Abstract
Global public health infrastructure is unprepared for emerging pathogen epidemics, in part because diagnostic tests are not developed in advance. The recent Zika, Ebola, and SARS-CoV-2 virus epidemics are cases in point. We demonstrate here that multicolored gold nanoparticles, when coupled to cross-reactive monoclonal antibody pairs generated from a single immunization regimen, can be used to create multiple diagnostics that specifically detect and distinguish related viruses. The multiplex approach for specific detection centers on immunochromatography with pairs of antibody-conjugated red and blue gold nanoparticles, coupled with clustering algorithms to detect and distinguish related pathogens. Cross-reactive antibodies were used to develop rapid tests for i) Dengue virus serotypes 1–4, ii) Zika virus, iii) Ebola and Marburg viruses, and iv) SARS-CoV and SARS-CoV-2 viruses. Multiplexed rapid antigen tests based on multicolored nanoparticles and cross-reactive antibodies and can be developed prospectively at low cost to improve preparedness for epidemic outbreaks.
Keywords: Nanoparticle, Nanosphere, Nanostar, Rapid antigen diagnostics, Pandemic preparedness, Infectious diseases, Antibody, Cross-reactive, Lateral flow chromatography
Graphical Abstract
Introduction
The past decade has seen the emergence of viruses that are serious human health threats; for example, SARS-CoV-2, Zika, Ebola, and now monkeypox. Deforestation, climate change, overpopulation, frequent air travel, and ineffective public health surveillance programs are determinants contributing to the spillover of pathogens into the human population [1]. Rapid diagnostics are useful both as point of care devices to detect and identify viral pathogens at the bedside, and also as surveillance devices to identify pathogens that are circulating in an environment. Rapid diagnostics have advantages of low cost, ease of use, transportability, and use in environments where power sources and reagent supplies are unpredictable. Today’s rapid diagnostics include paperfluidic lateral flow-type devices [2] as well as paper-based devices that use synthetic biology approaches [3], [4]. The selection of rapid diagnostic type depends on the application and the relative parameters of cost, simplicity, ease of use, and time to readout [5].
Among rapid point of care diagnostics, paper fluidic lateral flow devices offer low cost and ease of use without need for refrigeration, specialized chemicals, or specialized training. These devices are characteristically built from antibodies that are used in pairs, where one antibody is conjugated to a nanoparticle, while the other (“capture”) antibody is immobilized on a membrane. Sensitivity (true positive results) and specificity (true negative results) are measures of device performance. Developing an antibody-based paperfluidic diagnostic device requires at least six months of time, much of which is needed to generate and characterize antibodies, beginning with screening for binding affinity to the immunizing antigen, followed by testing in pairs to define differential antigen binding and cross reactivity. This 6–12-month process can cost many tens of thousands of dollars in salaries, animal costs, hybridoma preparation, and validation. Epidemics often appear suddenly and unexpectedly, and often without characterized antibodies that recognize the emerging pathogen. As a result, months pass before diagnostics are available, delaying care delivery to patients. This problem has the potential to repeat in the future as new emerging viruses spill over [6].
Viruses are classified according to genome type (RNA/DNA), genome organization (segmented/unsegmented), and replication strategy (nuclear/cytoplasmic) [7]. Viral proteins from members of the same virus family share amino acid sequence homologies, contributing to shared linear and conformational immunogenic epitopes that stimulate antibody responses. Embedded in and around the homologous regions are amino acid sequences that are unique to individual viral proteins; these regions can modulate protein structure and epitope presentation. Antibodies used for diagnostics are, in most cases, developed by immunizing rodents or camelids with individual viruses or their or purified proteins, although other technologies are being explored [8]. A central public health problem is that it is economically unrealistic to proactively develop individual diagnostics for dozens of pathogens. At the same time, several members of a virus family can be threats; for example, Dengue/Zika/St. Louis Encephalitis/Powassan (flaviviruses), Ebola/Marburg/ (filoviruses) and SAR-CoV/SARS-CoV-2/endemic coronaviruses. Public health would be served if it were possible to produce specific and sensitive diagnostics, covering a virus family, by using one or two cross reactive monoclonal antibody libraries. The approach would speed diagnostics development at lower cost.
In previous work, we developed rapid antigen tests to detect and distinguish the four dengue serotype viruses [2] using antibodies raised by immunizing mice individually with the NS1 protein from Dengue virus serotypes 1–4. Here, we demonstrate that the four Dengue virus serotypes can be detected and distinguished using antibodies raised to a single Dengue virus serotype NS1 protein (DV3), using red and blue gold nanoparticle conjugates to create signal colors and patterns that are deconvoluted by image analysis and data clustering. Further, we present evidence that pairing cross reactive with mono-reactive antibodies can yield effective rapid antigen tests, as shown with SARS/SARS-CoV-2 and Ebola/Marburg detection. The approaches and data presented here demonstrate the feasibility of using homologous epitope binding to prepare multiple rapid diagnostic tests per antigen immunization, limiting costs and enabling proactive, advance development of diagnostic tests for emerging human pathogens.
Results
Outline of the approach
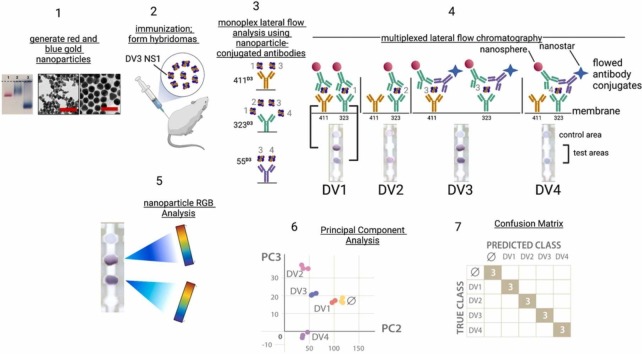
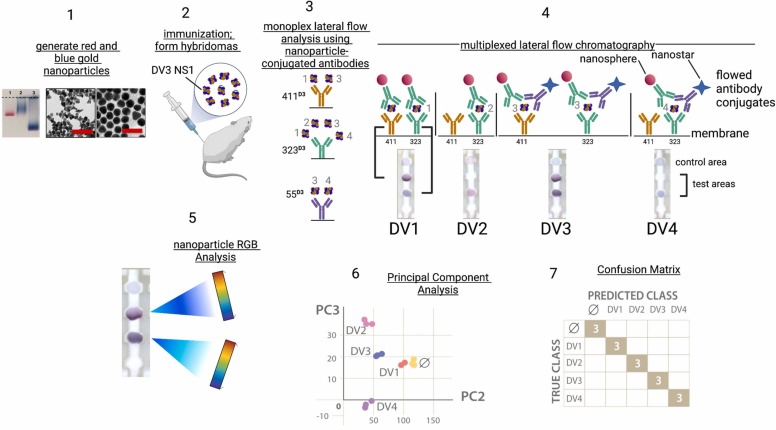
A graphic summary of the approach is presented in Fig. 1. In Step 1, blue and gold nanoparticles were generated from gold salts and characterized by electron microscopy, zeta potential, and hydrodynamic diameter analyses. In step 2, mice were immunized with the Dengue virus serotype 3 NS1 protein, followed by creating hybridomas from spleen or lymph node cells, and testing hybridoma supernatants containing monoclonal antibodies in an ELISA format. Step 3 of the workflow confirms ELISA antibody-antigen binding data using the lateral flow chromatography format. In the example of the Dengue 3 NS1 immunization, three monoclonal antibodies (323, 411, 55) bound differentially to the viral NS1 proteins, as indicated by the serotype number (1−4) bound to the immobilized antibody. In Step 4, optimal monoclonal antibody pairs are used to create multiplexed tests where flow phase antibodies are conjugated to red nanospheres or blue nanostars, and membrane-adsorbed antibodies are applied at the test areas of the strip membrane. Chromatographing the test strip with ligand generates signal patterns corresponding to the red and blue nanoparticle-antibody conjugates captured at the two test areas. By using two test areas and two colored nanoparticles, the theoretical total number of antigens that could be distinguiushed in the assays is 16 (red, blue, red/blue or no color) for each of the two test areas. In Step 5, the combinations of test area red and blue nanoparticle colors, representing antibody binding, are deconvoluted into their red/green/blue (RGB) color components using the open-source imaging software ImageJ, followed by principal component analysis and data clustering in Step 6 to distinguish the ligands. Step 7 is data analysis using a confusion matrix to assess the efficacy of the approach in detecting and distinguishing the NS1 proteins of Dengue virus serotypes 1–4, using antibodies raised to a single antigen, i.e. the NS1 proteins of Dengue virus serotype 3.
