Abstract
Cysteine proteinases expressed by schistosomes appear to play key roles in the digestion of host hemoglobin, the principal source of amino acid nutrients utilized by these parasites. We have shown previously that the predominant cysteine proteinase activity in soluble extracts and excretory/secretory (ES) products of adults of Schistosoma mansoni and S. japonicum is cathepsin L-like in its substrate specificity. However, biochemical analysis of the cathepsin L activity in extracts and ES products of schistosomes has been complicated by the presence of at least two distinct forms of schistosome cathepsin L, termed SmCL1 and SmCL2. We now report the purification and enzyme characteristics of active, recombinant SmCL1 which was obtained by transforming Saccharomyces cerevisiae with an expression plasmid encoding the preproenzyme of SmCL1. Recombinant SmCL1 was secreted by the transformed yeast into the culture media from which it was purified by gel filtration and ion-exchange chromatography. The purified enzyme exhibited substrate specificity against synthetic peptidyl substrates (e.g., Boc-Val-Leu-Lys-NHMec and Z-Phe-Arg-NHMec; kcat/Km = 17.25 and 6.24 mM−1 s−1, respectively) and against gelatin and hemoglobin, characteristic of cathepsin L. Immunoblot analysis using antiserum raised against recombinant SmCL1 demonstrated that native SmCL1 of 33 kDa was present in ES products and soluble extracts of S. mansoni. Using this antiserum and thin tissue sections, we localized the native SmCL1 to the gastrodermis and to the tegument of adult schistosomes. Recombinant SmCL1 was capable of degrading human hemoglobin at pH 4.0 to 4.5 but not higher, suggesting that denaturation of hemoglobin by low pH, as found in the cecum of the adult schistosome, may be necessary for its catalysis by cathepsin L and other gut-associated proteinases. Together, these results support a role for SmCL1 in the degradation of host hemoglobin within the gut of the schistosome.
Schistosomiasis afflicts more than 250 million people in tropical and subtropical regions. The disease is caused by blood flukes of the genus Schistosoma, and infection is acquired in contaminated water, where cercariae penetrate the skin. After migrating through the lungs and liver, the developing Schistosoma japonicum and S. mansoni parasites take up residence in the mesenteric veins, where male and female worms mature and reproduce. Each day, female schistosomes produce numerous eggs which move through the intestinal wall into the lumen of the bowel and are shed with the feces. The pathology associated with schistosomiasis caused by S. japonica and S. mansoni is primarily a consequence of inflammatory responses to eggs inadvertently carried to the liver and other sites (17).
Cysteine proteinases, including cathepsin L-like and cathepsin B-like proteinases, are considered important targets to which novel antischistosome chemotherapy and/or immunoprophylaxis could be directed (4, 9, 18). These enzymes appear to be involved in the degradation of host hemoglobin, the main source of nutrient used by schistosomules and adult worms (8, 14). Both activities are secreted by adult schistosomes (7), and tissue localization studies have indicated their presence in the gastrodermal cells lining the cecum of the parasite (25). Inhibitors of cysteine proteinases were shown to prevent hemoglobin digestion by schistosomula and decrease their viability in vitro (23, 25). Moreover, treatment of S. mansoni-infected mice with these inhibitors not only reduced worm burden but exhibited antifecundity effects (23).
Elucidation of the precise physiological role of the cathepsin L-like and cathepsin B-like proteinases of schistosomes has been hampered by the difficulty of obtaining homogeneous enzymes. The enzymes have similar molecular sizes and substrate specificities, and past biochemical studies appear to have been performed on enzyme mixtures (4). Furthermore, we and others have shown that schistosomes express at least two distinct cathepsin L proteinases, termed S. mansoni cathepsin L1 (SmCL1) and SmCL2 (10, 19, 21). Michel et al. have demonstrated that SmCL2 is expressed in the reproductive organs of S. mansoni (19), and thus we consider that it is unlikely to play a role in hemoglobin degradation in the gastrodermis or cecum of the schistosome.
We have recently described a system for obtaining functionally active cathepsin L proteinases of the digenean trematode Fasciola hepatica by expressing cDNAs encoding preprocathepsin L in the brewer’s yeast, Saccharomyces cerevisiae (12, 20). The recombinant F. hepatica proteinases were produced and processed by the yeast to their mature forms, thereby obviating the need for protein refolding and/or activation steps. Using the same approach, we now report the recombinant expression of SmCL1 cDNA (21). We purified recombinant SmCL1 from yeast culture supernatants and characterized its activity against a panel of synthetic substrates, gelatin, and human hemoglobin. Immunolocalization studies using antiserum raised against recombinant SmCL1 showed that native SmCL1 was present in the gastrodermal cells lining the cecum of adult worms and at other sites, and immunoblotting studies detected the enzyme in excretory/secretory (ES) products. Together, these results support a role for SmCL1 in the degradation of host hemoglobin within the gut of the schistosome.
MATERIALS AND METHODS
Synthetic peptidyl substrates and inhibitors.
Boc-Val-Leu-Lys-NHMec (Boc, t-butyloxycarbonyl; NHMec, 7-amino-4-methyl coumarin), benzyloxycarbonyl (Z)-Phe-Ala-diazomethylketone (CHN2), 1-3-carboxy-2-3-trans-epoxypropionyl-leucylamido(4-guanido)-butane (E-64), dithiothreitol, and l-cysteine were obtained from Sigma Chemical Co. Z-Phe-Arg-NHMec, Z-Phe-Val-Arg-NHMec, Z-Arg-Arg-NHMec, Z-Arg-NHMec, and tosyl (Tos)-Gly-Pro-Arg-NHMec were purchased from Bachem.
Schistosome extracts.
Soluble extracts of S. mansoni cercariae, separate-sex adults, and media containing cysteine proteinases secreted by cultured adult worms (ES products) were prepared as previously described (7, 8).
