Abstract
Treated wastewater is a major pathway by which antibiotic resistance genes (ARG) enter aquatic ecosystems. However, knowledge gaps remain concerning the dissemination of specific ARG and their association with bacterial hosts. Here, we employed shotgun metagenomics to track ARG and taxonomic markers in river biofilms along a gradient of fecal pollution depicted by crAssphage signatures. We found strong evidence for an impact of wastewater effluents on both community composition and resistomes. In the light of such simultaneity, we employed a model comparison technique to identify ARG–host relationships from nonassembled metagenomic DNA. Hereby, a major cause of spurious associations otherwise encountered in correlation-based ARG–host analyses was suppressed. For several families of ARG, namely those conferring resistance to beta-lactams, particular bacterial orders were identified as candidate hosts. The found associations of blaFOX and cphA with Aeromonadales or blaPER with Chromatiales support the outcome of independent evolutionary analyses and thus confirm the potential of the methodology. For other ARG families including blaIMP or tet, clusters of bacterial orders were identified which potentially harbor a major proportion of host species. For yet other ARG, like, for example, ant or erm, no particular host candidates were identifiable, indicating their spread across various taxonomic groups.
Keywords: antibiotic resistance, bacterial community, biofilm, river, metagenomics
Short abstract
Shotgun metagenomics allows for the identification of candidate bacterial hosts of antibiotic resistance genes in river biofilms.
Introduction
Antibiotic resistance in pathogenic bacteria poses a global problem of increasing concern.1 Successful management of the crisis of antibiotic resistance requires a holistic approach that recognizes the particular role of terrestrial and aquatic ecosystems.2,3 Environmental systems are, on the one hand, an original source of antibiotic resistance genes (ARG).4,5 On the other hand, they provide reservoirs and pathways for the spread of acquired ARG that emerge and proliferate in conjunction with antibiotic therapies in human healthcare and livestock farming.6−8
In particular, the role of treated wastewater emissions on the occurrence of ARG in surface waters was examined in different geographical settings and with a focus on varying resistance determinants.9−11 Moreover, a number of successful attempts were made to disentangle the impacts of wastewater disposal and other anthropogenic activities on freshwater resistomes.12−14 The vast majority of those case studies relied on qPCR technology to quantify selected ARG and, possibly, genetic elements involved in their proliferation like, for example, integrons. In recent years, shotgun metagenomics (SMG) is increasingly employed to capture ARG in environmental systems.15−17 In contrast to alternative methodologies, SMG yields information on the gene pool of entire bacterial communities without prior selection of targets. Thus, SMG potentially detects a broad spectrum of resistance genes, including those not being typically considered in surveillance.18 Moreover, metagenomic sequencing allows for the simultaneous extraction of information on resistome and community composition from one and the same set of DNA sequences. Finally, relative abundances obtained by SMG are believed to be unbiased since neither cultivation nor PCR are involved which could result in unequal target amplification. At the same time, the lack of amplification is considered the main drawback of SMG as it necessarily poses restrictions on sensitivity.
One of the biggest challenges of current antibiotic resistance research remains pinpointing the bacterial hosts of ARG in complex environmental communities. This issue is nowadays tackled by two complementary approaches. On the one hand, molecular methods are being developed to actually codetect ARG and phylogenetic markers in single cells via sophisticated PCR protocols,19,20 single-cell sequencing21 or the sequencing of metagenomic DNA that underwent cross-linking prior to extraction.22,23 On the other hand, there are attempts to link resistance and phylogenetic information exclusively in the computational domain. This includes the assembly of large contigs from fragmented metagenomic DNA24,25 and the study of empirical associations between the abundances of ARG and phylogenetic markers.16,26,27
With the assembly based approaches, reconstructed DNA fragments (contigs) are scanned for physical co-occurrences of ARG and phylogenetic markers on one and the same fragment such that, ideally, ARG–host relations can be deduced from individual samples. By contrast, the statistical approach necessarily compares multiple samples of different community composition to identify associations between the abundance of ARG and taxonomic markers. For the latter approach, ARG and taxonomic markers do not need to reside on identical DNA fragments. Considering that nonassembled fragments obtained from short-read sequencing typically comprise about 150–300 bp only, such co-occurrences actually represent very rare exceptions.
Experience with the different approaches to host identification was recently compiled in a review paper.28 Of the 21 case studies considered therein, the majority addressed microbiomes of wastewater or waste originating from livestock farming. Cultivation-independent studies on ARG–host associations in freshwater systems, however, are still scarce,16,29−31 with information being largely confined to eastern Asia.
The aim of the present study is to shed light on ARG–host associations in freshwater biofilms of a European river receiving treated wastewater from multiple plant effluents.
Here, we chose the statistical approach which compares ARG abundances in nonassembled metagenomic DNA across samples of variable community composition. In comparison to metagenome assembly, this approach is computationally cheap, and it consistently covers both chromosomal and plasmid-borne ARG. However, empirical ARG–host associations are prone to spurious correlations28 arising from hidden effects which simultaneously control resistome and community. The exposure of sampling sites to differing loads of treated wastewater is a typical example of such an effect. Consequently, we advanced the statistical inference of ARG–hosts relations from nonassembled metagenomic DNA by implementing a simple but novel model comparison technique. The latter accounts for simultaneous effects of wastewater disposal on resistomes and bacterial community composition and thus suppresses a major cause of spurious ARG–host relationships. The actual exposure of sampling sites to treated wastewater was assessed through the quantification of crAssphage sequences as proposed recently.32 To our knowledge, this study is the first of its kind addressing ARG–host relations in a river of central Europe. The outcomes are discussed in light of existing knowledge on the distribution of ARG across taxonomically defined bacterial groups.
Materials and Methods
Study Site and Sampling
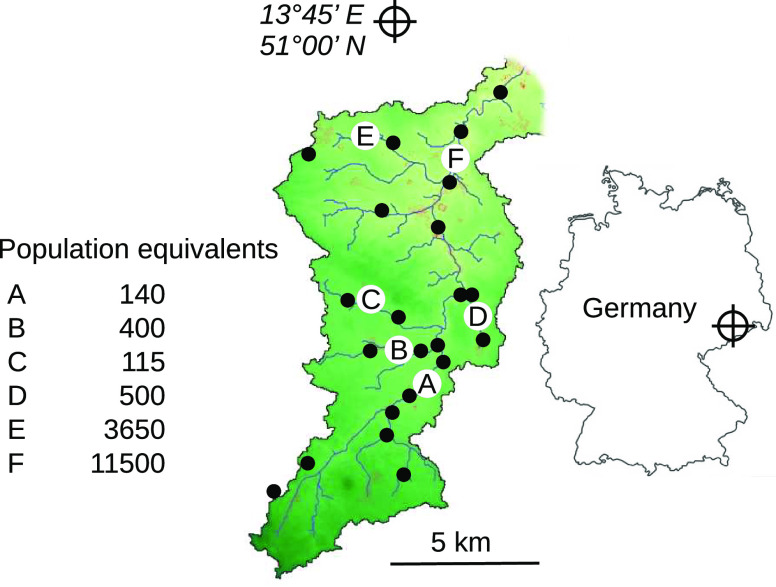
Samples were taken from the bottom sediment of a German mountain stream and its tributaries (Lockwitzbach River; Figure 1) in September 2017 and July 2018. The spatial distribution of sampling sites was chosen to reflect a gradient of anthropogenic pollution arising from the disposal of municipal wastewater. The latter is discharged into the river at six treatment plants (WWTP) with capacities ranging from about 100 to 12 000 population equivalents (Figure 1).
Figure 1.
Study catchment with sampling points (filled dots) and wastewater treatment plants (capital letters). Population equivalents reflect nominal treatment capacities reported by the environmental agency of Saxony (LfULG).