Fig. 1.
Graphic summary of rapid antigen diagnostics development using cross-reactive antibodies. Step 1: generate red nanospheres and blue nanostars from gold salts; Step 2: immunization; Step 3: Confirmation of ELISA binding using monoplex lateral flow chromatography. The naming standard (e.g. 411D3) identifies the antibody name (411) and the Dengue virus serotype antigen (D3). mAb 411 recognizes D1 and D3 NS1; mAb 323 recognizes D1-D4 serotype NS1 proteins; mAb 55 recognizes DV3 and DV4 serotype NS1 proteins. Step 4: multiplexed lateral flow chromatography, creating the test signal patterns that detect and distinguish the viral antigens. Four lateral flow chromatography strips are shown, each with mAb323D3 adsorbed at the lower test area, and mAb411D3 adsorbed at the upper test area. The control area is anti-mouse IgG. For the flowed antibody conjugates, mAb323D3 was conjugated to gold nanospheres, and mAb55D3 was conjugated to blue nanostars. Step 5: The distribution of red and blue nanoparticle colors in the test areas is determined by red/green/blue (RGB) analysis (ImageJ, NIH). Step 6: The data are clustered using principal component analysis. Step 7: A confusion matrix evaluates the performance of the tests in detecting and distinguishing the four Dengue virus serotype by comparing the predicted class with the true class. The number 3 indicates the number of tests that were run, and numbers falling on the diagonal represent a perfect correlation of predicted and true classes.
Multicolored nanoparticles and cross-reactive antibodies detect the four Dengue virus serotype NS1 proteins
ELISA data revealed that the Dengue virus serotype 3 (D3) NS1 protein immunization generated a mixture of antibodies recognizing not only DV3 NS1, but other dengue serotype NS1 proteins as well (Fig. 1, Step 2; Fig. S1) [2]. ELISA permits a relatively fast initial characterization of hybridoma antibody binding properties; however, it has been reported that, because of the distinct physical properties of the two assays, ELISA binding may not predict the results of the lateral flow format [2], [9] . Because our goal was to create lateral flow diagnostics, we performed subsequent binding analyses using the lateral flow immunochromatography format ( Fig. 2). The design of the lateral flow strips with the corresponding adsorbed and flow phase antibodies is shown in Fig. 2A. The anti-Fc test area served as a positive control for completed chromatography. Pairwise combinations of antibodies mAb411, mAb55 and mAb323, immobilized on nitrocellulose or conjugated to red gold nanospheres (NS) or blue gold nanostars (NSt), were chromatographed on “half strips” (Fig. 2A, 2B) [9] to identify pairs that detected and distinguished the dengue serotypes.
Fig. 2.
Detecting and distinguishing the four (1−4) Dengue serotypes using anti-NS1 antibodies from a Dengue serotype 3 NS1 immunization. A) Schematic of the lateral flow strips used for monoplex analysis of NS1 antibody binding pairs, using monoclonal antibodies 323, 55, and 411. The nomenclature NS-323DV3 refers to red nanospheres coupled to antibody 323, which was raised by immunizing mice with the DV3 NS1 protein antigen. Similarly, NSt-55DV3 refers to blue nanostars conjugated to antibody 55, which was raised using the DV3 NS1 protein antigen; B) Summary binding data from the monoplex antibody binding experiments; C-J) antibody-antigen binding isotherms; the dashed lines represent standard deviation. Panels C-F correspond to the 411/323 and 411/55 pairs (also Panel B, upper), while Panels G-J correspond to the 323/323 and 323/55 antibody pairs (also Panel B, lower). Panels C/G, D/H, E/I, and F/J used Dengue virus NS1 proteins from serotypes 1–4, respectively. K) schematic of the multiplexed rapid antigen test design, with monoclonal antibodies 411 and 323 adsorbed to different test areas of the nitrocellulose membrane; L) multiplexed analysis of the four dengue virus serotype NS1 proteins using serotype 3 NS1 monoclonal antibodies. The labels (D1, D2, etc.) refer to the Dengue virus serotype NS1 ligand present in the liquid phase chromatographed on the strips; M) principal component analysis and clustering of binding data; N) Confusion matrix of the linear discriminant assay. Data that fall on the diagonal are a perfect true class/predicted class fit. The number 3 in the shaded boxes refers to the number of test strips run per determination.
Chromatography was followed by image intensity analysis of the test areas (Fig. 1, Step 5) for quantification. The binding results are presented in Fig. 2B, where darker color reflects greater NS1 protein binding and corresponding signal intensities. The results (Fig. 2B) demonstrate that dengue virus 3 NS1 protein (D3) was detected in all four of the antibody combinations and orientations tested, as expected, because the antibodies were raised by immunizing mice with dengue serotype 3 NS1 antigen. The results also show that antibody pairs 411/323 and 411/55 failed to recognize D2 or D4 NS1. Differential binding was also observed with D1 NS1 where the 411/55 pair gave a weaker signal than 411/323. Because mAb 323 recognized dengue serotype NS1 proteins 1–4 (Fig. 2B), it could be used as a 323/323 “pan-dengue” and was the only pair among those tested that bound D2 NS1 protein (Fig. 2B). We speculate that the 323/323 pairs avoid binding interference because the NS1 proteins are dimeric or hexameric [10], providing multiple binding sites. The absence of binding is also a distinguishing feature; for example, monoclonal antibodies 411 and 55 did not recognize dengue serotypes 2 or 4 NS1 proteins (Fig. 2B) [2]). Together, the data presented in Fig. 2 demonstrate the differential binding properties of the 411/323/55 antibodies.
The data shown in Fig. 2B are initial single point concentration binding assays (1000 ng/ml NS1 antigen). To determine limits of detection and relative binding constants (Kd), we next performed lateral flow binding assays at a range of concentrations, as shown in panels 2 C-2 J. Langmuir isotherms were used to calculate the limits of detection and dissociation constants (Kd) of the antibody pairs from all four dengue serotypes (Table S1). Dengue virus serotype 3 NS1 protein was detected with the lowest limit of detection (3.7 ng/ml), a result that was expected because dengue virus serotype 3 NS1 protein was the immunizing antigen. In addition, dengue virus serotypes 2 and 4 detections showed the highest limits of detection (379 ng/ml and 370 ng/ml, respectively. Overall, limits of detection ranged from 4 to 400 ng/ml. Because the range of NS1 protein concentrations reported in infected patient sera can reach (15–50 μg/ml) [2], diagnostic tests using cross-reactive antibodies would exceed sensitivity requirements.
The data further demonstrate partial discrimination of the dengue virus serotype NS1 proteins. Positive signals with the 411/55 or 411/323 antibodies identify D1 or D3 serotype NS1 proteins because D2 and D4 serotype NS1 proteins are not recognized by these pairs. At the same time, D1 and D3 are not distinguished using the 411/55 and 411/323 pairs. A positive signal with the 323/323 pair without a corresponding signal in the 323/55 pair identified D2, while distinguishing it from D4. Taken together, dengue serotypes 2 and 4 can be detected and distinguished from others, while serotypes 1 and 3 can be detected, but not distinguished, by monoplex chromatography methods.
Multiplexed immunochromatography detects and distinguishes all dengue serotype NS1 proteins (1−4)
Multicolored gold nanoparticles and antibody conjugation
We used multiplexed immunochromatography with red and blue nanoparticles and anti-D3 NS1 mAb with differential binding properties. We hypothesized that specific detection of the dengue serotype NS1 proteins could be accomplished by combining multiplexed test strips with antibody-conjugated red and blue nanoparticles, representing orthogonal axes in the RBG (red-green-blue) color space [11], [12]. Lab-synthesized ∼13 ± 3 nm diameter blue gold nanostars with a length of 26 ± 5 nm (NSt), and 38 ± 4 nm nm diameter red gold nanospheres (Abcam) were characterized by dynamic light scattering, zeta potential, optical absorption and transmission electron microscopy to quantify particle shape, size, and monodispersity (Fig. 1 Step 1; Fig. S2). Antibodies mAb323 and mAb411 were immobilized on individual test areas of a nitrocellulose strip to create the multiplexed assay (Fig. 2K). For the liquid flow phase, mAb323 was conjugated to red nanospheres, and mAb55 was conjugated to blue nanostars (Fig. 2K). Ab coverage was 17.6 ± 1.2 Abs/NStar, and a coverage of 9.8 ± 0.7 Ab/nanosphere. Changes in hydrodynamic diameter (D H), zeta potential (ζ), and gel electrophoretic mobility were observed upon antibody conjugation, confirming formation of NS- and NStar-bioconjugates (Fig. S2).
