Abstract
Background
The mutations in SARS-CoV-2 variants of concern (VOC) facilitate the virus’ escape from the neutralizing antibodies induced by vaccines. However, the protection from hospitalization and death is not significantly diminished. Both vaccine boosters and infection improve immune responses and provide protection, suggesting that conserved and/or cross-reactive epitopes could be involved. While several important T- and B-cell epitopes have been identified, mainly in the S protein, the M and N proteins and their potential cross-reactive epitopes with other coronaviruses remain largely unexplored.
Aims
To identify and map new potential B- and T-cell epitopes within the SARS-CoV-2 S, M and N proteins, as well as cross-reactive epitopes with human coronaviruses.
Methods
Different bioinformatics tools were used to: i) Identify new and compile previously-reported B-and T-cell epitopes from SARS-CoV-2 S, M and N proteins; ii) Determine the mutations in S protein from VOC that affect B- and T-cell epitopes, and; iii) Identify cross-reactive epitopes with coronaviruses relevant to human health.
Results
New, potential B- and T-cell epitopes from S, M and N proteins as well as cross-reactive epitopes with other coronaviruses were found and mapped within the proteins’ structures.
Conclusion
Numerous potential B- and T-cell epitopes were found in S, M and N proteins, some of which are conserved between coronaviruses. VOCs present mutations within important epitopes in the S protein; however, a significant number of other epitopes remain unchanged. The epitopes identified here may contribute to augmenting the protective response to SARS-CoV-2 and its variants induced by infection and/or vaccination, and may also be used for the rational design of novel broad-spectrum coronavirus vaccines.
Key Words: SARS-CoV-2, B- and T-cell epitopes, Spike protein, Membrane protein, Nucleocapsid protein, Cross-reactivity, COVID-19 vaccines.Introduction
Coronaviruses are characterized by a high rate of genetic recombination and mutation, allowing them to colonize different ecological niches, resulting in the ability to infect and adapt to a wide range of hosts. Seasonal coronaviruses such as HKU1, NL63, 229E and OC43 are known to cause mild respiratory illnesses in humans. Additionally, other coronaviruses such as SARS-CoV, MERS-CoV and SARS-CoV-2 are able to cause severe respiratory illnesses with a high mortality rate. These last three viruses mentioned above have been responsible for major pandemics: SARS-CoV, in 2003, MERS-CoV in 2012 and the on-going pandemic caused by SARS-CoV-2, which began in 2019 (1). To this day, we are still battling to stop the pandemic caused by SARS-CoV-2, which had caused 597 million confirmed cases as of August 2022 and more than 6.4 million deaths worldwide (2,3). Although the effectiveness of SARS-CoV-2 vaccines to protect against infection has been shown to decrease over time, the protection against severe COVID-19 has been maintained (4). On the other hand, the virus is constantly evolving and new variants with increased ability to evade infection and vaccine-mediated immunity have and will continue to emerge (2). The changes appearing in the SARS-CoV-2 genome have affected mainly the gene encoding the spike protein (S) which is used in most of the approved vaccines in the world (5).
The Spike (S), membrane (M) and nucleocapsid (N) proteins are some of the most studied targets because they are the principal immunogens present in SARS-CoV-2 (6,7), and are also the most abundant proteins in the virus. Protein S has been the main target because it mediates the infection of host cells through angiotensin-converting enzyme 2 (ACE2) (8). For that reason, most of the vaccines approved by the World Health Organization (WHO) and the Food and Drug Administration (FDA) (9) target this antigen. In addition, the M protein is another key target for antibody responses, since it induces a dominant and long-lasting immune response in SARS-CoV-2 infected individuals (10). The N protein has played an important role in diagnosing SARS-CoV-2-positive patients, since it is used as a marker in the RT-PCR test (11., 12., 13.), and it has also been identified as an indicator of the progression of COVID-19 immunopathology (14).
Currently, the WHO recognizes five SARS-CoV-2 variants of concern (VOC) (Alpha, Beta, Gama, Delta and Omicron) and seven variants of interest (Epsilon, Eta, Iota, Kappa, Lambda, Theta and Zeta) (15), and this number is likely to increase in the future as long as the virus keeps actively circulating. Variant B.1.1.7 (Alpha) was first identified in the United Kingdom (UK) in September 2020. This variant is highly transmissible thanks to several mutations in the S protein that help the virus adhere more strongly to human cells and also help infected cells create new S proteins more efficiently (16,17). In addition, H69-V70 and Y144/145 deletions alter the shape of the S protein which may help the virus to evade certain antibodies (18). The effectiveness of the BNT162b2 vaccine (Pfizer/BioNTech) against the Alpha variant (89.5%), is similar to that observed in clinical trials (97.4%), showing a modest decrease in neutralizing activity (70%) of samples from vaccinated individuals, while maintaining the same level of protection against severe disease (19,20). Variant B.1.351 (Beta), which was first detected in South Africa, has shown approximately a 20 fold increase in affinity for ACE2 compared to the receptor-binding domain (RBD) of the Wuhan variant, making it more transmissible (21) and better able to escape neutralization by monoclonal antibodies due to the E484K mutation (22). Laboratory data has shown a moderately reduced efficacy against symptomatic disease with this variant, but high levels of efficacy (97%) against severe disease in individuals vaccinated with the BNT162b2 vaccine and 51% efficacy with the NVX-CoV2373 vaccine (Novavax) against this variant (20,23). Another extremely infectious variant, P.1 (Gamma), was first reported in Brazil in mid-2020, where it rapidly increased the number of infections to the point where it caused the country's healthcare system to collapse (24). However, high neutralizing antibody titers against this variant have been measured in serum collected from individuals vaccinated with the BNT162b2 vaccine (25). Variant B.1.617.2 (Delta) was first detected in India in December 2020. The mutations in this variant cause increased replication, leading to high viral loads and increased transmission. Two doses of the BNT162b2 vaccine or ChAdOx1 nCoV-19 (AstraZeneca) vaccine within the previous 39 days are effective in preventing (88 and 67%, respectively) symptomatic disease caused by the delta variant (26). Similarly, the B.1.427 and B.1.429 lineages of the Epsilon variant detected in California, United States, have significant mutations in the S protein (S13I, W152C, and D614G) and the RBD (L452R). Another variant, B.1.525 (Eta), first identified in the UK and Nigeria, presents several significant mutations in the S protein (A67V, 69del, 70del, 144del, D614G, Q677H, and F888L) and RBD (E484K). Some of these mutations are associated with increased infectivity, transmission, and a reduction in neutralization. The B.1.617.1 (Kappa) variant, circulating in India, has important mutations in the S protein (T95I, D614G, E154K, P681R, G142D, and Q1071H) as well as highly crucial mutations in the RBD (E484K and L452R). In the case of the P.2 (Zeta) and P.3 (Theta) variants, first identified in Brazil and Japan, respectively, they present some similar and important mutations in the S protein (D614G and V1176F) and the RBD (E484K), which cause a reduction of neutralization by antibodies and an increase in its ability to cause reinfection (27., 28., 29., 30.). Lastly, in November 2021 the Omicron variant (B.1.1.529) was first detected in South Africa, and it contains 30 mutations in the S protein, amongst them an insertion of three amino acids (EPE 214) and other changes such as V143/Y145 (31). Some of these mutations have been previously reported in the Alpha, Beta, Gamma, and Delta variants, as well as in the Kappa, Zeta, Lambda, and Mu variants, and have been associated with increased transmissibility and evasion of the immune responses (32). Omicron has been the variant in which the greatest number of mutations have been described so far: three within the M protein (D3G, Q19E and A63T) and three substitutions and one deletion of three residues in the N protein (31).
Protective immunity against SARS-CoV-2 involves both antibody and T-cell immune responses; adaptive immune responses mediated by CD8+ and CD4+ T-cells are essential to promote an efficient B-cell and antibody response as well as for the elimination of cells already infected by the SARS-CoV-2 virus, whereas B-cell and antibody responses are important for generating neutralizing antibodies to control the viral infection. Antibody and T-cell responses are directed at several SARS-CoV-2 components such as the M, N and S proteins, among others (33). The S protein has been used as the main target to induce protective antibody and T-cell responses. Conversely, the contribution of other antigens toward the immune response remains largely unexplored (34). It has been suggested that cross-reactivity from pre-existing CD4+ T-cells confers a certain degree of protection against COVID-19; Crotty et al., have proposed several scenarios to explain the role of these cross-reactive cells (35,36). In the first scenario, as the infection occurs, cross-reactive CD4+ T-cells would respond relatively slowly in comparison with the speed of viral replication, making it difficult to control the infection in the upper respiratory tract (URT). Nevertheless, this recall response could be fast enough to provide protection in the lung before the infection reaches this anatomical site, therefore helping to control disease severity. In the second scenario, memory follicular helper cross-reactive CD4+ T-cells would quickly get activated and help trigger a faster and more robust antibody response. This antibody response could then play an important role in controlling the virus, both in the URT and in the lung. Another scenario suggests that resident memory cross-reactive CD4+ T-cells, present in the URT, could restrict viral replication at the site of infection, allowing a controlled and efficient activation of the innate immune system that can successfully fight the infection. It is important to highlight that, although these hypotheses have not been proven, they are considered plausible. This is also supported by the observation that protection from severe disease conferred by vaccines with new variants has been maintained at constant levels despite the significant decrease in neutralizing antibody titers (36).
The aim of the study was to identify potential epitopes of SARS-CoV-2 and other coronaviruses of medical interest. Experimental evidence suggests that cross-reactive responses between coronaviruses occur among the population (37), but there is little information available on the particular epitopes that might drive these responses. Using web-based bioinformatic tools we have been able to build a repertoire of potential B- and T-cell epitopes that are found in SARS-CoV-2, and identify those epitopes that share high percentages of their identity with other coronaviruses relevant to human health. In addition, we present an analysis of mutations found in the S protein of SARS-CoV-2 variants and show that, while several potential epitopes are affected, the changes in immunogenicity are not necessarily detrimental, and in fact, there is an ample repertoire of epitopes that remain unchanged.
Materials and Methods
Epitope Prediction for T-and B-cells
The multiple reference sequences of SARS-CoV-2 S, M, and N proteins of the different seasonal and outbreak coronaviruses were obtained from the NCBI, Genbank. S protein QHR6390.2, M protein YP_009724393.1 and N protein QIC53221.1 sequences were used for the prediction of B- and T-cell epitopes.
