Abstract
In situ hybridization (ISH) is a powerful method for detecting specific RNAs at the cellular level. Although conventional ISH using hapten-labeled probes are useful for detecting multiple RNAs, the detection procedures are still complex and required longer time. Therefore, we introduced a new application of fluorescence resonance energy transfer (FRET)-based molecular beacon (MB) probes for ISH. MCF-7 cells and C57BL/6J mouse uterus were used for ISH. MB probes for ERα mRNA and 28S rRNA were labeled with Cy3/BHQ-2 and 6-FAM/DABCYL, and conventional probes were labeled with digoxigenin. Fluorescence measurements revealed that of more-rapid hybridization kinetics compared to conventional probes. In MCF-7 cells, 28S rRNA was detected in nucleolus and cytoplasm of all cells, whereas ERα mRNA was detected in some nucleolus. In the uterus, 28S rRNA was clearly detected using complementary MB probe, but there were no signals in control slides. Moreover, 28S rRNA was detected in all cells, whereas ERα mRNA was detected mainly in the epithelium. Fluorescence intensity of 28S rRNA was decreased significantly in 1 or 2 base-mismatched sequences, that indicates highly specific detection of target RNAs. In conclusion, the FRET-based MB probes are very useful for ISH, providing rapid hybridization, high sensitivity and specificity.
Keywords: in situ hybridization, FRET, molecular beacon probe, 28S rRNA, estrogen receptor alpha
I. Introduction
In situ hybridization (ISH) is an important molecular histochemical method for detecting the expression of specific genes in tissues and individual cells [22]. In principle, the expression of specific genes is detected based on the base pairing between a specific probe and a target nucleic acid sequence. Since ISH was first introduced in the field of biomedical research, various labels have been employed for probes, including radioisotopes, haptens, and fluorescent dyes. Radioisotope-labeled probes were initially used for ISH [8, 11]. In addition, a variety of next-generation nonradioactive probes have been developed, such as horseradish peroxidase (HRP)-labeled polyuridine as well as various hapten-based probes [22, 29]. The most common haptens currently used are digoxigenin (Dig) and biotin, and deoxynucleotides themselves can be haptenized by thymine-thymine dimer (T-T dimer) method [19–21]. Briefly, these nonradioactive probes labeled with haptens are detected by immunohistochemistry. Additionally, a technique utilizing fluorescent-labeled probes known as fluorescent in situ hybridization (FISH) was developed to enable detection of chromosomes and chromosomal abnormalities such as deletions, translocations, and amplifications [2, 31, 33]. Indeed, FISH has proven to be a powerful method for the analysis of specific DNA, but it is not suitable for analysis of RNA because of high background. Although great achievements were done in multiplex detection and signal amplification, the current procedures are still complex, time consuming, and require experienced personnel [14, 15, 17, 18, 28, 31, 34]. Therefore, simpler and more-rapid detection tools for ISH are needed.
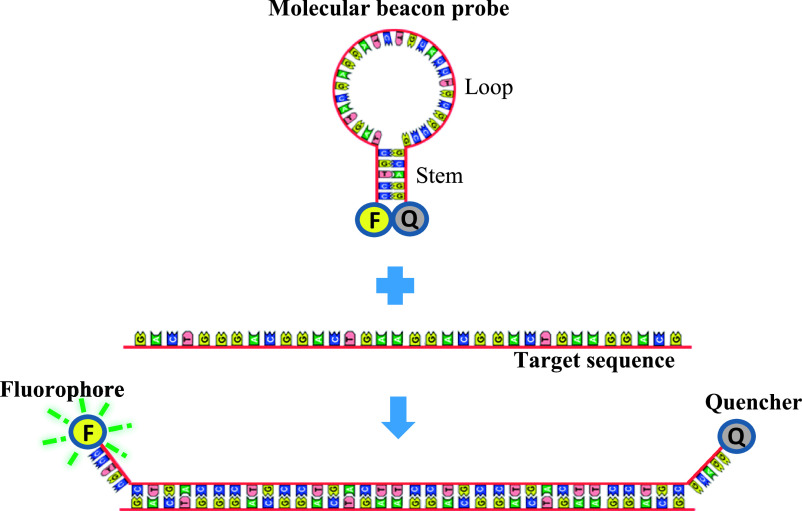
To simplify ISH protocols, we introduced a new detection system that uses a molecular beacon (MB) probe based on fluorescence resonance energy transfer (FRET) using paraffin-embedded tissue sections. The MB probes are a hairpin-shaped, single-stranded oligonucleotide sequence consisting of a loop, stem, fluorophore, and quencher [12]. The loop part of the probe is an oligonucleotide sequence that is complementary to the target mRNA, whereas the stem is formed by annealed complementary sequences at each end of the loop. Fluorophore and quencher molecules are labeled at the 5'- and 3'-ends. When the probe hybridizes with the target sequence, the fluorophore separates from the quencher molecule and produces a fluorescent signal based on the FRET principle. When the probe does not hybridize with the target sequence, the energy from the donor fluorophore is transferred to the acceptor quencher, which is in close proximity, thus preventing production of a fluorescent signal [38]. Also, the unique hairpin structure of the MB probe provides highly sensitive detection due to tiny probes and high-affinity binding to target nucleotides [4].
In practice, FRET-based MB probes are used in a variety of applications, including real-time PCR, genetic analyses, pathogen detection, analyses of nucleic acid–protein interactions, as well as various diagnostic clinical assays [1, 10, 24, 36, 37]. All of these methods provide an indication of the presence of target molecules in biological samples; however, in situ localization of target molecules has not been demonstrated yet. Therefore, we introduced a new application of FRET-based MB probes using cultured cells as well as paraffin-embedded tissues, which are the most widely used sample type in basic histochemical research and medical diagnostics due to good preservation of tissue morphology.
In the present study, a FRET-based MB probe was used for the simultaneous localization of ERα mRNA and 28S rRNAs by ISH. The sensitivity and specificity of the MB probe was confirmed using various control experiments, such as dot blot hybridization, competition and neutralization assays, RNase treatment, and mismatched sequences. We optimized the MB probe for ISH using cultured cells as well as paraffin-embedded tissues, and it was useful for rapid and simultaneous detection of different RNAs at the cellular and tissue levels.
II. Materials and Methods
Chemicals and biochemicals
β-Cyanoethyl phosphoramidites, 3'-DABCYL CPG (#20-5912), 5'-fluorescein phosphoramidite (#10-5901), and cyanine 3 CPG (#20-5913) were obtained from Glen Research (Sterling, VA, USA). Trizma base, bovine serum albumin (BSA), 3-aminopropyl-triethoxysilane, and Brij L23 were from Sigma Chemical Co. (St. Louis, MO, USA). Paraformaldehyde (PFA) was from Merck (Darmstadt, Germany). 3,3'-Diaminobenzidine–4 HCl (DAB) was from Dojindo Chemicals (Kumamoto, Japan). Deionized formamide was purchased from Nacalai Tesque (Kyoto, Japan). All other reagents used in this study were obtained from Fujifilm Wako Pure Chemicals (Osaka, Japan) and were of high analytical grade.
Cell culture
Human breast cancer cells (MCF-7) were grown in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum and maintained at 37°C in a humidified chamber with 5% CO2 [32]. Cells were attached to the slide glass pretreated with ethanol and UV irradiation, then fixed with 4% PFA/PBS for 15 min.