Multiplexed immunochromatography
We incubated mixtures of the conjugated nanospheres/nanostars with individual dengue serotype NS1 proteins and performed immunochromatography. Test area intensities and colors varied with each dengue serotype protein (Fig. 2L). For DV1 NS1, a blue signal formed at area 323, and a red/purple signal at area 411, indicating binding of NSt-mAb55 with mAb323 and both NS-mAb323 and NSt-mAb55 with mAb411. For DV2 NS1, membrane test area 323 showed a red signal, but area 411 did not show any signal, suggesting that Dengue 2 NS1 protein fails to bind to membrane-adsorbed mAb411, consistent with data in 2D). Purple signal, a combination of red and blue nanoparticles, appeared in tests run with D3 NS1 (Fig. 2L, strip D3) indicating that DV3 NS1 bound to both mAb323 and mAb411 on the membrane, using mAb323 conjugated to red nanospheres and mAb55 conjugated to blue nanostars. With D4V NS1, a blue signal appeared at area 323 (Fig. 2L; also, Fig. 2J), but there was no signal at area 411, indicating that NSt-mAb55 bound to immobilized mAb323 but not mAb411 (Fig. 2F). Nanoparticle signal was not visible at the no-protein control (NC) test area. Thus, the resulting test pattern and colors varied as a function of the NS1 serotype. The signal patterns for D1/D3 and D2/D4 were distinguishable by a trained observer, but less obvious to untrained observers.
Image analysis, linear discriminant analysis, and confusion matrixes
For objective discrimination in assigning test results, red, green and blue (RGB) color space values of all test areas were quantified from lateral flow strips, thereby defining the signal contribution from blue and red nanoparticles. Images of the test strips were analyzed to determine red-green-blue (RGB) values (ImageJ), followed by machine learning computational methods to deconvolute the signal color space data into clusters corresponding to specific viral proteins. Principal component analysis (PCA) separated clusters of the four serotypes and the blank (Fig. 2M). We trained a linear discriminant analysis (LDA) with six predictor variables (RGB values in antibody positions 323 and 411) to distinguish the four dengue serotypes. LDA results were transferred to a confusion matrix, where diagonal values show correctly classified responses, and off-diagonals show misclassified responses (Fig. 2N). The analysis demonstrates that LDA accurately identified and distinguished, with 100% classification accuracy, NS1 proteins from each of the four dengue serotypes.
Detecting and distinguishing Dengue disease from Zika virus disease
We next explored approaches to detect and distinguish the group of Dengue virus serotype NS1 proteins from Zika virus NS1 proteins. Zika and Dengue viruses co-circulate in the Americas [13], and their NS1 proteins have about 54% amino acid sequence identity [14] (Table S2). Cross reactivity of anti-dengue NS1 antibodies with Zika NS1 protein has been reported [15] and has been a major challenge to developing accurate/specific Dengue and zika diagnostic assays. We therefore designed a strategy to design a diagnostic test and distinguish Zika virus from Dengue virus NS1 proteins, using cross-reactive antibodies and multicolored nanoparticles. Matrix pairing results revealed that mAb136, prepared by immunizing mice with the Dengue virus serotype 1 NS1 protein, is a cross-reactive Dengue/zika antibody (Table S3) [2]. Monoplex chromatography was performed to evaluate mAb136 binding to Dengue and Zika NS1 proteins, using the test strip design strategy that is schematized in Fig. 3A. The design required liquid flow phase nanoparticle conjugates that would recognize the four Dengue serotype NS1 proteins as well as the Zika virus NS1 protein; therefore, we conjugated blue nanostars to a mixture of anti-dengue serotype NS1 antibodies, creating “PAN” nanoparticles decorated with anti-NS1 antibodies recognizing all Dengue serotype NS1 proteins, as well as the Zika virus NS1 protein (Fig. 3A, NSt-PANDV). In practice, we found that conjugating multiple antibodies to blue nanostars in a single reaction yielded greater lateral flow signal intensities than adding individually conjugated particles. mAb136 was conjugated to red nanospheres (Fig. 3A, NS-136DV1). The results of the monoplex binding experiments are presented graphically in Fig. 3B, where Dengue serotype 4 NS1 protein was recognized only by the 323/PAN pairs, while Zika virus NS1 was recognized by 136/136 and 136/PAN pairs (Fig. 3B, C), but not by 323/136 or 323/PAN pairs (Fig. 3B, F). A full set of isotherms (Fig. 3, Panels C-J) was run to define limits of detection for each of the antibody pairs with the four dengue serotype NS1 proteins and Zika virus NS1 protein (Table S4). The data in Table S4 demonstrate that the lateral flow tests detect Dengue NS1 proteins in the range of 30–100 ng/ml, while the concentration of circulating Dengue NS1 protein has been reported to reach 15–50 micrograms/ml [5]. The limit of detection for Zika virus NS1 protein is approximately 220 ng/ml (Table S4), while circulating Zika virus NS1 levels has been reported to reach about 2 micrograms/ml [16]. These sensitivity data demonstrate that the rapid antigen tests described in Fig. 3 and Table 4 would translate to diagnostic applications in detecting and distinguishing Zika from Dengue in patient infections.
Fig. 3.
Multiplexed lateral flow chromatography with red and blue nanoparticles to detect and distinguish the group of Dengue virus serotypes 1–4 NS1 proteins from Zika virus NS1. A) Design of the monoplex lateral flow tests used to characterize NS1 protein binding to mAbs 136 and 323, as well as the PAN blue nanostar particles, prepared by conjugating to mAbs 243DV2, 323DV3, 626DV4, and 271DV3; B) Monoplex antibody binding matrix; C-L) Monoplex antibody binding isotherms for Dengue virus serotypes 1–4 NS1 proteins and Zika virus NS1 protein; M) Design of the multiplexed rapid diagnostic test with mAbs 136 and 323 adsorbed to the nitrocellulose membrane, and chromatographed using NS-136DV1 and Nst-PANDV nanoparticles; N) Lateral flow immunochromatography using Dengue virus serotype1 NS1 and Zika virus NS1 proteins. Dengue virus serotype 1 NS1 protein alone is chromatographed on strip 1, while Zika virus NS1 protein alone is chromatographed on strip 5. Strips 2–4 represent mixtures of the Dengue and Zika virus NS1 proteins; O) Lateral flow immunochromatography using Dengue virus serotype 4 NS1 and Zika virus NS1 proteins. Dengue virus serotype 4 NS1 protein alone is chromatographed on strip 1, while Zika virus NS1 protein alone is chromatographed on strip 5. Strips 2–4 represent mixtures of the Dengue 4 and Zika virus NS1 proteins. P) principal component analysis of binding data; Q) Confusion matrix of the linear discriminant analysis.
