Abstract
A gold nanoparticle (AuNP) labeled CRISPR-Cas13a nucleic acid assay has been developed for sensitive solid-state nanopore sensing. Instead of directly detecting the translocation of RNA through a nanopore, our system utilizes non-covalent conjugates of AuNPs and RNA targets. Upon CRISPR activation, the AuNPs are liberated from the RNA, isolated, and passed through a nanopore sensor. Detection of the AuNPs can be observed as increasing ionic current in the chip. Each AuNP that is detected is enumerated as an event, leading to quantitative of molecular targets. Leveraging the high signal-to-noise ratio enabled by the AuNPs, a detection limit of 50 fM before front-end target amplification is achieved using SARS-CoV-2 RNA segments as a Cas13 target. Furthermore, a dynamic range of six orders of magnitude is demonstrated for quantitative RNA sensing. This simplified AuNP-based CRISPR assay is performed at the physiological temperature without relying on thermal cyclers. In addition, the nanopore reader is similar in size to a smartphone, making the assay system suitable for rapid and portable nucleic acid biomarker detection in either low-resource settings or hospitals.
Keywords: nanopore, CRISPR-Cas13a, point-of-care, biosensing, diagnostics
1. Introduction
In the past two decades, there has been an increasing frequency of emerging viral infections, including SARS, Ebola, and Zika, causing significant public health concerns.[1] The incredibly swift global spread of COVID-19 re-emphasizes the critical need for rapid and sensitive molecular diagnostics to combat current and future pandemics. Sensitive polymerase chain reaction (PCR) based tests are the gold standard for molecular diagnostics but rely on bulky and expensive instruments. Thus, they are not suitable for self-diagnosis or point-of-care (POC) settings.[2] High-throughput sequencing can decipher the entire genomic landscape of the pathogens but is time-consuming and requires bioinformatics for data interpretation.[3] Immunoassays, such as rapid antigen tests, are simple and rapid diagnostic methods but normally lack the sensitivity to reporting the low concentration biomarkers.[4] Assays with high limits of detection and low accuracy have been acceptable out of necessity, but there are many scenarios where a rapid, sensitive, POC device would be beneficial. Therefore, developing a simple to use, portable, and sensitive diagnostic platform is one important key to addressing the current challenges for molecular diagnostics.
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) are bacterial systems evolved to combat bacteriophage infections by recognizing specific nucleic acid sequences to activate nucleolytic cleavage activities. The trans-cleavage of RNA and DNA molecules by various CRISPR systems has been adapted for a variety of molecular uses, including the design of sensitive novel diagnostic platforms.[5] An enzyme turnover rate of ~1-10 molecules per second has been demonstrated with the CRISPR-Cas13a complex, thus significantly enhancing the sensitivity of RNA detection.[6] Further, through combination with either isothermal amplification or conventional PCR, the CRISPR platforms demonstrate unprecedented pathogen detection sensitivity.[7-9] Lateral flow strips based on CRISPR-Cas12a and -Cas13a have been developed for simple and sensitive detection of SARS-CoV-2 and other infectious diseases without a complicated sample extraction process.[10,11] However, even though paper-based strips are highly sensitive, they cannot be used for quantitative detection, which is crucial for monitoring disease progression in patients.[12]
In recent years, solid-state nanopore sensing has attracted increasing attention in detecting single molecules with many advantages, including controllable feature size, high sensitivity, simple readout, and label-free electronic sensing.[13,14] The technology has been applied to many areas such as DNA sequencing,[15,16] protein detection,[17] nanoparticle separation,[18] and energy conversion.[19] The principle behind nanopore detection is resistive pulse sensing. The ionic current either decreases due to the presence of an analyte which hinders the access of the ions to the nanopore volume or increases when metal nanoparticles are passing through the nanopore and enhance the ionic current due to the high conductivity.[20] Highly sensitive and quantitative detection can be achieved by analyzing the features of the current, such as duration time, amplitude, and signal frequency. CRISPR assays have shown outstanding competence in highly specific nucleic acid targeting and have been used in various nanopore sensing platforms. For instance, Nouri et al. developed a kind of solid-state CRISPR-Cas12a-assisted nanopores (SCAN) to specifically detect the HIV-1. The glass nanopore sensor is effective in monitoring the cleavage activity of the target DNA-activated Cas12a.[21] In addition, Nicole E. et al. demonstrated the use of a highly specific dCas9 probe to create unique barcodes on DNA that nanopore sensors can read. Multiple dCas9 probes are available to create characteristic structural patterns on DNA. These sequence-specific structures simultaneously recognize different DNA targets in a mixture, demonstrating an important step toward rapid DNA identification.[22]
Even though solid-state nanopores can directly detect DNA/RNA in solution,[23,24] a tagging molecule is often introduced to enhance the signal to noise ratio for the translocation.[25,26] In addition, a stringent pre-purification process must be included as the impurities present in the sample can disrupt the ionic current and cause false-positive signals. In the present study, we leverage the strong ionic signal pulse caused by gold nanoparticles (AuNPs) in the detection of viral RNA. A sensitive CRISPR-Cas13a assay has been developed to recognize the SARS-Cov-2 target RNA. After magnetic bead separation, the CRISPR-Cas13 cleaved AuNPs are introduced into a solid-state nanopore reader for detection. Here, we show that our strategy can differentiate between similar viral RNA strains (specificity) with a detection limit of 50 fM (sensitivity), covering six orders of magnitude (quantitative range). Our results established a key step towards the implementation of a portable, rapid, and highly quantitative nucleic acid detection technology in POC settings.
