Abstract
Triterpenoids are a class of natural products widely used in fields related to medicine and health due to their biological activities such as hepatoprotection, anti-inflammation, anti-viral, and anti-tumor. With the advancement in biotechnology, microorganisms have been used as cell factories to produce diverse natural products. Despite the significant progress that has been made in the construction of microbial cell factories for the heterogeneous biosynthesis of triterpenoids, the industrial production of triterpenoids employing microorganisms has been stymied due to the shortage of efficient enzymes as well as the low expression and low catalytic activity of heterologous proteins in microbes. Protein engineering has been demonstrated as an effective way for improving the specificity, catalytic activity, and stability of the enzyme, which can be employed to overcome these challenges. This review summarizes the current progress in the studies of Oxidosqualene cyclases (OSCs), cytochrome P450s (P450s), and UDP-glycosyltransferases (UGTs), the key enzymes in the triterpenoids synthetic pathway. The main obstacles restricting the efficient catalysis of these key enzymes are analyzed, the applications of protein engineering for the three key enzymes in the microbial synthesis of triterpenoids are systematically reviewed, and the challenges and prospects of protein engineering are also discussed.
Keywords: Oxidosqualene cyclase, Cytochrome P450, UDP-Glycosyltransferase, Triterpenoids, Protein engineering
1. Introduction
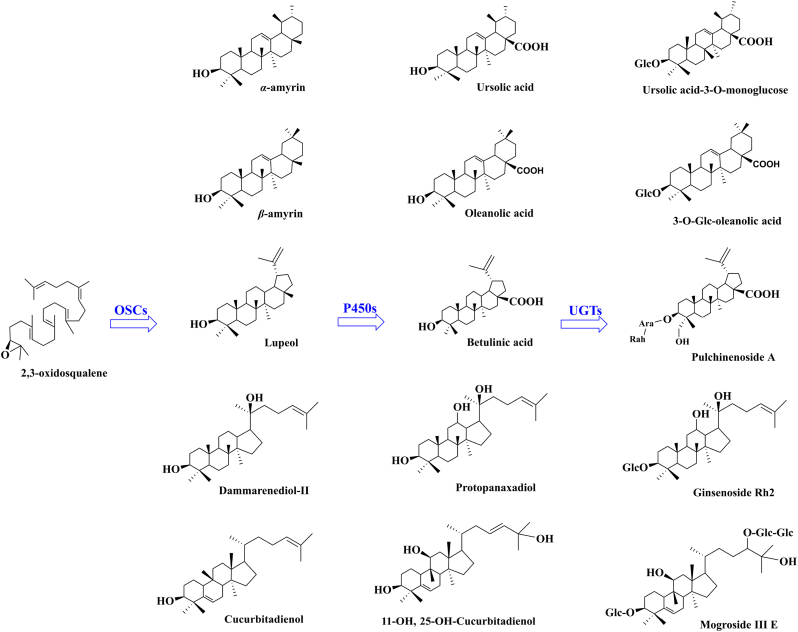
Triterpenoids are a class of secondary metabolites with diverse structures widely existing in plants. As the main active ingredients in traditional herbal medicines, they have pharmacological activities such as liver protection, anti-inflammation, anti-virus, and anti-tumor [[1], [2], [3]]. The structural diversity of triterpenoids makes them potential lead compounds for new drugs, and thus attracted the attention of the pharmaceutical industry [4]. Triterpenoids are mainly derived from the cyclization of 2,3-oxidosqualene, in which oxidosqualene cyclases catalyze the cyclization of 2,3-oxidosqualene to form triterpenoid skeletons, such as α-amyrin, β-amyrin, and lupeol, laying the foundation for the structural diversity of triterpenoids. Subsequently, the triterpenoid skeletons could be further mono-oxygenated by cytochrome P450s to produce different triterpenoid compounds such as ursolic acid, oleanolic acid and betulinic acid, etc. (Fig. 1). However, the clinical application of many triterpenoids is limited by their low solubility and bioavailability [5]. The glycosylation reaction mediated by UGTs can transfer sugar groups such as glucose and galactose from sugar donors to the hydroxyl or carboxyl groups of triterpenoids, therefore improving the water-solubility and bioavailability of triterpenoids. The products of the glycosylation reaction, triterpenoid saponins, thus have better pharmacological activities [6].
Fig. 1.
Biosynthetic pathways of triterpenoids.
In recent years, with the development of high-throughput sequencing technologies and web-based BLAST servers, as well as the publication of lots of plant transcriptome databases, more and more key enzymes for terpenoid synthesis have been mined. Meanwhile, the synthetic pathways of many terpenoids in plants have been analyzed and obtained [7]. Therefore, the synthesis of plant natural products using microbial cell factories becomes a research hotspot. For example, CYP72A63 has been identified to be a P450 enzyme involved in the synthesis of glycyrrhetinic acid (GA), and can significantly increase its yield [8,9]. When expressed in microbes, however, the plant-derived enzymes suffer from low catalytic activity and poor specificity, limiting the industrial production of natural products. Rational or semi-rational design based on crystal structure analysis or molecular dynamics simulations for protein engineering has been used to overcome these challenges [10,11]. Notably, the crystal structures of enzymes are of crucial importance in the rational or semi-rational design of protein engineering. While the known crystal structures for enzymes in the synthesis of plant triterpenoids are limited, indirect methods such as sequence alignment, protein homology modeling, and molecular simulation are employed in rational or semi-rational protein design. In addition, AlphaFold 2, a new tool in structural biology, can predict the structure of a typical protein in minutes with unprecedented accuracy. It can enable protein structure elucidation techniques to keep the pace of the genomic revolution in the future [12]. To date, key enzymes in several synthetic pathways have been modified via protein engineering, endowed with specific chemoselectivity, substrate selectivity, and catalytic activity [13,14]. These advancements facilitate the application of protein engineering in the microbial synthesis of triterpenoids and triterpenoid saponins. This review mainly introduces the current studies of key enzymes, OSCs, P450s, and UGTs, in the synthetic pathway of triterpenoids and triterpenoid saponins. An overview of the optimization of OSCs, P450s and UGTs by protein engineering is also provided.
2. Protein engineering
As the synthetic pathways of terpenoids in plants have been elucidated constantly, heterologous synthesis of triterpenoids in microorganisms has been achieved through various metabolic regulation strategies. Zhang et al. [15] overexpressed typical rate-limiting enzymes isopentenyl pyrophosphate isomerase (IDI), farnesyl-diphosphate synthase (FPPS), and squalene synthase (SQS) in Saccharomyces cerevisiae to enhance the supply of precursors and increase the yield of oleanane triterpenoids by 1.55-fold. Li et al. successfully synthesized the precursor 2,3-oxidosqualene in Escherichia coli, followed by the introduction of the enzymes with truncated N-terminal transmembrane domains, producing dammarenediol II with a final yield of 8.63 mg/L [16]. Akin to Saccharomyces cerevisiae, Yarrowia lipolytica is another excellent host for the production of triterpenoids. Li et al. [17] constructed a recombinant ginsenoside compound K pathway in Y. lipolytica which overexpressed MVA rate-limiting enzyme, employed a fused protein of P450 (PPDS) and NADPH-cytochrome P450 reductase (ATR1), leading to a final product of 30.4 mg/L. In order to increase the production of betulinic acid, cytochrome P450 oxidoreductases (CPR) from Medicago truncatula and CYP716A180 from Betula platyphylla were screened and paired by Jin et al. [18,19]. By co-expressing with ERG1, ERG9, and HMG1 in yeast, the titer of betulinic acid was increased to 51.87 ± 2.77 mg/L. Although the regulation and optimization of metabolic pathway can improve carbon flux to a certain extent, it is difficult to achieve large-scale production for most natural products. One of the bottlenecks is the poor catalytic activity and substrate specificity of key enzymes.
The emergence of protein engineering has greatly accelerated the industrialization of biosynthesis for triterpenoids and triterpenoid saponins. On the other hand, the update of many open source software and websites provides a theoretical basis and feasibility basis for the development of protein engineering. For example, the online BLAST tool can quickly match homologous templates for target enzymes from massive databases; The MODELER program in Discovery Studio v2.5, SWISS-MODEL, or AlphaFold2 [[20], [21], [22]] can use the method of homology modeling to predict the three-dimensional structure of a known sequence; The quality of the 3D model structure is evaluated by the software such as Seq Identity, GMQE, QMEAN, ProCheck, Molprobity, and Errat; The docking of the target protein and the catalytic product is simulated by Autodock Vina [23,24], which predicts the conformation of the ligand at the target binding site, calculates the binding energy, and predicts specific amino acid residues. The activity centers of key enzymes, catalytic products, and functional residues have been studied by researchers through computer-aided design combined with the crystal structures resolved. The section focuses on the application of protein engineering in the optimization of OSCs, P450s, and UGTs and the research progress in triterpenoid synthesis.
2.1. Protein engineering of OSCs
2.1.1. Crystal structure of OSCs
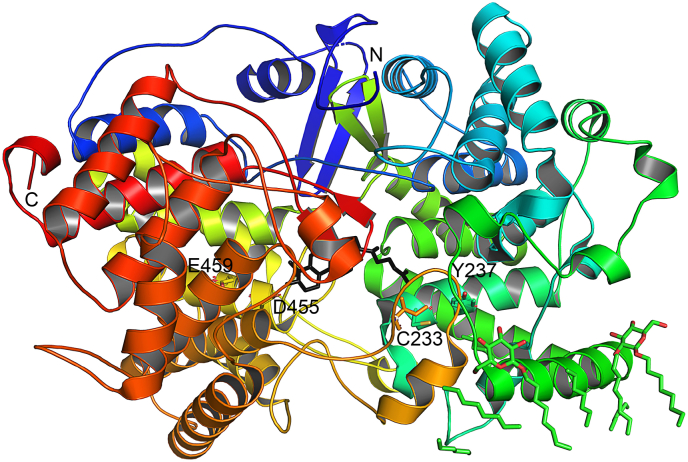
With the in-depth study of the three-dimensional structure of enzyme molecules and the rapid development of bioinformatics, rational design based on crystal structure followed by experimental analysis and computer simulation has become an important method for enzyme molecule modification. Therefore, protein engineering of OSCs from the perspective of structural biology and biochemistry is necessary to improve their performance and applicability. OSC is a membrane protein with a complex structure, which is difficult to be analyzed. To date, only two crystal structures of the human membrane protein OSC, namely OSC-lanosterol complex structure (PDB ID: 1W6K) and OSC-inhibitor Ro 48-8071 complex structure (PDB ID: 1W6J) have been resolved [25].
The crystal structure of OSC-lanosterol (Fig. 2) shows that one hydrogen bond is formed between the lanosterol 3-hydroxy group and the side chain of D455. This reveals the cyclization mechanism of 2,3-oxidosqualene, where D455 donates a proton to the epoxy group of the substrate to initiate the cyclization cascade. When a catalytic cycle is completed, the proton is transferred back from the final deprotonation step D455, or D455 recarbonylation through the water molecular chain and the carboxylic acid group of E459. In the crystal structure of human OSC containing the inhibitor Ro48-8071, the nitrogen atom of Ro48-8071 forms a charged hydrogen bond with the carboxylate of D455, and the bromine atom interacts with I524, Y237, and C233 residues near the channel constriction site, H232 terminates the cyclization of 2,3-oxysqualene. This is consistent with the catalytic mechanism of the OSC-lanosterol complex structure. Due to the difficulty in obtaining the crystal structure of OSCs enzyme, almost all the studies related to the molecular modification of OSCs in recent years have been carried out using human OSC as the template, such as ERG7 from S. cerevisiae, and AsHS1 from Avena strigosa [26,27].
Fig. 2.
Structure of human OSC in complex with Lanosterol.