After completing the monoplex assays (Fig. 3A-L), the multiplexed dipstick test configuration was prepared by immobilizing the Dengue-specific antibody mAb323 and the Dengue-Zika cross-reactive antibody mAb136 on separate test areas of a nitrocellulose strip (Fig. 3M). The monoplex binding data for Dengue serotypes 1–3 NS1 proteins revealed similar isotherms; however, these similar isotherms were distinct from Dengue serotype 4 binding (Fig. 3C-E and 3H-J). Because of the similarities in the D1-D3 isotherms, the examples of only Dengue serotype 1 NS1 binding and Dengue serotype 4 binding (Fig. 3N-Q) are shown, in isolation as well as in mixtures (Fig. 3N and O). Test dipsticks (Fig. 3M) were prepared and chromatographed with a mixture of conjugated red NS-mAb136 and conjugated blue NSt-PAN nanoparticles. When dengue virus serotype 1 NS1 protein was tested in isolation, signal appeared at areas 323 and 136, a result of binding of both blue NSt-PAN and NS-mAb136 (Fig. 3N, strip 1). The monoplex binding isotherm data demonstrated that Dengue 4 NS1 was not recognized mAb136, and Dengue 4 NS1 signal was observed only with the NS323/NSt-PAN pair (Fig. 3K). When ZIKV NS1 protein was tested in isolation, signal was observed only at test area 136 due to binding of only NSt-mAb136 to immobilized mAb136 (Fig. 3N, strip 5). Mixtures of D1 and ZIKV NS1 proteins showed intermediate signal intensities using D1 and D4 NS1 proteins (Fig. 3N and O, strips 2–4). The principal component analysis of the binding data is presented in Fig. 3P, and the confusion matrix in Fig. 3Q. The confusion matrix data demonstrate that the group of Dengue NS1 serotype NS1 proteins was detected and distinguished from Zika NS1 in 25/26 samples (>96%) classification accuracy. The same analysis was performed by including Dengue 1, Dengue 2, and Dengue 3 serotype NS1 proteins, showing a 60/64 (>94%) classification accuracy (Fig. S3). These data demonstrate that cross-reactive antibodies detected and distinguished Dengue and Zika virus NS1 proteins in a multiplex format, while the monoplex test described in Fig. 2 is the best approach for distinguishing the individual Dengue serotypes. The results presented in Fig. 3 are evidence that a multiplexed test using cross-reactive antibodies detects and multicolored nanoparticles distinguishes the group of Dengue 1 NS1 proteins from Zika virus NS1.
Detecting and distinguishing filoviruses and coronaviruses
To demonstrate applications beyond closely-related flaviviruses, we extended the approach toward detecting and distinguishing coronaviruses and filoviruses. The first goal was to detect the pandemic 2019 SARS-CoV-2 glycoprotein [17], [18] and to distinguish it from 2003 SARS-CoV [19], [20], using anti-spike protein antibodies raised against 2003 SARS-CoV. 2003 SARS-CoV and 2019 SARS-CoV-2 share 75% spike protein amino acid sequence identity [21]. Both viruses enter cells when the viral spike protein binds to the ACE2 receptor [22], and antibody cross-reactivity against the viral CoV-1 and CoV-2 spike proteins has been reported [23], [24]. A group of anti-SARS-CoV-1 monoclonal and polyclonal antibodies was screened by monoplex lateral flow chromatography, and the optimal pairs, and their limits of detection, are presented in Fig. 4A and Fig. S4. We immobilized anti-SARS-CoV spike protein antibody S18 or anti-SARS-CoV spike protein antibody S4 on nitrocellulose membrane test strips (Fig. 4A). Nanospheres were conjugated to anti-SARS spike protein antibody S6, while nanostars were conjugated to anti-SARS spike protein antibody S16. The data first show that an immobilized S18 antibody, paired with a mixture of S6 and S16 antibodies, detected both SARS and SARS-2 spike proteins (Fig. 4A, row S18, strips 2 and 3). The immobilized S4 antibody, when paired with S6 and S16 antibodies detected SARS-CoV protein (row S4, strip 3) but failed to detect SARS-CoV-2 spike protein (Fig. 4A, row S4, strip 2). The lateral flow strips were analyzed by RGB separation followed by principal component analysis (Fig. 4C). The linear discriminant analysis had 100% correlation when detecting and distinguishing the two coronavirus spike proteins (Fig. 4C and D). The results are evidence that the SARS-CoV and SARS CoV-2 spike glycoproteins were detected and distinguished using only anti-SARS-CoV-1 spike protein antibodies.
Fig. 4.
Detecting COVID19 and Ebola using SARS and Marburg antibodies, respectively. The abbreviations SARS-1 or CoV refer to 2003 SARS-CoV, while CoV-2 refers to 2019 SARS-CoV-2. A) schematic of the lateral flow strip designs showing antibodies S4 and S18 adsorbed to the test membrane, and antibodies S6 and S16 conjugated to red nanospheres and blue nanostars, respectively. B) Multiplexed detection of SARS-CoV (CoV) and SARS-CoV-2 (CoV-2) using SARS-CoV antibodies; C) principal component analysis of binding data for the two coronaviruses; D) Confusion matrix of the linear discriminant analysis (LDA) for the two coronaviruses; E) schematic of the lateral flow strip designs showing anti-Marburg glycoprotein antibodies 1G11 and 2G12 adsorbed to the test membrane, and antibodies 1G11 and 2G12 conjugated to red nanospheres and blue nanostars, respectively. F) Multiplexed detection of Ebola (E) and Marburg (M), as well as mixtures of the two viruses using Marburg antibodies. Marburg glycoprotein binding alone is shown on strip 1, while Ebola glycoprotein binding alone is shown on strip 5. Strips 2–4 represent mixtures of the Marburg and Ebola glycoproteins; (G) principal component analysis of binding data for the two filoviruses; H) Confusion matrix of the linear discriminant analysis for the two filoviruses glycoprotein binding data.
To evaluate the potential for using cross-reactive antibodies to detect viral proteins with lower amino acid homology scores, we turned to detecting the Ebola and Marburg virus glycoproteins (GP). Ebola and Marburg are co-circulating filoviruses whose envelope glycoproteins are limited to a 31% amino acid sequence identity [25]. Our hypothesis was that monoclonal antibodies raised against Marburg GP would detect and distinguish Marburg and Ebola glycoproteins. Pseudotyped viruses expressing the surface Marburg or Ebola glycoproteins were used in the binding assays. Baby hamster kidney (BHK) cells were infected with replication-competent vesicular stomatitis viruses wherein the Marburg or Ebola glycoproteins replaced the VSV surface glycoprotein. Supernatants from the infected cells contain infectious pseudoviruses that have the Marburg or Ebola envelope with the VSV rhabdovirus core [26]. Hybridomas producing anti-Marburg envelope glycoprotein antibodies were produced, and resulting monoclonal antibodies were tested to define cross-reactive and differential binding properties. mAb1G11 was adsorbed to the nitrocellulose membrane and also conjugated to red nanospheres, while mAb 2G12 was adsorbed to a second test area and conjugated to blue nanostars (Fig. 4E). The results of chromatographing the individual Marburg and Ebola pseudoviruses are shown on strips 1 and 5. The Marburg test signals are intense and purple, suggesting that both red and blue nanoparticles are captured. The Ebola pseudovirus test signals (strip 5), however, are less intense and also have greater red hue than purple, suggesting that binding is primarily the red nanospheres. Gradients of mixtures of the two pseudoviruses are presented in strips 2–4, showing the changes in test area color and intensity as the test protein levels changed. RGB and principal component analyses were performed (Fig. 4G), and the results were plotted as a confusion matrix (Fig. 4H). The data demonstrate that the Marburg glycoprotein, Ebola glycoprotein, and mixtures of the two proteins were distinguished with a 100% classification accuracy in the confusion matrix (Fig. 4H). These data demonstrate that viral proteins with only 31% amino acid identity can be detected and distinguished using cross-reactive antibodies. Taken together, the data presented in Fig. 2, Fig. 3, Fig. 4 are evidence that rapid diagnostics assembled using antibodies raised by immunization with a single viral antigen successfully detected and distinguished viral proteins from related pathogens (Dengue/Zika, Marburg/Ebola and SARS-CoV/SARS-CoV-2). These results have important implications for proactive and prospective preparation of diagnostic devices to detect emerging pathogens with pandemic potential.
Discussion
The broad goal of this work is to improve public health preparedness for emerging virus outbreaks. Global responses to pathogen outbreaks are best described today as reactive, meaning that action is taken to address an emerging pathogen outbreak only after it has spread. We propose alternate approaches that permit advance, proactive development of reagents and devices, at low cost, for emerging pathogens. The foundation of work described here is multicolored nanoparticles, as well as paperfluidic or lateral flow chromatography that requires only papers or membranes that are striped with antibodies. Lateral flow technologies [27] have been developed and validated over several decades, and used throughout the world for disease diagnosis and epidemiological surveillance [28]. The advantages of paperfluidic assays include simplicity, low cost, scaling, and use as point of care devices without power or instrumentation.
