Abstract
We cloned lbpB, encoding a predicted 80-kDa lipoprotein, upstream of lbpA. A nonpolar mutant (LbpB− LbpA+) had normal lactoferrin (LF) binding and grew normally with LF as an iron source, whereas LbpB− LbpA− and LbpB+ LbpA− strains had reduced binding of LF and did not grow with LF as an iron source. LbpB bound LF directly in an affinity purification, suggesting that LbpB might play a still-uncharacterized role in the LF iron utilization.
Most organisms use inorganic iron (Fe) as a cofactor in carrying out a variety of metabolic functions (34). In an oxidized environment, ferric iron is highly insoluble and relatively unavailable. Mammals solubilize and sequester extracellular Fe by binding to the glycoproteins transferrin (TF) in serum (34) and lactoferrin (LF) on mucosal surfaces (22). Since there is essentially no free Fe in humans, pathogens have evolved mechanisms to scavenge Fe from host proteins. Many bacteria synthesize and secrete siderophores, low-molecular-weight compounds which bind and transport Fe into the cell through specific siderophore receptors (16). Production of siderophores facilitates infection in several bacterial pathogens (26). Neisseria gonorrhoeae and several other gram-negative mucosal pathogens including Neisseria meningitidis do not produce siderophores (10, 23) but produce specific receptors for each of the glycoproteins TF and LF, as well as hemoglobin (5, 7, 10, 15, 21).
A receptor for LF, LbpA, was identified and the corresponding lbpA gene was cloned and characterized in N. gonorrhoeae (3) and N. meningitidis (29). Recently, a second LF receptor protein, LbpB, was identified in N. meningitidis (6, 20, 28). Here we report the cloning of lbpB from N. gonorrhoeae and characterization of the role of LbpB in gonococcal LF binding and Fe acquisition.
Cloning of lbpB.
We previously cloned the entire lbpA gene on four overlapping clones (3). pUNCH127 (Fig. 1) included 1,246 bp of DNA upstream of the 5′ end of the lbpA open reading frame (ORF). To test if the region upstream of lbpA was also involved in LF utilization, we inactivated it by inserting (31) an Ω cassette (32) into an AvaI site in pUNCH127, creating pUNCH130. Sequence analysis of pUNCH130 confirmed that the Ω fragment was inserted at the AvaI site. pUNCH130 DNA was introduced into wild-type gonococcal strain FA19 by transformation (4), resulting in a mutant strain, FA6839. The presence of the Ω fragment in the AvaI site in FA6839 was confirmed by Southern analysis with the use of an lbpB-specific oligonucleotide probe (positions 2451 to 2470 in the lbpB sequence in GenBank) or the 2-kb Ω fragment (32). The mutant FA6839 had a phenotype similar to that of lbpA mutant FA6775 (3), including loss of ability to (i) bind LF in a solid-phase assay, (ii) take up Fe from LF, (iii) utilize LF as an Fe source, and (iv) express LbpA. These results suggest that insertion of Ω upstream of lbpA had a polar effect on expression of LbpA.
FIG. 1.
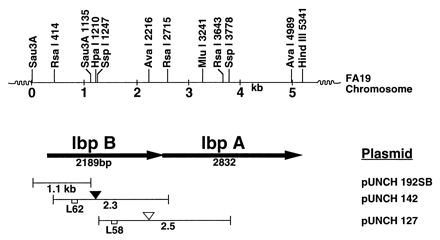
Physical map of the N. gonorrhoeae chromosome showing genetic organization of the lbpBA operon region. The top line represents the restriction map of the cloned FA19 chromosome fragment. The organization of lbpBA genes including the direction of transcription as indicated by arrows is shown in the next line. Plasmid pUNCH192SB is a 1.1-kb Sau3AI fragment. Plasmids pUNCH142 and pUNCH127 contain 2.3-kb RsaI and 2.5-kb SspI fragments, respectively. The solid inverted triangle denotes the position of the aphA3 insertion in strain FA6965(pUNCH142), and the open inverted triangle denotes the position of the Ω insertion in strain FA6839(pUNCH127). L58 and L62 refer to the oligonucleotides used as probes in isolating the clones pUNCH142 and pUNCH192SB, respectively.
