Abstract
Recently, Escherichia coli cytotoxic necrotizing factor 1 (CNF1) was shown to activate the low-molecular-mass GTPase RhoA by deamidation of Gln63, thereby inhibiting intrinsic and GTPase-activating protein (GAP)-stimulated GTPase activities (G. Schmidt, P. Sehr, M. Wilm, J. Selzer, M. Mann, and K. Aktories, Nature 387:725–729, 1997; G. Flatau, E. Lemichez, M. Gauthier, P. Chardin, S. Paris, C. Fiorentini, and P. Boquet, Nature 387:729–733, 1997). Here we report that in addition to RhoA, Cdc42 and Rac also are targets for CNF1 in vitro and in intact cells. Treatment of HeLa cells with CNF1 induced a transient formation of microspikes and formation of membrane ruffles. CNF1 caused a transient 10- to 50-fold increase in the activity of the c-Jun N-terminal kinase. Tryptic peptides of Cdc42 obtained from CNF1-treated cells by immunoprecipitation exhibited an increase in mass of 1 Da compared to control peptides, indicating the deamidation of glutamine 61 by the toxin. The same increase in mass was observed with the respective peptides obtained from CNF1-modified recombinant Cdc42 and Rac1. Modification of recombinant Cdc42 and Rac1 by CNF1 inhibited intrinsic and GAP-stimulated GTPase activities and retarded binding of 2′(3′)-O-(N-methylanthraniloyl)GDP. The data suggest that recombinant as well as cellular Cdc42 and Rac are substrates for CNF1.
The small GTPases of the Rho family (Rho, Rac, and Cdc42) are involved in the regulation of the actin cytoskeleton and act as molecular switches in various signaling processes. Rho induces formation of stress fibers and focal adhesions, Rac is involved in lamellipodia and membrane ruffle formation (37), and Cdc42 regulates microspike formation (21). Besides their role in the organization of the actin cytoskeleton, Rho proteins participate in the regulation of various signal transduction processes, including control of cell aggregation (48), integrin signaling (25, 49), and regulation of phosphatidylinositol-3-kinase (50), phosphatidylinositol-4-phosphate-5-kinase (4), and phospholipase D (22, 27, 47). Furthermore, Rho proteins are implicated in endocytosis (23, 41), secretion (36), control of transcription (28), cell cycle progression (33), and cell transformation (20).
Like other low-molecular-mass GTP-binding proteins, Rho proteins are regulated by a GTPase cycle which is controlled by at least three groups of Rho-interacting proteins. Whereas the inactive, GDP-bound forms of Rho proteins are stabilized and retained in the cytosol by a group of guanine nucleotide dissociation inhibitors (14), activation of Rho proteins is induced by guanine nucleotide exchange factors (9). GTP hydrolysis and thus inactivation of the Rho protein is accelerated by GTPase-activating proteins (GAPs) (31). Rho proteins are targets for various bacterial toxins. Clostridium botulinum exoenzyme C3 and related transferases ADP-ribosylate RhoA, -B, and -C (but not Rac1 and Cdc42) at Asn41, thereby inhibiting the biological activity of Rho (1, 16, 44). The large clostridial cytotoxins Clostridium difficile toxins A and B inactivate Rho GTPases by glucosylation at Thr35 or Thr37 (18, 19). All members of the Rho family are modified, including Rac and Cdc42. Clostridium sordellii lethal toxin glucosylates the small GTPases Rac1 and Cdc42 but not Rho (17, 35). In addition, Ras proteins are substrates for this toxin (17, 35). Finally, Clostridium novyi α-toxin uses UDP-GlucNAc to modify Rho, Rac1, and Cdc42 (45). All of these toxins cause inactivation of Rho proteins, thereby largely affecting the actin cytoskeleton and inhibiting Rho-dependent signal processes.
RhoA is also the target for cytotoxic necrotizing factor 1 (CNF1) and CNF2 from Escherichia coli (10, 34) and for the dermonecrotic toxin from Bordetella species. CNFs have been isolated from enteritis-affected children (3) and from various pathogenic E. coli strains from piglets and calves (6); however, little is known about their role in pathogenesis. In addition to their skin-necrotizing action, the toxins have been shown to be lethal for mice when injected intravenously (7). In cultured cells CNFs induce actin polymerization and inhibit cytokinesis, resulting in formation of multinucleated cells (8, 10, 34). It was shown that CNF1 modifies RhoA by deamidation of Gln63, thereby forming a glutamic acid residue at this position (12, 42). Because Gln63 of RhoA is essential for intrinsic and GAP-stimulated GTPase activity, the Rho protein is constitutively activated by CNF1 (39, 42). Here we studied whether Cdc42 and Rac also are targeted by CNF1 in vitro and in intact cells. We report that treatment of HeLa cells with CNF1 causes formation of membrane ruffles and microspikes and induces activation of the c-Jun N-terminal kinase (JNK). Concomitantly, deamidation of Cdc42 and Rac at Gln61 was detected, indicating that in addition to RhoA, Cdc42 and Rac are cellular targets for CNF1.
MATERIALS AND METHODS
2′(3′)-O-(N-methylanthraniloyl)GDP (mant-GDP) was synthesized, purified, and characterized as previously described (15). The Cdc42 plasmid was a gift from M. Aepfelbacher (Munich, Germany); the Rac1, RhoA, and GAP plasmids were from A. Hall (London, United Kingdom).
Cell culture.
HeLa cells were cultivated in Dulbecco’s modified Eagle’s medium (with 12 mM l-glutamine) supplemented with 10% fetal calf serum, 4 mM penicillin, and 4 mM streptomycin in a humidified atmosphere of 5% CO2 at 37°C. For CNF1 intoxication, the cells were treated with 500 ng of glutathione S-transferase (GST)–CNF1 per ml 1 day after seeding.
Actin staining.
