Abstract
Objective:
Copy number variants (CNVs) are strongly associated with neurodevelopmental and psychotic disorders. Early onset psychotic (EOP) illnesses, where symptoms appear before 18 years of age, are thought to be more strongly influenced by genetic factors than adult-onset psychotic disorders. However, the prevalence and effect of CNVs in EOP is unclear.
Methods.
We documented the prevalence of recurrent CNVs and the functional impact of deletions and duplications genome-wide in 137 children and adolescents with EOP compared to 5,540 individuals with autism spectrum disorders (ASD) and 16,504 population controls. Specifically, we compared the frequency of 47 recurrent CNVs previously associated with neurodevelopmental and neuropsychiatric illnesses in each cohort. Next, CNV risk scores (CRS), indices reflecting the dosage sensitivity for any gene across the genome that is encapsulated in a deletion or duplication separately, were compared between groups.
Results.
Prevalence of recurrent CNVs was higher in EOP than in ASD (OR=2.30, p=0.02) and controls (OR=5.06, p=3x10−5). However, the difference between the EOP and ASD was attenuated when EOP participants with co-occurring ASD were excluded. CRS was higher in the EOP group when compared to controls for both deletions (OR=1.30, p=9x10−8) and duplications (OR=1.09, p=0.02). In contrast, the EOP and ASD cohorts did not differ in terms of CRS.
Conclusions.
Given the high frequency of recurrent CNVs in EOP and comparable CRS in the EOP and ASD cohorts, our findings suggest that all children and adolescents with a psychotic diagnosis should undergo genetic screening, as is recommended in ASD.
Introduction
Rare copy number variants (CNVs) are deletions and duplications of genomic segments(1) with high relative risk for psychotic disorders like schizophrenia(2, 3). Recurrent CNVs are relatively common (1 in 10,000 or more) and generally occur due to non-allelic homologous recombination, resulting in similar or identical mutations in unrelated individuals(4, 5), and are found in approximately 2% of cases with adult-onset idiopathic schizophrenia(6). Individuals with childhood-onset schizophrenia whose symptoms begin prior to age 13 have a nearly 3-fold increase of recurrent CNVs relative to those with adult-onset illness, suggesting a greater genetic component in the childhood form of the disorder(7, 8). However, childhood-onset schizophrenia is rare(9) and few cohorts have been genetically characterized to date(7, 8). Moreover, only half of children and adolescents with a psychiatric diagnosis that includes prominent psychotic features meet the strict criteria for schizophrenia(10) and childhood diagnoses often change over the course of development(11). Consequently, there is considerable interest in understanding the genetic underpinnings of the more inclusive early onset psychosis (EOP) categorization, which captures psychotic symptomatology in various diagnoses. EOP, defined as any psychiatric diagnosis with pronounced psychotic symptoms with onset prior to 18 years of age, is associated with lower premorbid psychosocial function, more hospitalizations, poorer cognitive functioning, and worse overall prognosis than adult-onset illness(10, 12). Yet, functional outcomes are highly variable in EOP youth(10, 11) and CNV status has been shown to influence these outcomes(13). Although genomic information could help to disentangle the clinical heterogeneity in EOP, the genetic architecture of early onset psychosis is largely unknown.
Establishing the burden of recurrent CNVs in EOP is an important first step in characterizing the genetic architecture of this extreme phenotype(3). Documenting the frequencies of recurrent CNVs, mutations that occur at high enough rates in the population to foster their individual study, would facilitate the comparison of EOP with unaffected individuals and with other neurodevelopmental disorders. However, ~90% of CNVs identified in the clinic are non-recurrent and too rare (i.e., insufficient copies) for individual association studies to be practical(14). Recently, we developed a strategy to estimate an individual’s genome-wide CNV burden by deriving a single aggregate CNV Risk Score (CRS) that reflects the probability of intolerance to haploinsufficiency of each gene encapsulated by every CNV across the genome, regardless of the mutation’s population prevalence(15-17). The CRS is a scaler value that is broadly analogous to the polygenic risk score (PRS), except that while the PRS reflects an individual’s liability for an illness based on common genetic variation, the CRS reflects the dosage sensitivity for all genes across the genome that is encapsulated in a deletion (CRSdel) or duplication (CRSdup), separately. Here, the CRS was estimated with the loss-of-function observed/expected upper bound fraction (LOEUF) score(18). The LOEUF is calculated by comparing the observed and expected number of loss-of-function (LoF) mutations for a given gene in a reference population(18). Low LOEUF scores indicate strong selection against predicted LoF variation in a gene, while high LOEUF scores indicate relatively higher tolerance to inactivation. Thus, LOEUF scores provide a method for documenting the biological ramifications of individual genes and inferring pathobiology(15, 17, 19). Our CRS measure has been successfully used to model autism spectrum disorder (ASD)(17) and general intelligence(15, 16). Applying this method to an EOP sample enables us to model genome-wide dosage sensitivity in EOP and directly compare our index with ASD and unselected control cohorts, even in the context where the individual CNVs are extremely rare.
Findings that 5-15% of children with ASD carry a deleterious genetic mutation(20) have led organizations like the American Academy of Pediatrics to recommend that individuals presenting with ASD symptoms undergo genomic screening(21, 22). These established guidelines involve routine use of chromosomal microarrays to document CNVs(23). The burden of recurrent CNVs was similar for ASD and a small childhood onset schizophrenia cohort(7), suggesting that these disorders have comparable genetic architectures and should be subject to similar genetic screening approaches. Correspondingly, if the burden of recurrent CNVs is similar among children and adolescents with the broader EOP phenotype, this would strongly support the development of guidelines for genomic screening in this population. Such an approach could help aid diagnosis, therapeutic choices, and clinical staging of individuals with EOP, many of whom do not respond to first line treatments(24). However, there are currently no genomic screening guidelines for children or adults with psychotic disorders(25).
In the present manuscript we aim to 1) establish the prevalence of recurrent CNVs in our diagnostically heterogeneous EOP cohort (n=137) and compare this prevalence with that found in 5,540 individuals with ASD and 16,504 unselected population controls; and 2) compare the CRS observed among EOP probands to the ASD and control cohorts.
Methods
EOP Samples.
Unrelated EOP participants (N=137) were referred to the Developmental Neuropsychiatry Program at Boston Children’s Hospital (BCH). Clinical diagnoses were ascertained by a board-certified child psychiatrist (JGH) specializing in EOP. Diagnoses were subsequently confirmed via medical record with a DSM-5 checklist and a consensus diagnosis was reached (see Table 1). Inclusion criteria included having a DSM-5 diagnosis for current or lifetime Axis I psychotic disorder with onset prior to age 18. Exclusion criteria were 1) substance- or medication-induced psychosis; 2) psychosis secondary to a brain infection (e.g., encephalitis); 3) psychosis due to a neurodegenerative disorder such as Wilson’s disease, Dystonia muscular deformations, Huntington’s disease, Friedrich’s ataxia, Ataxia Telangiectasia, or Parkinson’s disorder; and 4) severe neurodevelopmental disorder or other impairment impacting ability to describe symptoms or provide other information required for this study. All EOP participants or guardians provided written informed consent (or assent for participants <18 years) on forms approved by the BCH IRB as part of the Manton Center for Orphan Disease Research. After providing consent/assent, each participant provided blood samples.
