Abstract
Angiosarcomas are rare, malignant soft tissue tumors in children that arise in a wide range of anatomical locations and have limited targeted therapies available. Here, we report a rare case of a pediatric angiosarcoma (pAS) with Li–Fraumeni syndrome (LFS) expressing a novel NOTCH1–ROS1 gene fusion. Although both NOTCH1 and ROS1 are established proto-oncogenes, our study is the first to describe the mechanistic role of NOTCH1–ROS1 fusion arising via intrachromosomal rearrangement. NOTCH1–ROS1 displayed potent neoplastic transformation propensity in vitro, and harbors tumorigenic potential in vivo, where it induced oncogenic activation of the MAPK, PI3K/mTOR, and JAK–STAT signaling pathways in a murine allograft model. We found an unexpected contribution of the NOTCH1 extracellular region in mediating NOTCH1–ROS1 activation and oncogenic function, highlighting the contribution of both NOTCH1 and ROS1 fusion partners in driving tumorigenicity. Interestingly, neither membrane localization nor fusion protein dimerization were found to be essential for NOTCH1–ROS1 fusion oncogenicity. To target NOTCH1–ROS1-driven tumors, we tested both NOTCH1-directed inhibitors and ROS1-targeted tyrosine kinase inhibitors (TKI) in heterologous models (NIH3T3, Ba/F3). Although NOTCH1 inhibitors did not suppress NOTCH1–ROS1-driven oncogenic growth, we found that oral entrectinib treatment effectively suppressed the growth of NOTCH-ROS1-driven tumors. Taken together, we report the first known pAS case with a novel NOTCH1–ROS1 alteration along with a detailed report on the function and therapeutic targeting of NOTCH1–ROS1. Our study highlights the importance of genomic profiling of rare cancers such as pAS to reveal actionable drivers and improve patient outcomes.
Keywords: metastatic angiosarcoma
INTRODUCTION
Angiosarcomas are vascular tumors that arise in cells lining blood or lymphatic vessels and display significant clinical heterogeneity in terms of anatomic distribution (Young et al. 2010). These soft tissue sarcomas are extremely rare in children, representing ∼0.3% of pediatric sarcomas overall, with greater prevalence in adults and elderly patients (Ferrari et al. 2002; Young et al. 2010). Pediatric angiosarcomas have a more aggressive clinical course than in adults, requiring multimodal therapies including surgical resection, chemotherapy, and/or radiation therapy, albeit with little success (Ayadi and Khabir 2010). Despite dismal outcomes in children, there have been no significant therapeutic advancements as the molecular and genetic drivers of pediatric angiosarcoma remain largely uncharacterized.
Much of our prior understanding of the genetic drivers of angiosarcoma come from analysis of adult cases. These studies have discovered oncogenic alterations that activate the phosphatidylinositol-3′-kinase/mammalian target of rapamycin pathway (PI3K/mTOR) (Italiano et al. 2012; Painter et al. 2020), leading to clinical testing of mTOR inhibitors in angiosarcoma patients that demonstrated partial responsiveness (Seki et al. 2012; Yoo et al. 2013). In addition to the PI3K/mTOR pathway, several sequencing studies have revealed frequent genetic mutations in TP53, KDR, PIK3CA, RAS, BRAF, MAPK1, and NF1, and amplifications in MAPK1/CRKL, CRAF, or BRAF driving activation of the mitogen activated protein kinase (MAPK) pathway in angiosarcomas (Garcia et al. 2000; Murali et al. 2015; Painter et al. 2020). These findings have supported successful preclinical testing of MEK inhibitor, trametinib in combination with either mTOR inhibitor, and everolimus in a genetic mouse model of adult angiosarcoma (Chadwick et al. 2018) or with pan-vascular endothelial growth factor receptor (VEGFR) inhibitor cediranib in angiosarcoma cell lines (Wagner et al. 2021). However, a single clinical report of a pediatric patient with KRAS-altered angiosarcoma showed no clinical benefit of treatment with MEK inhibitor (trametinib) therapy (Jeng et al. 2014), suggesting that the treatment modalities that are effective in preclinical models of adult angiosarcoma may not be translatable in the pediatric setting. Furthermore, pediatric angiosarcoma can occur in patients with germline TP53 mutations who have Li–Fraumeni syndrome (LFS), a rare familial cancer predisposition syndrome. LFS renders patients susceptible to different types of tumors including sarcomas in multiple locations, which may further differentiate the pediatric and adult angiosarcoma genetic landscape (Kamihara et al. 2014; Calvete et al. 2015). Hence, there is a need to specifically understand molecular pathogenesis and develop targeted therapeutics for children with LFS and pediatric angiosarcomas.
To address this, we performed genetic and molecular analysis of a rare case of LFS-related pediatric angiosarcoma. In addition to TP53 loss, our clinically validated, next-generation sequencing panel (Surrey et al. 2019) reported a unique translocation event between NOTCH1 and ROS1 that led to formation of a novel NOTCH1–ROS1 fusion product. Up to 20% of soft tissue sarcoma subtypes have chromosomal translocations (Nakano and Takahashi 2018) with rare cases of angiosarcomas harboring structural variants. We characterized the oncogenic profile of NOTCH1–ROS1 to understand the collaborative mechanisms of both NOTCH1 and ROS1 fusion partners. Furthermore, we sought to identify optimal targeted therapeutic options for NOTCH1–ROS1-driven pediatric angiosarcomas using orthotopic mouse models.
RESULTS
Clinical Presentation of a Pediatric Patient with Angiosarcoma Harboring NOTCH1–ROS1 Fusion
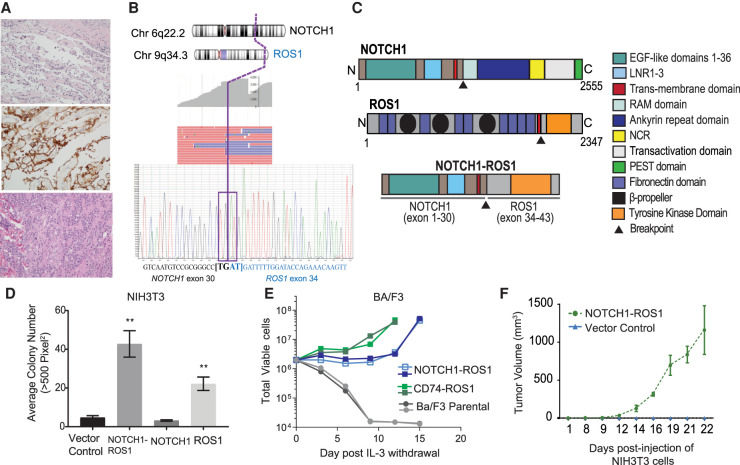
The 11-yr-old female patient presented to our institution for management of progressive Gorham–Stout disease (lymphatic malformation of the bones) with extensive bone resorption and chylothorax. A biopsy of the scapula performed 2 yr previously (age 9) was reviewed and had shown fragments of reactive bone and cartilage with an atypical vascular proliferation consisting of endothelial cells wrapping around individual muscle fibers (Fig. 1A, upper panel). The endothelial cells stained positively for vascular (CD31) and lymphatic (D240) markers (Fig. 1A, middle panel). There was no atypia or mitosis, and the differential diagnosis included lymphovascular malformation, lymphangiomatosis or a low-grade malignant vascular proliferation. For management of her progressive bone disease, the patient had received radiation to the chest and upper extremities ∼6 mo prior to presentation. At presentation to our institution (age 11), she was found to have soft tissue thickening of the left chest, shoulder, left upper extremity, and paraspinal region, which was positron emission tomography (PET)-avid, measuring more than 10 × 9 × 5 cm. Incisional biopsy of this lesion revealed an epithelioid angiosarcoma with prominent mitotic activity involving the dermis and subcutaneous tissue, with immunohistochemistry positive for endothelial markers CD31, von Willebrand factor, D240, and CD34 (Fig. 1A, bottom panel). Based on histologic features, this tumor was classified as Grade 3 under the French Federation of Cancer Centers Sarcoma Group (FNCLCC).
