Abstract
Fibroblast growth factor receptor inhibitors (FGFRi) were introduced into clinical trials on several cancer types and found to be particularly efficacious on urothelial cancer and cholangiocarcinoma. Although many enrolled patients responded well in clinical trials, there were some patients who did not respond to FGFRi even though their tumors carried the genomic changes that met the enrollment criteria. As already established, fibroblast growth factor receptor (FGFR) and epidermal growth factor receptor (EGFR) share the downstream signaling pathway of MAPK activation. Accordingly, it is conceivable that targeted inhibition of FGFR alone could leave the MAPK signaling unaffected when the signaling through EGFR is relatively strong. To test this hypothesis, we calculated here the FGFR to EGFR mRNA ratio (F/E for short) of biliary tract and urothelial cancer cell lines utilized in preclinical studies. In six biliary tract cancer cell lines, two responsive lines had an F/E of 9.5 and 9.0, whereas the F/E of four nonresponsive lines was 0.1–1.8. In 22 urothelial cancer cell lines, four of the five responsive lines showed an F/E of 2.8–4.9 (median, 3.6), whereas the F/E range of 17 nonresponsive lines was 0.01–2.7 (median, 0.6) (p = 0.004). We further investigated our 47 patient‐derived colorectal cancer‐stem cell spheroid lines. The 18 responsive lines showed relatively high F/E (median, 16.4), whereas 29 nonresponsive lines had low F/E (median, 9.2) (p = 0.0006). These results suggest that F/E is another strong predictor of responses to FGFRi that is as useful as the current genomic criteria based solely on the FGFR genomic changes.
Keywords: colorectal cancer, EGFR, EGFR inhibitor, FGFR, FGFR inhibitor
Fibroblast growth factor receptor (FGFR) inhibitors were introduced into clinical trials on several cancer types carrying FGFR genomic alterations. However, there were some patients who did not respond to FGFR inhibitors despite that their tumors carried the genomic changes that met the enrollment criteria. Here, we propose a novel parameter that helps predict the chemosensitivity of cancer patients to FGFR inhibitors.
![]()
1. INTRODUCTION
The fibroblast growth factor (FGF) family contains 22 ligands, whereas the FGF receptor (FGFR) has four receptor tyrosine kinase paralogs encoded by separate genes (FGFR1–FGFR4). 1 As FGF/FGFR signaling plays crucial roles in tumor cell proliferation, migration, and survival, they are considered as druggable therapeutic targets. 1
Several sets of preclinical data showed significant growth inhibition by small‐molecule FGFR inhibitors (FGFRi) on cancer cell lines or xenografts that had FGFR gene amplifications. 2 , 3 Accordingly, multiple clinical phase I trials of FGFRi were undertaken on patients with cancers carrying such genomic alterations. 4 , 5 , 6 However, FGFRi did not always improve patient survival compared with conventional treatments 4 , 5 except for urothelial cancer with FGFR3 mRNA overexpression. 6 Even in such a urothelial cancer trial, 6 approximately one quarter of enrolled patients with FGFR mRNA overexpression failed to respond to the drug. In contrast, a small subset of patients in these clinical trials showed significantly better (i.e., complete or partial) response to the FGFRi than others, regardless of the FGFR gene copy numbers or mRNA expression levels.
Although there are several reports about the effectiveness of FGFRi for colorectal cancer (CRC) cell lines, 7 there have been no clinical trials exclusively focusing on FGFRi for CRC patients. Using patient‐derived CRC tumor‐initiating cell or stem cell (SC) spheroids (or organoids) and their xenografts, we recently reported that some spheroid lines (7/25; 28%) responded significantly better to a pan‐FGFRi erdafitinib where growth effect index (GEI, indicating the relative growth rate to the no‐drug control) < 0.7 was defined as “responsive.” 8 Moreover, the combination of erdafitinib with an epidermal growth factor receptor (EGFR) inhibitor (EGFRi) erlotinib showed much stronger growth inhibition than either drug alone, as efficacy was observed in the majority of lines (21/25; 84%), including 56% (14/25) that was insensitive to erdafitinib alone. Consistently, the in vivo response of xenograft tumors to erdafitinib accurately reflected the above in vitro experiments. Surprisingly, we found little correlation between the sensitivity to the FGFRi and genetic/genomic alterations of CRC‐SC spheroids.
