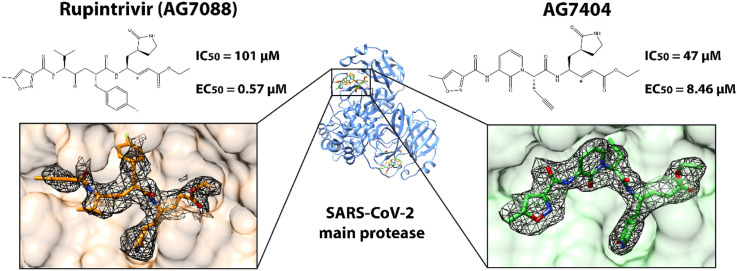
Abstract
Severe acute respiratory syndrome coronaviruses 1 and 2 (SARS-CoV-1 and SARS-CoV-2) pose a threat to global public health. The 3C-like main protease (Mpro), which presents structural similarity with the active site domain of enterovirus 3C protease, is one of the best-characterized drug targets of these viruses. Here we studied the antiviral activity of the orally bioavailable enterovirus protease inhibitor AG7404 against SARS-CoV-1 and SARS-CoV-2 from a structural, biochemical, and cellular perspective, comparing it with the related molecule rupintrivir (AG7800). Crystallographic structures of AG7404 in complex with SARS-CoV-1 Mpro and SARS-CoV-2 Mpro and of rupintrivir in complex with SARS-CoV-2 Mpro were solved, revealing that all protein residues interacting with the inhibitors are conserved between the two proteins. A detailed analysis of protein-inhibitor interactions indicates that AG7404 has a better fit to the active site of the target protease than rupintrivir. This observation was further confirmed by biochemical FRET assays showing IC50 values of 47 μM and 101 μM for AG7404 and rupintrivir, respectively, in the case of SARS-CoV-2 Mpro. Equivalent IC50 values for SARS-CoV-1 also revealed greater inhibitory capacity of AG7404, with a value of 29 μM vs. 66 μM for rupintrivir. Finally, the antiviral activity of the two inhibitors against SARS-CoV-2 was confirmed in a human cell culture model of SARS-CoV-2 infection, although rupintrivir showed a higher potency and selectivity index in this assay.
Keywords: Human coronavirus, SARS-CoV-1, SARS-CoV-2, Main protease Mpro, AG7404, Rupintrivir
Graphical abstract
Human coronaviruses (HCoVs) cause mild to severe respiratory tract illnesses in humans (Hu, 2019). The emergence in the last two decades of severe acute respiratory syndrome coronaviruses 1 and 2 (SARS-CoV-1 and SARS-CoV-2) and Middle East respiratory syndrome coronavirus (MERS-CoV) has revealed the extraordinary potential threat to global public health posed by HCoVs, with SARS-CoV-2 originating the Coronavirus Disease 2019(COVID-19) pandemic (Forman et al., 2022). In the search for drugs against coronavirus, the dimeric 3C-like main protease (Mpro) is one of the best-characterized drug targets (Anand et al., 2003; Yang et al., 2003; Linlin Zhang et al., 2020). Mpro is a key enzyme for viral replication since, together with the papain-like protease, it processes the newly synthesized polyprotein in functional proteins (Meyer et al., 2021).
Orally bioavailable compound AG7404 (also named compound 1) and the related molecule rupintrivir (AG7088) were developed against human rhinovirus, the etiological agent of the common cold. These drugs progressed to phase II/III and phase I of clinical trials, respectively (Hayden et al., 2003; Patick et al., 2005). We previously demonstrated that both compounds are active against the enterovirus B pathogen EV-93. By solving the crystal structures of the complexes of these two peptidomimetic compounds with the EV-93 3C protease (3Cpro), we showed that both imitate the P4 to P1’ peptide substrate, with an α,β-unsaturated ester at P1′ as a Michael acceptor to form an irreversible covalent bond with the active-site cysteine residue. Therefore, both compounds form a stable tetrahedral adduct that results in the irreversible inactivation of the protease. The inhibition of the proteolytic activity of 3Cpro was confirmed in biochemical assays and the activity of the two compounds against EV-93 was proved in vitro in infected cells (Costenaro et al., 2011). Although there is low sequence identity between picornavirus 3Cpro and coronavirus Mpro (<10%), the substrate specificity of both proteases is identical for P1 and P4 subsites. Moreover, even though Mpro domain III has no counterpart in 3Cpro, their active site is located in a cleft between domains I and II, which have a similar architecture to that of picornavirus 3Cpro. Given this structural similarity, it has been suggested that rhinovirus 3Cpro inhibitors may be modified and used for treating SARS (Anand et al., 2003). Therefore, we decided to test AG7404 and rupintrivir against SARS-CoV-2 and SARS-CoV-1.
Crystallographic structures of AG7404 bound to both SARS-CoV-2 Mpro and SARS-CoV-1 Mpro and rupintrivir bound to SARS-CoV-2 Mpro were solved at a resolution of 2.26, 2.53 and 2.26 Å, respectively (Table 1 ). All electron density maps showed clear densities corresponding to the inhibitors at the active sites, which allowed the building of one molecule of AG7404 or rupintrivir per protein monomer, forming a stable tetrahedral adduct (Fig. 1 , Supplementary Fig. 1). The molecular interactions of the inhibitors with Mpro were characterized (Supplementary Fig. 2, Supplementary Table 1). All protein residues interacting with the inhibitors are conserved between SARS-CoV-1 Mpro and SARS-CoV-2 Mpro, thereby suggesting that their efficacy as antivirals against both species might be similar.
Table 1.
Data collection and refinement statistics.
