Abstract
Abnormal RNA metabolism, regulated by various RNA binding proteins, can have functional consequences for multiple diseases. Heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) is an important RNA binding protein, that regulates various RNA metabolic processes, including transcription, alternative splicing of pre-mRNA, translation, miRNA processing and mRNA stability. As a potent splicing factor, hnRNP A1 can regulate multiple splicing events, including itself, collaborating with other cooperative or antagonistical splicing factors by binding to splicing sites and regulatory elements in exons or introns. hnRNP A1 can modulate gene transcription by directly interacting with promoters or indirectly impacting Pol II activities. Moreover, by interacting with the internal ribosome entry site (IRES) or 3′-UTR of mRNAs, hnRNP A1 can affect mRNA translation. hnRNP A1 can alter the stability of mRNAs by binding to specific locations of 3′-UTR, miRNAs biogenesis and Nonsense-mediated mRNA decay (NMD) pathway. In this review, we conclude the selective sites where hnRNP A1 binds to RNA and DNA, and the co-regulatory factors that interact with hnRNP A1. Given the dysregulation of hnRNP A1 in diverse diseases, especially in cancers and neurodegeneration diseases, targeting hnRNP A1 for therapeutic treatment is extremely promising. Therefore, this review also provides the small-molecule drugs, biomedicines and novel strategies targeting hnRNP A1 for therapeutic purposes.
Keywords: alternative splicing, hnRNP A1, RNA binding protein, RNA metabolism, splicing factor
Introduction
hnRNPs family consists of RNA-binding proteins, that includes at least 20 members named from A to U (1). hnRNP A1 belongs to the hnRNPA/B subfamily and is one of the most abundant and broadly expressed nuclear proteins. HnRNP A1 is first identified as one of the core proteins of ribonucleoprotein complexes (Choi and Dreyfuss, 1984; Lothstein et al., 1985; Wilk et al., 1985; Han et al., 2010). Subsequently, the RNA-binding ability (Schenkel et al., 1988) and the gene alternative splicing regulatory roles (Biamonti et al., 1989) of hnRNP A1 are observed. Intensive studies have revealed the role of hnRNP A1 in regulating normal physiological functions and pathologic processes (Roy et al., 2017; Clarke et al., 2021).
As a splicing factor, hnRNP A1 can modulate the splicing of crucial genes to produce specific protein variants, contributing to human diseases such as tumorigenesis and neurological diseases. Elevated hnRNP A1 levels in cancer cells attenuate cell apoptosis, by regulating the gene splicing process to generate specific protein variants (Kedzierska and Piekielko-Witkowska, 2017). hnRNP A1-mediated alternative splicing of genes in the brain causes severe mental disorders (Donev et al., 2007; Babic et al., 2013; Bruun et al., 2018; Beijer et al., 2021). hnRNP A1 also shows multiple physiological functions on cell proliferation (Yang et al., 2019), cell survival (Feng et al., 2018), cell cycle (Yu et al., 2015), cell migration, cell stemness, cellular senescence (Shimada et al., 2009), etc.
In addition to regulating mRNA alternative splicing events, hnRNP A1 involved in gene transcription, internal ribosomal entry sites (IRES)-dependent mRNA translation, mRNA transportation, mRNA stability and microRNA biogenesis. This review article describes our current understanding of hnRNP A1’s underlying mechanisms regulating RNA metabolism and provides existing approaches targeting hnRNP A1 or its functions.
The structure of hnRNP A1
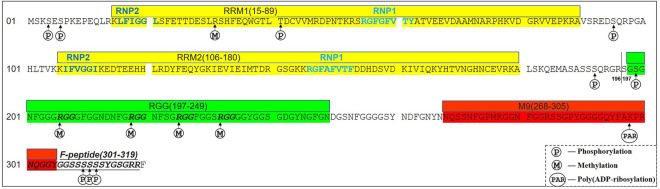
The hnRNP A1 gene is located at 12q13.13, consisting of 10 exons and 9 introns. The alternative splicing of pre-mRNA hnRNP A1 generates an alternate in-frame exon (exon 7b, 52 amino acids), resulting in an extended protein hnRNP A1B (372 amino acids, UniProt:P09651) compared to hnRNP A1 (320 amino acids, UniProt:P09651-2) (Hutchison et al., 2002). The hnRNP A1 isoform is much more abundant than hnRNP A1B isoform. The N-terminus domain (also named UP-1, 1–196 amino acids) of hnRNP A1 comprises two RNA recognition motifs (RRMs) with highly similar sequences, PRM1 and PRM2, which consist of four β-sheets, two α-helices (βαββαβ), conserved RNP1 octameric and RNP2 hexameric about 30 amino acid residues apart (Dreyfuss et al., 1988; Xu et al., 1997; Ding et al., 1999). The C-terminal of hnRNP A1 is a glycine-rich region (also called Glycine-rich domain, 197–320 amino acids), with an RGG box (197–249 amino acids) within four RGG repeats followed by nuclear localization signal (NLS) M9 domain (268–305 amino acids) and F peptide (301–319 amino acids) with six consecutive serines (S308-S313) (Figure 1) (Izaurralde et al., 1997; Allemand et al., 2005; Ghosh and Singh, 2018).
FIGURE 1.
A scheme of the hnRNP A1 primary amino acids sequence (UniProt:P09651-2). The amino acids (1–196) are the N-terminus (also named UP-1) of hnRNP A1, consisting of two RNA recognition motifs (RRM1 and RRM2, Yellow). The amino acids sequences of dark gray are the C-terminus (also named GRD,197–320), composed of RGG (197–249, Green) with four RGG motifs (Italic Bold), M9 (268–305, Red) and F-peptide (301–319, Italic Underline). The phosphorylation and poly (ADP-ribosylation) sites of hnRNP A1 are marked.
The two RRMs of hnRNP A1 are the common RNA-binding domains recognizing and binding to splicing regulatory elements to regulate gene alternative splicing events (Beusch et al., 2017). In addition, the two RBMs also contribute to RNA packaging and trafficking (Shamoo et al., 1995; Shan et al., 2000), and bind to single-stranded telomeric DNA (21). The Glycine-rich domain of hnRNP A1 mediates protein-protein interactions and RNA binding (Bekenstein and Soreq, 2013). The RGG box in Glycine-rich domain, which consists of four RGG repeats, has been shown to mediate self-interaction, and interaction with other hnRNPs or RNA binding proteins including serine-arginine (SR) proteins (Cartegni et al., 1996; Fisette et al., 2010). It can also recognize the telomere G-quadruplex DNA (22), as well as affect its internal ribosome entry site trans-acting factor (ITAF) activity (Wall and Lewis, 2017). M9 is a 38 amino acid-long motif, downstream of the RGG box, which is closely related to the cellular localization of hnRNP A1 and plays a key role in mRNA nuclear export and protein nuclear import (Siomi and Dreyfuss, 1995; Izaurralde et al., 1997), whereas hnRNP A1’s regulatory functions on the mRNA stability and translational activity are depended on its subcellular location. Furthermore, F-peptide is located at the C-terminal domain, adjacent to the M9 motif, and its phosphorylation status and can influence the rate of hnRNP A1 nuclear import (Allemand et al., 2005).
The function of hnRNP A1 in alternative splicing
Alternative splicing is a pre-mRNA process regulated by cis-acting elements and trans-acting factors. The cis-acting elements contain exonic splicing enhancer/silencer (ESE/ESS) and intronic splicing enhancer/silencer (ISE/ISS). These splicing enhancers or splicing silencers interact with trans-acting factors, conferring a positive or negative effect on splice site recognition by the spliceosome, ultimately influencing splicing outcome (Zhu et al., 2001). While the trans-acting factors comprise two major classes of splicing factors, hnRNPs and SR proteins (Busch and Hertel, 2012). hnRNP A1, one of the most important hnRNPs splicing factors, regulates alternative splicing in numerous mammalian genes including the caspase-2 gene, c-src, SMN2 gene, and even itself (Suzuki and Matsuoka, 2017). Moreover, the hnRNP A1 participates in the alternative splicing process of several genes in virus harboring Human Immunodeficiency Virus-1 (HIV-1) (Damgaard et al., 2002; Marchand et al., 2002; Zahler et al., 2004), Human papillomavirus (HPV) (Ajiro et al., 2016) and Human T-lymphotropic virus type 1(HTLV-1) (Princler et al., 2003), indicating that both viral RNA cis-elements and host splicing factors govern virus pre-mRNA alternative splicing (Kaur and Lal, 2020).
hnRNP A1 is associated with spliceosome assembly, then in the two consecutive transesterification reactions that lead to excision of the introns and joining of the exons (Jurica et al., 2002; Zhou et al., 2002). Finally, by establishing a complex with U2AF, hnRNP A1 causes the spliceosome to select functional 3′ splicing sites (Tavanez et al., 2012). hnRNP A1 can bind to exon splicing silencers (Del Gatto-Konczak et al., 1999), intron binding sites (Expert-Bezancon et al., 2004) or splice sites (Chiou et al., 2013) to repress exon splicing, suggesting that hnRNP A1 may act as a splicing repressor (Fisette et al., 2010). However, it can also have beneficial effects on exon splicing of several genes, including CDK2 (16), Fas (Oh et al., 2013) and IRF3(49). The alternative splicing regulatory functions of hnRNP A1, including how it interacts with the specific binding sites or other co-regulatory splicing factors, will be discussed in detail as following.
RNA specific binding sites of hnRNP A1
hnRNP A1 is an RNA-binding protein that binds to particular sequence, that Burd et al. (Burd and Dreyfuss, 1994) identified the high affinity hnRNP A1 binding sites, UAGGGA/U, using the systematic evolution of ligands by exponential enrichment (SELEX) experiment. Crosslinking immunoprecipitation followed by high throughput sequencing (CLIP-seq) analyses showed preferential binding of hnRNP A1 to UAGU sequence element (Huelga et al., 2012). Furthermore, the individual-nucleotide resolution crosslinking immunoprecipitation (iCLIP) identified UAGG as the hnRNP A1 binding motif (Bruun et al., 2016). The positions where hnRNP A1 interacts with its binding sequence may affect the consequences of the alternative splicing events. According to the study from Burd et al. (Burd and Dreyfuss, 1994), the ‘winner’ sequence containing a duplication of this UAGGGA/U sequence separated by two nucleotides showed the highest affinity for hnRNP A1, where the binding consensus resembled the 5′ and 3′ splice sites. The UAGGGC sequence at the E9 5′ splice site of the PKM gene has been identified as hnRNP A1’s binding site, facilitating the alternative splicing process that results in PKM2 isoform (David et al., 2010). In the alternative splicing process of the MAPT gene, hnRNP A1 binds to the sequence caaagGTGC at the 3′ splice site of exon 10 to promote exon 10 skipping (Liu et al., 2020). Another strong hnRNP A1 binding sequence (GAGGAAG) at 5′ splice site of exon 5 interacted with hnRNP A1, enabling Fas gene inclusion in distal exon 6 (Oh et al., 2013). These findings suggest that hnRNP A1 regulates the splicing by binding to a particular sequence in splice sites.
In addition to the splice sites, hnRNP A1 can bind splicing regulatory elements, such as splicing silencers in exons or introns, to regulate the alternative splicing of various genes. For exon splicing regulation, ESSs suppress exon inclusion by blocking the exon splice sites, while ESEs increase exon inclusion by recruiting SR splicing factors (Graveley, 2000). ISSs hinder exon inclusion by recruiting repressors, while ISEs antagonistically promote exon inclusion (Wang and Burge, 2008). However, the effects of a single regulatory element on alternative splicing events can have two opposing consequences depending on its locations and binding factors (Wang et al., 2012). hnRNP A1 regulates gene splicing by binding to splicing silencers in exons and introns. The binding of hnRNP A1 to the ESS sequence (TGCGGC) in Ron exon 12 is relevant for its ability to promote Ron exon 11 inclusion, contributing to mesenchymal-to-epithelial transition of cancer cells (Bonomi et al., 2013). Furthermore, the hnRNP A1 inhibits exon 7 inclusion during gene splicing by binding both ESSs at exon 7 and ISS N1 at intron 7 of SMN2, that suppressing the regulatory effects of hnRNP A1 on SMN2 alternative splicing leads to a functional protein with exon 7 inclusion beneficial in treating spinal muscular atrophy (Kashima et al., 2007a; Beusch et al., 2017). Besides promoting exon exclusion, hnRNP A1 can also promote exon inclusion by splicing. With binding to ESS sites in exon 12, hnRNP A1 increases exon 12 inclusion of ATP7B, that hnRNP A1 silencing promotes ATP7B exon12 exclusion, potentially attenuating the toxic effects of ATP7B exon 12 mutation in Wilson’s disease (Lin et al., 2015). Similarly, overexpression of hnRNP A1 causes CDK2 to be included in exon 5, promoting cell cycle progression of oral squamous cell carcinoma (Yu et al., 2015), Fas to be included in exon 6 protecting cells from apoptosis (Oh et al., 2013), and so on. The binding sites and modulated splicing events of hnRNP A1 are listed in Table 1.
TABLE 1.