DNA upstream of lbpA was cloned in two chromosome walking steps involving the use of oligonucleotide primers L58 and L62 (Fig. 1). Southern hybridization analysis showed that L58 hybridized to a 2.3-kb RsaI fragment (data not shown). We cloned the 2.3-kb RsaI fragment into the SmaI site of pUNCH615 (32), creating pUNCH142. Hybridization analysis with probe L62 identified a 1.1-kb Sau3A fragment of FA19 DNA. We ligated the 1.1-kb Sau3A fragment into the BamHI site of pMCL210 (27) harboring lacZ, allowing rapid color screening for inserts. Screening by colony hybridization produced two clones, pUNCH192 and pUNCH193, that differed in size. The clone pUNCH192 in Escherichia coli produced colonies on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and IPTG (isopropyl-β-d-thiogalactopyranoside) plates that had three phenotypes: slightly blue, white, and blue. Plasmid DNA from a colony belonging to each class was sequenced. Plasmid DNA from a slightly blue colony designated pUNCH192SB (Fig. 1) contained an 1,138-bp Sau3AI insert with a complete 5′ end of an apparent 2,189-bp-long ORF (Fig. 1). In contrast, the corresponding plasmid isolated from a white colony was 328 bp shorter than pUNCH192SB. The missing DNA included 21 bp at the 5′ end of the 2,189-bp ORF. Plasmid DNA from the blue colony contained primarily vector DNA. The clone pUNCH193 produced stable white colonies on similar media and was stable on subculture. Interestingly, pUNCH193 lacked 20 bp of DNA that included 5′-GTC GAA TCA ACG CCG ACC GC-3′ at positions 31 to 51 from the ATG codon start site of the 2,189-bp ORF in pUNCH192SB. To verify that the DNA in pUNCH192SB represented wild-type FA19, we PCR amplified and directly sequenced FA19 DNA flanking the deleted 20-bp region. A comparison of the sequences of FA19 DNA in the amplified product and the corresponding DNA in pUNCH192SB showed complete identity, confirming that the clone pUNCH192SB contained wild-type FA19 DNA. Thus, the set of three overlapping clones, pUNCH127, pUNCH142, and pUNCH192SB, represents 2,493 bp of contiguous FA19 DNA upstream of lbpA.
Nucleotide sequence analysis.
An examination of the sequences in clones pUNCH127, pUNCH142, and pUNCH192SB revealed a 2,189-bp ORF. The last 4 bp (ATGA) of the 2,189-bp ORF partially overlapped with the ATG initiation codon of the lbpA ORF. We designated the 2,189-bp ORF located upstream of lbpA as lbpB. Further upstream, we found 170 bp that were 56% identical to the 3′ end of an E. coli era gene that encodes a GTP binding protein (1). Immediately downstream of the era homolog there was a potential hairpin loop structure at positions 180 to 212. This structure has a ΔG (25°C) value of −20.8 kcal (33) and probably corresponds to a transcription terminator. In addition, a 10-bp inverted repeat at positions 184 to 208 and another 10-bp sequence at positions 1902 to 1911 matched gonococcal uptake sequence (12, 14).
Translation of the lbpB coding region predicted a protein of 728 amino acids with a molecular mass of 80 kDa and a pI value of 4.53. Analysis of the deduced N-terminal amino acid sequence of LbpB revealed an 18-amino-acid signal sequence with the lipoprotein modification consensus sequence LSAC at positions 16 to 19 (17). These data suggested that LbpB was a lipoprotein. Based on fluorographic analysis and palmitic acid labeling, Lewis et al. concluded that LbpB in N. meningitidis is a lipoprotein (20). LbpB was 31% identical to TbpB of the same gonococcal strain and 81% identical to the recently reported meningococcal LbpB (6, 28). Comparison of the predicted protein sequence of FA19 LbpB with that of meningococcal LbpB (6, 28) showed very similar features that differentiate each from the neisserial TbpB family. Both gonococcal and meningococcal LbpB proteins contained two stretches of negatively charged residues that were not present in TbpB family members: region one, residues 462 to 527, and region two, residues 689 to 708 (Fig. 2). These regions also exhibit considerable variation between meningococcal and gonococcal LbpB proteins. There was a deletion in region one in meningococcal LbpB relative to the gonococcal LbpB, and in region two, there was a deletion in gonococcal LbpB. Overall, these two charged domains were only about 50% identical, compared to 81% for the whole proteins. Each of the LbpB proteins contained an identical 21-amino-acid C terminus that was absent in all neisserial TbpB family members.