| SARS-CoV-2 Mpro- AG7404 (PDB 7ZQV) |
SARS-CoV-1 Mpro- AG7404 (PDB 7ZQW) |
SARS-CoV-2 Mpro- Rupintrivir (PDB 7P35) |
|
|---|---|---|---|
| Data collection statistics | |||
| Space group | P21 | P21212 | P21 |
| Unit-cell dimensions (Å, °) | a = 45.31, b = 54.08, c = 115.011, α = 90.00, β = 100.34, γ = 90.00 | a = 106.854, b = 45.22, c = 53.55, α = 90.00, β = 90.00, γ = 90.00 | a = 45.02, b = 54.02, c = 114.07, α = 90.00, β = 100.77, γ = 90.00 |
| Beamline | ID30A-3 (ESRF) | XALOC (ALBA) | XALOC (ALBA) |
| Wavelength (Å) | 0.96770 | 0.97918 | 0.97926 |
| Vm (Å/Da) | 2.06 | 1.91 | 2.03 |
| Solvent content (%) | 40.00 | 35.71 | 39.44 |
| Resolution range (Å) | 48.79–2.26 (2.45–2.26) | 47.87–2.53 (2.69–2.53) | 56.09–2.26 (2.29–2.26) |
| Number of observed reflections | 47,666 (2,657) | 32,205 (1,985) | 53,621 (2,639) |
| Number of unique reflections | 18,801 (941) | 7,634 (382) | 21,599 (1,085) |
| Rmergea | 0.096 (0.552) | 0.093 (1.495) | 0.060 (0.853) |
| CC1/2 | 0.985 (0.802) | 0.998 (0.431) | 0.998 (0.580) |
| Spherical completeness (%) | 72.7 (17.5) | 83.1 (24.5) | 84.4 (87.9) |
| Ellipsoidal completeness (%)b | 89.5 (56.8) | 91.9 (46.3) | |
| Mean I/σ (I) | 4.5 (1.4) | 10.1 (1.1) | 9.6 (1.4) |
| Multiplicity | 2.5 (2.8) | 4.2 (5.2) | 2.5 (2.4) |
| Refinement statistics | |||
| Rfactorc | 0.190 | 0.215 | 0.200 |
| Rfreec | 0.241 | 0.289 | 0.258 |
| Number of atoms (non-H) | 4,896 | 2,375 | 4,814 |
|
4,716 | 2,336 | 4,694 |
|
76 | 38 | 86 |
|
104 | 1 | 34 |
| r.m.s.d. for bond lengths (Å) | 0.006 | 0.006 | 0.006 |
| r.m.s.d. for bond angles (°) | 1.444 | 1.526 | 1.471 |
| Average B-factor (Å2) | 40.67 | 65.93 | 50.93 |
| Average B-factor for inhibitor molecules (Å2) | 60.92 (62.68 for chain A, 59.15 for chain B) | 82.69 | 94.59 (98.95 for chain A, 90.22 for chain B) |
| Ramachandran analysisd | |||
| Residues in allowed regions (%) | 99.67 | 99.67 | 99.34 |
| Residues in outlier regions (%) | 0.33 | 0.33 | 0.66 |
Values in parenthesis refer to the outer-most resolution shell.
Rmerge = ΣℎΣi∣Ii,ℎ−<Iℎ>∣/ΣℎΣiIi, where Ii,h is the ith-intensity measurement of reflection h and <Ih> is the average intensity for multiple measurements.
Ellipsoidal completeness was calculated with STARANISO (Tickle et al., 2018).
Rfactor and Rfree = Σ||Fo|−|Fc||/Σ|Fo|. Rfree was calculated for 5% of the reflections not used for refinement.
Ramachandran analysis was done with MolProbity (Chen et al., 2010).
Fig. 1.
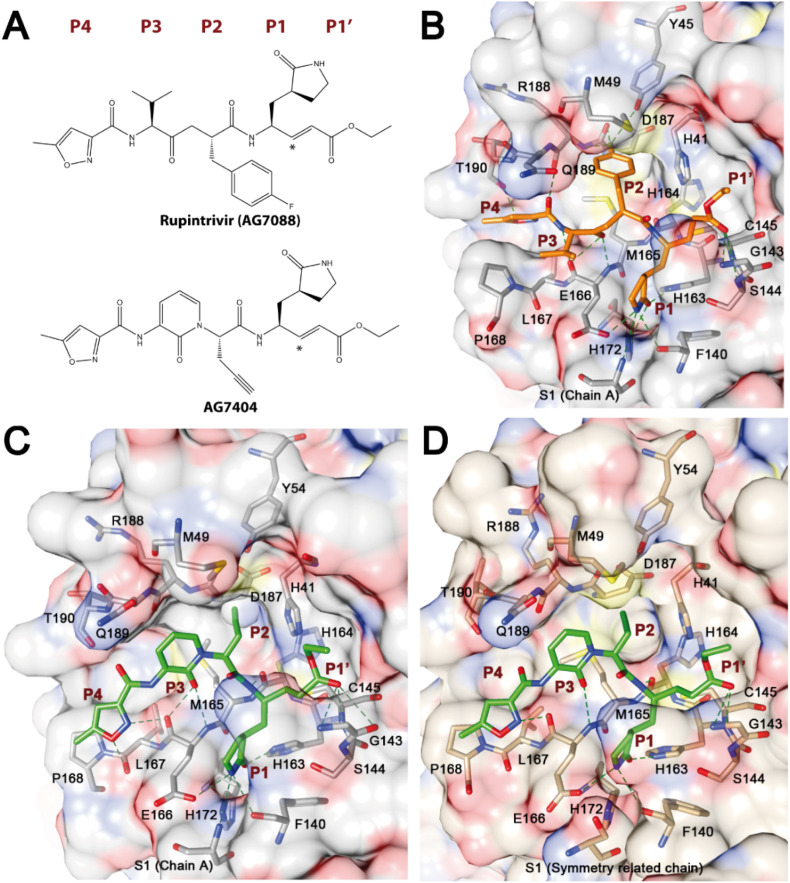
Structure of the inhibitors bound to Mproactive site. (A) Chemical structures of rupintrivir and AG7404. Asterisks indicate the carbon atoms that form irreversible covalent bonds with SARS-CoV-2 Mpro Cys145. (B) SARS-CoV-2 Mpro with rupintrivir. (C) SARS-CoV-2 Mpro with AG7404. (D) SARS-CoV-1 Mpro with AG7404. SARS-CoV-2 Mpro is shown in gray, SARS-CoV-1 Mpro in wheat, rupintrivir in orange and AG7404 in lime green. Proteins are shown as atom-colored surface, with atomic detail of the protein residues interacting with the compounds. Hydrogen bonds are marked with discontinuous dark green lines. Subsites from P4 to P1′ are also labeled.
P1′ and P1 moieties of the inhibitors are shared between the two compounds. In all the structures, P1’ ethyl ester group is positioned above the oxyanion hole formed by Gly143, Ser144 and Cys145 amide groups and makes hydrogen bonds with these residues. In the SARS-CoV-2 Mpro – AG7404 complex, inhibitor O3 forms an additional hydrogen bond with the side chain of catalytic His41. The P1 γ-lactam moiety was designed as a glutamine surrogate and is deeply embedded in the S1 pocket, forming hydrogen bonds with residues Phe140, Glu166 and Ser1 from the other chain of the dimer (Dragovich et al., 1999; Tan et al., 2013; Linlin Zhang et al., 2020, Zhang et al., 2020). Apart from the interactions in the γ-lactam ring, equivalent N8 of AG7404 and N12 of rupintrivir interacts with the carbonyl oxygen of His164.