As already established, FGFR and EGFR share the downstream signaling pathway of MAPK activation. Additionally, some reports showed that EGFR overexpression played a significant role in the proliferation of several types of cancer, including biliary, urothelial, and colorectal. 9 , 10 Accordingly, it is conceivable that targeted inhibition of FGFR alone might leave the MAPK signaling unaffected when the signaling through EGFR is relatively strong. In the present report, we reevaluated the preclinical data of biliary and urothelial cancers, followed by our own CRC‐SC spheroid data. Based on these results, we would like to propose a novel parameter, the FGFR to EGFR mRNA ratio (F/E), as a biomarker that helps predict cancer chemosensitivity to FGFRi.
2. MATERIALS AND METHODS
Human CRC samples were obtained from patients who underwent operations at the Department of Surgery, Kyoto University Hospital from 2014 to 2018. All materials and methods are described in Supporting Information.
3. RESULTS AND DISCUSSION
By scrutinizing the published data of FGFRi clinical trials on biliary and urothelial cancers, 11 , 12 we noticed an unresolved issue. Namely, most of these reports showed waterfall plots where tumors of some good fractions of enrolled patients responded well, demonstrating efficacy of the FGFRi. Importantly, however, there were sizable subsets in which tumors did not respond to the FGFRi despite the patients’ tumors carrying the genomic changes that met the enrollment criteria.
As already established, FGFR and EGFR share the downstream signaling pathway of MAPK activation as well as other pathways, 8 suggesting that targeted inhibition of FGFR alone might leave the MAPK signaling unaffected when the input through EGFR is relatively strong. To test such a possibility, we obtained data of FGFR1–4 and EGFR mRNA sequencing (Table S1) and genomic alterations such as gene mutations (Table S2) and fusions (Table S3) from the Broad Institute Cancer Cell Line Encyclopedia (https://portals.broadinstitute.org/ccle) of biliary tract and urothelial cancer cell lines for which FGFRi chemosensitivity had been published in preclinical studies. 13 , 14 , 15 Based on these mRNA expression data, we calculated the FGFR to EGFR mRNA ratio (F/E). We first summed up the expression levels of FGFR1–4 mRNAs by adding the transcripts per million of FGFR paralogs (sumFGFR). This is because human FGFR has four paralogs, FGFR1–4, and the particular paralog(s) that play the key role in tumorigenesis is different depending on the cancer type 6 as well as the individual case even in the same cancer type 16 (Figure S1). To take EGFR expression into consideration, we then divided this number with that of EGFR: F/E. We found no statistically significant associations between the FGFRi sensitivity and expression levels of the other ERBB family genes, ERBB2–4, and therefore excluded their expression levels from the F/E ratio (Figure S2 and Table S4).
In six biliary tract cancer cell lines tested, 13 two responsive cell lines had an F/E ratio of 9.5 and 9.0, whereas four nonresponsive ones had substantially lower ratios of 0.1–1.8 (Table 1). Not surprisingly, the two responsive cell lines had relatively high levels (148 and 214) of FGFR expression compared with four nonresponsive ones (5.6–83). The EGFR expression of the FGFRi‐responsive lines was relatively low (16 and 24), whereas that in the nonresponsive lines was at least twice as high (36–60). Accordingly, F/E helped to classify the cell lines’ sensitivity to FGFRi more clearly than the FGFR mRNA levels alone.
TABLE 1.
Correlation between mRNA expression ratio sumFGFR/EGFR (F/E) and sensitivity to fibroblast growth factor receptor (FGFR) inhibitors for six biliary tract cancer cell lines
| Cell line | mRNA expression level (TPM) | F/E | FGFR inhibitor sensitivity | ||
|---|---|---|---|---|---|
| sumFGFR | EGFR | ||||
| ICC13‐7 | 147.6 | 15.5 | 9.5 | R | |
| CCLP‐1 | 214.0 | 23.9 | 9.0 | R | |
| SNU1079 | 83.4 | 47.6 | 1.8 | NR | |
| SSP25 | 51.9 | 46.5 | 1.1 | NR | |
| GB2 | 9.6 | 35.8 | 0.3 | NR | |
| HUCCT1 | 5.6 | 59.6 | 0.1 | NR | |
Note: Rows for responsive cell lines are shaded.
Abbreviations: EGFR, epidermal growth factor receptor; NR, nonresponsive; R, responsive; TPM, transcripts per million.
