Abstract
The human brain vasculature is of vast medical importance: its dysfunction causes disability and death1, and the specialized structure it forms—the blood-brain barrier—impedes treatment of nearly all brain disorders2,3. Yet, we have no molecular map of the human brain vasculature. Here, we develop Vessel Isolation and Nuclei Extraction for Sequencing (VINE-seq) to profile the major human brain vascular and perivascular cell types through 143,793 single-nucleus transcriptomes from 25 hippocampus and cortex samples of 17 cognitively normal and Alzheimer’s disease (AD) subjects. We identify brain region- and species-enriched genes and pathways. We reveal molecular principles of human arteriovenous organization, recapitulating a gradual endothelial and punctuated mural cell continuum. We discover two subtypes of human pericytes, marked by solute transport and extracellular matrix (ECM) organization; and define perivascular versus meningeal fibroblast specialization. In AD, we observe selective vulnerability of ECM-maintaining pericytes and gene expression patterns implicating dysregulated blood flow. With an expanded survey of brain cell types, we find 30 of the top 45 AD GWAS genes expressed in the human brain vasculature, confirmed by immunostaining. Vascular GWAS genes map to endothelial protein transport, adaptive immune, and ECM pathways. Many are microglia-specific in mice, suggesting a partial evolutionary transfer of AD risk. Our work unravels the molecular basis of the human brain vasculature, informing our understanding of overall brain health, disease, and therapy.
Brain health depends on brain vascular health. The brain is one of the most highly perfused organs in the body, necessary to meet its unique metabolic needs4. Brain vascular dysfunction contributes to stroke1, congenital neurological disorders5 and age-related neurodegenerative disease5,6. The brain vasculature forms a special structure, the blood-brain barrier (BBB), that mediates selective and hemodynamically responsive movement of molecules between the blood and brain2,3. While necessary for optimal neuronal function4, the BBB frustrates the pharmacological treatment of nearly all brain disorders7,8, and extensive efforts are underway to discover targets on the human BBB to enhance drug delivery.
Specialized brain vascular properties arise from a complex community of interacting cell types9–11: endothelial cells, adjacent mural smooth muscle cells and pericytes, perivascular immune cells, and surrounding astrocytes that differ across brain regions and vary along an arteriovenous gradient12. Heterogeneity along this gradient produces functionally segmented circulatory, metabolic, and permeability properties necessary for brain health3,5.
Pioneering work has profiled the mouse brain vasculature12–15, but it remains unclear how conserved these findings are in humans. Single-nucleus studies have elucidated the cellular heterogeneity of the human brain in health and disease16–19. Yet, though vascular cell density20,21 (70,000 cells/mm3) approaches total glia density20, prior studies, to our knowledge, have depleted human brain vascular cells for unknown reasons. While human brain microvessels can be isolated for bulk assays, no method exists to extract nuclei from microvessels for analysis at single-cell resolution.
Given the scientific, medical, and pharmacological importance of the human brain vasculature, we set out to create a first single-cell atlas of the human brain vasculature of the hippocampus and cortex from control no cognitive impairment (NCI) and Alzheimer’s disease (AD) individuals.
Cells of the human brain vasculature
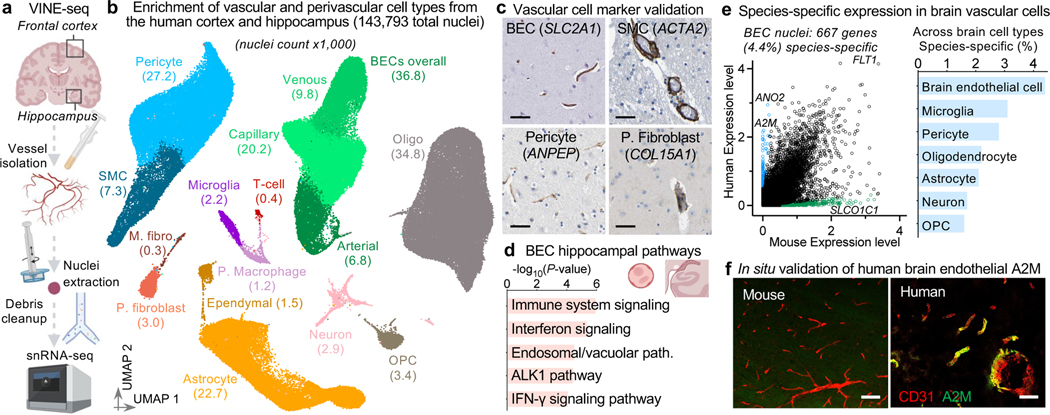
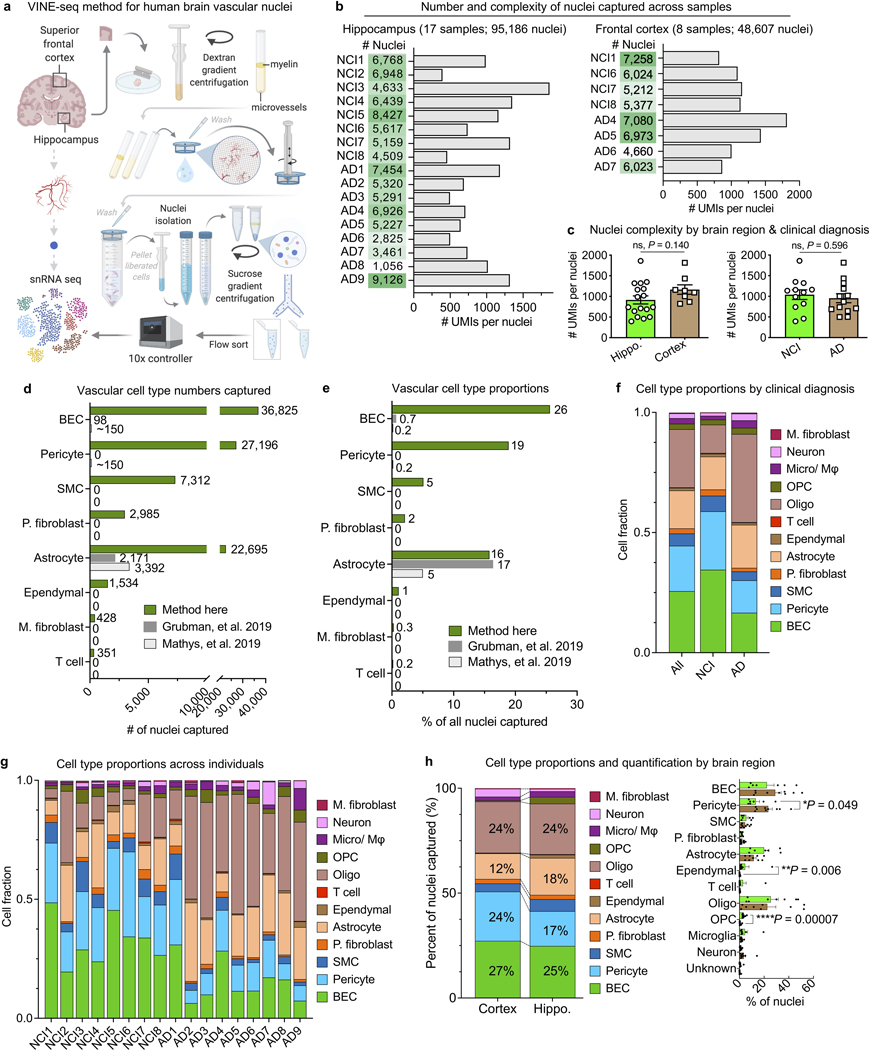
Extracting nuclei from brain microvessels is a challenge because the method must dissociate the vessel basement membrane to release nuclei without damaging the nuclei themselves. We systematically explored enzymatic, chemical, and physical approaches. Most resulted in damaged nuclei devoid of RNA reads. We found success adapting a gentle protocol for splenocyte isolation (Methods)—and combined it with extensive sucrose- and FACS-based cleanup to ensure high-quality data (Fig. 1a, Extended Data Fig. 1). We used our new method, VINE-seq (Vessel Isolation and Nuclei Extraction for Sequencing), to process 25 samples: the hippocampus of 9 AD and 8 age- and sex-matched NCI individuals, as well as the superior frontal cortex from a subset of 8 individuals (4 per group, Supplemental Table 1). Samples included a range of APOE genotypes, and NCI samples display no significant vascular pathologies in the studied hippocampus and cortex tissue (Supplemental Table 1, Extended Data Fig. 2a). After quality-control (Methods), we obtained 143,793 single-nucleus transcriptomes. Visualization in uniform manifold approximation and projection (UMAP) space separated nuclei into 15 major cell types (Fig. 1b), including all known vascular and perivascular cell types: endothelial cells (arterial, capillary, venous), smooth muscle cells, pericytes, astrocytes, macrophages, T cells, and both perivascular and meningeal fibroblasts. The number of cerebrovascular nuclei captured here exceed those in the literature by several hundred-fold (Supplemental Table 2, Extended Data Fig. 2b-c). Vascular cell type markers were validated in situ (Fig. 1c). Data can be browsed at https://twc-stanford.shinyapps.io/human_bbb.
Figure 1. Cells of the human brain vasculature.
a, VINE-seq method to enrich vascular nuclei from postmortem human brain samples.
b, Uniform Manifold Approximation and Projection (UMAP) of 143,793 nuclei from 25 human hippocampus and superior frontal cortex samples across 17 individuals, colored by cell type and labeled with nuclei numbers.
c, Immunohistochemical validation of cell type-specific gene markers. Scale bar = 50 microns. Image credit: Human Protein Atlas (http://www.proteinatlas.org)25.
d, Enriched biological pathways in BECs from the hippocampus compared to the superior frontal cortex, in not cognitive impairment (NCI) individuals (P value < 0.05, cumulative hypergeometric test).
e, Scatter plot (left) depicting mRNA expression levels (logCPM) of mouse and human genes with one-to-one orthologs in BECs. Divergently expressed genes are colored (>10-fold difference, minimum 0.5 log2CPM expression). Proportion of each brain cell type’s transcriptome that is specific to human versus mouse (same thresholds, right).
f, Immunohistochemical validation of A2M protein specifically in the human but not mouse vasculature. Scale bar = 50 microns.
VINE-seq captured three human brain immune cell types: microglia, macrophages, and T cells. Over 300 genes differentiated brain border macrophages and microglia, yielding new markers for human studies (Extended Data Fig. 2d-f). Analysis of canonical markers indicated our capture of brain memory CD8 cytotoxic and CD4 T cells (Extended Data Fig. 2g). Brain regional transcriptome differences were evident. Astrocyte transcriptional identity was the most influenced by brain region (Extended Data Fig. 2h-k). Moreover, hippocampal endothelial cells demonstrated greater baseline inflammation, such as IFN-γ signaling, than those in cortex (Fig. 1d). Such inflammatory signaling may inhibit hippocampal neurogenesis, and together with accelerated hippocampal pericyte loss (Extended Data Fig. 1h), provides a molecular hypothesis for hippocampal vasculature vulnerability and dysfunction22.
We next compared nuclei transcriptomes between human and mouse endothelial cells and pericytes. Using a strict cutoff (>10x difference, logCPM>0.5, Supplemental Table 4), we found hundreds of species-enriched genes in brain endothelial cells (BECs) and pericytes (Fig. 1e). Understanding species differences in neurons and glia has been subject to intense study. Together with microglia23, we find that BECs and pericytes exhibit the greatest transcriptional divergence (Fig. 1e, Extended Data Fig. 3). Several vascular solute transporters (e.g., GABA transporter SLC6A12) vary by species, suggesting differences in brain metabolism (Extended Data Fig. 3g). Several genes of disease and pharmacological importance also vary (Extended Data Fig. 3-4). Together, the VINE-seq method opens the human brain vasculature for molecular study and provides an important data resource for interrogating its diverse cell types.
Endothelial and mural cell organization
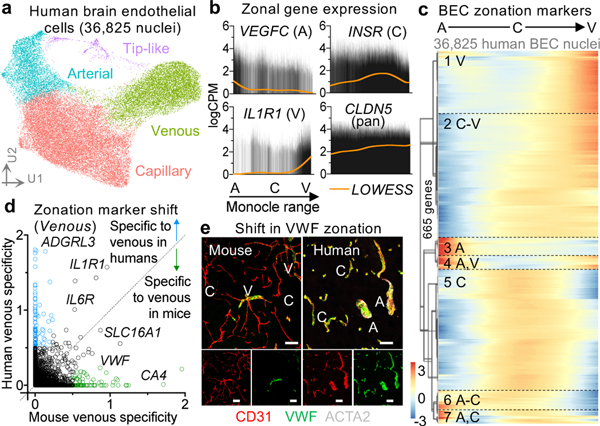
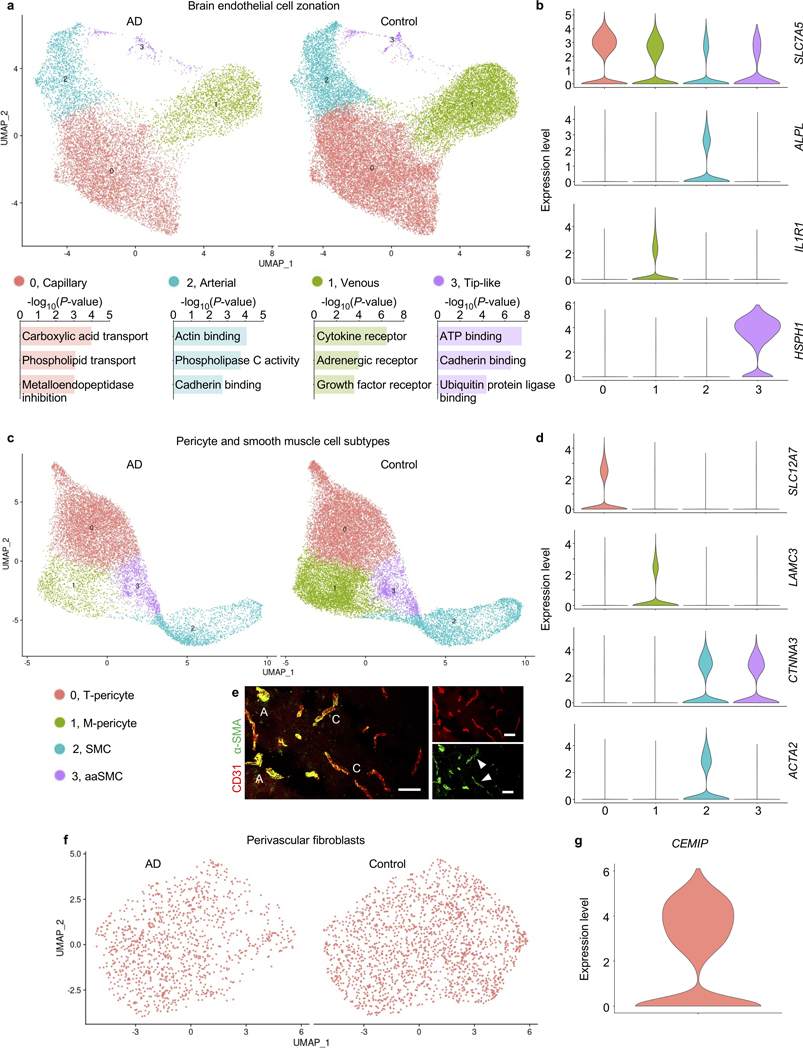
With our large-scale capture of vascular nuclei (>30x mouse12, >200x human16,17 prior studies), we sought to comprehensively characterize the molecular basis of endothelial and mural cell organization along the arteriovenous axis of the human brain vasculature. Changes along this axis are referred to as zonation12,15. Beginning with BECs, we clustered 36,825 nuclei into known arterial, capillary, and venous segments (Fig. 2a, Extended Data Fig. 5). Clusters were defined by established zonation markers, such as arterial VEGFC and ALPL; capillary MFSD2A and SLC7A5; and venous IL1R1 and NR2F212,15. While capillaries represent most (∼90%) of the endothelium, VINE-seq captured rarer arterial (at 19%) and venous (27%) BECs, because of better strainer retention or nuclei liberation. We noticed a small cluster (571 nuclei, ∼0.1%) outside conventional zonation. This cluster expressed genes characteristic of ‘tip’ cells (e.g., PLAUR and LAMB1) as well as ‘proteostatic’ heat shock proteins.
