Abstract
Sulfonylhydrazones are privileged structures with multifaceted pharmacological activity. Exploring the hypoglycemic properties of these organic compounds, we previously revealed a new series of N-sulfonylhydrazones (NSH) as antidiabetic drug candidates. Here, we evaluated the microsomal metabolism, chemical stability, and permeability profile of these NSH prototypes, focusing on the pharmacokinetic differences in N-methylated and non-N-methylated analogs. Our results demonstrated that the N-methylated analogs (LASSBio-1772 and LASSBio-1774) were metabolized by CYP, forming three and one metabolites, respectively. These prototypes exhibited chemical stability at pH 2.0 and 7.4 and brain penetration ability. On the other hand, non-N-methylated analogs (LASSBio-1771 and LASSBio-1773) were hydrolyzed in acid pH and could not cross the artificial blood–brain barrier. The cyano group in LASSBio-1771 was postulated as a possible site of interaction with the heme group, potentially inhibiting CYP enzymes. Moreover, prototypes with the methyl ester group were metabolized by carboxylesterase, and non-N-methylated analogs did not show oxidative metabolism. The prototypes (except LASSBio-1774) showed excellent gastrointestinal absorption. Altogether, our data support the idea that the methyl effect on NSH strongly alters their pharmacokinetic profile, enhances the recognition by CYP enzymes, promotes brain penetration, and plays a protective effect upon acid hydrolysis.
Introduction
Sulfonylhydrazones (SH) are organic compounds described as nonclassical isosteres of acylhydrazones (NA) and considered as privileged structures.1−3 The synthesis and applications of SH are widely explored in medicinal chemistry due to their different biological and pharmacological properties, such as antinociceptive,4−6 antiasthmatic,7 antioxidant,8 antibacterial,9,10 antidepressant,11 antineoplastic,12−14 and antidiabetic.15
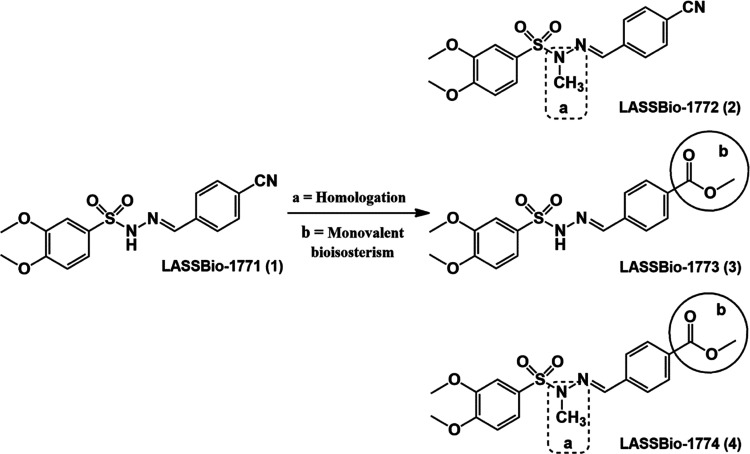
Previously, our group revealed a new series of N-sulfonylhydrazones (NSH) as antidiabetic drug candidates (Figure 1). These compounds demonstrated a very interesting hypoglycemic profile, decreasing neuropathic pain induced by diabetes and significantly reducing blood glucose levels in a murine streptozotocin-induced diabetes model.16,17 Although the in vitro plasma stability of these compounds has been studied,16 little is known about their metabolism and permeability profile.
Figure 1.
N-sulfonylhydrazone analogs (1–4) with hypoglycemic properties.
From this perspective and considering the effect of methyl groups in chemical conformation and drug metabolism,18 we investigate the differences in microsomal metabolism, chemical stability, and permeability profile of N-methylated and non-N-methylated sulfonylhydrazone analogs 1–4. Our data support the idea that methylation at the nitrogen in NSH plays a protective effect upon acid hydrolysis, promotes brain penetration, and intensely alters the recognition of N-sulfonylhydrazones in the CYP enzyme. To the best of our knowledge, this is the first study that evaluates the methyl effect on microsomal metabolism and permeability profile of N-sulfonylhydrazone prototypes.
Results and Discussion
Metabolism of N-Sulfonylhydrazone Prototypes (1–4)
We investigated phase I metabolism of N-sulfonylhydrazone prototypes 1–4 using liver microsomes from male Wistar rats. The CYP catalytic activity of microsomal fractions and the analytical method applied in this study were validated. The analytical parameters evaluated showed that the developed method presents specificity, linearity (R2 ≥ 0.99), sensitivity (0.5 μM), accuracy (RE ± 13%), intraday and interday precision (RSD < 9%), and good recovery (range 108–90%) (see the Supplementary Data).
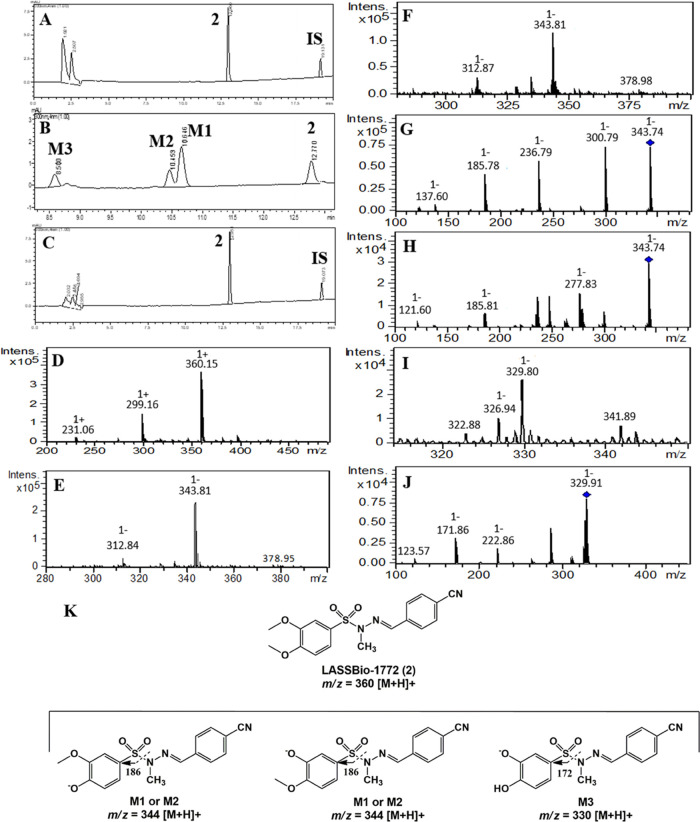
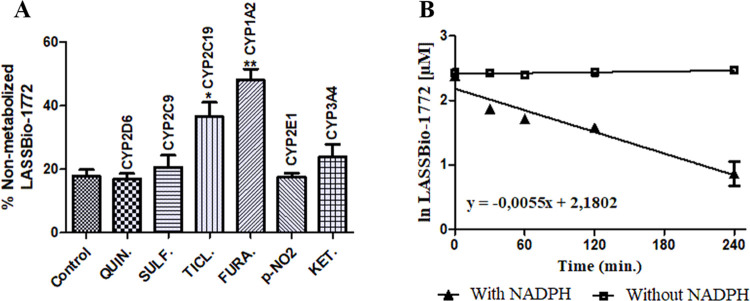
Our results revealed that compound 2 was only metabolized in the presence of NADPH-generating system and produced three major metabolites, identified as M1 (Rt = 10.6 min), M2 (Rt = 10.4 min), and M3 (Rt = 8.5 min). HPLC-MS analysis showed that M1 and M2 exhibit the same m/z value of 344 [M – H]−, consistent with the loss of 15 atomic units in comparison to compound 2 (m/z 360, [M + H]+), which might reflect O-demethylation in the 3,4-dimethoxyphenyl group (Figure 2A–F). Considering the steric and electronic effects, N-demethylation at the sulfonylhydrazone moiety is an unlikely process.19 However, to unequivocally identify the metabolites M1 and M2, the non-N-methylated analog 1 (LASSBio-1771) was coinjected in a metabolized sample and showed a different retention time (Rt = 10.1 min) of M1 and M2 (Figure S2). Furthermore, both metabolites present the fragment ions m/z 186 [M – H]− consistent with the S–N bond cleavage in the sulfonylhydrazone from O-demethylated metabolites. The metabolite M3 displayed m/z of 330 [M – H]−, in accordance with a double O-demethylation process in the 3,4-dimethoxyphenyl group and production of catechol metabolites. In a similar manner, the HPLC-MS/MS analysis of M3 showed a fragment ion m/z 172 [M – H]−, indicating the S–N bond cleavage in the sulfonylhydrazone of the catechol metabolite proposed as M3 (Figure 2G–K). The enzymatic inhibition study demonstrated that CYP1A2 and CYP2C19 metabolized LASSBio-1772 (2) since its biotransformation rate decreased by about 50 and 30% with the use of furafylline (40 μM) and ticlopidine (20 μM), respectively (Figure 3). A thermal inactivation study discarded the involvement of FMO on the metabolism of LASSBio-1772 (2) (Figure S3). Further studies using the human liver microsome and human-recombinant CYP are required to clarify whether the enzymes and compound 2 metabolites are compatible with our murine study.
Figure 2.
