Abstract
Generating a summary from findings has been recently explored (Zhang et al., 2018, 2020) in note types such as radiology reports that typically have short length. In this work, we focus on echocardiogram notes that is longer and more complex compared to previous note types. We formally define the task of echocardiography conclusion generation (EchoGen) as generating a conclusion given the findings section, with emphasis on key cardiac findings. To promote the development of EchoGen methods, we present a new benchmark, which consists of two datasets collected from two hospitals. We further compare both standard and state-of-the-art methods on this new benchmark, with an emphasis on factual consistency. To accomplish this, we develop a tool to automatically extract concept-attribute tuples from the text. We then propose an evaluation metric, FactComp, to compare concept-attribute tuples between the human reference and generated conclusions. Both automatic and human evaluations show that there is still a significant gap between human-written and machine-generated conclusions on echo reports in terms of factuality and overall quality1.
1. Introduction
Echocardiography (or echo) is a test that uses sound waves to produce live images of the heart (Mitchell et al., 2019). It has become routinely used to support the diagnosis, management, and follow-up of patients with suspected or known heart diseases. The echo report documents and communicates the evaluation of cardiac and vascular structures in the echocardiography study. As shown in Figure 1, a standard echo report usually consists of a demographic section, an echocardiographic evaluation section (also called the finding section), and a conclusion section (Gardin et al., 2002). In a typical workflow, consultants who interpret echocardiography provide the quantitative measurement and descriptive statements to describe pertinent findings, and then conclude.
Figure 1:
Echocardiography reports from the (a) EgMimic and (b) EgCLEVER datasets.
In this work, we formally study the task of echo conclusion generation (EchoGen), arising in clinical practice to relieve the clinician of tasks that may contribute to clinician burnout (Alsharqi et al., 2018). A practical system shall be able to generate statements that emphasize abnormal findings, and compare differences and similarities of the current study versus the previous one if available and relevant. We define EchoGen as a task of learning from the demographic and echocardiographic findings section and generating the conclusion section.
Neural network-based models (See et al., 2017; Lewis et al., 2020) are an attractive method for this task, but are difficult to apply without appropriate training data. To address this gap, we present a large-scale EchoGen benchmark, which consists of two datasets. Here we reply on one prexisting MIMIC-III dataset (EgMimic) and one newly collected dataset from the NewYork-Presbyterian Hospital (EgCLEVER) to cover different text genres, data sizes, and degrees of difficulty, and more importantly, highlight common challenges of EchoGen (Figure 1).
Beyond data, a second challenge for EchoGen is to evaluate the factual correctness of a generated conclusion. Automatic metrics such as ROUGE and METEOR only assess content selection but not other quality aspects, such as fluency, grammaticality, and coherence, and are not well-correlated with factuality, leading to the development of separate evaluation measures (Zhang et al., 2018; Falke et al., 2019; Kryscinski et al., 2020; Goyal and Durrett, 2021). This study proposes a new evaluation metric to measure factual consistency, called “FactComp” by considering both concept and their attributes in the fact equivalence criteria.
To better understand the challenge posed by EchoGen, we conducted experiments with five baselines: Tf-idf, RandSent, LexRank, FactExt, and Bart. We find that Bart exceeds other baselines by a large margin, but it has poor transferability when tested on cross-corpus settings. Further human evaluations indicate that there is still a significant gap between generated conclusions and human reference in terms of fluency and factual consistency.
In summary, our contributions can be summarized as follows. (1) We formally introduce the task of EchoGen. (2) We curate a large-scale benchmark from an existing representative dataset and a newly-collected dataset. (3) We introduce a new metric to measure the fact consistency for echo notes. (4) Our metric and human evaluations find that there is still a gap between human reference and generated conclusions for echo reports in terms of fluency and factual consistency.
2. Related works
While EchoGen has not been defined before, there are closely related tasks that were studied before: data-to-text generation, clinical report summarization, and evaluation.
Data-to-text Generation
Data-to-text generation is a task of generating text in natural language from non-linguistic input data such as tables and time series (Gatt and Krahmer, 2018; Wiseman et al., 2017; Gardent et al., 2017). Traditional approaches for data-to-text generation (Reiter and Dale, 2000) follow a pipeline of modules such as content selection, text structuring, and surface realization. Recent methods (Gehrmann et al., 2018; Harkous et al., 2020) generate text from data in an end-to-end fashion using the encoder-decoder approach. Data-to-text is also explored in healthcare (Pauws et al., 2019) to facilitate patient review.
Clinical report summarization
Clinical report summarization is a long-standing research problem (Adams et al., 2021). Both extractive and abstractive methods have been applied for summarization, covering cases from structured data to text, medical image to text, and history documents to text (Afantenos et al., 2005; Xiong et al., 2019; Pivovarov and Elhadad, 2015).