2. Results
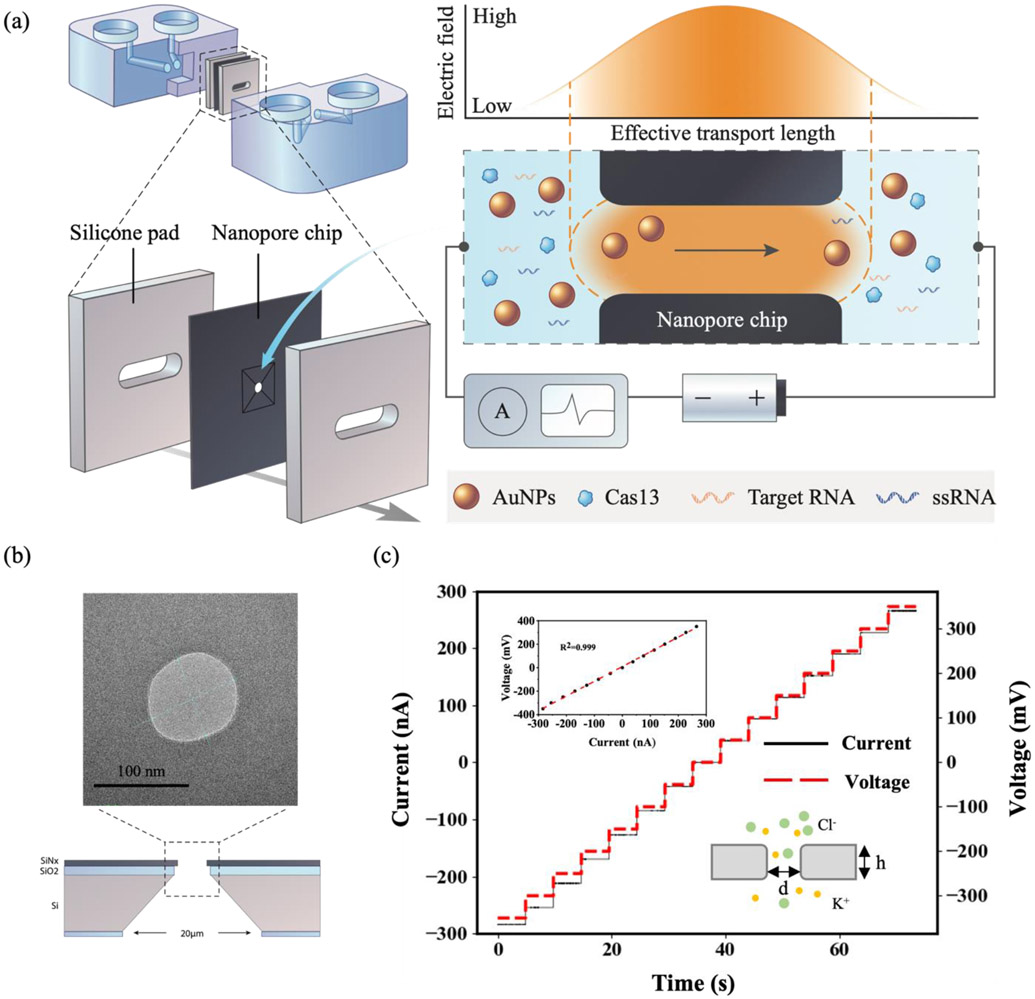
The approach for solid-state nanopore sensing is shown in Figure 1a. A SiNx nanopore chip is sandwiched between two microfluidic reservoirs shown in blue. Two AgCl electrodes are immersed in the reservoirs on both sides, and a constant voltage (−700 mV to 700 mV) is applied across the nanopore, causing a steady-state ionic-current flux at the picoamp (PA) level through the nanopore. The translocation of the free AuNPs changes the ionic current and is detected by the nanopore reader shown in the diagrams to the right. The free-standing SiNx membrane (thickness: 20 ± 3 nm) sits on a 200 μm thick silicon substrate with a frame size of 5 × 5 mm. A 60 nm thick SiO2 layer was deposited between the silicon substrate and the SiNx membrane to reduce the electrical noise. The nanopore (diameter: 90 nm) was prepared with a helium ion microscope (Figure 1b),[27] the pore is visible as a round opening of approximately 90 nm by transmission electron microscopy (TEM). Before introducing AuNPs into the chip, the open-pore current in 1 M KCl solution was analyzed to characterize the nanopore chip and the nanopore reader. As shown in Figure 1c, a linear relationship between the ionic current and the applied voltage in the range of −400 mA to 400 mA was recorded (R2 =0.999) in 1 M KCl with a corresponding pore conductance G, calculated to be 732.1 nS. The ionic conductance through an open solid-state nanopore can be calculated by
| (1) |
where σ is the solution conductivity, l is the membrane thickness, and d is the nanopore diameter.
Figure 1.
(a) Schematic of the solid-state nanopore sensing platform. The nanopore chip is sandwiched between two microfluidic reservoirs (blue) and silicone pads. The trans-cleavage of CRISPR-Cas13a releases the gold nanoparticles to pass through the solid-state nanopore and changes the ionic current. (b) TEM image of the nanopore chip with a pore diameter of 90 nm with diagram showing cross-section. (c) A representative current–voltage (I–V) calibration curve of a ∼90 nm nanopore in 1 M KCl. The inset shows the liner fits yield a conductance of 732.1 nS and the diagram of KCl buffer passing through the nanopore.
Here, l is 20 nm and σ is 10.5 S/m in 1 M KCl at 23°C.[28] The calculated nanopore diameter is 89.5 nm, which is consistent with the TEM characterization.
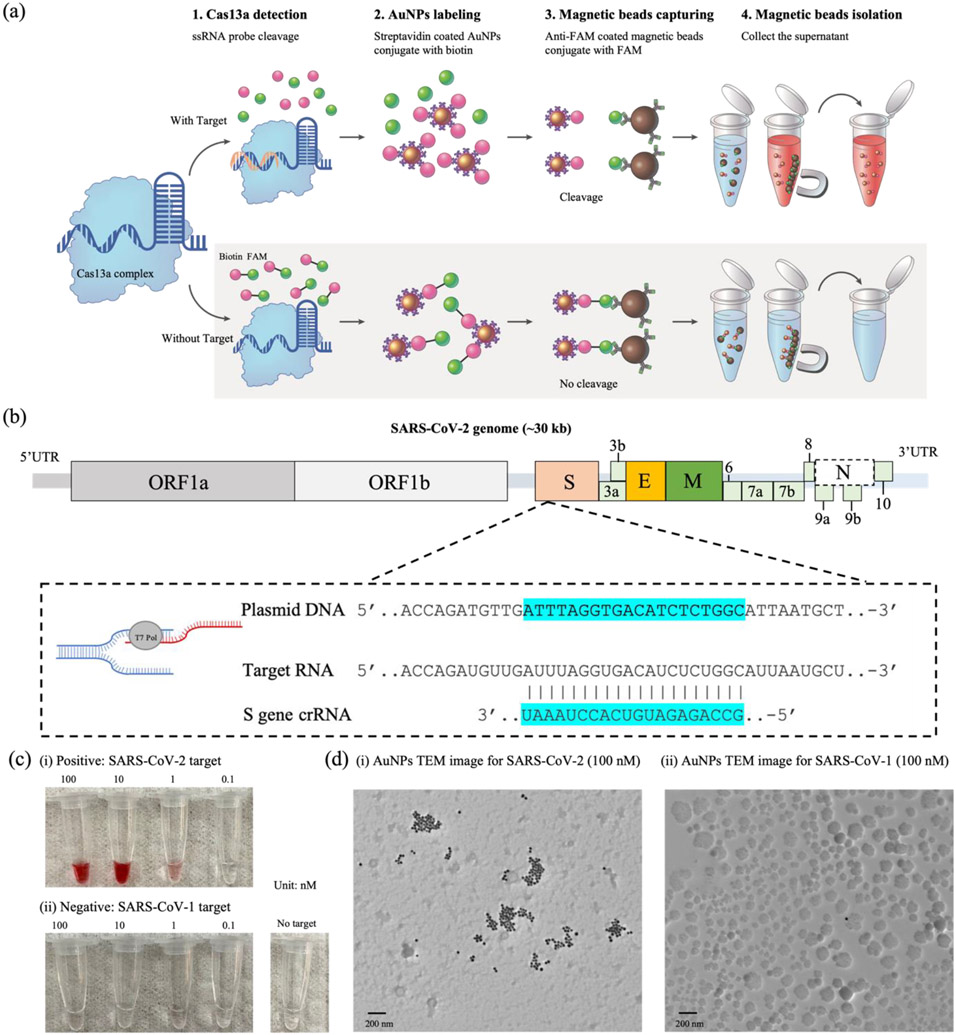
The schematic of the AuNP-Cas13 detection process is shown in Figure 2a. Lbu-Cas13a protein and crRNA are pre-mixed to form a Cas13a:crRNA complex. In this case, the crRNA is specific for a nucleotide sequence within the S reading frame of SARS-CoV-2. Then, biotin-FAM-labeled ssRNA probes are introduced into the mixture (step 1). Next, Streptavidin-coated AuNPs are added to the assay (step 2). The presence of the target RNA (SARS-CoV-2) activates the Cas13a:crRNA complex, cleaving the ssRNA probes in the solution. In the absence of the target RNA, the un-cleaved ssRNA probes are captured by anti-FAM coated magnetic beads (step 3) and isolated by a super-magnet (step 4). Since the ssRNA probe cleavage is correlated to the target concentration, the amount of AuNPs presented in the supernatant can be used for quantitative RNA target sensing. The target and the guide RNA sequences are listed in Figure 2b. The SARS-CoV-2 target (703 nts) was selected from the plasmid pUC57-SARS-CoV-2 (spike S gene). A 20-mer guide RNA was selected