The P2 part of the inhibitors differs, with an ethynyl group in AG7404 and a 1-fluorobenzen-4-yl group in rupintrivir. Hydrogen bonds are formed between rupintrivir F1 and Tyr54, Asp187 and Arg188 residues, although they might not be well established as electron density in this region was poorly defined. The ethynyl group in AG7404 is much smaller and stacks against His41, with residues Met165 and Asp187 also constraining its position. Electron density of this moiety is less defined when compared with the rest of the AG7404 compound, thus indicating that there is room for binding improvement in this area.
The major difference between the two inhibitors is found in the P3 region. In this regard, AG7404 has a 2-pyridon-1,3-diyl group that cycles with the following amine while rupintrivir has a valine amino acid-like moiety. N58 and O3 atoms in the peptidomimetic backbone of rupintrivir interact with residue Glu166, in a similar manner to O26 in AG7404. However, the electron density of the P3 moiety of AG7404 is much better defined than that of rupintrivir, thereby suggesting a better fit in the protease cavity.
Although the P4 moiety is common to both inhibitors, its position differs, probably because of a distinct orientation imposed by the position of the P3 groups (Supplementary Fig. 3A). While in rupintrivir weak electron density shows that the moiety interacts with Gln189 and Thr190 residues through O4 and O60, in AG7404 hydrogen bonds, both direct and water-mediated, are established with Glu166, Leu167 and Thr190 with O18, N17 and O22 inhibitor atoms. Such interactions are especially well defined in the structure of the SARS-CoV-2 Mpro – AG7404 complex.
Another X-ray structure of SARS-CoV-2 Mpro in complex with rupintrivir was recently published, showing a unique conformation of the compound in the protease active site, with the P2 moiety splitting up the catalytic His41 and Cys145 side chains (Lockbaum et al., 2021). However, in our structure, the P2 moiety is accommodated at the S2 subsite, as previously observed in enterovirus complexes, and the His41 side chain is in its functional position (Supplementary Fig. 3B).
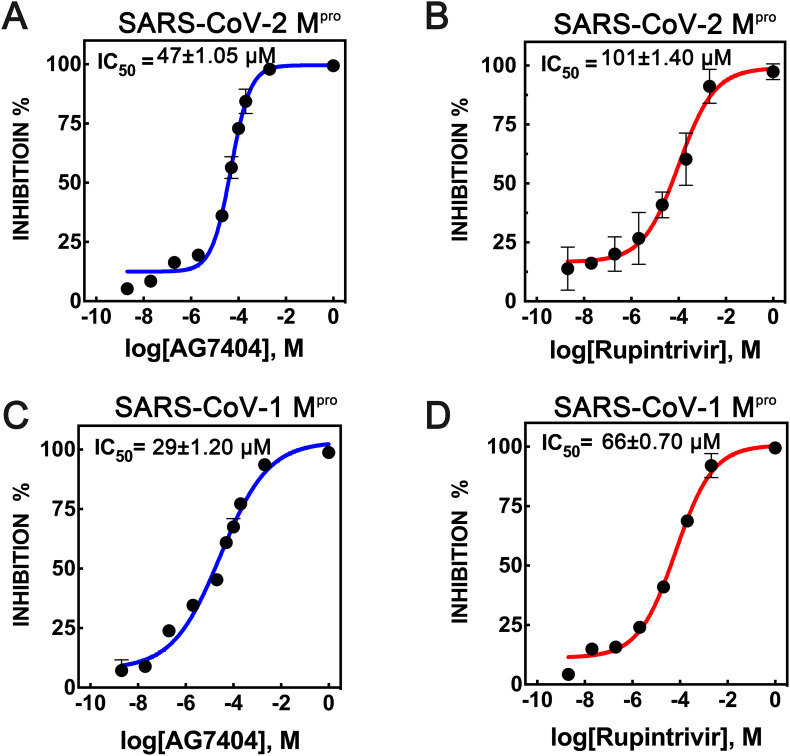
Previous biochemical studies have revealed that rupintrivir is a selective potent enterovirus 3C protease inhibitor, with IC50 values as low as 1.04 nM against rhinovirus HRV3C. It also inhibits SARS-CoV-2 Mpro but with much higher IC50 values of at least 68 μM (Liu et al., 2021; Vatansever et al., 2021). In our hands, rupintrivir also showed antiviral activity against the SARS-CoV Mpro enzymes, as shown by a fluorescence resonance energy transfer (FRET)-based inhibition assay, with IC50 values of 66 and 101 μM for SARS-CoV-1 and SARS-CoV-2, respectively (Fig. 2 ). The observation of two distinct binding modes of the inhibitor to HCoV Mpro (Lockbaum et al., 2021; this work) suggests a dynamic equilibrium of P2 orientations, which could explain the loss of the inhibitory capacity of rupintrivir against HCoV when compared to its potency against enterovirus.
Fig. 2.
Biochemical inhibition assays. (A) SARS-CoV-2 Mpro with AG7404. (B) SARS-CoV-2 Mpro with rupintrivir. (C) SARS-CoV-1 Mpro with AG7404. (D) SARS-CoV-1 Mpro with rupintrivir. The inhibition of Mpro activity was measured by FRET and the dose-response curves for IC50 values were determined by a non-linear regression model (four parameters). Triplicate experiments were performed for each data point n = 2, and the value is presented as mean ± standard deviation (SD).
The same assay was used to determine the activity of AG7404 against Mpro, which appears to be stronger than that of rupintrivir. The IC50 values of AG7404 against SARS-CoV-1 and SARS-CoV-2 are 29 μM and 47 μM, respectively. These results are in agreement with what we observe in the structural analysis of the complexes, with AG7404 having a more defined electron density, thus suggesting a better fit in the SARS Mpro active site.
To verify the antiviral activity of these molecules against SARS-CoV-2, in vitro cell-based infection assays were performed. To this end, an immunomicroscopy-based phenotypic assay was carried out in human lung adenocarcinoma A549 cells expressing SARS-CoV-2 human receptor ACE2 (A549-ACE2), which are susceptible to SARS-CoV-2 infection in culture.
