Abstract
Purpose
Aldo-keto reductase family 1 member C3 (AKR1C3) is important in prostate cancer progression, being a potential biomarker in metastatic castration-resistant prostate cancer (mCRPC). Previous explorations of AKR1C3 are mainly based on tissue samples. This study investigates using plasma-based liquid biopsy to validate the prognostic and predictive value of AKR1C3 in patients with mCRPC .
Materials and Methods
We prospectively recruited 62 patients with mCRPC. All patients received repeated prostate biopsies at the time of mCRPC diagnosis, and immunohistochemistry (IHC) staining was used to detect protein expression of AKR1C3 in the tissues. We took their blood simultaneously and performed digital droplet polymerase chain reaction (ddPCR) to measure expression levels of AKR1C3 in the exosomes. The detected plasma and tissue AKR1C3 expression levels were analyzed for patients’ overall survival (OS) and progression-free survival under first-line abiraterone use (ABI-PFS).
Results
All other baseline characteristics were balanced between the 2 groups. 15/62 (24.2%) and 25/62 (40.3%) patients showed AKR1C3-EXO positive (≥20 copies/20 μL) and AKR1C3-IHC positive, respectively. Positive AKR1C3-EXO expression was associated with decreased patients’ survival [ABI-PFS: 3.9 vs 10.1 months, P < .001; OS: 16.2 vs 32.5 months, P < .001]. AKR1C3-IHC positivity was also correlated with ABI-PFS and OS (P = .010, P = .016). In patients with worse baseline blood tests (including higher alkaline phosphatase (ALP) and lactate dehydrogenase (LDH) level and lower hemoglobin (HB) level), and lower ISUP/WHO group (<4), their OS was significantly worse when showing AKR1C3-EXO positive.
Conclusion
AKR1C3-EXO is associated with patient prognosis regarding OS and ABI-PFS and can be used as a biomarker in mCRPC.
Keywords: prostate cancer, metastatic castration-resistant prostate cancer, AKR1C3, exosome, small extracellular vesicles, prognosis, abiraterone
AKR1C3 is a potential biomarker of metastatic castration-resistant prostate cancer (mCRPC) and an important indicator in prostate cancer progression. However, the traditional detecting method of AKR1C3 relies on repeated prostate biopsies, which is not always achievable in clinical work. This study investigated the use of plasma-based liquid biopsy to validate the prognostic and predictive value of AKR1C3 in patients with mCRPC. This article finds that plasma exosomal AKR1C3 expression is associated with patient prognosis and can be used as a biomarker in patients with mCRPC.
Implications for Practice.
Plasma AKR1C3-EXO is associated with patient prognosis regarding overall survival and progression-free survival under first-line abiraterone use and can be used as a biomarker in metastatic castration-resistant prostate cancer.
Introduction
Androgen deprivation therapy (ADT) indisputably presents a rather high effective rate in treating prostate cancer. However, the wide application of AR antagonists has contributed to the progression to castration-resistant prostate cancer (CRPC) as well.1 CRPC is a lethal malignant disease prevalent in older men. As the end-stage of prostate cancer, many patients with CRPC have developed resistance to both classical and next-generation androgen-targeted drugs.
Aldo-keto reductase family 1 member C3 (AKR1C3), a critical enzyme in the de novo biosynthesis of steroids, is closely related to prostate cancer aggression.2 AKR1C3 belongs to the structurally similar AKR1C family, and its most known function is to participate in the oxidoreduction reactions to regulate androgen synthesis.3 At the same time, it is also a prostaglandin F synthase participating in cell proliferation hormone-independently.4 Moreover, AKR1C3 is also an androgen receptor (AR) enzymatic coactivator, directly and through binding Siah2 (an E3 ubiquitin ligase, which also increases AR transcriptional activity) to suppress itself degradative ubiquitination.5 It even participates in the epigenetic regulation and ferroptosis in prostate cancer.6,7 The expression of AKR1C3 is higher in metastatic and CRPC tissues than normal prostate tissues or benign prostatic hyperplasia, or localized prostate cancer.8-14 AKR1C3 upregulation could be a potential adaptive mechanism for ADT,11,12 leading to the pan-AR antagonist, chemotherapy, and radiotherapy resistance.15-18 Thus, AKR1C3 is likely a good candidate as a CRPC biomarker. However, a major bottleneck for novel biomarkers in CRPC, including AKR1C3, is the dependency on repeated biopsies. Many realistic factors contribute to the hurdle in retrieving repeated biopsy tissue, including the tumor positive rate of detection, difficulty in bone marrow biopsy, patients’ cooperation, etc.
Small extracellular vesicles (sEVs) are cell-derived compartments that participate in many biological processes. Cancer cell manufactured sEVs are critical mediators of intercellular communication, thus playing a significant role in tumor progression.19 SEVs are a broad concept and can be classified regarding their size, markers, and contents. The term exosomes in this manuscript refer to CD63+&CD9+&CD81+&TSG101+ small extracellular vesicles (sEVs) for conciseness. Exosomes can be detected in the early stage of disease, prior to PSA elevation, metastatic symptoms, or certain tissue marker positivity, in the case of mCRPC.19 Also, they are remarkably stable, making them a magnificent surrogate biomarker.