The sample material was obtained by pushing a transparent plastic tube vertically into the river bed and was representative of the top 5 cm of bottom sediments. The raw material mainly consisted of sand and silt mixed with varying proportions of gravel. Biofilm was growing on the surface of those particles, and a significant part of it was detached for DNA extraction (details below). Total organic matter accounted for approximately 2–5% of dry weight.33 For each site, the analyzed sample consisted of two pooled and thoroughly homogenized subsamples to accommodate for small-scale spatial heterogeneity. For reference purposes, effluent water (1 L per sample) was directly collected from the discharge pipe of the easily accessible treatment plant “F” (Figure 1). Glass bottles were used for intermediate storage of sediment material and effluent water at 4 °C before DNA extraction within 12 h after sampling. All glassware was pretreated with sodium hypochlorite to prevent a contamination of samples with foreign traces of DNA.
DNA Extraction and Sequencing
DNA was obtained from both liquid and sediment samples by means of a common extraction protocol. This was to avoid potential bias arising from the use of different protocols on subsets of samples. Consequently, sediment samples were preconditioned by rigorous shaking (450 rpm, 20 min) and the addition of sterile 0.1% sodiumdiphosphate (Na4P2O7) solution (20 mL per 100 g wet weight). According to experience,33 this detaches a significant proportion of bacteria from particle surfaces such that cells residing in the liquid phase are clearly representative of formerly attached biofilm. Liquid-phase material from preconditioned sediment (10 mL per sample) and WWTP effluent water (300 mL per sample) were filtered through 0.2 μm polycarbonate membranes (Whatman, Maidstone, Great Britain). DNA was extracted from filter residues by means of the PowerWaterKit (Qiagen, Hilden, Germany).
The obtained amounts of DNA ranged between 10 and 15 ng mL–1 for WWTP effluent water and between 50 and 150 ng DNA mL–1 for the sediment. The extracted DNA was stored at −20 °C until sequencing. All samples were shotgun-sequenced on a HiSeq device (2 × 150 bp paired end) by Eurofins Genomics Europe Sequencing GmbH, Konstanz, Germany.
Bioinformatics
We employed Trim Galore(34) to simultaneously remove adapter sequences, barcodes, and low quality information. Specifically, a Phread score of 28 (base call accuracy of 99.85%) was requested. Reads with a length of less than 100 bp after adapter cutting and quality filtering were discarded. Paired end reads were merged with the pandaseq assembler35 using its default settings. The merged sequences were aligned to the resfinder database of acquired ARG,36 version January 2020, available from the Center for Genomic Epidemiology (http://www.genomicepidemiology.org/) by means of blastn (http://blast.ncbi.nlm.nih.gov). Only high quality alignments were retained by requesting a maximum e-value of 1 × 10–20, a minimum match length of 80 bp, and a sequence identity greater or equal to 95%. A particular read often aligned well with multiple related ARG or ARG variants. Those cases were treated as a single hit and only the longest common name information was retained such that, for example, tet(M) and tet(O) collapsed into just tet. If the gene family could not be identified with certainty, the respective hits were dropped entirely.
The merged reads were further scanned for signatures of 16S rRNA genes with metaxa2(37) to identify the taxonomic composition of the bacterial community and to normalize resistance gene counts. Finally, the merged reads were scanned for crAssphage signatures as an indicator of human fecal pollution32 using the NCBI reference sequence NC_024711.138 cut into chunks of 500 bp. Only alignments with e-values ≤1 × 10–20 and ≥95% identity within a minimum of 80 bp were considered as hits. Basic properties of the samples are summarized in Table 1.
Table 1. Basic Properties of the Analyzed Materiala.
| sample material | samples | average number of merged sequences per sample | 16S rRNA gene copies per sample (average) |
|---|---|---|---|
| river biofilm | 62 | 2.5 × 10+7 | 3.1 × 10+4 |
| WWTP effluent | 8 | 2.2 × 10+7 | 2.5 × 10+4 |
The number of sequences was counted after merging forward and reverse reads; the average sequence length was 260 bp.
Data Analysis
The strict quality criteria applied in sequence processing and alignment were complemented by additional filter criteria. The latter guard against the misinterpretation of sequencing errors and they help to prevent frequent type II errors merely attributable to very small sample sizes. In particular, we discarded all ARG from analysis which did not reach a threshold frequency of 10 copies in at least one of the samples. Similarly, taxonomically defined bacterial groups were only considered if at least 50 copies of the respective 16S rRNA gene were detected in any of the samples. The number of crAssphage copies per sample ranged between 0 and 166, and the respective information was used without filtering.
Theoretically, the reliability of ARG detection could be compromised by a high degree of redundancy in the sense that most of the reads yielding a hit align to exactly the same short subsequence within a longer ARG fragment. Consequently, we analyzed the subject coverage for every ARG and each sample using an indicator α (eq 1; 0 < α ≤ 1).
| 1 |
where TS represents the total number of nucleotides in the subject sequence (length of gene fragment in resfinder database) and CS is the number of nucleotides of the subject sequence actually covered by hits. Finally, Cmax represents the upper limit of CS for a theoretical situation where all hits are fully independent without any overlaps. Consequently, α = 1 signals optimum conditions with all hits are nonredundant in the sense that they cover independent regions of the subject sequence. Lower values indicate partial redundancy, that is, overlaps between hits in terms of the covered nucleotides of the subject. For our data set, the “global” value of α aggregated over all samples and resistance genes was 0.86. Hence, only a minor fraction (≈15%) of the alignments counted as hits was actually subject to redundancy. At the level of individual genes, the values of α ranged between 0.77 (gene ant) and 0.91 (qnrS) which was considered acceptable.
To facilitate a comparison of ARG prevalence across samples, the number of ARG copies was generally divided by the corresponding number of 16S rRNA gene copies to yield relative abundances.39 The number of crAssphage copies found in a sample was normalized to the number of scanned reads reflecting the total amount of analyzed DNA. Models relating the abundance of ARG to the abundance of bacterial groups were fitted using raw gene counts to avoid artificial skill resulting from common normalization.40
Data analysis was conducted in R version 4.1.41 Empirical associations or differences between sample groups were tested for significance with nonparametric methods exclusively, namely “cor.test(method = “spearman”)” and “wilcox.test()”. Standard linear models were fitted with “lm()” and likelihood ratio tests were performed with “lrtest()”. Sorting of matrix rows by similarity was performed by hierarchical cluster analysis (“hclust()”, “cutree()”). In the context of multiple hypothesis testing, p values were adjusted with “p.adjust(method = “BH”)” to control the false discovery rate.42
Results and Discussion
General Properties of Samples
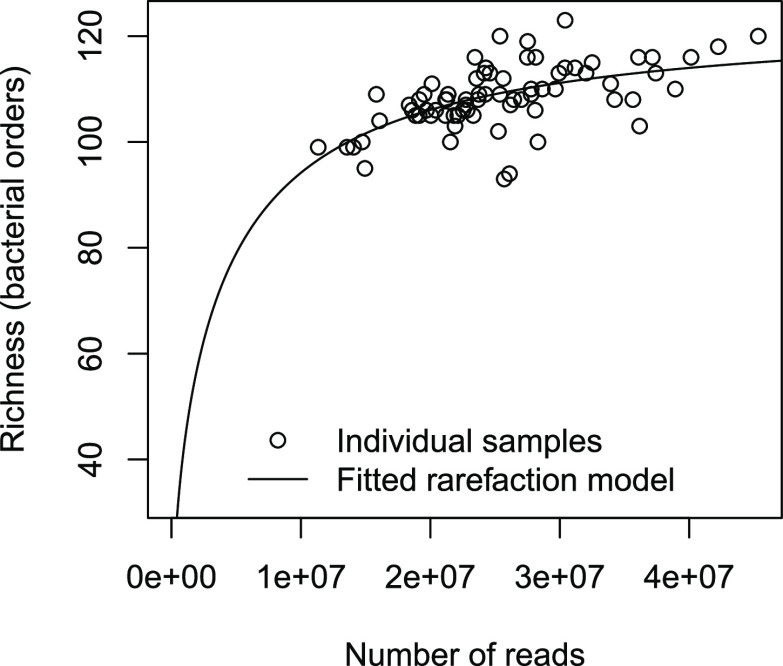
The average number of reads per sediment sample was 2.5 × 107 with only moderate variation (range: 1.1 × 107–4.5 × 107). At the same time, the samples did not vary substantially in terms of diversity based on the number of unique bacterial orders detected. In a rarefaction plot, all samples fell into the region where the curve levels off suggesting that diversity was adequately represented by the amount of sequenced material (Figure 2). No indication was found for a correlation between bacterial diversity and the amount of crAssphage signatures contained in samples (Spearman’s ρ = 0.07, p = 0.4).