Cloning of SmCL1 and yeast expression plasmid construction.
Isolation and characterization of the cDNA encoding the complete preprocathepsin SmCL1 (GenBank accession no. U07345) have been described previously (10, 21). The cDNA encoding the SmCL1 prepro enzyme was amplified by PCR using two primers designed to anneal to the 5′ and 3′ termini of the cDNA: SmCL1F (CGCAAGCTTATGCCTGTAAACCTCGAGTAC) and SmCL1R (CGCAAGCTTCCCCTAGTAGATCATCGCTGA). The primers included HindIII recognition sequences (underlined). The amplified fragments were cloned into pGemT (Promega). (Nucleotide sequencing of the recombinant plasmid verified that the sequence had not been mutated during the PCR.) The plasmid insert was excised with HindIII and ligated into the yeast expression plasmid pAAH5 (1, 20) (kindly provided by J. R. Dickinson, University of Wales, Cardiff, United Kingdom), linearized with HindIII. pAAH5 is a shuttle vector with the yeast replication region of the 2μm circle and the Escherichia coli replication region of pBR322. The HindIII cloning site is flanked at the 5′ side by the promoter and the untranslated leader of the yeast alcohol dehydrogenase gene ADC1 containing the ribosome-binding site (1). The SmCL1 insert provided the translation and termination codons. The signals required for posttranslational processing and intracellular sorting of the proenzyme are encoded by the prosegment-encoding sequences within the SmCL1 gene (7, 10, 21). An internal EcoRI site within the SmCL1 cDNA was used to determine the correct orientation of the SmCL1 insert within the vector. A clone with the correct orientation for expression was isolated and named pAAH5.SmCL1. E. coli MC1061 was used for propagating pAAH5 and recombinant constructs.
Transformation and culturing of S. cerevisiae.
S. cerevisiae DBY746 (Mata his3-D1-leu2-3 leu2-112 ura3-52 trp1-289a) (Yeast Genetic Stock Center, Department of Biophysics and Medical Physics, University of California, Berkeley) was routinely maintained in complex medium (YEPD); 10 g of yeast extract, 20 g of peptone, and 20 g of d-glucose per liter). S. cerevisiae cells were transformed with pAAH5.SmCL1 in the presence of lithium acetate (6). Yeast transformants were cultured in selective minimal medium (6.7 g of Bacto Yeast Nitrogen Base [lacking leucine but containing histidine and tryptophan; Difco], 10 g of d-glucose, and 20 mg of uracil, per liter in 0.1 M sodium citrate, pH 5.5). (The selection marker on pAAH5 is Leu2 [1].) For expression of recombinant yeast-expressed SmCL1 (ySmCL1), yeast cells were grown in an automative fermentor (New Brunswick model 101) in selective minimal medium at 30°C until the optical density at 600 nm reached 1.4. Yeast cells were removed by centrifugation, and the supernatant was stored at 4°C.
Purification of ySmCL1.
Five liters of pAAH5.SmCL1-transformed yeast supernatant was concentrated at 4°C to 20 ml in an Amicon 2000A concentrator, using an Amicon YM3 membrane (3,000-Da molecular mass cutoff). The concentrate was applied to a Sephacryl S200 HR (Pharmacia) gel filtration column (2.6 by 74.5 cm) equilibrated in 0.1 M Tris-HCl (pH 7.0) at 4°C. Proteins were eluted from the matrix with 0.1 M Tris-HCl (pH 7.0), and fractions (5 ml) containing cathepsin L activity, measured by using the fluorogenic substrate Z-Phe-Arg-NHMec (see below), were pooled. The pooled fractions were concentrated to 3 ml, dialyzed against 20 mM Tris-HCl (pH 7.0), and applied to a QAE-Sephadex A50 column (10 cm by 2.5 cm; Pharmacia), equilibrated in the same buffer. The column was washed with the equilibration buffer (375 ml), after which bound molecules were eluted on a 0 to 500 mM NaCl gradient. Fractions (5 ml) containing cathepsin L activity were pooled, concentrated as described above, and stored at −20°C.
Characterization of ySmCL1 proteinase activity.
Cathepsin L proteinase activity was characterized by using peptidyl-NHMec as substrates (below). These substrates were stored as a 1-mg/100-μl stock solution in dimethylformamide. Assays were carried out with a final concentration of 10 μM substrate in 0.1 M sodium phosphate buffer, pH 6.5, containing 1 mM dithiothreitol, in a volume of 1 ml. The solutions were incubated at 37°C for 1 h before the reaction was terminated by the addition of 0.2 ml of 1.7 M acetic acid. The amount of NHMec released was measured with a fluorimeter (370-nm excitation and 440-nm emission). One unit of enzyme activity was defined as that amount which catalyzed the release of 1 μmol of NHMec/min at 37°C.
Substrate specificity and kinetics of purified ySmCL1 were determined with the peptide substrates Z-Phe-Arg-NHMec, Z-Phe-Val-Arg-NHMec, Z-Arg-Arg-NHMec, Z-Arg-NHMec, Z-Gly-Pro-Arg-NHMec, and Boc-Val-Leu-Lys-NHMec. The kinetic constants, kcat and Km were obtained by nonlinear regression analysis using the Enzfitter program (15). Active-site titration using the cysteine proteinase inhibitor E-64 and the fluorogenic substrate Z-Phe-Arg-NHMec was performed to determine the molar concentration of active ySmCL1, using the method of Barrett et al. (2). For determination of the optimum pH of proteinase activity, the following buffers were used at a concentration of 50 mM: glycine, pH 2.5 to 3.0 and 9.1 to 10.0; sodium acetate, pH 3.5 to 5.5; sodium phosphate, pH 5.5 to 7.5; and Tris-HCl, pH 7.5 to 9.0. The ionic strength of each buffer was equalized to 100 mM by using NaCl.