Figure 1.
Histological and sequencing characteristics of a pediatric angiosarcoma with NOTCH1–ROS1 fusion that drives oncogenic cellular phenotypes. (A) A biopsy of the scapula 2 yr previous show a low-grade vascular lesion consisting of individual muscle fibers wrapped by a single layer of endothelial cells (hematoxylin and eosin [H&E], top panel) that are positive for lymphatic marker D240 (middle panel). The bottom panel shows 6 mo after radiation, incisional biopsy revealed a proliferation of malignant cells forming irregular vascular channels, consistent with angiosarcoma. All histology images are 20×. (B) Targeted RNA-seq results showing multiple next-generation sequencing (NGS) reads spanning the breakpoint (top) with confirmatory Sanger sequencing of cDNA exhibiting breakpoint (bottom). (C) NOTCH1–ROS1 fusion protein retains: NOTCH1 exons 1–30 encoding epidermal growth factor (EGF) domains 1–36, LNR 1–3, and transmembrane domain. ROS1 exons 34–43 encoding the transmembrane domain and complete tyrosine kinase. (D) Soft agar colony assays with stable NIH3T3 cell lines, quantification of colony counts shown for n = 5. (E) Ba/F3 proliferation assay performed in the absence of IL-3. (F) Flank xenograft tumor measurements of NSG mice engrafted with stable NIH3T3 cell lines, n = 5, P < 0.05.
Targeted RNA sequencing of the angiosarcoma identified a novel in-frame fusion between NOTCH1 exon 30 and ROS1 exon 34 (Fig. 1B). The fusion arises from a chromosomal translocation between Chromosome 9 (NOTCH1) and Chromosome 6 (ROS1). The targeted somatic next-generation sequencing (NGS) panel also identified additional DNA sequence and copy-number variants of clinical significance, including a TP53 c.331_365dup (p.T123Wrs*12) variant, whole chromosome gains of Chromosomes 7, 20, and 21, and loss of heterozygosity of Chromosome 17, containing TP53 (Table 1). For evaluation of germline TP53 status, the patient's peripheral blood was tested, and the same TP53 pathogenic variant (c.331_365dup) identified in the tumor was discovered as heterozygous germline variant. These data established a previously unknown diagnosis of LFS. Her family history is notable because her mother was deceased from an unknown type of brain tumor, and no further parental germline testing could be performed.
Table 1.
Clinically significant somatic tumor sequencing results
| Chromosome | Gene(s) | Type of alteration | Reference/Iioform | Nucleotide | Amino acid | Variant allele fraction | Comments |
|---|---|---|---|---|---|---|---|
| 17p | TP53 | Duplication | NM_001126114.2 | c.331_265dup | p.T123Wfs*12 | 0.427 | Also present in heterozygous state in patient's peripheral blood |
| Xp11.4-p11.22 | BCOR; KDM6A; ARAF; GATA1; KDM5C | Loss | n/a | n/a | n/a | n/a | |
| 1p36.22-q23.3 | MTOR; SPEN; SDHB; ARID1A; MPL; MUTYH; CDKN2C; JUN; JAK1 | Gain | n/a | n/a | n/a | n/a | |
| Chr 7 (whole chromosome) | Gain | n/a | n/a | n/a | n/a | ||
| Chr 17 (whole chromosome) | Loss of heterozygosity | n/a | n/a | n/a | n/a | Includes TP53 | |
| 17q24.2-q25.3 | PRKAR1A; FAM20A; RPTOR | Gain | n/a | n/a | n/a | n/a | |
| Chr 20 (whole chromosome) | Gain | n/a | n/a | n/a | n/a | ||
| Chr 21 (whole chromosome) | Gain | n/a | n/a | n/a | n/a |
The NOTCH1–ROS1 fusion contains the amino-terminal region of NOTCH1, which is a single-pass transmembrane receptor is fused to the carboxy-terminal of ROS1 tyrosine kinase. In NOTCH1–ROS1, NOTCH1 extracellular regions encoding the 36 epidermal growth factor (EGF)-like repeats. and transmembrane regions are retained but the intracellular regions, NOTCH1 ankyrin repeats and PEST domain, have been lost (Fig. 1C). Likewise, the amino-terminal region of ROS1 is lost in the fusion context, leaving only the ROS1 intracellular tyrosine kinase domain intact (Fig. 1C). Our observation highlights disruptions in both NOTCH1 and ROS1 in the NOTCH1–ROS1 fusion context, thereby suggesting that NOTCH1–ROS1 could be oncogenic through combinatorial effects of alterations in both NOTCH1 and ROS1. We therefore sought to characterize the oncogenic mechanism of action of both NOTCH1 and ROS1 in NOTCH1–ROS1-driven angiosarcoma.
NOTCH1–ROS1 Gene Fusion Drives Oncogenic Transformation in Heterologous Cell Models
Previous studies have shown gene fusions involving either NOTCH1 or ROS1 to be oncogenic driver mutations (Kumar-Sinha et al. 2015; Roskoski 2017). This led us to assess the transformation potential of NOTCH1–ROS1 fusion protein. Because of insufficient tumor tissue, we were unable to create an angiosarcoma patient–derived cell line. To pursue molecular characterization, we created NOTCH1–ROS1 stable expression in two independent heterologous cell model systems, NIH3T3 and Ba/F3, along with ROS1 full-length (FL), NOTCH1 FL, and vector control (Supplemental Figs. 1a,b, respectively). In soft agar colony formation assays, the NOTCH1–ROS1 expressing NIH3T3 cells show significant colony growth compared to vector control and NOTCH1 FL (Fig. 1D, P < 0.01). ROS1 FL expressing NIH3T3 exhibits significant growth compared to the vector control, but lower than seen with NOTCH1–ROS1, highlighting a proliferative advantage provided by the fusion context (Fig. 1D, P < 0.01). In the Ba/F3 cell model, we observed IL-3-independent proliferation in the NOTCH1–ROS1 and CD74-ROS1 (positive control) expressing cells but not in the ROS1 FL or parental cells (Fig. 1E). Next, we tested tumor forming capacity of NOTCH1–ROS1 in vivo by performing NIH3T3 flank allograft injections in immune-compromised NSG mice and observed tumor formation in only the NOTCH1–ROS1 fusion context (Fig. 1F), indicating that NOTCH1–ROS1 is an oncogenic driver.