Figure 2.
Number of bacterial orders detected in river biofilm in relation to sample size (number of reads). The structure of the fitted theoretical rarefaction model was adopted from Hess et al. 2019.39
Exposure of Sampling Sites to Treated Wastewater
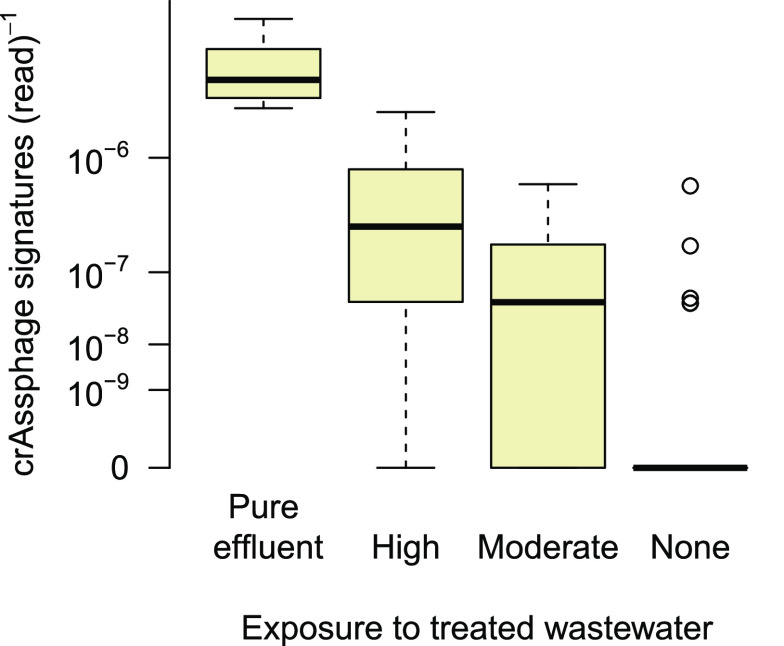
Wastewater effluents potentially affect both the resistome and the composition of bacterial communities in receiving waters. In statistical analyses targeted at the identification of ARG host, those simultaneous effects must be taken into account to avoid spurious correlations. However, the pollution status of a particular sampling site with regard to treated wastewater is difficult to quantify. This is due to uncertainty in estimated effluent rates, dilution effects, as well as possible in-stream retention all of which are subject to temporal variability. Recently, it has been suggested to employ the signature of crAssphage, a bacteriophage associated with the human gut flora38 as a tracer for fecal pollution.32 To verify the applicability of this approach, sampling sites were first grouped into ordinal classes according to the suspected exposure to wastewater: “None”’ for sites upstream of any WWTP, “High” for sites immediately downstream of WWTP where effluent and river have just mixed, and “Moderate” for sites in greater distance to upstream plants (≥2 km) where impacts of wastewater have potentially been attenuated by in-stream retention and dilution already. Then, all samples were scanned for crAssphage signatures and relative abundances were compared across the ordinal exposure classes (Figure 3). In fact, the number of crAssphage sequences per read differed significantly between sample classes with a continuous increase from unspoiled to highly exposed sites. In accordance with expectation, crAssphage abundance was highest in pure effluent water.
Figure 3.
Relative abundance of crAssphage signatures in pure WWTP effluent (very left) and river biofilms with different exposure to treated wastewater. Differences between adjacent groups are significant with all p < 0.02 (one-sided Wilcoxon rank sum test).
While the majority of samples collected upstream of any WWTP was free of crAssphage signatures, elevated signals were found at some sampling points (outliers in the rightmost box of Figure 3). The respective samples do not stand out from the rest, neither in sequencing depth (number of reads) nor in terms of bacterial diversity. Thus, very likely, these outliers indicate cases of illegal wastewater disposal. However, they could also reflect a lack of specificity since crAssphage signatures have recently been detected in feces of nonhuman origin.43
In conclusion, the number of crAssphage signatures per read appears to be a reasonable indicator for the exposure of sampling sites to treated wastewater. Since crAssphage information is continuous and quantitative, it was used as a proxy for wastewater exposure in all downstream statistics.
Impact of Wastewater on Bacterial Community Composition
Understanding the possible impact of wastewater disposal on the composition of in-river bacterial communities is necessary to properly interpret resistome information. In total, 71 bacterial orders were detected based on the chosen filter criteria. For about 20% of those, a positive association between their relative abundance and the exposure of the sampling sites to treated wastewater was established (Table 2). As expected, negative associations were observed as well, reflecting a linkage of certain bacterial groups with primary environmental habitats.
Table 2. Bacterial Orders Whose Relative Abundance in Riverbed Biofilms Was Positively Associated with crAssphage Contamination As an Indicator for Fecal Pollution.
| phylum | order | Spearman’s ρ | adj. p-value |
|---|---|---|---|
| Bacteroidetes | Flavobacteriales | 0.51 | 7.4 × 10–05 *** |
| Chlamydiae | Chlamydiales | 0.51 | 6.6 × 10–05 *** |
| Chloroflexi | Chloroflexales | 0.28 | 0.039 * |
| Cyanobacteria | Subsection I | 0.46 | 0.00033 *** |
| Subsection II | 0.43 | 9 × 10–04 *** | |
| Subsection III | 0.35 | 0.0078 ** | |
| Subsection V | 0.35 | 0.0068 ** | |
| Firmicutes | Lactobacillales | 0.53 | 6.1 × 10–05 *** |
| Fusobacteria | Fusobacteriales | 0.49 | 0.00014 *** |
| Proteobacteria | Bdellovibrionales | 0.47 | 0.00024 *** |
| Legionellales | 0.45 | 0.00053 *** | |
| Pseudomonadales | 0.37 | 0.0053 ** | |
| Rickettsiales | 0.66 | 3 × 10–08 *** | |
| Tenericutes | Mycoplasmatales | 0.38 | 0.0038 ** |
The bacterial groups listed in Table 2 form a heterogeneous set. The phyla Bacteroidetes, Firmicutes, Fusobacteria, and Proteobacteria, for example, are known to make up most of the human gut microbiome44,45 and a positive association is thus in agreement with expectation. Bacteroidetes and Proteobacteria have also been shown to majorly contribute to the composition of activated sludge microbiomes.46
By contrast, the correlation of crAssphage contamination with the abundance of Cyanobacteria is likely attributed to eutrophication caused by incomplete removal of nutrients in sewage treatment.47 These two examples illustrate how wastewater disposal can potentially shape river microbiomes via mechanisms of selection and invasion. The latter term, however, must be used with care since, depending on bacterial adaptive capabilities, we have to expect all forms of invasion ranging from only transient presence to the persistent integration into freshwater communities.
Although the observed relations between community composition and crAssphage abundance are plausible, the analyses are potentially compromised by the high level of aggregation. In particular, positive associations may remain unidentified due to the simultaneous occurrence of positive and negative association between crAssphage and taxonomic units at lower phylogenetic levels. For example, if “A” and “B” represent two bacterial families, a positive link between crAssphage and “A” may be “canceled out” by a negative link with “B” when the analysis is performed at order level.