Subconfluent HeLa cells grown on glass coverslips were washed three times with phosphate-buffered saline (PBS) and fixed with 4% formaldehyde–0.1% Triton X-100 for 10 min at room temperature. After being washed three times with PBS, the cells were incubated with rhodamine-conjugated phalloidin (6 μg/ml in PBS) for 1 h at room temperature. The cells were thoroughly washed with PBS and subjected to fluorescence microscopy.
Mutagenesis.
Q61E Cdc42 and Q61E Rac1 were constructed from pGEX2T-Cdc42 and pGEX2T-Rac1 by PCR in the presence of two primers (sense and corresponding antisense) carrying a single-base mismatch encoding the proper mutant (sense primer for Cdc42, 5′-CT GCA GGG GAA GAG GAT TAT GAC AG-3′; sense primer for Rac1, 5′-GG GAT ACA GCT GGA GAA GAA GAT TAT GAC-3′). The parental DNA was eliminated by using the restriction enzyme DpnI, which digests methylated DNA. The PCR products were transformed into Epicurean XL-2 Blue ultracompetent cells (Stratagene). After the mutation was verified by sequencing, the plasmids were transformed into E. coli BL21 for protein expression.
Protein expression.
For protein purification, E. coli strains carrying pGEX plasmids with the GTPase and CNF1 genes were grown in Luria-Bertani medium and induced with 0.2 mM isopropyl-β-d-thiogalactopyranoside at an optical density of 0.5. The cells were harvested at an optical density of 1.0, and the GST fusion proteins were purified with glutathione-Sepharose (Pharmacia). All GST fusion proteins except full-length CNF1, Rac1, and c-Jun were subjected to thrombin cleavage.
Nucleotide binding assay.
Control Cdc42/Rac, Q61E Cdc42/Rac, and CNF1-treated Cdc42/Rac (0.5 μM each) were incubated at 37°C in a buffer containing 150 mM NaCl, 2.5 mM MgCl2, and 10 mM triethanolamine (pH 7.5). After addition of 2 μM mant-GDP, the increase in light emission at 444 nm, due to the higher intensity of bound mant-GDP (three times stronger than that of free mant-GDP) excited at 357 nm, was monitored in a Perkin-Elmer LS 50B luminescence spectrometer.
Treatment of recombinant GTPases with CNF1.
The GTPases were incubated for 3 h at 37°C together with GST-CNF1 or ΔCNF1 (the active fragment of CNF1, consisting of amino acid residues 709 through 1014 of CNF1) (43) at a GTPase/toxin molar ratio of 5:1 in a buffer containing 20 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 1 mM dithiothreitol, and 1 mM EDTA.
GTPase activity.
The GTPases were loaded with [γ-32P]GTP by incubation for 5 min at 37°C in a solution containing 50 mM Tris-HCl (pH 7.4), 10 mM EDTA, and 2 mM dithiothreitol. After addition of MgCl2 and GTP to give concentrations of 30 and 1 mM, respectively, the samples were placed on ice. The GTPases were incubated at 37°C in the presence or absence of p50GAP at the concentrations indicated in the figure legends. The samples were filtered through nitrocellulose, and the filters were washed three times with 3 ml of ice-cold buffer containing 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, and 5 mM MgCl2. Subsequently, the radioactivity remaining on the filters was determined.
JNK assay.
After treatment with GST-CNF1, subconfluent HeLa cells grown on 10-cm-diameter petri dishes were lysed on ice for 15 min in 1 ml of lysis buffer (20 mM Tris-HCl [pH 7.4], 137 mM NaCl, 10% glycerol, 1% Triton X-100, 2 mM EDTA, 50 mM sodium β-glycerophosphate, 20 mM sodium pyrophosphate, 0.5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 5 μg of aprotinin per ml, 5 μg of leupeptin per ml, 5 mM benzamidine, and 1 mM sodium orthovanadate). The lysates were collected with a rubber policeman and cleared by centrifugation (10 min, 20,800 × g, 4°C). For determination of kinase activity, JNK was precipitated (2 h of end-over-end shaking at 4°C) from the cytosol with rabbit anti-JNK polyclonal antibody (Santa Cruz) bound to protein A-Sepharose (Pharmacia). In control experiments, the JNK antibody proved to be specific for JNK compared to preimmune serum. However, it did not distinguish between the different JNK subclasses (JNK1, JNK2, and JNK3) (not shown). The immunoprecipitates were washed two times in lysis buffer with 500 mM NaCl and once in kinase buffer (25 mM HEPES [pH 7.5], 10 mM MgCl2, 25 mM sodium β-glycerophosphate, 5 mM benzamidine, 1 mM sodium orthovanadate, and 0.5 mM dithiothreitol). Recombinant GST–c-Jun (2 μg), 100 μM ATP, and 0.5 μCi of [γ-32P]ATP were added, and the reaction mixture was incubated for 30 min at 30°C. The reaction was stopped with Laemmli sample buffer. The samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the proteins were transferred to nitrocellulose. The 32P-phosphorylated c-Jun was visualized by autoradiography, and subsequently the amount of precipitated JNK was determined by means of a primary rabbit anti-JNK antibody and a secondary horseradish peroxidase-coupled antirabbit antibody with enhanced chemiluminescence detection reagent (100 mM Tris-HCl, [pH 8.0], 1 mM luminol [Fluka], 0.2 mM p-coumaric acid [Fluka], 3 mM H2O2).
Immunoprecipitation.
Twenty micrograms of pMT2ST-Cdc42 DNA (with a hemagglutinin [HA] tag) was dissolved in 220 μl of buffer containing 10 mM Tris-HCl and 1 mM EDTA (pH 8.0) and coprecipitated with calcium phosphate according to the method of Sambrook et al. (40). Subconfluent HeLa cells growing on 10-cm-diameter dishes were incubated with the DNA precipitate overnight, and after a change to fresh medium, the cells were incubated for 16 h with 500 ng of CNF1 per ml. The cells were lysed in a buffer containing 20 mM triethanolamine (pH 7.4), 2 mM MgCl2, 2 mM EDTA, 1% Triton X-100, 0.5 mM phenylmethylsulfonyl fluoride, 0.5 mM benzamidine, 40 μg of aprotinin per ml, and 10 μg of leupeptin per ml. The lysates were centrifuged for 20 min (20,800 × g at 4°C), and the supernatants were incubated at 4°C with protein A-Sepharose (Pharmacia) in the presence of mouse monoclonal HA-antibody (Boehringer). The beads were washed three times with PBS, and thereafter the immunoprecipitated Cdc42 was digested for mass spectrometric analysis as described below.