This study investigates the prognostic and predictive value of AKR1C3 level in paired plasma exosomes and tissue samples from a group of prospectively recruited patients with mCRPC. Our goal is to validate the biomarker potential of AKR1C3 in mCRPC. We hope to provide a novel test method suggestion for AKR1C3, which can help clinicians choosing the most suitable therapy at the most appropriate time.
Methods
Patients
We prospectively recruited newly diagnosed patients with mCRPC with the intention for repeat prostate biopsies from 2014 to 2021. All candidates were required to receive repeat prostate biopsies with AKR1C3 immunohistochemistry (IHC) staining (AKR1C3-IHC hereafter; Sigma; A6229; 1:800 dilution). Samples with positive AKR1C3 immunostaining in tumor cells were considered positive. Exclusion criteria were accompanying other severe systemic diseases and the inability to adhere to regular follow-ups. We collect clinical and pathological data from the electronic medical record system. Three milliliters of blood were sampled from each enrolled patient at the time of inclusion. All patients were followed up once every 4 weeks until death or at the end of this study. The study received consent from each enrolled patient and was performed under the Declaration of Helsinki. The Medical Ethics Committee of West China Hospital, Sichuan University approved our study protocol.
Cell Culture and ddPCR System Set Up
Common prostate cancer cell lines (22Rv1, C4-2, LNCaP, PC-3, and DU-145) were cultured in 1640 with 50 U/mL penicillin-streptomycin and 10% fetal bovine serum (FBS; 10099141C, Gibco, ThermoFisher Scientific) at 37 °C. When cells reached 80% confluence, we changed to FBS-free medium 3 days before exosome extraction. Real-time quantitative reverse transcription PCR (Applied Biosystems A25743) results showed that 22Rv1 carried a much higher AKR1C3 expression than other common prostate cancer cell lines (Supplementary Fig. S1; primers: AKR1C3-forward: 5'-GCCTGTATTGGGATTTGGCACCTAT-3', AKR1C3-reverse: 5'-GCGGAACCCAGCTTCTATTGCTAA-3'; GAPDH-forward: 5'-GGAGCGAGATCCCTCCAAAAT-3', GAPDH-reverse: 5'-GGCTGTTGTCATACTTCTCATGG-3'). Thus, exosomes collected from the 22Rv1 cell supernatant set up the digital droplet polymerase chain reaction (ddPCR) system as described in detail before20; they were also used as the positive control in the following exosome validation procedures in grouping patients’ plasma exosomal AKR1C3 expression (AKR1C3-EXO hereafter).
Exosome Extraction
Cell culture supernatant (or fresh whole blood) was collected, centrifuged at 1600 × g at 4 ºC for 10 minutes, then 4000 × g at 4 ºC for 10 minutes (4000 × g, 4 ºC, 10 minutes for blood sample). After centrifugation, the cell supernatant (10 times diluted plasma with 10 mM PBS solution) was collected and filtered by a 0.22-μm filter. Then the cell supernatant was transferred into polycarbonate centrifuge tubes, and centrifuged at 120,000 × g for 2 h at 4 ºC (Beckman Coulter, Optima XE-90 Ultracentrifuge). Then we discard the supernatant and add 200 μL 10 mM PBS solution to resuspend the precipitate (exosomes).
Exosome Characterization
Immunoblotting was done with standard protocols using anti-CD9 antibody (Huabio, ET1601-9), anti-CD63 antibody (Immunoway, YT5525), anti-CD81 antibody (Cell Signal Technology, 56039), anti-TSG101 (Proteintech, 14497-1-AP) as primary antibodies. Exosomes were observed using transmission electron microscopy (JEOL, JEM-1400). We took 20 μL of exosome solution, dropped it on the ordinary carbon support membrane, incubated it at room temperature for 20 min, and then washed off the excess solution with filter paper. Then we added 20 μL 2% phosphotungstic acid solution and incubated it for 10 s at room temperature. Then we washed off the excess solution with filter paper. Finally, the transmission electron microscope parameter settings were acceleration voltage as 100 kV, exposure time as 0.5 s (JEM 1400, Japan Electronics). We took 10 μL exosome solution and diluted it to 1 mL with 10 mM PBS solution. After filtering the solution using a 0.22-μm filter, we used Nanosight NS300 (Nanosight Ltd., Amesbury, UK) for Nanoparticle tracking analysis (NTA) analyses. Each sample was tested for 30 s and was repeated 3 times.
Exosomal AKR1C3 mRNA Expression
QIAzol Lysis Reagent and miRNeasy Micro Kit (217804) were used to extract RNA from the exosome suspension. We used ddPCR to measure expression levels of AKR1C3 in the exosomal RNAs with One-Step RT-ddPCR kit, as we and others validated before.20,21 We reported the results as copies of the AKR1C3 mRNA per 20 µl. We repeated the procedures for every patient sample were for at least twice.
Statistics
This study used overall survival (OS, CRPC diagnosis to all-cause death) as the primary endpoint. Progression-free survival under first-line abiraterone (ABI-PFS) was used as the second study endpoint, defined as the time from abiraterone treatment to progression according to PCWG3 consensus. X-tile software was used to determine the best cutoff value.22 Chi-square test and rank-sum test were used for comparing baseline characteristic differences between groups. We used the Kaplan-Meier curve and log-rank test for survival curves. The Cox regression model was applied for the univariate and multivariate survival analyses. All tests were 2-sided. SPSS (version 25.0), X-tile (version 3.6.1), and R (version 4.0.5) were used in our analyses.