to recognize a 20-nt sequence within the SARS-CoV-2 target. To characterize the CRISPR assay, we utilized an established RNase Alert kit by using SARS-CoV-1 as a negative control (Figure S1). The photographs of the cleaved products contained within microtubes are shown in Figure 2c (i). The red color is observed for positive samples with a concentration above 0.1 nM. On the other hand, for negative samples, the collected supernatants do not show any color, regardless of the input target concentration (Figure 2c (ii)). The results were further confirmed by TEM imaging (Figure 2d). For the 100 nM positive sample (Figure 2d (i)), clusters of the AuNPs are easily observed, indicating that the ssRNA probes were cleaved, leaving the AuNPs in the supernatant. In the negative sample (Figure 2d (ii)), it was extremely difficult to observe AuNPs with TEM, indicating that most AuNPs are conjugated on the magnetic beads.
Figure 2.
(a) Schematic of the AuNP-Cas13-based nucleic acid detection strategy: Step 1. Cas13a/crRNA complex cleavage of ssRNA probes; Step 2. Streptavidin-coated AuNPs bind biotin-labeled ssRNA probes. Step 3. FAM-labeled ssRNA probes bind anti-FAM coated magnetic beads; Step 4. Magnetic bead isolation of non-cleaved ssRNA. (b) The SARS-CoV-2 gene locus, target site, and the guide RNA sequence. (c) Images of the AuNPs containing the indicated concentrations of (i) SARS-CoV-2 target RNA (positive) and (ii) SARS-CoV-1 target RNA (negative). (d) TEM images of the cleaved products of (i) 100 nM of SARS-CoV-2 target RNA and (ii) 100 nM of SARS-CoV-1 target RNA. Scale bars are 200 nm.
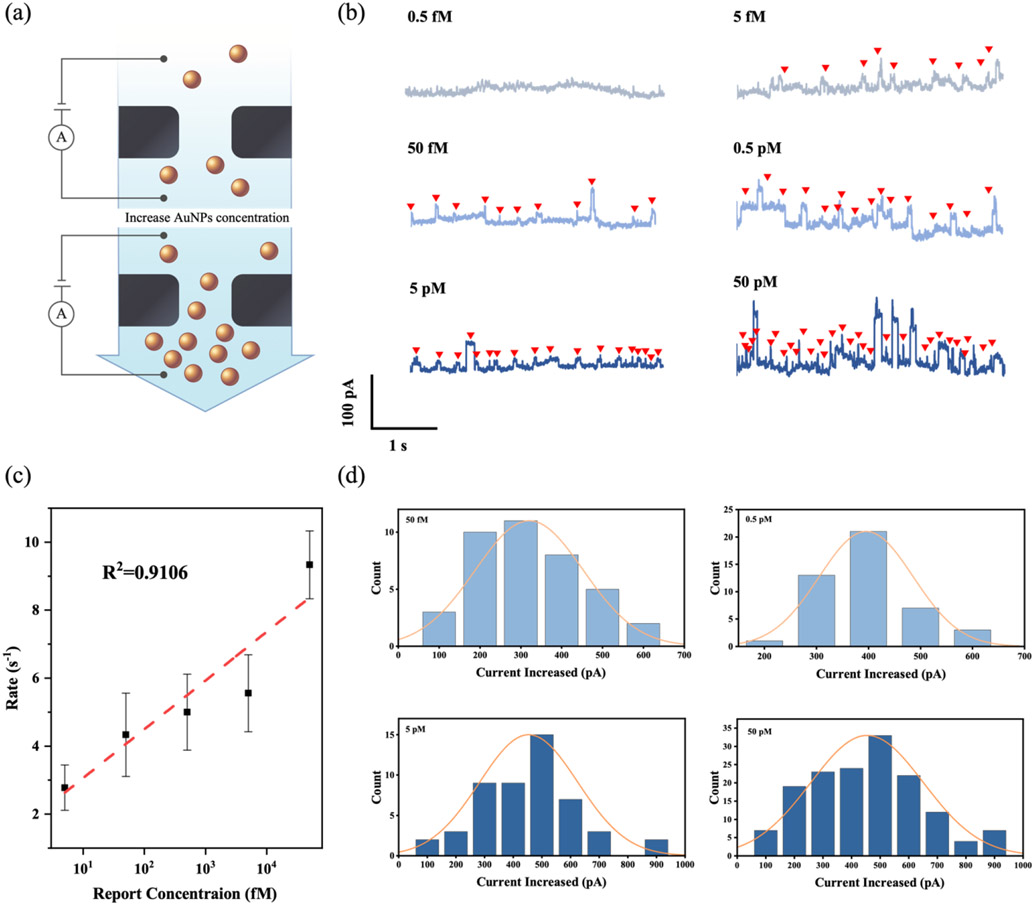
To validate that the nanopore event rate can be used as a quantitative readout for the AuNP and thus target RNA abundance, we performed nanopore counting with serially diluted Streptavidin-coated AuNPs in 1 M KCl solution (Figure 3a). Typical current traces of AuNPs with varying concentrations (0.5 fM to 50 pM) translocated through a 90 nm nanopore are shown in Figure 3b. The upward pulses along the time trace indicate that the negatively charged AuNPs bring new ions into the nanopore, resulting in a temporary increase in conductance.[29] The nanopore chip is small in size and occupies the largest resistance component in the whole circuit. Therefore, when the original solution (buffer solution) is replaced by a more conductive solution, the resistance of the circuit would decrease, resulting in the current increase. The current increases when adding more AuNPs to the solution since the conductivity of the AuNPs is higher than the buffer solution. A quick glance at these traces reveals that the events occur more frequently with the increase of the AuNPs concentration. The extracted event rate exponentially increases with the AuNPs concentration (R2 =0.9106), as shown in Figure 3c. The corresponding histogram of the current increase (ΔI) versus the AuNPs concentration is shown in Figure 3d. The ΔI value is 320.5 ± 132.1 pA for 50 fM AuNPs, 395.5 ± 90.3 pA for 0.5 pM AuNPs, 454.0 ± 171.7 pA for 5 pM AuNPs, and 454.3 ± 197.5 pA for 50 pM AuNPs, indicating that the ΔI increases with AuNP concentration. To further explore the current variation of the AuNPs, we extracted the dwell time and the distribution of event charge deficit (ECD) for the AuNPs with various concentrations (Figure S2). The calculated ECD value does not change with the different concentrations of the AuNPs.
