Abstract
Wnts, bone morphogenetic protein (BMP), and fibroblast growth factor (FGF) are paracrine signaling pathways implicated in the niche control of stem cell fate decisions. BMP-on and Wnt-off are the dominant quiescent niche signaling pathways in many cell types, including neural stem cells (NSCs). However, among the multiple inhibitory family members of the Wnt pathway, those with direct action after BMP4 stimulation in NSCs remain unclear. We examined 11 Wnt inhibitors in NSCs after BMP4 treatment. Wnt inhibitory factor 1 (Wif1) has been identified as the main factor reacting to BMP4 stimuli. RNA sequencing confirmed that Wif1 was markedly upregulated after BMP4 treatment in different gene expression analyses. Similar to the functional role of BMP4, Wif1 significantly decreased the cell cycle of NSCs and significantly inhibited cell proliferation (P < 0.05). Combined treatment with BMP4 and Wif1 significantly enhanced the inhibition of cell growth compared with the single treatment (P < 0.05). Wif1 expression was clearly lower in glioblastoma and low-grade glioma samples than in normal samples (P < 0.05). A functional analysis revealed that both BMP4 and Wif1 could decrease glioma cell growth. These effects were abrogated by the BMP inhibitor Noggin. The collective findings demonstrate that Wif1 plays a key role in quiescent NSC homeostasis and glioma cell growth downstream of BMP-on signaling. The functional roles of Wif1/BMP4 in glioma cells may provide a technical basis for regenerative medicine, drug discovery, and personal molecular therapy in future clinical treatments.
Keywords: BMP4, Wif1, neural stem cells, GBM, Wnt signaling pathway
Introduction
Undifferentiated neural stem cells (NSCs) can divide and differentiate to produce a variety of neural cell types, including neurons, astrocytes, and oligodendrocytes. Owing to their ability to expand in vitro, in vitro culture and induced differentiation of NSCs have been increasingly explored in recent years1. Glioblastoma (GBM) is a highly malignant brain tumor derived from glial and neuronal cells2. Therefore, it is essential to develop novel therapeutic options based on an understanding of neurodevelopment.
During the early developmental stages, bone morphogenetic protein (BMP), fibroblast growth factor (FGF), and Wnt act as key regulators to direct the developmental outcome of tissue organs3–5. The Wnt and BMP signaling pathways are involved in dynamic regulation and synergistically work through a variety of positive and negative regulators to ensure that cell proliferation and differentiation (which are essential for the development of tissue organs) occur. Although the morphology and function of organs vary across species, only a few signaling pathways control them. Among these pathways, the Wnt and BMP signaling molecules are highly conserved and often co-regulate multiple biological processes, such as growth, development, and stem cell maintenance6,7. This interaction mostly overlaps in time and space. Recent studies have shown that transcription factors that include Wnt, BMP, and FGF regulate NSC proliferation and differentiation. BMPs can affect the fate of NSCs and inhibit their proliferation8. Wnt signaling may play a role in the development of multiple cancers and diseases9,10. Although the regulation of cell fate and function by individual signaling pathways has been well described, it is unclear which of the multiple inhibition family members of the Wnt pathway acts directly on BMP4 stimulation in NSCs and neurologically related tumors, such as GBMs.
Cell fate can be determined by classical Wnt signaling pathways, and cell motility by non-classical Wnt signaling pathways11,12. The fate of NSCs is governed by the classic Wnt/β-catenin and BMP signaling pathways13–16. The Wif1 gene is located on chromosome 12q14.3 and is approximately 200 kilobase pairs (kbp) in length. Wif1 protein is abundant in human prostate, lung, and brain tissue. The Wnt ligand competes with Wif1 for binding to the coiled-coil protein receptor and is a downstream regulator of the Wnt/β-catenin signaling pathway that eventually blocks this signaling17. Tumors that have been studied by various researchers have downregulated Wif1 expression and altered Wnt signaling pathways18–20.
We studied two key signaling pathways intersecting at the molecular level. The results revealed that Wif1 is more active in Wnt inhibitors after BMP stimulation. A synergistic effect was observed between the two. Based on cell cycle, RNA sequencing, and cell function assays, we confirmed that Wif1 can effectively reduce the cell cycle of NSCs. The cycle of NSCs was controlled digitally. Growth of glioma cells was inhibited by both BMP4 and Wif1. These inhibitions were reversed by the BMP inhibitor Noggin. The collective findings identify Wif1 as a unique downstream mechanism of BMP signaling in the static homeostasis of NSCs and the growth of glioma cells. The findings also clarify the understanding of neural development and differentiation, and how diseases that include GBM develop and are treated.
Experimental Procedures
Cell Culture
NSCs were maintained in culture ware coated with poly-l-ornithine (P3655; Sigma-Aldrich, St. Louis, MO, USA) and mouse laminin (23017-015; Invitrogen, Grand Island, NY, USA) in Dulbecco’s Modified Eagle Medium (DMEM)/F12 (11320-033, Invitrogen) supplemented with N2 (17502-048, Invitrogen), l-glutamine (25030-081, Invitrogen), penicillin-streptomycin (15140-122, Invitrogen), and FGF2 (20 ng/mL, HZ-1271, Humanzyme, Rosemont, IL, USA). The quiescent-like state of the cells was induced using 50 ng/mL BMP4 (HZ-1045, Humanzyme). DMEM/F12 supplemented with 10% fetal bovine serum (FBS; 10099-141C, Gibco, Carlsbad, CA, USA) and 20 ng/mL penicillin-streptomycin (15140-122, Invitrogen) was used to culture glioma cell lines U87 and U251 purchased from the Cell Bank of the Chinese Academy of Science (Shanghai, China). All cells were cultured in an atmosphere of 5% CO2 at 37°C. Trypsin-EDTA (0.05%; 25300062, Invitrogen) and trypsin inhibitor (R007100, Invitrogen) were used for cell passage. HXX-6 is a primary cultured glioma stem cell line established from surgical specimens, as previously described21.
Plasmids and Cell Transfection
The Wif1 overexpression plasmid was constructed using the pLV-EF1α-EGFP (2A) Puro plasmid. Sequencing (1098 bp) was performed with forward primers (TTATCTAGAGCCACCATGGCCCGGAGAAGAGCCTTC) and reverse primers (TATGCGGCCGCTCACCAGATGTAATTGGATTCAG) into two digestion sites, XbaI and NotI. Plasmid transfection was performed following the standard protocol of the FuGENE HD Transfection Reagent (E2312, Promega, Madison, WI, USA).
Experimental Groups
Four groups of cells were designed as F5 (FGF 5 ng/mL), F20 (FGF 20 ng/mL), F5+B50 (FGF 5 ng/mL and BMP4 50 ng/mL), and F20+B50 (FGF 20 ng/mL and BMP4 50 ng/mL) to test the NSC culture system of FGF2 and BMP4. Both Wif1 protein (20 nM) and overexpression plasmid (2, 4, and 5 μg) were compared with the control group (FGF 20 ng/mL) to qualify the functional role of Wif1. Four groups were designed as FGF (FGF 20 ng/mL), FGF+BMP (FGF 20 ng/mL and BMP4 50 ng/mL), FGF+Wif1 (FGF 20 ng/mL and Wif1 20 nM), and FGF+BMP+Wif1 (FGF 20 ng/mL, BMP4 50 ng/mL, and Wif1 20 nM) to examine the differences of BMP4 and Wif1. Time series samples were collected every 24 h, based on the cell growth period.
