Abstract
Background:
Loco-regional treatment strategies of colorectal cancer (CRC) metastases are evolving, but biological markers that can benefit patients and assist physicians in clinical decisions are lacking. The primary objective of this systematic review and meta-analysis is to investigate the current knowledge on circulating DNA and its clinical utility in predicting outcomes in patients undergoing loco-regional treatment of CRC metastases.
Methods:
A systematic search of PubMed, Embase, and Cochrane Central Register of Controlled Trials was conducted on March 22, 2022. We included studies on patients undergoing loco-regional treatment of CRC metastases reporting the predictive or prognostic value of circulating DNA in the blood. Hazard ratios (HR) were pooled in separate random-effects meta-analyses to investigate if pre- or post-ablation measurements of circulating DNA were associated with survival. The risk of bias was assessed according to the Quality in Prognosis Studies tool.
Results:
Twenty-eight studies with 2868 patients were included, of which 16 studies were eligible for meta-analyses. As expected in this new research field, a majority of included studies (n = 21/28) had a high risk of bias in at least one domain. Circulating DNA above the cutoff in a plasma sample taken before loco-regional treatment was associated with a short recurrence-free survival [pooled HR = 2.8, 95% confidence interval (CI) 1.4–5.7, n = 162] and overall survival (pooled HR = 4.7, 95% CI 1.1–20.6, n = 105). Circulating DNA above the cutoff in a plasma sample taken after loco-regional treatment was associated with a short recurrence-free survival (pooled HR = 4.5, 95% CI 3.4–6.1, n = 569) and overall survival (pooled HR = 7.5, 95% CI 2.0–27.3, n = 161). There was limited data on the association between dynamics in circulating DNA and outcome.
Conclusions:
Measurements of circulating DNA can be valuable when selecting and monitoring patients undergoing loco-regional treatment of CRC metastases. Studies designed to investigate the true clinical utility of circulating DNA in the context of various ablation modalities are warranted.
The review has been registered at PROSPERO (ID: CRD42022320032)
Keywords: biomarker, cell-free DNA, circulating free DNA, circulating tumor DNA, loco-regional treatment, metastatic colorectal cancer
Introduction
Colorectal cancer (CRC) is the third most common cancer worldwide, and globally the second leading cause of cancer-related deaths.1 Approximately 20% of patients with CRC present with metastatic disease at the time of diagnosis, and around 50% of patients will eventually develop metastases. Metastatic colorectal cancer (mCRC) is a term that covers a continuum of conditions from one or a few metastases in one organ to widespread disease affecting many organs. Loco-regional treatment may be applied for different purposes in this continuum, for example, late-stage treatment of widespread disease, consolidation, or curation. The field of loco-regional therapies for managing mCRC is rapidly evolving, but biological markers that can assist in clinical decisions are lacking.
There is growing evidence that circulating DNA in blood can aid clinical decision-making in the care pathway of patients with both curable and incurable CRC.
Having minimal residual disease as detected by circulating tumor DNA (ctDNA) after curative treatment of CRC is associated with a very high risk of recurrence.2–6 A prospective study has shown that a ctDNA-guided approach after surgery for stage II CRC indeed can identify high-risk patients in need of adjuvant chemotherapy.7 In the setting of incurable mCRC, both the levels of total circulating cell-free DNA (cfDNA) and ctDNA have prognostic value, and the detection of specific mutations and their dynamics may provide additional predictive information relevant for tailored treatment approaches.8,9
Circulating DNA has the potential of assisting in selecting patients who may benefit from loco-regional treatment, optimizing neo-adjuvant and post-interventional adjuvant strategies, and improving follow-up.
The primary objective of this study is to present a systematic review and meta-analyses of circulating DNA as a biomarker and its clinical utility in predicting outcomes from loco-regional treatment of CRC metastases. With this review, we summarize the current knowledge and provide future perspectives on research in this field.
Methods
Prespecified eligibility criteria
Eligibility criteria include human studies on patients with metastatic colorectal adenocarcinoma receiving loco-regional treatment for metastases, where circulating DNA (including cfDNA and/or ctDNA) in the blood is associated with treatment response, recurrence, and/or a survival end point.
The review was registered with PROSPERO (CRD42022320032) and is reported in accordance with the PRISMA statement.10
Information sources and systematic search strategy
As of March 22, 2022, we searched PubMed, Embase, and Cochrane Central Register of Controlled Trials to identify relevant studies. The searches were not limited, that is, by language, year of publication, or type of publication. Specific search strategies and strings are specified in Supplementary Methods. Studies were managed using Covidence (Covidence systematic review software, Veritas Health Innovation, Melbourne, Australia, available at www.covidence.org).
Study evaluation and selection
Two researchers independently evaluated the studies through each phase of the review (i.e., screening, eligibility, and inclusion) (LBC, TT). Study screening for eligibility and data collection of relevant studies were performed using a predefined data extraction form. All studies were initially screened by title and abstract, followed by a full-text assessment. Consensus was reached by discussion with a third researcher if necessary (K-LGS). At least two researchers independently (LC, TT) tabulated core data items from included studies. We collected data on the study (author, year, design), the number of patients, loco-regional treatment modality, metastatic site, adjuvant therapy, circulating DNA marker/analytical method, time of blood sampling, evaluated cutoff(s), treatment response, recurrence, hazard ratio (HR) for survival end points including 95% confidence interval (CI) and p-value.
Risk of bias assessment
Risk of bias was assessed independently by two researchers (LC, TT) according to the Quality in Prognosis Studies (QUIPS) tool.11 Specifically, this included biases in the domains of study participation, study attrition, biomarker measurement, outcome measurement, study confounding, and statistical analysis and reporting. Each domain was scored according to the categories low, moderate, or high risk of bias. Consensus was reached by discussion (with a third researcher when necessary). An overall risk of bias judgment across domains was not performed.
Summary and synthesis of results
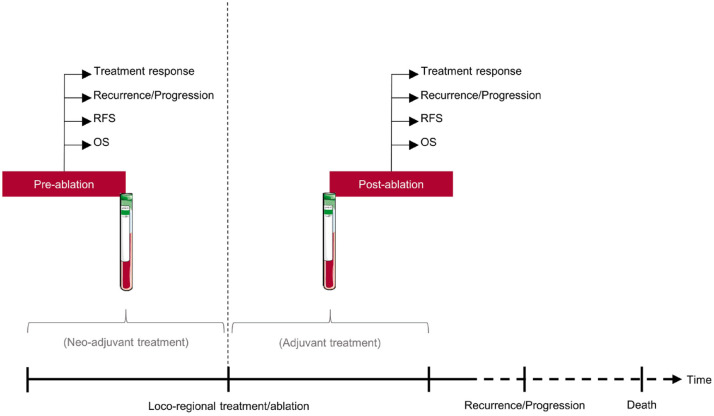
To illustrate different aspects of the clinical utility of circulating DNA as a biomarker in patients undergoing loco-regional treatment, we grouped studies based on the timing of blood sampling (i.e. liquid biopsies) (Figure 1).
Figure 1.
Schematic overview of clinical applications of liquid biopsies in patients undergoing loco-regional treatment of colorectal cancer metastases.
RFS, recurrence-free survival; OS, overall survival.
Pre-ablation circulating DNA was defined as a measurement of a given marker at a defined time-point before the planned loco-regional treatment/ablation. The time-point may be before, during, or after potential neo-adjuvant therapy. Post-ablation circulating DNA was defined as a measurement of a given marker at a time-point after loco-regional treatment/ablation. The time-point may be before, during, or after a potential post-interventional adjuvant therapy. Dynamics in circulating DNA was defined as a change in a circulating DNA marker from pre- to post-ablation.