Animals and tissue preparation
The uterus from 8-week-old C57BL/6J mice weighing 17–19 g were used in this study. Uterine tissue was obtained at proestrus stage, which is reported as highest level of ERα expression [40]. Mice were kept under constant 12-hr dark/12-hr light conditions and fed normal chow and drinking water ad libitum. The experimental protocol was approved by the Animal Ethics Review Committee of the University of Miyazaki (#2012-502-3). After animals were sacrificed, tissues were obtained and fixed in 4% PFA in PBS (pH 7.4) at room temperature for 24 hr and then embedded in paraffin [35].
DNA synthesis and purification
Probe oligomers were synthesized on a 1-μmol scale using an automatic DNA/RNA synthesizer (Nihon Techno Service, Saitama, Japan) and solid-phase phosphoramidite chemistry. After automated synthesis, the oligomers were cleaved from the support and deprotected by treatment with 28% ammonium hydroxide for 17 hr at 55°C. The crude oligomers were purified by denaturing polyacrylamide gel electrophoresis on a 15% gel. After ethanol precipitation, the purified oligomers were desalted using NAP10 columns (GE Healthcare, Buckinghamshire, UK) and identified by electrospray ionization mass spectrometry (ESI-MS) on a Q Exactive quadrupole-orbitrap mass spectrometer (Thermo Scientific, MA, USA).
Fluorescence measurement
Fluorescence was measured using a JASCO FP-8200 fluorescence spectroscopy (Tokyo, Japan), as described previously [16]. The complementary MB probe for 28S rRNA (1 ng/μl) was annealed by heating at 95°C for 3 min and then cooling (1.0°C/min) and storing at 4°C for 3 hr. MB probe and unlabeled homologous oligo-DNA for 28S rRNA (10 ng/μl) were added to the reaction buffer and incubated at 37°C. Reaction buffer contents were similar with hybridization medium of ISH that containing 10 mM Tris-HCl (pH 7.4), 1 mM EDTA, 0.6 M NaCl, 1× Denhardt’s solution, 250 μg/ml yeast tRNA, 125 μg/ml salmon sperm DNA, and 40% deionized formamide. Just after mixing, as well as 3, 6, 12 and 24 hr, fluorescence spectra of the mixture were measured. The excitation wavelength for the probe was 495 nm and the fluorescence emission was monitored at 520 nm. For each sample, at least two spectrum scans were accumulated over a wavelength range of 400 to 700 nm. For mismatch experiments, the complementary MB probe for 28S rRNA was incubated with excess amount of homologous and 1 or 2-base mismatched oligo-DNA, as well as unrelated sequences. After 3 hr of incubation at 37°C, the fluorescence intensity was measured.
Sequences and design of molecular beacon probes
The oligonucleotide probes used for ISH were listed in Table 1. 28S rRNA and ERα mRNA probe sequences were examined previously by Yoshii et al. and Hishikawa et al. [13, 42]. The loop sequence (target mRNA sequence) of MB was selected as a region with 35–65% GC content and a sequence with a predicted melting temperature (Tm) of ~65°C. As described previously, a five-base stem with a GC content of ~60% was attached respectively to the 3' and 5' ends of the loop sequence to prepare a stem-loop hairpin structure with a Tm of ~50°C, which can be determined using the online tool ‘DNA Mfold’ in the experimental condition used in this study (0.6 M NaCl) [41].
Table 1. .
Oligo-DNA probe sequences for ISH
| Target | Probe | Sequence and labeling |
|---|---|---|
| MB-28S rRNA | Complementary | FAM 5'-CCTGCtactaccaccaagatctgcacctgcggcgGCAGG-3' DABCYL |
| Homologous | FAM 5'-CCTGCcgccgcaggtgcagatcttggtggtagtaGCAGG-3' DABCYL | |
| MB-ERα mRNA | Antisense | Cy-3 5'-ATGGCgaggggctccagctcgttcccttggatctggtgcagcaagGCCAT-3' BHQ2 |
| Sense | Cy-3 5'-GAGGGatggccttgctgcaccagatccaagggaacgagctggagcCCCTC-3' BHQ2 | |
| Dig-28S rRNA | Complementary | 5'-tgctactaccaccaagatctgcacctgcggcggc-3' Dig |
| Homologous | 5'-gccgccgcaggtgcagatcttggtggtagtagca-3' Dig | |
| Mismatched probes in complementary sequence (28S rRNA) | ||
| 1-mismatch | FAM-5'-CCTGCtactaccaccaagacctgcacctgcggcgGCAGG-3'-DABCYL | |
| 2-mismatches | FAM-5'-CCTGCtactaccactaagatctgcgcctgcggcgGCACC-3'-DABCYL | |
| Mismatched homologous sequences (28S rRNA) | ||
| 1-mismatch | gccgccgcaggtgcaggtcttggtggtagtagca | |
| 2-mismatches | gccgccgcaggcgcagatcttagtggtagtagca | |
| Unrelated and unlabeled oligo-DNA | ccagtcctctgcgctcccccacgcctcatttgcgacagtcgcgtt | |
* For MB probe, stem and loop sequences are indicated as capital and small letters, respectively.
Dot blot hybridization
The procedures for dot blot hybridization were described previously [14, 19]. Two-microliter solution of homologous oligo-DNA for 28S rRNA were placed onto nitrocellulose membranes pretreated with 20× SSC (1× SSC = 0.15 M sodium chloride and 0.015 M sodium citrate, pH 7.0) in a series of spots at 1 pg to 10 ng per spot. After baking at 80°C for 2 hr, the membranes were incubated at 37°C for 2 hr with prehybridization medium containing 10 mM Tris-HCl (pH 7.4), 1 mM EDTA, 0.6 M NaCl, 1× Denhardt’s solution, 500 μg/ml yeast tRNA, 250 μg/ml salmon sperm DNA, and 50% deionized formamide. The membranes were then hybridized at 37°C for 3 hr with complementary and homologous MB probes in medium containing 10 mM Tris-HCl (pH 7.4), 1 mM EDTA, 0.6 M NaCl, 1× Denhardt’s solution, 250 μg/ml yeast tRNA, 125 μg/ml salmon sperm DNA, 10% dextran sulfate, and 40% deionized formamide. After successive washing, the membranes were observed using a Fujifilm LAS-3000 luminescent image analyzer (Fujifilm, Tokyo, Japan).
In situ hybridization
ISH followed a previously described standard protocol [6, 19]. Tissue sections were deparaffinized with toluene and rehydrated using a graded ethanol series, followed by treatment with 0.2 N HCl for 20 min and proteinase K digestion at 37°C for 15 min. After post-fixation with 4% PFA in PBS, the sections were immersed in 2 mg/ml glycine in PBS for 30 min and kept in 40% deionized formamide in 4× SSC until used for hybridization. Tissue sections were hybridized with MB probe for 3 hr at 37°C. Control experiments using Dig-labeled oligo-DNA probes were hybridized for overnight. The slides were washed three times with 2× SSC at 37°C and then twice with 2× SSC at room temperature. DAPI was used for nuclear counterstaining. Excess amounts of unlabeled complementary and homologous oligo-DNAs for 28S rRNA (100×) were used for competition and neutralization assays, respectively. Adjacent sections were digested with 100 μg/ml RNase prior to post-fixation for 1 hr at 37°C. In addition, an excess amount of unrelated unlabeled probe was applied together with the 28S rRNA probe. In parallel with MB probe, Dig-labeled 28S rRNA probe was examined in every experiment [42]. For mismatch experiment, mouse uterine tissue was hybridized with complementary MB for 28S rRNA, as well as 1 or 2 base-mismatched probes. The fluorescence intensity was measured in 100 cells in each groups and results were revealed as an average arbitrary fluorescence unit (ImageJ software, 1.53c NIH) [7, 32]. Microphotographs were taken using a laser scanning microscope (Zeiss LSM700), fluorescence microscope (Keyence BZ-X700), and light microscope (Olympus BX53) super resolution microscope (Olympus SpinSR10).