2.1.2. Regulation of product preference through protein engineering of OSCs
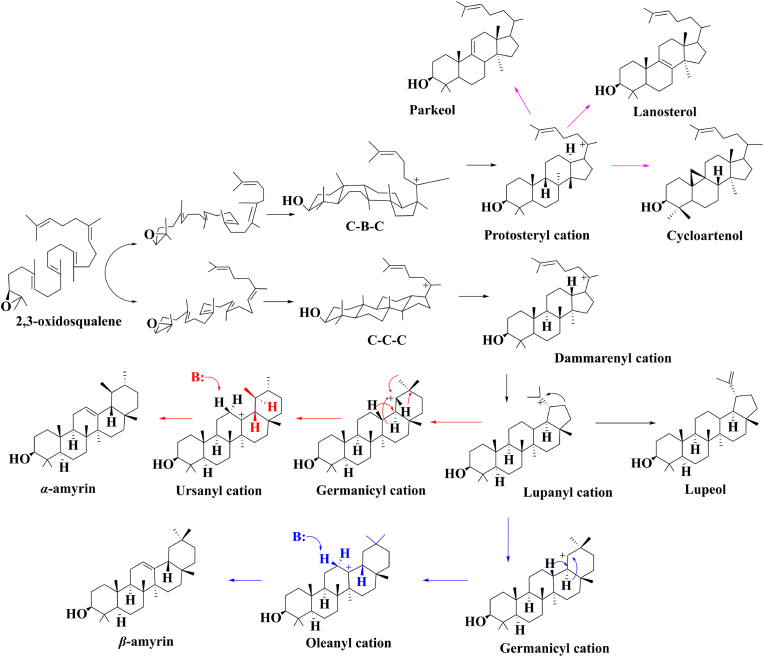
From the perspective of natural synthetic pathways, OSCs cyclize 2,3-oxidosqualene to generate a variety of compound skeletons for plant triterpenoid through a Chair-Boat-Chair (CBC) or a Chair-Chair-Chair (CCC) conformation and finally forms triterpenoids (e.g. cucurbitadienol, lupeol, and glutinol) (Fig. 3). At present, a considerable number of OSCs have been isolated, purified, cloned, and characterized through plant or transcriptome database mining. For example, Yu et al. [39] identified MdOSC1 from apple, which increased the production of α-amyrin in S. cerevisiae by 5.8-fold with a titer of 11.97 ± 0.61 mg/L. However, in addition to the target compound, there are small amounts of β-amyrin and lupin alcohol in the product. The MdOSC4 and MdOSC5 identified from apples by Andre et al. [40] also have product confounding, among which germanicol and lupeol are the main products. SlTTS2 from tomato catalyzed the production of triterpenoids with six compounds (e.g. α-, β- and δ-amyrin) [41]; KcMS from Kandelia candel was verified to be highly homologous to lupeol synthase, but the content of lupeol only accounted for half of the total product [42]. It can be concluded the plant-derived enzymes often have problems of poor specify when introduced into microbes. Therefore, protein modification of the key enzymes that have been mined is crucial, and the discovery of new enzymes provides a resource-rich database for protein engineering.
Fig. 3.
Formation of carbocation intermediates and triterpenoid skeletons under the catalysis of OSCs.
Combining advanced computational techniques and molecular biology methods, we can deeply understand the structural characteristics and functions of the enzyme. The correct docking of the substrate can provide important candidates for mutation, including active sites such as key catalytic or binding residues (Table 1). Liang et al. [27] discovered two novel hopane-type triterpenoid synthases, namely, AsHS1 and AsHS2, which produce hopenol B and hop-17(21)-en-3β-ol, respectively. AlphaFold2 modeling and QM/MM calculation showed that aromatic residues F257 and F729 of AsHS1 promoted the rearrangement reaction by interacting with hopyl C22 cations. The bulky side chain of Y410 affects the interaction between F257 and carbocation through steric hindrance, which is considered to be the key element to product differentiation. When Y410 was mutated to a small alanine (Y410 corresponds to A410 in AsHS2), the mutant mainly catalyzed the production of hop-17(21)-en-3β-ol, which was the same as AsHS2. This result reveals the possibility of changing product preference through amino acid substitution mutations. Eukaryotic OSCs specifically recognize (3S)-2,3-oxidosqualene as a substrate, rather than squalene or (3R)-2,3-oxidosqualene. Tsutomu et al. [43] compared the sequences of OSCs with some prokaryotic squalene-hopene cyclase (SHCs), and proposed that G600 was important for SHC's recognition of multiple substrates according to the fact that G600 was highly conserved in SHCs but not in OSCs. They knocked out G600 of squalene cyclase from Alicyclobacillus acidocaldarius, and the obtained mutant only took (3S)-2,3-oxidosqualene as the substrate. Conserved motifs 255MLCYCR260 and 258MWCYCR263 are present in lupeol synthase and β-amyrin synthase, respectively, and are actively involved in product differentiation. Kushiro et al. [33] changed the product from lupeol to β-amyrin by mutating L256 of lupeol synthase to L256W. Similarly, W259 of β-amyrin synthase is replaced by leucine, resulting in the termination of the reaction at the Lupenyl cation stage, and the product is dominated by lupeol. It can be seen that the unique active sites or functional domains of OSCs play an important role in stabilizing carbocation intermediates and product specificity. Interestingly, corresponding residues do not necessarily produce the same interactions. For example, the F474 residue of β-amyrin synthase (EtAS) from Euphorbia tirucalli corresponds to F365 of SHC, which stabilized the transient cation via cation–π interaction. But when F474 was mutated to aliphatic amino acids lacking π-electrons such as Val, Leu and Met, the polycyclic reaction still proceeded and produced pentacyclic triterpenoids without terminating at the bicyclic stage, which proved that SHC F365 has a distinct mode of action from EtAS F474 [29,30]. Another example is the catalytic efficiency of EtAS, which was significantly reduced after the mutation from phenylalanine to tryptophan at site 728. F728 promotes ring-expansion processes by stabilizing the cationic intermediates, while the steric bulk near the active site also affects enzyme-substrate binding, actively participating in the polycyclization pathway and the direction of the folded conformation [32].
Table 1.
Protein engineering of OSCs.
| OSC | GenBank | Substrates | Products | Key Residues and Mutants | Mechanism of action | Results | References |
|---|---|---|---|---|---|---|---|
| ItOSC2 | MT104569.1 | 2,3-oxidosqualene | α-amyrin | Y531/L256/L258 | formed the unusual Y-LL triplet at the active site | Yield greater than 87% | [28] |
| β-amyrin cyclase (E. tirucalli) | AB206470.1 | 2,3-oxidosqualene | β-amyrin | F474 | Appropriate volume of space near the B-ring formation site | F474L and F474 M increased yield by 60% and 82%, respectively | [29,30] |
| β-amyrin cyclase (E. tirucalli) | AB206470.1 | 2,3-oxidosqualene | lupeol | F413/Y259/W257 | Stabilization of intermediary cations via cation–π interactions | Generating lupeol synthase from the β-amyrin synthase | [31] |
| β-amyrin cyclase (E. tirucalli) | AB206470.1 | 2,3-oxidosqualene | triterpenoids | F728 | Stabilizes cationic intermediates and facilitates the ring expansion process | First demonstration of the criticality of cation-p interaction for OSCs | [32] |
| β-amyrin synthase (PNY) | AB009030.1 | 2,3-oxidosqualene | lupeol | W259L | W259 controls β-amyrin formation by stabilizing oleanyl cation | Produced lupeol as a major product | [33] |
| lupeol synthase (OEW) | AB025343.1 | 2,3-oxidosqualene | β-amyrin | L256W | L256 controls lupeol formation by stabilizing dammarenyl cation | Produced exclusively β-amyrin | [33] |
| AsHS1 | OM401324.1 | 2,3-oxidosqualene | hopenol B | F257/C366/Y410 | Promote rearrangement; Involved in the deprotonation process | Y410A mutant mainly produces hop-17(21)-en-3β-ol | [27] |
| AsHS2 | OM401325.1 | 2,3-oxidosqualene | hop-17(21)-en-3β-ol | A410 | The side chain size of site 410 leading to a difference in deprotonation positions | Could synthesize hopenol B alone | [27] |
| β-amyrin synthase (GsAS2) | KJ467352.1 | 2,3-oxidosqualene | β-amyrin | Y560 | Y560 and N610 together form a hydrophilic environment and improve the deprotonation of C13 | GsAS2 with Y560 had accumulated more oleanolic acid | [34] |
| SHC | AB007002.1 | 2,3-oxidosqualene | mono- and pentacyclic triterpene | D377C/V380E/V381A | The triple amino acids are responsible for the initiation of squalene cyclization | Cyclized 2,3-oxidosqualene to give mono- and pentacyclic triterpene | [35] |
| α-amyrin synthase (BfOSC3) | LC464980.1 | 2,3-oxidosqualene | α-amyrin | L258W | L258 mediates the lupeol specificity of OSCs | From mainly producing pentacyclic products to mainly producing tetracyclic products | [36] |
| OSC(OsOS) | MG932735.1 | 2,3-oxidosqualene | orysatinol | Y257 | The pi-electrons of Y257 stabilize the intermediary cation through cation-pi interaction | OsOSY257L,F,A resulted in complete loss of function | [37] |
| MdOSC1 | FJ032006.1 | 2,3-oxidosqualene | α-amyrin | N11I/P250H/P373S | Introduced hydrogen bonds and expand the substrate binding pocket | The yield of α-amyrin was increased 11-fold | [38] |
2.1.3. Improved product specificity through protein engineering of OSCs
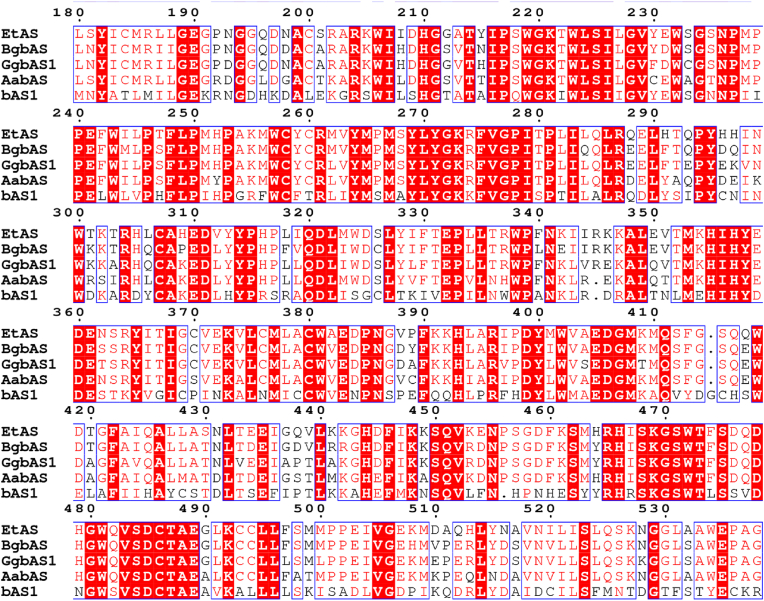
Although OSCs have generated sequence diversity during evolution, terpenoid synthases belonging to the same type have conserved motifs near the catalytic center (Fig. 4). For instance, β-amyrin synthase has a highly conserved MWCYCR motif, while lupine alcohol synthase keeps the conserved MLCYCR motif [28]. Commonly seen approaches based on the conserved motifs to improve the product specificity and catalytic activities of enzymes have been developed as follows: infrequent amino acids in conserved motifs are identified by comparing multiple amino acid sequences and are replaced with consensus amino acids or treated with site-directed mutations. Taking α-amyrin synthase as an example, there is seldom OSC that can specifically synthesize α-amyrin in nature. By comparing the amino acid sequences of ItOSC2 and representative plant OSCs, Wu et al. [28] identified Y531/L256/L258 as the three key active sites of ItOSC2. While amino acids L256 and L258 are close to lupanyl cation intermediates, preventing premature deprotonation of intermediates, Y531 is involved in the correct orientation of the lupanyl cation and other late intermediates, facilitating the deprotonation of the intermediates through the formation of hydrogen bond network between the hydroxyl group and water molecules. This results in highly specific α-amyrin products. However, site-directed mutagenesis based on sequence conservation analysis has certain limitations [44]. Since multiple amino acid mutations are prone to unpredictable epistasis, there are usually no more than 4 mutation sites, and the improvement in catalytic activity is limited to within 10-fold.