Figure 3.
(a) Schematic of AuNPs (40 nm) passing through a nanopore chip (90 nm) at different concentrations. (b) Typical current-time traces of AuNPs (0.5 fM, 5 fM, 50 fM, 0.5 pM, 5 pM, and 50 pM) passing through a 90 nm nanopore with arrows showing detection events. (c) Nanopore event rate versus AuNP concentration ranging from 0.5 fM to 50 nM. (d) The histograms of the distribution of current increase corresponding to the translocations of AuNPs with various concentrations, fitted to Gaussian curves.
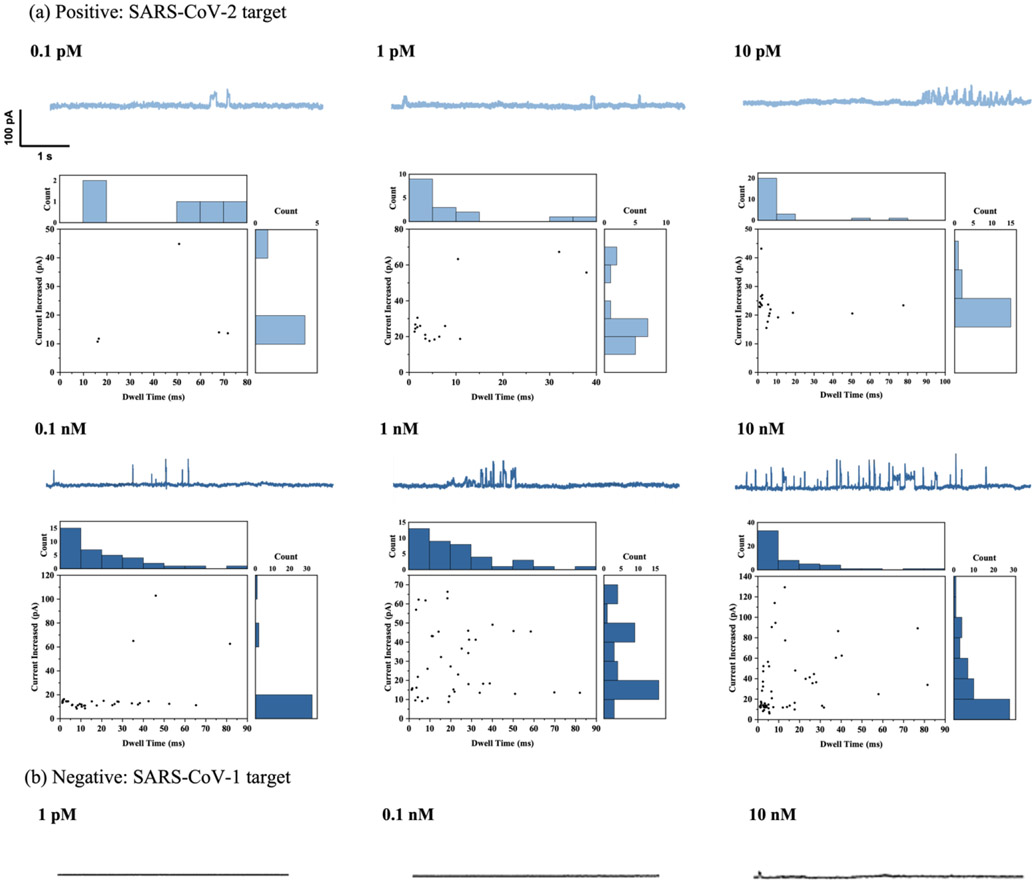
After verifying the relationship between the AuNPs and the nanopore event rate, RNA sensing was performed by varying the RNA concentration from 0.1 pM to 10 nM while setting the AuNPs concentration at 5.5 nM. The current traces were recorded using a single applied voltage (700 mV) for 5 min after the signal appeared. Multiple specific periods were analyzed for each concentration. Figure 4a shows the detected current trace of AuNPs with various SARS-CoV-2 RNA target concentrations and the corresponding ion current versus the observed translocation time. More current traces of AuNPs were detected with increasing target RNA concentrations. The current trace of the AuNPs for negative control (SARS-CoV-1) is shown in Figure 4b. A flat control signal was detected at most concentrations, and we only found a very low frequency of AuNPs in the 10 nM negative sample.
Figure 4.
(a) Ionic current vs. time trace for translocation events count, dwell time histograms, count and current histograms, and scatterplots of current and dwell time for different concentrations of SARS-CoV-2 target, labeled by AuNP-Cas13a assay. (b) Current trace of different concentrations of SARS-CoV-1 target (negative control).
