Abstract
Pancreatic ductal adenocarcinoma (PDAC) is a highly fatal malignancy with extremely poor prognosis. Gemcitabine resistance is a major challenge in the treatment of PDAC. Here, we showed that LINC00460 was associated with the response to gemcitabine both in PDAC patients and PDAC‐PDX. After knocking down LINC00460 in PDAC tumor cells, results of RNA sequencing followed by gene ontology analysis indicated that LINC00460 influenced the activity of growth factors and modified the extracellular matrix. FISH showed that LINC00460 is mostly located in the cytoplasm. Results of RNA pull‐down, LC–MS/MS, RIP, and immunoblotting confirmed that LINC00460 could directly bind to PDAP1. Furthermore, we demonstrated that LINC00460 mediated the cellular communication of PDAC tumor cells and CAFs by PDAP1/PDGFA/PDGFR signaling pathway and regulated the gemcitabine‐resistance function of CAFs, which could be reversed by treatment with a PDGFR inhibitor (crenolanib). PDAC‐PDX tumors with lower expression of LINC00460 showed a better response to gemcitabine plus crenolanib treatment. Our finding supported the application of LINC00460 in precision medicine that uses gemcitabine plus crenolanib to treat PDAC with low expression of LINC00460.
Keywords: cancer associated fibroblasts, gemcitabine‐resistance, LINC00460, pancreatic ductal adenocarcinoma, PDAP1
LINC00460 regulates the gemcitabine chemotherapy‐resistance function of CAFs via the PDAP1/PDGFA/PDGFR pathway, which could be attenuated by treatment with a PDGFR inhibitor (crenolanib).PDAC with low expressed LINC00460 is an indicator to apply treatment therapy of gemcitabine combine with crenolanib.
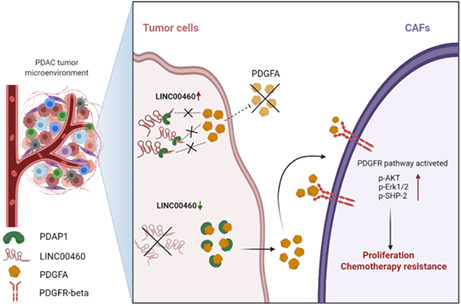
1. INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is a highly fatal malignancy with a 5‐year survival rate of 10%. At the time of diagnosis, 80%–85% of patients present with unresectable or metastatic disease. Even for a small subset of patients who are diagnosed with a localized, resectable tumor, the prognosis remains poor, with only 20% surviving for five years following surgery. 1 , 2 , 3 , 4
Gemcitabine was recommended as a first‐line chemotherapy for PDAC, but only a subset of patients benefited from it. 3 , 4 , 5 Published data have shown that patients who underwent a matched treatment based upon their actionable molecular alterations had a superior prognosis compared with those who underwent unmatched treatment. 6 Therefore, advanced indexes that could contribute to the selection of appropriate treatment are critical to making a clinically significant impact.
Pancreatic ductal adenocarcinoma is characterized by its special tumor microenvironment (TME), which accounts for approximately 90% of PDAC tumors. 7 Cancer‐associated fibroblasts (CAFs), immune cells, endothelial cells, and neurons are the predominant cell types within the TME. Cells in the TME create a unique network in which they interact with each other and produce extracellular matrix (ECM) components, which in turn present a complex signaling network with transformed tumor cells. 8 , 9 , 10 The function of the TME in PDAC remains debatable. Currently, there is no approved anti‐stromal therapy for PDAC.
Over the past decade, non‐coding RNAs have emerged as novel regulators in tumor progression, such as transcriptional regulation and cellular differentiation. Previous studies have demonstrated that long non‐coding RNAs (lncRNAs) could act as sponges to inhibit the expression of miRNAs, thereby suppressing the translation of mRNA targets. 11 , 12 , 13 Recently, an increasing number of studies have shown that lncRNAs could mediate the function of proteins by directly binding to them. 14 , 15 However, the molecular mechanisms underlying the direct binding of lncRNAs with proteins to mediate the cell function of CAFs remain unclear.
LINC00460 is a long non‐coding RNA that has been reported to play vital roles in various cancers. 16 In recent years, an increasing number of studies have evaluated the function and mechanism of lncRNAs in chemotherapy resistance. 17 , 18 However, the function of LINC00460 in the chemotherapy resistance in PDAC has not been reported. Here, we demonstrated that LINC00460 was associated with the response to gemcitabine of PDAC. LINC00460 mediated the cellular communication of tumor cells and CAFs during gemcitabine resistance by PDAP1/PDGFA/PDGFR signaling pathway. PDAC with low expressed LINC00460 showed a more positive response to gemcitabine plus PDGFR inhibitor (crenolanib). Thus, our findings indicated that gemcitabine plus crenolanib could be an option to treat PDAC with low expression of LINC00460.
2. MATERIALS AND METHODS
2.1. Patients' specimens
A total of 20 patients who received EUS‐guided fine‐needle aspiration biopsy and were confirmed as having PDAC by histology from June 2017 to March 2021 in the First Affiliated Hospital of Sun Yat‐sen University were enrolled in this study. All of these patients received gemcitabine neoadjuvant chemotherapy and had no prior chemotherapy or radiotherapy. The response assessment with imaging was assessed according to the Response Evaluation Criteria in Solid Tumors guidelines (RECIST, version1.1). Another 37 PDAC patients who underwent surgical resection at the First Affiliated Hospital of Sun Yat‐sen University were also enrolled in this study. This study was approved by the Ethical Committee of the First Affiliated Hospital of Sun Yat‐sen University ([2017]247). All patients provided written consent.
2.2. Primary CAFs and tumor cells isolation
Freshly collected tissue from the operation theater was transported in Hank's Balanced Salt Solution (#SH30588.01, Hyclone), cut into small pieces, and incubated with Collagenase I (200 U/mL; #LS004197, Worthington Biochem), DNAse I (10 μg/mL; #D5025‐15KU, Sigma‐Aldrich), and Collagenase P (0.5 mg/mL; #11213865001, Roche) in HBSS for 30 min at 37°C with 47 g. The solution was forced through a 100‐mm cell strainer followed by a 70‐mm cell strainer washed several times with FAST buffer (0.2% BSA and 2 mM EDTA in PBS). For isolating fibroblasts, the single cell suspension was incubated with anti‐fibroblast MicroBeads (130–050‐601, Miltenyi Biotec) in FAST buffer for 15 min at 4°C and then applied on LS columns (130–042‐401) in a magnetic field (QuadroMACS Separator; 130–090‐976). CAFs#1 were verified by immunofluorescence staining after MicroBeads sorting of cell suspensions of tissue specimens (Figure S2A). All experiments were performed up to five passages to avoid possible gross genomic changes during long‐term in vitro culture. Cells were verified by fibroblastic morphology in two dimensions: expression of stromal marker PDGFR (#ab32570, Abcam) and activated marker aSMA(#ab32575, Abcam) inimmunofluorescence assay. For isolating tumor cells, the Tumor Cell Isolation Kit was used according to the manufacturer's instructions (130–108‐339, Miltenyi Biotec).
2.3. Cell culture and transduction
Human PDAC cell lines PANC‐1 and CFPAC‐1 were purchased from Cellcook Cell Biotechnology (Guangzhou, China). PANC‐1 was cultured in DMEM‐high glucose pyruvate (Gibco, #11995065) supplemented with 10% FBS (Gibco, #A3160802), and CFPAC‐1 was cultured in L‐15 (Gibco, #11415056) supplemented with 10% FBS (Gibco, #A3160802). For the tumor cells/CAFs co‐culture system, cell culture inserts with 0.4 μm PC pore size in six well plates (Thermo Scientific 140,640) were applied in the co‐culture system. Then, 1 × 105 CAFs were seeded in the six well plate, and 5 × 104 pancreatic tumor cells were seeded in the inserts. For the gene knockdown and knockout assays, cells were infected with lentivirus encoding shRNA or sgRNA, respectively. LINC00460 expression was knocked down by transduction of pLKO.1‐Puro, and PDAP1 expression was knocked out by transduction with CRISPR‐Cas9‐V2. The target sequences used for LINC00460 shRNAs and PDAP1 sgRNAs are listed in Table S1.