Fig. 4.
Alignment of partial amino acid sequences of β-amyrin synthases.
2.2. Protein engineering of P450s
In general, P450 oxidase and its reductase can specifically oxidize different sites of the triterpenoid skeleton to produce triterpenoid compounds with various structures by introducing hydroxyl, carboxyl, or epoxyl groups. P450s have attracted much attention due to their wide range of biocatalyst activities. However, as membrane-localized proteins, plant P450s have low expression levels, low activity, and poor stability in microorganisms, which have been considered key bottlenecks in pathway engineering. In this section, protein engineering in improving the catalytic properties of P450 and increasing the yield of terpenoids is systematically reviewed, including optimization of redox chaperones, protein fusion, engineering of transmembrane regions, and rational engineering (Table 2).
Table 2.
Protein engineering of P450s.
| P450 | Gene ID | Source | Substrates | Products | Key Residues and Mutants | Mechanism of action | Results | References |
|---|---|---|---|---|---|---|---|---|
| CYP72A63 | 11410378 | Medicago truncatula | 11-oxo-β-amyrin | glycyrrhetol | L509I | Increases the hydrophobicity of the active pocket ends | Yield is 90.6% | [9] |
| CYP72A63 | 11410378 | Medicago truncatula | glycyrrhetol | glycyrrhetinic acid (GA) | T338S | Improve the hydrophilicity of the binding pocket | GA increased by 7.1-fold | [9] |
| CYP72A63 | 11410378 | Medicago truncatula | 11-oxo-β-amyrin | 29-OH-11-oxo-β-amyrin | L398I | Rotate the substrate 180° to reverse the relative positions of C-30 and C-29 | 100% regional selectivity | [9] |
| CYP72A63 | 11410378 | Medicago truncatula | 11-oxo-β-amyrin | GA | T338S/W205A | Eliminate limitations on proton transfer | 36.4 ± 3.0 mg/L | [9] |
| CYP72A154 | 371940465 | Glycyrrhiza uralensis | β-amyrin | 30-OH-β-amyrin | L149/L398 | L149 and L398 determined the substrate orientation, which controlled its product regioselectivity | Further enhance the production of high-value triterpenoid compounds | [4] |
| CYP72A62v2 | 11409246 | Medicago truncatula | β-amyrin | 30-OH-β-amyrin | V398L | L398 determined the substrate orientation, which controlled its product regioselectivity | Changed from being regioselective at C-29 to producing only C-30 oxidation products | [4] |
| CYP87D20 | 101213710 | cucumber | cucurbitadienol | 11-H- cucurbitadienol | E286A | E286A plays a role in shuttling protons into the active site | Decreases the production of 11C-Cuol and 11C-20H-Cuol | [45] |
| TwCYP712K1 | 2081630515 | Tripterygium wilfordii | friedelin | polpunonic acid | S110F | Maked the substrate closer to the reaction centre | S110F increased the production of polpunonic acid | [46] |
2.2.1. Enhanced triterpenoid production via protein fusion of CYP and CPR
P450 oxidoreductase (CPR) is responsible for the transfer of electrons from NAD(P)H to the heme center of Class II microsomal P450s, and is the redox partner of P450. It is considered to be an important factor affecting the catalytic activity of P450. Therefore, P450s mining and combining with CPRs for optimized electron transfer efficiency are important. Wang et al. [47] introduced CPR (GuCYB5) from Glycyrrhiza uralensis into the engineered yeast strain. This successfully reduced the accumulation of β-amyrin and increased the glycyrrhetic acid titer to 545 μg/L. Lodeiro et al. [48] compared the pairing effects of different CPRs with CYP71BA1 and found that the catalytic efficiency of AtCPR1 from Arabidopsis thaliana was 7.4-fold higher than that of RhCPR from Rhizopus oryzae. Jin et al. [18] sequentially expressed LiCPR from Lotus japonicus and MtCPR from M. truncatula in Y. lipolytica, and the production of betulinic acid increased from 25.62 mg/L to 32.33 mg/L.
To improve the efficiency of P450, the fusion protein is employed. The fusion of plant P450 and CPR has many advantages: (1) the fusion protein enhances the coupling between CYP and CPR, as well as improves the electron transfer efficiency; (2) the continuous reaction is accelerated to obtain higher products titer by reducing the physical distance between the two enzymes [49]; (3) the loss of electrons caused by the lack of inner membrane structure is avoided.
It has been reported that the uncoupling of P450 and CPR generates reactive oxygen species (ROS), which can reduce the efficiency of electron transfer and even lead to the death of heterologous host cells in severe cases [50,51]. For example, when protopanaxadiol synthase (PPDS) and CPR (ATR1) are overexpressed in the protopanaxadiol synthesis pathway, the coupling between the two is poor, and a large amount of reactive oxygen species is released, seriously threatening the health of the host S. cerevisiae. Linker of different lengths was used by Zhao et al. [52] to construct the self-sufficient PPDS-ATR1 fusion. Compared with the previous modification, the catalytic activity of all fusion enzymes increased by about 4.5-fold, and the conversion efficiency of dammarenediol-II was as high as 96%. Meanwhile, the growth of host cells was effectively improved. In order to enhance the electron transfer efficiency between CYP716A12 and ATR1, Li et al. [53] fused the two proteins employing the linker GSTSSG. This reduced the precursor β-amyrin and increased the oleanolic acid production in Y. lipolytica, reaching 540.7 mg/L. The fusion of opCYP706M1 and opt46AtCPR1 accelerated electron transfer from NADPH to CYP706M1, enhanced the oxidation reaction, and the yield of the target product was 6-fold higher [54]. In Prokaryotes where the sites for electron transfer such as the endoplasmic reticulum are lacking, the P450/CPR fusion is more needed. However, in the taxadiene production pathway of E. coli, this fixed impression was broken. The yield of taxadiene decreased after the fusion of CYP725A4 with CPR, even though the plasmid copy number and promoter were adjusted or replaced. Although its origin is unsettled, it is considered that the overexpression of CPR has a negative impact. Resource competition or inefficient NADPH utilization may be the reason [55,56].
In addition, the formation of triterpenoids by P450 oxidation may involve the continuous catalysis of multiple enzymes, i.e., the catalytic product of the previous enzyme is the reaction substrate of the next enzyme. Reducing the spatial distance between multiple enzymes through fusion proteins will help improve the transfer efficiency of substrates between them and minimize the loss of intermediates during transport. Enzyme cascade protein fusion and synthetic scaffold protein have been proven to be effective tools in reducing the distance [57]. For example, Moon et al. [58] constructed a peptide scaffold using the protein-protein interaction domain and co-located a variety of key enzymes to increase the effective concentration of inositol. As a result, the titer of d-glucaric acid was increased by about 5-fold. Baek et al. [59] utilized the synthetic protein scaffold to realize the spatial co-localizations of the heterogeneous pathway enzymes in E. coli, resulting in a 3-fold increase in butyrate production.
2.2.2. Improved expression of CYP and CPR by engineering the transmembrane domain
Both CPR and CYP are inner membrane proteins anchored to the endoplasmic reticulum through their N-terminal. In prokaryotes lacking organelle membranes (such as E. coli), these two enzymes cannot be expressed normally and mostly exist in the form of inclusion bodies [60]. Truncation or modification of the N-terminal transmembrane region of P450 oxidase is an effective strategy to regulate P450 activity and solubility in prokaryotes. A stretch of polar charged amino acid AKKTSSKGK was inserted into the N-terminal of CYP71A16 to generate a CYP71A16 mutant, resulting in up to ∼50 mg/L active monooxygenase [61]. To express the CPR from Candida apicola in a functional form in E. coli, Girhard et al. [62] truncated the N-terminal of CPR with different amino acid lengths, and the modified recombinant protein showed higher expression and stronger solubility. This is the first report of eukaryotic CPR to transfer electrons to bacterial P450s. Improving the compatibility between enzyme and host is an important condition for efficient heterologous expression of P450s. Thus, small libraries of N-terminal expression tag chimeras were developed to enhance the expression of hydrophobic P450 enzymes in cell factories. Diterpene synthase EpCYP76A1 has not previously been expressed in microbial hosts, but when its N-terminal was inserted with a tag, the expression of the EpCYP76A1 mutant in E. coli increased by 9-fold [63].
2.2.3. Crystal structure of P450s
As a low soluble membrane protein, the crystal structure analysis of P450 is also difficult to achieve. X-ray diffraction, cryo-electron microscopy, and nuclear magnetic resonance have been applied to the studies of the three-dimensional structure of proteins. To date, however, only several P450 crystal structures such as CYP116B46 heme domain (PDB ID, 6GII) [64], CYP4B1 (PDB ID: 5t6q. 1. A) [65], and P450BM3 [66] have been obtained. Without structures of P450s proteins involved in triterpenoid biosynthesis, analysis of structure-function relationships cannot be performed comprehensively. Fortunately, even though the sequence similarity of P450 oxidases is only 10%, their three-dimensional architecture is similar [67]. According to this feature, in the absence of high homology P450 oxidase crystal structure, modeling using the resolved crystal structure as a template can also meet the needs of the semi-rational design. Sun et al. [9] used the crystal structure of CYP4B1 with 26.99% and 23.83% homology to CYP72A63 and CYP72A154, respectively, as a template, and conducted homology modeling to study the transformation strategy and precisely tune its catalytic properties to achieve the selective synthesis of target products.
2.2.4. Improved regioselectivity of P450s through protein engineering
In order to solve the problems of low synthesis efficiency and complex by-products of triterpenoids, researchers attempted to obtain candidate enzymes by mining corresponding transcriptome database and comparative analysis. However, when plant P450s are expressed in microorganisms, they often show substrate heterogeneity or poor regioselectivity. As a result, some problems such as scattered metabolic flow in the synthetic pathway, low synthesis efficiency, and the coexistence of multiple by-products, often occur and thus limit the effective synthesis of target products. Meanwhile, it aggravates the difficulty of separation and purification of downstream products.
Protein engineering based on rational design or semi-rational design has been proved to be an effective way to improve the regioselectivity of P450s and achieve the specific synthesis of target products. CYP72A63 from M. truncatula has catalytic activity on the C-29 and C-30 positions of 11-oxo-β-amyrin, resulting in a mixed product of various structural analogs [9]. After the mutation from threonine to hydrophilic serine at site 338 of CYP72A63, the distance between glycyrrhetol C-30 OH and heme-Fe is drawn closer by hydrophilic interaction, and C-29 is pushed away from heme-Fe. Therefore, the regional selectivity of the mutant T338S to C-30 was improved, and the yield of glycyrrhetinic acid was increased 7.1-fold. When the leucine at position 398 was mutated to isoleucine, the substrate 11-oxo-β-amyrin rotated nearly 180° and the relative positions of C-30 and C-29 were reversed. At this point, the mutant generated a highly regioselective 29-OH-11-oxo-β-amyrin, an isomer of glycyrrhetol. CYP720B1 from Pinus taeda has a certain degree of substrate promiscuity, and its substrate binding site is almost entirely hydrophobic residues that may mediate interactions with lipophilic substrates. Ignea et al. [68] mutated these residues into amino acids with different volumes and sizes but still hydrophobicity. The shape of the binding cavity of the mutant and the way of binding with the substrate were changed. For example, the G359A mutant retained only the activity of oxidizing C-18 and produced 18-hydroxy-miltiradiene with a 2-fold increase in production efficiency.
Site-directed mutagenesis based on sequence conservation analysis is also beneficial to find important amino acid residues, particularly for those involved in substrate and product specificity. Fanani et al. [4] identified L149 and L398 as the key amino acids to regional selectivity of CYP72A63 to β-amyrin C-30 through structural analysis of the binding protein model based on amino acid sequence alignment. When V398 of CYP72A62v2 was replaced by leucine, the mutant changed from being regioselective at C-29 to producing only C-30 oxidation products such as 30-hydroxy-β-amyrin.
