Abstract
Domestic cats are susceptible to SARS-CoV-2 infection and can transmit the virus to other felines. A high number of COVID-19 human cases within the military personnel and a high density of stray cats living close to soldiers raised the need to perform active animal surveillance. We validated a novel quantitative serological microarray for use in cats, that enables simultaneous detection of IgG and IgM responses; in addition, molecular genetic SARS-CoV-2 detection was performed. Three out of 131 cats analyzed, showed IgG antibodies against SARS-CoV-2 RBD and S2P (2.3 %). None of cats were positive for SARS-CoV-2 RNA by RT-PCR. SARS-CoV-2 infection rate in soldiers ranged from 4.7 % to 16 % (average rate=8.9 %). Further investigations on a larger cohort are necessary, in the light of the emerging new viral variants in other animal species and in humans.
Keywords: COVID-19, One health, SARS-CoV-2, Stray cats, Military
1. Introduction
The human coronavirus disease 2019 (COVID-19) caused by the severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) has already affected more than 486 million people and killing 6.1 million humans worldwide (by the specified date of 3th April 2022) [1]. Israel was severely affected by the pandemic with over 2.6 million cases despite major efforts to control the outbreak. The close phylogenetic relationship of SARS-CoV-2 to a coronavirus from bats (Rhinolophus affinis) provides evidence that COVID-19 may have been caused by a virus spillover from bats to humans [2] directly or through another intermediate host which is yet to be identified [3]. Although, at present, the virus is transmitted almost exclusively from humans to humans, there are several reports of reverse zoonotic events, involving transmission from humans-to-dogs, humans-to-cats and humans-to-wild cats [4], [5], [6], [7]. Sustained natural transmission was documented in minks and in white-tailed deer [8], [9]. A human-to-animal and reverse animal to human transmission pathway was documented in a mink farm in the Netherlands, in Denmark [10], [11] and in two independent hamster-to-human transmission cycles in Hong Kong [12].
Domestic cats are more susceptible to infection with SARS-CoV-2 than other domestic animals such as dogs, pigs, chickens and ducks [13]. Experimentally infected cats with SARS-CoV-2 shed infectious virus through oral, nasal and fecal secretions, with almost no clinical signs, contrary to what was described in natural infected big cats [7] and can spread the virus to other cats through respiratory droplet transmission [13], [14]. Natural infection with SARS-CoV-2 in cats was reported in Spain, Italy, Netherlands, China, U.S.A, France, Belgium, Germany, Brazil, Japan and Argentina [5], [6], [8], [15], [16], [17], [18], [19], [20], [21], [22]. Most of the naturally infected cats acquired the infection from COVID-19 positive humans in their households; however, a few stray cats from Spain and Italy showed specific antibodies in sera proving that cat infection may also occur outdoors [15], [23].
Free roaming cats (FRC) in Israel are very abundant in urban locations and may impose a public human health hazard [24], [25]; in military bases encounters between FRC and soldiers in open spaces may occur, despite a military law that prohibits contact between soldiers and animals.
In order to control the dissemination of SARS-CoV-2 between soldiers, the army preventive medicine branch implemented a massive vaccination plan which resulted in more than 80 % of the soldiers immunized, and regularly executes epidemiological investigations and surveillance. In spite of the effort, 50,885 out of 100,000 of the military personnel were tested positive for COVID-19 by the end of February 2022. A combination of a high number of human cases and a high density of stray cats living in proximity with the soldiers, offers a unique environment for human-to-animal transmission. In addition, the emergence of variants (such Omicron) with ability to infect vaccinated human individuals in white-tailed deer [26], raises the need to perform active and passive surveillance in species of animals sensitive to viral transmission under a “One Health” approach [27]. The objectives of this study were to investigate natural infection of stray cats that live inside military bases in Israel by analyzing the presence of SARS-CoV-2 RNA and specific antibodies and to understand the epidemiological role cats may play in this unique ecosystem.
2. Materials and methods
2.1. Study area, data collection and sampling
The study was conducted in 11 military bases with varying levels of human COVID-19 incidence across Israel during February to July 2021. The study group included 132 cats captured during ‘trap-neuter-vaccine-return’ campaigns carried out to control stray cat populations. Cats were anaesthetized with a combination of intramuscular ketamine (Clorketam®, 5 mg/kg), xylazine (Chanazine®, 2 mg/kg) and butorphanol (Butomidor®, 0.4 mg/kg). After the neuter surgery, the cats received intramuscular dipyrone (Oftalgine® 20 mg/kg) and Amoxicillin (Clamoxil® L.A 15 mg/kg). A full physical examination was performed before the COVID-19 sampling and information regarding age, gender and health condition was recorded for all the animals. Blood was collected by venipuncture of the jugular vein into clot activator tubes for serology. Oropharyngeal swabs were collected by inserting the swab into the posterior pharynx and tonsillar areas. Nasal secretions were collected by inserting a swab into both nostrils and rotating it to absorb the nasal fluids. Rectal swabs were collected by inserting the swab 2 cm past the anal verge and rotating it. After collection, each swab was immersed in 3 ml viral transport medium (Biological Industries, Israel).
Sampling of animals for this study was approved by the Animal Ethics Committee of the Israeli Ministry of Defense (approval number 09–2021).