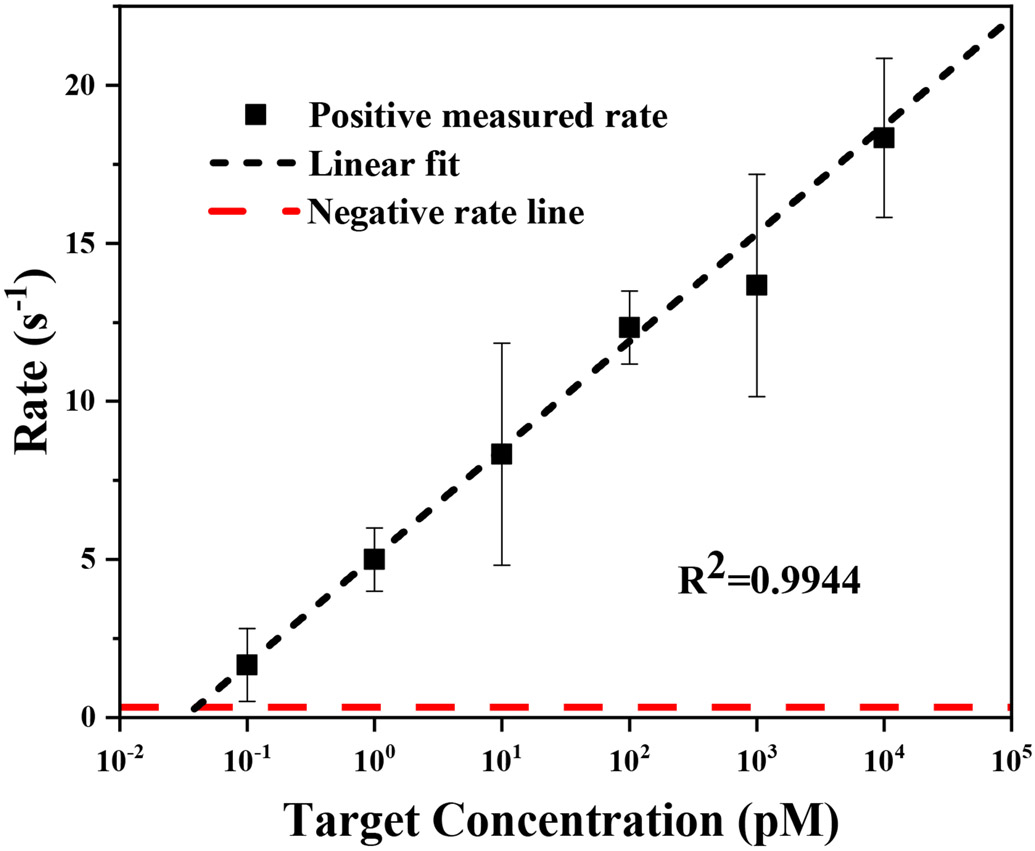
To quantify the target RNA concentration, a calibration curve of extracted rate versus AuNPs concentration was constructed (Figure 5). A linear relationship was found between the extracted rate and the logarithm of the AuNP concentration (R2=0.9944). The scattering intensity of the AuNPs increases with the increasing target RNA concentration (black curve). We are able to detect the signal from the AuNPs when the target concentration is low as 100 fM, thus achieving a highly sensitive detection. In contrast, the negative samples show a flat line and do not increase with the concentration. Extrapolating the positive measured rate and comparing it with the negative control suggests a detection limit of ~50 fM.
Figure 5.
Nanopore event rate versus RNA concentration (50 fM to 50 nM): SARS-CoV-2 (black); SARS-CoV-1 (red). (R2=0.9944)
3. Discussion
The CRISPR-enabled solid-state nanopore sensing established here shows great sensitivity and specificity for viral RNA sensing. Without front-end target amplification, a detection limit of 50 fM (30,000 copies/μL) is achieved by the unique trans-cleavage property of the CRISPR-Cas13a assay. This detection limit is close to the required sensitivity for rapid screening of SARS-CoV-2.[30] Moreover, our protocol can be integrated with established reverse transcription-loop-mediated isothermal amplification (RT-LAMP) and reverse transcription-recombinase polymerase amplification (RT-RPA) methods to extend the detection limit further by several orders of magnitude.[31,32]
Our solid-state nanopore sensing platform requires only a smartphone-sized nanopore reader designed for POC applications. Simple and inexpensive, CRISPR-based lateral flow strips have been developed to diagnose SARS-CoV-2, but these tests cannot provide quantitative results. On the other hand, qPCR (quantitative polymerase chain reaction) is ultra-sensitive and quantitative but relies on bulky instruments and cannot be used in POC settings or for frequent self-diagnosis.[33] Our strategy can differentiate SARS-CoV-2 and SARS-1 with femtomolar level sensitivity with a wide dynamic range of six orders of magnitude, ideally for accurate detection and frequent testing to monitor the viral load in the patients. Furthermore, since the nanopore chips are made by conventional microfabrication, numerous nanopores can be patterned on a single substrate. Coupling with the commercially available miniaturized multi-channel amplifiers, our system can enable a highly multiplexing diagnostic instrument. However, we realized that pore-to-pore variation might affect the detection limit. For future POC testing, all the nanopores will need to be calibrated with reference samples.
Due to the small feature size of the nanopores, the presence of debris and air bubbles can prevent the solution and target particles from flowing through the nanopore. Therefore, a pre-wetting and cleaning process is crucial for the stable translocation of the AuNPs. We found that treating the chip with a 5 min sonication, followed by washing with DI water (deionized water) and IPA (Isopropyl alcohol) is sufficient to wet and clean the chip. In our case, 40 nm AuNPs are chosen for this CRISPR assay and are passed through a 90 nm nanopore. The nanoparticle and nanopore size combination ensure the sensitive trace tracking of single AuNPs without causing nanopore blocking issues. With the same nanoparticle size, we determined that a larger nanopore can indeed reduce the blocking, but it also compromises the sensitivity. The mechanism of nanopore detection is to detect the concentration of nanoparticles by detecting the current change when nanoparticles pass through the nanopore at a certain time. When the nanopore size is too large in proportion to the size of the nanoparticles, most of the nanopore space is not affected when the nanoparticles pass through the nanopore. Therefore, the current change is not obvious, and it is difficult to distinguish it from the signal fluctuation caused by artefacts, such as current noise. For example, many translocation events were not distinguished from the background with the 150 nm nanopore but were apparent with the 90 nm nanopore (Figure S4).