In urothelial cancer cell lines, 14 , 15 four of the five responsive lines showed an F/E ranging from 2.8 to 4.9 (median, 3.6), whereas the range of 17 nonresponsive lines was from 0.01 to 2.7 (median, 0.6) (p = 0.004) (Table 2 and Figure S3), and the receiver operating characteristic (ROC) curve analysis showed an area under the ROC curve (AUC) of 0.94 (95% confidence interval [CI], 0.83–1.00). The only exception to our hypothesis here was SW780 that had the ratio of 1.0, and was responsive to FGFRi. Interestingly, this cell line carried an FGFR3‐BAIAP2L1 gene fusion that had oncogenic activity through MAPK signaling activation caused by ligand‐independent and constitutive dimerization. 17 These cancer cell lines also showed a similar tendency to the biliary tract cancer lines. Namely, the four responsive lines had relatively high FGFR expression (81–149) compared with 17 nonresponsive lines (5–50). In contrast, the EGFR mRNA levels of the FGFRi‐responsive lines were relatively low (21–36; median, 25), whereas those in the nonresponsive lines were slightly higher (6–98; median, 30). Accordingly, F/E reflected their sensitivity to FGFRi in a similar manner to the FGFR mRNA levels.
TABLE 2.
Correlation between mRNA expression ratio sumFGFR/EGFR (F/E) and sensitivity to fibroblast growth factor receptor (FGFR) inhibitors for 22 urothelial carcinoma cell lines
| Cell line | mRNA expression level (TPM) | F/E | FGFR inhibitor sensitivity | ||
|---|---|---|---|---|---|
| sumFGFR | EGFR | ||||
| JMSU1 | 103.5 | 21.3 | 4.9 | R | |
| RT112 | 149.0 | 30.7 | 4.9 | R | |
| RT4 | 80.5 | 22.2 | 3.6 | R | |
| UMUC14 | 100.6 | 35.8 | 2.8 | R | |
| J82 | 42.8 | 15.6 | 2.7 | NR | |
| SW1710 | 50.3 | 26.8 | 1.9 | NR | |
| BC3C | 11.0 | 6.3 | 1.8 | NR | |
| KMBC2 | 47.6 | 30.0 | 1.6 | NR | |
| TCCSUP | 22.7 | 17.3 | 1.3 | NR | |
| SW780 | 25.9 | 25.5 | 1.0 | R | |
| UMUC3 | 25.3 | 25.0 | 1.0 | NR | |
| BFTC905 | 28.2 | 28.6 | 1.0 | NR | |
| CAL29 | 43.6 | 59.8 | 0.7 | NR | |
| UMUC9 | 10.8 | 17.2 | 0.6 | NR | |
| T24 | 8.1 | 18.8 | 0.4 | NR | |
| KU1919 | 14.4 | 34.8 | 0.4 | NR | |
| HT1197 | 10.5 | 36.7 | 0.3 | NR | |
| HT1376 | 18.6 | 77.6 | 0.2 | NR | |
| VMCUB1 | 21.2 | 98.3 | 0.2 | NR | |
| 5637 | 10.6 | 50.1 | 0.2 | NR | |
| 647 V | 4.8 | 39.2 | 0.1 | NR | |
| UBLC1 | 8.0 | 779.9 | 0.0 | NR | |
Note: Rows for responsive cell lines are shaded.
Abbreviations: EGFR, epidermal growth factor receptor; NR, nonresponsive; R, responsive; TPM, transcripts per million.