We evaluated the predictive and prognostic significance of pre-ablation and post-ablation circulating DNA and dynamics where applicable. Data were summarized in tables. Meta-analyses were performed and data were summarized in forest plots. Studies were ordered according to publication year.
In the present description, further grouping according to treatment modality or intent was unfeasible due to the limited amount of studies and lack of details in the clinical setting.
Statistical analysis
HRs with corresponding 95% CIs reported in the studies were used to estimate the strength of the relationship between circulating DNA and survival. Only studies providing HR based on univariate analysis and corresponding 95% CI were included in the meta-analyses. Due to inter-study heterogeneity, study-specific results were pooled using a random-effects model. Funnel plots were generated to visually assess publication bias.
Heterogeneity was quantified by χ2 tests and inconsistency index (I2) tests statistics. A χ2 test of p < 0.10 or I2 > 50% indicated heterogeneity among studies. All analyses were performed with Stata software version 17.0 (Stata Corporation, College Station, TX, USA). A two-sided p-value below 0.05 was considered statistically significant.
When selecting results for the meta-analyses, the following applied: if multiple results for one outcome parameter (recurrence-free survival (RFS), overall survival (OS)) were available from an included study (e.g., different cutoffs, circulating DNA marker), all results were included if each result originated from separate study cohorts. The results with the most complete data (HR, 95% CI, p-value, n) were included when originating from the same study cohort. If the completeness was equal, we included the results that applied to the largest patient cohort.
Fisher’s exact test was applied to compare categorical variables.
Results
Eligibility assessment
A total of 876 study entries were identified and screened in abstract form according to the prespecified eligibility criteria. Only studies published in full-text versions were considered for inclusion.
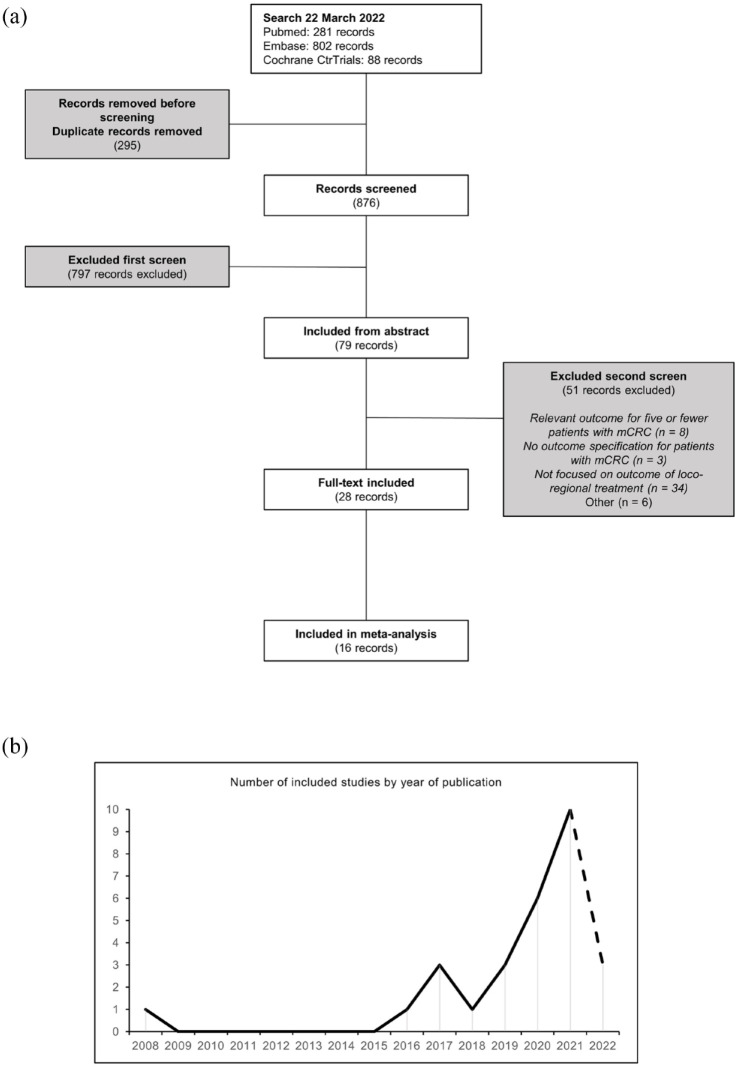
Twenty-eight publications were included in the final review, fulfilling all eligibility criteria. The eligibility assessment was summarized in a flow diagram (Figure 2(a)). An exponential increase in the number of eligible studies was seen from 2018 (Figure 2(b)). The studies included a total of 2868 patients and reported an association between circulating DNA and outcomes relevant to this systematic review in at least 864 patients.
Figure 2.
PRISMA diagram and eligible studies by year of publication. (a) PRISMA flow diagram of the study selection process and (b) number of included studies by year of publication.
Risk of bias
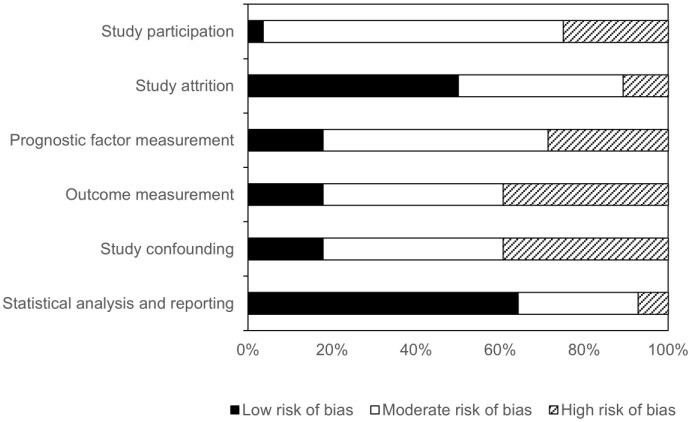
According to the risk of bias assessment, no studies had a low risk of bias in all six domains. A high risk of bias in at least one domain was seen in the majority of the studies (n = 21/28). A high risk of bias was most common in the domains ‘outcome measurement’ and ‘study confounding’ (Figure 3, Supplemental Table 1). The risk of bias in studies incorporated in the meta-analyses was comparable to the risk of bias across all studies included in the review (Supplemental Figure 1).
Figure 3.
Assessment of risk of bias using Quality in Prognosis Studies tool. The authors’ judgments regarding each risk-of-bias domain presented as percentages across all included studies.
Only one study12 reported to follow the REMARK guidelines.13
Characteristics of included studies
The studies were heterogeneous with regard to site of treated metastases, treatment modality, analytical method, circulating DNA marker, cutoff, time of sampling, and the number of included patients (Tables 1–3).
Table 1.