Quantitative Real-Time PCR (qPCR)
RNA was extracted from NSCs using TRIzol reagent (LS1000, Promega) according to the traditional protocol. The FastQuant RT Kit (KR106, TIANGEN, Beijing, China) was used to convert the RNA to cDNA. Quantitative RT-PCR was performed in the Bio-Rad T100 PCR system using SuperRealPreMix Plus (FP205, TIANGEN) and appropriate primers. The primer sequences are listed in Table 1. The effects of 18S rRNA and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) levels on the expression of target genes were quantitatively studied using the contrast CT method.
Table 1.
Primer Forward (F) and Reverse (R) Sequences.
| Genes | Sequence |
|---|---|
| 18sF | CATTCGAACGTCTGCCCTATC |
| 18sR | CCTGCTGCCTTCCTTGGA |
| GADPH F | TGACTCTACCCACGGCAAGTTCAA |
| GADPH R | ACGACATACTCAGCACCAGCATCA |
| BMP4 F | ACACAGGGACACACCAACCAT |
| BMP4 R | TGTGACCAGCTGTGTTCATCTTG |
| Noggin F | GCAGCTGCTTCAGTAAGCG |
| Noggin R | GCACTCGGAAATGATGGG |
| DKK1 F | TTGGAAGGGTGGGAATGTGAC |
| DKK1 R | ACCATTCTTCAGCATGTCAGAG |
| Cer1 F | CACCAAATTCACCACCACGC |
| Cer1 R | CGTCTTCACCATGCACTGAC |
| Wif1 F | TTGTACCTGTGGATCGACGC |
| Wif1 R | GGCTGGCATTCTTTGTTGGG |
| DKK2 F | ATGGGTTTTGTTGTGCTCGC |
| DKK2 R | CAGCCCATGTGAACCCTTCT |
| DKK3 F | GACACTCAGCACAAACTGCG |
| DKK3 R | TCTCCGTGTTGGTCTCGTTG |
| DKK4 F | GCGGAGGAAAACAGACCTCA |
| DKK4 R | AAATGGCGAGCACAGCAAAG |
| Sfrp1 F | ACGTGAGCTTCCAGTCGGAT |
| Sfrp1 R | CTTCTTGTAGCCCACGTTGTG |
| Sfrp2 F | ATCCTGGAGACAAAGAGCAAGACC |
| Sfrp2 R | TGACCAGATACGGAGCGTTGATG |
| Sfrp3 F | TCCGGGCTAAAGTTAAAGAGGT |
| Sfrp3 R | GACGGTATCCCTTGGAATGT |
| Sfrp4 F | TGAGGTCACAACTGTGGTCG |
| Sfrp4 R | ACACTGGCAGGAGGAATTGG |
| Sfrp5 F | TTGCCTCCTACTTGGACGAC |
| Sfrp5 R | CACCCGCATGGCTGGATATT |
| SMAD1 F | CCAAATGGCTAGTGGGTCTACA |
| SMAD1 R | GAAACACAGCAAAGTCCGACAC |
| CDK2 F | CTTTGCCGAAATGGTGACCC |
| CDK2 R | TAACTCCTGGCCAAACCACC |
Cell Cytometry
Cells (1 × 106) were collected in a 15-mL Corning tube after centrifugation at 250 × g for 3 min. The supernatant was discarded, and the cells were resuspended in 100 μL PBS (10010023, Gibco). The cells were fixed with 1 mL cold 75% ethanol and stored them at –20°C for further analysis. For cell cytometry, cell samples were stained with 400 μL PBS containing 500 ng/mL propidium iodide (15140122, Gibco) for 15 min at room temperature after centrifugation, and supernatant was discarded. The cell cytometry assay was performed on a BD FACSVerse machine (Franklin Lakes, NJ, USA) following standard procedures using the PE channel.
Immunofluorescence and Microscopy
The cells were fixed in 4% paraformaldehyde (PFA; E672002, BBI Life Sciences Corporation, Hong Kong, China) for 15 min at room temperature and permeabilised in PBS+0.1% Triton (X100, Sigma-Aldrich). The antibodies used were rabbit anti-Ki67 (1:1000; vpk451, Vector Laboratories Company, San Francisco, CA, USA) and DAPI (1:1000; D3571, Invitrogen). Photographs were taken using a Nikon ECLIPSE Ti microscope (Japan) and processed using Photoshop CS5.
RNA Sequencing
RNA purity was determined using a Qubit® 3.0 Fluorometer (Thermo Fisher Scientific, Waltham, England), and RNA integrity and concentration were assessed using an RNANano6000 Assay Kit (Agilent, Santa Clara, CA, USA) on a Bioanalyzer 2100 (Agilent). The total amount of RNA in each sample was 2 μg and was used as the input for RNA sample preparation. A sequencing library was created for the RNA library (#E7530L) using the NEBNext® Ultra RNA library for Illumina (Biolab, England), according to the manufacturer’s recommendations. An index code was added to the attribute sequences of each sample. The Qubit® RNA Assay Kit (Thermo Fisher Scientific) was used for the initial quantification and diluted to every microgram/microliter. The insert size was evaluated using an Agilent Bioanalyzer 2100 and accurately quantified using the StepOnePlus™ Real-Time PCR System (Thermo Fisher Scientific) with an effective library concentration >10 nM. Index-encoded sampling was performed using the HiSeq PE Cluster Kitv4-cBot-HS (Illumina) in the cBot cluster generation system as described by the manufacturer. After clustering was completed, sequence analysis was performed using Illumina Hiseq 4000, resulting in 150 bp paired-end data.
Genomic Analysis
Fragments per kilobase transcript mapped reads per million (FPKM) values were used. Pearson correlation analysis was performed using the R method. An R value > 90% indicated the consistency of the samples. Differentially expressed genes (DEGs) were detected using Morpheus online software (https://software.broadinstitute.org/morpheus).
Statistical Analysis
In the qPCR and immunohistochemistry (IHC) assays, receiver operating characteristic curves were used to determine the cut-off points. Cytology and IHC were analyzed using the Student’s t test. Data analysis was performed using SPSS software. In all experiments, P values < 0.05 were considered significant. All data are expressed as mean ± standard deviation.