To investigate the applicability of the F/E ratio to CRC treatment with FGFRi, we looked into our patient‐derived CRC‐SC spheroid lines. 8 We recently reported that seven (28%) of 25 RAS/RAF WT CRC‐SC lines responded to a pan‐FGFRi erdafitinib singly in culture, whereas 21 (84%) responded to erdafitinib in combination with an EGFRi erlotinib. 8 Moreover, we examined an additional 22 lines to determine erdafitinib and/or erlotinib sensitivities and F/E ratios, expanding our list to a total of 47 lines (Table 3). As anticipated, 18 (38%) singly‐responsive lines showed relatively high F/E, ranging from 7.2 to 866 (median, 16.4), whereas the 29 (62%) nonresponsive lines had a ratio range of 3.5–37 (median, 9.2) (p = 0.0006), with the ROC curve showing an AUC of 0.80 (95% CI, 0.67–0.93) for F/E as a predictor of erdafitinib response (Figure 1). Accordingly, we set the cut‐off F/E at 14 that divided responders from nonresponders more clearly than sumFGFR values. The four exceptional nonresponder cell lines (HC129T, HC185T, HC22T, and HC40T) suffered little in growth by the inhibitor (GEI of 0.80–0.95, where GEI <0.7 was defined as growth suppression) despite their F/E of 26–37, substantially higher than the threshold of 14. It is worth noting that two of these lines, HC129T and HC40T, were nonresponsive even to the combination of erdafitinib and erlotinib, which might be attributable to higher mRNA expression level of MET in HC129T cells or to PIK3CA‐activating E545K 18 and EGFR‐activating E114K 19 mutations in HC40T cells. Incidentally, the FGFR1 R820H mutation in HC129T cells did not appear to contribute to the refractoriness to erdafitinib because of the very low level of FGFR1 mRNA (Tables S5–S7). Five exceptions among the singly‐responsive lines were HC46T, HC28T, HC91T, HC183T, and HC72T with F/E ratios of 7–12, which were lower than the threshold of 14. Just in case, we undertook the Sashimi plot analysis 20 on these lines, and excluded the possibility of chromosome translocations that affected FGFR activity. The results indicated no obvious sign of translocations involving the FGFR3/4 genes (Figure S4) such as the one reported for the urothelial cancer cell line SW780 that contained an FGFR3‐BAIAP2L1 fusion and was responsive to FGFRi despite its low F/E of 1.0 (Tables 2 and S3). These results suggest that F/E can also be a useful biomarker for CRC chemosensitivity where the EGFR expression level fluctuates widely.
TABLE 3.
Correlation between mRNA expression ratio sumFGFR/EGFR (F/E) and sensitivity to fibroblast growth factor receptor (FGFR) inhibitor erdafitinib and/or epidermal growth factor receptor (EGFR) inhibitor erlotinib for 47 patient‐derived RAS/RAF WT colorectal cancer‐stem cell (CRC‐SC) spheroid lines
| Spheroid line | mRNA expression level (TPM) | F/E | Drug sensitivity (GEI) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sumFGFR | EGFR | Erdafitinib | Erlotinib | Erdafitinib/erlotinib combination | ||||||||
| HC137T a | 168.7 | 0.2 | 866.1 | R | (0.54) | (0.99) | NR | R | (0.54) | |||
| HC6T | 210.0 | 1.0 | 207.1 | R | (0.34) | (0.75) | NR | R | (0.26) | |||
| HC97T a | 193.9 | 2.3 | 82.7 | R | (0.35) | (0.88) | NR | R | (0.32) | |||
| HC129T | 265.2 | 7.2 | 36.8 | (0.90) | NR | (1.24) | NR | (0.74) | NR | |||
| HC67T | 259.4 | 7.6 | 34.3 | R | (0.59) | (1.17) | NR | R | (0.50) | |||