Results
Exosome Characterization
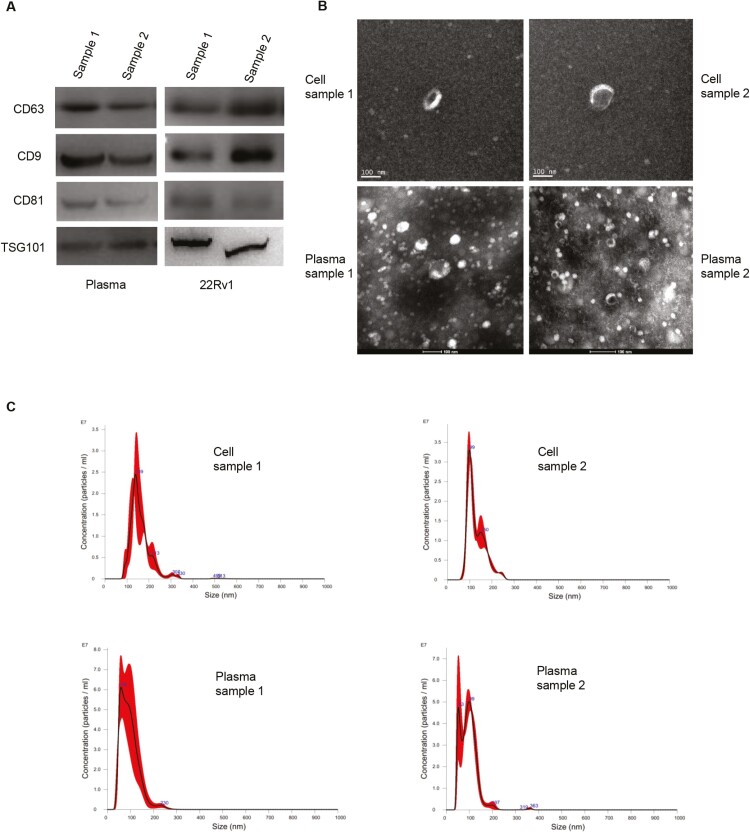
We characterized the exosomes by multiple complementary techniques, including NTA, western blot, and electron microscope, according to the recommendations of MISEV2018.23 Representative samples of each group showed positive expression of commonly used protein markers for exosome characterization: tetraspanins (CD63, CD81), TSPANs (CD9), and cytosolic proteins (TSG101) (Fig. 1A). Also, the scanning electron microscope (SEM) images of the exosomes were shown in Fig. 1B. Nanoparticle tracking analysis showed that most extracted particles were approximately 100 nm, consistent with the most commonly reported exosome size (Fig. 1C).
Figure 1.
Characterization of exosomes. Four selected samples were random representation of our patient’s plasma and cell culture supernatant. (A): Immunoblots for commonly used exosome-expressed protein markers: tetraspanins (CD63, CD81), TSPANs (CD9), and cytosolic proteins (TSG101). (B): Scanning electron microscopy images of extracted exosomes. (C): Nanoparticle tracking analysis for particle diameter.
Patient Baseline Characteristics
This study cohort contains 62 patients with mCRPC. The mean AKR1C3 exosome was 22.9 copies/20 μL. We used X-tile software to determine the cutoff value as 20 copies/20 μL since it was visualized as the best cut-point in creating subsets based on biomarker expression. X-tile is a bio-informatics tool using a graphical method to depict the presence of many tumor subpopulations and also display the association between a biomarker and outcome through a 2-dimensional projection of every possible subpopulation.22 Thus, AKR1C3-EXO was considered positive (≥20 copies/20 μL) or negative (<20 copies/20 μL), corresponding to 24.2% (15/62) and 75.8% (47/62) in this cohort, respectively. All other baseline characteristics, including age, baseline PSA, PSA at CRPC diagnosis, lactate dehydrogenase (LDH), alkaline phosphatase (ALP), hemoglobin (HB), CFS, and ISUP/WHO group were balanced between the 2 groups (Table 1). Regarding AKR1C3-IHC in biopsy samples, 40.3% (25/62) patients were positive. AKR1C3’s protein expression by IHC and the plasma exosomal mRNA expression showed a 67.7% concordance rate. Forty-four patients received abiraterone acetate as first-line therapy after CRPC diagnosis. The remaining 18 patients used docetaxel-based chemotherapy or other treatments. This cohort’s median OS and ABI-PFS were 29.9 and 8.6 months, respectively.
Table 1.