Results
BMP4 and FGF Induce Wif1 Activity in NSCs
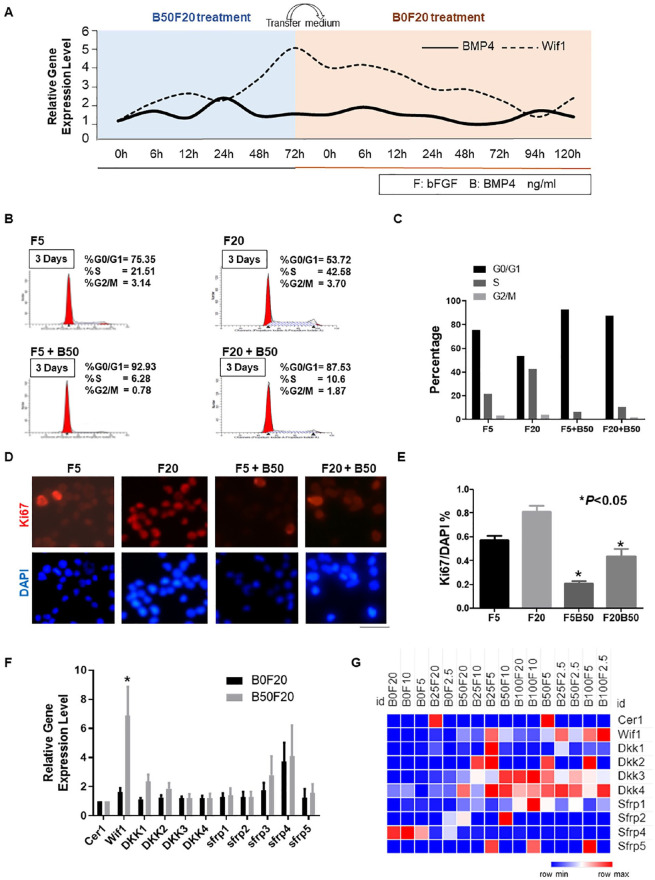
To test the expression trend of Wif1 after BMP4 treatment in NSCs, we collected RNA samples from samples treated using B50F20 (BMP4 50 ng/mL and FGF 20 ng/mL) and transferred to B0F20 (FGF 20 ng/mL) medium. The detailed time series results showed that Wif1 increased with BMP4 stimulation and decreased immediately after shift to the medium lacking BMP4 (Fig. 1A). In this study, two different FGF and BMP4 combinations were used to regulate the cell cycle of adult NSCs. Cell cycle status and cell proliferation rate were examined by staining with PI and Ki67 staining, respectively. The percentage of G0/G1 cells increased from 53.72% to 75.35%, while FGF dosage decreased from 20 to 5 ng/mL. Extra treatment with BMP4 (50 ng/mL) significantly increased the percentage of cells in the G0/G1 phase to 87.53% and 92.93%, respectively, compared with every single treatment with FGF (20 ng/mL and 5 ng/mL) (Fig. 1B and C). The ratio of DAPI-labeled Ki67-positive cells to DAPI-labeled cells indicates the proliferation rate of cells. Co-treatment with BMP (50 ng/mL) and FGF (20 ng/mL and 5 ng/mL) drastically decreased the NSC proliferation rate from 80% and 57% to approximately 40% and 21%, respectively (P < 0.05, Fig. 1D–E). Thus, we defined B50F20 as a quiescent-like status and B0F20 as an activated status for NSCs in our study.
Figure 1.
Wif1 displays a high expression after BMP4 stimulation. (A) qPCR using primers of BMP4 and Wif1 were performed within different time points samples from quiescent-like status (B0/B6/B12/B24/B48/B72) to proliferation (F0/F6/F12/F24/F48/F72/F96/F120). The gene expression of cells was normalized to the B0 cell and the 18S gene was used as an internal control. (B) PI staining cell cytometry test was performed to identify the distinct G0/G1, S, and G2/M phase status within the different treatments (FGF 20 ng/mL; FGF 20 ng/mL + BMP 50 ng/mL; FGF 5 ng/mL; FGF 5 ng/mL + BMP 50 ng/mL). (C) PI staining cell cytometry results were presented as a bar chart. (D and E) The ratio of Ki67 positive cells to DAPI labeling cells denotes the proliferation rate of the cells. BMP treatment significantly decreases the NSC proliferation rate (*P < 0.05). Three independent experiments have been performed, and the data are represented as mean ± SEM. (F) qPCR using primers of Wnt inhibitors was performed within Rattus NSCs cDNA (n = 3). The gene expression of proliferating cells was normalized to the expression of B50F20 cells, and the 18S gene was used as an internal control (*P < 0.05). (G) Expression patterns of Wnt inhibitors as FPKM values with different combinations of BMP4 and FGF.
Wnt signaling regulates stem cell quiescence22. However, among the multiple upstream inhibitors of Wnt signaling, the key factor regulating NSC quiescence is still unknown. To address this, qPCR was performed using primers for Cer1, Wif1, DKK family (DKK1-4), and sFRP family (sFRP1-5) in both activated and quiescent NSC cDNA samples. Wif1 and sFRP3/4 were significantly upregulated within quiescent samples compared with proliferation samples, with Wif1 displaying the most dramatic gene expression change (Fig. 1F). The FPKM values from the RNA sequencing results of the BMP/FGF digital control system were similar to the qPCR results. The FPKM value of Wif1 increased significantly with BMP4 concentration (Fig. 1G).
Wif1 Is the Most Responsive Gene to BMP4 in NSCs
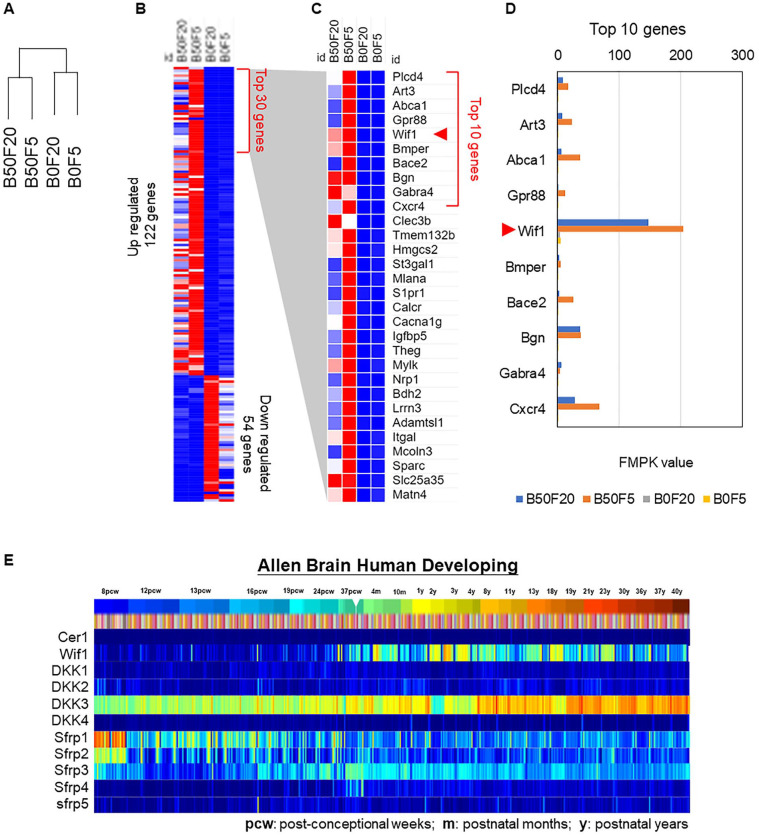
We performed RNA sequencing of NSCs with or without BMP4 treatment using different combinations of FGF and BMP4. The clustering results indicated that BMP4 was the main driver to group the samples as B50F20 together with B50F5 and B0F20 together with B0F5 (Fig. 2A). Identification of DEGs revealed 122 upregulated genes, of which 54 were BMP4-induced downregulated genes in NSCs (Fig. 2B). Of the top 30 upregulated genes from the whole genome transcriptional analysis, Wif1 was the fifth most upregulated gene (Fig. 2C). A bar plot of the FPKM values of the top 10 genes indicated that the gene expression of Wif1 was significantly higher (Fig. 2D). The findings indicate that Wif1 is the most responsive gene to BMP4 in NSCs. The expression pattern of all Wnt inhibitors in the human brain development period showed that Wif1 expression increased with age23, which indicates the importance of Wnt signaling during development and coincides with our results that Wif1 is highly expressed in quiescent-like NSCs (Fig. 2E).