The prediction of linear B-cell epitopes was performed through the Immune Epitope Database (IEDB) website (https://www.iedb.org/) using the Bepipred V1.0 and V2.0 linear epitope prediction algorithms (Supplementary 1) (38). Threshold values of 0.35 were used (corresponding to a specificity >0.49 and a sensitivity <0.75) and 0.55 (corresponding to a specificity >0.817 and sensitivity <0.292) with versions 1.0 and 2.0, respectively. Epitopes were chosen based on their IC50 binding values of <50 (high affinity) and <500 (average affinity). To determine whether the predicted B-cell epitopes were exposed on the surface of the protein, surface accessibility and secondary structure NetSurfP-2.0 (39) was used.
Prediction of conformational B-cell epitopes was performed using the ElliPro and DiscoTope 2.0 tools from the IEDB website (Supplementary Table 1) (39., 40., 41., 42., 43., 44., 45., 46., 47., 48., 49., 50.). The prediction of conformational epitopes using Ellipro is based on the protrusion index, which is defined as a range from 0–1, where the residues with higher scores being those that have greater accessibility to the solvent. This is already a default criterion on the server, based on the 3-dimensional (3D) structures of the SARS-CoV-2 S protein (PDB ID: 6VSB_1_1). On the other hand, discontinuous B-cell epitopes were predicted via the DiscoTope 2.0 server tool in IEDB with a default threshold of −3.7 (corresponding to a specificity >0.75 and a sensitivity <0.47), based on the 3D structures of the SARS-CoV-2 S protein (PDB ID: 6VSB_1_1, A chain).
For the prediction of the MHC-I-restricted T-cell epitopes, MHC-I binding predictions (NetMHCpan EL 4.0 method) (51) and Class I immunogenicity tools (52) from the IEDB website were used (Supplementary Table 1). Predictions were made for 6 HLA-A alleles (HLA-A * 01:01, HLA-A * 02:01, HLA-A * 03:01, HLA-A * 11:01, HLA-A * 23:01 and HLA-A * 24:02) and 6 HLA-B alleles ( HLA-B * 07:02, HLA-B * 08:01, HLA-B*35:01, HLA-B*40:01, HLA-B * 44:02 and HLA-B * 44:03) with a peptide length limit of 7 amino acids. The selection of T-cell epitopes was made based on IC50 affinity scores (<500) and immunogenicity scores (>-1 and <1). For the prediction of the MHC-II-restricted T-cell epitopes, we used the algorithm 2.22 recommended by IEDB and TepiTool (45) (Supplementary Table 1). Predictions were made for 3 HLA-DR alleles (HLA-DRB1 * 01: 01, HLA-DRB1 * 04: 01 and HLA-DRB1 * 07: 01), 8 HLA-DP alleles (HLA-DPA1 * 01: 03 / DPB1 * 03: 01, HLA-DPA1 * 02: 01 / DPB1 * 02: 01, HLA-DPA1 * 02: 02 / DPB1 * 02: 02 and HLA-DPA1 * 03: 01 / DPB1 * 23: 01) and 6 HLA-DQ alleles (HLA-DQA1 * 05: 01 / DQB1 * 03: 01, HLA-DQA1 * 01: 01 / DQB1 * 05: 01 and HLA-DQA1 * 03: 01 / DQB1 * 03: 02) with a peptide length limit of 15 amino acids and an average consensus percentile of the 20 prediction threshold. The selection of T-cell epitopes was made based on IC50 affinity scores (<500) (38,51., 52., 53.).
The chosen alleles were used for the predictions since they are present with a high frequency in the Mexican population and also have a significant frequency worldwide (54., 55., 56., 57.). These additional alleles were obtained from the allele frequency database (http://www.allelefrequencies.net/default.asp) (58) (Supplementary Table 1), using the region of North America as the search criteria, and ethnic Mexican groups present in all the State capitals of the Country.
Analysis of SARS-CoV-2 Mutations in Variants Being Monitored
Sequences of the SARS-CoV-2 genome isolated from different countries were accessed through the GISAID database (https://www.gisaid.org/) (59,60). Sequences were filtered from December 2019–December 30, 2021, we used 33 complete SARS-CoV-2 genomes obtained from different patients around the world (i.e., Australia, Germany, Italy, Mexico, Saudi Arabia, Taiwan, Vietnam, China, Belgium, Ecuador, Malaysia, New Zealand, Singapore, USA, Brazil, Estonia, South Africa, England, India, Botswana, and Hong Kong) with at least one sequence belonging to one of the 10 clades established by GISAID, and a representative sequence of each of the VOC sequences that had been reported. Only complete genomes (28000–30000 bps) were included in the analysis, and the reference sequence of NCBI from Wuhan (ID NC_045512.2) (Supplementary Table 2)
The Transeq EMBOSS tools (https://www.ebi.ac.uk/Tools/st/emboss_transeq/) (61,62) were used for sequence translation in the 6 nucleotide-to-amino acid reading frames. In order to find the translated protein within the reading frames and assemble the full protein sequences, we used a combination of Jalview, Clustal Omega and the NCBI protein BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (Supplementary Table 1) (48,49,61,63) using the SARS-CoV-2 reference sequences as the subject (S protein QHR63290.2, M protein YP_009724393.1 and N protein QIC53221.1). Once all the S, M and N protein sequences for the 33 genomes were obtained, we proceeded to make B and T-cell epitope predictions as described in the section “Epitope prediction for T- and B-cells”.
Analysis of Percentage of Identity
In addition to the reference sequences of SARS-CoV-2, we obtained the sequences of other seasonal and pandemic coronaviruses such as HCoV-HKU1 (HKU): S protein YP_173238.1; M protein AYN64564.1; N protein AMM42397.1, HcoV-NL63 (NL63): S protein YP_003767.1; M protein YP_003770.1; N protein AFV53152.1, HcoV-OC43 (OC43): S protein YP_009555241.1; M protein AAA45462.1; N protein QDH43730.1; MERS: S protein AKN11075.1; M protein YP_009047210.1; N protein QFQ59594.1; and, SARS-CoV: S protein AAU04646.1; M protein AAU04638.1; N protein ABI96968.1.
To determine the percentage of identity shared between SARS-CoV-2 and the other coronaviruses for each of the epitopes predicted and described in the section “Analysis of SARS-CoV-2 mutations and clinically significant variants” we used the EMBOSS Needle tool (https://www.ebi.ac.uk/Tools/psa/emboss_needle/) (64) (Supplementary Table 1). This tool can find similarities in the amino acid sequences of two proteins and determine a significant homology between the two (45). The threshold we established to consider a percentage of shared identity as significant was greater than 50% shared identity. This value was decided upon based on experimental data found in public databases (65,66), where cross-reactive epitopes can have percentages of shared identity ranging from as low as 38% to up to 100%. Thus, we chose a value within this range that could still provide some stringency to our analysis.
To generate a graphical representation of the epitopes, we used the full-length SARS-CoV-2 S protein structural model (ID: 6VSB_1_1) (67,68). Graphical representations of M (ID: QHD43419) and N (ID: QHD43423) S protein structures were generated through D-I-TASSER/ C-I-TASER (https://zhanggroup.org/) (69,70). The structures were analyzed using PyMOL software (Schrödinger LLC. Molecular Graphics System (PyMOL) Version 1.80 LLC, New York, NY 2015 (47). The basic online local alignment search tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (63) was used to evaluate the position of the predicted peptides in the protein sequences analyzed in this study.
Results
Prediction of B-cell Linear Epitopes
We conducted a literature review of studies that reported potential epitopes of the spike (S), membrane (M) and nucleocapsid (N) proteins of SARS-CoV-2. This review was done conducting an advanced search using the following words: SARS-CoV-2, epitopes of B and T-cells, S/spike protein, M/membrane protein and N/nucleocapsid protein, and establishing a publication time range in the search engine: PubMed https://pubmed.ncbi.nlm.nih.gov/ (67), Google Scholar https://scholar.google.com/ (71), ScienceDirect https://www.sciencedirect.com/ (72), and COVIDep https://covidep.ust.hk/ (73,74) databases. The review included reports generated between January 2020–January 2022. The initial search yielded a total of 1,361 publications, but only 47 of these were included in the analysis, given that other studies did not indicate the specific epitopes with which they worked.
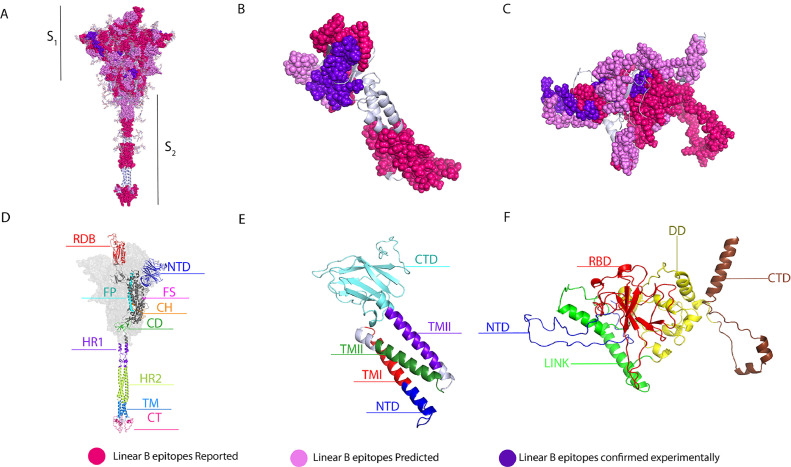
For the spike protein in particular, we were able to compile a total of 116 potential B-cell epitopes, of which 61 were epitopes previously reported in silico (ERIS) (73,75., 76., 77., 78., 79., 80.) by other research groups (Supplementary Table 3), and 19 were experimentally-confirmed epitopes (ECE) (56,78., 79., 80., 81., 82., 83., 84.) (Supplementary Table 4) also reported by others (Figure 1 ). Using BepiPred 1.0 and 2.0, our study predicted 36 epitopes that were selected based on their score and surface accessibility, and which are referred to as “epitopes predicted” in this study (EPITS). From these 36 EPITS, 29 had already been reported in studies by other research groups, and 7 were novel reported epitopes (NRE) (Supplementary Table 4). Epitopes from our predictions that overlapped with any ERIS, were fused into a single polypeptide, and marked in bold characters, and ECEs are shown in red (Supplementary Tables 3 and 4). To visualize the position of the compiled epitopes we used an optimized 3D model of the SARS-CoV-2 spike protein in its trimeric conformation (Figure 1A). ERIS are shown in magenta, EPITS are shown in pink and ECE are shown in purple. B-cell epitopes reported in silico and described in this study are distributed across all domains of the spike protein. The trimeric model shows several predicted epitopes, from this and other studies, that still lack experimental validation, highlighting the importance of further research on the immunogenicity of these predicted epitopes. It should be noted that experimentally confirmed epitopes are in the regions comprising the S1 polypeptide, which contains the N-terminal domain (NTD), the receptor-binding domain (RBD) and the receptor-binding motif (RBM) (Figure 1D) (67,85).