Mass spectrometry.
A saturated matrix solution of recrystallized 4-hydroxy-α-cyanocinnamic acid (Aldrich) and acetonitrile–aqueous 0.1% trifluoroacetic acid (1:1) was freshly prepared each day. The recombinant CNF1-treated and control proteins and the immunoprecipitated HA-Cdc42 were digested for 12 h at 30°C in 20 μl of 50 mM Tris-HCl (pH 7.8)–5 mM CaCl2 containing 0.1 μg of chymotrypsin (sequencing grade; Boehringer). Four microliters of the proteolytic peptide mixture was mixed with 1 μl of 10% trifluoroacetic acid, 2 μl of acetonitrile, 8 μl of saturated matrix, and 2 μl of marker peptides (5 pmol each of human adrenocorticotropin [amino acids 18 to 39] [molecular weight, 2,466] [Sigma] and human angiotensin II [molecular weight, 1,047] [Sigma]) for internal calibration. By using the dried-drop method of matrix crystallization, 1 μl of the sample-matrix solution was placed on the MALDI stainless-steel target and was allowed to air dry for several minutes at room temperature, resulting in a thin layer of fine granular matrix crystals. MALDI/time-of-flight mass spectrometry was performed on a Bruker Biflex mass spectrometer equipped with a nitrogen laser (λ = 337 nm) to desorb and ionize the samples. Mass spectra were recorded in the reflector positive mode in combination with delayed extraction. External calibration was routinely used, and internal calibration with two points which bracketed the mass range of interest was additionally performed to consolidate peptide masses further. The computer program MS-digest (from Peter Baker and Karl Clauser, University of California at San Francisco Mass Spectrometry Facility) was used for computer-assisted comparison of the tryptic peptide mapping data with the expected set of peptides.
Nucleotide sequence accession number.
The nucleotide sequence of the CNF1 gene is available from the EMBL data library under accession no. X70670.
RESULTS
Morphology of CNF1-treated HeLa cells.
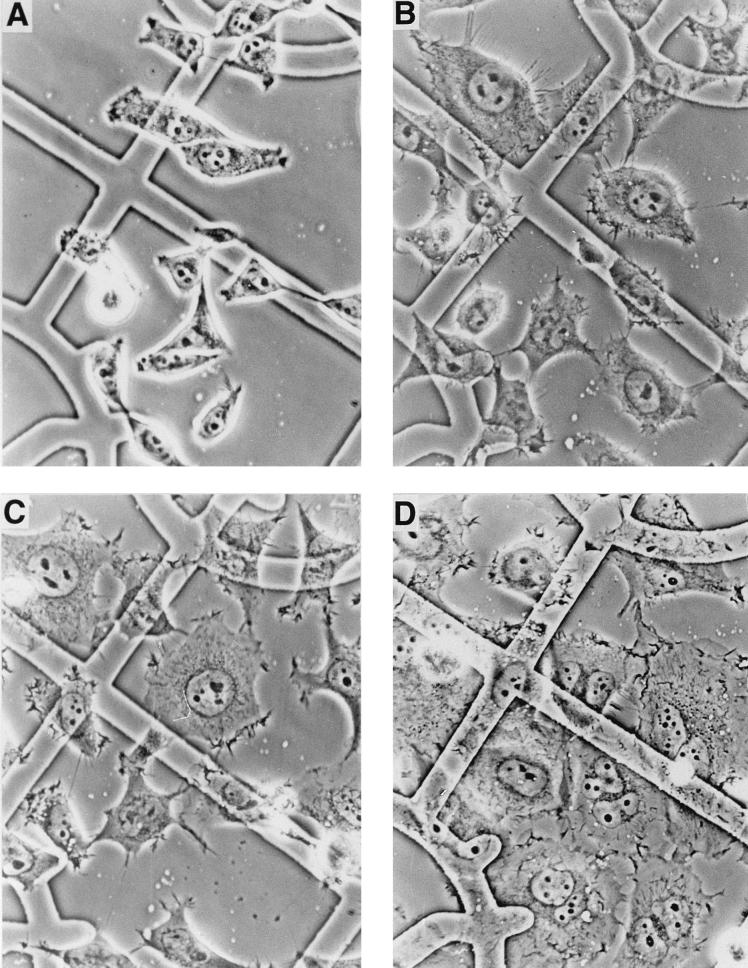
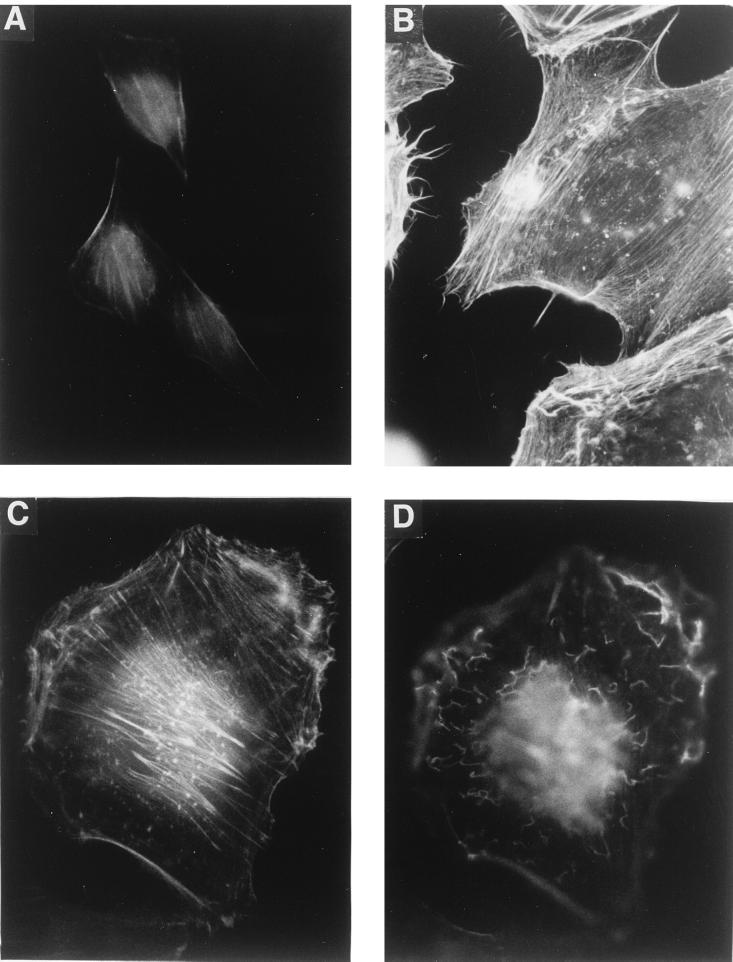
Treatment of HeLa cells with CNF1 induced dramatic morphological changes. Similar to results reported earlier (8, 10, 34), CNF1-treated cells increased in size and became multinucleated. In addition, we observed a transient formation of microspikes in subconfluent HeLa cells (Fig. 1). The microspikes appeared 3 to 6 h after addition of GST-CNF1 (500 ng/ml), suggesting a possible involvement of Cdc42 (see Discussion). At this time of toxin treatment, most cells were still mononuclear. After 24 h of GST-CNF1 treatment, the cells were enlarged and multinucleated, and the microspikes had almost completely disappeared. Membrane ruffles were visible in the phase-contrast microscope, suggesting an involvement of Rac as well (see Discussion). To visualize the actin cytoskeleton of HeLa cells after GST-CNF1 treatment, F-actin was stained with rhodamine-labeled phalloidin (Fig. 2). After 6 h of GST-CNF1-treatment, F-actin-containing membrane protrusions, stress fibers, and membrane ruffles of the cells were visible (Fig. 2B). After 24 h of GST-CNF1 treatment, only stress fibers (Fig. 2C) and membrane ruffles (Fig. 2D) appeared.