Microsomal metabolism of LASSBio-1772 (2) (10 μM) and its oxidative metabolites M1, M2, and M3 formed in the presence of NADPH. IS, internal standard (biphenyl l-4-carboxylate methyl, 10 μM). (A) Incubation at 0 min, compound 2, Rt = 12.9 min; (B) 240 min incubation time in the presence of NADPH, biotransformation in M1 (Rt = 10.6 min), M2 (Rt = 10.4 min), and M3 (Rt = 8.5 min); (C) no metabolization in the absence of NADPH, 240 min incubation time; (D) compound 2, HPLC-MS analysis, m/z 360 [M + H]+; (E) HPLC-MS analysis M1, m/z 344 [M – H]−; (F) HPLC-MS analysis M2, m/z 344 [M – H]−; (G) fragment ion m/z 186, HPLC-MS/MS analysis M1; (H) fragment ion m/z 186, HPLC-MS/MS analysis M2; (I) HPLC-MS analysis M3, m/z 330 [M – H]−; (J) fragment ion m/z 172, HPLC-MS/MS analysis M3. (K) Structure elucidation of fragment ions m/z 186 and 172.
Figure 3.
Metabolism inhibition and experimental half-life of compound 2. (A) Enzymatic inhibition study using specific inhibitors of CYP isoenzymes to establish the enzymes responsible for in vitro hepatic microsomal metabolism of compound 2: quinidine (6 μM, CYP2D6), sulfaphenazole (6 μM, CYP2C9), ticlopidine (20 μM, CYP2C19), furafylline (40 μM, CYP1A2), p-nitrophenol (100 μM, CYP2E1), and ketoconazole (1 μM, CYP3A4). The biotransformation rate decreased about 50 and 35% with furafylline and ticlopidine, respectively, showing that CYP1A2 and CYP2C19 were responsible for metabolizing LASSBio-1772 (2). (B) Determination of half-life (t1/2= 126 min) based on the straight-line equation achieved by the natural logarithm of LASSBio-1772 concentration vs incubation time in the presence of NADPH. Data represent the mean of triplicate, performed with three different microsome samples (n = 9) after 240 min of incubation at 37 °C. **p-value < 0.0003; *p-value < 0.005. One-way ANOVA followed by Dunnett’s post-test, using GraphPrism software (version 5.0).
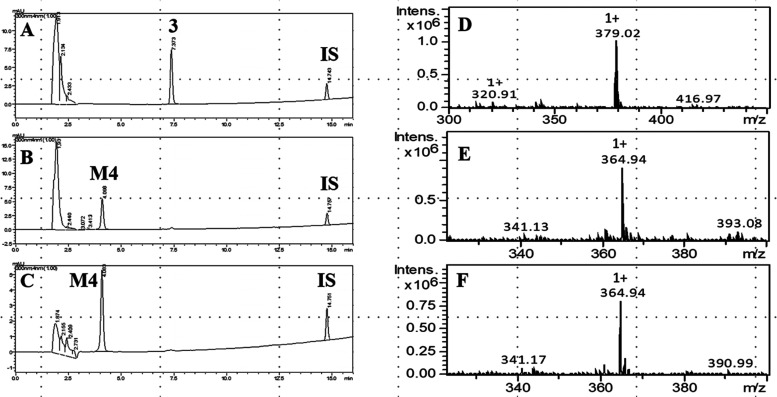
Additionally, we determined the experimental half-life (t1/2) for compound 2, which was 126 min, indicating high microsomal stability (Figure 3). On the other hand, compounds 3 (LASSBio-1773) and 4 (LASSBio-1774) were readily metabolized in the microsomal fraction in the presence and absence of NADPH. The biotransformation of compound 3 (t1/2 ∼5 min) generates a single metabolite, called M4 (Rt = 4.1 min), present in both experimental controls. The HPLC-MS analysis revealed that M4 shows the same m/z 365 [M + H]+ in the presence and absence of NADPH, consistent with the loss of 15 atomic units in comparison to compound 3 (m/z 379, [M + H]+). These findings suggest that compound 3 might be metabolized by carboxylesterase (CES), an NADPH-independent enzyme, and M4 represents a carboxylic acid derivative produced by the hydrolysis of methyl ester group present in LASSBio-1773 (3) (Figures 4 and 5A). Similar results were seen in our previous study, where this hydrolysis metabolite was revealed by an in vitro plasma stability assay.16 The involvement of CES was confirmed via incubation with bis-(p-nitrophenyl)phosphate (100 μM), where the metabolism of compound 3 was inhibited by almost 100% (Figure 5B). Finally, the hypothesis of methyl ester hydrolysis was validated with the coinjection of the proposed carboxylic acid derivative in previously metabolized samples. The increment in the chromatographic peak of M4 (Rt = 4.1 min) in both experimental controls confirmed that this metabolite corresponds to the suggested carboxylic acid derivative (Figure S4).
Figure 4.
Microsomal metabolism of LASSBio-1773 (3) (10 μM) and its hydrolysis metabolite M4, formed in the presence and absence of NADPH. IS, internal standard (biphenyl l-4-carboxylate methyl, 10 μM). (A) Incubation at 0 min, compound 3, Rt = 7.4 min; (B) 20 min incubation time in the presence of NADPH, biotransformation in M4 (Rt = 4.1 min); (C) 20 min incubation time in the absence of NADPH, biotransformation in M4 (Rt = 4.1 min); (D) compound 3, HPLC-MS analysis, m/z 379 [M + H]+; (E) HPLC-MS analysis M4, m/z 365 [M + H]+, formed in the presence of NADPH; (F) HPLC-MS analysis M4, m/z 365 [M + H]+, formed in the absence of NADPH.
Figure 5.
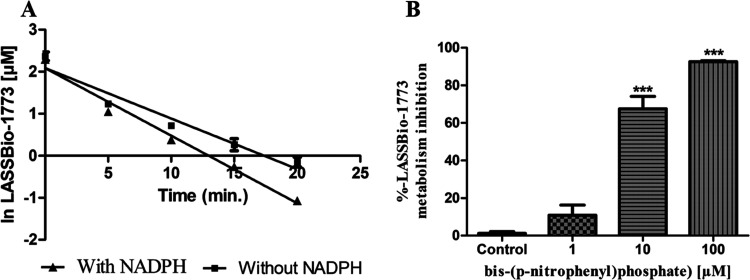
Experimental half-life and metabolism inhibition of compound 3. (A) Determination of half-life based on the straight-line equation achieved by the natural logarithm of LASSBio-1773 concentration vs incubation time in the presence and absence of cofactor (t1/2 = 4.3 min, presence of NADPH; t1/2 = 5.7 min, absence of NADPH). (B) Enzymatic inhibition study using bis-(p-nitrophenyl)phosphate at 1, 10, and 100 μM to inhibit CES. Data represent the mean of triplicate, performed with three different microsome samples (n = 9), after 20 min of incubation at 37 °C. ***p-value < 0.0001; one-way ANOVA followed by Dunnett’s post-test, using GraphPrism software (version 5.0).
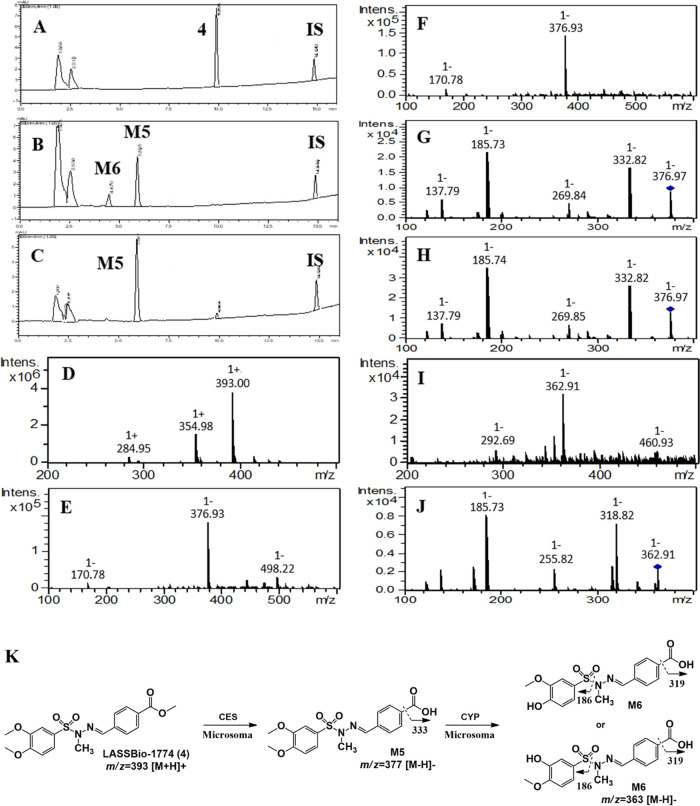
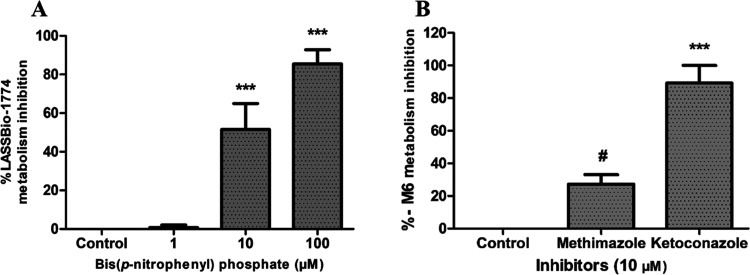
Similarly, compound 4 (m/z 393 [M + H]+, positive mode) showed a short experimental half-life (t1/2 < 5 min) and was biotransformed in two major metabolites identified as M5 (Rt = 5.9 min) and M6 (Rt = 4.5 min) (Figure 6A–C). The metabolite M5 was formed in the presence and absence of NADPH and displayed m/z 377 ([M – H]−, negative mode) and fragment ions m/z 333 in both controls, which is compatible with a carboxylic acid derivative created via methyl ester group hydrolysis promoted by CES (Figure 6D–H). The involvement of CES was confirmed by an enzymatic inhibition study in the same way as described for compound 3 (Figure 7A).
Figure 6.