Activated ROS1 Kinase in NOTCH1–ROS1 Triggers Oncogenic Activation of the MAPK, PI3K/mTOR, and JAK–STAT Pathways
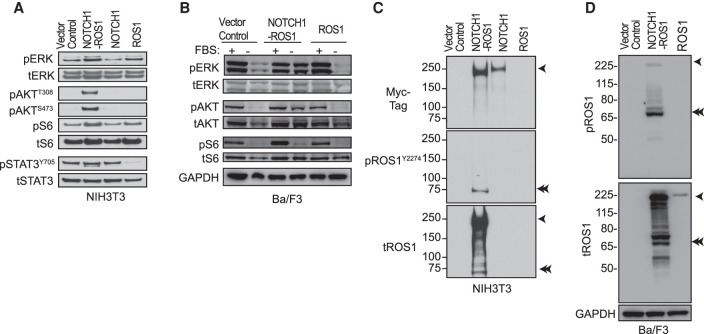
Because of the inclusion of the intact ROS1 tyrosine kinase domain in NOTCH1–ROS1, we investigated ROS1 downstream signaling pathway levels in our cell models. In serum-starved conditions, higher phosphorylation of ERK, AKT, S6, and STAT3 was observed, suggesting activation of the MAPK, PI3K/mTOR, and JAK-STAT pathways, respectively, in the NOTCH1–ROS1 expressing NIH3T3s compared to NOTCH1 FL and vector control (Fig. 2A). No apparent activation was observed in ROS1 FL expressing NIH3T3s compared to vector control (Fig. 2A). NOTCH1–ROS1-driven activation of MAPK, PI3K/mTOR, and JAK–STAT pathways was corroborated by our observations in Ba/F3 cells, where NOTCH-ROS1 expressing cells uniquely exhibited elevated phosphorylation of ERK, AKT, STAT3, and ribosomal S6 in a serum-independent manner (-FBS) as compared to ROS1 and control (Fig. 2B). Next, we assessed ROS1 kinase phosphorylation at Y2274, a site known to activate ROS1 kinase [20]. Within the NOTCH1–ROS1 expressing NIH3T3 and Ba/F3 cells, we noticed an unexpected, smaller band at ∼75 kDa with pROS1Y2274, suggesting a truncated, activated fragment of NOTCH1–ROS1 (Fig. 2C,D, respectively, denoted via double arrows). This cleaved NOTCH1–ROS1 fragment shows higher pROS1Y2274 compared to the full NOTCH1–ROS1 fusion, suggesting higher activation of ROS1 kinase domain in the cleaved fragment (Fig. 2C,D, respectively). We concluded that this cleaved NOTCH1–ROS1 protein could be a product of the wild-type NOTCH1 processing machinery acting on the fusion protein, suggesting functional involvement of ROS1 but also possibly NOTCH1 as fusion partners.
Figure 2.
Activated ROS1 kinase in NOTCH1–ROS1 drives downstream signaling via MAPK, PI3K/Mtor, and JAK–STAT pathways. Western blot analysis of (A) stable NIH3T3 and (B) Ba/F3 cell lines showing phosphorylated (p-) and total (t-) protein levels of MAPK, PI3K/Mtor, and JAK–STAT pathways. Western blot analysis to show activation of ROS1 kinase in (C) stable NIH3T3 and (D) Ba/F3 cell lines showing phosphorylated (p-) and total (t-) protein levels. High exposure tROS1 blot is included in Supplemental Figure 1a to show ROS1 levels in ROS1 NIH3T3 cell model.
NOTCH1 Extracellular Domain Regulates NOTCH1–ROS1 Function but Targeting with NOTCH Inhibitors Does Not Suppress Oncogenicity
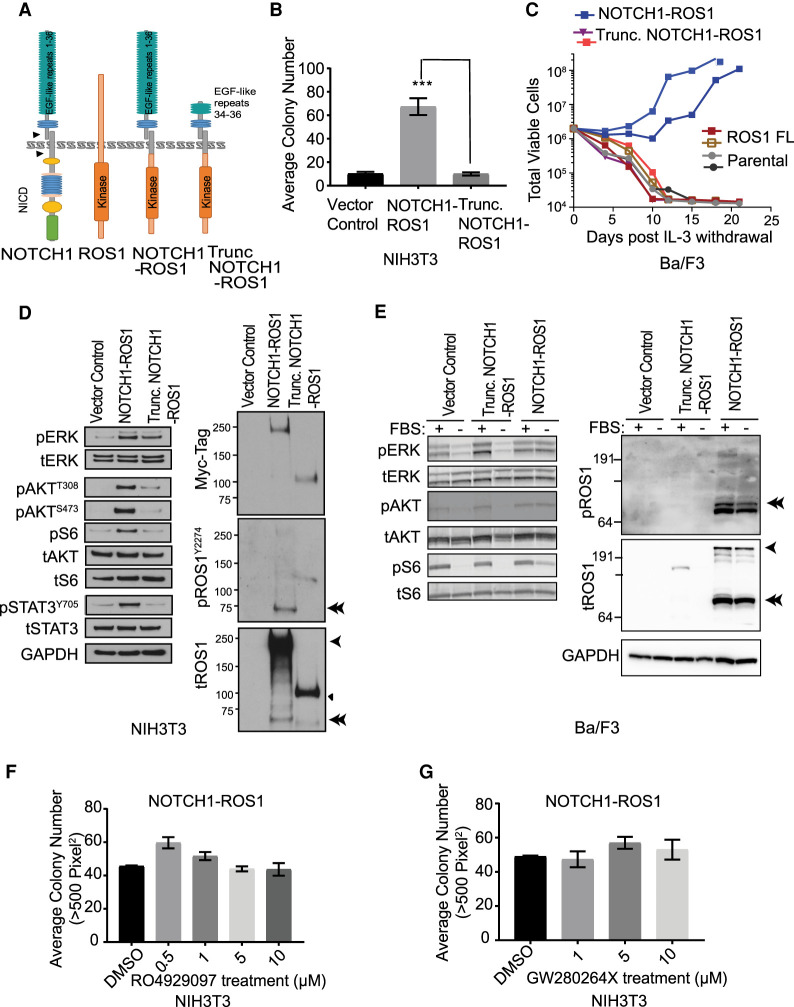
Under normal conditions, binding of ligands to the NOTCH1 receptor triggers downstream signaling through release of the NOTCH1 intracellular domain (NICD) that translocates to the nucleus and mediates transcriptional activation (Lieber et al. 1993). NOTCH1 processing is facilitated by two proteases, ADAM10 and γ-secretase (respective cleavage sites S2 and S3 shown by arrows in Fig. 3A), which are well-studied antitumor targets (Katoh and Katoh 2020). In NOTCH1–ROS1, the NICD is lost suggesting a loss of NOTCH1 transcriptional regulation as seen in our gene expression analysis in which NOTCH1–ROS1 expressing NIH3T3 cell models show significant difference in NOTCH1 pathway–regulated genes compared to NOTCH1 FL cells (Supplemental Fig. 2). This led us to investigate the contribution of the retained NOTCH1 extracellular region (EGF repeats 1–36) in NOTCH1–ROS1.
Figure 3.
Truncation of NOTCH1 extracellular domains causes loss of NOTCH1–ROS1 oncogenicity, despite lack of efficacy with existing preclinical NOTCH1 inhibitors. (A) Diagrammatic representation of NOTCH1, ROS1, NOTCH1–ROS1, and Trunc. NOTCH1–ROS1 predicted protein domains and localization. (B) Soft agar colony assays with stable NIH3T3 cells, quantification of colony counts shown for n = 5. (C) Ba/F3 proliferation assay performed in the absence of IL-3. Western blot analysis of (D) stable NIH3T3 and (E) Ba/F3 cell lines showing phosphorylated (p-) and total (t-) protein levels of MAPK, PI3K/mTOR, JAK–STAT pathways, and activation level of ROS1 kinase. fx1 represents NOTCH1–ROS1 (256 kDa); ◄ represents Trunc. NOTCH1–ROS1 (118 kDa); fx2 represents Cleaved NOTCH1–ROS1 (∼75 kDa). (F,G) Soft agar colony formation analysis with NIH3T3 cell models in presence of increasing doses of (F) gamma secretase inhibitor, RO4929097, and (G) ADAM 10/17 inhibitor, GW280264×. n = 10 each.