SARS-CoV-2 propagation was strongly inhibited by both rupintrivir and AG7404 in A549-ACE2 cells (Table 2 , Supplementary Fig. 4). Rupintrivir showed strong potency (EC50 0.57 μM; EC90 8.46 μM) and high selectivity (CC50 > 250 μM; SI50 > 438.6 μM), suggesting that this molecule inhibits SARS-CoV-2 infection as previously reported in these cells (Xie et al., 2020). AG7404 showed antiviral activity at higher doses (EC50 6.8 μM; EC90 90.90 μM; CC50 250 μM; SI50 36.8). Structural data and biochemical assays supported AG7404 as a better antiviral candidate against SARS-CoV-2 than rupintrivir, with the first showing tighter interactions with the target and a lower IC50. Nevertheless, cellular assays suggest that differences in additional factors in this experimental setup like cellular uptake, non-specific binding, compound solubility and stability, and/or compound biotransformation may reduce the expected effectiveness of AG7404 in the experimental system that was used (Hann and Simpson, 2014; Teuscher et al., 2017). Both compounds were found to be inactive against SARS-CoV-2 when using a Vero cell-based infection system (data not shown), as found previously with SARS-CoV-1 (Matthews et al., 2004). Moreover, a previous study showed that rupintrivir and AG7404 inhibited SARS-CoV-2 infection only at higher drug concentrations when tested on Vero E6 and Huh7 cells (Leike Zhang et al., 2020). This observation thus suggests that distinct cell lines may be required to study the potential of these and similar compounds. Vero cells are not an accurate model for human airway and lung epithelial cells. In contrast, A549-ACE2 cells are likely a closer mimicking model (Steuten et al., 2021). It has been found that some Mpro inhibitors cross-inhibit cell cathepsins involved in viral entry (Ma et al., 2022; Steuten et al., 2021). Cross-inhibition of cathepsins might play a synergic role in the case of rupintrivir, although AG7404 has not been assayed for this activity.
Table 2.
Cell-based inhibition assays with A549-ACE2 cell lines and SARS-CoV-2. Effective concentrations 50 and 90 (EC50 and EC90) and cytotoxic concentration 50 (CC50) are expressed in μM. Selectivity index 50 (SI50) was calculated as the ratio between the CC50 and the EC50. Remdesivir was used as a control.
| A549-ACE2 | ||||
|---|---|---|---|---|
| EC50 [95% C.I.] | EC90 [95% C.I.] | CC50 | SI50 | |
| AG7404 | 6.80 [3.20–13.51] | 90.90 [38.50–436.30] | 250 | 36.8 |
| Rupintrivir | 0.57 [0.33–0.87] | 8.46 [5.46–15.60] | >250 | >438.6 |
| Remdesivir | 0.033 [0.018–0.055] | 0.145 [0.082–0.432] | >250 | >7575.7 |
Since the beginning of the pandemic, both public research institutions and pharma companies made huge efforts to find clinical candidates that were active against COVID-19. Pfizer preclinical experiments showed that compound PF-00835231, previously developed as an inhibitor of SARS-CoV-1 Mpro, had suitable pharmaceutical properties as intravenous treatment with an EC50 which ranges from 158 to 422 nM (de Vries et al., 2021; Hoffman et al., 2020). More recently, the drug nirmatrelvir (PF-07321332), also developed by Pfizer with and EC50 of 16 nM, has been granted emergency use authorization by both the FDA and EMA for high-risk COVID-19 patients (Greasley et al., 2022; Hammond et al., 2022; Rai et al., 2022). In the medication known as Paxlovid, nirmatrelvir is administered together with ritonavir, a P450(CYP)3A inhibitor, to increase its effective concentration, since the cytochrome degrades the compound. Depletion of the cytochrome enzyme activity may increase the plasma concentration of other drugs. Therefore, Paxlovid has multiple contraindications according to its FDA and EMEA emergency use authorizations, because it interferes with other co-administered pharmacological products. Finding and developing new SARS-CoV inhibitors is thus still desirable, and various ongoing studies are addressing this objective using Mpro as a target (Agost-Beltrán et al., 2022; Liu et al., 2020).
Although our cell-based assays indicate that optimization of rupintrivir and AG7404 for improved cellular uptake or other factors is needed, their tight-fitting in the active site shown in the crystal structures, in particular of AG7404, makes the detailed structural data regarding the interaction of different chemical moieties of the inhibitors in SARS-CoV Mpro subsites beneficial to further develop additional compounds that bind specifically to SARS Mpro protease pockets. The structure of AG7404 in complex with SARS-CoV-2 Mpro was superposed with those of PF-00835231 and nirmatrelvir (Supplementary Figs. 3C and 3D). The three compounds share a γ-lactam moiety in P1 tightly fitted in the S1 pocket, which should be maintained in further inhibitor development initiatives. However, the warhead in PF-00835231 is a hydroxymethylketone, and in nirmatrelvir is a nitrile group, instead of the ethyl ester in rupintrivir and AG7404. Other parts of these compounds also differ, although the occupied sites are similar.
Taking into consideration the structural data above discussed, four new compounds were designed as potential compounds that could be synthesized and tested for antiviral activity (Supplementary Table 2). All of them share the already discussed γ-lactam moiety in P1 and a leucine-like group in P2 as in PF-00835231, since electron density in S2 subsite was not well defined in the AG7404 structure and there is enough space to accommodate this bigger hydrophobic group. Regarding the P3 chemical group the one present in AG7404 was maintained, as experimental structures show a well-defined electronic density that suggests a tight fitting in the protein subsite. The AG7404 P4 moiety was slightly modified in inhibitors 1 and 2, maintaining however the nitrogen atom that established a hydrogen bond in both our AG7404 structures. Oppositely, inhibitors 3 and 4 are smaller and were designed with a five-atom ring directly bound to the P3 six-atom ring, with the aim to check whether a smaller moiety could work better as the P4 experimental density in our structures is not that strong as that corresponding to P3 is. Finally, two of the compounds have a nitrile group as a warhead (inhibitors 1 and 3), while the other two have a hydroxymethylketone (inhibitors 2 and 4). This choice was based in Pfizer inhibitors which have proved potent and suitable from a pharmacological point of view. The four inhibitors were fit into the SARS-CoV-2 M pro active site by a covalent docking simulation (Supplementary Fig. 5, Supplementary Table 2). The values of the four binding energies verify that all four compounds behave as covalent inhibitors of SARS-CoV Mpro. Binding energy values suggest that the hydroxymethylketone warhead group produces a tighter binding than the nitrile one. Regarding the P3/P4 region, more favorable binding energies are obtained when the P4 ring moiety is maintained separated from the ring in P3, rather than fusing both. As a combination of both observations, inhibitor 2 presents the best binding affinity, with −10.1 kcal/mol.
Interestingly, the P3 moiety from inhibitors 3 and 4 appears in the docked models in a different disposition than that of P3 in AG7404, located outside the S3 pocket in a position closer to the catalytic residue His41 (Supplementary Figs. 5C and 5D). Therefore, docking results suggest that modifications of AG7404 with a smaller chemical group in the P3-P4 parts of the inhibitor do not guarantee the occupancy of S3 and S4 subsites of the protease active center.