FIG. 2.

Lineup of negatively charged residues in region one and region two of the gonococcal (NG) and meningococcal (NM) LbpB proteins. The numbers indicate the position in the unprocessed protein of the first amino acid listed. Amino acids identical between the two proteins are denoted by vertical lines; colons indicate highly conservative substitutions, and dots indicate less conservative substitutions.
lbpB gene product.
To study the function of lbpB, we disrupted lbpB by inserting a nonpolar aphA3 cassette (24) into an HpaI site in pUNCH142, creating pUNCH195 (Fig. 1; Table 1) and exchanged this mutant allele into the gonococcal strain FA19 as described above, creating FA6965. Sequence analysis confirmed that the cassette was inserted in the correct orientation and translational frame into pUNCH195. Southern hybridization with the lbpB-specific probe L66 (positions 1156 to 1176) or the 0.9-kb aphA3 fragment (24) confirmed the expected insertion in lbpB in FA6965. The lbpB gene product was identified with the use of polyclonal, affinity-purified antiserum raised against a synthetic 16-mer peptide, RTRDNGINLSGNGSTN, from the predicted C terminus of LbpB. We used isogenic strains FA6815, FA6985, and FA6986 (Table 1) lacking TbpB and TbpA in order to avoid potential problems due to production of proteins similar in size and structure to LbpB and LbpA. A Western blot probed with LbpB antibodies reacted with a protein of about 95 kDa in the parent strain FA6815 (Fig. 3A, lane 1). A protein of the same size also was produced by FA6985 (lbpA::mTn3 Cm) (Fig. 3A, lane 3) but not FA6986 (lbpB::aphA3) (Fig. 3A, lane 4). The latter strain did produce the 103-kDa LbpA, as expected (Fig. 3B, lane 4). FA6839 (lbpB::Ω) produced neither LbpA nor LbpB (Fig. 3, lanes 2). These data showed that the 95-kDa protein was the lbpB gene product and also provided experimental evidence that lbpB and lbpA were part of a single transcriptional unit.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Source or reference(s) |
|---|---|---|
| N. gonorrhoeae strains | ||
| FA19 | LF+ TF+ | 5, 23 |
| FA1090 | LF− TF+ | 8 |
| FA6775 | LF− Cmr (lbpA::mTn3 Cm) | 3 |
| FA6815 | TF− Strr Spcr (tbpB::Ω) | 2 |
| FA6839 | LF− TF+ Strr Spcr (lbpB::Ω); transformant, pUNCH130→FA19 | This study |
| FA6965 | LF+ Kanr (lbpB::aphA3); transformant, pUNCH195→FA19 | This study |
| FA6985 | LF− (lbpA::mTn3 Cm) TF− (tbpB::Ω), Cmr Strr Spcr; transformant, FA6775→FA6815 | This study |
| FA6986 | LF+ (lbpB::aphA3) TF− (tbpB::Ω) Kanr Strr Spcr; transformant, FA6965→FA6815 | This study |
| Plasmids | ||
| pHP45Ω | Source for the Ω fragment (Strr Spcr) | 32 |
| pMCL210 | Cloning vector, low copy; lacZα Cmr | 27 |
| pUC18K | Source for the aphA3 cassette (Kanr) | 24 |
| pUNCH127 | Apr Tetr; 2.5-kb FA19 lbpAB SspI fragment cloned into SspI site in pBR322 | 3 |
| pUNCH128 | Apr; FA19 insert in pUNCH127 cloned into BamHI site in pUNCH615 | 3 |
| pUNCH130 | Strr Spcr; Ω cassette inserted into AvaI site in pUNCH128 | This study |
| pUNCH142 | Apr; 2.3-kb FA19 RsaI fragment cloned into BamHI site in pUNCH615 | This study |
| pUNCH192 | Like pUNCH192SB but unstable | |
| pUNCH192SB | Cmr; 1.1-kb FA19 Sau3A fragment cloned into BamHI site in pMCL210 | This study |
| pUNCH193 | Like pUNCH192SB but deleted | This study |
| pUNCH195 | Kanr; aphA3 cassette inserted into HpaI site in pUNCH142 | This study |
| pUNCH615 | Apr; pBluescript II KS(−) with PstI site replaced with MluI | 32 |
FIG. 3.
