Abstract
Host genetics is a key determinant of COVID-19 outcomes. Previously, the COVID-19 Host Genetics Initiative genome-wide association study used common variants to identify multiple loci associated with COVID-19 outcomes. However, variants with the largest impact on COVID-19 outcomes are expected to be rare in the population. Hence, studying rare variants may provide additional insights into disease susceptibility and pathogenesis, thereby informing therapeutics development. Here, we combined whole-exome and whole-genome sequencing from 21 cohorts across 12 countries and performed rare variant exome-wide burden analyses for COVID-19 outcomes. In an analysis of 5,085 severe disease cases and 571,737 controls, we observed that carrying a rare deleterious variant in the SARS-CoV-2 sensor toll-like receptor TLR7 (on chromosome X) was associated with a 5.3-fold increase in severe disease (95% CI: 2.75–10.05, p = 5.41x10-7). This association was consistent across sexes. These results further support TLR7 as a genetic determinant of severe disease and suggest that larger studies on rare variants influencing COVID-19 outcomes could provide additional insights.
Author summary
COVID-19 clinical outcomes vary immensely, but a patient’s genetic make-up is an important determinant of how they will fare against the virus. While many genetic variants commonly found in the populations were previously found to be contributing to more severe disease by the COVID-19 Host Genetics Initiative, it isn’t clear if more rare variants found in less individuals could also play a role. This is important because genetic variants with the largest impact on COVID-19 severity are expected to be rarely found in the population, and these rare variants require different technologies to be studies (usually whole-exome or whole-genome sequencing). Here, we combined sequencing results from 21 cohorts across 12 countries to perform a rare variant association study. In an analysis comprising 5,085 participants with severe COVID-19 and 571,737 controls, we found that the gene for toll-like receptor 7 (TLR7) on chromosome X was an important determinant of severe COVID-19. Importantly, despite being found on a sex chromosome, this observation was consistent across both sexes.
Introduction
Despite successful vaccine programs, SARS-CoV-2 is still a major cause of mortality and widespread societal disruption [1,2]. While disease severity has correlated with well established epidemiological and clinical risk factors (e.g., advanced age, obesity, immunosuppression), these do not explain the wide range of COVID-19 presentations [3]. Hence, individuals without one of these known risk factors may have a genetic predisposition to severe COVID-19[4]. These genetic determinants to severe disease can, in turn, inform about the pathophysiology underlying COVID-19 severity and accelerate therapeutics development [5,6].
Previous work on COVID-19 host genetics using genome-wide association studies (GWASs) revealed 23 statistically robust genetic loci associated with either COVID-19 severity or susceptibility [7–11]. Given that most GWASs use genetic data obtained from genome-wide genotyping followed by imputation to measure the association between a phenotype and genetic variation, their reliability and statistical power declines as a variant’s frequency decreases, especially at allele frequencies of less than 1%[12]. Ascertainment of rare genetic variation can be improved with sequencing technology [13]. Rare variants are expected to be enriched for larger effect sizes, due to evolutionary pressure on highly deleterious variants, and may therefore provide unique insights into genetic predisposition to COVID-19 severity. Identifying such genes may highlight critical control points in the host response to SARS-CoV-2 infection.
Measuring the effect of rare genetic variants on a given phenotype (here COVID-19) is difficult. Specifically, while variants of large effect on COVID-19 are more likely to be rare, the converse is not true, and most rare variants are not expected to strongly impact COVID-19 severity [14]. Therefore, unless large sample sizes and careful statistical adjustments are used, most rare variant genetic associations studies risk being underpowered, and are at higher risk of false or inflated effect estimates if significant associations are found between COVID-19 and genetic loci. This is exemplified by the fact that several rare variant associations reported for COVID-19 have not been replicated in independent cohorts [15–17].
Here, we investigated the association of rare genetic variants on the risk of COVID-19 by combining gene burden test results from whole exome and whole genome sequencing. We build off recent work on exome-wide analyses [17] and include close to 5 times the number of severe cases, with a more genetically diverse cohort, to better study the effect of rare variants on COVID-19. To our knowledge, this is the first rare genetic variant burden test meta-analysis ever performed on a worldwide scale, including 21 cohorts, in 12 countries, including all main continental genetic ancestries.
Results
Study population and outcome
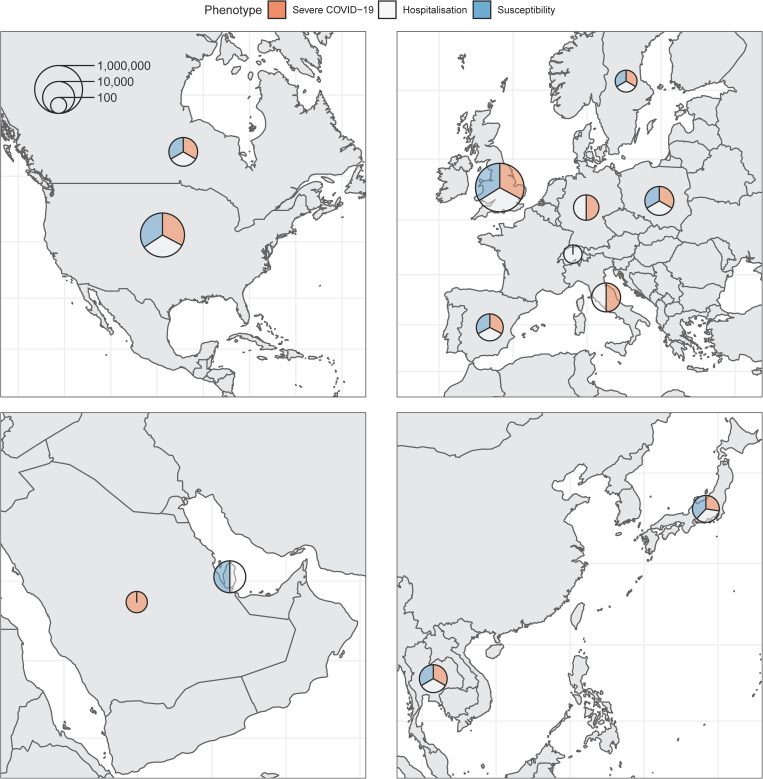
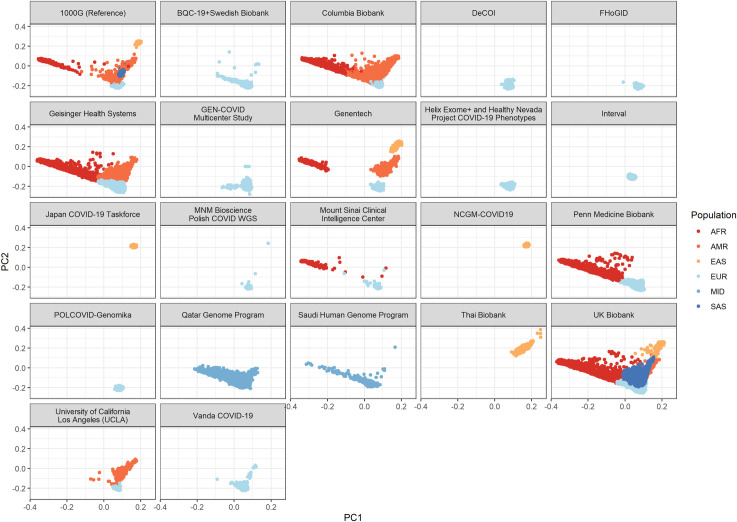
The final analysis included up to 28,159 individuals infected with SARS-CoV-2, and up to 597,165 controls from 21 cohorts in 12 countries (Fig 1). Most participants were of European genetic ancestry (n = 576,389), but the consortium also included participants of Admixed American (n = 4,529), African (n = 25,465), East Asian (n = 4,716), Middle Eastern (n = 4,977) and South Asian ancestries (n = 9,943). These resulted in a genetically diverse sample of participants (Fig 2). Participating cohorts enrolled patients based on local protocols, and both retrospective and prospective designs were used. Genetic sequencing was also performed locally, and cohorts were provided with a specific framework for quality control analyses, but each were allowed to deviate based on individual needs. Both exome (n = 11 cohorts) and genome sequencing (n = 10 cohorts) were included in the meta-analyses. The mean age of participants was 55.6 years, and 55.9% were females (S3 Table).
Fig 1. Maps of countries contributing data to the consortium.
Sample sizes (cases and controls) for each phenotype were added and represented on the logarithmic scale by each circle. Relative contribution to each phenotype is represented by the three colors. Maps obtained from https://www.naturalearthdata.com/.
Fig 2. Participant’s genome projection on the first and second genetic principal components of the 1000G reference panel.
AFR: African ancestry. AMR: admixed American ancestry. EAS: east Asian ancestry. EUR: European ancestry. MID: middle eastern ancestry. SAS: south Asian ancestry.
We studied three separate outcome phenotypes, as previously described by the COVID-19 Host Genetics Initiative (COVID-19 HGI)[8]. Briefly, the outcome cases were defined according to three standard COVID-19 HGI outcomes: A) severe disease: individuals with SARS-CoV-2 infection who died or required invasive respiratory support (extracorporeal membrane oxygenation, intubation with mechanical ventilation, high-flow oxygen support, or new bilevel or continuous positive airway pressure ventilation), B) hospitalisation: individuals with SARS-CoV-2 who died or required hospitalisation, and C) susceptibility to infection: any individual with SARS-CoV-2 infection. These are also referred to as A2, B2, and C2, respectively, in the COVID-19 HGI meta-analyses [8]. For all three phenotypes, controls were all individuals not classified as cases (including population controls with unknown COVID-19 status). The final meta-analyses included up to 5,085 cases and 571,737 controls for the severe disease outcome, 12,304 cases and 590,151 controls for the hospitalisation outcome, and 28,196 cases and 597,165 controls for the susceptibility outcome.