III. Results
Sensitivity of the MB probe
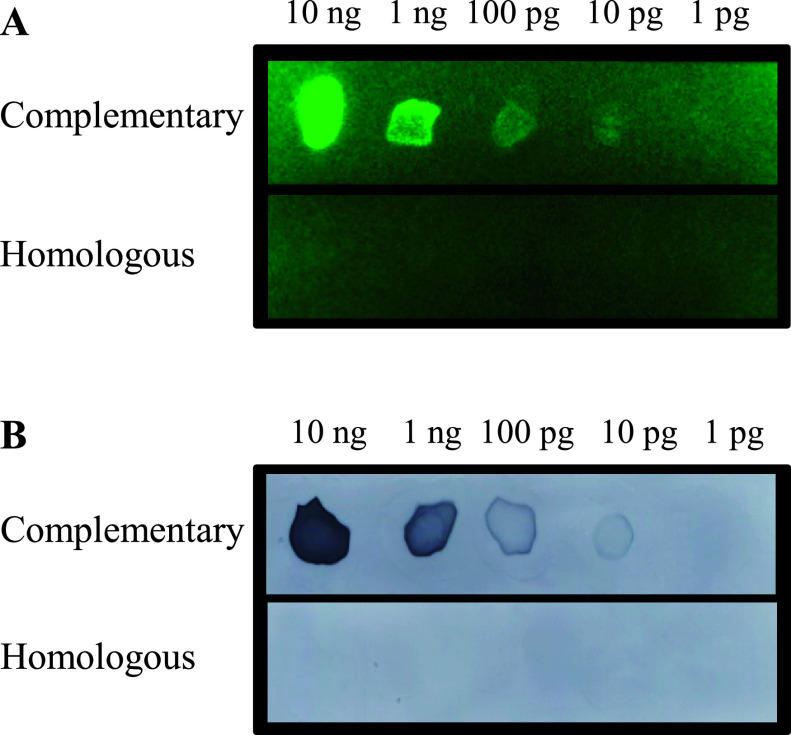
The principle of FRET-based MB probe was described in Figure 1. When the MB probe hybridizes with the target sequence, the fluorophore and the quencher molecules become distant and produces a fluorescent signal. The sensitivity of the MB probe was evaluated by dot blot hybridization. A total of 10 pg of oligo-DNA could be detected when the complementary MB probe for 28S rRNA was hybridized with unlabeled homologous oligo-DNA fixed on a nitrocellulose membrane (Fig. 2A). The homologous MB probe for 28S rRNA did not hybridized. In parallel with these experiments, 10 pg of oligo-DNA was detected using complementary oligo-DNA for 28S rRNA, but not homologous 28S rRNA probe (Fig. 2B). These findings indicated that the MB probes are highly sensitive probe.
Fig. 1.
Principle of in situ hybridization using the MB probe. The complementary sequence to target molecule is designed in the loop part of the probe. Stem part is formed by annealed complementary sequences and both the 5'- and 3'-ends were labeled with fluorophore and quencher molecules. After probe hybridization, the distance between the fluorophore and quencher molecules increases, enabling emission of fluorescence.
Fig. 2.
Dot blot hybridization. Various amount (1 pg to 10 ng per spot) of homologous oligo-DNA for 28S rRNA were spotted and then membranes were hybridized with complementary and homologous MB probes (A) and Dig-labeled (B) complementary and homologous oligo-DNA for 28S rRNA. 10 pg DNA were detected specifically.
Fluorescence intensity of the MB probe
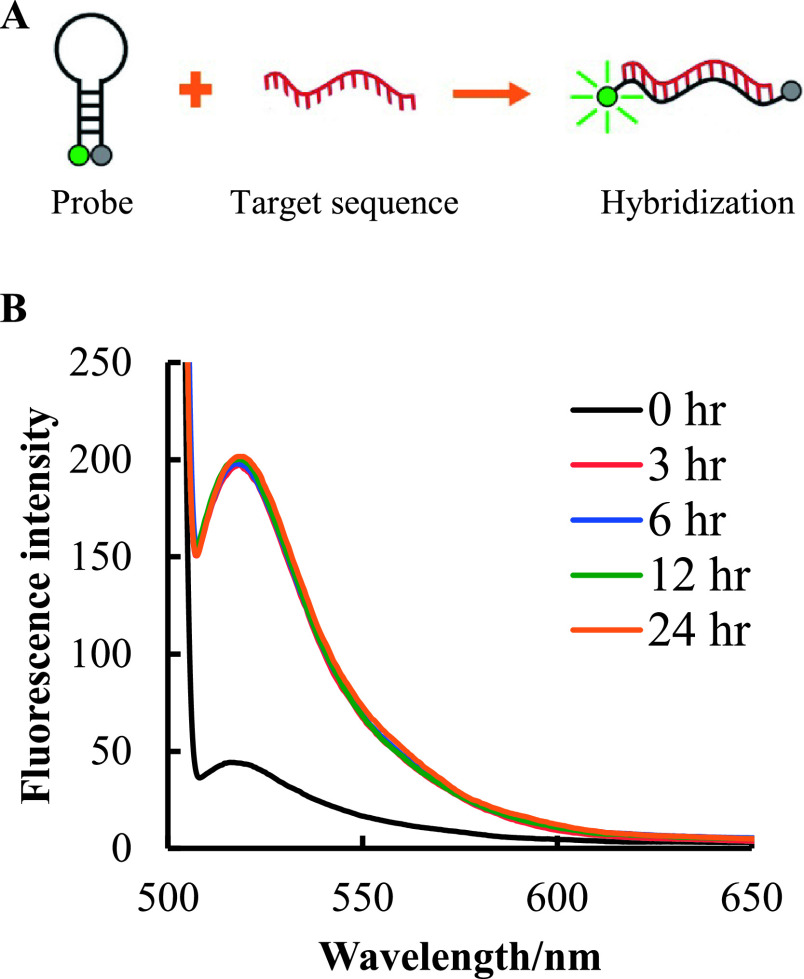
Fluorescence intensity of MB probe was measured by fluorescence spectroscopy at 0–24 hr after hybridization. After annealing, the complementary MB probe for 28S rRNA was gradually cooled-down to form a stem-loop conformation. Just after mixing with an excess amount of unlabeled homologous oligo-DNA (Fig. 3A), very low fluorescence intensity was observed (black line) (Fig. 3B). After 3 hr of incubation at 37°C to allow for hybridization of the MB probe with the target, a dramatic increase in fluorescence intensity was observed (red line). Interestingly, similar fluorescence intensity was observed at 3, 6, 12, and 24 hr after hybridization. These results indicate that once the MB probe hybridizes with the target sequence, it produces a stable fluorescent signal, regardless of incubation time.