The study described in this paper is conceptually distinct from prior lateral flow detection approaches; however, Flamand, Wiktor, and Kaprowski speculated forty years ago that panels of monoclonal antibodies could be used to distinguish among the Lyssavirus group [29]. Cross-reactive polyclonal antibodies, used in a chemical olfaction approach, have been reported for detection in a virus family [12]. Here, we have used monoclonal antibodies to detect not just a single pathogen (represented by the immunizing antigen), but also other pathogens whose homologous proteins share epitopes with the immunizing antigen. The data presented here demonstrate that antibodies directed against Dengue virus serotype 3, Ebola glycoprotein, and SARS CoV spike protein can be used to detect and distinguish Zika, Marburg, and SARS-CoV-2, respectively. By using this shared epitope approach, improved and timely patient care can be delivered while also reducing both the costs and time required to deliver screening or diagnostic tests in an emergency. Cost and time reductions stem from obviating the need to generate individual hybridomas for every pathogen.
Although antibody pairs and lateral flow diagnostics are well-established technologies, the physical and biological parameters underlying detection using complex human tissues or extracts are incompletely understood. Device development is in part an empirical process because detection is dependent upon many variables, including membrane porosity, pH, temperature, nanoparticle size/properties, flow rate, blocking non-specific binding, antibody specificity, and antibody affinities [28], [30]. At a minimum, rapid antigen detection requires pairs of monoclonal or polyclonal antibodies, where an optimal pair is composed of two antibodies that bind to antigen without interference. ELISA is routinely used for initial characterization of antigen binding because it is quantitative and can be used in a 96-well or 384-well format to achieve throughput. An issue, however, is that antibody binding and specificity measured in ELISA are not always predictive of behavior in lateral flow. Therefore, device developers recommend testing antibodies in the paired lateral flow format early in antibody characterization [9]. Initial pairs are selected using ELISA binding affinity data, complemented with the lateral flow matrix approach [2]. In this paper we present evidence that antibody cross reactivity is a positive attribute for detecting and distinguishing related pathogens.
At least three properties of antibody-antigen interactions are central to using antibodies to detect shared epitopes in homologous proteins: induced fit, conformational selection, and epitope interference. Induced fit differs from conformational selection in terms of whether a conformational change occurs before binding (conformational selection) or after binding (induced fit) [31]. Homologous viral proteins share common epitopes that are embedded in regions of non-homology; therefore, allosteric effects of nanoparticle-conjugated antibody binding to NS1 protein could change the structure of distant shared binding epitopes, affecting second antibody binding [32] (conformational selection). An implication of this proposed mechanism is that conjugated antibody binding to individual homologous viral proteins could cause subtle antigen conformational changes that are recognized differentially by the non-cognate antibodies, thereby distinguishing them [33]. In a possible example of induced fit, antigen binding to non-cognate antibodies could be accompanied by co-binding conformational changes (induced fit) that are distinct for individual related viral antigens because the shared epitopes are embedded in regions of sequence non-homology [34]. Again, the combination of both homologous and non-homologous regions in different antigens could influence structures that result from induced fit, and thereby affect second antibody binding. Third, either induced fit or conformational selection binding could increase or decrease antibody binding interference by bringing epitopes closer or separating them. Conformational selection and induced fit could also explain orientation dependency for the antibody pairs; that is, antigen binding by the conjugated antibody may alter antigen conformation to affect second antibody binding, and those changes may not duplicate if the nanoparticle-membrane orientation is reversed. Structural analysis of cognate and non-cognate antigen-antibody binding could be used to explore the importance of induced fit, conformational selection, and antibody binding interference.
Can detection by shared epitopes be applied widely, and is the approach generalizable to detecting many/most pathogens? The data presented here, showing detection of viruses in three different virus families, strongly suggest that the method is generalizable and can be applied to a range of pathogens. Antibodies used in the tests can be sourced from existing stocks, commercial products, or generated anew. In the example of generating new antibodies, mice would be immunized with a goal of creating multiple diagnostic tests from a single monoclonal antibody pool. Screening a large library of monoclonal antibodies to identify pairs is preferred over testing only a few commercial antibodies because of the greater diversity, increasing the probability of identifying effective pairs. Following the test runs, data capture can be accomplished by photography using mobile phones, including simple “flip” phones. Internet connectivity is desirable for downstream analysis, but not required at the time of image capture. RGB analysis, signal intensity quantification, and linear discriminant analysis can be performed on a mobile phone or, alternatively, uploaded for cloud analysis. The code used here runs on MatLab, a common engineering software package, and could be adapted for use on a mobile phone.
What are the main obstacles toward adopting this approach? The primary obstacle lies in reorienting public health attitudes from reactive tests to proactive and advance preparation of diagnostic and surveillance tools. Predicting which pathogen will be the next to spill over into the human population is challenging [1], but can be data-driven [35]. Events coinciding with spillover zoonoses have been defined and analyzed [36]. Ecologists, field biologists, government agencies and academic researchers are generating or collecting data that can be analyzed computationally to predict statistical trends that point to a possible epidemic [37], [38], [39]. Evidence of a successful prediction can be found in a warning about a possible outbreak of coronaviruses that appeared in 2007, twelve years before the SARS-CoV-2 pandemic [39]. These authors stated that: “The presence of a large reservoir of SARS-CoV-like viruses in horseshoe bats, together with the culture of eating exotic mammals in southern China, is a timebomb. The possibility of the reemergence of SARS and other novel viruses from animals or laboratories and therefore the need for preparedness, should not be ignored”. If that advice had been heeded, and diagnostic and surveillance tests developed in advance, the impact of the current pandemic might have been diminished.
Are there current examples of emerging viruses that may become threats to global human health; would public health benefit from advance diagnostics or surveillance screening? The list of potential virus threats is long, and it is expected that viruses will continue to spill over and threaten the human population [1], [40]. To begin, additional respiratory coronaviruses, as described by Cheng and Yuen [39] are positioned to spill over into the human population. With longer summers due to climate change, tick-borne viruses such as Powassan virus, which circulates in North America and causes a serious encephalitis, are emerging [41]. Brazil is undergoing climate change as well as Amazon deforestation, bringing humans into closer contact with arboviruses and intermediate hosts. Several flaviviruses [40], including Ilheus, Rocio, and Iguape, are being isolated from an expanding number of mosquito vectors, and could represent imminent threats for outbreaks or epidemics that could also threaten the United States [42]. There are additional examples of potential Arenavirus and Bunyavirus threats as well [43]. Few if any of these emerging threats have accompanying vaccines or surveillance/diagnostic tests to detect and distinguish pathogens.
Conclusions
Emerging and reemerging pathogens are serious health threats, and it is near certain that outbreaks of new diseases will appear in the future. There are thousands of known pathogens with potential to cause outbreaks, and success in addressing future threats requires accurate point of care diagnostics to detect and distinguish the infections. The recent outbreaks of Ebola, Zika, and SARS-CoV-2 are evidence that new approaches are needed for public health preparedness. The data presented here suggest that rapid diagnostics based on cross-reactive antibodies and multicolored nanoparticles to detect related pathogens can be prepared rapidly and at reduced costs, providing key early epidemiologic data as well as point-of-care use. A shared epitope approach, as described here, can circumvent the costly and time-consuming animal immunizations and monoclonal antibody characterizations, allowing identification of antibody pairs to detect and distinguish emerging pathogens during outbreaks.
Materials and methods
Reagents
Gold chloride trihydrate was purchased from Sigma-Aldrich (CAS: 16961-25-4). Bis(sulphatophenyl)phenyl-phosphine dihydrate (BPS), was purchased from Aldrich (CAS:308103–66–4). N-(2-Hydroxyethyl) piperazine-N’-(2-ethanesulphonic acid) (HEPES) was purchased from United States Biochemical Company (CAT: 16926) and 5 kD mPEG was from Nanocs. Fluorescent Goat anti-Mouse IgG (H+L) Secondary Antibody, DyLight 650 conjugate was purchased from Pierce.
Viruses
Replication-competent vesicular stomatitis viruses bearing the glycoprotein of Ebola virus (VSV-rGP-EBOV) or the glycoprotein of Marburg virus (VSV-rGP-MARV) were obtained from Dr. Sean Whelan. Virus was amplified in BHK (baby hamster kidney cells) and harvested from supernatant, followed by titering.