Single-variant analysis
We first performed an exome-wide association study using single variants with a MAF (minor allele frequency) higher than 0.1% and an allele count of 6 or more in at least one cohort, with the same additive model and covariates used in the COVID-19 HGI GWAS [8]. Analyses were performed separately by each cohort and each ancestry using Firth regression as applied in the Regenie software [18]. Firth regression is a penalized likelihood regression method that provides unbiased effect estimates even in highly unbalanced case-control analyses [19]. The summary statistics were then meta-analyzed with a fixed effect inverse-variance weighted model within each ancestry, and then with a DerSimonian-Laird random effect model across ancestries.
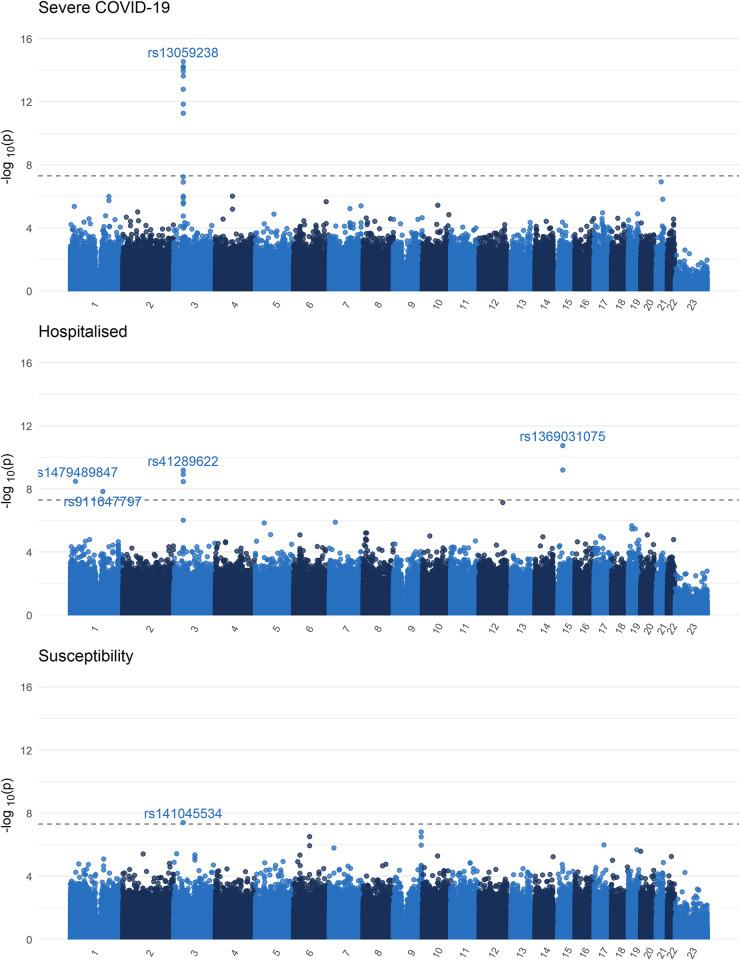
The previously described Neanderthal chromosome 3 locus associated with COVID-19 outcomes [8,20] was also found in all three phenotypes (Figs 3 and S1), with lead variants in the CXCR6 gene for the severe COVID-19 phenotype (rs13059238), and in FYCO1 in the hospitalisation phenotype (rs13069079), and for the LIMD1 gene in the susceptibility phenotype (rs141045534). Reassuringly, each cohort provided summary statistics in the chromosome 3 locus, suggesting that the QC process was working as intended (allowing for sample sizes and number of cases) (S2–S4 Figs).
Fig 3. Single variant exome-wide association study Manhattan plot (MAF>0.1%).
QQ-plot available in the S1 Fig. Black dashed line demarcates the genome-wide significance threshold (p < 5x10-8).
Three other loci were found for the hospitalization phenotypes. One at SRRM1 (rs1479489847, OR: 4.17, 95% CI 2.60–6.70, p = 3.25x10-9), IL6R (rs911647797, OR: 6.19, 95% CI: 3.29–11.6, p = 1.45x10-8), and another at cytoskeleton FRMD5 (rs1369031075, OR: 4.06, 95% CI: 2.70–6.11, p = 1.75x10-11). While these loci may hold biological plausibility (especially IL6R, given the use of IL-6 receptor inhibitors in the treatment of COVID-19), these associations were driven by two smaller cohorts (Genentech and Vanda, S5 Fig). However, the SRMM1 locus is located between two stretches of T nucleotides, while both the FRMD5 and the IL-6 loci are within GC rich regions, making variant calling difficult. Hence, these findings will require validation, despite the biological plausibility.
Finally, all genetic inflation factors were below 1 (S4 Table). Summary statistics for genome-wide significant variants can be found in S5 Table and QQ-plots can be found in S1 Fig.
Burden test definition
Given the expected paucity of large-effect size rare deleterious variants, strategies have been devised to increase statistical power to test associations between rare variants and biomedically-relevant outcomes. One such strategy is to use burden tests [21], where each variant is collapsed into larger sets of variants, and association is tested between groups of variants and an outcome. Here, we collapsed deleterious variants in each gene and devised the following burden test: for each gene, an individual received a score of 0 if they do not carry any deleterious variant, a score of 1 if they carry at least one non-homozygous deleterious variant, and a score of 2 if they carry at least 1 homozygous deleterious variant. As defined in previous studies on burden testing of rare variants[17,22] deleterious variants were chosen using three masks: 1) “M1" which uses only predicted loss of function variants, 2) “M3” which uses all variants in M1, as well as indels of moderate consequence as predicted by Ensembl [23], and missense variants classified as deleterious in 5 in-silico algorithms (see Methods), and 3) “M4”, which uses all variants in M1 and M3, and also adds all missense variants classified as deleterious in at least 1 of the in-silico algorithms.
The analyses were performed separately both for variants with MAF of less than 1%, and for variants of MAF less than 0.1%. We defined MAFs based on a combination of gnomAD [24] MAF annotations, and of cohort-specific common variant exclusion lists. These common variant exclusion lists included variants that achieved a MAF of >1% or >0.1% in at least one study population within the consortium. To reduce the effect of fluctuations due to sampling, a minor allele count (MAC) ≥ 6 in the corresponding study was required for inclusion in the common variant list. Such “blacklists” have been shown to increase statistical power by removing variants at lower risk of being highly deleterious, and it reduces the risk of having cohort-specific false-positive variants being retained on the overall analysis[25]. Hence for each MAF threshold, each cohort removed any variant with a MAF above the threshold in either gnomAD or the corresponding common variant exclusion list.
The resulting score (either 0, 1, or 2) for each mask was then regressed on each of our three phenotypes using logistic regression, controlling for age, age^2, sex, sex*age, sex*age2, and 10 common variant (MAF > 1%) genetic principal components (the same covariates as for COVID-19 HGI GWASs [7,8]). Additionally, given that population genetic structure and its confounding effect on phenotypes is different at the rare variant level [26], we also used the first 20 genetic principal components from rare variants (MAF<1%) as covariates in all our analyses. Analyses were otherwise done using the same approach as for single-variant analyses.
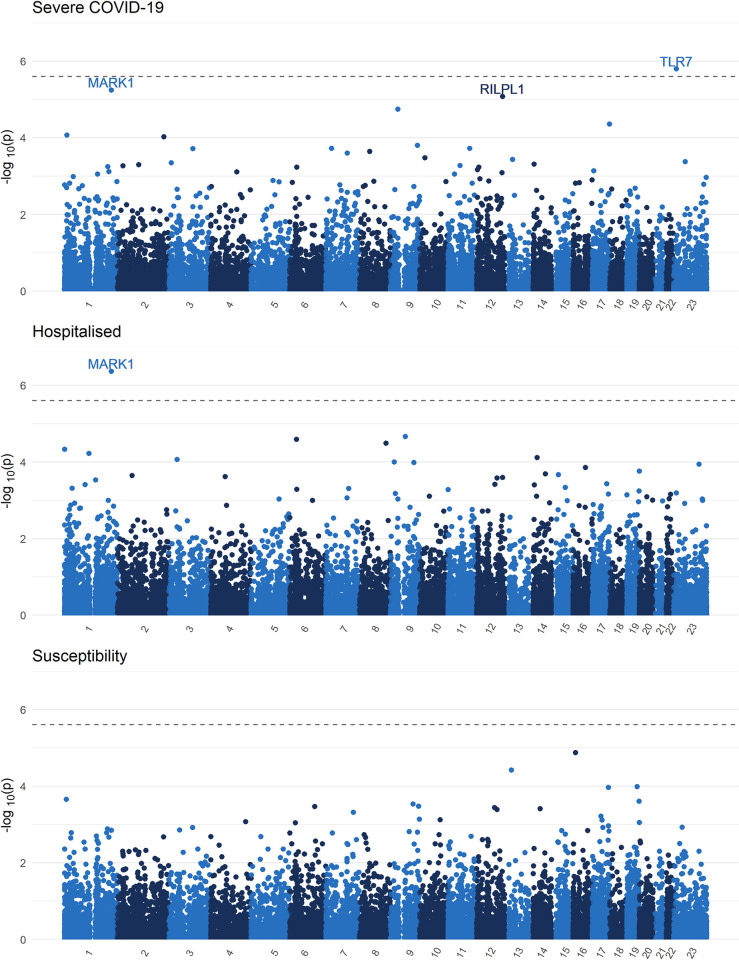
Exome-wide burden test analyses results
Our meta-analysis included a total of 18,883 protein-coding genes, and all burden test genetic inflation factors, for all masks, were less than 1 (S4 Table), suggesting that our results were not biased by population stratification and that Firth regression adequately adjusted for unbalanced case-control counts. Using an exome-wide significance p-value threshold of 0.05/20,000 = 2.5x10-6, we found 3 genes associated with one of the COVID-19 phenotypes in at least one mask in our meta-analyses (Table 1 and S6–S12 Figs). Of specific interest, we observed that carrying a predicted loss of function or in-silico highly deleterious missense variant (i.e., mask M3) in the toll-like receptor 7 (TLR7) gene was associated with a 5.3-fold increase (95% CI: 2.7–10.1, p = 5.41x10-7) in odds of severe COVID-19. TLR7 is an important part of the innate viral immunity, encoding a protein that recognizes coronaviruses and other single-stranded RNA viruses, leading to upregulation of the type-1 and type-2 interferon pathway [27]. Results from the severe COVID-19 outcome analyses of TLR7 with other masks also nearly reach our statistical significance threshold, with larger effects found in the M1 mask (OR: 13.6, 95% CI: 4.41–44.3, p = 1.64x10-5) and smaller effect in the M4 mask (OR: 3.12, 95% CI: 1.91–5.10, p = 5.30x10-6), though the latter was balanced by smaller standard errors due to the larger number of cases (3275 cases in M4 vs 1577 in M1), as expected. These findings further support previous reports of TLR7 errors of immunity underlying severe COVID-19 presentations [17,28–31].
