Abstract
Adoptive transfer of T cell receptor (TCR)‐engineered T cells targeting viral epitopes represents a promising approach for treating virus‐related cancers. However, the efficient identification of epitopes for T cells and the corresponding TCR remains challenging. Here, we report a workflow permitting the rapid generation of human papillomavirus (HPV)‐specific TCR‐T cells. Six epitopes of viral proteins belonged to HPV16 or HPV18 were predicted to have high affinity to A11:01 according to bioinformatic analysis. Subsequently, CTL induction were performed with these six antigen peptides separately, and antigen‐specific T cells were sorted by FACS. TCR clonotypes of these virus‐specific T cells were determined using next‐generation sequencing. To improve the efficiency of TCRαβ pair validation, a lentiviral vector library containing 116 TCR constructs was generated that consisted of predominant TCRs according to TCR repertoire analysis. Later, TCR library transduced T cells were simulated with peptide pool‐pulsed antigen‐presenting cells, then CD137‐positive cells were sorted and subjected to TCR repertoire analysis. The top‐hit TCRs and corresponding antigen peptides were deduced and validated. Through this workflow, a TCR targeting the E692–101 of HPV16 was identified. These HPV16‐specific TCR‐T cells showed high activity towards HPV16‐positive human cervical cancer cells in vitro and efficiently repressed tumor growth in a murine model. This study provides a HPV16‐specific TCR fitted to the HLA‐A11:01 population, and exemplifies an efficient approach that can be applied in large‐scale screening of virus‐specific TCRs, further encouraging researchers to exploit the therapeutic potential of the TCR‐T cell technique in treating virus‐related cancers.
Keywords: adoptive cell transfer, HPV, immunotherapy, TCR‐T cells, virus‐related cancers
This study provides a HPV16‐specific TCR fitted to HLA‐A11:01 population, and exemplifies an efficient approach which can be applied in large‐scale screen of virus‐specific TCRs, further encouraging researchers to exploit the therapeutic potential of TCR‐T cell technique in treating virus‐related cancers.
Abbreviations
- HLA
human leukocyte antigen
- HPV
human papillomavirus
- TCR‐T cell
T cell receptor‐engineered T cell
1. INTRODUCTION
Several viruses, such as the HPV, hepatitis B virus, and Epstein–Barr virus, have been well documented to induce human malignancies. 1 , 2 HPV‐related cancers, including cervical cancer, head and neck cancer, and vaginal cancer, account for 4.5% of cancer‐related death worldwide, especially in developing countries where the incidence rate is much higher due to poor public health administration. 3 , 4 The promotion of vaccination is gradually decreasing the overall infection rate, however, for patients with relapsing and refractory cancers, current treatments provide limited benefits and patients usually experience uncontrollable cancer development and die from metastasis.
HPV integrates into the genome of host cells, and forces host cells to constitutively express viral proteins such as E6 and E7, which are considered oncogenes driving the malignant transformation of host cells. 5 , 6 E6 and E7 are therefore recognized in the clinic as markers for the authentication of HPV infection. Interestingly, some parts of the HPV proteome, including E6 and E7, can be processed by the proteasome system of the host cell and presented as epitopes by MHC molecules. 7 , 8 Due to this characteristic, HPV‐infected cells can be recognized and eliminated by CTLs in an MHC‐dependent manner.
Recently, several studies have raised the possibility of TCR‐T cells in treating HPV‐related cancers. 9 HLA‐A02:01‐specific TCR‐T cells targeting E6 or E7 have been reported consecutively, 10 , 11 including a clinic trial initiated by the United States National Institutes of Health in 2019, 12 which preliminarily proved the safety and effectiveness of TCR‐T cells for the treatment of A02:01‐restricted metastatic HPV‐related cancers. Although A02:01 is one of the common HLA‐A types worldwide, the most prevalent HLA‐A type A11:01 among Asian populations (nearly 26%) and its matching TCR‐T cells has been rarely studied, encouraging researchers to explore candidate TCRs suitable for Asian populations.
In the present study, we intended to establish a workflow that enabled the rapid generation of TCR‐T cells targeting high‐risk HPV types. Furthermore, to improve the efficiency of TCRαβ pair validation, we introduced a TCR library‐based validation step in the traditional workflow. One TCR targeting the E6 protein of HPV16 was identified. The function of this HPV16‐specific TCR‐T cell was tested in vitro and in vivo, and showed specific killing on A11:01 restricted HPV16‐E6 positive tumors. Combining the TCR library‐based validation method with our previously published minigene library‐based epitopes screen method, 13 , 14 large‐scale screen of virus‐specific epitopes and establishment of corresponding TCR‐T cells will be possible.
2. MATERIALS AND METHODS
2.1. Patients and sample collection
Ninety‐eight patients with cervical lesions were enrolled from September 2018 to December 2019 at the Shenzhen People's Hospital. None of them had received chemotherapy or radiotherapy before sampling. Peripheral blood samples from all the patients were collected the and proceeded to HLA genotyping. Tumor tissues from 30 patients among the 98 patients were resected and the tissue‐infiltrating T cells from the tissues were subjected to TCR repertoire analysis; in addition, peripheral blood samples from 10 patients were collected and used for CTL induction. The clinicopathological characteristics are summarized in Tables 1, S1 and S2.
TABLE 1.