Fig. 3.
Time-dependent fluorescence intensity of the MB probe. The complementary MB probe for 28S rRNA was hybridized with the unlabeled homologous oligo-DNA for 28S rRNA (A) Green color represents fluorophore and red color represents target sequence. The signal was then measured after excitation at 495 nm (B). The black line indicates the fluorescence intensity at the beginning of incubation. The red, blue, green, and orange lines indicate 3, 6, 12, and 24 hr after incubation, respectively.
MB probe for cultured cell line
The usefulness of the MB probe was examined using cultured cells. In MCF-7 cells, the strong fluorescent signal of 28S rRNA was found in all nucleolus as well as throughout the cytoplasm of all cells (arrowheads indicate presence of 28S rRNA) (Fig. 4A). Interestingly, ERα mRNA was found in some nucleolus, but not all (arrows indicate absence in Fig. 4B). Moreover, ERα mRNA was localized mainly in the perinuclear region of the cytoplasm. No fluorescent signals were detected with the homologous MB probe for 28S rRNA (Fig. 4A inset) and sense MB probe for ERα mRNA (Fig. 4B inset). Moreover, the localization of 28S rRNA and ERα mRNA was confirmed by super resolution microscopy, and independent expressions were found in the cell nuclei (Fig. 4D–F). Collectively, these results indicated that the differential subcellular localization of target RNAs can be determined clearly by using ISH with MB probes.
Fig. 4.
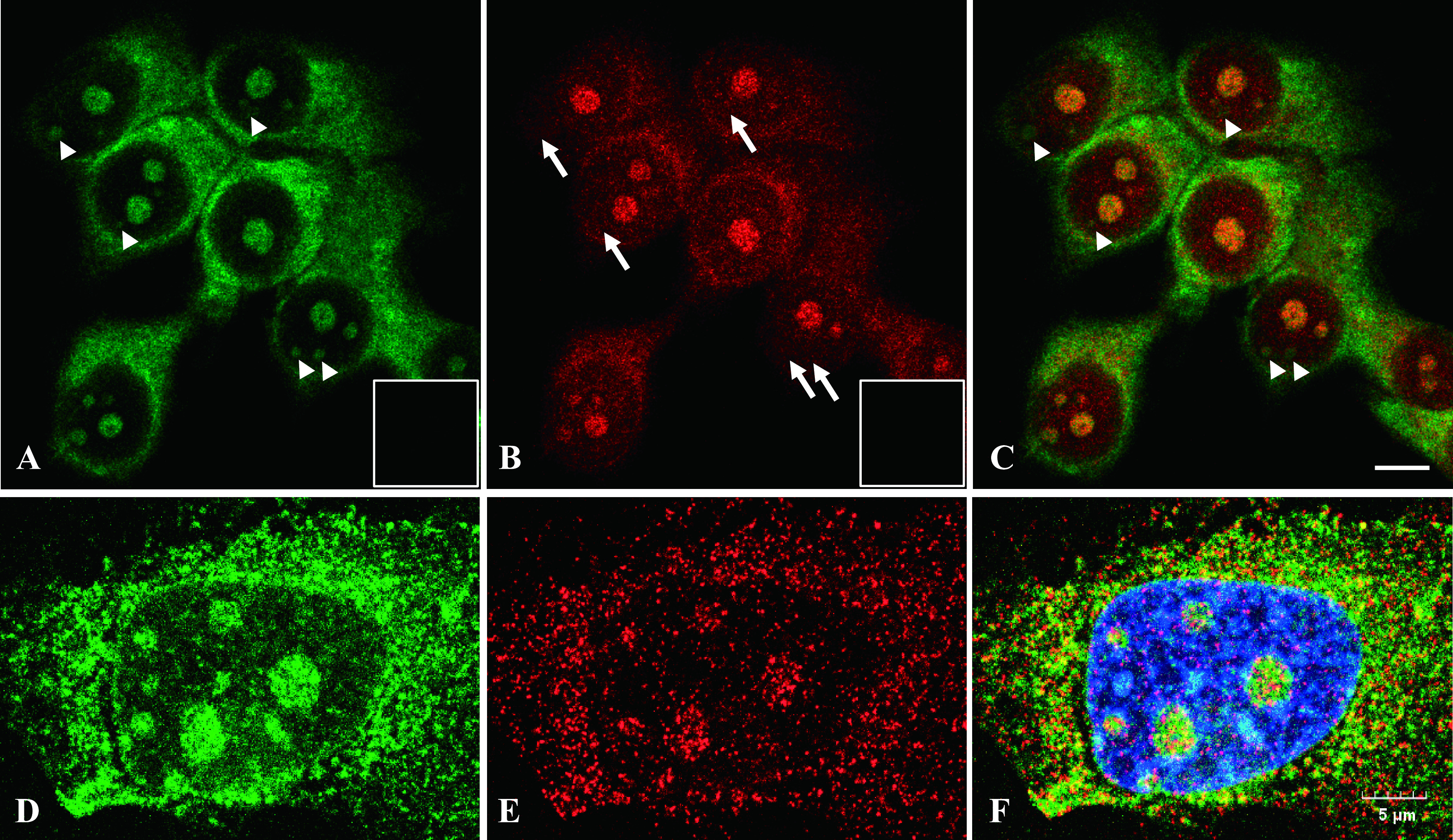
In situ detection of ERα mRNA and 28S rRNA in MCF-7 cells. Cells were attached onto a slide glass and reacted with complementary MB probe for 28S rRNA (green) (A) and antisense MB probe for ERα mRNA (red) (B). Both images were merged in (C). Arrowheads indicate presence of 28S rRNA; and arrows indicate absence of ERα mRNA in nucleoli. ERα mRNA was localized mainly in perinuclear region of MCF-7 cells. Another slide was reacted with homologous MB probe for 28S rRNA (inset in a) and sense MB probe for ERα mRNA (inset in B). Magnification ×630. Bar = 10 μm. Cells were analyzed by confocal microscope (A–B) and super resolution microscope (D–F). MB probe for 28S rRNA (D), ERα mRNA (E) and merged with DAPI (F).
Control experiments for the MB probe
Various control experiments were performed to confirm the specificity in the hybridization reaction with MB probe in ISH settings. As shown in Figure 5A, a strong fluorescent signal was observed in the cytoplasm as well as nucleolus in mouse uterus sections hybridized with the complementary MB probe for 28S rRNA, whereas no fluorescent staining was detected in sections hybridized with the homologous MB probe for 28S rRNA (Fig. 5B). Moreover, in competition and neutralization assays, no fluorescent signal was observed when the complementary probes for 28S rRNA were reacted with an excess amount of unlabeled complementary or homologous oligo-DNA for 28S rRNA, respectively (Fig. 5C, D). Further, a similar fluorescent signal to that with the complementary probe was observed in the cytoplasm and nucleolus in sections hybridized with the complementary oligo-DNA for 28S rRNA together with an excess amount of unrelated unlabeled oligo-DNA (Fig. 5E). In addition, no staining was detected when the slides were treated with RNase prior to hybridization with the complementary probes for 28S rRNA (Fig. 5F). Collectively, these results indicate that the MB probe enables specific in situ localization of target RNAs in the tissue sections. Parallel with the MB probe, control experiments were performed using the Dig-labeled oligo-DNA probe for 28S rRNA, and the results were consistent with those obtained with the MB probe (Fig. 5G–L).