In this work, the upper bound of the voltage setting is 700 mV, and the maximum current value that can be read is 200 nA. In the future, the sensitivity of solid-state nanopore sensing can be further improved by upgrading the electronic sensing circuit. Since the driving force of AuNPs movement comes from the electrical potential difference of the electrodes, increasing the voltage at both ends will increase the number and frequency of nanoparticles passing through the nanopore. In addition, increasing the current by either increasing the applied voltage or changing the buffer concentration can also increase the signal-to-noise ratio as the translocation signals from the background noise can be better identified.[34,35]
For the trace of the CRISPR-cleaved AuNPs, the histogram of the current increase does not fit a Gaussian distribution. There could be two reasons for this: 1) Impurities, such as CRISPR-Cas13 complexes and nucleic acid probes in the sample, can affect the translocation signal of the AuNPs. In contrast, the current signal of the pure AuNPs (Figure 3) fits the Gaussian distribution well because there are no impurities in the solution; 2) For CRISPR experiments, the AuNPs concentration in the solution is low. Since our data model will only fit the Gaussian distribution with a large amount of data, the statistical analysis is likely to deviate when the AuNP concentration is low. In the future, we will focus on improving our data model to interpret the data more effectively.[36]
One unique advantage of the CRISPR-Cas13a assay is that the guide RNA sequence can be programmed to detect a specific target RNA. In this work, we designed a guide sequence for SARS-CoV-2, and this guide showed great specificity against the SARS-CoV-1 negative control. This strategy can be extended to detecting other diseases, such as sepsis, cancer, and genetic disorders, by using nucleic acids as biomarkers.[37] The combination of the high specificity of the AuNP-based CRISPR assay with a sensitive solid-state nanopore sensor can provide an appealing alternative for nucleic acid detection methods. Finally, it is worth noting that the experimental setup presented here may be useful for studying the dynamics of microscopic particles, as well as the change of conductivity of microscopic particles in different solutions.[38]
4. Experimental Section
Purification of Lbu-Cas13a:
Purification of Lbu Cas13a was purified as previously described with some alterations.[39] Briefly, E. coli cells (Rosetta 2) were grown at 37°C with shaking at 180 rpm in LB broth supplemented with 0.5% glucose, 100 μg/mL ampicillin, and 5 μg/mL chloramphenicol, and induced with 0.5 mM IPTG at 0.8 OD. Cells were harvested after 16 hr of growth and resuspended in lysis buffer (50 mM HEPES pH 7.0, 500 mM NaCl, 5% glycerol, 1 mM DTT, 0.5 mM PMSF, and protease inhibitor). Cells were lysed by sonication and the resulting lysate was clarified by centrifugation at 4°C. Lbu-Cas13 was isolated through affinity chromatography using HisPur™ Ni-NTA Resin (Thermo) and the Cas13-containing eluate was dialyzed overnight at 4°C in dialysis buffer (50 mM HEPES pH 7.0, 250 mM NaCl, 5% glycerol, 1 mM DTT) and in the presence of TEV protease to cleave the His-MBP tag. Cas13 was further purified through cation exchange using a linear KCl gradient (0.25 M – 1 M) on a HiTrap SP HP column (GE) and ÄKTA pure FPLC system (GE). The Cas13-containing fractions were pooled, concentrated, and resolved by gel filtration on a Superdex™ 200 Increase 10/30 GL column (GE) using gel filtration buffer (20 mM HEPES pH 7.0, 200 mM KCl, 5% glycerol, 1 mM DTT). The purified protein was concentrated and stored at −80°C for subsequent experiments. Protein purity was assessed by SDS-PAGE and is shown in Figure S5.
Lbu-Cas13a Activity Assay:
Purified Lbu-Cas13a was tested for activity by a radioactive cleavage assay. Briefly, 100 or 1,000 nM Cas13a was incubated with 10% excess of guide RNA in cleavage buffer (20 mM HEPES pH 6.8, 50 mM KCl, 5 mM MgCl2, 10 μg/ BSA, 100 μg/mL yeast tRNA, 0.01% Igepal CA-630, 5% glycerol) and incubated at 37°C for 10 min to allow effector: guide complexing. Target RNA with a 5’ gamma ATP end label was then added at a final concentration of 2 nM, and the reactions were incubated at 37°C for 60 min. The reactions were terminated by the addition of 2x formamide loading buffer (95% deionized formamide, 20 mM EDTA pH 8.0, 0.05% w/v bromophenol blue, 0.05% w/v xylene cyanol), resolved by 15% urea-PAGE, and subsequently visualized with using a Typhoon™ 9410 Trio Phosphoimager (Cytiva). Activity assay results are presented in Figure S5.
RNA sequences:
Guide RNA and RNA reporters were purchased from IDT. Inc or synthesized in-house, and the sequences are listed in Table S1.
Preparation of SARS-CoV-2 and SARS-CoV-1 RNA segments:
The target SARS-CoV-2 (703 nucleotides) spike genes and negative control virus SARS-CoV-1 (660 nucleotides) (sequences are listed in Table S1) were amplified from plasmids pUC57-SARS-CoV-1 and pUC57-SARS-CoV-2 via PCR and TA cloned into vector TOPO-TA directly following a T7 promotor (Invitrogen, MA, USA). To generate linear template DNA for RNA synthesis, plasmids were digested at the 3’ terminus of the S coding fragments with HindIII (New England Biolabs, MA, USA), EtOH precipitated and then resuspended in dH2O. RNA fragments were synthesized using linear plasmid DNA as a template via T7 runoff reactions incubated for 4 hr at 37°C (RiboMAX, PROMEGA, WI, USA). Following RNA synthesis, reactions were DNase treated as per the manufacturer’s instructions. Proteins and excess nucleotides were removed by silica gel membrane column purification (RNeasy- QIAGEN, MD, USA). RNA size and initial quantitation was performed via agarose-TBE gel electrophoresis with RiboRuler RNA ladder (ThermoFisher, MD, USA) using densitometry. Quantitation was verified via UV absorption spectroscopy at 260 nm.
CRISPR Lbu-Cas13a trans-cleavage:
We first mixed CRISPR-Cas13a and guide RNA in the following order: RNase-free water (22μL), 6 μl of 5x Standard (STD) Buffer (250mM KCl, 100mM HEPES, 25 mM MgCl2, 5mM DTT, 25% Glycerol, pH 6.8), 1.67 μl of Lbu Cas13a in-stock solution (18 μM), 0.33 μl of guide RNA in-stock solution (100 μM) to a total volume of 30 μL. The mixture was incubated at 37°C for 2 min followed by 8 min at room temperature, then put on ice for later use. We then added 2 μL of the Cas13a-guide RNA complex into 11 μl of RNase-free water, 4 μl of 5x STD buffer, 2 μl of RNA target with different concentrations, and 1 μl of the biotin-fluorescein ssRNA reporter (10 μM) to a total volume of 20 μL. The reaction was incubated at 37°C for 30 min.
Immobilizing RNA reporters onto AuNPs:
Streptavidin-coated AuNPs with a diameter of 40 nm (1.1 nM) were purchased from nano Composix Inc. We concentrated the stock solution by a centrifuge process: we added 500 μL of stock AuNPs in a microcentrifuge tube and spin for 8 min (8000 rpm) and then removed 400 μl of the supernatant to achieve 100 μL (5.5 nM) of AuNPs. We took 10 μL of the concentrated AuNPs and added them into the 20 μL of Cas13-guide RNA-RNA reporter probe mixture. The sample was incubated on a rotary mixer at room temperature for 15 min.