Association of Wastewater Exposure with Antibiotic Resistance Genes
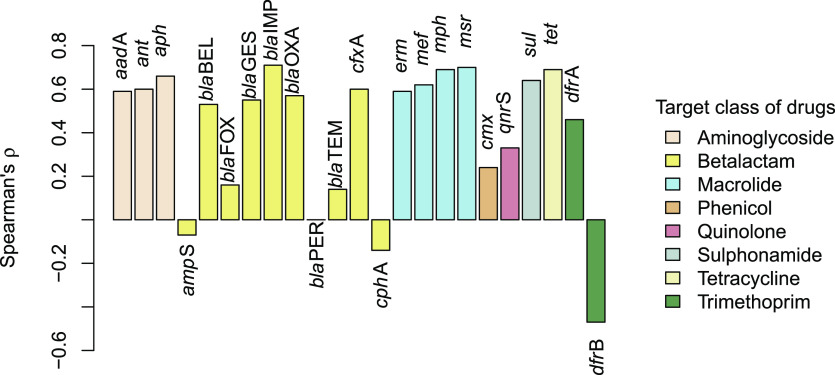
Relative ARG abundance was positively correlated with the exposure of sampling sites to treated wastewater for most of the genes (Figure 4, Supporting Information Table S1). This was especially true for genes conferring resistance to aminoglycosides, macrolides, sulfonamide, or tetracyclines. By contrast, there was no consistent pattern for genes providing resistance to beta-lactams or trimethoprim.
Figure 4.
Association of the relative abundance of ARG with the relative abundance of crAssphage signatures. All cases where |ρ|≥ 0.3 meet the significance criterion of p < 0.001 after correction for multiple testing.
In the case of gene families encoding for beta-lactamases, 5 out of 10 showed a positive association with crAssphage while the remaining five appeared to be unrelated. Between these two groups, no differences were found, neither with regard to the enzyme class nor in terms of the target spectrum of drugs. Specifically, genes coding for class A, B, and D beta-lactamases were present in both groups. Likewise, resistance to carbapenems, cephalosporins, cephamycins, monobactams, penams, or penems was not uniquely linked to either group, and most of the gene families—in both groups—are known to be effective against multiple beta-lactam antibiotics according to CARD records.48 Heterogeneous trends with regard to the occurrence of distinct beta-lactam resistance genes were also reported for other environments. For example, blaTEM genes were found to be ubiquitous in soil microbiomes, whereas the abundances of blaOXA or blaCTX correlated with anthropogenic impacts.49,50
Of particular interest is the result for the two variants of dihydrofolate reductase genes (dfr) conferring trimethoprim resistance. While the relative abundance of type I resistance (dfrA) was significantly increased in samples contaminated with crAssphage signatures, the opposite was true for type II (dfrB). Strikingly, dfrB was detected in the majority of riverbed biofilms, whereas it was not detected in any sample of treatment plant effluent which is unlikely to be by chance (p = 0.023; Fisher’s exact test).
On the one hand, this could point toward an environmental origin of the dfrB genes. On the other hand, genes of the dfrB family have already been detected in genomes of various clinically relevant strains51 with clear evidence for mobility.52 Hence, the detection of dfrB in environmental samples could be a result of resistance dissemination predominantly from nonpoint sources. In Germany, combined trimethoprim and sulfonamide treatment is not restricted to human infections but it is also a legal medication in a veterinary context, for example, for cattle, pigs, and pets. Within the catchment, seven larger facilities of commercial livestock farming were identified from aerial images and on-site inspection which could potentially serve as hot-spots of diffuse resistance dissemination. However, a statistical comparison of ARG abundances observed up- and downstream of any of those facilities did not indicate significant effects, irrespective of the gene family (Wilcoxon test; all adj. p-values >0.2).
Resistome data from sites located upstream of any treatment plant effluent underwent a separate analysis to better understand the outlier samples depicted as dots in Figure 3. No statistically significant difference in the relative abundance of ARG was found when comparing samples with and without elevated levels of crAssphage signatures. One straightforward conclusion could be that the crAssphage signal was misleading in the sense that it reflected some local contamination with feces of nonhuman origin. However, another plausible interpretation would be that elevated crAssphase levels indeed originated from illegal disposal of wastewater but the contributing population was very small and so was the likelihood of ARG dissemination from actually colonized individuals.
Association of ARG Abundance with Taxonomic Groups
In this work, we followed the statistics-based approach toward the problem of ARG–host identification from metagenomic information. Considering the study design along a gradient of pollution (Figure 1), mere correlations between the abundance of ARG and taxonomic markers are likely to yield spurious outcomes. This is due to the fact that both the abundance of ARG and bacterial groups are often simultaneously affected by wastewater effluents (Table 2, Figure 4). Consequently, we refrained from examining correlations but followed a model comparison approach. Specifically, we fitted linear models to predict the number of ARG copies in a sample, y, using two predictors: the presence of crAssphage signatures, x1, and the number of 16S rRNA gene copies associated with a particular taxonomic group of bacteria, x2 (eqs 2 and 3). To infer the possible value of bacterial abundance information, the two-predictor model (eq 3) was compared to the single-predictor version (eq 2) by means of a likelihood ratio test. Models with negative coefficients a2 were generally discarded since inverse relationship between ARG and bacterial abundances are not of interest in the context of host identification.
| 2 |
| 3 |
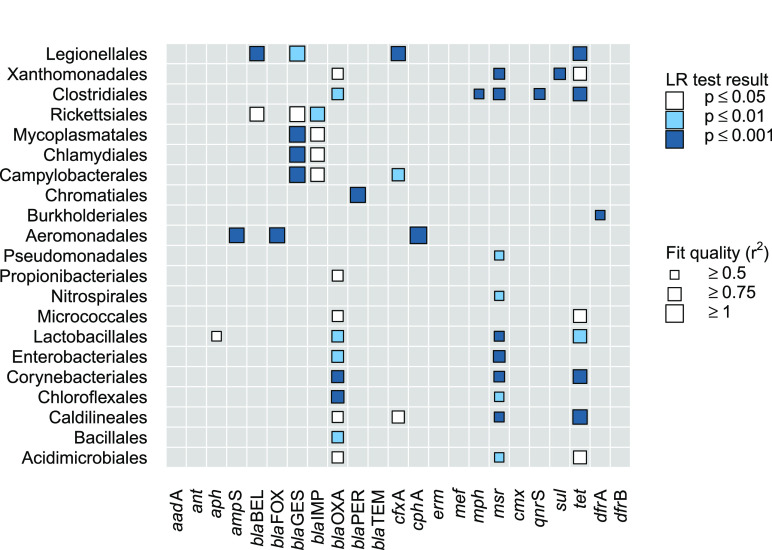
The model comparison approach disclosed a heterogeneous pattern (Figure 5).
Figure 5.
Performance of two-predictor models (eq 3) in relation to the single-predictor benchmarks (eq 2). Colors encode p-values of a likelihood ratio (LR) test with the null hypothesis being that community information does not contribute to predictive power; p-values were adjusted column-wise to account for the fact that all bacterial orders were considered as candidate predictors. Symbol size depicts the fraction of variance explained by eq 3; cases with r2 < 0.5 were suppressed for clarity. Row order was determined by cluster analysis.
For the majority of ARG, the incorporation of information on bacterial community composition allowed for an improved estimation of gene abundances. However, results differ substantially between particular ARG families. In the case of beta-lactam resistance genes like ampS, blaFOX, blaPER, or cphA, for example, only a single bacterial order contributed significantly to model performance. Thus, the respective orders are likely to play an actual role as host of the respective ARG.
By contrast, genes like blaGES, blaIMP, blaOXA, msr, or tet were found to be linked with numerous bacterial orders leaving more room for speculation. While all these orders could potentially serve as hosts, such outcomes are rather likely to reflect collinearity. The latter can arise from associations between bacterial community members due to, for example, common environmental preferences or even metabolic dependencies. Nevertheless, even those results are not entirely inconclusive since Figure 5 still depicts characteristic patterns. For example, the genes blaGES and blaIMP conferring resistance to carbapenems were found to be linked with a different set of bacterial orders than, for example, ARG of the blaOXA or tet group.