Fig. 5.
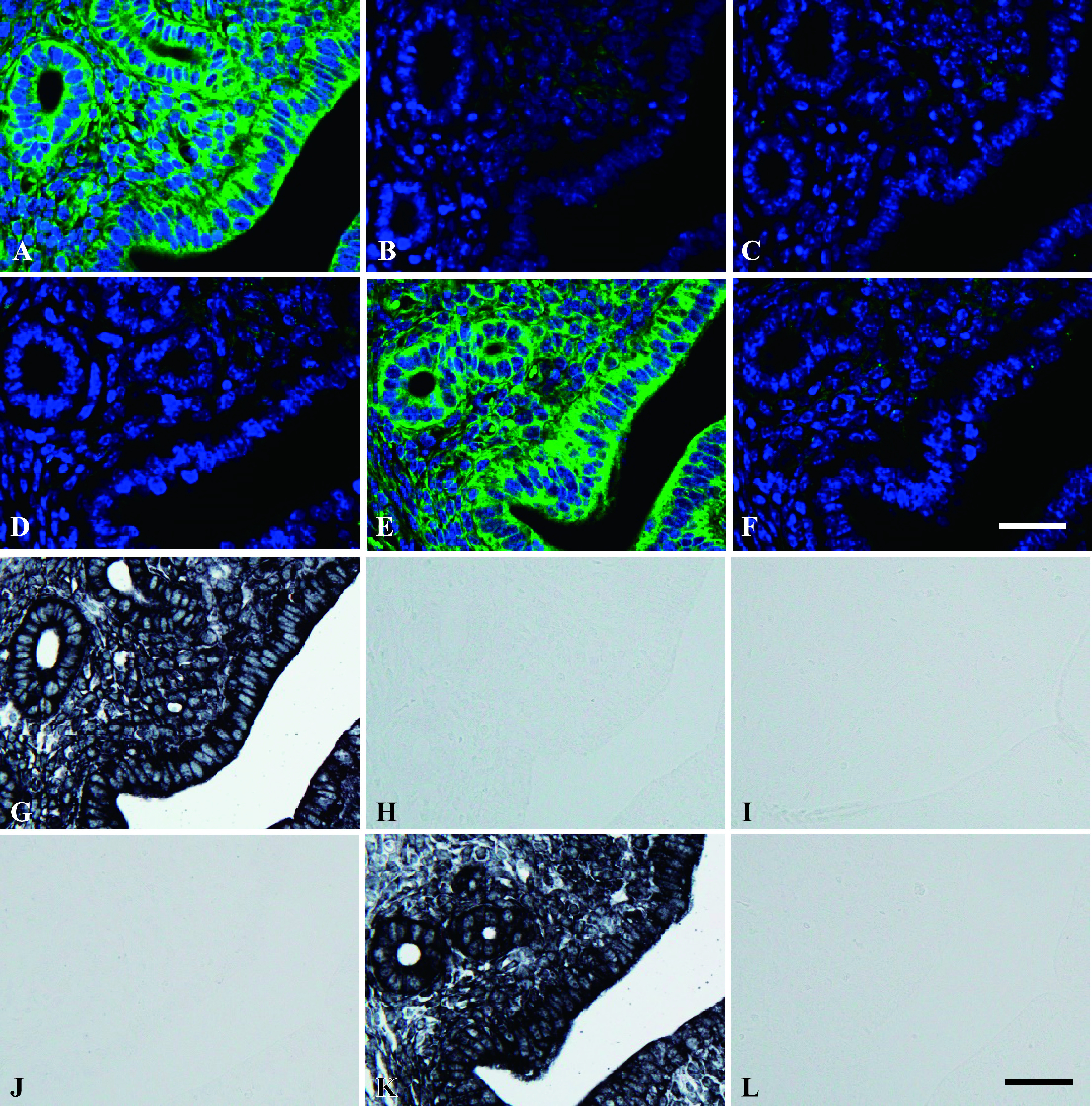
Control experiments for MB probes. Paraffin-embedded mouse uterus tissue was used for ISH. Uterine sections were hybridized with complementary MB probes for 28S rRNA (green) (A). Adjacent sections were hybridized with homologous (B) and complementary sequences in the presence of excess amounts of unlabeled complementary (C) and unlabeled homologous oligo-DNAs for 28S rRNA (D). Adjacent section was hybridized with the complementary oligo-DNA for 28S rRNA in the presence of an excess amount of unrelated unlabeled oligo-DNA (E). Tissue was treated with RNase prior to hybridization with MB probe for 28S rRNA (F). Uterine sections were hybridized with complementary oligo-DNA probe (G) homologous (H) and complementary sequences in the presence of excess amounts of unlabeled complementary (I) and unlabeled homologous oligo-DNAs (J) for 28S rRNA. Uterine sections were hybridized with the complementary 28S rRNA in the presence of an excess amount of unrelated unlabeled oligo-DNA (K). Tissue was treated with RNase prior to hybridization with complementary oligo-DNA probes for 28S rRNA (L). Magnification ×400. Bar = 20 μm.
Simultaneous detection of ERα mRNA and 28S rRNA using MB probes
In mouse uterus, ERα mRNA and 28S rRNA were detected simultaneously using MB probes. In mouse uterine sections hybridized with complementary MB probe for 28S rRNA, fluorescent signals were observed in all cells (Fig. 6A). Among them, the strongest fluorescent signals were observed in the glandular and luminal epithelium. On the same slide, the antisense MB probe for ERα mRNA was detected in glandular and luminal epithelial cells and some stromal cells (Fig. 6B). Merged image was shown in Figure 6E and arrows indicate co-localization. No signals from homologous MB probe for 28S rRNA (Fig. 6C) and sense MB probe for ERα mRNA (Fig. 6D) were detected in mouse uterine sections.
Fig. 6.