To assess this, we synthesized a shorter, experimental fusion construct, termed trunc. NOTCH1–ROS1 (trunc. denotes truncated) by deleting NOTCH1 exons 1–23 that encode EGF-like repeats 1–33 housing majority of NOTCH1 ligand-binding sites (Fig. 3A; Katoh and Katoh 2020). First, we investigated the subcellular localization of NOTCH1–ROS1 and trunc. NOTCH1–ROS1 as plasma membrane localization is essential for NOTCH1 receptor function. Furthermore, subcellular localization has been shown to modulate oncogenic signaling induced by ROS1 fusions (Neel et al. 2019). Using an in silico topological analysis tool (Dobson et al. 2015), we predicted both NOTCH1–ROS1 and trunc. NOTCH1–ROS1 to be membrane-anchored with a cytoplasmic ROS1 kinase domain (Fig. 3A). By subcellular fractionation, we observed localization of NOTCH1–ROS1 primarily at the cell membrane in NIH3T3 whereas trunc. NOTCH1–ROS1 was partially localized at the membrane with a portion of the total protein detectable in the cytosol (Supplemental Fig. 1c). Similar membrane localization of NOTCH1–ROS1 was observed in Ba/F3 cells that underwent biotinylation assays followed by neutravidin pulldown to represent proteins at the cellular membrane (Supplemental Fig. 3).
We next assessed the oncogenic profile of trunc. NOTCH1–ROS1 and observed loss of neoplastic transformation potential in both soft agar and IL-3 withdrawal assays in NIH3T3 and Ba/F3 expressing trunc. compared to full-length NOTCH1–ROS1 (Fig. 3B,C, respectively). Trunc. NOTCH1–ROS1 cells also showed lower MAPK phosphorylation and insignificant PI3K/mTOR and JAK–STAT pathway activation in NIH3T3s (Fig. 3D), correlating with lack of cellular transformation seen in soft agar assays. In Ba/F3 cells, no pathway activation was observed in serum-starved conditions with trunc. NOTCH1–ROS1 compared to NOTCH1–ROS1 (Fig. 3E). This shows that despite appropriate membrane localization, trunc. NOTCH1–ROS1 is not oncogenic. Interestingly, the cleaved NOTCH1–ROS1 fragment is seen only with the NOTCH1–ROS1 cells but not with trunc. NOTCH1–ROS1 (Fig. 3D,E; Supplemental Fig. 3, denoted via double arrows). This indicates that both NOTCH1–ROS1 and trunc. NOTCH1–ROS are trafficked to the plasma membrane but loss of NOTCH1 EGF repeats 1–33 in trunc. NOTCH-ROS1 likely leads to differential protein processing. These data indicate the relevance of NOTCH1 EGF repeats 1–33 in mediating oncogenic phenotype driven by NOTCH1–ROS1 via formation of the phosphorylated and cleaved NOTCH1–ROS1 fragment.
To further understand the mechanism of NOTCH1–ROS1 activation, we tested dimerization potential of NOTCH1–ROS1 as several RTKs are known to be activated via ligand-mediated dimerization (Kumar-Sinha et al. 2015). In coimmunoprecipitation assays, we observed homodimerization of NOTCH1–ROS1 (Supplemental Fig. 1d). To further dissect which fusion partner drives fusion dimerization, we tested heterodimerization of NOTCH1–ROS1 with both wild-type NOTCH1 and ROS1. Our results show protein–protein interaction in both cases with a stronger dimerization of ROS1 FL with NOTCH1–ROS1 compared to NOTCH1 FL (Supplemental Fig. 1e). However, we also observed trunc. NOTCH1–ROS1 interacting with NOTCH1 FL (Supplemental Fig. 1e). The absence of oncogenic phenotype with trunc. NOTCH1–ROS1 despite dimerization highlights that dimerization is not key for the oncogenic phenotype of the fusion.
Given our initial observations showing a role of NOTCH1 processing in NOTCH1–ROS1 oncogenicity, we assessed the response of NOTCH1-related targeted inhibitors on NIH3T3 cell models. We did not observe a decrease in soft agar growth of NIH3T3 expressing NOTCH1–ROS1 with increasing concentrations of γ-secretase inhibitor, RO4929097 (Fig. 3F) and ADAM 10/17 inhibitor, GW280264X (Fig. 3G) or any impact on downstream signaling (Supplemental Fig. 4).
Targeted Therapy with ROS1 Kinase Inhibitors Is Effective in Suppressing NOTCH1–ROS1-Driven Oncogenesis In Vitro and In Vivo
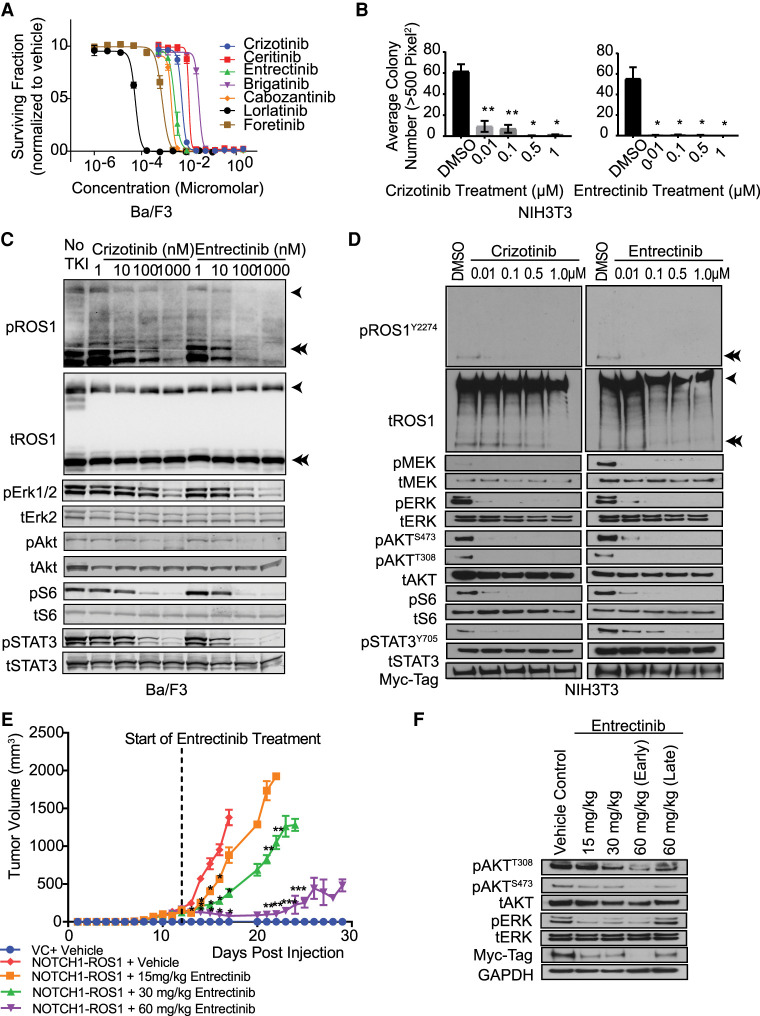
Tyrosine kinase inhibitors (TKIs) with activity against the ROS1-kinase domain have demonstrated noteworthy efficacy in patients suffering from cancers driven by ROS1-fusion oncogenes. Roughly 70% of ROS1-positive non-small-cell lung cancer patients exhibit an objective response to crizotinib, a TKI with ROS1/ALK activity, whereas median progression-free survival varies between 9 and 19.2 mo (Shaw et al. 2014; Mazieres et al. 2015).