Figure 1.
Linear B-cell epitopes in spike, membrane and nucleocapsid proteins. 3D graphical representations of A. Spike, B. Membrane and C Nucleocapsid proteins of SARS-CoV2. Linear B-cell epitopes reported by in silico studies found in the literature are shown in magenta color, epitopes predicted in this study are shown in pink, and predicted epitopes that have been experimentally confirmed to be immunogenic are shown in purple. The following domains and regions are also shown: D. S protein, in red the receptor binding region (RBD, 336-518), in dark blue the N-terminal domain (NTD, 16-291), in magenta the furin cleavage site (FS), in cyan the fusion peptide (FP, 817-834) (S1/S2), in orange the central helix (CH, 987-1034), in green the connecting domain (CD, 1080-1135), in purple and in green the heptad repeat domains 1 (HR1, 1047-1163) and 2 (HR2, 1163-1210) respectively, in light blue the transmembrane domain (TM, 1214-1234), and in pink the cytoplasmic tail (CT, 1235-1273); E. M protein, in cyan color the connecting domain (CTD, 101-222), the three transmembrane domains in red (TMI, 20-40), green (TMII, 51-71) and purple (TMIII, 80-100) respectively and in dark blue the N-terminal domain (NTD, 1-19 ) F. N protein, in red RNA binding domain (RBD, 50-174), in dark blue the N-terminal domain (NTD, 1-50), in yellow the dimerization domain (DD, 246-365), in brown the connection domain (CTD, 366-419) and in green central linker (LINK, 174-246). X-ray crystallography S protein ID: 6VSB_1_1, M protein ID: QHD43419 and N protein ID: QHD43423.
Following the same protocol that was used for the S protein, we performed the analysis for the membrane (M) and nucleocapsid (N) protein sequences. For the M protein, we were able to collect a total of 18 potential B-cell epitopes (Supplementary Table 3), of which 9 are classified as ERIS (56,73,77,78,81). In addition, we found 2 ECE (78) from the literature, as shown in Supplementary Tables 3 and 4. Using BepiPred 1.0 and 2.0, we predicted and selected 7 EPITS that were predicted both in this study and by other research groups. We did not find any new epitopes for this protein. For the representation of the compiled epitopes, we used a 3-D model of the M protein (Figure 1B), where we highlight the ERIS in magenta, the EPITS in pink, and the ECE are shown in purple. Additionally, we can observe that most of the epitopes reported, predicted and confirmed are located in the C-terminal and the N-terminal domains (Figure 1E) (86).
For the N protein (Figures 1C-1F), a total of 63 potential B-cell epitopes were collected initially; 35 of which were classified as ERIS (56,73,77,83), 5 as ECE (83) (Supplementary Tables 3 and 4), and 23 as EPITS. Of those 23 EPITS, 21 were predicted both in this study and by other research groups. The last 2 were NRE (Supplementary Table 3). The epitopes from our predictions that overlapped with ERIS, were fused into a single polypeptide highlighting in bold characters the EPITS and in red the ECE (Supplementary Table 3). Using the 3D model of the N protein (Figure 1C-1F), we were able to locate and visualize the position of each of the ERIS in magenta, EPITS in pink, and the ECE in purple.
Prediction of B-cell Discontinuous Epitopes of the S Protein
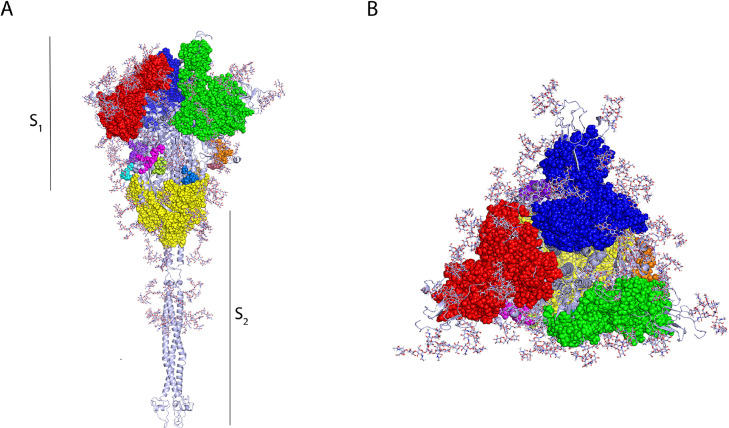
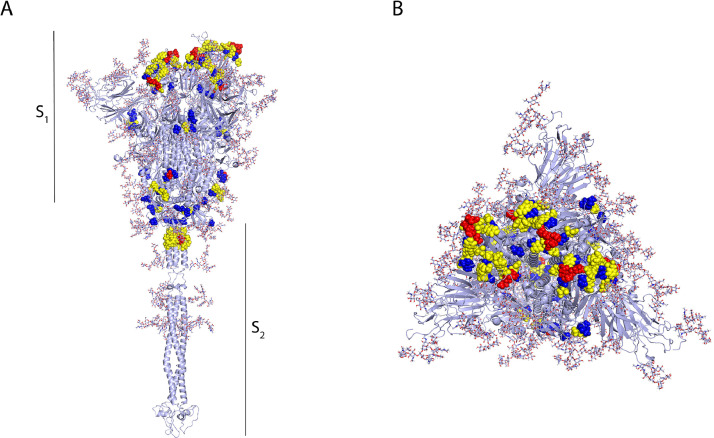
Using the ElliPro and DiscoTope tools from the IEDB website we predicted potential discontinuous epitopes on the SARS-CoV-2 spike glycoprotein. The prediction using ElliPro yielded 12 discontinuous epitopes (Figure 2 ) (Supplementary Table 5) 3 of which have residues belonging to the RBD and residues belonging to the NTD. A single epitope contained residues in the region of the fusion peptides (FP) and in HR1 (Figure 2). The remaining 8 epitopes had residues situated in the transitional region between the S1 and S2 subunits (Figure 2). Three epitopes predicted by ElliPro contained residues from 2 different monomers, these epitopes were shown in red, blue and green, respectively, in Figure 2A. In addition, one epitope shown in yellow in figure 2A, contained residues from each of the 3 monomers of the spike glycoprotein trimer, suggesting the presence of a highly antigenic region within the trimeric structure. Prediction of conformational epitopes using DiscoTope 2.0 resulted in 967 surface residues; of these, 55 were chosen based on a sensitivity of 0.47 and specificity of 0.75 (Supplementary Table 6). The selected residues are distributed mainly across 4 domains (Figure 3 ). In particular, one residue (N282) belongs to the NTD, and the RBD contains 28 of the predicted residues with 27 of these situated in the RBM (Figure 3). Within the S2 region, three residues (P793, I794 and P809) are located in the fusion peptide (FP) and another three residues (N914, Y917 and E918) fall within the HR1. The predicted residues are shown in Figure 3B, where predicted residues with a high specificity (91–100%) are shown in red, the residues with moderate specificity (86–90%) are shown in yellow and residues with a low specificity (75–85%) are shown in blue.
Figure 2.
Conformational B-cell epitopes of the SARS-CoV-2 spike glycoprotein displayed in trimer representation, A. Side and B. Top view. Predictions of conformational epitopes were obtained with the Ellipro tool. The major epitopes between chains A and B are shown in red, chains B and C are shown in green, and chains A and C are shown in yellow.
Figure 3.
Conformational B-cell epitopes of the SARS-CoV-2 spike glycoprotein displayed in monomer representation, A. Side and B. Top view. Predictions of conformational epitopes were obtained with the DiscoTope tool. The selected residues are mainly distributed in 4 domains. In particular, one residue (N282) belongs to the NTD, and the RBD contains 28 of the predicted residues, 27 of them located in the RBM. Within the S2 region, three residues (P793, I794 and P809) are located in the fusion peptide (FP) and three other residues (N914, Y917 and E918) are located within HR1. The predicted residues are shown in Figure 3B, where predicted residues with high specificity (91–100%) are shown in red, residues with moderate specificity (86–90%) are shown in yellow and residues with low specificity (75–85%) are shown in blue.
Prediction of T-cell Epitopes
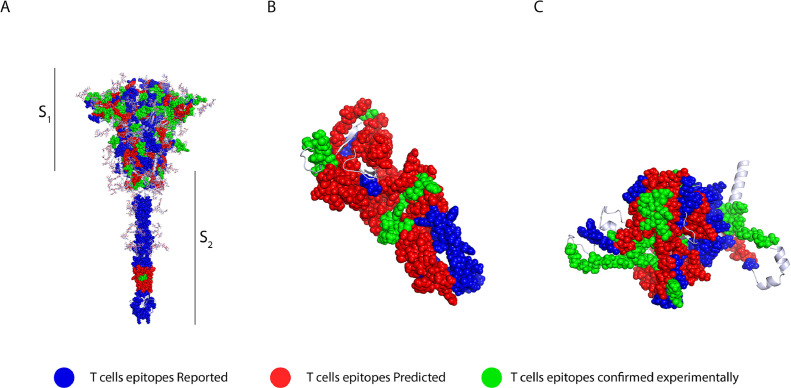
In total, for the S protein, 128 CD8+ and CD4+ T-cells epitopes were identified (Figure 4 A). 89 were identified for the M protein (Figure 4B), and 114 for the N protein (Figure 4C). From these, 73 ERIS were identified for the S protein (56,75,87., 88., 89., 90., 91., 92.), 77 for the M protein (80,81,93., 94., 95., 96., 97.) and 98 for the N protein (56,73,75,84,87., 88., 89., 90., 91., 92., 93., 94., 95., 96., 97.) (Supplementary Table 7). In addition, we were able to compile 9 ECE for the S protein (76,87,89., 90., 91., 92.,98), 3 for the M protein (87) and 5 for the N protein (76,84,87). In addition, 46 EPITS were detected for the S protein, 27 of which were NER: 10 restricted to MHC-I binding, 5 to MHC-II binding, and 12 were predicted to be promiscuous T-cell epitopes (presented to both CD4+ and CD8+ T-cells). For the M protein, 9 EPITS were identified, 2 of which were NER epitopes, one epitope was restricted for MHC-I and one for MHC-II, while no promiscuous peptides were found (Supplementary Table 7). Finally, for the N protein, 11 EPITS were identified, 2 of which were NER restricted to MCH-I, one NER was classified as promiscuous, and none were MHC-II restricted. For the visualization of epitopes, we used an optimized 3D model of the three proteins (Figure 4), the ERIS are shown in blue, while EPITS are shown in green and finally CD4+ or CD8+ T-cells ECE appear in red. The epitopes described in the literature can be found throughout the structure of spike protein, while the predicted epitopes are present throughout the structure, except for a notable gap in the transmembrane domain (TD) (Figure 4A). In the case of the M protein (Figure 4B), the EPITS are evenly distributed throughout the protein, while ECE are mainly situated in the N-terminal and C-terminal domains. Finally, the N protein 3D model shows that the distribution of the epitopes is across the entire protein, while the ECE were mapped mainly in the RNA-binding domain, the binding site and within the dimerization domain (Figure 4C).