Microsomal metabolism of LASSBio-1774 (4) (10 μM). IS, internal standard (biphenyl l-4-carboxylate methyl, 10 μM). (A) Incubation at 0 min, compound 4, Rt = 9.9 min; (B) 5 min incubation time in the presence of NADPH, biotransformation in M5 (Rt = 5.9 min) and M6 Rt = 4.5 min; (C) 5 min incubation time in the absence of NADPH, biotransformation in M5 (Rt = 5.9 min); (D) compound 4, HPLC-MS analysis positive mode, m/z 393 [M + H]+; (E) HPLC-MS analysis M5 negative mode, m/z 377 [M – H]−, formed in the presence of NADPH; (F) HPLC-MS analysis M5 negative mode, m/z 377 [M – H]−, formed in the absence of NADPH; (G) fragment ion m/z 333 [M – H]−, HPLC-MS/MS analysis M5 in the presence of NADPH; (H) fragment ion m/z 333 [M – H]−, HPLC-MS/MS analysis M5 in the absence of NADPH; (I) HPLC-MS analysis M6 negative mode, m/z 363 [M – H]−, formed in the presence of NADPH; and (J) structure elucidation of fragment ions from M5 and M6.
Figure 7.
Metabolism inhibition study of compound 4 and its metabolite M6. (A) Enzymatic inhibition study using bis-(p-nitrophenyl)phosphate at 1, 10, and 100 μM to inhibit CES. (B) Enzymatic inhibition study to identify the oxidative class of enzymes responsible for M6 metabolite formation: the inhibition study was performed in the presence of NADPH-generating system using ketoconazole (unspecific CYP inhibitor at 10 μM) and methimazole, an FMO inhibitor. The values of AUC from M6 in the positive control were compared with the AUC in the presence of ketoconazole and methimazole. Data represent the mean of triplicate, performed with three different microsome samples (n = 9), after 15 min of incubation at 37 °C. ***p-value < 0.0003; #p-value > 0.05. One-way ANOVA followed by Dunnett’s post-test, using GraphPrism software (version 5.0).
On the other hand, the metabolite M6 was formed only in the presence of NADPH and shows m/z 363 ([M – H]−, negative mode) and fragment ions m/z 186 and 319, suggesting an O-demethylation process from the metabolite M5 (m/z 377, [M–H]−, negative mode) (Figure 6I,J). Furthermore, this metabolite was not formed in the presence of bis-(p-nitrophenyl)phosphate at 100 μM, reinforcing that M6 is a metabolite of M5 (experiment performed in the presence of NADPH). Although unlikely, the possibility of N-demethylation at the sulfonylhydrazone moiety was discarded by coinjecting the non-N-methylated analog of M5 (Rt = 4.1 min), which shows a different retention time of M6 (Figure S5). The participation of CYP in producing M6 was confirmed by an enzymatic inhibition study using CYP and FMO inhibitors. After the incubation with ketoconazole (10 μM), the formation of metabolite M6 was inhibited by almost 90%, showing that the production of M6 is dependent on the catalytic action of CYP isozymes (Figure 7B).
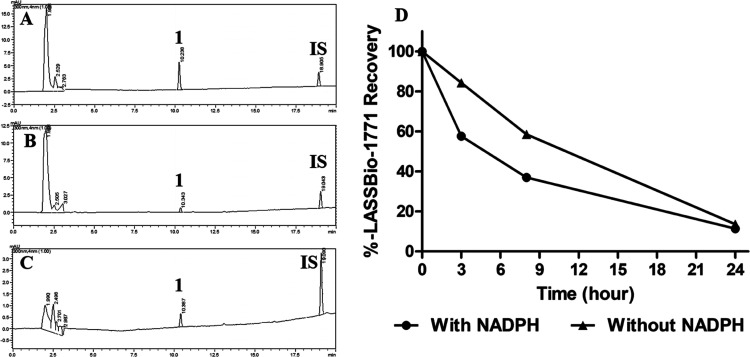
Interestingly, the microsomal metabolism analysis of compound 1 showed atypical results compared to its N-sulfonylhydrazone analogs (2–3). Our study showed that although there was an apparent consumption of this prototype, no metabolites were detected. As shown in Figure 8, the peak area of compound 1 (Rt = 10.3 min) declined while the incubation time increased in the presence and absence of NADPH. However, no metabolite chromatographic peaks were identified. After 24 h of incubation at 37 °C, the recovery rate of LASSBio-1771 (1) was lower than 20% for both experimental controls (Figure 8D). Among the hypotheses formulated to explain such behavior (see the Supplementary Data), the one that attributes compound 1 to the ability to inhibit CYP, involving its nitrile subunit, seems the most plausible. Considering that the consumption of compound 1 increased with and without NADPH, we hypothesize that the inhibition occurs by a competitive and reversible CYP inhibition through a bind between the heme iron atom and the cyano group. In this sense, the inhibition may occur without the involvement of NADPH as a cofactor once this mechanism of inhibition does not require the previous metabolism of the prototype.20 From this perspective, it is important to consider that even though the consumption of LASSBio-1771 occurs in both controls, it looks like the kinetics happen differently. After 3 h of incubation, the consumption of compound 1 was around 50% in the presence of NADPH. In contrast, without NADPH, the consumption was only approximately 15% (Figure 8B). These results indicate that LASSBio-1771 could also act as a time-dependent inhibitor, where the CYP inhibition is time-, concentration-, and NADPH-dependent.21
Figure 8.
Microsomal profile of LASSBio-1771 (1) (10 μM). IS, internal standard (biphenyl l-4-carboxylate methyl, 10 μM). (A) Incubation at 0 min, compound 1, Rt = 10.3 min; (B) 24 h incubation at 37 °C in the presence of NADPH, no metabolite peaks were identified; (C) 24 h incubation at 37 °C in the absence of NADPH, no metabolite peaks were identified; (D) recovery rate of compound 1 at 0, 3, 8, and 24 h of incubation in the presence and absence of NADPH.
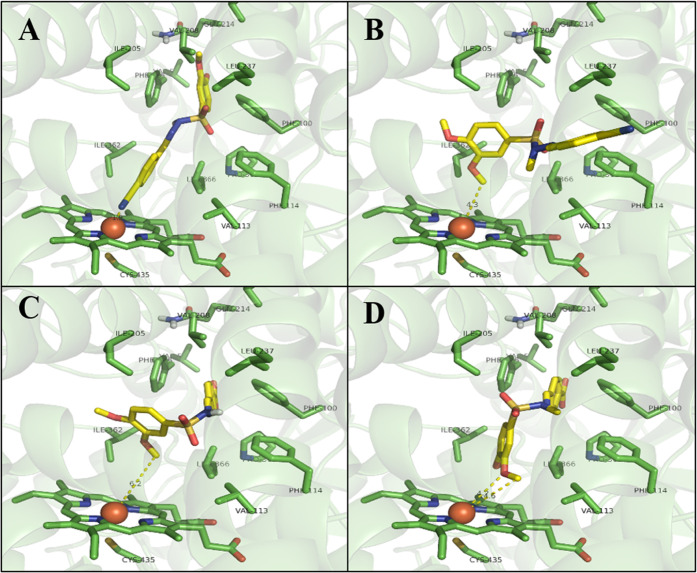
To explore the possibility of LASSBio-1771 (1) inhibiting a CYP enzyme, we conducted a molecular modeling analysis, where compound 1 and its N-methylated analog 2 were docked into the CYP2C19 isozyme. Our results showed that LASSBio-1771 (1) performs a coordination interaction with the iron ion through the nitrogen lone pair of the cyano group, which was not observed for the interaction mode of LASSBio-1772 (2), in which the methoxy group in the meta position was close to the iron atom (Figure 9A,B). In this way, the docking results point out compound 1 as a possible CYP inhibitor, which agrees with the experimental data. To deeply understand the strength of the coordination interaction, we performed a bond order analysis using the PM7 semiempirical method with MOPAC2016 software. Additionally, we carried out docking studies in the PBD 4GQS with common pharmacophoric groups for CYP inhibition, such as pyridine and triazole rings, to compare with the coordination interaction performed by the cyano group in the bond order analysis. The analysis indicated a bond order of 0.5 for the cyano group, comparable with the coordination interactions of the lone nitrogen pairs in pyridine (bond order = 0.4) and triazole (bond order = 0.5) rings. These results suggest that LASSBio-1771 (1) is a potential CYP inhibitor. However, additional studies are required to clarify whether CYP2C19 or another microsomal enzyme could be inhibited by compound 1in vivo. Considering that electrophilicity and steric access to the cyano group are similar between LASSBio-1771 (1) and LASSBio-1772 (2), we hypothesized that conformational differences promoted the methyl effect may influence the affinity and intrinsic activity of these prototypes by the oxidative enzymes. This hypothesis agrees with our previous study that revealed conformational differences between LASSBio-1773 (3) and LASSBio-1774 (4).22
Figure 9.
Molecular modeling study of N-sulfonylhydrazone prototypes (1–4). (A) LASSBio-1771 (1) docked into the CYP2C19 isozyme, coordination interaction with the iron ion through the nitrogen lone pair of the cyan group. (B) LASSBio-1772 (2) docked into the CYP2C19 isozyme, demonstrating the interaction of the methoxy group in the meta position with the iron atom. (C) M4 metabolite from LASSBio-1773 (3) docked into the CYP2C19 isozyme, showing a 6.2 Å distance from the methoxy groups and the iron ion. (D) M5 metabolite from LASSBio-1774 (4) docked into the CYP2C19 isozyme, showing a 4.6 Å distance from the methoxy groups and iron ion. CYP2C19 isozyme (PBD: 4GQS).