Magnetic bead isolation:
Dynabeads™ MyOne™ Streptavidin C1 with a diameter of 1 μm (10 mg/ml) were purchased from Thermo Fisher Scientific Inc. Biotinylated anti-fluorescein antibody (1 mg/ml) was purchased from Vector Laboratories Inc. Before conjugation, streptavidin-coated magnetic beads were washed three times with 1x PBS buffer. Ten microliters of biotinylated anti-fluorescein antibody (1 mg/ml) were added to the 10 μL Dynabeads solution, followed by incubation at room temperature using a rotary mixer 30 min. After incubation, the beads were washed three times with 1x PBS buffer to remove any unbound anti-fluorescein antibody. Thirty microliters of AuNPs labeled Cas13a reaction products from the last step were then added. The reaction tube was incubated at 37°C for 30 min. After the reaction, the magnetic beads were isolated by a magnet and left the supernatant for nanopore experiments. Alternatively, the Cas13a-based detection reactions could be detected using fluorescence detection assay. Reactions were performed as above, with minor modifications: Briefly, each Cas13-based fluorescence detection reaction was prepared in a 0.6 ml microcentrifuge tube with the following components: 32.5 μl of RNase-free water, 13 μl of 5x STD buffer, 6.5 μl of RNA target with different concentration (positive target: SARS-CoV-2, negative target: SARS-CoV-1), 6.5 μl of Cas13a/crRNA complex, and 6.5 μl of RNase alert substrate (5 μM). The results were collected by a spectrofluorometer (FP-8550 Spectrofluorometer, JASCO Inc.)
SiNx Nanopore Fabrication and Preparation:
The 90 nm diameter SiNx nanopore chips were provided by Norcada Inc. Before each experiment, the nanopore chip was sonicated for 5 min to remove the debris. DI water (deionized water) and IPA (Isopropyl alcohol) were used to wash the chip and dried the sample with high-pressure gas.
Nanopore Sensing and Data Analysis:
The portable nanopore reader (100 kHz bandwidth) was purchased from Elements Inc. The nanopore flow-cell was constructed with two translucent parts made by Delrin [PolyOxyMethylene (POM)]. Each part has a channel and a reservoir to hold the electrolyte. The two translucent parts sandwiched the nanopore chip in the middle and then were assembled with a 15 × 25 mm2 PCB board with integrated Ag/AgCl electrodes. Two microliters AuNPs samples were added into 90 μL of buffer (1M KCl) on each side of the reservoirs. Constant voltage (−700 mV to 700 mV) was applied across the portable nanopore reader in this study. The Elements data reader collected the current data with a frequency of 20k. The Elements data analyzer (data analysis program developed by the Elements. Inc) and a customized Python program was used to extract the single molecule translocation events, the ionic current dip, and the molecule dwell time. Specifically, the data analyzer detects the events by setting the baseline threshold, which is the average value of the noise floor,[40] and the high threshold, which is at least five standard deviations (5σ) above the noise floor. The dwell time of the events which are lower than 0.1 ms or longer than 100 ms is considered not regular dwell time, and the events are discarded.
TEM imaging:
JEOL (JEM-2010, Japan) was used to image the AuNPs (HV=200.0 kV, direct Mag=6 k×).
Supplementary Material
Acknowledgments
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R35GM142763 (to KD) and R35GM133462 (to MRO), UNYTE Pipeline-to-Pilot grant (37161; to KD and MRO), and Shenzhen Ruhan Genetech (37189). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors thank Xian Boles and the RIT PHT180 for the schematic design.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Conflict of Interest
M.R.O is an inventor on patent applications related to CRISPR-Cas systems and uses thereof. M.R.O is a member of the scientific advisory boards for Dahlia Biosciences and LocanaBio, and an equity holder in Dahlia Biosciences and LocanaBio.
References
- [1].Global Disease Detection Timeline, https://www.cdc.gov/globalhealth/infographics/global-health-security/global-disease-detection-timeline.html (accessed: December 2021).
- [2].Vogels CBF, Brito AF, Wyllie AL, Fauver JR, Ott IM, Kalinich CC, Petrone ME, Casanovas-Massana A, Catherine Muenker M, Moore AJ, Klein J, Lu P, Lu-Culligan A, Jiang X, Kim DJ, Kudo E, Mao T, Moriyama M, Oh JE, Park A, Silva J, Song E, Takahashi T, Taura M, Tokuyama M, Venkataraman A, Weizman O-E, Wong P, Yang Y, Cheemarla NR, White EB, Lapidus S, Earnest R, Geng B, Vijayakumar P, Odio C, Fournier J, Bermejo S, Farhadian S, Dela Cruz CS, Iwasaki A, Ko AI, Landry ML, Foxman EF, Grubaugh ND, Nat. Microbiol 2020, 5, 1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Villamor DEV, Ho T, Al Rwahnih M, Martin RR, Tzanetakis IE, Phytopathology® 2019, 109, 716. [DOI] [PubMed] [Google Scholar]
- [4].Klumpp-Thomas C, Kalish H, Drew M, Hunsberger S, Snead K, Fay MP, Mehalko J, Shunmugavel A, Wall V, Frank P, Denson J-P, Hong M, Gulten G, Messing S, Hicks J, Michael S, Gillette W, Hall MD, Memoli MJ, Esposito D, Sadtler K, Nat. Commun 2021, 12, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bao M, Chen Q, Xu Z, Jensen EC, Liu C, Waitkus JT, Yuan X, He Q, Qin P, Du K, ACS Sens. 2021, 6, 2497. [DOI] [PubMed] [Google Scholar]
- [6].Ramachandran A, Santiago JG, Anal. Chem 2021, 93, 7456. [DOI] [PubMed] [Google Scholar]