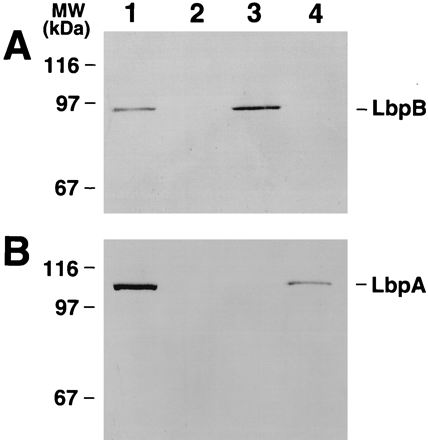
Expression of LbpB and LbpA in N. gonorrhoeae. Shown are Western blots containing total membranes prepared from Fe-starved cells probed with affinity-purified LbpB polyclonal antibody (A) and monoclonal LbpA antibody (B) (3). Lanes: 1, parent strain FA6815 (LbpB+ LbpA+); 2, FA6839 (LbpB− LbpA−); 3, FA6985 (LbpB+ LbpA−); 4, FA6986 (LbpB− LbpA+).
We noticed that the nonpolar mutant FA6986 (lbpB::aphA3) expressed reduced amounts of LbpA (Fig. 3B, lane 4) compared to the parent strain FA6815 (lbpB+ lbpA+) (Fig. 3B, lane 1). We confirmed this by comparing the intensities of the LbpA protein bands in linear range of detection in Western blots. We estimated that the reduction was approximately fivefold from the level for the parent strain (data not shown). We concluded that the aphA3 insertion in lbpB in FA6986 caused reduced production of the downstream lbpA gene product. The reason for reduced expression of LbpA in the lbpB::aphA3 mutant is not understood. Insertion of the aphA3 cassette in lbpB clearly did allow transcriptional read-through into lbpA but may have attenuated transcription downstream of the insertion.
LF binding.
To determine whether LbpB participated in LF binding, we performed dot blot assays of LF binding to whole cells. Bound LF was detected with horseradish peroxidase-conjugated anti-human LF antibody (Accurate Chemical & Scientific Corp., Westbury, N.Y.) (3). The results demonstrated that the LbpB− LbpA+ mutant FA6986 bound approximately as much LF as the LbpB+ LbpA+ parent strain FA6815 (data not shown). In contrast, no binding was detected for cells lacking LbpA, regardless of whether LbpB was present. Therefore, the solid-phase binding assay failed to detect LF binding by gonococci expressing LbpB alone. LbpB-specific binding was not detected under slightly different conditions (6, 28) for the solid-phase assay (data not shown).
To better quantitate LF binding, we measured the amount of radiolabeled LF bound by living cells in an equilibrium-phase binding assay developed by Cornelissen and Sparling (9). The specific activity of iodinated LF was 2.9 × 105 cpm/μg of LF. Approximately 1 × 107 to 5 × 107 CFU of gonococcal cells grown in chelexed defined medium with no added Fe (CDM-0) to induce Fe stress (5) were mixed with 2 to 100 nM 125I-LF in the presence of 1% bovine serum albumin and 5 μM unlabeled LF in individual wells of a Multi-Screen microtiter dish (0.45-μm-pore-size filter; MAHV N45; Millipore, Cambridge, Mass.). After being allowed to bind for 20 min at room temperature, unbound LF was removed by filtration, followed by five washes with CDM-0. Filters were dried, punched out, and counted. Specific binding was the difference between total binding without cold LF and binding that occurred in the presence of at least a 50-fold-excess concentration of cold LF. All four strains studied exhibited specific, saturable LF binding (Fig. 4). The wild-type strain bound significantly more LF than did each of the other strains. An apparent trend toward an intermediate level of LF binding in FA6965 (LbpB− LbpA+) was noted, but the differences were not statistically significant by the t test in conjunction with the Bonferroni procedure (19).
FIG. 4.