To the best of our knowledge, few clinical summarization datasets are available. MEDIQA 2021 ST provides a task of generating radiology impression statements from textual clinical findings in radiology reports (Ben Abacha et al., 2021) collected from the indiana University dataset and Stanford Health Care. CLIP is a dataset on discharge notes, where the authors’ task was to extract the follow-up action items from notes (Mullenbach et al., 2021). This dataset is more suitable for developing information extraction (IE) systems or extractive summarization methods. Adams et al. (2021) developed a dataset CLINSUM from Columbia University Irving Medical Center, focusing on discharge summary notes. While they identified the complex, multi-document summarization task, the dataset is not public to promote the model development by other researchers.
In comparison, our EchoGen is a completely new task on a new note type – echocardiograms. More importantly, the benchmark covers a diverse range of text genres from two resources. We expect that the models that perform better on both datasets will be more robust in real-word settings.
Evaluation on clinical text
Evaluation of clinical text generation or summarization is a challenging research area. Existing methods include automatic approaches and human judgments. For example, commonly used ROUGE-based evaluation metrics measure the overlapping n-grams or longest common sub-sequence between the reference and generated summaries. BERTScore (Zhang et al., 2019) (or HOLMS) is an alternative that accounts for lexical variations by comparing the similarity of semantic representations encoded via BERT (Devlin et al., 2019). However, human evaluations show that these metrics do not always correlate well with factual consistency measurement. Hence, many research works focus on developing automatic consistency metrics that correlate better with human evaluations.
Goodrich et al. (2019) measure the factual consistency as the ratio of overlap between relation triplets under fixed schema extracted from the reference and the generated summary. Kryscinski et al. (2020) propose an entailment-based model FactCC to check whether the source text entails each sentence in the generated summary. Wang et al. (2020) and Durmus et al. (2020) propose QA-based methods that measure the amount of information in the generated summary supported by the source. However, these evaluation approaches often consist of auxiliary modules trained on external or artificial datasets, which is prohibitively expensive and time-consuming to collect. In addition, these modules are hardly generalizable to other clinical settings. Our proposed fact extractor FACTEXT instead relies on linguistic knowledge and is shown to have higher generalizability.
3. EchoGen
3.1. Task definition
We first formulate the EchoGen task. Let x = {x1, …, xm} be the demographics and findings section of an echo report, the goal is to generate a conclusion y = {y1, …, yn}, where m and n are the length of the source section and the generated section of an echo report, respectively. In this work, x is the finding section of a report. We leave leveraging the correlations, if any, between demographic values and generated conclusions into future works.
3.2. Dataset construction
The EchoGen benchmark contains two corpora (Table 1. Here, we reply on one prexisting dataset because it is widely used in the clinical NLP community and one newly collected dataset to cover different text styles and levels of difficulties.
Table 1:
Statistics for the EchoGen benchmark.
| EgMimic | EgCLEVER | |
|---|---|---|
| Notes | 44,085 | 13,000 |
| Train | 41,164 | 10,081 |
| Dev | 1,447 | 1,406 |
| Test | 1,474 | 1,513 |
| Source sentences | 19 | 19 |
| Conclusion sentences | 14 | 12 |
| Source tokens | 173 | 219 |
| Conclusion tokens | 150 | 72 |
EgMimic
The first dataset was sampled from the MIMIC-III dataset (Medical Information Mart for Intensive Care III) (Johnson et al., 2016). MIMIC-III is a de-identified clinical database composed of over 40,000 patients admitted in the ICUs at Beth Israel Deaconess Medical Center. Of those, we collected echo reports from the noteevents table, whose category is “Echo”.
We applied the RadText tool 2 to split the notes into a sequence of sections. It uses a rule-based matching algorithm with default rules adapted from SecTag with reported recall of 99% (Denny et al., 2008). We then selected the “Findings” section as the input and the “Conclusion” section as the human reference. We sampled a collection of 41,164, 1,447, and 1,474 reports for training, development, and test, respectively (Table 1). Note that we sampled the echo notes at the patient level. This strategy will ensure that no participant was in more than one group to avoid cross-contamination between the training and test datasets.
EgCLEVER
The second dataset is a collection of echo notes in English for heart failure patients from the “PrediCtion of EarLy REadmissions in Patients with CongestiVE HeaRt Failure” (CLEVER) cohort at NewYork-Presbyterian Hospital (called EgCLEVER). The patients were admitted and discharged with billing codes ICD-9 Code 428 or ICD-10 Code I50 from January 2008 and July 2018. The study was reviewed and approved by the NewYork-Presbyterian Hospital Institutional Review Board.
We used the same method to preprocess EgCLEVER and sampled a collection of 10,081, 1,406, and 1,513 reports for training, development, and test, respectively.