Finally, Figure 5 also highlights a few ARG families whose relative abundance was not significantly associated with any bacterial order (e.g., aadA, ant, erm). Such an outcome would be expected for ARG that are widely spread across taxonomic groups possibly facilitated through horizontal transfer of plasmids with a broad host range. In fact, genes like aadA or erm have been detected in chromosomal and plasmid DNA of numerous bacterial hosts according to current CARD48 records, including Gram-positive and negative species. Furthermore, the particular role of horizontal transfer for the dissemination of these genes has been highlighted earlier.53,54
Several of the bacterial orders highlighted in Figure 5 harbor potentially pathogenic species with drug resistance being clinically relevant. This includes, for example, Aeromonadales, Clostridiales, or Enterobacteriales, while some of the other bacterial groups have not been in the focus of drug resistance research so far. Unfortunately, a validation of the empirically identified relationships suggested by Figure 5 is not easily accomplished. On the one hand, case studies on ARG–host relationships in surface waters are not only limited in number but also with regard to geographical coverage.16,29−31 On the other hand, whole genome-based information on ARG prevalence found in repositories like CARD48 is potentially strongly biased toward culturable bacteria of clinical relevance.
Nevertheless, some of the ARG–host relations proposed here are strongly supported by previous studies on the phylogeny of resistance genes.55,56 This is especially so for ARG that were positively linked to the order Aeromonadales. For instance, in agreement with Figure 5, the fish pathogen Aeromonas allosaccharophila was recently identified as the original host of blaFOX genes.57 Similarly, the cphA gene was previously found to be linked with the host Aeromonas hydrophila.58
The association of blaPER beta-lactamases with the bacterial order Chromatiales implied by Figure 5 represents another case supported by external evaluation. Only recently, it was suggested that blaPER genes were originally acquired by the genus Pararheinheimera, a member of Chromatiales, long before the era of antibiotics59 with the occurrence in human pathogens being a result of later horizontal transmission.
At a higher level, the candidate associations proposed here support the outcome of previous research on ARG–host associations28 according to which the majority of hosts falls into the groups of Proteobacteria and Firmicutes. These two phyla contribute the majority of bacterial orders depicted in Figure 5.
The cases of blaFOX, blaPER, and cphA clearly illustrate that the statistical inference of ARG hosts from nonassembled metagenomic data is actually feasible. However, challenges remain with regard to several aspects. First of all, it seems necessary to apply the approach to even larger data sets exhibiting considerable variation in the composition of bacterial communities. On the one hand, this would allow for an improved detection of false positives by, for example, cross-validation methods. On the other hand, it may help to further disentangle some of the clusters depicted in Figure 5 where a particular set of ARG was associated with multiple bacterial groups and host relations thus remain inconclusive.
Second, it seems necessary to further increase resolution, especially of the taxonomic data as pointed out recently.28 This is due to considerable diversity at higher taxonomic levels like orders or phyla which may obscure ARG–host relations manifested at genera or species level. In this respect, progress could be made through sequencing technologies yielding longer reads.60
Third, the statistics-based approach to the identification of ARG can certainly profit from independent validation data produced with complementary techniques like, for example, metagenome assembly61 or Hi-C sequencing.23 A sole validation against information on ARG prevalence observed in isolates (many of which having a clinical background) can hardly suffice. Even worse, it could result in the false rejection of identified ARG–host relationships in environmental systems for the reason for apparent implausibility.
To possibly validate the ARG–host associations reported here, we applied the memory-efficient MEGAHIT software62 to assemble six of the metagenomes. About 1/5 of the ARG and 1/10 of the 16S rRNA-based taxonomic markers detected on the original reads were recovered from contigs. However, among the 8 × 106 generated contigs there were only three instances with simultaneous information on resistance and host. All three cases were related to the macrolide resistance gene msrE with Comamonadaceae (order Burkholderiales; 2×) and Microoscillaceae (order Cytophagales; 1×) being reported as the candidate hosts. This amount of information is clearly insufficient for serious validation. In our case, the major limitation appeared to be the insufficient length of the assembled contigs. While a few long sequences of up to about 900 kbp were obtained, median contig lengths ranged between 590 and 800 bp only and the codetection of ARG and specific 16S rRNA information was thus unlikely. Moreover, one needs to keep in mind that assembly based strategies are not suited to the identification of ARG host when the resistance is plasmid-borne. In fact, the full consideration of plasmid-borne ARG is one of the major advantages of the statistical approach to host identification.
A successful identification of ARG–host relations by the statistical approach requires a number of conditions to be met. First of all, multiple metagenomic samples must be analyzed each contributing millions of reads. A high number of reads per sample is the key to the detection and quantification of less abundant bacterial groups and ARG. Likewise, statistical significance of empirical ARG–host relationships is more likely to be established the more individual samples are available. However, a high number of deeply sequenced samples alone does not guarantee success. Rather, it is crucial that individual samples differ with regard to community composition. Without variance in community composition, the statistics-based inference of ARG–host relations would clearly be infeasible.
Finally, it needs to be stressed that any ARG–host relations proposed by statistical analyses should be treated with the usual caution. First, false positives cannot be avoided entirely and a small proportion of the identified associations may thus reflect coincidence. This is especially so when the number of multiple tests becomes large, for example, in response to a fine-grained decomposition of the bacterial community. To guard against false positives, obtained p-values should undergo rigorous adjustment as in the preparation of Figure 5. Second but more important, candidate bacterial groups must not be regarded as the “exclusive hosts”. While they potentially play an outstanding role, many ARG families have invaded multiple branches of the phylogenetic tree by horizontal transfer and certainly persist in multiple bacterial orders or phyla.63,64
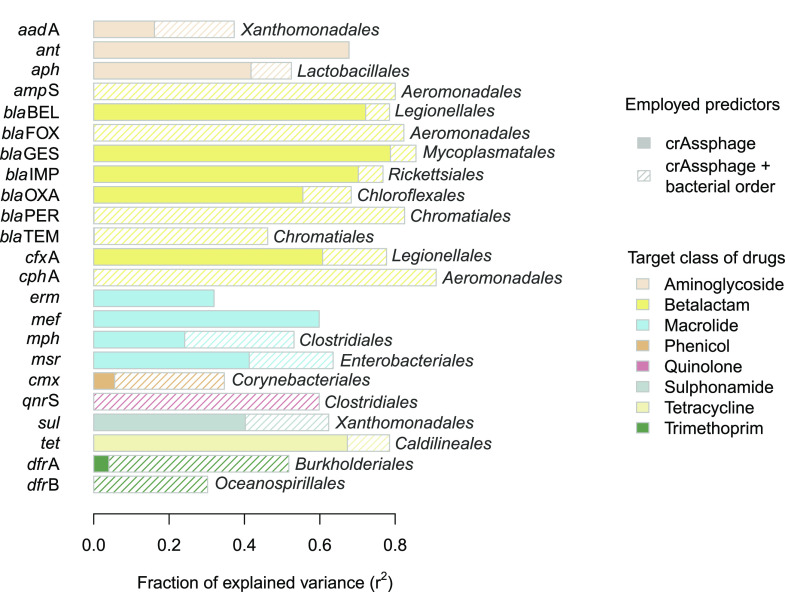
To better illustrate the possible contribution of taxonomic information to the prediction of ARG prevalence, the matrix from Figure 5 was filtered to those bacterial orders that allowed for the best fit of eq 3. Furthermore, the fraction of explained variance was decomposed so as to highlight the contribution of community information in relation to crAssphage information (Figure 6). For more than 3/4 of the gene families, crAssphage data combined with information on a single bacterial order explained over 50% of the observed variance in ARG abundance. For genes conferring resistance to beta-lactam antibiotics, up to about 80% of the variance were explained.
Figure 6.
Performance of linear models predicting the relative abundance of ARG families from crAssphage abundance (eq 2; solid bars) or crAssphage abundance combined with information on bacterial community composition (eq 3; total bar length). For each ARG family, the indicated bacterial order is the one that allowed for the best fit of eq 3 and shaded parts represent the corresponding improvement of the model. If no order name is given, community information had no added value whatsoever.