2.4. Xenograft Assay and Tumor Stroma Measurement
Four‐week‐old female BALB/c‐nu/nu mice (obtained from the Laboratory Animal Center of SUN YAT‐SEN University) were kept in the Animal Room F zone of Zhongshan School of Medicine; 1 × 106 CFPAC‐1 or PANC‐1 cells with 2 × 106 CAFs#1 cells in 150 μL PBS were inoculated subcutaneously into the left flank of nude mice. Tumors were measured every 3 days. Tumor volume was calculated with the formula: V = 0.5 × length × width 2. The tumor‐bearing mice were killed 33 days after inoculation, and the tumors were subsequently removed for further study. Tumor slices were stained using a Masson Trichrome Staining Kit (LEAGENE, #DC0033). Digital microphotographs (200× and 400×) were taken using a NIKON Eclipse Ci microscope, and three microscopic fields were randomly selected. The images were analyzed using ImageJ software (version 1.8), and the average stromal proportions were calculated. The detailed operating protocol using ImageJ software can be found in Section 2.5.
2.5. Patient‐derived xenograft subcutaneous tumor model
We set the Patient‐derived xenograft (PDX) tumor without gemcitabine treatment as the first passage (P1). The LINC00460 expression level was quantified using the PDX(P1) tumors. PDX(P1) were divided into high and low LINC00460 expression groups according to the Ct value of LINC00460. We pass the PDX(P1) tumors in BALB/C nude mice with the treatment of gemcitabine to gain the following passages: P2, P3 and P4. Tumors of equal volume (3 × 3 × 3 mm) were grafted subcutaneously into the left flank of BALB/c mice. Drugs ((1) 1%DMSO saline, (2) gemcitabine 50 mg.kg−1 twice weekly, (3) crenolanib 20 mg kg−1 daily, and (4) combined drug regimen 2 + 3) were injected intraperitoneally until the tumor volume was >150 mm3. The termination of the experiment was four weeks after drug injection. The PDAC patients' specimen utilization and animal experiments were carried out with the approval of the Ethical Committee of the First Affiliated Hospital of Sun Yat‐sen University ([2019]124, [2020]005).
2.6. Subcellular fraction location
The separation of nucleus and cytoplasm fractions was performed with NE‐PER nuclear and cytoplasmic extraction reagents (Thermo, #78833, USA) according to the manufacturer's instructions. Afterwards, the qRT‐PCR assay was performed to measure percentages of LINC00460, U6, and GAPDH in the nucleus and cytoplasm, respectively.
2.7. RNA FISH
RNA FISH assay was performed with the Ribo FISH Kit (Ribo, China) according to the manufacturer's instructions. The lncRNA probe mix of LINC00460 and positive nucleus control probe U6 and positive cytoplasm control probe 18S were designed and purchased from Ribo. Images of cultured cells were obtained using the ZEISS Laser Scanning Confocal Microscope LSM710 with oil ×67 magnification lenses, and ZEN 2.3 software was used.
2.8. RNA immunoprecipitation qRT‐PCR assays
In LINC00460 RNA immunoprecipitation‐qPCR experiments, a total of 1 × 107 PANC‐1 or CFPAC‐1 cells were harvested in IP lysis buffer 150 mM KCl, 0.5 mM DTT, 5 mM EDTA, 0.5% NP‐40, 25 mM Tris, pH 7.4. Each lysate was further divided into two groups: anti‐PDAP1 and anti‐IgG (Input). Normal IgG (#A7007, Beyotime) was used as the negative control. Subsequently, the RBP captured, together with the bound RNA, was collected using Dynabeads (#10002D/10004D, Thermo Fisher). After washing off the unbound material, the RBP was digested using Proteinase K, and the RNA bound to immunoprecipitated RBP was purified and reverse transcribed into cDNA. Then, a qPCR assay was performed to measure the percentage input of LINC00460 in each group. The primer sequences used for RIP‐qPCR analysis are listed in Table S2.
2.9. RNA pull‐down and Label‐free quantitative proteomics by LC–MS/ MS
The pcDNA3.1(−) plasmids of LINC00460 transcripts (NR_034119.2) were constructed, respectively, followed by restriction digestion, agarose electrophoresis, and gel extraction. Reverse transcription of template DNA and streptavidin labeling was performed using the TranscriptAid T7 High Yield Transcription Kit (#K0441, Thermo). The protein lysates were harvested from CFPAC‐1, PANC‐1, and BXPC3 cells. Binding of magnetic beads labeled RNA streptavidin was performed using the Pierce Magnetic RNA‐Protein Pull‐Down Kit (#20164, Thermo). Then, peptide samples were separated and analyzed by liquid chromatograph‐mass spectrometer/mass spectrometer (LC–MS/MS). The full mass and subsequent MS/MS analyses were performed in an Orbitrap analyzer with a resolution of 70,000 for MS1 (at 200 mass/charge ratio [m/z]) and 17,500 for MS2, respectively. The automatic gain control target for MS1 was set to 3.0 × 10 + 6, with a maximum injection time (IT) of 50 ms and 5.0 × 10 + 4 for MS2 with a maximum IT of 100 ms. The top 20 most intense ions were fragmented by higher‐energy collisional dissociation (HCD) with normalized collision energy of 27% and an isolation window of 2 m/z. The dynamic exclusion time window was 30 s. The analysis results were processed using MaxQuant (1.5.6.0). The protein database is from the UNIPROT database (Uniprot_human_2016_09). Finally, statistical analysis was performed on the standardized quantitative results to obtain corresponding differentially expressed proteins.
2.10. Western blot analysis
The cell sample protein was harvested using the Pierce lysis buffer, and the tissue sample protein was harvested by using the Total Protein Extraction Kit (Invent, SD‐001), which resolved on 4%–12% SDS‐PAGE (ACE, Biotech) and transferred for 90 minutes at 280 mA to PVDF membranes (Millipore, USA). The membranes were blocked for 1 h in Tris‐buffered saline‐Tween 20 (TBST) with 5% BSA. Thereafter, immunoblottings were performed with below primary antibodies against PDAP1 (#15081‐1‐AP, Proteintech), PDGFA (#203911, Abcam), PDGFR (#32570, Abcam), a PDGFR Activation Antibody Sampler Kit (#12651, CST), β‐actin (#4970, CST), and GAPDH (#60004‐1‐Ig, Proteintech) in TBST buffer containing 5% BSA overnight. After incubation with the corresponding secondary antibody against rabbit IgG‐HRP‐linked (#7071, CST) for 1 h, the signals were finally visualized using Immobilon Western Blotting substrate (Millipore, USA).
2.11. ELISA
The expression level of PDGFA in cell supernatant medium was evaluated using an ELISA Kit (SEA528Hu, Cloud‐Clone Corp) according to the manufacturer's instructions.
2.12. RNA secondary structure and RNA–Protein Interaction prediction
The secondary structure of LINC00460 was predicted by the RNA fold web server: http://rna.tbi.univie.ac.at/cgi‐bin/RNAWebSuite/RNAfold.cgi. The LINC00460 sequence was uploaded onto the server, and the secondary structure of LINC00460 was calculated based on the recommended settings and parameters. For fold algorithms and basic options, “minimum free energy and partition function” and “avoid isolated base pairs” were chosen. The prediction of LINC00460‐PDAP1 interaction probability was performed using the online software from RPIsea website (http://pridb.gdcb.iastate.edu/RPISeq/).
2.13. Statistical analysis
The experiment was set up to use a minimum of three independent experiments. Results are reported as means ± SD. All statistical analyses were performed using GraphPad Prism 9.0. Comparisons were performed using a two‐tailed paired Student's t‐test or one‐way analysis of variance (ANOVA), as indicated in individual figures. Differences were considered statistically significant at two‐tailed p < 0.05 (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001).
3. RESULTS
3.1. LINC00460 was associated with the gemcitabine resistance in pancreatic ductal adenocarcinoma
To explore the role of lncRNAs in the occurrence of gemcitabine resistance in patients with PDAC, we analyzed the clinical data of 20 patients with unresectable locally advanced PDAC. Patients' clinical characteristics are shown in Table 1. All of these 20 patients underwent fine‐needle aspiration biopsy followed by treatment with gemcitabine. Patients were divided into high and low LINC00460 expression groups according to the relative expression level of LINC00460. The median of △CT was defined as the threshold. In the group with a higher level of LINC00460, a significantly high proportion of patients achieved a partial response and a complete response, while a low proportion of patients had stable and progressive disease (partial response + complete response: 5 vs 0; stable disease + progressive disease: 5 vs 10; p = 0.0098) compared with the group with a lower level of LINC00460 (Figure 1A,B). These results highlighted that LINC00460 might be associated with the sensitivity to gemcitabine.