The alternative splicing events regulated by hnRNP A1.
| Genes | Binding sites | Isoforms | Co-regulatory factors | Diseases/Tissues/Cells/Virus | Reference |
|---|---|---|---|---|---|
| APOL1 | a consensus cis-acting element in exon 4 | Promoting exon 4 exclusion | — | Human glomerular and tubular cells | Cheatham et al. (2018) |
| APP | Alu element in introns 6 and 8 | Promoting exons 7 and 8 skipping | SRSF2(Cooperative) | NT2N neuronal cells | Donev et al. (2007) |
| AR | UAGGGA in splice sites | Promoting AR-V7 expression | — | Prostate cancer | Nadiminty et al. (2015) |
| ATP7B | ESS sites in exon12 | Promoting exon 12 inclusion | — | Wilson’s diseases | Lin et al. (2015) |
| ATM | Alu-derived Intronic Splicing enhancer (ISE) in intron 20 | Promoting cryptic exon exclusion | DAZAP1 (Antagonistic) | HeLa cell | Pastor and Pagani, (2011) |
| Bcl-x | 5′ splice site | Promoting Bcl-x(S) expression | (hnRNP) F/H/Sam68 (Cooperative) | HEK293 cell | Cloutier et al. (2018) |
| beta-tropomyosin | G-rich intronic sequence (S3) downstream of exon 6B | Promoting exon 6B exclusion | SRSF1, SRSF2 (Antagonistic); hnRNP F/H (Cooperative) | HeLa cell | Expert-Bezancon et al. (2004) |
| CCDC50 | — | Promoting exon 6 skipping | — | Clear cell renal cell carcinoma | Sun et al. (2020) |
| CD44 | Splice regulatory elements in exon v5 | Promoting exon v5 exclusion | — | CB3 and NIH-3T3 cells | Matter et al. (2000) |
| CD44 | — | Increasing c5v6v7v8v9v10c6 and c5v6v8v9v10c6; inhibiting c5v6c6 | — | MCF7, MCF10A and MDA-MB-231 cells | Loh et al. (2015) |
| CDK2 | GUAGUAGU in intron 4 | Promoting exon 5 inclusion | — | Oral squamous cell carcinoma | Yu et al. (2015) |
| CEACAM1 | 3′ to exon 7 | Promoting exon 7 exclusion | hnRNP L (Cooperative) | ZR75 and MDA-MB-468 cells | Dery et al. (2011) |
| hnRNP M (Antagonistic) | |||||
| CFTR | ISS of intron 9 | Promoting exon 9 skipping | SRSF1, SRSF5, SRSF6, SRSF4 (Cooperative) | Hep3B cell | Pagani et al. (2000) |
| c-H-ras | intronic silencer sequence (rasISS1) of intron D2 | Promoting intron D1 exclusion | SRSF2, SRSF5 (Antagonistic) | HeLa cell | Guil et al. (2003) |
| c-src | 3′ splice site of exon N1 | Promoting exon N1 exclusion | SRSF1, SRSF2 (Antagonistic) hnRNP I (Cooperative) | HeLa and WERI-1 cells | Rooke et al. (2003) |
| Fas | GAGGAA at 5′ splice site of exon 5 | Promoting exon 6 inclusion | — | MDA-MB-231, HeLa and HCT116 cells | Oh et al. (2013) |
| FGFR2 | K-SAM ESS | Promoting K-SAM exon skipping | TIA-1 (Antagonistic) | HEK293 cell | Gesnel et al. (2009) |
| Gαs | — | Promoting exon 3 skipping | SRSF1(Antagonistic) | Myometrial smooth muscle cells | Pollard et al. (2002) |
| GLA | ESS overlapping the 5′ splice site | Preventing pseudoexon inclusion | hnRNP A2/B1 (Cooperative) | HeLa and HepG2 cells | Palhais et al. (2016) |
| GRIN1 gene | exonic UAGGs and the 5′-splice-site proximal GGGG motif | Promoting CI cassette exon (exon 19) skipping | hnRNP H (Antagonistic) | Rat cortical culture; PC12 and C2C12 cells | (Han et al., 2005; An and Grabowski, 2007) |
| HER2 | — | Promoting intron 8 retention | — | SKBR3 cell | Silipo et al. (2017) |
| HIV-1 rev/tat | intronic splicing silencer (ISS), a novel UAG motif in the exon splicing enhancer (ESE), and the exon splicing silencer (ESS3) | Inhibiting the second intron exclusion | SRSF1, SRSF2(Antagonistic) | HIV-1 | (Damgaard et al., 2002; Marchand et al., 2002; Zahler et al., 2004) |
| HN1 | — | Promoting alternative polyadenylation (APA) -3′ UTR shortening | — | HEK293, HUVEC and A549 cells | Jia et al. (2019) |
| HMGCR | — | Promoting exon 13 skipping | — | HepG2, Hep3B and Huh7 cells | Yu et al. (2014) |
| HPV18 | ESS at the E7 | Preventing HPV18 233^416 splicing in the E6 ORF | — | HPV | Ajiro et al. (2016) |
| HipK3 | HipK3-T purine-rich region | Promoting HipK3-T exclusion | Tra2β-1 (Antagonistic) | Testis | Venables et al. (2005) |
| hnRNP A1 | — | Promoting intron 10 inclusion | Autoregulation | HeLa and NSC34 cells | Suzuki and Matsuoka, (2017) |
| hnRNP A1 | the CE1a and CE4 elements | Promoting exon 7B skipping | hnRNP A2 (Cooperative) | HeLa, CB3C7, and CB3C7-20 cells | Hutchison et al. (2002) |
| IKBKAP | ISS of intron 20; ESS1 and ESS2 of exon 20 | Promoting exon 20 skipping | — | Familial dysautonomia | Bruun et al. (2018) |
| IRF-3 | (UAGGGA) binding motifs in intron 1 | Promoting exons 2 and 3 inclusion | SRSF1 (Cooperative) | A549 and Calu-6 cells | Guo et al. (2013) |
| Ich-1 | — | Promoting exon 9 (61bp) inclusion | SRSF1, SRSF2(Antagonistic) | HeLa cell | Jiang et al. (1998) |
| INSR | AGGGA sites in intron 10 | Promoting exon 11 skipping | hnRNP F (Antagonistic) | HeLa, HepG2 and HEK293 cells | Talukdar et al. (2011) |
| LOXL4 | — | Promoting exon 9 skipping | — | ES-2 and MDA-MB-231 cells | Sebban et al. (2013) |
| Mag | UAGGU at the 5′ splice site of Mag exon 12 | Promoting (S-MAG) exon 12 skipping | — | Mouse brainstem; HeLa and CG4 cells | (Zhao et al., 2010; Zearfoss et al., 2013) |
| MLCK | UAGGGA in Intron 10 | Promoting exon 11 skipping | — | Human pulmonary artery endothelial cells | Mascarenhas et al. (2018) |
| Max | intronic region in intron 4 | Promoting exon 5 inclusion | — | Glioblastoma | Babic et al. (2013) |
| Mdm2 | — | Promoting exon3-10 skipping | — | HaCaT cell | Feng et al. (2016) |
| PKM | Intronic UAGGGC sequence flanking exon 9 | Repressing the use of exon 9 to generate PKM2 (exons 9 and 10 mutually splicing) | hnRNP A2, hnRNP I (Cooperative) | brain and glioma samples; NIH-3T3, C2C12 cells | David et al. (2010) |
| PKM | — | Repressing the use of exon 9 to generate PKM2 (exons 9 and 10 mutually splicing) | hnRNP A2, hnRNP I (Cooperative) | HeLa, HEK293, U-118MG, A-172, SK-N-BE and C2C12 cells | Clower et al. (2010) |
| PKM | Intronic UAGGGC sequence flanking exon 9 | Repressing the use of exon 9 to generate PKM2 (exons 9 and 10 mutually splicing) | SAM68(Cooperative) | Lung adenocarcinoma | Zhu et al. (2021) |
| PKM | Exon9-Intron9; Exon9; Exon10 | Repressing the use of exon 9 to generate PKM2 (exons 9 and 10 mutually splicing) | SRSF3(Cooperative) | Colon cancer DLD-1 and WiDr Cells | Kuranaga et al. (2018) |
| PKM | — | Repressing the use of exon 9 to generate PKM2 (exons 9 and 10 mutually splicing) | NEK2(Cooperative) | Multiple Myeloma Cells | Gu et al. (2017) |
| PKM | — | Repressing the use of exon 9 to generate PKM2 (exons 9 and 10 mutually splicing) | HIF1 (Cooperative) | Mouse cardiac muscle | Williams et al. (2018) |
| PKM | — | Repressing the use of exon 9 to generate PKM2 (exons 9 and 10 mutually splicing) | RBM4 (Antagonistic) | Neuronal differentiation of MSCs | Su et al. (2017) |
| PKM | Intronic UAGGGC sequence flanking exon 9 | Repressing the use of exon 9 to generate PKM2 (exons 9 and 10 mutually splicing) | RBMX (Antagonistic) | Bladder cancer | Yan et al. (2021) |
| p53-inducible gene 3 (PIG3) | ESS in exon 4 | Promoting exon 4 skipping | — | HeLa and MCF7 cells | Nicholls and Beattie, (2008) |
| RAGE | — | Inhibiting alternative inclusion of part of intron 9 and removal of exon 10 to form flRAGE | Tra2β-1 (Antagonistic) | SH-SY5Y cell | Liu et al. (2015) |
| Rac1 | UAAAGA within exon 3b | Promoting exon 3b exclusion | — | SCp2 and EpH4 cells | Pelisch et al. (2012) |
| Ron | Splicing silencer of exon 12 | Inhibiting Ron exon 11 skipping (Δ Ron) | SRSF1(Antagonistic) | HeLa, KATOⅢ and MDA-MB-435S cells | Bonomi et al. (2013) |
| SMN2 | intronic splicing silencer ISS-N1 at the beginning of intron 7 | Promoting exon 7 skipping | Tra2β-1 (Antagonistic) hnRNP I (Cooperative) | Spinal muscular atrophy | (Kashima and Manley, 2003; Kashima et al., 2007b; Beusch et al., 2017) |
| Smad2 | — | Promoting exon 9 skipping | Rpl22(Cooperative) | Zebrafish embryos | Zhang et al. (2017) |
| tau | 3′ splice site of exon 10 | Promoting exon 10 skipping; intron 9 exclusion | — | SH-SY5Y and HEK293T cells | Liu et al. (2020) |
| Tid1 | — | Promoting exon 11 skipping | hnRNP A2 (Cooperative) | Non-small cell lung cancer | Chen et al. (2016) |
| TIMP1 | 3′ splice site of exon 4 | Promoting intron 3 retention | — | HCT116 cell | Flodrops et al. (2020) |
| TRA2B | — | Promoting exon 2 inclusion | hnRNP U (Antagonistic) | HCT116 and HCEC-1CT cells | Nishikawa et al. (2019) |
hnRNP A1 binding to both the splicing sites and splicing regulatory elements is required for the splicing of some certain genes, such as Insulin Receptor (INSR) gene. In the alternative splicing of INSR gene, hnRNP A1 binds to both the 5’ splice site of intron 11 and the ISE site of intron 10, preventing the inclusion of exon 11 in INSR gene splicing, finally influencing the glucose metabolism (Talukdar et al., 2011). Besides, the alternative splicing regulatory activities of hnRNP A1 were dependent not only on the RNA-binding properties but also on protein-protein interactions with other splicing factors (Cartegni et al., 1996). As mentioned above, the selective binding is mostly mediated by the Gly-rich C-terminal region. The hnRNP A1-interacted proteins responsible for alternative splicing regulation can be sorted into two categories: cooperative and antagonistic splicing factors.
Alternative splicing regulation by hnRNP A1 with cooperative splicing factors
hnRNP A1 participates in spliceosome assembly, and can interact with other splicing factors and RBPs to regulate alternative splicing of genes cooperatively (Jurica et al., 2002; Zhou et al., 2002). Firstly, the homotypic interactions of hnRNP A1 via its UP1 domain have been documented using the bioluminescence resonance energy transfer (BRET) technology (Ding et al., 1999; Fisette et al., 2010). Like hnRNP A1, the longer isoform hnRNP A1B also exhibits homotypic interactions (Gueroussov et al., 2017; Gagne et al., 2021). Other RBPs, such as hnRNP C and TDP-43, have been shown to dimerize, suggesting that dimerization could affect their functions (Shiina et al., 2010; Cienikova et al., 2015). Secondly, hnRNP A1 cooperates with other hnRNPs family members to modulate gene alternative splicing. hnRNP A1B-hnRNP A1 heterodimer was observed in neurons (Gagne et al., 2021). The heterotypic interactions between hnRNP H and hnRNP A1 in live cells can be captured in live cells through BRET signals mediated by the C-terminal of hnRNP A1 and H (Fisette et al., 2010). hnRNP A1 and hnRNP H can collaborate in modulating 5′ splice site selection to further regulate alternative splicing of genes containing Bcl-x (Cloutier et al., 2018), beta-tropomyosin (Expert-Bezancon et al., 2004), and viral genes (Stoltzfus and Madsen, 2006). By regulating the alternative splicing of PKM gene, hnRNP A1, hnRNP A2 and hnRNPI (also known as PTB) worked together to enhance PKM2 expression (David et al., 2010). SAM68’s 351–443 aa region binds to the RGG motif of hnRNP A1, causing hnRNP A1-dependent PKM splicing to promote oncogenic PKM2 isoform formation while inhibiting PKM1 isoform formation. For c-src exon N1 splicing regulation, hnRNP A1 collaborates with hnRNP I to play key regulatory roles for exon exclusion by binding to 3’splice site of exon N1. The removal of hnRNP I binding sites in N1 Exon inhibits its effects of splicing repression but does not influence hnRNP A1-modulated N1 exon skipping. Thus, the impact of hnRNP A1 and hnRNP I on c-src splicing regulation are cumulative rather than synergistic (Rooke et al., 2003). hnRNPs such as hnRNP F (66), hnRNP A2/B1 (69) and others, as shown in Table 1 have been determined to collaborate with hnRNP A1 modulating gene alternative splicing. Thirdly, several SRs could cooperate with hnRNP A1 in regulating splicing events, although SR proteins often compete with hnRNPs (Table 1). These findings suggest that the regulation of alternative splicing events by hnRNP A1 still requires the involvement of multiple splicing factors including hnRNPs and SRs.