Synthesis
We found strong evidence for an impact of wastewater effluents on both the taxonomic composition of river bacterial biofilms and their enrichment with ARG. With regard to the latter, the strongest effects were observed for gene families conferring a reduced susceptibility to aminoglycosides, macrolides, tetracyclines, and selected beta-lactam antibiotics including carbapenems. However, the abundance of several other ARG families coding for beta-lactamases (e.g., blaTEM, ampS) or type II trimethoprim resistance (dfrB) was apparently unrelated to wastewater disposal. Hence, the respective genes are either naturally present in river microbiomes independent of human impacts or they are linked with anthropogenic nonpoint emissions.
The statistics-based approach to the identification of ARG hosts yielded promising results. In several cases, the relative abundance of a particular ARG proved to be significantly associated with the occurrence of just one or two specific bacterial orders. For several ARG families mediating beta-lactam resistance (blaFOX, blaPER, cphA) the proposed host relationships strongly support recent external evidence rooted in evolutionary analyses. However, even cases where a particular ARG is found to be associated with multiple bacterial groups (or no group at all) are not without relevance. In fact, those cases may reflect an efficient horizontal transfer of antibiotic resistance through highly mobile genetic elements and call for further examination. Overall, community composition proved to be a valuable input to statistical resistome models. Specifically, for 20 out of 23 resistance gene families, information on the abundance of particular bacterial orders significantly improved the performance of linear models predicting ARG abundance.
Acknowledgments
We thank Ulrike Mogck and Anna-Sophia Reichelt for their help with DNA isolation as well as Uli Klümper, Tamar Kohn, and three anonymous reviewers for valuable comments on an earlier version of the manuscript. This work has received funding from the German Ministry of Education and Research, BMBF (Grant Nos. 01KI1909A (EMBARK) and 16LC1904A (ANTIVERSA)) and from the German Research Foundation, DFG (Grant No. BE2299/14-1). The costs of sequencing were covered by the German Research Foundation (DFG) via the TU Dresden “support the best” program.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.est.2c00370.
Table S1: Relative abundance of antibiotic resistance genes (ARG copies/16S rRNA gene copies) in river biofilms sampled downstream of wastewater effluents (“highly exposed”) in comparison to values observed upstream of any treatment plant (“not exposed”). Reported p-values refer to a Wilcoxon rank sum test and underwent adjustment for multiple testing. Stars indicated significance according to the usual convention (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Laxminarayan R.; Duse A.; Wattal C.; Zaidi A. K. M.; Wertheim H. F. L.; Sumpradit N.; Vlieghe E.; Hara G. L.; Gould I. M.; Goossens H.; Greko C.; So A. D.; Bigdeli M.; Tomson G.; Woodhouse W.; Ombaka E.; Peralta A. Q.; Qamar F. N.; Mir F.; Kariuki S.; Bhutta Z. A.; Coates A.; Bergstrom R.; Wright G. D.; Brown E. D.; Cars O. Antibiotic resistance - the need for global solutions. Lancet Infectious Diseases 2013, 13, 1057–1098. 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- Tiedje J. M.; Wang F.; Manaia C. M.; Virta M.; Sheng H.; Ma L.; Zhang T.; Topp E. Antibiotic Resistance Genes in the Human-Impacted Environment: A One Health Perspective. Pedosphere 2019, 29, 273–282. 10.1016/S1002-0160(18)60062-1. [DOI] [Google Scholar]
- Berendonk T.; Manaia C.; Merlin C.; Fatta-Kassinos D.; Cytryn E.; Walsh F.; Bürgmann H.; Sørum H.; Norström M.; Pons M.-N.; Kreuzinger N.; Huovinen P.; Stefani S.; Schwartz T.; Kisand V.; Baquero F.; Martinez J. Tackling antibiotic resistance: the environmental framework. Nature Reviews Microbiology 2015, 13, 310–317. 10.1038/nrmicro3439. [DOI] [PubMed] [Google Scholar]
- D’Costa V. M.; King C. E.; Kalan L.; Morar M.; Sung W. W. L.; Schwarz C.; Froese D.; Zazula G.; Calmels F.; Debruyne R.; Golding G. B.; Poinar H. N.; Wright G. D. Antibiotic resistance is ancient. Nature 2011, 477, 457–461. 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- Canteón R. Antibiotic resistance genes from the environment: a perspective through newly identified antibiotic resistance mechanisms in the clinical setting. Clinical Microbiology and Infection 2009, 15, 20–25. 10.1111/j.1469-0691.2008.02679.x. [DOI] [PubMed] [Google Scholar]
- Pärnänen K. M. M.; Narciso-da-Rocha C.; Kneis D.; Berendonk T. U.; Cacace D.; Do T. T.; Elpers C.; Fatta-Kassinos D.; Henriques I.; Jaeger T.; Karkman A.; Martinez J. L.; Michael S. G.; Michael-Kordatou I.; O’Sullivan K.; Rodriguez-Mozaz S.; Schwartz T.; Sheng H.; Sørum H.; Stedtfeld R. D.; Tiedje J. M.; Giustina S. V. D.; Walsh F.; Vaz-Moreira I.; Virta M.; Manaia C. M. Antibiotic Resistance in European Wastewater Treatment Plants Mirrors the Pattern of Clinical Antibiotic Resistance Prevalence. Science Advances 2019, 5, eaau9124 10.1126/sciadv.aau9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.; Yang Q. E.; Zhou X.; Wang F.-H.; Muurinen J.; Virta M. P.; Brandt K. K.; Zhu Y.-G. Antibiotic resistome in the livestock and aquaculture industries: Status and solutions. Critical Reviews in Environmental Science and Technology 2021, 51, 2159–2196. 10.1080/10643389.2020.1777815. [DOI] [Google Scholar]
- He Y.; Yuan Q.; Mathieu J.; Stadler L.; Senehi N.; Sun R.; Alvarez P. J. J. Antibiotic resistance genes from livestock waste: occurrence, dissemination, and treatment. npj Clean Water 2020, 3, 1–11. 10.1038/s41545-020-0051-0. [DOI] [Google Scholar]
- LaPara T. M.; Burch T. R.; McNamara P. J.; Tan D. T.; Yan M.; Eichmiller J. J. Tertiary-Treated Municipal Wastewater is a Significant Point Source of Antibiotic Resistance Genes into Duluth-Superior Harbor. Environ. Sci. Technol. 2011, 45, 9543–9549. 10.1021/es202775r. [DOI] [PubMed] [Google Scholar]
- Sabri N. A.; Schmitt H.; Van der Zaan B.; Gerritsen H. W.; Zuidema T.; Rijnaarts H. H. M.; Langenhoff A. A. M. Prevalence of antibiotics and antibiotic resistance genes in a wastewater effluent-receiving river in the Netherlands. Journal of Environmental Chemical Engineering 2020, 8, 102245. 10.1016/j.jece.2018.03.004. [DOI] [Google Scholar]
- Cacace D.; Fatta-Kassinos D.; Manaia C. M.; Cytryn E.; Kreuzinger N.; Rizzo L.; Karaolia P.; Schwartz T.; Alexander J.; Merlin C.; Garelick H.; Schmitt H.; de Vries D.; Schwermer C. U.; Meric S.; Ozkal C. B.; Pons M.-N.; Kneis D.; Berendonk T. U. Antibiotic resistance genes in treated wastewater and in the receiving water bodies: A pan-European survey of urban settings. Water Res. 2019, 162, 320–330. 10.1016/j.watres.2019.06.039. [DOI] [PubMed] [Google Scholar]
- Storteboom H.; Arabi M.; Davis J. G.; Crimi B.; Pruden A. Tracking Antibiotic Resistance Genes in the South Platte River Basin Using Molecular Signatures of Urban, Agricultural, And Pristine Sources. Environ. Sci. Technol. 2010, 44, 7397–7404. 10.1021/es101657s. [DOI] [PubMed] [Google Scholar]