Number of active-duty soldiers as well as COVID-19 annual and monthly incidence in each military base were obtained from the clinical report records of the information branch of the IDF medical corps, between May 2020 until May 2021. COVID-19 testing in soldiers during the study period was performed when soldiers showed clinical signs or were in close contact with a positive individual. Positive soldiers were sent to a recovery center (‘The recovery village-Ashkelon’) and suspected soldiers were quarantined in their homes.
2.2. Serological analysis
2.2.1. Microarray analysis
A quantitative multi-component serological array detecting simultaneously IgG and IgM targeting SARS-CoV-2 spike protein subunit 2 (S2P), receptor-binding domain (RBD) and nucleocapsid (N) developed by Fisher et al., was standardized and validated for use in cat serological surveillance [28]. SARS-CoV-2 recombinant proteins RBD, S2P, and N were designed, expressed, and purified as described previously [[29], [30]]. The array consisted of SARS CoV-2 pre-fusion stabilized spike (S2P), RBD and N proteins, that were spotted (18 spots) on 16 sun-arrays nitrocellulose-coated slides (Grace Bio Labs, GBL, Bend, OR) using a non-contact Piezo dispensing microarray spotter (Scienion Inc., Berlin, Germany). Slides were pre-blocked to avoid non-specific binding with blocking buffer (0.1 % Tween 20, 3.3 % Bovine Serum Albumin (BSA) in PBS) for 30 min at room temperature, washed (Tween 20 0.1 %, 5 % BSA in PBS), dried and stored desiccated until use. Cats’ sera were diluted (1:200) in array buffer (0.1 % Tween 20, 1 % BSA in PBS) and loaded separately on one of 16 sub-arrays. Following an incubation of 30 min at room temperature, the slides were washed twice with 100 μL of PBT (Tween 20 0.1 % in PBS) and probed with goat anti-cat IgG coupled to Alexa fluor® 647 conjugate (Jackson ImmunoResearch, USA) and goat anti-cat IgM coupled with Fluorescein isothiocyanate conjugated (FITC) (abcam, USA). Following an additional wash step, the resulting fluorescent signal was scanned at two wavelengths (625, 470 nm) using a SciReader FL2 system (Scienion Inc., Germany). The average median fluorescent intensity (MFI) values for each antigen (18 spots) were collected at different exposure times (6–400 ms) and recorded using Scienion scan Array software (Scienion Inc., Germany). Calculated MFI values (“raw crude") were generated from boxed values for each sample. Each slide contains 16 sub-arrays, allowing analysis of up to 16 samples. Three of these sub-arrays are dedicated to controls: SARS-CoV-2 positive serum (P), negative control (naïve pre-pandemic sera), and reagent control (Blank), each loaded in a single sub-array spotted with recombinant S2P, RBD, and N. For the evaluation of seroconversion, we determined several parameters of the multi-component array, including acceptance ranges, linearity, lower limits of detection (LLOD) and quantitation (LLOQ) following previous publication [28].
Cat control samples used for validation of the system were: two sera samples of SPF (specific pathogen free) Israeli cats obtained before the pandemic as negative controls, two positive feline coronavirus (FCoV) sera tested for potential cross-reactive antibodies; two SARS-CoV-2-naturally infected cats from Italy (University of Bari), 2 sentinel SARS-CoV-2-infected cats from U.S.A (Kansas State University) and one experimentally infected cat from U.S.A (Colorado State University) were used as positive control.
2.2.2. FCoV ELISA
All seropositive samples for SARS-CoV-2 were tested for presence of FCoV IgG antibodies using an ELISA commercial kit (Biogal, Israel) following the instructions.
2.3. Polymerase chain reaction
RNA extraction and real-time PCR assays targeting the SARS-CoV-2 E and N gene was performed as described previously [31]. The primers and probes used are described in the appendix (material and methods section).
2.4. Statistical analysis
The Pearson correlation coefficient was calculated to determine correlation between COVID-19 in soldiers and number of soldiers; and between MFI values of the different microarray antigen assays (S2P, RBD and N). Statistical analysis was performed by using SPSS version 22 (IBM, USA).
3. Results
3.1. Sample and data collection
The study included samples of 132 stray cats (seventy male cats and sixty-two females) from eleven military bases collected during February to July 2021 ( Table 1). In total, 360 swab
Table 1.
Number of cats sampled, collection time and presence of respiratory signs.
| Location | Date of sample collection | Male cats | Female cats | Cats with respiratory signs* |
|---|---|---|---|---|
| Glilot | Feb-21 | 3 | 11 | 1 |
| Tzrifin | Mar/May-2021 | 6 | 12 | 1 |
| Kfir Brigade | Mar-2021 | 16 | 8 | 2 |
| The recovery village-Ashkelon | Apr-21 | 4 | 3 | 1 |
| Re`im Camp | May-21 | 15 | 9 | 0 |
| Camp 80 | May-21 | 4 | 5 | 1 |
| Menashe Brigade | May-21 | 2 | 4 | 0 |
| Biranit | May-21 | 6 | 4 | 0 |
| Training base 1 officer school | Jun-21 | 11 | 1 | 0 |
| Military collage Glilot | Jul-21 | 1 | 3 | 0 |
| Jerusalem | Jul-21 | 2 | 2 | 0 |
Ocular or nasal secretions.
samples (120 nasal swabs, 120 oropharyngeal swabs and 120 rectal swabs) and 131 serum samples were collected. One hundred and nineteen cats had both swab and serum samples, twelve cats had only serum samples and one cat had only swab samples ( Table 2).
Table 2.