Expression of recombinant SmCL1 in E. coli and preparation of rabbit anti-SmCL1 serum.
A cDNA encoding the mature SmCL1 was ligated into the E. coli expression vector pQE30 (Qiagen, Chatsworth, Calif.) and used to transform E. coli M15 as previously described by Dalton et al. (7). LB medium containing ampicillin (100 μg/ml) and kanamycin (25 μg/ml) was inoculated with transformed cells and incubated at 37°C with shaking until the optical density at 600 nm reached 0.8. Expression of recombinant bacterium-expressed SmCL1 (bSmCL1) was induced by addition of isopropyl-1-thio-β-d-galactopyranoside to 1 mM, and the cells were harvested 5 h later by centrifugation. The cell pellet was resuspended in 0.1 M sodium phosphate–0.01 M Tris-HCl (pH 8.0) containing 6 M guanidine hydrochloride at 5 ml per g of cell pellet and sonicated for 8 min (duty cycle, 25%; output, 2.5) (Branson Sonifier 250; Branson Ultrasonics) to disrupt bacterial cells. The extract was centrifuged at 14,000 × g for 30 min, and the supernatant was incubated with 2 ml of Ni-nitrilotriacetic acid (NTA) Superflow resin (Qiagen) for 1 h at room temperature. The resin was packed into a column and washed with 5 volumes of 0.1 M sodium phosphate–0.01 M Tris-HCl (pH 7.2) containing 8 M urea. Recombinant bSmCL1 was eluted with a linear gradient of imidazole, prepared at 250 mM in the last buffer, at 0.5 ml/min over 50 ml. One-milliliter fractions were collected and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting with a monoclonal antibody specific for the R-G-S-H-H-H-H epitope (Qiagen) engineered onto the C terminus of the recombinant protein expressed in pQE30. Purified bSmCL1 was used as antigen to raise an anti-bSmCL1 serum in a New Zealand White rabbit. The rabbit was immunized five times with 20 μg of bSmCL1 in QuilA adjuvant (Superfos Biosector, Frederikssund, Denmark) with intervals of 3 weeks between boosts. Antibodies in the serum of these immunized rabbits reacted with recombinant SmCL1 but not SmCL2 in immunoblotting experiments (data not shown).
SDS-PAGE analysis, zymography, immunoblotting, and glycosylation studies.
Native and recombinant schistosome proteins were analyzed by SDS-PAGE (12% gel) under reducing conditions as described by Dalton et al. (7). Both zymographic analysis using gels containing copolymerized gelatin and immunoblotting were performed with anti-bSmCL1 serum as previously described (7, 10, 11, 22). Glycoproteins were detected by using a DIG Glycan detection kit (Boehringer, Mannheim, Germany) in which transferrin and creatinase were used as positive and negative controls, respectively. Protein concentrations were measured by using a DC protein assay kit from Bio-Rad.
Hemoglobin proteolysis.
Hemoglobin was prepared as described previously (3). Hemoglobin (150 μg) was incubated with ySmCL1 (20 μg) at 37°C for 18 h in the presence of 1 mM dithiothreitol. Digestions were carried out in the following buffers: 0.1 M sodium acetate, pH 4.0 and 4.5; 0.1 M sodium citrate, pH 5.0 and 5.5; and 0.1 M sodium phosphate, pH 6.0 and 6.5. The ionic strength of each buffer was equalized to 100 mM by using NaCl. Following the incubation, the reaction products were analyzed by SDS-PAGE (15% gel) under nonreducing conditions and staining with Coomassie brilliant blue R250.
Immunolocalization of SmCL1 in adult worms.
Mixed-sex adult worms were perfused from mice and then embedded in Tissue-Tek O.C.T. medium (Sakura Finetek, Torrance, Calif.), after which 10-μm sections were cut with a cryostat microtome. The sections were mounted on glass slides and air dried for 4 h. Sections were fixed in ice-cold acetone for 2 min, washed in phosphate-buffered saline (PBS), and incubated in goat normal serum diluted 1:5 in PBS for 30 min to inhibit nonspecific background with the secondary antibody (below). After being washed in PBS, sections were incubated in rabbit anti-bSmCL1 or control (preimmunization) serum diluted 1:20 in PBS–1% bovine serum albumin for 1 h. Sections were washed in PBS and incubated in goat anti-rabbit IgG conjugated to fluorescein isothiocyanate (Calbiochem, La Jolla, Calif.) diluted 1:100 in PBS–1% bovine serum albumin. (The conjugated antibody had been preadsorbed to bovine, horse, human, and mouse sera by the manufacturer.) All incubations were performed at room temperature. After further washing in PBS, sections were mounted in Crystal/Mount medium (Biomeda, Foster City, Calif.), viewed under UV light on an Olympus BX60 microscope, and photographed.
RESULTS
Recombinant SmCL1 expressed in yeast and bacteria.
Ten positive yeast clones transformed with pAAH5.SmCL1 were selected and tested for secretion of proteinase activity into culture media with Z-Phe-Arg-NHMec as the substrate (see below). All 10 secreted proteinase at similar levels, but the clone producing most enzyme activity was used for subsequent fermentations. By contrast, medium in which yeast transformed with nonrecombinant pAAH5 were cultured did not contain Z-Phe-Arg-NHMec-cleaving activity (data not shown). The concentrate from a 5-liter fermentation using the positive clone (above) exhibited potent activity against Z-Phe-Arg-NHMec. This activity was enhanced by dithiothreitol and completely blocked by E-64 (5 μM) and by the cathepsin L and cathepsin B-specific inhibitor Z-Phe-Ala-CHN2 (5 μM) (data not shown). These results showed that the pAAH5.SmCL1-transformed yeast secreted cathepsin L-like cysteine proteinase activity. The specific activity of this proteinase in the culture supernatant was 0.06 U/mg of protein (Table 1).