- Pruden A.; Arabi M.; Storteboom H. N. Correlation Between Upstream Human Activities and Riverine Antibiotic Resistance Genes. Environ. Sci. Technol. 2012, 46, 11541–11549. 10.1021/es302657r. [DOI] [PubMed] [Google Scholar]
- Amos G. C.; Gozzard E.; Carter C. E.; Mead A.; Bowes M. J.; Hawkey P. M.; Zhang L.; Singer A. C.; Gaze W. H.; Wellington E. M. H. Validated predictive modelling of the environmental resistome. ISME Journal 2015, 9, 1467–1476. 10.1038/ismej.2014.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson-Palme J.; Larsson D. G. J.; Kristiansson E. Using metagenomics to investigate human and environmental resistomes. J. Antimicrob. Chemother. 2017, 72, 2690–2703. 10.1093/jac/dkx199. [DOI] [PubMed] [Google Scholar]
- Li B.; Yang Y.; Ma L.; Ju F.; Guo F.; Tiedje J. M.; Zhang T. Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME Journal 2015, 9, 2490–2502. 10.1038/ismej.2015.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baral D.; Dvorak B. I.; Admiraal D.; Jia S.; Zhang C.; Li X. Tracking the Sources of Antibiotic Resistance Genes in an Urban Stream during Wet Weather using Shotgun Metagenomic Analyses. Environ. Sci. Technol. 2018, 52, 9033–9044. 10.1021/acs.est.8b01219. [DOI] [PubMed] [Google Scholar]
- Kneis D.; Berendonk T.; Heß S. High prevalence of colistin resistance genes in German municipal wastewater. Sci. Total Environ. 2019, 694, 133454. 10.1016/j.scitotenv.2019.07.260. [DOI] [PubMed] [Google Scholar]
- Spencer S. J.; Tamminen M. V.; Preheim S. P.; Guo M. T.; Briggs A. W.; Brito I. L.; A Weitz D.; Pitkänen L. K.; Vigneault F.; Virta M. P.; Alm E. J. Massively parallel sequencing of single cells by epicPCR links functional genes with phylogenetic markers. ISME Journal 2016, 10, 427–436. 10.1038/ismej.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultman J.; Tamminen M.; Pärnänen K.; Cairns J.; Karkman A.; Virta M. Host range of antibiotic resistance genes in wastewater treatment plant influent and effluent. FEMS Microbiology Ecology 2018, 94, fiy038. 10.1093/femsec/fiy038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourcy C. F. A. d.; Vlaminck I. D.; Kanbar J. N.; Wang J.; Gawad C.; Quake S. R. A Quantitative Comparison of Single-Cell Whole Genome Amplification Methods. PLoS One 2014, 9, e105585 10.1371/journal.pone.0105585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitel C. W.; Froenicke L.; Lang J. M.; Korf I. F.; Michelmore R. W.; Eisen J. A.; Darling A. E. Strain- and plasmid-level deconvolution of a synthetic metagenome by sequencing proximity ligation products. PeerJ. 2014, 2, e415 10.7717/peerj.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder T.; Press M. O.; Sullivan S.; Liachko I.; Top E. M. Linking the resistome and plasmidome to the microbiome. ISME journal 2019, 13, 2437–2446. 10.1038/s41396-019-0446-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.-X.; Anantharaman K.; Shaiber A.; Eren A. M.; Banfield J. F. Accurate and complete genomes from metagenomes. Genome Res. 2020, 30, 315–333. 10.1101/gr.258640.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers R. M.; Kyrpides N. C.; Stepanauskas R.; Harmon-Smith M.; Doud D.; Reddy T. B. K.; Schulz F.; Jarett J.; Rivers A. R.; Eloe-Fadrosh E. A.; Tringe S. G.; Ivanova N. N.; Copeland A.; Clum A.; Becraft E. D.; Malmstrom R. R.; Birren B.; Podar M.; Bork P.; Weinstock G. M.; Garrity G. M.; Dodsworth J. A.; Yooseph S.; Sutton G.; Glöckner F. O.; Gilbert J. A.; Nelson W. C.; Hallam S. J.; Jungbluth S. P.; Ettema T. J. G.; Tighe S.; Konstantinidis K. T.; Liu W.-T.; Baker B. J.; Rattei T.; Eisen J. A.; Hedlund B.; McMahon K. D.; Fierer N.; Knight R.; Finn R.; Cochrane G.; Karsch-Mizrachi I.; Tyson G. W.; Rinke C.; Lapidus A.; Meyer F.; Yilmaz P.; Parks D. H.; Murat Eren A.; Schriml L.; Banfield J. F.; Hugenholtz P.; Woyke T. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 2017, 35, 725–731. 10.1038/nbt.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J.-Q.; An X.-L.; Li B.; Chen Q.-L.; Gillings M. R.; Chen H.; Zhang T.; Zhu Y.-G. Metagenomics of urban sewage identifies an extensively shared antibiotic resistome in China. Microbiome 2017, 5, 84. 10.1186/s40168-017-0298-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y.-G.; Zhao Y.; Li B.; Huang C.-L.; Zhang S.-Y.; Yu S.; Chen Y.-S.; Zhang T.; Gillings M. R.; Su J.-Q. Continental-scale pollution of estuaries with antibiotic resistance genes. Nature Microbiology 2017, 2, 16270. 10.1038/nmicrobiol.2016.270. [DOI] [PubMed] [Google Scholar]
- Rice E. W.; Wang P.; Smith A. L.; Stadler L. B. Determining Hosts of Antibiotic Resistance Genes: A Review of Methodological Advances. Environmental Science & Technology Letters 2020, 7, 282–291. 10.1021/acs.estlett.0c00202. [DOI] [Google Scholar]
- Jia S.; Zhang X.-X.; Miao Y.; Zhao Y.; Ye L.; Li B.; Zhang T. Fate of antibiotic resistance genes and their associations with bacterial community in livestock breeding wastewater and its receiving river water. Water Res. 2017, 124, 259–268. 10.1016/j.watres.2017.07.061. [DOI] [PubMed] [Google Scholar]
- Qiu W.; Sun J.; Fang M.; Luo S.; Tian Y.; Dong P.; Xu B.; Zheng C. Occurrence of antibiotics in the main rivers of Shenzhen, China: Association with antibiotic resistance genes and microbial community. Sci. Total Environ. 2019, 653, 334–341. 10.1016/j.scitotenv.2018.10.398. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Lu S.; Liu X.; Chen J.; Han M.; Wang Z.; Guo W. Profiles of antibiotic resistance genes in an inland salt-lake Ebinur Lake, Xinjiang, China: The relationship with antibiotics, environmental factors, and microbial communities. Ecotoxicology and Environmental Safety 2021, 221, 112427. 10.1016/j.ecoenv.2021.112427. [DOI] [PubMed] [Google Scholar]
- Karkman A.; Pärnänen K.; Larsson D. G. J. Fecal pollution can explain antibiotic resistance gene abundances in anthropogenically impacted environments. Nat. Commun. 2019, 10, 80. 10.1038/s41467-018-07992-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heß S.; Berendonk T. U.; Kneis D. Antibiotic resistant bacteria and resistance genes in the bottom sediment of a small stream and the potential impact of remobilization. FEMS Microbiology Ecology 2018, 94, fiy128. 10.1093/femsec/fiy128. [DOI] [PubMed] [Google Scholar]
- Krueger F.A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files, 2015. https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ [Google Scholar]
- Masella A. P.; Bartram A. K.; Truszkowski J. M.; Brown D. G.; Neufeld J. D. PANDAseq: paired-end assembler for illumina sequences. BMB Bioinformatics 2012, 13, 31. 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E.; Hasman H.; Cosentino S.; Vestergaard M.; Rasmussen S.; Lund O.; Aarestrup F. M.; Larsen M. V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson-Palme J.; Hartmann M.; Eriksson K. M.; Pal C.; Thorell K.; Larsson D. G. J.; Nilsson R. H. Improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Molecular Ecology Resources 2015, 15, 1403–1414. 10.1111/1755-0998.12399. [DOI] [PubMed] [Google Scholar]