Comparison
The task of EchoGen varies with the data source, which may depend on the individual hospital. Figure 1 shows one echo report from EgMimic and one from EgCLEVER. The EgMimic report more closely resembles the task of data-to-text generation (Gatt and Krahmer, 2018; Pauws et al., 2019), where the finding section consists of structured data (here, noun phrases in a key-value format), and the conclusion section is written by selecting important findings and expanding them to coherent natural language text. Since data-to-text often has a more complex tabular structure, the result here is somewhere in between pure data and natural language as the tabular structure is not explicit. Therefore, even though the number of tokens in the input is not much shorter than the conclusion section, the conclusion does contain less information than the input.
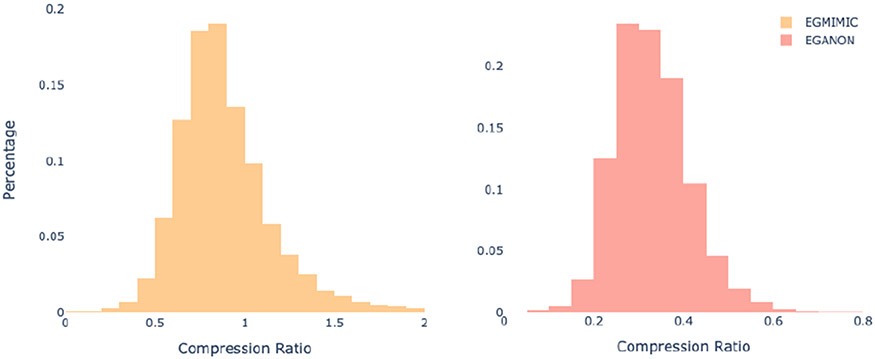
On the other hand, the conclusion section of our collected dataset EgCLEVER involves more heavily selecting and summarizing content from unstructured text input. The distribution of word compression ratio for both datasets further confirms our observations (Figure 2). The compression ratio is centered around 0.8 for EgMimic and 0.3 for EgCLEVER.
Figure 2:
Distribution of word compression ratio on EgMimic and EgCLEVER. The ratio defined as the quotient of number of tokens in the reference and that in the source.
3.3. Evaluation Metrics
ROUGE
First, we use the standard ROUGE scores (Lin, 2004), and report the F1 scores for ROUGE-1, ROUGE-2, and ROUGE-L, which compare the word-level unigram, bigram, and longest common sequence overlap between the generated and the human reference conclusion, respectively.
Factual Consistency
For Factual Consistency evaluation, we define a Factual F1 score, inspired by (Zhang et al., 2020). Specifically, we first extract and represent the facts f as a list of “(Concept, Attribute)” pairs ⟨f1, … fn⟩. For example, in the sentence “Right ventricular chamber size and free wall motion are normal”, the fact list is ⟨(right ventricular chamber size, normal), (free wall motion, normal)⟩.
The evaluation is then carried out by comparing the list f from the human reference to the list of facts from a generated conclusion. This requires that a concept and its attributes be extracted correctly to count as one fact.
Finally, the evaluation results are reported using the standard Precision, Recall, and F1-score metrics.
Here, FE is the factual equivalence criteria and can be defined in various modes.
Strict matching
The strict matching mode requires exact matching, and it holds when both the concept and attribute are the same. .
BERTScore matching
This mode uses greedy matching to maximize the matching similarity. Each fact is matched to the most similar fact in the human reference. Here, we concatenate the attribute with the concept to form a factual noun phrase, and used the BERTScore to measure the similarity between two phrases (Zhang et al., 2019). .
However, both modes have flaws. For example, strict matching does not consider lexical variation and semantic equivalence. On the other hand, since concept-attribute pairs are supposed to be independent, aligning each fact from the generated conclusion to the most similar one in the reference via BERTScore matching is less meaningful if they are two different facts. Therefore, we relax the definition of these modes and propose approximate matching.
Approximate matching
This mode combines strict matching and BERTScore matching. Specifically, a predicted fact is equivalent to a reference fact if their BERTScore is above a threshold t3. .
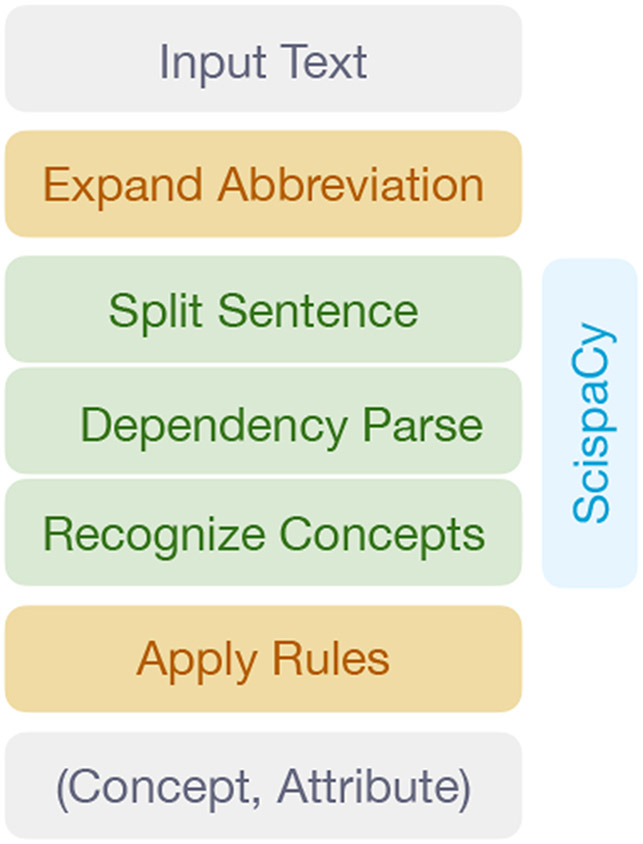
To extract the facts from the text, we develop a rule-based fact extraction system FactExt (Figure 3). The tool first splits the text into sentences, and then obtains the universal dependencies (de Marneffe et al., 2021) from the sentences. It further detects UMLS© concepts mentioned in the sentence. Here we focused on the common 55 concepts in the echo notes identified in the data driven way4. We used the ScispaCy model (Neumann et al., 2019) trained on MedMentions (Mohan and Li, 2018) to process the text.