TABLE 1.
Chromatographic purification of recombinant ySmCL1 on gel filtration and ion-exchange matrices
| Prepn | Protein (mg) | Activity (U) | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Culture medium | 53.1 | 3.24 | 0.06 | 100 | 1 |
| S200 HR | |||||
| Peak I | 12.52 | 2.12 | 0.169 | 65.4 | 2.82 |
| Peak II | 2.03 | 1.12 | 0.552 | 34.6 | 30.2 |
| QAE-Sepadex poola | 0.18 | 0.45 | 2.5 | 14.0 | 42.0 |
Only proteinase activity in the S200 HR peak II was applied to the QAE-Sephadex column.
The mature form of SmCL1 expressed in E. coli was isolated from inclusion bodies under denaturing conditions by affinity chromatography on nickel chelate (Ni-NTA) resin. SDS-PAGE analysis indicated that a protein which migrated at ∼24 kDa was eluted from the Ni-NTA resin and was >90% homogeneous (data not shown). The recombinant protein reacted strongly on immunoblots with the monoclonal antibody to the polyhistidine ligand (not shown), demonstrating, based on this reactivity and its predicted size of 24 kDa, that it was the recombinant, mature form of SmCL1 (not shown). Recombinant bSmCL1 was used as the antigen to prepare a monospecific rabbit antiserum to SmCL1, which strongly recognized bSmCL1 at a dilution of 1:2,000 in immunoblots (not shown).
Purification of ySmCL1.
ySmCL1 was purified from yeast culture media by gel filtration followed by ion-exchange chromatography. Z-Phe-Arg-NHMec-cleaving activity resolved as two peaks on the S200 HR matrix. Fractions corresponding to both peaks were separately pooled. Although the total activity was greater in the first peak (peak I) than in the second peak (peak II), further purification was performed with the enzyme pool of peak II since it contained proteinase with much higher specific activity (peak I, 0.169 U/mg; peak II, 0.552 U/mg) (Table 1). When the peak II activity was applied to the ion-exchange QAE-Sephadex matrix, the majority of the proteolytic activity failed to bind to the resin and was collected in the run-through fractions. Little proteolytic activity was subsequently eluted on the NaCl gradient (data not shown). The proteinase in the run-through from QAE-Sephadex exhibited a specific activity of 2.5 U/mg, which represented a 42-fold enrichment compared to the fermentation supernatant (Table 1).
We divided the run-through into three pools based on elution time from QAE-Sephadex and then examined the protein profile of the pools, along with the concentrated culture media, and peak II from S200 HR by Coomassie staining after SDS-PAGE (Fig. 1A). The gel demonstrated that we had enriched a protein of 45 kDa close to purity (Fig. 1A, lane 5) on the gel filtration followed by anion-exchange resins. Immunoblot analysis of the same preparations demonstrated that this 45-kDa protein reacted very strongly on immunoblots with the anti-bSmCL1 serum but did not react with control (preimmunization) serum, verifying its identity as recombinant SmCL1 (Fig. 1B and C). This preparation (Fig. 1A, lane 5) was used for the characterization of ySmCL1 detailed below.
FIG. 1.
Chromatographic purification of ySmCL1 on gel filtration and anion-exchange matrices. Ten to 20 μg of protein of concentrated culture medium (lane 1), S200 HR peak II (lane 2), and QAE-Sephadex run-through pool I (lane 3), pool II (lane 4), and pool III (lane 5) were separated by SDS-PAGE (12% gel) under reducing conditions. Gels were either stained with Coomassie brilliant blue R (A) or transferred to nitrocellulose and probed with rabbit anti-bSmCL1 serum (B) or control serum (C).
The molecular size of ySmCL1 was greater than the size predicted for the mature SmCL1 (24.1 kDa) or the proenzyme (35 kDa) (21). Since the SmCL1 sequence contains three potential N-linked glycosylation sites, the purified protein was tested for the presence of N-linked sugar residues by using an enzyme immunoassay-based glycan detection system. The recombinant protein showed a positive reaction for the presence of glucan (data not shown). Glycosylation of ySmCL1 may explain its slow migration on gels.
Purified ySmCL1 and soluble extracts of adult S. mansoni were analyzed for cysteine proteinase activity by gelatin-substrate PAGE under native conditions. Two prominent gelatinolytic bands were observed in extracts of adult S. mansoni parasites but were apparent only under reducing conditions when the gels were incubated in the presence of cysteine (Fig. 2A). We have previously attributed these activities in schistosome extracts to cathepsin L-like cysteine proteinases (7, 10). The proteinases are known to be also expressed by S. mansoni cercariae and schistosomules and to occur in ES products from cultures of adult schistosomes (7, 8). ySmCL1 resolved as a single band which migrated more slowly than the two proteinases in the schistosome extracts. The slower migration may be due to hyperglycosylation, as discussed above. Like the activities in the schistosome extracts, activity of ySmCL1 was enhanced markedly by the reducing agent, cysteine (Fig. 2B). By contrast, extracts and supernatants of nontransformed yeast did not exhibit gelatinolytic activity (data not shown). The optimum pH for the gelatinolytic activity of ySmCL1 was pH 6.5 (data not shown).
FIG. 2.
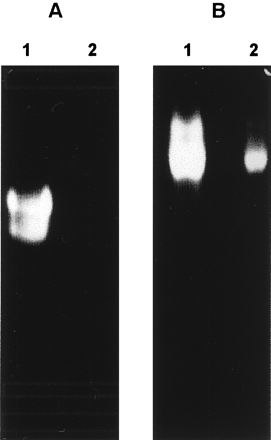
Gelatinolytic activity of ySmCL1. Soluble extracts of adult S. mansoni (A) and ySmCL1 (B) were analyzed by 10% gelatin-substrate PAGE (zymography) at pH 6.5 in the presence (lanes 1) and absence (lanes 2) of 10 mM cysteine.