FIG. 1.
Phase-contrast micrographs of HeLa cells treated with CNF1. HeLa cells grown on gridded glass coverslips were treated with GST-CNF1 (500 ng/ml) and photographed after 0 (A), 6 (B), 12 (C), and 24 (D) h (40× objective).
FIG. 2.
Actin staining of CNF1-treated HeLa cells. HeLa cells were treated with GST-CNF1 (500 ng/ml) for 0 (A), 6 (B), and 24 (C and D) h. Thereafter, the cells were fixed and the F-actin was stained with rhodamine-phalloidine. (C and D) Micrographs from the same cell, with focusing below the apical cell surface (C) and on the apical surface of the cell (D). For all panels, a 100× oil immersion objective was used.
Activation of JNK in CNF1-treated HeLa cells.
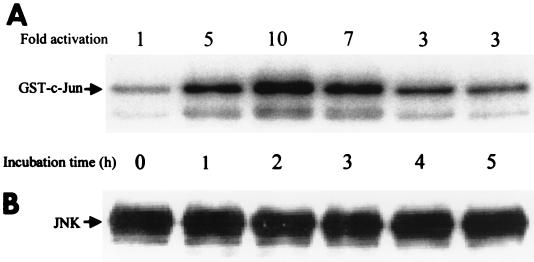
Cdc42 and Rac are known to activate JNK (2, 5, 28). To examine whether JNK is activated in CNF1-treated cells, we immunoprecipitated the kinase from lysates of HeLa cells treated for up to 5 h with GST-CNF1 (500 ng/ml) and tested its activity with GST–c-Jun as a substrate in the kinase assay. Figure 3A shows the radiolabeling of GST–c-Jun after 32P phosphorylation by JNK. As early as 1 h after treatment of cells with CNF1, JNK activity was increased, and after 2 h of treatment, a maximal (10- to 50-fold) activation was observed. After 3 h of CNF1 treatment, the JNK activity was reduced again but remained above control levels. The immunoblot in Fig. 3B shows that equal amounts of JNK were precipitated in each sample.
FIG. 3.
(A) Time course of the CNF1-induced activation of JNK. HeLa cells were incubated for 0 to 5 h with 500 ng of GST-CNF1 per ml. After lysis, the endogenous JNK was immunoprecipitated from the clear lysates. The activity of the JNK was determined in a kinase assay, in which the immunocomplexed JNK was incubated for 30 min at 30°C with [γ-32P]ATP and GST–c-Jun as substrates. The samples were subjected to SDS-PAGE and Western blotting. Phosphorylation of GST–c-Jun was quantified by the Image Quant program of the Phosphorimager (Molecular Dynamics). (B) The amount of precipitated JNK was determined by enhanced chemiluminescence.
Mass spectrometric analysis of CNF1-treated Cdc42 and Rac.
Since the morphological changes of HeLa cells and the activation of JNK induced by CNF1 suggested the involvement of Cdc42 and Rac in the action of the toxin (see Discussion), we attempted to verify deamidation of these GTPases by CNF1 in intact cells. Therefore, HeLa cells transfected with HA-tagged Cdc42 were treated with GST-CNF1 for 16 h. Thereafter, HA-Cdc42 was precipitated with anti-HA antibodies, digested with chymotrypsin, and subjected to mass spectrometric analysis as described in Materials and Methods. We found that a 2,683.3-Da peptide of low intensity, which corresponds to amino acid residues 47 through 70 of Cdc42, increased in mass by 1 Da after treatment of the HeLa cells with GST-CNF1 (not shown). An identical increase in mass of 1 Da was observed in CNF1-treated recombinant Cdc42 and Rac1 (peptides 52 through 70 and 57 through 70, respectively) (Fig. 4). Both isoforms of Cdc42 (brain and placenta [29, 46]) were substrates of CNF1 (not shown).