| HC185T a | 286.4 | 8.9 | 32.3 | (0.75) | NR | (0.82) | NR | R | (0.64) | |||
| HC22T | 354.4 | 11.4 | 31.0 | (0.80) | NR | (0.90) | NR | R | (0.59) | |||
| HC40T | 138.7 | 5.4 | 25.8 | (0.95) | NR | (0.93) | NR | (0.86) | NR | |||
| HC9T | 96.7 | 3.8 | 25.1 | R | (0.48) | (0.73) | NR | R | (0.21) | |||
| HC24T a | 245.5 | 12.6 | 19.5 | R | (0.68) | (0.74) | NR | R | (0.56) | |||
| HC128T a | 87.9 | 5.0 | 17.6 | R | (0.48) | (0.81) | NR | R | (0.32) | |||
| HC179T a | 196.0 | 11.7 | 16.8 | R | (0.62) | (0.72) | NR | R | (0.48) | |||
| HC90T a | 197.4 | 12.0 | 16.4 | R | (0.54) | (1.11) | NR | R | (0.46) | |||
| HC80T | 191.2 | 11.7 | 16.4 | R | (0.50) | (0.99) | NR | R | (0.32) | |||
| HC172T a | 153.2 | 9.8 | 15.6 | R | (0.66) | (0.90) | NR | R | (0.60) | |||
| HC93T | 127.6 | 8.6 | 14.9 | R | (0.52) | (1.22) | NR | R | (0.28) | |||
| HC20T | 137.2 | 9.2 | 14.8 | R | (0.46) | (0.72) | NR | R | (0.44) | |||
| HC16T | 143.5 | 10.2 | 14.0 | (0.76) | NR | (1.13) | NR | R | (0.58) | |||
| HC7T | 107.9 | 8.0 | 13.5 | (0.89) | NR | R | (0.44) | R | (0.30) | |||
| HC10T | 126.4 | 10.1 | 12.6 | (0.82) | NR | R | (0.67) | R | (0.31) | |||
| HC102T a | 136.4 | 11.1 | 12.3 | (0.73) | NR | R | (0.70) | R | (0.42) | |||
| HC46T a | 143.6 | 11.7 | 12.2 | R | (0.39) | (0.79) | NR | R | (0.17) | |||
| HC146T | 99.0 | 8.5 | 11.7 | (0.81) | NR | (0.83) | NR | R | (0.37) | |||
| HC28T | 78.6 | 7.0 | 11.3 | R | (0.67) | (1.07) | NR | R | (0.26) | |||
| HC193T a | 258.2 | 24.5 | 10.5 | (0.74) | NR | (0.89) | NR | R | (0.35) | |||
| HC1T | 149.6 | 14.4 | 10.4 | (0.80) | NR | (0.88) | NR | R | (0.46) | |||
| HC106T | 110.0 | 10.9 | 10.1 | (1.01) | NR | (1.13) | NR | (1.15) | NR | |||
| HC195T a | 201.7 | 20.8 | 9.7 | (0.73) | NR | (0.92) | NR | R | (0.57) | |||
| HC178T a | 147.7 | 15.7 | 9.4 | (0.81) | NR | (0.72) | NR | R | (0.51) | |||
| HC94T a | 136.2 | 14.7 | 9.2 | (0.82) | NR | (0.77) | NR | R | (0.57) | |||
| HC120T | 227.8 | 24.9 | 9.2 | (0.92) | NR | (0.82) | NR | (0.71) | NR | |||
| HC91T a | 76.0 | 9.0 | 8.4 | R | (0.63) | R | (0.69) | R | (0.30) | |||
| HC183T a | 73.3 | 8.9 | 8.2 | R | (0.39) | (0.87) | NR | R | (0.23) | |||
| HC117T | 69.7 | 8.8 | 7.9 | (1.02) | NR | R | (0.63) | R | (0.20) | |||
| HC72T a | 130.8 | 18.1 | 7.2 | R | (0.51) | (0.99) | NR | R | (0.47) | |||
| HC73T | 123.7 | 17.1 | 7.2 | (0.86) | NR | R | (0.66) | R | (0.40) | |||
| HC127T a | 160.2 | 22.3 | 7.2 | (0.90) | NR | (0.75) | NR | R | (0.42) | |||
| HC21T | 122.2 | 17.1 | 7.1 | (0.77) | NR | (0.85) | NR | R | (0.35) | |||
| HC8T | 73.2 | 10.3 | 7.1 | (0.96) | NR | (0.71) | NR | R | (0.32) | |||
| HC149T a | 201.4 | 29.2 | 6.9 | (0.95) | NR | (0.73) | NR | R | (0.50) | |||
| HC74T | 91.2 | 13.7 | 6.7 | (0.99) | NR | (0.92) | NR | R | (0.68) | |||
| HC108T | 83.2 | 14.8 | 5.6 | (0.80) | NR | R | (0.57) | R | (0.31) | |||
| HC35T a | 49.2 | 11.3 | 4.3 | (0.90) | NR | (0.99) | NR | (0.98) | NR | |||
| HC142T | 71.9 | 18.7 | 3.8 | (1.00) | NR | (1.08) | NR | R | (0.35) | |||
| HC122T a | 75.6 | 19.9 | 3.8 | (1.02) | NR | (0.78) | NR | (0.76) | NR | |||
| HC11T | 95.8 | 27.6 | 3.5 | (0.85) | NR | (0.78) | NR | R | (0.41) | |||
| HC83T a | 66.5 | 19.3 | 3.5 | (0.76) | NR | R | (0.51) | R | (0.37) | |||
Note: Shaded rows indicate spheroid lines responsive to erdafitinib alone.