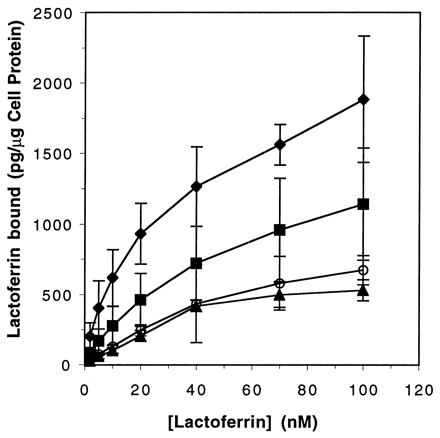
Isotherms for binding of LF to lbp mutants. The curves represent the amount of specifically bound LF as a function of LF concentration. Each point represents the mean of four individual experiments. Symbols: ⧫, FA19 (LbpB+ LbpA+); ■, FA6965 (LbpB− LbpA+); ○, FA6775 (LbpB+ LbpA−); ▴, FA6839 (LbpB− LbpA−).
The data obtained in the liquid-phase LF-binding experiments were analyzed with the computer program Receptor Fit Saturation Two-Site (Lundon Software, Inc., Cleveland Heights, Ohio). This program generates Kd and copy number estimates based on the best fit of observed data to progressively more complex models. Table 2 contains the best estimates of Kd and copy numbers generated by this program. Considerable variability was observed for LF binding to all strains; thus, these estimates are best considered comparisons of LF binding to isogenic strains rather than absolute indicators of intrinsic receptor affinity or copy number. Isogenic strains expressing LbpA exhibited complex binding phenomena consistent with the presence of at least two populations of binding sites, with Kds ranging from 5 to 500 nM. In contrast, the isogenic mutants that did not express LbpA bound LF to a more homogeneous population of receptors, with a single apparent Kd of approximately 45 nM. This was true of both LbpB+ and LbpB− strains, suggesting that LbpB played no detectable role in LF binding under the conditions employed in this assay. Somewhat surprisingly, the mutant that did not express either of the two identified LF-binding proteins (LbpB and LbpA) retained the ability to bind LF. This observation suggested that there were uncharacterized LF-binding components on the gonococcal cell surface.
TABLE 2.
Kd and copy number estimates for LF receptors expressed by isogenic mutants
| Strain | Phenotype | Kd (nM) | Copy number (M) |
|---|---|---|---|
| FA19 | LbpA+ LbpB+ | 4.8 (site 1) | 9.3 × 10−12 (N1) |
| 550 (site 2) | 6.9 × 10−11 (N2) | ||
| FA6965 | LbpA+ LbpB− | 5.0 (site 1) | 2.3 × 10−12 (N1) |
| 95 (site 2) | 2.0 × 10−11 (N2) | ||
| FA6775 | LbpA− LbpB+ | 45 (site 1) | 1 × 10−11 (N1) |
| FA6839 | LbpA− LbpB− | 45 (site 1) | 1 × 10−11 (N1) |
LF affinity isolation of LbpB.
To determine whether LbpB could bind LF in the absence of LbpA, total membranes prepared from Fe-starved FA6815 (LbpB+ LbpA+) and its isogenic derivatives, FA6985 (LbpB+ LbpA−) and FA6986 (LbpB− LbpA+), were subjected to LF-agarose affinity purification (7, 11). Total membrane proteins were dissolved in 2% Zwittergent 3, 14 (Calbiochem) and rocked with a 50% slurry of LF-agarose overnight at 4°C. The unbound material was removed by centrifugation, and the LF-binding protein was eluted in a Laemmli sample buffer for analytical sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting. Results (Fig. 5) showed an excellent recovery of LbpB in the absence of LbpA, indicating that LbpB was capable of binding LF under these conditions. LbpA also could be isolated by this procedure from total membranes in the absence of LbpB. The amount of LbpA in membrane preparations from the two strains was roughly the same (lanes A and C), but less LbpA was purified from the LbpB− LbpA+ strain than from wild type (lanes D and F). This suggested that LbpA alone did not bind to LF as avidly as it did in the presence of LbpB.
FIG. 5.
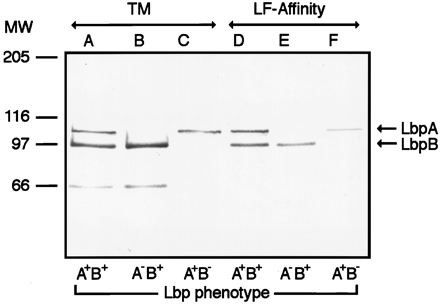
Western blot of LF-agarose affinity-purified LbpB and LbpA from total membranes of Fe-starved FA6815 (LbpB+ LbpA+) (lanes A and D), FA6985 (LbpB+ LbpA−) (lanes B and E), and FA6986 (LbpB− LbpA+) (lanes C and F). The blot was probed with antibodies to LbpA and then LbpB. The anti-LbpB antibody used here was raised against an LbpB His-tagged fusion protein. The 66-kDa band present in lanes A and B was recognized by antiserum elicited against full-length recombinant LbpB but not against an LbpB-specific peptide (Fig. 3 and data not shown). This protein band probably represents a breakdown product of holo-LbpB. Lanes A to C, total membranes (TM); lanes D to F, LF-agarose affinity-purified proteins. MW, molecular weight in thousands.