Clinical characteristics of 98 patients with cervical lesions
| Clinicopathologic parameters | ||
|---|---|---|
| Age | Median (years) | 47 |
| HPV status | Positive | 84 (84/98, 85.71%) |
| HPV16 | 45 (45/84, 53.57%) | |
| Pure 16 | 37 (37/84, 44.05%) | |
| Mix 16 | 8 (8/84, 9.52%) | |
| HPV18 | 10 (10/84, 11.90%) | |
| Pure 18 | 9 (9/84, 10.71%) | |
| Mix 18 | 1 (1/84, 1.19%) | |
| Others | 36 (36/84, 42.86%) | |
| Negative | 4 (4/98, 4.08%) | |
| Not available | 10 (10/98, 10.20%) | |
| Pathological type | CIN | 45 (45/98, 45.92%) |
| Cervical squamous cell carcinoma | 41(41/98, 41.84%) | |
| Cervical adenocarcinoma | 11(11/98, 11.22%) | |
| Clear cell carcinoma | 1 (1/98, 1.02%) | |
| Stage | CIN | 45 (45/98, 45.92%) |
| Tis | 2 (2/98, 2.04%) | |
| I | 39 (39/98, 39.80%) | |
| II | 12 (12/98, 12.24%) | |
| III | 0 (0/98, 0.00%) | |
| HLA‐A alleles | IV | 0 (0/98, 0.00%) |
| 11:01 | 51 (51/196, 26.02%) | |
| 24:02 | 30 (30/196, 15.31%) | |
| 02:07 | 25 (25/196, 12.76%) | |
| 33:03 | 21 (21/196, 10.71%) | |
| 02:01 | 12 (12/196, 6.12%) | |
| 11:02 | 10 (10/196, 5.10%) | |
| 02:06 | 8 (8/196, 4.08%) | |
| 02:03 | 7 (7/196, 3.57%) | |
| 31:01 | 6 (6/196, 3.06%) | |
| 01:01 | 6 (6/196, 3.06%) | |
| 26:01 | 5 (6/196, 2.55%) | |
| 30:01 | 5 (5/196, 2.55%) | |
| 32:01 | 3 (3/196, 1.53%) | |
| 29:01 | 2 (2/196, 1.02%) | |
| 30:04 | 1 (1/196, 0.51%) | |
| 24:20 | 1 (1/196, 0.51%) | |
| 68:01 | 1 (1/196, 0.51%) | |
| 29:02 | 1 (1/196, 0.51%) | |
| 03:01 | 1 (1/196, 0.51%) | |
2.2. Cell lines
The HLA‐A11:01‐homozygous immortalized cell line C1R‐A11, which is commonly used as antigen‐presenting cells, was kindly provided by OncoTherapy Science, Inc. (Kawasaki) and was maintained in RPMI‐1640 medium (HyClone) supplied with 10% FBS (Gibco, USA) and 100 U/ml penicillin/streptomycin (HyClone). The cervical cancer cell line CaSki was purchased from Procell Life Science & Technology (China) and cultured in RPMI‐1640 medium supplemented with 10% FBS and 100 U/ml penicillin/streptomycin.
2.3. Bioinformatic analysis of epitopes with high affinity to HLA‐A11:01
The amino acid sequences of E6 and E7 were obtained from the NCBI database. The NetMHC v4.0 platform was used to predict the binding affinity of all possible 8‐mer to 11‐mer epitopes; epitopes for which binding affinity to A11:01 was less than 500 nM were considered as candidate epitopes. Peptides of candidate epitopes were synthesized by CSBio (China). All peptides were reconstituted in DMSO to 20 μg/μl and stored at −80°C.
2.4. Induction of antigen peptide‐specific CTLs
The induction of antigen peptide‐specific T cells was performed as described previously. 15 A known epitope of a cytomegalovirus restricted to HLA‐A11:01 was used as a positive control. Briefly, on the 1st day, 1 × 108 human PBMCs were isolated from 50 ml of fresh peripheral blood using Ficoll gradient centrifugation. CD8+ T cells were isolated using the Dynabeads CD8 Positive Isolation Kit (Thermo Fisher Scientific). The remaining CD8− cells were used to generate monocyte‐derived dendritic cells (DCs) using a plastic adherence method. CD8− cells were cultured in CellGro DC (Cellgenix) medium containing 1% human AB serum (ABS, Thermo Fisher Scientific), 500 U/ml of IL‐4 (Thermo Fisher Scientific), and 1000 U/ml of GM‐CSF (Thermo Fisher Scientific) for 72 h in six‐well plates (Thermo Fisher Scientific) at 37°C, 5% CO2. On the 4th day, in total, 5.0 × 105 matured DCs were seeded into one well of a 24‐well plate and pulsed with antigen peptide (20 μg/ml) for 16 h. On the 5th day, pulsed DCs were inactivated with 30 μg/ml of mitomycin C (Sigma) for 30 min, then 1.0 × 105 DCs were co‐cultured with 5 × 105 CD8+ T cells in CellGro DC/5% ABS containing 30 ng/ml of IL‐21 (Thermo Fisher Scientific) in a 48‐well plate. Next, 10 ng/ml IL‐7 (R&D Systems) and IL‐15 (Novoprotein) were supplied on the 8th, 10th, and 12th day. On the 15th day, antigen peptide‐specific T cells were stained with peptide–MHC dextramers (Immudex) separately and analyzed using FACS. CD8+/Dextramer+ T cells were sorted using FACS and subjected to TCR repertoire analysis.
2.5. TCR repertoire analysis
TCR clonotypes of the T cells were analyzed by next‐generation sequencing, as described previously. 16 Briefly, total RNA was extracted from tumors using the AllPrep DNA/RNA mini kit (Qiagen), or RNA from sorted CD8+/Dextramer+ T cells was extracted using the PicoPure RNA Isolation Kit (Life Technologies) according to the manufacturer's instructions. cDNAs with common 5′‐RACE adapters were synthesized from total RNA using the SMART library construction kit (Clontech Laboratories). TCRα and TCRβ cDNAs were amplified by PCR using a forward primer for the SMART adapter and reverse primers corresponding to the constant regions of each TCRα and TCRβ chain. Illumina index sequences with barcodes were added using the Nextera XT Index kit (Illumina), then the library was sequenced using 300‐bp paired‐end reads on an Illumina MiSeq system using the MiSeq Reagent v3 600‐cycles kit (Illumina, USA). Tcrip software was used to analyze the sequence data.
2.6. Construction of TCR library and TCR‐engineered T cells
After bulk TCR sequencing, top rank alpha chains and beta chains were selected and randomly paired to obtain a library of 116 candidate TCR constructs. Detailed information on the library can be found in Table S3. The TCR library was cloned into lentiviral vectors for T‐cell transduction. Multiplicity of infection (MOI) was controlled at 0.2 to ensure single lentivirus integration into the genome of each transduced T cell.