Abbreviations: GEI, growth effect index; NR, nonresponsive (GEI ≥ 0.7); R, responsive (GEI < 0.7).
New data in addition to those of 25 CRC‐SC lines published in our previous report.
FIGURE 1.
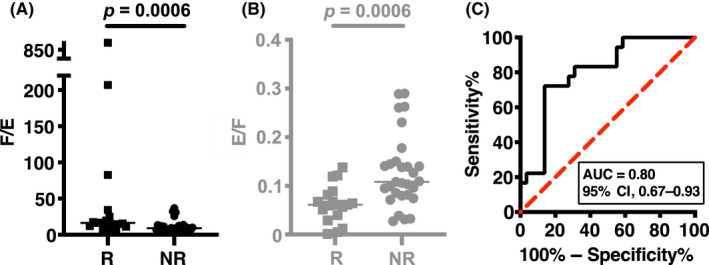
(A) Mann–Whitney U‐test of sumFGFR/EGFR (F/E) regarding fibroblast growth factor receptor inhibitor (FGFRi) sensitivity in 47 colorectal cancer‐stem cell (CRC‐SC) spheroid lines. (B) Mann–Whitney U‐test of EGFR/sumFGFR (E/F) regarding FGFRi sensitivity in 47 CRC‐SC spheroid lines. Note that this is just to visualize high F/E samples more easily in a reciprocal plot. (C) Receiver operating characteristic (ROC) curve analysis of F/E regarding FGFRi sensitivity in 47 CRC‐SC spheroid lines. Horizontal bar shows the median. AUC, area under the ROC curve; CI, confidence interval; R, responsive; NR, nonresponsive
Together, these results show that F/E is another strong predictor of responses to FGFRi that is as useful as the current genomic criteria that are based solely on the FGFR genomic changes.
AUTHOR CONTRIBUTIONS
All three authors designed the study, acquired and analyzed the data, and wrote the manuscript.
DISCLOSURE
The authors declare no competing interests.
ETHICAL APPROVAL
Approval of the research protocol by an institutional review board: The study protocol was approved by the Ethics Committee of Kyoto University, Approval Nos. R0915 and R0857, FY2015–2022.
Informed consent: Written informed consent was obtained from all patients
Registry and registration no. of the study/trial: N/A.
Animal studies: N/A.
Supporting information
Figures S1–S4
Tables S1–S7
Appendix S1
ACKNOWLEDGMENTS
We thank H. Miyoshi, F. Kakizaki, T. Morimoto, H. Matsubara, and T. Aoyama for help and discussions, and the members of Division of Gastrointestinal Surgery at Department of Surgery, Kyoto University Hospital for help in collecting colon cancer specimens. We also thank S. Youssefian for comments on the manuscript.
Kitano S, Yamamoto T, Taketo MM. Novel parameter for cancer chemosensitivity to fibroblast growth factor receptor inhibitors. Cancer Sci. 2022;113:4005‐4010. doi: 10.1111/cas.15523
DATA AVAILABILITY STATEMENT
mRNA expression data files of 47 RAS/RAF WT patient‐derived CRC‐SC spheroid lines have been deposited in the Gene Expression Omnibus under accession code GSE205787.