Number and type of sample collected from cats in each location.
| Location name | Serum | Swaps |
||||
|---|---|---|---|---|---|---|
| Pharyngeal swab | Nasal swab | Rectal swab | Total swabs | Total | ||
| Glilot | 14 | 14 | 14 | 14 | 42 | 84 |
| Tzrifin | 18 | 10 | 10 | 10 | 30 | 60 |
| Kfir Brigade | 24 | 24 | 24 | 24 | 72 | 144 |
| The recovery village-Ashkelon | 7 | 7 | 7 | 7 | 21 | 42 |
| Re`im Camp | 23 | 24 | 24 | 24 | 72 | 144 |
| Camp 80 | 9 | 9 | 9 | 9 | 27 | 54 |
| Menashe Brigade | 6 | 6 | 6 | 6 | 18 | 36 |
| Biranit | 10 | 10 | 10 | 10 | 30 | 60 |
| Training base 1 officer school | 12 | 12 | 12 | 12 | 36 | 72 |
| Military collage Glilot | 4 | 0 | 0 | 0 | 0 | 0 |
| Jerusalem | 4 | 4 | 4 | 4 | 12 | 24 |
| Total | 131 | 120 | 120 | 120 | 360 | 851 |
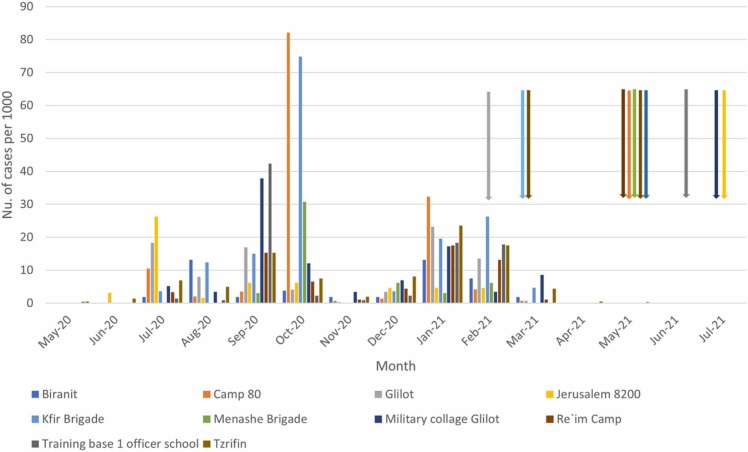
Respiratory signs (nasal and/or ocular discharge) were observed in six animals from five different locations (Table 1). The number of cats trapped and sampled from each base ranged between 4 and 24 ( Table 2, Table 3). Human annual COVID-19 incidence in the surveyed military bases ranged between 4.7 % and 16 % (mean rate: 8.9%, SE= 1.2) from May 2020 until May 2021 (period that includes the first three waves of COVID-19 in Israel) (Table 3). Monthly numbers of new cases per 1000 soldiers are described in Fig. 1, Fig. 2.
Table 3.
Covid-19 incidence and rate of seropositive cats.
| Location | Human Covid-19 incidence (%) | total nu. of cats | Seropositive cats (%) |
|---|---|---|---|
| Glilot | 8.9 | 14 | 0 |
| Tzrifin | 9.3 | 18 | 1 (5.5) |
| Kfir Brigade | 16 | 24 | 0 |
| The recovery village-Ashkelon | ND | 7 | 0 |
| Re`im Camp | 6.2 | 24 | 2 (8.3) |
| Camp 80 | 13.7 | 9 | 0 |
| Menashe Brigade | 4.9 | 6 | 0 |
| Biranit | 4.7 | 10 | 0 |
| Training base 1 officer school | 8.6 | 12 | 0 |
| Military collage Glilot | 9.8 | 4 | 0 |
| Jerusalem | 5.7 | 4 | 0 |
ND: Not determinable since all soldiers were COVID-19 recovery patients, thus it was not possible to calculate the prevalence.
Fig. 1.
COVID-19 cases per month in soldiers in each location. Arrows indicate the month of cat trapping and sampling, and the color shows the location.
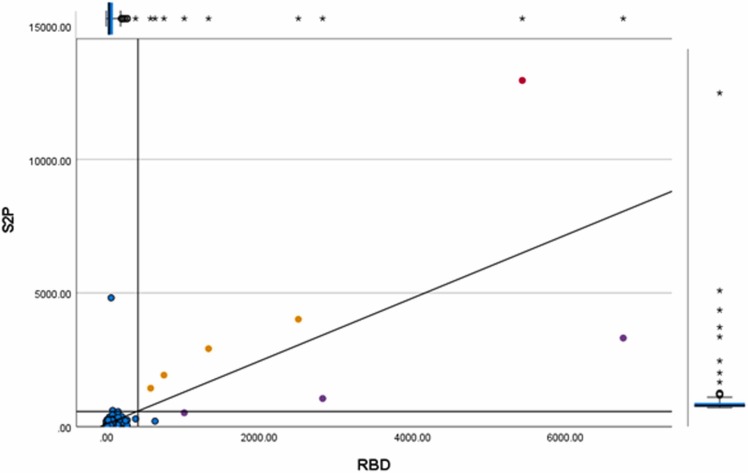
Fig. 2.
S2P and RBD MFI values of the samples and controls tested in the serological microarray. The red circle indicates the positive control sample of an experimentally infected cat. The yellow circles indicate the positive control samples of naturally infected cats. The purple circles indicate the positive cat samples from the cohort study. The blue circles indicate the negative samples. The parallel axis show the cutoff (LLOQ) of RBD and S2P. The unparallel axis show the trend line.
