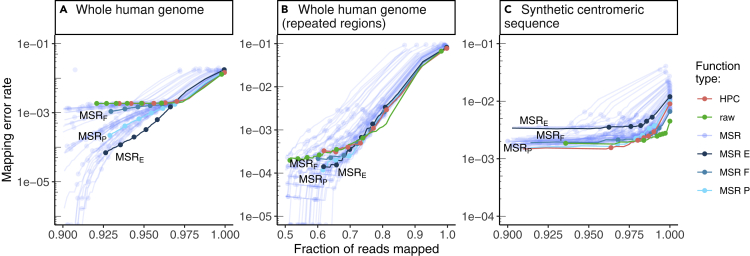
Figure 5.
Performance of our 58 selected mapping-friendly sequence reductions across genomes on reads simulated by nanosim
(Panel A) shows the whole human genome assembly,
(B and C) the subset of mapped reads from panel B that originate from repetitive regions, and C) the “TandemTools” synthetic centromeric reference sequence. We highlighted the best-performing mapping-friendly sequence reductions as MSR E, F, and P, respectively, in terms of cumulative mapeval mapping error rate, fraction of reads mapped, and percentage of better thresholds than HPC. Each point on a line represents, from left to right, the mapping quality thresholds 60, 50, 40, 30, 20, 10, and 0. For the first point of each line, only reads of mapping quality 60 are considered, and the y value represents the rate of these reads that are not correctly mapped, the x value represents the fraction of reads that are mapped at this threshold. The next point is computed for all reads of mapping quality , etc. The rightmost point on any curve represents the mapping error rate and the fraction of mapped reads for all primary alignments. The x-axes are clipped for lower mapped read fractions to better differentiate HPC, raw and MSRs E, F, and P. See Also Figure S7.