Figure 2. Organizing principles of human brain endothelial cells.
a, UMAP of 36,825 human brain endothelial cell (BEC) nuclei, colored by zonation.
b, Zonal expression of transcripts across human BECs ordered by Monocle pseudotime. LOWESS regression line (orange) and density of black lines (counts) correspond with expression levels. A = arterial, C = capillary, and V = venous.
c, Heatmap of zonation-dependent gene expression in human BECs.
d, Scatter plot depicting the specificity of transcripts for venous BECs in mice12 versus humans. Venous specificity score = avg(logFC(vein/cap), logFC(vein/art)). For example, VWF is predicted to be more specific to venous BECs in mice than it is in humans. See Extended Data Fig. 9 for arterial and capillary specificity plots.
e, Immunohistochemical validation of VWF specificity to venous BECs in mice but not in humans. Scale bar = 50 microns.
We next ordered endothelial nuclei along a one-dimensional pseudotime24 axis to recapitulate the anatomical arteriovenous axis. Arterial and venous markers peaked at opposite ends, with capillary markers in between (Fig. 2b). We used the 665 most significantly variable cluster genes to unbiasedly order endothelial nuclei and observed seven gradually changing gene expression patterns, representing arterial, capillary, or venous markers, and combinations thereof (Fig. 2c). We confirmed that the Monocle range represented a cell order matching anatomical arteriovenous zonation via immunostaining25 (Extended Data Fig. 6a). Our patterns recapitulate the gradual/seamless zonation continuum described in mice—but interestingly, this similar overall continuum arises from significantly different component zonation markers (Supplemental Table 2, 5).
We thus wondered whether established zonation markers in mice would be conserved in humans. We calculated a score (Methods) measuring each gene’s specificity to a given zonation (e.g., arterial, capillary, venous). Indeed, we observed in each vessel segment a significant number of markers that lost their zonation specificity between species (Fig. 2d, Extended Data Fig. 6). For example, the blood clotting gene von Willebrand factor (VWF) is largely expressed in mouse venous endothelial cells12,15. However, VWF is expressed throughout the human cerebrovasculature, even in small diameter capillaries (Fig. 2d-e). VWF abundance predisposes for ischemic stroke26, raising implications for translational studies of stroke.
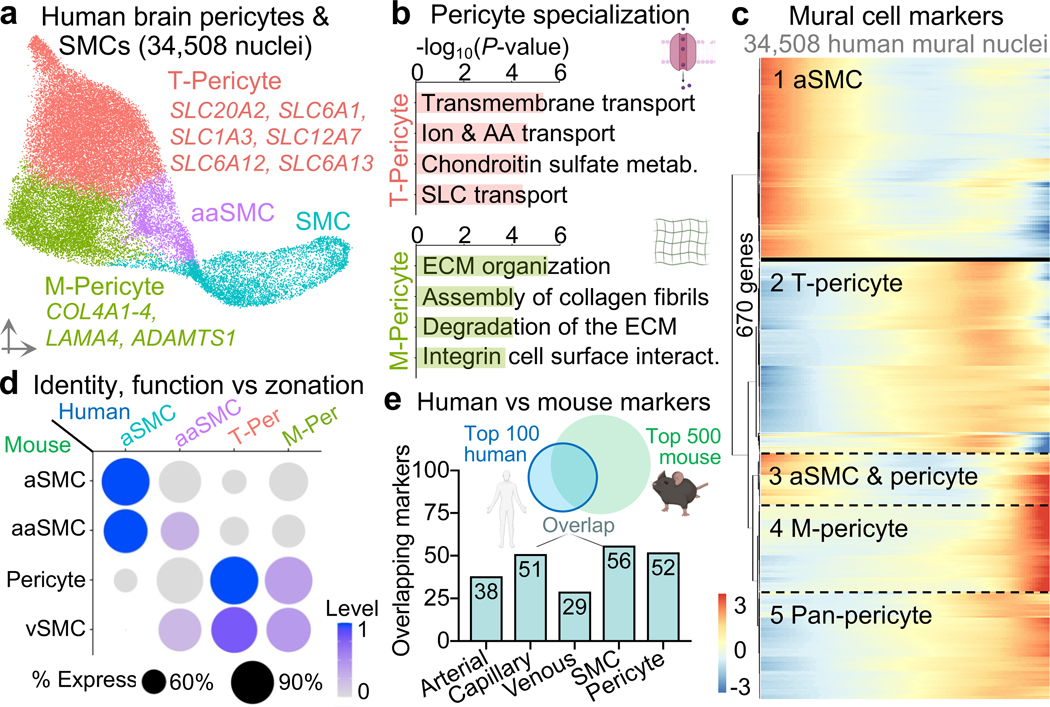
Next with mural cells, we clustered 34,508 nuclei into arterial smooth muscle cells (aSMCs), arteriolar SMCs (aaSMCs), and interestingly, two subclusters of pericytes (Fig. 3a-b). One pericyte subcluster enriched for small molecule transmembrane transport, which we call T-pericytes (for transport); while the other for extracellular matrix (ECM) organization, M-pericytes (for matrix). Thus, function rather than anatomical location may be the major driver of human pericyte transcriptional identity. M-pericytes may thus contribute to small vessel diseases like CADASIL, CARASIL, and Collagen IV deficiencies for which perturbations in the cerebrovascular ECM cause disease27. Moreover, human T-pericytes—but not mouse pericytes—express transporters like the GABA transporter SLC6A1 (involved in epilepsy) and glutamate transporter SLC1A3 (Extended Data Fig. 3g). Because recent mouse pericyte datasets have cautioned against endothelial contamination, we assessed and found no such contamination in our human pericyte nuclei (Extended Data Fig. 2c).
Figure 3. Organizing principles of human brain mural cells.
a, UMAP of 34,508 human pericyte and smooth muscle cell nuclei, colored by cell subtype. aSMC = arterial smooth muscle cell (aSMC), aaSMC = arteriole SMCs, T-Pericyte = solute transport pericytes, and M-Pericyte = Extracellular matrix regulating pericytes.
b, Enriched biological pathways in T- and M-pericytes compared to remaining SMC and pericyte populations (P value < 0.05, cumulative hypergeometric test).
c, Heatmap of gene expression in human SMCs and pericytes. Solid line delineating aaSMC/aSMCs from pericytes reflects an abrupt transcriptomic transition. Note that unlike BECs, mural cell ordering does not reflect anatomical arteriovenous ordering.
d, Mapping expression of mouse mural cell markers onto human mural cell types. The top 500 mouse markers were aggregated into four distinct modules (‘aSMCs’, ‘aaSMCs’, capillary ‘pericytes’, venous smooth muscle cells ‘vSMCs’)12 and their expression assessed in the four transcriptionally distinct human mural cell types.
e, Overlap between the top 100 human endothelial and mural cell subtype markers and those identified in mice. A more lenient set of 500 mouse markers12 was used for comparison for robust results. Note that species-conservation of a cell type marker depends on species-specific changes in the given cell type and changes among remaining cell types.
To study mural cell zonation, we used the 670 most significantly variable and robustly expressed cluster genes to order all mural cell nuclei and observed the expected order of aSMC markers on one end (e.g., ACTA2, TAGLN), followed by aaSMC (e.g., SLIT3, CTNNA3); and pericyte markers on the other end (e.g., ABCC9, PTN), confirmed in situ (Fig. 3c, Extended Data Fig. 6d). Recapitulating patterns described in mice12—and as opposed to the gradual zonation pattern in BECs—, human mural cells exhibited an abrupt gene expression transition between SMCs and pericytes (Fig. 3c). To determine whether T- and M-pericyte ordering (Fig. 3c) reflects anatomical arteriovenous order, we mapped them to mouse mural cells markers12 and found no differences in their expression of mouse capillary ‘Pericyte’ versus venous mural cell ‘vSMC’ markers (Fig. 3d). This suggests that T- and M-pericytes do not segregate by arteriovenous segment. Immunostaining confirmed that T- and M-pericyte markers are each present across both small and large diameter brain vessels (Extended Data Fig. 6d).
Together, these data suggest that human pericytes transcriptionally identify more by functional specialization than by arteriovenous location; and that while aSMCs and aaSMCs reside on arterial and arteriolar vessels respectively, T- and M-pericytes intermix along the capillary and venous vasculature. As with BECs, we find that only a minority of top mouse SMC and pericyte markers retain their predictive value in humans (Fig 3e, Extended Data Fig. 3b, 6d). Because proteins encoded by zonation marker genes perform a variety of important functions at defined arteriovenous locations, species-specific endothelial and mural cell differences likely reflect fundamental differences in brain vascular properties that can now be studied.
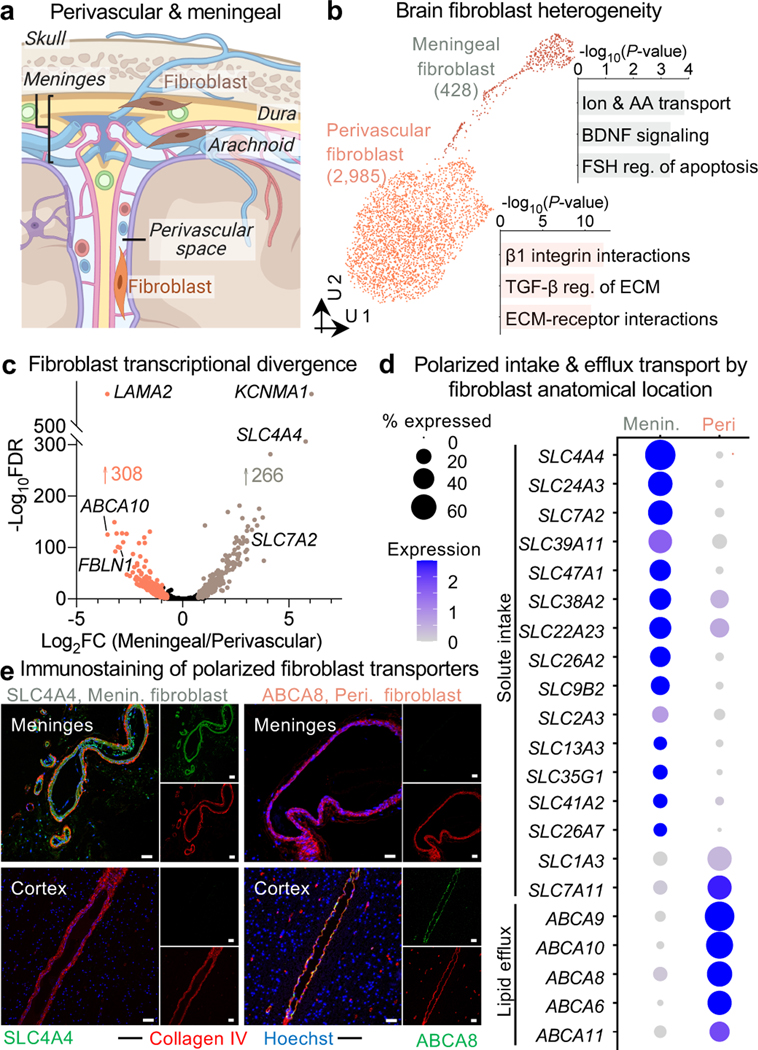
Brain fibroblasts molecularly defined
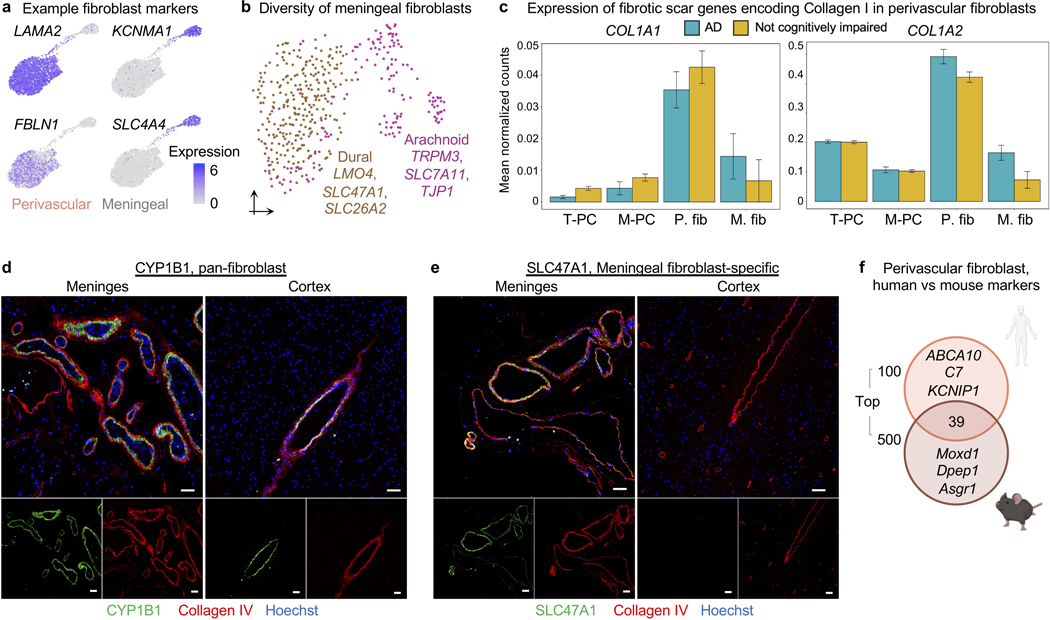
Using annotations from recent mouse studies12,28, we noticed fibroblasts from both the human brain vascular and meningeal barriers (Fig. 4a). Cooperating with the vascular BBB, the recently (re)discovered meningeal lymphatics plays important roles in waste clearance and neuroimmune surveillance29,30. Fibroblasts transcriptionally segregated according to anatomical location: vascular versus meningeal (Fig. 4a-b), but also separated according to meningeal layers (Extended Data Fig. 7a-b). Pathway enrichment analysis of marker genes revealed specialized fibroblast functions (Fig. 4b): perivascular fibroblasts showed enriched expression for ECM structural components or its modifiers/receptors (e.g., “TGF-β regulation of ECM”), while meningeal fibroblasts enriched for solute transporters. Fibrotic scars arise from the pathological deposition of Collagen I (COL1)-rich ECM. Consistent with elegant mouse lineage tracing studies31, our work indicates that human perivascular fibroblasts are the main producers of COL1 in the brain and thus likely form fibrotic scars after brain injury (Extended Data Fig. 7c).