Figure 3:
The pipeline of the fact extractor FactExt.
Afterward, we applied rules to all identified concepts and subsequently found the attributes that describe the concept. We include negation as an attribute but not uncertainty words as they rarely show up in the text. in this work, we utilized the universal dependency graph to define rules (Chambers et al., 2007). Therefore, the rules take advantage of linguistic knowledge so that the search of attributes is not limited to fixed word distance. The comprehensive rules can be found at our released code. The performance of FactExt is discussed in Section 4.
3.4. Baseline models for benchmarking
We consider 5 baseline models.
Tf-idf
Given a source x, Tf-idf first searches for the most similar source x′ over all training data based on TF-IDF features and then chooses corresponding conclusion y′ as a conclusion for the source x.
RandSent
We randomly select k = 12 sentences from a source as its conclusion, where k is determined according to the average number of conclusion sentences in two collected datasets.
LexRank
LexRank constructs a graph representation of the course, where nodes are sentences and edges are similarities between sentences (Erkan and Radev, 2004). It then applies the PageRank algorithm on the graph to extract top k = 12 most relevant sentences from the source.
FactExt
We first extract all facts f from a source and then construct a conclusion by concatenating them together. We next convert (Concept, Attribute) pairs into noun phrases by attaching attributes to the beginning of concepts. For example. (right ventricular chamber size, normal) converts to “normal right ventricular chamber size”.
Bart
BART (Lewis et al., 2020) is a pretrained language model that recently demonstrates the state-of-the-art performance in text summarization. It models the conditional likelihood p(y∣x) = Σt p(yt∣y<t, x), where y<t denotes generated tokens before time step t. We fine-tune a pretrained BART initialized with facebook/bart-large-xsum on both datasets.
4. Benchmark results and discussion
Rule-based system
Table 2 shows the performance of FactExt on randomly sampled 25 examples from two datasets. Two authors of the work manually annotated all (Concept, Attribute) tuples of sampled examples for evaluation. We obtain Cohen’s kappa κ = 0.81, which indicates a strong agreement. We observe that the system has high precision in all settings but with a drop in recall. This indicates that most (Concept, Attribute) pairs can be correctly identified with a few pairs missed. Further analysis demonstrates that the “Findings” section in EgMimic is more well structured than in EgCLEVER. Therefore, FactExt on the former setting achieves higher recall and F1.
Table 2:
The performance of FactExt on 25 randomly sampled Echo notes from the validation set of EgMimic (13) and EgCLEVER (12). Each report consists of one “Findings” section and one “Conclusion” section. All statistics are obtained by averaging scores from each report.
| EgMimic |
EgCLEVER |
Overall |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | P | R | F1 | |
| Findings | 94.3 | 83.4 | 88.3 | 88.8 | 73.1 | 79.9 | 91.7 | 78.5 | 84.3 |
| Conclusion | 91.2 | 76.1 | 82.6 | 96.7 | 93.5 | 95.0 | 93.8 | 84.4 | 88.5 |
| Overall | 92.8 | 79.8 | 85.5 | 92.8 | 83.3 | 87.4 | 92.8 | 81.5 | 86.4 |
Baseline Comparisons
Table 3 and 4 show the results of baseline approaches on the EgMimic and EgCLEVER datasets.
