To the Editor: More than 2 years into the coronavirus disease 2019 (Covid-19) pandemic, the global population carries heterogeneous immune histories derived from various exposures to infection, viral variants, and vaccination.1 Evidence at the level of binding and neutralizing antibodies and B-cell and T-cell immunity suggests that a history of infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can have a negative effect on subsequent protective immunity.1 In particular, the immune response to B.1.1.529 (omicron) subvariants could be compromised by differential immune imprinting in persons who have had a previous infection with the original virus or the B.1.1.7 (alpha) variant.1
We investigated the epidemiologic evidence for immune imprinting in persons with specific immune histories related to natural infection. We evaluated the incidence of repeat reinfection in the national cohort of persons in Qatar who had had a documented omicron BA.1 or BA.2 reinfection after a primary infection with non-omicron SARS-CoV-2 (the “double-primed” cohort) as compared with the incidence of reinfection in the national cohort of persons who had had a documented primary infection with omicron BA.1 or BA.2 (the “omicron-primed” cohort).2 This analysis was performed as a matched retrospective cohort study (Section S1 in the Supplementary Appendix, available with the full text of this letter at NEJM.org).
Data on SARS-CoV-2 laboratory testing, clinical infection, vaccination, and demographic characteristics were extracted from the Qatar national SARS-CoV-2 databases. Persons in both cohorts were exactly matched in a 1:3 ratio according to sex, 10-year age group, nationality, number of coexisting conditions, and calendar week of the omicron subvariant infection. The follow-up period started at 90 days after documentation of the omicron subvariant infection. Vaccinated persons were excluded. Associations were estimated with the use of Cox proportional-hazards regression models. Hazard ratios were adjusted for the factors used for matching.
Figure S1 in the Supplementary Appendix shows the population selection process, and Table S1 shows the baseline characteristics of the full and matched cohorts. The matched cohorts included 7873 persons in the double-primed cohort and 22,349 persons in the omicron-primed cohort. The study population was representative of the unvaccinated population of Qatar with respect to demographic characteristics and histories of SARS-CoV-2 infection (Table S2).
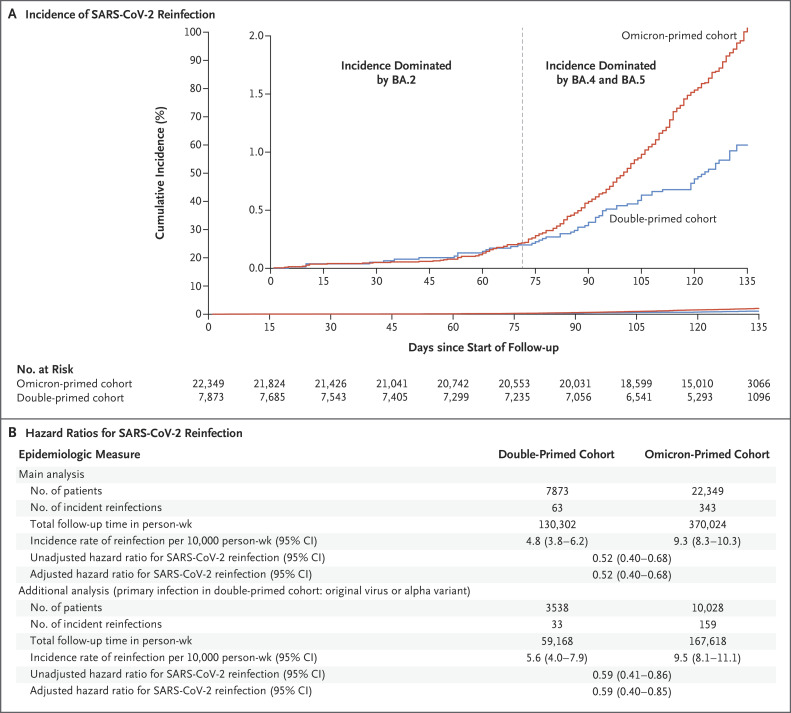
During follow-up, 63 reinfections occurred in the double-primed cohort and 343 occurred in the omicron-primed cohort; none of the infections progressed to severe, critical, or fatal Covid-19 (Fig. S1). At 135 days after the start of follow-up, the cumulative incidence of reinfection was 1.1% (95% confidence interval [CI], 0.8 to 1.4) in the double-primed cohort and 2.1% (95% CI, 1.8 to 2.3) in the omicron-primed cohort (Figure 1A). In the comparison of the full matched double-primed cohort with the omicron-primed cohort, the adjusted hazard ratio for reinfection was 0.52 (95% CI, 0.40 to 0.68). In an analysis involving the subgroup of persons in the double-primed cohort whose primary infection was with the original virus or the alpha variant as compared with the omicron-primed cohort, the adjusted hazard ratio for infection was 0.59 (95% CI, 0.40 to 0.85) (Figure 1B).