Studies reporting on association between pre-ablation circulating DNA and treatment response, recurrence, and/or survival.
| First author | Year | Metastatic site | Treatment modality | Analytical method | Circulating DNA marker | Cutoff | Time of sampling | n | Circulating DNA marker statistically significantly associated with | Ref | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genetic mutation unless otherwise specified | Treatment response | Recurrence | RFS | OS | |||||||||
| Diehl | 2008 | Liver, lung, oment, stomach | Resection, RFA | BEAMing | Tumor-informed (APC, KRAS, PIK3CA, TP53) | Detection | On the day of surgery | 16 | NR | D | NR | NR | 14 |
| Shin | 2017 | NR | Resection | MassArray | Tumor specific KRAS mutation | Detection | On the day of surgery | NR | NR | NR | NR | Yes | 40 |
| Boysen | 2018 | Liver | DEBIRI-TACE | DFA | cfDNA-level | 75th percentile | Baseline | NR | No | NR | NR | D | 38 |
| Narayan | 2019 | Liver | Resection | Targeted sequencing | 9 genes/cfDNA level | Detection/levelc | During surgery before resection | 59 | NR | NR | No | No/Yesh | 17 |
| Bidard | 2019 | Liver | Resection | ddPCR | KRAS | Detection | Before neo-adjuvant CT | NR | NR | NR | NR | Yes | 12 |
| After 1 month of neo- adjuvant CT | NR | NR | NR | No | |||||||||
| Before liver surgery | NR | NR | NR | Yes | |||||||||
| He | 2020 | Liver | Resection | Targeted sequencing | 41 genes/cfDNA level | High versus low levelc | Within 7 days before surgery | 19 | NR | NR | Yes | NR | 18 |
| Beagan | 2020 | Peritoneal, liver, lnn | CRS-HIPEC, resection | ddPCR | Tumor-informed (48 genes) and KRAS | Detection | During surgery before resection | 24 | NR | Noe | Yesf/Nog | NR | 19 |
| Polivka | 2020 | Liver | Resection | ddPCR | KRAS | Detection/FA = 3.33%/percentage increment | Preoperative | 30 | NR | NR | No | No/Yesh | 20 |
| Boysen | 2020B | Liver | HAI | DFA | cfDNA-level | 75th percentile | Before first HAI | NR | Yes | NR | NR | Yes | 16 |
| Kobayashi | 2021 | Liver | Resection | Targeted sequencing | 74 genes | Detection | Within a month of Liver resection | 40 | NR | Yes | Yes | No | 21 |
| Nakamura | 2021 | Liver, lung, lnn | PBT, SBRT | Targeted sequencing | 18 genes | Detection/MaxAF = 1%/clonal versus multiclonal | Before ablative radiotherapy | 20 | NR | NR | No/Yesh | NR | 22 |
| Tie | 2021 | Liver | Resection | Targeted sequencing | Tumor-informed (15 genes) | Detection | Before possible neo-adjuvant CT and surgery | 54 | NR | NR | No | No | 23 |
| Lee | 2021 | Liver, lung, peritoneum, lnn, rectum | Resection | Targeted sequencing | Tumor-informed | Detection | Before resection | 58 | NR | D | No | No | 24 |
| Wang | 2021 | Liver | Resection | Targeted sequencing | 451 genes | Detection | At diagnosis before treatment | NR | NR | NR | No | NR | 25 |
| Before resection | NR | NR | No | NR | |||||||||
| Pellini | 2021 | Liver and others | Resection | Targeted sequencing | 197 genes | Detection | On the day of surgery | 24 | NR | Noe | No | NR | 26 |
| Parikh | 2021 | NR | NR | Targeted sequencing | Multiple analytical features | Detection | Preoperatively | 15 | NR | Noe | NR | NR | 27 |
| Reinerta | 2022 | Liver | Resection | ddPCR | Tumor-informed (APC, BRAF, KRAS, NRAS, PIK3CA, TP53)b | Detection | Prior to liver resection | 68 | NR | No | NR | NR | 28 |
| Øgaarda | 2022 | Liver | Resection | ddPCR | TriMeth | Positived | Prior to liver resection | NR | NR | No | No | NR | 29 |
Studies included in at least one meta-analysis are indicated in bold type. For some outcomes, n may be lower.
Same study group.
Preoperative plasma samples from two patients, where no liver metastasis tissue was available.
No cutoff reported.
Samples were classified as ‘TriMeth positive’ if two out of three TriMeth markers showed >1 positive droplet.
Fisher’s exact test applied.
All patients.
Patients without liver metastases.
Different associations for different markers of circulating DNA.
cfDNA, circulating cell-free DNA; CRS-HIPEC, cytoreductive surgery and hyperthermic intraperitoneal chemotherapy; CT, chemotherapy; D, descriptively with no statistical test; ddPCR, droplet digital polymerase chain reaction; DEBIRI-TACE, transarterial chemoembolization with irinotecan loaded beads; DFA, direct flourescent assay; FA, fractional abundance; HAI, hepatic artery infusion; lnn, lymph nodes; MaxAF, maximum allele frequency; n, number of patients with reported association between pre-ablation circulating DNA and at least one outcome; NR, not reported; PBT, proton beam therapy; Ref, reference; RFA, radiofrequency ablation; SBRT, stereotactic body radiation therapy; OS, overall survival; RFS, recurrence-free survival.
Table 2.
Studies reporting on association between post-ablation circulating DNA and treatment response, recurrence, and/or survival.
| First author | Year | Metastatic site | Treatment modality | Analytical Method | Circulating DNA marker | Cutoff | Time of sampling | n | Circulating DNA marker statistically significantly associated with | Ref | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genetic mutation unless otherwise specified | Treatment response | Recurrence | RFS | OS | |||||||||
| Diehl | 2008 | Liver, lung, oment, stomach | Resection, RFA | BEAMing | Tumor-informed (APC, KRAS, PIK3CA, TP53) | Detection | Postoperative | 16 | NR | D | NR | NR | 14 |
| Yamada | 2016 | Liver, lung, lnn | Resection | Peptide nucleic acid clamping | KRAS | Detection | Within 1 month of resection | 7 | NR | Nog | NR | NR | 30 |
| Schøler* | 2017 | Liver | Resection, RFA | ddPCR | Tumor-informed (KRAS, NRAS, BRAF) | Detection | Within 3 months of resection | 23 | NR | Nog | Yes | NR | 31 |
| Benešová | 2019 | Liver, lung, lnn, muscle, peritoneum | Resection | PCR with the formation of heteroduplexes and subsequent separation using DCE | Tumor-informed (KRAS, TP53, APC, PIK3CA, BRAF, CTNNB1) | Detection | 1 week after resection | 27 | NR | D | NR | NR | 32 |
| He | 2020 | Liver | Resection | Targeted sequencing | 41 genes/cfDNA level | High versus low levele | Within 7 days of resection | 19 | NR | NR | No | NR | 18 |
| Boysen | 2020A | Liver, lung | Resection, RFA, SBRT | ddPCR | Variousb | Detection | 2 weeks after ablation | 35 | NR | Nog | Yes | NR | 2 |
| Beagan | 2020 | Peritoneal, liver, lnn | CRS-HIPEC, resection | ddPCR | Tumor-informed (48 genes) and KRAS | Detection | Within a month of CRS-HIPEC | 24 | NR | Nog | NR | NR | 19 |
| Boysen | 2020B | Liver | HAI | DFA | cfDNA-level | 75th percentile | At the end of HAI | NR | NR | NR | NR | No | 15 |
| Mason | 2021 | Liver, lung, lnn, peritoneum, ovary, adrenal gland | Resection | Targeted sequencing | 70 genes | Detection | Postoperative | 63 | NR | No | NR | Yes | 33 |
| Tie | 2021 | Liver | Resection | Targeted sequencing | Tumor-informed (15 genes) | Detection | Postoperative (before ACT) | 49 | NR | Yesg | Yes | Yes | 23 |
| End of treatment (resection and possible ACT) | NR | Yesg | Yes | Yes | |||||||||
| Lee | 2021 | Liver, lung, peritoneum, lnn, rectum | Resection | Targeted sequencing | Tumor-informed | Detection | 3–4 weeks after resection | 58 | NR | D | NR | NR | 24 |
| Wang | 2021 | Liver | Resection | Targeted sequencing | 451 genes | Detection | Postoperative (before ACT) | NR | NR | Yesg | Yes | NR | 25 |
| After completion of ACT | NR | Yesg | Yes | NR | |||||||||
| Loupakis | 2021 | Liver, lung, peritoneum, others | Resection | mPCR-NGS | Tumor-informed | Detection | Postoperative (before ACT) | 112 | NR | Yesg | Yes | Yes | 34 |
| Bolhuis | 2021 | Liver | Resection, local ablative therapy | ddPCR | RAS hotspot mutations | Detection | Within 100 days of ablation | 23 | NR | Nog | Yes | NR | 35 |
| Parikh | 2021 | NR | NR | Targeted sequencing | Multiple analytical features | Detection | App. 1 month after surgery and possible ACT | 16 | NR | Nog | Yes | NR | 27 |
| Reinert a | 2022 | Liver | Resection | ddPCR | Tumor-informed (APC, BRAF, KRAS, NRAS, PIK3CA, TP53)c | Detection | Day 30 (±14 days) after surgery and prior to ACT | 40 | NR | Yes | Yes | NR | 28 |
| Øgaard a | 2022 | Liver | Resection | ddPCR | TriMeth | Positivef | Within 3 months after resection prior to possible ACT | NR | NR | Yes | Yes | NR | 29 |
| Within 3 months after resection (non-ACT treated patients) | NR | Yes | Yes | NR | |||||||||
| After completion of ACT | NR | Yes | Yes | NR | |||||||||
| Nishioka | 2022 | Liver | Resection | Targeted sequencing | 23/70 genesd | Detection | Within 180 days after resection | 105 | NR | Yes | Yes | Yes | 36 |
Studies included in at least one meta-analysis are indicated in bold type. For some outcomes, n may be lower.