Figure 4.
T-cell epitopes in spike, membrane and nucleocapsid proteins. 3D graphical representations of A. Spike, B. Membrane and C. Nucleocapsid proteins of SARS-CoV2. T-cell epitopes reported by in silico studies found in the literature are shown in blue, epitopes predicted in this study are shown in green, and predicted epitopes that have been experimentally confirmed to be immunogenic are shown in red. The epitopes described in the literature are found throughout the spike protein structure; the predicted epitopes are present throughout the structure, except for a notable gap in the transmembrane domain (TD). In the M protein, EPITS are evenly distributed throughout the protein, whereas ECEs are mainly located in the N-terminal and C-terminal domains. On the other hand, the 3D model of the N protein shows that the distribution of epitopes is throughout the entire protein. In contrast, the ECEs were mapped mainly in the RNA-binding domain, the binding site and the dimerization domain.
X-ray crystallography S protein ID: 6VSB_1_1, M protein ID: QHD43419 and N protein ID: QHD43423.
Mutations Present in SARS-CoV-2 Variants of Concern and in Variants Being Monitored
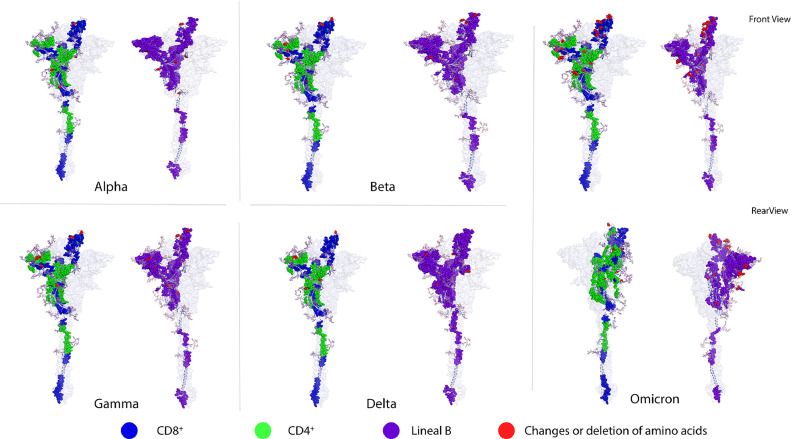
To map the mutations reported for the Alpha, Beta, Gamma, Delta and Omicron variants of concern, we used an optimized 3D model of the S protein, where the mutations that have been reported are shown in red. The model also depicted the EPITS, CD8+ T-cell epitopes which are shown in blue, the CD4+ T-cell epitopes in green, and the linear B-cell epitopes in purple (Figure 5 ). This model shows that mutations in these variants are mainly located in the S1 polypeptide region, leaving the epitopes that are positioned in other regions of the S protein intact.
Figure 5.
Amino acid changes in SARS-CoV-2 variants being monitored (Alpha, Beta and Gamma) and variants of concern (Delta and Omicron). 3D graphical representations of the spike protein of SARS-CoV2 where changes or deletions in specific amino acids are shown in red for Alpha, Beta, Gamma, Delta and Omicron variants. CD8+ T-cell epitopes are shown in blue, CD4+ T-cell epitopes are shown in green, and B-cell epitopes are shown in purple. All epitopes compiled in this work (ER, EP and ED) are shown. RBD, NTD, and S1 incision sites are the main regions of the protein that present amino acid deletions and substitutions. Front and rear views of the Omicron variant are included to highlight the high number of amino acid changes along the protein sequence. X-ray crystallography S protein ID: 6VSB_1_1.
To evaluate epitope modifications in different SARS-CoV2 isolates from different geographical regions, a multiple sequence alignment analysis of the spike, membrane and the nucleocapsid proteins of SARS-CoV-2 was performed (Supplementary Tables 2, 8, 9 and 10). This made it possible to identify epitopes that vary from the reference sequence with respect to other selected sequences. We detected modifications in the sequences of the 3 evaluated proteins (S, M and N). In total we compiled changes in 31 potential linear B-cell epitopes: 15 for S, 2 for M and 14 for N protein, ranging from one amino acid change to displacement of the reading frame from one to two positions (Supplementary Table 8). In the case of peptides presented by HLA-I, we detected alterations in 72 epitopes, 20 for the S protein, 3 for the M protein and 49 for the N protein (Supplementary Table 9). In particular, for the epitopes that are presented by HLA-I, it was noted that even one change in the amino acid sequence, either by substitution or displacement of the reading frame, modified the immunogenicity index, as well as the specificity for the HLA (Supplementary Table 9). For peptides restricted to HLA-II, 57 epitopes were identified among the variants that showed sequence modifications, 36 belonging to the S protein, 6 belonging to the M protein and 15 to the N protein (Supplementary Table 10).
Identity Analysis Between SARS-CoV-2 Proteins and Other Human Coronaviruses
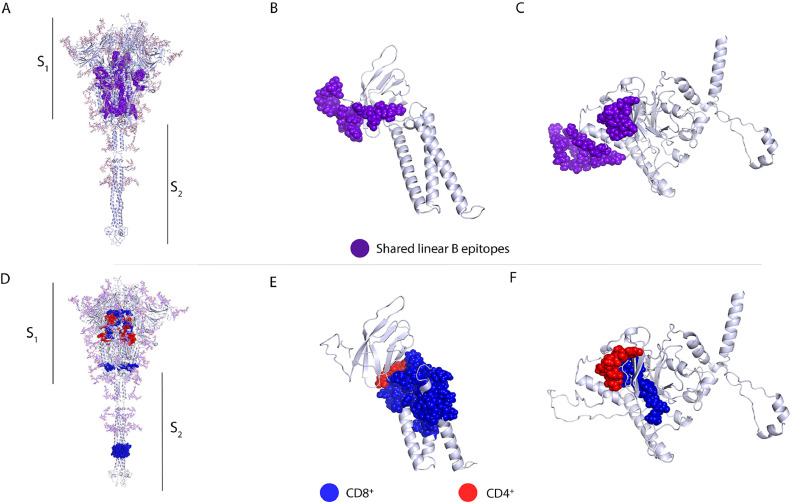
To extend our epitope analysis to other human coronaviruses, a multiple sequence alignment of the S protein from SARS-CoV-2 and other human coronaviruses was performed. We identified 25 linear B-cell epitopes (Figure 6 A) (Supplementary Table 11) that shared a percentage of identity between the S protein of SARS-CoV-2 protein and other human coronaviruses (SARS-CoV, MERS-CoV, HKU1, NL63, OC43, and 229E). This analysis made it possible to identify conserved epitopes among these coronaviruses that could be targets for cross-reactive antibodies. We identified epitope GQSKRVDFC which was the only one preserved among the 6 coronaviruses, with more than 50% shared identity. The same analysis was performed for the M and N proteins. For the M protein, 3 epitopes were identified (Supplementary Table 11) that have more than 50% shared identity between the M protein of SARS-CoV-2 and the M protein of the viruses described above, all preserved epitopes are in the C-terminal domain of the protein (Figure 4B) (80). For the nucleocapsid protein, 7 epitopes with more than 50% shared identity were identified mainly with SARS-CoV, while the shared identity with other coronaviruses varied (Supplementary Table 11). These conserved epitopes were in the N-terminal domain and the RNA-binding domain (Figure 6C) (99).
Figure 6.
Conserved linear B-cell and T-cell epitopes in spike, membrane and nucleocapsid proteins of human coronaviruses. 3D graphical representations of the A, and D. Spike, B, and E. Membrane and C, and F. Nucleocapsid proteins of SARS-CoV2. Epitopes that have above 50% shared identity with epitopes from seasonal coronaviruses (HKU1, NL63, 229E and OC43), SARS-CoV and MERS are highlighted in different colors. Linear B-cell epitopes are shown in purple A–C. CD8+ T-cell epitopes are represented in blue, CD4+ T-cell epitopes are shown in red. All epitopes (ERIS, EPITS and EEC) were included in the analysis. X-ray crystallography S protein ID: 6VSB_1_1, M protein ID: QHD43419 and N protein ID: QHD43423.
The strategy described above using the EMBOSS Needle, was also used to determine conserved CD8+ or /and CD4+ T-cell epitopes. For the S protein, 10 different MHC-I-restricted epitopes were identified (Figure 6 in blue) that have a certain shared identity with the seven human coronaviruses (Supplementary Table 12). Most conserved epitopes are located in fusion peptides (FP), the central helical region (CH), the C-terminal domain (CTD), and in the heptad repeat 2 (HR2) protein regions (Figure 6D). Supplementary Table 12 shows the conserved peptide sequences with more than 50% shared identity between the SARS-CoV-2 sequence and that of the other six coronaviruses. For epitopes restricted to MHC-II (red), we identified 9 conserved epitopes located in the same protein regions as described above for MHC-I-restricted epitopes. For the M protein, we identified one epitope restricted to MHC-I (blue) and one restricted to MHC-II (red), with >50% shared identity between SARS-CoV-2 and the other 6 human coronaviruses (Figure 6E) (Supplementary Table 12). In the case of the N protein, 3 conserved MHC-I epitopes (blue) that shared more than 50% shared identity with other human coronaviruses were identified; these epitopes were located between the RNA-binding domain and the binding site. For MHC-II restricted epitopes (red), only one epitope in the N protein was identified (Supplementary Table 12, and Figure 6F). SARS-CoV and SARS-CoV-2 epitope sequences shared the highest percentage of identity. This was expected given that SARS-CoV is the closest evolutionary virus to SARS-CoV-2 amongst all the human coronaviruses (100,101). In the case of epitopes that are considered promiscuous (that can be recognized by both CD8+ and CD4+ T-cells) 14 were found in the S protein, none of which had more than 50% shared identity between SARS-CoV-2 and all the other human coronaviruses. For the M protein, 6 promiscuous epitopes were identified, while for the N protein, 7 promiscuous epitopes were found (Supplementary Table 12).