3.2. Polymerase chain reaction
All samples tested, including cats showing respiratory signs, were negative for SARS-CoV-2 RNA by RT-PCR.
3.3. Microarray serological analysis
Results are presented as MFI, and the binding of each animal's serum antibodies (IgG and IgM) to all the spotted antigens is obtained simultaneously from a single subarray. All 5 positive control samples presented full IgG-specific seroconversion on all three antigens. In contrast, IgM in two of the controls, reacted with none of the antigens, and thus, IgM was not suitable as seroconversion indicator. The sensitivity of the method, determined as a reaction with the full antigen panel, was 100 % and 60 % for IgG and IgM respectively.
Both negative SPF cats tested negative for all IgG antigen panel. One of the two FCoV control samples reacted with SARS-CoV-2 S2P and the N antigen, showing cross-reaction. The second FCoV control sample reacted with the N antigen. Altogether, despite some unspecific interactions, none of the negative control samples (SPF cats and FCoV samples) reacted with all three antigens (100 % specificity).
Since N showed non-specific results, with many cats expressing N background signals that exceeded the determined LLOQ, it was not considered as a parameter for positivity. MFI values of SARS-CoV-2 S2P and RBD showed a strong correlation with each other (R=0.746, p < 0.001); therefore, we defined a seropositive sample as any sample being positive for both SARS-CoV-2 S2P and RBD. Following this criterion, from a total of 131 cats tested against SARS-CoV-2 S2P, RBD and N antigens, three cat samples (two males and one female) were positive by SARS-CoV-2 S2P and RBD (2.3 %) (Table 3) and all tested negative for FCoV ELISA. IgG MFI values against S2P, RBD and N antigens of the controls and cohort samples, as well as LLOD and LLOQ values, are described in the Apendix (results section Table A.1 and table A. 2).
Of the seropositive cats, two males belonged to Re`im camp and one seropositive female cat belonged to the Tzrifin camp. All animals were adults, with good physical condition and no visible respiratory signs.
3.4. Statistical analysis
No correlation was found between COVID-19 incidence and the number of soldiers in each location (R=0.416, p = 0.231).
4. Discussion
In this study we evaluated the presence of SARS-CoV-2 RNA in cats and modified a clinical serological assay [28] to detect antibodies against SARS-CoV-2 in 131 stray cats living within the limits of 11 IDF military bases. This is the first study to demonstrate natural exposure of stray cats to SARS-CoV-2 in Israel and in a military environment worldwide.
The quantitative multi-component serological microarray used in this study employs an algorithm to calculate the exposure-induced fluorescence, which is related to the serum’s antibodies concentration and affinity against fixed SARS-CoV-2 antigens, enabling simultaneous detection of IgG and IgM responses [28].
We defined seropositivity on the basis of the results of the SARS-CoV-2 positive control samples obtained from infected cats in U.S.A and Italy. Results indicate that IgG antibodies enable sensitive and specific discrimination between negative and SARS-CoV-2 positive animals. Analysis of positive animal samples showed seroconversion with all array antigens. Some naivë individuals had N background signals, probably due to cross-reacting antibodies as a consequence of previous exposure to circulating FCoV; therefore, N was discarded as a serologic screening antigen. Although S2P showed cross-reaction with one of the FCoV control samples, it showed strong correlation with RBD (R=0.75, p < 0.001); therefore, we defined seropositivity when both, the S2P and RBD microarray results were positive. Afterwards, antibodies against FCoV were excluded in SARS-CoV-2 seropositive samples by performing an ELISA commercial test (Biogal, Israel). IgM did not react with any of the microarray antigens in two of the positive controls. In a previous study, a decrease in IgM seropositivity against RBD was reported in cats after 28 days post infection; in addition, IgM responses were less robust than IgG [14].
Soldiers that were positive or suspected with COVID-19 were quarantined (either in their homes or in a recovery center); hence, the exposure to cats was possible before the symptoms onset; time when one out of five positive individuals shed infectious virus [32]. From the cohort study, three cats (2.3 %) were found seropositive against S2P and RBD. Positive cats belonged to the Re`im and Tzrifin military bases where human COVID-19 annual incidences were 6.2 % and 9.3 % respectively, between May 2020 and May 2021. Due to the small sample size, we were not able to perform a correlation test between rate of COVID-19 in soldiers and rate of seropositive cats with enough statistical power. Nevertheless, due to the low number of positive animals, it may be concluded that in military bases, cats do not have an important epidemiological role in SARS-CoV-2 transmission cycle and confirms the assumption that testing FRC cannot be used as a sentinel species of COVID-19 in a specific territory [33].
Our results are in concordance with other studies performed on stray cats where seroprevalence was 0.4 %, 1 % and 3.5 % in the Netherlands, Italy and Spain, respectively [15], [23], [34]. However, it is important to point out that SARS-CoV-2 prevalence may be influenced by various factors, such as the SARS-CoV-2 epidemic condition in the geographical area examined, the different serological tests implemented and the criteria chosen to define seropositivity [23], [35].