Lentiviruses were generated using the pMD2.G and pSPAX2 envelope plasmids in HEK293T cells. Freshly isolated PBMCs were activated with anti‐CD3 and anti‐CD28 (Miltenyi Biotec) overnight, and proceeded to virus infection. Viral supernatant mixed with 400 U/ml IL‐2 (R&D System, USA) and 8 μg/ml protamine (Sigma Aldrich) was added onto peripheral blood lymphocytes (PBLs) and centrifuged at 800 g and 25°C for 1.5 h. Transduced T cells were maintained in GT551‐H3 medium (TaKaRa) supplied with 5% human serum (GemCell), 1% penicillin/streptomycin (HyClone), and 400 IU/ml IL‐2. The expression of TCR genes was routinely examined using a mouse TCRβ‐specific antibody (BD Biosciences) by FACS.
2.7. Functional analysis of TCR‐T cells
The numbers of interferon (IFN)‐γ secreted T cells were detected using enzyme‐linked immunospot (ELISPOT) kit (Mabtech) according to the manufacturer's instruction. Briefly, on the 1st day, C1R‐A11 cells were pulsed with antigen peptide and TCR‐T cells were rested overnight. On the 2nd day, 2 × 104 pulsed C1R‐A11 cells were co‐cultured with 5 × 104 TCR‐T cells in a 96‐well plate for 20 h. On the 3rd day, spots were captured using an automated ELISPOT reader ImmunoSpot S4 (Cellular Technology Ltd.) and analyzed using the ImmunoSpot Professional Software package (Version 5.1, Cellular Technology Ltd).
The levels of secreted IFN‐γ, IL2, and TNFα of TCR‐T cells were determined using enzyme‐linked immunosorbent assay (ELISA) assay. Supernatants of TCR‐T cells co‐cultured with antigen peptide‐pulsed or nonpulsed C1R‐A11 cells were collected and analyzed as the manufacturer's instruction suggested.
The killing abilities of TCR‐T cells were examined using the CytoTox 96 Non‐Radioactive Cytotoxicity Assay kit (Promega). Briefly, C1R‐A11 cells were pulsed with antigen peptide (10−6 M) for 16 h, then co‐cultured with TCR‐T cells at different effector:target ratios for 4 h. The supernatant containing lactate dehydrogenase (LDH) that were released from the damaged C1R‐A11 cells was measured following the manufacturer's instruction. Maximum LDH released from target cells was measured by lysing C1R‐A11 cells with 0.9% Triton X‐100 reagent. The percentage of killing was quantified as: % Cytotoxicity = [(Experimental – Effector Spontaneous – Target Spontaneous)/(Target Maximum – Target Spontaneous)] × 100.
2.8. Generation of E692 –101‐expressing cervical cancer cell line
The sequences of E692–101 (GTTLEQQYNK) and HLA‐A11:01 open‐reading frames were cloned into a lentiviral vector and transduced into a cervical cancer cell line CaSki. E692–101 and A11:01 stably expressing CaSki‐E692–101 was selected with 2 μg/ml puromycin for 7 days until transfection efficiency was confirmed by FACS.
2.9. Murine model of adoptive T cell therapy
An immunodeficient mouse strain NCG (GemPharmatech) that lacks mature T, B and natural killer (NK) cells was used in this study. Next, 4‐week‐old NCG mice were inoculated subcutaneously with 1 × 107 CaSki‐E692–101 cells. Mice were divided randomly into three groups: the TCR‐T cell‐treated group, the control T cell‐treated group, and the tumor group (n = 5). Control T and TCR‐T cells were established from PBL from the same donor and maintained under the same culture conditions. T cells were cultured in vitro for 14 days and then used for in vivo infusion. Next, 3 × 107 T cells were injected intravenously to the mice on the 10th day after tumor inoculation. Here, 400,000 IU IL‐2 (Kingsley Pharmaceutical) was given intraperitoneally for the first 3 days. Tumor size was recorded using vernier caliper measurement or in vivo bioluminescence measurement using an IVIS imaging system (PerkinElmer). Briefly, luciferin (200 mg/kg, Promega) was injected intraperitoneally into mice; 15 min later, mice were anesthetized and bioluminescence images were captured in the lateral position on the IVIS apparatus. Photons emitted from the tumor were quantified using Living Image Software (PerkinElmer).
2.10. Statistical analysis
All experiments were repeated independently three times under identical conditions. Data are presented as the mean ± SEM. Student's t‐test or one‐way ANOVA with Tukey's honestly significant difference (HSD) test was used to analyze the statistical differences. Correlation analysis were done using GraphPad Prism version 8.0 (GraphPad Software). A p‐value <0.05 was considered statistically significant.
3. RESULTS
3.1. Epidemiology of HPV‐related cervical lesions in an east Asian population
To investigate the prevalent HLA‐A types and HPV types in east Asia, 98 patients living in the Shenzhen of Guangdong Province were enrolled in this study. The age of these patients ranged from 27 to 76; patients had been diagnosed with different cervical lesions: 45 of the 98 cases were cervical intraepithelial neoplasm, 41 cases were cervical squamous carcinoma, 11 cases were cervical adenocarcinoma, and one case was clear cell carcinoma. Among the 98 patients, a high proportion were diagnosed as infection with HPV16 (53.57%) or HPV18 (11.90%), suggesting the prevalence of these two high‐risk HPV types. Further analysis of the abundance of cervical tissue‐infiltrating T cells from 30 patients using TCR repertoire analysis revealed that the diversity of T cell clonotypes was much higher in HPV16‐ or HPV18‐related cancers than in other HPV type‐related cancers (Figure 1A,B), suggesting a more immune‐suppressive tumor microenvironment modified by HPV16 and HPV18. In addition, according to HLA genotyping, A11:01 was the most prevalent type (26.02%) in this cohort, which was consistent with other study in which A11:01 was found to be the most prevalent type in the Asian population, 17 so A11:01 was chosen for the following study.
FIGURE 1.