2.2.5. Enhanced catalytic activity of P450s through protein engineering
The proton delivery pathway is important for the catalytic activity of P450s. Molecular docking results of CYP72A63 with the substrate 11-oxo-β-amyrin suggest that K236 has the function of transferring protons from solvent to alcohol-acid pair, while the bulky side chain of W205 may hinder proton transfer, thereby limiting the catalytic activity of the enzyme. After the mutation of tryptophan into alanine with a small volume, the restriction of steric hindrance on proton transfer was removed, and the double mutant CYP72A63 (T338S/W205A) resulted in the yield of glycyrrhetinic acid reaching 36.4 ± 3.0 mg/L [9]. Huang et al. remodeled the CYPs microenvironment by increasing NADPH supply based on overexpressing CYP72A63 (T338S/W205A) to achieve de novo synthesis of glycyrrhetic acid 3-O-mono-β-D-glucuronide (GAMG) in S. cerevisiae, and the yield was increased from 0.02 μg/L to 92.00 μg/L [69]. Thus, unblocking the restriction that impedes proton transport is an effective way to increase specific P450 activity.
Seifert et al. conducted systematic computational analysis for the crystal structures of 29 P450s and the sequences of 6379 CYPs. Their study showed that two amino acids adjacent to the heme center were both hydrophobic, a phenomenon that existed in most of the analyzed P450s. F87 and A328 of P450BM3 were indicated to be the corresponding two hydrophobic residues. By replacing them with nonpolar amino acids (e.g. Ala, Val, Phe, Leu, and lle), the mutants showed similar or higher activity. In addition, the large volume amino acid Phe87 located near the binding pocket is also considered to be the key active site of P450BM3 [70]. Dietrich et al. [71] mutated phenylalanine into small volume alanine (F87A), which enhanced the coupling efficiency of the epoxidation of uscatadiene, confirming that steric hindrance does affect the binding of the enzyme to the substrate.
2.3. Protein engineering of UGTs
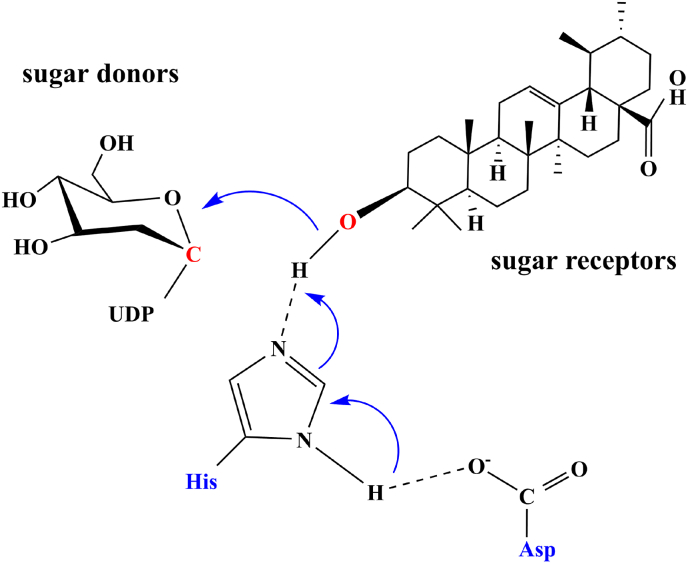
UDP-glycosyltransferase mediates the glycosylation reaction of triterpenoids. The resulting compounds are called triterpenoid saponins, which usually have better physicochemical properties and are easier to be stored and transported in plants. From the perspective of structural characteristics, UDP-glycosyltransferase has two binding domains. One domain is located in a highly conserved region consisting of 44 amino acids at the C-terminus, known as the plant secondary product glycosyltransferase (PSPG) motif, involved in the recognition and binding of sugar donors. The other is the sugar receptor binding domain, which is mainly involved in the recognition and binding of sugar receptors. The amino acid with high N-terminal variability is an important reason for the poor substrate selectivity and specificity of glycosyltransferases. The highly conserved histidine, located between the binding domain of sugar donor and sugar receptor, is the catalytic center of glycosyltransferase, and the neighboring aspartic acid plays a conformational and charge balancing role. The mutation of histidine and/or aspartic acid leads to the loss of enzyme activity [72], suggesting that the catalytic pair of His-Asp is an indispensable key amino acid residue (Fig. 5). Glycosylation promotes the structural and functional diversity of natural products and provides many possibilities and opportunities for the development of the food and pharmaceutical industries. For example, while most of the basic glycosides in ginseng, such as protopanaxadiol and protopanaxatriol, have diverse pharmacological activities, ginsenosides (such as ginsenoside Rh2 and ginsenoside Rg3) are the main bioactive ingredients of ginseng and have better applications in the pharmaceutical industry [73]. The glycosyltransferase UGT73C11 was used to catalyze the C-3 hydroxyl of GA to GAMG, which has a sweetness of 941-fold higher than that of sucrose and has better solubility and wider pharmacological activity [74,75]. Precise glycosylation of C-3 OH of triterpenoids can efficiently produce higher value-added derivatives such as GA-3-O-glucose, calenduloside E, and oleanolic acid 3-O-glucose.
Fig. 5.
Mechanism of SN2 bimolecular nucleophilic substitution reaction of UGTs.
Even though the microbial synthesis of some terpenoid saponins has been achieved, it is still a huge challenge to realize their industrialization due to the following difficulties in the application of reported UGTs. Firstly, the low regioselectivity of UGT inhibits the efficient production of target products. For example, UGT73C33 from licorice shows high activity towards C-3 OH and C-30 COOH of pentacyclic triterpenoids but produced three glycosides due to poor substrate selectivity [76]. The glycosyltransferase UGT73C11 from Barbarea vulgaris displays catalytic activity for oleanolic acid C-3 OH and C-28 COOH, resulting in mixed products, which increases the cost of downstream separation and purification [77]. Secondly, the number of introduced sugar groups affecting the pharmacological activity is difficult to control [78,79]. The C-3 OH of GA is more likely to be introduced into two glucuronic acid groups to generate glycyrrhizic acid (GL) than to introduce one to generate GAMG. Meanwhile, the bioavailability of GL is much lower than that of GAMG [77]. Thirdly, the low catalytic activity of UGT leads to the low yield of terpenoid saponins in microbes and thus limits the industrial applications of microbial synthesis. The glucuronyltransferase OAGT identified by Tang et al. [80] from the transcriptome data of Panax zingiberensis showed the catalytic activity of oleanolic acid C-3 OH after recombinant expression in E. coli. However, the target product, calenduloside E, was only obtained at a very low yield. The glycosyltransferase GuGT14 identified in licorice by bioinformatics can specifically synthesize GAMG, but the conversion rate is only 40% [81]. UGT74AG5 expressed in S. cerevisiae showed high regioselectivity for ursolic acid C-28 COOH, and synthesized a rare ursolic acid 28-O-β-d-glucopyranoside, but the yield was very low [82]. Fourthly, the high specificity of UGT for sugar donors restricts its applications. For example, soyasapogenol glucuronosyl-transferase (UGASGT), isolated from the seeds of Glycine max, synthesizes oleanolic acid 3-O-β-glucuronide using UDP-glucuronic acid rather than UDP-glucose as the sugar donor [83]. The last one is the substrate heterogeneity of UGT. With UDP-Glc, UDP-Rha, UDP-Gal, UDP-Ara, UDP-Xyl as sugar donors, the corresponding sugar groups can be introduced into glycyrrhetinic acid through UGT73C33 and UGT73F24, but the catalytic activity of UGT towards different sugar donors is distinct [76]. GmSGT2 from G. max catalyzes the production of triterpenoid saponins from UDP-Gal, UDP-Glc, UDP-Ara and UDP-Xyl, of which UGT has the highest catalytic activity towards UDP-Gal [84]. In general, the above five problems still limit the synthesis of terpenoid saponins in microbial cell factories. As shown in Table 3, protein engineering is a helpful strategy to address these problems and achieve high regioselectivity and efficient glycosylation of UGT.
Table 3.
Protein engineering of UGTs.
| UGT | Substrates | Products | Key Residues and Mutants | Mechanism of action | Results | References |
|---|---|---|---|---|---|---|
| UGT73F24 | GA | GA-3-O-glucose | I23G/L84 N | Reduce steric hindrance; Expanded binding pocket | Activity increased by 4.1‐fold | [76] |
| UGTSr | stevioside | rebaudioside A | H25/D124 | His25 abstracts a proton from the acceptor; Asp124 stabilizes His25 by forming hydrogen bonds | Stability and specificity increased | [85] |
| UGTPg71A29 | Rh1 | Rg1 | Q283 | Affects conformation through hydrophobic interactions | 80% reduction in activity after mutation | [86] |
| UGT74AC1 | mogrol | mogroside IE | T79Y/L48 M/R28H/L109I/S15A/M76L/H47R | The hydrophobic interactions between enzyme and substrate that play a critical role in anchoring triterpenoids at the active center | Catalytic ability increased by 102–104 folds | [72] |
| UGT51 | protopanoxadiol | ginsenoside Rh2 | S81A/L82A/V84A/K92A/E96K/S129A/N172D | Residues were 9–20 Å away from the docked substrate and involved in a significant conformational change upon substrate binding | Catalytic efficiency (kcat/Km) increased by ∼ 1800-fold | [87] |
| UGTPg45 | protopanoxadiol | ginsenoside Rh2 | Q222H/A322V | Enhanced binding to PPD; Affects the configuration of the binding pocket | Rh2 increased by 70% | [88] |
| UGT73P12 | GAMG | gycyrrhizin | R32 | Generating salt Bridges and hydrogen bonds; Improves affinity with substrates | Identified Arg32 as the essential residue of UGT73P12 | [89] |
2.3.1. Crystal structure of UGTs
Significantly improving the catalytic efficiency of plant UGT remains a challenge due to the lack of sufficient crystal templates for structural modeling of enzymes and in-depth understanding of their reaction mechanisms. At present, lots of glycosyltransferases (GTs) have been excavated and identified [90]. However, only 13 plant UGT crystal structures have been analyzed and included in Protein Data Bank Database (PDB: https://www.rcsb.org/), such as UGT74F2 from Arabidopsis [91], UGT76G1 from Stevia Rebaudiana [92] and PtUGT1 from Polygonum tinctorium [93]. A smaller number of UGT crystals were identified in the database as responsible for triterpenoid glycosylation, including MtUGT71G1 from M. truncatula [94], ScUGT51 from S. cerevisiae [95], and UGT74AC1 from Siraitia grosvenorii [72]. The crystal structure is important for protein engineering of UGTS. UGT74AC1 from Siraitia grosvenorii (SgUGT74AC1) has a high region-specific but low catalytic activity for glycosylation of mogrol C3-OH. In order to understand the molecular basis of enzyme and substrate binding, Li et al. [72] used X-ray crystallography to elucidate the crystal structure of SgUGT74AC1 (PDB ID: 6L90) and its complex with substrate UDP-glucose (PDB ID: 6L8Z). In addition, error-prone polymerase chain reaction (EP-PCR) and sequence alignment combined with semi-rational design were used to generate mutants to improve the catalytic efficiency of UGT74AC1, which significantly increased the catalytic capacity of triterpenoid glycosylation by 102-104 times. Finally, they revealed that the enhanced catalytic activity of UGT mutants was attributed to the improved hydrophobic interaction between enzyme and substrate, which promoted the anchoring of triterpenoids at the active center. Currently, the crystal structures of most enzymes have not been resolved, but three-dimensional structures obtained by homology modeling can be used for molecular dynamics simulations to rationally modify proteins.
