Abstract
Introduction
MicroRNAs (miR or miRNA) are short regulatory RNAs, which modulate post-transcriptional gene expression. Dysregulation of these molecules contributes to pathogenicity of autoimmune disorders, such as multiple sclerosis (MS).
Aims
This study was conducted to investigate changed expression pattern of miRNA-145 and miRNA-155 in MS.
Methods
We collected blood samples of 75 patients with relapsing-remitting MS patients and 75 healthy controls. Ficoll-Hypaque density gradient method was used to isolate peripheral blood mononuclear cells. Also, total RNA was extracted and subjected to RT-PCR analysis. We used the Mann–Whitney U test to evaluate the differences in expression levels of target miRNAs between the groups.
Results
We found that expression of miRNA-145 (P = 0.012) and miRNA-155 (P = 0.005) were partly reduced in patients with relapse-remitting MS in comparison with healthy controls. The miRNA-145 had an area under curve (AUC) of 0.621 (P = 0.01) and miRNA-155 levels had an AUC of 0.625 (P = 0.008).
Conclusion
Decreased expression of miRNA-145 and miRNA-155 contributes to development of relapse-remitting MS, while further large scale observational studies and meta-analyses are required.
Keywords: microRNA, miR-145, miR-155, Real-time PCR, Relapsing-remitting MS, Multiple sclerosis
Introduction
Multiple sclerosis (MS) is a multifactorial, immune-mediated disorder of the central nervous system (CNS) [1]. Its complex pathological processes lead to inflammatory demyelination and axonal damage of the CNS neurons, finally giving rise to neurological disability [1, 2]. The clinical course of MS can exhibit changing patterns over the time [3], but the usual course is characterized by a primary relapsing-remitting (RR) stage followed by the secondary progressive (SP) phase, and finally, the primary progressive (PP) phase, which is observed in only a small group of patients [4, 5, 6]. Similar to the clinical course and manifestations of MS, its response to therapeutic interventions, as well as the underlying pathogenicity mechanisms seems to be heterogeneous [7]. Biomarkers are considered as objective measures, which could be evaluated as indicators of regular biological processes, pathological processes or responses to therapeutic interventions [8]. Identification of valid biomarkers for MS bears a potential for a better diagnosis of MS, monitoring of the disease progress, and assessment of therapeutic responses [9].
MicroRNAs (miR or miRNA) are a class of short, widely-expressed, regulatory RNA molecules that modulate gene expression at the post-transcriptional stage [1]. Evidence suggests that these small molecules play a significant role in several cellular and physiological functions. Therefore, dysregulation of miRNA expression results in various pathological conditions, including autoimmunity and inflammation [10]. Recent studies have indicated that miRNAs circulate in a considerably stable form in human blood, supporting the strong potential for using them as promising diagnostic biomarkers [11, 12]. Investigations of miRNA expression profiles in RRMS, SPMS and PPMS patients indicate the differential expression of miRNAs in whole blood [13], peripheral blood mononuclear cells (PBMCs) [9, 10], B- and T-cell subsets [14–16], and active MS lesions [17]. These investigations have also proven that dysregulation of various miRNAs is closely related to development of MS [14–16].
Previous studies have suggested that the altered expression of miRNA-145 [18, 19] and miRNA-155 [20, 21, 22] contributes to the development of MS, and these molecules could serve as diagnostic biomarkers of the disease. However, the level of expression of the biomarkers in patients with MS in Iran have not been recently evaluated. So, in the present study, we compared the expression pattern of the mentioned candidate miRNAs in blood samples of patients with RRMS and healthy controls, in order to investigate their potential to be considered as diagnostic target for RRMS.
Methods
Study design and sample collection
In this case-control study, 75 patients with RRMS and 75 healthy controls were obtained from the Imam Reza Hospital, Tabriz University of Medical Sciences, Tabriz, Iran. Blood samples of the eligible participants were recruited between 2019 and 2020. The Expanded Disability Status Scale (EDSS) were calculated for the included patients with RRMS by an expert neurologist. The inclusion criteria were a diagnosis of MS according to the McDonald Diagnostic criteria [23] and being at least 18 years of age. The control participants should not have any history of neurodegenerative or mental disorders.
Written informed consent was obtained from all of the participants at the beginning of the study. The study protocol was approved by the ethics committee of Tabriz University of Medical Sciences (Ethics number
IR.TBZMED.REC.1400.433). All methods were performed in accordance with the national guidelines and regulations and the ethical standards of the Declaration of Helsinki 1964.