TABLE 1.
Clinical characteristics of 20 PDAC patients
| Features | High expression of LINC00460 | Low expression of LINC00460 | p‐values |
|---|---|---|---|
| Age, years, median (range) | 55.0 (41.0–70.0) | 51.5 (40.0–80.0) | 0.890 |
| Sex | 1.000 | ||
| Male | 5 | 5 | |
| Female | 5 | 5 | |
| Tumor location | 0.628 | ||
| Head and neck of pancreas | 6 | 8 | |
| Body and tail of pancreas | 4 | 2 | |
| Imaging T category | 0.645 | ||
| T1 | 0 | 0 | |
| T2 | 3 | 4 | |
| T3 | 1 | 2 | |
| T4 | 6 | 4 | |
| Imaging N category | 0.582 | ||
| N0 | 3 | 1 | |
| N1, N2 | 7 | 9 | |
| Imaging M category | 1.000 | ||
| M0 | 9 | 9 | |
| M1 | 1 | 1 | |
| Imaging stage | 0.657 | ||
| IA | 0 | 0 | |
| IB | 1 | 0 | |
| IIA | 0 | 0 | |
| IIB | 3 | 5 | |
| III | 5 | 4 | |
| IV | 1 | 1 | |
| Drug response | 0.033 | ||
| CR | 0 | 0 | |
| PR | 5 | 0 | |
| SD | 4 | 7 | |
| PD | 1 | 3 |
Abbreviations: CR, complete response; PD, progressive disease; PDAC, pancreatic ductal adenocarcinoma; PR, partial response; SD, stable disease.
FIGURE 1.
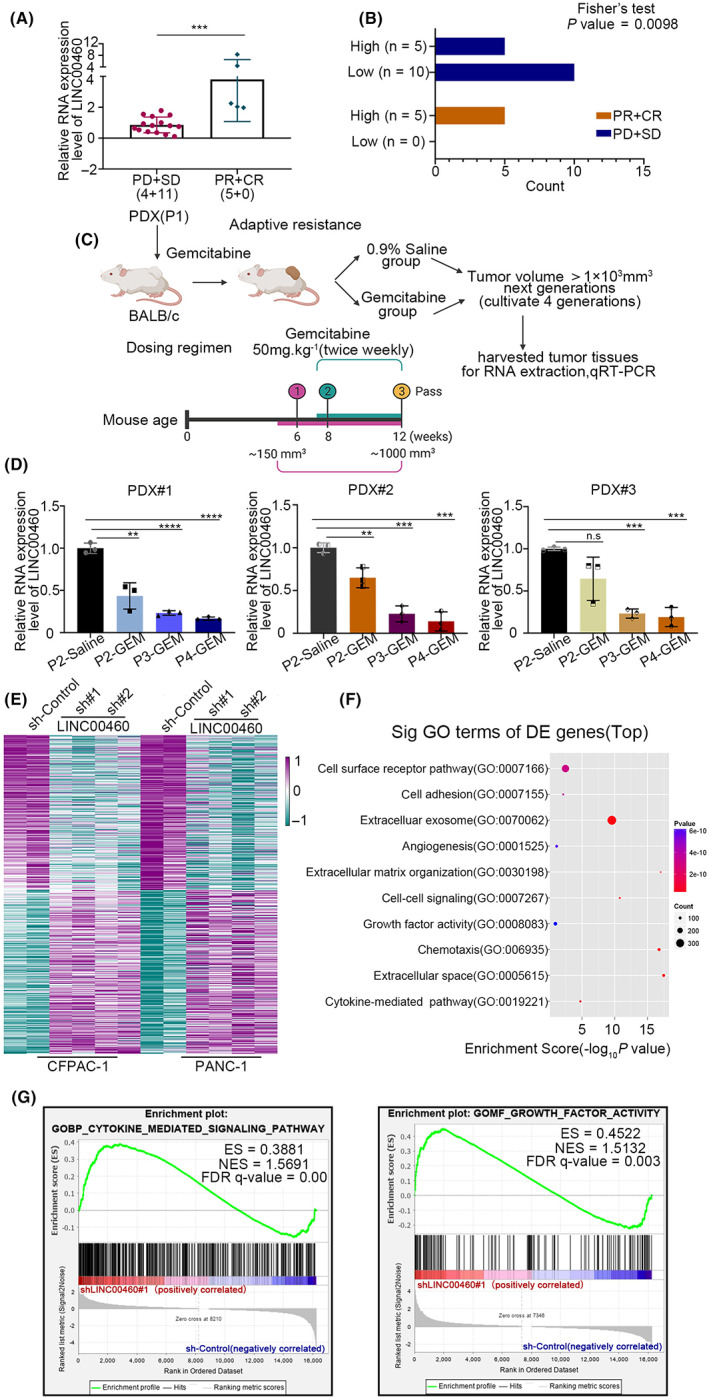
LINC00460 was associated with the gemcitabine resistance in pancreatic ductal adenocarcinoma cells (PDAC). (A) Relative expression level of LINC00460 was detected by qRT‐PCR (local chemotherapy cohort, n = 20; drug measure of response: PD (progressive disease); SD (stable disease); PR (partial response); CR (complete response)). (B) The correlation between high/low LINC00460 level and gemcitabine response was analyzed using Fisher's exact test. (C) Schematic to generate gemcitabine‐resistant primary human PDAC for PDX studies. PDX (P1) tumors were grafted subcutaneously to BALB/c mice for the gemcitabine‐resistance model (P2–P4). Once tumor volume reached 150 mm3, mice were treated with gemcitabine 50 mg/kg by intraperitoneal injection twice weekly until tumor growth relapsed (approximately 6 weeks), indicating drug resistance. (D) Relative expression level of LINC00460 in PDX tumor cells of the gemcitabine‐resistant/control model was detected by qRT‐PCR. (E) Heatmap of 734 differentially expressed genes in shControl (cyan) versus shLINC00460 (purple) (356 downregulated and 378 upregulated (n = 4 and 8; all absolute fold change >2 or ≤2, p‐value < 0.01). (F) GO annotation using genes whose differential expression is significant. (G) GSEA results showed that cytokine‐mediated signaling pathway and growth factors were activated when LINC00460 was knocked down.
To verify this hypothesis, we established a gemcitabine‐resistant PDX mouse model (Figure 1C). We set the PDX tumor without gemcitabine treatment as the first passage (P1). We passed the PDX (P1) tumors in BALB/C nude mice with the treatment of gemcitabine to gain the following passages: (P2, P3, and P4). We isolated tumor cells from gemcitabine‐resistant/control PDX tumors and tested the expression level of LINC00460. Results showed that the expression level of LINC00460 decreased gradually after gemcitabine treatment, but there was no significant change in normal saline treatment (Figures 1D, S1A). These results indicated that a lower level of LINC00460 was related to resistance to gemcitabine.
3.2. LINC00460 influences the activity of growth factors and modifies the extracellular matrix by binding to PDAP1
To elucidate the function and mechanism of LINC00460 in PDAC, we knocked down (KD) LINC00460 by shRNAs in two PDAC cell lines (Figure S1B,C) and performed RNA sequencing (RNAseq). The heatmap showed the differentially expressed genes after LINC00460 was knocked down (Figure 1E). GO analysis revealed the probable function of LINC00460 (Figure 1F). Interestingly, we found that LINC00460 might play a role in influencing the activity of growth factors and modifying the extracellular matrix, and this has not been previously reported. GSEA results showed that cytokine‐mediated signaling pathway and growth factors were activated when LINC00460 was knocked down (Figures 1G, S1D). Based on these results, we hypothesized that LINC00460 might play its role by regulating CAFs through growth factors and then progressing to modify the extracellular matrix.
To verify our hypothesis, we sent gemcitabine‐resistant/control PDX tumors for Masson trichrome staining to test the proportion of tumor cells (red) and stroma (blue). The results showed that the proportion of stroma in gemcitabine‐resistant PDX tumors was significantly increased compared to the control (Figure 2A,B). We also attempted to test the expression and correlation between LINC00460 and FAP (fibroblast activation protein alpha, a marker of CAFs) in human PDAC tumor tissues. We found that LINC00460 was negatively correlated with FAP (Figure 2C,D). As the expression level of LINC00460 decreased gradually after gemcitabine treatment, these results indicated that LINC00460 might have the function of regulating CAFs and modifying the extracellular matrix.