Table 1:
Diagnostic and Demographic Breakdown of EOP Cohort
| Characteristic | Full EOP Sample (N=137) |
% | EOP without ASD or ID (n=89) |
% | EOP without Schizophrenia (n=99) |
% | EOP Onset <13 Years (n=101) |
% |
|---|---|---|---|---|---|---|---|---|
| Sex | ||||||||
| Male | 88 | 64.2 | 50 | 56.2 | 63 | 63.6 | 65 | 64.4 |
| Female | 49 | 35.8 | 39 | 43.8 | 36 | 36.4 | 36 | 35.6 |
| Race | ||||||||
| Asian or Pacific Islander | 3 | 2.2 | 3 | 3.4 | 2 | 2.0 | 2 | 2.0 |
| Black/African American | 21 | 15.3 | 15 | 16.9 | 16 | 16.2 | 12 | 11.9 |
| Two or more races | 2 | 1.5 | 2 | 2.2 | 0 | 0.0 | 2 | 2.0 |
| White | 90 | 65.7 | 56 | 62.9 | 65 | 65.7 | 67 | 66.3 |
| Unknown/Not available | 21 | 15.3 | 13 | 14.6 | 16 | 16.2 | 18 | 17.8 |
| Ethnicity | ||||||||
| Hispanic/Latino | 22 | 16.1 | 15 | 16.9 | 16 | 16.2 | 15 | 14.9 |
| Not Hispanic/Latino | 91 | 66.4 | 62 | 69.7 | 66 | 66.7 | 67 | 66.3 |
| Unknown/Not available | 24 | 17.5 | 12 | 13.5 | 17 | 17.2 | 19 | 18.8 |
| Age at Psychosis Symptom Onset | ||||||||
| <8 years | 38 | 27.7 | 23 | 25.8 | 27 | 27.3 | 38 | 37.6 |
| 8-12 years old | 63 | 46.0 | 43 | 48.3 | 43 | 43.4 | 63 | 62.4 |
| 13-18 years old | 36 | 26.3 | 23 | 25.8 | 29 | 29.3 | -- | 0.0 |
| Primary Psychosis Spectrum Disorder | ||||||||
| Schizophrenia | 38 | 27.7 | 21 | 23.6 | -- | 0.0 | 31 | 30.7 |
| Affective Psychosis | ||||||||
| Schizoaffective Disorder | 11 | 8.0 | 6 | 6.7 | 11 | 11.1 | 8 | 7.9 |
| (Bipolar Type) | ||||||||
| Schizoaffective Disorder | 8 | 5.8 | 8 | 9.0 | 8 | 8.1 | 5 | 5.0 |
| (Depressed Type) | ||||||||
| MDD Psychotic Features | 17 | 12.4 | 15 | 16.9 | 17 | 17.2 | 10 | 9.9 |
| BP Psychotic Features | 17 | 12.4 | 13 | 14.6 | 17 | 17.2 | 12 | 11.9 |
| Schizophreniform | 2 | 1.5 | 0 | 0.0 | 2 | 2.0 | 2 | 2.0 |
| Disorder | ||||||||
| Brief Psychotic Disorder | 1 | 0.7 | 0 | 0.0 | 1 | 1.0 | 0 | 0.0 |
| Other Specified or | 43 | 31.3 | 26 | 29.2 | 43 | 43.4 | 33 | 32.7 |
| Unspecified | ||||||||
| Schizophrenia Spectrum | ||||||||
| and Other Psychotic | ||||||||
| Disorder | ||||||||
| Co-Occurring Diagnoses | ||||||||
| Autism Spectrum | 47 | 34.2 | ||||||
| Disorders (ASD) | ||||||||
| Intellectual Disability | 17 | 12.4 | -- | 0.0 | 10 | 10.1 | 11 | 10.9 |
| History of Seizures | 24 | 17.5 | 12 | 13.5 | 15 | 15.2 | 19 | 18.8 |
ASD and Unselected Populations.
We compared the EOP participants to two pooled ASD cohorts: 2,585 children from the Simons Simplex Collection (SSC)(26) and 3,171 probands from the MSSNG database(27). We also compared EOP participants to individuals from three pooled unselected community-based cohorts: IMAGEN (N=1,802)(28), Generation Scotland (GS)(29) (N=14,160), and the Lothian Birth Cohort(30) (LBC) (N=554) (see Figure 1). Studies for each cohort were reviewed by local institutional review boards and are described elsewhere(26-30).
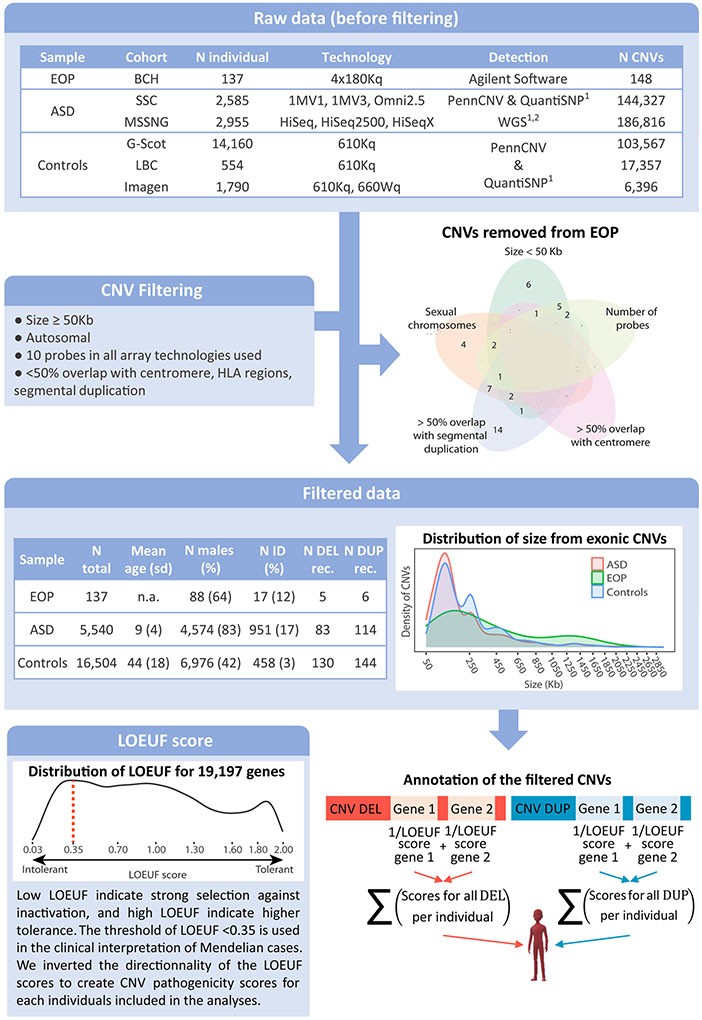
Figure 1: Methodological pipeline for CNV filtering and annotation.
The first table describes CNVs identified in the EOP, ASD and unselected cohorts before filtering. The Venn diagram represents distribution of the EOP CNVs discarded in function of the filtering criteria to which they belong: size is >50 Kb; fewer than 10 probes in at least one of the technologies used; overlap with a centromere or a segmental duplication (<50%); or positioned on a sexual chromosome. The second table indicates the prevalence of filtered CNVs, demographic and comorbidity data for EOP, ASD and controls samples. The first density plot presents the distribution of the size of the genome-wide CNVs included in the analyses across the different samples (EOP in green, ASD in red, unselected population in blue). The CNV sizes on the x axis are represented with a square root transformation. The final density plot represents the distribution of LOEUF score across 19,197 coding genes. A LOEUF score ≤ 0.35 is the defined clinical threshold for intolerant genes. For CNV annotation, the coding gene totally encompassed in deletions and duplications were identified and the LOEUF score of each gene was attributed. For each individual, the number of gene encompassed and the 1/LOEUF scores for deletions and duplications were summed separately. CNV: copy number variant; N: number; EOP: early-onset psychosis; BCH: Boston children hospital; ASD: autism spectrum disorders; SSC: Simons Simplex Collection; Controls: unselected populations; G-Scot: Generation Scotland; LBC: Lothian birth cohort; ID: intellectual disability; N DEL/DUP rec: Number of recurrent deletions/duplications previously associated with neurodevelopmental or neuropsychiatric disorders; LOEUF: loss-of-function observed/expected upper bound fraction.