Table 1. Exome-wide significant findings, as well as other TLR7 results (for the severe phenotype only).
Note that for Masks M1, all deleterious variants had a MAF<0.1%, and hence both burden tests (MAF<1% and 0.1%) gave the same results. Full results available in S4 Table.
| Gene | Mask | Phenotype | MAF | Beta | Standard Error | Odds Ratio | 95% Confidence Interval | P-value | Heterogeneity p-value | N Cases 0|1|2 Burden Test | N Controls 0|1|2 Burden Test |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Meta-Analysis Across Ancestries | |||||||||||
| MARK1 | M1 | Severe COVID-19 | <0.1% | 3.17 | 0.67 | 23.9 | 6.5–88.2 | 1.89x10-6 | 0.883 | 1935|4|0 | 540031|92|0 |
| MARK1 | M1 | Severe COVID-19 | <1% | 3.17 | 0.67 | 23.9 | 6.5–88.2 | 1.89x10-6 | 0.883 | 1935|4|0 | 540031|92|0 |
| MARK1 | M1 | Hospitalisation | <0.1% | 2.51 | 0.48 | 12.3 | 4.8–31.2 | 1.43x10-7 | 0.893 | 6132|8|0 | 547943|93|0 |
| MARK1 | M1 | Hospitalisation | <1% | 2.51 | 0.48 | 12.3 | 4.8–31.2 | 1.43x10-7 | 0.893 | 6132|8|0 | 547943|93|0 |
| RILPL1 | M1 | Severe COVID-19 | <0.1% | 3.01 | 0.64 | 20.2 | 5.8–70.7 | 2.42x10-6 | 0.941 | 1745|4|0 | 558448|121|0 |
| TLR7 | M3 | Severe COVID-19 | <0.1% | 1.66 | 0.33 | 5.25 | 2.75–10.05 | 5.41x10-7 | 0.755 | 3101|2|5 | 519047|83|47 |
| TLR7 | M3 | Severe COVID-19 | <1% | 1.63 | 0.33 | 5.10 | 2.67–9.72 | 7.48x10-7 | 0.760 | 3275|2|5 | 519834|85|47 |
| Other TLR7 results for severe phenotype | |||||||||||
| TLR7 | M1 | Severe COVID-19 | <0.1% | 2.61 | 0.60 | 13.6 | 4.14–44.4 | 1.64x10-5 | 0.820 | 1577|0|2 | 508987|13|11 |
| TLR7 | M1 | Severe COVID-19 | <1% | 2.61 | 0.60 | 13.6 | 4.14–44.4 | 1.64x10-5 | 0.820 | 1577|0|2 | 508987|13|11 |
| TLR7 | M4 | Severe COVID-19 | <0.1% | 1.14 | 0.25 | 3.12 | 1.91–5.10 | 5.30x10-6 | 0.854 | 3275|3|7 | 519616|210|139 |
|
TLR7 |
M4 |
Severe COVID-19 |
<1% |
1.11 |
0.24 |
3.03 |
1.90–4.85 |
3.43x10-6 |
0.956 |
3273|5|8 |
521166|221|144 |
In the meta-analyses, we also found that pLoFs (M1) in MARK1 were associated with a 23.9-fold increase in the odds of severe COVID-19 (95% CI: 6.5–88.2, p = 1.89x10-6), and a 12.3-fold increase in the odds of hospitalisation due to COVID-19 (95% CI: 4.8–31.2, p = 1.43x10-7). While the number of MARK1 pLoFs (M1) found in severe and hospitalized cases was small (MAC = 4 and MAC = 8, respectively), the signal was consistent in our three largest cohorts: UK Biobank, Penn Medicine, and Geisinger Health Services (S9–S12 Figs). MARK1 is a member of the microtubule affinity-regulating kinase family, and is involved in multiple biological processes, chief among which is the promotion of microtubule dynamics [32]. MARK1 has previously been shown to interact with the SARS-CoV-2 ORF9b protein [33], further supporting its potential role in COVID-19. Lastly, our meta-analyses also found marginal evidence for an association between severe COVID-19 and pLoFs (M1) in RILPL1 (OR: 20.2, 95% CI: 5.8–70.7, p = 2.42x10-6), a gene that, like MARK1, is associated with microtubule formation and ciliopathy [34].
We then meta-analyzed p-values using the aggregated Cauchy association test [35] (ACAT). ACAT accounts for correlation between test statistics (as is expected here) by treating p-values as Cauchy random variables, and taking their weighted average, which also is Cauchy distributed. With ACAT, the association between TLR7 and severe COVID-19 (p = 1.58x10-6), and between MARK1 and hospitalisation (p = 4.30x10-7) remained exome-significant (Fig 4). Full summary statistics are available in S6 Table.
Fig 4. Exome burden test ACAT p-value meta-analysis Manhattan plots and QQ plots.
QQ-plot available in the S6 Fig. Black dashed line demarcates the Bonferroni significance threshold (p < 0.5/20,000).
Finally, we note that for both TLR7 and MARK1, the signal was driven by European ancestry participants. Further, while the larger biobanks contributed to these findings, smaller prospective cohorts also provided cases with rare variants at both genes, highlighting the importance of study design in rare variant association testing (S6–S12 Figs).
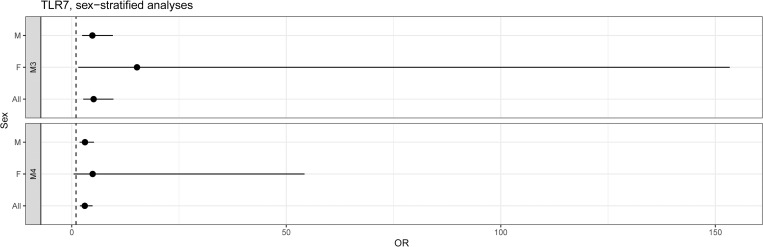
TLR7 sex stratified analyses
Given that TLR7 is located on the X chromosome, we performed sex-stratified analyses of the severe disease phenotype to determine if the effect was also observed in females. These could only be done for the M3 and M4 masks due to very low number of M1 mask qualifying variants (Fig 5). In both we still see a clear effect among males with a 4.81-fold increase in the odds of severe COVID-19 in M3-variant carriers (95% CI: 2.41–9.59, 5 case carriers, 47 control carriers), and a 3.08-fold increase in M4-variant carriers (95% CI: 1.83–5.20, 7 case carriers, 143 control carriers). In females, we still observed a nominally significant signal in the M3 mask, with a 15.2-fold in odds of severe disease in M3-variant carriers (95% CI: 1.51–153.4). However, an M3-variant was observed in only one female with severe disease (heterozygous) in these analyses (compared to 76 heterozygous controls). In M4 variant, the analyses included 2 female heterozygous carriers (and 203 heterozygous controls), with a 4.86-fold in odds of severe disease (95% CI: 0.43–54.3).
Fig 5. Sex-stratified TLR7 analyses.
Rare variants in interferon-related genes and at previously reported genome-wide significant loci
Despite a 7.7-fold increase in number of cases, and a 1,069-fold increase in number of controls, the previously reported associations of genes in the interferon pathway with COVID-19 outcomes [15,16] could not be replicated with either our exome-wide significance threshold (S7 Table) or a more liberal one of p = 0.05/10 = 0.005 (based on Bonferroni correction by the number of genes in the interferon pathway defined in a previous study[15]).
We also tested for rare variant associations between GWAS candidate genes from genome-wide significant loci in the COVID-19 HGI GWAS meta-analyses, but observed no exome-wide significant associations (S8 Table). However, at a more liberal Bonferroni threshold of p = 0.05/46 = 0.001 (correcting for the 46 genes in the COVID-19 HGI GWAS associated loci), we observed an increased burden of pLoF (M1) or missense variants (M3 mask) in ABO gene among those susceptible to SARS-CoV-2 infection (Table 2). For example, individuals carrying a pLoF (M1) with MAF<0.1% in ABO were at a 2.34-fold higher risk of having a positive SARS-CoV-2 infection (95% CI: 1.50–3.64, p = 1.6x10-4). The ABO results were driven mainly by European and African ancestry participants (S13 Fig). Note that deleterious variants in ABO often lead to blood groups A and B [36,37], which is consistent with the epidemiological association that non-type-O individuals are at higher risk of COVID-19[38]. However, more work is required to better understand the genetics of this locus as it relates to COVID-19 outcomes. Lastly, missense variants in NSF (mask M4, MAF<1%) were also associated with higher susceptibility to SARS-CoV-2 (OR: 1.48, 95% CI: 1.21–1.82, p = 1.4x10-4), but this association was not present in other masks (S9 Table).
Table 2. Results of burden tests at genes identified from common variants GWAS in the COVID-19 HGI.