Table 3:
Results on EgMimic. ROUGE-1/2/L represent the ROUGE-F1 scores. FC represents Factual Consistency using the approximate matching.
| ROUGE-1 |
ROUGE-2 |
ROUGE-L |
FC |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | P | R | F1 | P | R | F1 | |
| Tf-idf | 47.7 | 47.2 | 44.9 | 27.4 | 27.1 | 25.9 | 39.2 | 38.7 | 36.9 | 40.2 | 41.0 | 38.8 |
| RandSent | 58.3 | 49.2 | 51.4 | 34.0 | 29.7 | 30.5 | 47.9 | 41.0 | 42.6 | 49.6 | 45.8 | 45.9 |
| LexRank | 60.5 | 51.5 | 53.8 | 37.0 | 32.3 | 33.3 | 49.9 | 43.1 | 44.7 | 53.6 | 47.5 | 48.3 |
| FactExt | 69.1 | 51.7 | 57.4 | 40.0 | 30.0 | 33.2 | 63.8 | 47.6 | 52.9 | 48.8 | 66.0 | 54.9 |
| Bart | 65.5 | 67.4 | 69.5 | 55.5 | 57.2 | 55.5 | 65.5 | 67.4 | 65.5 | 72.0 | 66.4 | 67.9 |
Table 4:
Results on EgCLEVER. ROUGE-1/2/L represent the ROUGE-F1 scores. FC represents Factual Consistency using the approximate matching.
| ROUGE-1 |
ROUGE-2 |
ROUGE-L |
FC |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | P | R | F1 | P | R | F1 | |
| Tf-idf | 58.1 | 57.0 | 55.5 | 40.8 | 40.3 | 39.1 | 52.5 | 51.5 | 50.2 | 59.8 | 60.2 | 57.8 |
| RandSent | 37.2 | 57.7 | 44.3 | 17.0 | 26.9 | 20.4 | 28.7 | 44.9 | 34.2 | 33.9 | 34.3 | 32.5 |
| LexRank | 40.2 | 58.7 | 46.6 | 18.0 | 27.2 | 21.2 | 30.8 | 45.5 | 35.8 | 33.4 | 36.5 | 33.1 |
| FactExt | 49.1 | 49.7 | 48.3 | 25.3 | 25.9 | 25.0 | 47.4 | 47.9 | 46.6 | 35.1 | 50.6 | 40.4 |
| Bart | 76.1 | 72.4 | 73.3 | 63.5 | 60.5 | 61.2 | 73.0 | 69.5 | 70.4 | 85.8 | 73.4 | 78.3 |
Overall, Bart achieves superior performance over other baselines by a large margin, showing the promising result of using abstractive summarization models.
RandSent and LexRank have similar performances on both datasets. The result is reasonable because LexRank relies on inter-sentence similarity to select sentences, but similarities between conclusion sentences are limited in clinical notes.
The Tf-idf baseline has contrary performance on two datasets. Recall that this approach copies the reference directly from the report with the most similar source in the training data. Since the “Conclusion” section is written as structured noun phrases in EgCLEVER and as complete sentences in EgMimic, Tf-idf is more likely to achieve a higher ROUGE score in EgCLEVER, which has fewer lexical variations in the “Conclusion” section.
Information Extraction v.s. Text Summarization
To tackle the summarization of echocardiography reports as an information extraction (IE) task, we provide our rule-based fact extractor FactExt as a performance lower bound. As shown in Table 3 and 4, the rule-based system falls short of performance in both evaluation metrics. Since FactExt concatenates all (Concept, Attribute) pairs as noun phrases to form a generated conclusion section, it fails to distill the key information of the source. Further, since the importance of a concept in one report depends on the overall levels of importance of other concepts, external human annotations are required. However, it is hard to reach a consensus on the importance of concepts between domain experts on our dataset (See Human Evaluation below). Therefore, these annotations are deemed to have limited usability, and an iE model trained on them may not be transferable to other clinical datasets.
Alternatively, machine learning based models outperform FactExt by a large margin in terms of both ROUGE scores and factual consistency evaluation. This suggests that summarization models can approximate the capability of an IE system and identify more critical facts.
Extractive Summarization v.s. Abstractive Summarization
FactExt is a strong extractive baseline that selects all concept and attribute pairs f as an extractive conclusion. However, the low recall under our defined evaluation metric indicates that (1) f is not capable of describing all the information in the reference; and (2) domain knowledge is required to generate novel information. The low precision score of FactExt, on the other hand, shows that the reference is highly selective of the source text as the majority of facts are excluded from the reference.
Transferability of the model across datasets
We intentionally designed the test set to be partially from a hospital system different from the training set (out-of-domain) to test the generalizability of the models. Results are shown in Table 5. As expected, the performance drops significantly in both datasets and is worse than all baselines in Table 3 and 4. The low FC scores indicate that organizations do not share a unified consensus of important information.
Table 5:
Cross-corpus results of models trained on EchoMIMIC and EgCLEVER using BART. R-1, R-2, R-L represent the ROUGE-F1 scores. FC represents Factual Consistency using the approximate matching. Numbers in parenthesis indicates the performance of the model on the dataset it trained on.
| Training corpus |
EgMimic |
EgCLEVER |
||||||
|---|---|---|---|---|---|---|---|---|
| R-1 | R-2 | R-L | FC | R-1 | R-2 | R-L | FC | |
| EgMimic | (69.5) | (55.2) | (65.5) | (67.9) | 39.9 | 13.9 | 24.2 | 28.1 |
| EgCLEVER | 32.6 | 14.2 | 23.9 | 24.9 | (73.1) | (60.8) | (70.2) | (78.3) |
5. Human Evaluation
To compare the quality of generated text against a human reference, we conduct a human evaluation following Zhang et al. (2020). We randomly sampled 50 echo reports from the development set of EgMimic. For each example, we presented echo findings to two Neurologist and Pulmonary Critical Care physicians along with the human references and summaries generated from BART in random order. We asked the physicians to compare them in three dimensions (1) fluency, (2) factual consistency, and (3) overall quality. For each metric, we asked the physicians to select the better one, with ties allowed.