Genotyping and CNV Calling.
For the EOP sample, genomic DNA from blood was extracted using standard protocols. Dye-swap array-CGH experiments were performed according to the experimental procedures described by Agilent Technologies (Santa Clara, CA) using standard 4×180K Surescan arrays and analyzed with the Agilent Cytogenomics Software. Probe sequences and locations are based on Genome Reference Consortium build 37 (GRCh37/hg19). Three criteria were used to determine the presence of a CNV: 1) at least 7 consecutive probes in the same direction; 2) 1.5-fold average difference between test and reference DNA; and 3) CNV not present in the within-slide control DNA sample. We applied a previously published pipeline to data from the ASD and control cohorts(15-17). To harmonize the samples, CNVs were filtered by discarding: CNVs <50kb; CNVs that appeared on sex chromosomes; CNVs with >50% overlap with segmental duplication or centromere; and CNVs with <10 probes across all detection technologies used in all included cohorts (see Figure 1).
Recurrent CNV Analyses.
We identified 47 loci and genes previously associated with neurodevelopmental or neuropsychiatric disorders (see Supplementary Table 1) to document the frequency of recurrent CNVs in EOP. These loci were defined by >40% overlap with a specific deletion or duplication or if the genes were disrupted by the CNVs(31, 32). Recurrent CNVs share a common size, similar breakpoints, and recur in multiple individuals in a population(3, 4). Recurrent CNVs generally occur due to non-allelic homologous recombination (NAHR), which is typically mediated by low-copy repeats (LCRs), resulting in recombination hotspots, gene conversion and apparent minimal efficient processing segments(5). In contrast, nonrecurrent CNVs are defined as structural variants with dissimilar endpoints or junctions that do not coincide with LCRs but tend to occur in the vicinity of regions that are rich in LCRs, resulting in complex regional genomic architecture. LCRs do not mediate but may stimulate nonrecurrent events(5). Although, non-recurrent CNVs are of different sizes in each patient, they can share a small region of overlap whose change in copy number may result in shared clinical features among different patients(4). Yet, given differences in breakpoints and genes effected, nonrecurrent CNVs, such as private CNVs, are almost impossible to study individually. However, the overall impact of non-recurrent CNVs can be indexed using genome-wide scores like the CRS, as described below. To test for differences in the prevalence of recurrent CNVs between EOP, ASD, and controls, two-sided Fisher’s exact tests, computed using the fisher.test() function in R, were employed. The Benjamin-Hochberg correction for false discovery rate (FDR) were used to adjust for multiple comparisons.
CRS: Genome-Wide Dosage Sensitivity Analyses.
The CNV Risk Score (CRS) reflects the probability of intolerance to haploinsufficiency of each gene in the genome that is encapsulated in every CNV identified in an individual, regardless of the population prevalence of the mutation(15-17). In this study, the CRS was calculated as the sum of 1/LOEUF for deletions (CRSdel) and duplications (CRSdup) separately using our previously published annotation pipeline (see Figure 1). Briefly, each coding gene with all isoforms fully encompassed in filtered CNVs was identified using ENSEMBL map (hg19)(33) and was annotated using the inverse LOEUF (1/LOEUF) score (gnomAD version 2.1.1)(18), which is available for 19,197 genes and ranges from 0.5 (gene tolerant to haploinsufficiency) to 33.3 (gene intolerant to haploinsufficiency). A score of 0 was assigned to individuals with no coding genes encompassed in any CNV. We tested the genome-wide CNV burden with logistic regression models:
β0, β1, β2 and β3 are the vectors of coefficients for fixed effects. The logistic regression models were computed using the glm() function in R.
Sensitivity Analyses.
We performed sensitivity analyses to investigate the robustness of our main results. Specifically, we ran a series of analyses after excluding EOP participants (1) with co-occurring ASD, (2) with co-occurring Intellectual Disability (ID), (3) with a diagnosis of schizophrenia, or (4) with illness onset prior to 13 years of age. Each of these sensitivity analyses used statistical models identical to those described above, with smaller sample sizes (see Table 1) and were designed to refine our appreciation of the CNV burden in EOP relative to the ASD and unselected cohorts.
Results
EOP Cohort.
A total of 137 EOP patients (88 (64.2%) male) were included in this study (see Table 1). Mean age of psychosis symptom onset was 9.8 years (range 4-17 years old), with 101 (72.3%) patients having psychosis onset before age 13 years. Thirty-eight (28%) of individuals with EOP had co-occurring ASD, 17 (12%) had ID, and 7 (5%) had both ASD and ID. Thirty-eight (28%) individuals met criteria DSM-V for schizophrenia. Sixty-nine CNVs meeting quality control criteria were identified in 55 individuals from the EOP cohort (see Figure 1).
Prevalence of Recurrent CNVs in EOP, ASD and Unselected Population Cohorts.
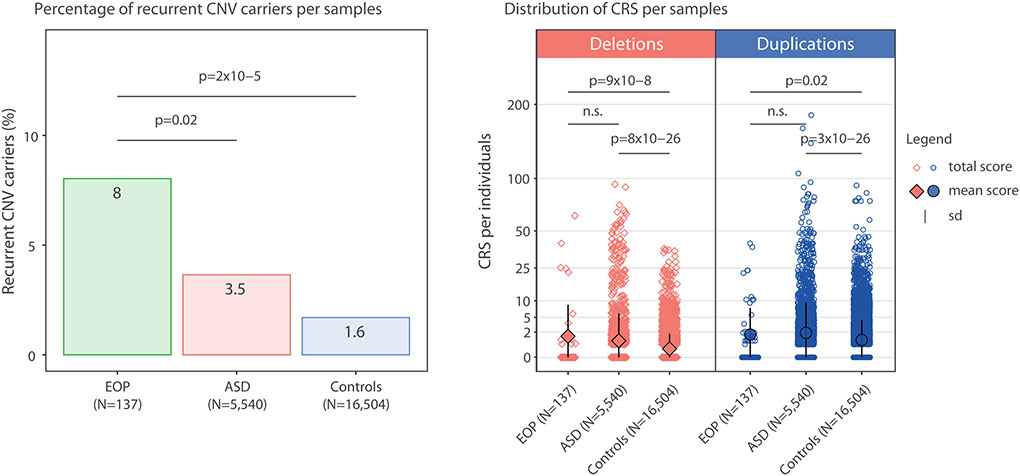
When focusing on recurrent CNVs previously associated with neurodevelopmental or neuropsychiatric disorders (see Supplementary Table 1), we found 11 (8.0% of sample) recurrent CNV carriers in the EOP cohort (see Figure 2A, Table 2 and Supplementary Table 2). In contrast, 193 (3.5%) individuals from the ASD cohort and 273 (1.6%) individuals from the unselected population were recurrent CNV carriers. Thus, the prevalence of recurrent CNVs in children and adolescents with EOP was double that observed in ASD (OR=2.42, 95% CI=1.16-4.57, p=0.02) and five times the rate in unselected populations (OR=5.19, 95% CI=2.50-9.75, p=2 × 10−5) (see Table 2).
Figure 2: CNV Burden in EOP, ASD and Unselected Populations.