FIG. 4.
Mass spectrometric analysis of in vitro-modified Cdc42 (A) and Rac1 (B). Purified recombinant Cdc42 and Rac1 were digested with chymotrypsin after incubation with CNF1 (molar ratio of GTPase to toxin, 5:1) and, as a control, without the toxin. The masses of the resultant peptides were determined in a mass spectrometer. aa, amino acid.
Biochemical properties of recombinant Cdc42 and Rac1 modified by CNF1.
Recently, it was reported that Gln63 of RhoA is deamidated by CNF1 (12, 42). Thus, toxin-modified RhoA possesses the same biochemical properties as the mutant Q63E RhoA (42). To test whether this is also true for CNF1-treated Cdc42 and Rac1, we changed the equivalent glutamine residue (Gln61) in Cdc42 and Rac1 to glutamic acid and compared the biochemical properties of the mutants with CNF1-treated Cdc42 and Rac1.
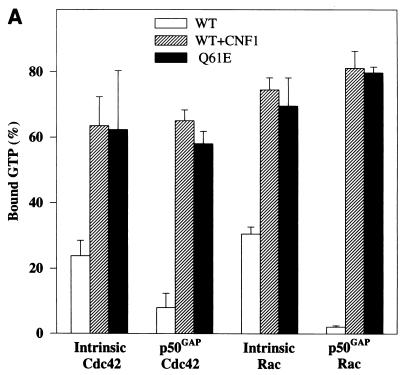
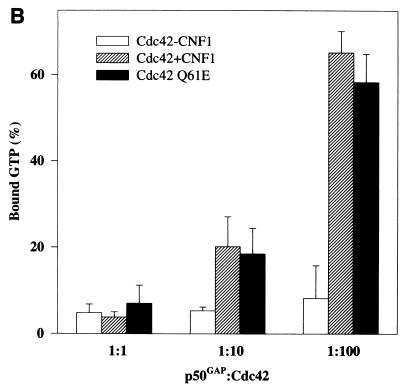
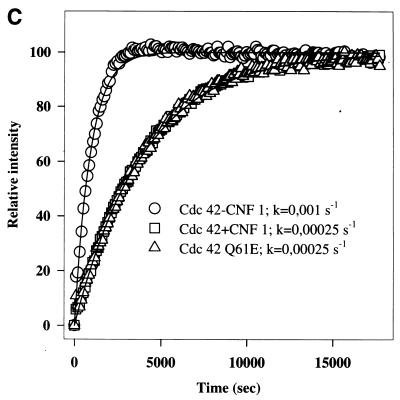
Figure 5A shows that the intrinsic and GAP-stimulated GTPase activities of CNF1-treated Cdc42 and Rac1 were inhibited to the same extent as those of the Q61E mutants. At high concentrations of GAP (molar ratio of p50GAP to Cdc42 of 1:1), however, the inhibition of the GTPase activities of CNF1-treated Cdc42 and of the Q61E mutant was overcome, and bound GTP was completely hydrolyzed (Fig. 5B). Furthermore, CNF1-treated Cdc42 and the Q61E mutant exhibited exactly the same retardation in mant-GDP binding (Fig. 5C). The mant-GDP binding assay reflects the release of prebound nucleotide, which was approximately 90% GDP in all samples. Essentially the same retardation in mant-GDP binding was observed with CNF1-treated Rac1 and Q61E Rac (not shown).
FIG. 5.
Effects of CNF1 treatment on GTPase activities and nucleotide binding of Cdc42 and Rac1. (A and B) Wild-type (WT) Cdc42, wild-type Rac1, and the respective Q61E mutants were incubated without (WT and Q61E) and with (WT) ΔCNF1 (molar ratio of GTPase to toxin, 5:1) for 3 h, and thereafter the GTPases were loaded with [γ-32P]GTP. Data are given as means and standard deviations (n = 3). (A) Loaded GTPases were incubated without (intrinsic) and with p50GAP (molar ratio of p50GAP to Cdc42/Rac, 1:50) for 4 min at 37°C. The samples were then filtered through nitrocellulose, and the radioactivity remaining on the filters was counted. (B) GTPase activity was determined with Cdc42 at different p50GAP/Cdc42 ratios. (C) Wild-type Cdc42, Q61E Cdc42, and CNF1-treated Cdc42 (0.5 μM each) were incubated with 2 μM mant-GDP, and the increase in fluorescence at 444 nm (due to the higher intensity of bound mant-GDP excited at 357 nm) was recorded. The k values are the dissociation rates for GDP.
DISCUSSION
Recently, we and others reported that CNF1 causes deamidation of Gln63 of RhoA, resulting in inhibition of GTP hydrolysis (12, 42). Since CNF1-modified RhoA is constitutively active, the deamidation is a plausible explanation for the CNF1-induced formation of stress fibers. It has been proposed that the effect of CNF1 is specific for RhoA and that other members of the Rho family, i.e., Cdc42 and Rac, are not modified by the toxin (11, 26). Preliminary studies performed in our laboratory, however, suggested that recombinant Cdc42 and Rac are substrates for the toxin. Therefore, we were prompted to investigate in more detail whether Cdc42 and Rac also serve as substrates for CNF1 in intact cells. A first hint of the involvement of Cdc42 in the action of CNF1 was the finding that treatment of HeLa cells with GST-CNF1 caused a marked formation of microspikes, which are known to be regulated by Cdc42 (32). This formation of microspikes or filopodia occurred rather early in the intoxication process and was transient. The reason for the transiency is not clear; however, formation of microspikes in cells induced by microinjection of constitutive active Cdc42 also appears to be transient (14). As previously reported for Hep-2 cells (8), another striking morphological feature of CNF1-treated cells is the occurrence of membrane ruffles. We found membrane ruffles in CNF1-treated HeLa cells visualized by actin staining and scanning electron microscopy (not shown), which suggests that Rac is activated by CNF1 as well. Although Rho may also participate in membrane ruffling in some cell types (30), Rac is believed to be a major regulator of formation of lamellipodia with subsequent membrane ruffling (37). However, the present data do not exclude the possibility that Rac is activated by CNF1-modified Cdc42 through the activation cascade described for these proteins, in which Cdc42 is capable of activating Rac (32). Microinjection of constitutively active Cdc42 induces Rac-dependent formation of lamellipodia in quiescent Swiss 3T3 cells (32).