Figure 2.
Wif1 is identified with genomic analysis in NSCs after BMP4 treatment. (A) Gene clustering from B50F20, B50F5, B0F20, and B0F5 for the NSCs RNA sequencing results. (B) Identification of significantly upregulated (122 genes) and downregulated (54 genes) genes. (C) Expression patterns of top 30 upregulated genes. (D) Bar chart of FPKM values from top 10 upregulated genes. (E) Expression trends of Wnt inhibitors during human brain development.
Upregulation of Wif1 Suppresses the Cell Cycle of NSCs
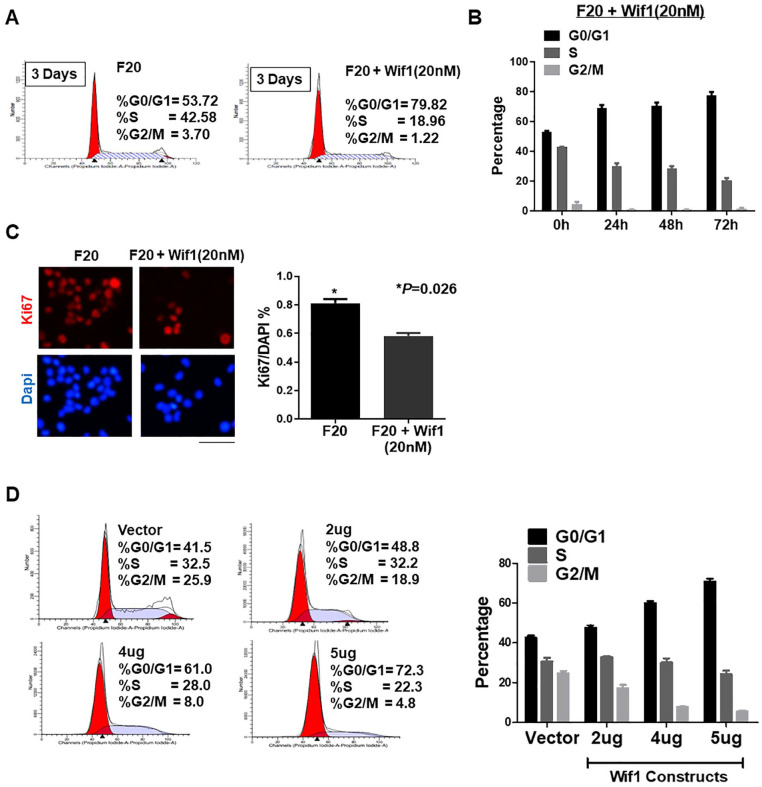
To investigate the functional roles of Wif1, PI staining cytometry assay and Ki67 staining IHC analysis were performed using commercial Wif1 factors (novoproteins). Extra treatment with Wif1 (20 nM) significantly increased the percentage of G0/G1 cells compared with the single treatment of FGF (20 ng/mL) from 53.72 to 79.82 (Fig. 3A and B). Co-treatment with Wif1 (20 nM) and FGF (20 ng/mL) apparently decreased the NSC proliferation rate from 80% to approximately 60% (P = 0.026, Fig. 3C). To confirm the importance of Wif1, we constructed a Wif1 overexpression plasmid. Cell cycle analysis was performed using PI staining. The Wif1 plasmid increased the percentage of G0/G1 cells in a dose-dependent manner compared with the vector from approximately 40% to nearly 80% (Fig. 3D). Thus, upregulation of Wif1 significantly blocked the cell cycle of NSCs and decreased cell proliferation.
Figure 3.
Wif1 regulates the cell cycle of NSCs. (A) PI staining cell cytometry test was performed to identify the distinct G0/G1, S, and G2/M phase status within the treatments of FGF (20 ng/mL) control and FGF (20 ng/mL) plus Wif1 (50 nM). Wif1 successfully blocks the cells at G0/G1 phase at 79.82%. (B) PI staining cell cytometry results from different time points were presented as a bar chart. (C) The ratio of Ki67 positive cells to DAPI labeling cells stands for the proliferation rate of the cells. Wif1 (*P = 0.026) significantly decreased cell proliferation. (D) PI staining cell cytometry results were presented as a bar chart. After transfection of the Wif1 construct, the percentage of G0/G1 was increased dose-dependently (2 μg/4 μg/5 μg; n = 3; *P < 0.05).
Wif1 and BMP4 Synergistically Regulate the Cell Cycle of NSCs
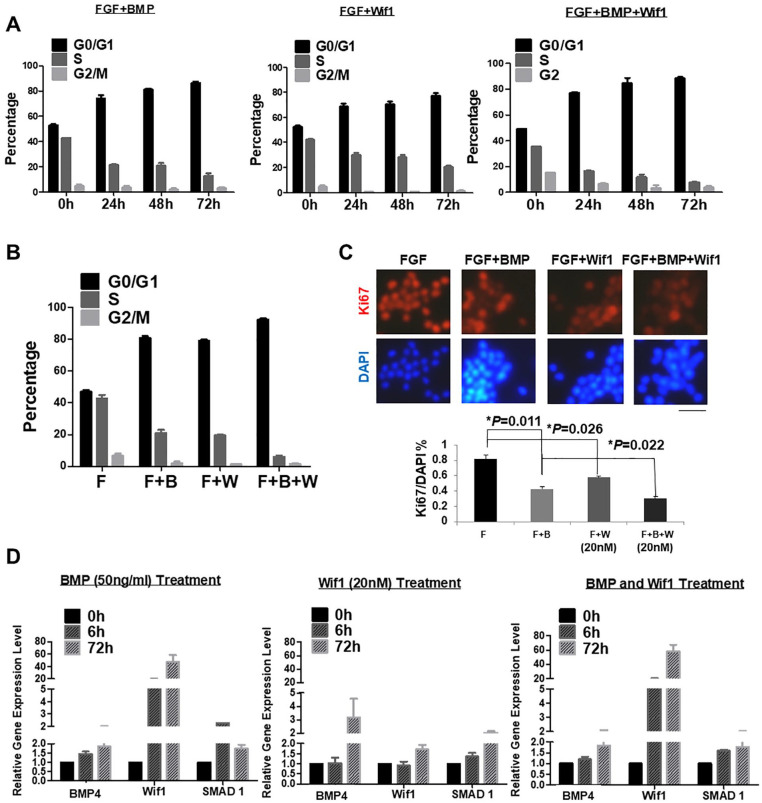
BMP functions in the entry of NSCs to quiescence24. Whether BMP can be replaced or acts synergistically with other factors is unknown. Since Wif1 has similar roles as BMP4 in causing a quiescence-like state for NSCs, we performed functional experiments with either a single treatment of BMP4 or Wif1, or co-treatment to examine their joint action. PI staining was used to detect the cell cycle in samples every 24 h. Together with the presence of FGF (20 ng/mL), sole treatment with either Wif1 (20 nM) or BMP4 (50 ng/mL) significantly increased the percentage of G0/G1 cells from under 50% to approximately 80%. Co-treatment with both Wif1 (20 nM) and BMP4 (50 ng/mL) drastically increased the percentage of G0/G1 cells up to 90% (Fig. 4A). If we only focused on the 72 h results, it was clear that the co-treatment with Wif1 and BMP4 enhanced the blocking effect of G0/G1 by close to 90% compared with their single treatment (Fig. 4B). To confirm the synergistic regulation of Wif1 and BMP4, Ki67 staining IHC analysis was performed. A bar chart plot revealed that the combination of Wif1 and BMP4 drastically decreased cell proliferation from 80% to approximately 30%, and Wif1 alone significantly downregulated the cell proliferation rate to 50% as compared with the BMP single treatment at 40% (P = 0.022) (Fig. 4C). qPCR was performed to detect relative gene expression with Wnt and BMP signaling. Interestingly, BMP4 could upregulate the expression of Wif1 and Wif1 treatment also apparently activated the BMP pathway by increasing the expression of BMP4 and Smad1 (Fig. 4D).