A. Prevalence of Recurrent CNVs. Rates of disease-related recurrent CNVs in individuals with early onset psychosis (EOP), autism spectrum disorder (ASD) probands and controls from the unselected population. B. Genome-Wide Gene Dossage Effects (CRS). Scores for deletions and duplications per individual are represented in red diamonds and blue circles, respectively. Individuals without a CNV or with a non-coding CNV have a score of 0. Coding CNVs have scores ranging from 0.5 to approximately 180. Y axis: CRS value (root squared of the sum of 1/LOEUF of all genes encompassed in CNVs identified in each individual). The largest diamonds/circles and black bars represent the mean and standard deviation of each group. EOP: early-onset psychosis; ASD: autism spectrum disorder; Controls: unselected population; DEL: deletions; DUP: duplications; n.s.: non-significant; sd: standard deviation.
Table 2:
Enrichment of recurrent CNVs in EOP relative to ASD and Unselected Control Cohorts
| ASD Contrast | ASD carriers | EOP carriers | OR | 95%CI | P-value |
|---|---|---|---|---|---|
| EOP (N=137) | 193 | 11 | 2.42 | 1.16-4.57 | 0.02 |
| EOP without ASD (N=90) | 193 | 6 | 1.79 | 0.63-4.11 | 0.16 |
| EOP without ID (N=120) | 193 | 7 | 1.72 | 0.67-3.72 | 0.20 |
| EOP without schizophrenia (N=99) | 193 | 8 | 2.43 | 1.01-5.10 | 0.02 |
| EOP < 13 years old (N=101) | 193 | 6 | 1.75 | 0.62-4.02 | 0.17 |
| Control Contrast | Control carriers | EOP carriers | OR | 95%CI | P-value |
| EOP (N=137) | 273 | 11 | 5.19 | 2.50-9.75 | 2 × 10−5 |
| EOP without ASD (N=90) | 273 | 6 | 3.83 | 1.36-8.77 | 6 × 10−3 |
| EOP without ID (N=120) | 273 | 7 | 3.68 | 1.43-7.94 | 4 × 10−3 |
| EOP without schizophrenia (N=99) | 273 | 8 | 5.22 | 2.17-10.88 | 3 × 10−4 |
| EOP < 13 years old (N=101) | 273 | 6 | 3.75 | 1.33-8.58 | 7 × 10−3 |
Legend: Odds ratios are computed using Fisher’s exact test. Significant p-values are in bold (≤ 0.05). Sensitivity analysis involved excluding individuals in the EOP sample: 1) with co-occurring ASD; 2) with co-occurring ID; 3) with a diagnosis of schizophrenia; or 5) with psychosis onset prior to age 13. EOP: early-onset psychosis; ASD: autism spectrum disorder (N=5,540); Controls: unselected population (N=16,504); ID: intellectual disability; N: number of individuals; OR: Odds ratio; 95% CI: 95% confidence intervals.
Three recurrent CNVs were individually enriched in EOP participants relative to unselected controls after FDR correction (see Supplementary Table 3): 1q21.1 duplication (pFDR=6x10−4, see Supplementary Table 4 for a description of 1q21.1 patients), 16p13.11 deletion (pFDR=0.01) and 22q11.2 proximal deletion (pFDR=0.02). These same loci were enriched in the EOP cohort relative to the ASD cohort, but none survived FDR correction.
CRS in EOP, ASD and Unselected Population Cohorts.
CRS were higher in EOP participants relative to the unselected cohort for both deletions (CRSdel 1.39 vs. 0.23, OR=1.30, 95% CI=1.26-1.35, p=9 × 10−8) and duplications (CRSdup 1.63 vs. 0.94, OR=1.09, 95% CI=1.06-1.12, p=0.02) (see Figure 2B and Table 3). Similar results were found when comparing ASD participants to the unselected cohort (CRSdel 0.86 vs. 0.23, OR=1.24, 95% CI=1.22-1.26, p=8 × 10−26; CRSdup 1.88 vs. 0.94, OR=1.12, 95% CI=1.11-1.13, p= 3 × 10−26). However, CRS did not differ significantly between the EOP and ASD cohorts (CRSdel 1.39 vs. 0.86, OR=1.03, 95% CI=1.01-1.06, p=0.33; CRSdup 1.63 vs. 1.88 OR=0.98; 95% CI=0.96-1.01 p=0.61). No sex differences (49 females, 88 males) were observed for the CRS for deletions (OR=1.01, 95% CI= 0.96-109, p=0.70) or duplications (OR=0.97, 95% CI= 0.91-103, p=0.34).
Table 3:
Genome-wide CRS Differences in EOP relative to ASD and Unselected Control Cohorts
| Deletions | Duplications | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ASD Contrast | ASD CRS |
EOP CRS |
OR | 95%CI | p-value | ASD CRS |
EOP CRS |
OR | 95%CI | p-value |
| EOP (N=137) | 0.86 | 1.39 | 1.03 | 1.01-1.06 | 0.33 | 1.88 | 1.63 | 0.98 | 0.96-1.01 | 0.61 |
| EOP without ASD (N=90) | 0.86 | 1.00 | 1.01 | 0.97-1.04 | 0.89 | 1.88 | 1.42 | 0.96 | 0.93-1.00 | 0.48 |
| EOP without ID (N=120) | 0.86 | 0.53 | 0.94 | 0.89-0.99 | 0.42 | 1.88 | 1.63 | 0.98 | 0.95-1.01 | 0.64 |
| EOP without schizophrenia (N=99) | 0.86 | 1.59 | 1.04 | 1.02-1.07 | 0.24 | 1.88 | 1.76 | 0.99 | 0.96-1.02 | 0.78 |
| EOP < 13 years old (N=101) | 0.86 | 1.25 | 1.03 | 1.00-1.06 | 0.56 | 1.88 | 1.34 | 0.95 | 0.92-0.99 | 0.42 |
| Control Contrast | Control CRS |
EOP CRS |
OR | 95%CI | p-value | Control CRS |
EOP CRS |
OR | 95%CI | p-value |
| EOP (N=137) | 0.23 | 1.39 | 1.30 | 1.26-1.35 | 9x10−8 | 0.94 | 1.63 | 1.09 | 1.06-1.12 | 0.02 |
| EOP without ASD (N=90) | 0.23 | 1.00 | 1.24 | 1.19-1.30 | 3x10−4 | 0.94 | 1.42 | 1.07 | 1.04-1.11 | 0.17 |
| EOP without ID (N=120) | 0.23 | 0.53 | 1.19 | 1.12-1.27 | 0.04 | 0.94 | 1.63 | 1.09 | 1.06-1.12 | 0.03 |
| EOP without schizophrenia (N=99) | 0.23 | 1.59 | 1.31 | 1.27-1.36 | 3 × 10−7 | 0.94 | 1.76 | 1.10 | 1.07-1.13 | 0.02 |
| EOP < 13 years old (N=101) | 0.23 | 1.25 | 1.34 | 1.28-1.39 | 4 × 10−7 | 0.94 | 1.34 | 1.06 | 1.03-1.10 | 0.25 |
Legend: Effect of gene dosage on EOP risk. Odds ratios are computed using logistic regressions including the CNV risk score (CRS; sum of 1/LOEUF score) for genes totally encompassed in deletions and duplications as the two main explanatory variables. OR represents the mean risk conferred by a deletion or a duplication including 1 intolerant gene (a LOEUF ≤ 0.35). All models were adjusted for sex. CRS presented in the table are the mean score per samples. Sensitivity analysis involved excluding individuals in the EOP sample: 1) with co-occurring ASD; 2) with co-occurring ID; 3) with a diagnosis of schizophrenia; or 5) with psychosis onset prior to age 13. EOP: early-onset psychosis; ASD: autism spectrum disorder cohort (n=5,540); ID: intellectual disability; Controls: unselected population (n=16,50); LOEUF: Loss-of-function observed/expected upper fraction; OR: Odds ratio; 95%CI: 95% Confidence interval
Sensitivity Analyses: Prevalence of Recurrent CNVs in EOP Subgroups.
