Abstract
Background
Vaccination against COVID-19 was implemented very quickly, but the emergence of new variants that can evade the previous acquired immunological protection highlights the importance of understanding the mechanisms involved in the immune response generated after SARS-CoV-2 infection or vaccination.
Objectives
Since most of our knowledge on the humoral immunity generated against SARS-CoV-2 has been obtained from studies with infected patients before vaccination, our goal here was to evaluate seroconversion and its correlation with the titers of neutralizing antibodies (NAbs) in individuals who received the complete initial recommended vaccination schedule with three different vaccines.
Study design
We analyzed serum IgG, IgA and total NAbs against the trimeric SARS-CoV-2 Spike (S) protein or its receptor binding domain (RBD) in blood samples collected from 118 healthy individuals without known previous infection, before and after receiving the first and the second dose of CoronaVac (n = 18), ChAdOx-1 (n = 68) or BNT162b2 (n = 32) vaccines.
Results
We found that although IgG titers were high in all sera collected after the two doses of these vaccines, NAbs amounts varies among the groups. In contrast, serum NAbs concentrations were much more comparable to the IgA levels, indicating that these antibodies would have a major neutralizing capacity against SARS-CoV-2.
Conclusions
Altogether our data suggest that quantification of serum anti-S or anti-RBD IgA, rather than IgG, may be a valuable tool to screen NAbs and may be considered for surveillance of vaccine coverage.
Keywords: SARS-CoV-2, Vaccination, IgG, IgA, Neutralizing antibodies
Abbreviations
- S
Spike protein;
- RBD
receptor binding domain;
- Abs
antibodies;
- NAbs
neutralizing antibodies;
- ELISA
Enzyme-Linked Immunosorbent Assay.
1. Background
The lack of effective treatments for COVID-19 control led to the rapid development of several types of immunizing agents against SARS-CoV-2 early in the pandemic [1]. In April 2022, ten different vaccines were approved for large scale immunization worldwide [2] and currently 199 vaccine candidates are in the preclinical phase and 172 in clinical development [3]. Presently, about 68% of the world's population received at least one vaccine dose [4], but there are still many challenges to achieve a full protection against the disease and/or infection, including the development of strategies to induce long-term immunization and protection against new variants [5,6].
The quantification of serum antibodies (Abs), usually IgG, is an important tool for monitoring infection [7] and vaccination coverage [8], but not all Abs induced by the immunizing agent are neutralizing antibodies (NAbs) [9]. Thus, NAbs’ quantification represents the most appropriate way to verify the humoral protection against infection [10], but the assay costs make them inconvenient for serological surveillance.
2. Study design
We analyzed NAbs and IgG and IgA responses against SARS-CoV-2 Spike (S) protein or its receptor binding domain (RBD) in a cohort of 118 healthy individuals before and after receiving the first and the second doses of either CoronaVac (n = 18), ChAdOx-1 (n = 68) or BNT162b2 (n = 32) vaccines. A total of 250 blood samples were collected 15 to 30 days before vaccination (T0), 15 to 20 days after the first dose (T1), and/or 15 to 60 days after the second dose (T2) of each vaccine, according to the scheme represented in Fig. 1 . The participants completed an informed consent form and answered a survey providing data on demographics, medical history, and vaccine information, approved by the local ethics committees (CEP approvals HUCFF/UFRJ #35,303,120.5.0000.5257 and IOC/Fiocruz CAAE #56246022.1.0000.5248). Participants with previous SARS-CoV-2 infection were excluded.
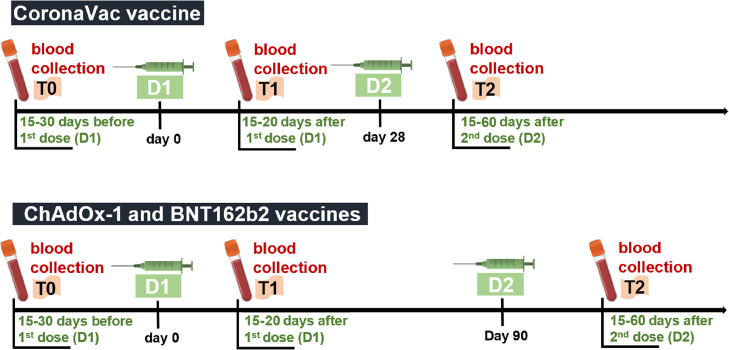
Fig. 1.
Diagram representing the blood collection scheme. A total of 250 blood samples were collected from 118 healthy individuals 15 to 30 days before vaccination (T0), 15 to 20 days after receiving the first dose (T1), and/or 15 to 60 days after receiving the second dose (T2) of either CoronaVac (n = 18), ChAdOx-1 (n = 68) or BNT162b2 (n = 32) vaccines. It is important to consider that not all participants underwent the three blood collections. We highlight that the participants vaccinated with CoronaVac received the second dose 28 days after receiving the first dose, while those vaccinated with ChAdOx-1 or BNT162b2 received the second dose 90 days after the first dose, according to the immunization protocols adopted by the Brazilian Ministry of Health. Blood samples were collected from 2020 to 2022 in the Laboratório de Análises Clínicas of the Faculdade de Farmácia at the Universidade Federal do Rio de Janeiro and in the Laboratório de Biotecnologia e Fisiologia das Infecções Virais of the Instituto Oswaldo Cruz. All samples were heat-inactivated at 56 °C for 30 min, aliquoted, and stored at −80 °C until further analysis.