Discussion
The SARS-CoV-2 S protein interacts with the host cell receptor, ACE2, to mediate virus infection. For this reason, the S protein, or its RBD domain, have been used to develop most of the anti-COVID-19 vaccines that are currently in use around the globe. As the pandemic progressed, several mutations within this protein have been identified in different SARS-CoV-2 variants of concern. At different levels, these mutations render the virus resistant to neutralization mediated by antibodies generated from a previous infections or vaccination, resulting in a reduced immunity against infection with these variants. However, the level of protection against serious disease, hospitalization and death has remained constant (102,103).
Through the literature review and epitope prediction exercises carried out in this work, we were able to compile 116 linear B-cell epitopes within the S protein, reporting 7 new epitopes, most of them located at the RBD, the junction of S1/S2 and the furin site (Figure 1A). Recent studies identified neutralizing antibodies directed at these sites. In that sense, the new epitopes reported here are potential targets for neutralizing antibodies (78,104). Several regions of the S protein have been shown to be targets for neutralizing antibodies. Of these, the RBD is one of the most important binding sites (Figure 1A). It has been reported that the epitope GDEVRQIAPGQTGKIADYNYKLPDD, which overlaps with one of the epitopes predicted in this work (Supplementary Table 3), generates neutralizing antibodies in mice (105). In addition, other studies have shown that different monoclonal antibodies that interact with multiple residues at this site, exhibit neutralizing activity (96,106). The epitopes ASYQTQTNSPRRARSVASQ and IIAYTMSLGAENSVAYSNN (Supplementary Table 3), predicted in our study, have also been described by Polyiam et al. within the SYQTQTNSPRRARSVASQSIIAYTMSLGAENSVAYSN polypeptide. These epitopes are located at the incision site of S1/S2 (Figure 1A), where the RRAR sequence is recognized and cleaved by the furin protease, resulting in the separation of the S1 and S2 domains during virus assembly. Antibodies directed at this immunodominant region could block the cleavage of the S protein during the viral invasion process (78). Both the Alpha and Beta SARS-CoV-2 variants exhibit amino acid substitutions in these epitopes, and we could also identify these changes in sequences from viruses isolated in several countries such as Estonia, South Africa and Singapore (Supplementary Table 8). Of particular importance, the Beta variant presents a change in the IIAYTMSLGAENSVAYSNN peptide, where the A701V substitution affects this epitope at the cleavage site of cathepsin L (107).
The peptides ILPDPSKPSKRS, FIEDLLFNKVTLADAGFFIKQYGDCLG and PSKPSKRSFIEDLLFNKV are neutralizing epitopes located at the S2 excision site (residues 805-842). Antibody binding at this region results in the inhibition of the molecule excision. Additionally, we located an epitope at the cytoplasmic site of the S protein (CKFDEDDSEPVLKGVKLHYT-1234-1273) (Figure 1A) (Supplementary Table 3), which has been identified as an immunodominant epitope (94,108). This epitope has not been shown to generate neutralizing antibodies against the SARS-CoV-2 virus. However, a study with the porcine epidemic diarrhea virus (PEDV) (an alpha coronavirus) reported B-cell epitopes for neutralizing antibodies located at the cytoplasmic region of the S protein (109). These results suggest that this region could be another important targets for neutralizing antibodies within the SARS-CoV-2 S protein.
Recent studies have reported individuals who recovered from SARS-CoV infection that have neutralizing antibodies against the virus but are not able to neutralize SARS-CoV-2. However, when these subjects were vaccinated with one or two doses of the BNT162b2 mRNA vaccine, they produced high-level, broad-spectrum antibody responses that could effectively neutralize all of the SARS-CoV-2 variants of interest and seasonal human coronaviruses (28,110). Given that the NTD and RBD are the main targets for neutralization, we searched for epitopes conserved between SARS-CoV and SARS-CoV-2 in these regions that could explain cross-neutralization (29). We could not find any conserved linear B-cell epitopes that had at least 50% shared identity between the two coronaviruses, suggesting that neutralization at these sites is mainly mediated by conformational epitopes.
As was the case for the S protein, immuno-dominant epitopes have also been reported for the SARS-CoV-2 M and N proteins. In patients with severe disease treated in the Intensive Care Unit (ICU), high IgG titers specific to the S protein linear B-cell epitopes TESNKKFLPFQQFGRDIA, PSKPSKRSFIEDLLFNK and to the N protein epitope NNAAIVLQLPQGTTLPKG have been found (80). Additionally, the N protein epitope mentioned above has been associated with lymphopenia in patients with COVID-19 (14). These S and N epitopes have a low mutation rate (<2%) and could be used as markers for COVID-19 induced immunopathology (14).
Even though neutralizing antibody responses are involved in protection against COVID-19 induced by SARS-CoV-2 infection or vaccination, T-cell immune responses have also been identified as an extremely important component of the immunity against COVID-19. A study in patients with mild and severe COVID-19 showed the presence of effector and central memory SARS-CoV-2-specific T-cells. In particular, mild cases generated higher levels of cytokine-producing CD8+ T-cells (111). Strong memory-specific T-cell responses to SARS-CoV-2 have also been detected in individuals who had mild and asymptomatic infections, in some cases even in the absence of an antibody response (112). Another study reported T-cell responses specific to SARS-CoV-2 peptide stimulation in pre-pandemic samples, which suggests T-cell cross-reactivity with seasonal coronaviruses (56). Although pending experimental confirmation, our work provides a panel of 33 T-cell epitopes that could potentially be involved in cross-reactive T-cell responses to different coronaviruses (Supplementary Table 12).
Even though ElliPro and DiscoTope yielded the prediction of epitopes in similar regions of the spike glycoprotein, there are limitations in the accuracy of these predictions given the nature of conformational epitopes. As more experimental data is generated, predictions of conformational epitopes for the SARS-CoV-2 spike glycoprotein will become more refined and precise. Despite the limitations, we believe that investigating potential neutralizing antibodies against the predicted residues should be pursued. Given that coronaviruses have a latent pandemic potential, we were interested to see if amongst the epitopes compiled in this study there were conserved epitopes between the SARS-CoV-2 spike glycoprotein and other relevant human coronaviruses. We identified only 7 linear B-cell epitopes that shared a certain identity between the SARS-CoV-2 spike glycoprotein and that of the other coronaviruses. The percentage of shared identity between peptides ranged from 19–100%, with only one epitope with at least 60% shared identity between SARS-CoV-2 and all the other coronaviruses. Recent studies have shown that anti-spike antibodies generated in response to SARS-CoV infection recognize the spike glycoprotein of SARS-CoV-2 and vice versa, suggesting antigenic similarities between the spike glycoprotein of these two viruses. However, these cross-reactive antibodies did not show any neutralizing activity against any other virus except the one that caused the infection (113). Given that SARS-CoV and SARS-CoV-2 share the highest similarity, it is highly unlikely that these antibodies could have neutralizing activity against any of the other human coronaviruses. Another study showed that memory B-cells from convalescent patients once infected with SARS-CoV, produce a repertoire of monoclonal antibodies that cross-neutralize SARS-CoV-2, while they showed no binding affinity for the OC43 or MERS spike glycoproteins (114). A different study showed that antibody responses against infection with seasonal human coronaviruses elicits neutralizing antibodies against SARS-CoV-2. However, the responses were measured shortly after infection occurred and therefore there is no information about the potential longevity of such responses (115). All these observations show that cross-reactive humoral responses between coronaviruses are plausible. However, it is still not clear whether they are robust enough to provide protection against infection and if they are long-lasting.
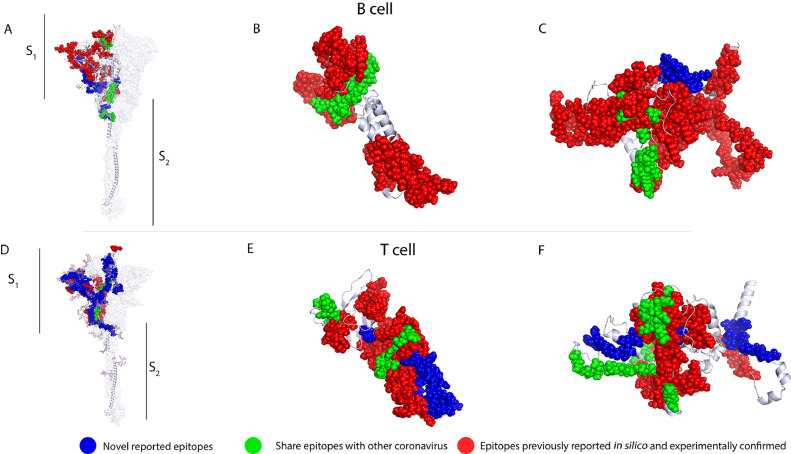
In vitro studies have shown that specific CD4+ and CD8+ lymphocytes from convalescent patients are activated by S protein peptides (37,116,117). In addition, a study with the BNT162b2 vaccine reported that before vaccination, SARS-CoV-2 S protein specific T-cells were found in some unexposed individuals, suggesting these clones were induced by exposure to seasonal coronaviruses (110). The presence of cross-reactive T-cells has been associated with better humoral and cellular responses to vaccination, and the authors also reported the presence of Th1 CD4+ and polyclonal CD8+ T-cells as well as central memory CD4+ and CD8+ T-cells for 6 months after immunization, suggesting long-lasting T-cell responses induced by vaccination (118). These cross-reactive epitopes are located in the S2 region, which, compared to the S1 and the RBD in particular, is less polymorphic (Supplementary Table 12), supporting the use of these epitopes for the design of broad-spectrum vaccines (Figures 7 A-7F, and Supplementary Table 13).
Figure 7.
The locations of proposed epitopes set for a possible broad-spectrum T and B-cell vaccine candidate, as well as potential cross-reactive epitopes, including, newly reported epitopes in blue, shared epitopes with other coronavirus are shown in green and epitopes previously reported in silico and experimentally confirmed are shown in red. 3D graphical representations of spike A and D. membrane B and E. and nucleocapsid C and F proteins of SARS-CoV-2.