1. Materials and methods
1.1. SARS-CoV-1 and SARS-CoV-1 Mpro expression and purification
Synthetic genes codifying for both proteins were purchased from GeneArt (Thermo Fisher Scientific) with sequences optimized for Escherichia coli codon usage. The DNA sequence codifying for the protein was cloned using the InFusion™ method into a pOPINS vector, which allows expression of target proteins with N-terminal His-SUMO tags (Berrow et al., 2007). The proteins were expressed in E. coli BL21(DE3) cells grown at 37 °C. After induction at an optical density of ∼0.6 with 0.4 mM of isopropyl β-D-1-thiogalactopyranoside (IPTG), cells were kept overnight at 25 °C. They were then resuspended in 50 mM Tris-HCl pH 7.6, 250 mM NaCl, 30 mM imidazole and 40 μg/mL DNase I, and lysis was performed using a cell disruptor (Constant Systems, Ltd.) at 20 kPsi. Next, samples were clarified by centrifugation for 45 min at 30,000×g and purified by a three-step chromatography protocol. First, proteins were loaded onto a HisTrap HP column (Cytiva) equilibrated in binding buffer (50 mM Tris-Hcl pH 7.6, 250 mM NaCl, 30 mM imidazole) and eluted with a linear gradient with elution buffer (50 mM Tris-HCl pH 7.6, 250 mM NaCl, 500 mM imidazole). His-SUMO-Mpro fusion proteins were digested with ULP1 SUMO protease overnight at a 1:500 protease:substrate molar ratio while dializing against a buffer containing 20 mM Tris pH 7.6, 250 mM NaCl buffer. Protein samples were then loaded again onto a HisTrap HP column to perform reverse affinity chromatography with the same buffers as in the previous step. Protein samples eluted in the non-bound fraction were pooled and loaded onto a Superdex75 Increase 10/300 column (Cytiva). Samples were concentrated with 10,000 Da molecular weight cutoff Amicon® Ultra Centrifugal Filters (Merck Millipore).
1.2. Complex preparation
AG7404 ((E)-(S)-4-((S)-2-{3-[(5-methyl-isoxazole-3-carbonyl)-amino]-2-oxo-2H-pyridin-1-yl}-pent-4-ynoylamino)-5-((S)-2-oxo-pyrrolidin-3-yl)-pent-2-enoic acid ethyl ester) was provided by Pfizer under a Pure Compound Grant. Rupintrivir or AG7088 (4-{2-(4-fluoro-benzyl)-6-methyl-5-[(5-methyl-isoxazole-3-carbonil)-amino]-4-oxo-heptanoylamino}-5-(2-oxo-pyrrolidin-3-yl)-pentanoicacidethylester) was purchased from Tocris Bioscience. Both compounds were dissolved in 100% DMSO. The SARS-CoV-2 Mpro– AG7404 complex was prepared at a final protein concentracion of 3.5 mg/mL, the SARS-CoV-1 Mpro– AG7404 complex at 4 mg/mL and the SARS-CoV-2 Mpro– rupintrivir complex at 3.1 mg/mL. In all cases, a 30-fold molar excess of the inhibitor was added, and samples were kept overnight at 4 °C to allow the formation of the covalent complex before conducting further experiments.
1.3. Crystallization and data collection
All crystals were prepared at 20 °C by the hanging drop vapor diffusion technique.SARS-CoV-2 M pro – AG7404 crystals grew in drops where the complex was mixed with a reservoir that contained 0.2M sodium chloride, 0.1M HEPES pH 7 and 25% w/v PEG 3350. SARS-CoV-1Mpro – AG7404 crystals grew in drops where the complex was mixed with a reservoir solution that contained 0.2M lithium sulfate monohydrate, 0.1M HEPES pH 8 and 27% PEG 3350. SARS-CoV-2 Mpro – rupintrivir crystals grew in drops where the complex was mixed with a reservoir that contained 0.2M triammonium citrate and 18% w/v PEG 3350. All crystals were mounted in loops and flash-frozen in liquid nitrogen using a cryoprotective buffer containing the reservoir solution plus 30% glycerol (SARS-CoV-2 Mpro – AG7404, SARS-CoV-2 Mpro – rupintrivir) or 20% glycerol (SARS-CoV-1 Mpro – AG7404). X-ray diffraction data for the SARS-CoV-1 Mpro – AG7404 and SARS-CoV-2 Mpro – rupintrivir complexes were collected at the XALOC beamline at ALBA Synchrotron in Cerdanyola del Vallès (Spain), while the data for the SARS-CoV-2 Mpro – AG7404 complex were collected at ID30A-3 beamline at the European Synchrotron Radiation Facility (ESRF) in Grenoble (France).
1.4. Structure determination
Diffraction data were processed with XDS (Kabsch, 2010) using the autoPROC pipeline (Vonrhein et al., 2011). For the SARS-CoV-2 Mpro – AG7404 and SARS-CoV-1 Mpro – AG7404 datasets, STARANISO autoprocessing was used (Tickle et al., 2018). Structures were solved by molecular replacement with Phaser (McCoy et al., 2007) using previously deposited structures of SARS-CoV-1 and SARS-CoV-2 Mpro with PDB accession codes 1UJ1 and 6Y2E, respectively, as search models (Yang et al., 2003; Linlin Zhang et al., 2020). Inhibitors were manually fitted in the active site electron densities with Coot, and manual model rebuilding alternated with refinement using REFMAC5 was performed (Emsley et al., 2010; Murshudov et al., 2011). JLigand was used to generate the restraints for the covalent bonds between the proteins and the inhibitors (Lebedev et al., 2012). Structures were validated with Molprobity (Chen et al., 2010). Atomic coordinates and structure factors for the reported crystal structure have been deposited in the Protein Data Bank under accession codes 7ZQV (SARS-CoV-2 Mpro - AG7404 complex), 7ZQW (SARS-CoV-1 Mpro - AG7404 complex) and 7P35 (SARS-CoV-2 Mpro - rupintrivir complex). Diagrams detailing the interactions between the inhibitors and the proteins were obtained with Ligplot+ (Laskowski and Swindells, 2011).
1.5. Biochemical inhibition assays
Fluorescence resonance energy transfer (FRET)-based inhibition assays were performed with a fluorescent peptide substrate comprising a cleavage site of SARS-CoV-1 and SARS-CoV-2 Mpro (Dabcyl-KTSAVLQ↓SGFRKM-E(Edans)-NH2); GenScript) and a buffer made up of 20 mM TRIS, 100 mM NaCl, 1 mM EDTA, and 1 mM DTT, pH 7.3 (Zhu et al., 2011). Stock solutions of the inhibitors were prepared with 100% DMSO. For determination of the IC50, 1.5 μM of the proteases was incubated with rupintrivir or AG7404 at concentrations ranging from 20 nM to 2 mM overnight at 4 °C and for 30 min at 37 °C. Afterwards, 50 μL of the complex at a final protein concentration of 750 nM was pipetted into a 96-well plate. The reaction was then started by addition of 50 μL of the substrate dissolved in the reaction buffer at a final concentration of 20 μM. The fluorescence signal of the Edans caused by the cleavage of the substrate was monitored at 460 nm after exciting at 360 nm using a Synergy H1 Hybrid Multimode Plate Reader (Agilent). Reactions without FRET substrate and without protease were performed as a control for all conditions. IC50 values were calculated using the GraphPad Prism 6.0 software (GraphPad). All measurements were performed in triplicate and are presented as the mean ± standard deviation.