Same study group.
Depending on how well the observed mutations in the tissue had been characterized, ctDNA was analyzed either by singleplex or multiplex ddPCR assays.
Preoperative plasma samples from two patients, where no liver metastasis tissue was available.
Depending on the time period.
No cutoff reported.
Samples were classified as ‘TriMeth positive’ if two out of three TriMeth markers showed >1 positive droplet.
Fisher’s exact test applied.
ACT, adjuvant chemotherapy; cfDNA, circulating cell-free DNA; CRS-HIPEC, cytoreductive surgery and hyperthermic intraperitoneal chemotherapy; D, descriptively with no statistical test; ddPCR, droplet digital polymerase chain reaction; DCE, Denaturing capillary electrophoresis; HAI, hepatic arterial infusion; lnn, lymph nodes; mPCR-NGS, tumor-informed multiplex PCR next-generation sequencing assay; n, number of patients with reported association between post-ablation circulating DNA and at least one outcome; NR, not reported; Ref, reference; RFA, radiofrequency ablation; SBRT, stereotactic body radiation therapy; OS, overall survival; RFS, recurrence-free survival.
Table 3.
Studies reporting on correlation between dynamics in circulating DNA and treatment response, recurrence, and/or survival.
| First author | Year | Metastatic site | Treatment modality | Analytical method | Circulating DNA marker | Time of sampling | Groups compared | n | Circulating DNA marker statistically significantly associated with | Ref | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Treatment response | Recurrence | RFS | OS | ||||||||||
| Janowski | 2017 | Liver | SIRT | qPCR | Fragmentation index | 1. Immediately prior to SIRT | Decrease versusincrease | NR | NR | NR | Noc | Nod/Yese | 37 |
| 2. 2–4 weeks after SIRT | |||||||||||||
| Boysen | 2018 | Liver | DEBIRI-TACE | DFA | cfDNA level | 1. Baseline | Decline above the median value versus decline below the median value | NR | NR | NR | NR | D | 38 |
| 2. During the 8 weeks of treatment | |||||||||||||
| Iwai | 2019 | Liver | Resection | RTqPCR | cfDNA level/integrity/cLBR | 1. On the day of surgery. | Decrease versus increase | 41 | NR | No/Yesa,b | No/Yesa | NR | 39 |
| 2. 1 month after surgery | |||||||||||||
For some outcomes, n may be lower.
Different association for different ‘circulating free DNA’ marker.
Fisher’s exact test applied.
Progression-free survival in the liver.
Overall survival since SIRT.
Overall survival since diagnosis.
cfDNA, circulating cell-free DNA; cLBR, long cfDNA fragment/β-globin ratio; D, descriptively with no statistical test; DEBIRI-TACE, transarterial chemoembolization with irinotecan loaded beads; DFA, direct fluorescent assay; n, number of patients with reported association between dynamics in circulating DNA and at least one outcome; NR, not reported; qPCR, quantitative polymerase chain reaction; Ref, reference; RTqPCR, real-time qPCR; SIRT, selective internal radiation therapy OS, overall survival; RFS, recurrence-free survival.
The majority of studies investigated treatment of liver metastases by resection (n = 14/28)12,17,18,20,21,23,25,28,29,31,33,35,36,39 (for some patients in combination with local ablative therapy31,35 or resection of non-liver metastases33), repeated transarterial chemoembolization (TACE) with irinotecan loaded beads (DEBIRI-TACE) (n = 1/28),38 hepatic artery infusion (HAI) (n = 1/28),15 or selective internal radiation therapy (SIRT) (n = 1/28).37 Other studies investigated treatment of metastases in various sites by resection (n = 6/28),14,24,26,30,32,34 ablative radiotherapy (n = 1/28),22 or various treatment modalities [i.e., resection and/or RFA and/or stereotactic body radiation therapy (SBRT)] (n = 1/28).2 One study investigated treatment of peritoneal carcinomatosis by cytoreductive surgery and hyperthermic intraperitoneal chemotherapy (CRS-HIPEC) (for some patients in combination with resection of liver metastases) (n = 1/28).19 The remaining studies had insufficient reporting regarding metastatic site (n = 1/28)40 or metastatic site and treatment modality (n = 1/28).27
The analytical methods used included polymerase chain reaction (PCR) based methods (e.g., BEAMing, ddPCR) (n = 13/28),2,12,14,19,20,28–32,35,37,39 targeted sequencing (n = 11/28),17,18,21–27,33,36 direct fluorescent assay (DFA) (n = 2/28),38,15 mass spectrometry (n = 1/28),40 or combined PCR and targeted sequencing (n = 1/28).34
The majority of studies chose a tumor-agnostic marker of ctDNA, that is, assays covering mutations in RAS/BRAF (n = 6/28),2,12,20,30,35,40 large gene panels covering various numbers of genes (n = 6/28),21,22,25,26,33,36 or epigenetic markers (n = 2/28).29,27 Less frequently a tumor-informed approach was applied (n = 7/28)14,23,24,28,31,32,34 or both a tumor-agnostic and a tumor-informed marker (n = 1/28)19 or a tumor-agnostic marker and cfDNA level (n = 2/28).17,18 For the remaining studies, cfDNA level and/or fragment length were evaluated (n = 4/28).15,37–39
The limit of detection was the most frequently chosen cutoff (n = 22/28).2,12,14,17,19–28,30–36,40
Only a few studies reported on the coordination between blood sampling and neo-adjuvant and/or post-interventional adjuvant therapy (n = 4/28).12,23,25,29,27
Pre-ablation circulating DNA and outcome
A total of 18 studies reported on the correlation between pre-ablation circulating DNA and treatment response, recurrence, or survival (Table 1).