Only genes with p<0.05/46 are shown here. Full results available in S8 Table.
| Gene | Mask | Phenotype | MAF | Beta | Standard Error | Odds Ratio | 95% Confidence Interval | P-value | Heterogeneity p-value | N Cases 0|1|2 Burden Test | N Controls 0|1|2 Burden Test |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NSF | M4 | Susceptibility | <1% | 0.395 | 0.104 | 1.484 | 1.21–1.82 | 1.44x10-4 | 0.866 | 25752|127|2 | 585642|1907|5 |
| ABO | M1 | Susceptibility | <0.1% | 0.851 | 0.226 | 2.341 | 1.50–3.65 | 1.68x10-4 | 0.498 | 22778|27|0 | 572310|296|0 |
| ABO | M1 | Susceptibility | <1% | 0.784 | 0.209 | 2.19 | 1.45–3.30 | 1.75x10-4 | 0.826 | 23460|34|0 | 574608|364|0 |
| ABO | M3 | Susceptibility | <1% | 0.729 | 0.195 | 2.073 | 1.41–3.04 | 1.89x10-4 | 0.869 | 24455|42|0 | 575051|434|0 |
| ABO | M1 | Hospitalisation | <0.1% | 1.33 | 0.395 | 3.78 | 1.74–8.20 | 7.56x10-4 | 0.542 | 7859|12|0 | 561642|291|0 |
| ABO | M1 | Susceptibility | <0.1% | 0.736 | 0.222 | 2.088 | 1.35–3.23 | 9.35x10-4 | 0.512 | 23779|29|0 | 572799|320|0 |
Replication in GenOMICC
Data for the M1 mask for TLR7 and MARK1 in the severe COVID-19 phenotype was then replicated with the GenOMICC cohort [11], a prospective study enrolling critically ill individuals with COVID-19, with controls selected from the 100,000 genomes cohort[39]. Results are shown in Table 3. For TLR7, European ancestry individuals with a pLoF (M1) had a 4.70-fold increase in odds of severe disease (95% CI: 1.58 to 14.0, p = 0.005). In the sample of South Asian ancestry individuals, a pLoF (M1) was associated with a 1.90-fold increase in odds of severe disease, but the 95% confidence interval crossed the null (0.23 to 15.6, p = 0.55), which was likely due to a much smaller sample size than in the European ancestry subgroup (1,202 vs 10,645). Of interest, in both Europeans and South Asians, no pLoFs were observed in either of the control groups.
Table 3. Replication of M1 mask, severe COVID-19, MARK1 and TLR7 results in the GenOMICC cohort. Note that the same variants were included in both the MAF<1% and MAF<0.1% replication, and the same results were obtained (shown here).
| Gene | Ancestry | Beta | Standard Error | Odds Ratio | 95% Confidence Interval | P-value | N Cases 0|1|2 Burden Test | N Controls 0|1|2 Burden Test |
|---|---|---|---|---|---|---|---|---|
| MARK1 | EUR | 0.195 | 1.42 | 1.21 | 0.075–19.7 | 0.891 | 5988|1|0 | 4655|1|0 |
| MARK1 | SAS | 1.44 | 2.19 | 4.21 | 0.058–307.6 | 0.511 | 787|1|0 | 414|0|0 |
| TLR7 | EUR | 1.55 | 0.555 | 4.70 | 1.58–14.0 | 0.005 | 5980|1|8 | 4566|0|0 |
| TLR7 | SAS | 0.640 | 1.08 | 1.90 | 0.230–15.6 | 0.552 | 786|0|2 | 414|0|0 |
On the other hand, we could not replicate an effect from MARK1, which demonstrated an OR of 1.21 in European ancestry participants (95% CI 0.075 to 19.7, p = 0.89) and an OR of 4.21 in South Asian ancestry individuals (95% CI 0.058 to 307, p = 0.51).
Discussion
Whole genome and whole exome sequencing can provide unique insights into genetic determinants of COVID-19, by uncovering associations between rare genetic variants and COVID-19. Specifically, gene burden tests can be particularly helpful, because they test for coding variants, thereby pointing directly to a causal gene and often suggesting a direction of effect. However, such studies require careful control for population stratification and an adapted analysis method such as burden testing, in order to have enough statistical power to find those associations. In our study, we observed that individuals with rare deleterious variants at TLR7 are at increased risk of severe COVID-19 (up to 13.1-fold increase in odds in those with pLoFs). Although this association was suggested by previous studies [28–30], our study provides the most definitive evidence for the role of TLR7 in COVID-19 pathogenesis, with exome-wide significance for this gene in the discovery phase followed by strong replication in a large independent cohort. TLR7 is a well-studied part of the antiviral immunity cascade and stimulates the interferon pathway after recognizing viral pathogen-associated molecular patterns. Given its location on the X chromosome, it has been hypothesis that it could partly explain the observed COVID-19 outcome differences between sexes [40–42], and to our knowledge, this is the first study to show that even in heterozygous females, this gene can potentially play a role in severe disease. Further, this our results suggest that TLR7 mediated genetic predisposition to severe COVID-19 may be a dominant or co-dominant trait, an observation that cannot be made in cohorts limited to male participants[28,30].
We also uncovered a potential role for cellular microtubule disruption in the pathogenesis of COVID-19 and the microtubule network is known to be exploited by other viruses during infections [43]. Indeed, the MARK1 protein has been shown to interact with SARS-CoV-2 in previous in-vitro experiments [33]. Nevertheless, these findings at MARK1 were not replicated in the GenOMICC cohort and will need to be tested in larger cohorts, especially given the small number of highly deleterious variants that we found in our consortium. Lastly, we found single variant associations at IL6R, SRRM1, and FRMD5. While IL6R is is already a therapeutic target [44,45] for COVID-19, and SRRM1 has been reported in a previous pre-print [46], these were found in smaller cohorts and will require replication.
To our knowledge, this is the first time a rare variant burden test meta-analysis has been attempted on such a large scale. Our framework allowed for easy and interpretable summary statistics results, while at the same time preventing participant de-identification or any breach of confidentiality that stems from sharing results of rare genetic variant analyses [47]. It also provides important insights into how these endeavours should be planned in the future. First, our burden test operated under the assumption that the effect of any of the deleterious variants on the phenotype would be in the same direction and did not account for compound deleterious variant heterozygosity. This allowed for easier meta-analysis across cohorts, but may have decreased statistical power. Other methods may be needed in future analysis to soften this assumption, though some of these cannot be easily meta-analyzed across multiple cohorts directly from summary statistics (e.g., SKAT-O [48]). Similarly, methods that combine both rare and common variants might also provide additional insights into disease outcomes [31,49]. Second, our results highlight the importance at looking at different categories of variants through different masks to increase sensitivity and specificity of our burden tests. Third, while the largest biobanks contributed the most to the signal observed at TLR7 and MARK1, many of our smaller prospective COVID-19 specific cohorts also contributed to the signal. This further highlights the importance of robust study design to improve statistical power, especially with rare variant associations. Lastly, work remains to be done to standardize sequencing and annotation pipelines to allow comparisons of results easily across studies and cohorts. Here, we provided a pipeline framework to every participating cohort, but there remains room for process harmonization. While the decentralized approach to genetic sequencing, quality control, and analyses allowed for more rapid generation of results, it may come at the cost of larger variance in our estimates. In the future, more sophisticated approaches may be required to increase statistical power of exome-wide rare variant association studies [50].
Our study had limitations. First, even if this is one of the world’s largest consortia using sequencing technologies for the study of rare variants, we remain limited by a relatively small sample size. For example, in a recent analyses of UK Biobank exomes, many of the phenotypes for which multiple genes were found using burden tests had a much higher number of cases than in our analyses (e.g. blonde hair colour, with 48,595 cases) [22]. Further, rare variant signals were commonly found in regions enriched in common variants found in GWASs. The fact that ABO and NSF were the only genes from the COVID-19 HGI GWAS that were also identified in our burden test (albeit using a more liberal significance threshold), also suggests a lack of statistical power. Similarly, GenOMICC, a cohort of similar size, was also unable to find rare variant associations using burden tests [11]. However, their analysis methods were different from ours, making further comparisons difficult. Nevertheless, this provides clear guidance that smaller studies looking at the effect of rare variants across the genome are at considerable risk of finding both false positive and false negative associations. Second, many cohorts used population controls, which may have decreased statistical power given that some controls may have been misclassified. However, given that COVID-19 critical illness remains a rare phenomenon [51], our severe disease phenotype results are unlikely to be strongly affected by this. Finally, the use of population control is a long-established strategy in GWAS burden tests [7,8,11,22,52], and the statistical power gain from increasing our sample size is likely to have counter-balanced the misclassification bias.
In summary, we reproduced an exome-wide significant association with severe COVID-19 outcomes in carriers of rare deleterious variants at TLR7, for both sexes. Our results also suggest an association between the cellular microtubule network and severe disease, which requires further validation. More importantly, our results underline the fact that future genome-wide studies of rare variants will require considerably larger sample size, but our work provides a roadmap for such collaborative efforts.
Methods
Ethics statement
Each cohort had the following statement to make on ethics:
BQC-19
Each participant or their legal representative (if the participant was incapable to consent) provided informed consent to the biobank. If a participant regained capacity to give consent, informed consent was obtained again directly from the participant. The study was approved by the Jewish General Hospital and Centre Hospitaler de l’Université de Montréal institutional review boards.
Columbia Biobank: Recruitment and sequencing of participants from the Columbia COVID-19 Biobank were approved by the Columbia University Institutional Review Board (IRB) protocol AAAS7370 and the genetic analyses were approved under protocol AAAS7948. A subset of patients was included under a public health crisis IRB waiver of consent specifically for COVID-19 studies if patients were deceased, not able to consent, or if the study team was unable to contact them as per IRB protocol AAAS7370.
DeCOI
Informed consent was obtained from each participant or the legal representative. DeCOI received ethical approval by the Ethical Review Board (ERB) of the participating hospitals/centres (Technical University Munich, Munich, Germany; Medical Faculty Bonn, Bonn, Germany; Medical Board of the Saarland, Germany; University Duisburg-Essen, Germany; Medical Faculty Duesseldorf, Duesseldorf, Germany)
FHoGID
Each participant or their legal representative provided informed consent to the biobank. FHoGID received ethical approval by the Commission cantonale d’éthique de la recherche sur l’être humain.
GEN-COVID multicenter study: The patients were informed of this research and agreed to it through the informed consent process. The GEN-COVID is a multicentre academic observational study that was approved by the Internal Review Boards (IRB) of each participating centre (protocol code 16917, dated March 16, 2020 for GEN-COVID at the University Hospital of Siena).