Alternative splicing regulation by hnRNP A1 with antagonistic splicing factors
Numerous splicing factors have been identified as hnRNP A1 antagonists in regulating alternative splicing. SR proteins are non-snRNP prtoteins involved in both constitutive and the controlled splicing processes (Graveley et al., 2001). SR splicing factors are the most predominant splicing factors antagonistically interacting with hnRNP A1 to modulate the splicing process. SRSF1 (ASF/SF2) is a well-established hnRNP A1 antagonist (Mayeda et al., 1993). According to the study from Mayeda et al. (Mayeda and Krainer, 1992), when alternative 5′ splice sites are present in pre-mRNAs, the relative concentration of hnRNP A1 and the crucial splicing factor SRSF1 influence the selection of 5′ splice site. Instead of having substrate-specific effects, these splicing factors have a shared effect on the polarity of alternative 5′ splice-site selection. When the concentration of SRSF1 is higher than hnRNP A1, it will lead to activation of proximal 5′ splice sites. While when the hnRNP A1 expression level is higher than SRSF1, it will benefit the activation of distal 5’ splice sites (Mayeda and Krainer, 1992). In exon 3 HIV-1 tat pre-mRNA, researchers investigated that hnRNP A1 and SR proteins participate in the antagonistic effects between ESE and ESS(33). hnRNP A1 inhibits tat23 splicing by cooperating along the exon, beginning at ESS3. SRSF1 reverses the process and leads to exon recognition, while SRSF2 (SC35) is ineffective (Zhu et al., 2001). The SRSF2-dependent splicing could be specifically blocked and mediated by the hnRNP A1 binding motif UAGUGAA in ESS3(33). Several other SRs and SR-like splicing factors that serve as hnRNP A1 antagonists have also been discovered (Jiang et al., 1998; Pollard et al., 2002; Guil et al., 2003; Rooke et al., 2003; Expert-Bezancon et al., 2004; Venables et al., 2005; Bonomi et al., 2013; Liu et al., 2015).
Furthermore, under certain circumstances, several hnRNPs splicing factors may have the opposite effect on hnRNP A1 on alternative splicing regulation. By binding to a particular motif positioned centrally and 3′ to exon 7, hnRNP A1 promotes exon 7 exclusion of CEACAM1 gene, while overexpression of hnRNP M causes exon 7 inclusion (Dery et al., 2011). Similarly, hnRNP H (79, 80) and hnRNP F (61) antagonize the activity of hnRNP A1 on the exon skipping in the splicing events of GRIN1 and INSR genes. Interestingly, hnRNP A1 promotes exon 2 inclusion of Tra2β, while hnRNP U facilitates its skipping, and further ectopic overexpression or deletion experiments reveal that hnRNP A1 and hnRNP U also influence the transcription of TRA2B, suggesting important roles of hnRNP A1 and hnRNP U in the coupling between gene transcription and alternative splicing, as they have both DNA- and RNA-binding abilities (Nishikawa et al., 2019).
There is antagonism between various types of splicing factors and hnRNP A1 in addition to the hnRNPs and SRs proteins. RBMX can competitively bind to the RGG motif of hnRNP A1, blocking the binding of hnRNP A1 to the sequences flanking PKM exon 9, suppressing the formation of PKM2 (82). RNA binding protein TIA-1 promotes K-SAM exon inclusion of FGFR2 through binding to the downstream intron, while hnRNP A1 represses the exon splicing by binding to the exon (Gesnel et al., 2009).
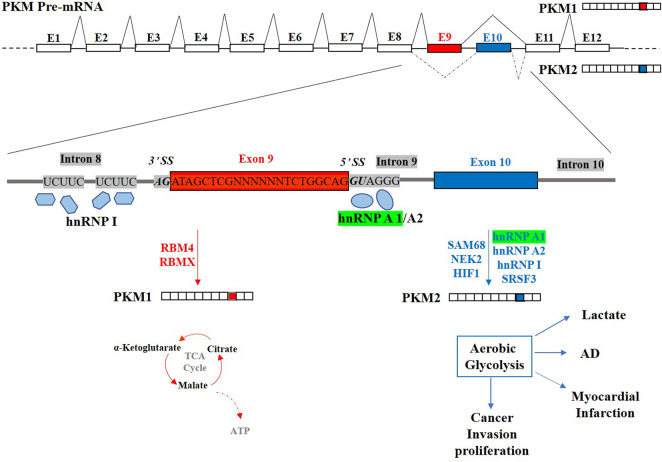
Altogether, hnRNP A1 is an essential regulator in alternative splicing. HnRNP A1 can collaborate with other factors to cooperatively and antagonistically regulate alternative splicing by interacting with specific sites, splicing regulatory elements in exons or introns, and specific protein domains. Consider the hnRNP A1-regulated alternative splicing of PKM gene, which is a canonical mutually exclusive alternative splicing (Figure 2). Alternative splicing of the PKM gene to skip exon 9 and include exon 10 generates PKM2 in cancer cells through hnRNP A1/A2 binding to UAGGG at 5’ SS in intron 9 and hnRNP I interacting with two UCUUC in intron 8 (Clower et al., 2010). Besides, RBM4 (85) and HIF1 (Williams et al., 2018) exert regulatory role in alternative splicing of PKM in the brain and heart. Other factors such as SRSF3(87), SAM68(88), NEK2 (89) and RBMX (82) are also involved in the alternative splicing process of the PKM gene via interacting with hnRNP A1. The isoform PKM2, results in aerobic glycolysis (the Warburg effect) with high lactate production, contributing to AD, myocardial infarction and cancer progression. The use of exon 9 to generate the PKM1 isoform is crucial for the tricarboxylic acid cycle (TCA cycle) and oxidative phosphorylation producing maximum ATP. hnRNP A1 cooperates with cooperative factors (hnRNP A2, hnRNP I, SRSF3, SAM68, NEK2 and HIF1) and antagonistic factors (RBMX and RBM4) to regulate alternative splicing of PKM gene. The hnRNP A1 can regulate the alternative splicing process of multiple target genes and correspondingly, the alternative splicing process of one single gene is regulated by multiple splicing factors.
FIGURE 2.
Schematic representation of the human pre-PKM gene and the regulation of its mutually exclusive alternative splicing. Exon9 (E9) and Exon10 (E10) of PKM gene are two mutually exclusive exons. Generation of the PKM1 isoform using E9 is crucial for tricarboxylic acid cycle (TCA cycle) and oxidative phosphorylation producing maximum ATP. PKM2, including E10 but not E9, results in aerobic glycolysis (the Warburg effect) with high lactate production, contributing to AD, myocardial infarction and cancer progression. hnRNP A1 (Highlighted in Green) suppresses the E9 inclusion by binding to 5′ SS downstream E9 and co-regulation with cooperative factors (hnRNP A2, hnRNP I, SRSF3, SAM68, NEK2 and HIF1) and antagonistic factor (RBMX and RBM4).
In addition to canonical linear splicing of pre-mRNA, the back-splicing has been recently well studied to produce circular RNAs (circRNAs) by the joint of a downstream 5′ splice site and an upstream 3′ splice site. Both hnRNP and SR proteins could regulate circRNA biogenesis (Kramer et al., 2015). Particularly, the characteristics of those RNA binding proteins to bind the introns flanking circRNA and form a dimmer, effectively contribute to the biogenesis of circRNAs (Conn et al., 2015). Accordingly, the engineering circRNA regulators (ECRRs) has been proposed recently by Qi et al. (Qi et al., 2021), to specifically promote circular RNA production by combining sequence-specific RNA binding motifs of human Pumilio 1 (PUF domain) with functional domains that could form dimerization, including the UP1 (RRM) domain of HNRNP A1. The PUF-hnRNP A1 ECRR could significantly promote the circRNA production of the exogenous circRNA minigene reporter circGFP, indicating an important cricRNA biogenesis regulatory role of hnRNP A1. Given the importance of circRNAs in various physiological and pathologic conditions, the effects of hnRNP A1 on back-splicing control and specific circRNA generation are worthy of further exploration.
hnRNP A1 regulates mRNA transcription and translation
Although hnRNP A1 is a powerful RNA binding protein, its role in transcription regulation has been suggested as it could prevent the transcription elongation factor phospho-TEFb from interacting with its repressor molecule, the 7SK RNA (Barrandon et al., 2007; Van Herreweghe et al., 2007). The hnRNP A1 UP1 domain bound specifically to the SL3 domain of 7SK snRNA, forming the 7SK-hnRNP A1 complex, further influenced the 7SK snRNA bioactivity and P-TEFb availability, affecting the initial transcription and elongation processes (Luo et al., 2021). Downregulation of hnRNP A1 causes the promoter-proximal pausing of RNA polymerase II (Pol II) on TEFb-dependent genes, as well as transcriptional repression, suggesting that hnRNP A1 has a role in controlling transcription elongation by Pol II (Lemieux et al., 2015). Along with the eukaryotic gene, hnRNP A1 can bind to the complementary (-)-strand of the leader RNA and intergenic sequence of mouse hepatitis virus (MHV) RNA, enhancing the viral RNA transcription (Li et al., 1997). Besides, N6-methyladenosine (m6A) modification of RNA, an addition of a methyl group at position N6 of adenosine, can influence RNA transcription, alternative splicing, degradation, and translation. The m6A modification is catalyzed by methyltransferase (“writers”), removed by demethylase (“erasers”) and recognized by m6A binding protein (“readers”). As one of the “readers”, HnRNPs including hnRNPA2/B1 (Alarcon et al., 2015) and hnRNP C (Huang et al., 2021) can recognize the methylation sequence on RNAs. Recently, Kumar et al. (Kumar et al., 2021) discovered that hnRNP A1 could be recruited to the m6A-modified SARS-CoV-2 RNA, and act as a m6A “reader” to promote the transcription.
Also, hnRNP A1 has been shown to possess DNA binding activity, by interacting with G-quadruplex structure in the promoter of genes to promote the gene transcription (Cogoi et al., 2017). hnRNP A1 firmly binds the quadruplex-forming GC-elements upstream of the primary transcription start sites and their higher i-motif conformations, increasing HRAS gene transcription (Miglietta et al., 2015). Table 2 lists the genes transcriptionally regulated by hnRNP A1. Interestingly, hnRNP A1 exerts regulating alternative splicing of some certain genes, such as TRA2B, concomitantly influencing its transcription activity, indicating that hnRNP A1 probably involve in the co-transcriptional splicing modulation (Nishikawa et al., 2019).
TABLE 2.
The transcription and translation of genes enhanced by hnRNP A1.
| Genes | Binding sites | Functions | Reference |
|---|---|---|---|
| Acta2 | Promoter | Enhancing Transcription | Huang et al. (2013) |
| APOE | -219T site in the promoter | Enhancing Transcription | Campillos et al. (2003) |
| ANXA7 | Promoter | Enhancing Transcription | Torosyan et al. (2010) |
| mouse hepatitis virus (MHV) RNA | MHV(-)-strand leader and IG sequences | Enhancing Transcription | Li et al. (1997) |
| Tagln | Promoter | Enhancing Transcription | Huang et al. (2013) |
| TRA2B | G-quadruplex | Enhancing Transcription | Nishikawa et al. (2019) |
| c-myc | IRES | Enhancing Translation | (Jo et al., 2008; Shi et al., 2016) |
| cyclin D1 | IRES | Enhancing Translation | Jo et al. (2008) |
| egr2 | IRES | Enhancing Translation | Rubsamen et al. (2012) |
| EV71 | IRES | Enhancing Translation | Leong et al. (2015) |
| FGF-2 | IRES | Enhancing Translation | Bonnal et al. (2005) |
| HIF1a | IRES | Enhancing Translation | (Gao et al., 2017; Zeng et al., 2021) |
| HRAS | GC-elements | Enhancing Translation | Miglietta et al. (2015) |
| KRAS | G-quadruplex | Enhancing Translation | Paramasivam et al. (2009) |
| MELOE-1 | IRES | Enhancing Translation | Charpentier et al. (2021) |
| Nfil3 | IRES | Enhancing Translation | Kim et al. (2017) |
| RON | G-quadruplex in 5′-UTR | Enhancing Translation | Cammas et al. (2016) |
| sST2 | IRES | Enhancing Translation | Kunze et al. (2016) |
| VRK1 | 3′-UTR | Enhancing the translation | Ryu et al. (2021) |
hnRNP A1 is primarily distributed in the nucleus as RNA binding protein, however, it may translocate to the cytoplasm with specific phosphorylation when expose to growth factors (Kunze et al., 2016) and stress stimuli such as osmotic shock or UV irradiation (van der Houven van Oordt et al., 2000; Feng et al., 2018). Translocation is crucial for translational regulatory functions of hnRNP A1 as it can bind to the internal ribosome entry site (IRES) of mRNA. hnRNP A1 acts as an ITAF, affecting the mRNA translation process, regulating the sterol-regulatory-element-binding protein 1a (SREBP-1a) expression in hepatocytes and hepatoma cells (Damiano et al., 2013), and hence modulating the expression of several enzymes involved in lipid synthesis. In addition to the eukaryotic genes, hnRNP A1 enhances the translation of EV71 and HRV-2 by binding to the IRES (128, 129). In hnRNP A1 driven IRES-dependent EV71 RNA translation process, UP1 domain of hnRNP A1 interacts specifically with the stem-loop II (SLII) of the IRES, required for the next translation (Levengood et al., 2013).
In Drosophila, hnRNP A1 binds to the 3′ untranslated region (UTR) of Nanos mRNA, inhibiting Nanos translation, whereas poly (ADP-ribosylation) of hnRNP A1 relieves the translation repression (Ji and Tulin, 2016). hnRNP A1 can bind to the IRES of cyclin D1 and c-Myc to promote gene translation, however, phosphorylation of hnRNP A1 on serine 199 modulated by Akt suppresses the IRES activity (Jo et al., 2008). PRMT5 facilitates the interaction of hnRNP A1 with IRES by methylating R218 and R225 by to promote IRES-dependent translation of cyclin D1 and c-Myc (Gao et al., 2017). These findings suggest that the ITAF activity of hnRNP A1 is influenced not only by its expression level but also by its post-translational modifications (PTMs) status, and the PTMs of hnRNP A1 are crucial regulators of ITAF bioactivities.
In addition to its translation promoting activities, the translational repression effects of hnRNP A1 via interaction with IRES have also been reported (Table 3). For example, hnRNP A1 binds to the IRES of cellular apoptotic peptidase activating factor 1 (apaf-1) mRNA to repress its translation (Li et al., 2019). hnRNP A1 interacts with the IRES of X-linked inhibitor of apoptosis (XIAP), suppressing its translation (Lewis et al., 2007). These findings show that hnRNP A1, along with subcellular localization, plays a vital role in regulating IRES-dependent translation.