Antigens and antibodies
Dengue and Zika NS1 proteins were obtained from Native Antigen Company (UK). SARS-CoV-1 antibodies as well as SARS-CoV-1 and SARS-CoV-2 Spike proteins were obtained from Sino Biologicals. DENV anti-NS1 and Marburg anti-GP monoclonal antibodies were produced in mice (Covance, Inc; Denver, PA; and Precision Antibody, respectively), following an approved animal care protocol. BALB-c mice were immunized with purified recombinant DENV NS1 or Marburg virus GP expressed in mammalian cells (Native Antigen, UK or IBT Biosciences, respectively). From each of the immunized groups, one seroconverted animal was used for cell fusion to generate hybridomas.
Nanoparticles
Blue gold nanostars were synthesized by mixing 400 μl of 140 mM HEPES (pH 7.4) with 600 μl of 18 MΩ deionized (MilliQ) water, followed by the addition of 16 μl of 25 mM gold chloride trihydrate and further vortexing. After vortexing, the solution sat undisturbed for 1 h, during which the NSt formed. Next, ∼ 0.5 mg BPS was added for NSt stabilization, and the solution was briefly vortexed and left undisturbed for 1 h. For antibody conjugation, the NSt were separated from excess reagents by centrifugation at 12000 rcf for 12 min. The resulting NSt pellet was resuspended in 400 μl of 10 mM HEPES at pH 7.7. Next, 10 μl of 1 mg/ml antibody was added and the mixture was vortexed, and further agitated overnight, during which time the antibodies were able to bind to the nanostars. After the overnight incubation, in order to avoid nonspecific binding on the nanostars, 10 μl of 0.1 mM 5kD mPEG was added, and the solution was briefly vortexed and further agitated in a rocker for 15 min, during which time mPEG passivated any bare gold surfaces. Finally, nanoparticles were centrifuged for 12 min at 10000 rcf to separate excess reagents. The NP pellets were resuspended in MilliQ water at OD 20 (520 nm wavelength) for use in the lateral flow tests.
Bioconjugation of nanospheres
Innova-coat nanoparticles (20 OD, Mini Kit, 40 nm diameter) were conjugated as indicated by the manufacturer. In short, antibody was diluted to 0.1 mg/ml in the supplied dilution buffer and mixed with 42 μl of reaction buffer. 45 μl of this mixture was used to suspend the lyophilized nanoparticles, the mixture was incubated for 15 min at room temperature, followed by the addition of 5 μl of the proprietary quencher solution.
Nanoparticle characterization
Optical absorption spectra of the NP were obtained on a Cary 100 UV Vis from Agilent Technologies. NP morphology was characterized with a FEI Tecnai G2 transmission electron microscope at 120 kV. ImageJ was used to process the images and measure the NP dimensions. In addition, a Zetasizer Nano ZS (Malvern Instruments) was used to measure the hydrodynamic diameter (DH) and the zeta potential (ζ) of the NP. Agarose gel electrophoresis was used to confirm the antibody and mPEG binding on the nanoparticles. In short, 1% agarose gels were prepared and NPs were loaded by mixing 8 μl of concentrated NPs with 4 μl of 50% glycerol in MilliQ water. Fluorescence spectroscopy was used to quantify antibody loading per nanoparticle, using a supernatant-loss method. Antibody coverage was measured by attaching fluorescent antibodies to the nanoparticles.
Running the immunochromatography assays
Lateral flow half-strips (9) were used for rapid prototyping (2). Antibodies were immobilized on nitrocellulose membranes (EMD Millipore HF18002XSS) by manually pipetting 0.3 μl of a 2 mg/ml solution of antibodies onto the nitrocellulose membrane followed by drying for at least 30 min. Strips were attached to a wick (GB003 Gel Blot Paper) with adhesive paper (MIBA-010 Backing Card, 0.020" thickness; DCN Diagnostics, Carlsbad CA). For the positive control area, 0.3 μl of anti-mouse Fc antibody (EMD Millipore AQ127) at 1 mg/ml was spotted on the control line. Dipsticks were dropped into a solution containing: 8 μl of 1% Tween-20 in PBS and 4 μl of 50% sucrose in water, 5 μl of a mixture of NSt and Innova NS and 30 μl of the analyte, typically diluted in filtered human serum (Sigma). Supernatants from cells infected with the vesicular stomatitis virus (VSV) pseudotypes were used for Ebola and Marburg test development. The tests were run by letting the solution migrate through the strip via capillary action.
Image quantification and analysis
After drying, the finished tests were photographed or scanned, and test area intensities were quantified with ImageJ, using the measure mean grayscale value function. Both RGB and grayscale data were recorded. Limits of detection were measured using serially diluted recombinant NS1 (Dengue and Zika), or serially diluted infected cell supernatants as proteins antigens (Ebola, Marburg), or serially diluted recombinant Spike protein (SARS and SARS-CoV-2). The signal intensities were quantified (ImageJ), normalized by the intensity at the highest concentration, following: . Where grayo is the measured gray value of the blank, graymax is the gray value of the highest concentration point (at saturation), and grayn is the gray value at each concentration. After normalization, gray values were plotted and fitted in a Langmuir equation of the form: . Where [antigen] is the concentration of antigen present in the 30 μl of sample in the solution, and KD eff represents the effective binding constant in a Langmuir-like system. The limit of detection was calculated from the curve fit as the concentration found at the intersection with a line representing the value of the blank plus 5-times the standard deviation of blank intensity.
Image analysis of the nitrocellulose strips
RGB information was extracted during manual ImageJ analysis by splitting the image data of the strips into red, green and blue components. Matlab was used to train a Linear Discriminant Analysis (LDA) with 5-fold cross-validation that used the six predictor variables (red, blue and green intensities of two test bands in each assay) to distinguish the clusters where the data aggregated. Confusion matrices were plotted by comparing the expected true results from LDA (True class), versus the LDA results (Predicted class). The resulting confusion matrix shows the performance of the Values in the diagonal represented correctly-classified tests, while values out of diagonal represented incorrectly-classified results. The classifier accuracy was measured by using bootstrap methods and was always over 80%, typically around 95%. Principal component analysis was performed in Matlab and used to plot the results from six to two dimensions to visualize the separate clusters.
Statistical analysis
Prism 8 was used to calculate and graph the data. Unpaired, two-tailed t test or one-way ANOVA was performed to test for statistical significance, as indicated in the figure legends.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Lee Gehrke reports financial support was provided by National Institutes of Health (R33AI100190 and AI151807). Helena de Puig reports financial support was provided by Broshy Foundation. Helena de Puig reports financial support was provided by Tata Trusts. Lee Gehrke reports equipment, drugs, or supplies was provided by US Food and Drug Administration. Lee Gehrke reports a relationship with IDx20 that includes: consulting or advisory and equity or stocks. Irene Bosch reports a relationship with IDx20 that includes: board membership, employment, and equity or stocks. Nol Salcedo reports a relationship with IDx20 that includes: employment. Helena de Puig reports a relationship with IDx20 that includes: consulting or advisory. James Collins reports a relationship with Sherlock Biosciences that includes: board membership, consulting or advisory, and equity or stocks. Lee Gehrke has patent #9488613 issued to Massachusetts Institute of Technology. Irene Bosch, Kimberly Hamad-Schifferli, Helena de Puig has patent #9488613 issued to Massachusetts Institute of Technology. Lee Gehrke has patent #10551381 with royalties paid to Massachusetts Institute of Technology. Irene Bosch, Helena de Puig, Kimberly Hamad-Schifferli has patent #10551381 with royalties paid to Massachusetts Institute of Technology.
Acknowledgments
Financial support was provided by National Institutes of Health (R33AI100190 and AI151807). We thank Brian Baker and Patrick Regan of the Winchester Engineering and Analytic Center, FDA, for encouraging the development of this project and for access to equipment and materials. We thank Dr. Sean Whelan (Washington University) for providing the vesicular stomatitis virus-pseudotyped Marburg and Ebola viruses. The authors would like to thank CMSE at MIT for the use of facilities. Viruses were obtained from BEI Resources, NIAID, NIH. We thank Megan Hiley, Bobby Brooke Herrera and members of MilliporeSigma: Martin Zillmann, Kimberly Mann, Melissa Holstein, Meghan Higson, Christopher Gillespie, and Patricia Kumpey for assistance with the production and purification of detection antibodies used in this study. We thank Cristina Rodriguez Quijada and Marc Carré for experimental assistance. BioRender was used to prepare Fig. 1, Fig. 2, Fig. 3, Fig. 4.