In many cell types, regulation of JNK activity by extracellular stimuli involves Cdc42 and Rac but not Rho (2, 5, 28). Thus, the finding that CNF1 treatment of HeLa cells increases JNK activity is in line with the notion that CNF1 modifies Cdc42 and/or Rac in intact cells. Interestingly, as with the formation of microspikes, we observed a transient effect of CNF1 on JNK activity. However, the activation of JNK and the formation of microspikes peak at 2 and 6 h after addition of CNF1 to the cells, respectively (not shown). It remains to be investigated whether the reductions of JNK activity and of microspike formation depend on the same regulatory mechanism.
Direct evidence for the modification of Cdc42 by CNF1 was obtained by mass spectrometry of immunoprecipitated HA-tagged Cdc42 from CNF1-treated HeLa cells. We detected an increase in mass of 1 Da for the peptide consisting of the amino acid residues 47 to 70, indicating a deamidation of Gln61 of Cdc42 (deamidation results in a mass shift of 1 Da). Gln61 is equivalent to Gln63 of RhoA, which was shown to be deamidated by CNF1 (12, 42). Our attempts to precipitate overexpressed Rac1 were not successful. The increase in mass of 1 Da was consistently observed in peptides formed by digestion of CNF1-treated recombinant Cdc42 and Rac1.
The biochemical properties of recombinant Cdc42 and Rac1 modified by CNF1 were identical to those of the corresponding mutants (Q61E). First, in nondenaturing gels but not in SDS-PAGE, the CNF1-treated Cdc42 and Q61E Cdc42 exhibited an identical mobility shift (not shown). Second, the GTPase activities of toxin-treated Cdc42 and Rac1 were inhibited to the same extent as those of the Q61E mutants. Finally, mant-GDP binding to CNF1-treated Cdc42 and Rac1 was retarded similarly to that to the Q61E mutants. Previously we reported that in contrast to RhoA, treatment of recombinant Cdc42 and Rac1 by CNF1 causes only partial inhibition of their GTPase activities (42). The reasons for this discrepancy are most likely a different efficiency of p50GAP in activating the various subtypes of Rho GTPases (24, 38) and the fact that p50GAP was used at rather high concentrations in the previous study. The finding that CNF1-treated Cdc42 and Rac1 shared all properties with the Q61E mutants corroborates the notion that Gln61 is deamidated by CNF1. Moreover, even repetitive microinjection of constitutively active RhoA is not sufficient to cause the same phenotype as induced by treatment of cells with CNF1 (unpublished observation), indicating the existence of further targets for CNF1. Taken together, the data in the present study indicate that not only RhoA but also Cdc42 can be deamidated in intact cells, and they strongly suggest that Rac can also be targeted by CNF1 in vivo. Thus, CNF1 may be a useful tool as general activator of the Rho family GTPases in signal transduction research.
ACKNOWLEDGMENTS
We thank Volker Speth for electron micrographs and Iris Misicka for technical assistance.
This work was supported by Sonderforschungsbereich (SFB) 366 and DFG grant RA 642/3-2.
REFERENCES
- 1.Aktories K, Koch G. Clostridium botulinum ADP-ribosyltransferase C3. In: Aktories K, editor. Bacterial toxins: tools in cell biology and pharmacology. Weinheim, Germany: Chapman & Hall; 1997. pp. 61–69. [Google Scholar]
- 2.Bagrodia S, Dérijard B, Davis R J, Cerione R A. Cdc42 and PAK-mediated signaling leads to Jun kinase and p38 mitogen-activated protein kinase activation. J Biol Chem. 1995;270:27995–27998. doi: 10.1074/jbc.270.47.27995. [DOI] [PubMed] [Google Scholar]
- 3.Caprioli A, Falbo V, Roda L G, Ruggeri F M, Zona C. Partial purification and characterization of an Escherichia coli toxic factor that induces morphological cell alterations. Infect Immun. 1983;39:1300–1306. doi: 10.1128/iai.39.3.1300-1306.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chong L D, Traynor-Kaplan A, Bokoch G M, Schwartz M A. The small GTP-binding protein Rho regulates a phosphatidylinositol 4-phosphate 5-kinase in mammalian cells. Cell. 1994;79:507–513. doi: 10.1016/0092-8674(94)90259-3. [DOI] [PubMed] [Google Scholar]
- 5.Coso O A, Chiariello M, Yu J-C, Teramoto H, Crespo P, Xu N, Miki T, Gutkind J S. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 6.de Rycke J, Guillot J F, Boivin R. Cytotoxins in non-enterotoxigenic strains of Escherichia coli isolated from feces of diarrheic calves. Vet Microbiol. 1987;15:137–150. doi: 10.1016/0378-1135(87)90139-8. [DOI] [PubMed] [Google Scholar]
- 7.de Rycke J, Phan-Thanh L, Bernard S. Immunochemical identification and biological characterization of cytotoxic necrotizing factor from Escherichia coli. J Clin Microbiol. 1997;27:983–988. doi: 10.1128/jcm.27.5.983-988.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falzano L, Fiorentini C, Donelli G, Michel E, Kocks C, Cossart P, Cabanié L, Oswald E, Boquet P. Induction of phagocytic behaviour in human epithelial cells by Escherichia coli cytotoxic necrotizing factor type 1. Mol Microbiol. 1993;9:1247–1254. doi: 10.1111/j.1365-2958.1993.tb01254.x. [DOI] [PubMed] [Google Scholar]
- 9.Feig L A. Guanine-nucleotide exchange factors: a family of positive regulators of Ras and related GTPases. Curr Opin Cell Biol. 1994;6:204–211. doi: 10.1016/0955-0674(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 10.Fiorentini C, Donelli G, Matarrese P, Fabbri A, Paradisi S, Boquet P. Escherichia coli cytotoxic necrotizing factor 1: evidence for induction of actin assembly by constitutive activation of the p21 Rho GTPase. Infect Immun. 1995;63:3936–3944. doi: 10.1128/iai.63.10.3936-3944.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fiorentini C, Fabbri A, Flatau G, Donelli G, Matarrese P, Lemichez E, Falzano L, Boquet P. Escherichia coli cytotoxic necrotizing factor 1 (CNF1), a toxin that activates the Rho GTPase. J Biol Chem. 1997;272:19532–19537. doi: 10.1074/jbc.272.31.19532. [DOI] [PubMed] [Google Scholar]
- 12.Flatau G, Lemichez E, Gauthier M, Chardin P, Paris S, Fiorentini C, Boquet P. Toxin-induced activation of the G protein p21 Rho by deamidation of glutamine. Nature. 1997;387:729–733. doi: 10.1038/42743. [DOI] [PubMed] [Google Scholar]
- 13.Fukumoto Y, Kaibuchi K, Hori Y, Fujioka H, Araki S, Ueda T, Kikuchi A, Takai Y. Molecular cloning and characterization of a novel type of regulatory protein (GDI) for the rho proteins, ras p21-like small GTP-binding proteins. Oncogene. 1990;5:1321–1328. [PubMed] [Google Scholar]
- 14.Hall, A. Personal communication.