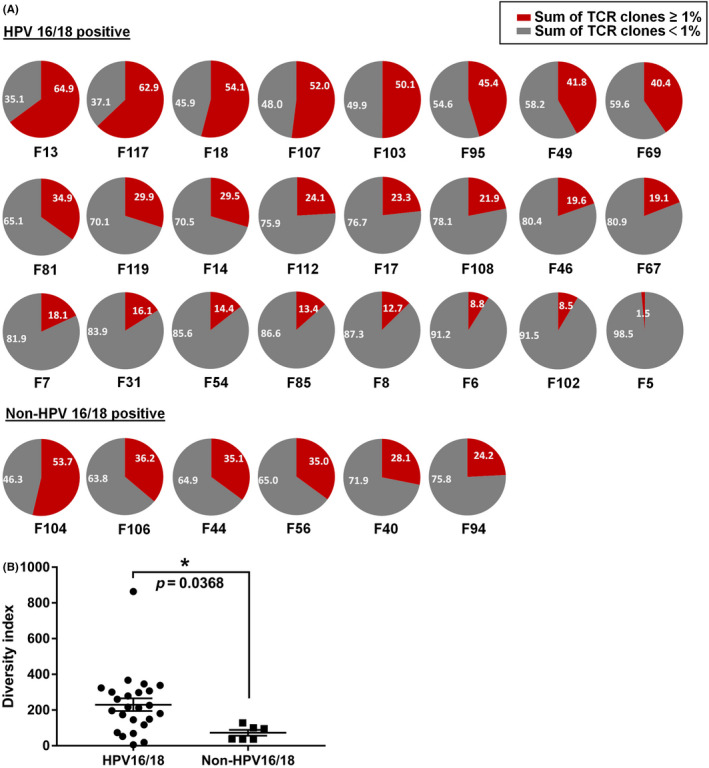
Diversity of T cell clonotypes in the tumor microenvironment of cervical cancers infected with different HPV types. Tumor issues were collected from 30 patients with cervical cancers, and RNA of which were proceeded to TCR repertoire analysis using next‐generation sequencing. (A) Pie charts depict the frequency distribution of unique TCR clone. The gray portion of each pie chart represents the sum of clones with frequencies <1%, and the red portion of each pie chart represents the sum of clones with frequencies >1%. (B) The diversity index calculated for TCRβ in HPV16‐ or HPV18‐positive cervical cancers (n = 24) and other HPV type‐infected cervical cancers (n = 6). Asterisk indicates significant difference between the two groups
3.2. Induction of E6 and E7 specific cytotoxic T cells and identification of their TCR clonotypes
Three epitopes of HPV16 and three epitopes of HPV18 that were predicted to have high affinity for HLA‐A11:01 were chosen for CTL induction (Table 2). Five of the six epitopes (except HPV18‐E641–50) successfully induced antigen‐specific T cells (Figure 2A). After FACS‐based sorting and subsequent TCR repertoire analysis using next‐generation sequencing, the clonotypes of antigen‐specific cytotoxic T cells were determined. The dominant TCRαβ pair existed in HPV16‐E692–101‐induced or HPV18‐E759–67‐induced cytotoxic T cells, other antigen peptide‐induced T cells consisted of multiple TCRαβ clones (Figure 2B), making the following individual TCR verification difficult. To improve the efficiency of TCR validation, we decided to select between two and four of the top‐ranking frequent TCRα or TCRβ from each antigen peptide‐induced T cell, and paired them randomly, to finally construct a lentiviral vector library containing 116 unique TCRαβ pairs. The TCR library were transduced into T cells from heathy donors; the MOI was controlled at 0.2 to ensure one virus transduced into a single T cell. Subsequently, the TCR library transduced T cells were stimulated with a peptide pool (all five antigen peptides) of pulsed or mock‐pulsed antigen‐presenting cells, and reactive T cells were further enriched by CD137 positivity (Figure 3A,B). Unstimulated TCR library transduced T cells and sorted CD137+ T cells were subjected to TCR repertoire analysis, and clonal TCRs that were significantly enriched compared with unstimulated or mock counterpart were considered as antigen‐reactive TCRs.
TABLE 2.
Epitopes predicted to be bound to HLA‐A*11:01
| Epitopes | HPV type | Sequence | Affinity to HLA‐A (IC50, nM) |
|---|---|---|---|
| E633–41 | HPV16 | IILECVYCK | 43 |
| E692–101 | HPV16 | GTTLEQQYNK | 126.8 |
| E789–97 | HPV16 | IVCPICSQK | 48.7 |
| E684–92 | HPV18 | SVYGDTLEK | 13.4 |
| E641–50 | HPV18 | LTEVFEFAFK | 32 |
| E759–67 | HPV18 | HTMLCMCCK | 61.3 |
FIGURE 2.
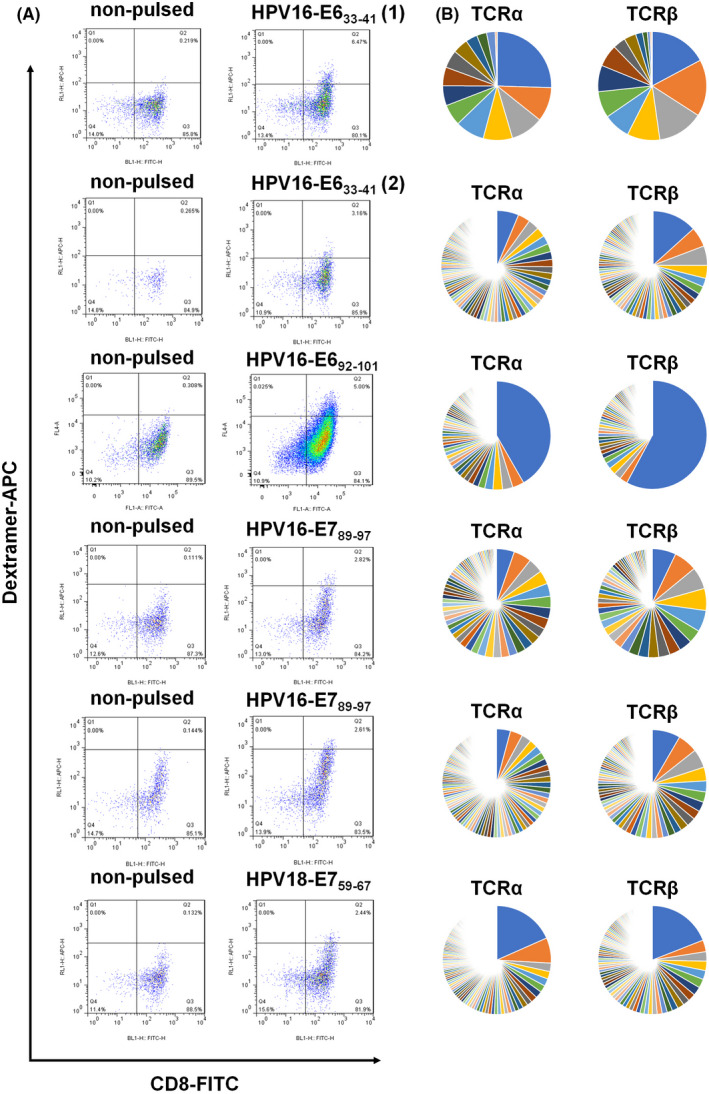
Identification of HPV‐specific cytotoxic T cells. (A) Cytotoxic T cell induction was performed with individual antigen peptide and specific T cells were enriched with the peptide–MHC‐dextramer fluorescent probe by FACS (inductions for HPV16‐E633–41 were performed twice). (B) RNAs of enriched T cells were isolated separately and subjected to TCR repertoire analysis using next‐generation sequencing. The frequencies of TCRA, TCRB, and their unique CDR3 sequences are shown
FIGURE 3.