Biographies
 Helena de Puig, Ph.D. completed her graduate studies in Lee Gehrke’s laboratory and received the Ph.D. from the Department of Mechanical Engineering at the Massachusetts Institute of Technology. She is currently a postdoctoral fellow in James Collins’ laboratory at the Wyss Institute for Biologically Inspired Engineering at Harvard University. Helena uses and edits Nature’s toolbox of proteins/DNA and materials to give new capabilities to materials, cells, and proteins, toward lowering healthcare costs.
Helena de Puig, Ph.D. completed her graduate studies in Lee Gehrke’s laboratory and received the Ph.D. from the Department of Mechanical Engineering at the Massachusetts Institute of Technology. She is currently a postdoctoral fellow in James Collins’ laboratory at the Wyss Institute for Biologically Inspired Engineering at Harvard University. Helena uses and edits Nature’s toolbox of proteins/DNA and materials to give new capabilities to materials, cells, and proteins, toward lowering healthcare costs.
 Irene Bosch is a Ph.D. graduate of the Harvard School of Public Health and a former Professor of Medicine at UMass Medical School, in Worcester, MA. She has more than twenty years of experience in the areas of infectious diseases, including SARS, flaviviruses and retroviruses. Dr. Bosch is a founder and CEO of IDx20, which aims to develop technologies and solutions to predict and diagnose infectious disease outbreaks.
Irene Bosch is a Ph.D. graduate of the Harvard School of Public Health and a former Professor of Medicine at UMass Medical School, in Worcester, MA. She has more than twenty years of experience in the areas of infectious diseases, including SARS, flaviviruses and retroviruses. Dr. Bosch is a founder and CEO of IDx20, which aims to develop technologies and solutions to predict and diagnose infectious disease outbreaks.
 Nol Salcedo, M.S. is a graduate of the Universidad de Los Andes, Mérida, Venezuela. Ms. Salcedo has performed research for thirteen years in the field of rapid diagnostics for infectious diseases. Her experience also includes studies of sexually transmitted diseases, respiratory diseases, SARS, and flaviviruses. Ms. Salcedo was among the lead contributors to an MIT-industry project involving the development rapid antigen tests for dengue, zika, chikungunya, and powassan viruses. Her current position is Laboratory director for diagnostics development at IDx20.
Nol Salcedo, M.S. is a graduate of the Universidad de Los Andes, Mérida, Venezuela. Ms. Salcedo has performed research for thirteen years in the field of rapid diagnostics for infectious diseases. Her experience also includes studies of sexually transmitted diseases, respiratory diseases, SARS, and flaviviruses. Ms. Salcedo was among the lead contributors to an MIT-industry project involving the development rapid antigen tests for dengue, zika, chikungunya, and powassan viruses. Her current position is Laboratory director for diagnostics development at IDx20.
 Kimberly Hamad-Schifferliis a professor in the Department of Engineering and the School for the Environment at the University of Massachusetts Boston. Her research is focused on using nanomaterials for biological applications, including low-cost diagnostics for infectious diseases and other pathogens, food safety, and water quality.
Kimberly Hamad-Schifferliis a professor in the Department of Engineering and the School for the Environment at the University of Massachusetts Boston. Her research is focused on using nanomaterials for biological applications, including low-cost diagnostics for infectious diseases and other pathogens, food safety, and water quality.
 James Collins, Ph.D. is the Termeer Professor of Bioengineering in the Department of Biological Engineering and Institute for Medical Engineering & Science at the Massachusetts Institute of Technology. His research group works in synthetic biology and systems biology, with a particular focus on using network biology approaches to study antibiotic action, bacterial defense mechanisms, and the emergence of resistance.
James Collins, Ph.D. is the Termeer Professor of Bioengineering in the Department of Biological Engineering and Institute for Medical Engineering & Science at the Massachusetts Institute of Technology. His research group works in synthetic biology and systems biology, with a particular focus on using network biology approaches to study antibiotic action, bacterial defense mechanisms, and the emergence of resistance.
 Lee Gehrke, Ph.D. is the Hermann L.F. von Helmholtz Professor of Health Sciences and Technology at MIT and Professor of Microbiology at Harvard Medical School. His laboratory has studied the structure and function of viral RNA-protein complexes, host-protein interactions and pathogenesis of positive-sense single stranded RNA viruses, as well as the development of rapid antigen diagnostics for RNA viruses.
Lee Gehrke, Ph.D. is the Hermann L.F. von Helmholtz Professor of Health Sciences and Technology at MIT and Professor of Microbiology at Harvard Medical School. His laboratory has studied the structure and function of viral RNA-protein complexes, host-protein interactions and pathogenesis of positive-sense single stranded RNA viruses, as well as the development of rapid antigen diagnostics for RNA viruses.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.nantod.2022.101669.
Appendix A. Supplementary material
Supplementary material
.
Data Availability
Data will be made available on request.
References
- 1.Howard C.R., Fletcher N.F. Emerging virus diseases: can we ever expect the unexpected. Emerg. Microbes Infect. 2012;1 doi: 10.1038/emi.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosch I., de Puig H., Hiley M., Carré-Camps M., Perdomo-Celis F., Narváez C.F., Salgado D.M., Senthoor D., O’Grady M., Phillips E., Durbin A., Fandos D., Miyazaki H., Yen C.-W., Gélvez-Ramírez M., Warke R.V., Ribeiro L.S., Teixeira M.M., Almeida R.P., Muñóz-Medina J.E., Ludert J.E., Nogueira M.L., Colombo T.E., Terzian A.C.B., Bozza P.T., Calheiros A.S., Vieira Y.R., Barbosa-Lima G., Vizzoni A., Cerbino-Neto J., Bozza F.A., Souza T.M.L., Trugilho M.R.O., de Filippis A.M.B., de Sequeira P.C., Marques E.T.A., Magalhaes T., Díaz F.J., Restrepo B.N., Marín K., Mattar S., Olson D., Asturias E.J., Lucera M., Singla M., Medigeshi G.R., de Bosch N., Tam J., Gómez-Márquez J., Clavet C., Villar L., Hamad-Schifferli K., Gehrke L. Rapid antigen tests for dengue virus serotypes and Zika virus in patient serum. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aan1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pardee K., Green A.A., Ferrante T., Cameron D.E., DaleyKeyser A., Yin P., Collins J.J. Paper-based synthetic gene networks. Cell. 2014;159:940–954. doi: 10.1016/j.cell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pardee K., Green A.A., Takahashi M.K., Braff D., Lambert G., Lee J.W., Ferrante T., Ma D., Donghia N., Fan M., Daringer N.M., Bosch I., Dudley D.M., O’Connor D.H., Gehrke L., Collins J.J. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell. 2016;165:1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- 5.Muller D.A., Depelsenaire A.C.I., Young P.R. Clinical and laboratory diagnosis of Dengue virus infection. J. Infect. Dis. 2017;215:S89–S95. doi: 10.1093/infdis/jiw649. [DOI] [PubMed] [Google Scholar]
- 6.Ebel G.D. Update on Powassan virus: emergence of a North American tick-borne flavivirus. Annu. Rev. Entomol. 2010;55:95–110. doi: 10.1146/annurev-ento-112408-085446. [DOI] [PubMed] [Google Scholar]
- 7.Baltimore D. Expression of animal virus genomes. Bacteriol. Rev. 1971;35:235–241. doi: 10.1128/br.35.3.235-241.1971. 〈https://www.ncbi.nlm.nih.gov/pubmed/4329869〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McMahon C., Baier A.S., Pascolutti R., Wegrecki M., Zheng S., Ong J.X., Erlandson S.C., Hilger D., Rasmussen S.G.F., Ring A.M., Manglik A., Kruse A.C. Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nat. Struct. Mol. Biol. 2018;25:289–296. doi: 10.1038/s41594-018-0028-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D. Wild, The Immunoassay Handbook, in: Theory and Applications of Ligand Binding, ELISA and Related Techniques, Fourth, Elsevier, The Boulevard, Langford Lane, Kidlington, Oxford, OX5 1GB, UK, 2013.