- 15.Hiratsuka T. New ribose-modified fluorescent analogs of adenine and guanine nucleotides available as substrates for various enzymes. Biochim Biophys Acta. 1983;742:496–508. doi: 10.1016/0167-4838(83)90267-4. [DOI] [PubMed] [Google Scholar]
- 16.Just I, Mohr C, Schallehn G, Menard L, Didsbury J R, Vandekerckhove J, van Damme J, Aktories K. Purification and characterization of an ADP-ribosyltransferase produced by Clostridium limosum. J Biol Chem. 1992;267:10274–10280. [PubMed] [Google Scholar]
- 17.Just I, Selzer J, Hofmann F, Green G A, Aktories K. Inactivation of Ras by Clostridium sordellii lethal toxin-catalyzed glucosylation. J Biol Chem. 1996;271:10149–10153. doi: 10.1074/jbc.271.17.10149. [DOI] [PubMed] [Google Scholar]
- 18.Just I, Selzer J, Wilm M, Von Eichel-Streiber C, Mann M, Aktories K. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature. 1995;375:500–503. doi: 10.1038/375500a0. [DOI] [PubMed] [Google Scholar]
- 19.Just I, Wilm M, Selzer J, Rex G, Von Eichel-Streiber C, Mann M, Aktories K. The enterotoxin from Clostridium difficile (ToxA) monoglucosylates the Rho proteins. J Biol Chem. 1995;270:13932–13936. doi: 10.1074/jbc.270.23.13932. [DOI] [PubMed] [Google Scholar]
- 20.Khosravi-Far R, Solski P A, Clark G J, Kinch M S, Der C J. Activation of Rac1, RhoA, and mitogen-activated protein kinases is required for Ras transformation. Mol Cell Biol. 1995;15:6443–6453. doi: 10.1128/mcb.15.11.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozma R, Ahmed S, Best A, Lim L. The Ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol Cell Biol. 1995;15:1942–1952. doi: 10.1128/mcb.15.4.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuribara H, Tago K, Yokozeki T, Sasaki T, Takai Y, Morii N, Narumiya S, Katada T, Kanaho Y. Synergistic activation of rat brain phospholipase D by ADP-ribosylation factor and rhoA p21, and its inhibition by Clostridium botulinum C3 exoenzyme. J Biol Chem. 1995;270:25667–25671. doi: 10.1074/jbc.270.43.25667. [DOI] [PubMed] [Google Scholar]
- 23.Lamaze C, Chuang T H, Terlecky L J, Bokoch G M, Schmid S L. Regulation of receptor-mediated endocytosis by Rho and Rac. Nature. 1996;382:177–179. doi: 10.1038/382177a0. [DOI] [PubMed] [Google Scholar]
- 24.Lancaster C A, Taylor-Harris P M, Self A J, Brill S, Van Erp H E, Hall A. Characterization of rhoGAP. A GTPase-activating protein for rho-related small GTPases. J Biol Chem. 1994;269:1137–1142. [PubMed] [Google Scholar]
- 25.Laudanna C, Campbell J J, Butcher E C. Role of Rho in chemoattractant-activated leukocyte adhesion through integrins. Science. 1996;271:981–983. doi: 10.1126/science.271.5251.981. [DOI] [PubMed] [Google Scholar]
- 26.Lemichez E, Flatau G, Bruzzone M, Boquet P, Gauthier M. Molecular localization of the Escherichia coli cytotoxic necrotizing factor CNF1 cell-binding and catalytic domains. Mol Microbiol. 1997;24:1061–1070. doi: 10.1046/j.1365-2958.1997.4151781.x. [DOI] [PubMed] [Google Scholar]
- 27.Malcolm K C, Ross A H, Qiu R-G, Symons M, Exton J H. Activation of rat liver phospholipase D by the small GTP-binding protein RhoA. J Biol Chem. 1994;269:25951–25954. [PubMed] [Google Scholar]
- 28.Minden A, Lin A, Claret F-X, Abo A, Karin M. Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 29.Munemitsu S, Innis M A, Clark R, McCormick F, Ullrich A, Polakis P. Molecular cloning and expression of a G25K cDNA, the human homolog of the yeast cell cycle gene CDC42. Mol Cell Biol. 1990;10:5977–5982. doi: 10.1128/mcb.10.11.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishiyama T, Sasaki T, Takaishi K, Kato M, Yaku H, Araki K, Matsuura Y, Takai Y. rac p21 is involved in insulin-induced membrane ruffling and rho p21 is involved in hepatocyte growth factor- and 12-O-tetradecanoylphorbol-13-acetate-induced membrane ruffling in KB cells. Mol Cell Biol. 1994;14:2447–2456. doi: 10.1128/mcb.14.4.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nobes C, Hall A. Regulation and function of the Rho subfamily of small GTPases. Curr Opin Genet Dev. 1994;4:77–81. doi: 10.1016/0959-437x(94)90094-9. [DOI] [PubMed] [Google Scholar]
- 32.Nobes C D, Hall A. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 33.Olson M F, Ashworth A, Hall A. An essential role for Rho, Rac, and Cdc42 GTPases in cell cycle progression through G1. Science. 1995;269:1270–1272. doi: 10.1126/science.7652575. [DOI] [PubMed] [Google Scholar]