Genentech
The protocol was reviewed by the institutional review board or ethics committee at each site. Written informed consent was obtained from all the patients or, if written consent could not be provided, the patient’s legally authorized representative could provide oral consent with appropriate documentation by the investigator. Details on institutional review boards are provided in S9 Table.
GenOMICC
GenOMICC was approved by the appropriate research ethics committees (Scotland, 15/SS/0110; England, Wales and Northern Ireland, 19/WM/0247). Informed consent was obtained for all participants.
Geisinher Health Systems: All subjects consented to participation and the analysis was approved by the Geisinger Institutional Review Board under project number 2006–0258.
Helix Exome+ and Healthy Nevada Project COVID-19 Phenotypes: informed consent was obtained for all participants. The Healthy Nevada Project study was reviewed and approved by the University of Nevada, Reno Institutional Review Board (IRB, project 956068–12)
Thai Biobank ()
Informed consent was obtained for each participant via the biobank. The study was approved by the Institutional Review Board of the Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand (COA No. 691/2021).
Japan COVID-19 Task Force: Each participant or their legal representative (if the participant was incapable to consent) provided informed consent to the biobank. Study was approved by the ethical committees of Keio University School of Medicine, Osaka University Graduate School of Medicine, and affiliated institutes.
Interval WGS
After reading study leaflets and participating in a discussion with donor carer staff, eligible donors were asked to complete the trial consent form before giving a blood donation. The National Research Ethics Service (United Kingdom) approved this study.
MNM Diagnostics (Polish Covid WGS)
All participants, or their guardians/parents for the participants under 18), provided their informed consent before collecting their blood samples. The study was approved by the Institutional Ethics Committee of the Central Clinical Hospital of the Ministry of Interior and Administration in Warsaw, Poland (decision nr: 41/2020 from 03.04.2020 and 125/2020 from 1.07.2020).
MSCIC
This research protocol was reviewed and approved by the Icahn School of Medicine at Mount Sinai Institutional Review Board (IRB) (STUDY-20-00341). During the height of the SARS-CoV-2 pandemic in New York City, all patients admitted to the Mount Sinai Health System were made aware of the research study by a notice included in their admission paperwork. The notice outlined details of the planned research, potential specimen collection and the opportunity to opt-out of research. Flyers announcing the study were also posted throughout the health system. Given the monumental hurdles of consenting sick and infectious patients in isolation rooms, the IRB allowed for specimen collection to occur prior to obtaining research consent at the time of clinical blood collection. Patients and/or their legally authorized representative provided consent to the research study, including genetic profiling for research and data sharing on an individual level. In a subset of individuals, who were unreachable following hospital discharge, we were unable to obtain written informed consent. In these cases, data cannot be share further. All data used these these analyses were anonymized same as above.
Penn medicine
Recruitment of PMBB participants was approved under IRB protocol 813913 and supported by Perelman School of Medicine at University of Pennsylvania.
POLCOVID-Genomika
All study participants provided written informed consent and received detailed information on the study and associated risk before enrollment. The study was approved by the Bioethics Committee of the Medical University of Bialystok.
Qatar Genome Program
All QBB participants signed an Informed Consent Form prior to their participation; QBB study protocol ethical approval was obtained from the Hamad Medical Corporation Ethics Committee in 2011 and continued with QBB Institutional Review Board (IRB) from 2017 onwards and it is renewed on an annual basis
Saudi human genome program
Informed Consent was provided to each participant or their legal guardian (if the participant could not consent) by the corresponding institute. This study was approved the IRB of each participating hospitals, and the IRB at King Abdullah International Medical Research Centre, Ministry of National Guard–Health Affairs, Riyadh, Ministry of Health, and King Fahad Medical City.
Swedish Biobank
Informed consent was obtained for all study participants. The study was approved by the National Ethical Review Agency (Sweden) (No. 2020–01623).
UK Biobank
All subjects consented to participation. The UK Biobank was approved by the North West Multi-centre Research Ethics Committee (United Kingdom) (11/NW/0382). The work described herein was approved by the UK Biobank under application no. 26041.
University of California, Los Angeles biobank
Each participant or their legal representative (if the participant was incapable to consent) provided informed consent to the biobank. If a participant regained capacity to give consent, informed consent was obtained again directly from the participant. This study was considered human subjects research exempt because all genetic and electronic health records were de-identified. This study was approved by the UCLA Health Institutional Review Board.
Vanda COVID-19
All participants consented to WGS. The study was reviewed and approved by Advarra IRB; Pro00043096.
COVID-19 outcome phenotypes
For all analyses, we used three case-control definitions: A) Severe COVID-19, where cases were those who died, or required either mechanical ventilation (including extracorporeal membrane oxygenation), high-flow oxygen supplementation, new continuous positive airway pressure ventilation, or new bilevel positive airway pressure ventilation, B) Hospitalized COVID-19, where cases were all those who died or were admitted with COVID-19, and C) Susceptibility to COVID-19, where cases are anyone who tested positive for COVID-19, self-reported an infection to SARS-CoV-2, or had a mention of COVID-19 in their medical record. For all three, controls were individuals who did not match case definitions, including population controls for which case status was unknown (given that most patients are neither admitted with COVID-19, nor develop severe disease [53]). These three analyses are also referred to as analyses A2, B2, and C2 by the COVID-19 Host Genetics Initiative [8], respectively.
Cohort inclusion criteria and genetic sequencing
Any cohort with access to genetic sequencing data and the associated patient level phenotypes were allowed in this study. Specifically, both whole-genome and whole-exome sequencing was allowed, and there were no limitations in the platform used. There were no minimal number of cases or controls necessary for inclusion. However, the first step of Regenie, which was used to perform all tests (see below), uses a polygenic risk score which implicitly requires that a certain sample size threshold be reached (which depends on the phenotype and the observed genetic variation). Hence, cohorts were included if they were able to perform this step. All cohorts obtained approval from their respective institutional review boards, and informed consent was obtained from all participants. More details on each cohort’s study design and ethics approval can be found in the S3 and S1 Tables.
Variant calling and quality control
Variant calling was performed locally by each cohort, with the pre-requisite that variants should not be joint-called separately between cases and controls. Quality control was also performed individually by each cohort according to individual needs. However, a general quality control framework was made available using the Hail software [54]. This included variant normalization and left alignment to a reference genome, removal of samples with call rate less than 97% or mean depth less than 20. Genotypes were set to unknown if they had genotype quality less than 20, depth less than 10, or poor allele balance (more than 0.1 for homozygous reference calls, less than 0.9 for homozygous alternative calls, and either below 0.25 or above 0.75 for heterozygous calls. Finally, variants were removed from if the mean genotype quality was less than 11, mean depth was less than 6, mean call rate less than or equal to 0.8, and Hardy-Weinberg equilibrium p-value less than or equal to 5x10-8 (10−16 for single variant association tests). Details on variant calling and quality control is described for each cohort in the S3 Table.
Single variant association tests
We performed single variant association tests using a GWAS additive model framework, with the following covariates: age, age2, sex, age*sex, age2*sex, 10 genetic principal components obtained from common genetic variants (MAF>1%). Each cohort performed their analyses separately for each genetic ancestry, but also restricted their variants to those with MAF>0.1% and MAC>6. Summary statistics were then meta-analyzed using a fixed effect model within each ancestry and using a DerSimonian-Laird random effect model across ancestries with the Metal package [55] and its random effect extension [56]. Lastly, given that multiple technologies were used for sequencing, and that whole-exome sequencing can provide variant calls of worse quality in its off-target regions [57], we used the UKB, GHS, and Penn Medicine whole-exome sequencing variants as our “reference panel” for whole-exome sequencing. Hence, only variants reported in at least one of these biobanks were used in the final single-variant analyses.
Variant exclusion list
For the burden tests, we also compiled a list of variants that had a MAF > 1% or > 0.1% in any of the participating cohorts. This list was used to filter out variants that were less likely to have a true deleterious effect on COVID-19, even if they were considered rare in other cohorts, or in reference panels [25]. We created two such variant exclusion lists: one to be used in our burden test with variants of MAF less than 1%, and the other for the analysis with MAF less than 0.1%. In any cohort, if a variant had a minor allele count of 6 or more, and a MAF of more than 1% (or 0.1%), this variant was added to our exclusion list. This list was then shared with all participating cohorts, and all variants contained were removed from our burden tests.
Gene burden tests
The following analyses generally followed the methods used by recent literature on large-scale whole-exome sequencing [22] and the COVID-19 HGI [8].
The burden tests were performed by pooling variants in three different variant sets (called masks), as described in recent UK Biobank whole-exome sequencing papers by Backman et al.[22] and Kosmicki et al.[17].: “M1” which included loss of functions as defined by high impact variants in the Ensembl database[23] (i.e. transcript ablation, splice acceptor variant, splice donor variant, stop gained, frameshift variant, stop lost, start lost, transcript amplification), “M3” which included all variants in M1 as well as moderate impact indels and any missense variants that was predicted to be deleterious based on all of the in-silico pathogenicity prediction scores used, and “M4” which included all variants in M3 as well as all missense variants that were predicted to be deleterious in at least one of the in-silico pathogenicity prediction scores used. For in-silico prediction, we used the following five tools: SIFT [58], LRT [59], MutationTaster[60], PolyPhen2[61] with the HDIV database, and PolyPhen2 with the HVAR database. Protein coding variants were collapsed on canonical gene transcripts.
Once variants were collapsed into genes in each participant, for each mask, genes were given a score of 0 if the participant had no variants in the mask, a score of 1 if the participant had one or more heterozygous variant in this mask, and a score of 2 if the participant had one or more homozygous variant in this mask. These scores were used as regressors in logistic regression models for the three COVID-19 outcomes above. These regressions were also adjusted for age, age2, sex, age*sex, age2*sex, 10 genetic principal components obtained from common genetic variants (MAF>1%), and 20 genetic principal components obtained from rare genetic variants (MAF<1%). The Regenie software [18] was used to perform all burden tests, and generate the scores above. Regenie uses Firth penalized likelihood to adjust for rare or unbalanced events, providing unbiased effect estimates.