- 10.Muller D.A., Young P.R. The flavivirus NS1 protein: molecular and structural biology, immunology, role in pathogenesis and application as a diagnostic biomarker. Antivir. Res. 2013;98:192–208. doi: 10.1016/j.antiviral.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 11.de Puig H., Tam J.O., Yen C.-W., Gehrke L., Hamad-Schifferli K. The extinction coefficient of gold nanostars. J. Phys. Chem. C. Nanomater. Interfaces. 2015;119:17408–17415. doi: 10.1021/acs.jpcc.5b03624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodriguez-Quijada C., Gomez-Marquez J., Hamad-Schifferli K. Repurposing old antibodies for new diseases by exploiting cross-reactivity and multicolored nanoparticles. ACS Nano. 2020;14:6626–6635. doi: 10.1021/acsnano.9b09049. [DOI] [PubMed] [Google Scholar]
- 13.Villamil-Gómez W.E., Rodríguez-Morales A.J., Uribe-García A.M., González-Arismendy E., Castellanos J.E., Calvo E.P., Álvarez-Mon M., Musso D. Zika, dengue, and chikungunya co-infection in a pregnant woman from Colombia. Int. J. Infect. Dis. 2016;51:135–138. doi: 10.1016/j.ijid.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 14.Shan C., Xie X., Barrett A.D.T., Garcia-Blanco M.A., Tesh R.B., da P.F., Vasconcelos C., Vasilakis N., Weaver S.C., Shi P.-Y. Zika Virus: Diagnosis, Therapeutics, and Vaccine. ACS Infect. Dis. 2016;2:170–172. doi: 10.1021/acsinfecdis.6b00030. [DOI] [PubMed] [Google Scholar]
- 15.Gyurech D., Schilling J., Schmidt-Chanasit J., Cassinotti P., Kaeppeli F., Dobec M. False positive dengue NS1 antigen test in a traveller with an acute Zika virus infection imported into Switzerland. Swiss Med. Wkly. 2016;146:w14296. doi: 10.4414/smw.2016.14296. [DOI] [PubMed] [Google Scholar]
- 16.Beddingfield B.J., Hartnett J.N., Wilson R.B., Kulakosky P.C., Andersen K.G., Robles-Sikisaka R., Grubaugh N.D., Aybar A., Nunez M.-Z., Fermin C.D., Garry R.F. Zika Virus Non-Structural Protein 1 Antigen-Capture Immunoassay. Viruses. 2021;13 doi: 10.3390/v13091771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M.A., Weldon W.C., Biggs H.M., Uyeki T.M., Pillai S.K. Washington State 2019-nCoV case investigation team, first case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munster V.J., Koopmans M., van Doremalen N., van Riel D., de Wit E. A Novel Coronavirus Emerging in China — key questions for impact assessment. N. Engl. J. Med. 2020;382:692–694. doi: 10.1056/nejmp2000929. [DOI] [PubMed] [Google Scholar]
- 19.Marra M.A., Jones S.J.M., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S.N., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 20.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.-E., Humphrey C.D., Shieh W.-J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.-Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. SARS Working Group, A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 21.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 23.Yuan M., Wu N.C., Zhu X., Lee C.-C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu Y., Yu D., Han Y., Yan H., Chong H., Ren L., Wang J., Li T., He Y. Cross-reactive neutralization of SARS-CoV-2 by serum antibodies from recovered SARS patients and immunized animals. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abc9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Babirye P., Musubika C., Kirimunda S., Downing R., Lutwama J.J., Mbidde E.K., Weyer J., Paweska J.T., Joloba M.L., Wayengera M. Identity and validity of conserved B cell epitopes of filovirus glycoprotein: towards rapid diagnostic testing for Ebola and possibly Marburg virus disease. BMC Infect. Dis. 2018;18 doi: 10.1186/s12879-018-3409-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whelan S.P., Ball L.A., Barr J.N., Wertz G.T. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc. Natl. Acad. Sci. USA. 1995;92:8388–8392. doi: 10.1073/pnas.92.18.8388. 〈http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=7667300〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plotz C.M., Singer J.M. The latex fixation test. I. Application to the serologic diagnosis of rheumatoid arthritis. Am. J. Med. 1956;21:888–892. 〈https://www.ncbi.nlm.nih.gov/pubmed/13372565〉 [PubMed] [Google Scholar]
- 28.Wong R., Tse H. & Business Media. Springer Science; 2008. Lateral Flow Immunoassay.〈https://play.google.com/store/books/details?id=D4-capuCoRUC〉 [Google Scholar]
- 29.Flamand A., Wiktor T.J., Koprowski H. Use of hybridoma monoclonal antibodies in the detection of antigenic differences between rabies and rabies-related virus proteins. I. The nucleocapsid protein. J. Gen. Virol. 1980;48:97–104. doi: 10.1099/0022-1317-48-1-97. [DOI] [PubMed] [Google Scholar]
- 30.de Puig H., Bosch I., Gehrke L., Hamad-Schifferli K. Challenges of the nano-bio interface in lateral flow and dipstick immunoassays. Trends Biotechnol. 2017;35:1169–1180. doi: 10.1016/j.tibtech.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paul F., Weikl T.R. How to distinguish conformational selection and induced fit based on chemical relaxation rates. PLoS Comput. Biol. 2016;12 doi: 10.1371/journal.pcbi.1005067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cepica A., Yason C., Ralling G. The use of ELISA for detection of the antibody-induced conformational change in a viral protein and its intermolecular spread. J. Virol. Methods. 1990;28:1–13. doi: 10.1016/0166-0934(90)90082-q. 〈https://www.ncbi.nlm.nih.gov/pubmed/2112149〉 [DOI] [PubMed] [Google Scholar]
- 33.Dekker E.L., Dore I., Porta C., van Regenmortel M.H. Conformational specificity of monoclonal antibodies used in the diagnosis of tomato mosaic virus. Arch. Virol. 1987;94:191–203. doi: 10.1007/BF01310713. [DOI] [PubMed] [Google Scholar]
- 34.Rini J.M., Schulze-Gahmen U., Wilson I.A. Structural evidence for induced fit as a mechanism for antibody-antigen recognition. Science. 1992;255:959–965. doi: 10.1126/science.1546293. [DOI] [PubMed] [Google Scholar]
- 35.Johnson C.K., Hitchens P.L., Pandit P.S., Rushmore J., Evans T.S., Young C.C.W., Doyle M.M. Global shifts in mammalian population trends reveal key predictors of virus spillover risk. Proc. Biol. Sci. 2020;287:20192736. doi: 10.1098/rspb.2019.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plowright R.K., Parrish C.R., McCallum H., Hudson P.J., Ko A.I., Graham A.L., Lloyd-Smith J.O. Pathways to zoonotic spillover. Nat. Rev. Microbiol. 2017;15:502–510. doi: 10.1038/nrmicro.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venkatramanan S., Lewis B., Chen J., Higdon D., Vullikanti A., Marathe M. Using data-driven agent-based models for forecasting emerging infectious diseases. Epidemics. 2018;22:43–49. doi: 10.1016/j.epidem.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muellner U., Fournié G., Muellner P., Ahlstrom C., Pfeiffer D.U. epidemix-An interactive multi-model application for teaching and visualizing infectious disease transmission. Epidemics. 2018;23:49–54. doi: 10.1016/j.epidem.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 39.Cheng V.C.C., Lau S.K.P., Woo P.C.Y., Yuen K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin. Microbiol. Rev. 2007;20:660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pierson T.C., Diamond M.S. The continued threat of emerging flaviviruses. Nat. Microbiol. 2020 doi: 10.1038/s41564-020-0714-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fatmi S.S., Zehra R., Carpenter D.O. Powassan Virus-A new reemerging tick-borne disease. Front Public Health. 2017;5:342. doi: 10.3389/fpubh.2017.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cunha M.S., Luchs A., da Costa A.C., de G., Ribeiro O., Dos Santos F.C.P., Nogueira J.S., Komninakis S.V., Marinho R.D.S.S., Witkin S.S., Villanova F., Deng X., Sabino E.C., Delwart E., Leal É., Nogueira M.L., Maiorka P.C. Detection and characterization of Ilheus and Iguape virus genomes in historical mosquito samples from Southern Brazil. Acta Trop. 2020;205 doi: 10.1016/j.actatropica.2020.105401. [DOI] [PubMed] [Google Scholar]
- 43.Elliott R.M., Brennan B. Emerging phleboviruses. Curr. Opin. Virol. 2014;5:50–57. doi: 10.1016/j.coviro.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data will be made available on request.