Activity of ySmCL1 against synthetic peptides and hemoglobin.
The substrate specificity of the ySmCL1 was characterized by using fluorogenic peptide substrates (Table 2). Initial studies showed that the enzyme efficiently cleaved the cathepsin L- and cathepsin B-specific substrate Z-Phe-Arg-NHMec but exhibited minimal activity against Z-Arg-Arg-NHMec, a substrate diagnostic of cathepsin B, and against Z-Arg-NHMec, a cathepsin B and cathepsin H substrate (not shown). Analysis of reaction kinetics demonstrated that the enzyme cleaved Boc-Val-Leu-Lys-NHMec with greater efficiency (kcat/Km) than any of the other substrates examined, including Z-Phe-Arg-NHMec (Table 2). This observation is consistent with our earlier report of the substrate specificity of cathepsin L-like activity in soluble extracts of schistosomes (7–10, 21). In com-parison to Z-Phe-Arg-NHMec, Z-Phe-Val-Arg-NHMec, a substrate diagnostic of cathepsin S, was cleaved much less efficiently by recombinant SmCL1. ySmCL1 also cleaved Tos-Gly-Pro-Arg-NHMec, a substrate which we have shown can distinguish different classes of F. hepatica cathepsin L (11, 12, 20, 22), although the efficiency of cleavage was not as high as reported for F. hepatica cathepsin L2 (11, 12). ySmCL1 exhibited activity over a wide pH range (pH 3.5 to 10.0), although it exhibited a pH optimum for activity of 6.5 against the peptide substrates Z-Phe-Arg-NHMec and Boc-Val-Leu-Lys-NHMec (Fig. 3).
TABLE 2.
Reaction kinetics of recombinant ySmCL1 on peptide substrates
| Substrate | Km (μM) | kcat (s−1) | kcat/Km (mM−1 s−1) |
|---|---|---|---|
| Z-Phe-Arg-NHMec | 8.5 | 0.053 | 6.24 |
| Boc-Val-Leu-Lys-NHMec | 10.2 | 0.176 | 17.25 |
| Z-Phe-Val-Arg-NHMec | 9.8 | 0.02 | 1.32 |
| Tos-Gly-Pro-Arg-NHMec | 15.1 | 0.04 | 2.72 |
FIG. 3.
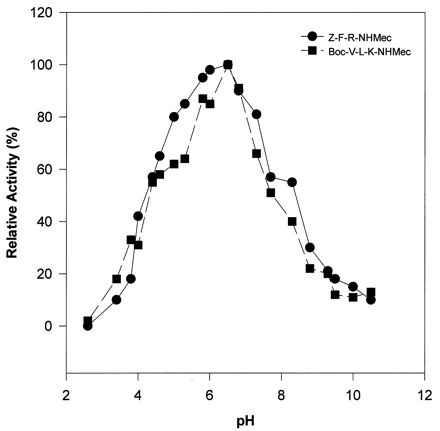
pH profile of activities of ySmCL1 against peptide substrates. The activities of ySmCL1 against Z-Phe-Arg-NHMec (Z-F-R-NHMec) and Z-Val-Leu-Lys-NHMec (Z-V-L-K-NHMec) were measured at different pHs. Points represent the means of duplicate experiments and are plotted as relative activity.
ySmCL1 cleaved human hemoglobin and, based on the smeared appearance of the digested products, cleaved this substrate at more than one site. In contrast to peptide substrates and gelatin, where it showed a pH optimum for activity of pH 6.5, ySmCL1 most efficiently cleaved hemoglobin at pH 4.5. Indeed, hemoglobin was not digested at pH 5.0 or above (Fig. 4).
FIG. 4.
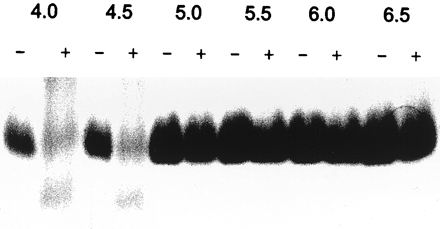
Hydrolysis of native human hemoglobin by ySmCL1. Native human hemoglobin was incubated with (+) and without (−) purified ySmCL1. Reactions were carried out at 37°C, at different pHs and in the presence of 1 mM dithiothreitol. After incubation for 18 h, the reaction products were resolved by SDS-PAGE (15% gel) under nonreducing conditions, and the gels were stained with Coomassie blue.
Identification of native SmCL1 in soluble extracts and ES products of schistosomes.
Soluble extracts of female and male adults, cercariae, and ES products of S. mansoni were separated by SDS-PAGE (12% gel), transferred to nitrocellulose, and probed with rabbit anti-bSmCL1 serum. Each lane was loaded with 10 μg of protein. We identified in the male and female adult S. mansoni extracts and ES products an antigen of ∼33 kDa that appeared to represent mature, native SmCL1 (Fig. 5). Based on the intensity of the signal, this protein was present at a higher concentration in female than male extract. We identified in male, female, and cercarial extracts a second antigen of ∼43 kDa that likely represents the proenzyme form of SmCL1. Also evident were several weakly staining bands of 40 to 35 kDa, possibly breakdown products or differentially glycosylated isoforms of SmCL1. The 33-kDa band was very prominent in ES products, whereas the 43-kDa band was not present, indicating that SmCL1 is processed to its active form before being secreted into the gut. No bands were visualized on replicate blots probed with the control (preimmunization) rabbit serum (not shown).
FIG. 5.