1.6. Cell-based inhibition assays
An immunomicroscopy-based phenotypic assay was performed in A549 cells expressing SARS-CoV-2 receptor ACE2 (A549-ACE2) (Ginex et al., 2021). A549 cells were kindly provided by Dr. Juan Ortín (CNB-CSIC). The cells were transduced with a retroviral vector enabling the expression of ACE2 in a di-cistronic cassette conferring resistance to blasticidine. Transduced populations were selected using 2.5 μg/mL of blasticidine. All cell cultures were kept in complete media (DMEM) supplemented with 10 mM HEPES, 1X non-essential amino acids (Gibco), 100 U/mL penicillin-streptomycin (GIBCO) and 10% fetal bovine serum (FBS; heat-inactivated at 56 °C for 30 min). All experiments were performed at 37 °C in a CO2 incubator (5% CO2), in the presence of 2% FBS and in the absence of selection antibiotics. Inhibitors were dissolved in 100% DMSO at a concentration of 100 mM and subsequently diluted in cell culture media.
The toxicity of the compounds was evaluated by studying cell confluency, as well as metabolic activity using a MTT-formazan assay. Cell monolayers were seeded in 96-well plates. The day after, cells were treated with concentrations ranging from 250 μM to 0.1 μM and 48 h later they were subjected to MTT assays using standard procedures. The CC50 values were graphically interpolated from dose-response curves obtained with three biological replicates.
SARS-CoV-2 (Orthocoronavirinae; Alphacoronavirus; Sarbecovirus; strain NL/2020) was kindly provided by Dr. R. Molenkamp, Erasmus University Medical Center Rotterdam. SARS-CoV-2 stocks were produced and titrated in Vero E6 cells as described previously (Ginex et al., 2021). A549-ACE2 cells were seeded onto 96-well plates as described above and infected at low multiplicity of infection (MOI = 0.01) in the presence of the indicated compound concentrations. Forty-eight hours post-infection, cells were fixed for 20 min at room temperature with a 4% formaldehyde solution in PBS, washed twice with PBS, and kept in incubation buffer (3% BSA; 0.3% Triton X100 in PBS) for 1 h. A rabbit monoclonal antibody against the N protein was diluted in the incubation buffer (1:2000, v/v; Genetex HL344) and incubated with the cells for 1 h; after this time, cells were washed with PBS and subsequently incubated with a 1:500 (v/v) dilution of a goat anti-rabbit conjugated to Alexa 488 (Invitrogen-Carlsbad, CA). To control for unexpected toxicity of the compounds, nuclei were stained with DAPI (Life Technologies) during the secondary antibody incubation as recommended by the manufacturer. Cells were washed with PBS and imaged using an automated multimode reader (TECAN Spark Cyto; Austria). All the infection experiments were performed by mixing the virus and compound dilutions 1:1 (v/v) before addition to the target cells. Remdesivir (Santa Cruz) was used as a control (Pruijssers et al., 2020; Xie et al., 2020).
1.7. Covalent docking of designed compounds
Covalent dockings were performed with AutoDockFR (Ravindranath et al., 2015). Chemical structures of the designed compounds were obtained with ChemDraw (RRID:SCR_016768). SMILES strings were used to obtain a PDB file in Chimera (Pettersen et al., 2004) which was then converted to PDBQT format by OpenBabel (O'Boyle et al., 2011). Ligands were prepared in the product form, including carbon backbone and sidechain atoms from Cys145, which are shared with the receptor. Correspondence between atoms in the root section in PDBQT format and backbone, alpha and beta carbons of cysteine were established using the “-R” option from the prepare_ligand4.py script from MGLTools (Morris et al., 2009). Crystal structure of the SARS-CoV-2 Mpro in complex with AG7404 without the inhibitor was prepared and used to generate the target file for docking with AutoDockFR and AutoGridFR programs. A cube with length of 30 Å centered in AG7404 was used as grid box, alpha and beta carbons were defined as covalently bonded atoms, the nitrogen atom from Cys145 was the tensor atom and Cys145 was set as the residue allowed to form a covalent bond with the receptor. Docking was performed with the AutoDockFR program with carbon backbone, alpha carbon and beta carbon atoms from Cys defined as overlapping atoms between the receptor and the ligand.