RNA extraction
Following the preparation of blood samples from participants, PBMCs were isolated using a Ficoll-Hypaque density gradient. Following that, approximately 200 µl of PBMC samples were used for RNA extraction using Trizol reagent (Roche) according to the manufacturer’s instructions. Following the evaluation of the quality and quantity of extracted RNAs using the NanoDrop Spectrophotometer (NanoDrop ND-2000 C, Thermo Fisher Scientific, Massachusetts, USA), 1000 ng of the RNAs were reverse transcribed to complementary DNA (cDNA) using the microRNA First Strand cDNA Synthesis kit (EXIQON) as directed by the manufacturer.
Genotyping
Real-time polymerase chain reaction (RT-PCR) assays were performed in the LightCycler® 96 Instrument using the BioFACTTM 2X Real-Time PCR Master Mix to evaluate the expression levels of target microRNAs. According to Schmittgen and Livak [24], the expression of target miRNAs was normalized to the U6 reference gene and evaluated using the 2 −∆CT approach. Each reaction was carried out three times in a total volume of 10 µl, with 5 µl of Master Mix, 2 µl of DEPC-treated water, 1 µl of Primer Mix, and 2 µl of cDNA. Table 1 shows the primer sequences. The reaction conditions were represented in Table 2.
Table 1.
Target sequences for real-time polymerase chain reaction (RT-PCR) and primer sequences.
| Micro-RNA | Stem loop | Forward primer | Reverse primer |
|---|---|---|---|
| miRNA-155 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGAAACCCC | CGTGCTCATTTTAATGCTAATC | CCAGTGCAGGGTCCGAGGTA |
| miRNA-145 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGAAGGGAT | CGTGCTCACGTCCAGTTTTC | CCAGTGCAGGGTCCGAGGTA |
| U6 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAAATAT | GCTTCGGCAGCACATATACTAAAAT | CGCTTCACGAATTTGCGTGTCAT |
Table 2.
Conditions for the real-time polymerase chain reaction (RT-PCR).
| Step | Tm | Time | Cycle namber |
|---|---|---|---|
| Pre-incubation | 94c | 600s | 1 |
| Denaturation | 94c | 10s | 45 |
| Annealing and Extension | 60c | 60s | |
| Melting | |||
Statistical methods
The Mann–Whitney U test was used to assess the differences in target miRNA expression levels between the two groups. Furthermore, a receiver operating characteristic (ROC) analysis was performed to establish the discriminative capacity of miRNA levels. All data were expressed as mean and standard deviation (SD) of experiments, with a statistical significance level of P < 0.05. All of the graphics were created using the GraphPad Prism version 6.00 program (Graph Pad, San Diego, CA, USA).
Results
The present study included a total of 150 participants, made up of 75 RRMS cases and 75 controls. There were 14 (19%) and 16 (21%) men among cases and controls, respectively. The mean age of participants was 34.5 (range: 22–75) in cases and 34.3 (range: 20–75) in controls. Among the cases, the age of onset of the disease was 27.7 ± 14.5 years old and time of diagnosis until sample collection was 17.3 ± 10.7 years. Moreover, the EDSS in patients were 4.12 ± 0.88.
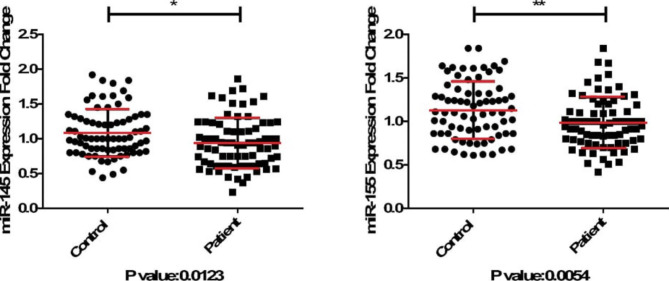
RT-PCR analysis demonstrated that expression of miRNA-145 and miRNA-155 were considerably lower in RRMS specimens in comparison with healthy controls (P = 0.012 and P = 0.005, respectively) (Fig. 1).
Fig. 1.
The relative expression of miRNA-145 and miRNA-155 in relapsing-remitting multiple sclerosis samples compared to healthy controls.
ROC curve analysis was performed to assess the potential diagnostic value of the studied miRNAs and a notable level in separating RRMS and controls showed that miRNA-145 had an area under the curve (AUC) of 0.621 (P = 0.01) and miRNA-155 levels had an AUC of 0.625 (P = 0.008), suggesting that both of these miRNAs could be applied as clinical biomarkers for evaluation of MS (Fig. 2).
Fig. 2.