Although a small number of cats were seropositive, all animals tested negative by RT-PCR. Absence of RT-PCR positive results is not unexpected since the cat sampling was perfomed at the end of third COVID-19 wave in Israel when few positive soldiers were diagnosed positve; this finding is in accordance with a previous epidemiological study on SARS-CoV-2 in cats where the prevalence of seropositive cats was low (0.9 %) and all cats were RT-PCR negative [5], [23]. This suggests virus shed in cats is very limited and only transient, as reported in experimentally infected cats that stopped shedding infectious virus at day 10 [14], [36]. Naturally infected cats also showed shorter duration of SARS-CoV-2 RNA shedding in nasal and sputum samples compared to their human owners [37]. SARS-CoV-2 IgG antibodies can be identified as early as 7 days post infection in cats and may remain detectable for up to 110 days and 10 months in experimentally and naturally infected cats, respectively [14], [16], [36], [38]. We are not able to determine the time and source of infection of the seropositive cats in our study. We can estimate a period of 39–69 days since the last human case of SARS-CoV-2 infection was reported in March 2021 (Fig. 1) during the third wave of COVID-19 in Israel, until seropositive cats were sampled in Re`im (09.05.2021). In Tzrifin, the last human COVID-19 case was reported in May 2021 (Fig. 1), during which a seropositive cat was sampled from this location (27.05.2021). However, besides humans, other possible sources of infection in cats are other infected cats or other intermediate hosts; a SARS-CoV-2 contaminated environment was not infective suggesting low feasibility for indirect fomite transmission [14], [39]. In any case, we have proven that cats living in closed military bases with almost no contact with outside cats were exposed to SARS-CoV-2 and seroconverted.
5. Conclusions
Since Israel has one of the largest colony of cats in the world, with thousands of permanent ‘feeding areas’, sero-surveillance studies of stray cats together with molecular detection are crucial to further understand the cats’ epidemiological role in the transmission of SARS-CoV-2 in Israel and to detect potential new variants. In order to achieve this, further surveillance should be performed during a COVID-19 human wave when SARS-CoV-2 is actively spreading. A mink-associated variant identified in Denmark due to a mutation in the spike protein probably represented a virus adaptation which may have increased its transmissibility between minks and allowed zoonotic transmission to humans [11], [40]. Another virus adaptation associated to confer escape to monoclonal antibodies in humans was found in a cat model [41]. If a variant with improved binding to the cat ACE2 receptor should emerge, it is possible that this would impact cat-to-cat transmission and increase the potential for cats to serve as reservoirs of the virus with severe repercussions for public health. Further studies on larger cohorts are warranted, especially in the light of the emerging new viral variants, using a One Health approach.
Funding
This research was funded by the Israel Defense Forces (IDF) Medical Corps and Directorate of Defense Research & Development, Israeli Ministry of Defense (IMOD DDR&D). J.A.R work was funded through grants from the National Bio and Agro-Defense Facility (NBAF) Transition Fund from the State of Kansas , the AMP Core of the Center of Emerging and Zoonotic Infectious Diseases (CEZID) from National Institute of General Medical Sciences (NIGMS) under award number P20GM130448, and the NIAID supported Center of Excellence for Influenza Research and Response (CEIRR, contract number 75N93021C00016).
Institutional review board statement
The animal study protocol was approved by the Animal Ethics Committee of the Israeli Ministry of Defense (approval number 09–2021).
CRediT authorship contribution statement
The J.A.R laboratory received support from Tonix Pharmaceuticals, Genus plc, Xing Technologies and Zoetis, outside of the reported work. J.A.R is inventor of patents and patent applications on the use of antivirals and vaccines for the treatment and prevention of virus infections, owned by Kansas State University, KS.
Conflict of Interest
The J.A.R laboratory received support from Tonix Pharmaceuticals, Genus plc, Xing Technologies and Zoetis, outside of the reported work. J.A.R is inventor of patents and patent applications on the use of antivirals and vaccines for the treatment and prevention of virus infections, owned by Kansas State University, KS.
Acknowledgments
We would like to thank Dr. Morly Fisher and Dr. Adva Mechaly from the Israeli Institute for Biological Research for the analysis of the samples, and Dr. Angela Bosco-Lauth from the University of Colorado (U.S.A) for sending us a positive control of an experimentally infected cat with SARS-CoV-2.
Data availability statement
Data are contained within the article and the supplementary material.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.cimid.2022.101905.
Appendix A. Supplementary material
Supplementary material.
.
References
- 1.W.H.O.. Coronavirus (COVID-19), global situation. available online: https:// covid19.who.int/ (accessed on 3th April 2022).
