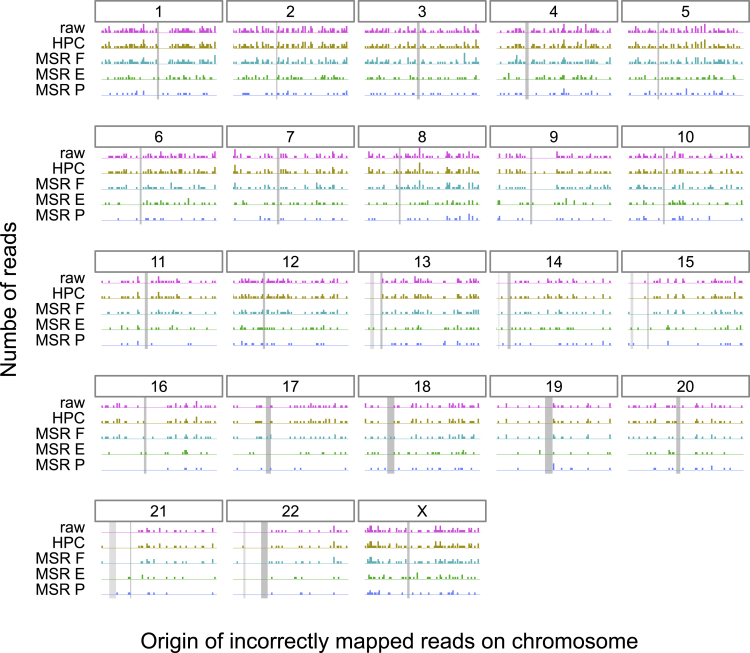
Figure 6.
Histogram of the original simulated positions for the incorrectly mapped reads using minimap2 at high mapping qualities across the whole human genome, for several transformation methods
For a given chromosome, each row represents the number of simulated reads starting at that particular region. The dark gray rectangle represents the position of the centromere for that chromosome, obtained from annotations provided by the T2T consortium (http://t2t.gi.ucsc.edu/chm13/hub/t2t-chm13-v1.1/). Similarly, for chromosomes 13, 14, 15, 21, and 22, a lighter gray rectangle represents the position of the “stalk” satellites also containing repetitive regions. For HPC and raw reads only alignments of mapping quality 60 were considered. To provide a fair comparison, alignments with mapping qualities were considered for MSRs E, F, and P. See also Figures S1–S5.