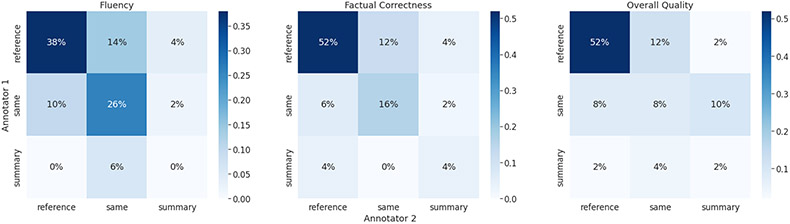
Since it is difficult to reach an agreement between physicians, we show the human evaluation result as confusion matrices in Figure 4. Across all three dimensions, both physicians agree that human reference is better among half of the selected samples (the upper-left cell of each figure). Further, most of the percentages fall into the top left two-by-two sub-matrices, with the main diagonal being the most frequent. This indicates that physicians have a consensus that generated conclusion is less preferred. There are also uncertainties about whether a reference is better or tied with a generated conclusion (around 20% at off-diagonal). Overall, model-generated summaries are still undesired compared to human reference in terms of fluency, factual consistency, and overall quality.
Figure 4:
Confusion matrices of human evaluation results on 50 randomly sampled echo notes from EgMimic. Results are shown in percentage and “same” means there is a tie between a reference and a generated conclusion.
6. Limitations
While our conducted human evaluation suggests that generated summaries from Bart tend to have more factual errors than human reference, the accuracy of factuality comparison between Bart and other baselines is still limited by the quality of our proposed system FactExt. Its performance, especially recall, depends on the accuracy of the ScispaCy model we use and the number of common concepts we focus on (55 in this work). For example, we can integrate the recommended phrases that echocardiographers may choose to use to describe pertinent findings by the American Society of Echocardiography (Gardin et al., 2002). We leave continually designing a more robust information extraction system or learning-based models, which both (1) rely less on domain-specific concepts; and (2) generalize to other types of notes, to future works.
7. Conclusion
In this study, we introduce EchoGen, a new benchmark for evaluating and analyzing models for echocardiography report conclusion generation. We systematically analyze the performance of several baseline methods with our proposed evaluation metric and conclude that there is still a gap between human reference and generated conclusions for echo reports in terms of fluency and factual consistency. Detailed analysis shows that our benchmarking can be used to evaluate the capacity of the models to understand the clinical text and, moreover, to shed light on the future directions for developing clinical text generation and summarization systems.
Acknowledgements
This work was supported by the National Library of Medicine under Award No. 4R00LM013001 and the Amazon Web Services Diagnostic Development Initiative. This work was partially supported by a gift from Amazon and a gift from Salesforce.
A. Echo Concepts
aneurysm, anular, aorta, apex, appendage, arch, arteriosus, artery, atheroma, atrial, atrium, calcification, cava, cavity size, chamber size, chordae, color doppler, defect, disease, effusion, ejection fraction, excursion, foramen, hypertension, hypertrophy, inflammation, leaflet, mitral, muscles, ovale, pad, pericardium, pressure, prolapse, prosthesis, regurgitation, ring, root, septum, shortening, sinus, space, stenosis, structure, tamponade, thicknesses, thrombus, tricuspid, valve, vegetation, velocities, velocity, ventricle, ventricular, wall motion.
Footnotes
Ethical considerations
The research has been designated by IRB at NewYork-Presbyterian Hospital as Not Human Subject Research. The Protocol Number is 20-10022833.
Code for data construction and model evaluation is available at https://github.com/bionlplab/echo_summarization.
We set threshold t = 0.85 in this study based on the performance on the validation set.
Specific concepts are shown in Appendix A.