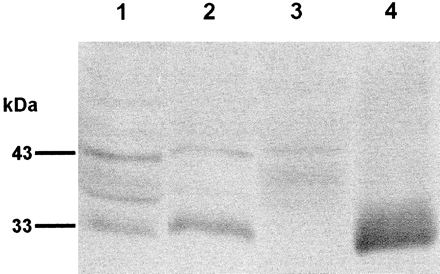
Immunoblot analysis of S. mansoni soluble extracts and ES products. Extracts of adult male schistosomes (lane 1), adult females (lane 2), mixed-sex cercariae (lane 3), and ES products from mixed-sex adults (lane 4) were separated by SDS-PAGE, transferred to nitrocellulose, and probed with anti-bSmCL1 serum. Replicate filters probed with control (preimmunization) serum showed no reactivity (data not shown).
Immunolocalization of native SmCL1 in adult worms.
Using rabbit preimmunization and anti-bSmCL1 sera, we probed thin sections of adult male and female S. mansoni to determine the site of expression and/or activity of SmCL1. A series of 10-μm-thick longitudinal, diagonal, and transverse sections were examined by light microscopy. No specific reactivity was observed on sections probed with preimmunization serum (Fig. 6A). By contrast, immunofluorescent labeling was observed on sections probed with anti-bSmCL1 serum (i) at the tegument of adult worms of both sexes, with more prominent reactivity at and immediately below the tegument on the ventral surface of male worms (Fig. 6B), and (ii) in the gastrodermal cells lining the lumen of the schistosome gut (Fig. 6C). No reactivity was evident at other sites or organs in the adult worms.
FIG. 6.

Immunolocalization of native SmCL1 in adult schistosomes. Longitudinal sections of male worms were probed with preimmunization (A; scale bar = 5 μm) or rabbit anti-bSmCL1 (B; scale bar = 10 μm) serum followed by labeling with anti-rabbit antibody-fluorescein conjugate. No specific labeling was observed with preimmunization serum (A), but intense labeling was observed in the tegument, particularly on the ventral surface, with anti-bSmCL1 (B). Labeling was also observed with anti-bSmCL1 in the gastrodermal cells lining the gut, as shown in the transverse sections of female adult worms (C; scale bar = 2.5 μm). VT, ventral tegument; DT, doral tegument; P, parenchyma; LU, gut lumen; GA, gastrodermis.
DISCUSSION
Given the difficulty in obtaining large numbers of schistosomes, it is not practical to isolate the enzymes directly from schistosomes for analysis of their biochemical activities or physiological roles. cDNAs encoding two discrete forms of cathepsin L from adult S. mansoni (SmCL1 and SmCL2) and adult S. japonicum (SjCL1 and SjCL2) have been reported (10, 19, 21). In a previous attempt to obtain functionally active schistosome cathepsins, we expressed the cognate S. mansoni cDNAs in E. coli; unfortunately, the recombinant proteins were compartmentalized by the bacteria into inclusion bodies from which we have been unable to isolate correctly folded, active proteinases (7). Accordingly, we have now used a eukaryotic expression system, S. cerevisiae transformed with the expression plasmid pAAH5. (We have successfully used this system to produce substantial quantities of each of two forms of cathepsin L from the related trematode parasite F. hepatica [12, 20]). By transforming yeast with pAAH5 encoding the full preproenzyme sequence of SmCL1, we obtained functional expression of active, recombinant schistosome cathepsin L. Manipulations to denature, refold, and activate the recombinant enzyme were not necessary.
Purification of ySmCL1 from the culture medium was achieved by using gel filtration followed by ion-exchange chromatography. The enzyme resolved as two peaks in gel filtration chromatography, although we subjected only the second peak, which contained the enzyme in higher specific activity, to purification by ion-exchange chromatography. The exclusion of the first peak resulted in loss of the much of the available enzyme, as it contained 65% of the total proteolytic activity. We observed a similar elution profile of F. hepatica cathepsin L proteinases on gel filtration, where it appears that aggregation of the recombinant enzyme to yeast proteins causes the protein to resolve in separate peaks (12, 20). Nevertheless, we obtained from a 5-liter fermentation sufficient ySmCL1 for analysis of the substrate specificity, enzyme kinetics, and hemoglobinolysis studies.
ySmCL1 exhibited a molecular size of 45 kDa, greater than the predicted sizes for the mature enzyme (24.1 kDa) and the proenzyme (35 kDa) (21). Glycosylation of ySmCL1 by the yeast cells may have contributed to its retarded migration in gels. Mature SmCL1 has three potential glycosylation sites (7, 21), and S. cerevisiae is known to hyperglycosylate recombinant proteins (5). The molecular sizes for the native schistosome SmCL1 (33 kDa) and the proenzyme (43 kDa), identified in immunoblots, are also greater than the predicted sizes, which may indicate that the enzymes are naturally glycosylated. Hyperglycosylation by yeast can result in an inactive recombinant protein; to avoid this problem, Lipps et al. (16) used a mutant cathepsin B Sm31 cDNA from which glycosylation sites had been deleted. Notwithstanding these mutations, the yeast-expressed recombinant cathepsin B required exogenous pepsin for its activation. By contrast, ySmCL1 was enzymatically active, and its activity was enhanced at acidic pH and by reducing conditions, as expected for a cathepsin L cysteine proteinase. The activity was also completely inhibited by the general cysteine proteinase inhibitor E-64 as well as the specific cathepsin L inhibitor Z-Phe-Ala-CHN2.