FIGURE 2.
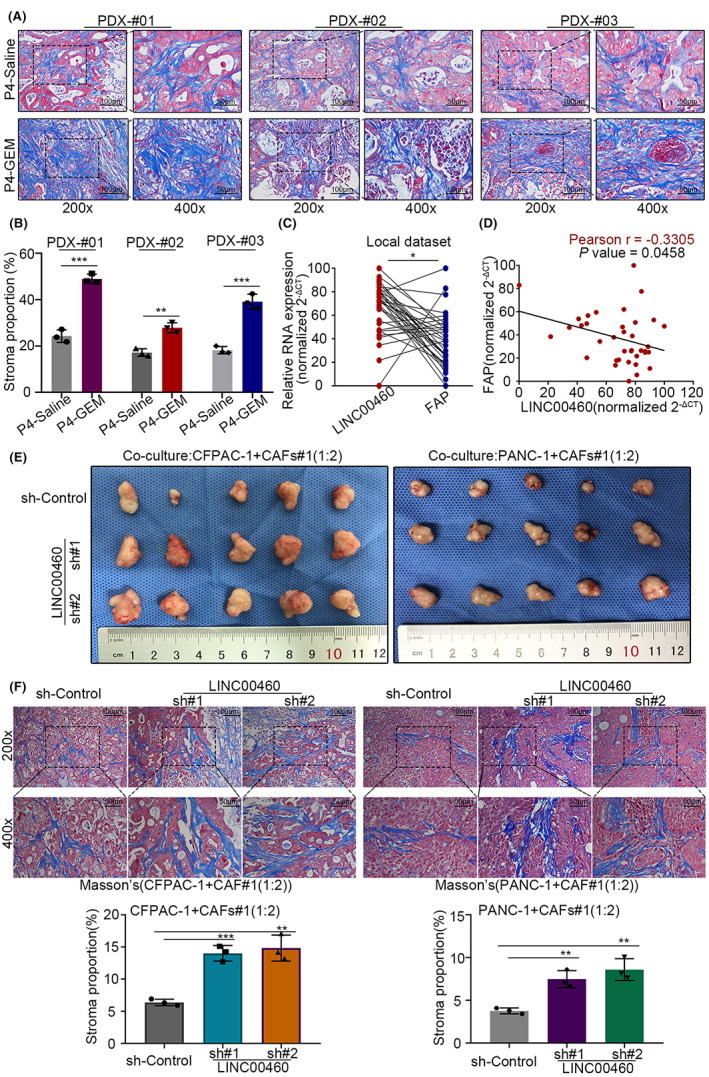
LINC00460 could modify the extracellular matrix. (A) Masson trichrome staining of PDX tumor samples. (B) Histogram plot showed the stroma proportion of the PDX tumors in (A). (C) Paired dot plot showed the relative expression level, measured by qRT‐PCR, and the correlation of LINC00460 and FAP in human PDAC tissues. (D) Plot showed the direct correlation between LINC00460 and FAP, based on the result of (C). (E) Xenograft tumors from respective groups are shown. (F) Masson trichrome staining of xenograft tumor samples with sh‐LINC00460 and sh‐Control, histogram plot showed the stroma proportion of the xenograft tumors.
Then we investigated the biological function of LINC00460. Results of the CCK‐8 assay showed that when PDAC tumor cells were cultured alone, the growth curve of LINC00460 KD groups was not statistically different from the control group (Figure S2B). Subsequently, we performed a xenograft assay with a mixed injection of PDAC tumor cells (LINC00460 KD/control groups separately) and CAFs (WT). Results showed that after co‐culturing PDAC tumor cells with CAFs, the tumor volume of LINC00460 KD groups was significantly higher than the control group (Figures 2E, S2C). Furthermore, when the xenograft tumors were subjected to Masson trichrome staining, we found that the proportion of stroma was significantly increased in LINC00460 KD groups (Figure 2F). These results indicated that LINC00460 modified the extracellular matrix with the presence of CAFs.
To gain deeper insight into the effects of LINC00460, RNA FISH and subcellular fractionation location assays were performed to determine the location of LINC00460 in PDAC cells (Figure 3A,B). The results showed that LINC00460 is mainly located in the cytoplasm. LncRNA can directly bind with proteins and influence the activity of the protein. 14 , 15 As LINC00460 is mainly located in the cytoplasm, we hypothesized that LINC00460 interacts with certain proteins. Therefore, an RNA pull‐down was performed to obtain proteins that could bind with LINC00460, and the proteins were subjected to LC–MS/MS for identification. PDAP1, a RNA‐binding protein, 19 , 20 , 21 , 22 was among the proteins with high rich scores (Figure 3C, D). PDAP1 was reported as a phosphoprotein that upregulates the PDGFA‐stimulated growth of fibroblasts by binding to PDGFA. The RNA immunoprecipitation (RIP) results verified that LINC00460 could bind to PDAP1 (Figure 3E). To determine the detailed sequence of LINC00460 that interacts with PDAP1, different fragments of the functional regions of LINC00460 were generated based on its secondary structure predicted using an RNA fold software (Figure 3F). By performing an RNA pull‐down assay, we identified the region (238–747 nt) that was essential for binding to PDAP1 (Figure 3G). PDAC is an extremely “hard” tumor due to its unique and abundant TME, which accounts for approximately 90% of the tumor space. CAFs are one of the major cell types in the TME. CAFs were reported to regulate chemotherapy resistance in PDAC. 23 , 24 , 25 , 26 Taken together, the above results and previous reports indicated that LINC00460 might bind to PDAP1, regulate the growth of CAFs and, thus, mediate the gemcitabiner esistance in PDAC.
FIGURE 3.
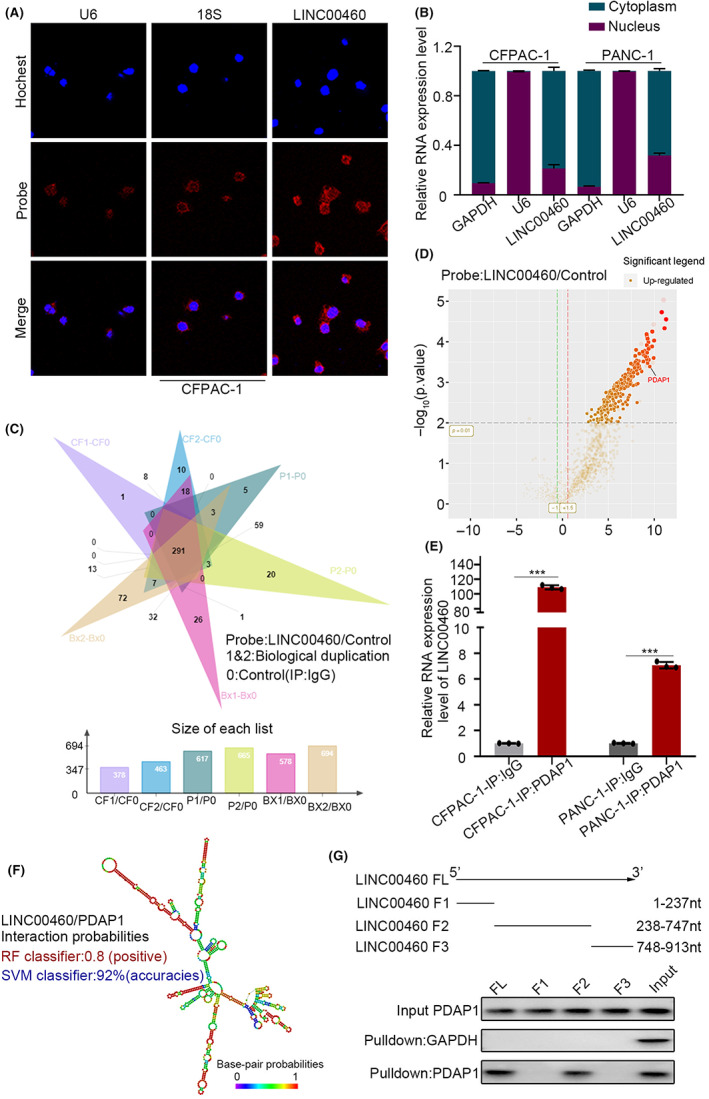
LINC00460 could directly bind with PDAP1. (A) FISH assay showed the sub‐cellular location of LINC00460 in CFPAC‐1. (U6: nucleus location positive control, 18S: cytoplasm control) The subcellular location of LINC00460 is mainly in cytoplasm, followed by the nucleus. (B) LINC00460 expression in the nucleus and cytoplasm fractions by qRT‐PCR were consistent with FISH results. Data from three independent experiments were expressed as mean ± SD. (C) RNA‐pull‐down‐MS results showed 291 proteins have the possibility of binding to LINC00460 in the intersection of three PDAC cell lines. (D) Volcano plot of RNA‐pull‐down‐MS revealed that PDAP1 (Log10[iBAQ] = 9.10) interacts with LINC00460. The red font indicates PDAP1. (E) RNA immunoprecipitation (RIP)‐qPCR assay detects the coupling of PDAP1 and LINC00460. (F) Predicted structure of the LINC00460 determined by RNAfold software and the prediction of LINC00460/PDAP1 interaction probabilities using RPIseq website. (G) Immunoblot of PDAP1 in RNA pull‐down extracts with different LINC00460 fragment sequences (1–237, 238–747, and 748–913).