References
- Adams Griffin, Alsentzer Emily, Ketenci Mert, Zucker Jason, and Noémie Elhadad. 2021. What’s in a Summary? Laying the Groundwork for Advances in Hospital-Course Summarization. In Proceedings of the 2021 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, pages 4794–4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afantenos Stergos, Karkaletsis Vangelis, and Stamatopoulos Panagiotis. 2005. Summarization from medical documents: a survey. Artificial Intelligence in Medicine, 33(2):157–177. [DOI] [PubMed] [Google Scholar]
- Alsharqi M, Woodward WJ, Mumith JA, Markham DC, Upton R, and Leeson P. 2018. Artificial intelligence and echocardiography. Echo Research and Practice, pages R115–R125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abacha Asma Ben, Mrabet Yassine, Zhang Yuhao, Shivade Chaitanya, Langlotz Curtis, and Demner-Fushman Dina. 2021. Overview of the MEDIQA 2021 Shared Task on Summarization in the Medical Domain. In Proceedings of the 20th Workshop on Biomedical Language Processing, pages 74–85, Online. Association for Computational Linguistics. [Google Scholar]
- Chambers Nathanael, Cer Daniel, Grenager Trond, Hall David, Kiddon Chloe, MacCartney Bill, De Marneffe Marie-Catherine, Ramage Daniel, Yeh Eric, and Manning Christopher D.. 2007. Learning alignments and leveraging natural logic. In Proceedings of the ACL-PASCAL Workshop on Textual Entailment and Paraphrasing, pages 165–170. [Google Scholar]
- de Marneffe Marie-Catherine, Manning Christopher D., Nivre Joakim, and Zeman Daniel. 2021. Universal Dependencies. Computational Linguistics, pages 1–54. [Google Scholar]
- Denny Joshua Charles, Miller Randolph A., Johnson Kevin B., and Spickard Anderson. 2008. Development and evaluation of a clinical note section header terminology. AMIA … Annual Symposium proceedings. AMIA Symposium, pages 156–60. [PMC free article] [PubMed] [Google Scholar]
- Devlin Jacob, Chang Ming-Wei, Lee Kenton, and Toutanova Kristina. 2019. BERT: pre-training of deep bidirectional transformers for language understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers), pages 4171–4186, Minneapolis, Minnesota. Association for Computational Linguistics. [Google Scholar]
- Durmus Esin, He He, and Diab Mona. 2020. FEQA: A question answering evaluation framework for faithfulness assessment in abstractive summarization. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics. Association for Computational Linguistics. [Google Scholar]
- Erkan Günes and Radev Dragomir R.. 2004. Lexrank: Graph-based lexical centrality as salience in text summarization. Journal of artificial intelligence research, 22:457–479. [Google Scholar]
- Falke Tobias, Ribeiro Leonardo F. R., Utama Prasetya Ajie, Dagan Ido, and Gurevych Iryna. 2019. Ranking generated summaries by correctness: An interesting but challenging application for natural language inference. In Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics. Association for Computational Linguistics. [Google Scholar]
- Gardent Claire, Shimorina Anastasia, Narayan Shashi, and Perez-Beltrachini Laura. 2017. The WebNLG challenge: Generating text from RDF data. In Proceedings of the 10th International Conference on Natural Language Generation, pages 124–133, Santiago de Compostela, Spain. Association for Computational Linguistics. [Google Scholar]
- Gardin Julius M., Adams David B., Douglas Pamela S., Feigenbaum Harvey, Forst David H., Fraser Alan G., Grayburn Paul A., Katz Alan S., Keller Andrew M., Kerber Richard E., Khandheria Bijoy K., Klein Allan L., Lang Roberto M., Pierard Luc A., Quinones Miguel A., Schnittger Ingela, and American Society of Echocardiography. 2002. Recommendations for a standardized report for adult transthoracic echocardiography: A report from the American Society of Echocardiography’s Nomenclature and Standards Committee and Task Force for a Standardized Echocardiography Report. Journal of the American Society of Echocardiography: Official Publication of the American Society of Echocardiography, 15(3):275–290. [DOI] [PubMed] [Google Scholar]
- Gatt Albert and Krahmer Emiel. 2018. Survey of the state of the art in natural language generation: Core tasks, applications and evaluation. J. Artif. Int. Res, 61(1):65–170. [Google Scholar]
- Gehrmann Sebastian, Dai Falcon, Elder Henry, and Rush Alexander. 2018. End-to-end content and plan selection for data-to-text generation. In Proceedings of the 11th International Conference on Natural Language Generation, pages 46–56, Tilburg University, The Netherlands. Association for Computational Linguistics. [Google Scholar]
- Goodrich Ben, Rao Vinay, Liu Peter J., and Saleh Mohammad. 2019. Assessing the factual accuracy of generated text. In Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, KDD ‘19, page 166–175, New York, NY, USA. Association for Computing Machinery. [Google Scholar]
- Goyal Tanya and Durrett Greg. 2021. Annotating and Modeling Fine-grained Factuality in Summarization. In Proceedings of the 2021 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, pages 1449–1462, Online. Association for Computational Linguistics. [Google Scholar]