Kinetic studies showed that ySmCL1 preferred substrates with a hydrophobic residue in the P2 position, including Boc-Val-Leu-Lys-NHMec and Z-Phe-Arg-NHMec. By contrast, the enzyme showed minimal activity toward the cathepsin B substrates Z-Arg-NHMec and Z-Arg-Arg-NHMec. It is noteworthy that, and consistent with our earlier findings on the cathepsin L-like activities in extracts and ES products of schistosomes (7, 10), ySmCL1 exhibited a marked preference for Boc-Val-Leu-Lys-NHMec over Z-Phe-Arg-NHMec. Earlier studies by Dowd et al. (11, 12) showed that purified cathepsin L’s from the related trematode F. hepatica have a similar substrate preference for Boc-Val-Leu-Lys-NHMec. The presence of an additional residue in the P3 position, Val in this case, may increase the overall binding energy of the substrate in the active site of the enzyme, resulting in more efficient hydrolysis. Nevertheless, these observations indicate that Boc-Val-Leu-Lys-NHMec may be a more sensitive substrate for measuring cathepsin L-like activity in helminth parasites than Z-Phe-Arg-NHMec, which has classically been used to demonstrate cathepsin L in mammalian tissues (2).
ySmCL1 showed higher pH optima for activity against gelatin and synthetic peptidyl substrates than for hemoglobin. While ySmCL1 was most active against Boc-Val-Leu-Lys-NHMec at pH 6.5, it was inactive against hemoglobin at pH 5.0 and higher. This finding indicates that denaturation of the hemoglobin by acidic pH may be required before it can be digested by SmCL1, and this may reflect the physicochemical environment of the schistosome gut, which appears to be acidic (4). Earlier studies by us and others showed that both cathepsin L- and cathepsin D-like proteinases were secreted by adult S. mansoni and that both enzymes were involved in the degradation of hemoglobin (3, 7, 13). The present results demonstrating the presence of SmCL1 in the gastrodermal cells lining the gut (at higher levels in female than in male parasites), its presence in ES products, and its ability to digest hemoglobin at acidic pH signal the probable biological function of this schistosome cathepsin. Together, they indicate that SmCL1 plays a role in proteolysis of hemoglobin within the schistosome gut. If this is so, SmCL1 has a role discrete from that of SmCL2, which is located in the reproductive organs (19). It is noteworthy that SmCL1 is located in the tegument of male schistosomes in addition to the digestive tract. Other enzymes, such as schistosome legumain and cathepsin D, that are associated with the digestive tract have also been located in the tegument (9, 26). These enzymes may function in intracellular protein turnover or in membrane biogenesis, in addition to playing roles in the degradation of hemoglobin.
If SmCL1 plays a central role in hemoglobin proteolysis as the present results indicate, it represents a potential target for antischistosomal therapies. In view of the sequence differences between schistosome and human cathepsin L, including divergence in their active site residues (9) and differential sensitivity to diazomethanes (10), it is feasible that inhibitors that selectively inhibit the schistosome cathepsin L’s could be developed. Indeed, the potential antischistosomal effects of drugs targeted at cysteine proteinases has been demonstrated by Wasilewski et al. (23), using morpholinourea-Phe-Ala-CHN2 and analogues. While these drugs would be inhibitors of both cathepsin L and cathepsin B, they produced dramatic reductions in schistosome worm loads and fecundity in infected mice and were lethal to cultured schistosomula. Since it is now clear that schistosome cathepsins, including SmCL1, can be produced in sufficient quantities in yeast, development of specific inhibitors of these proteinases can now be addressed.
ACKNOWLEDGMENTS
We thank Malcolm Jones and Michael Walsh for help with immunolocalization and Mary Duke for maintenance of the schistosome life cycle.
Ciaran Brady was a recipient of a grant from the Cavan County Council and Forbairt, Ireland. We are grateful for the financial support provided by Dublin City University and the Australian National Health & Medical Research Council. Andrew J. Dowd is a recipient of an award from the Higher Education Authority, Ireland.
REFERENCES
- 1.Ammerer G. Expression of genes in yeast using the ADC1 promoter. Methods Enzymol. 1983;101:192–201. doi: 10.1016/0076-6879(83)01014-9. [DOI] [PubMed] [Google Scholar]
- 2.Barrett A J, Kembhavi A A, Brown M A, Kirschke H, Knight C G, Tamai M, Hanada K. l-trans-Epoxysuccinyl-leucyl-amido(4-guanidino)butane (E-64) and its analogues and inhibitors of cysteine proteinases including cathepsins B, H and L. Biochem J. 1982;201:189–198. doi: 10.1042/bj2010189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker M M, Harrop S A, Dalton J P, Kalinna B B, McManus D P, Brindley P J. Cloning and characterization of the Schistosoma japonicum aspartic proteinase involved in hemoglobin degradation. J Biol Chem. 1995;270:24496–24501. doi: 10.1074/jbc.270.41.24496. [DOI] [PubMed] [Google Scholar]
- 4.Brindley P J, Kalinna B M, Dalton J P, Day S R, Wong J Y M, Smythe M L, McManus D P. Proteolytic degradation of host hemoglobin by schistosomes. Mol Biochem Parasitol. 1997;89:1–9. doi: 10.1016/s0166-6851(97)00098-4. [DOI] [PubMed] [Google Scholar]