Figure 4.
Wif1 and BMP4 synergistically regulate the NSC cell cycle. (A) PI staining cell cytometry results presented as a bar chart. The treatments with FGF/BMP, FGF/Wif1, and FGF/BMP/Wif1 significantly increased the percentage of G0/G1 phase time-dependently (n = 3) (*P < 0.05). (B) PI staining cell cytometry results were presented as a bar chart. The single treatment with either BMP4 or Wif1 accompanied with FGF significantly increased the percentage of the G0/G1 phase, while co-treatment of FGF, BMP4, and Wif1 could enhance the blocking effect of the NSC cell cycle. (C) The ratio of Ki67 positive cells to DAPI labeling cells denotes the proliferation rate of the cells. The single treatment with either BMP (*P = 0.011) or Wif1 (*P = 0.026) accompanied with FGF significantly decreased the cell proliferation, while co-treatment of FGF, BMP, and Wif1 enhanced the inhibition effect (*P = 0.022). (D) qPCR using primers of BMP4, Wif1, and Smad1 were performed within different time points (0 h/6 h/72 h) samples of the above treatments. The gene expression of cells was normalized to the 0 h sample, and the 18S gene was used as an internal control.
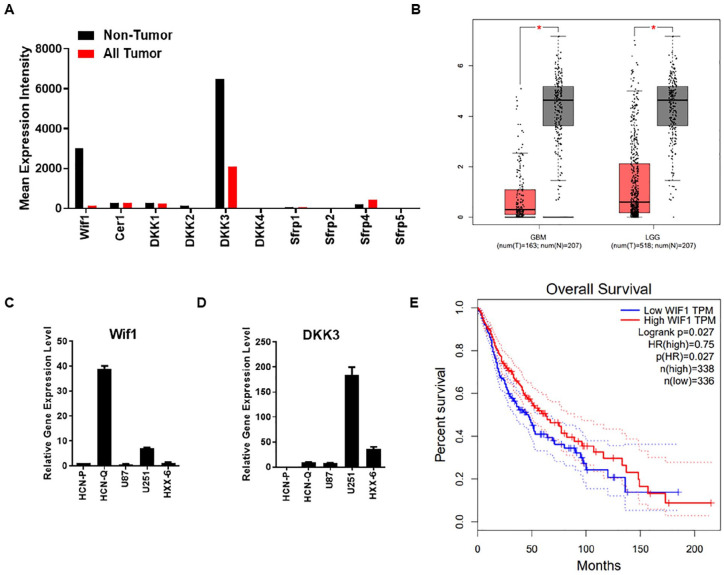
Wif1 Is Expressed at a High Level in Non-tumor Samples
Except for NSCs, the expression of Wif1 in brain malignant tumors is still important. We collected data from The Cancer Genome Atlas and Genotype-Tissue Expression (GTEx) and analyzed them using Gene Expression Profiling Interactive Analysis 2 (GEPIA2)25. Both Wif1 and DKK3 showed significantly higher expression in non-tumor samples compared with all tumor samples (Fig. 5A). When we restricted our analysis to the database of low-grade glioma (LGG) and malignant glioma (GBM), the expression of Wif1 was also lower in tumor samples (Fig. 5B). The different patterns of Wif1 in tumor and non-tumor samples suggest that Wif1 may play an essential functional role in tumourigenesis. Malignant glioma is a brain cancer with high lethality and recurrence rate26. Glioma stem cells (GSCs) may be the most important reason for the high recurrence27–29. Identifying the differences between normal NSCs and GSCs is important for selecting potential therapeutic targets. Here, we included the GSCs cell line HXX-6 we established before21 to observe the expression of Wif1 and DKK3 in active NSCs, quiescent-like NSCs, glioma cells (U87 and U251), and GCSs (HXX-6). Wif1 was highly expressed within quiescent-like NSCs (Fig. 5C), while DKK3 was highly expressed in glioma cells U251 (Fig. 5D). The overall survival analysis also indicated that high Wif1 expression correlated with longer survival periods (Fig. 5E), which may suggest its potential suppressive role in tumor treatment.
Figure 5.
Wif1 is presented with a high expression in non-tumor samples. (A) Gene expression of Wnt inhibitors in non-tumor and tumor samples. (B) Gene expression of Wif1 in GBM and LGG database. The red box stands for tumor samples, and the gray box stands for non-tumor samples. (C and D) qPCR using primers of Wif1 and DKK3 was performed within different samples as NSCs, glioma cell lines, and primary cultured glioma stem cells. (E) Overall survival results correlated with Wif1 level in GBM and LGG.