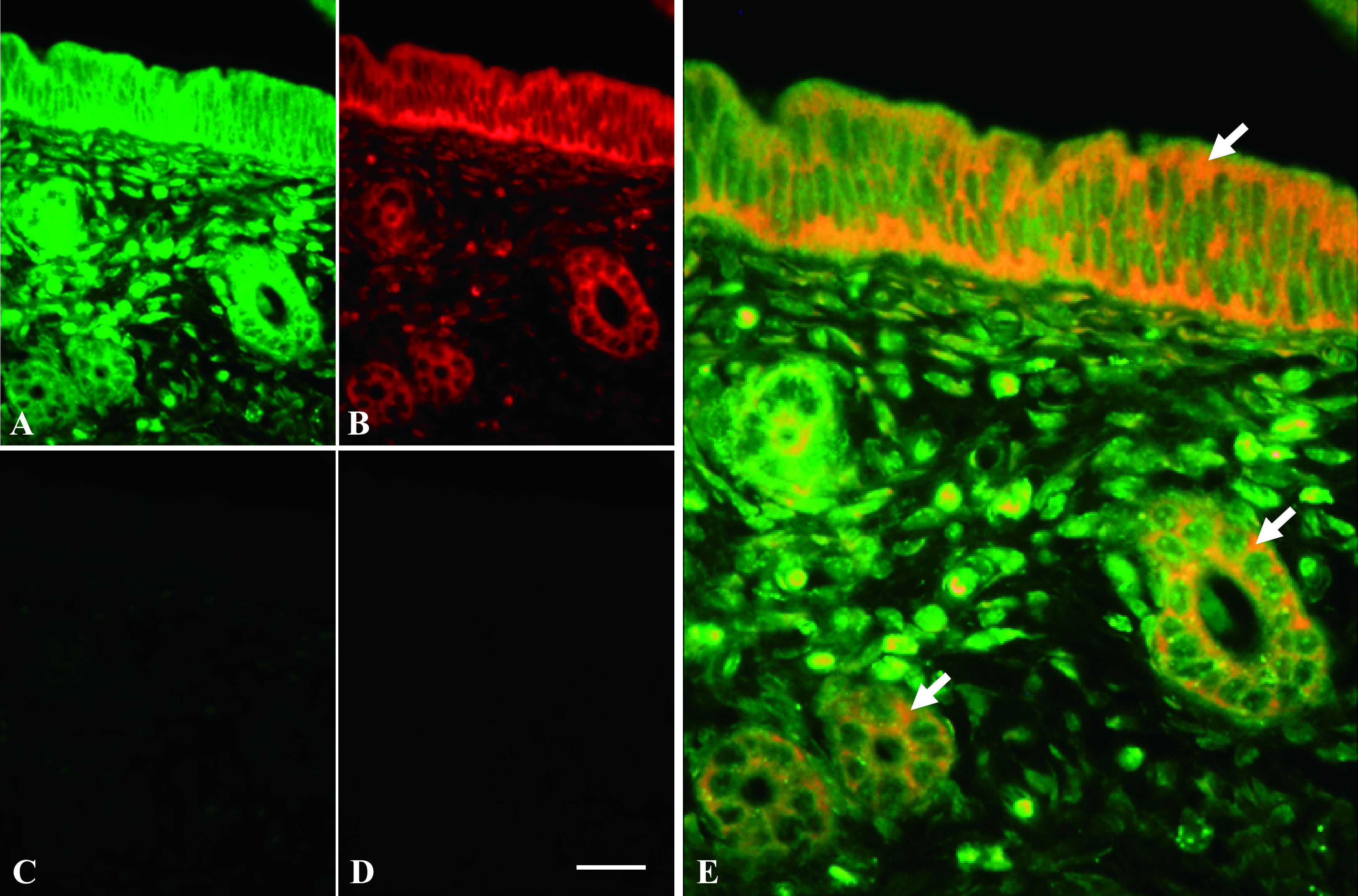
ERα mRNA and 28S rRNA expression in mouse uterine tissue using ISH. Uterine tissue was hybridized with complementary MB probe for 28S rRNA (green) (A) and antisense MB probe for ERα mRNA (red) (B). Both images were merged in (E), and arrows indicate co-localization of ERα mRNA and 28S rRNA expressions. Adjacent uterine section was hybridized with homologous MB probe for 28S rRNA (C) and sense MB probe for ERα mRNA (D). Magnification ×400. Bar = 20 μm.
Discrimination of mismatched transcripts by MB probes
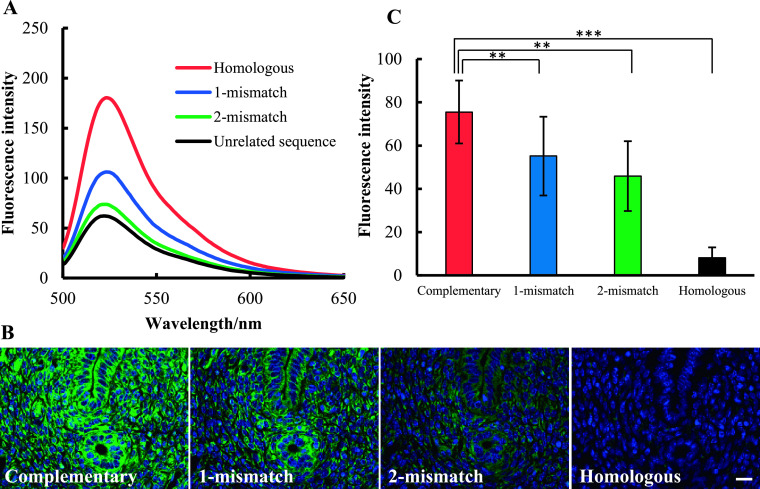
The mismatch discrimination ability of MB probe was tested both in medium and tissue sections. First, mismatch discrimination of MB probe was tested in medium. In this experiment, complementary MB probe for 28S rRNA was mixed with homologous sequence that has 1 or 2 base-mismatches (Table 1) (Fig. 7A). The fluorescent intensity was decreased in 1 or 2 base-mismatched sequences. Minimal fluorescence intensity was found in tube that mixed with unrelated sequence. Then, mismatch discrimination of MB probe was examined in tissue sections. As shown in Figure 7B, a strong fluorescent signal was observed in complementary MB probe for 28S rRNA. However, the fluorescent signal was gradually decreased with 1 or 2 base-mismatched MB probes. No fluorescent signal was detected in sections hybridized with the homologous MB probe for 28S rRNA. When the staining intensity was measured by ImageJ, significant decreases were confirmed between complementary and 1, 2 base-mismatched sequences (Fig. 7C). All these findings demonstrate that the MB probe could discriminate target sequence based on 1 or 2 base-mismatches in both medium and tissue sections.
Fig. 7.
Mismatch discrimination of MB probe. Complementary MB probe for 28S rRNA was hybridized with fully homologous, 1 or 2 base-mismatched sequences, then the fluorescence intensity was measured by fluorescence spectroscopy (A). For control experiment, the unrelated sequence was used. Mouse uterus sections were reacted with complementary MB probe and 1, 2 base-mismatched probes (B). For control experiment, the homologous MB probe was used. Magnification ×400. Bar = 20 μm. Fluorescence signal intensity was measured by ImageJ software and results were revealed as bar-graph (C).
IV. Discussion
In this study, we introduced a FRET-based MB probe to localize target RNAs in cultured cells and tissue by ISH. Pathological and basic research samples are generally preserved as formalin-fixed paraffin-embedded tissues, and banks of archived tissues preserved in this manner are useful resources for biomedical research [26]. Our results demonstrate that MB probes can be very useful for ISH, providing high sensitivity, specificity, and simpler compared to conventional probes for ISH.
The FRET-based MB probe enabled detection of 28S rRNA with high sensitivity and specificity. In our study, dot blot hybridization could detect as small as 10 pg of oligo-DNA, indicating that the MB probes are sufficiently sensitive for use in ISH, with sensitivity similar to that of Dig-labeled oligo-DNA probes [14]. In order to examine the hybridization kinetics of the MB probes, we measured the fluorescence intensity at 0, 3, 6, 12 and 24 hr using fluorescence spectroscopy. A strong fluorescent signal was observed with 3 hr of incubation, and there was no difference between 3 hr and overnight incubation. Conventional ISH protocols using T-T dimers or Dig-labeled probes require 2 to 3 days to complete, whereas analyses using an MB probe can be completed within 1 day due to the more-rapid hybridization, and an immunodetection system is not necessary for MB probes [21]. Recently, the fast FISH protocol is reported that only 1 hr is required for hybridization, however it is useful for DNA detection, but not for RNA [8, 9]. Therefore, a major advantage of using MB probes is the ability to rapidly detect multiple RNAs due to the comparatively faster hybridization.