TABLE 3.
The transcription and translation of genes repressed by hnRNP A1.
| Genes | Binding sites | Functions | Reference |
|---|---|---|---|
| human thymidine kinase (htk) | an ATTT sequence motif in CCRU of the promoter | Repressing Transcription | Lau et al. (2000) |
| apaf-1 | IRES | Repressing Translation | Li et al. (2019) |
| Nanos | 3′ UTR | Repressing Translation | Ji and Tulin, (2016) |
| XIAP | IRES | Repressing Translation | Lewis et al. (2007) |
hnRNP A1 influences RNA stability
hnRNP A1 influences mRNA stability in addition to alternative splicing, transcription and translation. The mRNA stability and degradation regulatory pathways have been extensively reviewed by Clarke et al. (Clarke et al., 2021). AU-rich elements (ARE) in 3′ untranslated region (UTR) of mRNA function as a potent mRNA destabilizing element (Chen and Shyu, 1995). By binding to the reiterated AUUUA sequence of the 3′-UTR of the lymphokine, c-myc and c-fos proto-oncogene mRNA in human T lymphocytes, hnRNP A1 enhances the mRNA stability (Hamilton et al., 1993). Furthermore, hnRNP A1 is identified as an ARE-binding protein in 3′UTR of cIAP1, (crucial member of the apoptosis inhibitor family), increasing its mRNA stability under cytotoxic conditions such as UV radiation (Zhao et al., 2009). These findings are suggestive of a cancer-promoting effect of hnRNP A1 through influencing specific mRNA stability. Further research shows that ARE binding ability of hnRNP A1 is negatively correlated with its serine-threonine phosphorylation status (Hamilton et al., 1997). Besides binding to ARE, hnRNP A1 can bind to a putative hairpin-loop region in the 3′ UTR of CYP2A5 mRNA, enhancing the mRNA stability (Glisovic et al., 2003).
miRNAs are endogenous single-stranded RNAs that negatively regulates the targeted mRNAs. The RNase III Drosha enzyme catalyzes the biogenesis of miRNAs, forming the stem-loop precursors, which are then processed by type III ribonuclease Dicer, producing the mature miRNAs. The hnRNP A1 participates in this process and regulates the biogenesis of miRNAs. HnRNP A1 binds to the conserved terminal loop of pri-let-7a-1 and inhibits its processing by Drosha, inhibiting let-7a biogenesis (Michlewski and Caceres, 2010). In contrast, hnRNP A1 promotes miRNA-18a biogenesis mediated by identifying the terminal loop RNA and therefore creating a favorable cleavage site for Drosha, which could be a potential general principle of miRNA biogenesis and regulation (Michlewski et al., 2008; Kooshapur et al., 2018). Furthermore, hnRNP A1 serves as a powerful loading protein for microRNAs in small extracellular vesicle (sEV-miRNAs), facilitating tumor proliferation and migration of non-small cell lung cancer (Li et al., 2021). hnRNP A1 has also shown to mediate the package of miR-196a into cancer-associated fibroblasts (CAF)-derived exosomes targeting CDKN1B and ING5 to endow cisplatin resistance of head and neck cancer (Qin et al., 2019). As a result, based on the contribution of miRNAs to the stability and translation of mRNAs (Fabian et al., 2010), hnRNP A1 can modulate mRNA stability and translation indirectly mediated by miRNAs.
Nonsense-mediated mRNA decay (NMD) is a conserved mRNA quality control mechanism for ensuring the fidelity of gene expression, which is another mRNA degrading pathway targeting the mRNAs that lack of the proper arrangement of translational signals. A large percentage of alternatively spliced transcripts that harbor a premature termination codon (PTC) can be degraded by NMD pathway (Lareau et al., 2007). Alternative polyadenylation (APA) is an alternative splicing process that produces transcript 3′-UTRs with distinct sequences, lengths and stabilities. Alternative polyadenylation of 3′UTR in hnRNP A2/B1 pre-mRNA is induced by increased hnRNP A1, resulting in mRNA degradation via the NMD pathway (Patry et al., 2003; McGlincy et al., 2010). However, the role of hnRNP A1 in NMD is still largely unknown, requiring further investigation to identify the underlying mechanisms.
Available approaches targeting hnRNP A1
The crucial roles of hnRNP A1 have been indicated in cancers (Roy et al., 2017) and neurodegenerative diseases (Clarke et al., 2021). hnRNP A1 selectively regulates mRNA splicing processes, promoting expression of specific protein variants linked to tumorigenesis and cancer progression, and also modulates the transcription and translation of several oncogenes or anticancer genes (Roy et al., 2017). The pathogenesis of neurodegenerative diseases including amyotrophic lateral sclerosis, multiple sclerosis, Alzheimer’s disease, and Huntington’s disease may be influenced by hnRNP A1 dysregulation (Clarke et al., 2021). Therefore, targeting hnRNP A1 for the treatments of the relevant cancers and neurological disorders may be encouraging. Here, we have reviewed the reported approaches targeting hnRNP A1 (Table 4).
TABLE 4.
Available Approaches Targeting hnRNP A1.
| Candidates | Targeting process affected/Checked | Reference |
|---|---|---|
| VPC-80051 | targeting the RNA-binding domain (RBD) of hnRNP A1 | Carabet et al. (2019) |
| Camptothecin | bind directly to hnRNP A1 and inhibit the hnRNP A1/ topoisomerase I (top I) interaction | Manita et al. (2011) |
| Riluzole | bind directly to hnRNP A1 and inhibit its ITAF activity | Benavides-Serrato et al. (2020) |
| Compound 11 | block hnRNP A1 from interacting with IRES of c-MYC and cyclin D1 | Holmes et al. (2016) |
| Quercetin | binds to the C-terminal region of hnRNPA1, causing it to be retained in the cytoplasm | (Ko et al., 2014; Tummala et al., 2017) |
| Idarubicin | impairing the binding between EV71 IRES RNA and hnRNP A1 | Hou et al. (2016) |
| Tetracaine hydrochloride | reduced protein stability of hnRNP A1 | Huang et al. (2022) |
| miR-18a | mRNA degradation | Fujiya et al. (2014) |
| miR-490 | mRNA degradation | Zhou et al. (2016) |
| miR-206 | mRNA degradation | Fu et al. (2020) |
| miR-424 | mRNA degradation | Otsuka et al. (2018) |
| miR-503 | mRNA degradation | Otsuka et al. (2018) |
| miR-135a-5p | mRNA degradation | Sokol et al. (2018) |
| miR-149–5p | mRNA degradation | |
| miR-137 | mRNA degradation | Sun et al. (2012) |
| lncRNA RP11-81H3.2 | directly interacts with miR-339, weakening the repression from miRNA to mRNA of hnRNP A1 | Chen et al. (2020) |
| lncRNA ANCR | sponge miR-140–3p to inhibit hnRNP A1 degradation | Wen et al. (2020) |
| lncRNA XIST | sponge miR-326 to inhibit hnRNP A1 degradation | Ding et al. (2021) |
| BC15 | an hnRNP A1-specific single-stranded DNA aptamer, used as hnRNP A1 inhibitor | Li et al. (2012) |
| ASO (SMN) | target the hnRNP A1 binding ISS of SMN intron 7 enhances SMN2 exon 7 inclusion | Beusch et al. (2017) |
| SSO (MTRR) | block the hnRNP A1 binding ESEs created by c.903 + 469T>C MTRR mutation correcting the splicing and restoring protein activity | Palhais et al. (2015) |
With a computer-aided drug discovery approach, Carabet et al. (Carabet et al., 2019) developed an inhibitor named VPC-80051 to target the RNA-binding domain (RBD) of hnRNP A1. The compound could effectively suppress both c-Myc transcription and an alternative splice variant of androgen receptor (AR-V7) generation modulated by hnRNP A1. Camptothecin (CPT) is an anti-tumor natural product, that can bind directly to hnRNP A1 and inhibit the hnRNP A1/ topoisomerase I (top I) interaction (Manita et al., 2011), wherein top I is essential for maintaining DNA helical structure and is important for cancer progression. Benavides-Serrato et al. (Benavides-Serrato et al., 2020) discovered that riluzole could bind directly to hnRNP A1 and inhibit its ITAF activity in glioblastoma via a riluzole-bead coupled binding assay. Holmes et al. (Holmes et al., 2016) identified a drug named compound 11 (C11) that blocks hnRNP A1 from interacting with IRES of c-MYC and cyclin D1, in glioblastoma cells. Using affinity chromatography, mass spectrometry and in vitro binding experiments, it was discovered that quercetin, a flavonoid, interact directly with hnRNP A1. Quercetin binds to the C-terminal region of hnRNPA1, causing it to be retained in the cytoplasm, and subsequently exerts its anti-cancer effects on prostate cancer cells (Ko et al., 2014; Tummala et al., 2017). Idarubicin, an anthracycline compound used for cancer therapy, has been identified as a broad-spectrum enterovirus replication inhibitor that selectively inhibits impairing the binding between EV71 IRES RNA and hnRNP A1 (168). Our recent work, that tetracaine hydrochloride, a local anesthetic, was found to induce the melanoma cell cycle by downregulating hnRNP A1. We discovered that tetracaine hydrochloride treatment reduced protein stability of hnRNP A1, however the underlying molecular mechanism needs further investigation (Huang et al., 2022). These findings suggest that some small-molecule chemical drugs can inhibit hnRNP A1 activity. However, the application of these drugs targeting hnRNP A1 are facing challenges with targeting specificity, side effects and drug delivery efficiency.
hnRNP A1 activities can be inhibited by various biopharmaceutical approaches. miR-18a is reported to target hnRNP A1 in colon cancer cells, because miRNAs bind to mRNAs and downregulate the transcripts of target genes based on sequence complementarity (Fujiya et al., 2014). The miR-490 binds directly to the 3′-UTR of hnRNPA1 mRNA repressing its translation in gastric cancer cells (Zhou et al., 2016). The tumor suppressor miR-206 directly targets hnRNPA1 to attenuate the Warburg effect and proliferation of colon cancer cells (Fu et al., 2020). In breast cancer, resveratrol induces tumor-suppressive miRNAs miR-424 and miR-503, which suppress breast cancer cell proliferation by downregulating the hnRNP A1 expression (Otsuka et al., 2018). As per the reports, miR-135a-5p, miR-149–5p, miR-137, miR-339 are the other miRNAs that target hnRNP A1 (174–176). Long noncoding RNA (lncRNA) can regulate hnRNP A1 expression level via a competing endogenous RNA (CeRNA) mechanism. In gastric cancer, lncRNA RP11-81H3.2 directly interacts with miR-339, weakening the repression from miRNA to mRNA of hnRNP A1 (176). The lncRNA ANCR can sponge miR-140–3p to inhibit hnRNP A1 degradation promoting hepatocellular carcinoma metastasis (Wen et al., 2020). Similarly, lncRNA XIST upregulates hnRNP A1 in Multiple sclerosis (MS) through the XIST-miR-326-HNRNPA1 signaling axis (Ding et al., 2021). However, one miRNA targets more than one mRNA, and one single mRNA can be targeted by multiple miRNAs, translation of miRNAs to clinical practice remains challenging.
Additionally, more approaches targeting hnRNP A1 have been developed. BC15, an hnRNP A1-specific single-stranded DNA aptamer, is used as hnRNP A1 inhibitor, eliciting a strong anticancer effect on the proliferation of cultured hepatoma cells (Li et al., 2012). Anti-sense oligonucleotides (ASO) that target the hnRNP A1 binding ISS of SMN intron 7 enhances SMN2 exon 7 inclusion, reflecting advantages in neuromuscular disease spinal muscular atrophy (Beusch et al., 2017). A similar splice-shifting oligonucleotide (SSO) is utilized to block the hnRNP A1 binding ESEs created by c.903 + 469T>C MTRR mutation correcting the splicing and restoring protein activity (Palhais et al., 2015). Proteolysis targeting chimeras (PROTACs) have recently been proposed for in vivo protein degradation by recruiting E3 ubiquitin ligases with high-affinity ligands (Bondeson et al., 2015). In addition, Ghidini et al. (Ghidini et al., 2021) have introduced an updated PROTAC named RNA-PROTACs for effectively degrading RBPs using small RNA mimics docking the RNA-binding site of the RBP, which is a very promising direction for rapidly and selectively targeting RBPs in various diseases.
Conclusion
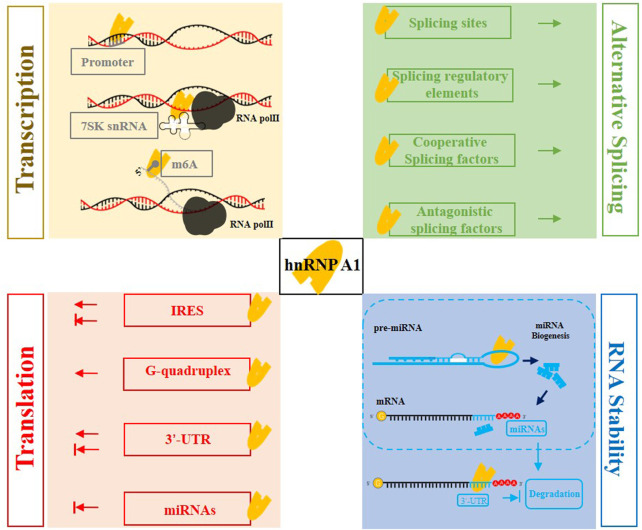
hnRNP A1 belongs to the hnRNP subfamily and is among the most abundant and widely expressed nuclear proteins. It has multiple functions including participation in transcription regulation, alternative splicing, mRNA translation, miRNA processing and mRNA stability according to its RNA and DNA binding ability (Figure 3). hnRNP A1 can influence the transcription process by directly interacting with the G-quadruplex structure in the promoter, as well as indirectly controlling the transcription of TEFb-dependent genes by influencing transcription elongation by Pol II. hnRNP A1 regulates alternative splicing of multiple genes, collaborating with other cooperative or/and antagonistical splicing factors by binding to splicing sites, splicing regulatory elements of exons or introns, and specific protein domains. HnRNP A1 may exert as ITAF, influencing the IRES-dependent mRNA translation and gene translation by binding to the 3′-UTR of mRNAs. hnRNP A1 can effectively affect the stability of mRNAs via binding to specific sites of 3′-UTR, miRNAs biogenesis and NMD pathway. Finally, the targeting hnRNP A1 approaches are reviewed including traditional chemical drugs and biomedicines. According to the evidence, hnRNP A1 plays a crucial role in regulating RNA metabolism, and its dysregulation has been linked to diverse diseases, including cancers and neurodegeneration diseases. As a result, more in-depth exploration in the functions and underlying molecular mechanisms of hnRNP A1 is required, as well as the development of the rapid, selective and highly effective approaches targeting hnRNP A1.