We quantified IgG and IgA antibodies against the S and RBD proteins (Wuhan strain) in all sera samples at T0, T1 and T2, using an in-house ELISA assay previously developed and validated by our group [11], and NAbs concentration in T2 using the immunoenzimatic GenScript cPass™ SARS-CoV-2 assay. Demographic information of the participants is shown in Table 1 .
Table 1.
| Subject demographics and clinical characteristics.
| Characteristics | CoronaVac (n = 18) | ChAdOx-1 (n = 68) | BTN162b2 (n = 32) | |
|---|---|---|---|---|
| Age, average (range) | 38 (22 - 60) | 34 (22–58) | 39 (21 - 60) | |
| Sex,% (n) | Female | 89 (16) | 72 (49) | 56 (18) |
| Male | 11 (2) | 28 (19) | 44 (14) | |
| Comorbidities,% (n) | Diabetes mellitus | 0 | 4.4 (3) | 3.1 (1) |
| Hypertension | 5.5 (1) | 8.8 (6) | 12.5 (4) | |
| Lung diseases | 0 | 4.4 (3) | 3.1 (1) | |
| Autoimmune disease | 0 | 5.8 (4) | 0 | |
| Other comorbidities | 5.5 (1) | 2.9 (2) | 3.1 (1) |
3. Results
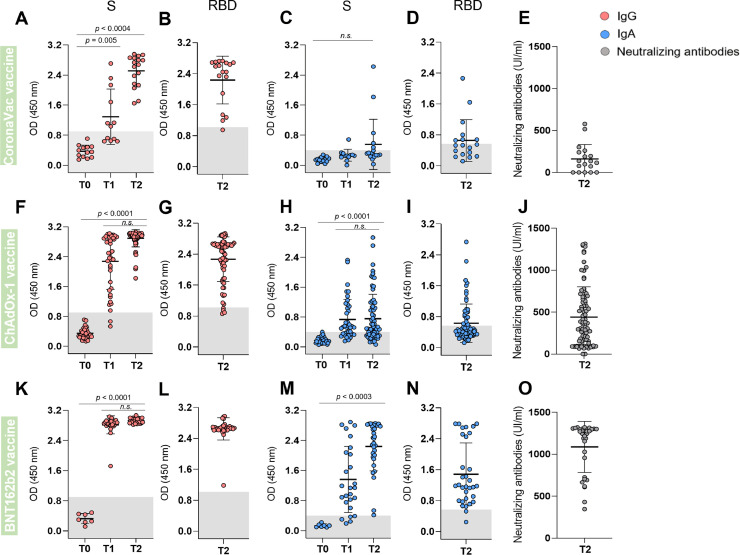
After the complete initial recommended vaccination schedule (two doses; T2), the three vaccines induced high anti-S (Fig. 2 A,F,K) and anti-RBD (Fig. 2B,G,L) IgG titers. Individuals vaccinated with ChAdOx-1 and BNT162b2 also presented high levels of anti-S IgG after the first dose (T1) (Fig. 2F,K). Regarding IgA response, we observed that CoronaVac did not induce a significant increase in either anti-S or anti-RBD IgA, even after the second dose (Fig. 2C,D), while some of participants vaccinated with ChAdOx-1 presented anti-S and anti-RBD IgA reactivity in T2 (Fig. 2H,I). BNT162b2 vaccine induced high anti-S IgA titers even in the T1, with a remarkable increase after the second dose (T2). In this case the anti-S IgA, the titers were slightly higher than those of anti-RBD IgA (Fig. 2M,N). Variations in our sample regarding the time span upon vaccination (15 to 60 days after T2), the age range of the participants (from 18 to 60 years) or the difference in the ratio of females and males for the three vaccination regiments did not affect the serum Abs profile. (Suppl. Fig. 1).
Fig. 2.