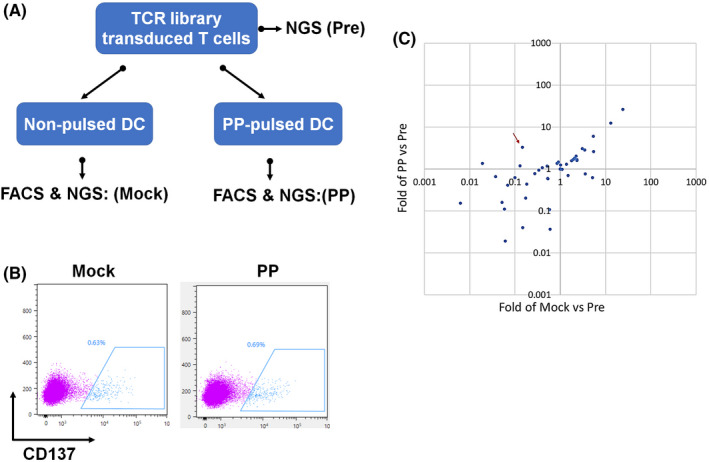
Strategy to discriminate ‘real’ antigen‐reactive TCRs. (A) A lentiviral vector library containing 116 TCRs was constructed and transduced into T cells and the MOI was controlled at 0.2 to ensure a single T cell was infected by one lentivirus. The TCR library transduced T cells were then stimulated with the peptide pool or mock‐pulsed autologous monocyte‐derived dendritic cells (MoDC). (B) Reactive T cells were further enriched by CD137 positivity. (C) Unstimulated TCR library transduced T cells and sorted CD137‐positive T cells were subjected to TCR repertoire analysis. The red arrow points to the antigen‐reactive clonal TCR051 that was significantly enriched compared with unstimulated or its mock counterpart
3.3. E692 –101 specific TCR‐T cells showed antigen‐specific killing activities in vitro
This workflow identified TCR051 as an antigen‐reactive TCR, which was 3.3‐fold enriched compared with its unstimulated counterpart and 22.7‐fold enriched compared with the mock counterpart, and was deduced to belong to E692–101‐induced cytotoxic T cells (Figure 3C). Furthermore, the validation screen was also performed with an artificial cell line (HLA‐null K562 transduced with A11:01), in which TCR051 was 7.0‐fold or 3.4‐fold enriched compared with its unstimulated or mock counterpart (Figure S1), indicating that the choice of antigen‐presenting cell is flexible. To further confirm the reactivity of TCR051, a lentiviral vector encoding TCR051 was constructed and individually transduced into T cells. FACS‐confirmed TCR051 efficiently recognized an artificial peptide–MHC mimic probe (Figure 4A,B). Furthermore, the functional analysis of TCR051 was performed in an in vitro tumor‐killing model. IFN‐γ ELISPOT assay and ELISA revealed that TCR051 transduced T cells were activated by E692–101‐pulsed antigen‐presenting cells in a dose‐dependent manner, which had no response on control peptide‐pulsed antigen‐presenting cells (Figure 5A–E). Similarly, TCR051 transduced T cells showed antigen‐specific killing on E692–101‐pulsed antigen‐presenting cells, whereas it only had slight toxicity for control peptide‐pulsed antigen‐presenting cells, which might be an TCR‐independent inhibition of T cells on tumor cell lines (Figure 5F).
FIGURE 4.
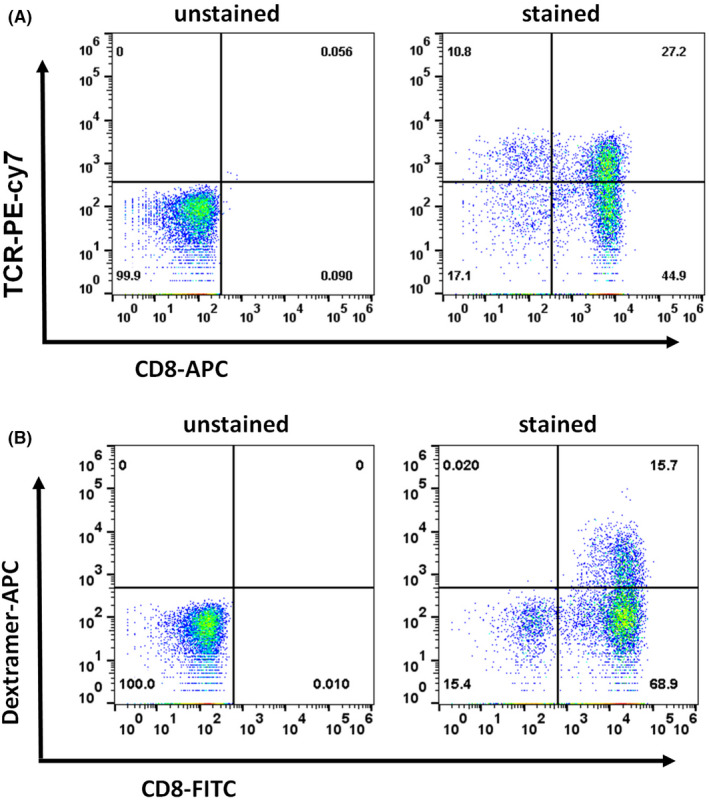
Construction of E692–101 specific TCR‐T cells. A lentiviral vector carrying TCR051 was transduced into peripheral blood lymphocytes (PBL) from a healthy donor. (A) Expression of exogenous TCRs was detected by FACS with an antibody against the mouse constant region of TCRβ. (B) The specificity of these TCR‐T cells was determined by FACS using an E692–101‐MHC‐dextramer fluorescent probe
FIGURE 5.