Figure 4. Molecular definitions for brain perivascular and meningeal fibroblasts.
a, Anatomical reference of the human meninges (dura and arachnoid) and perivascular space, each with a resident fibroblast population.
b, UMAP of 2,985 human perivascular fibroblast-like nuclei and 428 meningeal fibroblast nuclei. Enriched biological pathways derived from respective fibroblast cell type markers (Supplemental Table 2, P value < 0.05, cumulative hypergeometric test).
c, Differentially expressed genes between perivascular (left, orange) and meningeal (right, brown) fibroblasts (MAST, Benjamini Hochberg correction; FDR < 0.01 and logFC>0.5 [log2FC>0.72] to be colored significant).
d, Expression of all differentially expressed (from (c)) SLC and ABC family members across perivascular and meningeal fibroblasts.
e, Immunostaining validation of polarized meningeal and perivascular fibroblast transporter expression: the meningeal fibroblast-specific influx pump SLC4A4 (left, green) stains selectively in the meninges but not cortical vasculature (red); and vice versa for the perivascular-specific efflux pump ABCA8 (right, green). Scale = 50 μM.
Closer comparison of differentially expressed genes (DEGs) between fibroblast populations revealed a polarization of solute influx and efflux pumps: meningeal fibroblasts specifically expressed SLC influx solute transporters, while perivascular fibroblasts exclusively expressed ABC efflux pumps (Fig. 4d). We confirmed this polarized expression in situ (Fig. 4e, Extended Data Fig. 7d-e). Perivascular fibroblasts reside in the Virchow-Robin space, and thus like meningeal fibroblasts, bathe in the cerebrospinal fluid (CSF). This cooperative circuit of polarized transporters suggests fibroblast regulation of solute exchange between the brain and CSF32. Finally, as in endothelial and mural cells, perivascular fibroblast-like cell markers varied by species (Extended Data Fig. 7f). Together, these data provide a first characterization of human brain fibroblast diversity, revealing the molecular basis of their anatomical specialization and a cooperative circuit for CSF solute exchange.
Vascular cell type perturbations in AD
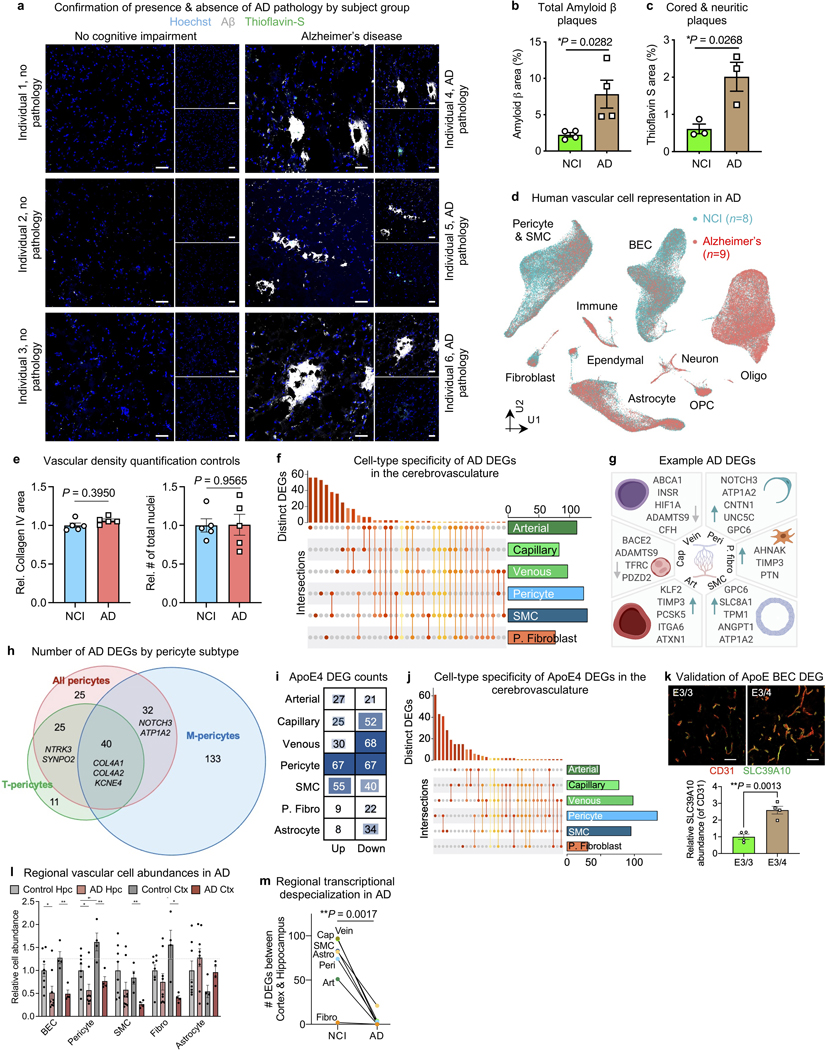
AD is a neurodegenerative disorder with progressive impairment of cognitive function leading to dementia. Impairment arises from complex perturbations in cell composition and gene expression. We thus sought to profile changes in the AD human brain vasculature at single-cell resolution. We defined our patient groups by clinical diagnosis and confirmed the presence of β-amyloid plaques in the hippocampus and cortex (Extended Data Fig. 8a-c).
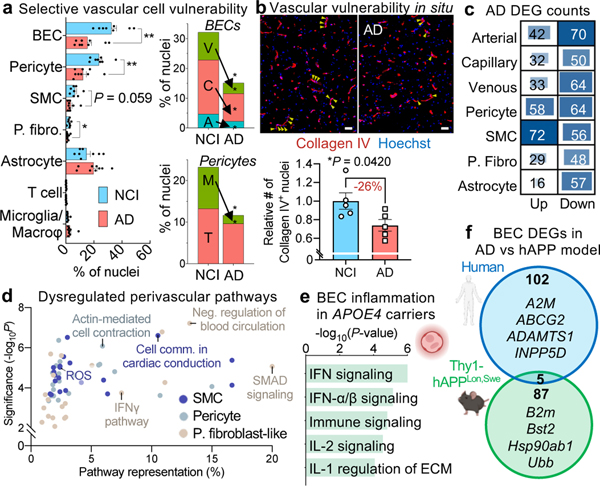
Recent studies have identified context-dependent, disease associated glial subpopulations16,17,33. We did not observe new vascular cell subclusters emerge in AD (Fig. 5a, Extended Data Fig. 8d). In contrast, we found a strong loss of brain vascular nuclei—across endothelial, SMC, pericyte, and fibroblast-like cells (Fig. 5a-b). Thus, beyond focal cerebrovascular damage, vascular loss may be widespread across the arteriovenous axis. Among pericytes, M-pericytes involved in ECM organization exhibited selective vulnerability (Fig. 5b), providing a molecular hypothesis for structural BBB breakdown reported in AD22. Yet, FACS and snRNA-seq approaches may not always yield reliable cell quantifications. Thus, we stained hippocampal tissue to quantify the number of Hoechst+ vascular cells within Collagen IV+ cerebrovasculature. While we found no significant differences in the area of Collagen IV+ vessels or total number of Hoechst+ nuclei, we found a significant decrease with AD in the number of Hoechst+ nuclei within Collagen IV+ vessels, consistent with reports of “string vessels” in disease34 (Fig. 5b, Extended Data Fig. 8d-e). In short, we find selective vulnerability of specific vascular cell subpopulations in AD.
Figure 5. Vascular cell-type specific perturbations in Alzheimer’s disease.
a, Proportion of cell types captured in Alzheimer’s disease (AD) and no cognitive impairment individuals (NCI) (left). Proportion of BEC and pericyte subpopulations in AD and NCI (right) (n = 8 NCI, n = 9 AD, two-sided t-test; mean +/− s.e.m.). **BEC P = 0.002, **Pericyte P = 0.003, *P. fibroblast P = 0.0461.
b, Immunohistochemical validation of a loss of vascular cell density in AD (number of Hoechst+ nuclei within Collagen IV+ vasculature). Scale bar = 50 microns (n = 5 NCI and AD, nested two-sided t-test; mean +/− s.e.m.). Yellow arrows denote example vascular nuclei.
c, Differentially expressed gene (DEG) counts for each cell type in AD. The intensity of the blue color and the size of the squares are proportional to entry values.
d, Enriched biological pathways from AD differentially expressed genes in pericytes, smooth muscle cells, and perivascular fibroblast-like cells, plotted by Pathway Representation (in a given pathway, what proportion of all members are DEGs) and Significance (-log10P) of pathway enrichment.
e, Enriched biological pathways from genes upregulated in AD APOE4 carriers in capillary and venous endothelial cells (P value < 0.05, cumulative hypergeometric test).
f, Venn diagram comparing DEG BECs in human AD samples compared to those from the Thy1-hAPP T41BLon,Swe amyloidosis mouse model37. Note that only genes with human-mouse orthologs are shown, and that the absolute logFC threshold for calling DEGs in mouse APP BECs was lowered to 0.15 (by half) to ensure claims of limited overlap with human BECs were robust.
We next examined cell type-specific gene expression changes in AD (Methods). We identified 463 unique differentially expressed genes using more stringent thresholds (DEGs, Methods, Fig. 5c, Supplemental Table 6). Overall, mural cells exhibited the strongest changes, and other cell types exhibited a signature of gene repression: 61–78% of DEGs were downregulated (Fig. 5c). DEGs were robustly detected across different levels of expression (Supplemental Table 6). Most DEGs were cell type- and zonation-specific (Extended Data Fig. 8f), suggesting a heterogenous response to AD pathology across the vasculature. Intriguingly, several DEGs are risk genes implicated in AD and small vessel disease GWAS studies (Extended Data Fig. 8g-h). At the pathway level, DEGs in mural cells and fibroblasts implicated dysregulated vasoconstriction and compromised blood flow (Fig. 5d). This provides a potential molecular mechanism for cerebral hypoperfusion discernible in MRI of living AD patients35.
APOE4 carriers may exhibit accelerated BBB breakdown before cognitive impairment36. We found dramatic interferon inflammation in the endothelium of APOE4 carriers (Fig. 5e, Extended Data Fig. 8i-k, Supplemental Table 7). Next, we compared AD DEGs in BECs with those in mouse models of AD. Such models have facilitated mechanistic study of β-amyloid pathology, but recent reports describe significant species differences in various cell types, like microglia23. We isolated BECs from 12–14 month old Thy1-hAPPLon,Swe mice37 (and littermate wild-type controls) and processed them for single-cell sequencing. Surprisingly, we observed minimal overlap between human AD and mouse hAPP BEC DEGs (Fig. 5f). Finally, because AD pathology begins and spreads via a strikingly consistent regional pattern, we assessed the impact of AD on brain regional vascular specialization. We found regional differences in cell density and transcriptional profiles largely erased in AD (Extended Data Fig. 8l-m)—suggesting impairments in brain region-specific vascular functions.
Together, these findings show that AD patients exhibit heterogeneous cell type-, zonation-, region-, and species-specific vulnerabilities and perturbations across the brain vasculature that require single-cell approaches to profile.