All analyses were performed separately for each of six genetic ancestries (African, Admixed American, East Asian, European, Middle Eastern, and South Asian). Summary statistics were meta-analyzed as for the single variant analysis. Participant assignment to genetic ancestry was done locally by each cohort, more details on the methods can be found in the S3 Table.
Lastly, we used ACAT [35] to meta-analyze p-values across masks, within each phenotype separately. ACAT is not affected by lack of independence between tests. These values were used to draw Manhattan and QQ plots in Fig 2.
Supporting information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
Note that all ancestries are shown together. Black dashed line represents the nominal statistical significance threshold (p = 0.05).
(DOCX)
Note that all ancestries are shown together. Black dashed line represents the nominal statistical significance threshold (p = 0.05)
(DOCX)
Note that all ancestries are shown together. Black dashed line represents the nominal statistical significance threshold (p = 0.05).
(DOCX)
(DOCX)
(DOCX)
X-axes are cut at -10 and 10.
(DOCX)
X-axes are cut at -10 and 10.
(DOCX)
X-axes are cut at -10 and 10.
(DOCX)
X-axes are cut at -10 and 10.
(DOCX)
X-axes are cut at -10 and 10.
(DOCX)
X-axes are cut at -10 and 10.
(DOCX)
X-axis cut at 50. Figures show odds ratios and 95% confidence intervals.
(DOCX)
Acknowledgments
We thank the patients who volunteered to all participating cohorts, and the researchers and clinicians who enrolled them into the respective studies. A full list of acknowledgments can be found in S1 and S2 Tables.
Data Availability
Code availability Code guidance is available at https://github.com/DrGBL/WES.WGS. Data availability The exome-wide burden test summary statistics are available in the manuscript's supporting information files. The single variant association studies summary statistics are available openly on the GWAS Catalog[62] (study numbers GCST90132193, GCST90132194, and GCST90132195). Participant level data from each corresponding cohorts may be accessed according to the rules of each cohort’s data sharing policies. Specifically, we refer readers to the following resources: - Biobanque Québecoise de la COVID-19 (BQC-19): https://www.bqc19.ca/ - Sedish biobank: https://swecovid.org/ - Columbia Biobank: https://www.vagelos.columbia.edu/research/researchers/core-and-shared-facilities/new-instruments-and-facilities/columbia-university-biobank - Geisinger Health Systems: https://www.geisinger.edu/research - Helix Exome+ and Healthy Nevada Project: https://healthynv.org/ - Penn Medicine Biobank: https://pmbb.med.upenn.edu/ - GEN-COVID Multicenter Study: https://sites.google.com/dbm.unisi.it/gen-covid - Qatar Genome Program: https://www.qatargenome.org.qa/ - Deutsche COVID-19 OMICS Initiative (DeCOI): https://decoi.eu/ - POLCOVID-Genomika: Medical University of Bialystok ethics board. - FHoGID: Commission cantonale d'éthique de la recherche sur l'être humain (CER-VD, https://www.cer-vd.ch/) - Interval: https://www.intervalstudy.org.uk/ - Saudi Human Genome Program: https://shgp.kacst.edu.sa/index.en.html - Genentech: https://www.gene.com/ - Mount Sinai Clinical Intelligence Center: https://labs.icahn.mssm.edu/minervalab/resources/data-ark/mscic-covid-19-biobank/ - Vanda COVID-19: https://www.vandapharma.com/ - University of California Los Angeles: https://www.uclahealth.org/precision-health/research - Japan COVID-19 Taskforce: https://www.covid19-taskforce.jp/en/home/ - Thai Biobank: Institutional Review Board of the Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand (COA No. 691/2021) - MNM Biosience Polish COVID WGS: https://mnmbioscience.com/ - UK Biobank: https://www.ukbiobank.ac.uk/.
Funding Statement
Genome sequencing of Biobanque Québec COVID-19 was funded by the CanCOGeN HostSeq project, with contribution from Fonds de Recherche Québec Santé (FRQS), Génome Québec, and the Public Health Agency of Canada. The Richards group is supported by the Canadian Institutes of Health Research (CIHR), the Lady Davis Institute of the Jewish General Hospital, the Canadian Foundation for Innovation, the NIH, Cancer Research UK, and FRQS. The Richards research group is supported by the Canadian Institutes of Health Research (CIHR: 365825; 409511, 100558, 169303), the McGill Interdisciplinary Initiative in Infection and Immunity (MI4), the Lady Davis Institute of the Jewish General Hospital, the Jewish General Hospital Foundation, the Canadian Foundation for Innovation, the NIH Foundation, Cancer Research UK, Genome Québec, the Public Health Agency of Canada, McGill University, Cancer Research UK [grant number C18281/A29019] and the Fonds de Recherche Québec Santé (FRQS). JBR is supported by a FRQS Mérite Clinical Research Scholarship. Support from Calcul Québec and Compute Canada is acknowledged. GBL is supported by FRQS and CIHR fellowships. The Columbia COVID-19 Biobank is supported by the Vagelos College of Physicians & Surgeons Office for Research, Precision Medicine Resource, and Biomedical Informatics Resource of the Columbia University Irving Institute for Clinical and Translational Research (CTSA). Columbia CTSA is funded by the National Center for Advancing Translational Sciences (UL1TR001873). The Columbia University COVID-19 Biobank was supported by Columbia University and the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (NIH), through grant no. UL1TR001873. DeCOI NGS was supported by funding from the German Research Foundation (DFG; INST 217/1011-1) and were performed at the DFG-funded NGS Competence Center Tübingen (INST 37/1049-1) – Project-ID 286/2020B01 – 428994620 and the DFG-funded West German Genome Center (INST 216/981-1) - Project Number 407493903. Funding for this work was further received from the Stiftung Universitätsmedizin Essen, Germany. The COMRI cohort is funded through in-house institutional funding of the Technical University of Munich, Munich, Germany. Individual grants are as follows: AS is supported by the BONFOR program of the Medical Faculty, University of Bonn (O-149.0134). KUL is supported by the Emmy-Noether programm of the German Research Foundation (DFG; LU 1944/3-1). RB was supported by the State of Saarland and the Dr. Rolf M. Schwiete Foundation. ECS is supported by the Munich Clinician Scientist Programm (MCSP) and the DFG (SCHU2419/2-1). JR received funding from DFG, RY159/3-1; DFG SFB1403; BMBF COVIM 01KX2021. MA and PS were supported by Netzwerk-Universitaetsmedizin-COVIM: (NaFoUniMedCovid19, FKZ: 01KX2021) and the BMFB (Idepico). For FhOGID: P-YB is supported by the Swiss National Science Foundation (31CA30_196036, 33IC30_179636 and 314730_192616), the Leenaards Foundation, the Santos-Suarez Foundation as well as grants allocated by Carigest. CR is supported by the Swiss National Science Foundation (31CA30_196036, 31003A_176097, and 310030_204285). The GEN-COVID Multicenter Study (Italy) was funded by the MIUR project “Dipartimenti di Eccellenza 2018-2020” to Department of Medical Biotechnologies University of Siena, Italy (Italian D.L. n.18 March 17, 2020), private donors for COVID-19 research, “Bando Ricerca COVID-19 Toscana” project to Azienda Ospedaliero-Universitaria Senese, charity fund 2020 from Intesa San Paolo dedicated to the project N. B/2020/0119 “Identificazione delle basi genetiche determinanti la variabilità clinica della risposta a COVID-19 nella popolazione italiana”, the Italian Ministry of University and Research for funding within the “Bando FISR 2020” in COVID-19 and the Istituto Buddista Italiano Soka Gakkai for funding the project “PAT-COVID: Host genetics and pathogenetic mechanisms of COVID-19” (ID n. 2020-2016_RIC_3). The GEN-COVID (Spain) study received support from Instituto de Salud Carlos III (ISCIII): GePEM (PI16/01478/Cofinanciado FEDER; A.S.), DIAVIR (DTS19/00049/Cofinanciado FEDER, A.S.), Resvi-Omics (PI19/01039/Cofinanciado FEDER, A.S.), ReSVinext (PI16/01569/Cofinanciado FEDER, F.M.T.), Enterogen (PI19/01090/Cofinanciado FEDER, F.M.T.); Agencia Gallega para la Gestión del Conocimiento en Salud (ACIS): BI-BACVIR (PRIS-3, A.S.), and CovidPhy (SA 304 C, A.S.); Agencia Gallega de Innovación (GAIN): Grupos con Potencial de Crecimiento (IN607B 2020/08, A.S.), GEN-COVID (IN845D 2020/23, F.M.T.); Framework Partnership Agreement between the Consellería de Sanidad de la XUNTA de Galicia and GENVIP-IDIS - 2021-2024 (SERGAS-IDIS march 2021); and consorcio Centro de Investigación Biomédica en Red de Enfermedades Respiratorias (CB21/06/00103; F.M.T.). For Genentech: The COVACTA study was supported by F. Hoffmann-La Roche Ltd and, in part, by federal funds received from the U.S. Department of Health and Human Services, Office of the Assistant Secretary for Preparedness and Response, and Biomedical Advanced Research and Development Authority, under grant number HHSO100201800036C. For Helix+ and Healthy Nevada Project: Funding was provided to Desert Research Institute (DRI) by the Nevada Governor's Office of Economic Development. Funding was provided to the Renown Institute for Health Innovation by Renown Health and the Renown Health Foundation. Thai Biobank (Host genetic factors in COVID-19 patients in relation to disease susceptibility, disease severity and pharmacogenomics) funding was obtained from the following sources: 1.Ratchadapiseksompotch Fund, Faculty of Medicine, Chulalongkorn University (RA(PO) 003/63 and 764002-HE01). 2.Grant for the Healthcare-associated Infection Research Group STAR (Special Task Force for Activating Research), Chulalongkorn University (STF 6100430002-1). 3.Grant for Development of New Faculty Staff, Ratchadaphiseksomphot Endowment Fund (DNS 64_002_30_001_2). 4.The e-ASIA Joint Research Program (e-ASIA JRP) as administered by the National Science and Technology Development Agency. 