- 2.Zhou P., Xing-Lou Y., Xian-Guang W., Ben H., Lei Z., Wei S., Hao-Rui Y., Zhu L., Bei H., Chao-Lin H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Xiao-Shuang Zheng X.S.X.S., Zhao K., Chen Qj, Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao J., Wei C., Bao-Ping T. The potential intermediate hosts for SARS-CoV-2. Front Microbiol. 2020 doi: 10.3389/fmicb.2020.580137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrs V.R., Peiris M., Tam K.W.S., Law P.Y.T., Brackman C.J., To E.M.W., Yu V.Y.T., Chu D.K.W., Perera R.A.P.M., Sit T.H.C. SARS-CoV-2 in quarantined domestic cats from covid-19 households or close contacts, Hong Kong, China. Emerg. Infect. Dis. 2020;26:3071–3074. doi: 10.3201/eid2612.202786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patterson E.I., Elia G., Grassi A., Giordano A., Desario C., Medardo M., Smith S.L., Anderson E.R., Prince T., Patterson G.T., Lorusso E., Lucente M.S., Lanave G., Lauzi S., Bonfanti U., Stranieri A., Martella V., Solari Basano F., Barrs V.R., Radford A.D., Agrimi U., Hughes G.L., Paltrinieri S., Decaro N. Evidence of exposure to SARS-CoV-2 in cats and dogs from households in Italy. Nat. Commun. 2020 doi: 10.1038/s41467-020-20097-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newman A., Smith D., Ghai R.R., Wallace R.M., Torchetti M.K., Loiacono C., Murrell L.S., Carpenter A., Moroff S., Rooney J.A., Behravesh C.B. First reported cases of SARS-CoV-2 infection in companion animals - New York, March-April 2020. MMWR Morb. Mortal. Wkly Rep. 2020;69:710–713. doi: 10.15585/mmwr.mm6923e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McAloose D., Laverack M., Wang L., Killian M.L., Caserta L.C., Yuan F., Mitchell P.K., Queen K., Mauldin M.R., Cronk B.D., Bartlett S.L., Sykes J.M., Zec S., Stokol T., Ingerman K., Delaney M.A., Frederikson R., Ivancicic M., Jenkins-Moore M., Mozingo K., Franzen K., Kuzmin I., Bergeson N.H., Goodman L., Wang H., Tong S., Fang Y., Olmstead C., McCann C., Thomas P., Goodrich E., Elvinger F., Smith D.C., Slavinski S., Calle P.C., Terio K., Torchetti M.K., Diel D.G. From people to Panthera: natural SARS-CoV-2 in tigers and lions at the Bronx Zoo. mBio. 2020 doi: 10.1128/mBio.02220-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oreshkova N., Molenaar R.J., Vreman S., Harders F., Munnink B.B.O., Hakze-van der Honing R.W., Gerhards N., Tolsma P., Bouwstra R., Sikkema R.S., Tacken M.G., de Rooij M.M., Weesendorp E., Engelsma M.Y., Bruschke C.J., Smit L.A., Koopmans M., van der Poel W.H., Stegeman A. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Eur. Surveill. 2020 doi: 10.2807/1560-7917.ES.2020.25.23.2001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hale V.L., Dennis P.M., Bride D.S., Nolting J.M., Madden C., Huey D., Ehrlich M., Grieser J., Winston J., Lombardi D., Gibson S., Saif L., Killian M.L., Lantz K., Tell R.M., Torchetti M., Robbe-Austerman S., Nelson M.I., Faith S.A., Bowman A.S. SARS-CoV-2 infection in free-ranging white-tailed deer. Nature. 2022;602:481–486. doi: 10.1038/s41586-021-04353-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munnink B.B.O., Sikkema R.S., Nieuwenhuijse D.F., Molenaar R.J.M., Munger E., Molenkamp R., van der Spek A., Tolsma P., Rietveld A., Brouwer M., Bouwmeester-Vincken N., Harders F., Hakze-van der Honing R., Wegdam-Blans M.C.A., Bouwstra R.J., GeurtsvanKessel C., van der Eijk A.A., Velkers F.C., Smit L.A.M., Stegeman A., van der Poel W.H.M., Koopmans M.P.G. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;8:172–177. doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammer A.S., Quaade M.L., Rasmussen T.B., Fonager J., Rasmussen M., Mundbjerg K., Lohse L., Strandbygaard B., Jørgensen C.S., Alfaro-Núñez A., Rosenstierne M.W., Boklund A., Halasa T., Fomsgaard A., Belsham G.J., Bøtner A. SARS-CoV-2 transmission between mink (Neovison vison) and humans, Denmark. Emerg. Infect. Dis. 2021;27:547–551. doi: 10.3201/eid2702.203794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yen H., Sit T.H.C., Brackman C.J., Chuk S.S.Y., Gu H., Tam K.W.S., Law P.Y.T., Leung G.M., Peiris M., Poon L.L.M. Transmission of SARS-CoV-2 delta variant (AY.127) from pet hamsters to humans, leading to onward human-to-human transmission: a case study. Lancet. 2022;399:1070–1078. doi: 10.1016/S0140-6736(22)00326-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z., Zhao Y., Liu P., Liang L., Cui P., Wang J., Zhang X., Guan Y., Tan W., Wu G., Chen H., Bu Z. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bosco-Lauth A.M., Hartwig A.E., Porter S.T., Gordy P.W., Nehring M., Byas A.D., VandeWoude S., Ragan I.K., Maison R.M., Bowen R.A. Experimental infection of domestic dogs and cats with SARS-CoV-2: pathogenesis, transmission, and response to reexposure in cats. Proc. Natl. Acad. Sci. U. S. A. 2020;117:26382–26388. doi: 10.1073/pnas.2013102117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villanueva-Saz S., Giner J., Tobajas A.P., Pérez M.D., González-Ramírez A.M., Macías-León J., González A., Verde M., Yzuel A., Hurtado-Guerrero R., Pardo J., Santiago L., Paño-Pardo J.R., Ruíz H., Lacasta D.M., Sánchez L., Marteles D., Gracia A.P., Fernández A. Serological evidence of SARS-CoV-2 and co-infections in stray cats in Spain. Transbound. Emerg. Dis. 2021;69:1056–1064. doi: 10.1111/tbed.14062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Q., Zhang H., Gao J., Huang K., Yang Y., Hui X., He X., Li C., Gong W., Zhang Y., Peng C., Gao X., Chen H., Zou Z., Shi Z.L., Jin M. A serological survey of SARS-CoV-2 in cat in Wuhan. Emerg. Microbes. Infect. 2020;9:2013–2019. doi: 10.1080/22221751.2020.1817796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sailleau C., Dumarest M., Vanhomwegen J., Delaplace M., Caro V., Kwasiborski A., Hourdel V., Chevaillier P., Barbarino A., Comtet L., Pourquier P., Klonjkowski B., Manuguerra J.C., Zientara S., Le Poder S. First detection and genome sequencing of SARS-CoV-2 in an infected cat in France. Transbound. Emerg. Dis. 2020;67:2324–2328. doi: 10.1111/tbed.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garigliany M., Van Laere A., Clercx C., Giet D., Escriou N., Huon C., van der Werf S., Eloit M., Desmecht D. SARS-CoV-2 natural transmission from human to cat, Belgium, March 2020. Emerg. Infect. Dis. 2020;26:3069–3071. doi: 10.3201/eid2612.202223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michelitsch A., Schön J., Hoffmann D., Beer M., Wernike K. The second wave of SARS-CoV-2 circulation-antibody detection in the domestic cat population in Germany. Viruses. 2021;13:1009. doi: 10.3390/v13061009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calvet A.G., Pereira S.A., Ogrzewalska M., Pauvolid-Corrêa A., Resende P.C., de Souza Tassinari W., de Pina Costa A., Oliveira Keidel L., Sampaio Barreto da Rocha A., Borges da Silva M.F., dos Santos S.A., Machado Lima A.B., Campos Vargas de Moraes I., Velho Mendes Junior A.A., das Chagas Souza T., Martins E.B., R.Orsini Ornellas M., Lopes Corrêa I.M., da Silva Antonio L., Guaraldo F., Fernando do Couto Motta P., Brasil M., Mendonça Siqueira I.D., Ferreira Gremião R., Caldas Menezes Investigation of SARS-CoV-2 infection in dogs and cats of humans diagnosed with COVID-19 in Rio de Janeiro, Brazil. PLoS One. 2021 doi: 10.1371/journal.pone.0250853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imanishi I., Asahina R., Hayashi S., Uchiyama J., Hisasue M., Yamasaki M., Murata Y., Morikawa S., Mizutani T., Sakaguc M. Serological study of SARS-CoV-2 antibodies in Japanese cats: analysis of risk factors among cat lifestyles. Vet. Res. 2022 doi: 10.21203/rs.3.rs-1113354/v1. (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pecora A., Malacari D.A., Mozgovoj M.V., Díaz M., Peralta A.V., Cacciabue M., Puebla A.F., Carusso C., Mundo S.L., Gonzalez Lopez Ledesma M.M. Anthropogenic infection of domestic cats with SARS-CoV-2 alpha variant b.1.1.7 lineage in Buenos Aires. Front Vet. Sci. 2022;9 doi: 10.3389/fvets.2022.790058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spada E., Vitale F., Bruno F., Castelli G., Reale S., Perego R., Baggiani L., Proverbio D. A pre- and during pandemic survey of Sars-Cov-2 infection in stray colony and shelter cats from a high endemic area of Northern Italy. Viruses. 2021;13 doi: 10.3390/v13040618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gunther I., Raz T., Klement E. Association of neutering with health and welfare of urban free-roaming cat population in Israel, during 2012-2014. Prev. Vet. Med. 2018;157:26–33. doi: 10.1016/j.prevetmed.2018.05.018. [DOI] [PubMed] [Google Scholar]
- 25.Seimenis A., Tabbaa D. Stray animal populations and public health in the South Mediterranean and the Middle East regions. Vet. Ital. 2014;50:131–136. doi: 10.12834/VetIt.48.134.3. [DOI] [PubMed] [Google Scholar]
- 26.Vandegrift K.J., Yon M., Surendran-Nair M., Gontu A., Amirthalingam S., Nissly R.H., Levine N., Stuber T., DeNicola A.J., Boulanger J.R., Kotschwar N., Aucoin S.G., Simon R., Toal K., Olsen R.J., Davis J.J., Bold D., Gaudreault N.N., Richt J.A., Musser J.M., Hudson P.J., Kapur V., Kuchipudi S.V. Detection of SARS-CoV-2 Omicron variant (B.1.1.529) infection of white-tailed deer. bioRxiv. 2022 doi: 10.1101/2022.02.04.479189. [DOI] [Google Scholar]
- 27.Davis M.F., Innes G.K. The cat's in the bag: despite limited cat-to-cat severe acute respiratory syndrome coronavirus 2 transmission, one health surveillance efforts are needed. J. Infect. Dis. 2021;223:1309–1312. doi: 10.1093/infdis/jiab106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fisher M., Manor A., Abramovitch H., Fatelevich E., Afrimov Y., Bilinsky G., Lupu E., Ben-Shmuel A., Glinert T., Madar-Balakirski N., Marcus H., Mechaly A. A novel quantitative multi-component serological assay for SARS-CoV-2 vaccine evaluation. Anal. Chem. 2022;94:4380–4389. doi: 10.1021/acs.analchem.1c05264. [DOI] [PubMed] [Google Scholar]