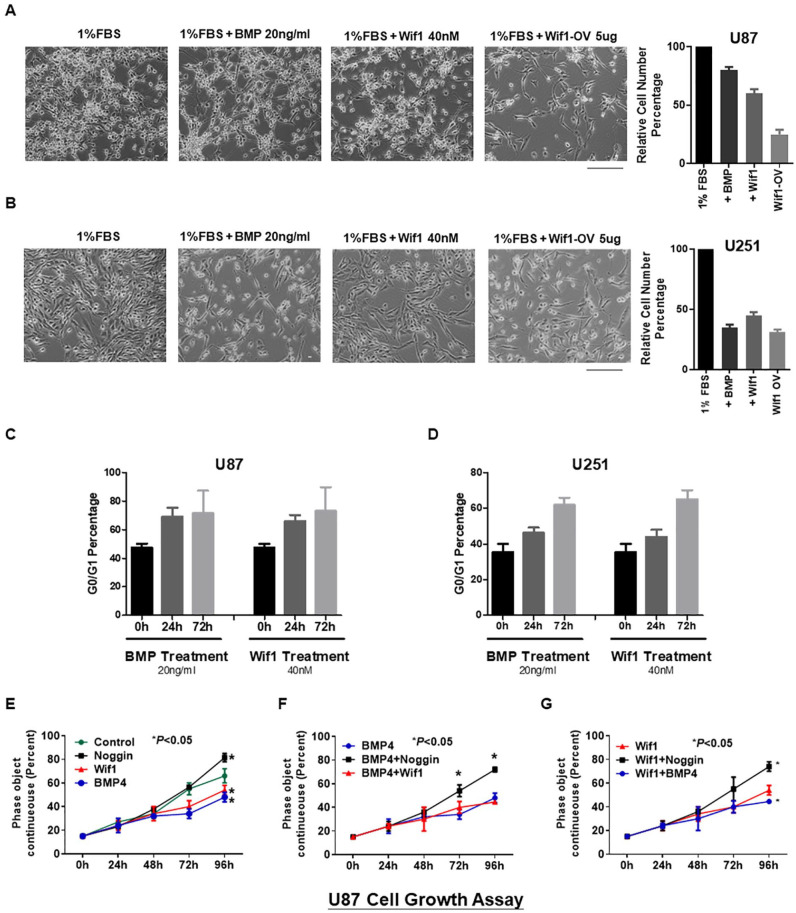
Wif1 Depresses Growth of Glioma Cells Synergistically With BMP Signaling
To study whether Wif1 could inhibit glioma growth, we included glioma cell lines U87 and u251 for functional validation. We seeded the same number of cells on day 0 and observed cell density on day 3. Since glioma cell lines grow too fast with 10% FBS, to better understand the functions of BMP4 and Wif1, we reduced the concentration of FBS to 1%. Our results showed that upregulation of both BMP4 and Wif1 successfully decreased the cell density in U87 and U251 (Fig. 6A and B). The PI staining cytometry assay showed a time-course increase in G0/G1 blocking after BMP4 (20 ng/mL) and Wif1 (40 nM) treatment in these two cell lines (Fig. 6C and D). We then performed a cell growth assay to detect the interaction between BMP signaling and Wif1 in U87 cells. Single treatment with BMP4 and Wif1 significantly decreased cell number at 96 h, while the BMP inhibitor Noggin increased cell number (Fig. 6E). When combined with BMP4, Noggin successfully upregulated the growth curve at 72 h and 96 h, as compared with BMP4 alone or along with BMP4 (Fig. 6F). At the same time, Noggin could reverse the inhibition of a single Wif1 treatment (Fig. 6G). Thus, inhibition of BMP signaling may affect downstream Wif1 and abrogate their effects on cell growth.
Figure 6.
Wif1 depresses growth of glioma cell lines synergistically with BMP signaling. (A and B) The cell density of control, BMP4 treatment (20 ng/mL), Wif1 treatment (40 nM), and Wif1 overexpression plasmid in 1% FBS medium after 72 h of treatment in glioma cell lines U87 and U251. (C and D) The G0/G1 phase status of PI staining cell cytometry results was presented as a bar chart. Both BMP4 and Wif1 could block the cell cycle at G0/G1 phase in U87 and U251. (E–G) A cell growth curve was drawn based on the cell density of U87 cells recorded every 24 h from start to 96 h. Wif1 and BMP4 decreased the cell density at 96 h while BMP inhibitor Noggin increased the cell density. Co-treatment with BMP4 and Wif1 enhanced the depression of cell density.
Discussion
The findings demonstrate that Wif1 plays an important role in the quiescence and activation of NSCs evoked in response to BMP4 stimulation. The significant upregulation of Wif1 upon BMP4 treatment was confirmed by RNA sequencing results. IHC results showed that Wif1 can significantly inhibit the cell cycle of NSCs in a manner similar to that of BMP4. We achieved digital regulation of the NSC cycle by manipulating different concentrations and combinations of BMP4 and Wif1. The expression of Wif1 was significantly reduced in GBM and LGG. Functional studies demonstrated that both BMP4 and Wif1 inhibited the growth of glioma cells and that this inhibition was released by the BMP inhibitor Noggin. The collective results show that Wif1 has an important role in the BMP signaling pathway maintenance of the stability of NSCs and the growth of glioma cells. The effect of Wif1/BMP4 on gliomas lays a technical foundation for the development of new drugs and personalized treatment.
Mesenchymal stem cells (MSCs) play an important role in neural development, have the potential to regenerate and differentiate, and are also closely related to the generation and recurrence of brain tumors. Owing to the presence of a large number of quiescent NSCs, the cells can generate new neurons through numerous intermediate progenitor cells. Activation of quiescent NSCs can generate progenitor cells and influence progenitor cell proliferation and neuronal fate. Cell transfer and differentiation are important processes in neural cell formation. Local signals in the microenvironment of NSCs can regulate these signals at various stages. Among the above signals, those in the Wnt, BMP, and FGF families may have important regulatory functions30. Although the Wnt pathway is involved in the formation of adult nerves, the relationship between Wnt and other signaling pathways has not yet been elucidated31. Wnt signaling regulates multiple developmental processes in the embryonic brain and regulates the differentiation and proliferation of progenitor cells in various adult tissues, such as hair follicles, intestinal tissue, blood, bone, and the nervous system12. Among the Wnt signaling pathways, the upstream inhibitors contain multiple proteins, including the DKK family (DKK1–DKK4), sFRP family (sFRP1–sFRP5), Cer1, and Wnt inhibitory factor 1 (Wif1). Wif1 is a secreted Wnt inhibitor that binds to any Wnt agonist and blocks the interaction between Wnt and its specific receptor, thereby inhibiting Wnt signaling32. The “Wnt-off” theory regulating the stem cell quiescence has been proved by many previous works with different choices of the upstream inhibitors22. In the mature dentate granule niche, sFRP3 is an important inhibitory factor that regulates NSC quiescence33. The expression of Wif1 was clearly higher than that of any other Wnt inhibitors in BMP4-induced quiescent-like Rattus mature NSCs. Further functional experiments suggested that Wif1 could decrease the NSC cell cycle grade and maintain them in a quiescent-like state with the help of FGF, but independent of BMP.
Among the proteins that bind directly to Wnt ligands during Wnt signaling regulation, Wif123 received the least recognition. This molecule is involved in the development of several organs34–36. Its important roles in neurogenesis, axonal extension, stem cell maintenance, and tumors have been reported37,38. Wif1 is a typical negative regulator of Wnt/β-linked protein signaling. Dysregulation of Wnt pathway components has been associated with tumor development and recurrence39,40. Among these, Wif1 is downregulated in a variety of cancers17. Recent studies have confirmed the downregulation of Wif1 expression in human gliomas and astrocytomas41. In addition, the expression of Wif1 protein was also significantly reduced in high-grade gliomas20. Our results further elucidate the Wif1/BMP interaction in glioma cells. This provides a new direction for therapeutic options for gliomas.