Baseline characteristics of the total cohort and the AKR1C3-EXO negative and positive groups.
| Total cohort (n = 62) | AKR1C3-EXO negative (n = 47) | AKR1C3-EXO positive (n = 15) | P-value | |
|---|---|---|---|---|
| AKR1C3-IHC (%) | ||||
| Negative, n (%) | 37 (59.68) | 32 (68.09) | 5 (33.33) | .037 |
| Positive, n (%) | 25 (40.32) | 15 (31.91) | 10 (66.67) | |
| Age, years, median [IQR] | 69.000 [62.250, 72.000] | 69.000 [61.000, 72.000] | 69.000 [64.500, 73.500] | .537 |
| ≤70, n (%) | 33 (53.23) | 27 (52.94) | 6 (54.55) | 1.000 |
| >70, n (%) | 29 (46.77) | 24 (47.06) | 5 (45.45) | |
| Baseline PSA, ng/mL, median [IQR] | 100.100 [78.735, 159.250] | 100.100 [67.270, 186.150] | 100.100 [100.100, 100.100] | .905 |
| ≤100, n (%) | 19 (30.65) | 16 (31.37) | 3 (27.27) | 1.000 |
| >100, n (%) | 43 (69.35) | 35 (68.63) | 8 (72.73) | |
| PSA at CPRC, ng/mL, median [IQR] | 10.645 [3.425, 35.837] | 9.210 [3.275, 28.150] | 30.030 [9.450, 100.100] | .053 |
| ≤100, n (%) | 30 (48.39) | 26 (50.98) | 4 (36.36) | .584 |
| >100, n (%) | 32 (51.61) | 25 (49.02) | 7 (63.64) | |
| Baseline LDH, IU/L, median [IQR] | 210.000 [174.000, 247.500] | 210.000 [174.500, 253.000] | 205.000 [161.000, 232.000] | .439 |
| ≤300, n (%) | 53 (85.48) | 40 (85.11) | 13 (86.67) | 1.000 |
| >300, n (%) | 9 (14.52) | 7 (14.89) | 2 (13.33) | |
| Baseline ALP, IU/L, median [IQR] | 105.000 [77.500, 167.250] | 102.000 [73.500, 173.000] | 124.000 [110.000, 134.000] | .637 |
| ≤160, n (%) | 48 (77.42) | 34 (72.34) | 14 (93.33) | .181 |
| >160, n (%) | 14 (22.58) | 13 (27.66) | 1 (6.67) | |
| Baseline HB, IU/L, median [IQR] | 135.000 [121.500, 144.250] | 132.000 [117.500, 146.000] | 138.000 [135.000, 141.000] | .646 |
| ≤120, n (%) | 23 (37.10) | 16 (34.04) | 7 (46.67) | .566 |
| >120, n (%) | 39 (62.90) | 31 (65.96) | 8 (53.33) | |
| CFS months, median, [IQR] | 11.533 [7.042, 23.342] | 13.233 [7.017, 22.950] | 10.033 [7.050, 21.600] | .639 |
| ≤12, n (%) | 33 (53.23) | 23 (48.94) | 10 (66.67) | .368 |
| >12, n (%) | 29 (46.77) | 24 (51.06) | 5 (33.33) | |
| ISUP/WHO group | ||||
| ≤4, n (%) | 7 (11.29) | 5 (10.64) | 2 (13.33) | 1.000 |
| >4, n (%) | 55 (88.71) | 42 (89.36) | 13 (86.67) |
Abbreviations: AKR1C3, aldo-keto reductase family 1 member C3; PSA, prostate-specific antigen; CRPC, castration-resistant prostate cancer; LDH, lactate dehydrogenase; ALP, alkaline phosphatase; HB, hemoglobin; ECOG, eastern cooperative oncology Group; CFS, castration-resistance-free survival; ABI: abiraterone.
Survival Analyses
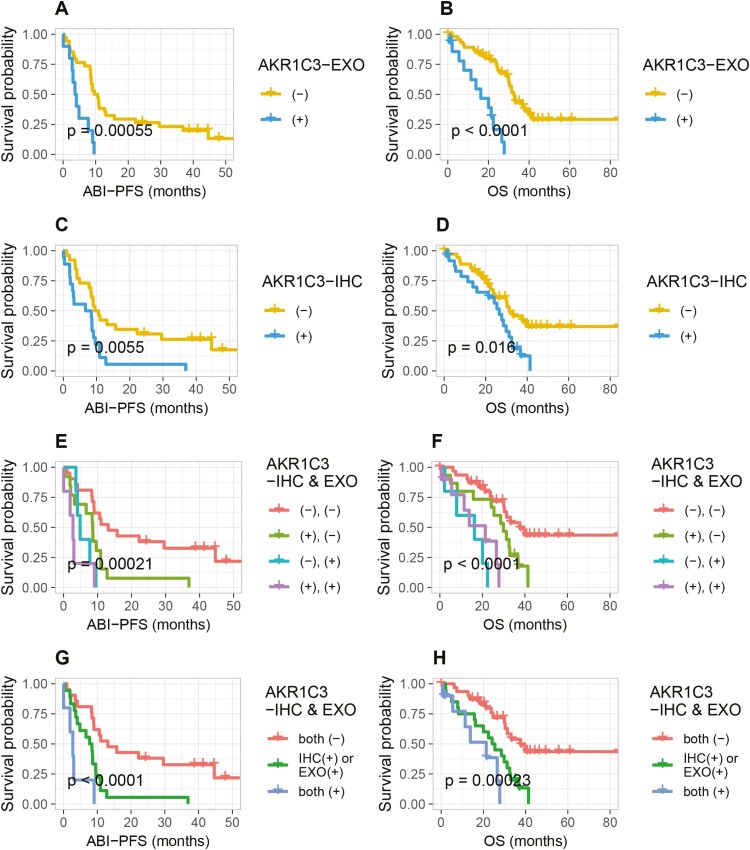
The presence of positive AKR1C3-EXO expression sharply decreased patients’ survival, both in univariate and multivariate analyses [AKR1C3-EXO (+) vs AKR1C3-EXO (–): ABI-PFS: 3.9 vs 10.1 months, HR = 3.81 (1.69-8.58), P = .001; OS: 16.2 vs 32.5 months, HR = 5.41 (2.44-12.01), P < .001, Fig. 2A, B, Table 2]. Similarly, AKR1C3-IHC positivity was also correlated with ABI-PFS and OS, with a lower HR value [HR = 2.5 (1.28-4.86), P = .010; HR = 2.16 (1.14-4.1), P = .016, Fig. 2C, D]. Then we further focused on patients with differential AKR1C3 expression regarding exosome and IHC. Patients harboring both AKR1C3-EXO and AKR1C3-IHC positivity and those without any positivity displayed the shortest and longest survival, respectively. The survival of others (harboring AKR1C3-EXO or AKR1C3-IHC positivity) were in between [both (+) vs IHC (+) or EXO (+) vs both (–): ABI-PFS: 2.8 vs 8.1 vs 13.1 months; OS: 21.4 vs 24.8 vs 37.6 months]. Notably, within group with only one positivity, patients with only EXO (+) seemed to have worse survival than patients with only IHC (+) [only EXO (+) vs only IHC (+): ABI-PFS: 4.9 vs 8.6 months; OS: 16.2 vs 29.9 months] (Fig. 2E-H).
Figure 2.
Kaplan-Meier curves. (A, B): AKR1C3-EXO expression in predicting ABI-PFS and OS. (C, D): AKR1C3-IHC expression in predicting ABI-PFS and OS. (E-H): combined expression of AKR1C3-EXO and AKR1C3-IHC in predicting ABI-PFS and OS. Abbreviations: AKR1C3-EXO, plasma exosomal AKR1C3 expression (positive: ≥20 copies/20 μL; negative: <20 copies/20 μL); AKR1C3-IHC, AKR1C3 IHC staining in repeat prostate biopsies; ABI-PFS, progression-free survival under first-line abiraterone (the time from abiraterone treatment to progression); OS, overall survival (CRPC diagnosis to all-cause death).
Table 2.
Univariate and multivariate survival analyses of AKR1C3-EXO and other clinicopathological factors in predicting OS and ABI-PFS.
| OS | ABI-PFS | |||||||
|---|---|---|---|---|---|---|---|---|
| Univariate analyses | Multivariate analyses | Univariate analyses | Multivariate analyses | |||||
| HR (lower limit, upper limit) | P | HR (lower limit, upper limit) | P | HR (lower limit, upper limit) | P | HR (lower limit, upper limit) | P | |
| AKR1C3-EXO(+) | 5.41 (2.44-12.01) | <.001 | 4.51 (1.97-10.36) | <.001 | 3.81 (1.69-8.58) | .001 | 3.67 (1.62, 8.29) | .002 |
| AKR1C3-IHC(+) | 2.16 (1.14-4.1) | .019 | 1.62 (0.82, 3.20) | .169 | 2.5 (1.28-4.86) | .007 | 2.27 (1.03, 4.99) | .042 |
| Baseline PSA >100 ng/mL | 1.14 (0.57-2.25) | .712 | — | — | 1.29 (0.62-2.66) | .498 | — | — |
| PSA at CRPC diagnosis >10 ng/mL | 1.15 (0.61-2.18) | .664 | — | — | 0.75 (0.39-1.45) | .389 | — | — |
| LDH >300 IU/L | 0.55 (0.19-1.56) | .263 | — | — | 0.45 (0.14-1.49) | .191 | — | — |
| ALP >160 IU/L | 0.67 (0.32-1.43) | .303 | — | — | 0.63 (0.3-1.35) | .235 | — | — |
| HB >120 g/L | 0.9 (0.45-1.78) | .764 | — | — | 0.79 (0.4-1.58) | .508 | — | — |
| CFS >12 months | 0.61 (0.31-1.19) | .147 | — | — | 0.55 (0.28-1.08) | .083 | 0.863 (0.383, 1.944) | .722 |
| ISUP/WHO group >4 | 1.25 (0.38-4.07) | .714 | — | — | 1.38 (0.42-4.53) | .594 | — | — |
| Age >70 years | 0.75 (0.39-1.43) | .383 | — | — | 1.02 (0.53-1.95) | .952 | — | — |
Abbreviations: AKR1C3-EXO, plasma exosomal AKR1C3 expression; PSA, prostate-specific antigen; CRPC, castration-resistant prostate cancer; LDH, lactate dehydrogenase; ALP, alkaline phosphatase; HB, hemoglobin; CFS, castration-resistance free survival; HR, hazard ratio.