Figure 1. Incidence of SARS-CoV-2 Reinfection in the Double-Primed and Omicron-Primed Cohorts.
The double-primed cohort included persons with a documented reinfection with B.1.1.529 (omicron) subvariant BA.1 or BA.2 after a primary infection with pre-omicron severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and the omicron-primed cohort included persons with a documented primary infection with an omicron BA.1 or BA.2 subvariant. The inset in Panel A shows the same data on an expanded y axis. The main analysis included the full matched cohorts; in an additional analysis (Panel B), the double-primed cohort included only persons whose primary infection had been with the original virus or the B.1.1.7 (alpha) variant. Hazard ratios were adjusted for the factors used for matching. This study was conducted in Qatar between December 19, 2021, and August 15, 2022. Follow-up started 90 days after documentation of reinfection. The median duration of follow-up was 125 days (interquartile range, 114 to 132) in each cohort.
In the first 70 days of follow-up, when infections were dominated by the BA.2 subvariant,2,3 the adjusted hazard ratio for infection was 0.92 (95% CI, 0.51 to 1.65). However, the cumulative incidence curves diverged when the BA.4 and BA.5 subvariants were introduced and subsequently dominated4 (adjusted hazard ratio, 0.46; 95% CI, 0.34 to 0.62) (Figure 1A).
Limitations of the study are discussed in Section S1. One potential limitation was the difference in the frequencies of testing between the two cohorts, but a sensitivity analysis with adjustment for these differences showed results similar to those in the main analysis.
Omicron infection induces strong protection against a subsequent omicron infection.2,4 In the present cohort study, an additional, earlier infection with non-omicron SARS-CoV-2 was found to strengthen this protection against a subsequent omicron infection. The earlier pre-omicron infection may have broadened the immune response against a future reinfection challenge.
Supplementary Appendix
Disclosure Forms
This letter was published on October 12, 2022, at NEJM.org.
Footnotes
Supported by the Biomedical Research Program and the Biostatistics, Epidemiology, and Biomathematics Research Core at Weill Cornell Medicine–Qatar; the Qatar Ministry of Public Health; Hamad Medical Corporation; and Sidra Medicine. The Qatar Genome Program and Qatar University Biomedical Research Center supported viral genome sequencing.
Disclosure forms provided by the authors are available with the full text of this letter at NEJM.org.
References
- 1.Reynolds CJ, Pade C, Gibbons JM, et al. Immune boosting by B.1.1.529 (omicron) depends on previous SARS-CoV-2 exposure. Science 2022;377(6603):eabq1841-eabq1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chemaitelly H, Ayoub HH, Coyle P, et al. Protection of omicron sub-lineage infection against reinfection with another omicron sub-lineage. Nat Commun 2022;13:4675-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altarawneh HN, Chemaitelly H, Ayoub HH, et al. Effects of previous infection and vaccination on symptomatic omicron infections. N Engl J Med 2022;387:21-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altarawneh HN, Chemaitelly H, Ayoub HH, et al. Protective effect of previous SARS-CoV-2 infection against omicron BA.4 and BA.5 subvariants. N Engl J Med. DOI: 10.1056/NEJMc2209306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.