As can be seen in Table 2, the frequencies of recurrent CNVs in the various EOP subgroups (excluding ASD, excluding ID, and excluding schizophrenia) did not differ from those observed in the ASD cohort. However, when excluding EOP participants with later symptom onset, the subgroup had statistically fewer recurrent CNVs than expected when compared with the ASD cohort (p=0.02, see Table 2). In contrast, each EOP subgroup had a higher prevalence of recurrent CNVs when compared to the unselected control cohort.
Sensitivity Analyses: CRS in EOP Subgroups.
As in the full sample, we observed no significant differences in dosage sensitivity between any EOP subgroup and the ASD cohort for deletions or duplications (see Table 3). In contrast, the CRS index was elevated for deletions in every EOP subgroup relative to the unselected control cohort. For duplications, the CRS was significantly increased in the EOP subgroups when excluding individuals with ID (p=0.03) and those with schizophrenia (p=0.02; see Table 3).
Discussion
Prevalence of recurrent CNVs in children and adolescents with various early onset psychosis (EOP) diagnoses was far higher than in unselected population controls. In contrast, the EOP sample showed a similar prevalence of recurrent CNVs to a large ASD cohort. Prevalence of recurrent CNVs in our EOP cohort was also in line with previous reports of individuals with the more restrictive childhood onset schizophrenia diagnosis(7), even when individuals with a schizophrenia diagnosis were excluded from our cohort. Initially, we selected and analyzed recurrent CNVs that were previously associated with neurodevelopmental(34) and psychotic(2) illnesses(6) (see Supplementary Table 1). However, these recurrent CNVs represent only a fraction of all CNVs observed in the population. Thus, we also tested for group differences in CRS, an index of genome-wide dosage sensitivity for deletions and duplications(15). We found higher CRSdel and CRSdup in the EOP sample relative to the control population. In contrast, CRS was comparable in the EOP and ASD cohorts. This general pattern of results held even when individuals in the EOP sample with co-occurring ASD or co-occurring ID were excluded, suggesting that these co-occurring disorders were not completely responsible for the observed CNV burden. Our results indicate that EOP is associated with a substantial CNV burden, strongly suggesting that systematic genetic screening in EOP is clinically warranted.
Given the success of genetic screening in ASD(35), our findings suggest that all children and adolescents with a psychotic diagnosis could substantially benefit from chromosomal microarray testing, with the potential for further testing contingent upon family history and/or clinical features. Universal genetic screening(25) could help disentangle the clinical heterogeneity among EOP youth(11), potentially leading to specific treatment regiments. As detailed in Moreno-De-Luca et al.(36), genetic diagnoses allow clinicians to communicate more effectively with patients and families, and facilitate genetic counseling. Genetic information could also connect families to additional resources and networks, such as other families with the same CNV. Forming cohorts of patients with the same CNV has yielded valuable information about comorbidities, the range of possible phenotypes, and disease progression in other areas of medicine(37-39). Furthermore, children and adolescents with EOP who carry recurrent CNVs associated with serious non-psychiatric medical conditions (e.g., cardiovascular abnormalities in 22q11.2 deletion syndrome or the high incidence of hypotonia and epilepsy in 15q11.2 duplication carriers) could be more carefully monitored. Finally, information derived from genetic screening is often invaluable to families of children with ASD(35), helping parents appreciate the biological nature of the illness. Similar genetic information would no doubt be well received by EOP families too. Overall, genetic screening in EOP has the potential to bring us one step closer to true precision medicine in pediatric psychiatry.
Among the recurrent CNVs previously associated with neurodevelopmental and neuropsychic illness, we documented an enrichment for three mutations: 22q11.2 proximal deletion (due to a lack of carriers in the population control), 16p13.11 deletion and 1q21.1 duplication. Each of these CNVs was reported in the prior childhood onset schizophrenia study(7) and in ASD cohorts(17). These specific CNVs could be particularly informative to the pathobiology of psychosis and neurodevelopment, particularly since these mutations were commonly observed in a large sample of individuals with idiopathic adult-onset schizophrenia(2). Since the number of CNVs observed is directly related to sample size, it is likely that with a larger EOP cohort, evidence for additional recurrent CNVs will emerge. Thus, it is difficult to speculate about the genetic architecture of EOP and whether child and adolescent psychosis is more strongly influenced by an enumerable set of rare mutations of large effect or by countless common mutations of small effect (e.g., polygenic model), currently the favored model for idiopathic adult-onset psychosis. Further investigations with much larger EOP samples are needed.
Since recurrent CNVs reflect only a fraction of deletions and duplications observed in neurodevelopmental and neuropsychiatric illnesses, we also used the CRS to index dosage sensitivity across the genome. We demonstrated a higher overall burden of genes intolerant to mutation in EOP compared to unselected samples. Moreover, larger effect sizes were observed for deletions than duplications. Interestingly, effect-sizes observed for EOP were in the same range as those found when comparing ASD to unselected populations(17), suggesting a major contribution of haploinsufficiency in both EOP and ASD. Additional work is needed to document the relative strength of our CRS score in EOP compared to adult-onset psychotic disorders, as well as to other neurodevelopmental disorders.
A series of sensitivity analyses determined the impact association of co-occurring neurodevelopmental disorders, psychosis diagnosis and symptom onset with CNV burden. These analyses documented few differences between the EOP subgroups and the ASD cohort in terms of the frequencies of recurrent CNVs or with regards to genome-wide dosage sensitivity. In contrast, regardless of the portion of the EOP sample studied, we found that individuals with early onset psychosis had increased prevalence of recurrent CNVs and increased CRS for deletions relative to the unselected control cohort, with significant findings for the CRS for duplications for specific subgroups. The sensitivity analyses excluded co-occurring ASD or co-occurring ID demonstrated that the CNV burden observed in EOP is not simply due to the presence of other neurodevelopmental disorders in the EOP sample. To assess psychiatric diagnostic specificity, one sensitivity analysis excluded EOP participants with a schizophrenia diagnosis. The prevalence and CRS for this subgroup of individuals with affective and other psychoses remained relatively unchanged, suggesting that the specific diagnosis of schizophrenia did not drive the CNV burden observed in the full EOP cohort. Finally, to compare our findings more directly with those reported in childhood onset schizophrenia(7), we excluded EOP individuals whose psychosis symptoms manifest at 13 years or older. However, excluding individuals with later symptom onset did not substantively change the pattern of results. Given that these sensitivity analyses included relatively small sample sizes, we have limited power to detect group differences and larger samples are needed to fully address these issues.
Our study has several strengths, such as the use of a unique sample of EOP patients with a range of comorbidities, which is highly representative of children and adolescents with psychotic disorders. Moreover, we were able to compare CNV burden in EOP to ASD and general population controls. However, our EOP sample is small, which is to be anticipated given the rarity of psychosis in children and adolescents. Future studies with larger samples might reveal additional recurrent CNVs or stronger effects of genome-wide duplications, similar to findings in larger ASD or schizophrenia samples(2, 17). Nonetheless, the high frequency of CNVs in our EOP cohort suggests that routine screening for CNVs should be made available to EOP patients and could have important implications for genetic counseling and patient management. These relatively high penetrance risk alleles are also promising targets for biological research aimed at developing animal and cellular models to identify novel disease mechanisms and drug targets for psychotic disorders.