FIGURE 3.
The multifaceted roles of hnRNP A1 in mRNA metabolism. hnRNP A1 affects gene transcription through binding to 7SK snRNA or directly binding to the gene promoter; hnRNP A1 can be recruited to the m6A-modification, and act as a m6A “reader” to promote the transcription. hnRNP A1 enhances mRNA stability through binding to specific sites of 3′-UTR. hnRNP A1 also contributes to the biogenesis of miRNAs, and then the specific miRNAs suppress the mRNA translation or targeting the mRNA decreasing the stability. hnRNP A1 regulates alternative splicing events by binding to specific splice sites and splicing regulatory elements, as well as interacting with cooperative or antagonistic splicing factors. hnRNP A1 increases gene translation by binding to G-quadruplex, and shows positive or negative effects when using its ITAF activity binding to IRES sequence or binding to 3′-UTR of mRNAs.
Author contributions
The first draft of the manuscript was written by JF, JZ, and YL. JF and WH instructed, supervised and finalized the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by National Natural Science Foundation of China (No. 81902796), China Postdoctoral Science Foundation (No. 2019M662985) and the Science and Technology Department of Sichuan Province (No. 2022NSFSC1594).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Ajiro M., Tang S., Doorbar J., Zheng Z. M. (2016). Serine/arginine-rich splicing factor 3 and heterogeneous nuclear ribonucleoprotein A1 regulate alternative RNA splicing and gene expression of human papillomavirus 18 through two functionally distinguishable cis elements. J. Virol. 90 (20), 9138–9152. 10.1128/JVI.00965-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon C. R., Goodarzi H., Lee H., Liu X., Tavazoie S., Tavazoie S. F. (2015). HNRNPA2B1 is a mediator of m(6)a-dependent nuclear RNA processing events. Cell 162 (6), 1299–1308. 10.1016/j.cell.2015.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allemand E., Guil S., Myers M., Moscat J., Caceres J. F., Krainer A. R. (2005). Regulation of heterogenous nuclear ribonucleoprotein A1 transport by phosphorylation in cells stressed by osmotic shock. Proc. Natl. Acad. Sci. U. S. A. 102 (10), 3605–3610. 10.1073/pnas.0409889102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An P., Grabowski P. J. (2007). Exon silencing by UAGG motifs in response to neuronal excitation. PLoS Biol. 5 (2), e36. 10.1371/journal.pbio.0050036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babic I., Anderson E. S., Tanaka K., Guo D., Masui K., Li B., et al. (2013). EGFR mutation-induced alternative splicing of Max contributes to growth of glycolytic tumors in brain cancer. Cell Metab. 17 (6), 1000–1008. 10.1016/j.cmet.2013.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrandon C., Bonnet F., Nguyen V. T., Labas V., Bensaude O. (2007). The transcription-dependent dissociation of P-TEFb-HEXIM1-7SK RNA relies upon formation of hnRNP-7SK RNA complexes. Mol. Cell. Biol. 27 (20), 6996–7006. 10.1128/MCB.00975-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beijer D., Kim H. J., Guo L., O'Donovan K., Mademan I., Deconinck T., et al. (2021). Characterization of HNRNPA1 mutations defines diversity in pathogenic mechanisms and clinical presentation. JCI Insight 6 (14), 148363. 10.1172/jci.insight.148363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekenstein U., Soreq H. (2013). Heterogeneous nuclear ribonucleoprotein A1 in health and neurodegenerative disease: From structural insights to post-transcriptional regulatory roles. Mol. Cell. Neurosci. 56, 436–446. 10.1016/j.mcn.2012.12.002 [DOI] [PubMed] [Google Scholar]
- Benavides-Serrato A., Saunders J. T., Holmes B., Nishimura R. N., Lichtenstein A., Gera J. (2020). Repurposing potential of riluzole as an ITAF inhibitor in mTOR therapy resistant glioblastoma. Int. J. Mol. Sci. 21 (1), E344. 10.3390/ijms21010344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beusch I., Barraud P., Moursy A., Clery A., Allain F. H. (2017). Tandem hnRNP A1 RNA recognition motifs act in concert to repress the splicing of survival motor neuron exon 7. Elife 6, e25736. 10.7554/eLife.25736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biamonti G., Buvoli M., Bassi M. T., Morandi C., Cobianchi F., Riva S. (1989). Isolation of an active gene encoding human hnRNP protein A1. Evidence for alternative splicing. J. Mol. Biol. 207 (3), 491–503. 10.1016/0022-2836(89)90459-2 [DOI] [PubMed] [Google Scholar]
- Bondeson D. P., Mares A., Smith I. E., Ko E., Campos S., Miah A. H., et al. (2015). Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat. Chem. Biol. 11 (8), 611–617. 10.1038/nchembio.1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnal S., Pileur F., Orsini C., Parker F., Pujol F., Prats A. C., et al. (2005). Heterogeneous nuclear ribonucleoprotein A1 is a novel internal ribosome entry site trans-acting factor that modulates alternative initiation of translation of the fibroblast growth factor 2 mRNA. J. Biol. Chem. 280 (6), 4144–4153. 10.1074/jbc.M411492200 [DOI] [PubMed] [Google Scholar]
- Bonomi S., di Matteo A., Buratti E., Cabianca D. S., Baralle F. E., Ghigna C., et al. (2013). HnRNP A1 controls a splicing regulatory circuit promoting mesenchymal-to-epithelial transition. Nucleic Acids Res. 41 (18), 8665–8679. 10.1093/nar/gkt579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruun G. H., Bang J. M. V., Christensen L. L., Broner S., Petersen U. S. S., Guerra B., et al. (2018). Blocking of an intronic splicing silencer completely rescues IKBKAP exon 20 splicing in familial dysautonomia patient cells. Nucleic Acids Res. 46 (15), 7938–7952. 10.1093/nar/gky395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruun G. H., Doktor T. K., Borch-Jensen J., Masuda A., Krainer A. R., Ohno K., et al. (2016). Global identification of hnRNP A1 binding sites for SSO-based splicing modulation. BMC Biol. 14, 54. 10.1186/s12915-016-0279-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C. G., Dreyfuss G. (1994). RNA binding specificity of hnRNP A1: Significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 13 (5), 1197–1204. 10.1002/j.1460-2075.1994.tb06369.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A., Hertel K. J. (2012). Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip. Rev. RNA 3 (1), 1–12. 10.1002/wrna.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammas A., Lacroix-Triki M., Pierredon S., Le Bras M., Iacovoni J. S., Teulade-Fichou M. P., et al. (2016). hnRNP A1-mediated translational regulation of the G quadruplex-containing RON receptor tyrosine kinase mRNA linked to tumor progression. Oncotarget 7 (13), 16793–16805. 10.18632/oncotarget.7589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campillos M., Lamas J. R., Garcia M. A., Bullido M. J., Valdivieso F., Vazquez J. (2003). Specific interaction of heterogeneous nuclear ribonucleoprotein A1 with the -219T allelic form modulates APOE promoter activity. Nucleic Acids Res. 31 (12), 3063–3070. 10.1093/nar/gkg435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabet L. A., Leblanc E., Lallous N., Morin H., Ghaidi F., Lee J., et al. (2019). Computer-aided discovery of small molecules targeting the RNA splicing activity of hnRNP A1 in castration-resistant prostate cancer. Molecules 24 (4), E763. 10.3390/molecules24040763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L., Maconi M., Morandi E., Cobianchi F., Riva S., Biamonti G. (1996). hnRNP A1 selectively interacts through its Gly-rich domain with different RNA-binding proteins. J. Mol. Biol. 259 (3), 337–348. 10.1006/jmbi.1996.0324 [DOI] [PubMed] [Google Scholar]
- Charpentier M., Dupre E., Fortun A., Briand F., Maillasson M., Com E., et al. (2021). hnRNP-A1 binds to the IRES of MELOE-1 antigen to promote MELOE-1 translation in stressed melanoma cells. Mol. Oncol. 16, 594–606. 10.1002/1878-0261.13088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheatham A. M., Davis S. E., Khatua A. K., Popik W. (2018). Blocking the 5' splice site of exon 4 by a morpholino oligomer triggers APOL1 protein isoform switch. Sci. Rep. 8 (1), 8739. 10.1038/s41598-018-27104-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. Y., Jan C. I., Pi W. C., Wang W. L., Yang P. C., Wang T. H., et al. (2016). Heterogeneous nuclear ribonucleoproteins A1 and A2 modulate expression of Tid1 isoforms and EGFR signaling in non-small cell lung cancer. Oncotarget 7 (13), 16760–16772. 10.18632/oncotarget.7606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. Y., Shyu A. B. (1995). AU-Rich elements: Characterization and importance in mRNA degradation. Trends biochem. Sci. 20 (11), 465–470. 10.1016/s0968-0004(00)89102-1 [DOI] [PubMed] [Google Scholar]
- Chen F. R., Sha S. M., Wang S. H., Shi H. T., Dong L., Liu D., et al. (2020). RP11-81H3.2 promotes gastric cancer progression through miR-339-HNRNPA1 interaction network. Cancer Med. 9 (7), 2524–2534. 10.1002/cam4.2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou N. T., Shankarling G., Lynch K. W. (2013). hnRNP L and hnRNP A1 induce extended U1 snRNA interactions with an exon to repress spliceosome assembly. Mol. Cell 49 (5), 972–982. 10.1016/j.molcel.2012.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. D., Dreyfuss G. (1984). Isolation of the heterogeneous nuclear RNA-ribonucleoprotein complex (hnRNP): A unique supramolecular assembly. Proc. Natl. Acad. Sci. U. S. A. 81 (23), 7471–7475. 10.1073/pnas.81.23.7471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cienikova Z., Jayne S., Damberger F. F., Allain F. H., Maris C. (2015). Evidence for cooperative tandem binding of hnRNP C RRMs in mRNA processing. Rna 21 (11), 1931–1942. 10.1261/rna.052373.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J. P., Thibault P. A., Salapa H. E., Levin M. C. (2021). A comprehensive analysis of the role of hnRNP A1 function and dysfunction in the pathogenesis of neurodegenerative disease. Front. Mol. Biosci. 8, 659610. 10.3389/fmolb.2021.659610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier A., Shkreta L., Toutant J., Durand M., Thibault P., Chabot B. (2018). hnRNP A1/A2 and Sam68 collaborate with SRSF10 to control the alternative splicing response to oxaliplatin-mediated DNA damage. Sci. Rep. 8 (1), 2206. 10.1038/s41598-018-20360-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clower C. V., Chatterjee D., Wang Z., Cantley L. C., Vander Heiden M. G., Krainer A. R. (2010). The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc. Natl. Acad. Sci. U. S. A. 107 (5), 1894–1899. 10.1073/pnas.0914845107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoi S., Rapozzi V., Cauci S., Xodo L. E. (2017). Critical role of hnRNP A1 in activating kras transcription in pancreatic cancer cells: A molecular mechanism involving G4 DNA. Biochim. Biophys. Acta. Gen. Subj. 1861 (5), 1389–1398. 10.1016/j.bbagen.2016.11.031 [DOI] [PubMed] [Google Scholar]
- Conn S. J., Pillman K. A., Toubia J., Conn V. M., Salmanidis M., Phillips C. A., et al. (2015). The RNA binding protein quaking regulates formation of circRNAs. Cell 160 (6), 1125–1134. 10.1016/j.cell.2015.02.014 [DOI] [PubMed] [Google Scholar]
- Damgaard C. K., Tange T. O., Kjems J. (2002). hnRNP A1 controls HIV-1 mRNA splicing through cooperative binding to intron and exon splicing silencers in the context of a conserved secondary structure. Rna 8 (11), 1401–1415. 10.1017/s1355838202023075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiano F., Rochira A., Tocci R., Alemanno S., Gnoni A., Siculella L. (2013). hnRNP A1 mediates the activation of the IRES-dependent SREBP-1a mRNA translation in response to endoplasmic reticulum stress. Biochem. J. 449 (2), 543–553. 10.1042/BJ20120906 [DOI] [PubMed] [Google Scholar]
- David C. J., Chen M., Assanah M., Canoll P., Manley J. L. (2010). HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 463 (7279), 364–368. 10.1038/nature08697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Gatto-Konczak F., Olive M., Gesnel M. C., Breathnach R. (1999). hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol. Cell. Biol. 19 (1), 251–260. 10.1128/mcb.19.1.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dery K. J., Gaur S., Gencheva M., Yen Y., Shively J. E., Gaur R. K. (2011). Mechanistic control of carcinoembryonic antigen-related cell adhesion molecule-1 (CEACAM1) splice isoforms by the heterogeneous nuclear ribonuclear proteins hnRNP L, hnRNP A1, and hnRNP M. J. Biol. Chem. 286 (18), 16039–16051. 10.1074/jbc.M110.204057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J., Hayashi M. K., Zhang Y., Manche L., Krainer A. R., Xu R. M. (1999). Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 13 (9), 1102–1115. 10.1101/gad.13.9.1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Li T., Yan X., Cui M., Wang C., Wang S., et al. (2021). Identification of hub lncRNA ceRNAs in multiple sclerosis based on ceRNA mechanisms. Mol. Genet. Genomics. 296 (2), 423–435. 10.1007/s00438-020-01750-1 [DOI] [PubMed] [Google Scholar]