2.3.2. Enhanced catalytic activity of UGTs through protein engineering
Biochemical or amino acid sequence information is important for selecting mutated amino acids. The key amino acids in the active pocket usually determine or directly affect the catalytic activity and substrate specificity of the enzyme. Zhang et al. [76] improved the activity of UGT73F24 towards glycyrrhetinic acid C3-OH by molecular docking and mutation of key residues, leading to the de novo synthesis of GAMG in engineered yeast. They found that L84 is located on a flexible loop at the substrate pocket entrance, which has a certain steric hindrance effect. After L84 is mutated to asparagine, the steric hindrance decreased and the active pocket increased. The combined mutation of L84 and I23 enhanced the activity of UGT73F24 (I23G/L84N) by 4.1-fold [76]. Through nine rounds of iterative saturation mutation of amino acid residues in the glycosyltransferase-receptor complex model, UGT51 was engineered into a glycosyltransferase for the efficient synthesis of ginsenoside Rh2. The catalytic efficiency (kcat/Km) was improved by about 1800 times [87]. In the 3D structure of UGTSL2 from S. rebaudiana, N358 is close to the substrate channel and interacts with UDPG through two hydrogen bonds. Mutation of N358 to phenylalanine in the UGTSL2 (N358F) multi-enzyme reaction system increased the production of rebaudioside D by 60% [96]. Despite the key residus interact directly with the substrate, the catalytic results of the triterpenoid glycosyltransferase mutant UGT74AC1 (T79Y/L48M/R28H/L109I) showed that even the amino acid residues with a long distance from the substrate can also affect the glycosyltransferase activity. Moreover, the mutation of S15A/M76L/H47R enhanced the hydrophobic interaction with the substrate and the binding affinity. As a result, the catalytic activity of mutant UGT74AC1 (T79Y/L48M/R28H/L109I/S15A/M76L/H47R) was 4.17 104 -fold higher than that of wild type [72]. Therefore, the disclosure of key sites is more conducive to the analysis of enzyme catalytic mechanism and protein engineering. Lu et al. [86] constructed a protein model of UGTPg71A29 using the crystal structure of MtUGT71G1 as a template, and speculated that key sites played a role in the catalytic process. Among them, Q283 affected the spatial conformation of UGTPg71A29 by forming a hydrophobic interaction with the substrate, and promoted the conversion of Rd to Rb1. Moreover, G18, M22, Q251 and S253 are also important for maintaining the catalytic activity since the catalytic activity of glycosyltransferase decreases when any of the four sites are mutated.
Activity-based sequence conservation analysis (ASCA) is also an effective method to improve the selection specificity and catalytic activity of UGT. UGT74AC1 was used for sequence alignment with 12 plant UGTs that catalyze mogrosol by Li et al. [72]. They performed combinatorial mutations of four residues (S15A, L109I, M76L and H47R) that were shown to be present at high frequency. As a result, the catalytic activity of the UGT74AC1 mutant was increased 96-fold. Both UGTPg45 and UGTPn50 have the function of catalyzing protopanaxadiol (PPD) to ginsenoside Rh2. By comparing the amino acid sequences of them, it was found that two amino acids were missing in UGTPn50, corresponding to A322 and E323 of UGTPg45. The insertion of corresponding amino acids into UGTPn50 was carried out followed by the modification according to beneficial mutations (Q222H and A322V) of UGTPg45. As a result, the catalytic activity of mutant UGTPn50 was 75.4% higher than that of wild type [88]. Yin et al. [97] identified conservative amino acid residues and mutated them through sequence comparison of methionine adenosyltransferase (MAT) from different sources. The mutant MAT (I303V) showed higher catalytic activity than the wild type. These results suggest that analogizing beneficial mutations by sequence comparison is effective to improve the catalytic efficiency of wild-type glycosyltransferase.
2.3.3. Expanded sugar-donor selectivity through protein engineering
Most UGTs are highly specific for the selection of sugar donors, which not only increase the production cost of compounds, but also limit the structural diversity of saponins. UDP-glucose is the sole sugar donor for UGT92G1 and UGT92G2 from G. max [98]. UGT73P12 from licorice prefers to use UDP-glucuronic acid rather than UDP-glucose or UDP-galactose as the sugar donor [89]. Although the chassis host has been modified and optimized, it is still difficult to provide sufficient and diverse glycosylated donors. Modification of glycosyltransferase through the underlying mechanism of substrate selectivity and specificity of glycosylation can effectively solve this problem. Chen et al. [99] analyzed the structural features of UGT78H2 and found that the 23rd residue of the plant secondary product glycosyltransferase (PSPG) motif, which corresponds to the 360th residue in UGT78H2, influences the interactions of enzyme and ligand by forming hydrogen bonds with the donor. When lysine was mutated to asparagine at site 360 and combined with the beneficial mutation N340P, the catalytic activity of the double mutant for UDP-galactose was increased by 23%, and the selectivity of sugar donors was also broadened. In addition to using UDP-glucuronic acid and UDP-galactose, it could also use UDP-glucose as a substrate. Similarly, R350 in the PSPG conserved motif of UGT88D7 is a key amino acid for recognizing UDP-glucuronic acid. When arginine was replaced by tryptophan, the utilization efficiency of UDP-glucuronic acid was significantly reduced. Interestingly, the mutant obtained novel UDP-glucose transfer activity [100]. The residue R25 side chain of the N-terminal in UGT94B1 is positively charged and directed towards the carboxylic acid group of the UDP-GlcUA sugar donor. Osmani et al. [101] mutated R25 as the active site to produce mutant proteins R25S, R25G, R25K and R25P, which significantly reduced the catalytic activity of the sugar donor UDP-GlcUA. All the other mutants except R25P showed a preference for UDP-Glc, and the catalytic activity was increased 3–4 fold. When D374 of UGT76E2 was mutated to glutamic acid, the amino acid side chain at position 374 was extended and closer to the –OH groups of the sugar, which promoted the formation of hydrogen bonds between the mutant and UDP-Gal. And UGT76E2 (D374E) showed UDP-Gal activity that was not found in the wild type [102]. The glycosyltransferase UGT73P12 is responsible for the last step in the de novo synthesis of glycyrrhizic acid, which function is to introduce a glucuronic acid group on the basis of GAMG. The positively charged side chain of R32 is considered to be the key amino acid to the specific recognition of UDP-glucuronic acid by UGT73P12. The reason is that under the combined influence of the resultant salt bridge and electrostatic interaction, the positively charged side chain of R32 is oriented toward the negatively charged carboxyl group in UDP-glucuronic acid that provides high affinity for it. Following mutation of arginine to serine, the sugar-donor substrate of UGT73P12 (R32S) was changed from UDP-glucuronic acid to UDP-glucose and UDP-galactose [89].
3. Perspective
Metabolic engineering in recombinant microbial cell factories shows great potential for the production of various triterpenoids. At the same time, candidate genes are mined according to the sequence libraries and information on gene expression profiles generated by high-throughput sequencing technologies and the databases provided by the PhytoMetaSyn project [103], the 1 KP project [104] and the Medicinal Plant Genomics Resource project. Although some progress has been made in the heterologous synthesis of plant triterpenoids by microorganisms, great challenges still exist for their commercial production. The lack of efficient key enzymes is one of the most critical limiting factors, especially the key enzymes in the triterpenoid synthetic pathway such as OSCs, P450s, and UGTs. Recently, the discovery of cellulose synthase-like (CSLs) enzymes as triterpenoid glucuronyltransferases break the traditional view that glycosylation is only catalyzed by UDP-dependent glycosyltransferases and fill a knowledge gap in the biosynthetic pathways of triterpenoid saponins. Jozwiak et al. [105] successfully catalyzed medicagenic acid to medicagenic acid 3-O-glucuronide using cellulose synthase-like G (CSLG) belonging to GT family 2. At the same time, they performed site-directed mutagenesis of CSLG, revealing that the active site amino acids S442 and K483 are important for the enzyme to specifically recognize UDP-GlcA. Two other cellulose synthase-like M-subfamily enzymes (AeCSLMs) derived from Aralia elata have been reported to catalyze oleanolic acid and echinocystic acid to produce calenduloside E and echinocystic acid 3-O-glucuronopyranoside, respectively, and have been applied to the de novo synthesis of oleanane-type pentacyclic triterpenes in yeast [106]. The cellulose synthase superfamily-derived glycosyltransferase (CSyGT) can introduce glucuronic acid at C-3 of Glycyrrhetinic acid. Therefore, it has been used in the de novo synthesis of glycyrrhizin [107]. Although the catalytic mechanism of the CSyGTs is still unclear, Soo et al. hypothesise that the transmembrane topology of CSyGTs is critical for triterpene glycosylation. More novel enzymes have been discovered in triterpenoid synthesis. Tao et al. [108] discovered two fungal chimeric class I triterpene synthases that use dimethylallyl diphosphate and isopentenyl diphosphate or hexaprenyl diphosphate as substrates to synthesize triterpenoids, which overturns the long-held belief that all triterpenoids are synthesized exclusively from squalene. The discovery and mining of enzymes in the synthesis of triterpenoids provides more candidates that can be employed in the microbial production.
Despite the obtain of novel efficient key enzymes, the modifications of known enzymes are alternative ways for efficient production. Protein engineering of the key enzymes have attracted much attention of the researchers. In recent years, rational design of enzymes involved in triterpenoid biosynthesis has become a trend by means of sequence alignment, molecular evolutionary analysis and protein homology modeling. Successful examples include ItOSC2 (Y531/L256/L258) [28], UGT71G1 (Y202A) [109], CYP72A63 (T338S/W205A) [9], etc. Significant advances in protein engineering have accelerated the improvements in enzyme specificity, activity, and stability. Moreover, the protein engineering of key enzymes could lead to novel reactions that do not occur in nature and thus generate different types of specialized metabolites. The crystal structure of enzymes is the basis for the elucidation of enzyme-substrate binding and interactions, which helps to optimize the catalytic properties of enzymes through protein design. Even though the docking of enzyme-substrate active sites can be studied by indirect methods such as homology modelling and molecular simulation, the semi-rational design lacking crystallographic data is still challenging. Therefore, accelerating the acquisition of the three-dimensional structure of enzyme molecules is one of the main problems need to be solved in protein engineering [110].
In recent years, the technologies for obtaining the three-dimensional structure of enzymes mainly include nuclear magnetic resonance and X-ray crystallography. Padyana et al. [111] obtained the crystal structure of human squalene epoxide (SQLE), a key rate-limiting enzyme in cholesterol biosynthesis, through the most classical X-ray crystallography. The key conformational rearrangement necessary for the binding of inhibitors including NB-598 and compound-4″ was determined, and the structural basis of catalytic epoxidation of SQLE was clarified. However, there are many difficulties in the crystallization process for some proteins, such as membrane proteins. Fortunately, the improvement of single-particle electron cryomicroscopy (CRYo-EM) resolution makes up for this deficiency. Yao et al. [112] performed high-resolution cryoEM structural analysis of AcrB reconstituted into liposomes by cryo-EM to obtain the membrane protein structure in physiological state. Gong et al. [113] measured the three-dimensional structure of human cholesterol intracellular transporter Niemann-pick C1 (NPC1) and its complex with EboV-GPCL by CRYO-EM. Qian et al. [114] analyzed the near-atomic resolution cryo-EM structure of the full-length human ABCA1 protein, which laid an important foundation for understanding the mechanism of ABCA1-mediated lipid export and the pathogenesis of related diseases. AlphaFold 2, a representative combination of artificial intelligence and biophysical ideas, will become an important tool for structural biology. It can provide an initial structural model for X-ray crystal diffraction, avoiding the embarrassment that the structural model cannot be established due to lack of phase information. More targeted mutation sites to stabilize protein structures can also be provided. AlphaFold 2 can also guide the classification of 2D images in single-particle cryo-EM structure analysis. Obviously, the development of structural biology will be driven by the combination of AlphaFold 2 and experimental structural biology.
Different from structure-guided protein engineering, the enzyme gene functional modification method based on Rosetta and the amino acid co-evolution information calculated by GREMLIN is suitable for enzyme molecules with rare crystal structures, which have been used to optimize high-resolution protein structures [115]. Ma et al. changed the function of cucurbitadienol synthase from cucumber by Rosetta, resulting in a conversion of catalytic activity from tetracyclic products to pentacyclic products [116]. Li et al. [117] transformed the multifunctional P450 enzyme CYP87D20 involved in the synthesis of cucurbitacin C into a mono-oxygenase, which has the function of directionally catalyzing cucurbitadienol to generate 11-hydroxyl cucurbitadienol, the precursor of mogrol. Zhang et al. performed protein surface engineering based on the CYP87D20 mutant, and combined with amino acid site mutations in the active region to further improve the catalytic activity of 11-hydroxycucurbitadienol synthase [45].