Supplementary Material
Acknowledgments:
We gratefully acknowledge the patients and families who participated in this study.
Disclosures:
Author JGH holds equity in and is founding head of the scientific advisory board for Mightier/Neuromotion Labs, a company making emotional regulation training video games and has received consulting income from Alkermes, Inc., Neurocrine, and Sunovion pharmaceutical companies. The remaining authors have no conflicts. This work was funded by the Tommy Fuss Center for Neuropsychiatric Research, the Boston Children’s Hospital Intellectual and Developmental Disabilities Research Center Molecular Genetics Core Facility supported by P50HD105351 from the NIH Eunice Kennedy Shriver National Institute of Child Health and Human Development, and NIH/NIMH U01 MH119690, The Stanley Center of the Broad Institute of MIT and Harvard, the Jonathan and Robin Klein and Anne and Paul Marcus families.
References
- 1.Girirajan S, Rosenfeld JA, Coe BP, Parikh S, Friedman N, Goldstein A, Filipink RA, McConnell JS, Angle B, Meschino WS, Nezarati MM, Asamoah A, Jackson KE, Gowans GC, Martin JA, Carmany EP, Stockton DW, Schnur RE, Penney LS, Martin DM, Raskin S, Leppig K, Thiese H, Smith R, Aberg E, Niyazov DM, Escobar LF, El-Khechen D, Johnson KD, Lebel RR, Siefkas K, Ball S, Shur N, McGuire M, Brasington CK, Spence JE, Martin LS, Clericuzio C, Ballif BC, Shaffer LG, Eichler EE. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. The New England journal of medicine. 2012;367:1321–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marshall CR, Howrigan DP, Merico D, Thiruvahindrapuram B, Wu W, Greer DS, Antaki D, Shetty A, Holmans PA, Pinto D, Gujral M, Brandler WM, Malhotra D, Wang Z, Fajarado KVF, Maile MS, Ripke S, Agartz I, Albus M, Alexander M, Amin F, Atkins J, Bacanu SA, Belliveau RA, Bergen SE, Bertalan M, Bevilacqua E, Bigdeli TB, Black DW, Bruggeman R, Buccola NG, Buckner RL, Bulik-Sullivan B, Byerley W, Cahn W, Cai G, Cairns MJ, Campion D, Cantor RM, Carr VJ, Carrera N, Catts SV, Chambert KD, Cheng W, Cloninger CR, Cohen D, Cormican P, Craddock N, Crespo-Facorro B, Crowley JJ, Curtis D, Davidson M, Davis KL, Degenhardt F, Del Favero J, DeLisi LE, Dikeos D, Dinan T, Djurovic S, Donohoe G, Drapeau E, Duan J, Dudbridge F, Eichhammer P, Eriksson J, Escott-Price V, Essioux L, Fanous AH, Farh KH, Farrell MS, Frank J, Franke L, Freedman R, Freimer NB, Friedman JI, Forstner AJ, Fromer M, Genovese G, Georgieva L, Gershon ES, Giegling I, Giusti-Rodríguez P, Godard S, Goldstein JI, Gratten J, de Haan L, Hamshere ML, Hansen M, Hansen T, Haroutunian V, Hartmann AM, Henskens FA, Herms S, Hirschhorn JN, Hoffmann P, Hofman A, Huang H, Ikeda M, Joa I, Kähler AK, Kahn RS, Kalaydjieva L, Karjalainen J, Kavanagh D, Keller MC, Kelly BJ, Kennedy JL, Kim Y, Knowles JA, Konte B, Laurent C, Lee P, Lee SH, Legge SE, Lerer B, Levy DL, Liang KY, Lieberman J, Lönnqvist J, Loughland CM, Magnusson PKE, Maher BS, Maier W, Mallet J, Mattheisen M, Mattingsdal M, McCarley RW, McDonald C, McIntosh AM, Meier S, Meijer CJ, Melle I, Mesholam-Gately RI, Metspalu A, Michie PT, Milani L, Milanova V, Mokrab Y, Morris DW, Müller-Myhsok B, Murphy KC, Murray RM, Myin-Germeys I, Nenadic I, Nertney DA, Nestadt G, Nicodemus KK, Nisenbaum L, Nordin A, O'Callaghan E, O'Dushlaine C, Oh SY, Olincy A, Olsen L, O'Neill FA, Van Os J, Pantelis C, Papadimitriou GN, Parkhomenko E, Pato MT, Paunio T, Perkins DO, Pers TH, Pietiläinen O, Pimm J, Pocklington AJ, Powell J, Price A, Pulver AE, Purcell SM, Quested D, Rasmussen HB, Reichenberg A, Reimers MA, Richards AL, Roffman JL, Roussos P, Ruderfer DM, Salomaa V, Sanders AR, Savitz A, Schall U, Schulze TG, Schwab SG, Scolnick EM, Scott RJ, Seidman LJ, Shi J, Silverman JM, Smoller JW, Söderman E, Spencer CCA, Stahl EA, Strengman E, Strohmaier J, Stroup TS, Suvisaari J, Svrakic DM, Szatkiewicz JP, Thirumalai S, Tooney PA, Veijola J, Visscher PM, Waddington J, Walsh D, Webb BT, Weiser M, Wildenauer DB, Williams NM, Williams S, Witt SH, Wolen AR, Wormley BK, Wray NR, Wu JQ, Zai CC, Adolfsson R, Andreassen OA, Blackwood DHR, Bramon E, Buxbaum JD, Cichon S, Collier DA, Corvin A, Daly MJ, Darvasi A, Domenici E, Esko T, Gejman PV, Gill M, Gurling H, Hultman CM, Iwata N, Jablensky AV, Jönsson EG, Kendler KS, Kirov G, Knight J, Levinson DF, Li QS, McCarroll SA, McQuillin A, Moran JL, Mowry BJ, Nöthen MM, Ophoff RA, Owen MJ, Palotie A, Pato CN, Petryshen TL, Posthuma D, Rietschel M, Riley BP, Rujescu D, Sklar P, St Clair D, Walters JTR, Werge T, Sullivan PF, O'Donovan MC, Scherer SW, Neale BM, Sebat J, Consortium PEI, Consortium CaSWGotPG. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nature genetics. 2017;49:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148:1223–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu W, Zhang F, Lupski JR. Mechanisms for human genomic rearrangements. Pathogenetics. 2008;1:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergen SE, Ploner A, Howrigan D, O'Donovan MC, Smoller JW, Sullivan PF, Sebat J, Neale B, Kendler KS, Consortium CAGatSWGotPG. Joint Contributions of Rare Copy Number Variants and Common SNPs to Risk for Schizophrenia. The American journal of psychiatry. 2019;176:29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahn K, Gotay N, Andersen TM, Anvari AA, Gochman P, Lee Y, Sanders S, Guha S, Darvasi A, Glessner JT, Hakonarson H, Lencz T, State MW, Shugart YY, Rapoport JL. High rate of disease-related copy number variations in childhood onset schizophrenia. Mol Psychiatry. 2014;19:568–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez A, Drozd MM, Thümmler S, Dor E, Capovilla M, Askenazy F, Bardoni B. Childhood-Onset Schizophrenia: A Systematic Overview of Its Genetic Heterogeneity From Classical Studies to the Genomic Era. Frontiers in Genetics. 2019;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicolson R, Rapoport JL. Childhood-onset schizophrenia: rare but worth studying. Biological psychiatry. 1999;46:1418–1428. [DOI] [PubMed] [Google Scholar]