- Donev R., Newall A., Thome J., Sheer D. (2007). A role for SC35 and hnRNPA1 in the determination of amyloid precursor protein isoforms. Mol. Psychiatry 12 (7), 681–690. 10.1038/sj.mp.4001971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G., Philipson L., Mattaj I. W. (1988). Ribonucleoprotein particles in cellular processes. J. Cell Biol. 106 (5), 1419–1425. 10.1083/jcb.106.5.1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Expert-Bezancon A., Sureau A., Durosay P., Salesse R., Groeneveld H., Lecaer J. P., et al. (2004). hnRNP A1 and the SR proteins ASF/SF2 and SC35 have antagonistic functions in splicing of beta-tropomyosin exon 6B. J. Biol. Chem. 279 (37), 38249–38259. 10.1074/jbc.M405377200 [DOI] [PubMed] [Google Scholar]
- Fabian M. R., Sonenberg N., Filipowicz W. (2010). Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79, 351–379. 10.1146/annurev-biochem-060308-103103 [DOI] [PubMed] [Google Scholar]
- Feng J., Li L., Tong L., Tang L., Wu S. (2016). The involvement of splicing factor hnRNP A1 in UVB-induced alternative splicing of hdm2. Photochem. Photobiol. 92 (2), 318–324. 10.1111/php.12564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Liao Y., Xu X., Yi Q., He L., Tang L. (2018). hnRNP A1 promotes keratinocyte cell survival post UVB radiation through PI3K/Akt/mTOR pathway. Exp. Cell Res. 362 (2), 394–399. 10.1016/j.yexcr.2017.12.002 [DOI] [PubMed] [Google Scholar]
- Fisette J. F., Toutant J., Dugre-Brisson S., Desgroseillers L., Chabot B. (2010). hnRNP A1 and hnRNP H can collaborate to modulate 5' splice site selection. Rna 16 (1), 228–238. 10.1261/rna.1890310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flodrops M., Dujardin G., Busson A., Trouve P., Ka C., Simon B., et al. (2020). TIMP1 intron 3 retention is a marker of colon cancer progression controlled by hnRNPA1. Mol. Biol. Rep. 47 (4), 3031–3040. 10.1007/s11033-020-05375-w [DOI] [PubMed] [Google Scholar]
- Fu R., Yang P., Amin S., Li Z. (2020). A novel miR-206/hnRNPA1/PKM2 axis reshapes the Warburg effect to suppress colon cancer growth. Biochem. Biophys. Res. Commun. 531 (4), 465–471. 10.1016/j.bbrc.2020.08.019 [DOI] [PubMed] [Google Scholar]
- Fujiya M., Konishi H., Mohamed Kamel M. K., Ueno N., Inaba Y., Moriichi K., et al. (2014). microRNA-18a induces apoptosis in colon cancer cells via the autophagolysosomal degradation of oncogenic heterogeneous nuclear ribonucleoprotein A1. Oncogene 33 (40), 4847–4856. 10.1038/onc.2013.429 [DOI] [PubMed] [Google Scholar]
- Gagne M., Deshaies J. E., Sidibe H., Benchaar Y., Arbour D., Dubinski A., et al. (2021). hnRNP A1B, a splice variant of HNRNPA1, is spatially and temporally regulated. Front. Neurosci. 15, 724307. 10.3389/fnins.2021.724307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G., Dhar S., Bedford M. T. (2017). PRMT5 regulates IRES-dependent translation via methylation of hnRNP A1. Nucleic Acids Res. 45 (8), 4359–4369. 10.1093/nar/gkw1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesnel M. C., Del Gatto-Konczak F., Breathnach R. (2009). Combined use of MS2 and PP7 coat fusions shows that TIA-1 dominates hnRNP A1 for K-SAM exon splicing control. J. Biomed. Biotechnol. 2009, 104853. 10.1155/2009/104853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghidini A., Clery A., Halloy F., Allain F. H. T., Hall J. (2021). RNA-PROTACs: Degraders of RNA-binding proteins. Angew. Chem. Int. Ed. Engl. 60 (6), 3163–3169. 10.1002/anie.202012330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M., Singh M. (2018). RGG-box in hnRNPA1 specifically recognizes the telomere G-quadruplex DNA and enhances the G-quadruplex unfolding ability of UP1 domain. Nucleic Acids Res. 46 (19), 10246–10261. 10.1093/nar/gky854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisovic T., Ben-David Y., Lang M. A., Raffalli-Mathieu F. (2003). Interplay between hnRNP A1 and a cis-acting element in the 3' UTR of CYP2A5 mRNA is central for high expression of the gene. FEBS Lett. 535 (1-3), 147–152. 10.1016/s0014-5793(02)03893-0 [DOI] [PubMed] [Google Scholar]
- Graveley B. R., Hertel K. J., Maniatis T. (2001). The role of U2AF35 and U2AF65 in enhancer-dependent splicing. Rna 7 (6), 806–818. 10.1017/s1355838201010317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley B. R. (2000). Sorting out the complexity of SR protein functions. Rna 6 (9), 1197–1211. 10.1017/s1355838200000960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Xia J., Xu H., Frech I., Tricot G., Zhan F. (2017). NEK2 promotes aerobic glycolysis in multiple myeloma through regulating splicing of pyruvate kinase. J. Hematol. Oncol. 10 (1), 17. 10.1186/s13045-017-0392-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueroussov S., Weatheritt R. J., O'Hanlon D., Lin Z. Y., Narula A., Gingras A. C., et al. (2017). Regulatory expansion in mammals of multivalent hnRNP assemblies that globally control alternative splicing. Cell 170 (2), 324–339. 10.1016/j.cell.2017.06.037 [DOI] [PubMed] [Google Scholar]
- Guil S., Gattoni R., Carrascal M., Abian J., Stevenin J., Bach-Elias M. (2003). Roles of hnRNP A1, SR proteins, and p68 helicase in c-H-ras alternative splicing regulation. Mol. Cell. Biol. 23 (8), 2927–2941. 10.1128/mcb.23.8.2927-2941.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R., Li Y., Ning J., Sun D., Lin L., Liu X. (2013). HnRNP A1/A2 and SF2/ASF regulate alternative splicing of interferon regulatory factor-3 and affect immunomodulatory functions in human non-small cell lung cancer cells. PloS one 8 (4), e62729. 10.1371/journal.pone.0062729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton B. J., Burns C. M., Nichols R. C., Rigby W. F. (1997). Modulation of AUUUA response element binding by heterogeneous nuclear ribonucleoprotein A1 in human T lymphocytes. The roles of cytoplasmic location, transcription, and phosphorylation. J. Biol. Chem. 272 (45), 28732–28741. 10.1074/jbc.272.45.28732 [DOI] [PubMed] [Google Scholar]
- Hamilton B. J., Nagy E., Malter J. S., Arrick B. A., Rigby W. F. (1993). Association of heterogeneous nuclear ribonucleoprotein A1 and C proteins with reiterated AUUUA sequences. J. Biol. Chem. 268 (12), 8881–8887. 10.1016/s0021-9258(18)52955-0 [DOI] [PubMed] [Google Scholar]
- Han K., Yeo G., An P., Burge C. B., Grabowski P. J. (2005). A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol. 3 (5), e158. 10.1371/journal.pbio.0030158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S. P., Tang Y. H., Smith R. (2010). Functional diversity of the hnRNPs: Past, present and perspectives. Biochem. J. 430 (3), 379–392. 10.1042/BJ20100396 [DOI] [PubMed] [Google Scholar]
- Holmes B., Lee J., Landon K. A., Benavides-Serrato A., Bashir T., Jung M. E., et al. (2016). Mechanistic target of rapamycin (mTOR) inhibition synergizes with reduced internal ribosome entry site (IRES)-mediated translation of cyclin D1 and c-MYC mRNAs to treat glioblastoma. J. Biol. Chem. 291 (27), 14146–14159. 10.1074/jbc.M116.726927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H. Y., Lu W. W., Wu K. Y., Lin C. W., Kung S. H. (2016). Idarubicin is a broad-spectrum enterovirus replication inhibitor that selectively targets the virus internal ribosomal entry site. J. Gen. Virol. 97 (5), 1122–1133. 10.1099/jgv.0.000431 [DOI] [PubMed] [Google Scholar]
- Huang X., Chen Y., Yi J., Yi P., Jia J., Liao Y., et al. (2022). Tetracaine hydrochloride induces cell cycle arrest in melanoma by downregulating hnRNPA1. Toxicol. Appl. Pharmacol. 434, 115810. 10.1016/j.taap.2021.115810 [DOI] [PubMed] [Google Scholar]
- Huang X. T., Li J. H., Zhu X. X., Huang C. S., Gao Z. X., Xu Q. C., et al. (2021). HNRNPC impedes m(6)A-dependent anti-metastatic alternative splicing events in pancreatic ductal adenocarcinoma. Cancer Lett. 518, 196–206. 10.1016/j.canlet.2021.07.016 [DOI] [PubMed] [Google Scholar]
- Huang Y., Lin L., Yu X., Wen G., Pu X., Zhao H., et al. (2013). Functional involvements of heterogeneous nuclear ribonucleoprotein A1 in smooth muscle differentiation from stem cells in vitro and in vivo . Stem Cells 31 (5), 906–917. 10.1002/stem.1324 [DOI] [PubMed] [Google Scholar]
- Huelga S. C., Vu A. Q., Arnold J. D., Liang T. Y., Liu P. P., Yan B. Y., et al. (2012). Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep. 1 (2), 167–178. 10.1016/j.celrep.2012.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison S., LeBel C., Blanchette M., Chabot B. (2002). Distinct sets of adjacent heterogeneous nuclear ribonucleoprotein (hnRNP) A1/A2 binding sites control 5' splice site selection in the hnRNP A1 mRNA precursor. J. Biol. Chem. 277 (33), 29745–29752. 10.1074/jbc.M203633200 [DOI] [PubMed] [Google Scholar]
- Izaurralde E., Jarmolowski A., Beisel C., Mattaj I. W., Dreyfuss G., Fischer U., et al. (1997). A role for the M9 transport signal of hnRNP A1 in mRNA nuclear export. J. Cell Biol. 137 (1), 27–35. 10.1083/jcb.137.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y., Tulin A. V. (2016). Poly(ADP-Ribosyl)ation of hnRNP A1 protein controls translational repression in Drosophila. Mol. Cell. Biol. 36 (19), 2476–2486. 10.1128/MCB.00207-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Q., Nie H., Yu P., Xie B., Wang C., Yang F., et al. (2019). HNRNPA1-mediated 3' UTR length changes of HN1 contributes to cancer- and senescence-associated phenotypes. Aging (Albany NY) 11 (13), 4407–4437. 10.18632/aging.102060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z. H., Zhang W. J., Rao Y., Wu J. Y. (1998). Regulation of Ich-1 pre-mRNA alternative splicing and apoptosis by mammalian splicing factors. Proc. Natl. Acad. Sci. U. S. A. 95 (16), 9155–9160. 10.1073/pnas.95.16.9155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo O. D., Martin J., Bernath A., Masri J., Lichtenstein A., Gera J. (2008). Heterogeneous nuclear ribonucleoprotein A1 regulates cyclin D1 and c-myc internal ribosome entry site function through Akt signaling. J. Biol. Chem. 283 (34), 23274–23287. 10.1074/jbc.M801185200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica M. S., Licklider L. J., Gygi S. R., Grigorieff N., Moore M. J. (2002). Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. Rna 8 (4), 426–439. 10.1017/s1355838202021088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima T., Manley J. L. (2003). A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat. Genet. 34 (4), 460–463. 10.1038/ng1207 [DOI] [PubMed] [Google Scholar]
- Kashima T., Rao N., David C. J., Manley J. L. (2007). hnRNP A1 functions with specificity in repression of SMN2 exon 7 splicing. Hum. Mol. Genet. 16 (24), 3149–3159. 10.1093/hmg/ddm276 [DOI] [PubMed] [Google Scholar]
- Kashima T., Rao N., Manley J. L. (2007). An intronic element contributes to splicing repression in spinal muscular atrophy. Proc. Natl. Acad. Sci. U. S. A. 104 (9), 3426–3431. 10.1073/pnas.0700343104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur R., Lal S. K. (2020). The multifarious roles of heterogeneous ribonucleoprotein A1 in viral infections. Rev. Med. Virol. 30 (2), e2097. 10.1002/rmv.2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedzierska H., Piekielko-Witkowska A. (2017). Splicing factors of SR and hnRNP families as regulators of apoptosis in cancer. Cancer Lett. 396, 53–65. 10.1016/j.canlet.2017.03.013 [DOI] [PubMed] [Google Scholar]
- Kim H. J., Lee H. R., Seo J. Y., Ryu H. G., Lee K. H., Kim D. Y., et al. (2017). Heterogeneous nuclear ribonucleoprotein A1 regulates rhythmic synthesis of mouse Nfil3 protein via IRES-mediated translation. Sci. Rep. 7, 42882. 10.1038/srep42882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko C. C., Chen Y. J., Chen C. T., Liu Y. C., Cheng F. C., Hsu K. C., et al. (2014). Chemical proteomics identifies heterogeneous nuclear ribonucleoprotein (hnRNP) A1 as the molecular target of quercetin in its anti-cancer effects in PC-3 cells. J. Biol. Chem. 289 (32), 22078–22089. 10.1074/jbc.M114.553248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooshapur H., Choudhury N. R., Simon B., Muhlbauer M., Jussupow A., Fernandez N., et al. (2018). Structural basis for terminal loop recognition and stimulation of pri-miRNA-18a processing by hnRNP A1. Nat. Commun. 9 (1), 2479. 10.1038/s41467-018-04871-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer M. C., Liang D., Tatomer D. C., Gold B., March Z. M., Cherry S., et al. (2015). Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 29 (20), 2168–2182. 10.1101/gad.270421.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Khandelwal N., Chander Y., Nagori H., Verma A., Barua A., et al. (2021). S-adenosylmethionine-dependent methyltransferase inhibitor DZNep blocks transcription and translation of SARS-CoV-2 genome with a low tendency to select for drug-resistant viral variants. Antivir. Res. 197, 105232. 10.1016/j.antiviral.2021.105232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze M. M., Benz F., Brauss T. F., Lampe S., Weigand J. E., Braun J., et al. (2016). sST2 translation is regulated by FGF2 via an hnRNP A1-mediated IRES-dependent mechanism. Biochim. Biophys. Acta 1859 (7), 848–859. 10.1016/j.bbagrm.2016.05.005 [DOI] [PubMed] [Google Scholar]