Protein engineering is expected to be developed as a useful approach for the commercial production of useful triterpenoids. This strategy also opens a new perspective for synthetic biology by constructing mutant libraries of key enzymes to generate triterpenoid synthases with novel structures and prominent functions, as well as more and more functional triterpenoid compounds. In future, enzyme catalysts for any chemical reaction could be designed as machine learning, multi-site mutagenesis, high-throughput biosensors, and other technologies are more and more matured. The employment of protein engineering in microbial synthesis of plant triterpenoids will greatly accelerate their industrial application.
Author contributions
Yan Luo: Writing - original draft, Writing - review & editing, Visualization; Yaozhu Jiang: Partial writing; Linhao Chen: Investigation, Visualization; Chun Li: Supervision; Ying Wang: Project administration; Supervision; Validation; Writing - review & editing.
Declaration of competing interest
The authors declare no competing interest.
Acknowledgements
The authors would like to acknowledge the funding support from the National Key Research and Development Program of China (2019YFA0905700, 2018YFA0901800), the National Natural Science Foundation of China (22078020) and Young Elite Scientists Sponsorship Program by CAST (2019QNRC001).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Xu G.B., Xiao Y.H., Zhang Q.Y., Zhou M., Liao S.G. Hepatoprotective natural triterpenoids. Eur J Med Chem. 2018;145:691–716. doi: 10.1016/j.ejmech.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 2.Dembitsky V.M., Gloriozova T.A., Poroikov V.V. Antitumor profile of carbon-bridged steroids (cbs) and triterpenoids. Mar Drugs. 2021;19 doi: 10.3390/md19060324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsui S., Matsumoto H., Sonoda Y., Ando K., Aizu-Yokota E., Sato T., et al. Glycyrrhizin and related compounds down-regulate production of inflammatory chemokines il-8 and eotaxin 1 in a human lung fibroblast cell line. Int Immunopharm. 2004;4:1633–1644. doi: 10.1016/j.intimp.2004.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fanani M.Z., Fukushima E.O., Sawai S., Tang J., Ishimori M., Sudo H., et al. Molecular basis of C-30 product regioselectivity of legume oxidases involved in high-value triterpenoid biosynthesis. Front Plant Sci. 2019;10:1520. doi: 10.3389/fpls.2019.01520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou M., Zhang R.-H., Wang M., Xu G.-B., Liao S.-G. Prodrugs of triterpenoids and their derivatives. Eur J Med Chem. 2017;131:222–236. doi: 10.1016/j.ejmech.2017.03.005. https://doi.org/10.1016/j.ejmech.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Huang G., Lv M., Hu J., Huang K., Hong X. Glycosylation and activities of natural products. Mini Rev Med Chem. 2016;16:1013–1016. doi: 10.2174/138955751612160727164559. [DOI] [PubMed] [Google Scholar]
- 7.Seki H., Tamura K., Muranaka T. P450s and ugts: key players in the structural diversity of triterpenoid saponins. Plant Cell Physiol. 2015;56:1463–1471. doi: 10.1093/pcp/pcv062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun W., Qin L., Xue H., Yu Y., Ma Y., Wang Y., et al. Novel trends for producing plant triterpenoids in yeast. Crit Rev Biotechnol. 2019;39:618–632. doi: 10.1080/07388551.2019.1608503. [DOI] [PubMed] [Google Scholar]
- 9.Sun W., Xue H., Liu H., Lv B., Yu Y., Wang Y., et al. Controlling chemo- and regioselectivity of a plant P450 in yeast cell toward rare licorice triterpenoid biosynthesis. ACS Catal. 2020;10:4253–4260. doi: 10.1021/acscatal.0c00128. [DOI] [Google Scholar]
- 10.Guo J., Zhou Yongjin J., Matthew L Hillwig, Shen Y., Yang L., Wang Y., et al. CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proc Natl Acad Sci USA. 2013;110:12108–12113. doi: 10.1073/pnas.1218061110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo J., Ma X., Cai Y., Ma Y., Zhan Z., Zhou Y.J., et al. Cytochrome p450 promiscuity leads to a bifurcating biosynthetic pathway for tanshinones. New Phytol. 2016;210:525–534. doi: 10.1111/nph.13790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with alphafold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maldonado M.R., Alnoch R.C., de Almeida J.M., Santos L A d, Andretta A.T., Ropaín R. d P.C., et al. Key mutation sites for improvement of the enantioselectivity of lipases through protein engineering. Biochem Eng J. 2021;172 doi: 10.1016/j.bej.2021.108047. [DOI] [Google Scholar]
- 14.Werner N., Ramirez-Sarmiento C.A., Agosin E. Protein engineering of carotenoid cleavage dioxygenases to optimize β-ionone biosynthesis in yeast cell factories. Food Chem. 2019;299 doi: 10.1016/j.foodchem.2019.125089. [DOI] [PubMed] [Google Scholar]
- 15.Zhang G., Cao Q., Liu J., Liu B., Li J., Li C. Refactoring β-amyrin synthesis in Saccharomyces cerevisiae. AIChE J. 2015;61:3172–3179. doi: 10.1002/aic.14950. [DOI] [Google Scholar]
- 16.Li D., Zhang Q., Zhou Z., Zhao F., Lu W. Heterologous biosynthesis of triterpenoid dammarenediol-II in engineered Escherichia coli. Biotechnol Lett. 2016;38:603–609. doi: 10.1007/s10529-015-2032-9. [DOI] [PubMed] [Google Scholar]
- 17.Li D., Wu Y., Zhang C., Sun J., Zhou Z., Lu W. Production of triterpene ginsenoside compound k in the non-conventional yeast Yarrowia lipolytica. J Agric Food Chem. 2019;67:2581–2588. doi: 10.1021/acs.jafc.9b00009. [DOI] [PubMed] [Google Scholar]
- 18.Jin C.-C., Zhang J.-L., Song H., Cao Y.-X. Boosting the biosynthesis of betulinic acid and related triterpenoids in Yarrowia lipolytica via multimodular metabolic engineering. Microb Cell Factories. 2019;18:77. doi: 10.1186/s12934-019-1127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muhammad A., Feng X., Rasool A., Sun W., Li C. Production of plant natural products through engineered Yarrowia lipolytica. Biotechnol Adv. 2020;43 doi: 10.1016/j.biotechadv.2020.107555. [DOI] [PubMed] [Google Scholar]
- 20.Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., et al. Swiss-model: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., et al. Swiss-model: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tunyasuvunakool K., Adler J., Wu Z., Green T., Zielinski M., Zidek A., et al. Highly accurate protein structure prediction for the human proteome. Nature. 2021;596:590–596. doi: 10.1038/s41586-021-03828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seeliger D., de Groot B.L. Ligand docking and binding site analysis with pymol and autodock/vina. J Comput Aided Mol Des. 2010;24:417–422. doi: 10.1007/s10822-010-9352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trott O., Olson A.J. Autodock vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thoma R., Schulz-Gasch T., D 'Arcy B., Benz J., Aebi J., Dehmlow H., et al. Insight into steroid scaffold formation from the structure of human oxidosqualene cyclase. Nature. 2004;432:118–122. doi: 10.1038/nature02993. [DOI] [PubMed] [Google Scholar]
- 26.Wu T.-K., Liu Y.-T., Chang C.-H., Yu M.-T., Wang H.-J. Site-saturated mutagenesis of histidine 234 of Saccharomyces cerevisiae oxidosqualene-lanosterol cyclase demonstrates dual functions in cyclization and rearrangement reactions. J Am Chem Soc. 2006;128:6414–6419. doi: 10.1021/ja058782p. [DOI] [PubMed] [Google Scholar]
- 27.Liang M., Zhang F., Xu J., Wang X., Wu R., Xue Z. A conserved mechanism affecting hydride shifting and deprotonation in the synthesis of hopane triterpenes as compositions of wax in oat. Proc Natl Acad Sci USA. 2022;119 doi: 10.1073/pnas.2118709119. https://doi.org/doi:10.1073/pnas.2118709119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu S., Zhang F., Xiong W., Molnár I., Liang J., Ji A., et al. An unexpected oxidosqualene cyclase active site architecture in the iris tectorum multifunctional α-amyrin synthase. ACS Catal. 2020;10:9515–9520. doi: 10.1021/acscatal.0c03231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito R., Masukawa Y., Nakada C., Amari K., Nakano C., Hoshino T. Β-amyrin synthase from Euphorbia tirucalli. Steric bulk, not the π-electrons of phe, at position 474 has a key role in affording the correct folding of the substrate to complete the normal polycyclization cascade. Org Biomol Chem. 2014;12:3836–3846. doi: 10.1039/C4OB00064A. [DOI] [PubMed] [Google Scholar]
- 30.Morikubo N., Fukuda Y., Ohtake K., Shinya N., Kiga D., Sakamoto K., et al. Cation−π interaction in the polyolefin cyclization cascade uncovered by incorporating unnatural amino acids into the catalytic sites of squalene cyclase. J Am Chem Soc. 2006;128:13184–13194. doi: 10.1021/ja063358p. [DOI] [PubMed] [Google Scholar]
- 31.Ito R., Nakada C., Hoshino T. β-amyrin synthase from Euphorbia tirucalli L. Functional analyses of the highly conserved aromatic residues Phe413, Tyr259 and Trp257 disclose the importance of the appropriate steric bulk, and cation–π and CH–π interactions for the efficient catalytic action of the polyolefin cyclization cascade. Org Biomol Chem. 2016;15:177–188. doi: 10.1039/c6ob02539k. [DOI] [PubMed] [Google Scholar]
- 32.Ito R., Hashimoto I., Masukawa Y., Hoshino T. Effect of cation–π interactions and steric bulk on the catalytic action of oxidosqualene cyclase: a case study of phe728 of β-amyrin synthase from Euphorbia tirucalli L. Chem Eur J. 2013;19:17150–17158. doi: 10.1002/chem.201301917. [DOI] [PubMed] [Google Scholar]
- 33.Kushiro T., Shibuya M., Masuda K., Ebizuka Y. Engineering lupeol synthase into β-amyrin synthase. J Am Chem Soc. 2000;122:6816–6824. doi: 10.1021/ja0010709. [DOI] [Google Scholar]
- 34.Liu Y., Zhao Z., Xue Z., Wang L., Cai Y., Wang P., et al. An intronless β-amyrin synthase gene is more efficient in oleanolic acid accumulation than its paralog in gentiana straminea. Sci Rep. 2016;6 doi: 10.1038/srep33364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dang T., Prestwich G.D. Site-directed mutagenesis of squalene-hopene cyclase: altered substrate specificity and product distribution. Chem Bio. 2000;7:643–649. doi: 10.1016/S1074-5521(00)00003-X. [DOI] [PubMed] [Google Scholar]
- 36.Srisawat P., Fukushima E.O., Yasumoto S., Robertlee J., Suzuki H., Seki H., et al. Identification of oxidosqualene cyclases from the medicinal legume tree Bauhinia forficata: a step toward discovering preponderant α-amyrin-producing activity. New Phytol. 2019;224:352–366. doi: 10.1111/nph.16013. [DOI] [PubMed] [Google Scholar]
- 37.Xue Z., Tan Z., Huang A., Zhou Y., Sun J., Wang X., et al. Identification of key amino acid residues determining product specificity of 2,3-oxidosqualene cyclase in oryza species. New Phytol. 2018;218:1076–1088. doi: 10.1111/nph.15080. [DOI] [PubMed] [Google Scholar]
- 38.Yu Y., Rasool A., Liu H., Lv B., Chang P., Song H., et al. Engineering Saccharomyces cerevisiae for high yield production of α-amyrin via synergistic remodeling of α-amyrin synthase and expanding the storage pool. Metab Eng. 2020;62:72–83. doi: 10.1016/j.ymben.2020.08.010. [DOI] [PubMed] [Google Scholar]