LF utilization.
To assess the contribution of LbpB in utilization of Fe from LF, we compared growth in CDM supplemented with LF of strains that carried either intact lbpB or inactivated lbpB. On agar plates or in CDM broth culture (5), the nonpolar lbpB mutant FA6965 (LbpB− LbpA+) failed to show a growth defect, growing to a level equal to that of wild-type FA19 (data not shown).
55Fe-LF uptake.
To detect smaller effects of LbpB in LF utilization, we measured the amount of 55Fe-LF taken up by the use of established procedures (2, 5). Results indicated that the nonpolar lbpB::aphA3 mutant FA6965 (LbpB− LbpA+) acquired 55Fe from LF at about 60% of wild-type levels (P < 0.03) (data not shown). In contrast, LbpA− mutants, FA6775 and FA6839, failed to take up a significant amount of 55Fe from LF (P < 0.01). We could not determine whether reduced Fe uptake from LF in FA6965 was due to loss of LbpB or to decreased expression of LbpA.
Conclusions.
We cloned an N. gonorrhoeae lbpB gene, providing genetic evidence for the existence of a second LF receptor protein in the gonococcus, similar to that described recently for meningococci (6, 20, 28). Despite difficulties encountered in isolating the 5′ end of the gonococcal lbpB gene, we succeeded in isolating a clone (pUNCH192SB) with an intact 5′ end of lbpB. Attempts to clone the 5′ end of the lbpB gene in N. meningitidis were unsuccessful (6, 20, 28).
A two-gene lbpB lbpA transcriptional unit was implicated by the observations that the 3′ end of lbpB overlapped the 5′ end of lbpA, and creation of a polar mutation in lbpB resulted in simultaneous loss of expression of the lbpB and lbpA genes. Recent experiments by Lewis et al. (20) showed that the lbpB and lbpA genes in meningococci are organized in a single transcriptional unit.
Both the gonococcal and meningococcal LbpB proteins contain two strongly acidic domains, notably absent in TbpB proteins, which are rich in aspartic acid and glutamic acid. This suggests that these regions could be involved in binding of LF, which is highly cationic (34). These regions are quite dissimilar in primary sequence in the known examples in gonococci and meningococci, suggesting that these also could be antigenic domains recognized by the host immune response. Future experiments hopefully will answer whether these domains are involved in binding LF and are immunogenic.
The role of LbpB in mediating Fe acquisition from LF is unclear. The LbpB− mutant (LbpB− LbpA+) retained the ability to bind LF, and this was reflected in its capacity to utilize LF as an Fe source. We demonstrated that LbpB, in the absence of LbpA, could be isolated by LF-agarose affinity, which is the best evidence that LbpB is an LF-binding protein.
We found that the LbpB− LbpA+ mutant used LF as an Fe source approximately as well as did the parent, as evidenced by similar growth patterns. The growth patterns of similar meningococcal LbpB− LbpA+ mutants varied, but all mutants grew to some extent (28). Differences in growth exhibited by strains in these reports might be explained on the basis of varying levels of LbpA expression in the LbpB− LbpA+ strain. For example, negligible growth noted by Lewis et al. might have resulted from a more than 10-fold reduction in LbpA expression (20). Near-normal growth exhibited by the mutant used by Pettersson et al. was correlated with LbpA expression similar to that of LbpB (28). The general conclusion is that LbpA is essential for LF-Fe utilization, whereas LbpB is not essential.
The question of how important LF is to pathogenesis remains. Many pathogens and nonpathogens can survive on the mucosal surface without being able to bind LF or to use LF as a source of Fe. For example, all Haemophilus influenzae strains are LF− (18, 30), and about one-half of gonococci are LF− (3, 25). Indeed, gonococcal strain FA1090 is LF− due to a large lbpBA deletion (references 13 and 20 and data not shown), yet it causes urethritis readily in male human volunteers (8). Future work is required to clarify the role of LF in bacterial pathogenesis and the roles of LbpB, in particular in utilization of LF.