- Kuranaga Y., Sugito N., Shinohara H., Tsujino T., Taniguchi K., Komura K., et al. (2018). SRSF3, a splicer of the PKM gene, regulates cell growth and maintenance of cancer-specific energy metabolism in colon cancer cells. Int. J. Mol. Sci. 19 (10), E3012. 10.3390/ijms19103012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau L. F., Brooks A. N., Soergel D. A., Meng Q., Brenner S. E. (2007). The coupling of alternative splicing and nonsense-mediated mRNA decay. Adv. Exp. Med. Biol. 623, 190–211. 10.1007/978-0-387-77374-2_12 [DOI] [PubMed] [Google Scholar]
- Lau J. S., Baumeister P., Kim E., Roy B., Hsieh T. Y., Lai M., et al. (2000). Heterogeneous nuclear ribonucleoproteins as regulators of gene expression through interactions with the human thymidine kinase promoter. J. Cell. Biochem. 79 (3), 395–406. [DOI] [PubMed] [Google Scholar]
- Lemieux B., Blanchette M., Monette A., Mouland A. J., Wellinger R. J., Chabot B. (2015). A function for the hnRNP A1/A2 proteins in transcription elongation. PloS one 10 (5), e0126654. 10.1371/journal.pone.0126654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong S. Y., Ong B. K., Chu J. J. (2015). The role of Misshapen NCK-related kinase (MINK), a novel Ste20 family kinase, in the IRES-mediated protein translation of human enterovirus 71. PLoS Pathog. 11 (3), e1004686. 10.1371/journal.ppat.1004686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levengood J. D., Tolbert M., Li M. L., Tolbert B. S. (2013). High-affinity interaction of hnRNP A1 with conserved RNA structural elements is required for translation and replication of enterovirus 71. RNA Biol. 10 (7), 1136–1145. 10.4161/rna.25107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S. M., Veyrier A., Hosszu Ungureanu N., Bonnal S., Vagner S., Holcik M. (2007). Subcellular relocalization of a trans-acting factor regulates XIAP IRES-dependent translation. Mol. Biol. Cell 18 (4), 1302–1311. 10.1091/mbc.e06-06-0515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. P., Zhang X., Duncan R., Comai L., Lai M. M. (1997). Heterogeneous nuclear ribonucleoprotein A1 binds to the transcription-regulatory region of mouse hepatitis virus RNA. Proc. Natl. Acad. Sci. U. S. A. 94 (18), 9544–9549. 10.1073/pnas.94.18.9544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. L., Lin J. Y., Chen B. S., Weng K. F., Shih S. R., Calderon J. D., et al. (2019). EV71 3C protease induces apoptosis by cleavage of hnRNP A1 to promote apaf-1 translation. PloS one 14 (9), e0221048. 10.1371/journal.pone.0221048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Wang W., Ding H., Xu H., Zhao Q., Li J., et al. (2012). Aptamer BC15 against heterogeneous nuclear ribonucleoprotein A1 has potential value in diagnosis and therapy of hepatocarcinoma. Nucleic Acid. Ther. 22 (6), 391–398. 10.1089/nat.2012.0363 [DOI] [PubMed] [Google Scholar]
- Li Y., Zhang J., Li S., Guo C., Li Q., Zhang X., et al. (2021). Heterogeneous nuclear ribonucleoprotein A1 loads batched tumor-promoting MicroRNAs into small extracellular vesicles with the assist of caveolin-1 in A549 cells. Front. Cell Dev. Biol. 9, 687912. 10.3389/fcell.2021.687912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. J., Ho T. J., Lin T. H., Hsu W. Y., Huang S. M., Liao C. C., et al. (2015). P-coumaric acid regulates exon 12 splicing of the ATP7B gene by modulating hnRNP A1 protein expressions. Biomed. (Taipei) 5 (2), 10. 10.7603/s40681-015-0010-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. Y., Li H. L., Su J. B., Ding F. H., Zhao J. J., Chai F., et al. (2015). Regulation of RAGE splicing by hnRNP A1 and Tra2β-1 and its potential role in AD pathogenesis. J. Neurochem. 133 (2), 187–198. 10.1111/jnc.13069 [DOI] [PubMed] [Google Scholar]
- Liu Y., Kim D., Choi N., Oh J., Ha J., Zhou J., et al. (2020). hnRNP A1 regulates alternative splicing of tau exon 10 by targeting 3' splice sites. Cells 9 (4), E936. 10.3390/cells9040936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh T. J., Moon H., Cho S., Jang H., Liu Y. C., Tai H., et al. (2015). CD44 alternative splicing and hnRNP A1 expression are associated with the metastasis of breast cancer. Oncol. Rep. 34 (3), 1231–1238. 10.3892/or.2015.4110 [DOI] [PubMed] [Google Scholar]
- Lothstein L., Arenstorf H. P., Chung S. Y., Walker B. W., Wooley J. C., LeStourgeon W. M. (1985). General organization of protein in HeLa 40S nuclear ribonucleoprotein particles. J. Cell Biol. 100 (5), 1570–1581. 10.1083/jcb.100.5.1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L., Chiu L. Y., Sugarman A., Gupta P., Rouskin S., Tolbert B. S. (2021). HnRNP A1/A2 proteins assemble onto 7SK snRNA via context dependent interactions. J. Mol. Biol. 433 (9), 166885. 10.1016/j.jmb.2021.166885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manita D., Toba Y., Takakusagi Y., Matsumoto Y., Kusayanagi T., Takakusagi K., et al. (2011). Camptothecin (CPT) directly binds to human heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) and inhibits the hnRNP A1/topoisomerase I interaction. Bioorg. Med. Chem. 19 (24), 7690–7697. 10.1016/j.bmc.2011.09.059 [DOI] [PubMed] [Google Scholar]
- Marchand V., Mereau A., Jacquenet S., Thomas D., Mougin A., Gattoni R., et al. (2002). A Janus splicing regulatory element modulates HIV-1 tat and rev mRNA production by coordination of hnRNP A1 cooperative binding. J. Mol. Biol. 323 (4), 629–652. 10.1016/s0022-2836(02)00967-1 [DOI] [PubMed] [Google Scholar]
- Mascarenhas J. B., Tchourbanov A. Y., Danilov S. M., Zhou T., Wang T., Garcia J. G. N. (2018). The splicing factor hnRNPA1 regulates alternate splicing of the MYLK gene. Am. J. Respir. Cell Mol. Biol. 58 (5), 604–613. 10.1165/rcmb.2017-0141OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter N., Marx M., Weg-Remers S., Ponta H., Herrlich P., Konig H. (2000). Heterogeneous ribonucleoprotein A1 is part of an exon-specific splice-silencing complex controlled by oncogenic signaling pathways. J. Biol. Chem. 275 (45), 35353–35360. 10.1074/jbc.M004692200 [DOI] [PubMed] [Google Scholar]
- Mayeda A., Helfman D. M., Krainer A. R. (1993). Modulation of exon skipping and inclusion by heterogeneous nuclear ribonucleoprotein A1 and pre-mRNA splicing factor SF2/ASF. Mol. Cell. Biol. 13 (5), 2993–3001. 10.1128/mcb.13.5.2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A., Krainer A. R. (1992). Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell 68 (2), 365–375. 10.1016/0092-8674(92)90477-t [DOI] [PubMed] [Google Scholar]
- McGlincy N. J., Tan L. Y., Paul N., Zavolan M., Lilley K. S., Smith C. W. (2010). Expression proteomics of UPF1 knockdown in HeLa cells reveals autoregulation of hnRNP A2/B1 mediated by alternative splicing resulting in nonsense-mediated mRNA decay. BMC Genomics 11, 565. 10.1186/1471-2164-11-565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlewski G., Caceres J. F. (2010). Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat. Struct. Mol. Biol. 17 (8), 1011–1018. 10.1038/nsmb.1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlewski G., Guil S., Semple C. A., Caceres J. F. (2008). Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol. Cell 32 (3), 383–393. 10.1016/j.molcel.2008.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miglietta G., Cogoi S., Pedersen E. B., Xodo L. E. (2015). GC-elements controlling HRAS transcription form i-motif structures unfolded by heterogeneous ribonucleoprotein particle A1. Sci. Rep. 5, 18097. 10.1038/srep18097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadiminty N., Tummala R., Liu C., Lou W., Evans C. P., Gao A. C. (2015). NF-κB2/p52:c-Myc:hnRNPA1 pathway regulates expression of androgen receptor splice variants and enzalutamide sensitivity in prostate cancer. Mol. Cancer Ther. 14 (8), 1884–1895. 10.1158/1535-7163.MCT-14-1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls C. D., Beattie T. L. (2008). Multiple factors influence the normal and UV-inducible alternative splicing of PIG3. Biochim. Biophys. Acta 1779 (12), 838–849. 10.1016/j.bbagrm.2008.08.009 [DOI] [PubMed] [Google Scholar]
- Nishikawa T., Kuwano Y., Takahara Y., Nishida K., Rokutan K. (2019). HnRNPA1 interacts with G-quadruplex in the TRA2B promoter and stimulates its transcription in human colon cancer cells. Sci. Rep. 9 (1), 10276. 10.1038/s41598-019-46659-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H., Lee E., Jang H. N., Lee J., Moon H., Sheng Z., et al. (2013). hnRNP A1 contacts exon 5 to promote exon 6 inclusion of apoptotic Fas gene. Apoptosis 18 (7), 825–835. 10.1007/s10495-013-0824-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka K., Yamamoto Y., Ochiya T. (2018). Regulatory role of resveratrol, a microRNA-controlling compound, in HNRNPA1 expression, which is associated with poor prognosis in breast cancer. Oncotarget 9 (37), 24718–24730. 10.18632/oncotarget.25339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani F., Buratti E., Stuani C., Romano M., Zuccato E., Niksic M., et al. (2000). Splicing factors induce cystic fibrosis transmembrane regulator exon 9 skipping through a nonevolutionary conserved intronic element. J. Biol. Chem. 275 (28), 21041–21047. 10.1074/jbc.M910165199 [DOI] [PubMed] [Google Scholar]
- Palhais B., Dembic M., Sabaratnam R., Nielsen K. S., Doktor T. K., Bruun G. H., et al. (2016). The prevalent deep intronic c. 639+919 G>A GLA mutation causes pseudoexon activation and Fabry disease by abolishing the binding of hnRNPA1 and hnRNP A2/B1 to a splicing silencer. Mol. Genet. Metab. 119 (3), 258–269. 10.1016/j.ymgme.2016.08.007 [DOI] [PubMed] [Google Scholar]
- Palhais B., Praestegaard V. S., Sabaratnam R., Doktor T. K., Lutz S., Burda P., et al. (2015). Splice-shifting oligonucleotide (SSO) mediated blocking of an exonic splicing enhancer (ESE) created by the prevalent c.903+469T>C MTRR mutation corrects splicing and restores enzyme activity in patient cells. Nucleic Acids Res. 43 (9), 4627–4639. 10.1093/nar/gkv275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paramasivam M., Membrino A., Cogoi S., Fukuda H., Nakagama H., Xodo L. E. (2009). Protein hnRNP A1 and its derivative Up1 unfold quadruplex DNA in the human KRAS promoter: Implications for transcription. Nucleic Acids Res. 37 (9), 2841–2853. 10.1093/nar/gkp138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor T., Pagani F. (2011). Interaction of hnRNPA1/A2 and DAZAP1 with an Alu-derived intronic splicing enhancer regulates ATM aberrant splicing. PloS one 6 (8), e23349. 10.1371/journal.pone.0023349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patry C., Bouchard L., Labrecque P., Gendron D., Lemieux B., Toutant J., et al. (2003). Small interfering RNA-mediated reduction in heterogeneous nuclear ribonucleoparticule A1/A2 proteins induces apoptosis in human cancer cells but not in normal mortal cell lines. Cancer Res. 63 (22), 7679–7688. [PubMed] [Google Scholar]
- Pelisch F., Khauv D., Risso G., Stallings-Mann M., Blaustein M., Quadrana L., et al. (2012). Involvement of hnRNP A1 in the matrix metalloprotease-3-dependent regulation of Rac1 pre-mRNA splicing. J. Cell. Biochem. 113 (7), 2319–2329. 10.1002/jcb.24103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard A. J., Krainer A. R., Robson S. C., Europe-Finner G. N. (2002). Alternative splicing of the adenylyl cyclase stimulatory G-protein G alpha(s) is regulated by SF2/ASF and heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) and involves the use of an unusual TG 3'-splice Site. J. Biol. Chem. 277 (18), 15241–15251. 10.1074/jbc.M109046200 [DOI] [PubMed] [Google Scholar]
- Princler G. L., Julias J. G., Hughes S. H., Derse D. (2003). Roles of viral and cellular proteins in the expression of alternatively spliced HTLV-1 pX mRNAs. Virology 317 (1), 136–145. 10.1016/j.virol.2003.09.010 [DOI] [PubMed] [Google Scholar]