Two studies evaluated the association between circulating DNA and treatment response (defined as complete or partial response according to RECIST).41 In one study, circulating DNA level below the cutoff was associated with treatment response to HAI.15 In contrast, the other study did not find any statistically significant association with treatment response to DEBIRI-TACE.38
Eight studies evaluated the association between circulating DNA and recurrence.14,19,21,28,29,24,26,27 One study reported a statistically significant association, with a higher risk of recurrence in patients with circulating DNA above the cutoff (n=1/8).21 The remaining studies were either descriptive (n = 2/8)14,24 or reported no significant association (n = 5/8).19,26,27,28,29
Eleven studies evaluated the association between circulating DNA and RFS . Four studies reported a statistically significantly shorter RFS in patients with circulating DNA above the cutoff (n = 4/11).18,19,21,22 Of note, the results were ambiguous in two studies depending on the selected cutoff,22 or which study participants were eligible for the analysis.19 The remaining studies reported no significant association (n = 7/11).17,20,23–26,29
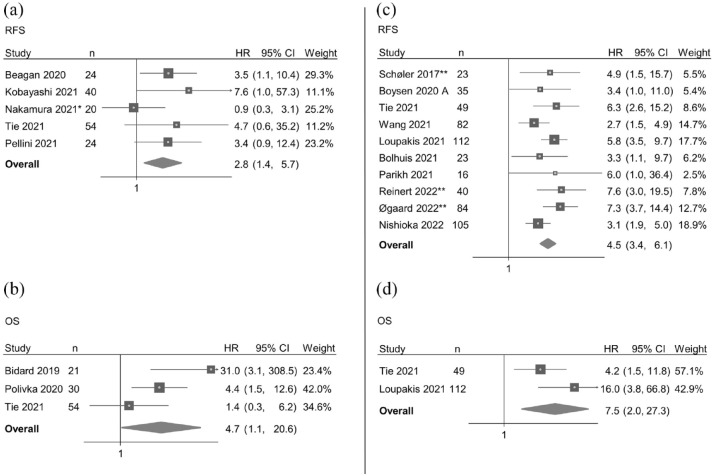
Meta-analysis of the association between pre-ablation circulating DNA and RFS included five eligible studies.19,21–23,26 Circulating DNA above the cutoff in a pre-ablation sample was associated with a shorter RFS (pooled HR = 2.8, 95% CI 1.4–5.7, n = 162) (Figure 4(a)), and the funnel plot indicated no major publication bias (Supplemental Figure 2). Minimal heterogeneity was observed among the reports (χ2 = 4.69, df = 5, p = 0.32, I2 = 20.3%).
Figure 4.
Forest plots of the association between circulating DNA and survival. Forest plots of the association between circulating DNA above the cutoff in a pre-ablation sample and RFS (a) and OS (b); between circulating DNA above the cutoff in a post-ablation sample and RFS (c) and OS (d); all under the random-effects model. Studies ordered according to publication date.
CI, confidence interval; HR, hazard ratio; n, number of patients included in the analysis; OS, overall survival; RFS, recurrence-free survival.
*More than one result with complete data due to test of different circulating DNA markers. Results from the analysis with circulating tumor DNA detectability as marker are included in the meta-analysis. **Same research group.
Nine studies evaluated the association between circulating DNA and OS. Five studies reported a statistically significantly shorter OS in patients with circulating DNA above the cutoff (n = 5/9).12,15,17,20,40
Of note, the results were ambiguous in three studies depending on the selected circulating DNA marker,17 cutoff,20 or time of sampling.12 The remaining studies were either descriptive (n = 1/9) 38 or reported no significant association (n = 3/9).21,23,24 The remaining studies evaluated the association without statistically significant results (n = 3/18),21,23,24 described the association without statistical tests (n = 1/18),38 or did not report on the association between circulating DNA and OS (n = 9/18). 14,18,19,22,25–29
Meta-analysis of the association between pre-ablation circulating DNA and OS included three eligible studies.12,20,23 Circulating DNA above the cutoff in a pre-ablation sample was associated with a shorter OS (pooled HR = 4.7, 95% CI 1.1–20.6, n = 105) (Figure 4(b)), and the funnel plot indicated no major publication bias (Supplemental Figure 2). Some heterogeneity was observed among the reports (χ2 = 5.0, df = 3, p = 0.08, I2 = 63.9%).
Despite inter-study heterogeneity and lack of systematic analysis of the prognostic value, results indicate that pre-ablation circulating DNA could have prognostic value across treatment modalities and methods.
Post-ablation circulating DNA and outcome
A total of 18 studies reported on the correlation between post-ablation circulating DNA and recurrence or survival (Table 2).
No studies investigated the association between post-ablation circulating DNA and treatment response. Sixteen studies evaluated the association between circulating DNA and recurrence. Six of the studies reported a statistically significant association, with a higher risk of recurrence in patients with circulating DNA above the cutoff (n = 6/16).23,25,28,29,34,36 The remaining studies were either descriptive (n=3/16) 14,24,32 or reported no significant association (n = 7/16).2,19,27,30,31,33,35
Eleven studies evaluated the association between circulating DNA and RFS. A majority of studies reported a statistically significantly shorter RFS in patients with circulating DNA above the cutoff (n = 10/11).2,23,27–29,31,34–36 The remaining study reported no significant association n = 1/11).18
Meta-analysis of the association between post-ablation circulating DNA and RFS included 10 eligible studies.2,23,25,27–29,31,34,35,36 Circulating DNA above the cutoff in a post-ablation sample was associated with a shorter RFS (pooled HR = 4.5, 95% CI 3.4–6.1, n = 569) (Figure 4(c)), and the funnel plots indicated no major publication bias (Supplemental Figure 2). Minimal heterogeneity was observed (χ2 = 10.5, df = 10, p = 0.31, I2 = 28.5%).
Five studies evaluated the association between circulating DNA and OS. Four studies reported a statistically significantly shorter OS in patients with circulating DNA above the cutoff (n = 4/5).23,33,34,36 The remaining study reported no significant association (n = 1/5).15
Meta-analysis of the association between post-ablation circulating DNA and OS included two eligible studies.23,34 Circulating DNA above the cutoff in a post-ablation sample was associated with a shorter OS (pooled HR = 7.5, 95% CI 2.0–27.3, n = 161) (Figure 4(d)). Some heterogeneity was observed among the reports (χ2 = 2.21, df = 2, p = 0.14, I2 = 54.8%).
Despite inter-study heterogeneity and lack of systematic analysis of the prognostic value, results were uniform and indicate that post-ablation circulating DNA has prognostic value across treatment modalities and methods.
Post-resection circulating DNA and outcome
Of the 18 studies reporting on the correlation between post-ablation circulating DNA and treatment recurrence or survival, 11 studies evaluated solely resection as loco-regional treatment modality.18,23–25,28–30,32–34,36 All studies reporting a statistically significant association between circulating DNA above the cutoff and a higher risk of recurrence23,25,28,29,34,36 and/or shorter OS 23,33,34,36 was among these 11 studies.
Dynamics in circulating DNA and outcome
Only three studies reported on the correlation between dynamics in circulating DNA and recurrence or survival (Table 3). None of the studies investigated dynamics in circulating DNA in relation to treatment response.
Iwai and colleagues reported on the association between quantitative changes in circulating DNA and recurrence.39 The results were ambiguous depending on the selected circulating DNA marker. There were statistically significant higher risk of recurrence and shorter RFS in patients with decreasing ratio compared to patients with an increasing ratio. A similar association was not observed when evaluating dynamics in integrity or cfDNA level.39
Janowski and colleagues evaluated the association between dynamics in circulating DNA and RFS without significant results. However, they reported a statistically significant shorter OS (counted from the time of diagnosis) in patients with a fragmentation index <1 compared to patients with a fragmentation index >1.37
Boysen and colleagues described an association between dynamics in circulating DNA and OS but did not perform any statistical tests.38
Due to the limited number of studies and diverging results, no definite conclusions can be drawn on the correlation between dynamics in circulating DNA and prognosis.