3.3. LINC00460 regulates the proliferation of CAFs via the PDAP1/PDGFA/PDGFR pathway
To further clarify how LINC00460 interacts with PDAP1, we tested the expression and content of PDAP1 and PDGFA using qRT‐PCR and western blot. The results of qRT‐PCR showed that after LINC00460 knockdown, the expression levels of PDAP1 and PDGFA did not change significantly (Figure 4A). However, the western blot results showed that even though the protein content of PDAP1 remained unchanged, the protein content of PDGFA decreased conspicuously (Figure 4B). As PDGFA is a secreted protein, ELISA was performed to determine the amount of PDGFA in the culture supernatant. The ELISA results showed that PDGFA significantly increased in the culture supernatant of LINC00460 knockdown cells (Figure 4C). These results indicate that LINC00460 could regulate the secretion of PDGFA. To verify whether LINC00460 mediates the secretion of PDGFA by interacting with PDAP1, PDAP1 knockout (PDAP1‐KO) cell lines were generated (Figure 4D). After knocking down LINC00460 in PDAP1‐KO cell lines, the western blot and ELISA results showed that the protein content of PDGFA in cells and culture supernatant remained unchanged (Figure 4E,F). These results confirmed that LINC00460 regulates PDGFA secretion by binding to PDAP1. PDGFA is a PDGFR ligand. PDGFR is highly expressed in fibroblasts. Our results validated that in PDAC, PDGFR was rarely expressed in tumor cells but was highly expressed in CAFs (Figure 4G). After co‐culturing CAFs and tumor cells, we found that CAFs showed enhanced proliferation ability and activated the PDGFR pathway when co‐cultured with LINC00460 knockdown tumor cells. This effect could be reversed by treatment with a PDGFR inhibitor (Figure 4H–K); crenolanib is an effective and selective PDGFR inhibitor. Taken together, we demonstrated that LINC00460 regulates the proliferation of CAFs by binding to PDAP1.
FIGURE 4.
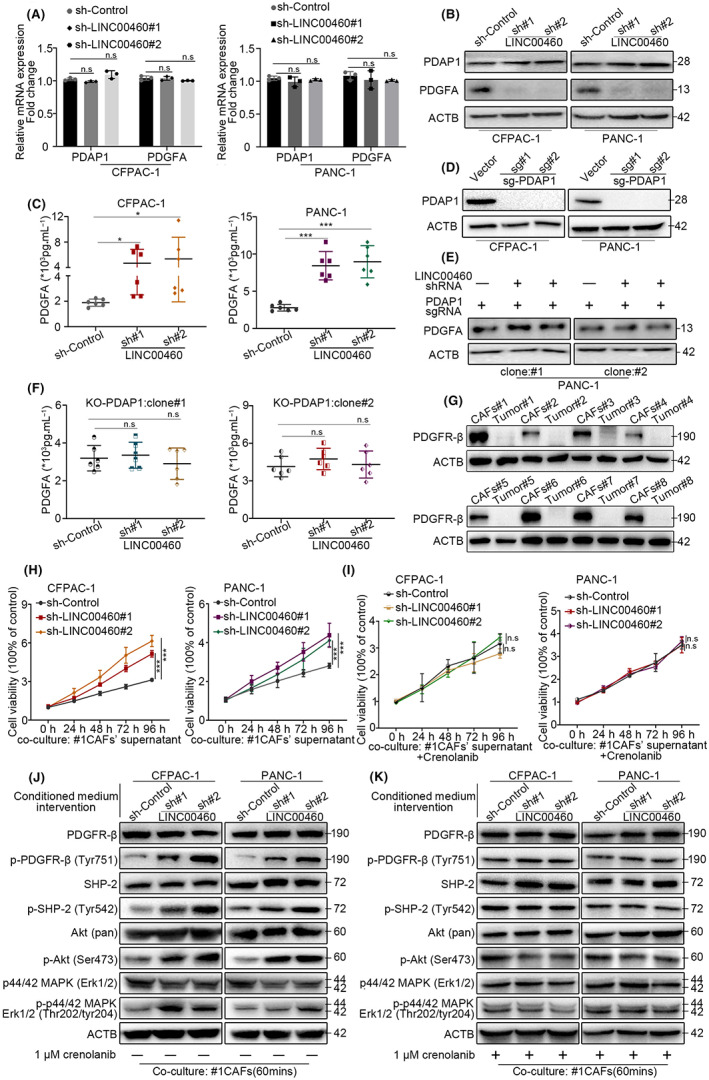
LINC00460 could regulate the proliferation of CAFs via the PDAP1/PDGFA/PDGFR pathway. (A) qRT‐PCR assay detected the mRNA level of PDAP1 and PDGFA after transduced LINC00460 shRNAs. (B) Western blot assay detected the protein level of PDAP1 and PDGFA after transduced LINC00460 shRNAs. (C) The concentration of PDGFA in the medium supernatant of CFAPC‐1 and PANC‐1 after silencing LINC00460 was measured by ELISA assay; values represent means ± SD of six replicate samples in each group. (D) Western blot assay validated KO‐PDAP1 cell lines. (E) Western blot assay showed the expression level of PDGFA in PANC‐1 cell line (KO‐PDAP1) after transduction of LINC00460 shRNAs. (F) ELISA assay showed the concentration of PDGFA in medium supernatant of PANC‐1 cell line (KO‐PDAP1) after transduction of LINC00460 shRNAs. Values represent means ± SD of six replicate samples in each group. (G) Western blot assay showed PDGFR‐β expression in eight pairs of PDAC CAFs and tumor cells. (H) Cell viability showed the different proliferation rate of CAFs after co‐cultured with the supernatant of CFPAC‐1 and PANC‐1. (I) Cell viability showed that crenolanib can reverse the proliferation of CAFs caused by silencing LINC00460. (J) Western blot analysis of the PDGFR‐β pathway activated genes in after transduced LIINC00460 shRNAs whose co‐cultured with CFPAC‐1 or PANC‐1 supernatant. (K) Western blot shows crenolanib can reverse PDGFR‐β pathway activation.
3.4. LINC00460 mediates resistance of pancreatic ductal adenocarcinoma cells to gemcitabine by regulating CAFs
To explore how LINC00460 mediates cell resistance to gemcitabine in PDAC, we measured the IC50 value of gemcitabine in LINC00460 knockdown cells. The results showed no significant differences in the IC50 values of gemcitabine between LINC00460 knockdown cells and control cells (Figure 5A,E). In co‐cultured CAFs and PDAC cells, LINC00460 knockdown cells displayed more than two‐fold higher IC50 values than control cells (Figure 5C,G). We then added a PDGFR inhibitor to the co‐culture system, and the results showed that the PDGFR inhibitor efficiently decreased the IC50 values in the LINC00460 knockdown group. These results indicate that LINC00460 mediates the resistance of PDAC cells to gemcitabine by regulating CAFs (Figure 5B,D,F,H,I).
FIGURE 5.