Altogether, our data support the idea that the methyl effect, introduced by sulfonylhydrazone N-methylation, influences the recognition of CYP enzymes since only N-methylated analogs (i.e., compounds 2 and 4) exhibited oxidative metabolites formed by CYP activity. In this regard, we carried out docking studies involving the metabolites M4 and M5 formed by the biotransformation of compounds 3 and 4, respectively. This analysis demonstrated that both metabolites adopted similar interactions in the active site of CYP2C19, showing a folded conformation, where the rings with the methoxy groups face the HEME group. However, there is a clear difference in the angulation and, consequently, in the distance between the methoxy groups of each compound in relation to the iron ion. The metabolite M5 from LASSBio-1774 (4) is positioned closer to the iron ion (4.6 Å distance), which may be an indication that this compound is more susceptible to undergoing dealkylation reactions in the methoxy groups in relation to the metabolite M4 from LASSBio-1773 (3) (6.2 Å distance from the iron ion). These data are in agreement with the experimental observations (Figure 9C,D). Furthermore, our results demonstrated that N-methylation might cause an angulation difference in the dihedral angle between the nitrogen of sulfonylhydrazone (ArSO2NRN=CHAr) and the phenyl ring substituted with the methoxy groups. In the M4 metabolite of compound 3, the dihedral angle H-N–S-C is equal to 176.4°, while in the M5 metabolite of compound 4, the dihedral angle C-N–S-C is equal to 95.8° (Figure S7). These findings agree with our previous study22 and support the idea that N-methylation in N-sulfonylhydrazones might influence the recognition of these prototypes upon CYP enzymes. Additionally, our data emphasize how a simple molecular modification could modulate the pharmacokinetics properties, highlighting the importance of metabolite profile investigation in drug discovery.23
Considering that the CYP1A2 isozyme also plays an essential role in LASSBio-1772 (2) oxidative metabolism, we performed a new series of docking studies using PDB 2HI4. The results do not point out the possibility of a coordination interaction with the iron ion, and any significant differences in the interaction mode of LASSBio-1771 compared with LASSBio-1772 or with LASSBio-1773 and LASSBio-1774 were observed, indicating that this isoform is not the one inhibited by LASSBio-1771 (Figure S13).
Chemical Stability
The chemical stability of N-sulfonylhydrazone prototypes (1–4) was evaluated in buffer solution with pH 2 and pH 7.4 to mimic their stability in biological environments, considering the stomach and plasma pH and taking into account the presence of the labile sites for hydrolysis, i.e., ester and imine subunits (Figure 1).
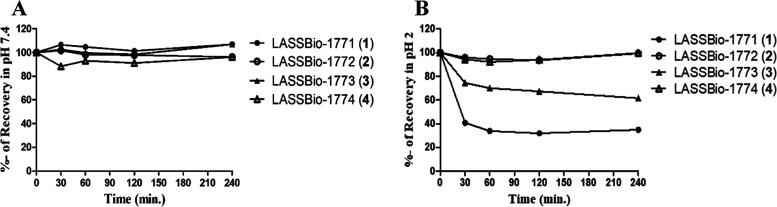
As shown in Figure 10, all compounds (1–4) are stable at pH 7.4. On the other hand, the HPLC-PDA analyses demonstrated that only the N-methylated prototypes, LASSBio-1772 (2) and LASSBio-1774 (4), were stable at pH 2, with a recovery of 99-93%. The non-N-methylated analogs, LASSBio-1771 (1) and LASSBio-1773 (3), were unstable at pH 2, with hydrolysis rates of about 60 and 40%, respectively. The HPLC-PDA results showed that after incubation at 37 °C, hydrolysis products (1a/1b and 3a/3b) were detected for compounds 1 and 3 but were not observed for N-methylated prototypes 2 and 4 even after 4 h of incubation (Figure S9). As expected, the hydrolysis products were formed breaking the imine bond, generating a sulfonylhydrazide and the corresponding aldehyde products. These products were identified by spiking samples after incubation time (pH 2, at 37 °C). For LASSBio-1771 (1), the samples were spiked with [(3,4-dimethoxyphenyl)sulfonyl]hydrazide (1a) and 4-formylbenzonitrile (1b), and for LASSBio-1773 (3), the samples were spiked with [(3,4-dimethoxyphenyl)sulfonyl]hydrazide (3a) and methyl-4-formylbenzoate (3b). The area under the curve (AUC) of hydrolysis products increases substantially after spiking (Figure S10).
Figure 10.
Chemical stability profile of compounds 1, 2, 3, and 4. (A) Analysis at pH 7.4. (B) Analyses at pH 2, showing that non-N-methylated analogs, LASSBio-1771 (1) and LASSBio-1773 (3), were unstable in pH 2. Experiments were performed in triplicate, and the average values are represented.
Also, the LUMO orbitals and atomic coefficient of imine carbon were calculated from the lowest energy conformer of each N-sulfonylhydrazone (1–4) (Spartan 14′, Wavefunction Inc.). This analysis demonstrated that non-N-methylated prototypes (1 and 3) present the LUMO orbital at a lower energy level than their N-methylated analogs (2 and 4), making them more reactive toward nucleophilic attack by water. Besides, the atomic coefficient of imine carbon in compounds 1 and 3 is slightly higher than those from their N-methylated analogs (2 and 4), which also corroborates the experimental findings since it indicates greater participation of imine carbon for the formation of LUMO in compounds 1 and 3, facilitating the nucleophilic attack by water (Table 1). Similar results were seen in a study, where N-methylation in an N-acylhydrazone derivative seems to promote higher stability in pH 2 than non-N-methylated analogs.24
Table 1. Energy Levels of LUMO Orbitals and Atomic Coefficient Values of Iminic Carbon from N-sulfonylhydrazone Prototypes (1–4) Calculated by the SPARTAN 14′ Program (Wavefunction Inc.).
| compounds | atomic coefficient | LUMO (eV) | molar refractivity |
|---|---|---|---|
| 1 | 0.09 | 1.42 | 33.23 |
| 2 | 0.07 | 1.54 | 35.42 |
| 3 | 0.11 | 1.58 | 36.34 |
| 4 | 0.07 | 1.83 | 42.09 |
The extrapolation of these data to the eventual behavior of biophase indicates that all prototypes might show chemical stability at plasma pH and reveal the instability of compounds 1 and 3 at gastric pH, suggesting attention to using both prototypes by oral administration. Furthermore, these findings might reflect a protective effect of the N-methyl group in N-sulfonylhydrazones for acid hydrolysis.
Permeability Profile
Gastrointestinal absorption and brain penetration of compounds 1–4 were predicted by a parallel artificial membrane permeability assay (PAMPA).23,25−27 All experiments were validated by quality control (QC) standards, and the samples were analyzed in triplicate at two different times.
The prediction of brain penetration (PAMPA-BBB) employed seven QC standards, which were applied in the linear equation to classify the permeability profile of the N-sulfonylhydrazone prototypes (1–4). The X-axis represents the known permeability coefficient (Pe) reported in the literature for QC standards, and the Y-axis represents the Pe experimental values. In this study, a good correlation between the experimental results and the literature data was achieved [Pe (exp.) = 11,459 (liter.) −0.7211 (R2 = 0,993)] (Figure S11). Compounds with Pe values <1.57 were classified as nonpermeable (BBB-), Pe values >3.86 were correlated with brain penetration ability (BBB+), and Pe values between 1.57 and 3.86 were considered borderline. Our results of PAMPA-BBB indicate that the non-N-methylated compounds, LASSBio-1771 (1) and LASSBio-1773 (3), are nonpermeable, while their N-methylated analogs, compound 2 and compound 4, may penetrate in the brain (Pe > 3.86). The pKa values of compounds 1 and 3 might influence their inability to penetrate the brain since they assume an ionized form at pH 7.4 (Table 2).16 Furthermore, the presence of the N-methyl group promotes the enhanced lipophilicity of compounds 2 and 4, which might impact the permeability profile of these prototypes.28
Table 2. Permeability Coefficient of N-sulfonylhydrazone Prototypes (1–4) and Quality Control Standards Determined by the PAMPA-BBB Assay.
| compounds | literature permeability (10–6 cm/s)a | experimental permeability (10–6 cm/s) | classificationc | pKab |
|---|---|---|---|---|
| atenolol | 0.8 | 0.10 ± 0.21 | BBB– | |
| caffeine | 1.3 | 0.77 ± 0.58 | BBB– | |
| enoxacin | 0.9 | 0.57 ± 0.12 | BBB– | |
| hydrocortisone | 1.9 | 1.26 ± 0.78 | BBB– | |
| ofloxacin | 0.8 | 0.29 ± 0.43 | BBB– | |
| testosterone | 17 | 19.9 ± 0.53 | BBB+ | |
| verapamil | 16 | 16.3 ± 1.74 | BBB+ | |
| LASSBio-1771 (1) | 0.43 ± 0.54 | BBB– | 6.35 | |
| LASSBio-1772 (2) | 4.18 ± 0.03 | BBB+ | ND | |
| LASSBio-1773 (3) | 0.45 ± 0.63 | BBB– | 6.35 | |
| LASSBio-1774 (4) | 9.01 ± 0.09 | BBB+ | ND |
On the other hand, all prototypes, except compound 4, showed a high rate of fraction absorption (Fa%), indicating that these compounds must be well absorbed across the gastrointestinal tract and, consequently, can be orally administrated. In this analysis, compound 4 could not be evaluated by PAMPA-GIT since it was not soluble in the solution PBS pH 6.6-10 mM/DMSO (95:5) (Table 3). These results emphasize that N-methylation in N-sulfonylhydrazones can promote brain penetration but not affect gastrointestinal absorption.