- 5.Buckholz R G, Gleeson M A G. Yeast systems for the production of heterologous proteins. Bio/Technology. 1991;9:1067–1072. doi: 10.1038/nbt1191-1067. [DOI] [PubMed] [Google Scholar]
- 6.Carter B L A, Irani M, McKay V L, Seale R L, Sledziewski A V, Smith R A. Expression and secretion of foreign genes in yeast. In: Glover D M, editor. DNA cloning. III. Oxford, England: IRL Press; 1987. pp. 141–161. [Google Scholar]
- 7.Dalton J P, Clough K A, Jones M K, Brindley P J. Characterization of the cathepsin-like cysteine proteinases of Schistosoma mansoni. Infect Immun. 1996;64:1328–1334. doi: 10.1128/iai.64.4.1328-1334.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalton J P, Clough K A, Jones M K, Brindley P J. The cysteine proteinases of Schistosoma mansoni cercariae. Parasitology. 1997;114:105–112. doi: 10.1017/s003118209600830x. [DOI] [PubMed] [Google Scholar]
- 9.Dalton J P, Smith A M, Clough K A, Brindley P J. Digestion of haemoglobin by schistosomes: 35 years on. Parasitol Today. 1995;11:299–303. doi: 10.1016/0169-4758(95)80045-x. [DOI] [PubMed] [Google Scholar]
- 10.Day S R, Dalton J P, Clough K A, Leonardo L, Tiu W U, Brindley P J. Characterization and cloning of the cathepsin L proteinases of Schistosoma japonicum. Biochem Biophys Res Commun. 1995;217:1–9. doi: 10.1006/bbrc.1995.2737. [DOI] [PubMed] [Google Scholar]
- 11.Dowd A J, Smith A M, McGonigle S, Dalton J P. Purification of a second cathepsin L proteinase secreted by the parasitic trematode Fasciola hepatica. Eur J Biochem. 1994;223:91–98. doi: 10.1111/j.1432-1033.1994.tb18969.x. [DOI] [PubMed] [Google Scholar]
- 12.Dowd A J, Tort J, Roche L, Ryan T, Dalton J P. Isolation of a cDNA encoding Fasciola hepatica cathepsin L2 and functional expression in Saccharomyces cerevisiae. Mol Biochem Parasitol. 1997;88:241–246. doi: 10.1016/s0166-6851(97)00090-x. [DOI] [PubMed] [Google Scholar]
- 13.Ghoneim H, Klinkert M-Q. Biochemical properties of purified cathepsin B from Schistosoma mansoni. Int J Parasitol. 1995;25:1515–1519. doi: 10.1016/0020-7519(95)00079-8. [DOI] [PubMed] [Google Scholar]
- 14.Lawrence J D. The ingestion of red blood cells by Schistosoma mansoni. J Parasitol. 1973;59:60–63. [PubMed] [Google Scholar]
- 15.Leatherbarrow R J. Enzfitter. Cambridge, England: Elsevier Biosoft; 1987. [Google Scholar]
- 16.Lipps G, Fullkrug R, Beck E. Cathepsin B of Schistosoma mansoni. Purification and activation of the recombinant proenzyme secreted by Saccharomyces cerevisiae. J Biol Chem. 1996;271:1717–1725. doi: 10.1074/jbc.271.3.1717. [DOI] [PubMed] [Google Scholar]
- 17.Mahmood A A F, Wahab M F A. Schistosomiasis. In: Warren K S, Mahmood A A F, editors. Tropical and geographical medicine. 2nd ed. New York, N.Y: McGraw-Hill; 1990. pp. 458–473. [Google Scholar]
- 18.McKerrow J H, Doenhoff M J. Schistosome proteinases. Parasitol Today. 1988;4:334–340. doi: 10.1016/0169-4758(88)90002-6. [DOI] [PubMed] [Google Scholar]
- 19.Michel A, Ghoneim H, Resto M, Klinkert M-Q, Kunz W. Sequence, characterisation and localisation of a cysteine proteinase cathepsin L in Schistosoma mansoni. Mol Biochem Parasitol. 1995;73:7–18. doi: 10.1016/0166-6851(95)00092-f. [DOI] [PubMed] [Google Scholar]
- 20.Roche L, Dowd A J, Tort J, McGonigle S, McSweeney A, Curley G P, Ryan T, Dalton J P. Functional expression of Fasciola hepatica cathepsin L1 in Saccharomyces cerevisiae. Eur J Biochem. 1997;245:373–380. doi: 10.1111/j.1432-1033.1997.t01-1-00373.x. [DOI] [PubMed] [Google Scholar]
- 21.Smith A M, Dalton J P, Clough K A, Kilbane C L, Harrop S A, Hole N, Brindley P J. Adult Schistosoma mansoni express cathepsin L proteinase activity. Mol Biochem Parasitol. 1994;67:11–19. doi: 10.1016/0166-6851(94)90091-4. [DOI] [PubMed] [Google Scholar]
- 22.Smith A M, Dowd A J, McGonigle S, Keegan P S, Brennan G, Trudgett A, Dalton J P. Purification of a cathepsin L-like proteinase secreted by adult Fasciola hepatica. Mol Biochem Parasitol. 1993;62:1–8. doi: 10.1016/0166-6851(93)90171-s. [DOI] [PubMed] [Google Scholar]
- 23.Wasilewski M M, Lim K C, Phillips J, McKerrow J H. Cysteine proteinase inhibitors block schistosome haemoglobin degradation in vitro and decrease worm burden and egg production in vivo. Mol Biochem Parasitol. 1996;81:179–189. doi: 10.1016/0166-6851(96)02703-x. [DOI] [PubMed] [Google Scholar]
- 24.Yoshino T P, Lodes M J, Rege A A, Chappell C L. Proteinase activity in miracidia, transformation excretory-secretory products, and primary sporocysts of Schistosoma mansoni. J Parasitol. 1993;79:23–31. [PubMed] [Google Scholar]
- 25.Zerda K S, Dresden M H, Chappell C L. Schistosoma mansoni: expression and role of cysteine proteinases in developing schistosomula. Exp Parasitol. 1988;67:238–246. doi: 10.1016/0014-4894(88)90071-9. [DOI] [PubMed] [Google Scholar]
- 26.Zhong C, Skelly P J, Leaffer D, Cohn R G, Caulfield J P, Shoemaker C B. Immunolocalisation of a Schistosoma mansoni facilitated diffusion glucose transporter to the basal, but not the apical, membranes of the surface synticum. Parasitology. 1995;110:383–394. doi: 10.1017/s0031182000064726. [DOI] [PubMed] [Google Scholar]