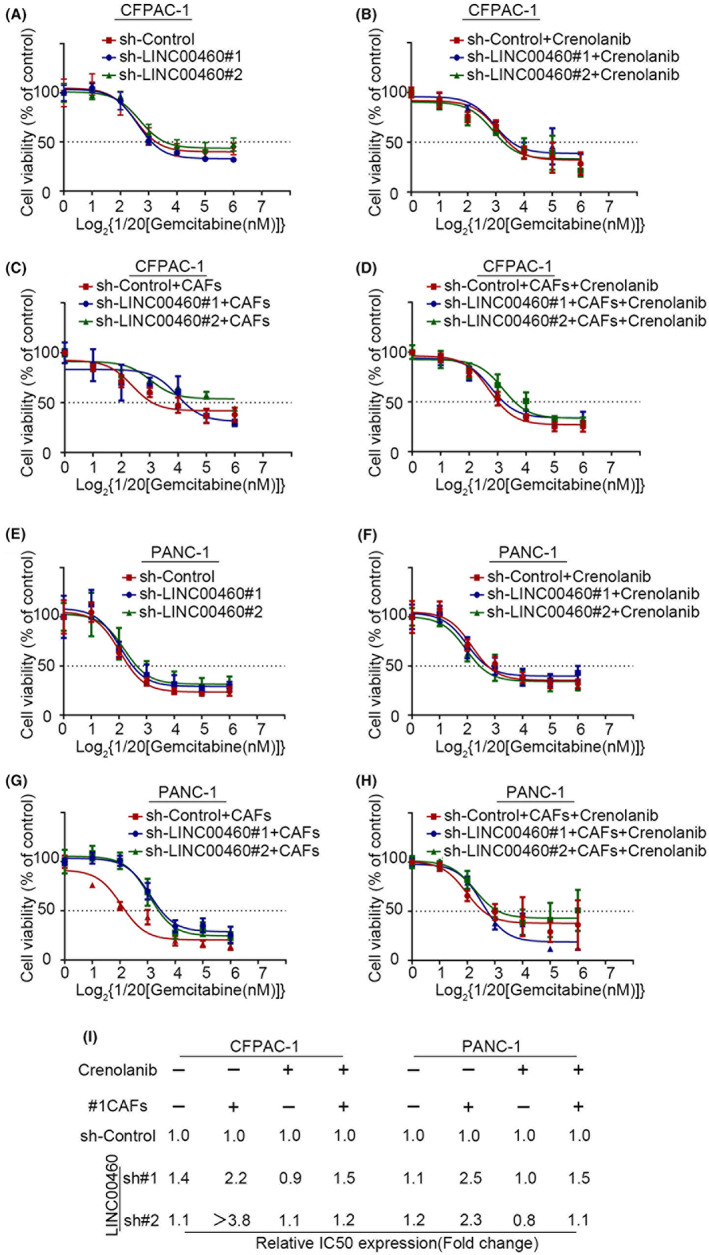
LINC00460 mediated the chemotherapy resistance to gemcitabine by regulating CAFs. (A–H) The cell viability of pancreatic cancer cells 72 hours after adding different concentrations of gemcitabine. The fitting curves showed the IC50 value of gemcitabine in LINC00460 knockdown/control pancreatic cancer cells (A‐D: CFPAC‐1; E‐H: PANC‐1; A,E, pancreatic cells alone; B,F, pancreatic cells with crenolanib; C,G, pancreatic cells with CAFs; D,H, pancreatic cells with CAFs and crenolanib). (I) The table shows the exact IC50 value of gemcitabine in A–H.
3.5. Pancreatic ductal adenocarcinoma cells with low expression of LINC00460 is an indicator for use of PDGFR inhibitor treatment
We demonstrated that LINC00460 mediates resistance to gemcitabine by regulating CAFs via the PDGFR pathway. Furthermore, we wondered whether tumors with lower expression of LINC00460 showed higher sensitivity to PDGFR inhibitors. We then adopted the PDX model to prove this hypothesis (Figure 6A). We gave gemcitabine or PDGFR inhibitor (crenolanib) or two drugs combined to the mice and measured the tumor volume. After giving gemcitabine, the tumor volume of PDX with higher expressed LINC00460 (PDX‐H) decreased more than PDX with lower expressed LINC00460 (PDX‐L) (Figure 6C). These results corresponded with our thesis that PDAC with high expression of LINC00460 was more sensitive to gemcitabine. After giving gemcitabine combined with crenolanib, compared with the gemcitabine group, PDX‐L showed significantly decreased tumor volume than PDX‐H (Figure 6B,C). This result indicated that tumors with low expression of LINC00460 could benefit more from PDGFR inhibitor. Thus, PDAC with low expression of LINC00460 can be an indicator for the use of PDGFR inhibitor treatment.
FIGURE 6.
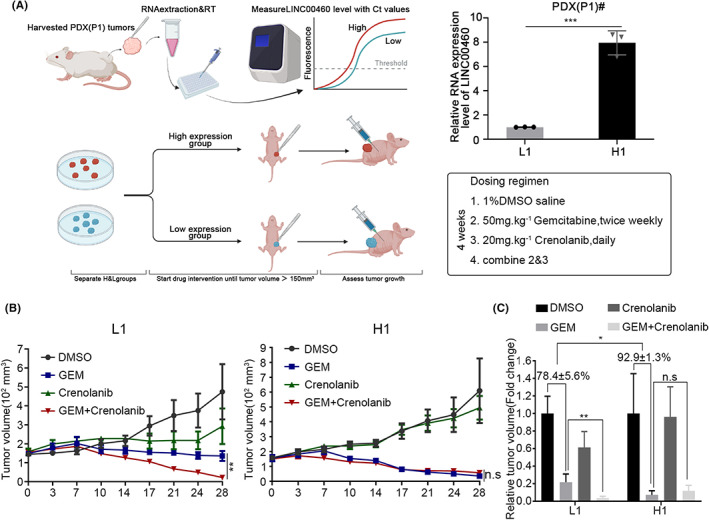
Pancreatic cancer with low expressed LINC00460 is an indicator to use PDGFR inhibitor. (A) qRT‐PCR results showed the expression level of LINC00460 in PDX tumors. PDX were divided to two groups (L, H) based on the expression level of LINC00460. (B) The tumor volume of PDX tumors after given DMSO/gemcitabine/crenolanib/gemcitabine+crenolanib. (C) The relative tumor volume of PDX tumors in B (the average volume of DMSO in each group were set as 100%); values represent means ± SD of five replicate samples in each group.
4. DISCUSSION
Gemcitabine is recommended as a first‐line chemotherapy for PDAC, but only a subset of patients are sensitive to this treatment. Inspired by this, we explored the biological characteristics of PDAC tumors that were resistant to gemcitabine. We established a PDX mouse model of gemcitabine resistance. The experimental mice were continuously treated with gemcitabine for four passages. LncRNAs are involved in several pathophysiological processes in pancreatic cancer, breast cancer, and other malignancies. 24 , 25 , 27 In recent years, an increasing number of studies have evaluated the function and mechanism of lncRNAs in chemotherapy resistance. 17 , 18 However, the role of lncRNAs in gemcitabine chemotherapy resistance still needs to be elucidated. Our results showed that PDAC with high expression of LINC00460 was more sensitive to gemcitabine. Therefore, we speculated that LINC00460 might be associated with tumor resistance to gemcitabine. To verify this hypothesis, we tested the expression levels of LINC00460 in every passage of the gemcitabine‐resistant PDX model. Consistent with our hypothesis, with passages past and increased gemcitabine resistance, the expression level of LINC00460 decreased. LINC00460 regulates the proliferation, migration, and invasion of tumor cells, but its role in chemotherapy resistance has rarely been studied. 16 This prompted us to elucidate the mechanism of action of LINC00460 in the occurrence of chemotherapy resistance.
By knocking down LINC00460 in two PDAC cell lines and performing RNAseq, we examined the probable function of LINC00460 by gene ontology (GO) analysis. After confirming its relationship with chemotherapy resistance, we found that LINC00460 regulates cell–cell communication (including growth factor activity, cell surface receptor signaling pathway, cell–cell signaling, and cytokine‐mediated signaling pathway) and modifies the extracellular matrix (including extracellular space, extracellular exosome, and ECM organization). The location of lncRNAs could indicate their function. Results of RNA FISH and subcellular fractionation location assays showed that LINC00460 was mainly located in the cytoplasm. LncRNAs have mostly been reported to function as sponges, inhibit the expression of siRNAs, and promote the translation of target mRNAs. Recently, an increasing number of studies have shown that lncRNAs could mediate the function of proteins by directly binding to them. 14 , 15 LINC00460 sponges miR‐320a and miR‐485‐5p and mediates the proliferation, migration, and invasion of tumor cells. 28 , 29 However, its role in mediating chemotherapy resistance has not been well studied. Therefore, we speculated that LINC00460 might mediate chemotherapy resistance by binding to proteins. RNA pull‐down and LC–MS/MS were performed to detect proteins that could directly bind to LINC00460. After combining the results of GO analysis, which showed that LINC00460 regulated the cell–cell communication and modified the extracellular matrix, we found that PDAP1 acts as an RNA‐binding protein. This phosphoprotein could upregulate the PDGFA‐stimulated growth of fibroblasts by binding to PDGFA. 30 PDGFA and its receptor PDGFR are involved in the occurrence of drug resistance in patients with breast cancer and glioblastoma. 31 , 32 Our results demonstrated that PDAP1 could bind to 238‐747 nt of LINC00460. Hence, we hypothesized that LINC00460 might mediate chemotherapy by activating the PDAP1 pathway.