Table 3. Permeability Coefficient and Fraction Absorption of N-sulfonylhydrazone Prototypes (1–3) and Quality Control Standards Determined by the PAMPA-GIT Assayc.
| compounds | literature fraction absorption (%)a | experimental permeability (10–6 cm/s) | experimental fraction absorption (%)b | classificationb |
|---|---|---|---|---|
| acyclovir | 21 | 0.1 | 8.62 | low |
| atenolol | 52 | 0.1 | 15.81 | low |
| ceftriaxone | 1 | 0.3 | 23.70 | intermediate |
| coumarin | 100 | 27.9 | 100.00 | high |
| diclofenac | 100 | 14 | 100.00 | high |
| hydrocortisone | 91 | 6.7 | 100.00 | high |
| norfloxacin | 35 | 0.8 | 51.39 | intermediate |
| ranitidine | 55 | 0.4 | 49.75 | intermediate |
| sulfasalazine | 12 | 0.1 | 8.62 | low |
| verapamil | 98 | 7.3 | 99.86 | high |
| LASSBio-1771 (1) | 18.2 | 100.00 | high | |
| LASSBio-1772 (2) | 13.1 | 100.00 | high | |
| LASSBio-1773 (3) | 23.2 | 100.00 | high | |
| LASSBio-1774 (4) | NA | NA | NA |
Zhu et al.29
Fraction absorption classification: High, 70–100% Fa; Intermediate, 30–69% Fa; and Low, 0–29% Fa.
NA = nonanalyzed. Compound 4 could not be evaluated by PAMPA-GIT since it was not soluble in the solution PBS pH 6.6–10 mM: DMSO (95:5).
Conclusions
N-methylation at the sulfonylhydrazone framework strongly alters the recognition of NSH derivatives upon the CYP enzyme, which might be associated with conformational differences promoted by the methyl effect. The presence of the cyano group in a non-N-methylated analog was pointed to as a possible site of coordination interaction with the iron atom of the heme group, which can potentially inhibit CYP enzymes. Furthermore, only the N-methylated analogs exhibited oxidative metabolites formed by CYP action, reinforcing the influence of N-methylation on the metabolism profile in N-sulfonylhydrazones. Additionally, the presence of the methyl group promotes brain penetration and may play a protective function against acid hydrolysis. On the other hand, gastrointestinal absorption was not affected by the methyl effect.
Experimental Section
Chemicals and Reagents
N-sulfonylhydrazone prototypes, LASSBio-1771 (1), LASSBio-1772 (2), LASSBio-1773 (3), LASSBio-1774 (4), and the internal standard (IP), biphenyl l-4-carboxylate methyl, were synthesized at the Laboratório de Avaliação e Síntese de Substâncias Bioativas (LASSBio, UFRJ, Brazil) with ≥98% pure by HPLC (see the Supplemental Data). HPLC grade solvents and reagents, i.e., acetonitrile, methanol, and formic acid (96%), were purchased from Tedia-Brazil. Type I grade water produced by an ultrapure water purification system (Master System MS2000, Gehaka, Brazil) was used in experiments and HPLC analysis. Bis(p-nitrophenyl) phosphate sodium salt, D-glucose-6-phosphate, glucose-6-phosphate dehydrogenase, β-nicotinamide adenine dinucleotide phosphate (NADP+), magnesium chloride hexahydrate, quinidine, sulfaphenazole, furafylline, p-nitrophenol, ketoconazole, bicinchoninic acid (BCA) protein assay kit, and salts used for buffer preparation were purchased from Sigma-Aldrich (St. Louis, MO).
Chromatographic Equipment
A Shimadzu Prominence HPLC system (Shimadzu, Japan) with a photodiode array detector (SPD-M20A) was used for analytical analysis. The HPLC system was equipped with a binary pump (LC-20AD), a degasser (DGU-20A5), and an autosampler (SIL-20A).
The HPLC-MS and HPLC-MS/MS analysis were carried out in the HPLC system (Shimadzu, Japan) coupled with a mass spectrometer with electrospray ionization, amaZon SL model Esquire 6000—ESI Ion Trap Msn System (Bruker Daltonics). The mass spectrometry and ionization parameters were set as follows: range of mass-to-charge ratio (m/z) 100-2000, capillary voltage 3500 V, nitrogen gas flow 6.0 L/min, temperature 250 °C, nebulizer pressure 4.0 psi, and collision energy at 25 eV using helium as collision gas.
Chemical Stability Study
The study of chemical hydrolysis of the N-sulfonylhydrazone prototypes (1–4) was performed at pH 2 and pH 7.4 to mimic their stability in biological environments, considering the stomach and plasma pH. Each prototype (100 μM) was mixed in a buffer solution of pH 2 (HCl 0.2 M) and pH 7.4 (phosphate buffer, PBS) and incubated under stirring in a water bath at 37 °C for 0, 30, 60, 120, and 240 min. At the end of the incubation, the samples in acid medium (pH 2) were neutralized by phosphate buffer (PBS, pH 8.5). Compound extraction was performed using 1 mL of ice-cold acetonitrile. The organic phase was separated, filtered through PVDF syringe filters (Millex: 0.22 μm pore size), and analyzed by HPLC-PDA.24,30,31 The stability profile was evaluated based on the comparison with samples not incubated at 37 °C.
Chromatographic separation was carried out employing a C18 column (Kromasil C18, 250 mm × 4.6 mm, 5 μm) preceded by a guard column (Phenomenex C18, 4 × 3 mm2). The column was held at room temperature (20 ± 2 °C). The mobile phase was constituted of solution A (type I grade water) and organic solution B (acetonitrile). The chemical stability analysis of LASSBio-1771 (1) and LASSBio-1773 (3) prototypes was performed with an isocratic method (1 mL·min–1) over 12 min (A: B, 1:1, v/v). For their N-methylated analogs, LASSBio-1772 (2) and LASSBio-1774 (4), the isocratic method (1 mL·min–1) was set as follows: 60% B: 40% A, 0–10 min. A volume of 20 μL of each sample was injected, and the quantification was carried out at 254 and 300 nm wavelengths.
Metabolism Study of N-Sulfonylhydrazone Prototypes (1–4)
This study investigated phase I microsomal metabolism (oxidative and hydrolytic) of N-sulfonylhydrazone prototypes (1–4) using rat liver microsomal fraction obtained from male Wistar rats (280–330 g). The microsomal fractions were prepared by the centrifugal differential process following the method described by Cabrera and co-workers.32 The BCA protein assay kit was used for protein quantification of liver microsomes.33
Samples were prepared based on a standard mixture, including 1 mg protein/mL of rat microsomal fraction, NADPH-generating system (0.4 mM NADP+, 1.3 mM MgCl2, 3.5 mM glucose-6-phosphate and 0.5 U/mL glucose-6-phosphate dehydrogenases), compounds 1–4 (10 μM, from stock solutions in DMSO), and PBS (pH 7.4) in a sufficient volume to complete 250 μL. The samples were incubated under stirring in a water bath at 37 °C for different times. After incubation, samples from compounds 1 and 3 were acidified to pH 5 with formic acid (HPLC grade, 98%). Then, 500 μL of methanol and 500 μL of acetonitrile were added at ice-cold temperature to all samples, which were vortexed for 10 s, kept at 4 °C for protein precipitation, and centrifuged at 13,500 rpm for 15 min at 4 °C. The organic phase was collected, and 10 μM IS was added. Finally, all of the samples were filtered through PVDF syringe filters (Millex: 0.22 μm pore size) and then analyzed by HPLC-PDA and HPLC-MS/MS in positive and negative modes. All of the reactions were performed in triplicate, and each prototype (1–4) was analyzed in a separate system. The DMSO final concentration was 0.5%.34,35
Chromatographic separation was carried out employing a C18 column (Kromasil C18, 250 mm × 4.6 mm, 5 μm) preceded by a guard column (Phenomenex C18, 4 × 3 mm2), and the mobile phase constituted of aqueous solution A (0.1% formic acid) and organic solution B (acetonitrile). The following gradient method (1 mL·min–1) was used for LASSBio-1771 (1) and LASSBio-1772 (2): 0-16 min, linear gradient from 40 to 90% B; 16-20 min, 90% B. Additionally, for LASSBio-1773 (3) and LASSBio-1774 (4), a gradient method (1 mL·min–1) was applied over 16 min as follows: 0–12 min, linear gradient from 50 to 90% B; 12–16 min, 90% B. The column was held at room temperature (20 ± 2 °C), and a volume of 20 μL of each sample was injected and quantified at a 300 nm wavelength. The metabolites’ structural analysis derived from the N-sulfonylhydrazone prototypes (1–4) was inspected by HPLC-MS and HPLC-MS/MS in positive or negative mode, considering the metabolically labile sites of these molecules.
In this study, all experiments were accompanied by three assay controls corresponding to (i) blank sample, in the absence of the prototype (1–4), to analyze chromatographic peaks related to the biological matrix; (ii) control in the presence of NADPH-generating system to investigate the activity of NADPH-dependent enzymes, i.e., isoforms of cytochrome P450 (CYP) and/or flavin-containing monooxygenase (FMO); and (iii) control in the absence of NADPH-generating system to verify carboxylesterase (CES) action, i.e., NADPH-independent enzyme.
The experimental half-life (t1/2) was determined by applying the equation: t1/2 = 0.693/a, where a corresponding to the slope of the straight-line equation of natural logarithm of sample concentration vs incubation time.34 A calibration curve was used to establish the prototype concentration in the buffered medium. The following incubation times were applied to the microsomal metabolism study: 0, 3, 8, and 24 h to compound 1; 30, 60, 120, and 240 min to compound 2; 0, 5, 10, 15, and 20 min to compounds 3 and 4.
All experimental procedures were approved by the Animal Care and Use Committee from the Federal University of Rio de Janeiro (protocol number: 028/15). The analytical methods were validated following guidelines from Food and Drug Administration (FDA)36 and the Brazilian Regulatory Agency (ANVISA).37 The analytical parameters analyzed were linearity, sensitivity, specificity, accuracy, recovery, and intraday and interday precision (see the Supplementary Data).