Discussion
Loco-regional treatment of CRC metastases is a fast-growing therapeutic field, where eligible patients typically present with oligometastatic/limited disease. There is an increasing number of patients being offered loco-regional therapies, using various ablative modalities.16 Loco-regional treatment can potentially improve survival and enhance the quality of life for patients with an otherwise poor prognosis.42,43 To continue the development of this field, prognostic and predictive biomarkers are needed to assist in more optimal selection for individual modalities and to optimize the most relevant time-points for response evaluation and treatment adaption. Circulating DNA might offer the chance to improve the selection of patients with oligometastatic disease who are eligible for loco-regional treatment, sparing patients unnecessary therapies and saving money for health-care systems.
Summary of main results
Twenty-eight studies were included in the systematic review, and 16 of those were eligible for an investigation on the prognostic impact of circulating DNA in a pre- and/or post-ablation plasma sample across different treatment modalities. Based on these publications, we were able to demonstrate a clear prognostic value of post-ablation circulating DNA. The prognostic value of pre-ablation circulating DNA was less clear and needs to be elucidated from adequately designed prospective studies. None of the included studies were designed to investigate if circulating DNA is a predictive biomarker and the role of dynamics in circulating DNA remains only very briefly investigated.
Overall completeness and applicability of evidence
Most studies investigated if circulating DNA in a pre- and/or post-ablation sample could predict prognosis. Few studies evaluated if a change in circulating DNA comparing pre- and post-ablation samples could predict treatment response and/or prognosis. The study setups were heterogeneous, for example, in terms of metastatic site, treatment intent, and treatment modality. Interestingly, some studies did not report on metastatic site and/or treatment modality, which hampers generalizability of the results.
Although all studies used plasma as the source of circulating DNA according to current recommendations,44 there were variations in pre-analytical and analytical procedures, which may confound the identified associations between circulating DNA and outcome. For example, the processing steps (i.e., centrifugation, storage), and thereby the risk of contamination with leucocytes, varied. Furthermore, the studies utilized varying volumes of plasma, from which the circulating DNA was extracted. Low sample volumes can potentially limit the detection of ctDNA and subsequently affect test sensitivity. Within the past years, significant work has been conducted to reach a consensus on the standardization of pre-analytical and analytical steps. This will hopefully improve the generalizability and applicability of future research results.
The optimal time-point for blood sampling is not known. From the studies included in this review, it is not possible to identify the optimal time period between loco-regional treatment and blood sampling. We observed heterogeneity in the studies included both with regard to timing with neo-adjuvant or post-interventional adjuvant therapies. We believe timing may depend on disease stages and ablative modalities, as each modality may give rise to different biological reactions and affect circulating DNA in its own way. Resection induces cfDNA increase, which may persist for weeks and could be reflected in the measurement of circulating DNA.45 Less is known about how circulating DNA is affected by other treatment modalities. An example is SBRT of cranial and extra cranial primary or metastatic tumors which has shown excellent outcomes with high local control rates and low probability of normal tissue toxicity.46,47 Traditional radiobiological studies demonstrated radiation-induced DNA damage and cell kill via apoptosis induction and reproductive cell death. Experiments in immune radiobiology clearly show that the cell killing effect of radiation is largely dependent on tumor-infiltrating cytotoxic CD8+ T cells. Radiation interacts with immunological activation cascades at different levels. Radiation thus not only impacts tumor cells but has multiple effects on immune cells within the tumor tissue and on normal cells of the tumor microenvironment.48 These biological factors could all influence the pattern of circulating DNA release into the blood following the procedures, and much relevant knowledge can be expected from prospective studies of circulating DNA dynamics after SBRT. Similar complex biological mechanisms can be expected following SIRT, also frequently used in liver-dominant metastatic disease.
Equally important when searching for the optimal time-point for blood sampling is the clinical question at hand. A pre-ablation sample could be valuable when deciding whether the patient will benefit from loco-regional treatment or not, thereby limiting the number of unnecessary treatments. Alternatively, evaluating the effect of neo-adjuvant chemotherapy prior to loco-regional treatment. A post-ablation sample could be valuable in guiding post-interventional adjuvant chemotherapy and follow-up (i.e., intensified in patients with circulating DNA above the cutoff and de-escalated in patients circulating DNA below the cutoff). Standard of care adjuvant chemotherapy, administered after loco-regional treatment of CRC metastases, can clear ctDNA from the circulation, but recurrence of ctDNA after the termination of chemotherapy may indicate the need of intensified post-interventional adjuvant chemotherapy.2 On the other hand, patients without detectable ctDNA after treatment of oligometastatic CRC may not benefit from treatment with post-interventional adjuvant chemotherapy. This is being evaluated in an ongoing randomized study by our group (OPTIMISE, NCT04680260) where ctDNA-guided post-interventional adjuvant therapy is evaluated against standard of care.49 The purpose of an intensified follow-up would be to detect recurrence in a less advanced stage with a potentially higher chance of cure.
In order to gain further insight into the optimal time-point for blood sampling in the context of different loco-regional modalities, we believe it is of utmost importance to evaluate this in individual prospective clinical trials with consecutive post-ablation samples. The trials should carefully take into consideration pre-analytical/analytical standardization and alignment of sampling, in addition to the cfDNA biology of the ablation modality and the clinical questions at hand. Furthermore, we believe it will be important to integrate biological information from circulating DNA with advanced imaging modalities (i.e., diffusion-weighted magnetic resonance imaging in patients treated with TACE, SIRT, or SBRT).
Quality of the evidence
The body of evidence suggests that pre- and post-ablation circulating DNA above a chosen cutoff confer a poor prognosis for patients undergoing loco-regional treatment of CRC metastases. However, it is still unclear to what degree circulating DNA can replace/complement the current standard of care (i.e., radiological assessment) since there was no consensus among studies as to the optimal method, marker, cutoff, or time of sampling.
Circulating DNA as a prognostic marker in the loco-regional treatment of CRC metastases is a novel research field. Consequently, studies included in this review tended to be descriptive, exploratory, retrospective, and with few included participants. Even though well conducted, studies often had a high risk of bias in one or more domains as seen from the QUIPS bias assessment. The risk of bias in individual studies could have been reduced by the systematic use of the REMARK guidelines.13
Limitations
Our review has several limitations. Firstly, we described the results both qualitatively and quantitatively depending on the data availability due to great inter-study heterogeneity. Consequently, only 16 out of 28 studies had sufficient data for inclusion in the meta-analyses. Secondly, all studies providing sufficient data were included in the meta-analyses despite great variance in quality and risk of bias, which could lead to imprecisions of the combined effect measures. Thirdly, we did not search unpublished material for inclusion. This could introduce publication bias and subsequently overestimate combined effect measures. Of note, we did not see any major tendencies of publication bias when visually assessing the funnel plots. Finally, due to the great heterogeneity among studies, it was not possible to further investigate whether discrepancies in results were attributed to specific study characteristics.
Future perspectives
In conclusion, circulating DNA has a clear prognostic potential in patients undergoing loco-regional treatment of CRC metastases. Prospective clinical trials with standardized methodologies are needed to evaluate if circulating DNA can be a cost-effective tool that add clinical value (i.e., improved survival and quality of life) in the care pathway of this patient group.