- Qi Y., Han W., Chen D., Zhao J., Bai L., Huang F., et al. (2021). Engineering circular RNA regulators to specifically promote circular RNA production. Theranostics 11 (15), 7322–7336. 10.7150/thno.56990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X., Guo H., Wang X., Zhu X., Yan M., Wang X., et al. (2019). Exosomal miR-196a derived from cancer-associated fibroblasts confers cisplatin resistance in head and neck cancer through targeting CDKN1B and ING5. Genome Biol. 20 (1), 12. 10.1186/s13059-018-1604-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooke N., Markovtsov V., Cagavi E., Black D. L. (2003). Roles for SR proteins and hnRNP A1 in the regulation of c-src exon N1. Mol. Cell. Biol. 23 (6), 1874–1884. 10.1128/mcb.23.6.1874-1884.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R., Huang Y., Seckl M. J., Pardo O. E. (2017). Emerging roles of hnRNPA1 in modulating malignant transformation. WIREs RNA 8 (6), 1. 10.1002/wrna.1431 [DOI] [PubMed] [Google Scholar]
- Rubsamen D., Blees J. S., Schulz K., Doring C., Hansmann M. L., Heide H., et al. (2012). IRES-dependent translation of egr2 is induced under inflammatory conditions. Rna 18 (10), 1910–1920. 10.1261/rna.033019.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H. G., Jung Y., Lee N., Seo J. Y., Kim S. W., Lee K. H., et al. (2021). HNRNP A1 promotes lung cancer cell proliferation by modulating VRK1 translation. Int. J. Mol. Sci. 22 (11), 5506. 10.3390/ijms22115506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel J., Sekeris C. E., Alonso A., Bautz E. K. (1988). RNA-binding properties of hnRNP proteins. Eur. J. Biochem. 171 (3), 565–569. 10.1111/j.1432-1033.1988.tb13825.x [DOI] [PubMed] [Google Scholar]
- Sebban S., Golan-Gerstl R., Karni R., Vaksman O., Davidson B., Reich R. (2013). Alternatively spliced lysyl oxidase-like 4 isoforms have a pro-metastatic role in cancer. Clin. Exp. Metastasis 30 (1), 103–117. 10.1007/s10585-012-9514-0 [DOI] [PubMed] [Google Scholar]
- Shamoo Y., Abdul-Manan N., Williams K. R. (1995). Multiple RNA binding domains (RBDs) just don't add up. Nucleic Acids Res. 23 (5), 725–728. 10.1093/nar/23.5.725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan J., Moran-Jones K., Munro T. P., Kidd G. J., Winzor D. J., Hoek K. S., et al. (2000). Binding of an RNA trafficking response element to heterogeneous nuclear ribonucleoproteins A1 and A2. J. Biol. Chem. 275 (49), 38286–38295. 10.1074/jbc.M007642200 [DOI] [PubMed] [Google Scholar]
- Shi Y., Yang Y., Hoang B., Bardeleben C., Holmes B., Gera J., et al. (2016). Therapeutic potential of targeting IRES-dependent c-myc translation in multiple myeloma cells during ER stress. Oncogene 35 (8), 1015–1024. 10.1038/onc.2015.156 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Shiina Y., Arima K., Tabunoki H., Satoh J. (2010). TDP-43 dimerizes in human cells in culture. Cell. Mol. Neurobiol. 30 (4), 641–652. 10.1007/s10571-009-9489-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada N., Rios I., Moran H., Sayers B., Hubbard K. (2009). p38 MAP kinase-dependent regulation of the expression level and subcellular distribution of heterogeneous nuclear ribonucleoprotein A1 and its involvement in cellular senescence in normal human fibroblasts. RNA Biol. 6 (3), 293–304. 10.4161/rna.6.3.8497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silipo M., Gautrey H., Satam S., Lennard T., Tyson-Capper A. (2017). How is Herstatin, a tumor suppressor splice variant of the oncogene HER2, regulated? RNA Biol. 14 (5), 536–543. 10.1080/15476286.2016.1267074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H., Dreyfuss G. (1995). A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 129 (3), 551–560. 10.1083/jcb.129.3.551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol E., Kedzierska H., Czubaty A., Rybicka B., Rodzik K., Tanski Z., et al. (2018). microRNA-mediated regulation of splicing factors SRSF1, SRSF2 and hnRNP A1 in context of their alternatively spliced 3'UTRs. Exp. Cell Res. 363 (2), 208–217. 10.1016/j.yexcr.2018.01.009 [DOI] [PubMed] [Google Scholar]
- Stoltzfus C. M., Madsen J. M. (2006). Role of viral splicing elements and cellular RNA binding proteins in regulation of HIV-1 alternative RNA splicing. Curr. HIV Res. 4 (1), 43–55. 10.2174/157016206775197655 [DOI] [PubMed] [Google Scholar]
- Su C. H., Hung K. Y., Hung S. C., Tarn W. Y. (2017). RBM4 regulates neuronal differentiation of mesenchymal stem cells by modulating alternative splicing of pyruvate kinase M. Mol. Cell. Biol. 37 (3), e004666–e004716. 10.1128/MCB.00466-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G., Zhou H., Chen K., Zeng J., Zhang Y., Yan L., et al. (2020). HnRNP A1 - mediated alternative splicing of CCDC50 contributes to cancer progression of clear cell renal cell carcinoma via ZNF395. J. Exp. Clin. Cancer Res. 39 (1), 116. 10.1186/s13046-020-01606-x [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sun Y., Zhao X., Zhou Y., Hu Y. (2012). miR-124, miR-137 and miR-340 regulate colorectal cancer growth via inhibition of the Warburg effect. Oncol. Rep. 28 (4), 1346–1352. 10.3892/or.2012.1958 [DOI] [PubMed] [Google Scholar]
- Suzuki H., Matsuoka M. (2017). hnRNPA1 autoregulates its own mRNA expression to remain non-cytotoxic. Mol. Cell. Biochem. 427 (1-2), 123–131. 10.1007/s11010-016-2904-x [DOI] [PubMed] [Google Scholar]
- Talukdar I., Sen S., Urbano R., Thompson J., Yates J. R., 3rd, Webster N. J. (2011). hnRNP A1 and hnRNP F modulate the alternative splicing of exon 11 of the insulin receptor gene. PloS one 6 (11), e27869. 10.1371/journal.pone.0027869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavanez J. P., Madl T., Kooshapur H., Sattler M., Valcarcel J. (2012). hnRNP A1 proofreads 3' splice site recognition by U2AF. Mol. Cell 45 (3), 314–329. 10.1016/j.molcel.2011.11.033 [DOI] [PubMed] [Google Scholar]
- Torosyan Y., Dobi A., Glasman M., Mezhevaya K., Naga S., Huang W., et al. (2010). Role of multi-hnRNP nuclear complex in regulation of tumor suppressor ANXA7 in prostate cancer cells. Oncogene 29 (17), 2457–2466. 10.1038/onc.2010.2 [DOI] [PubMed] [Google Scholar]
- Tummala R., Lou W., Gao A. C., Nadiminty N. (2017). Quercetin targets hnRNPA1 to overcome enzalutamide resistance in prostate cancer cells. Mol. Cancer Ther. 16 (12), 2770–2779. 10.1158/1535-7163.MCT-17-0030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Houven van Oordt W., Diaz-Meco M. T., Lozano J., Krainer A. R., Moscat J., Caceres J. F. (2000). The MKK(3/6)-p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. J. Cell Biol. 149 (2), 307–316. 10.1083/jcb.149.2.307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Herreweghe E., Egloff S., Goiffon I., Jady B. E., Froment C., Monsarrat B., et al. (2007). Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J. 26 (15), 3570–3580. 10.1038/sj.emboj.7601783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables J. P., Bourgeois C. F., Dalgliesh C., Kister L., Stevenin J., Elliott D. J. (2005). Up-regulation of the ubiquitous alternative splicing factor Tra2beta causes inclusion of a germ cell-specific exon. Hum. Mol. Genet. 14 (16), 2289–2303. 10.1093/hmg/ddi233 [DOI] [PubMed] [Google Scholar]
- Wall M. L., Lewis S. M. (2017). Methylarginines within the RGG-motif region of hnRNP A1 affect its IRES trans-acting factor Activity and are required for hnRNP A1 stress granule localization and formation. J. Mol. Biol. 429 (2), 295–307. 10.1016/j.jmb.2016.12.011 [DOI] [PubMed] [Google Scholar]
- Wang Y., Ma M., Xiao X., Wang Z. (2012). Intronic splicing enhancers, cognate splicing factors and context-dependent regulation rules. Nat. Struct. Mol. Biol. 19 (10), 1044–1052. 10.1038/nsmb.2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Burge C. B. (2008). Splicing regulation: From a parts list of regulatory elements to an integrated splicing code. Rna 14 (5), 802–813. 10.1261/rna.876308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z., Lian L., Ding H., Hu Y., Xiao Z., Xiong K., et al. (2020). LncRNA ANCR promotes hepatocellular carcinoma metastasis through upregulating HNRNPA1 expression. RNA Biol. 17 (3), 381–394. 10.1080/15476286.2019.1708547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk H. E., Werr H., Friedrich D., Kiltz H. H., Schafer K. P. (1985). The core proteins of 35S hnRNP complexes. Characterization of nine different species. Eur. J. Biochem. 146 (1), 71–81. 10.1111/j.1432-1033.1985.tb08621.x [DOI] [PubMed] [Google Scholar]
- Williams A. L., Khadka V., Tang M., Avelar A., Schunke K. J., Menor M., et al. (2018). HIF1 mediates a switch in pyruvate kinase isoforms after myocardial infarction. Physiol. Genomics 50 (7), 479–494. 10.1152/physiolgenomics.00130.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R. M., Jokhan L., Cheng X., Mayeda A., Krainer A. R. (1997). Crystal structure of human UP1, the domain of hnRNP A1 that contains two RNA-recognition motifs. Structure 5 (4), 559–570. 10.1016/s0969-2126(97)00211-6 [DOI] [PubMed] [Google Scholar]
- Yan Q., Zeng P., Zhou X., Zhao X., Chen R., Qiao J., et al. (2021). RBMX suppresses tumorigenicity and progression of bladder cancer by interacting with the hnRNP A1 protein to regulate PKM alternative splicing. Oncogene 40 (15), 2635–2650. 10.1038/s41388-021-01666-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Zhu R., Zhao X., Liu L., Zhou Z., Zhao L., et al. (2019). Sirtuin-mediated deacetylation of hnRNP A1 suppresses glycolysis and growth in hepatocellular carcinoma. Oncogene 38 (25), 4915–4931. 10.1038/s41388-019-0764-z [DOI] [PubMed] [Google Scholar]
- Yu C., Guo J., Liu Y., Jia J., Jia R., Fan M. (2015). Oral squamous cancer cell exploits hnRNP A1 to regulate cell cycle and proliferation. J. Cell. Physiol. 230 (9), 2252–2261. 10.1002/jcp.24956 [DOI] [PubMed] [Google Scholar]
- Yu C. Y., Theusch E., Lo K., Mangravite L. M., Naidoo D., Kutilova M., et al. (2014). HNRNPA1 regulates HMGCR alternative splicing and modulates cellular cholesterol metabolism. Hum. Mol. Genet. 23 (2), 319–332. 10.1093/hmg/ddt422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler A. M., Damgaard C. K., Kjems J., Caputi M. (2004). SC35 and heterogeneous nuclear ribonucleoprotein A/B proteins bind to a juxtaposed exonic splicing enhancer/exonic splicing silencer element to regulate HIV-1 tat exon 2 splicing. J. Biol. Chem. 279 (11), 10077–10084. 10.1074/jbc.M312743200 [DOI] [PubMed] [Google Scholar]
- Zearfoss N. R., Johnson E. S., Ryder S. P. (2013). hnRNP A1 and secondary structure coordinate alternative splicing of Mag. Mag. Rna. 19 (7), 948–957. 10.1261/rna.036780.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J., Xu H., Huang C., Sun Y., Xiao H., Yu G., et al. (2021). CD46 splice variant enhances translation of specific mRNAs linked to an aggressive tumor cell phenotype in bladder cancer. Mol. Ther. Nucleic Acids 24, 140–153. 10.1016/j.omtn.2021.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., O'Leary M. N., Peri S., Wang M., Zha J., Melov S., et al. (2017). Ribosomal proteins Rpl22 and Rpl22l1 control morphogenesis by regulating pre-mRNA splicing. Cell Rep. 18 (2), 545–556. 10.1016/j.celrep.2016.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Mandler M. D., Yi H., Feng Y. (2010). Quaking I controls a unique cytoplasmic pathway that regulates alternative splicing of myelin-associated glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 107 (44), 19061–19066. 10.1073/pnas.1007487107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T. T., Graber T. E., Jordan L. E., Cloutier M., Lewis S. M., Goulet I., et al. (2009). hnRNP A1 regulates UV-induced NF-kappaB signalling through destabilization of cIAP1 mRNA. Cell Death Differ. 16 (2), 244–252. 10.1038/cdd.2008.146 [DOI] [PubMed] [Google Scholar]
- Zhou B., Wang Y., Jiang J., Jiang H., Song J., Han T., et al. (2016). The long noncoding RNA colon cancer-associated transcript-1/miR-490 axis regulates gastric cancer cell migration by targeting hnRNPA1. IUBMB Life 68 (3), 201–210. 10.1002/iub.1474 [DOI] [PubMed] [Google Scholar]
- Zhou Z., Licklider L. J., Gygi S. P., Reed R. (2002). Comprehensive proteomic analysis of the human spliceosome. Nature 419 (6903), 182–185. 10.1038/nature01031 [DOI] [PubMed] [Google Scholar]
- Zhu J., Mayeda A., Krainer A. R. (2001). Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell 8 (6), 1351–1361. 10.1016/s1097-2765(01)00409-9 [DOI] [PubMed] [Google Scholar]
- Zhu S., Chen W., Wang J., Qi L., Pan H., Feng Z., et al. (2021). SAM68 promotes tumorigenesis in lung adenocarcinoma by regulating metabolic conversion via PKM alternative splicing. Theranostics 11 (7), 3359–3375. 10.7150/thno.51360 [DOI] [PMC free article] [PubMed] [Google Scholar]