Nucleotide sequence accession number.
The GenBank accession number for the 2,189-bp ORF is AF072890.
Acknowledgments
This work was supported by Public Health Service grants AI31496 and AI26837 from the National Institute of Allergy and Infectious Diseases.
We are grateful to Chris Elkins for his assistance with preparation of antipeptide sera and affinity isolation procedures. We also thank Chris Thomas for his expert computer assistance. We acknowledge the generosity of P. K. Sen for advice on statistical analysis and Rishikesh Chakravorty for analyzing the LF binding data. We are thankful to Bill Shafer for providing plasmid pUC18K and the UNC-CH Automated DNA Sequencing Facility for DNA sequencing.
REFERENCES
- 1.Ahnn J, March P E, Takiff H E, Inouye M. A GTP binding protein of Escherichia coli has homology to yeast RAS proteins. Proc Natl Acad Sci USA. 1986;83:8849–8853. doi: 10.1073/pnas.83.23.8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson J E, Sparling P F, Cornelissen C N. Gonococcal transferrin-binding protein 2 facilitates but is not essential for transferrin utilization. J Bacteriol. 1994;176:3162–3170. doi: 10.1128/jb.176.11.3162-3170.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biswas G D, Sparling P F. Characterization of lbpA, the structural gene for a lactoferrin receptor in Neisseria gonorrhoeae. Infect Immun. 1995;63:2958–2967. doi: 10.1128/iai.63.8.2958-2967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biswas G D, Burnstein K L, Sparling P F. Linearization of donor DNA during plasmid transformation in Neisseria gonorrhoeae. J Bacteriol. 1986;168:756–761. doi: 10.1128/jb.168.2.756-761.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanton K J, Biswas G D, Tsai J, Adams J, Dyer D W, Davis S M, Koch G G, Sen P K, Sparling P F. Genetic evidence that Neisseria gonorrohoeae produces specific receptors for transferrin and lacteroferrin. J Bacteriol. 1990;172:5225–5235. doi: 10.1128/jb.172.9.5225-5235.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonnah R A, Schryvers A B. Preparation and characterization of Neisseria meningitidis mutants deficient in production of the human lactoferrin binding proteins LbpA and LbpB. J Bacteriol. 1998;180:3080–3090. doi: 10.1128/jb.180.12.3080-3090.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen C-J, Sparling P F, Lewis L A, Dyer D W, Elkins C. Identification and purification of a hemoglobin-binding outer membrane protein from Neisseria gonorrhoeae. Infect Immun. 1996;64:5008–5014. doi: 10.1128/iai.64.12.5008-5014.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornelissen C N, Kelley M, Hobbs M M, Anderson J E, Sparling P F. The transferrin receptor expressed by gonococcal strain FA1090 is required for the experimental infection of human volunteers. Mol Microbiol. 1998;27:611–616. doi: 10.1046/j.1365-2958.1998.00710.x. [DOI] [PubMed] [Google Scholar]
- 9.Cornelissen C N, Sparling P F. Binding and surface-exposure characteristics of the gonococcal transferrin receptor are dependent on both transferrin-binding proteins. J Bacteriol. 1996;178:1437–1444. doi: 10.1128/jb.178.5.1437-1444.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornelissen C N, Sparling P F. Iron piracy: acquisition of transferrin-bound iron by bacterial pathogens. Mol Microbiol. 1994;14:843–850. doi: 10.1111/j.1365-2958.1994.tb01320.x. [DOI] [PubMed] [Google Scholar]
- 11.Elkins, C. Unpublished observations.
- 12.Elkins C, Thomas C E, Seifert H S, Sparling P F. Species-specific uptake of DNA by gonococci is mediated by a 10-base-pair sequence. J Bacteriol. 1991;173:3911–3913. doi: 10.1128/jb.173.12.3911-3913.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonococcal Genome Sequence Database.http://dna1.chem.uoknor.edu/.