- 34.Oswald E, Sugai M, Labigne A, Wu H C, Fiorentini C, Boquet P, O’Brien A D. Cytotoxic necrotizing factor type 2 produced by virulent Escherichia coli modifies the small GTP-binding proteins Rho involved in assembly of actin stress fibers. Proc Natl Acad Sci USA. 1994;91:3814–3818. doi: 10.1073/pnas.91.9.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Popoff M R, Chaves O E, Lemichez E, Von Eichel-Streiber C, Thelestam M, Chardin P, Cussac D, Chavrier P, Flatau G, Giry M, Gunzburg J, Boquet P. Ras, Rap, and Rac small GTP-binding proteins are targets for Clostridium sordellii lethal toxin glucosylation. J Biol Chem. 1996;271:10217–10224. doi: 10.1074/jbc.271.17.10217. [DOI] [PubMed] [Google Scholar]
- 36.Prepens U, Just I, Von Eichel-Streiber C, Aktories K. Inhibition of FcɛRI-mediated activation of rat basophilic leukemia cells by Clostridium difficile toxin B (monoglucosyltransferase) J Biol Chem. 1996;271:7324–7329. doi: 10.1074/jbc.271.13.7324. [DOI] [PubMed] [Google Scholar]
- 37.Ridley A J, Paterson H F, Johnston C L, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 38.Ridley A J, Self A J, Kasmi F, Paterson H F, Hall A, Marshall C J, Ellis C. rho family GTPase activating proteins p190, bcr and rhoGAP show distinct specificities in vitro and in vivo. EMBO J. 1993;12:5151–5160. doi: 10.1002/j.1460-2075.1993.tb06210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rittinger K, Walker P A, Eccleston J F, Smerdon S J, Gamblin S J. Structure at 1.65 A of RhoA and its GTPase-activating protein in complex with a transition-state analogue. Nature. 1997;389:758–762. doi: 10.1038/39651. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 41.Schmalzing G, Richter H P, Hansen A, Schwarz W, Just I, Aktories K. Involvement of the GTP binding protein Rho in constitutive endocytosis in Xenopus laevis oocytes. J Cell Biol. 1995;130:1319–1332. doi: 10.1083/jcb.130.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt G, Sehr P, Wilm M, Selzer J, Mann M, Aktories K. Deamidation of Gln63 of Rho induced by Escherichia coli cytotoxic necrotizing factor 1. Nature. 1997;387:725–729. doi: 10.1038/42735. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt G, Selzer J, Lerm M, Aktories K. The Rho-deamidating cytotoxic-necrotizing factor CNF1 from Escherichia coli possesses transglutaminase activity—cysteine-866 and histidine-881 are essential for enzyme activity. J Biol Chem. 1998;273:13669–13674. doi: 10.1074/jbc.273.22.13669. [DOI] [PubMed] [Google Scholar]
- 44.Sekine A, Fujiwara M, Narumiya S. Asparagine residue in the rho gene product is the modification site for botulinum ADP-ribosyltransferase. J Biol Chem. 1989;264:8602–8605. [PubMed] [Google Scholar]
- 45.Selzer J, Hofmann F, Rex G, Wilm M, Mann M, Just I, Aktories K. Clostridium novyi a-toxin-catalyzed incorporation of GlcNAc into Rho subfamily proteins. J Biol Chem. 1996;271:25173–25177. doi: 10.1074/jbc.271.41.25173. [DOI] [PubMed] [Google Scholar]
- 46.Shinjo K, Koland J G, Hart M J, Narasimhan V, Johnson D I, Evans T, Cerione R A. Molecular cloning of the gene for the human placental GTP-binding protein Gp (G25K): identification of this GTP-binding protein as the human homolog of the yeast cell-division-cycle protein CDC42. Proc Natl Acad Sci USA. 1990;87:9853–9857. doi: 10.1073/pnas.87.24.9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siddiqi A R, Smith J L, Ross A H, Qiu R-G, Symons M, Exton J H. Regulation of phospholipase D in HL60 cells. J Biol Chem. 1995;270:8466–8473. doi: 10.1074/jbc.270.15.8466. [DOI] [PubMed] [Google Scholar]
- 48.Tominaga T, Sugie K, Hirata M, Morii N, Fukata J, Uchida A, Imura H, Narumiya S. Inhibition of PMA-induced, LFA-1-dependent lymphocyte aggregation by ADP-ribosylation of the small molecular weight GTP binding protein, rho. J Cell Biol. 1993;120:1529–1537. doi: 10.1083/jcb.120.6.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Udagawa T, McIntyre B W. ADP-ribosylation of the G protein Rho inhibits integrin regulation of tumor cell growth. J Biol Chem. 1996;271:12542–12548. doi: 10.1074/jbc.271.21.12542. [DOI] [PubMed] [Google Scholar]
- 50.Zhang J, King W G, Dillon S, Hall A, Feig L, Rittenhouse S E. Activation of platelet phosphatidylinositide 3-kinase requires the small GTP-binding protein Rho. J Biol Chem. 1993;268:22251–22254. [PubMed] [Google Scholar]