AD GWAS variants in human BBB cell types
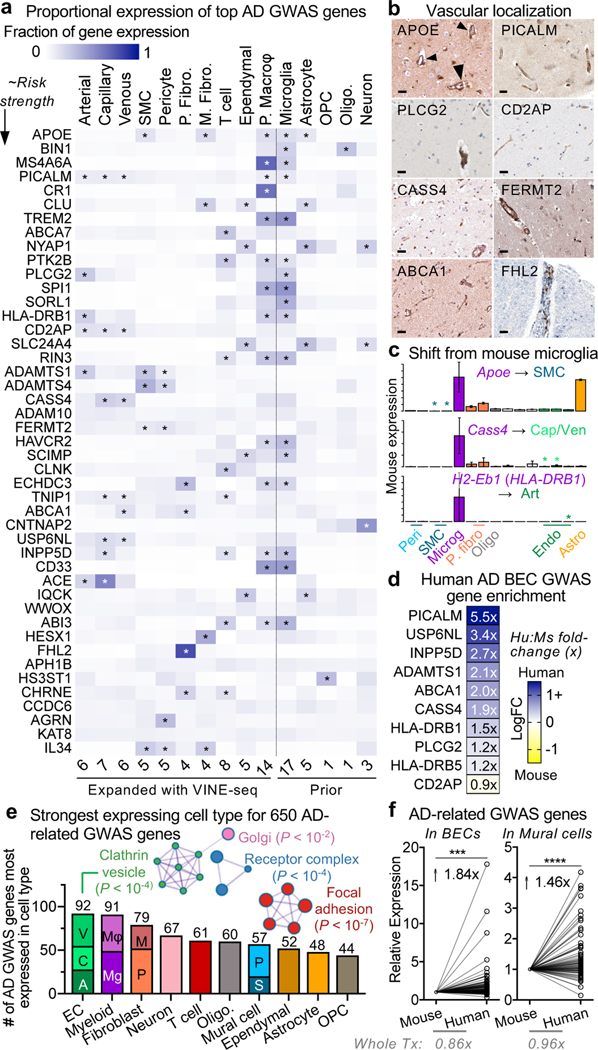
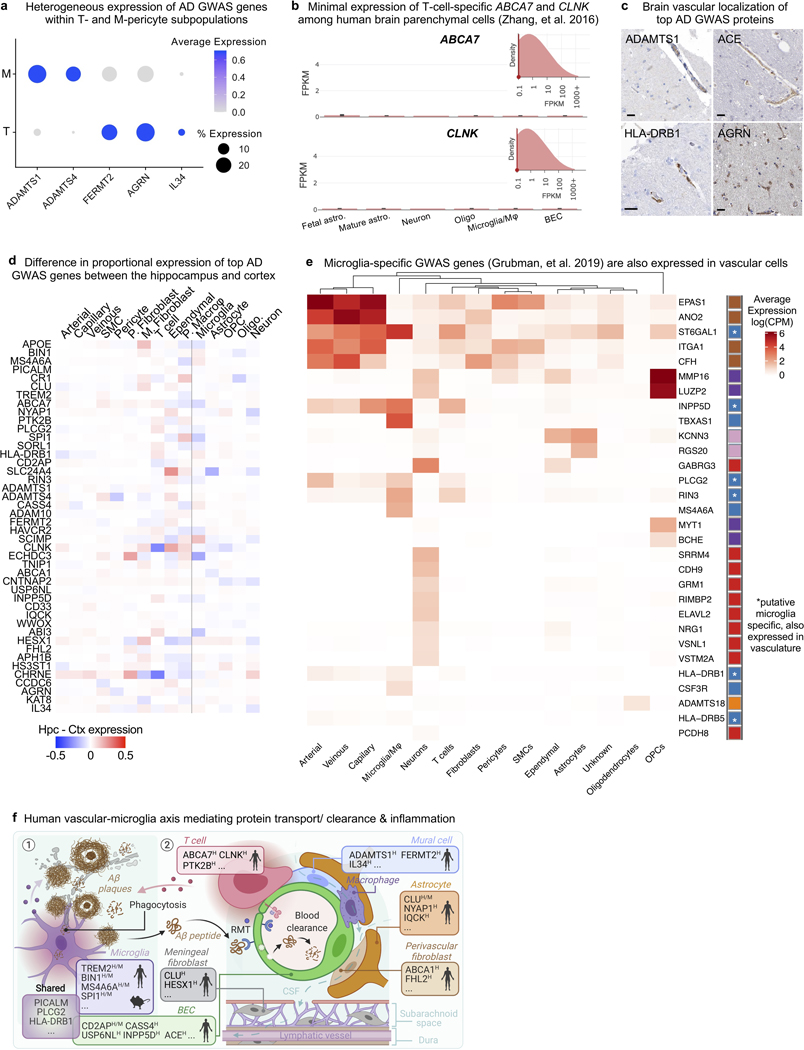
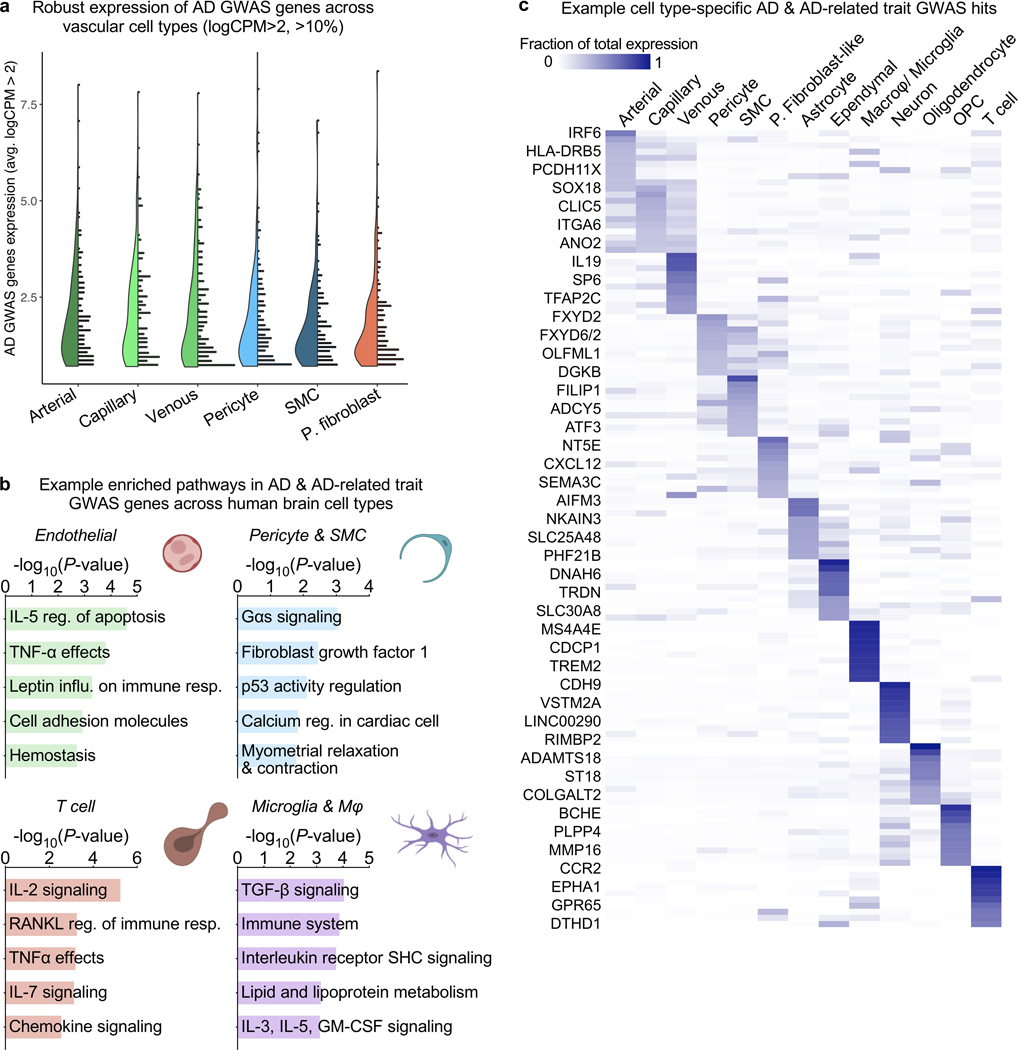
A major goal of biomedical research is to understand how genetic variation contributes to disease. GWAS studies have nominated genes contributing to AD risk, though their cell type contexts were unknown. Recent studies strongly implicate microglia as the major AD GWAS-expressing cell type23,38–41. We wondered, however, whether the unintended depletion of brain vascular cells prematurely dismissed their potential contributions. We curated AD GWAS studies39–41 to order the top 45 risk genes. With our expanded survey of brain cell types, we calculated the cell type proportional expression for each GWAS gene using Expression Weighted Cell Type Enrichment (EWCE)42. We indeed observed among brain parenchymal cells a specific microglial signature for top AD GWAS genes such as TREM2, MS4A6A, CR1, and SPI1 that are now under intense mechanistic study (Fig. 6a, right).
Figure 6. GWAS disease variants are enriched in the human brain vasculature.
a, Proportional expression of the top 45 AD GWAS genes across all major brain cell types. Expression values for a given gene sums to 1 across cell types using the EWCE method42. Genes ordered in approximate risk strength39–41,51. Asterisks denote strongest expressing cell types. Cells to the left of dashed line are from the vasculature, newly added here; to the right, parenchymal cells captured before. Numbers on the bottom summarize the number of GWAS genes enriched in a given cell type.
Note: MSA46A represents the average expression of MS4A46A, MS4A4A, and MS4A4E; likewise, HLADRB1 averages HLA-DRB1 and HLA-DRB5. EPHA1 was not robustly detected.
b, Immunohistochemical confirmation of vascular localization of proteins encoded by top AD GWAS genes from (a). Scale bar = 25 microns. Arrowheads in APOE point to signal around larger diameter vessels, consistent with SMC expression. Image credit: Human Protein Atlas (http://www.proteinatlas.org)25.
c, Examples of genes expressed specifically in mouse microglia but then also expressed in human brain vascular cell types (n of ∼3,500 whole cell mouse transcriptomes12, mean value +/− SEM).
d, BEC heatmap of top AD GWAS genes colored by logFC(human/mouse) and labeled by the linear foldchange (human/mouse) value.
e, Quantification of the number of AD and AD-related trait GWAS genes17 most expressed in a given cell type. 383 of 651 genes (59%) mapped to vascular or perivascular cell types. PPI network of GO Cellular Components (P value < 0.05, cumulative hypergeometric test).
A = arterial, C = capillary, V = venous endothelial cell (EC). Mg = microglia, and Mφ = macrophage. In fibroblasts, M = meningeal and P = perivascular. In mural cells, S = SMC and P = pericyte.
f, Human enrichment of AD-related trait GWAS genes17 highest expressed in BECs (left) and mural cells (right). In contrast to GWAS genes, the ratio of human to mouse expression across the overall transcriptome is less than or ∼1 for both cell types (bottom, paired two-sided t-test, ****P < 0.0001 and ***P = 0.0002).
Intriguingly, we noticed several GWAS genes were strongly expressed in human brain vascular and perivascular cell types (Fig. 6a, left, Extended Data Fig. 9). This included two genes previously implicated in the mouse vasculature, PICALM and CD2AP43,44. But this also included surprising genes, such as the immune-related PLCG2 and HLA-DRB1/5 in arterial cells, endocytic INPP5D and USP6NL in capillaries, and ECM-related ADAMTS1, ADAMTS4, FERMT2, and AGRN in SMCs and pericytes (Fig. 6a). Within pericytes, expression varied across M- and T-pericyte subtypes (Extended Data Fig. 9a). APOE, linked to myeloid cells and astrocytes, was robustly expressed in human SMCs and meningeal fibroblasts. Several GWAS genes like ABCA7 and CLNK enrich in T cells, and independent datasets show minimal expression in other brain cell types (Extended Data Fig. 9b). Several genes like ABCA1, FHL2, HESX1, and IL34 enrich in fibroblasts. Importantly, we confirmed our findings via immunostaining of proteins encoded by vascular GWAS genes, such as CASS4, FERMT2, PLCG2, and FHL2 (Fig. 6b, Extended Data Fig. 9c). Most GWAS genes were expressed similarly between the hippocampus and cortex (Extended Data Fig. 9d). In total, at least 30 of the top 45 AD GWAS genes are enriched in cells of the human brain vasculature (not including those solely in perivascular macrophages), suggesting thorough vascular and perivascular involvement in AD.
We next wondered whether these genes are expressed in different cell types between mice and humans. Many genes like APOE, CASS4, INPP5D, and HLA-DRB1 were predominately microglial in mice12 but then also exhibited vascular expression in humans (Fig. 6c, Extended Data Fig. 9e). Nearly every top AD GWAS gene expressed in BECs showed greater expression in humans (Fig. 6d). Together, these data suggest a partial evolutionary transfer of AD risk genes and pathways from microglia to the vasculature from mice to humans (Extended Data Fig. 9f).
We broadened our scope to hundreds of GWAS genes for AD and AD-related traits17. We observed robust, cell type-specific vascular and perivascular expression (Extended Data Fig. 10). For each gene, we assigned the cell type with the strongest expression, discovering surprisingly, that BECs harbored the most AD-related GWAS genes, followed by microglia/ macrophages (Fig. 6e, Supplemental Table 8). Within BECs, AD-related GWAS genes enriched in protein endo- and transcytosis components, such as receptor and clathrin vesicle components (Fig. 6d-e). We recently demonstrated a decline in BEC clathrin-mediated transcytosis8 with age, suggesting one mechanism by which aging and risk genes converge to increase AD risk. In total, over half of AD-related GWAS genes mapped to vascular or perivascular cell types (383 of 651). As with top GWAS genes, we observed enhanced human over mouse expression of AD-related genes in BECs and pericytes (Fig. 6f). Importantly, this human-enhanced expression is not observed for the whole transcriptome.
Together, these data provide a more comprehensive understanding of the diverse cell types contributing to AD risk. We suggest that an evolutionarily expanded vascular-microglia axis underlies the genetic risk for AD via shared protein clearance (BEC-microglia) and inflammatory pathways (BEC-T cell-microglia) (Fig. 6e, Extended Data Fig. 9f).
Discussion
We report here 143,793 single-cell, genome-wide quantitative transcriptomes from the human brain vasculature in health and AD. We molecularly define the principal vascular cell types; their differences by brain region and species; and the organizational principles of endothelial, mural, and fibroblast-like cells. We reveal selective vulnerability of vascular subpopulations and transcriptomic perturbations associated with clinically-diagnosed AD; and the unexpected expression of AD GWAS genes across human brain vascular cell types, confirmed in situ. Data are available to browse at https://twc-stanford.shinyapps.io/human_bbb.
How do human vascular GWAS genes fit into established AD pathways? Current understanding implicates β-amyloid metabolism, cholesterol/lipid dysfunction, innate immunity, and endocytosis43. We confirm these pathways and expand the cell types involved, for example, debris clearance via BEC clathrin-mediated endocytosis—and adaptive T cell in addition to innate immunity. We propose that the expansion of the human brain, brain activity, and activity byproducts (like β-amyloid45) necessitate enhanced clearance mechanisms and neuroimmune surveillance. In this model, microglia are still frontline participants in AD pathogenesis. But more than in mice, human vascular and perivascular cells partake. For example, microglial clearance functions can become overwhelmed46, diverting debris to BECs. Indeed, microglial depletion results in cerebral amyloid angiopathy47. But unlike microglia, vascular cells do not proliferate efficiently. Thus, constant vascular exposure to debris like β-amyloid triggers cell death, dysfunction, and impaired blood and CSF flow. Recent work identified a CD8 TEMRA population clonally expanded in AD CSF48, potentially influenced by T cell GWAS genes. Together, we suggest an intertwined microglia-vascular axis expanded in humans, with vascular cells playing an auxiliary role via shared endocytosis and inflammatory pathways. We note though the likelihood of additional human brain vascular contributions, as evidenced by SMC, pericyte, and fibroblast-enriched GWAS genes of unclear function. In general, disease risk variants enrich in gene expression-regulating enhancer regions49 that undergo accelerated species divergence50, providing one potential explanation for human-specific expression and dysregulation of AD risk genes in cerebrovascular cell types.