REFERENCES
- 1. Babina IS, Turner NC. Advances and challenges in targeting FGFR signalling in cancer. Nat Rev Cancer. 2017;17:318‐332. [DOI] [PubMed] [Google Scholar]
- 2. Guagnano V, Kauffmann A, Wohrle S, et al. FGFR genetic alterations predict for sensitivity to NVP‐BGJ398, a selective pan‐FGFR inhibitor. Cancer Discov. 2012;2:1118‐1133. [DOI] [PubMed] [Google Scholar]
- 3. Zhang J, Zhang L, Su X, et al. Translating the therapeutic potential of AZD4547 in FGFR1‐amplified non‐small cell lung cancer through the use of patient‐derived tumor xenograft models. Clin Cancer Res. 2012;18:6658‐6667. [DOI] [PubMed] [Google Scholar]
- 4. Van Cutsem E, Bang Y‐J, Mansoor W, et al. A randomized, open‐label study of the efficacy and safety of AZD4547 monotherapy versus paclitaxel for the treatment of advanced gastric adenocarcinoma with FGFR2 polysomy or gene amplification. Ann Oncol. 2017;28:1316‐1324. [DOI] [PubMed] [Google Scholar]
- 5. Paik PK, Shen R, Berger MF, et al. A phase Ib open‐label multicenter study of AZD4547 in patients with advanced squamous cell lung cancers. Clin Cancer Res. 2017;23:5366‐5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schuler M, Cho BC, Sayehli CM, et al. Rogaratinib in patients with advanced cancers selected by FGFR mRNA expression: a phase 1 dose‐escalation and dose‐expansion study. Lancet Oncol. 2019;20:1454‐1466. [DOI] [PubMed] [Google Scholar]
- 7. Manchado E, Weissmueller S, Morris JP, et al. A combinatorial strategy for treating KRAS‐mutant lung cancer. Nature. 2016;534:647‐651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yamamoto T, Miyoshi H, Kakizaki F, et al. Chemosensitivity of patient‐derived cancer stem cells identifies colorectal cancer patients with potential benefit from FGFR inhibitor therapy. Cancer. 2020;12:2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Philip PA, Mahoney MR, Allmer C, et al. Phase II study of erlotinib in patients with advanced biliary cancer. J Clin Oncol. 2006;24:3069‐3074. [DOI] [PubMed] [Google Scholar]
- 10. Rebouissou S, Bernard‐Pierrot I, de Reyniès A, et al. EGFR as a potential therapeutic target for a subset of muscle‐invasive bladder cancers presenting a basal‐like phenotype. Sci Transl Med. 2014;6:244ra291. [DOI] [PubMed] [Google Scholar]
- 11. Javle M, Lowery M, Shroff RT, et al. Phase II study of BGJ398 in patients with FGFR‐altered advanced cholangiocarcinoma. J Clin Oncol. 2018;36:276‐282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Loriot Y, Necchi A, Park SH, et al. Erdafitinib in locally advanced or metastatic urothelial carcinoma. N Engl J Med. 2019;381:338‐348. [DOI] [PubMed] [Google Scholar]
- 13. Goyal L, Shi L, Liu LY, et al. TAS‐120 overcomes resistance to ATP‐competitive FGFR inhibitors in patients with FGFR2 fusion–positive intrahepatic cholangiocarcinoma. Cancer Discov. 2019;9:1064‐1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kikuchi A, Suzuki T, Nakazawa T, et al. ASP5878, a selective FGFR inhibitor, to treat FGFR3‐dependent urothelial cancer with or without chemoresistance. Cancer Sci. 2017;108:236‐242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Perera TPS, Jovcheva E, Mevellec L, et al. Discovery and pharmacological characterization of JNJ‐42756493 (erdafitinib), a functionally selective small‐molecule FGFR family inhibitor. Mol Cancer Ther. 2017;16:1010‐1020. [DOI] [PubMed] [Google Scholar]
- 16. Sánchez‐Guixé M, Hierro C, Jiménez J, et al. High FGFR1–4 mRNA expression levels correlate with response to selective FGFR inhibitors in breast cancer. Clin Cancer Res. 2022;28:137‐149. [DOI] [PubMed] [Google Scholar]
- 17. Nakanishi Y, Akiyama N, Tsukaguchi T, et al. Mechanism of oncogenic signal activation by the novel fusion kinase FGFR3–BAIAP2L1. Mol Cancer Ther. 2015;14:704‐712. [DOI] [PubMed] [Google Scholar]
- 18. Wang Q, Shi Y‐L, Zhou K, et al. PIK3CA mutations confer resistance to first‐line chemotherapy in colorectal cancer. Cell Death Dis. 2018;9:739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim N, Cho D, Kim H, et al. Colorectal adenocarcinoma‐derived EGFR mutants are oncogenic and sensitive to EGFR‐targeted monoclonal antibodies, cetuximab and panitumumab. Int J Cancer. 2020;146:2194‐2200. [DOI] [PubMed] [Google Scholar]
- 20. Katz Y, Wang ET, Silterra J, et al. Quantitative visualization of alternative exon expression from RNA‐seq data. Bioinformatics. 2015;31:2400‐2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1–S4
Tables S1–S7
Appendix S1
Data Availability Statement
mRNA expression data files of 47 RAS/RAF WT patient‐derived CRC‐SC spheroid lines have been deposited in the Gene Expression Omnibus under accession code GSE205787.