The results of RNAseq did not show significant changes in the expression of PDAP1 in LINC00460 knockdown cells, which was confirmed by conducting qRT‐PCR and western blot. This indicated that LINC00460 did not act on the DNA or mRNA of PDAP1 and thus verified that LINC00460 could bind to PDAP1. As PDAP1 functions based on the status of the PDGFA, we evaluated the expression and location of PDGFA. Our results showed that LINC00460 did not regulate the expression levels of PDGFA, but the level of PDGFA expression in the extracellular matrix increased after knockdown of LINC00460. LINC00460 could not regulate the secretion of PDGFA in PDAP1 KO cells. This finding verified that LINC00460 regulated the secretion of PDGFA, which was mediated by PDAP1. PDGFA is a secreted protein that functions by stimulating the PDGFR expression (including PDGFR‐α and PDGFR‐β). PDGFR is a biomarker of fibroblast formation. PDGFR‐α and PDGFR‐β are expressed at similar levels in fibroblasts. PDGFR‐β participates in the regulation of the cell function of fibroblasts, and PDGFR‐β+ CAFs are associated with a worse prognosis in pancreatic cancer. 33 , 34 , 35 , 36 , 37 , 38 In line with these reports, we found that PDGFR was highly expressed in CAFs but was rarely expressed in PDAC tumor cells. Further results showed that knockdown of LINC00460 increased the proliferation of CAFs and activated the PDGFR signaling pathway, and this effect could be reversed by treatment with crenolanib (PDGFR inhibitor). To explore the relationship between CAFs and gemcitabine chemotherapy resistance, we determined the IC50 value of PDAC tumor cells. The results showed that knockdown of LINC00460 could increase the IC50 value of PDAC tumor cells when they were co‐cultured with CAFs but not when co‐cultured without CAFs. This effect was reversed by treatment with crenolanib. Taken together, these results indicate that LINC00460 regulates the gemcitabine chemotherapy‐resistance function of CAFs by directly binding to PDAP1 and mediating the PDGFA/PDGFR signaling pathway (Figure 7).
FIGURE 7.
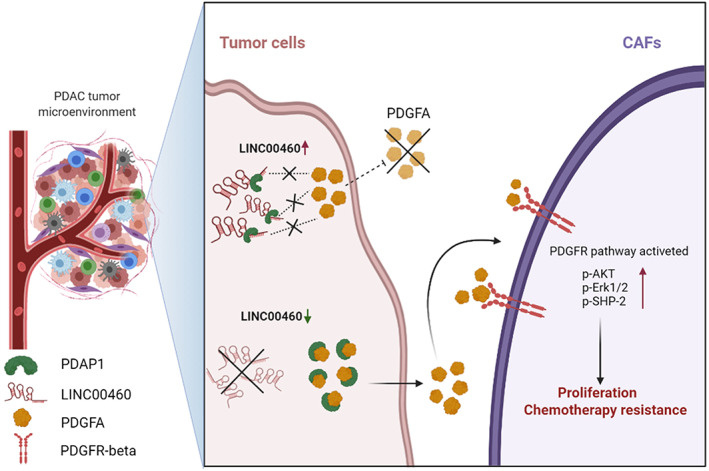
Proposed schematic model illustrating the interaction between LINC00460 and PDAP1, leading to the increase of CAF proliferation, which could cause chemotherapy resistance.
The results also indicated that crenolanib could reverse the gemcitabine chemotherapy‐resistance effect induced by LINC00460 low‐expressing cells. This inspired us to determine whether gemcitabine could be combined with crenolanib in the treatment of LINC00460 low‐expressing tumors. To verify this hypothesis, we adopted the PDX model and tested its response to gemcitabine/crenolanib/gemcitabine + crenolanib. In line with our results, when treated with gemcitabine alone, tumors with lower expression of LINC00460 were less sensitive compared with tumors with higher expression of LINC00460. However, when combined with gemcitabine and crenolanib, tumors with lower expression of LINC00460 showed a better response. Therefore, we concluded that PDAC with low expression of LINC00460 is an indicator for the use of PDGFR inhibitor treatment combined with gemcitabine.
In conclusion, we have demonstrated that LINC00460 regulates the gemcitabine chemotherapy‐resistance function of CAFs by directly binding to PDAP1 and mediating the PDGFA/PDGFR signaling pathway. PDAC with low expression of LINC00460 is an indicator for use of PDGFR inhibitor treatment combined with gemcitabine.
AUTHOR CONTRIBUTIONS
Xiao‐Xu Zhu: Conceptualization, Data curation, Writing ‐ original draft; Jian‐Hui Li: Data curation, Formal analysis, Writing ‐ original draft; Xuhao Ni: Methodology, Project administration; Xiao Wu: Methodology, Validation; Xun Hou: Investigation, Validation; Ya‐Xiong Li: Software, Data curation; Shi‐Jin Li: Investigation, Software; Wei Zhao: Funding acquisition, Writing ‐ review and editing; Xiao‐Yu Yin: Writing ‐ review and editing, Supervision.
ACKNOWLEDGMENT
This study was supported by the National Natural Science Foundation of China (Grant No. 81772522, No.82072644, and No. 31771630).
DISCLOSURE
The authors have no conflicts of interest to disclose.
ETHICS STATEMENT
Approval of the research protocol: This study was approved by the Ethical Committee of the First Affiliated Hospital of Sun Yat‐sen University.
Informed consent: All samples in this study were collected from the First Affiliated Hospital of Sun Yat‐sen University. All patients provided signed informed consent based on recognized guidelines.
Registry and the Registration No. of the study/trial: [2017]247.
Animal Studies: All experiments involving animals and human patient specimens were conducted in accordance with the Declaration of Helsinki and were approved by the Ethical Committee of the First Affiliated Hospital of Sun Yat‐sen University ([2019]124, [2020]005).
Supporting information
Figure S1 Cytokine fluctuations and growth factor activity when LINC00460 was knocked down
Figure S2. LINC00460 does not affect the proliferation of PDAC
Table S1. sgRNAs and shRNAs used in this study.
Table S2. Sequences of primers used in this study.
Supplemental Materials and Methods
Zhu X‐X, Li J‐H, Ni X, et al. Pancreatic ductal adenocarcinoma cells regulated the gemcitabine‐resistance function of CAFs by LINC00460 . Cancer Sci. 2022;113:3735‐3750. doi: 10.1111/cas.15547
Xiao‐Xu Zhu and Jian‐Hui Li contributed equally to this work.
Contributor Information
Wei Zhao, Email: zhaowei23@mail.sysu.edu.cn.
Xiao‐Yu Yin, Email: yinxy@mail.sysu.edu.cn.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in “RNA fold” at http://rna.tbi.univie.ac.at/cgi‐bin/RNAWebSuite/RNAfold.cgi and “RPIsea” at http://pridb.gdcb.iastate.edu/RPISeq/.