Subset Analyses
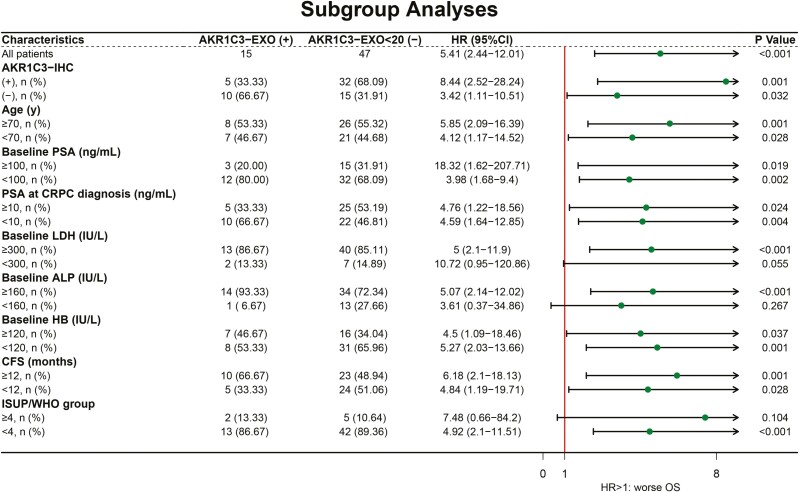
To explore further whether certain subset groups of people could benefit more from the prognostic value of the AKR1C3-EXO test, we did a subgroup survival analysis with cutoff values based on the medians for continuous variables (Fig. 3). The results suggest that in patients with worse baseline blood tests (including higher ALP and LDH level and lower HB level), and lower ISUP/WHO group (<4), their OS was significantly worse when showing AKR1C3-EXO positive. The prognostic value of AKR1C3-EXO was independent of AKR1C3-IHC, age, baseline PSA, PSA at CRPC diagnosis, CFS.
Figure 3.
The prognostic value of AKR1C3-EXO in different clinicopathological subgroups. Abbreviations: AKR1C3-EXO, plasma exosomal AKR1C3 expression (positive: ≥20 copies/20 μL; negative: <20 copies/20 μL); PSA, prostate-specific antigen; CRPC, castration-resistant prostate cancer; LDH, lactate dehydrogenase; ALP, alkaline phosphatase; HB, hemoglobin; CFS, castration-resistance-free survival; HR, hazard ratio.
Discussion
This study investigated the clinical value of AKR1C3 regarding its 2 detection methods: traditional IHC staining on tissue samples and liquid biopsy method for exosomal AKR1C3 mRNA expression in the blood, in a prospective mCRPC cohort. Our results suggest that positive AKR1C3-EXO expression is a robust prognostic predictor and an indicator for abiraterone use in patients with mCRPC. Furthermore, the AKR1C3-EXO test is likely to be more informative in patients with worse baseline blood test results, and lower ISUP/WHO group (<4), despite their AKR1C3-IHC status, age, baseline PSA, PSA at CRPC diagnosis, CFS.
Previous studies displayed the association of AKR1C3 expression and PSA-PFS after radical prostatectomy.13 Our prior publications indicated that AKR1C3 protein expression in tissue biopsies related to early resistance and glucocorticoid choice for ABI treatment in patients with mCRPC.24,25 These observations acknowledge the importance of evaluating AKR1C3 in different stages of prostate cancer. Nevertheless, repeated biopsies are not always feasible and cost-effective in progressed patients, not to mention that patients who are often not in good condition at this stage have to suffer from the biopsies with possible complications. Also, tissue biopsy can only represent the part being sampled out, limiting our understanding of tumor aggressiveness’s comprehensive status.
On top of that, prostate cancer is a well-known example of polyclonal origin, which imposes further restrictions on tissue biopsy use, especially in a stage when tumors show intense heterogeneity. Cancer-derived exosomes have been studied for a decade due to their remarkable accessibility and stability. This current study echos with the importance of AKR1C3 regarding not only therapy-guiding value, but also direct prognostic significance in patients with mCRPC. Most importantly, it offers us an alternative way: using plasma instead of biopsy tissues can also provide valuable information. In fact, its prognostic and predictive value is not inferior, if not superior, to AKR1C3-IHC positivity in tissue samples since AKR1C3-EXO shows a higher HR value.
Our subgroup analyses also show some interesting results. Being a solid prognosticator, AKR1C3-EXO is associated with worse survival regardless of AKR1C3-IHC, age, baseline PSA, PSA at CRPC diagnosis, CFS. This reminds the clinicians that performing an AKR1C3-EXO is beneficial no matter they have repeated biopsy AKR1C3-IHC results or not. Also, patients’ PSA status at diagnosis or CRPC does not affect the necessity of this test either. However, we do notice that patients with worse baseline blood test (including higher ALP and LDH level and lower HB level), and lower ISUP/WHO group (<4) are strongly recommended to perform the AKR1C3-EXO test, since these subgroups demonstrate significantly shorter OS.
PSA and its derivatives are indeed the cornerstones of prostate cancer biomarkers. At the same time, its value in managing the advanced disease is not as satisfying as that in earlier disease stages, especially with the emergence of highly aggressive CRPC variants with low PSA (eg, neuroendocrine prostate cancer).1 Consequently, discovering novel biomarkers to stratify and provide therapy advice for patients with advanced disease has been a field of ongoing research. Despite rapid advances in technology, there is still a lack of efficient biomarkers to provide prognostic indications or identify patients who are unlikely to respond well to androgen-targeted drugs, especially for patients in the CRPC stage. Current validated prognostic or predictive molecules still heavily rely on tissue samples or high-throughput next-generation sequencing. These are certainly rather exciting explorations, while there is a long way to go before really translate them into clinical utility.