Enzymatic Inhibition Study
The enzymatic pathway involved in the microsomal metabolism of the N-sulfonylhydrazone prototypes (2–4) was identified by in vitro experiments using an irreversible inhibitor of oxidative and hydrolytic enzymes (i.e., CYP, FMO, and CES). Ketoconazole at 10 μM was applied for nonspecific inhibition of CYP isoenzymes34 and bis-(p-nitrophenyl)phosphate (1, 10, and 100 μM)38,39 and methimazole (10 μM)40,41 for CES and FMO inhibition, respectively. The involvement of FMO catalytic action was also verified by thermal inactivation at 50 °C for 1.5 min.42 To identify the CYP isoenzymes responsible for oxidative metabolism, the following specific inhibitors were applied: quinidine (6 μM, CYP2D6); sulfaphenazole (6 μM, CYP2C9); ticlopidine (20 μM, CYP2C19);43 furafylline (40 μM, CYP1A2);44p-nitrophenol (100 μM, CYP2E1);34,45 and ketoconazole (1 μM, CYP3A4).45−47
The inhibitors were preincubated for 30 min at 37 °C with rat liver microsomes in the presence or absence of an NADPH-generating system. Then, 10 μM N-sulfonylhydrazone prototype (2-4) was added, and the mixture was incubated at 37 °C. The effect of microsomal inhibition was measured by comparison with positive controls in the absence of inhibitors. All reactions were performed in triplicate and analyzed in three different rat liver microsomal fractions (n = 9).
Molecular Modeling
N-sulfonylhydrazones (1–4) were constructed with a Spartan 14′ (Wavefunction Inc.) and submitted for conformational analysis by the molecular mechanics method MMFF. The most stable conformer of each compound was optimized using the semiempirical method PM6, and a geometry reoptimization of the lowest energy conformer was performed using the Hartree–Fock quantum method (6-31G*). The LUMO orbitals and atomic coefficient of iminic carbon were calculated from the lowest energy conformer to assist in interpreting experimental data from the chemical stability study. These analyses were performed in a theoretical vacuum environment.
The docking studies were performed with a CYP2C19 isoenzyme (PBD: 4GQS) through CCDC software GOLD’v.2021.1.0 with the GoldScore function available in GOLD, which was chosen based on the obtained RMSD values (0.8 Å) for the redocking analysis of the cocrystallized ligand (Figure S12).48 In the next step, LASSBio-1771 (1), LASSBio-1772 (2), and the M1 (X, X) metabolites of LASSBio-1773 (3) and LASSBio-1774 (4) were docked into the CYP2C19 isozyme (PBD: 4GQS). Docking studies were also performed in the CYP1A2 isoenzyme (PBD: 2HI4) following the same methodology described above. The redocking analysis of the cocrystallized ligand of CYP1A2 (PBD: 2HI4) also pointed out the GoldScore function as best for docking studies performance, based on the obtained RMSD values (0.6 Å).
Permeability Assay
Prediction of the gastrointestinal absorption (PAMPA-GIT) and brain penetration (PAMPA-BBB) was evaluated by a parallel artificial membrane permeability assay (PAMPA) using donor and acceptor 96-well plates, which work as passive transcellular permeability compartments.25−27 Donor plates were coated with porcine brain lipid [25] and L-α-phosphatidylcholine from soybeans23 (20 mg·mL–1, in dodecane) for PAMPA-BBB and PAMPA-GIT, respectively.
Permeability assay of gastrointestinal absorption was performed in a similar manner as previously described.23,25 The following compounds were used as quality control (QC) standards: acyclovir, atenolol, ceftriaxone, coumarin, diclofenac, hydrocortisone, norfloxacin, ranitidine, sulfasalazine, and verapamil.29 A stock solution of N-sulfonylhydrazones (1–4) and QC standards was prepared (DMSO, 10 mM), and 250 μL of these solutions was diluted 20 times in PBS pH 6.6–10 mM. The solutions were filtered (Millex: 0.45 μm pore size) and added to the donor plate after impregnating it with 5 μL of L-α-phosphatidylcholine from soybeans (20 mg mL–1, in dodecane). One-hundred eighty microliters of solution PBS pH 7.4:DMSO (95:5) was added to the acceptor 96-well plate, and the donor plate was gently put on the acceptor plate to form the permeability compartments, which was left under stirring (50 rpm) for 8h at room temperature (23 ± 2 °C). In the end, the assembled donor–acceptor plates were separated, and the compounds 1–4 and QC standards were quantified by a multiwavelength UV plate reader (SpectraMax 5, Molecular Devices) at 250–450 nm. Optical density values were used to determine the fraction absorption (Fa%)49 using the program JMP version 10.0. Blank samples were prepared with 180 μL of solution PBS pH 7.4/DMSO (95:5) and analyzed in the same manner. The tested compounds were classified based on their Fa% values as well-absorbed compounds, 70–100% Fa; intermediate-absorbed compounds, 30–69% Fa; and low absorption, 0–29% Fa.50
The prediction of brain penetration (PAMPA-BBB) was evaluated in the same way as Pérez and cols. (2012) as previously described. From this perspective, 1 mg of test compounds (1–4) and QC standards (i.e., atenolol, caffeine, enoxacin, hydrocortisone, ofloxacin, testosterone, and verapamil) were dissolved in 1.5 mL of ethanol, diluted with 3.5 mL of PBS pH 7.4, and filtered (Millex: 0.45 μm pore size). The donor plate was impregnated with 5 μL of porcine brain lipid (20 mg mL–1, in dodecane), and 180 μL of PBS pH 7.4:etanol (70:30) solution was added to the acceptor 96-well plates. Then, 180 μL of each compound solution was added to the donor plate, and the receptor plate was carefully placed into the donor plate. The permeability system was left undisturbed for 2 h and 45 min at room temperature (23 ± 2 °C). Then, the donor and acceptor plates were separated, and the compounds 1–4 and QC standards were quantified by a multiwavelength UV plate reader (SpectraMax 5, Molecular Devices) at 250–450 nm. Optical density values were applied in a linear equation to determine the permeability coefficient (Pe) through the artificial membrane.26,27 The compounds were classified as permeable (BBB+) or nonpermeable (BBB−).
The PAMPA-GIT and PAMPA-BBB were performed in triplicate and at two different times (n = 2). The Pe and Fa% represent the average of the six runs analyzed.
Statistical Analysis
Statistical analyses were performed using GraphPrism software (version 5.0). Group comparisons were carried out by one-way analysis of variance (ANOVA) followed by Dunnett’s post-test. P-value ≤ 0.05 was considered significant for all assays.
Acknowledgments
The authors thank INCT-INOFAR, CNPq, FAPERJ, and CAPES for their support.
Glossary
Abbreviations Used
- SH
sulfonylhydrazone
- NA
acylhydrazone
- NSH
N-sulfonylhydrazone
- CYP
cytochrome P450
- HPLC-MS
high-performance liquid chromatography coupled with mass spectrometry
- HPLC-MS/MS
high-performance liquid chromatography coupled with tandem mass spectrometry
- FMO
flavin-containing monooxygenase
- CES
carboxylesterase
- NADPH
reduced nicotinamide adenine dinucleotide phosphate
- QC
quality control standards
- PAMPA
parallel artificial membrane permeability assay
- PAMPA-BBB
parallel artificial membrane permeability assay to predict brain penetration
- PAMPA-GIT
parallel artificial membrane permeability assay to predict gastrointestinal absorption
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c04368.
Chromatograms of metabolism study of N-sulfonylhydrazone prototypes (1–4); CYP catalytic activity validation and chemical stability; linear correlation of QC standards in PAMPA-BBB; and data of analytical method validation (PDF)
Author Contributions
J.S.G. and L.M.L. designed the metabolism study and chemical stability experiments. J.S.G. drafted the manuscript. J.S.G. performed chemical stability analysis and metabolism study. J.S.G. and T.R.C. performed the PAMPA analysis. P.S.M.P., C.A.M.F., and C.M.R.S. performed the molecular modeling study. T.R.C., P.S.M.P., C.A.M.F., C.M.R.S., E.J.B., and L.M.L. revised the paper. L.M.L. and E.J.B. provided funding acquisition. J.S.G., T.R.C., P.S.M.P., C.A.M.F., C.M.R.S., E.J.B., and L.M.L. provided the scientific oversight of the study.
The authors declare no competing financial interest.