- Harkous Hamza, Groves Isabel, and Saffari Amir. 2020. Have your text and use it too! end-to-end neural data-to-text generation with semantic fidelity. In Proceedings of the 28th International Conference on Computational Linguistics, pages 2410–2424, Barcelona, Spain (Online). International Committee on Computational Linguistics. [Google Scholar]
- Johnson Alistair E. W., Pollard Tom J., Shen Lu, Lehman LiWei H., Feng Mengling, Ghassemi Mohammad, Moody Benjamin, Szolovits Peter, Celi Leo Anthony, and Mark Roger G.. 2016. MIMIC-III, a freely accessible critical care database. Scientific data, 3:160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryscinski Wojciech, McCann Bryan, Xiong Caiming, and Socher Richard. 2020. Evaluating the factual consistency of abstractive text summarization. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP). Association for Computational Linguistics. [Google Scholar]
- Lewis Mike, Liu Yinhan, Goyal Naman, Ghazvininejad Marjan, Mohamed Abdelrahman, Levy Omer, Stoyanov Veselin, and Zettlemoyer Luke. 2020. BART: Denoising Sequence-to-Sequence Pretraining for Natural Language Generation, Translation, and Comprehension. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, pages 7871–7880, Online. Association for Computational Linguistics. [Google Scholar]
- Lin Chin-Yew. 2004. ROUGE: A package for automatic evaluation of summaries. In Text Summarization Branches out: Proceedings of the ACL-04 Workshop, volume 8, pages 1–8. Barcelona, Spain. [Google Scholar]
- Mitchell Carol, Rahko Peter S., Blauwet Lori A., Canaday Barry, Finstuen Joshua A., Foster Michael C., Horton Kenneth, Ogunyankin Kofo O., Palma Richard A., and Velazquez Eric J.. 2019. Guidelines for Performing a Comprehensive Transthoracic Echocardiographic Examination in Adults: Recommendations from the American Society of Echocardiography. Journal of the American Society of Echocardiography: Official Publication of the American Society of Echocardiography, 32(1):1–64. [DOI] [PubMed] [Google Scholar]
- Mohan Sunil and Li Donghui. 2018. MedMentions: A Large Biomedical Corpus Annotated with UMLS Concepts. In Automated Knowledge Base Construction (AKBC). [Google Scholar]
- Mullenbach James, Pruksachatkun Yada, Adler Sean, Seale Jennifer, Swartz Jordan, McKelvey Greg, Dai Hui, Yang Yi, and Sontag David. 2021. CLIP: A Dataset for Extracting Action Items for Physicians from Hospital Discharge Notes. In Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing (Volume 1: Long Papers), pages 1365–1378, Online. Association for Computational Linguistics. [Google Scholar]
- Neumann Mark, King Daniel, Beltagy Iz, and Ammar Waleed. 2019. ScispaCy: Fast and Robust Models for Biomedical Natural Language Processing. In Proceedings of the 18th BioNLP Workshop and Shared Task, pages 319–327, Florence, Italy. Association for Computational Linguistics. [Google Scholar]
- Pauws Steffen, Gatt Albert, Krahmer Emiel, and Reiter Ehud. 2019. Making Effective Use of Healthcare Data Using Data-to-Text Technology, pages 119–145. Springer International. [Google Scholar]
- Pivovarov Rimma and Elhadad Noémie. 2015. Automated methods for the summarization of electronic health records. Journal of the American Medical Informatics Association: JAMIA, 22(5):938–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter Ehud and Dale Robert. 2000. Building Natural Language Generation Systems. Cambridge University Press. [Google Scholar]
- See Abigail, Liu Peter J., and Manning Christopher D.. 2017. Get To The Point: Summarization with Pointer-Generator Networks. In Proceedings of the 55th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), pages 1073–1083, Vancouver, Canada. Association for Computational Linguistics. [Google Scholar]
- Wang Alex, Cho Kyunghyun, and Lewis Mike. 2020. Asking and answering questions to evaluate the factual consistency of summaries. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics. Association for Computational Linguistics. [Google Scholar]
- Wiseman Sam, Shieber Stuart, and Rush Alexander. 2017. Challenges in data-to-document generation. In Proceedings of the 2017 Conference on Empirical Methods in Natural Language Processing, pages 2253–2263, Copenhagen, Denmark. Association for Computational Linguistics. [Google Scholar]
- Xiong Y, Tang B, Chen Q, Wang X, and Yan J. 2019. A study on automatic generation of chinese discharge summary. In 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pages 1681–1687. [Google Scholar]
- Zhang Tianyi, Kishore Varsha, Wu Felix, Weinberger Kilian Q., and Artzi Yoav. 2019. BERTScore: Evaluating Text Generation with BERT. In International Conference on Learning Representations. [Google Scholar]
- Zhang Yuhao, Yi Ding Daisy, Qian Tianpei, Manning Christopher D., and Langlotz Curtis P.. 2018. Learning to summarize radiology findings. In EMNLP 2018 workshop on health text mining and information analysis. [Google Scholar]
- Zhang Yuhao, Merck Derek, Tsai Emily, Manning Christopher D., and Langlotz Curtis. 2020. Optimizing the factual correctness of a summary: A study of summarizing radiology reports. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics. Association for Computational Linguistics. [Google Scholar]