In our study, we were able to detect several CD4+ and CD8+ T-cell epitopes, as well as promiscuous epitopes, most of which are located in the S2 region (Supplementary Table 7). The study of epitopes of this region could be also relevant to study the memory T-cell compartment induced by SARS-CoV-2 infection or vaccination. In addition to the S protein epitopes reported here, we found 83 and 105 epitopes within the M and N proteins, respectively. In the case of the M protein, only one MHC-I and one MHC-II epitope were found; whereas for the N protein, only two MHC-I epitopes were found (Figures 4B and 4C). In a previous study, 34 participants with severe and mild COVID-19 responded to M protein peptides, 11 responded to GAVILRLRGHLRIAGHHLGR, 16 to TSRTLSYYKLGASQRVA and 3 to LLESELVIGAVILRGHLR (Supplementary Table 7). The first two epitopes activated CD4+ T-cells and the later CD8+ T-cells (83). These peptides were also found by our analysis. Furthermore, the M protein epitope LRGHLRIAGHHLGRCDIKDL has previously been described as a highly conserved epitope. A study conducted by Heide et al. reported that this peptide was recognized by 12 out of 34 patients with COVID-19, inducing CD4+ T-cells to polarize to an effector memory phenotype. These data suggest that M protein epitopes could also be relevant in the induction of immunity against SARS-CoV-2 (119).
As for the N protein, a study reported that in 19 out of 37 donors of peripheral blood cells who had not been exposed to SARS-CoV or to SARS-CoV-2, the presence of SARS-CoV-2-specific CD4+ IFNγ T-cells was detected (119), again suggesting potential cross-reactive responses with seasonal coronaviruses. In addition, T-cell responses against NSP7 and NSP13 non-structural proteins have been identified in donors with no previous exposure to SARS-CoV or SARS-CoV-2 (111), whereas donor samples from COVID-19 and SARS recovered patients reacted preferentially to the N protein. In addition, the characterization of specific responses to the N protein in a donor without prior exposure to SARS-CoV and SARS-CoV-2, identified the CD4+ and CD8+ T-cell epitope MKDLSPRWFYYLGTGPEAG, considered to be a promiscuous epitope by Peng et al.; this epitope was also recognized by 12 out of 34 patients who recovered from mild and severe COVID-19 (83). Part of this epitope, from position 104–113 (Supplementary Table 7), was also found by our analysis, and is located within an N-protein region with a high degree of similarity to the MERS-CoV, OC43, and HKU1 N proteins. Therefore, we consider this epitope could be relevant for broadly protective vaccines.
An important issue is whether the immunity generated by infection or vaccination with the SARS-CoV-2 Wuhan strain, and with its S protein, respectively, could provide protection against infection, symptomatic mild or severe disease or death caused by the variants of concern of this virus. Mapping the amino acid changes within the S protein from the variants of concern together with the B- and T-cell epitopes reported here, revealed that the RBD, NTD and S1 incision site (Figure 5), are the main regions of the protein that presented amino acid deletions and substitutions. These changes were found in B- and T-cell epitopes and might alter their antigenicity. Such is the case with several B-cell epitopes (GDEVRQIAPGQTGKIADYNYKLPDD, YQAGSTPCNGV, and YGFQPTNGVGYQ) and T-cells epitopes (NATRFASVYAWNRK, CVADYSVLYNSASFSKCYGVSPTKLN, DLCFTNVYADSFVI, RQIAPGQTGKIA, TPCNGVEGFNCY, LQSYGFQPTNGVG, and YGYQPYRVVLSF) reported in this study. Additionally, epitope DPFLGVYYHKNNKSWMESEFRVYSSANNCTFEYVSQPFLM is recognized by the monoclonal antibody 4A8 (120), where a deletion in the alpha variant Y144/14 and a substitution in the gamma variant R190S have also been reported (Supplementary Table 8).
Data gathered involving the Omicron variant has revealed that the N440K, G446S, G496S, and Q498R mutations confer the ability to escape antibody responses. It was also found that the Q498S and N501S mutations are involved in immune evasion mechanisms through improving the binding to the ACE2 receptor (121) (Figure 5) (Supplementary Tables 8-10). This might have an impact on vaccine effectiveness, since it has been reported that the sera from patients recovered from infection with the Wuhan strain, or vaccinated with the original strain S protein vaccine, present a reduction on neutralizing antibody titres against the variants of concern. The neutralizing capacity of antibodies against Omicron is importantly reduced in healthy and COVID-19 convalescent individuals who completed their vaccination scheme with BNT162b2 (Pfizer/BioNTech) or mRNA-1273 (Moderna-mRNA) vaccines, as well as in COVID-19 convalescent individuals (not vaccinated). This suggests that, despite the vaccination status, protection could also be affected. In particular, epidemiological data shows that previously acquired immunity is not highly protective against symptomatic infection with other variants (122., 123., 124.). However, we found multiple cross-reactive B- and T-cell epitopes that are not altered within the different variants, suggesting that protection against symptomatic or severe disease could be maintained in these individuals although the protection against infection may be compromised. It has been noted that the majority of patients with a third vaccine dose did not present severe symptoms against different variants including Omicron, suggesting that vaccine boosters do amplify the neutralizing capacity of antibodies. In addition, we hypothesize that conserved epitopes could be involved in protection against severe disease or death with novel variants (122,124).
In conclusion, the results of this work and the experimental evidence that emerges daily, support the theory that cross-reactive B- and T-cell responses to coronaviruses’ common epitopes, could play a key role in the immunity to SARS-CoV-2 and its variants. We found new B- and T-cell potential epitopes in SARS-CoV-2 S, M and N proteins, and we identified T- and B-cell epitopes with a high percentage of shared identity with other human coronaviruses. We noted that T-cell epitopes have higher shared identity percentages compared to B-cell epitopes. In addition, we analyzed the mutations present within the S protein of SARS-CoV-2 and its variants, and observed that most epitope changes are located on the S1 region. Furthermore, we found that the greater number of changes located within total epitopes are found in Delta, followed by Beta, Gamma, Omicron and Alpha. Nevertheless, an important number of epitopes remained unchanged among these variants, suggesting that these conserved S, M and N protein epitopes could mediate the cross-protection induced by infection and might be involved in the protection against new SARS-CoV-2 variants. Taken together, this knowledge could be useful for the rational design of new and broad-spectrum SARS-CoV-2 vaccines.
Conflicts of Interest
All authors declare no conflicts of interest.
Acknowledgments
We would like to thank Guillermo Ramón Torres for kindly facilitating the use of PyMOLⓇ software in order to build the 3D models presented in this manuscript. Project was sponsored by the Consejo Nacional de Ciencia y Tecnología (CONACYT) Mexico Grant No. 313494 awarded to CLM. The sponsors played no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. SSR-H CVU-921827 and DP-O CVU-890789 receive grants from Consejo Nacional de Ciencia y Tecnología (CONACyT) and SNI III research assistant
Archives of Medical Research 53 (2022) x–x
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.arcmed.2022.10.007.
Appendix. Supplementary materials
References
- 1.Yang H, Rao Z. Structural biology of SARS-CoV-2 and implications for therapeutic development. Nat Rev Microbiol. 2021;19:685–700. doi: 10.1038/s41579-021-00630-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.ArcGIS Dashboards 2020. Available from: https://www.arcgis.com/apps/dashboards/bda7594740fd40299423467b48e9ecf6. (Accessed November 22, 2021).
- 3.Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ssentongo P, Ssentongo AE, Voleti N, et al. SARS-CoV-2 vaccine effectiveness against infection, symptomatic and severe COVID-19: a systematic review and meta-analysis. BMC Infect Dis. 2022;22:439. doi: 10.1186/s12879-022-07418-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farmacovigilancia de vacunas para COVID-19 - Catálogo. Farmacovigilancia de vacunas para COVID-19 2022. Available from: https://covid-19pharmacovigilance.paho.org/(Accessed August 10, 2022).