Author contributions
MC conceived and supervised the study; MFF and AHM prepared the protein samples; MFF, AHM, SMG and JPS performed the structural experiments; MFF, AHM and JPS performed the biochemical experiments; PB, VC, UG and PG performed the cellular experiments; MFF performed the covalent docking simulations; MFF, AHM, SMG, JPS, UG, PG and MC analyzed the results, and MFF, AHM, SMG, UG, PG and MC wrote or contributed to the writing of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was supported by the Ministry of Science and Innovation of Spain (grant numbers BFU2017-83720-P and PID-2020-120141GB-I00 to MC) and Spanish National Research Council (grant numbers 2020AEPP116 to MC and PIE-RD-COVID-19 ref. E202020E079 to the CNB-CSIC). We acknowledge institutional funding from the Ministry of Science and Innovation of Spain through the Severo Ochoa Award of Excellence (SEV-2015-0500 and CEX2019-000913-S to IRB Barcelona) and Maria de Maeztu Award (MDM-2014-0435 to IBMB-CSIC Structural Biology Unit), and from the Catalan Government's CERCA Programme (grant to IRB Barcelona). This research work was also funded by the European Commission – NextGenerationEU (Regulation EU 2020/2094), through CSIC's Global Health Platform (PTI Salud Global). AHM was supported by National Agency for Research and Development (ANID)/Scholarship Program POSTDOCTORADO BECAS CHILE/2019–74200135. SMG was supported by an FPI fellowship from the Ministry of Science and Innovation of Spain. AG7404 ((E)-(S)-4-((S)-2-{3-[(5-methyl-isoxazole-3-carbonyl)-amino]-2-oxo-2H-pyridin-1-yl}-pent-4-ynoylamino)-5-((S)-2-oxo-pyrrolidin-3-yl)-pent-2-enoic acid ethyl ester) was provided by Pfizer under Pure Compound Grant 63236753. The authors wish to thank the personnel of the following facilities for help during crystallization and X-ray data acquisition: Automated Crystallization Platform at IBMB; XALOC beamline at the ALBA Synchrotron, Cerdanyola del Vallès, Spain, and ID30A-3 beamline at the European Synchrotron Radiation Source (ESRF), Grenoble, France.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2022.105458.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Agost-Beltrán L., de la Hoz-Rodríguez S., Bou-Iserte L., Rodríguez S., Fernández-de-la-Pradilla A., González F.V. Advances in the development of SARS-CoV-2 Mpro inhibitors. Molecules. 2022;27:2523. doi: 10.3390/molecules27082523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) Structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. 80-. [DOI] [PubMed] [Google Scholar]
- Berrow N.S., Alderton D., Sainsbury S., Nettleship J., Assenberg R., Rahman N., Stuart D.I., Owens R.J. A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 2007;35 doi: 10.1093/nar/gkm047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V.B., Arendall W.B., Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costenaro L., Kaczmarska Z., Arnan C., Janowski R., Coutard B., Solà M., Gorbalenya A.E., Norder H., Canard B., Coll M. Structural basis for antiviral inhibition of the main protease, 3C, from human enterovirus 93. J. Virol. 2011;85:10764–10773. doi: 10.1128/jvi.05062-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries M., Mohamed A.S., Prescott R.A., Valero-Jimenez A.M., Desvignes L., O'Connor R., Steppan C., Devlin J.C., Ivanova E., Herrera A., Schinlever A., Loose P., Ruggles K., Koralov S.B., Anderson A.S., Binder J., Dittmann M. A comparative analysis of SARS-CoV-2 antivirals characterizes 3CL pro inhibitor PF-00835231 as a potential new treatment for COVID-19. J. Virol. 2021;95 doi: 10.1128/jvi.01819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragovich P.S., Zhou R., Skalitzky D.J., Fuhrman S.A., Patick A.K., Ford C.E., Meador J.W., Worland S.T. Solid-phase synthesis of irreversible human rhinovirus 3C protease inhibitors. Part 1: optimization of tripeptides incorporating N-terminal amides. Bioorg. Med. Chem. 1999;7:589–598. doi: 10.1016/S0968-0896(99)00005-X. [DOI] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman R., Azzopardi-muscat N., Kirkby V., Lessof S., Limaro N., Pastorino G., Permanand G., Ci M., Schalkwyk V., Torbica A., Busse R., Figueras J., Mckee M., Mossialos E. Health policy Drawing light from the pandemic : rethinking strategies for health policy and beyond. Health Pol. 2022;126:1–6. doi: 10.1016/j.healthpol.2021.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginex T., Garaigorta U., Ramírez D., Castro V., Nozal V., Maestro I., García-Cárceles J., Campillo N.E., Martinez A., Gastaminza P., Gil C. Host-directed FDA-approved drugs with antiviral activity against SARS-CoV-2 identified by hierarchical in silico/in vitro screening methods. Pharmaceuticals. 2021;14 doi: 10.3390/ph14040332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greasley S.E., Noell S., Plotnikova O., Ferre R., Liu W., Bolanos B., Fennell K., Nicki J., Craig T., Zhu Y., Stewart A.E., Steppan C.M. Structural basis for the in vitro efficacy of nirmatrelvir against SARS-CoV-2 variants. J. Biol. Chem. 2022;298 doi: 10.1016/j.jbc.2022.101972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond J., Leister-Tebbe H., Gardner A., Abreu P., Bao W., Wisemandle W., Baniecki M., Hendrick V.M., Damle B., Simón-Campos A., Pypstra R., Rusnak J.M. Oral nirmatrelvir for high-risk, nonhospitalized adults with covid-19. N. Engl. J. Med. 2022;1–12 doi: 10.1056/nejmoa2118542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann M.M., Simpson G.L. Intracellular drug concentration and disposition - the missing link? Methods. 2014;68:283–285. doi: 10.1016/j.ymeth.2014.05.009. [DOI] [PubMed] [Google Scholar]
- Hayden F.G., Turner R.B., Gwaltney J.M., Chi-burris K., Gersten M., Hsyu P., Patick A.K., Iii G.J.S., Zalman L.S., Al H.E.T. Phase II , randomized , double-blind , placebo-controlled studies of ruprintrivir nasal spray 2-percent suspension for prevention and treatment of experimentally induced rhinovirus colds in. Healthy Volunteers. 2003;47:3907–3916. doi: 10.1128/AAC.47.12.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman R.L., Kania R.S., Brothers M.A., Davies J.F., Ferre R.A., Gajiwala K.S., He M., Hogan R.J., Kozminski K., Li L.Y., Lockner J.W., Lou J., Marra M.T., Mitchell L.J., Murray B.W., Nieman J.A., Noell S., Planken S.P., Rowe T., Ryan K., Smith G.J., Solowiej J.E., Steppan C.M., Taggart B. Discovery of ketone-based covalent inhibitors of coronavirus 3CL proteases for the potential therapeutic treatment of COVID-19. J. Med. Chem. 2020;63:12725–12747. doi: 10.1021/acs.jmedchem.0c01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2019 doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., Swindells M.B. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- Lebedev A.A., Young P., Isupov M.N., Moroz O.V., Vagin A.A., Murshudov G.N. JLigand: a graphical tool for the CCP4 template-restraint library. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012;68:431–440. doi: 10.1107/S090744491200251X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Boland S., Scholle M.D., Bardiot D., Marchand A., Chaltin P., Blatt L.M., Beigelman L., Symons J.A., Raboisson P., Gurard-Levin Z.A., Vandyck K., Deval J. Dual inhibition of SARS-CoV-2 and human rhinovirus with protease inhibitors in clinical development. Antivir. Res. 2021;187 doi: 10.1016/j.antiviral.2021.105020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liang C., Xin L., Ren X., Tian L., Ju X., Li H., Wang Yongbo, Zhao Q., Liu H., Cao W., Xie X., Zhang D., Wang Yu, Jian Y. The development of Coronavirus 3C-Like protease (3CLpro) inhibitors from 2010 to 2020. Eur. J. Med. Chem. 2020;206 doi: 10.1016/j.ejmech.2020.112711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockbaum G.J., Henes M., Lee J.M., Timm J., Nalivaika E.A., Thompson P.R., Kurt Yilmaz N., Schiffer C.A. Pan-3C protease inhibitor rupintrivir binds SARS-CoV-2 main protease in a unique binding mode. Biochemistry. 2021;60:2925–2931. doi: 10.1021/acs.biochem.1c00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X.R., Alugubelli Y.R., Ma Y., Vatansever E.C., Scott D.A., Qiao Y., Yu G., Xu S., Liu W.R. MPI8 is potent against SARS-CoV-2 by inhibiting dually and selectively the SARS-CoV-2 main protease and the host cathepsin L. ChemMedChem. 2022;17:1–8. doi: 10.1002/cmdc.202100456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews D.A., Patick A.K., Baker R.O., Brothers M.A., Dragovich P.S., Hartmann C.J., Johnson T.O., Mucker E.M., Reich S.H., Rejto P.A.J., Rose P.W., Zwiers S.H., Huggins J. In: Institute of Medicine (US) Forum on Microbial Threats. Learning from SARS: Preparing for the Next Disease Outbreak: Workshop Summary. Knobler S., Mahmoud A., Lemon S., et al., editors. National Academies Press (US); Washington (DC): 2004. In vitro antiviral activity of human rhinovirus 3C protease inhibitors against the SARS coronavirus; pp. 186–193. [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B., Chiaravalli J., Gellenoncourt S., Brownridge P., Bryne D.P., Daly L.A., Grauslys A., Walter M., Agou F., Chakrabarti L.A., Craik C.S., Eyers C.E., Eyers P.A., Gambin Y., Jones A.R., Sierecki E., Verdin E., Vignuzzi M., Emmott E. Characterising proteolysis during SARS-CoV-2 infection identifies viral cleavage sites and cellular targets with therapeutic potential. Nat. Commun. 2021;12:1–16. doi: 10.1038/s41467-021-25796-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;3:2785–2791. doi: 10.1002/jcc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G.N., Skubák P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open Babel. J. Cheminform. 2011;3:1–14. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patick A.K., Brothers M.A., Maldonado F., Binford S., Maldonado O., Fuhrman S., Petersen A., Iii G.J.S., Zalman L.S., Burns-naas L.A., Tran J.Q. In vitro antiviral activity and single-dose pharmacokinetics in humans of a novel. Orally Bioavailable Inhibitor of Human Rhinovirus 3C Protease. 2005;49:2267–2275. doi: 10.1128/AAC.49.6.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pruijssers A.J., George A.S., Schäfer A., Leist S.R., Gralinksi L.E., Dinnon K.H., Yount B.L., Agostini M.L., Stevens L.J., Chappell J.D., Lu X., Hughes T.M., Gully K., Martinez D.R., Brown A.J., Graham R.L., Perry J.K., Du Pont V., Pitts J., Ma B., Babusis D., Murakami E., Feng J.Y., Bilello J.P., Porter D.P., Cihlar T., Baric R.S., Denison M.R., Sheahan T.P. Remdesivir inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.107940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai D.K., Yurgelonis I., McMonagle P., Rothan H.A., Hao L., Gribenko A., Titova E., Kreiswirth B., White K.M., Zhu Y., Anderson A.S., Cardin R.D. 2022. Nirmatrelvir, an Orally Active Mpro Inhibitor, Is a Potent Inhibitor of SARS-CoV-2 Variants of Concern. bioRxiv. [DOI] [Google Scholar]
- Ravindranath P.A., Forli S., Goodsell D.S., Olson A.J., Sanner M.F. AutoDockFR: advances in protein-ligand docking with explicitly specified binding site flexibility. PLoS Comput. Biol. 2015;11:1–28. doi: 10.1371/journal.pcbi.1004586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuten K., Kim H., Widen J.C., Babin B.M., Onguka O., Lovell S., Bolgi O., Cerikan B., Neufeldt C.J., Cortese M., Muir R.K., Bennett J.M., Geiss-Friedlander R., Peters C., Bartenschlager R., Bogyo M. Challenges for targeting SARS-CoV-2 proteases as a therapeutic strategy for COVID-19. ACS Infect. Dis. 2021;7:1457–1468. doi: 10.1021/acsinfecdis.0c00815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J., George S., Kusov Y., Perbandt M., Anemuller S., Mesters J.R., Norder H., Coutard B., Lacroix C., Leyssen P., Neyts J., Hilgenfeld R. 3C protease of enterovirus 68: structure-based design of Michael acceptor inhibitors and their broad-spectrum antiviral effects against picornaviruses. J. Virol. 2013;87:4339–4351. doi: 10.1128/jvi.01123-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuscher K.B., Zhang M., Ji H. A versatile method to determine the cellular bioavailability of small-molecule inhibitors. J. Med. Chem. 2017;60:157–169. doi: 10.1021/acs.jmedchem.6b00923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tickle I.J., Flensburg C., Keller P., Paciorek W., Sharff A., Vonrhein C., Bricogne G. Phasing Ltd; Cambridge, United Kingdom Glob: 2018. STARANISO. [Google Scholar]
- Vatansever E.C., Yang K.S., Drelich A.K., Kratch K.C., Cho C.C., Kempaiah K.R., Hsu J.C., Mellott D.M., Xu S., Tseng C.T.K., Liu W.R. Bepridil is potent against SARS-CoV-2 in vitro. Proc. Natl. Acad. Sci. U.S.A. 2021;118:1–8. doi: 10.1073/pnas.2012201118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonrhein C., Flensburg C., Keller P., Sharff A., Smart O., Paciorek W., Womack T., Bricogne G. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Muruato A.E., Zhang X., Lokugamage K.G., Fontes-Garfias C.R., Zou J., Liu J., Ren P., Balakrishnan M., Cihlar T., Tseng C.T.K., Makino S., Menachery V.D., Bilello J.P., Shi P.Y. A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19. Nat. Commun. 2020;11:1–11. doi: 10.1038/s41467-020-19055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., Gao G.F., Anand K., Bartlam M., Hilgenfeld R., Rao Z. 2003. The Crystal Structures of Severe Acute Respiratory Syndrome Virus Main Protease and its Complex with an Inhibitor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Linlin, Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. 80-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Leike, Liu J., Cao R., Xu M., Wu Y., Shang W., Wang X., Zhang H., Jiang X., Sun Y., Hu H., Li Y., Zou G., Zhang M., Zhao L., Li W., Guo X., Zhuang X., Yang X. Lou, Shi Z.L., Deng F., Hu Z., Xiao G., Wang M., Zhong W. Comparative antiviral efficacy of viral protease inhibitors against the novel SARS-CoV-2 in vitro. Virol. Sin. 2020;35:776–784. doi: 10.1007/s12250-020-00288-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., George S., Schmidt M.F., Al-Gharabli S.I., Rademann J., Hilgenfeld R. Peptide aldehyde inhibitors challenge the substrate specificity of the SARS-coronavirus main protease. Antivir. Res. 2011;92:204–212. doi: 10.1016/j.antiviral.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.