To target ROS1 kinase–driven oncogenic signaling, we tested several ROS1 TKIs in the NOTCH1–ROS1 onco-addicted Ba/F3 cells. All ROS1 inhibitors used in the screen were able to inhibit ROS1 signaling (Fig. 4A). The U.S. Food and Drug Administration (FDA)-approved ROS1 inhibitors, crizotinib and entrectinib, were chosen for further evaluation (Facchinetti and Friboulet 2019). Both crizotinib and entrectinib induced significant, dose-dependent suppression of NIH3T3 colony growth in soft agar assays (Fig. 4B, P < 0.05) and inhibited MAPK, PI3K/mTOR, and JAK–STAT effector pathways in both Ba/F3 and NIH3T3 cell model systems (Fig. 4C,D, respectively). To test entrectinib in vivo, NOTCH1–ROS1 expressing NIH3T3 cells were injected as flank allografts in NSG mice. Oral-gavage with entrectinib resulted in significant dose-dependent decrease in tumor volume (Fig. 4E). The entrectinib steady state trough is reported as a benchmark for simulating the study's effectiveness in replicating true drug bioavailability in patients (Supplemental Fig. 5b). Treatment of NIH3T3 NOTCH1-allograft tumors with entrectinib significantly increased in survival time as compared to vehicle for both the 30- and 60-mg/kg treatment groups (Supplemental Fig. 5a, P < 0.0001). Immunoblotting of lysates prepared from tumor tissue after oral treatment showed inhibition of AKT and ERK activation concordant with NIH3T3 NOTCH1–ROS1 cells in vitro (Fig. 4F). Thus, entrectinib therapy prolonged overall survival and decreased tumor growth of NOTCH1–ROS1 tumors in vivo.
Figure 4.
ROS1-directed targeted inhibitors target NOTCH1–ROS1-driven signaling and in vitro and in vivo oncogenic growth. (A) Ba/F3 proliferation assay using clinical ROS1-directed inhibitors in BA/F3 cell growth. (B) Soft agar colony formation analysis with NIH3T3 cell models in presence of increasing doses of entrectinib and crizotinib. n = 10, * denotes P-value ≤0.05 and ** denotes P-value ≤0.01. Western blot analysis of (C) Ba/F3 model and (D) NIH3T3 model upon treatment with crizotinib and entrectinib. (E) Tumor volume plot from in vivo flank xenograft of NOTCH1–ROS1 expressing NIH3T3 and vector control cells in NSG mice and bi-daily oral gavage treatment with entrectinib. n = 8 Significant decrease compared to * NOTCH1–ROS1 + Vehicle, ** NOTCH1–ROS1 + 15 mg/kg Entrectinib, *** NOTCH1–ROS1 + 30 mg/kg Entrectinib. (F) Western blot analysis of mouse tumor lysates to show targeting of MAPK, PI3K, and JAK/STAT and expression of fusion protein in the myc-tag blot.
DISCUSSION
Pediatric angiosarcomas are extremely rare entities, and although there is some understanding of the molecular pathophysiology or iatrogenic etiology for these patients, no effective therapies exist. Our study describes a novel oncogenic mechanism of action and therapeutic targetability of the NOTCH1–ROS1 fusion found in a rare case of pediatric angiosarcoma. This is the first report, to our knowledge, describing the critical role of NOTCH1 extracellular region in mediating membrane localization and dimerization of a therapeutically targetable fusion protein in angiosarcomas. Genetic heterogeneity is pervasive across angiosarcomas, with PI3K/mTOR and MAPK pathways being commonly affected, and some adult cases have also been shown to harbor mutually exclusive MYC amplifications or TP53 alterations and CDKN2A loss (Murali et al. 2015). In our study, the pediatric patient had LFS and several clinically relevant copy-number variants. It is likely that radiation therapy to the low-grade vascular lesion induced chromosomal breakage with subsequent transformation to angiosarcoma in the setting of unrecognized LFS. She rapidly succumbed to disease before targeted therapy could be initiated. An ongoing patient-partnered approach, called the Angiosarcoma Project (Painter et al. 2020), has begun elucidating the etiology for rare adult angiosarcomas leading to potential therapeutic opportunities. Such an effort in the pediatric setting is lacking and could provide more biological insight into the etiology of angiosarcoma in children.
Although several studies have previously shown gene fusions involving either NOTCH1 or ROS1 to be oncogenic driver mutations across cancers (Kumar-Sinha et al. 2015; Roskoski 2017), ours is the first to define the mechanism of the unique combination of amino-terminal NOTCH1 exons 1–30 to carboxy-terminal ROS1 exons 34–43 in a pediatric angiosarcoma. Interestingly, NOTCH1 has been implicated as both an oncogene (in T-cell acute lymphoblastic leukemia [ALL], medulloblastoma, osteosarcoma) and a tumor suppressor (in B-cell ALL, and acute myeloid leukemia [AML]) in pediatric cancers (Lieber et al. 1993; Kumar-Sinha et al. 2015). Normally, Notch1 receptor protein is activated by cell–cell interactions and regulates cell fate during development (Katoh and Katoh 2020). When acting as an oncogene, NOTCH1 has been shown to harbor a hotspot of activating mutations in the PEST domain, and as a 3′ gene fusion partner in constructs both dependent and independent of γ-secretase cleavage (Katoh and Katoh 2020). In the context of a tumor suppressor, NOTCH1-mediated signaling has also been demonstrated to be absent in neuroendocrine tumors, whereas activation of NOTCH1 in vitro significantly reduced tumor growth (Kunnimalaiyaan and Chen 2007). These studies highlight the diverse functionality of NOTCH1 across the cancer landscape, and our finding provides a novel mechanism via which the NOTCH1 amino-terminal domain contributes to fusion biology.
ROS1 is a receptor tyrosine kinase (RTK) driving growth signaling via activation of the PI3K/AKT, MAPK/ERK, and JAK–STAT pathways (Xiong et al. 1996; Zong et al. 1998; Biskup et al. 2004). ROS1 is an orphan receptor, implying that its ligand is currently unknown (Drilon et al. 2021). The ROS1 kinase domain has been detected in more than 50 different fusions as the 3′ partner in both adult and pediatric cancers (Drilon et al. 2021). However, it has not been previously reported as a fusion partner in pediatric angiosarcoma. GOPC–ROS1 and CEP85L–ROS1 fusions have been previously reported in adult angiosarcomas (Giacomini et al. 2013; Marks et al. 2019). ROS1 fusions drive oncogenesis via ligand-independent, constitutive activation of canonical signaling pathways, partly driven by amino-terminal partner-mediated intracellular protein localization (Neel et al. 2019). Interestingly, our study demonstrates that NOTCH1–ROS1 has NOTCH1-mediated membrane localization and dimerization as well as ROS1-mediated dimerization, suggesting that in the context of this fusion, canonical dimerization might be required for ROS1 kinase fusion activation at the membrane. This contrasts with the intracellular ROS1 fusions that show dimerization-independent activity (Neel et al. 2019). This brings up the possibility that NOTCH1–ROS1 may function as a different mechanistic class of membrane-bound ROS1 oncogenic fusion.
The pathological diversity and rare occurrence of angiosarcomas makes it challenging to determine optimal treatment strategies. The standard of care treatment options for angiosarcomas include surgery, chemotherapy, and/or radiotherapy, which are often toxic among pediatric patients. In adults, a few clinical trials with tyrosine kinase inhibitors have shown partial responsiveness such as the use of sorafenib against vascular sarcomas (Penel et al. 2014). The aberrant expression of programmed death ligand 1 (PD-L1) in angiosarcomas (Botti et al. 2017) has led to the successful use of anti-PD1 immunotherapy in an older patient (Sindhu et al. 2017). However, such clinical studies are lacking for pediatric angiosarcomas. Our study highlights the preclinical efficacy of ROS1 inhibitors in targeting NOTCH1–ROS1-driven tumors, which can be tested clinically as a potent targeted therapy in the arsenal against angiosarcomas.