Here we present an easy, non-invasive, cost-effective detection method on a molecule that is acknowledged to play an essential role in prostate cancer. A simple blood-based test of exosomal AKR1C3 can provide significant clues for patient management and prognosis. Moreover, the development of AKR1C3 inhibitors is thriving,26-28 and the AKR1C3-EXO test can be a bona fide marker for those too. This study is limited by a relatively small cohort; future studies with larger cohorts are needed to validate our findings.
Conclusion
Plasma AKR1C3-EXO is associated with patient prognosis regarding OS and ABI-PFS and can be used as a biomarker in mCRPC.
Supplementary Material
Contributor Information
Sha Zhu, Department of Urology, Institute of Urology, West China Hospital, Sichuan University, Chengdu, Sichuan, People’s Republic of China.
Yuchao Ni, Department of Urology, Institute of Urology, West China Hospital, Sichuan University, Chengdu, Sichuan, People’s Republic of China.
Zilin Wang, Department of Urology, Institute of Urology, West China Hospital, Sichuan University, Chengdu, Sichuan, People’s Republic of China.
Xingming Zhang, Department of Urology, Institute of Urology, West China Hospital, Sichuan University, Chengdu, Sichuan, People’s Republic of China.
Yaowen Zhang, Department of Urology, Institute of Urology, West China Hospital, Sichuan University, Chengdu, Sichuan, People’s Republic of China.
Fengnian Zhao, Department of Urology, Institute of Urology, West China Hospital, Sichuan University, Chengdu, Sichuan, People’s Republic of China.
Jindong Dai, Department of Urology, Institute of Urology, West China Hospital, Sichuan University, Chengdu, Sichuan, People’s Republic of China.
Zhipeng Wang, Department of Urology, Institute of Urology, West China Hospital, Sichuan University, Chengdu, Sichuan, People’s Republic of China.
Xudong Zhu, Department of Urology, Institute of Urology, West China Hospital, Sichuan University, Chengdu, Sichuan, People’s Republic of China.
Junru Chen, Department of Urology, Institute of Urology, West China Hospital, Sichuan University, Chengdu, Sichuan, People’s Republic of China.
Jinge Zhao, Department of Urology, Institute of Urology, West China Hospital, Sichuan University, Chengdu, Sichuan, People’s Republic of China.
Yuhao Zeng, Department of Urology, Institute of Urology, West China Hospital, Sichuan University, Chengdu, Sichuan, People’s Republic of China.
Ni Chen, Department of Pathology, Institute of Urology, West People’s Republic of China Hospital, Sichuan University, Chengdu, Sichuan, People’s Republic of China.
Peng Zeng, 3D Medicines Inc., Shanghai, People’s Republic of China.
Pengfei Shen, Department of Urology, Institute of Urology, West China Hospital, Sichuan University, Chengdu, Sichuan, People’s Republic of China.
Guangxi Sun, Department of Urology, Institute of Urology, West China Hospital, Sichuan University, Chengdu, Sichuan, People’s Republic of China.
Hao Zeng, Department of Urology, Institute of Urology, West China Hospital, Sichuan University, Chengdu, Sichuan, People’s Republic of China.
Funding
The Natural Science Foundation of China (NSFC 81902577), The Research Foundation for the Postdoctoral Program of Sichuan University (2021SCU12014).
Conflict of Interest
The authors declared no conflicts of interest.
Author Contributions
Conception/design: S.Z., G.S., H.Z. Provision of study material or patients: Yaowen Zhang, F.Z., J.D., Z.W., Xingming Zhang, J.C., J.Z., Yuhao Zeng, Xudong Zhu, N.C., P.S. Collection and/or assembly of data: Y.N., Z.W. Data analysis and interpretation: P.Z. Manuscript writing: S.Z., H.Z. Final approval of manuscript: All authors.
Data Availability
The data underlying this article will be shared on reasonable request to the corresponding author.