Supplemental Material
Supplemental material, sj-docx-1-tam-10.1177_17588359221133171 for Circulating DNA in patients undergoing loco-regional treatment of colorectal cancer metastases: a systematic review and meta-analysis by Louise B. Callesen, Tana Takacova, Julian Hamfjord, Florian Würschmidt, Karl J. Oldhafer, Roland Brüning, Dirk Arnold and Karen-Lise G. Spindler in Therapeutic Advances in Medical Oncology
Acknowledgments
We would like to thank Karen Sigaard, Librarian, Aarhus University, for help in developing the search string.
Footnotes
ORCID iDs: Louise B. Callesen  https://orcid.org/0000-0003-2799-1492
https://orcid.org/0000-0003-2799-1492
Tana Takacova  https://orcid.org/0000-0002-9974-4282
https://orcid.org/0000-0002-9974-4282
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Louise B. Callesen, Department of Experimental Clinical Oncology, Aarhus University Hospital, Palle Juul-Jensens Boulevard 99, Aarhus N 8200, Denmark; Institute of Clinical Medicine, Aarhus University, Aarhus, Denmark.
Tana Takacova, Asklepios Tumorzentrum Hamburg, Hamburg, Germany; Department of Oncology and Palliative Care with Sections Hematology and Rheumatology, AK Altona, Hamburg, Germany.
Julian Hamfjord, Department of Oncology, Oslo University Hospital, Oslo, Norway; Department of Cancer Genetics, Institute for Cancer Research, Oslo University Hospital, Oslo, Norway; Institute of Clinical Medicine, Faculty of Medicine, University of Oslo, Oslo, Norway.
Florian Würschmidt, Radiologische Allianz, Hamburg, Germany.
Karl J. Oldhafer, Asklepios Tumorzentrum Hamburg, Hamburg, Germany Department of Surgery, Division of Hepato-biliary and Pancreatic (HBP) Surgery, AK Barmbek, Hamburg, Germany; Faculty of Medicine, Semmelweis University Budapest, Asklepios Campus Hamburg, Hamburg, Germany.
Roland Brüning, Asklepios Tumorzentrum Hamburg, Hamburg, Germany; Department of Radiology and Neuroradiology, AK Barmbek, Hamburg, Germany.
Dirk Arnold, Asklepios Tumorzentrum Hamburg, Hamburg, Germany; Department of Oncology and Palliative Care with Sections Hematology and Rheumatology, AK Altona, Hamburg, Germany.
Karen-Lise G. Spindler, Department of Experimental Clinical Oncology, Aarhus University Hospital, Aarhus, Denmark Institute of Clinical Medicine, Aarhus University, Aarhus Denmark; Asklepios Tumorzentrum Hamburg, Hamburg, Germany.
Declarations
Ethics approval and consent to participate: Not applicable.
Consent for publication: Not applicable.
Author contribution(s): Louise B. Callesen: Conceptualization; Formal analysis; Investigation; Methodology; Project administration; Visualization; Writing – original draft; Writing – review & editing.
Tana Takacova: Investigation; Writing – review & editing.
Julian Hamfjord: Conceptualization; Methodology; Writing – review & editing.
Florian Würschmidt: Conceptualization; Writing – review & editing.
Karl J. Oldhafer: Conceptualization; Writing – review & editing.
Roland Brüning: Conceptualization; Writing – review & editing.
Dirk Arnold: Conceptualization; Writing – review & editing.
Karen-Lise G. Spindler: Conceptualization; Funding acquisition; Investigation; Methodology; Supervision; Writing – review & editing.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: LBC was supported by DCCC ctDNA Research Center – The Danish Research Center for Circulating Tumor DNA Guided Cancer Management, Danish Cancer Society (grant no. R257-A14700), and Danish Comprehensive Cancer Centers. K-LGS was supported by Health Research Foundation of Central Denmark Region (grant no. A1602). The remaining authors received no specific funding for this work.
The authors declare that there is no conflict of interest.
Availability of data and materials: The full text of all included studies was retrieved from the online databases PubMed, Embase, and Cochrane Central Register of Controlled Trials. The data of this systematic review and meta-analyses are all public and available from PubMed, Embase, and Cochrane Central Register of Controlled Trials. Template data collection forms, data extracted from included studies, data used for analyses, and analytic code used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- 1. Ferlay J, Colombet M, Soerjomataram I, et al. Cancer statistics for the year 2020: an overview. Int J Cancer 2021; 149: 778–789. [DOI] [PubMed] [Google Scholar]
- 2. Boysen AK, Pallisgaard N, Andersen CSA, et al. Circulating tumor DNA as a marker of minimal residual disease following local treatment of metastases from colorectal cancer. Acta Oncol 2020; 59: 1424–1429. [DOI] [PubMed] [Google Scholar]
- 3. Wang Y, Li L, Cohen JD, et al. Prognostic potential of circulating tumor DNA measurement in postoperative surveillance of nonmetastatic colorectal cancer. JAMA Oncol 2019; 5: 1118–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reinert T, Scholer LV, Thomsen R, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 2016; 65: 625–634. [DOI] [PubMed] [Google Scholar]
- 5. Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 2016; 8: 346ra92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reinert T, Henriksen TV, Christensen E, et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol 2019; 5: 1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tie J, Cohen JD, Lahouel K, et al. Circulating tumor DNA analysis guiding adjuvant therapy in stage II colon cancer. N Engl J Med 2022; 386: 2261–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Spindler KG, Boysen AK, Pallisgård N, et al. Cell-free DNA in metastatic colorectal cancer: a systematic review and meta-analysis. Oncologist 2017; 22: 1049–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Callesen LB, Hamfjord J, Boysen AK, et al. Circulating tumour DNA and its clinical utility in predicting treatment response or survival in patients with metastatic colorectal cancer: a systematic review and meta-analysis. Br J Cancer 2022; 127: 500–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Page MJ, McKenzie JE, Bossuyt PM, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 2021; 10: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hayden JA, Côté P, Bombardier C. Evaluation of the quality of prognosis studies in systematic reviews. Ann Intern Med 2006; 144: 427–437. [DOI] [PubMed] [Google Scholar]
- 12. Bidard F-C, Kiavue N, Ychou M, et al. Circulating tumor cells and circulating tumor DNA detection in potentially resectable metastatic colorectal cancer: a prospective ancillary study to the unicancer prodige-14 trial. Cells 2019; 8: 516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McShane LM, Altman DG, Sauerbrei W, et al. REporting recommendations for tumour MARKer prognostic studies (REMARK). Eur J Cancer 2005; 41: 1690–1696. [DOI] [PubMed] [Google Scholar]
- 14. Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008; 14: 985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boysen AK, Schou JV, Jensen BV, et al. Prognostic and predictive value of circulating DNA for hepatic arterial infusion of chemotherapy for patients with colorectal cancer liver metastases. Mol Clin Oncol 2020; 13: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boysen AK, Spindler K-LL, Høyer M, et al. Metastasis directed therapy for liver and lung metastases from colorectal cancer–A population-based study. Int J cancer 2018; 143: 3218–3226. [DOI] [PubMed] [Google Scholar]