- Dutilh B. E.; Cassman N.; McNair K.; Sanchez S. E.; Silva G. G. Z.; Boling L.; Barr J. J.; Speth D. R.; Seguritan V.; Aziz R. K.; Felts B.; Dinsdale E. A.; Mokili J. L.; Edwards R. A. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat. Commun. 2014, 5, 4498. 10.1038/ncomms5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heß S.; Kneis D.; Österlund T.; Li B.; Kristiansson E.; Berendonk T. Sewage from airplanes exhibits high abundance and diversity of antibiotic resistance genes. Environ. Sci. Technol. 2019, 53, 13898–13905. 10.1021/acs.est.9b03236. [DOI] [PubMed] [Google Scholar]
- Pearson K. Mathematical contributions to the theory of evolution. – On a form of spurious correlation which may arise when indices are used in the measurement of organs. Proceedings of the Royal Society of London 1897, 60, 489–498. 10.1098/rspl.1896.0076. [DOI] [Google Scholar]
- R Core Team R.: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Benjamini Y.; Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B 1995, 57, 289–300. 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- Li Y.; Gordon E.; Shean R. C.; Idle A.; Deng X.; Greninger A. L.; Delwart E. CrAssphage and its bacterial host in cat feces. Sci. Rep. 2021, 11, 815. 10.1038/s41598-020-80076-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson M. J.; Cusack S.; O’Sullivan O.; Greene-Diniz R.; de Weerd H.; Flannery E.; Marchesi J. R.; Falush D.; Dinan T.; Fitzgerald G.; Stanton C.; van Sinderen D.; O’Connor M.; Harnedy N.; O’Connor K.; Henry C.; O’Mahony D.; Fitzgerald A. P.; Shanahan F.; Twomey C.; Hill C.; Ross R. P.; O’Toole P. W. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 4586–4591. 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida A.; Mitchell A. L.; Boland M.; Forster S. C.; Gloor G. B.; Tarkowska A.; Lawley T. D.; Finn R. D. A new genomic blueprint of the human gut microbiota. Nature 2019, 568, 499–504. 10.1038/s41586-019-0965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J.; Bu Y.; Zhang X.-X.; Huang K.; He X.; Ye L.; Shan Z.; Ren H. Metagenomic analysis of bacterial community composition and antibiotic resistance genes in a wastewater treatment plant and its receiving surface water. Ecotoxicology and Environmental Safety 2016, 132, 260–269. 10.1016/j.ecoenv.2016.06.016. [DOI] [PubMed] [Google Scholar]
- Hendriks A. T. W. M.; Langeveld J. G. Rethinking Wastewater Treatment Plant Effluent Standards: Nutrient Reduction or Nutrient Control?. Environ. Sci. Technol. 2017, 51, 4735–4737. 10.1021/acs.est.7b01186. [DOI] [PubMed] [Google Scholar]
- Alcock B. P.; Raphenya A. R.; Lau T. T. Y.; Tsang K. K.; Bouchard M.; Edalatmand A.; Huynh W.; Nguyen A.-L. V.; Cheng A. A.; Liu S.; Min S. Y.; Miroshnichenko A.; Tran H.-K.; Werfalli R. E.; Nasir J. A.; Oloni M.; Speicher D. J.; Florescu A.; Singh B.; Faltyn M.; Hernandez-Koutoucheva A.; Sharma A. N.; Bordeleau E.; Pawlowski A. C.; Zubyk H. L.; Dooley D.; Griffiths E.; Maguire F.; Winsor G. L.; Beiko R. G.; Brinkman F. S. L.; Hsiao W. W. L.; Domselaar G. V.; McArthur A. G. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatica J.; Yang K.; Pagaling E.; Jurkevitch E.; Yan T.; Cytryn E. Resistance of Undisturbed Soil Microbiomes to Ceftriaxone Indicates Extended Spectrum ß-Lactamase Activity. Frontiers in Microbiology 2015, 6, 1233. 10.3389/fmicb.2015.01233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampouris I. D.; Agrawal S.; Orschler L.; Cacace D.; Kunze S.; Berendonk T. U.; Klümper U. Antibiotic resistance gene load and irrigation intensity determine the impact of wastewater irrigation on antimicrobial resistance in the soil microbiome. Water Res. 2021, 193, 116818. 10.1016/j.watres.2021.116818. [DOI] [PubMed] [Google Scholar]
- Lemay-St-Denis C.; Diwan S.-S.; Pelletier J. N. The Bacterial Genomic Context of Highly Trimethoprim-Resistant DfrB Dihydrofolate Reductases Highlights an Emerging Threat to Public Health. Antibiotics 2021, 10, 433. 10.3390/antibiotics10040433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulouse J. L.; Edens T. J.; Alejaldre L.; Manges A. R.; Pelletier J. N. Integron-Associated DfrB4, a Previously Uncharacterized Member of the Trimethoprim-Resistant Dihydrofolate Reductase B Family, Is a Clinically Identified Emergent Source of Antibiotic Resistance. Antimicrob. Agents Chemother. 2017, 61, e02665–16. 10.1128/AAC.02665-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popowska M.; Krawczyk-Balska A. Broad-host-range IncP-1 plasmids and their resistance potential. Frontiers in Microbiology 2013, 4, 44. 10.3389/fmicb.2013.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L.; Brinas L.; Verlinde A.; Ide L.; Nordmann P. BEL-1, a Novel Clavulanic Acid-Inhibited Extended-Spectrum ß-Lactamase, and the Class 1 Integron In120 in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2005, 49, 3743–3748. 10.1128/AAC.49.9.3743-3748.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebmeyer S.; Kristiansson E.; Larsson D. G. J. A framework for identifying the recent origins of mobile antibiotic resistance genes. Communications Biology 2021, 4, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaly T.; Tetu S.; Gillings M. Predicting the taxonomic and environmental sources of integron gene cassettes using structural and sequence homology of attC sites. Communications Biology 2021, 4, 946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebmeyer S.; Kristiansson E.; Larsson D. G. J. The mobile FOX AmpC beta-lactamases originated in Aeromonas allosaccharophila. Int. J. Antimicrob. Agents 2019, 54, 798–802. 10.1016/j.ijantimicag.2019.09.017. [DOI] [PubMed] [Google Scholar]
- Massidda O.; Rossolini G. M.; Satta G. The Aeromonas hydrophila cphA gene: molecular heterogeneity among class B metallo-beta-lactamases. J. Bacteriol. 1991, 173, 4611–4617. 10.1128/jb.173.15.4611-4617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebmeyer S.; Kristiansson E.; Larsson D. G. J. PER extended-spectrum β-lactamases originate from Pararheinheimera spp. Int. J. Antimicrob. Agents 2019, 53, 158–164. 10.1016/j.ijantimicag.2018.10.019. [DOI] [PubMed] [Google Scholar]
- Suzuki Y.; Nishijima S.; Furuta Y.; Yoshimura J.; Suda W.; Oshima K.; Hattori M.; Morishita S. Long-read metagenomic exploration of extrachromosomal mobile genetic elements in the human gut. Microbiome 2019, 7, 119. 10.1186/s40168-019-0737-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.; Klümper U.; Liu Y.; Yang Y.; Wei Q.; Lin J.-G.; Gu J.-D.; Li M. Metagenomic and metatranscriptomic analyses reveal activity and hosts of antibiotic resistance genes in activated sludge. Environ. Int. 2019, 129, 208–220. 10.1016/j.envint.2019.05.036. [DOI] [PubMed] [Google Scholar]
- Li D.; Liu C.-M.; Luo R.; Sadakane K.; Lam T.-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- Barlow M.; Hall B. Phylogenetic Analysis Shows That the OXA β-Lactamase Genes Have Been on Plasmids for Millions of Years. Journal of Molecular Evolution 2002, 55, 314–321. 10.1007/s00239-002-2328-y. [DOI] [PubMed] [Google Scholar]
- Klümper U.; Riber L.; Dechesne A.; Sannazzarro A.; Hansen L. H.; Sørensen S. J.; Smets B. F. Broad host range plasmids can invade an unexpectedly diverse fraction of a soil bacterial community. ISME Journal 2015, 9, 934–945. 10.1038/ismej.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.