- 29.Noy-Porat T., Makdasi E., Alcalay R., Mechaly A., Levy Y., Bercovich-Kinori A., Zauberman A., Tamir H., Yahalom-Ronen Y., Israeli M., Epstein E., Achdout H., Melamed S., Chitlaru T., Weiss S., Peretz E., Rosen O., Paran N., Yitzhaki S., Shapira S.C., Israely T., Mazor O., Rosenfeld R. A panel of human neutralizing mAbs targeting SARS-CoV-2 spike at multiple epitopes. Nat. Commun. 2020;11:4303. doi: 10.1038/s41467-020-18159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barlev-Gross M., Weiss S., Ben-Shmuel A., Sittner A., Eden K., Mazuz N., Glinert I., Elad Bar-David R., Puni S., Amit O., Kriger O., Schuster R., Alcalay E., Makdasi E., Epstein T., Noy-Porat R., Rosenfeld H., Achdout O., Mazor T., Israely H., Levy A., Mechaly Spike vs nucleocapsid SARS-CoV-2 antigen detection: application in nasopharyngeal swab specimens. Anal. Bioanal. Chem. 2021;413:3501–3510. doi: 10.1007/s00216-021-03298-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eur. Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hakki S., Zhou J., Jonnerby J., Singanayagam A., Barnett J.L., Madon K.J., C A.K., Kelly H., Houston S., Nevin J., Fenn R., Kundu M.A., Crone S., Ahmad N., Derqui-Fernandez E., Conibear P.S., Freemont G.P., Taylor N., Ferguson M., Zambon W.S., Barclay J., Dunning A., Lalvani Lancet Respir. Med. 2022 doi: 10.1016/S2213-2600(22)00226-0. [DOI] [Google Scholar]
- 33.Stranieri A., Lauzi S., Giordano A., Galimberti L., Ratti G., Nicola Decaro N., Brioschi F., Lelli D., Gabba S., Amarachi N.L., Lorusso E., Moreno A., Trogu T., Paltrinieri S. Absence of SARS-CoV-2 RNA and anti-SARS-CoV-2 antibodies in stray cats. Transbound. Emerg. Dis. 2021 doi: 10.1111/tbed.14200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao S., Schuurman N., Li W., Wang C., Smit L.A.M., Broens E.M., Wagenaar J.A., van Kuppeveld F.J.M., Bosch B.J., Egberink E. Serologic screening of severe acute respiratory syndrome coronavirus 2 infection in cats and dogs during first coronavirus disease wave, the Netherlands. Emerg. Infect. Dis. 2021;27:1362–1370. doi: 10.3201/eid2705.204055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fritz M., Rosolen B., Krafft E., Becquart P., Elguero E., Vratskikh O., Denolly S., Boson B., Vanhomwegen J., Gouilh M.A., Kodjo A., Chirouze C., Rosolen S.G., Legros V., Leroy E.M. High prevalence of SARS-CoV-2 antibodies in pets from COVID-19+ households. One Health. 2021 doi: 10.1016/j.onehlt.2020.100192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaudreault N.N., Trujillo J.D., Carossino M., Meekins D.A., Morozov I., Madden D.W., Indran S.V., Bold D., Balaraman V., Kwon T., Artiaga B.L., Cool K., García-Sastre A., Ma W., Wilson W.C., Henningson J., Balasuriya U.B.R., Richt J.A. SARS-CoV-2 infection, disease and transmission in domestic cats. Emerg. Microbes Infect. 2020;9:2322–2332. doi: 10.1080/22221751.2020.1833687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neira V., Brito B., Belén B., Berrios F., Valdés V., Gutierrez A., Ariyama N., Espinoza P., Retamal P., Holmes E.C. A household case evidences shorter shedding of SARS-CoV-2 in naturally infected cats compared to their human owners. Emerg. Microbes Infect. 2021;10:376–383. doi: 10.1080/22221751.2020.1863132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Decaro N., Grassi A., Lorusso E., Patterson E.I., Lorusso A., Desario C., Anderson E.R., Vasinioti V., Wastika C.E., Hughes G.L., Valleriani F., Colitti B., Ricci D., Buonavoglia D., Rosati S., Cavaliere N., Paltrinieri S., Lauzi S., Elia G., Buonavoglia C. Long-term persistence of neutralizing SARS-CoV-2 antibodies in pets. Transbound. Emerg. Dis. 2021 doi: 10.1111/tbed.14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ben-Shmuel A., Brosh-Nissimov T., Glinert I., Bar-David E., Sittner A., Poni R., Cohen R., Achdout H., Tamir H., Yahalom-Ronen Y., Politi B., Melamed S., Vitner E., Cherry L., Israeli O., Beth-Din A., Paran N., Israely T., Yitzhaki S., Levy H., Weiss S. Detection and infectivity potential of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) environmental contamination in isolation units and quarantine facilities. Clin. Microbiol. Infect. 2020;26:1658–1662. doi: 10.1016/j.cmi.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Welkers M.R.A., Han A.X., Reusken C.B.E.M., Eggink D. Possible host-adaptation of SARS-CoV-2 due to improved ACE2 receptor binding in mink. Virus Evol. 2021;7 doi: 10.1093/ve/veaa094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Braun K.M., Moreno G.K., Halfmann P.J., Hodcroft E.B., Baker D.A., Boehm E.C., Weiler A.M., Haj A.K., Hatta M., Chiba S., Maemura T., Kawaoka Y., Koelle K., O'Connor D.H., Friedrich T.C. Transmission of SARS-CoV-2 in domestic cats imposes a narrow bottleneck. PLoS Pathog. 2021 doi: 10.1371/journal.ppat.1009373. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Data Availability Statement
Data are contained within the article and the supplementary material.