- 39.Yu Y., Chang P., Yu H., Ren H., Hong D., Li Z., et al. Productive amyrin synthases for efficient α-amyrin synthesis in engineered Saccharomyces cerevisiae. ACS Synth Biol. 2018;7:2391–2402. doi: 10.1021/acssynbio.8b00176. [DOI] [PubMed] [Google Scholar]
- 40.Andre C.M., Legay S., Deleruelle A., Nieuwenhuizen N., Punter M., Brendolise C., et al. Multifunctional oxidosqualene cyclases and cytochrome P450 involved in the biosynthesis of apple fruit triterpenic acids. New Phytol. 2016;211:1279–1294. doi: 10.1111/nph.13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z., Guhling O., Yao R., Li F., Yeats T.H., Rose J.K., et al. Two oxidosqualene cyclases responsible for biosynthesis of tomato fruit cuticular triterpenoids. Plant Physiol. 2011;155:540–552. doi: 10.1104/pp.110.162883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Basyuni M., Oku H., Inafuku M., Baba S., Iwasaki H., Oshiro K., et al. Molecular cloning and functional expression of a multifunctional triterpene synthase cdna from a mangrove species kandelia candel (L.) druce. Phytochemistry (Oxf) 2006;67:2517–2524. doi: 10.1016/j.phytochem.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 43.Tsutomu, Hoshino, Kunio, Shimizu Tsutomu, Sato Deletion of the Gly600 residue ofalicyclobacillus acidocaldarius squalene cyclase alters the substrate specificity into that of the eukaryotic-type cyclase specific to (3S)-2,3-oxidosqualene. Angew Chem. 2004;43:6700–6703. doi: 10.1002/ange.200461523. [DOI] [PubMed] [Google Scholar]
- 44.Khersonsky O., Lipsh R., Avizemer Z., Ashani Y., Goldsmith M., Leader H., et al. Automated design of efficient and functionally diverse enzyme repertoires. Mol Cell. 2018;72:178–186. doi: 10.1016/j.molcel.2018.08.033. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X., Luo W., Yao Y., Luo X., Han C., Zhong Y., et al. Enhanced chemoselectivity of a plant cytochrome P450 through protein engineering of surface and catalytic residues. aBIOTECH. 2021;2:215–225. doi: 10.1007/s42994-021-00056-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou J., Hu T., Liu Y., Tu L., Song Y., Lu Y., et al. Cytochrome P450 catalyses the 29-carboxyl group formation of celastrol. Phytochemistry. 2021;190 doi: 10.1016/j.phytochem.2021.112868. [DOI] [PubMed] [Google Scholar]
- 47.Wang C., Su X., Sun M., Zhang M., Wu J., Xing J., et al. Efficient production of glycyrrhetinic acid in metabolically engineered Saccharomyces cerevisiae via an integrated strategy. Microb Cell Factories. 2019;18:95. doi: 10.1186/s12934-019-1138-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lodeiro S., Xiong Q., Wilson W.K., Kolesnikova M.D., Onak C.S., Matsuda S.P. An oxidosqualene cyclase makes numerous products by diverse mechanisms: a challenge to prevailing concepts of triterpene biosynthesis. J Am Chem Soc. 2007;129:11213–11222. doi: 10.1021/ja073133u. [DOI] [PubMed] [Google Scholar]
- 49.Jensen M.K., Keasling J.D. Recent applications of synthetic biology tools for yeast metabolic engineering. FEMS Yeast Res. 2015;15:1–10. doi: 10.1111/1567-1364.12185. [DOI] [PubMed] [Google Scholar]
- 50.Mellor S.B., Vinde M.H., Nielsen A.Z., Hanke G.T., Abdiaziz K., Roessler M.M., et al. Defining optimal electron transfer partners for light-driven cytochrome P450 reactions. Metab Eng. 2019;55:33–43. doi: 10.1016/j.ymben.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y., Ma L., Su P., Huang L., Gao W. Cytochrome P450s in plant terpenoid biosynthesis: discovery, characterization and metabolic engineering. Crit Rev Biotechnol. 2021:1–21. doi: 10.1080/07388551.2021.2003292. [DOI] [PubMed] [Google Scholar]
- 52.Zhao F., Bai P., Liu T., Li D., Zhang X., Lu W., et al. Optimization of a cytochrome P450 oxidation system for enhancing protopanaxadiol production in Saccharomyces cerevisiae. Biotechnol Bioeng. 2016;113:1787–1795. doi: 10.1002/bit.25934. [DOI] [PubMed] [Google Scholar]
- 53.Li D., Wu Y., Wei P., Gao X., Li M., Zhang C., et al. Metabolic engineering of Yarrowia lipolytica for heterologous oleanolic acid production. Chem Eng Sci. 2020;218 doi: 10.1016/j.ces.2020.115529. [DOI] [Google Scholar]
- 54.Guo X., Sun J., Li D., Lu W. Heterologous biosynthesis of (+)-nootkatone in unconventional yeast. Yarrowia lipolytica. Biochem Eng J. 2018;137:125–131. doi: 10.1016/j.bej.2018.05.023. [DOI] [Google Scholar]
- 55.Urlacher V.B., Girhard M. Cytochrome P450 monooxygenases in biotechnology and synthetic biology. Trends Biotechnol. 2019;37:882–897. doi: 10.1016/j.tibtech.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 56.Bradley W Biggs, Lim Chin G., Sagliani K., Shankar S., Stephanopoulos G., De Mey M., et al. Overcoming heterologous protein interdependency to optimize P450-mediated taxol precursor synthesis in Escherichia coli. Proc Natl Acad Sci USA. 2016;113:3209–3214. doi: 10.1073/pnas.1515826113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones J.A., Toparlak O.D., Koffas M.A. Metabolic pathway balancing and its role in the production of biofuels and chemicals. Curr Opin Biotechnol. 2015;33:52–59. doi: 10.1016/j.copbio.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 58.Moon T.S., Dueber J.E., Shiue E., Prather K.L.J. Use of modular, synthetic scaffolds for improved production of glucaric acid in engineered E. Coli. Metab Eng. 2010;12:298–305. doi: 10.1016/j.ymben.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 59.Baek J.M., Mazumdar S., Lee S.W., Jung M.Y., Lim J.H., Seo S.W., et al. Butyrate production in engineered Escherichia coli with synthetic scaffolds. Biotechnol Bioeng. 2013;110:2790–2794. doi: 10.1002/bit.24925. [DOI] [PubMed] [Google Scholar]
- 60.Schlegel S., Klepsch M., Gialama D., Wickstrm D., Slotboom D.J., Gier J.D. Revolutionizing membrane protein overexpression in bacteria. Microb Biotechnol. 2010;3 doi: 10.1111/j.1751-7915.2009.00148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kranzfinger S., Mahmoud O., Ricklefs E., Ditz N., Bakkes P.J., Urlacher V.B. Insights into the functional properties of the marneral oxidase CYP71A16 from Arabidopsis thaliana. BBA Proteins Proteom. 2017:1866. doi: 10.1016/j.bbapap.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 62.Girhard M., Tieves F., Weber E., Smit M.S., Urlacher V.B. Cytochrome P450 reductase from Candida apicola: versatile redox partner for bacterial P450s. Appl Microbiol Biotechnol. 2013;97:1625–1635. doi: 10.1007/s00253-012-4026-z. [DOI] [PubMed] [Google Scholar]
- 63.Vazquez-Albacete, D. , Cavaleiro, A. M. , Christensen, U. , Susanna Seppala, Birger Lindberg Moeller, et al. An expression tag toolbox for microbial production of membrane bound plant cytochromes P450. Biotechnol Bioeng 2016; 114: 751-760. https://doi.org/10.1002/bit.26203. [DOI] [PubMed]
- 64.Tavanti M., Porter J.L., Levy C.W., Gómez Castellanos J.R., Flitsch S.L., Turner N.J. The crystal structure of P450-tt heme-domain provides the first structural insights into the versatile class VII P450s. Biochem Biophys Res Commun. 2018;501:846–850. doi: 10.1016/j.bbrc.2018.05.014. https://doi.org/10.1016/j.bbrc.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 65.Hsu M.H., Ba Er B.R., Rettie A.E., Johnson E.F. The crystal structure of cytochrome P450 4B1 (CYP4B1) monooxygenase complexed with octane discloses several structural adaptations for ω-hydroxylation. J Biol Chem. 2017;292:5610–5621. doi: 10.1074/jbc.m117.775494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ravichandran Kurumbail G., Boddupalli Sekhar S., Charles A Hasermann, Peterson Julian A., Deisenhofer J. Crystal structure of hemoprotein domain of p450BM-3, a prototype for microsomal P450's. Science. 1993;261:731–736. doi: 10.1126/science.8342039. [DOI] [PubMed] [Google Scholar]
- 67.Chen J., Fan F., Qu G., Tang J., Xi Y., Bi C., et al. Identification of absidia orchidis steroid 11beta-hydroxylation system and its application in engineering Saccharomyces cerevisiae for one-step biotransformation to produce hydrocortisone. Metab Eng. 2020;57:31–42. doi: 10.1016/j.ymben.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 68.Ignea C., Ioannou E., Georgantea P., Loupassaki S., Trikka F.A., Kanellis A.K., et al. Reconstructing the chemical diversity of labdane-type diterpene biosynthesis in yeast. Metab Eng. 2015;28:91–103. doi: 10.1016/j.ymben.2014.12.001. https://doi.org/10.1016/j.ymben.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 69.Huang Y., Jiang D., Ren G., Yin Y., Sun Y., Liu T., et al. De novo production of glycyrrhetic acid 3-O-mono-beta-D-glucuronide in Saccharomyces cerevisiae. Front Bioeng Biotechnol. 2021;9 doi: 10.3389/fbioe.2021.709120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seifert A., Pleiss J. Identification of selectivity-determining residues in cytochrome Pp450 monooxygenases: a systematic analysis of the substrate recognition site 5. Proteins. 2009;74:1028–1035. doi: 10.1002/prot.22242. [DOI] [PubMed] [Google Scholar]
- 71.Dietrich J.A., Yoshikuni Y., Fisher K.J., Woolard F.X., Ockey D., McPhee D.J., et al. A novel semi-biosynthetic route for artemisinin production using engineered substrate-promiscuous P450BM3. ACS Chem Biol. 2009;4:261–267. doi: 10.1021/cb900006h. [DOI] [PubMed] [Google Scholar]
- 72.Li J., Yang J., Mu S., Shang N., Liu C., Zhu Y., et al. Efficient O-glycosylation of triterpenes enabled by protein engineering of plant glycosyltransferase UGT74AC1. ACS Catal. 2020;10:3629–3639. doi: 10.1021/acscatal.9b05232. [DOI] [Google Scholar]
- 73.Dai Z., Wang B., Liu Y., Shi M., Wang D., Zhang X., et al. Producing aglycons of ginsenosides in bakers' yeast. Sci Rep. 2014;4:3698. doi: 10.1038/srep03698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mizutani K., Kuramoto T., Tamura Y., Ohtake N., Doi S., Nakaura M., et al. Sweetness of glycyrrhetic acid 3-O-β-D-monoglucuronide and the related glycosides. Biosci Biotechnol Biochem. 1994;58:554–555. doi: 10.1271/bbb.58.554. [DOI] [PubMed] [Google Scholar]
- 75.Liu X., Zhang L., Feng X., Lv B., Li C. Biosynthesis of glycyrrhetinic acid-3-O-monoglucose using glycosyltransferase UGT73C11 from Barbarea vulgaris. Ind Eng Chem Res. 2017;56:14949–14958. doi: 10.1021/acs.iecr.7b03391. [DOI] [Google Scholar]
- 76.Zhang L., Ren S., Liu X., Liu X., Guo F., Sun W., et al. Mining of UDP-glucosyltrfansferases in licorice for controllable glycosylation of pentacyclic triterpenoids. Biotechnol Bioeng. 2020;117:3651–3663. doi: 10.1002/bit.27518. [DOI] [PubMed] [Google Scholar]