REFERENCES
- 1. Pereira SP, Oldfield L, Ney A, et al. Early detection of pancreatic cancer. Lancet Gastroenterol Hepatol. 2020;5(7):698‐710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cazes A, Betancourt O, Esparza E, et al. A MET Targeting antibody–drug conjugate overcomes gemcitabine resistance in pancreatic cancer. Clin Cancer Res. 2021;27(7):2100‐2110. [DOI] [PubMed] [Google Scholar]
- 3. Mizrahi JD, Surana R, Valle JW, Shroff RT. Pancreatic cancer. Lancet. 2020;395(10242):2008‐2020. [DOI] [PubMed] [Google Scholar]
- 4. Dell'Aquila E, Fulgenzi C, Minelli A, et al. Prognostic and predictive factors in pancreatic cancer. Oncotarget. 2020;11(10):924‐941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zeng S, Pottler M, Lan B, Grutzmann R, Pilarsky C, Yang H. Chemoresistance in pancreatic cancer. Int J Mol Sci. 2019;20(18):4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pishvaian MJ, Blais EM, Brody JR, et al. Overall survival in patients with pancreatic cancer receiving matched therapies following molecular profiling: a retrospective analysis of the know your tumor registry trial. Lancet Oncol. 2020;21(4):508‐518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dougan SK. The pancreatic cancer microenvironment. Cancer J. 2017;23(6):321‐325. [DOI] [PubMed] [Google Scholar]
- 8. Zhang Y, Recouvreux MV, Jung M, et al. Macropinocytosis in cancer‐associated fibroblasts is dependent on CaMKK2/ARHGEF2 signaling and functions to support tumor and stromal cell fitness. Cancer Discov. 2021;11:1808‐1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang S, Li Y, Xing C, et al. Tumor microenvironment in chemoresistance, metastasis and immunotherapy of pancreatic cancer. Am J Cancer Res. 2020;10(7):4561‐4567. [PMC free article] [PubMed] [Google Scholar]
- 10. Norton J, Foster D, Chinta M, Titan A, Longaker M. Pancreatic cancer associated fibroblasts (CAF): Under‐explored target for pancreatic cancer treatment. Cancers (Basel). 2020;12(5):1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Reese M, Dhayat SA. Small extracellular vesicle non‐coding RNAs in pancreatic cancer: molecular mechanisms and clinical implications. J Hematol Oncol. 2021;14(1):141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Farooqi AA, Nayyab S, Martinelli C, et al. Regulation of hippo, TGFbeta/SMAD, Wnt/beta‐catenin, JAK/STAT, and NOTCH by long non‐coding RNAs in pancreatic cancer. Front Oncol. 2021;11:657965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Song P, Yang F, Jin H, Wang X. The regulation of protein translation and its implications for cancer. Signal Transduct Target Ther. 2021;6(1):68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu L, Chen Y, Huang Y, et al. Long non‐coding RNA ANRIL promotes homologous recombination‐mediated DNA repair by maintaining ATR protein stability to enhance cancer resistance. Mol Cancer. 2021;20(1):94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lou M, Tang X, Wang G, et al. Long noncoding RNA BS‐DRL1 modulates the DNA damage response and genome stability by interacting with HMGB1 in neurons. Nat Commun. 2021;12(1):4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ghafouri‐Fard S, Khoshbakht T, Taheri M, Hajiesmaeili M. Long intergenic non‐protein coding RNA 460: review of its role in carcinogenesis. Pathol Res Pract. 2021;225:153556. [DOI] [PubMed] [Google Scholar]
- 17. Zhou C, Yi C, Yi Y, et al. LncRNA PVT1 promotes gemcitabine resistance of pancreatic cancer via activating Wnt/beta‐catenin and autophagy pathway through modulating the miR‐619‐5p/Pygo2 and miR‐619‐5p/ATG14 axes. Mol Cancer. 2020;19(1):118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xiong G, Liu C, Yang G, et al. Long noncoding RNA GSTM3TV2 upregulates LAT2 and OLR1 by competitively sponging let‐7 to promote gemcitabine resistance in pancreatic cancer. J Hematol Oncol. 2019;12(1):97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Iadevaia V, Wouters MD, Kanitz A, Matia‐Gonzalez AM, Laing EE, Gerber AP. Tandem RNA isolation reveals functional rearrangement of RNA‐binding proteins on CDKN1B/p27(Kip1) 3'UTRs in cisplatin treated cells. RNA Biol. 2020;17(1):33‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Castello A, Fischer B, Frese CK, et al. Comprehensive identification of RNA‐binding domains in human cells. Mol Cell. 2016;63(4):696‐710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Castello A, Fischer B, Eichelbaum K, et al. Insights into RNA biology from an atlas of mammalian mRNA‐binding proteins. Cell. 2012;149(6):1393‐1406. [DOI] [PubMed] [Google Scholar]
- 22. Baltz AG, Munschauer M, Schwanhausser B, et al. The mRNA‐bound proteome and its global occupancy profile on protein‐coding transcripts. Mol Cell. 2012;46(5):674‐690. [DOI] [PubMed] [Google Scholar]
- 23. Menezes S, Okail MH, Jalil S, Kocher HM, Cameron A. Cancer‐associated fibroblasts in pancreatic cancer: new subtypes, new markers, new targets. J Pathol. 2022;257:526‐544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang LZ, Yang JE, Luo YW, Liu FT, Yuan YF, Zhuang SM. A p53/lnc‐Ip53 negative feedback loop regulates tumor growth and chemoresistance. Adv Sci (Weinh). 2020;7(21):2001364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xie W, Chu M, Song G, et al. Emerging roles of long noncoding RNAs in chemoresistance of pancreatic cancer. Semin Cancer Biol. 2022;83:303‐318. [DOI] [PubMed] [Google Scholar]
- 26. Chen F, Chen J, Yang L, et al. Extracellular vesicle‐packaged HIF‐1alpha‐stabilizing lncRNA from tumour‐associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat Cell Biol. 2019;21(4):498‐510. [DOI] [PubMed] [Google Scholar]
- 27. Chen F, Chen J, Yang L, et al. Extracellular vesicle‐packaged HIF‐1alpha‐stabilizing lncRNA from tumor‐associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat Cell Biol. 2019;21(4):498‐510. [DOI] [PubMed] [Google Scholar]
- 28. Li G, Kong Q. LncRNA LINC00460 promotes the papillary thyroid cancer progression by regulating the LINC00460/miR‐485‐5p/Raf1 axis. Biol Res. 2019;52(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang Y, Wang R, Feng L, Ma H, Fang J. LINC00460 promotes cell proliferation, migration, invasion, and epithelial‐mesenchymal transition of head and neck squamous cell carcinoma via miR‐320a/BGN Axis. Onco Targets Ther. 2021;4:2279‐2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fischer WH, Schubert D. Characterization of a novel platelet‐derived growth factor‐associated protein. J Neurochem. 1996;66(5):2213‐2216. [DOI] [PubMed] [Google Scholar]
- 31. Chou CW, Huang YM, Chang YJ, Huang CY, Hung CS. Identified the novel resistant biomarkers for taxane‐based therapy for triple‐negative breast cancer. Int J Med Sci. 2021;18(12):2521‐2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu T, Ma W, Xu H, et al. PDGF‐mediated mesenchymal transformation renders endothelial resistance to anti‐VEGF treatment in glioblastoma. Nat Commun. 2018;9(1):3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Strell C, Folkvaljon D, Holmberg E, et al. High PDGFRb expression predicts resistance to radiotherapy in DCIS within the SweDCIS randomized trial. Clin Cancer Res. 2021;27(12):3469‐3477. [DOI] [PubMed] [Google Scholar]
- 34. Hu G, Huang L, Zhong K, et al. PDGFR‐beta (+) fibroblasts deteriorate survival in human solid tumors: a meta‐analysis. Aging (Albany NY). 2021;13(10):13693‐13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nitta T, Tsutsumi M, Nitta S, et al. Fibroblasts as a source of self‐antigens for central immune tolerance. Nat Immunol. 2020;21(10):1172‐1180. [DOI] [PubMed] [Google Scholar]
- 36. Wilhelm A, Aldridge V, Haldar D, et al. CD248/endosialin critically regulates hepatic stellate cell proliferation during chronic liver injury via a PDGF‐regulated mechanism. Gut. 2016;65(7):1175‐1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gargett CE, Schwab KE, Deane JA. Endometrial stem/progenitor cells: the first 10 years. Hum Reprod Update. 2016;22(2):137‐163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Primac I, Maquoi E, Blacher S, et al. Stromal integrin alpha11 regulates PDGFR‐beta signaling and promotes breast cancer progression. J Clin Invest. 2019;129(11):4609‐4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Cytokine fluctuations and growth factor activity when LINC00460 was knocked down
Figure S2. LINC00460 does not affect the proliferation of PDAC
Table S1. sgRNAs and shRNAs used in this study.
Table S2. Sequences of primers used in this study.
Supplemental Materials and Methods
Data Availability Statement
The data that support the findings of this study are openly available in “RNA fold” at http://rna.tbi.univie.ac.at/cgi‐bin/RNAWebSuite/RNAfold.cgi and “RPIsea” at http://pridb.gdcb.iastate.edu/RPISeq/.