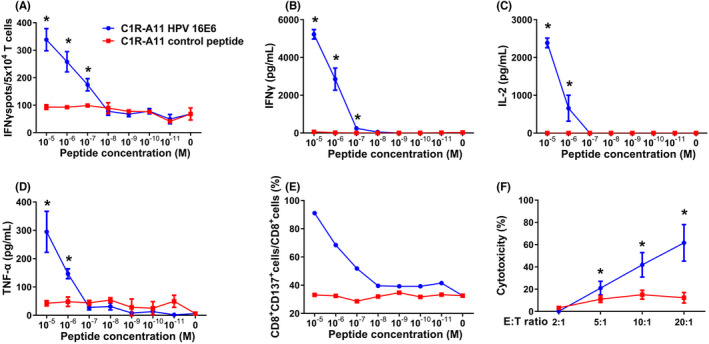
Functional assay of E692–101‐specific TCR‐T cells. TCR051 transduced T cells were co‐cultured with C1R‐A11 cells pulsed with different concentrations of E692–101 or mock peptide. (A) The number of IFNγ‐secreted T cells was determined using IFN‐γ ELISPOT assay. (B–D) The amount of secreted IFNγ, IL‐2 and TNFα was detected by ELISA. (E) The proportion of CD137+ T cells was analyzed by FACS. TCR051 transduced T cells were co‐cultured with E692–101 or mock peptide‐pulsed C1R‐A11 cells at different effector:target (E:T) ratios. (F) Killing ability was determined by lactate dehydrogenase (LDH) leakage of target cells. Asterisks indicate significant difference (p < 0.05) between the two groups
3.4. E692 –101 specific TCR‐T cells efficiently inhibited the growth of E692 –101‐carrying cervical cancers in vivo
To further confirm the in vivo tumor‐killing ability and safety of TCR051 transduced T cells, a cervical cancer cell line stably expressing E692–101, HLA‐A11:01, and a luciferase reporter was established, and transplanted subcutaneously (s.c.) into immunodeficient mice. When the tumor cell graft reached 5 mm, TCR051 transduced T cells or mock‐transduced T cells were adoptively transferred intravenously (i.v.), and tumor size was continuously recorded. Using in vivo imaging, tumors were seen to began to regress in TCR‐T cells treated mice at the 14th day after treatment, and showed continuous tumor control during the whole 30‐day treatment, whereas nontreated or control T cells failed to control the tumor growth (Figure 6B‐D). In addition, a high proportion of TCR‐T cells was detectable in the peripheral blood of mice at the final stage of treatment (Figure 6E,F), suggesting that antigen induced a strong proliferation of the TCR‐T cells.
FIGURE 6.
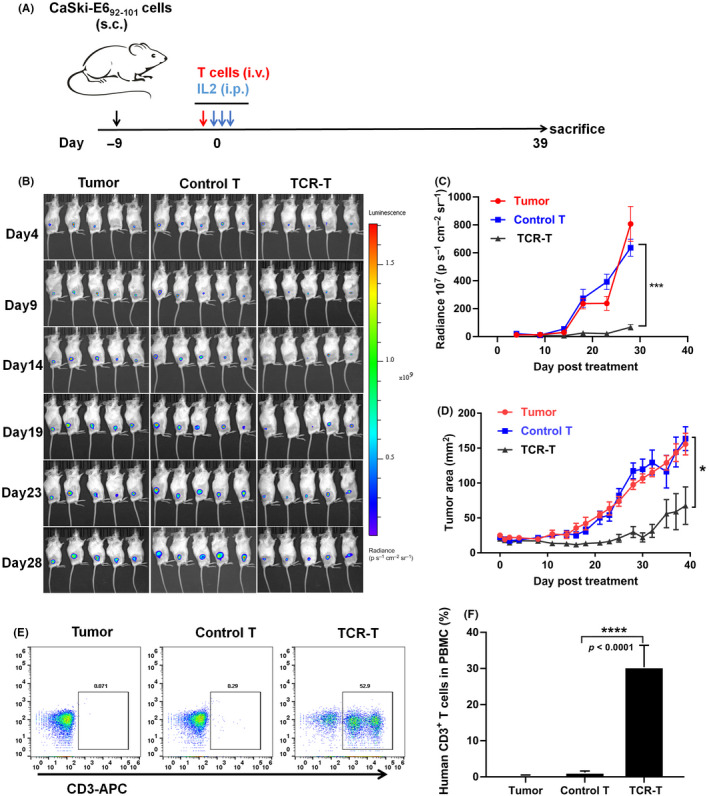
Evaluation of the tumor‐killing abilities of E692–101‐specific TCR‐T cells in a murine model of adoptive therapy. (A) Immunodeficient mice were subcutaneously (s.c.) implanted with human cervical tumor cells CaSki that expressed E692–101 epitopes and a luciferase reporter for 9 days, and were intravenously (i.v.) treated with TCR051 transduced T cells (TCR‐T) or mock‐transduced T cells (Control T). Then, 400,000 IU systemic IL‐2 was given for the first 3 days to sustain activities of T cells. (B) Bioluminescence images of tumors at 4–28 days after adoptive T cell transfer. (C) Quantitation of the luminescence signals. (D) Tumor areas at the surface of tumors were measured over time. (E, F) Proportion of human CD3+ T cells in the peripheral blood of mice at the end of this experiment was recorded. Data are presented as mean ± SEM (n = 5 per group)
4. DISCUSSION
Virus‐related cancers were commonly characterized as highly immunosuppressive. 18 , 19 , 20 During the procedure of chronic virus infections, the tumor microenvironment becomes highly suppressive, rendering the exclusion of T cell infiltration. This might partly explain why some virus‐related cancers are cold tumors and are poorly responsive to conventional therapy or immune checkpoint therapy. Therefore, adoptive cell transfer supplying additional tumor‐specific T cells to help patients eradicate tumors is a reasonable approach for treating late‐stage recurrent virus‐related cancers.