Supplementary Material
References
- Cunha M. R.; Tavares M. T.; Carvalho C. F.; Silva N. A. T.; Souza A. D. F.; Pereira G. J. V.; Ferreira F. F.; Parise-Filho R. Environmentally Safe Condition for the Synthesis of Aryl and Alkyl Sulfonyl Hydrazones via One-Pot Reaction. ACS Sustainable Chem. Eng. 2016, 4, 1899–1905. 10.1021/acssuschemeng.6b00193. [DOI] [Google Scholar]
- Lima L.; Barreiro E. Bioisosterism: A Useful Strategy for Molecular Modification and Drug Design. Curr. Med. Chem. 2005, 12, 23–49. 10.2174/0929867053363540. [DOI] [PubMed] [Google Scholar]
- Lima L. M.; Barreiro E. J.. Beyond Bioisosterism: New Concepts in Drug Discovery. In Comprehensive Medicinal Chemistry III, Chackalamannil S.; Rotella D.; Ward S. E., Eds.; Elsevier, 2017; pp 186–210. [Google Scholar]
- Lima P. C.; Lima L. M.; Silva K. C. M.; Léda P. H. O.; Miranda A. L. P.; Fraga C. A. M.; Barreiro E. J. Synthesis and Analgesic Activity of Novel N-Acylarylhydrazones and Isosters, Derived from Natural Safrole. Eur. J. Med. Chem. 2000, 35, 187–203. 10.1016/S0223-5234(00)00120-3. [DOI] [PubMed] [Google Scholar]
- Lima L. M.; Amarante E. G.; Miranda A. L. P.; Fraga C. A. M.; Barreiro E. J. Synthesis and Antinociceptive Profile of Novel Acidic Sulphonylhydrazone Derivatives From Natural Safrole. Pharm. Pharmacol. Commun. 1999, 5, 673–678. 10.1211/146080899128734370. [DOI] [Google Scholar]
- de Oliveira K. N.; Souza M. M.; Sathler P. C.; Magalhães U. O.; Rodrigues C. R.; Castro H. C.; Palm P. R.; Sarda M.; Perotto P. E.; Cezar S.; de Brito M. A.; Ferreira A. S. S. R.; Cabral L. M.; Machado C.; Nunes R. J. Sulphonamide and Sulphonyl-Hydrazone Cyclic Imide Derivatives: Antinociceptive Activity, Molecular Modeling and In Silico ADMET Screening. Arch. Pharm. Res. 2012, 35, 1713–1722. 10.1007/s12272-012-1002-1. [DOI] [PubMed] [Google Scholar]
- Nunes I. K. d. C.; de Souza E. T.; Martins I. R. R.; Barbosa G.; Moraes Junior M. O. d.; Medeiros M. d. M.; Silva S. W. D.; Balliano T. L.; da Silva B. A.; Silva P. M. R.; Carvalho V.; de F.; Martins M. A.; Lima L. M. Discovery of Sulfonyl Hydrazone Derivative as a New Selective PDE4A and PDE4D Inhibitor by Lead-Optimization Approach on the Prototype LASSBio-448: In Vitro and in Vivo Preclinical Studies. Eur. J. Med. Chem. 2020, 204, 112492 10.1016/j.ejmech.2020.112492. [DOI] [PubMed] [Google Scholar]
- Karaman N.; Oruç-Emre E. E.; Sıcak Y.; Çatıkkaş B.; Karaküçük-İyidoğan A.; Öztürk M. Microwave-Assisted Synthesis of New Sulfonyl Hydrazones, Screening of Biological Activities and Investigation of Structure–Activity Relationship. Med. Chem. Res. 2016, 25, 1590–1607. 10.1007/s00044-016-1592-0. [DOI] [Google Scholar]
- Popiołek Ł.; Szeremeta S.; Biernasiuk A.; Wujec M. Novel 2,4,6-Trimethylbenzenesulfonyl Hydrazones with Antibacterial Activity: Synthesis and In Vitro Study. Materials 2021, 14, 2723. 10.3390/ma14112723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gündüzalp A. B.; Özmen Ü. Ö.; Çevrimli B. S.; Mamaş S.; Çete S. Synthesis, Characterization, Electrochemical Behavior, and Antimicrobial Activities of Aromatic/Heteroaromatic Sulfonylhydrazone Derivatives. Med. Chem. Res. 2014, 23, 3255–3268. 10.1007/s00044-013-0907-7. [DOI] [Google Scholar]
- de Oliveira K. N.; Costa P.; Santin J. R.; Mazzambani L.; Bürger C.; Mora C.; Nunes R. J.; de Souza M. M. Synthesis and Antidepressant-like Activity Evaluation of Sulphonamides and Sulphonyl-Hydrazones. Bioorg. Med. Chem. 2011, 19, 4295–4306. 10.1016/j.bmc.2011.05.056. [DOI] [PubMed] [Google Scholar]
- Yang K.; Yang J.-Q.; Luo S.-H.; Mei W.-J.; Lin J.-Y.; Zhan J.-Q.; Wang Z.-Y. Synthesis of N-2(5H)-Furanonyl Sulfonyl Hydrazone Derivatives and Their Biological Evaluation in Vitro and in Vivo Activity against MCF-7 Breast Cancer Cells. Bioorg. Chem. 2021, 107, 104518 10.1016/j.bioorg.2020.104518. [DOI] [PubMed] [Google Scholar]
- Şenkardeş S.; Han M. İ.; Kulabaş N.; Abbak M.; Çevik Ö.; Küçükgüzel İ.; Küçükgüzel Ş. G. Synthesis, Molecular Docking and Evaluation of Novel Sulfonyl Hydrazones as Anticancer Agents and COX-2 Inhibitors. Mol. Diversity 2020, 24, 673–689. 10.1007/s11030-019-09974-z. [DOI] [PubMed] [Google Scholar]
- Wei D.-C.; Pan Y.; Wang H.; Xu W.-J.; Chen C.; Zheng J.-H.; Cai D. Synthesis of Substituted Aromatic Heterocyclic Sulfonyl Hydrazone Compounds and in Vitro Anti-Hepatoma Activity: Preliminary Results. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 4720–4729. 10.26355/eurrev_201807_15532. [DOI] [PubMed] [Google Scholar]
- Zapata-Sudo G.; Lima L. M.; Pereira S. L.; Trachez M. M.; Costa F. P.; Souza B. J.; Monteiro C. E. S.; Romeiro N. C.; D’Andrea E. D.; Sudo R. T.; Barreiro E. J. Docking, Synthesis and Anti-Diabetic Activity of Novel Sulfonylhydrazone Derivatives Designed as PPAR-Gamma Agonists. Curr. Top. Med. Chem. 2012, 12, 2037–2048. 10.2174/156802612804910205. [DOI] [PubMed] [Google Scholar]
- Lima L. M.; Zapata-Sudo G.; da Costa Nunes I. K.; Segundo Chaves de Araujo J.; da Silva J.; Manhães Trachez M.; Fernandes da Silva T.; P da Costa F.; Sudo R.; Barreiro E. Synthesis, Solubility, Plasma Stability, and Pharmacological Evaluation of Novel Sulfonylhydrazones Designed as Anti-Diabetic Agents. Drug Des., Dev. Ther. 2016, Volume 10, 2869–2879. 10.2147/DDDT.S108327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima L. M.; Trachez M. M.; Araujo J. S. C.; Silva J. S.; Amaral D. N.; Sudo R. T.; Barreiro E. J.; Zapata-Sudo G. Novel Partial Agonist of PPAR-Gamma for Treatment of Diabetic Neuropathy in Rats. J. Diabetes Metab. 2014, 05, 2. 10.4172/2155-6156.1000392. [DOI] [Google Scholar]
- Barreiro E. J.; Kümmerle A. E.; Fraga C. A. M. The Methylation Effect in Medicinal Chemistry. Chem. Rev. 2011, 111, 5215–5246. 10.1021/cr200060g. [DOI] [PubMed] [Google Scholar]
- Meunier B.; de Visser S. P.; Shaik S. Mechanism of Oxidation Reactions Catalyzed by Cytochrome P450 Enzymes. Chem. Rev. 2004, 104, 3947–3980. 10.1021/cr020443g. [DOI] [PubMed] [Google Scholar]
- Deodhar M.; Al Rihani S. B.; Arwood M. J.; Darakjian L.; Dow P.; Turgeon J.; Michaud V. Mechanisms of CYP450 Inhibition: Understanding Drug-Drug Interactions Due to Mechanism-Based Inhibition in Clinical Practice. Pharmaceutics 2020, 12, 846. 10.3390/pharmaceutics12090846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana E.; Dansette P.; Poli S. Cytochrome P450 Enzymes Mechanism Based Inhibitors: Common Sub-Structures and Reactivity. Curr. Drug Metab. 2005, 6, 413–451. 10.2174/138920005774330639. [DOI] [PubMed] [Google Scholar]
- Ibiapino A. L.; de Figueiredo L. P.; Lima L. M.; Barreiro E. J.; Punzo F.; Ferreira F. F. Structural and Physicochemical Characterization of Sulfonylhydrazone Derivatives Designed as Hypoglycemic Agents. New J. Chem. 2017, 41, 6464–6474. 10.1039/C7NJ00074J. [DOI] [Google Scholar]
- Fortuna A.; Alves G.; Soares-Da-Silva P.; Falcão A. Optimization of a Parallel Artificial Membrane Permeability Assay for the Fast and Simultaneous Prediction of Human Intestinal Absorption and Plasma Protein Binding of Drug Candidates: Application to Dibenz[b,f]Azepine-5-Carboxamide Derivatives. J. Pharm. Sci. 2012, 101, 530–540. 10.1002/jps.22796. [DOI] [PubMed] [Google Scholar]
- Rodrigues D. A.; Ferreira-Silva G. À.; Ferreira A. C. S.; Fernandes R. A.; Kwee J. K.; Sant’Anna C. M. R.; Ionta M.; Fraga C. A. M. Design, Synthesis, and Pharmacological Evaluation of Novel N -Acylhydrazone Derivatives as Potent Histone Deacetylase 6/8 Dual Inhibitors. J. Med. Chem. 2016, 59, 655–670. 10.1021/acs.jmedchem.5b01525. [DOI] [PubMed] [Google Scholar]
- Sugano K.; Hamada H.; Machida M.; Ushio H.; Saitoh K.; Terada K. Optimized Conditions of Bio-Mimetic Artificial Membrane Permeation Assay. Int. J. Pharm 2001, 228, 181–188. 10.1016/S0378-5173(01)00845-6. [DOI] [PubMed] [Google Scholar]