METHODS
Clinical Sequencing
Tumor tissue was sequenced using the Children's Hospital of Philadelphia Comprehensive Solid Tumor Panel as previously described (Surrey et al. 2019). Briefly, this custom panel interrogates 238 genes for DNA sequence alteration and copy-number variation using capture-based NGS. The panel also includes targeted RNA-seq for 110 additional genes using anchored multiplex polymerase chain technology followed by NGS to identify known and novel gene fusions. Supplemental Table 1 contains sequencing coverage information for the NOTCH1–ROS1 fusion gene breakpoint. Fusion gene confirmation was performed by nested polymerase chain reaction (PCR) and Sanger sequencing on reverse-transcribed RNA from tumor tissue. Sanger sequencing of peripheral blood leukocytes was used to confirm the TP53 constitutional variant with custom-designed primers.
Cell Culture
NIH3T3 and HEK293 cells were obtained from ATCC and maintained in 1× DMEM with 10% Donor Bovine Serum (DBS) and Fetal Bovine Serum (FBS), respectively. Ba/F3 cells were maintained in 1× RPMI with 10% FBS with or without addition of 2 µg/mL recombinant murine IL-3. Serum starvation of Ba/F3 cells was accomplished by suspending cells in 1× RPMI with 1 mg/mL BSA. Cell lines were routinely tested for mycoplasma infection.
Cloning and Generation of Heterologous Cell Lines
NOTCH1–ROS1 and Trunc. NOTCH1–ROS1 constructs were synthesized as gateway compatible entry clones by GenScript. Full-length NOTCH1 and full-length ROS1 were purchased as gateway entry clones from GenScript and PlasmID/Dana-Farber/Harvard Cancer Center DNA Resource Core, respectively. Subcloning was conducted to integrate NOTCH1–ROS1, Trunc. NOTCH1–ROS1, NOTCH1, and ROS1 into Gateway-compatible N-MYC-tagged pMX-Puro Retroviral Vector (Cell Biolabs). NIH3T3 cells were transduced using a previously described infection protocol (Jain et al. 2017). Ba/F3 cells were transduced with virus generated from the Plat-E retroviral packaging cell line (Cell Biolabs). Prior to infection, cells were treated with 10 µg/mL polybrene. Virus was harvested at 48 h and filtered with a 0.45 µm syringe filter before addition. Upon viral infection and addition of 10 mM HEPES, cells were inoculated for 90 min and then incubated for 72 h. An amount of 2 µg/mL puromycin was then utilized to select stably transduced cells. All constructs were subcloned into gateway destination vectors with amino-terminal Myc and Flag tags Invitrogen). Expression of tagged proteins was detected using anti-Myc HRP Antibody (Invitrogen R951-25; 1:5000) and anti-Flag HRP antibody (Sigma-Aldrich A8592; 1:5000).
Western Blotting and Antibodies
NOTCH1–ROS1 expressing NIH3T3 and HEK293 cells were lysed in RIPA buffer and normalized using a Pierce 660-nm Protein Assay. Lysates were run on NuPAGE precast gels (4%–12% Bis-Tris or 3%–8% Tris-Acetate; Invitrogen). For MAPK, PI3K, and JAK/STAT pathway analysis, pMEK (#9154), tMEK (#4694), pERK (#4370), pERK (#9101), tERK (#4695), tERK (#4696), pAKT Thr308 (#4056), pAKT Ser473 (#9271), pAKT Ser473 (#4060), tAKT (#2920), pS6 (#4858), tS6(#2317), pSTAT3 (#9145), tSTAT3 (#9139), pROS1 Tyr2274 (#3078), tROS1 (#15027), and GAPDH (#3683) antibodies were purchased from Cell Signaling (1:1000).
Soft Agar Colony–Forming Assays
Anchorage independent growth capability of stably expressing NIH3T3 cells was evaluated via soft agar colony formation assay. NIH3T3 cells expressing NOTCH1–ROS1, Trunc. NOTCH1–ROS1, NOTCH1, ROS1, and retroviral vector control were plated in 0.7% agar with DMEM and DBS in cell-repellent 96-well plates in n = 5 and n = 3 for baseline and drug studies, respectively. Cell colonies could form for 2 wk with media changed every 3 d and imaged. Image analysis was performed with ImageJ in which colonies of >500 pixels were included in the count.
IL-3 Withdrawal Assay
Oncogenicity of Notch-ROS1 fusion was assessed via IL-3 withdrawal assay performed in Ba/F3 cells, as described previously (Marks et al. 2019). Briefly, cells expressing no transgene, NOTCH-ROS1, Trunc. Notch-ROS1, CD74-ROS1, or ROS1 were suspended at 500,000 cells/mL in RPMI + 10% FBS without addition of IL-3. Cells were manually counted every 3–4 d until cells either died out or grew 100-fold relative to starting cell number.
Coimmunoprecipitation
Protein–protein interactions of NOTCH1–ROS1 with itself, wild-type NOTCH1, and wild-type ROS1 were investigated via cotransfections of Myc and Flag-tagged constructs into HEK293 using manufacturer defined protocol for Lipofectamine 3000 (Invitrogen). Anti-Flag antibody coated beads (Millipore-Sigma M8823) were used to immunoprecipitate the tagged proteins during rotation at 2 h at 4°C. Three 15-min washes using RIPA buffer were performed at 4°C followed by a PBS wash and elution using 2× LDS (lithium dodecyl sulfate). Following elution, samples were heated for 10 min to 70°C and analyzed through western blotting.
Nanostring Pathway Analysis
Nanostring pathway analysis was conducted with the nCounter PanCancer Pathways Panel CodeSet (XT-CSO-PATH1-12, Nanostring) containing 770 human genes from 13 pathways commonly associated with cancer. Multiplex gene expression analysis was performed according to published protocol for using isolated RNA. Isolated RNA was obtained from stably expressing NIH3T3 cells utilizing the RNeasy Plus Mini Kit (QIAGEN 74134) and was analyzed for RNA integrity using a bioanalyzer before use. Expression data was analyzed using the nSolver Data Analysis Software Suite with heatmaps being generated through the advanced analysis add-on.
Subcellular Fractionation Assay
Subcellular fractionation was performed using the Subcellular Protein Fractionation Kit for Cultured Cells (Thermo Scientific 78840) according to manufacturer's protocol with noted changes. Wash step with cytoplasmic extraction buffer was performed 2× after initial extraction of cytoplasmic fraction to reduce contamination of the membrane fraction with residual cytoplasmic extract. Samples were analyzed through western blotting using tROS1 (#15027), HSP90 (#4877), and Na,K-ATPase (#3010) antibodies purchased from Cell Signaling (all 1:1000), in addition to the anti-Myc Antibody from Invitrogen (R951-25, 1:5000).
Biotinylation Assays
Biotinylation was performed using the EZ-Link Sulfo-NHS-SS-Biotin reagent (Thermo Scientific 21331) adhering to the manufacturer's protocol with minor changes. Cells were incubated with biotin at 4°C to reduce cellular internalization of reagent. An initial wash with 25 mM Tris-HCl (pH 8.0) was also performed postbiotinylation to quench unreacted biotin reagent. Pierce NeutrAvidin Agarose resin was then used to bind biotinylated cell surface proteins and washed to remove unbound intracellular proteins. Samples were analyzed through western blotting using tROS1 (Cell Signaling15027; 1:1000) and anti-GAPDH (Origene TA802519; 1:7000).