- 6.Kim D, Lee JY, Yang JS, et al. The Architecture of SARS-CoV-2 Transcriptome. Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas S. The Structure of the Membrane Protein of SARS-CoV-2 Resembles the Sugar Transporter SemiSWEET. Pathog Immun. 2020;5:342–363. doi: 10.20411/pai.v5i1.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pérez-Abeledo M, Sanz Moreno JC. [SARS-CoV-2 variants, a still unfinished story] Vacunas. 2021;22:173–179. doi: 10.1016/j.vacun.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karamloo F, König R. SARS-CoV-2 immunogenicity at the crossroads. Allergy. 2020;75:1822–1824. doi: 10.1111/all.14360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolgin E. Pan-coronavirus vaccine pipeline takes form. Nat Rev Drug Discov. 2022;21:324–326. doi: 10.1038/d41573-022-00074-6. [DOI] [PubMed] [Google Scholar]
- 11.Thieme CJ, Anft M, Paniskaki K, et al. Robust T-cell Response Toward Spike, Membrane, and Nucleocapsid SARS-CoV-2 Proteins Is Not Associated with Recovery in Critical COVID-19 Patients. Cell Rep Med. 2020;1 doi: 10.1016/j.xcrm.2020.100092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hikichi T, Sakamoto M, Harada M, et al. Identification of cytotoxic T-cells and their T-cell receptor sequences targeting COVID-19 using MHC class I-binding peptides. J Hum Genet. 2022;67:411–419. doi: 10.1038/s10038-022-01013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu HS, Hsieh YC, Su IJ, et al. Early detection of antibodies against various structural proteins of the SARS-associated coronavirus in SARS patients. J Biomed Sci. 2004;11:117–126. doi: 10.1007/BF02256554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan BE, Chong VCL, Chan SSW, et al. Hematologic parameters in patients with COVID-19 infection. Am J Hematol. 2020;95:E131–E134. doi: 10.1002/ajh.25774. [DOI] [PubMed] [Google Scholar]
- 15.Chakraborty C, Bhattacharya M, Sharma AR. Present variants of concern and variants of interest of severe acute respiratory syndrome coronavirus 2: Their significant mutations in S-glycoprotein, infectivity, re-infectivity, immune escape and vaccines activity. Rev Med Virol. 2022;32 doi: 10.1002/rmv.2270. E2270. [DOI] [Google Scholar]
- 16.Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Virological. 2020 https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 Available from: (Accessed September 6, 2022) [Google Scholar]
- 17.Shrock E, Fujimura E, Kula T, et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020;370 doi: 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomson EC, Rosen LE, Shepherd JG, et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184:1171–1187. doi: 10.1016/j.cell.2021.01.037. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdool Karim SS, de Oliveira T. New SARS-CoV-2 Variants - Clinical, Public Health, and Vaccine Implications. N Engl J Med. 2021;384:1866–1868. doi: 10.1056/NEJMc2100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abu-Raddad LJ, Chemaitelly H, Butt AA. National Study Group for COVID-19 Vaccination. Effectiveness of the BNT162b2 Covid-19 Vaccine against the B.1.1.7 and B.1.351 Variants. N Engl J Med. 2021;385:187–189. doi: 10.1056/NEJMc2104974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang P, Nair MS, Liu L, et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 22.Zhou D, Dejnirattisai W, Supasa P, et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184:2348–2361. doi: 10.1016/j.cell.2021.02.037. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shinde V, Bhikha S, Hoosain Z, et al. Efficacy of NVX-CoV2373 Covid-19 Vaccine against the B.1.351 Variant. N Engl J Med. 2021;384:1899–1909. doi: 10.1056/NEJMoa2103055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faria NR, Mellan TA, Whittaker C, et al. Genomics and epidemiology of a novel SARS-CoV-2 lineage in Manaus, Brazil. medRxiv. 2021 doi: 10.1101/2021.02.26.21252554. (Accessed September 23, 2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parry H, Tut G, Bruton R, et al. mRNA vaccination in people over 80 years of age induces strong humoral immune responses against SARS-CoV-2 with cross neutralization of P.1 Brazilian variant. Elife. 2021;10:e69375. doi: 10.7554/eLife.69375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez Bernal J, Andrews N, Gower C, et al. Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant. N Engl J Med. 2021;385:585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng X, Garcia-Knight MA, Khalid MM, et al. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell. 2021;184:3426–3437. doi: 10.1016/j.cell.2021.04.025. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jangra S, Ye C, Rathnasinghe R, Personalized Virology Initiative study group SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. Lancet Microbe. 2021;2:e283–e2834. doi: 10.1016/S2666-5247(21)00068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yadav PD, Sapkal GN, Abraham P, et al. Neutralization Potential of Covishield Vaccinated Individuals Sera Against B.1.617.1. Clin Infect Dis. 2022;74:558–559. doi: 10.1093/cid/ciab483. [DOI] [PubMed] [Google Scholar]
- 30.Mansbach RA, Chakraborty S, Nguyen K, et al. The SARS-CoV-2 Spike variant D614G favors an open conformational state. Sci Adv. 2021;7:eabf3671. doi: 10.1126/sciadv.abf3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He X, Hong W, Pan X, et al. SARS-CoV-2 Omicron variant: Characteristics and prevention. MedComm (2020) 2021;2:838–845. doi: 10.1002/mco2.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilhelm A, Widera M, Grikscheit K, et al. Reduced neutralization of SARS-CoV-2 Omicron variant by vaccine Sera and monoclonal antibodies. bioRxiv. 2021 https://www.medrxiv.org/content/10.1101/2021.12.07.21267432v4 Available from: (Accessed January 5, 2022) [Google Scholar]
- 33.Prompetchara E, Ketloy C, Palaga T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. 2020;38:1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- 34.Jeyanathan M, Afkhami S, Smaill F, et al. Immunological considerations for COVID-19 vaccine strategies. Nat Rev Immunol. 2020;20:615–632. doi: 10.1038/s41577-020-00434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loyal L, Braun J, Henze L, et al. Cross-reactive CD4+ T-cells enhance SARS-CoV-2 immune responses upon infection and vaccination. Science. 2021;374:eabh1823. doi: 10.1126/science.abh1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lipsitch M, Grad YH, Sette A, et al. Cross-reactive memory T-cells and herd immunity to SARS-CoV-2. Nat Rev Immunol. 2020;20:709–713. doi: 10.1038/s41577-020-00460-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grifoni A, Weiskopf D, Ramirez SI, et al. Targets of T-cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501. doi: 10.1016/j.cell.2020.05.015. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vita R, Mahajan S, Overton JA, et al. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 2019;47:D339–S343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klausen MS, Jespersen MC, Nielsen H, et al. NetSurfP-2.0: Improved prediction of protein structural features by integrated deep learning. Proteins. 2019;87:520–527. doi: 10.1002/prot.25674. [DOI] [PubMed] [Google Scholar]
- 40.Ponomarenko J, Bui HH, Li W, et al. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics. 2008;9:514. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kringelum JV, Lundegaard C, Lund O, et al. Reliable B-cell epitope predictions: impacts of method development and improved benchmarking. PLoS Comput Biol. 2012;8 doi: 10.1371/journal.pcbi.1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clark K, Karsch-Mizrachi I, Lipman DJ, et al. GenBank. Nucleic Acids Res. 2016;44:D67–D72. doi: 10.1093/nar/gkv1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fleri W, Paul S, Dhanda SK, et al. The Immune Epitope Database and Analysis Resource in Epitope Discovery and Synthetic Vaccine Design. Front Immunol. 2017;8:278. doi: 10.3389/fimmu.2017.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dhanda SK, Mahajan S, Paul S, et al. IEDB-AR: immune epitope database-analysis resource in 2019. Nucleic Acids Res. 2019;47:W502–W506. doi: 10.1093/nar/gkz452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 46.Zheng W, Zhang C, Li Y, et al. Folding non-homologous proteins by coupling deep-learning contact maps with I-TASSER assembly simulations. Cell Rep Methods. 2021;1 doi: 10.1016/j.crmeth.2021.100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chaudhari R, Li Z. PyMine: a PyMOL plugin to integrate and visualize data for drug discovery. BMC Res Notes. 2015;8:517. doi: 10.1186/s13104-015-1483-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sievers F, Higgins DG. Clustal omega. Curr Protoc Bioinformatics. 2014;48 doi: 10.1002/0471250953.bi0313s48. 3.13.1–16. [DOI] [PubMed] [Google Scholar]
- 49.Waterhouse AM, Procter JB, Martin DMA, et al. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gonzalez-Galarza FF, McCabe A, Santos EJMD, et al. Allele frequency net database (AFND) 2020 update: gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res. 2020;48:D783–D788. doi: 10.1093/nar/gkz1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jurtz V, Paul S, Andreatta M, et al. NetMHCpan-4.0: Improved Peptide-MHC Class I Interaction Predictions Integrating Eluted Ligand and Peptide Binding Affinity Data. J Immunol. 2017;199:3360–3368. doi: 10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Calis JJA, Maybeno M, Greenbaum JA, et al. Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Comput Biol. 2013;9 doi: 10.1371/journal.pcbi.1003266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paul S, Sidney J, Sette A, et al. TepiTool: A Pipeline for Computational Prediction of T-cell Epitope Candidates. Curr Protoc Immunol. 2016;114 doi: 10.1002/cpim.12. 18.19.1–18.19.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greenbaum J, Sidney J, Chung J, et al. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics. 2011;63:325–335. doi: 10.1007/s00251-011-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zanotto PM, Gould EA, Gao GF, et al. Population dynamics of flaviviruses revealed by molecular phylogenies. Proc Natl Acad Sci USA. 1996;93:548–553. doi: 10.1073/pnas.93.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grifoni A, Sidney J, Zhang Y, et al. A Sequence Homology and Bioinformatic Approach Can Predict Candidate Targets for Immune Responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–680. doi: 10.1016/j.chom.2020.03.002. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huhn O, Chazara O, Ivarsson MA, et al. High-Resolution Genetic and Phenotypic Analysis of KIR2DL1 Alleles and Their Association with Pre-Eclampsia. J Immunol. 2018;201:2593–2601. doi: 10.4049/jimmunol.1800860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.The Allele Frequency Net Database - Allele, haplotype and genotype frequencies in Worldwide Populations 2021. Available from: http://www.allelefrequencies.net/default.asp. (Accessed November 22, 2021).
- 59.Delaune D, Hul V, Karlsson EA, et al. A novel SARS-CoV-2 related coronavirus in bats from Cambodia. Nat Commun. 2021;12:6563. doi: 10.1038/s41467-021-26809-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.GISAID - gisaid.org 2021. Available from: https://www.gisaid.org/(Accessed November 22, 2021).
- 61.Madeira F, Park YM, Lee J, et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.EMBL-EBI. EMBOSS transeq 2021. Available from: https://www.ebi.ac.uk/Tools/st/emboss_transeq/(Accessed August 10, 2021).
- 63.Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 64.EMBL-EBI. EMBOSS Needle 2021. Available from: https://www.ebi.ac.uk/Tools/psa/emboss_needle/(Accessed August 10, 2021).
- 65.Ivanciuc O. SDAP: Structural database of allergenic proteins 2022. Available from: https://fermi.utmb.edu/(Accessed August 20, 2022).
- 66.Ivanciuc O, Schein CH, Braun W. SDAP: database and computational tools for allergenic proteins. Nucleic Acids Res. 2003;31:359–362. doi: 10.1093/nar/gkg010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woo H, Park SJ, Choi YK, et al. Developing a Fully Glycosylated Full-Length SARS-CoV-2 Spike Protein Model in a Viral Membrane. J Phys Chem B. 2020;124:7128–7137. doi: 10.1021/acs.jpcb.0c04553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jo S, Kim T, Iyer VG, et al. CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 69.Zhang C, Zheng W, Huang X, et al. Protein Structure and Sequence Reanalysis of 2019-nCoV Genome Refutes Snakes as Its Intermediate Host and the Unique Similarity between Its Spike Protein Insertions and HIV-1. J Proteome Res. 2020;19:1351–1360. doi: 10.1021/acs.jproteome.0c00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Modeling of the SARS-COV-2 Genome using D-I-TASSER 2021. Available from: https://zhanggroup.org//COVID-19/(Accessed January 18, 2021).
- 71.PubMed. Choice. 2021;43(06) https://pubmed.ncbi.nlm.nih.gov 43–3422 –43–3422. Available from: (Accessed February 02, 2022) [Google Scholar]
- 72.Noll CE ScienceDirect. Adv Space Res. 2010;45:1440. https://www.sciencedirect.com/ Available from: (Accessed February 02, 2022) [Google Scholar]
- 73.Ahmed SF, Quadeer AA, McKay MR. COVIDep: a web-based platform for real-time reporting of vaccine target recommendations for SARS-CoV-2. Nat Protoc. 2020;15:2141–2142. doi: 10.1038/s41596-020-0358-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.COVIDep. Available from: https://covidep.ust.hk/(Accessed February 2, 2022).