The field now has a near complete census of human brain cell types. This atlas expands by orders of magnitude the number of human brain vascular cell type and zonation markers to inform research, such as identifying vessels, validating organoid fidelity, or deconvoluting bulk RNA-seq datasets. Our work also facilitates translational opportunities. This dataset provides targets for antibodies and other modalities to deliver therapeutics to the brain. VINE-seq enables study of brain vascular contributions to various diseases, such as stroke, multiple sclerosis, and COVID-19. But as with recent snRNA-seq studies16–19, it will be important to distinguish which vascular transcriptional perturbations respond to versus drive disease, clarify their links to clinical and pathologic traits, and dissect the mechanisms by which vascular-expressed AD variants confer disease risk. Overall, VINE-seq and the ensuing single-cell atlas provide a blueprint for studying the molecular basis of the human brain vasculature.
Methods
Isolation of vascular nuclei from frozen post-mortem brain tissue
Post-mortem fresh-frozen hippocampus and superior frontal cortex tissue were obtained from the Stanford/ VA/ NIA Aging Clinical Research Center (ACRC) with approval from local ethics committees and patient consent. Group characteristics are presented in Supplemental Table 1. Note, individuals were grouped by clinical diagnosis, with two of the NCI individuals exhibiting amyloid beta plaque staining in the hippocampus, though not to a sufficient degree for an expert pathologist to diagnose Alzheimer’s disease by histopathological criteria. Clinical instead of pathologic diagnosis was chosen because of potentially vascular contributions to AD independent of the well-known hallmarks of AD, β-amyloid and tau pathophysiology6. All procedures were carried out on ice in a 4°C cold room as rapidly as possible. 0.3 grams or more of brain tissue was thawed on ice for 5 minutes with 5 ml of nuclei buffer (NB): 1% BSA containing 0.2 U μl−1 RNase inhibitor (Takara, 2313A) and EDTA-free Protease Inhibitor Cocktail (Roche, 11873580001). Tissue was quickly minced and homogenized with 7 ml glass douncers (357424, Wheaton) until no visible chunks of debris remained. Similar to before53, homogenates were transferred into 50 ml tubes containing 35 ml of chilled 32% dextran (D8821, Sigma) in HBSS. Samples were vigorously mixed before centrifugation at 4,400g for 20 minutes with no brake. After centrifugation, samples separate into a top myelin layer, middle parenchymal layer, and vascular-enriched pellet. The myelin layer was aspirated, tips changed, and the parenchymal layer carefully removed without disturbing the pellet. Pellets were resuspended in 8 ml of 32% dextran, transferred to 15 ml falcon tubes, and centrifuged again. Vascularenriched pellets were gently resuspended in 1ml of NB and added to pre-wetted 40 μm strainer sitting atop 50 ml falcon tubes. From here diverging from prior protocol, strainers were washed with 10 ml of cold 0.32 M sucrose in PBS and 90 ml of PBS until flow through the strainers was unimpeded to deplete contaminating parenchymal cells from trapped microvessels. At this step, retained microvessels turn white in color, indicating removal of circulating blood cells. Strainers were switched to new collection 50 ml falcon tubes. Various techniques were tested and optimized to extract vascular cells from the isolated microvessels (e.g., enzymatic digestion, TisssueRuptor, sonication, etc.), but nearly all resulted in loss of nuclei integrity or low nuclei complexity (<50 median genes/ nuclei). Eventually, adapting a method for the isolation of murine splenocytes proved successful: vascular fragments were mashed four times through the cell strainer using the plunger end of a 3 ml syringe, with intermittent elution via 10 ml of 0.32 sucrose and 40 ml of PBS. Liberated vascular cells were pelleted at 500g for 10 minutes and resuspended in 1.5 ml of EZ Prep Lysis Buffer (Sigma, NUC101) spiked with 0.2 U μl−1 RNase inhibitor (Takara, 2313A) and EDTA-free Protease Inhibitor Cocktail (Roche, 11873580001). Nuclei were homogenized with 2 ml glass douncers (D8938, Sigma) 20 times with pestle B (pestle A optional). Spiked EZ lysis buffer was added to samples up to 4 ml and incubated on ice for 5 minutes before pelleting at 500g for 6 minutes. This incubation step was repeated. Debris was depleted via a sucrose gradient before flow cytometry isolation of nuclei. Briefly, pelleted nuclei were resuspended in 0.5ml of NB before the addition of 0.9 ml of 2.2 M sucrose in PBS. This mixture was layered atop 0.5 ml of 2.2 M and samples were centrifuged at 14,000g for 45 minutes at 4°C, with no brake. Pellets were aspirated in 1ml of NB, filtered through a 40 μm strainer (Flowmi), transferred to FACS tubes, stained with Hoechst 3342 (1:2000, Thermo) and rabbit monoclonal anti-NeuN Alexa Fluor® 647 (1:500, Abcam, ab190565), and nuclei collected on a SH800S Cell Sorter into chilled tubes containing 1 ml of NB without protease inhibitor. In pilot runs, we noticed the cytometer overestimated nuclei counts by ∼3.4x, and thus we sorted ∼34,000 nuclei to target ∼10,000 nuclei per sample. Sorted samples were inspected for lack of debris on a brightfield microscope. We note that an iodixanol gradient54 can substitute for the 2.2 M sucrose, but that unfortunately with either gradient, flow sorting is required—unlike parenchymal myelin debris, vascular debris is not sufficiently removed by gradient centrifugation alone. Vascular debris will confound downstream cDNA traces with higher background and low molecular weight peaks.
Droplet-based snRNA-sequencing
For droplet-based snRNA-seq, libraries were prepared using the Chromium Single Cell 3ʹ v3 according to the manufacturer’s protocol (10x Genomics), targeting 10,000 nuclei per sample after flow sorting (Sony SH800S Cell Sorter). 15 PCR cycles were applied to generate cDNA before 16 cycles for final library generation. Generated snRNA-seq libraries were sequenced on S4 lanes of a NovaSeq 6000 (150 cycles, Novogene).
snRNA-seq quality control
Gene counts were obtained by aligning reads to the hg38 genome (refdata-gex-GRCh38–2020-A) using CellRanger software (v.4.0.0) (10x Genomics). To account for unspliced nuclear transcripts, reads mapping to pre-mRNA were counted. As previously published, a cut-off value of 200 unique molecular identifiers (UMIs) was used to select single nuclei for further analysis16,55. As initial reference, the entire dataset was projected onto two-dimensional space using Uniform Manifold Approximation and Projection (UMAP) on the top 30 principal components56. Three approaches were combined for strict quality control: (1) outliers with a high ratio of mitochondrial (>5%, <200 features) relative to endogenous RNAs and homotypic doublets (> 5000 features) were removed in Seurat57; (2) after scTransform normalization and integration, doublets and multiplets were filtered out using DoubletFinder58; and (3) after DoubletFinder, nuclei were manually inspected using known cell type-specific marker genes, with nuclei expressing more than one cell type-specific marker further filtered16,18,58–62. For example, BEC nuclei containing any reads for the following cell type markers were subsequently filtered: MOBP, MBP, MOG, SLC38A11, LAMA2, PDGFRB, GFAP, SLC1A2, and AQP4. We note that the vascular nuclei in prior human single cell datasets exhibit contamination with other cell type-specific gene markers, potentially confounding downstream analysis. After applying these filtering steps, the dataset contained 143,793 high-quality, single nuclei.
Cell annotations & differential gene expression analysis
Seurat’s Integration function was used to align data with default settings. Genes were projected into principal component (PC) space using the principal component analysis (RunPCA). The first 30 dimensions were used as inputs into Seurat’s FindNeighbors, FindClusters (at 0.2 resolution) and RunUMAP functions. Briefly, a shared-nearest-neighbor graph was constructed based on the Euclidean distance metric in PC space, and cells were clustered using the Louvain method. RunUMAP functions with default settings was used to calculate 2-dimensional UMAP coordinates and search for distinct cell populations. Positive differential expression of each cluster against all other clusters (MAST) was used to identify marker genes for each cluster63. We annotated cell-types using previously published marker genes16,19,62,64. For brain endothelial cells, zonation specificity scores for each gene were calculated separately for arterial, capillary, and venous segments as in the following example for a given gene in capillaries:
Differential gene expression of genes comparing Alzheimer’s disease, ApoE4, and NCI samples—or comparing cell type sub-cluster markers—was done using the MAST63 algorithm, which implements a two-part hurdle model. Seurat natural log (fold change) > 0.5 (absolute value), adjusted P value (Bonferroni correction) < 0.01, and expression in greater than 10% of cells in both comparison groups were required to consider a gene differentially expressed for subcluster analysis and natural log (fold change) > 0.3 (absolute value), adjusted P value (Bonferroni correction) < 0.01, and expression in greater than 10% of cells in both comparison groups for Alzheimer’s disease and ApoE4 comparisons, both more stringent than the default Seurat settings. We incorporated age, gender, and batch as covariates in our model. A more lenient threshold of the above but with natural log (fold change) > 0.2 (absolute value) was used for brain region (i.e., hippocampus vs cortex. Biological pathway and gene ontology enrichment analysis was performed using Enrichr65 or Metascape66 with input species set to Homo sapiens66. UpSet plots were generated using identified differentially expressed genes as inputs using the R package UpSetR67. Diagrams were created with BioRender.
Monocle trajectory analysis
Monocle was used to generate the pseudotime trajectory analysis in brain endothelial and mural cells24. Cells were clustered in Seurat and cluster markers used as input into Monocle to infer arteriovenous relationships within endothelial cells and pericytes. Specifically, UMAP embeddings and cell sub-clusters generated from Seurat were converted to a cell_data_set object using SeuratWrappers (v.0.2.0) and then used as input to perform trajectory graph learning and pseudo-time measurement through Independent Component Analysis (ICA) with Monocle. Cluster marker genes identified in Seurat were used to generate a pseudotime route and plotted using the ‘plot_pseudotime_heatmap’ function. For mural cells, variable genes were limited to those with log[Average Expression] >1 (Seurat) for even more robust cell ordering.
Cell-cell communication
Cell-cell interactions based on the expression of known ligand-receptor pairs in different cell types were inferred using CellChatDB68 (v.0.02). Briefly, we followed the official workflow and loaded the normalized counts into CellChat and applied the preprocessing functions identifyOverExpressedGenes, identifyOverExpressedInteractions, and projectData with standard parameters set. As database we selected the Secreted Signaling pathways and used the pre-compiled human Protein-Protein-Interactions as a priori network information. For the main analyses the core functions computeCommunProb, computeCommunProbPathway, and aggregateNet were applied using standard parameters and fixed randomization seeds. Finally, to determine the senders and receivers in the network the function netAnalysis_signalingRole was applied on the netP data slot.
Animals
Aged C57BL/6 male mice (19-months old) were obtained from the National Institute on Aging rodent colony. Young male C57BL/6 mice (3-months old) were obtained from Jackson Laboratories or Charles River Laboratories. Thy1-hAPPLon,Swe male mice and littermate wild-type control37 mice were raised until 12–14 months of age. All mice were kept on a 12-h light/dark cycle and provided ad libitum access to food and water. All animal care and procedures complied with the Animal Welfare Act and were in accordance with institutional guidelines and approved by the V.A. Palo Alto Committee on Animal Research and the institutional administrative panel of laboratory animal care at Stanford University.
Mouse wild-type and APP T41B BEC single-cell and nuclei sequencing
Whole cell isolation from the CNS followed previously described methods69–71. Briefly, cortices and hippocampi were microdissected, minced, and digested using the Neural Dissociation Kit (Miltenyi). Suspensions were filtered through a 100 µm strainer and myelin removed by centrifugation in 0.9 M sucrose. The remaining myelin-depleted cell suspension was blocked for ten minutes with Fc preblock (CD16/ CD32, BD 553141) on ice and stained for 20 minutes with antibodies to distinguish brain endothelial cells (CD31+/ CD45-). Brain endothelial cells from 12–14 month old Thy1-hAPPLon,Swe mice and littermate wild-type control37 mice (pool of 4–6 mice per group) were sorted into PBS with 0.1% BSA. Nuclei isolation from 4–6 month-old mouse hippocampi followed protocols adapted from previous studies16,17,54,62,72. Briefly, tissue was homogenized using a glass douncer in 2 ml of ice-cold EZ PREP buffer (Sigma, N3408) and incubated on ice for 5 min. Centrifuged nuclei were resuspended in 1% BSA in PBS with 0.2 U μl−1 RNase inhibitor and filtered through a 40 μm cell strainer. Cells or nuclei were immediately counted using a Neubauer haemocytometer and loaded on a Chromium Single-Cell Instrument (10x Genomics, Pleasanton, CA, USA) to generate single-cell GEMs. The 10x-Genomics v3 libraries were prepared as per the manufacturer’s instructions. Libraries were sequenced on an Illumina NextSeq 550 (paired-end; read 1: 28 cycles; i7 index: 8 cycles, i5 index: 0 cycles; read 2: 91 cycles). De-multiplexing was performed using the Cellranger toolkit (v3.0.0) “cellranger mkfastq” command and the “cellranger count” command for alignment to the mouse transcriptome, cell barcode partitioning, collapsing unique-molecular identifier (UMI) to transcripts, and gene-level quantification. ∼70% sequencing saturation (>20,000 reads per cell) was achieved, for a median of ∼2,000 genes detected per cell and ∼16,500 genes detected in total. Downstream analysis using the Seurat package (v3)73 was performed as previously described12, applying standard algorithms for cell filtration, feature selection, and dimensionality reduction. Samples with fewer than 1,000 and more than 4,000 unique feature counts, samples with more than 15% mitochondrial RNA, samples with more than 15% small subunit ribosomal genes (Rps), and counts of more than 10,000 were excluded from the analysis. Genes were projected into principal component (PC) space using the principal component analysis (RunPCA). The first 30 dimensions were used as inputs into Seurat’s FindNeighbors and RunTsne functions. Briefly, a shared-nearest-neighbor graph was constructed based on the Euclidean distance metric in PC space, and cells were clustered using the Louvain method. RunTsne functions with default settings was used to calculate 2-dimensional tSNE coordinates and search for distinct cell populations. Cells and clusters were then visualized using 3-D t-distributed Stochastic Neighbor embedding on the same distance metric. Differential gene expression analysis was done by applying the Model-based Analysis of Single-cell Transcriptomics (MAST). Significant differentially expressed genes in Thy1-hAPPLon,Swe BECs were called by Log (fold change) > 0.15 (absolute value), adjusted P value (Bonferroni correction) < 0.01. This lowered Log (fold change) was to ensure our claims of limited overlap with human AD BECs were robust.