- 77.Augustin J.M., Drok S., Shinoda T., Sanmiya K., Nielsen J.K., Khakimov B., et al. UDP-glycosyltransferases from the UGT73C subfamily in barbarea vulgaris catalyze sapogenin 3-O-glucosylation in saponin-mediated insect resistance. Plant Physiol. 2012;160:1881–1895. doi: 10.1104/pp.112.202747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Itkin M., Davidovich-Rikanati R., Cohen S., Portnoy V., Doron-Faigenboim A., Oren E., et al. The biosynthetic pathway of the nonsugar, high-intensity sweetener mogroside V from Siraitia grosvenorii. Proc NatI Acad Sci. 2016;113:E7619–E7628. doi: 10.1073/pnas.1604828113. https://doi.org/doi:10.1073/pnas.1604828113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Olsson K., Carlsen S., Semmler A., Simón E., Mikkelsen M.D., Møller B.L. Microbial production of next-generation stevia sweeteners. Microb Cell Factories. 2016;15:207. doi: 10.1186/s12934-016-0609-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tang Q.-Y., Chen G., Song W.-L., Fan W., Wei K.-H., He S.-M., et al. Transcriptome analysis of panax zingiberensis identifies genes encoding oleanolic acid glucuronosyltransferase involved in the biosynthesis of oleanane-type ginsenosides. Planta. 2019;249:393–406. doi: 10.1007/s00425-018-2995-6. [DOI] [PubMed] [Google Scholar]
- 81.Chen K., Hu Z-m, Song W., Wang Z-l, He J-b, Shi X-m, et al. Diversity of O-glycosyltransferases contributes to the biosynthesis of flavonoid and triterpenoid glycosides in glycyrrhiza uralensis. ACS Synth Biol. 2019;8:1858–1866. doi: 10.1021/acssynbio.9b00171. [DOI] [PubMed] [Google Scholar]
- 82.Ji X., Lin S., Chen Y., Liu J., Yun X., Wang T., et al. Identification of α-amyrin 28-carboxylase and glycosyltransferase from ilex asprella and production of ursolic acid 28-O-β-d-glucopyranoside in engineered yeast. Front Plant Sci. 2020;11 doi: 10.3389/fpls.2020.00612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kurosawa Y., Takahara H., Shiraiwa M. UDP-glucuronic acid:Soyasapogenol glucuronosyltransferase involved in saponin biosynthesis in germinating soybean seeds. Planta. 2002;215:620–629. doi: 10.1007/s00425-002-0781-x. [DOI] [PubMed] [Google Scholar]
- 84.Gao Y., Zhang L., Feng X., Liu X., Guo F., Lv B., et al. Galactosylation of monosaccharide derivatives of glycyrrhetinic acid by UDP-glycosyltransferase gmsgt2 from Glycine max. J Agric Food Chem. 2020;68:8580–8588. doi: 10.1021/acs.jafc.0c03842. [DOI] [PubMed] [Google Scholar]
- 85.Madhav H., Bhasker S., Chinnamma M. Functional and structural variation of uridine diphosphate glycosyltransferase (UGT) gene of stevia rebaudiana–ugtsr involved in the synthesis of rebaudioside a. Plant Physiol Biochem. 2013;63:245–253. doi: 10.1016/j.plaphy.2012.11.029. [DOI] [PubMed] [Google Scholar]
- 86.Lu J., Yao L., Li J.-X., Liu S.-J., Hu Y.-Y., Wang S.-H., et al. Characterization of UDP-glycosyltransferase involved in biosynthesis of ginsenosides Rg1 and Rb1 and identification of critical conserved amino acid residues for its function. J Agric Food Chem. 2018;66:9446–9455. doi: 10.1021/acs.jafc.8b02544. [DOI] [PubMed] [Google Scholar]
- 87.Zhuang Y., Yang G.-Y., Chen X., Liu Q., Zhang X., Deng Z., et al. Biosynthesis of plant-derived ginsenoside Rh2 in yeast via repurposing a key promiscuous microbial enzyme. Metab Eng. 2017;42:25–32. doi: 10.1016/j.ymben.2017.04.009. https://doi.org/10.1016/j.ymben.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 88.Wang P., Wei W., Ye W., Li X., Zhao W., Yang C., et al. Synthesizing ginsenoside Rh2 in Saccharomyces cerevisiae cell factory at high-efficiency. Cell Discov. 2019;5 doi: 10.1038/s41421-018-0075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nomura Y., Seki H., Suzuki T., Ohyama K., Mizutani M., Kaku T., et al. Functional specialization of UDP-glycosyltransferase 73P12 in licorice to produce a sweet triterpenoid saponin, glycyrrhizin. Plant J. 2019;99:1127–1143. doi: 10.1111/tpj.14409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kurze E., Wust M., Liao J., McGraphery K., Hoffmann T., Song C., et al. Structure-function relationship of terpenoid glycosyltransferases from plants. Nat Prod Rep. 2022;39:389–409. doi: 10.1039/d1np00038a. [DOI] [PubMed] [Google Scholar]
- 91.George Thompson A.M., Iancu C.V., Neet K.E., Dean J.V., Choe J.Y. Differences in salicylic acid glucose conjugations by UGT74F1 and UGT74F2 from Arabidopsis thaliana. Sci Rep. 2017;7 doi: 10.1038/srep46629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang T., Zhang J., Ke D., Yang W., Tang M., Jiang J., et al. Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations. Nat Commun. 2019;10:3214. doi: 10.1038/s41467-019-11154-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hsu T.M., Welner D.H., Russ Z.N., Cervantes B., Prathuri R.L., Adams P.D., et al. Employing a biochemical protecting group for a sustainable indigo dyeing strategy. Nat Chem Biol. 2018;14:256–261. doi: 10.1038/nchembio.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shao H., He X., Achnine L., Blount J.W., Dixon R.A., Wang X. Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from medicago truncatula. Plant Cell. 2005;17:3141–3154. doi: 10.1105/tpc.105.035055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen L., Zhang Y., Feng Y. Structural dissection of sterol glycosyltransferase UGT51 from Saccharomyces cerevisiae for substrate specificity. J Struct Biol. 2018;204:371–379. doi: 10.1016/j.jsb.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 96.Chen L., Cai R., Weng J., Li Y., Jia H., Chen K., et al. Production of rebaudioside d from stevioside using a UGTSL2 Asn358Phe mutant in a multi-enzyme system. Microb Biotechnol. 2020;13:974–983. doi: 10.1111/1751-7915.13539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yin C., Zheng T., Chang X. Biosynthesis of s-adenosylmethionine by magnetically immobilized Escherichia coli cells highly expressing a methionine adenosyltransferase variant. Molecules. 2017;22 doi: 10.3390/molecules22081365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rehman H.M., Nawaz M.A., Shah Z.H., Chung G., Yang S.H. Molecular elucidation of two novel seed specific flavonoid glycosyltransferases in soybean. J Plant Biol. 2018;61:320–329. doi: 10.1007/s12374-018-0103-x. [DOI] [Google Scholar]
- 99.Chen Q., Liu X., Hu Y., Wang Y., Sun B., Chen T., et al. Broaden the sugar donor selectivity of blackberry glycosyltransferase UGT78H2 through residual substitutions. Int J Biol Macromol. 2021;166:277–287. doi: 10.1016/j.ijbiomac.2020.10.184. [DOI] [PubMed] [Google Scholar]
- 100.Noguchi A., Horikawa M., Fukui Y., Fukuchi-Mizutani M., Iuchi-Okada A., Ishiguro M., et al. Local differentiation of sugar donor specificity of flavonoid glycosyltransferase in lamiales. Plant Cell. 2009;21:1556–1572. doi: 10.1105/tpc.108.063826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Osmani S.A., Bak S., Imberty A., Ller O.M. Catalytic key amino acids and udp-sugar donor specificity of a plant glucuronosyltransferase, UGT94B1: molecular modeling substantiated by site-specific mutagenesis and biochemical analyses. Plant Physiol. 2008;148:1295–1308. doi: 10.1104/pp.108.128256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Akere A., Chen S.H., Liu X., Chen Y., Haider S. Structure-based enzyme engineering improves donor-substrate recognition of arabidopsis thaliana glycosyltransferases. Biochem J. 2020;477 doi: 10.1042/bcj20200477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xiao M., Zhang Y., Chen X., Lee E.J., Barber C.J., Chakrabarty R., et al. Transcriptome analysis based on next-generation sequencing of non-model plants producing specialized metabolites of biotechnological interest. J Biotechnol. 2013;166:122–134. doi: 10.1016/j.jbiotec.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 104.Matasci N., Hung L.H., Yan Z., Carpenter E.J., Wickett N.J., Mirarab S., et al. Data access for the 1,000 plants (1kp) project. GigaScience. 2014;3:17. doi: 10.1186/2047-217X-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jozwiak A., Sonawane P.D., Panda S., Garagounis C., Papadopoulou K.K., Abebie B., et al. Plant terpenoid metabolism co-opts a component of the cell wall biosynthesis machinery. Nat Chem Biol. 2020;16:740–748. doi: 10.1038/s41589-020-0541-x. [DOI] [PubMed] [Google Scholar]
- 106.Wang Y., Zhang H., Ri H.C., An Z., Wang X., Zhou J.-N., et al. Deletion and tandem duplications of biosynthetic genes drive the diversity of triterpenoids in Aralia elata. Nat Commun. 2022;13:2224. doi: 10.1038/s41467-022-29908-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chung S.Y., Seki H., Fujisawa Y., Shimoda Y., Hiraga S., Nomura Y., et al. A cellulose synthase-derived enzyme catalyses 3-O-glucuronosylation in saponin biosynthesis. Nat Commun. 2020;11:5664. doi: 10.1038/s41467-020-19399-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tao H., Lauterbach L., Bian G., Chen R., Hou A., Mori T., et al. Discovery of non-squalene triterpenes. Nature. 2022;606:414–419. doi: 10.1038/s41586-022-04773-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.He X.-Z., Wang X., Dixon R.A. Mutational analysis of the medicago glycosyltransferase UGT71G1 reveals residues that control regioselectivity for (iso)flavonoid glycosylation. J Biol Chem. 2006;281:34441–34447. doi: 10.1074/jbc.M605767200. [DOI] [PubMed] [Google Scholar]
- 110.Espinoza R.V., Sherman D.H. Exploring the molecular basis for selective C-H functionalization in plant P450s. Synth Syst Biotechnol. 2020;5:97–98. doi: 10.1016/j.synbio.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Padyana A.K., Gross S., Jin L., Cianchetta G., Narayanaswamy R., Wang F., et al. Structure and inhibition mechanism of the catalytic domain of human squalene epoxidase. Nat Commun. 2019;10:97. doi: 10.1038/s41467-018-07928-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yao X., Fan X., Yan N. Cryo-em analysis of a membrane protein embedded in the liposome. Proc Natl Acad Sci U S A. 2020;117:18497–18503. doi: 10.1073/pnas.2009385117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gong X., Qian H., Zhou X., Wu J., Wan T., Cao P., et al. Structural insights into the niemann-pick C1 (NPC1)-mediated cholesterol transfer and ebola infection. Cell. 2016:1467–1478. doi: 10.1016/j.cell.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Qian H., Zhao X., Cao P., Lei J., Yan N., Gong X. Structure of the human lipid exporter abca1. Cell. 2017;169:1228–1239. doi: 10.1016/j.cell.2017.05.020. [DOI] [PubMed] [Google Scholar]
- 115.Song Y., DiMaio F., Wang Ray Y-R, Kim D., Miles C., Brunette T.J., et al. High-resolution comparative modeling with rosettacm. Structure. 2013;21:1735–1742. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ma Y., Zhou Y., Ovchinnikov S., Greisen P., Huang S., Shang Y. New insights into substrate folding preference of plant oscs. Sci Bull. 2016;61:1407–1412. doi: 10.1007/s11434-016-1103-1. [DOI] [Google Scholar]
- 117.Li D., Ma Y., Zhou Y., Gou J., Zhong Y., Zhao L., et al. A structural and data-driven approach to engineering a plant cytochrome P450 enzyme. Sci China Life Sci. 2019;62:873–882. doi: 10.1007/s11427-019-9538-3. [DOI] [PubMed] [Google Scholar]