- 10.Stentebjerg-Olesen M, Pagsberg AK, Fink-Jensen A, Correll CU, Jeppesen P. Clinical Characteristics and Predictors of Outcome of Schizophrenia-Spectrum Psychosis in Children and Adolescents: A Systematic Review. Journal of child and adolescent psychopharmacology. 2016;26:410–427. [DOI] [PubMed] [Google Scholar]
- 11.Castro-Fornieles J, Baeza I, de la Serna E, Gonzalez-Pinto A, Parellada M, Graell M, Moreno D, Otero S, Arango C. Two-year diagnostic stability in early-onset first-episode psychosis. J Child Psychol Psychiatry. 2011;52:1089–1098. [DOI] [PubMed] [Google Scholar]
- 12.Patel PK, Leathem LD, Currin DL, Karlsgodt KH. Adolescent Neurodevelopment and Vulnerability to Psychosis. Biol Psychiatry. 2021;89:184–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hubbard L, Rees E, Morris DW, Lynham AJ, Richards AL, Pardinas AF, Legge SE, Harold D, Zammit S, Corvin AC, Gill MG, Hall J, Holmans P, O'Donovan MC, Owen MJ, Donohoe G, Kirov G, Pocklington A, Walters JTR. Rare Copy Number Variants Are Associated With Poorer Cognition in Schizophrenia. Biol Psychiatry. 2021;90:28–34. [DOI] [PubMed] [Google Scholar]
- 14.Zarrei M, Burton CL, Engchuan W, Young EJ, Higginbotham EJ, MacDonald JR, Trost B, Chan AJS, Walker S, Lamoureux S, Heung T, Mojarad BA, Kellam B, Paton T, Faheem M, Miron K, Lu C, Wang T, Samler K, Wang X, Costain G, Hoang N, Pellecchia G, Wei J, Patel RV, Thiruvahindrapuram B, Roifman M, Merico D, Goodale T, Drmic I, Speevak M, Howe JL, Yuen RKC, Buchanan JA, Vorstman JAS, Marshall CR, Wintle RF, Rosenberg DR, Hanna GL, Woodbury-Smith M, Cytrynbaum C, Zwaigenbaum L, Elsabbagh M, Flanagan J, Fernandez BA, Carter MT, Szatmari P, Roberts W, Lerch J, Liu X, Nicolson R, Georgiades S, Weksberg R, Arnold PD, Bassett AS, Crosbie J, Schachar R, Stavropoulos DJ, Anagnostou E, Scherer SW. A large data resource of genomic copy number variation across neurodevelopmental disorders. NPJ Genom Med. 2019;4:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huguet G, Schramm C, Douard E, Tamer P, Main A, Monin P, England J, Jizi K, Renne T, Poirier M, Nowak S, Martin CO, Younis N, Knoth IS, Jean-Louis M, Saci Z, Auger M, Tihy F, Mathonnet G, Maftei C, Léveillé F, Porteous D, Davies G, Redmond P, Harris SE, Hill WD, Lemyre E, Schumann G, Bourgeron T, Pausova Z, Paus T, Karama S, Lippe S, Deary IJ, Almasy L, Labbe A, Glahn D, Greenwood CMT, Jacquemont S. Genome-wide analysis of gene dosage in 24,092 individuals estimates that 10,000 genes modulate cognitive ability. Molecular psychiatry. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huguet G, Schramm C, Douard E, Jiang L, Labbe A, Tihy F, Mathonnet G, Nizard S, Lemyre E, Mathieu A, Poline J-B, Loth E, Toro R, Schumann G, Conrod P, Pausova Z, Greenwood C, Paus T, Bourgeron T, Jacquemont S, Consortium ftI. Measuring and Estimating the Effect Sizes of Copy Number Variants on General Intelligence in Community-Based Samples. JAMA psychiatry. 2018;75:447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Douard E, Zeribi A, Schramm C, Tamer P, Loum MA, Nowak S, Saci Z, Lord MP, Rodríguez-Herreros B, Jean-Louis M, Moreau C, Loth E, Schumann G, Pausova Z, Elsabbagh M, Almasy L, Glahn DC, Bourgeron T, Labbe A, Paus T, Mottron L, Greenwood CMT, Huguet G, Jacquemont S. Effect Sizes of Deletions and Duplications on Autism Risk Across the Genome. The American journal of psychiatry. 2021;178:87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, Gauthier LD, Brand H, Solomonson M, Watts NA, Rhodes D, Singer-Berk M, England EM, Seaby EG, Kosmicki JA, Walters RK, Tashman K, Farjoun Y, Banks E, Poterba T, Wang A, Seed C, Whiffin N, Chong JX, Samocha KE, Pierce-Hoffman E, Zappala Z, O'Donnell-Luria AH, Minikel EV, Weisburd B, Lek M, Ware JS, Vittal C, Armean IM, Bergelson L, Cibulskis K, Connolly KM, Covarrubias M, Donnelly S, Ferriera S, Gabriel S, Gentry J, Gupta N, Jeandet T, Kaplan D, Llanwarne C, Munshi R, Novod S, Petrillo N, Roazen D, Ruano-Rubio V, Saltzman A, Schleicher M, Soto J, Tibbetts K, Tolonen C, Wade G, Talkowski ME, Neale BM, Daly MJ, MacArthur DG The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wainberg M, Merico D, Huguet G, Zarrei M, Jacquemont S, Scherer SW, Tripathy SJ. Deletion of Loss-of-Function–Intolerant Genes and Risk of 5 Psychiatric Disorders. JAMA Psychiatry. 2022;79:78–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tammimies K, Marshall CR, Walker S, Kaur G, Thiruvahindrapuram B, Lionel AC, Yuen RKC, Uddin M, Roberts W, Weksberg R, Woodbury-Smith M, Zwaigenbaum L, Anagnostou E, Wang Z, Wei J, Howe JL, Gazzellone MJ, Lau L, Sung WWL, Whitten K, Vardy C, Crosbie V, Tsang B, D’Abate L, Tong WWL, Luscombe S, Doyle T, Carter MT, Szatmari P, Stuckless S, Merico D, Stavropoulos DJ, Scherer SW, Fernandez BA. Molecular Diagnostic Yield of Chromosomal Microarray Analysis and Whole-Exome Sequencing in Children With Autism Spectrum Disorder. Jama. 2015;314:895–903. [DOI] [PubMed] [Google Scholar]
- 21.Council on Children With D, Section on Developmental Behavioral P, Bright Futures Steering C, Medical Home Initiatives for Children With Special Needs Project Advisory C. Identifying infants and young children with developmental disorders in the medical home: an algorithm for developmental surveillance and screening. Pediatrics. 2006;118:405–420. [DOI] [PubMed] [Google Scholar]
- 22.Hyman SL, Levy SE, Myers SM, Council On Children With Disabilities SOD, Behavioral P. Identification, Evaluation, and Management of Children With Autism Spectrum Disorder. Pediatrics. 2020;145. [DOI] [PubMed] [Google Scholar]
- 23.Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, Church DM, Crolla JA, Eichler EE, Epstein CJ, Faucett WA, Feuk L, Friedman JM, Hamosh A, Jackson L, Kaminsky EB, Kok K, Krantz ID, Kuhn RM, Lee C, Ostell JM, Rosenberg C, Scherer Sw, Spinner NB, Stavropoulos DJ, Tepperberg JH, Thorland EC, Vermeesch JR, Waggoner DJ, Watson MS, Martin CL, Ledbetter DH. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. American journal of human genetics. 2010;86:749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sikich L, Frazier JA, McClellan J, Findling RL, Vitiello B, Ritz L, Ambler D, Puglia M, Maloney AE, Michael E, De Jong S, Slifka K, Noyes N, Hlastala S, Pierson L, McNamara NK, Delporto-Bedoya D, Anderson R, Hamer RM, Lieberman JA. Double-blind comparison of first- and second-generation antipsychotics in early-onset schizophrenia and schizo-affective disorder: findings from the treatment of early-onset schizophrenia spectrum disorders (TEOSS) study. Am J Psychiatry. 2008;165:1420–1431. [DOI] [PubMed] [Google Scholar]