- [7].Wang Y, Zhang Y, Chen J, Wang M, Zhang T, Luo W, Li Y, Wu Y, Zeng B, Zhang K, Deng R, Li W, Anal. Chem 2021, 93, 3393. [DOI] [PubMed] [Google Scholar]
- [8].Chaijarasphong T, Thammachai T, Itsathitphaisarn O, Sritunyalucksana K, Suebsing R, Aquaculture 2019, 512, 734340. [Google Scholar]
- [9].Zhang WS, Pan J, Li F, Zhu M, Xu M, Zhu H, Yu Y, Su G, Anal. Chem 2021, 93, 4126. [DOI] [PubMed] [Google Scholar]
- [10].Mukama O, Wu J, Li Z, Liang Q, Yi Z, Lu X, Liu Y, Liu Y, Hussain M, Makafe GG, Liu J, Xu N, Zeng L, Biosens. Bioelectron 2020, 159, 112143. [DOI] [PubMed] [Google Scholar]
- [11].Rauch JN, Valois E, Solley SC, Braig F, Lach RS, Audouard M, Ponce-Rojas JC, Costello MS, Baxter NJ, Kosik KS, Arias C, Acosta-Alvear D, Wilson MZ, J. Clin. Microbiol 59, e02402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhu X, Wang X, Han L, Chen T, Wang L, Li H, Li S, He L, Fu X, Chen S, Xing M, Chen H, Wang Y, Biosens. Bioelectron 2020, 166, 112437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yusko EC, Bruhn BR, Eggenberger OM, Houghtaling J, Rollings RC, Walsh NC, Nandivada S, Pindrus M, Hall AR, Sept D, Li J, Kalonia DS, Mayer M, Nat. Nanotechnol 2017, 12, 360. [DOI] [PubMed] [Google Scholar]
- [14].Nouri R, Jiang Y, Tang Z, Lian XL, Guan W, Nano Lett. 2021, 21, 8393. [DOI] [PubMed] [Google Scholar]
- [15].Larkin J, Henley RY, Jadhav V, Korlach J, Wanunu M, Nat. Nanotechnol 2017, 12, 1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sethi K, Dailey GP, Zahid OK, Taylor EW, Ruzicka JA, Hall AR, ACS Nano 2021, 15, 8474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Luo Y, Wu L, Tu J, Lu Z, Int. J. Mol. Sci 2020, 21, 2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Volarić M, Veseljak D, Mravinac B, Meštrović N, Despot-Slade E, Genes 2021, 12, 1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cao L, Wen Q, Feng Y, Ji D, Li H, Li N, Jiang L, Guo W, Adv. Funct. Mater 2018, 28, 1804189. [Google Scholar]
- [20].Xue L, Yamazaki H, Ren R, Wanunu M, Ivanov AP, Edel JB, Nat. Rev. Mater 2020, 5, 931. [Google Scholar]
- [21].Nouri R, Jiang Y, Lian XL, Guan W, ACS Sens. 2020, 5, 1273. [DOI] [PubMed] [Google Scholar]
- [22].Weckman NE, Ermann N, Gutierrez R, Chen K, Graham J, Tivony R, Heron A, Keyser UF, ACS Sens. 2019, 4, 2065. [DOI] [PubMed] [Google Scholar]
- [23].Wanunu M, Dadosh T, Ray V, Jin J, McReynolds L, Drndić M, Nat. Nanotechnol 2010, 5, 807. [DOI] [PubMed] [Google Scholar]
- [24].Gao Y, Liu X, Wu B, Wang H, Xi F, Kohnen MV, Reddy ASN, Gu L, Genome Biol. 2021, 22, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yang W, Restrepo-Pérez L, Bengtson M, Heerema SJ, Birnie A, van der Torre J, Dekker C, Nano Lett. 2018, 18, 6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sze JYY, Ivanov AP, Cass AEG, Edel JB, Nat. Commun 2017, 8, 1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Nanopore Products, https://www.norcada.com/products/nanopore-products/ (accessed: December 2021).
- [28].Wu H, Liu H, Tan S, Yu J, Zhao W, Wang L, Liu Q, J. Phys. Chem. C 2014, 118, 26825. [Google Scholar]
- [29].Pal S, R. B., Jugade S, Rao A, Naik A, Chakraborty B, Varma MM, Sens. Actuators B Chem 2020, 325, 128785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Fozouni P, Son S, Díaz de León Derby M, Knott GJ, Gray CN, D’Ambrosio MV, Zhao C, Switz NA, Kumar GR, Stephens SI, Boehm D, Tsou C-L, Shu J, Bhuiya A, Armstrong M, Harris AR, Chen P-Y, Osterloh JM, Meyer-Franke A, Joehnk B, Walcott K, Sil A, Langelier C, Pollard KS, Crawford ED, Puschnik AS, Phelps M, Kistler A, DeRisi JL, Doudna JA, Fletcher DA, Ott M, Cell 2021, 184, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Patchsung M, Jantarug K, Pattama A, Aphicho K, Suraritdechachai S, Meesawat P, Sappakhaw K, Leelahakorn N, Ruenkam T, Wongsatit T, Athipanyasilp N, Eiamthong B, Lakkanasirorat B, Phoodokmai T, Niljianskul N, Pakotiprapha D, Chanarat S, Homchan A, Tinikul R, Kamutira P, Phiwkaow K, Soithongcharoen S, Kantiwiriyawanitch C, Pongsupasa V, Trisrivirat D, Jaroensuk J, Wongnate T, Maenpuen S, Chaiyen P, Kamnerdnakta S, Swangsri J, Chuthapisith S, Sirivatanauksorn Y, Chaimayo C, Sutthent R, Kantakamalakul W, Joung J, Ladha A, Jin X, Gootenberg JS, Abudayyeh OO, Zhang F, Horthongkham N, Uttamapinant C, Nat. Biomed. Eng 2020, 4, 1140. [DOI] [PubMed] [Google Scholar]
- [32].Mahas A, Wang Q, Marsic T, Mahfouz MM, ACS Synth. Biol 2021, 10, 2541. [DOI] [PubMed] [Google Scholar]
- [33].Yun NR, Kim C-M, Kim DY, Seo J-W, Kim D-M, Sci. Rep 2021, 11, 14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Tang Y, Cao L, Zhan K, Xie Y, Sun D, Hou X, Chen S, Sens. Actuators B Chem 2019, 286, 315. [Google Scholar]
- [35].Nouri R, Tang Z, Guan W, ACS Sens. 2019, 4, 3007. [DOI] [PubMed] [Google Scholar]
- [36].Arima A, Tsutsui M, Washio T, Baba Y, Kawai T, Anal. Chem 2021, 93, 215. [DOI] [PubMed] [Google Scholar]
- [37].Palaz F, Kalkan AK, Can Ö, Demir AN, Tozluyurt A, Özcan A, Ozsoz M, ACS Synth. Biol 2021, 10, 1245. [DOI] [PubMed] [Google Scholar]
- [38].Antaw F, Anderson W, Wuethrich A, Trau M, Langmuir 2021, 37, 4772. [DOI] [PubMed] [Google Scholar]
- [39].East-Seletsky A, O’Connell MR, Knight SC, Burstein D, Cate JHD, Tjian R, Doudna JA, Nature 2016, 538, 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Rivas F, Zahid OK, Reesink HL, Peal BT, Nixon AJ, DeAngelis PL, Skardal A, Rahbar E, Hall AR, Nat. Commun 2018, 9, 1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.