- 14.Goodman S D, Scocca J J. Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1988;85:6982–6986. doi: 10.1073/pnas.85.18.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray-Owen S, Schryvers A B. Bacterial transferrin and lactoferrin receptors. Trends Microbiol. 1996;5:185–191. doi: 10.1016/0966-842x(96)10025-1. [DOI] [PubMed] [Google Scholar]
- 16.Guerinot M L. Microbial iron transport. Annu Rev Microbiol. 1994;48:743–772. doi: 10.1146/annurev.mi.48.100194.003523. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi S, Wu H C. Lipoproteins in bacteria. J Bioenerg Biomembr. 1990;22:451–471. doi: 10.1007/BF00763177. [DOI] [PubMed] [Google Scholar]
- 18.Herrington D A, Sparling P F. Haemophilus influenzae can use human transferrin as a sole source for required iron. Infect Immun. 1985;48:248–251. doi: 10.1128/iai.48.1.248-251.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hochberg Y, Tamhane A. Multiple comparison procedures. 1987. p. 30. and 363. John Wiley & Sons, Inc., New York, N.Y. [Google Scholar]
- 20.Lewis L A, Rhode K H, Behrens B, Gray E, Toth S I, Roe B A, Dyer D. Identification and molecular analysis of lbpBA, which encodes the two-component meningococcal lactoferrin receptor. Infect Immun. 1998;66:3017–3023. doi: 10.1128/iai.66.6.3017-3023.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis L A, Gray E, Wang Y-P, Roe B A, Dyer D. Molecular characterization of hpuAB, the haemoglobin-heptaglobin-utilization operon of Neisseria meningitidis. Mol Microbiol. 1997;23:737–749. doi: 10.1046/j.1365-2958.1997.2501619.x. [DOI] [PubMed] [Google Scholar]
- 22.Masson P L, Heremans J F, Dive C H. An iron-binding protein common to many external secretions. Clin Chim Acta. 1966;14:735–739. [Google Scholar]
- 23.McKenna W R, Mickelsen P A, Sparling P F, Dyer D W. Iron uptake from lactoferrin and transferrin by Neisseria gonorrhoeae. Infect Immun. 1988;56:785–791. doi: 10.1128/iai.56.4.785-791.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menard R, Sansonetti P J, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mickelsen P A, Blackman E, Sparling P F. Ability of Neisseria gonorrhoeae, Neisseria meningitidis, and commensal Neisseria species to obtain iron from lactoferrin. Infect Immun. 1982;35:915–920. doi: 10.1128/iai.35.3.915-920.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mietzner T A, Morse S A. The role of iron binding proteins in the survival of pathogenic bacteria. Annu Rev Nutr. 1994;14:471–493. doi: 10.1146/annurev.nu.14.070194.002351. [DOI] [PubMed] [Google Scholar]
- 27.Nakano Y, Yoshida Y, Yamashita Y, Koga T. Construction of a series of pACYC-derived plasmid vectors. Gene. 1995;162:157–158. doi: 10.1016/0378-1119(95)00320-6. [DOI] [PubMed] [Google Scholar]
- 28.Pettersson A, Prinz T, Umar A, Biezen J, Tommassen J. Molecular characterization of lbpB, the second lactoferrin-binding protein of Neisseria meningitidis. Mol Microbiol. 1998;27:599–610. doi: 10.1046/j.1365-2958.1998.00707.x. [DOI] [PubMed] [Google Scholar]
- 29.Pettersson A, Maas A, Tommassen J. Identification of the iroA gene product of Neisseria meningitidis as a lactoferrin receptor. J Bacteriol. 1994;176:1764–1766. doi: 10.1128/jb.176.6.1764-1766.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pidcock K A, Wooten J A, Daley B A, Stull T L. Iron acquisition by Haemophilus influenzae. Infect Immun. 1988;56:721–725. doi: 10.1128/iai.56.4.721-725.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Thomas C E, Sparling P F. Identification and cloning of a fur homologue from Neisseria meningitidis. Mol Microbiol. 1994;11:725–737. doi: 10.1111/j.1365-2958.1994.tb00350.x. [DOI] [PubMed] [Google Scholar]
- 33.Tinoco I, Jr, Borer P N, Dengler B, Levine M D, Uhlenbeck O C, Crothers D M, Gralla J. Improved estimation of secondary structure in ribonucleic acids. Nature (London) New Biol. 1973;246:40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- 34.Welch S. The chemistry and biology of iron. In: Welch S, editor. Transferrin: the iron carrier. Boca Raton, Fla: CRC Press, Inc.; 1992. pp. 41–59. [Google Scholar]