Minimal non-specific staining is one of the major advantages of using MB probes, especially when using appropriate fluorophore and quencher molecules [17]. Indeed, the fluorescent dye and quencher molecules should be selected carefully to avoid overlap in the emission spectrum, which could affect fluorescence intensity and background staining [25]. We therefore chose FAM/DABCYL and Cy3/BHQ-2 pairs, which exhibit minimal spectral overlap. Indeed, it is difficult to evaluate background staining for 28S rRNA due to its ubiquitous expression throughout the cell. However, various control experimental results were clearly indicated that MB probe has less background staining in tissue sections. Furthermore, MB probes are also comparable to locked nucleic acid (LNA) probes, which are useful for detecting mRNA and miRNA. The high binding affinity of LNA probes confers both advantages and disadvantages, such as better detection of target sequences but also revealed strong non-specific staining [23]. Also, the major disadvantage of LNA probe is the difficulty to establish control experiments. Therefore, based on the FRET principle, MB probes provide the major advantage of decreased background staining in comparison to LNA, conventional haptens, and fluorescent-labeled probes.
In general, conventional linear probes have two possible states: unbound and bound to the target. However, MB probes have three possible states: free with a stem-loop conformation; free as a random coil; and bound to the target [38]. Based on free energy, the stem-loop conformation is the most stable state and does not produce fluorescence. Although the random coil state can produce fluorescence, unbound probes are washed out after the hybridization step. Therefore, fluorescent signals from the random coil state can be eliminated from tissues. Thus, in tissue sections, fluorescent signals will be produced only by MB probes that have hybridized with target RNAs, which ensures high specificity and minimal background staining [3, 39]. Most importantly, MB probes produce fluorescent signals directly when they bind to the target, without using additional immunohistochemical detection steps, which often cause non-specific staining in conventional ISH. Furthermore, fluorescent labeled probes were often used for FISH, which also uses as direct method. However, FISH is mainly used for DNA detection, but not for RNA. Detection of RNA, such as gene transcripts in cellular and tissue level is important to understand biological processes. Therefore, MB probes are beneficial for specific detection of target RNAs.
In addition, the reason why non-specific binding is reduced and consequently specific staining achieved with the use of MB probes might be that a higher free-energy barrier exists in the hairpin-opening process. A previous report described the potential and free-energy profiles for structured DNA (molecular beacon) and random-coil DNA [5]. Compared to the hybridization of unstructured single-stranded DNA (ssDNA; used in conventional ISH), the hybridization of structured ssDNA (used in this study) must overcome one more energy barrier, which represents the hairpin opening of the MB stem. Overcoming this energy barrier can be achieved with a perfectly matched target. Indeed, MB probes can be used to detect single-nucleotide polymorphisms based on the ability to discriminate single base differences in PCR [27, 30]. In our study, the fluorescence intensity decreased dramatically in 1 or 2 base-mismatch probes in both medium and tissue sections. These findings indicate that MB probes are highly specific for the detection of target RNAs.
In conclusion, ISH using a FRET-based MB probe is a highly sensitive and specific tool that enables simple and rapid detection of specific RNAs in cultured cells and paraffin-embedded tissues.
V. Conflicts of Interest
We have no conflict of interest to declare.
VI. Acknowledgments
This study was supported in part by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (No. 16K08471, 21K06738 to Y. Hishikawa, No. 19K16477 to N. Choijookhuu). We gratefully acknowledge the Frontier Science Research Center at the University of Miyazaki for allowing us to use their facilities.
VII. References
- 1.Alan, L., Zelenka, J., Jezek, J., Dlaskova, A. and Jezek, P. (2010) Fluorescent in situ hybridization of mitochondrial DNA and RNA. Acta Biochim. Pol. 57; 403–408. [PubMed] [Google Scholar]
- 2.Bauman, J. G., Wiegant, J., Borst, P. and van. Duijn, P. (1980) A new method for fluorescence microscopical localization of specific DNA sequences by in situ hybridization of fluorochromelabelled RNA. Exp. Cell Res. 128; 485–490. [DOI] [PubMed] [Google Scholar]
- 3.Bonnet, G., Tyagi, S., Libchaber, A. and Kramer, F. R. (1999) Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl. Acad. Sci. U S A 96; 6171–6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bratu, D. P., Catrina, I. E. and Marras, S. A. (2011) Tiny MBs for in vivo mRNA detection. Methods Mol. Biol. 714; 141–157. [DOI] [PubMed] [Google Scholar]
- 5.Chen, C., Wang, W., Wang, Z., Wei, F. and Zhao, X. S. (2007) Influence of secondary structure on kinetics and reaction mechanism of DNA hybridization. Nucleic Acids Res. 35; 2875–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choijookhuu, N., Sato, Y., Nishino, T., Endo, D., Hishikawa, Y. and Koji, T. (2012) Estrogen-dependent regulation of sodium/hydrogen exchanger-3 (NHE3) expression via estrogen receptor beta in proximal colon of pregnant mice. Histochem. Cell Biol. 137; 575–587. [DOI] [PubMed] [Google Scholar]
- 7.Cizkova, K., Foltynkova, T., Gachechiladze, M. and Tauber, Z. (2021) Comparative Analysis of Immunohistochemical Staining Intensity Determined by Light Microscopy, ImageJ and QuPath in Placental Hofbauer Cells. Acta Histochem. Cytochem. 54; 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coghlan, J. P., Aldred, P., Haralambidis, J., Niall, H. D., Penschow, J. D. and Tregear, G. W. (1985) Hybridization histochemistry. Anal. Biochem. 149; 1–28. [DOI] [PubMed] [Google Scholar]
- 9.Duncan, D. J., Vandenberghe, M. E., Scott, M. L. J. and Barker, C. (2019) Fast fluorescence in situ hybridisation for the enhanced detection of MET in non-small cell lung cancer. PLoS One 14; e0223926. doi: 10.1371/journal.pone.0223926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gago, S., Buitrago, M. J., Clemons, K. V., Cuenca-Estrella, M., Mirels, L. F. and Stevens, D. A. (2014) Development and validation of a quantitative real-time PCR assay for the early diagnosis of coccidioidomycosis. Diagn. Microbiol. Infect. Dis. 79; 214–221. [DOI] [PubMed] [Google Scholar]
- 11.Gall, J. G. and Pardue, M. L. (1969) Formation and detection of RNA-DNA hybrid molecules in cytological preparations. Proc. Natl. Acad. Sci. U S A 63; 378–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goel, G., Kumar, A., Puniya, A. K., Chen, W. and Singh, K. (2005) MB: a multitask probe. J. Appl. Microbiol. 99; 435–442. [DOI] [PubMed] [Google Scholar]