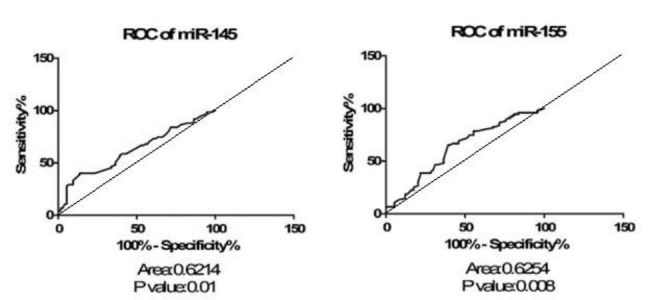
ROC curve of relapsing-remitting multiple sclerosis sample sets analyzed for relative expression level of miRNA-145 and miRNA-155. A diagonal line cutting the coordinates 0,0 and 100,100 with an AUC of 0.5 has been drawn. The difference between this line and the ROC curve of the proposed biomarker is the graphical representation of the quality of the diagnostic biomarker and could be helpful to easily evaluate the given ROC analysis.
Discussion
Findings of the present study showed that there was a significant difference between patients with RRMS and controls in terms of miRNA-145 and miRNA-155 expression which was in favor of the control group.
The miRNA-155 is an inflammation regulator and plays roles in MS development by different mechanisms [22]. In this regard, it can lead to blood-brain barrier disruption, promote demyelination processes and neuropathic pain which can be associated with neuropsychiatric complications like depression and anxiety in patients with MS [22]. In the article by Aljawadi and colleagues miRNA-155 was significantly higher expressed in peripheral blood leukocytes of 25 untreated MS patients compared to 25 healthy controls (p < 0.05) [25]. A previous study conducted by Paraboschit et al. on 10 patients with RRMS and 10 age- and sex-matched controls showed a close to significant results of higher expression of miRNA-155 in cases than controls (p = 0.053) [26]. Despite the findings of the study, we found that miRNA-155 was significantly higher presented in controls, not patients with MS (p = 0.005). It should be noted that the abovementioned study revealed a close to significant level and also different populations and measurements can lead to the variations between the findings. Moreover, the higher level of significance or lower p-values can be adjusted by larger sample size in our study.
The miRNA-145 is an miRNA which its down- or up-regulations are associated with different cancers (e.g. breast and gastric cancers), non-cancerous disorders (e.g. anemia, asthma, biliary atresia, diabetic nephropathy, and hepatopulmonary syndrome), and MS [18, 27]. A case-control study on 37 patients with MS, including 18 with RRMS and 19 with secondary progressive MS and 23 healthy controls showed a significantly higher expression of miRNA-145 in MS patients than controls (0.028 vs. 0.013; p = 0.028) [19]. The study also found an AUC of 0.67 (p = 0.028) which is close to our findings (AUC = 0.62), and represents that it could be used for discrimination of MS from healthy subjects [19]. Another study conducted by Keller and colleagues on 20 patients with RRMS and 19 controls revealed a significantly upregulated of has-miRNA-145 in MS patients (p < 0.0001) [9]. Moreover, it was the most remarkable dysregulated miRNA out of 866 assessed miRNAs which had an AUC value of 0.96 [9]. The higher AUC values of the study compared with our study could be due to different methodologies and objectives of the two study. The study by Aljawadi et al. also evaluated the expression levels of miRNA-145 among Iraqi patients with MS and revealed no significant difference between cases and controls in terms of miRNA-145 expression (p > 0.05) [25]. Another study also showed upregulation of miRNA-145 and highlighted the potential roles of this miRNA as diagnostic biomarker in PBMCs of MS patients (AUC value = 0.785; p = 0.0004) [1]. On the other hand, we found a remarkable higher expression of the miRNA-145 in controls. The differences in sample sizes and inclusion criteria for included patients could lead to the variations in the results.
It is one of the first and most recent studies which evaluated the expression of the two circulating miRNAs involved in MS development among the Iranian populations. Nevertheless, we acknowledge that our study has several limitations. Firstly, some clinical data such as administered treatments for the cases were not reported. Secondly, the findings could not be representative for other populations and regions since we collected data from residents of one of the cities in Iran. Therefore, sampling of other races or ethnicities might lead to different results. Thirdly, we used plasma serum of participants as a non-invasive method to measure plasma serum levels of participants instead of measuring the levels of miRNAs expression in different cell populations. Fourthly, the effects of genetic polymorphisms in the development of MS and its differences between patients with MS and healthy controls have not been evaluated, as a cohort on Egyptian patients with MS revealed that TT genotype and T allele in miRNA-155 were associated with elevated risk of MS [21]. Fifthly, we only included patients with RRMS, while other subtypes of MS were not included in the present study, so we cannot evaluate the roles of miRNA on distinguishing different MS subtypes. In this regard, the study by Gandhi et al. represented that miRNA-145-5p is significantly high regulated in RRMS than secondary progressive MS (p = 0.01) [28].