- 75.Denis M, Vandeweerd V, Verbeke R, et al. Covipendium : information available to support the development of medical countermeasures and interventions against COVID-19. Transdisciplinary Insights. 2021;4:1–296. https://biblio.ugent.be/publication/8710735 Available from: (Accessed September 6, 2022) [Google Scholar]
- 76.Kumar S. Drug and vaccine design against novel Coronavirus (2019-nCoV) spike protein through computational approach. Preprints. 2020 https://www.preprints.org/manuscript/202002.0071/v1 Available from: (Accessed September 6, 2022) [Google Scholar]
- 77.Singhal T. A Review of Coronavirus Disease-2019 (COVID-19) Indian J Pediatr. 2020;87:281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Polyiam K, Phoolcharoen W, Butkhot N, et al. Immunodominant linear B-cell epitopes in the spike and membrane proteins of SARS-CoV-2 identified by immunoinformatics prediction and immunoassay. Sci Rep. 2021;11:20383. doi: 10.1038/s41598-021-99642-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Le Bert N, Tan AT, Kunasegaran K, et al. SARS-CoV-2-specific T-cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 80.Amrun SN, Lee CYP, Lee B, et al. Linear B-cell epitopes in the spike and nucleocapsid proteins as markers of SARS-CoV-2 exposure and disease severity. EBioMedicine. 2020;58 doi: 10.1016/j.ebiom.2020.102911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ud-Din M, Albutti A, Ullah A, et al. Vaccinomics to Design a Multi-Epitopes Vaccine for Acinetobacter baumannii. Int J Environ Res Public Health. 2022;19:19. doi: 10.3390/ijerph19095568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.ul Qamar MT, F Shahid, Ashfaq UA, et al. Structural modeling and conserved epitopes prediction against SARS-COV-2 structural proteins for vaccine development. Research Square. 2020 doi: 10.1089/vim.2016.0033. https://www.researchsquare.com/article/rs-14534/v1 Available from. (Accessed September 6, 2022) [DOI] [PubMed] [Google Scholar]
- 83.Peng Y, Mentzer AJ, Liu G, et al. Broad and strong memory CD4+ and CD8+ T-cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kar T, Narsaria U, Basak S, et al. A candidate multi-epitope vaccine against SARS-CoV-2. Sci Rep. 2020;10:10895. doi: 10.1038/s41598-020-67749-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang Y, Yang C, Xu XF, et al. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. https://www.nature.com/articles/s41401-020-0485-4 Available from: (Accessed September 6, 2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mahtarin R, Islam S, Islam MJ, et al. Structure and dynamics of membrane protein in SARS-CoV-2. J Biomol Struct Dyn. 2022;40:4725–4738. doi: 10.1080/07391102.2020.1861983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shomuradova AS, Vagida MS, Sheetikov SA, et al. SARS-CoV-2 Epitopes Are Recognized by a Public and Diverse Repertoire of Human T-cell Receptors. Immunity. 2020;53:1245–1257. doi: 10.1016/j.immuni.2020.11.004. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kumar S, Maurya VK, Prasad AK, et al. Structural, glycosylation and antigenic variation between 2019 novel coronavirus (2019-nCoV) and SARS coronavirus (SARS-CoV) Virusdisease. 2020;31:13–21. doi: 10.1007/s13337-020-00571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stamatakis G, Samiotaki M, Mpakali A, et al. Generation of SARS-CoV-2 S1 Spike Glycoprotein Putative Antigenic Epitopes in Vitro by Intracellular Aminopeptidases. J Proteome Res. 2020;19:4398–4406. doi: 10.1021/acs.jproteome.0c00457. [DOI] [PubMed] [Google Scholar]
- 90.Chukwudozie OS, Gray CM, Fagbayi TA, et al. Immuno-informatics design of a multimeric epitope peptide based vaccine targeting SARS-CoV-2 spike glycoprotein. PLoS One. 2021;16 doi: 10.1371/journal.pone.0248061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hisham Y, Ashhab Y, Hwang SH, et al. Identification of Highly Conserved SARS-CoV-2 Antigenic Epitopes with Wide Coverage Using Reverse Vaccinology Approach. Viruses. 2021;13:787. doi: 10.3390/v13050787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.ASANLAATK epitope - Immune Epitope Database (IEDB) 2021. Available from: http://www.iedb.org/epitope/4321. (Accessed January 17, 2022).
- 93.Slathia P, Sharma P. Prediction of T and B-cell Epitopes in the Proteome of SARS-CoV-2 for Potential Use in Diagnostics and Vaccine Design. ChemRxiv. 2020 https://chemrxiv.org/engage/chemrxiv/article-details/60c749deee301c4145c79b4f Available from: (Accessde September 6, 2022) [Google Scholar]
- 94.Li Y, Lai DY, Zhang HN, et al. Linear epitopes of SARS-CoV-2 spike protein elicit neutralizing antibodies in COVID-19 patients. Cell Mol Immunol. 2020;17:1095–1097. doi: 10.1038/s41423-020-00523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kiyotani K, Toyoshima Y, Nemoto K, et al. Bioinformatic prediction of potential T-cell epitopes for SARS-Cov-2. J Hum Genet. 2020;65:569–575. doi: 10.1038/s10038-020-0771-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lu S, Xie XX, Zhao L, et al. The immunodominant and neutralization linear epitopes for SARS-CoV-2. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2020.108666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fast E, Altman RB, Chen B. Potential T-cell and B-cell Epitopes of 2019-nCoV. bioRxiv. 2020 https://www.biorxiv.org/content/10.1101/2020.02.19.955484v2 Available from: (Accessde September 6, 2022) [Google Scholar]
- 98.Schreibing F, Hannani M, Ticconi F, et al. Dissecting CD8+ T-cell pathology of severe SARS-CoV-2 infection by single-cell epitope mapping. bioRxiv. 2021 doi: 10.3389/fimmu.2022.1066176. https://biorxiv.org/cgi/content/short/2021.03.03.432690 Available from: (Accessde September 6, 2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cubuk J, Alston JJ, Incicco JJ, et al. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat Commun. 2021;12:1936. doi: 10.1038/s41467-021-21953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jaimes JA, André NM, Chappie JS, et al. Phylogenetic Analysis and Structural Modeling of SARS-CoV-2 Spike Protein Reveals an Evolutionary Distinct and Proteolytically Sensitive Activation Loop. J Mol Biol. 2020;432:3309–3325. doi: 10.1016/j.jmb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Choi A, Koch M, Wu K, et al. Safety and immunogenicity of SARS-CoV-2 variant mRNA vaccine boosters in healthy adults: an interim analysis. Nat Med. 2021;27:2025–2031. doi: 10.1038/s41591-021-01527-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nasreen S, Chung H, He S, et al. Effectiveness of mRNA and ChAdOx1 COVID-19 vaccines against symptomatic SARS-CoV-2 infection and severe outcomes with variants of concern in Ontario. bioRxiv. medRxiv. 2021 doi: 10.1038/s41564-021-01053-0. https://www.medrxiv.org/content/10.1101/2021.06.28.21259420v3 Available from: (Accessed October 17, 2021) [DOI] [PubMed] [Google Scholar]
- 104.Poh CM, Carissimo G, Wang B, et al. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nat Commun. 2020;11:2806. doi: 10.1038/s41467-020-16638-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li L, Zhao Z, Yang X, et al. Identification of four linear B-cell epitopes on the SARS-CoV-2 spike protein able to elicit neutralizing antibodies. bioRxiv. 2020 https://www.biorxiv.org/content/10.1101/2020.12.13.422550v1 Available from: (Accessed September 6, 2022) [Google Scholar]
- 106.Wu F, Wang A, Liu M, et al. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. bioRxiv. medRxiv. 2020 https://www.medrxiv.org/content/10.1101/2020.03.30.20047365v2 Available from: (Accessed August 8, 2020) [Google Scholar]
- 107.Xia X. Domains and Functions of Spike Protein in Sars-Cov-2 in the Context of Vaccine Design. Viruses. 2021;13:109. doi: 10.3390/v13010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yi Z, Ling Y, Zhang X, et al. Functional mapping of B-cell linear epitopes of SARS-CoV-2 in COVID-19 convalescent population. Emerg Microbes Infect. 2020;9:1988–1996. doi: 10.1080/22221751.2020.1815591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cruz DJM, Kim CJ, Shin HJ. Phage-displayed peptides having antigenic similarities with porcine epidemic diarrhea virus (PEDV) neutralizing epitopes. Virology. 2006;354:28–34. doi: 10.1016/j.virol.2006.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tan CW, Chia WN, Young BE, et al. Pan-Sarbecovirus Neutralizing Antibodies in BNT162b2-Immunized SARS-CoV-1 Survivors. N Engl J Med. 2021;385:1401–1406. doi: 10.1056/NEJMoa2108453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bertoletti A, Le Bert N, Qui M, et al. SARS-CoV-2-specific T-cells in infection and vaccination. Cell Mol Immunol. 2021;18:2307–2312. doi: 10.1038/s41423-021-00743-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sekine T, Perez-Potti A, Rivera-Ballesteros O, et al. Robust T-cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell. 2020;183:158–168. doi: 10.1016/j.cell.2020.08.017. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lv H, Wu NC, Tsang OTY, et al. Cross-reactive Antibody Response between SARS-CoV-2 and SARS-CoV Infections. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.107725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pinto D, Park YJ, Beltramello M, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 115.Ng OW, Chia A, Tan AT, et al. Memory T-cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine. 2016;34:2008–2014. doi: 10.1016/j.vaccine.2016.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sette A, Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 118.Guerrera G, Picozza M, D'Orso S, et al. BNT162b2 vaccination induces durable SARS-CoV-2-specific T-cells with a stem cell memory phenotype. Sci Immunol. 2021;6:eabl5344. doi: 10.1126/sciimmunol.abl5344. [DOI] [PubMed] [Google Scholar]
- 119.Heide J, Schulte S, Kohsar M, et al. Broadly directed SARS-CoV-2-specific CD4+ T-cell response includes frequently detected peptide specificities within the membrane and nucleoprotein in patients with acute and resolved COVID-19. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chi X, Yan R, Zhang J, et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Miller NL, Clark T, Raman R, et al. Insights on the mutational landscape of the SARS-CoV-2 Omicron variant. bioRxiv. 2021 doi: 10.1101/2021.12.06.471499. (Accessed December 28, 2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Carreño JM, Alshammary H, Tcheou J, et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 2022;602:682–688. doi: 10.1038/s41586-022-04399-5. [DOI] [PubMed] [Google Scholar]
- 123.Schmidt F, Muecksch F, Weisblum Y, et al. Plasma Neutralization of the SARS-CoV-2 Omicron Variant. N Engl J Med. 2022;386:599–601. doi: 10.1056/NEJMc2119641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pulliam JRC, van Schalkwyk C, Govender N, et al. Increased risk of SARS-CoV-2 reinfection associated with emergence of Omicron in South Africa. bioRxiv. 2021 doi: 10.1126/science.abn4947. https://www.medrxiv.org/content/10.1101/2021.11.11.21266068v3 Available from: (Accessed March 10, 2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.