Cellular Drug Assays
Entrectinib was provided by Ignyta, whereas crizotinib and RO4929097 were purchased from Selleck Chemicals. GW280264X was purchased from AOBIUS Inc. Aderbaib was purchased from Astatech Inc. Cells were plated at 1 × 106 cells/well in a six-well plate and serum starved for 24 h followed by exposure to specific drug for 1 h. All drugs were dissolved in dimethyl sulfoxide and stored at −20°C. Ba/F3 cell viability assays were performed as described previously (Davare et al. 2015). Briefly, inhibitors were loaded at a 2× concentration onto 384-well plates using a D300 Digital Dispenser (HP) and then NOTCH-ROS1 cells post-IL-3 withdrawal were seeded onto the drug-printed plates using a Multidrop Combi Reagent Dispenser (Thermo Scientific). Plates were incubated for 72 h at 37°C; 5% CO2 and then viability was assessed via MTS (BioVision), in which absorbance was quantified using a Biotek Synergy H1 microplate reader. IC50 determination and nonlinear curve fitting were subsequently performed with GraphPad Prism software. Experiments were performed at minimum two separate times in triplicate.
Animal Studies
The Children's Hospital of Philadelphia Institutional Animal Care and Use Committee approved all associated animal protocols. Homozygous NSG mice were purchased from Jackson Laboratories and housed under aseptic conditions. Mice used were 5-wk-old and randomized for treatment with equal sex representation and no investigator blinding.
Mouse flank xenograft studies with NOTCH1–ROS1 fusion expressing NIH3T3 cells: NIH3T3 cell lines were injected subcutaneously into NSG mouse flanks (baseline growth study n = 4 per cell line; entrectinib study n = 8 per treatment arm). Tumor growth was measured daily and tumor volume calculated using the formula
volume = (π/6) × (L × W × H/3)3.
Quantification of Entrectinib in Plasma (Pharmacokinetic Analysis)
Entrectinib was extracted from mouse plasma samples with an isotopically labeled internal standard (entrectinib-d8; Ignyta) by liquid–liquid extraction using acetonitrile. Extracts were injected onto an Xbridge C18 column (50 × 2.1 mm, 2.5 mm; Waters Corporation), maintained at 40°C, and eluted with a binary mobile phase gradient using 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B) with a constant flow rate of 0.4 mL/min. The initial mobile phase condition of 90:10, A:B, was held until 0.1 min, when the composition changed to 5:95, A:B gradually until 0.5 min. From 0.5 min to 2.0 min, the mobile phase held at 5:95, A:B, then reverted back to the initial conditions (90:10, A:B) at 2.15 min, then held at these conditions until 6 min. Positive ion tandem mass spectrometry (MS/MS) with multiple reaction monitoring (m/z 561.2 to m/z 302.1 for entrectinib) was used for analyte detection. The lower limit of quantification (LLQ) for entrectinib was 2 ng/mL for a 15-mL aliquot of mouse plasma with a higher limit of quantification (HLQ) of 1000 ng/mL.
ADDITIONAL INFORMATION
Data Deposition and Access
Our genomic findings have been submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession number VCV001710089.1.
Ethics Statement
Review of patient data was approved by the Children's Hospital of Pennsylvania (CHOP) institutional review board (17-013802)
Acknowledgments
We are thankful to the patient and her family for their participation in this study and the Children's Brain Tumor Network for their support of this study.
Author Contributions
P.J., A.J.W., A.R., and M.D. conceptualized the design of the study. P.J., J.S., and S.I. conducted the in vitro investigations, and P.J., J.S., and T.S. conducted in vivo experiments. M.L, L.F.S., and J.P. provided a pathologic review of the study. C.B. and E.F. conducted the pharmacokinetic (PK) study. P.J., J.S., H.H., and S.I. performed data interpretation and analysis. P.J., J.S., M.D., and S.I. wrote and edited the manuscript. All authors reviewed, contributed to, and approved the final manuscript.
Funding
This research was supported by funds from the Children's Brain Tumor Network at the Children's Hospital of Philadelphia, a St. Baldrick's Summer Fellow, Award, and funds from the OHSU Doernbecher Children's Hospital Foundation.
Competing Interest Statement
The authors have declared no competing interest.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Ayadi L, Khabir A. 2010. Pediatric angiosarcoma of soft tissue: a rare clinicopathologic entity. Arch Pathol Lab Med 134: 481–485. 10.5858/134.3.481 [DOI] [PubMed] [Google Scholar]
- Biskup C, Böhmer A, Pusch R, Kelbauskas L, Gorshokov A, Majoul I, Lindenau J, Benndorf K, Böhmer FD. 2004. Visualization of SHP-1-target interaction. J Cell Sci 117: 5165–5178. 10.1242/jcs.01397 [DOI] [PubMed] [Google Scholar]
- Botti G, Scognamiglio G, Marra L, Pizzolorusso A, Di Bonito M, De Cecio R, Cantile M, De Chiara A. 2017. Programmed death ligand 1 (PD-L1) expression in primary angiosarcoma. J Cancer 8: 3166–3172. 10.7150/jca.19060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvete O, Martinez P, Garcia-Pavia P, Benitez-Buelga C, Paumard-Hernández B, Fernandez V, Dominguez F, Salas C, Romero-Laorden N, Garcia-Donas J, et al. 2015. A mutation in the POT1 gene is responsible for cardiac angiosarcoma in TP53-negative Li–Fraumeni-like families. Nat Commun 6: 8383. 10.1038/ncomms9383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick ML, Lane A, Thomas D, Smith AR, White AR, Davidson D, Feng Y, Boscolo E, Zheng Y, Adams DM, et al. 2018. Combined mTOR and MEK inhibition is an effective therapy in a novel mouse model for angiosarcoma. Oncotarget 9: 24750–24765. 10.18632/oncotarget.25345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davare MA, Vellore NA, Wagner JP, Eide CA, Goodman JR, Drilon A, Deininger MW, O'Hare T, Druker BJ. 2015. Structural insight into selectivity and resistance profiles of ROS1 tyrosine kinase inhibitors. Proc Natl Acad Sci 112: E5381–E5390. 10.1073/pnas.1515281112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson L, Remenyi I, Tusnady GE. 2015. CCTOP: a Consensus Constrained TOPology prediction web server. Nucleic Acids Res 43: W408–W412. 10.1093/nar/gkv451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drilon A, Jenkins C, Iyer S, Schoenfeld A, Keddy C, Davare MA. 2021. ROS1-dependent cancers: biology, diagnostics and therapeutics. Nat Rev Clin Oncol 18: 35–55. 10.1038/s41571-020-0408-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchinetti F, Friboulet L. 2019. Profile of entrectinib and its potential in the treatment of ROS1-positive NSCLC: evidence to date. Lung Cancer (Auckl) 10: 87–94. 10.2147/LCTT.S190786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari A, Casanova M, Bisogno G, Cecchetto G, Meazza C, Gandola L, Garaventa A, Mattke A, Treuner J, Carli M. 2002. Malignant vascular tumors in children and adolescents: a report from the Italian and German Soft Tissue Sarcoma Cooperative Group. Med Pediatr Oncol 39: 109–114. 10.1002/mpo.10078 [DOI] [PubMed] [Google Scholar]