Conclusion
The presented data revealed that miRNA-145 and miRNA-155 expression is partly reduced in RRMS. Since our study only includes patients with RRMS and healthy controls, further investigations and large scale observational studies are needed to evaluate the expression of mentioned miRNAs in other MS subtypes. Also, the underlying mechanism of these miRNAs in MS pathogenesis should be explained in future studies.
Acknowledgements
The authors are grateful of patients, healthy individuals, and their family who contributed to accomplishment of this research.
List of abbreviations
- MS
Multiple sclerosis.
- CNS
central nervous system.
- RR
relapsing-remitting.
- SP
secondary progressive.
- PP
primary progressive.
- miR
MicroRNA.
- mi-RNA
MicroRNA.
- PBMC
peripheral blood mononuclear cell.
- EDSS
Expanded Disability Status Scale.
- cDNA
complementary DNA.
- RT-PCR
Real-time polymerase chain reaction.
- ROC
receiver operating characteristic.
- SD
standard deviation.
- AUC
area under the curve.
Authors’ contributions
S.S. designed the study. S.A.A., M.A., D.S., S.S.E., and S.S.: patient data acquisition, statistical analysis, and interpretation of data. A.F., S.A.N., and S.S.: preparation and critically revision of the manuscript. All the authors have read and approved the final version of the manuscript.
Funding
None.
Data Availability
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
Declarations
Ethics approval and consent to participate
Written informed consent was obtained from all of the participants at the beginning of the study. The study protocol was approved by the ethics committee of Tabriz University of Medical Sciences (Ethics number: IR.TBZMED.REC.1400.433). All methods were performed in accordance with the national guidelines and regulations and the ethical standards of the Declaration of Helsinki 1964.
Consent for publication
Not applicable.
Competing interests
The authors have declared that no competing financial interests or personal relationships exists in this paper.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Søndergaard HB, Hesse D, Krakauer M, Sørensen PS, Sellebjerg F. Differential microRNA expression in blood in multiple sclerosis. Multiple Scler J. 2013;19(14):1849–57. doi: 10.1177/1352458513490542. [DOI] [PubMed] [Google Scholar]
- 2.Baulina N, Kulakova O, Kiselev I, Osmak G, Popova E, Boyko A, et al. Immune-related miRNA expression patterns in peripheral blood mononuclear cells differ in multiple sclerosis relapse and remission. J Neuroimmunol. 2018;317:67–76. doi: 10.1016/j.jneuroim.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 3.Lublin FD, Reingold SC. Defining the clinical course of multiple sclerosis: results of an international survey. National Multiple Sclerosis Society (USA) Advisory Committee on Clinical Trials of New Agents in Multiple Sclerosis. Neurology. 1996;46(4):907–11. doi: 10.1212/WNL.46.4.907. [DOI] [PubMed] [Google Scholar]
- 4.Mandia D, Ferraro OE, Nosari G, Montomoli C, Zardini E, Bergamaschi R. Environmental factors and multiple sclerosis severity: a descriptive study. Int J Environ Res Public Health. 2014;11(6):6417–32. doi: 10.3390/ijerph110606417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inojosa H, Proschmann U, Akgün K, Ziemssen T. A focus on secondary progressive multiple sclerosis (SPMS): challenges in diagnosis and definition. J Neurol. 2021;268(4):1210–21. doi: 10.1007/s00415-019-09489-5. [DOI] [PubMed] [Google Scholar]
- 6.Thompson AJ, Polman CH, Miller DH, McDonald WI, Brochet B, Filippi MMX, et al. Primary progressive multiple sclerosis. Brain. 1997;120(Pt 6):1085–96. doi: 10.1093/brain/120.6.1085. [DOI] [PubMed] [Google Scholar]
- 7.Compston A, Coles A. Multiple sclerosis. Lancet. 2008;372(9648):1502–17. doi: 10.1016/S0140-6736(08)61620-7. [DOI] [PubMed] [Google Scholar]
- 8.Bielekova B, Martin R. Development of biomarkers in multiple sclerosis. Brain. 2004;127(Pt 7):1463–78. doi: 10.1093/brain/awh176. [DOI] [PubMed] [Google Scholar]