5.Health Systems Research Institute, TSRI Fund (CU_FRB640001_01_30_10) and Thailand Research Fund (DPG6180001). Further, PC is supported by Ratchadapiseksompotch Fund, Faculty of Medicine,Chulalongkorn University, Bangkok, Thailand, Grant number RA(PO) 003/63. VN is supported by Ratchadapiseksompotch Fund, Faculty of Medicine,Chulalongkorn University, Bangkok, Thailand, Grant number RA(PO) 001/63. NH is supported by The e-ASIA Joint Research Program (e-ASIA JRP) as administered by the National Science and Technology Development Agency. VS is supported by Health Systems Research Institute (64-132) and the Ratchadapisek Sompoch Endowment Fund, Chulalongkorn University (764002-HE01), Bangkok, Thailand. Interval was funded by the NHS Blood and Transplant, the National Institute for Health Research, the UK Medical Research Council, and the British Heart Foundation Japan COVID-19 Task Force acknowledges the contribution of Japan Agency for Medical Research and Development (AMED) and Japan Science and Technology Agency (JST). The Japan NCGM-COVID-19 study was supported in part by Grants-in-Aid for Research from the National Center for Global Health and Medicine (20A2009) and the Agency for Medical Research and Development (AMED) (JP20fk0108416 and JP20fk0108104). MNM Diagnostics (Polish COVID WGS) partially supported by the Polish National Science Centre grant No. SZPITALE-JEDNOIMIENNE/2/2020 and by the Medical Research Agency grant No 2020/ABM /COVID19/0022. The Penn Medicine Biobank is supported by Perelman School of Medicine at University of Pennsylvania, a gift from the Smilow family, and the National Center for Advancing Translational Sciences of the National Institutes of Health under CTSA award number UL1TR001878. The POLCOVID-Genomika study was financially supported by the Polish Medical Research Agency (ABM) grant no. 2020/ABM/COVID19/0001. The Qatar Genome Program and Qatar Biobank are both Research, Development & Innovation entities within Qatar Foundation for Education, Science and Community Development. The Saudi COVID-19 acknowledges the Saudi Ministry of Health and King Abdulaziz City for Science and Technology (KACST). The Swedish Biobank received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement No 824110 (EASI-Genomics). Sequencing was performed by the National Genomics Infrastructure SNP&SEQ facility, which is supported by Science for Life Laboratory, the Swedish Research Council, and the Knut and Alice Wallenberg Foundation. UCLA acknowledges OCRC, Microsoft COVID Compute Funding, Illumina in-kind donation. We thank the UCLA COVID-19 Oversight Research Committee, Microsoft COVID Compute Funding, Illumina in-kind donation, and UCLA David Geffen School of Medicine - Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research Award Program" for funding for this project under award #20-10 ("COVID-19 Host Genomics Registry at UCLA" PI:Pasaniuc). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cai Y, Kwek S, Tang SSL, Saffari SE, Lum E, Yoon S, et al. Impact of the COVID-19 pandemic on a tertiary care public hospital in Singapore: Resources and economic costs. J Hosp Infect. 2021. doi: 10.1016/j.jhin.2021.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mulholland RH, Wood R, Stagg HR, Fischbacher C, Villacampa J, Simpson CR, et al. Impact of COVID-19 on accident and emergency attendances and emergency and planned hospital admissions in Scotland: an interrupted time-series analysis. J R Soc Med. 2020;113: 444–453. doi: 10.1177/0141076820962447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395: 497–506. doi: 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakanishi T, Pigazzini S, Degenhardt F, Cordioli M, Butler-Laporte G, Maya-Miles D, et al. Age-dependent impact of the major common genetic risk factor for COVID-19 on severity and mortality. J Clin Invest. 2021;131: e152386. doi: 10.1172/JCI152386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou S, Butler-Laporte G, Nakanishi T, Morrison DR, Afilalo J, Afilalo M, et al. A Neanderthal OAS1 isoform protects individuals of European ancestry against COVID-19 susceptibility and severity. Nat Med. 2021;27: 659–667. doi: 10.1038/s41591-021-01281-1 [DOI] [PubMed] [Google Scholar]
- 6.Gaziano L, Giambartolomei C, Pereira AC, Gaulton A, Posner DC, Swanson SA, et al. Actionable druggable genome-wide Mendelian randomization identifies repurposing opportunities for COVID-19. Nat Med. 2021;27: 668–676. doi: 10.1038/s41591-021-01310-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.COVID-19 Host Genetics Initiative. A first update on mapping the human genetic architecture of COVID-19. Nature. 2022. Aug; 608(7921):E1–E10. Epub 2022 Aug 3. doi: 10.1038/s41586-022-04826-7 ; PMCID: PMC9352569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.COVID-19 Host Genetics Initiative, Niemi MEK, Karjalainen J, Liao RG, Neale BM, Daly M, et al. Mapping the human genetic architecture of COVID-19. Nature. 2021. doi: 10.1038/s41586-021-03767-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellinghaus D, Degenhardt F, Bujanda L, Buti M, Albillos A, Invernizzi P, et al. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N Engl J Med. 2020;383: 1522–1534. doi: 10.1056/NEJMoa2020283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pairo-Castineira E, Clohisey S, Klaric L, Bretherick AD, Rawlik K, Pasko D, et al. Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591: 92–98. doi: 10.1038/s41586-020-03065-y [DOI] [PubMed] [Google Scholar]
- 11.Kousathanas A, Pairo-Castineira E, Rawlik K, Stuckey A, Odhams CA, Walker S, et al. Whole genome sequencing reveals host factors underlying critical Covid-19. Nature. 2022. doi: 10.1038/s41586-022-04576-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tam V, Patel N, Turcotte M, Bossé Y, Paré G, Meyre D. Benefits and limitations of genome-wide association studies. Nat Rev Genet. 2019;20: 467–484. doi: 10.1038/s41576-019-0127-1 [DOI] [PubMed] [Google Scholar]
- 13.Taliun D, Harris DN, Kessler MD, Carlson J, Szpiech ZA, Torres R, et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590: 290–299. doi: 10.1038/s41586-021-03205-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganna A, Satterstrom FK, Zekavat SM, Das I, Kurki MI, Churchhouse C, et al. Quantifying the Impact of Rare and Ultra-rare Coding Variation across the Phenotypic Spectrum. Am J Hum Genet. 2018;102: 1204–1211. doi: 10.1016/j.ajhg.2018.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qian Z, Paul B, Zhiyong L, Jérémie LP, Marcela M-V, Jie C, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science (80-). 2020;370: eabd4570. doi: 10.1126/science.abd4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Povysil G, Butler-Laporte G, Shang N, Wang C, Khan A, Alaamery M, et al. Rare loss-of-function variants in type I IFN immunity genes are not associated with severe COVID-19. J Clin Invest. 2021;131. doi: 10.1172/JCI147834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kosmicki JA, Horowitz JE, Banerjee N, Lanche R, Marcketta A, Maxwell E, et al. Pan-ancestry exome-wide association analyses of COVID-19 outcomes in 586,157 individuals. Am J Hum Genet. 2021;108: 1350–1355. doi: 10.1016/j.ajhg.2021.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mbatchou J, Barnard L, Backman J, Marcketta A, Kosmicki JA, Ziyatdinov A, et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat Genet. 2021;53: 1097–1103. doi: 10.1038/s41588-021-00870-7 [DOI] [PubMed] [Google Scholar]
- 19.Wang X. Firth logistic regression for rare variant association tests. Frontiers in Genetics. 2014. Available: https://www.frontiersin.org/article/10.3389/fgene.2014.00187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeberg H, Pääbo S. The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nature. 2020;587: 610–612. doi: 10.1038/s41586-020-2818-3 [DOI] [PubMed] [Google Scholar]
- 21.Cirulli ET. The Increasing Importance of Gene-Based Analyses. PLOS Genet. 2016;12: e1005852. Available: doi: 10.1371/journal.pgen.1005852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Backman JD, Li AH, Marcketta A, Sun D, Mbatchou J, Kessler MD, et al. Exome sequencing and analysis of 454,787 UK Biobank participants. Nature. 2021;599: 628–634. doi: 10.1038/s41586-021-04103-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howe KL, Achuthan P, Allen J, Allen J, Alvarez-Jarreta J, Amode MR, et al. Ensembl 2021. Nucleic Acids Res. 2021;49: D884–D891. doi: 10.1093/nar/gkaa942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581: 434–443. doi: 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maffucci P, Bigio B, Rapaport F, Cobat A, Borghesi A, Lopez M, et al. Blacklisting variants common in private cohorts but not in public databases optimizes human exome analysis. Proc Natl Acad Sci. 2019;116: 950 LP– 959. doi: 10.1073/pnas.1808403116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathieson I, McVean G. Differential confounding of rare and common variants in spatially structured populations. Nat Genet. 2012;44: 243–246. doi: 10.1038/ng.1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petes C, Odoardi N, Gee K. The Toll for Trafficking: Toll-Like Receptor 7 Delivery to the Endosome. Frontiers in Immunology. 2017. p. 1075. Available: https://www.frontiersin.org/article/10.3389/fimmu.2017.01075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Made CI, Simons A, Schuurs-Hoeijmakers J, van den Heuvel G, Mantere T, Kersten S, et al. Presence of Genetic Variants Among Young Men With Severe COVID-19. JAMA. 2020;324: 663–673. doi: 10.1001/jama.2020.13719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fallerini C, Daga S, Mantovani S, Benetti E, Picchiotti N, Francisci D, et al. Association of Toll-like receptor 7 variants with life-threatening COVID-19 disease in males: findings from a nested case-control study. van de Veerdonk FL, van der Meer JWM, editors. Elife. 2021;10: e67569. doi: 10.7554/eLife.67569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asano T, Boisson B, Onodi F, Matuozzo D, Moncada-Velez M, Maglorius Renkilaraj MRL, et al. X-linked recessive TLR7 deficiency in ~1% of men under 60 years old with life-threatening COVID-19. Sci Immunol. 2021;6. doi: 10.1126/sciimmunol.abl4348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mantovani S, Daga S, Fallerini C, Baldassarri M, Benetti E, Picchiotti N, et al. Rare variants in Toll-like receptor 7 results in functional impairment and downregulation of cytokine-mediated signaling in COVID-19 patients. Genes Immun. 2022;23: 51–56. doi: 10.1038/s41435-021-00157-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drewes G, Ebneth A, Preuss U, Mandelkow EM, Mandelkow E. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell. 1997;89: 297–308. doi: 10.1016/s0092-8674(00)80208-1 [DOI] [PubMed] [Google Scholar]