GWAS analysis
For calculation of proportional cell type-specific gene expression, we followed the expression weighted cell type enrichment (EWCE) method described by Skene et al.42, and used previously on human snRNA-seq data17. For Alzheimer’s disease (AD) analysis, we compiled a list of top GWAS risk genes from Lambert et al.39, Kunkle et al.40, and Jansen et al.41, sorted descending by approximate P-value. Each gene’s expression sums to 1 across the cell types, with each heatmap cell showing the fraction of total gene expression as determined from EWCE analysis. The set of 720 AD and AD-related trait GWAS genes were obtained from Grubman, et al.17, and using EWCE analysis, the strongest expressing cell type was determined for each gene. Note that the original list was slightly parsed to 720, as several genes were not detected as expressed in our dataset.
Immunohistochemistry
Fresh-frozen NCI and AD human brain tissue (hippocampus and superior frontal cortex adjacent to tissue processed for snRNA-seq as well as meninges) was subjected to immunohistochemistry (IHC). 10 µm sections mounted on SuperFrost Plus glass slides were fixed with 4% paraformaldehyde (Electron Microscopy Services, 15714S) diluted in PBS at 4°C for 15 minutes before dehydration via an ethanol series or air drying. Sections were blocked in TBS++ (TBS + 3% donkey serum (130787, Jackson ImmunoResearch) + 0.25% Triton X-100 (T8787, Sigma-Aldrich)) for 1.5 hours at room temperature. Sections were incubated with primary antibodies at 4°C overnight: goat polyclonal anti-Collagen Type IV (1:200, AB769, Sigma), rabbit polyclonal anti-CYP1B1 (1:100, HPA026863, Atlas Antibodies), rabbit polyclonal anti-SLC4A4 (1:100, HPA035628, Atlas Antibodies), rabbit polyclonal anti-SLC47A1 (1:100, HPA021987, Atlas Antibodies), rabbit polyclonal anti-ABCA8 (1:100, HPA044914, Atlas Antibodies), mouse monoclonal anti-CD31 (1:100, JC70A, Dako), rabbit polyclonal anti-VWF (1:100, GA527, Dako), rabbit polyclonal anti-SLC39A10 (1:100, HPA066087, Atlas Antibodies), rabbit polyclonal anti-ALPL (1:100, HPA007105, Atlas Antibodies), rabbit polyclonal anti-A2M (1:100, HPA002265, Atlas Antibodies), rabbit monoclonal anti-β-Amyloid (1:500, clone D54D2 XP, CST), and mouse monoclonal anti-Actin, α-Smooth Muscle - Cy3 (1:100, clone 1A4, Sigma). Sections were washed, stained with Alexa Fluor-conjugated secondary antibodies (1:250) and Hoechst 33342 (1:2000, H3570, Thermo), mounted and coverslipped with ProLong Gold (Life Technologies) or VECTASHIELD (Vector Laboratories before imaging on a confocal laser scanning microscope (Zeiss LSM880). Age-related autofluorescence was quenched prior to mounting with Sudan Black B, as before8,71. National Institutes of Health ImageJ software was used to quantify the number of Hoechst+ nuclei per image, the percentage of vasculature (Collagen IV), the number of Hoechst+ nuclei within Collagen IV+ vasculature, or the predicted DEG SLC39A10 among CD31+ vasculature, following previously described protocols8,15,74. In short, at least five images were stained per patient, and imaging and analyses were performed by a blinded observer.
Statistics and Reproducibility
Immunostaining validation experiments were repeated independently at least twice with similar results. As indicated in the legends, some immunostaining images come from the Human Protein Atlas25,75 and are available at v21.proteinatlas.org.
Extended Data
Extended Data Fig. 1. Enhanced capture and characterization of human brain vascular nuclei.
a, Detailed schematic of the VINE-seq method to capture human brain vascular and immune cell types for single-nucleus sequencing.
b, Total number of nuclei, median number of unique molecular identifiers (UMI), and median number of genes for each human sample sequenced from hippocampus and superior frontal cortex.
c, Quantification of the median number of genes detected per nuclei across subject groups (n=17 hippocampus and n=8 cortex; n=8 NCI and n=9 AD, two-sided t-test; mean +/− s.e.m.).
d, e, Quantification of the number (d) and proportion (e) of cerebrovascular cell types captured via the VINE-seq method introduced here compared to recent snRNA-seq studies16,17.
f, Summary quantification of the proportion of captured cell types by NCI and AD individuals.
g, Quantification of the proportion of captured cell types across individuals.
h, Summary (left) and quantification (right) of the proportion of captured cell types by brain region (n=17 hippocampus and n=8 cortex; n=8 NCI and n=9 AD, two-sided t-test; mean +/− s.e.m.).
Extended Data Fig. 2. Diversity and heterogeneity of human brain vascular cell types.
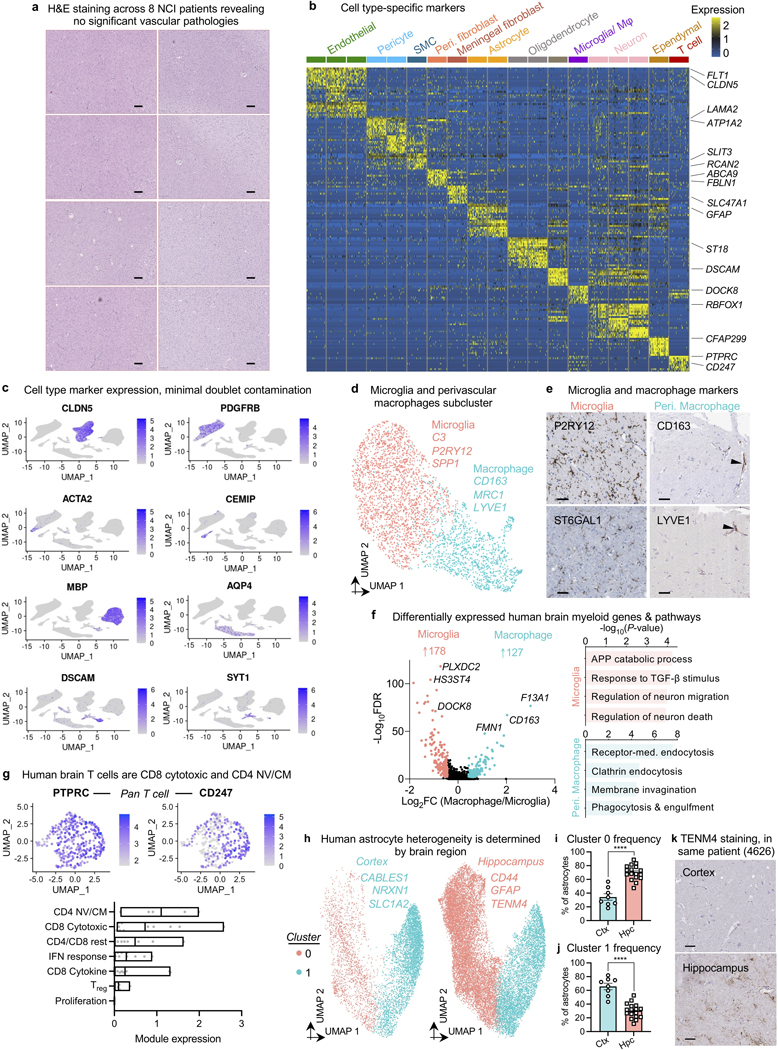
a, Representative H&E images used by two neuropathologists to evaluate NCI cortical and hippocampal tissue for vascular pathology. No significant vascular pathology was observed. Scale bar = 200 μM.
b, Discovery of the top cell type-specific marker genes across the major classes of cells captured. The color bar indicates gene expression from low (blue) to high (yellow).
c, Validation of cell type annotations and confirmation of minimal doublet contamination using established cell type markers.
d, UMAP projection of captured myeloid cells, forming two distinct clusters corresponding to parenchymal microglia and brain barrier macrophages. Example marker genes listed.
e, Immunohistochemical validation of microglial and perivascular macrophage markers. Scale bar = 50 microns. Image credit: Human Protein Atlas (http://www.proteinatlas.org)25,75.
f, Global view of differentially expressed genes comparing human brain macrophages and microglia (left, MAST, Benjamini Hochberg correction; FDR < 0.01 and logFC>0.5 [log2FC>0.72] to be colored significant). Pathways enriched in microglia versus macrophages (right), recapitulating interesting biology such as the unique TGF-β molecular signature in microglia76.
g, Expression of top gene markers for various T cell subtypes (top), and quantification of their expression as a module (bottom)77. Brain T cells exhibit highest expression of markers corresponding to CD8 cytotoxic and CD4 Naive/Central memory (NV/CM) T cells.
h, UMAP projection of captured astrocytes, forming two distinct clusters, and split by brain region. Example marker genes listed.
i-j, Quantification of astrocyte cluster 0 (b) and 1 (c) frequency in the cortex and hippocampus (n=8 cortex and n=17 hippocampus, Mann-Whitney t-test; mean +/− s.e.m.).
k, Immunohistochemical validation of the brain region-specific astrocyte marker TENM4. Scale bar = 50 microns.
Extended Data Fig. 3. Species-specific gene expression across brain cell types and their pharmacological relevance.
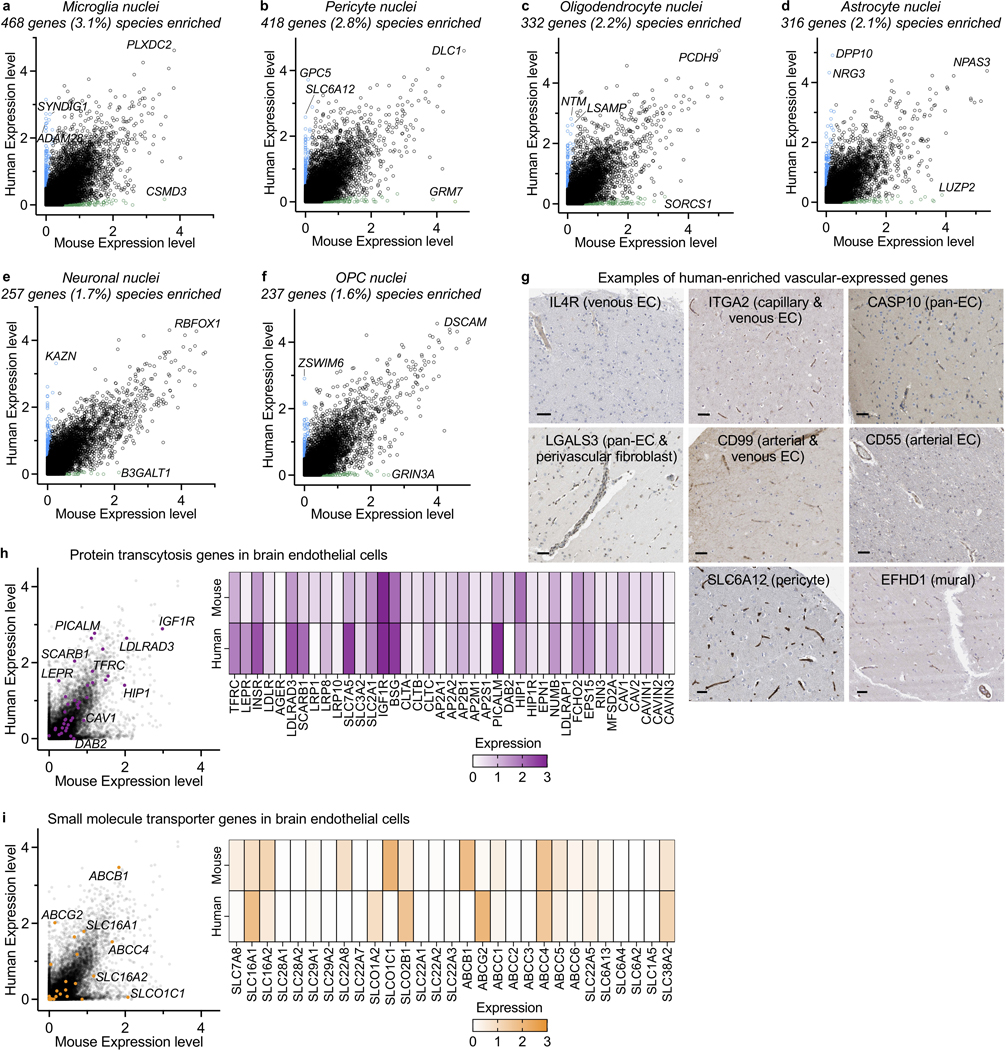
a-f, Identification of species-specific genes. Both mouse and human transcriptomes were generated and analyzed similarly via single-nucleus RNA-sequencing. Mice were 19 months of age to match the average age of our human cohort. Species-specific/enriched are colored.
g, Immunohistochemical confirmation of genes predicted to be enriched or specific to human cerebrovascular cells compared to mouse (isolated mouse nuclei and per Vanlandewijck, et al., 2018)12, in terms of overall expression or zonation. In parenthesis is the cell type predicted to be uniquely or exhibiting enriched expressed in human over mouse. Scale bar = 50 microns. Image credit: Human Protein Atlas (http://www.proteinatlas.org)25,75.
h-i, Mouse and human brain endothelial cell expression of genes mediating protein transcytosis (h) and small molecule influx and efflux (i).