- 9.Keller A, Leidinger P, Lange J, Borries A, Schroers H, Scheffler M, et al. Multiple sclerosis: microRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PLoS ONE. 2009;4(10):e7440. doi: 10.1371/journal.pone.0007440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otaegui D, Baranzini SE, Armañanzas R, Calvo B, Muñoz-Culla M, Khankhanian P, et al. Differential micro RNA expression in PBMC from multiple sclerosis patients. PLoS ONE. 2009;4(7):e6309. doi: 10.1371/journal.pone.0006309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 13.Cox MB, Cairns MJ, Gandhi KS, Carroll AP, Moscovis S, Stewart GJ, et al. MicroRNAs miR-17 and miR-20a inhibit T cell activation genes and are under-expressed in MS whole blood. PLoS ONE. 2010;5(8):e12132. doi: 10.1371/journal.pone.0012132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, et al. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10(12):1252–9. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- 15.Lindberg RL, Hoffmann F, Mehling M, Kuhle J, Kappos L. Altered expression of miR-17-5p in CD4 + lymphocytes of relapsing-remitting multiple sclerosis patients. Eur J Immunol. 2010;40(3):888–98. doi: 10.1002/eji.200940032. [DOI] [PubMed] [Google Scholar]
- 16.De Santis G, Ferracin M, Biondani A, Caniatti L, Rosaria Tola M, Castellazzi M, et al. Altered miRNA expression in T regulatory cells in course of multiple sclerosis. J Neuroimmunol. 2010;226(1–2):165–71. doi: 10.1016/j.jneuroim.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 17.Junker A, Krumbholz M, Eisele S, Mohan H, Augstein F, Bittner R, et al. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain. 2009;132(Pt 12):3342–52. doi: 10.1093/brain/awp300. [DOI] [PubMed] [Google Scholar]
- 18.Kadkhoda S, Ghafouri-Fard S. Function of miRNA-145–5p in the pathogenesis of human disorders. Pathol - Res Pract. 2022;231:153780. doi: 10.1016/j.prp.2022.153780. [DOI] [PubMed] [Google Scholar]
- 19.Sharaf-Eldin WE, Kishk NA, Gad YZ, Hassan H, Ali MAM, Zaki MS, et al. Extracellular miR-145, miR-223 and miR-326 expression signature allow for differential diagnosis of immune-mediated neuroinflammatory diseases. J Neurol Sci. 2017;383:188–98. doi: 10.1016/j.jns.2017.11.014. [DOI] [PubMed] [Google Scholar]
- 20.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A. 2007;104(5):1604–9. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ali MA, Shaker OG, Eid HM, Mahmoud EE, Ezzat EM, Gaber SN. Relationship between miR–155 and miR–146a polymorphisms and susceptibility to multiple sclerosis in an Egyptian cohort. Biomed Rep. 2020;12(5):276–84. doi: 10.3892/br.2020.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maciak K, Dziedzic A, Miller E, Saluk-Bijak J. miR-155 as an Important Regulator of Multiple Sclerosis Pathogenesis. A Review. International Journal of Molecular Sciences. 2021;22(9). [DOI] [PMC free article] [PubMed]
- 23.McDonald WI, Compston A, Edan G, Goodkin D, Hartung H-P, Lublin FD, et al. Recommended diagnostic criteria for multiple sclerosis: Guidelines from the international panel on the diagnosis of multiple sclerosis. Ann Neurol. 2001;50(1):121–7. doi: 10.1002/ana.1032. [DOI] [PubMed] [Google Scholar]
- 24.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3(6):1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 25.Aljawadi ZA, Al-Derzi AR, Abdul-Majeed BA, Almahdawi AM. MicroRNAs (20a, 146a, 155, and 145) expressions in a sample of Iraqi patients with multiple sclerosis. J Fac Med Baghdad. 2016;58(4):371–7. [Google Scholar]
- 26.Paraboschi EM, Soldà G, Gemmati D, Orioli E, Zeri G, Benedetti MD, et al. Genetic Association and Altered Gene Expression of Mir-155 in Multiple Sclerosis Patients. International Journal of Molecular Sciences. 2011;12(12). [DOI] [PMC free article] [PubMed]
- 27.Mansoor SR, Ghasemi-Kasman M, Yavarpour-Bali H. The role of microRNAs in multiple sclerosis. Int Rev Immunol. 2022;41(2):57–71. doi: 10.1080/08830185.2020.1826474. [DOI] [PubMed] [Google Scholar]
- 28.Gandhi R, Healy B, Gholipour T, Egorova S, Musallam A, Hussain MS, et al. Circulating MicroRNAs as biomarkers for disease staging in multiple sclerosis. Ann Neurol. 2013;73(6):729–40. doi: 10.1002/ana.23880. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.