Because the channel-to-channel crosstalk pattern may be direct, indirect, unidirectional, or bidirectional, it generally occurs in feedback loops. The pattern of integration of these signals may vary for different cell types, developmental stages, physiological/pathological conditions, and protein cell locations42. Through the synergy of BMP and Wnt signaling, neural cell growth can be promoted during embryonic neural development43 but antagonistically interacts to maintain adult stem cell quiescence22. In telogen hair follicle stem cells, ablation of BMPRI significantly reduces the Wnt inhibitor DKK3 and induces the expression of Wnt 7a, 7B genes44. In mouse MSCs, BMP impedes Wnt-induced cell proliferation through the interaction of downstream Smad1 and disheveled segment polarity protein 1 (DVL-1) in the cytoplasm45. Recently, a protein called Hopx was found to cooperate with the BMP pathway and react with activated Smad proteins to inhibit the activation of Wnt signaling46. Overall, these findings consistently indicate that BMP and Wnt signaling antagonistically interact in adult stem cell quiescent niches.
Cell cycle length is an important factor that affects the differentiation potential and tendency of stem cells. Strict temporal control of signaling activity is required during the development and maintenance of homeostasis. Accurate modulation of the cell cycle is important for homeostasis of tissues and organs. The disruption of the cell cycle is a key feature of malignancy. Identification of key factors regulating the cell cycle during neurodevelopment could help precisely control the NSC development process and provide reliable therapeutic options for neurological diseases. This study confirmed the role of Wif1 in regulating the cell cycle of NSCs.
Conclusion
This study provides a first demonstration that Wif1 is involved in the crosstalk between the BMP and Wnt signaling pathways. The cell cycle can be exquisitely regulated by the kinetics of BMP4 and Wif1. As such, Wif1 plays an important role in maintaining the state of BMP4 signaling, keeping neural and brain tumor stem cells at bay (quiescence). The study findings lay the foundation for further understanding of the interaction between BMP/Wnt/FGF and the development of individualized therapeutic strategies. Further functional tests and transcriptional analysis with multiple glioma cell lines and/or primary tissues from clinical patients would help to better understand molecular mechanisms underlying the interaction and crosstalk among diverse pathways such as BMP/Wnt/FGF signaling.
Acknowledgments
We thank all the colleges in our lab and Editage (www.editage.com) for English language editing.
Footnotes
Author Contributions: XCD, GZL, and CCH conceived and designed the experiments; XCD, CCH, FYT, and HXY performed the experiments; CCH, GZL, and LZ analyzed the data; XCD, LZ, and CCH wrote the manuscript; FYT, CCH, and GZL provided financial support. All the authors approved the final manuscript.
Availability of Data and Materials: All data generated or analyzed during this study are included in this article.
Ethical Approval: This study was approved by our institutional review board.
Statement of Human and Animal Rights: This article does not contain any studies with human or animal subjects.
Statement of Informed Consent: There are no human subjects in this article and informed consent is not applicable.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by funds from the National Key R&D Program of China (2019YFA0110300) and the National Natural Science Foundation of China (31600819 to CHC, 81773302 to YTF, and 31571058 and 32070862 to ZLG).
ORCID iD: Chunhui Cai  https://orcid.org/0000-0002-8700-4383
https://orcid.org/0000-0002-8700-4383
References
- 1. Kalies S, Heinemann D, Schomaker M, Murua Escobar H, Heisterkamp A, Ripken T, Meyer H. Plasmonic laser treatment for Morpholino oligomer delivery in antisense applications. J Biophotonics. 2014;7(10):825–33. [DOI] [PubMed] [Google Scholar]
- 2. Szopa W, Burley TA, Kramer-Marek G, Kaspera W. Diagnostic and therapeutic biomarkers in glioblastoma: current status and future perspectives. Biomed Res Int. 2017;2017:8013575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hirabayashi Y, Itoh Y, Tabata H, Nakajima K, Akiyama T, Masuyama N, Gotoh Y. The Wnt/beta-catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development. 2004;131(12):2791–2801. [DOI] [PubMed] [Google Scholar]
- 4. Muroyama Y, Kondoh H, Takada S. Wnt proteins promote neuronal differentiation in neural stem cell culture. Biochem Biophys Res Commun. 2004;313(4):915–21. [DOI] [PubMed] [Google Scholar]
- 5. ten Berge D, Brugmann SA, Helms JA, Nusse R. Wnt and FGF signals interact to coordinate growth with cell fate specification during limb development. Development. 2008;135(19):3247–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297(5580):365–69. [DOI] [PubMed] [Google Scholar]
- 7. Cunningham TJ, Kumar S, Yamaguchi TP, Duester G. Wnt8a and Wnt3a cooperate in the axial stem cell niche to promote mammalian body axis extension. Dev Dyn. 2015;244(6):797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu A, Niswander LA. Bone morphogenetic protein signaling and vertebrate nervous system development. Nat Rev Neurosci. 2005;6(12):945–54. [DOI] [PubMed] [Google Scholar]
- 9. Almario RU, Karakas SE. Roles of circulating WNT-signaling proteins and WNT-inhibitors in human adiposity, insulin resistance, insulin secretion, and inflammation. Horm Metab Res. 2015;47(2):152–57. [DOI] [PubMed] [Google Scholar]
- 10. Ding SL, Yang ZW, Wang J, Zhang XL, Chen XM, Lu FM. Integrative analysis of aberrant Wnt signaling in hepatitis B virus-related hepatocellular carcinoma. World J Gastroenterol. 2015;21(20):6317–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kühl M, Sheldahl LC, Park M, Miller JR, Moon RT. The Wnt/Ca2+ pathway: a new vertebrate Wnt signaling pathway takes shape. Trends Genet. 2000;16(7):279–83. [DOI] [PubMed] [Google Scholar]
- 12. Mao Y, Ge X, Frank CL, Madison JM, Koehler AN, Doud MK, Tassa C, Berry EM, Soda T, Singh KK, Biechele T, et al. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3beta/beta-catenin signaling. Cell. 2009;136(6):1017–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149(6):1192–205. [DOI] [PubMed] [Google Scholar]
- 14. Cui Y, Han J, Xiao Z, Chen T, Wang B, Chen B, Liu S, Han S, Fang Y, Wei J, Wang X, et al. The miR-20-Rest-Wnt signaling axis regulates neural progenitor cell differentiation. Sci Rep. 2016;6:23300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mimura S, Suga M, Okada K, Kinehara M, Nikawa H, Furue MK. Bone morphogenetic protein 4 promotes craniofacial neural crest induction from human pluripotent stem cells. Int J Dev Biol. 2016;60(1–3):21–28. [DOI] [PubMed] [Google Scholar]