- Di L.; Kerns E. H.; Fan K.; McConnell O. J.; Carter G. T. High Throughput Artificial Membrane Permeability Assay for Blood–Brain Barrier. Eur. J. Med. Chem. 2003, 38, 223–232. 10.1016/S0223-5234(03)00012-6. [DOI] [PubMed] [Google Scholar]
- Pérez D. I.; Pistolozzi M.; Palomo V.; Redondo M.; Fortugno C.; Gil C.; Felix G.; Martinez A.; Bertucci C. 5-Imino-1,2-4-Thiadiazoles and Quinazolines Derivatives as Glycogen Synthase Kinase 3β (GSK-3β) and Phosphodiesterase 7 (PDE7) Inhibitors: Determination of Blood–Brain Barrier Penetration and Binding to Human Serum Albumin. Eur. J. Pharm. Sci. 2012, 45, 677–684. 10.1016/j.ejps.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Miller R. R.; Madeira M.; Wood H. B.; Geissler W. M.; Raab C. E.; Martin I. J. Integrating the Impact of Lipophilicity on Potency and Pharmacokinetic Parameters Enables the Use of Diverse Chemical Space during Small Molecule Drug Optimization. J. Med. Chem. 2020, 63, 12156–12170. 10.1021/acs.jmedchem.9b01813. [DOI] [PubMed] [Google Scholar]
- Zhu C.; Jiang L.; Chen T.-M.; Hwang K.-K. A Comparative Study of Artificial Membrane Permeability Assay for High Throughput Profiling of Drug Absorption Potential. Eur. J. Med. Chem. 2002, 37, 399–407. 10.1016/S0223-5234(02)01360-0. [DOI] [PubMed] [Google Scholar]
- Perry M. d. J.; Carvalho E.; Rosa E.; Iley J. Towards an Efficient Prodrug of the Alkylating Metabolite Monomethyltriazene: Synthesis and Stability of N-Acylamino Acid Derivatives of Triazenes. Eur. J. Med. Chem. 2009, 44, 1049–1056. 10.1016/j.ejmech.2008.06.022. [DOI] [PubMed] [Google Scholar]
- Alves M. A.; de Queiroz A. C.; Alexandre-Moreira M. S.; Varela J.; Cerecetto H.; González M.; Doriguetto A. C.; Landre I. M.; Barreiro E. J.; Lima L. M. Design, Synthesis and in Vitro Trypanocidal and Leishmanicidal Activities of Novel Semicarbazone Derivatives. Eur. J. Med. Chem. 2015, 100, 24–33. 10.1016/j.ejmech.2015.05.046. [DOI] [PubMed] [Google Scholar]
- Cabrera M.; Lavaggi M. L.; Hernández P.; Merlino A.; Gerpe A.; Porcal W.; Boiani M.; Ferreira A.; Monge A.; de Cerain A. L.; González M.; Cerecetto H. Cytotoxic, Mutagenic and Genotoxic Effects of New Anti-T. Cruzi 5-Phenylethenylbenzofuroxans. Contribution of Phase I Metabolites on the Mutagenicity Induction. Toxicol. Lett. 2009, 190, 140–149. 10.1016/j.toxlet.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Smith P. K.; Krohn R. I.; Hermanson G. T.; Mallia A. K.; Gartner F. H.; Provenzano M. D.; Fujimoto E. K.; Goeke N. M.; Olson B. J.; Klenk D. C. Measurement of Protein Using Bicinchoninic Acid. Anal. Biochem. 1985, 150, 76–85. 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Costa Nunes I.; Tinoco L.; Martins-Junior H.; Rezende C.; Barreiro E.; Lima L. In Vitro Microsomal Hepatic Metabolism of Antiasthmatic Prototype LASSBio-448. Curr. Top. Med. Chem. 2014, 14, 1388–1398. 10.2174/1568026614666140506123718. [DOI] [PubMed] [Google Scholar]
- Fraga A. G. M.; da Silva L. L.; Fraga C. A. M.; Barreiro E. J. CYP1A2-Mediated Biotransformation of Cardioactive 2-Thienylidene-3,4-Methylenedioxybenzoylhydrazine (LASSBio-294) by Rat Liver Microsomes and Human Recombinant CYP Enzymes. Eur. J. Med. Chem. 2011, 46, 349–355. 10.1016/j.ejmech.2010.11.024. [DOI] [PubMed] [Google Scholar]
- Food and Drug Administration, Bioanalytical Method Validation Guidance for Industry Online. 2018, https://www.fda.gov/files/drugs/published/Bioanalytical-Method-Validation-Guidance-for-Industry.pdf (accessed 15 Sep, 2021).
- Ministério da Saúde, RESOLUÇÃO - RDC No. 27, DE 17 DE MAIO DE 2012 online. Brazil: ANVISA-Ministério da Saúde; 2012-2021, 2012, https://bvsms.saude.gov.br/bvs/saudelegis/anvisa/2012/rdc0027_17_05_2012.html (accessed 15 Sep, 2021).
- Büch H.; Buzello W.; Heymann E.; Krisch K. Inhibition of Phenacetin- and Acetanilide-Induced Methemoglobinemia in the Rat by the Carboxylesterase Inhibitor Bis-[p-Nitrophenyl] Phosphate. Biochem. Pharmacol. 1969, 18, 801. 10.1016/0006-2952(69)90050-1. [DOI] [PubMed] [Google Scholar]
- Kim M.-J.; Jeong E. S.; Park J.-S.; Lee S.-J.; Ghim J. L.; Choi C.-S.; Shin J.-G.; Kim D.-H. Multiple Cytochrome P450 Isoforms Are Involved in the Generation of a Pharmacologically Active Thiol Metabolite, Whereas Paraoxonase 1 and Carboxylesterase 1 Catalyze the Formation of a Thiol Metabolite Isomer from Ticlopidine. Drug Metab. Dispos. 2014, 42, 141–152. 10.1124/dmd.113.053017. [DOI] [PubMed] [Google Scholar]
- Taniguchi-Takizawa T.; Shimizu M.; Kume T.; Yamazaki H. Benzydamine N-Oxygenation as an Index for Flavin-Containing Monooxygenase Activity and Benzydamine N-Demethylation by Cytochrome P450 Enzymes in Liver Microsomes from Rats, Dogs, Monkeys, and Humans. Drug Metab. Pharmacokinet. 2015, 30, 64–69. 10.1016/j.dmpk.2014.09.006. [DOI] [PubMed] [Google Scholar]
- Hamman M. A.; Haehner-Daniels B. D.; Wrighton S. A.; Rettie A. E.; Hall S. D. Stereoselective Sulfoxidation of Sulindac Sulfide by Flavin-Containing Monooxygenases. Biochem. Pharmacol. 2000, 60, 7–17. 10.1016/S0006-2952(00)00301-4. [DOI] [PubMed] [Google Scholar]
- Kot M.; Daniel W. A. Relative Contribution of Rat Cytochrome P450 Isoforms to the Metabolism of Caffeine: The Pathway and Concentration Dependence. Biochem. Pharmacol. 2008, 75, 1538–1549. 10.1016/j.bcp.2007.12.017. [DOI] [PubMed] [Google Scholar]
- Bocato M. Z.; de Lima Moreira F.; de Albuquerque N. C. P.; de Gaitani C. M.; de Oliveira A. R. M. In Vitro Enantioselective Human Liver Microsomal Metabolism and Prediction of in Vivo Pharmacokinetic Parameters of Tetrabenazine by DLLME-CE. J. Pharm. Biomed. Anal. 2016, 128, 528–537. 10.1016/j.jpba.2016.06.028. [DOI] [PubMed] [Google Scholar]
- Yamagata S.; Ohmori S.; Suzuki N.; Yoshino M.; Hino M.; Ishii I.; Kitada M. Metabolism of Dacarbazine by Rat Liver Microsomes Contribution of CYP1A Enzymes to Dacarbazine N-Demethylation. Drug Metab. Dispos. 1998, 26, 379–382. [PubMed] [Google Scholar]
- Madan A.; Fisher A.; Jin L.; Chapman D.; Bozigian H. P. In Vitro Metabolism of Indiplon and an Assessment of Its Drug Interaction Potential. Xenobiotica 2007, 37, 736–752. 10.1080/00498250701433054. [DOI] [PubMed] [Google Scholar]
- Zhou H.; Shi R.; Ma B.; Ma Y.; Wang C.; Wu D.; Wang X.; Cheng N. CYP450 1A2 and Multiple UGT1A Isoforms Are Responsible for Jatrorrhizine Metabolism in Human Liver Microsomes: METABOLISM OF JATRORRHIZINE IN HUMAN LIVER MICROSOMES. Biopharm. Drug Dispos. 2013, 34, 176–185. 10.1002/bdd.1835. [DOI] [PubMed] [Google Scholar]
- Jing W.-H.; Song Y.-L.; Yan R.; Wang Y.-T. Identification of Cytochrome P450 Isoenzymes Involved in Metabolism of (+)-Praeruptorin A, a Calcium Channel Blocker, by Human Liver Microsomes Using Ultra High-Performance Liquid Chromatography Coupled with Tandem Mass Spectrometry. J. Pharm. Biomed. Anal. 2013, 77, 175–188. 10.1016/j.jpba.2013.01.023. [DOI] [PubMed] [Google Scholar]
- Reynald R. L.; Sansen S.; Stout C. D.; Johnson E. F. Structural Characterization of Human Cytochrome P450 2C19. J. Biol. Chem. 2012, 287, 44581–44591. 10.1074/jbc.M112.424895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano K.; Hamada H.; Machida M.; Ushio H. High Throughput Prediction of Oral Absorption: Improvement of the Composition of the Lipid Solution Used in Parallel Artificial Membrane Permeation Assay. SLAS Discovery 2001, 6, 189–196. 10.1177/108705710100600309. [DOI] [PubMed] [Google Scholar]
- Kansy M.; Senner F.; Gubernator K. Physicochemical High Throughput Screening: Parallel Artificial Membrane Permeation Assay in the Description of Passive Absorption Processes. J. Med. Chem. 1998, 41, 1007–1010. 10.1021/jm970530e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.