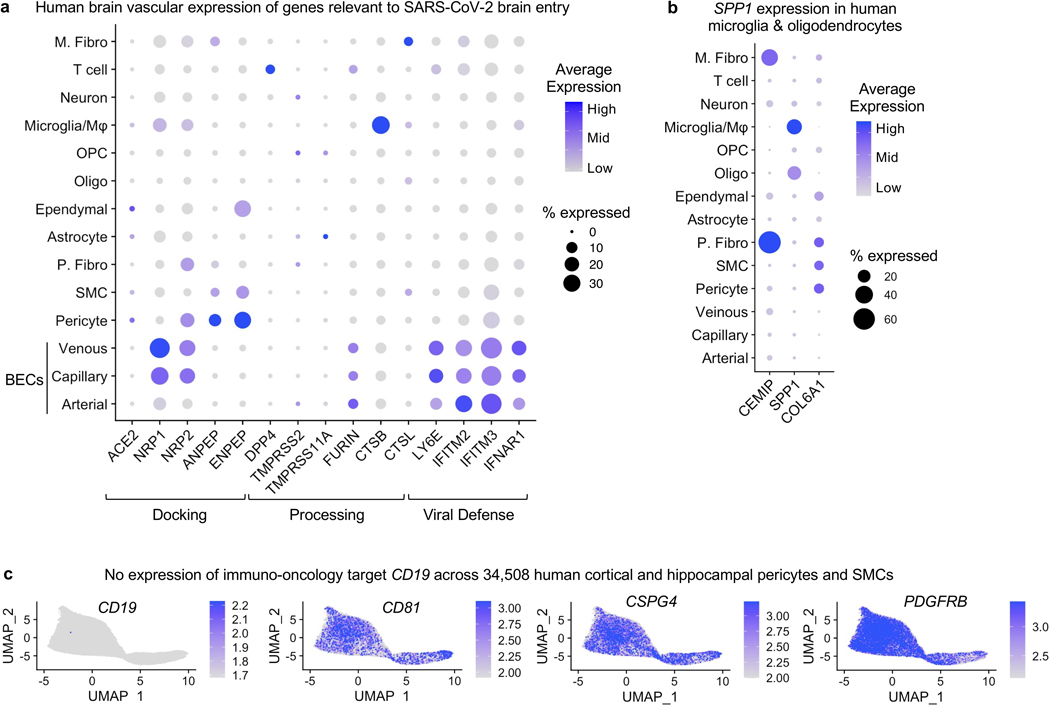
Extended Data Fig. 4. Human brain vascular expression of genes relevant to disease.
a, Brain vascular expression of genes relevant to SARS-CoV-2 brain entry, as summarized in Iadecola, et al. 202078.
b, Expression of the mouse perivascular fibroblast-like gene Spp1 is instead specifically expressed in human myeloid cells and oligodendrocytes (SPP1, top)79.
c, No expression of the immuno-oncology target CD19 and its chaperone CD81 across human adult brain pericytes and smooth muscle cells80. Note: cells with any finite expression are ordered to the front to ensure all expression is visible, but this carries the potential to visually overestimate average expression
Extended Data Fig. 5. Brain endothelial and mural cell zonation and subpopulations.
a, UMAP projection of captured brain endothelial cells, organizing by arteriovenous zonation. Bottom, tip cell markers expressed in the tip-like/ proteostatic EC cluster.
b, Validation of brain endothelial cell zonation clusters using established zonation markers12. Violin plots are centered around the median, with their shape representing cell distribution.
c, d, As in (a-b) but for pericytes and smooth muscle cells. Note that the anatomical locations of pericyte 0 and 1 have not yet been determined.
e, Immunohistochemical validation of ACTA2 (α-SMA) expression in human smooth muscle cells and less so in capillary pericytes. A denotes arterial and C denotes capillary. Arrowheads specify capillary pericytes expressing ACTA2. Scale bar = 50 microns.
f, g, As in (a-b) but for perivascular fibroblast-like cells, as recently discovered in mice12.
Extended Data Fig. 6. Brain endothelial zonation and mural cell subtype markers.
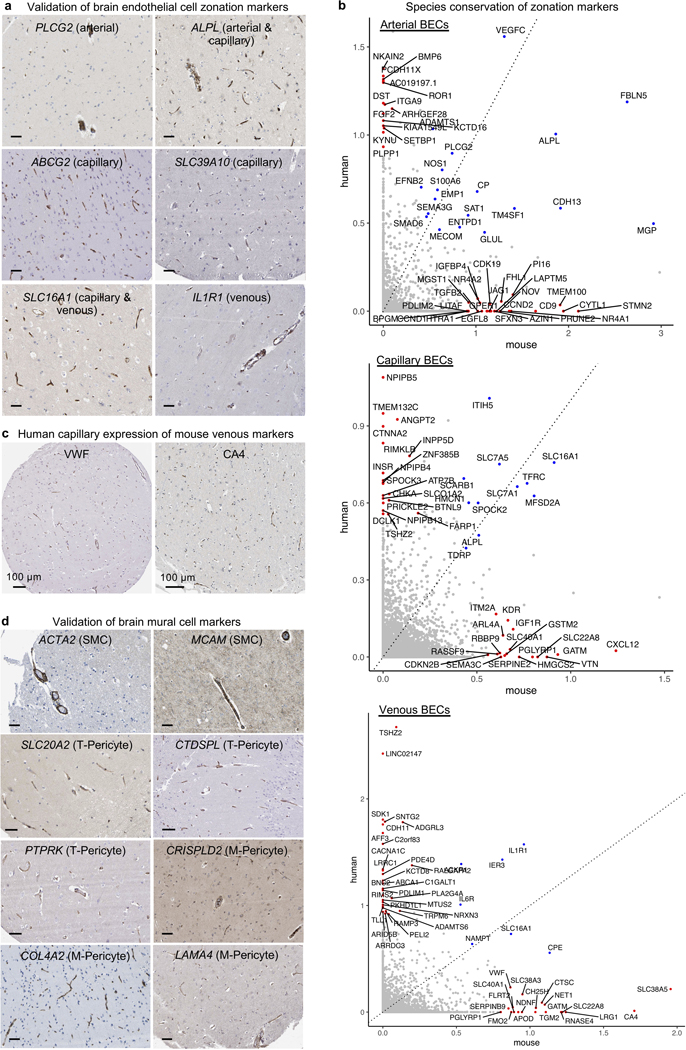
a, Immunohistochemical validation of zonation and cell subtype markers in brain endothelial cells. Scale bar = 50 microns. Image credit: Human Protein Atlas (http://www.proteinatlas.org)25,75.
b, Comparison of the zonal specificity of genes in arterial, capillary, and venous cells. Axis plot a specificity score, as defined in the Methods. For example, specificity score for capillaries = avg(logFC(cap/ven), logFC(cap/art)).
c, Immunohistochemical validation of capillary expression in human brains of the mouse venous-specific marker VWF and CA4, with similar patterns observed across multiple primary antibody clones. Scale bar = 100 microns. Image credit: Human Protein Atlas (http://www.proteinatlas.org)25,75.
d, Immunohistochemical validation of zonation and cell subtype markers in brain SMCs and pericytes. Scale bar = 50 microns. Image credit: Human Protein Atlas (http://www.proteinatlas.org)25,75.
Extended Data Fig. 7. Specialization and functions of human brain fibroblasts.
a, Expression of example markers demarcating perivascular from meningeal fibroblasts.
b, UMAP of 428 meningeal fibroblast nuclei, subclustering into anatomically segregated dural and arachnoid space fibroblasts.
c, Expression of the genes constituting the major fibrotic scar component Collagen I in pericytes and fibroblasts. Collagen I is composed of two components, COL1A1 and COL1A2. Column annotations: T-PC = solute transport pericyte and M-PC = Extracellular matrix regulating pericyte, P. FB = Perivascular fibroblast, and M. FB = Meningeal fibroblast.
d-e, Protein immunostaining validation of polarized expression of human brain meningeal and perivascular fibroblast pumps: the common marker CYP1B1 (a, serves as a control) and the meningeal fibroblast-specific influx pump SLC47A1 (b). Scale = 50 μM.
f, Overlap between the top 100 perivascular fibroblast-like cell markers and those identified in mice. A more lenient set of 500 (instead of 100) mouse markers52 were used for comparison to ensure claims of species-specificity were robust. Note: the species-conservation of a cell type marker depends on speciesspecific changes in the given cell type and changes amongst the remaining background cell types.
Extended Data Fig. 8. Vascular cell-type specific perturbations in AD patients and ApoE4 carriers.
a, Immunohistochemistry with anti-β-amyloid antibody (D54D2, white), Thioflavin S (green), and Hoechst (blue) in the hippocampus of NCI and AD individuals. Scale bar = 40 microns.
b, Quantification of β-amyloid immunostaining in (a) for overall β-amyloid (n=4 NCI and AD, two-sided t-test; mean +/− s.e.m.).
c, As in (b) but for cored and neuritic β-amyloid plaques (n=3 NCI and AD, two-sided t-test; mean +/− s.e.m.).
d, UMAP of 143,793 nuclei captured from 17 human hippocampus and superior frontal cortex samples, colored by Alzheimer’s disease (AD) diagnosis.
e, Quantification controls for Fig. 5b. Quantification of Collagen IV+ vasculature (left) and number of total (regardless of Collagen IV+ overlap) Hoechst+ nuclei (n = 5 NCI and AD, nested two-sided t-test; mean +/− s.e.m.).
f, Matrix layout for intersections of AD DEGs shared across and specific to each cell type. Circles in the matrix indicate sets that are part of the intersection, showing that most DEGs are cell type-specific.
g, Example differentially expressed genes (DEGs) in AD: arterial (Art), capillary (Cap), venous (Vein), pericyte (Peri), perivascular fibro blast-like cell (P. fibro), and smooth muscle cell (SMC). Blue arrow indicates upregulated and grey arrow downregulated genes.
h, Summary of the number of AD DEGs by pericyte class: T-, M-, and all pericytes combined to evaluate DEGs that could arise due to a disproportionate loss of M-pericytes in AD.
i, Differentially expressed gene (DEG) counts for each cell type in ApoE4 carriers (n = 5 ApoE3/3, n = 11 ApoE3/4 or ApoE4/4): arterial (Art), capillary (Cap), venous (Vein), pericyte (Peri), perivascular fibro blast-like cell (P. fibro), and smooth muscle cell (SMC). The intensity of the blue color and the size of the squares are proportional to entry values.
j, Matrix layout for intersections of ApoE4 DEGs shared across and specific to each cell type. Circles in the matrix indicate sets that are part of the intersection, showing that most DEGs are cell type-specific.
k, Immunohistochemical validation of the predicted upregulated anti-inflammatory DEG SLC39A10 in venous BECs of ApoE4 carriers. Scale bar = 50 microns (n = 4 ApoE3/3 and ApoE4 carriers, nested two-sided t-test; mean +/− s.e.m.).
l, Among patients with both hippocampus and superior frontal cortex profiled (n=4 NCI and n=4 AD), quantification of the relative abundance of major vascular cell types (NCI hippocampus set as reference, unpaired two-sided t-test; mean +/− s.e.m.). *BEC P = 0.0260, **BEC P = 0.0023, *Pericyte P (left) = 0.0357, *Pericyte P (mid) = 0.0237, **Pericyte P = 0.0077, **SMC P = 0.0075, *Fibroblast P = 0.0109, *Astrocyte P = 0.0357
m, As in (l), but comparison of the number of DEGs between brain regions for each cerebrovascular cell type. Analysis done separately for NCI and AD samples (n=7 cell types, unpaired two-sided t-test; mean +/− s.e.m.).
Extended Data Fig. 9. Re-evaluation and characterization of top AD GWAS genes expressed in the human brain vasculature.
a, Heterogeneous expression of AD GWAS genes across T- and M-pericyte subtypes.
b, RNA-seq data of the predicted T cell-specific AD GWAS genes EPHA1 and ABCA7 in an independent dataset81, corroborating minimal expression across resident/ parenchymal brain cells.
c, Immunohistochemical confirmation of vascular localization of proteins encoded by 12 top AD GWAS genes from (a). Scale bar = 25 microns. Arrowheads in APOE point to signal around larger diameter vessels, consistent with predicted SMC expression. Image credit: Human Protein Atlas (http://www.proteinatlas.org)25,75.
d, Heatmap comparing expression patterns of top AD GWAS genes in the hippocampus and superior frontal cortex: e.g., several microglia-expressed GWAS genes like APOE, MS4A4A, and TREM2 are more highly expressed in hippocampal compared to cortical microglia/ macrophages.
e, GWAS genes found to be expressed specifically in microglia among cells captured using the conventional nuclei isolation process (from Grubman, Chew, Ouyang, et al. 201917) are also expressed in vascular cells (asterisks).
f, Summary of AD GWAS genes enriched in microglia and vascular cells mediating common pathways in protein clearance and inflammation. Mouse and human superscripts denote whether expression has been confirmed in that species for a given gene. Proposed model is described in Discussion.
Extended Data Fig. 10. Brain vascular and perivascular expression of AD and AD-related GWAS genes.
a, Expression of Alzheimer’s disease (AD) and AD-related GWAS risk genes (from Grubman, Chew, Ouyang, et al. 2019)17 across human vascular cells.
b, Enriched biological pathways amongst AD and AD-related trait GWAS genes expressed in each cell type.
c, For each cell type, the top 10 most specifically expressed AD and AD-related trait GWAS genes.
Supplementary Material
Acknowledgments
We thank T. Iram, E. Tapp, N. Lu, M. Haney, O. Hahn, M.J. Estrada, S.M. Shi, and other members of the Wyss-Coray lab for feedback and support; Hansruedi Mathys, David A. Bennett, and participants in the CSHL BBB 2021 meeting for valuable advice; and H. Zhang and K. Dickey for laboratory management. This work was funded by the NOMIS Foundation (T.W.-C.), the National Institute on Aging (T32-AG0047126 to A.C.Y., 1RF1AG059694 to T.W.-C), Nan Fung Life Sciences (T.W.-C.), the Bertarelli Brain Rejuvenation Sequencing Cluster (an initiative of the Stanford Wu Tsai Neurosciences Institute), the Stanford Alzheimer’s Disease Research Center (P30 AG066515). This work was supported by a grant from the Simons Foundation Award # 811253TWC. A.C.Y was supported by a Siebel Scholarship. F.K. and A.K. are a part of the CORSAAR study supported by the State of Saarland, the Saarland University, and the Rolf M. Schwiete Stiftung. This study was supported by the AHA-Allen Initiative in Brain Health and Cognitive Impairment: 19PABHI34580007. The statements in this work are solely the responsibility of the authors and do not necessarily represent the views of the American Heart Association (AHA) or the Paul G. Allen Frontiers Group. Graphics were created with BioRender.com.
Footnotes
Competing interests
T.W.-C. is a co-founder and scientific advisor of Alkahest Inc. A.C.Y., R.V., and T.W.-C. are co-founders and scientific advisors of Qinotto Inc.
Data Availability.
Raw sequencing data is deposited under NCBI GEO: GSE163577. Data is also available to explore via an interactive web browser: https://twc-stanford.shinyapps.io/human_bbb.