HPV16 and HPV18 are the two of most prevalent HPV types, accounting for 70% of HPV‐related cancers. 21 HPV16 and HPV18 are defined as high‐risk HPV types due to their highly malignant transformation abilities. By analyzing the TCR clonotypes of tissue‐infiltrating T cells between HPV16‐ or HPV18‐positive cervical cancers and other HPV type‐related cervical cancers, the T cell diversity in HPV16‐ or HPV18‐positive cancers was significantly more than for other HPV types, suggesting that HPV16 or HPV18 inhibits T cell proliferation. Although the conclusion is not confirmed, as the number of other HPV type‐related cases in this cohort is small, this supports the idea that HPV16‐ and HPV18‐postitive cancers might be more immunosuppressive and might require more adoptive T cell therapy.
The E6 and E7 proteins are two oncogenic proteins in the HPV proteome and are biomarkers for HPV infection. 22 In addition, these proteins are exogenous and are not expressed by host cells, making them attractive targets for TCR‐T cells. Therefore, E6 and E7 were chosen as the targets for TCR‐T cells in this study. Certainly, other components of the HPV proteome are predicted to show high affinity to A11:01, such as E2284–292 (NTTPIVHLK), E257–66 (QVVPTLAVSK), E2103–112 (LTAPTGCIKK), or E281–91 (TIYNSQYSNEK), and could also be potential targets of TCR‐T cell design, and this requires further validation.
The core processes of TCR‐T cell development are screening CTL‐specific epitopes and the corresponding TCRs in an efficient and accurate manner. We previously established a six‐step workflow allowing the generation of tumor neoantigen‐specific TCR‐T cells within 2 months. 13 Furthermore, to accelerate the recognition of CTL epitopes, we updated the workflow using a tandem minigene library and exploited tumor infiltrating T cells as an alternative for autologous peripheral blood. These modifications enabled a narrowing down of the neoantigen candidates and broadened the sources of tumor reactive T cells. 14 Although these modifications permitted rapidly determination of CTL‐specific epitopes, hurdles exist in the process of TCR validation. How to dig out the correct antigen‐corresponding TCR from numerous TCRαβ pairs during TCR repertoire analysis remains difficult. Here, we exemplify a TCR library‐based validation that could examine hundreds of TCRs and several antigen peptides at once. This method largely improves validation efficiency and reduces expense.
Although the HPV‐specific TCR‐T cells showed good performance in antigen‐specific activation in vitro and tumor inhibition in vivo, we noticed that it was not able to fully eradicate tumors. This might be due to cancer cells downregulating epitopes or losing MHC expression in the in vivo environment or, as we deduced, that majorities of TCR isolated from peripheral blood were of moderate affinity. 14 Therefore, upgrading TCR affinity for example through genetic mutation to obtain higher‐affinity TCR, 23 or adjuvant therapy such as synergism with radiotherapy, 24 , 25 might be required to achieve better tumor control. Several studies have also highlighted the importance of CD4+ T cells in adoptive cell therapy. 26 , 27 We are currently establishing CD4+ T cell‐based TCR‐T cells, and will evaluate whether the combination of CD4+ and CD8+ TCR‐T cells could achieve better tumor eradication.
The real human tumors are heterogeneous, indicating that some epitopes might not be shared by all tumor cells. Therefore, there is a need to determine the most shared epitopes or to apply multiple target TCR‐T cells to avoid immune escape of those tumor cells not expressing specific antigens. The rapid development of biotechnology will make the large‐scale screening of TCRs possible. Several solutions have been raised, such as high‐throughout TCR synthesis at the single cell level. 28 Combined with our previous techniques, this study exemplifies a technical route that could be applied for large‐scale epitopes and the corresponding TCR screening by combining TCR library‐based validation with a minigene library‐based epitopes screen. Certainly, this approach will not be limited to HPV‐related cancers, but also applicable for other virus‐related cancers such as HBV, EBV, or neoantigen screen.
FUNDING INFORMATION
This project is supported by the Shenzhen Natural Science Foundation (JCYJ20190807150615224, JCYJ20210324114009026), Shenzhen International Collaborative Innovation Program (GJHZ20190821162003794, GJHZ20210705142209028), the National Natural Science Foundation of China (82002956), the Guangdong Basic and Applied Basic Research Foundation (2019A1515110149).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
APPROVAL OF THE RESEARCH PROTOCOL
The study protocol was approved by the Institutional Ethics Review Board of the Shenzhen People's Hospital (approval number [2019] 007) and conducted in compliance with the declaration of Helsinki.
INFORMED CONSENT
All patients were enrolled in this study after informed consent was obtained.
REGISTRY AND THE REGISTRATION NO. OF THE STUDY/TRIAL
N/A.
ANIMAL STUDIES
Animal experiments were performed under the approval of the Ethics Committee of Shenzhen People's Hospital of China (approval number LL‐KY‐2020033).
Supporting information
Figure S1
Table S1
Table S2
Table S3
ACKNOWLEDGMENTS
This work was supported by the Shenzhen Natural Science Foundation (grant number: JCYJ20190807150615224, JCYJ20210324114009026), Shenzhen International Collaborative Innovation Program (grant number: GJHZ20190821162003794, GJHZ20210705142209028), the National Natural Science Foundation of China (grant number: 82002956), the Guangdong Basic and Applied Basic Research Foundation (grant number: 2019A1515110149).
Jiang J, Xia M, Zhang L, et al. Rapid generation of genetically engineered T cells for the treatment of virus‐related cancers. Cancer Sci. 2022;113:3686‐3697. doi: 10.1111/cas.15528
Jinxing Jiang and Ming Xia authors contributed equally.
Contributor Information
Teng Wei, Email: weiteng.shenzhen@outlook.com.