- 25.Martin CL, Wain KE, Oetjens MT, Tolwinski K, Palen E, Hare-Harris A, Habegger L, Maxwell EK, Reid JG, Walsh LK, Myers SM, Ledbetter DH. Identification of Neuropsychiatric Copy Number Variants in a Health Care System Population. JAMA Psychiatry. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischbach GD, Lord C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron. 2010;68:192–195. [DOI] [PubMed] [Google Scholar]
- 27.C Yuen RK, Merico D, Bookman M, L Howe J, Thiruvahindrapuram B, Patel RV, Whitney J, Deflaux N,Bingham J, Wang Z, Pellecchia G, Buchanan JA, Walker S, Marshall CR, Uddin M, Zarrei M, Deneault E, D'Abate L, Chan AJ, Koyanagi S, Paton T, Pereira SL, Hoang N, Engchuan W, Higginbotham EJ, Ho K, Lamoureux S, Li W, MacDonald JR, Nalpathamkalam T, Sung WW, Tsoi FJ, Wei J, Xu L, Tasse AM, Kirby E, Van Etten W, Twigger S, Roberts W, Drmic I, Jilderda S, Modi BM, Kellam B, Szego M, Cytrynbaum C, Weksberg R, Zwaigenbaum L, Woodbury-Smith M, Brian J, Senman L, Iaboni A, Doyle-Thomas K, Thompson A, Chrysler C, Leef J, Savion-Lemieux T, Smith IM, Liu X, Nicolson R, Seifer V, Fedele A, Cook EH, Dager S, Estes A, Gallagher L, Malow BA, Parr JR, Spence SJ, Vorstman J, Frey BJ, Robinson JT, Strug LJ, Fernandez BA, Elsabbagh M, Carter MT, Hallmayer J, Knoppers BM, Anagnostou E, Szatmari P, Ring RH, Glazer D, Pletcher MT, Scherer SW. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nature neuroscience. 2017;20:602–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schumann G, Loth E, Banaschewski T, Barbot A, Barker G, Büchel C, Conrod PJ, Dalley JW, Flor H, Gallinat J, Garavan H, Heinz A, Itterman B, Lathrop M, Mallik C, Mann K, Martinot JL, Paus T, Poline JB, Robbins TW, Rietschel M, Reed L, Smolka M, Spanagel R, Speiser C, Stephens DN, Ströhle A, Struve M, the Ic. The IMAGEN study: reinforcement-related behaviour in normal brain function and psychopathology. Molecular psychiatry. 2010;15:1128–1139. [DOI] [PubMed] [Google Scholar]
- 29.Smith BH, Campbell A, Linksted P, Fitzpatrick B, Jackson C, Kerr SM, Deary IJ, Macintyre DJ, Campbell H, McGilchrist M, Hocking LJ, Wisely L, Ford I, Lindsay RS, Morton R, Palmer CN, Dominiczak AF, Porteous DJ, Morris AD. Cohort Profile: Generation Scotland: Scottish Family Health Study (GS:SFHS). The study, its participants and their potential for genetic research on health and illness. International journal of epidemiology. 2013;42:689–700. [DOI] [PubMed] [Google Scholar]
- 30.Deary IJ, Gow AJ, Pattie A, Starr JM. Cohort profile: the Lothian Birth Cohorts of 1921 and 1936. International journal of epidemiology. 2012;41:1576–1584. [DOI] [PubMed] [Google Scholar]
- 31.Kasak L, Rull K, Sõber S, Laan M. Copy number variation profile in the placental and parental genomes of recurrent pregnancy loss families. Scientific reports. 2017;7:45327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsuang DW, Millard SP, Ely B, Chi P, Wang K, Raskind WH, Kim S, Brkanac Z, Yu CE. The effect of algorithms on copy number variant detection. PloS one. 2010;5:e14456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, Barnes I, Berry A, Bignell A, Carbonell Sala S, Chrast J, Cunningham F, Di Domenico T, Donaldson S, Fiddes IT, García Girón C, Gonzalez JM, Grego T, Hardy M, Hourlier T, Hunt T, Izuogu OG, Lagarde J, Martin FJ, Martínez L, Mohanan S, Muir P, Navarro FCP, Parker A, Pei B, Pozo F, Ruffier M, Schmitt BM, Stapleton E, Suner MM, Sycheva I, Uszczynska-Ratajczak B, Xu J, Yates A, Zerbino D, Zhang Y, Aken B, Choudhary JS, Gerstein M, Guigó R, Hubbard TJP, Kellis M, Paten B, Reymond A, Tress ML, Flicek P. GENCODE reference annotation for the human and mouse genomes. Nucleic acids research. 2019;47:D766–d773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kendall KM, Bracher-Smith M, Fitzpatrick H, Lynham A, Rees E, Escott-Price V, Owen MJ, O'Donovan MC, Walters JTR, Kirov G. Cognitive performance and functional outcomes of carriers of pathogenic copy number variants: analysis of the UK Biobank. The British journal of psychiatry : the journal of mental science. 2019;214:297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeste SS, Geschwind DH. Disentangling the heterogeneity of autism spectrum disorder through genetic findings. Nat Rev Neurol. 2014;10:74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreno-De-Luca D, Ross ME, Ross DA. Leveraging the Power of Genetics to Bring Precision Medicine to Psychiatry: Too Little of a Good Thing? Biological psychiatry. 2018;83:e45–e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, Chu SH, Moreau MP, Gupta AR, Thomson SA, Mason CE, Bilguvar K, Celestino-Soper PB, Choi M, Crawford EL, Davis L, Wright NR, Dhodapkar RM, DiCola M, DiLullo NM, Fernandez TV, Fielding-Singh V, Fishman DO, Frahm S, Garagaloyan R, Goh GS, Kammela S, Klei L, Lowe JK, Lund SC, McGrew AD, Meyer KA, Moffat WJ, Murdoch JD, O'Roak BJ, Ober GT, Pottenger RS, Raubeson MJ, Song Y, Wang Q, Yaspan BL, Yu TW, Yurkiewicz IR, Beaudet AL, Cantor RM, Curland M, Grice DE, Günel M, Lifton RP, Mane SM, Martin DM, Shaw CA, Sheldon M, Tischfield JA, Walsh CA, Morrow EM, Ledbetter DH, Fombonne E, Lord C, Martin CL, Brooks AI, Sutcliffe JS, Cook EH Jr., Geschwind D, Roeder K, Devlin B, State MW. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahmad A, Bhattacharya S, Sridhar A, Iqbal AM, Mariani TJ. Recurrent copy number variants associated with bronchopulmonary dysplasia. Pediatric research. 2016;79:940–945. [DOI] [PubMed] [Google Scholar]
- 39.Glassford MR, Purcell RH, Pass S, Murphy MM, Bassell GJ, Mulle JG. Caregiver Perspectives on a Child's Diagnosis of 3q29 Deletion: "We Can't Just Wish This Thing Away". J Dev Behav Pediatr. 2022;43:e94–e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.