References
- 1.Feigin VL et al. Global and regional burden of stroke during 1990–2010: Findings from the Global Burden of Disease Study 2010. Lancet 383(9913), 245–255. (2014). doi: 10.1016/S0140-6736(13)61953-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chow BW & Gu C. The Molecular Constituents of the Blood-Brain Barrier. Trends Neurosci. 38, 598–608 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Profaci CP, Munji RN, Pulido RS & Daneman R. The blood-brain barrier in health and disease: Important unanswered questions. J. Exp. Med. 217(4). (2020). doi: 10.1084/jem.20190062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Obermeier B, Daneman R & Ransohoff RM. Development, maintenance and disruption of the blood-brain barrier. Nature Medicine 19(12), 1584–1596. (2013). doi: 10.1038/nm.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sweeney MD, Zhao Z, Montagne A, Nelson AR & Zlokovic BV. Blood-brain barrier: From physiology to disease and back. Physiological Reviews 99(1), 21–78. (2019). doi: 10.1152/physrev.00050.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iadecola C. The Pathobiology of Vascular Dementia. Neuron 80(4), 844–866. (2013). doi: 10.1016/j.neuron.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pardridge WM. Drug transport across the blood-brain barrier. Journal of Cerebral Blood Flow and Metabolism 32(11), 1959–1972. (2012). doi: 10.1038/jcbfm.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang AC et al. Physiological blood–brain transport is impaired with age by a shift in transcytosis. Nature 583(7816), 425–430. (2020). doi: 10.1038/s41586-020-2453-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daneman R, Zhou L, Kebede AA & Barres BA. Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature 468, 562–566 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Armulik A et al. Pericytes regulate the blood-brain barrier. Nature 468, 557–561 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Janzer RC & Raff MC. Astrocytes induce blood-brain barrier properties in endothelial cells. Nature 325(6101), 253–257. (1987). doi: 10.1038/325253a0 [DOI] [PubMed] [Google Scholar]
- 12.Vanlandewijck M et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Sabbagh MF et al. Transcriptional and epigenomic landscapes of CNS and non-CNS vascular endothelial cells. Elife 7, e36187. (2018). doi: 10.7554/elife.36187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalucka J et al. Single-Cell Transcriptome Atlas of Murine Endothelial Cells. Cell 180(4), 764–779. (2020). doi: 10.1016/j.cell.2020.01.015 [DOI] [PubMed] [Google Scholar]
- 15.Chen MB et al. Brain Endothelial Cells Are Exquisite Sensors of Age-Related Circulatory Cues. Cell Rep. 30(13), 4418–4432. (2020). doi: 10.1016/j.celrep.2020.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathys H et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570(7761), 332–337. (2019). doi: 10.1038/s41586-019-1195-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grubman A et al. A single-cell atlas of entorhinal cortex from individuals with Alzheimer’s disease reveals cell-type-specific gene expression regulation. Nat. Neurosci. 22(12), 2087–2097. (2019). doi: 10.1038/s41593-019-0539-4 [DOI] [PubMed] [Google Scholar]
- 18.Jäkel S et al. Altered human oligodendrocyte heterogeneity in multiple sclerosis. Nature 566(7745), 543–547. (2019). doi: 10.1038/s41586-019-0903-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Velmeshev D et al. Single-cell genomics identifies cell type–specific molecular changes in autism. Science 364(6441), 685–689. (2019). doi: 10.1126/science.aav8130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keller D, Erö C & Markram H. Cell densities in the mouse brain: A systematic review. Frontiers in Neuroanatomy 12, 83. (2018). doi: 10.3389/fnana.2018.00083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niedowicz DM et al. Obesity and diabetes cause cognitive dysfunction in the absence of accelerated β-amyloid deposition in a novel murine model of mixed or vascular dementia. Acta Neuropathol. Commun. 2(1), 1–17. (2014). doi: 10.1186/2051-5960-2-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montagne A et al. Blood-Brain barrier breakdown in the aging human hippocampus. Neuron 85, 296–302 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geirsdottir L et al. Cross-Species Single-Cell Analysis Reveals Divergence of the Primate Microglia Program. Cell 179(7), 1609–1622. (2019). doi: 10.1016/j.cell.2019.11.010 [DOI] [PubMed] [Google Scholar]
- 24.Trapnell C et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. (2014). doi: 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uhlén M et al. Tissue-based map of the human proteome. Science 347(6220). (2015). doi: 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- 26.De Meyer SF, Stoll G, Wagner DD & Kleinschnitz C. Von Willebrand factor: An emerging target in stroke therapy. Stroke (2012). doi: 10.1161/STROKEAHA.111.628867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mao M, Alavi MV, Labelle-Dumais C & Gould DB. Type IV Collagens and Basement Membrane Diseases: Cell Biology and Pathogenic Mechanisms. Curr. Top. Membr. 43(2), 599–606. (2015). doi: 10.1016/bs.ctm.2015.09.002 [DOI] [PubMed] [Google Scholar]
- 28.DeSisto J et al. Single-Cell Transcriptomic Analyses of the Developing Meninges Reveal Meningeal Fibroblast Diversity and Function. Dev. Cell 54(1), 43–59. (2020). doi: 10.1016/j.devcel.2020.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Louveau A et al. Structural and functional features of central nervous system lymphatic vessels. Nature 523(7560), 337–341. (2015). doi: 10.1038/nature14432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aspelund A et al. A dural lymphatic vascular system that drains brain interstitial fluid and macromolecules. J. Exp. Med. 212(7), 991–999. (2015). doi: 10.1084/jem.20142290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dorrier CE et al. CNS fibroblasts form a fibrotic scar in response to immune cell infiltration. Nat. Neurosci. 24(2), 234–244. (2021). doi: 10.1038/s41593-020-00770-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iliff JJ et al. A paravascular pathway facilitates CSF flow through the brain parenchyma and the clearance of interstitial solutes, including amyloid β. Sci. Transl. Med. 4(147), 147ra111–147ra111. (2012). doi: 10.1126/scitranslmed.3003748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keren-Shaul H et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 169(7), 1276–1290. (2017). doi: 10.1016/j.cell.2017.05.018 [DOI] [PubMed] [Google Scholar]
- 34.Brown WR. A review of string vessels or collapsed, empty basement membrane tubes. Journal of Alzheimer’s Disease 21(3), 725–739. (2010). doi: 10.3233/JAD-2010-100219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roher AE et al. Cerebral blood flow in Alzheimer’s disease. Vasc. Health Risk Manag. 8, 599. (2012). doi: 10.2147/VHRM.S34874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montagne A et al. APOE4 leads to blood–brain barrier dysfunction predicting cognitive decline. Nature 581(7806), 71–76. (2020). doi: 10.1038/s41586-020-2247-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rockenstein E, Mallory M, Mante M, Sisk A & Masliaha E. Early formation of mature amyloid-β protein deposits in a mutant APP transgenic model depends on levels of Aβ142. J. Neurosci. Res. 66(4), 573–582. (2001). doi: 10.1002/jnr.1247 [DOI] [PubMed] [Google Scholar]
- 38.Nott A et al. Brain cell type–specific enhancer–promoter interactome maps and disease-risk association. Science 366(6469), 1134–1139. (2019). doi: 10.1126/science.aay0793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lambert JC et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 45(12), 1452–1458. (2013). doi: 10.1038/ng.2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kunkle BW et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 51(3), 414–430. (2019). doi: 10.1038/s41588-019-0358-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jansen IE et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 51(3), 404–413. (2019). doi: 10.1038/s41588-018-0311-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skene NG & Grant SGN. Identification of vulnerable cell types in major brain disorders using single cell transcriptomes and expression weighted cell type enrichment. Front. Neurosci. 10, 16. (2016). doi: 10.3389/fnins.2016.00016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karch CM & Goate AM. Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biological Psychiatry 77(1), 43–51. (2015). doi: 10.1016/j.biopsych.2014.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao Z et al. Central role for PICALM in amyloid-β blood-brain barrier transcytosis and clearance. Nat. Neurosci. 18(7), 978–987. (2015). doi: 10.1038/nn.4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cirrito JR et al. Synaptic activity regulates interstitial fluid amyloid-β levels in vivo. Neuron 48(6), 913–922. (2005). doi: 10.1016/j.neuron.2005.10.028 [DOI] [PubMed] [Google Scholar]
- 46.Safaiyan S et al. Age-related myelin degradation burdens the clearance function of microglia during aging. Nat. Neurosci. 19(8), 995–998. (2016). doi: 10.1038/nn.4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spangenberg E et al. Sustained microglial depletion with CSF1R inhibitor impairs parenchymal plaque development in an Alzheimer’s disease model. Nat. Commun. 10(1), 1–21. (2019). doi: 10.1038/s41467-019-11674-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gate D et al. Clonally expanded CD8 T cells patrol the cerebrospinal fluid in Alzheimer’s disease. Nature 577(7790), 399–404. (2020). doi: 10.1038/s41586-019-1895-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Farh KKH et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518(7539), 337–343. (2015). doi: 10.1038/nature13835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Villar D et al. Enhancer evolution across 20 mammalian species. Cell 160(3), 554–566. (2015). doi: 10.1016/j.cell.2015.01.006 Additional/ Method References [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wightman DP, Jansen IE, Savage JE, Shadrin AA, Bahrami S, Rongve A, ... & 23andMe Research Team. (2020). Largest GWAS (N= 1,126,563) of Alzheimer’s Disease Implicates Microglia and Immune Cells. medRxiv. [Google Scholar]
- 52.Vanlandewijck M, He L, Mäe MA, Andrae J, Ando K, Del Gaudio F, ... & Betsholtz C (2018). A molecular atlas of cell types and zonation in the brain vasculature. Nature, 554(7693), 475–480. [DOI] [PubMed] [Google Scholar]
- 53.Lee YK, Uchida H, Smith H, Ito A, & Sanchez T (2019). The isolation and molecular characterization of cerebral microvessels. Nature protocols, 14(11), 3059–3081. [DOI] [PubMed] [Google Scholar]
- 54.Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, ... & Chang HY (2017). An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nature methods, 14(10), 959–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang AC, Kern F, Losada PM, Agam MR, Maat CA, Schmartz GP, ... & Wyss-Coray T (2021). Dysregulation of brain and choroid plexus cell types in severe COVID19. Nature, 595(7868), 565–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McInnes L, Healy J, & Melville J (2018). Umap: Uniform manifold approximation and projection for dimension reduction. arXiv preprint arXiv:1802.03426. [Google Scholar]
- 57.Satija R, Farrell JA, Gennert D, Schier AF, & Regev A (2015). Spatial reconstruction of single-cell gene expression data. Nature biotechnology, 33(5), 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McGinnis CS, Murrow LM, & Gartner ZJ (2019). DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell systems, 8(4), 329337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, Van Der Zwan J, ... & Linnarsson S (2018). Molecular architecture of the mouse nervous system. Cell, 174(4), 999–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zeisel A, Muñoz-Manchado AB, Codeluppi S, Lönnerberg P, La Manno G, Juréus A, ... & Linnarsson S (2015). Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science, 347(6226), 1138–1142. [DOI] [PubMed] [Google Scholar]
- 61.Yang AC, Kern F, Losada PM, Agam MR, Maat CA, Schmartz GP, ... & Wyss-Coray T (2021). Dysregulation of brain and choroid plexus cell types in severe COVID19. Nature, 595(7868), 565–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou Y, Song WM, Andhey PS, Swain A, Levy T, Miller KR, ... & Colonna M (2020). Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nature medicine, 26(1), 131142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, ... & Gottardo R (2015). MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome biology, 16(1), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lake BB, Chen S, Sos BC, Fan J, Kaeser GE, Yung YC, ... & Zhang K (2018). Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nature biotechnology, 36(1), 70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, ... & Ma’ayan A (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC bioinformatics, 14(1), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, ... & Chanda SK (2019). Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nature communications, 10(1), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Conway JR, Lex A, & Gehlenborg N (2017). UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, ... & Nie Q (2021). Inference and analysis of cell-cell communication using CellChat. Nature communications, 12(1), 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tabula Muris Consortium. (2018). Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature, 562(7727), 367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zuchero YJY, Chen X, Bien-Ly N, Bumbaca D, Tong RK, Gao X, ... & Watts RJ (2016). Discovery of novel blood-brain barrier targets to enhance brain uptake of therapeutic antibodies. Neuron, 89(1), 70–82. [DOI] [PubMed] [Google Scholar]
- 71.Yousef H, Czupalla CJ, Lee D, Chen MB, Burke AN, Zera KA, ... & WyssCoray T (2019). Aged blood impairs hippocampal neural precursor activity and activates microglia via brain endothelial cell VCAM1. Nature medicine, 25(6), 988–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, ... & Zhang F (2015). In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nature biotechnology, 33(1), 102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Butler A, Hoffman P, Smibert P, Papalexi E, & Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature biotechnology, 36(5), 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang AC, Du Bois H, Olsson N, Gate D, Lehallier B, Berdnik D, ... & Wyss-Coray T (2018). Multiple click-selective tRNA Synthetases expand mammalian cell-specific proteomics. Journal of the American Chemical Society, 140(23), 7046–7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thul PJ, Åkesson L, Wiking M, Mahdessian D, Geladaki A, Blal HA, ... & Lundberg E. (2017). A subcellular map of the human proteome. Science, 356(6340). [DOI] [PubMed] [Google Scholar]
- 76.Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, ... & Weiner HL (2014). Identification of a unique TGF-β–dependent molecular and functional signature in microglia. Nature neuroscience, 17(1), 131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Szabo PA, Levitin HM, Miron M, Snyder ME, Senda T, Yuan J, ... & Sims PA (2019). Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease. Nature communications, 10(1), 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Iadecola C, Anrather J, & Kamel H (2020). Effects of COVID-19 on the nervous system. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Månberg A, Skene N, Sanders F, Trusohamn M, Remnestål J, Szczepińska A, ... & Lewandowski SA (2021). Altered perivascular fibroblast activity precedes ALS disease onset. Nature medicine, 27(4), 640–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Parker KR, Migliorini D, Perkey E, Yost KE, Bhaduri A, Bagga P, ... & Satpathy AT (2020). Single-cell analyses identify brain mural cells expressing CD19 as potential off-tumor targets for CAR-T immunotherapies. Cell, 183(1), 126–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, ... & Barres BA (2016). Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron, 89(1), 37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data is deposited under NCBI GEO: GSE163577. Data is also available to explore via an interactive web browser: https://twc-stanford.shinyapps.io/human_bbb.