- 17. Narayan RR, Goldman DA, Gonen M, et al. Peripheral circulating tumor dna detection predicts poor outcomes after liver resection for metastatic colorectal cancer. Ann Surg Oncol 2019; 26: 1824–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. He Y, Ma X, Chen K, et al. Perioperative circulating tumor DNA in colorectal liver metastases: concordance with metastatic tissue and predictive value for tumor burden and prognosis. Cancer Manag Res 2020; 12: 1621–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Beagan JJ, Sluiter NR, Bach S, et al. Circulating tumor DNA as a preoperative marker of recurrence in patients with peritoneal metastases of colorectal cancer: a clinical feasibility study. J Clin Med 2020; 9: 1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Polivka J, Windrichova J, Pesta M, et al. The level of preoperative plasma KRAS mutations and CEA predict survival of patients undergoing surgery for colorectal cancer liver metastases. Cancers 2020; 12: 2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kobayashi S, Nakamura Y, Taniguchi H, et al. Impact of preoperative circulating tumor DNA status on survival outcomes after hepatectomy for resectable colorectal liver metastases. Ann Surg Oncol 2021; 28: 4744–4755. [DOI] [PubMed] [Google Scholar]
- 22. Nakamura M, Kageyama S-I, Seki M, et al. Liquid biopsy cell-free DNA biomarkers in patients with oligometastatic colorectal cancer treated by ablative radiotherapy. Anticancer Res 2021; 41: 829–834. [DOI] [PubMed] [Google Scholar]
- 23. Tie J, Wang Y, Cohen J, et al. Circulating tumor DNA dynamics and recurrence risk in patients undergoing curative intent resection of colorectal cancer liver metastases: A prospective cohort study. PLoS Med 2021; 18: e1003620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee S, Park Y-S, Chang W-J, et al. Clinical implication of liquid biopsy in colorectal cancer patients treated with metastasectomy. Cancers 2021; 13: 2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang D-S, Yang H, Liu X-Y, et al. Dynamic monitoring of circulating tumor DNA to predict prognosis and efficacy of adjuvant chemotherapy after resection of colorectal liver metastases. Theranostics 2021; 11: 7018–7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pellini B, Pejovic N, Feng W, et al. ctDNA MRD detection and personalized oncogenomic analysis in oligometastatic colorectal cancer from plasma and urine. JCO Precis Oncol 2021; 5: 378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Parikh AR, Van Seventer EE, Siravegna G, et al. Minimal residual disease detection using a plasma-only circulating tumor DNA assay in patients with colorectal cancer. Clin Cancer Res 2021; 27: 5586–5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reinert T, Petersen LMS, Henriksen TV, et al. Circulating tumor DNA for prognosis assessment and postoperative management after curative-intent resection of colorectal liver metastases. Int J Cancer 2022; 150: 1537–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Øgaard N, Reinert T, Henriksen TV, et al. Tumour-agnostic circulating tumour DNA analysis for improved recurrence surveillance after resection of colorectal liver metastases: A prospective cohort study. Eur J Cancer 2022; 163: 163–176. [DOI] [PubMed] [Google Scholar]
- 30. Yamada T, Iwai T, Takahashi G, et al. Utility of KRAS mutation detection using circulating cell-free DNA from patients with colorectal cancer. Cancer Sci 2016; 107: 936–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schøler LV, Reinert T, Orntoft MW, et al. Clinical implications of monitoring circulating tumor DNA in patients with colorectal cancer. Clin Cancer Res 2017; 23: 5437–5445. [DOI] [PubMed] [Google Scholar]
- 32. Benešová L, Hálková T, Ptácˇková R, et al. Significance of postoperative follow-up of patients with metastatic colorectal cancer using circulating tumor DNA. World J Gastroenterol 2019; 25: 6939–6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mason MC, Tzeng C-WD, Tran Cao HS, et al. Preliminary analysis of liquid biopsy after hepatectomy for colorectal liver metastases. J Am Coll Surg 2021; 233: 82.e1–89.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Loupakis F, Sharma S, Derouazi M, et al. Detection of molecular residual disease using personalized circulating tumor DNA assay in patients with colorectal cancer undergoing resection of metastases. JCO Precis Oncol 2021; 5: 1166–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bolhuis K, van ’t Erve I, Mijnals C, et al. Postoperative circulating tumour DNA is associated with pathologic response and recurrence-free survival after resection of colorectal cancer liver metastases. EBioMedicine 2021; 70: 103498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nishioka Y, Chun YS, Overman MJ, et al. Effect of co-mutation of RAS and TP53 on postoperative ctDNA detection and early recurrence after hepatectomy for colorectal liver metastases. J Am Coll Surg 2022; 234: 474–483. [DOI] [PubMed] [Google Scholar]
- 37. Janowski E, Timofeeva O, Chasovskikh S, et al. Yttrium-90 radioembolization for colorectal cancer liver metastases in KRAS wild-type and mutant patients: clinical and ccfDNA studies. Oncol Rep 2017; 37: 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boysen AK, Jensen M, Nielsen DT, et al. Cell-free DNA and chemoembolization in patients with liver metastases from colorectal cancer. Oncol Lett 2018; 16: 2654–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Iwai T, Yamada T, Takahashi G, et al. Circulating cell-free long DNA fragments predict post-hepatectomy recurrence of colorectal liver metastases. Eur J Surg Oncol 2020; 46: 108–114. [DOI] [PubMed] [Google Scholar]
- 40. Shin SJ, Chun SM, Kim TI, et al. Feasibility of multiplexed gene mutation detection in plasma samples of colorectal cancer patients by mass spectrometric genotyping. PLoS One 2017; 12: e0176340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009; 45: 228–247. [DOI] [PubMed] [Google Scholar]
- 42. Lehtomäki K, Stedt HP, Osterlund E, et al. Health-related quality of life in metastatic colorectal cancer patients treated with curative resection and/or local ablative therapy or systemic therapy in the finnish RAXO-study. Cancers 2022; 14: 1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Osterlund P, Salminen T, Soveri LM, et al. Repeated centralized multidisciplinary team assessment of resectability, clinical behavior, and outcomes in 1086 Finnish metastatic colorectal cancer patients (RAXO): a nationwide prospective intervention study. Lancet Reg Heal-Eur 2021; 3: 100049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. ISO. ISO 20186-3:2019 - Molecular in vitro diagnostic examinations — Specifications for pre-examination processes for venous whole blood — Part 3: Isolated circulating cell free DNA from plasma. https://www.iso.org/standard/69800.html (2019, accessed 19 January 2022).
- 45. Henriksen TV, Reinert T, Christensen E, et al. The effect of surgical trauma on circulating free DNA levels in cancer patients—implications for studies of circulating tumor DNA. Mol Oncol 2020; 14: 1670–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Buergy D, Würschmidt F, Gkika E, et al. Stereotactic or conformal radiotherapy for adrenal metastases: patient characteristics and outcomes in a multicenter analysis. Int J Cancer 2021; 149: 358–370. [DOI] [PubMed] [Google Scholar]
- 47. Palma DA, Olson R, Harrow S, et al. Stereotactic ablative radiotherapy versus standard of care palliative treatment in patients with oligometastatic cancers (SABR-COMET): a randomised, phase 2, open-label trial. Lancet 2019; 393: 2051–2058. [DOI] [PubMed] [Google Scholar]
- 48. Lippitz BE, Harris RA. A translational concept of immuno-radiobiology. Radiother Oncol 2019; 140: 116–124. [DOI] [PubMed] [Google Scholar]
- 49. Callesen LB, Hansen TF, Andersen RF, et al. OPTIMISE: optimisation of treatment selection and follow-up in oligometastatic colorectal cancer–a ctDNA-guided phase II randomised approach. Study protocol. Acta Oncol. Epub ahead of print September 2022. DOI: 10.1080/0284186X.2022.2116728. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-tam-10.1177_17588359221133171 for Circulating DNA in patients undergoing loco-regional treatment of colorectal cancer metastases: a systematic review and meta-analysis by Louise B. Callesen, Tana Takacova, Julian Hamfjord, Florian Würschmidt, Karl J. Oldhafer, Roland Brüning, Dirk Arnold and Karen-Lise G. Spindler in Therapeutic Advances in Medical Oncology