References
- 1. Boumahdi S, de Sauvage F.J.. The great escape: tumour cell plasticity in resistance to targeted therapy. Nat Rev Drug Discov. 2020;19(1):39-56. [DOI] [PubMed] [Google Scholar]
- 2. Liu Y, et al. Overview of AKR1C3: inhibitor achievements and disease insights. J Med Chem. 2020;63(20):11305-11329. 10.1021/acs.jmedchem.9b02138. [DOI] [PubMed] [Google Scholar]
- 3. Powell K, et al. ERG/AKR1C3/AR constitutes a feed-forward loop for AR signaling in prostate cancer cells. Clin Cancer Res. 2015;21(11):2569-2579. 10.1158/1078-0432.CCR-14-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Penning TM. Aldo-keto reductase (AKR) 1C3 inhibitors: a patent review. Expert Opin Ther Pat. 2017;27(12):1329-1340. 10.1080/13543776.2017.1379503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yepuru M, et al. Steroidogenic enzyme AKR1C3 is a novel androgen receptor-selective coactivator that promotes prostate cancer growth. Clin Cancer Res. 2013;19(20):5613-5625. 10.1158/1078-0432.CCR-13-1151. [DOI] [PubMed] [Google Scholar]
- 6. Doig CL, et al. Knockdown of AKR1C3 exposes a potential epigenetic susceptibility in prostate cancer cells. J Steroid Biochem Mol Biol. 2016;155(Pt A):47-55. 10.1016/j.jsbmb.2015.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stockwell BR, et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell 2017;171(2):273-285. 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hamid AR, et al. Aldo-keto reductase family 1 member C3 (AKR1C3) is a biomarker and therapeutic target for castration-resistant prostate cancer. Mol Med. 2013;18(1):1449-1455. 10.2119/molmed.2012.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mitsiades N, et al. Distinct patterns of dysregulated expression of enzymes involved in androgen synthesis and metabolism in metastatic prostate cancer tumors. Cancer Res. 2012;72(23):6142-6152. 10.1158/0008-5472.CAN-12-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stanbrough M, et al. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res. 2006;66(5):2815-2825. 10.1158/0008-5472.CAN-05-4000. [DOI] [PubMed] [Google Scholar]
- 11. Hofland J, et al. Evidence of limited contributions for intratumoral steroidogenesis in prostate cancer. Cancer Res. 2010;70(3):1256-1264. 10.1158/0008-5472.CAN-09-2092. [DOI] [PubMed] [Google Scholar]
- 12. Pfeiffer MJ, et al. Steroidogenic enzymes and stem cell markers are upregulated during androgen deprivation in prostate cancer. Mol Med. 2011;17(7-8):657-664. 10.2119/molmed.2010.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miyazaki Y, et al. Consecutive prostate cancer specimens revealed increased Aldo⁻keto reductase family 1 member C3 expression with progression to castration-resistant prostate cancer. J Clin Med. 2019;8(5):601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu C, et al. Intracrine androgens and AKR1C3 activation confer resistance to enzalutamide in prostate cancer. Cancer Res. 2015;75(7):1413-1422. 10.1158/0008-5472.CAN-14-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hertzog JR, et al. AKR1C3 mediates pan-AR antagonist resistance in castration-resistant prostate cancer. Prostate 2020;80(14):1223-1232. 10.1002/pros.24049. [DOI] [PubMed] [Google Scholar]
- 16. Liu C, et al. Inhibition of AKR1C3 activation overcomes resistance to abiraterone in advanced prostate cancer. Mol Cancer Ther. 2017;16(1):35-44. 10.1158/1535-7163.MCT-16-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sun SQ, et al. Overexpression of AKR1C3 significantly enhances human prostate cancer cells resistance to radiation. Oncotarget 2016;7(30):48050-48058. 10.18632/oncotarget.10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gurioli G, et al. Circulating tumor cell gene expression and plasma AR gene copy number as biomarkers for castration-resistant prostate cancer patients treated with cabazitaxel. BMC Med. 2022;20(1):48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lorenc T, et al. , Exosomes in prostate cancer diagnosis, prognosis and therapy. Int J Mol Sci . 2020;21(6):2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhu S, et al. Exosomal TUBB3 mRNA expression of metastatic castration-resistant prostate cancer patients: association with patient outcome under abiraterone. Cancer Med 2021;10(18):6282-6290. 10.1002/cam4.4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Del Re M, et al. The detection of androgen receptor splice variant 7 in plasma-derived exosomal RNA strongly predicts resistance to hormonal therapy in metastatic prostate cancer patients. Eur Urol. 2017;71(4):680-687. 10.1016/j.eururo.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 22. Camp RL, Dolled-Filhart M, Rimm DL.. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10(21):7252-7259. 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 23. Théry C, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 2018;7(1):1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhao J, et al. AKR1C3 expression in primary lesion rebiopsy at the time of metastatic castration-resistant prostate cancer is strongly associated with poor efficacy of abiraterone as a first-line therapy. Prostate 2019;79(13):1553-1562. 10.1002/pros.23875. [DOI] [PubMed] [Google Scholar]
- 25. Ni YC, et al. , Predictors of efficacy of corticosteroid switching from abiraterone plus prednisone to dexamethasone in patients with metastatic castration-resistant prostate cancer. Asian J Androl. 2022;24(2):154-160. 10.4103/aja202143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Endo S, et al. Development of Novel AKR1C3 inhibitors as new potential treatment for castration-resistant prostate cancer. J Med Chem. 2020;63(18):10396-10411. 10.1021/acs.jmedchem.0c00939. [DOI] [PubMed] [Google Scholar]
- 27. Verma K, et al. AKR1C3 inhibitor KV-37 exhibits antineoplastic effects and potentiates enzalutamide in combination therapy in prostate adenocarcinoma cells. Mol Cancer Ther. 2018;17(9):1833-1845. 10.1158/1535-7163.MCT-17-1023. [DOI] [PubMed] [Google Scholar]
- 28. Graham LS, et al. Targeting backdoor androgen synthesis through AKR1C3 inhibition: A presurgical hormonal ablative neoadjuvant trial in high-risk localized prostate cancer. Prostate 2021;81(7):418-426. 10.1002/pros.24118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.