- 13.Hishikawa, Y., Damavandi, E., Izumi, S. and Koji, T. (2003) Molecular histochemical analysis of estrogen receptor alpha and beta expressions in the mouse ovary: in situ hybridization and Southwestern histochemistry. Med. Electron Microsc. 36; 67–73. [DOI] [PubMed] [Google Scholar]
- 14.Hishikawa, Y., Koji, T., Dhar, D. K., Kinugasa, S., Yamaguchi, M. and Nagasue, N. (1999) Metallothionein expression correlates with metastatic and proliferative potential in squamous cell carcinoma of the oesophagus. Br. J. Cancer 81; 712–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huber, D., Voithenberg, L. V. and Kaigala, G. V. (2018) Fluorescence in situ hybridization (FISH): History, limitations and what to expect from micro-scale FISH? Micro Nano Engineer. 1; 15–24. [Google Scholar]
- 16.Ishizuka, T., Liu, H. S., Ito, K. and Xu, Y. (2016) Fluorescence imaging of chromosomal DNA using click chemistry. Sci. Rep. 6; 33217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johansson, M. K. (2006) Choosing reporter-quencher pairs for efficient quenching through formation of intramolecular dimers. Methods Mol. Biol. 335; 17–29. [DOI] [PubMed] [Google Scholar]
- 18.Köhler, A., Lauritzen, B. and van, Noorden, C. J. F. (2000) Signal amplification in immunohistochemistry at the light microscopic level using biotinylated tyramide and nanogold–silver staining. J. Histochem. Cytochem. 48; 933–941. [DOI] [PubMed] [Google Scholar]
- 19.Koji, T. and Brenner, R. M. (1993) Localization of estrogen receptor messenger ribonucleic acid in rhesus monkey uterus by nonradioactive in situ hybridization with digoxigenin-labeled oligodeoxynucleotides. Endocrinology 132; 382–392. [DOI] [PubMed] [Google Scholar]
- 20.Koji, T., Marcio, C., Rubin, J. S., Slayden, O. D., Csaky, K. G., Aaronson, S. A., et al. (1994) Progesterone-dependent expression of keratinocyte growth factor mRNA in stromal cells of the primate endometrium: keratinocyte growth factor as a progestomedin. J. Cell Biol. 125; 393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koji, T. and Nakane, P. K. (1990) Localization in situ of specific mRNA using thymine-thymine dimerized DNA probes. Sensitive and reliable non-radioactive in situ hybridization. Acta Pathol. Jpn. 40; 793–807. [DOI] [PubMed] [Google Scholar]
- 22.Koji, T. and Nakane, P. K. (1996) Recent advances in molecular histochemical techniques: in situ hybridization and southwestern histochemistry. J. Electron Microsc. 45; 119–127. [DOI] [PubMed] [Google Scholar]
- 23.Kubota, K., Ohashi, A., Imachi, H. and Harada, H. (2006) Improved in situ hybridization efficiency with locked-nucleic-acid-incorporated DNA probes. Appl. Environ. Microbiol. 72; 5311–5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makristathis, A., Riss, S. and Hirschl, A. M. (2014) A novel fluorescence in situ hybridization test for rapid pathogen identification in positive blood cultures. Clin. Microbiol. Infect. 20; O760–O763. doi: 10.1111/1469-0691.12561. [DOI] [PubMed] [Google Scholar]
- 25.Marras, S. A. (2006) Selection of fluorophore and quencher pairs for fluorescent nucleic acid hybridization probes. Methods Mol. Biol. 335; 3–16. [DOI] [PubMed] [Google Scholar]
- 26.McDonald, S. A. (2010) Principles of Research Tissue Banking and Specimen Evaluation from the Pathologist’s Perspective. Biopreserv. Biobank. 8; 197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mhlanga, M. M. and Malmberg, L. (2001) Using MBs to detect single-nucleotide polymorphisms with real-time PCR. Methods 25; 463–471. [DOI] [PubMed] [Google Scholar]
- 28.Murakami, M., Kawakami, R., Niko, Y., Tsuda, T., Mori, H., Yatsuzuka, K., et al. (2020) High-quality Fluorescence Imaging of the Human Acrosyringium Using a Transparency: Enhancing Technique and an Improved, Fluorescent Solvatochromic Pyrene Probe. Acta Histochem. Cytochem. 53; 131–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakane, P. K. and Wilson, M. B. (1975) Cytochemical localization of polyadenylic acid (poly A). J. Cell Biol. 67; 302a. [Google Scholar]
- 30.Nguyen, L. V., Giannetti, S., Warren-Smith, S., Cooper, A., Selleri, S., Cucinotta, A., et al. (2014) Genotyping single nucleotide polymorphisms using different MB multiplexed within a suspended core optical fiber. Sensors (Basel) 14; 14488–14499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rudkin, G. T. and Stollar, B. D. (1977) High resolution detection of DNA-RNA hybrids in situ by indirect immunofluorescence. Nature 265; 472–473. [DOI] [PubMed] [Google Scholar]
- 32.Shiroto, Y., Saga, R., Yoshino, H., Hosokawa, Y., Isokawa, K. and Tsuruga, E. (2021) Matrix Metalloproteinase-2 Activated by Ultraviolet-B Degrades Human Ciliary Zonules In Vitro. Acta Histochem. Cytochem. 54; 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singer, R. H. and Ward, D. C. (1982) Actin gene expression visualized in chicken muscle tissue culture by using in situ hybridization with a biotinated nucleotide analog. Proc. Natl. Acad. Sci. U S A 79; 7331–7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Speicher, M. R., Gwyn Ballard, S. and Ward, D. C. (1996) Karyotyping human chromosomes by combinatorial multi-fluor FISH. Nat. Genet. 12; 368–375. [DOI] [PubMed] [Google Scholar]
- 35.Sugita, N., Choijookhuu, N., Yano, K., Lee, D., Ikenoue, M., Fidya, et al. (2021) Depletion of high-mobility group box 2 causes seminiferous tubule atrophy via aberrant expression of androgen and estrogen receptors in mouse testis. Biol. Reprod. 105; 1510–1520. [DOI] [PubMed] [Google Scholar]
- 36.Szuhai, K., Sandhaus, E., Kolkman-Uljee, S. M., Lemaitre, M., Truffert, J. C., Dirks, R. W., et al. (2001) A novel strategy for human papillomavirus detection and genotyping with SybrGreen and MB polymerase chain reaction. Am. J. Pathol. 159; 1651–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tyagi, S. and Kramer, F. R. (1996) MBs: probes that fluoresce upon hybridization. Nat. Biotechnol. 14; 303–308. [DOI] [PubMed] [Google Scholar]
- 38.Tyagi, S., Marras, S. A. and Kramer, F. R. (2000) Wavelength-shifting MBs. Nat. Biotechnol. 18; 1191–1196. [DOI] [PubMed] [Google Scholar]
- 39.Vargas, D. Y., Raj, A., Marras, S. A., Kramer, F. R. and Tyagi, S. (2005) Mechanism of mRNA transport in the nucleus. Proc. Natl. Acad. Sci. U S A 102; 17008–17013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, H., Eriksson, H. and Sahlin, L. (2000) Estrogen receptors alpha and beta in the female reproductive tract of the rat during the estrous cycle. Biol. Reprod. 63; 1331–1340. [DOI] [PubMed] [Google Scholar]
- 41.Wile, B. M., Ban, K., Yoon, Y. S. and Bao, G. (2014) Molecular beacon-enabled purification of living cells by targeting cell type-specific mRNAs. Nat. Protoc. 9; 2411–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshii, A., Koji, T., Ohsawa, N. and Nakane, P. K. (1995) In situ localization of ribosomal RNAs is a reliable reference for hybridizable RNA in tissue sections. J. Histochem. Cytochem. 43; 321–327. [DOI] [PubMed] [Google Scholar]