- 16. Varela-Nallar L, Inestrosa NC. Wnt signaling in the regulation of adult hippocampal neurogenesis. Front Cell Neurosci. 2013; 7:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wissmann C, Wild PJ, Kaiser S, Roepcke S, Stoehr R, Woenckhaus M, Kristiansen G, Hsieh JC, Hofstaedter F, Hartmann A, Knuechel R, et al. WIF1, a component of the Wnt pathway, is down-regulated in prostate, breast, lung, and bladder cancer. J Pathol. 2003;201(2):204–12. [DOI] [PubMed] [Google Scholar]
- 18. Queimado L, Obeso D, Hatfield MD, Yang Y, Thompson DM, Reis AM. Dysregulation of Wnt pathway components in human salivary gland tumors. Arch Otolaryngol Head Neck Surg. 2008;134(1):94–101. [DOI] [PubMed] [Google Scholar]
- 19. Deng X, Hou C, Wang H, Liang T, Zhu L. Hypermethylation of WIF1 and its inhibitory role in the tumor growth of endometrial adenocarcinoma. Mol Med Rep. 2017;16(5):7497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lambiv WL, Vassallo I, Delorenzi M, Shay T, Diserens AC, Misra A, Feuerstein B, Murat A, Migliavacca E, Hamou MF, Sciuscio D, et al. The Wnt inhibitory factor 1 (WIF1) is targeted in glioblastoma and has a tumor suppressing function potentially by induction of senescence. Neuro Oncol. 2011;13(7): 736–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Han XX, Cai C, Yu LM, Wang M, Hu DY, Ren J, Zhang MH, Zhu LY, Zhang WH, Huang W, He H, et al. A fast and efficient approach to obtaining high-purity glioma stem cell culture. Front Genet. 2021;12:639858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327(5965):542–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shen EH, Overly CC, Jones AR. The Allen Human Brain Atlas: comprehensive gene expression mapping of the human brain. Trends Neurosci. 2012;35(12):711–14. [DOI] [PubMed] [Google Scholar]
- 24. Mira H, Andreu Z, Suh H, Lie DC, Jessberger S, Consiglio A, San Emeterio J, Hortiguela R, Marques-Torrejon MA, Nakashima K, Colak D, et al. Signaling through BMPR-IA regulates quiescence and long-term activity of neural stem cells in the adult hippocampus. Cell Stem Cell. 2010;7(1):78–89. [DOI] [PubMed] [Google Scholar]
- 25. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu S, Mischel PS. Same script, different cast: different cell origins shape molecular features and therapeutic response in glioblastoma. Cancer Cell. 2020;38(3):311–13. [DOI] [PubMed] [Google Scholar]
- 27. Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, Richman AR, Silverbush D, Shaw ML, Hebert CM, Dewitt J, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178(4):835–49.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Couturier CP, Ayyadhury S, Le PU, Nadaf J, Monlong J, Riva G, Allache R, Baig S, Yan X, Bourgey M, Lee C, et al. Single-cell RNA-seq reveals that glioblastoma recapitulates a normal neurodevelopmental hierarchy. Nat Commun. 2020;11(1):3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhu Z, Mesci P, Bernatchez JA, Gimple RC, Wang XX, Schafer ST, Wettersten HI, Beck S, Clark AE, Wu QL, Prager BC, et al. Zika virus targets glioblastoma stem cells through a SOX2-Integrin αvβ5 axis. Cell Stem Cell. 2020;26(2):187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Choe Y, Pleasure SJ, Mira H. Control of adult neurogenesis by short-range morphogenic-signaling molecules. Cold Spring Harb Perspect Biol. 2015;8(3):a018887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Armenteros T, Andreu Z, Hortiguela R, Lie DC, Mira H. BMP and WNT signaling cooperate through LEF1 in the neuronal specification of adult hippocampal neural stem and progenitor cells. Sci Rep. 2018;8(1):9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kawano Y, Kypta R. Secreted antagonists of the Wnt signaling pathway. J Cell Sci. 2003;116(Pt 13):2627–34. [DOI] [PubMed] [Google Scholar]
- 33. Jang MH, Bonaguidi MA, Kitabatake Y, Sun J, Song J, Kang E, Jun H, Zhong C, Su Y, Guo JU, Wang MX, et al. Secreted frizzled-related protein 3 regulates activity-dependent adult hippocampal neurogenesis. Cell Stem Cell. 2013;12(2):215–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hunter DD, Zhang M, Ferguson JW, Koch M, Brunken WJ. The extracellular matrix component WIF-1 is expressed during, and can modulate, retinal development. Mol Cell Neurosci. 2004;27(4):477–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nakaya N, Lee HS, Takada Y, Tzchori I, Tomarev SI. Zebrafish olfactomedin 1 regulates retinal axon elongation in vivo and is a modulator of Wnt signaling pathway. J Neurosci. 2008;28(31):7900–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xu B, Chen C, Chen H, Zheng SG, Bringas P, Jr, Xu M, Zhou X, Chen D, Umans L, Zwijsen A, Shi W. Smad1 and its target gene Wif1 coordinate BMP and Wnt signaling activities to regulate fetal lung development. Development. 2011;138(5):925–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lim FT, Ogawa S, Smith AI, Parhar IS. Proteomics identification of potential candidates involved in cell proliferation for early stage of brain regeneration in the adult zebrafish. Zebrafish. 2017;14(1):10–22. [DOI] [PubMed] [Google Scholar]
- 38. Nayak S, Mahenthiran A, Yang Y, McClendon M, Mania-Farnell B, James CD, Kessler JA, Tomita T, Cheng SY, Stupp SI, Xi G. Bone morphogenetic protein 4 targeting glioma stem-like cells for malignant glioma treatment: latest advances and implications for clinical application. Cancers (Basel). 2020;12(2):516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nusse R, Clevers H. Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169(6):985–99. [DOI] [PubMed] [Google Scholar]
- 40. Tai D, Wells K, Arcaroli J, Vanderbilt C, Aisner DL, Messersmith WA, Lieu CH. Targeting the WNT signaling pathway in cancer therapeutics. Oncologist. 2015;20(10):1189–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang Z, Wang Y, Fang J, Chen F, Liu J, Wu J, Wang Y, Song T, Zeng F, Rao Y. Downregulation of WIF-1 by hypermethylation in astrocytomas. Acta Biochim Biophys Sin (Shanghai). 2010;42(6):418–25. [DOI] [PubMed] [Google Scholar]
- 42. Guo X, Wang XF. Signaling cross-talk between TGF-beta/BMP and other pathways. Cell Res. 2009;19(1):71–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Urban N, Guillemot F. Neurogenesis in the embryonic and adult brain: same regulators, different roles. Front Cell Neurosci. 2014;8:396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kandyba E, Leung Y, Chen YB, Widelitz R, Chuong CM, Kobielak K. Competitive balance of intrabulge BMP/Wnt signaling reveals a robust gene network ruling stem cell homeostasis and cyclic activation. Proc Natl Acad Sci U S A. 2013; 110(4):1351–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu Z, Tang Y, Qiu T, Cao X, Clemens TL. A dishevelled-1/Smad1 interaction couples WNT and bone morphogenetic protein signaling pathways in uncommitted bone marrow stromal cells. J Biol Chem. 2006;281(25):17156–63. [DOI] [PubMed] [Google Scholar]
- 46. Jain R, Li D, Gupta M, Manderfield LJ, Ifkovits JL, Wang Q, Liu F, Liu Y, Poleshko A, Padmanabhan A, Raum JC, et al. HEART DEVELOPMENT. Integration of Bmp and Wnt signaling by Hopx specifies commitment of cardiomyoblasts. Science. 2015;348(6242):aaa6071. [DOI] [PMC free article] [PubMed] [Google Scholar]