- Garcia JM, Gonzalez R, Silva JM, Dominguez G, Vegazo IS, Gamallo C, Provencio M, España P, Bonilla F. 2000. Mutational status of K-ras and TP53 genes in primary sarcomas of the heart. Br J Cancer 82: 1183–1185. 10.1054/bjoc.1999.1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomini CP, Sun S, Varma S, Shain AH, Giacomini MM, Balagtas J, Sweeney RT, Lai E, Del Vecchio CA, Forster AD, et al. 2013. Breakpoint analysis of transcriptional and genomic profiles uncovers novel gene fusions spanning multiple human cancer types. PLoS Genet 9: e1003464. 10.1371/journal.pgen.1003464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Italiano A, Chen CL, Thomas R, Breen M, Bonnet F, Sevenet N, Longy M, Maki RG, Coindre JM, Antonescu CR. 2012. Alterations of the p53 and PIK3CA/AKT/mTOR pathways in angiosarcomas: a pattern distinct from other sarcomas with complex genomics. Cancer 118: 5878–5887. 10.1002/cncr.27614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain P, Fierst TM, Han HJ, Smith TE, Vakil A, Storm PB, Resnick AC, Waanders AJ. 2017. CRAF gene fusions in pediatric low-grade gliomas define a distinct drug response based on dimerization profiles. Oncogene 36: 6348–6358. 10.1038/onc.2017.276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeng MR, Fuh B, Blatt J, Gupta A, Merrow AC, Hammill A, Adams D. 2014. Malignant transformation of infantile hemangioma to angiosarcoma: response to chemotherapy with bevacizumab. Pediatr Blood Cancer 61: 2115–2117. 10.1002/pbc.25067 [DOI] [PubMed] [Google Scholar]
- Kamihara J, Rana HQ, Garber JE. 2014. Germline TP53 mutations and the changing landscape of Li–Fraumeni syndrome. Hum Mutat 35: 654–662. 10.1002/humu.22559 [DOI] [PubMed] [Google Scholar]
- Katoh M, Katoh M. 2020. Precision medicine for human cancers with Notch signaling dysregulation (review). Int J Mol Med 45: 279–297. 10.3892/ijmm.2019.4418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar-Sinha C, Kalyana-Sundaram S, Chinnaiyan AM. 2015. Landscape of gene fusions in epithelial cancers: seq and ye shall find. Genome Med 7: 129. 10.1186/s13073-015-0252-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunnimalaiyaan M, Chen H. 2007. Tumor suppressor role of Notch-1 signaling in neuroendocrine tumors. Oncologist 12: 535–542. 10.1634/theoncologist.12-5-535 [DOI] [PubMed] [Google Scholar]
- Lieber T, Kidd S, Alcamo E, Corbin V, Young MW. 1993. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev 7: 1949–1965. 10.1101/gad.7.10.1949 [DOI] [PubMed] [Google Scholar]
- Marks EI, Pamarthy S, Dizon D, Birnbaum A, Yakirevich E, Safran H, Carneiro BA. 2019. ROS1-GOPC/FIG: a novel gene fusion in hepatic angiosarcoma. Oncotarget 10: 245–251. 10.18632/oncotarget.26521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazières J, Zalcman G, Crino L, Biondani P, Barlesi F, Filleron T, Dingemans AM, Lena H, Monnet I, Rothschild SI, et al. 2015. Crizotinib therapy for advanced lung adenocarcinoma and a ROS1 rearrangement: results from the EUROS1 cohort. J Clin Oncol 33: 992–999. 10.1200/JCO.2014.58.3302 [DOI] [PubMed] [Google Scholar]
- Murali R, Chandramohan R, Möller I, Scholz SL, Berger M, Huberman K, Viale A, Pirun M, Socci ND, Bouvier N, et al. 2015. Targeted massively parallel sequencing of angiosarcomas reveals frequent activation of the mitogen activated protein kinase pathway. Oncotarget 6: 36041–36052. 10.18632/oncotarget.5936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Takahashi S. 2018. Translocation-related sarcomas. Int J Mol Sci 19: 3784. 10.3390/ijms19123784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel DS, Allegakoen DV, Olivas V, Mayekar MK, Hemmati G, Chatterjee N, Blakely CM, McCoach CE, Rotow JK, Le A, et al. 2019. Differential subcellular localization regulates oncogenic signaling by ROS1 kinase fusion proteins. Cancer Res 79: 546–556. 10.1158/0008-5472.CAN-18-1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter CA, Jain E, Tomson BN, Dunphy M, Stoddard RE, Thomas BS, Damon AL, Shah S, Kim D, Gómez Tejeda Zañudo J, et al. 2020. The Angiosarcoma Project: enabling genomic and clinical discoveries in a rare cancer through patient-partnered research. Nat Med 26: 181–187. 10.1038/s41591-019-0749-z [DOI] [PubMed] [Google Scholar]
- Penel N, Ray-Coquard I, Bal-Mahieu C, Chevreau C, Le Cesne A, Italiano A, Bompas E, Clisant S, Baldeyrou B, Lansiaux A, et al. 2014. Low level of baseline circulating VEGF-A is associated with better outcome in patients with vascular sarcomas receiving sorafenib: an ancillary study from a phase II trial. Target Oncol 9: 273–277. 10.1007/s11523-013-0299-0 [DOI] [PubMed] [Google Scholar]
- Roskoski R Jr. 2017. ROS1 protein-tyrosine kinase inhibitors in the treatment of ROS1 fusion protein-driven non–small cell lung cancers. Pharmacol Res 121: 202–212. 10.1016/j.phrs.2017.04.022 [DOI] [PubMed] [Google Scholar]
- Seki Y, Yamamoto N, Tamura Y, Goto Y, Shibata T, Tanioka M, Asahina H, Nokihara H, Yamada Y, Shimamoto T, et al. 2012. Phase I study for ridaforolimus, an oral mTOR inhibitor, in Japanese patients with advanced solid tumors. Cancer Chemother Pharmacol 69: 1099–1105. 10.1007/s00280-011-1788-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AT, Ou SH, Bang YJ, Camidge DR, Solomon BJ, Salgia R, Riely GJ, Varella-Garcia M, Shapiro GI, Costa DB, et al. 2014. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med 371: 1963–1971. 10.1056/NEJMoa1406766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindhu S, Gimber LH, Cranmer L, McBride A, Kraft AS. 2017. Angiosarcoma treated successfully with anti-PD-1 therapy: a case report. J Immunother Cancer 5: 58. 10.1186/s40425-017-0263-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surrey LF, MacFarland SP, Chang F, Cao K, Rathi KS, Akgumus GT, Gallo D, Lin F, Gleason A, Raman P, et al. 2019. Clinical utility of custom-designed NGS panel testing in pediatric tumors. Genome Med 11: 32. 10.1186/s13073-019-0644-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner MJ, Lyons YA, Siedel JH, Dood R, Nagaraja AS, Haemmerle M, Mangala LS, Chanana P, Lazar AJ, Wang W-L, et al. 2021. Combined VEGFR and MAPK pathway inhibition in angiosarcoma. Sci Rep 11: 9362. 10.1038/s41598-021-88703-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Q, Chan JL, Zong CS, Wang LH. 1996. Two chimeric receptors of epidermal growth factor receptor and c-Ros that differ in their transmembrane domains have opposite effects on cell growth. Mol Cell Biol 16: 1509–1518. 10.1128/MCB.16.4.1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo C, Lee J, Rha SY, Park KH, Kim TM, Kim YJ, Lee HJ, Lee KH, Ahn JH. 2013. Multicenter phase II study of everolimus in patients with metastatic or recurrent bone and soft-tissue sarcomas after failure of anthracycline and ifosfamide. Invest New Drugs 31: 1602–1608. 10.1007/s10637-013-0028-7 [DOI] [PubMed] [Google Scholar]
- Young RJ, Brown NJ, Reed MW, Hughes D, Woll PJ. 2010. Angiosarcoma. Lancet Oncol 11: 983–991. 10.1016/S1470-2045(10)70023-1 [DOI] [PubMed] [Google Scholar]
- Zong CS, Zeng L, Jiang Y, Sadowski HB, Wang LH. 1998. Stat3 plays an important role in oncogenic Ros- and insulin-like growth factor I receptor-induced anchorage-independent growth. J Biol Chem 273: 28065–28072. 10.1074/jbc.273.43.28065 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Our genomic findings have been submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession number VCV001710089.1.