Lili Ren, Email: ren.lili@szhospital.com.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Zapatka M, Borozan I, Brewer DS, et al. The landscape of viral associations in human cancers. Nat Genet. 2020;52:320‐330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. de Martel C, Georges D, Bray F, Ferlay J, Clifford GM. Global burden of cancer attributable to infections in 2018: a worldwide incidence analysis. Lancet Glob Health. 2020;8:e180‐e190. [DOI] [PubMed] [Google Scholar]
- 3. de Martel C, Plummer M, Vignat J, Franceschi S. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int J Cancer. 2017;141:664‐670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maxwell JH, Grandis JR, Ferris RL. HPV‐associated head and neck cancer: unique features of epidemiology and clinical management. Annu Rev Med. 2016;67:91‐101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McBride AA, Warburton A. The role of integration in oncogenic progression of HPV‐associated cancers. PLoS Pathog. 2017;13:e1006211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hoppe‐Seyler K, Bossler F, Braun JA, Herrmann AL, Hoppe‐Seyler F. The HPV E6/E7 oncogenes: key factors for viral carcinogenesis and therapeutic targets. Trends Microbiol. 2018;26:158‐168. [DOI] [PubMed] [Google Scholar]
- 7. Ramos CA, Narala N, Vyas GM, et al. Human papillomavirus type 16 E6/E7‐specific cytotoxic T lymphocytes for adoptive immunotherapy of HPV‐associated malignancies. J Immunother. 2013;36:66‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tsang KY, Fantini M, Fernando RI, et al. Identification and characterization of enhancer agonist human cytotoxic T‐cell epitopes of the human papillomavirus type 16 (HPV16) E6/E7. Vaccine. 2017;35:2605‐2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fakhr E, Modic Ž, Cid‐Arregui A. Recent developments in immunotherapy of cancers caused by human papillomaviruses. Immunology. 2021;163:33‐45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Draper LM, Kwong ML, Gros A, et al. Targeting of HPV‐16+ epithelial cancer cells by TCR gene engineered T cells directed against E6. Clin Cancer Res. 2015;21:4431‐4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jin BY, Campbell TE, Draper LM, et al. Engineered T cells targeting E7 mediate regression of human papillomavirus cancers in a murine model. JCI Insight. 2018;3:e99488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Doran SL, Stevanović S, Adhikary S, et al. T‐cell receptor gene therapy for human papillomavirus‐associated epithelial cancers: a first‐in‐human, phase I/II study. J Clin Oncol. 2019;37:2759‐2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ren L, Leisegang M, Deng B, et al. Identification of neoantigen‐specific T cells and their targets: implications for immunotherapy of head and neck squamous cell carcinoma. Oncoimmunology. 2019;8:e1568813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wei T, Leisegang M, Xia M, et al. Generation of neoantigen‐specific T cells for adoptive cell transfer for treating head and neck squamous cell carcinoma. Oncoimmunology. 2021;10:1929726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Matsuda T, Leisegang M, Park JH, et al. Induction of neoantigen‐specific cytotoxic T cells and construction of T‐cell receptor‐engineered T cells for ovarian cancer. Clin Cancer Res. 2018;24:5357‐5367. [DOI] [PubMed] [Google Scholar]
- 16. Fang H, Yamaguchi R, Liu X, et al. Quantitative T cell repertoire analysis by deep cDNA sequencing of T cell receptor α and β chains using next‐generation sequencing (NGS). Oncoimmunology. 2014;3:e968467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gonzalez‐Galarza FF, McCabe A, Santos E, et al. Allele frequency net database (AFND) 2020 update: gold‐standard data classification, open access genotype data and new query tools. Nucleic Acids Res. 2020;48:D783‐D788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shamseddine AA, Burman B, Lee NY, Zamarin D, Riaz N. Tumor immunity and immunotherapy for HPV‐related cancers. Cancer Discov. 2021;11:1896‐1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen Y, Tian Z. HBV‐induced immune imbalance in the development of HCC. Front Immunol. 2019;10:2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bauer M, Jasinski‐Bergner S, Mandelboim O, Wickenhauser C, Seliger B. Epstein‐Barr virus‐associated malignancies and immune escape: the role of the tumor microenvironment and tumor cell evasion strategies. Cancers (Basel). 2021;13:5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Braaten KP, Laufer MR. Human papillomavirus (HPV), HPV‐related disease, and the HPV vaccine. Rev Obstet Gynecol. 2008;1:2‐10. [PMC free article] [PubMed] [Google Scholar]
- 22. Ara R, Khatun S, Pervin S, et al. Role of molecular biomarker human papilloma virus (HPV) E6 oncoprotein in cervical cancer screening. Gynecol Oncol. 2020;158:590‐596. [DOI] [PubMed] [Google Scholar]
- 23. Liu Q, Tian Y, Li Y, et al. In vivo therapeutic effects of affinity‐improved‐TCR engineered T‐cells on HBV‐related hepatocellular carcinoma. J Immunother Cancer. 2020;8:e001748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Minn I, Rowe SP, Pomper MG. Enhancing CAR T‐cell therapy through cellular imaging and radiotherapy. Lancet Oncol. 2019;20:e443‐e451. [DOI] [PubMed] [Google Scholar]
- 25. Watanabe N, McKenna MK, Rosewell Shaw A, Suzuki M. Clinical CAR‐T cell and oncolytic virotherapy for cancer treatment. Mol Ther. 2021;29:505‐520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moeller M, Haynes NM, Kershaw MH, et al. Adoptive transfer of gene‐engineered CD4+ helper T cells induces potent primary and secondary tumor rejection. Blood. 2005;106:2995‐3003. [DOI] [PubMed] [Google Scholar]
- 27. Wang LX, Shu S, Disis ML, Plautz GE. Adoptive transfer of tumor‐primed, in vitro‐activated, CD4+ T effector cells (TEs) combined with CD8+ TEs provides intratumoral TE proliferation and synergistic antitumor response. Blood. 2007;109:4865‐4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Spindler MJ, Nelson AL, Wagner EK, et al. Massively parallel interrogation and mining of natively paired human TCRαβ repertoires. Nat Biotechnol. 2020;38:609‐619. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Table S1
Table S2
Table S3
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