- 33.Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583: 459–468. doi: 10.1038/s41586-020-2286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaub JR, Stearns T. The Rilp-like proteins Rilpl1 and Rilpl2 regulate ciliary membrane content. Mol Biol Cell. 2013;24: 453–464. doi: 10.1091/mbc.E12-08-0598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Chen S, Li Z, Morrison AC, Boerwinkle E, Lin X. ACAT: A Fast and Powerful p Value Combination Method for Rare-Variant Analysis in Sequencing Studies. Am J Hum Genet. 2019;104: 410–421. doi: 10.1016/j.ajhg.2019.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ying Y, Hong X, Xu X, Chen S, He J, Zhu F, et al. Molecular Basis of ABO Variants Including Identification of 16 Novel ABO Subgroup Alleles in Chinese Han Population. Transfus Med Hemotherapy. 2020;47: 160–166. doi: 10.1159/000501862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hult AK, Yazer MH, Jørgensen R, Hellberg Å, Hustinx H, Peyrard T, et al. Weak A phenotypes associated with novel ABO alleles carrying the A2-related 1061C deletion and various missense substitutions. Transfusion. 2010;50: 1471–1486. doi: 10.1111/j.1537-2995.2010.02670.x [DOI] [PubMed] [Google Scholar]
- 38.Zietz M, Zucker J, Tatonetti NP. Associations between blood type and COVID-19 infection, intubation, and death. Nat Commun. 2020;11: 5761. doi: 10.1038/s41467-020-19623-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.100,000 Genomes Pilot on Rare-Disease Diagnosis in Health Care—Preliminary Report. N Engl J Med. 2021;385: 1868–1880. doi: 10.1056/NEJMoa2035790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spiering AE, de Vries TJ. Why Females Do Better: The X Chromosomal TLR7 Gene-Dose Effect in COVID-19. Frontiers in Immunology. 2021. Available: https://www.frontiersin.org/article/10.3389/fimmu.2021.756262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arnold CG, Libby A, Vest A, Hopkinson A, Monte AA. Immune mechanisms associated with sex-based differences in severe COVID-19 clinical outcomes. Biol Sex Differ. 2022;13: 7. doi: 10.1186/s13293-022-00417-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Solanich X, Vargas-Parra G, van der Made CI, Simons A, Schuurs-Hoeijmakers J, Antolí A, et al. Genetic Screening for TLR7 Variants in Young and Previously Healthy Men With Severe COVID-19. Front Immunol. 2021;12: 719115. doi: 10.3389/fimmu.2021.719115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simpson C, Yamauchi Y. Microtubules in Influenza Virus Entry and Egress. Viruses. 2020. doi: 10.3390/v12010117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.RECOVERY Collaborative Group. Tocilizumab in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial. Lancet (London, England). 2021;397: 1637–1645. doi: 10.1016/S0140-6736(21)00676-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Interleukin-6 Receptor Antagonists in Critically Ill Patients with Covid-19. N Engl J Med. 2021;384: 1491–1502. doi: 10.1056/NEJMoa2100433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts GHL, Park DS, Coignet M V, McCurdy SR, Knight SC, Partha R, et al. AncestryDNA COVID-19 Host Genetic Study Identifies Three Novel Loci. medRxiv. 2020; 2020.10.06.20205864. doi: 10.1101/2020.10.06.20205864 [DOI] [Google Scholar]
- 47.Sankararaman S, Obozinski G, Jordan MI, Halperin E. Genomic privacy and limits of individual detection in a pool. Nat Genet. 2009;41: 965–967. doi: 10.1038/ng.436 [DOI] [PubMed] [Google Scholar]
- 48.Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, et al. Optimal Unified Approach for Rare-Variant Association Testing with Application to Small-Sample Case-Control Whole-Exome Sequencing Studies. Am J Hum Genet. 2012;91: 224–237. doi: 10.1016/j.ajhg.2012.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wainschtein P, Jain D, Zheng Z, Aslibekyan S, Becker D, Bi W, et al. Assessing the contribution of rare variants to complex trait heritability from whole-genome sequence data. Nat Genet. 2022. doi: 10.1038/s41588-021-00997-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warnat-Herresthal S, Schultze H, Shastry KL, Manamohan S, Mukherjee S, Garg V, et al. Swarm Learning for decentralized and confidential clinical machine learning. Nature. 2021;594: 265–270. doi: 10.1038/s41586-021-03583-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williamson EJ, Walker AJ, Bhaskaran K, Bacon S, Bates C, Morton CE, et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature. 2020;584: 430–436. doi: 10.1038/s41586-020-2521-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cirulli ET, White S, Read RW, Elhanan G, Metcalf WJ, Tanudjaja F, et al. Genome-wide rare variant analysis for thousands of phenotypes in over 70,000 exomes from two cohorts. Nat Commun. 2020;11: 542. doi: 10.1038/s41467-020-14288-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mutambudzi M, Niedzwiedz C, Macdonald EB, Leyland A, Mair F, Anderson J, et al. Occupation and risk of severe COVID-19: prospective cohort study of 120 075 UK Biobank participants. Occup Environ Med. 2021;78: 307 LP– 314. doi: 10.1136/oemed-2020-106731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hail Team. Hail 0.2. 2021. Available: https://github.com/hail-is/hail
- 55.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010/07/08. 2010;26: 2190–2191. doi: 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hemani G. random-metal. In: GitHub repository [Internet]. 2017 [cited 15 Mar 2022]. Available: https://github.com/explodecomputer/random-metal
- 57.Guo Y, Long J, He J, Li C-I, Cai Q, Shu X-O, et al. Exome sequencing generates high quality data in non-target regions. BMC Genomics. 2012;13: 194. doi: 10.1186/1471-2164-13-194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC. SIFT missense predictions for genomes. Nat Protoc. 2016;11: 1–9. doi: 10.1038/nprot.2015.123 [DOI] [PubMed] [Google Scholar]
- 59.Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19: 1553–1561. doi: 10.1101/gr.092619.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7: 575–576. doi: 10.1038/nmeth0810-575 [DOI] [PubMed] [Google Scholar]
- 61.Adzhubei I, Jordan DM, Sunyaev SR. Predicting Functional Effect of Human Missense Mutations Using PolyPhen-2. Curr Protoc Hum Genet. 2013;76: 7.20.1–7.20.41. doi: 10.1002/0471142905.hg0720s76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res, 2019, Vol. 47 (Database issue): D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
Note that all ancestries are shown together. Black dashed line represents the nominal statistical significance threshold (p = 0.05).
(DOCX)
Note that all ancestries are shown together. Black dashed line represents the nominal statistical significance threshold (p = 0.05)
(DOCX)
Note that all ancestries are shown together. Black dashed line represents the nominal statistical significance threshold (p = 0.05).
(DOCX)
(DOCX)
(DOCX)
X-axes are cut at -10 and 10.
(DOCX)
X-axes are cut at -10 and 10.
(DOCX)
X-axes are cut at -10 and 10.
(DOCX)
X-axes are cut at -10 and 10.
(DOCX)
X-axes are cut at -10 and 10.
(DOCX)
X-axes are cut at -10 and 10.
(DOCX)
X-axis cut at 50. Figures show odds ratios and 95% confidence intervals.
(DOCX)
Data Availability Statement
Code availability Code guidance is available at https://github.com/DrGBL/WES.WGS. Data availability The exome-wide burden test summary statistics are available in the manuscript's supporting information files. The single variant association studies summary statistics are available openly on the GWAS Catalog[62] (study numbers GCST90132193, GCST90132194, and GCST90132195). Participant level data from each corresponding cohorts may be accessed according to the rules of each cohort’s data sharing policies. Specifically, we refer readers to the following resources: - Biobanque Québecoise de la COVID-19 (BQC-19): https://www.bqc19.ca/ - Sedish biobank: https://swecovid.org/ - Columbia Biobank: https://www.vagelos.columbia.edu/research/researchers/core-and-shared-facilities/new-instruments-and-facilities/columbia-university-biobank - Geisinger Health Systems: https://www.geisinger.edu/research - Helix Exome+ and Healthy Nevada Project: https://healthynv.org/ - Penn Medicine Biobank: https://pmbb.med.upenn.edu/ - GEN-COVID Multicenter Study: https://sites.google.com/dbm.unisi.it/gen-covid - Qatar Genome Program: https://www.qatargenome.org.qa/ - Deutsche COVID-19 OMICS Initiative (DeCOI): https://decoi.eu/ - POLCOVID-Genomika: Medical University of Bialystok ethics board. - FHoGID: Commission cantonale d'éthique de la recherche sur l'être humain (CER-VD, https://www.cer-vd.ch/) - Interval: https://www.intervalstudy.org.uk/ - Saudi Human Genome Program: https://shgp.kacst.edu.sa/index.en.html - Genentech: https://www.gene.com/ - Mount Sinai Clinical Intelligence Center: https://labs.icahn.mssm.edu/minervalab/resources/data-ark/mscic-covid-19-biobank/ - Vanda COVID-19: https://www.vandapharma.com/ - University of California Los Angeles: https://www.uclahealth.org/precision-health/research - Japan COVID-19 Taskforce: https://www.covid19-taskforce.jp/en/home/ - Thai Biobank: Institutional Review Board of the Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand (COA No. 691/2021) - MNM Biosience Polish COVID WGS: https://mnmbioscience.com/ - UK Biobank: https://www.ukbiobank.ac.uk/.