Serum antibody levels before and after vaccination. Serum samples from subjects who received CoronaVac (A-E), ChAdOx-1 (F-J), or BNT162b2 (K-O) vaccines were analyzed 15 to 30 days before vaccination (T0); 15 to 20 days after receiving the first dose of one vaccine (T1); and 15 to 60 days after receiving the second vaccine dose (T2). Reactivity of IgG (A, B, F, G, K, L – red symbols) or IgA (C, D, H, I, M, N – blue symbols) against S (A, C, F, H, K, M)) and RBD (B, D, G, I, L, N) of SARS-CoV-2 (Wuhan strain) was analyzed by ELISA. Briefly, sera samples diluted 1:50 in a 1% BSA solution in PBS-T were incubated for 2 h in 96 well-plates previously coated overnight, at 4 °C, with each of the antigens (50 μl of a 4 μg/ml solution), and blocked for 1 h with 3% BSA in PBS-T. The samples were incubated for 1 h with the detection antibodies (pan anti-IgG or pan anti-IgA, which recognizes all Ig isotypes) and reactivity was quantified spectrophotometrically at 450 nm after the addition of the chromogenic substrate (3,3′,5,5′-tetramethylbenzidine dihydrochloride, TMB). The gray band in the graphs represents the cut-off of each analysis, calculated as the mean + 3 SD of the absorbance values of 42 pre-pandemic sera. Semi-quantification of NAbs (E, J, O – dark gray symbols) was conducted using the cPass™ kit (GenScript). The NAbs titers were determined using a calibrator based on a standard neutralizing curve (WHO-GenScript). The inhibition rate of RBD binding to the ACE2 was the product of the interpolated titer from the standard curve and the sample dilution factor required to achieve the OD450 value that falls within the linear range. NAbs’ titers were significantly different between CoronaVac x ChAdOx-1 (p = 0.0026), CoronaVac x BNT162b2 (p<0.0001) or ChAdOx-1 x BNT162b2 (p<0.0001). Data were analyzed using ANOVA test followed by Sidak test using the GraphPad Prism V.8 program.
To better understand whether the different antibody responses generated by the vaccines could be associated with the protection they provide, we evaluated the percentage of NAbs in T2. On average, high NAbs’ levels were found in participants who received the BNT162b2 (1086.4 UI/ml), while lower levels were observed in individuals who received ChAdOx-1 (439.5 UI/ml) and even lower for CoronaVac (162.2 UI/ml) (Fig. 2E,J,O). NAbs levels were not affected by demographic aspects such as sex, age and presence of previous comorbidities (Suppl. Fig. 2).
Although the three vaccines induced high levels of IgG, serum NAbs concentrations were much more comparable to the IgA levels (Suppl. Fig. 3). When the individuals showing high IgG titers were grouped according to their NAbs levels, they appear equally distributed among the groups. In contrast, most of the individuals with high IgA titers exhibited high (60%) NAbs levels, while 32% presented medium and only a marginal group (8%) presented low NAbs levels. In the IgA negative group, in turn, only 5% individuals presented high NAbs levels. These results suggest that individuals having high IgA levels are more likely to have also more NAbs.
4. Discussion
Vaccination against SARS-CoV-2 was implemented very quickly, but most of our knowledge on the humoral immunity against the virus has been obtained with infected patients before vaccination. These studies showed that the post-infection levels of IgG and IgM produced against S and N (nucleocapsid) proteins waned in a few months [12], [13], [14], [15]. On the other hand, the dynamics of serum IgA against SARS-CoV-2 proteins are still poorly understood. Some studies showed a long-term decay of IgA titers [16,17], but more recent studies with vaccinated individuals showed that both serum IgA and IgG waned in a short period [18], [19], [20]. As IgA is the predominant serum immunoglobulin at the onset of disease and its titers correlated with the disease severity [21], it is reasonable to hypothesize that it plays a role in the infection control. Furthermore, IgAs produced in the mucosa are potent NAbs that efficiently stop SARS-CoV-2 infection [22].
NAbs produced after infection or vaccination are essential for protection against new infections [23,9]. During SARS-CoV-2 infection, the majority neutralizing antibodies are produced against RBD [24], suggesting these antibodies as the choice for serological surveillance [25]. However, a recent study showed that plasma IgA from >60% of a cohort of early convalescent COVID-19 subjects inhibited the interaction between RBD and ACE2 [26]. Therefore, monitoring the serum IgA response in the context of vaccination is critical to address its protective role in the polyclonal antibody response. Accordingly, our study shows a high correspondence between NAbs levels and the IgA response after two doses of SARS-CoV-2 vaccines, particularly ChAdOx-1 or BNT162b2. CoronaVac did not induce anti-S or anti-RBD IgA and generated low NAbs titers. However, the importance of the cellular immune response should also be considered [27,28], as data from subjects vaccinated with CoronaVac confirmed its safety and efficacy [29]. Indeed, its use in the pandemic early stages offered good protection against reinfection, even before the booster dose [30,31]. Moreover, the high level of serum IgG generated by CoronaVac could mediate the antibody-dependent phagocytosis, contributing to the virus clearance [26].
The immune response generated by ChAdOx-1 [32] and especially by BNT162b2 [33] has been shown to be quite effective in producing NAbs, corroborating the data presented here. Moreover, we showed that despite the high S and RBD-specific IgG production induced for all three vaccines, the levels of this immunoglobulin did not correlate to the NAbs. Nonetheless, it is important to note that we used here a high sensitivity ELISA, which allows most samples to be classified far above the cutoff. In addition, the cPass kit does not quantify all neutralizing antibodies, only those capable of binding and inhibit the interaction between RBD and ACE2.
In summary, our data showed that the serum IgA profile in vaccinated subjects is very similar to that of NAbs, which agrees with findings highlighting the robust IgA neutralizing capacity after infection by SARS-CoV-2 [34,35]. In conclusion, we suggest that quantification of serum anti-S or anti-RBD IgA, rather than IgG, may be a valuable tool to screen NAbs and may be considered for surveillance of vaccine coverage.
Funding
This work was supported by Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), Brazil [grant number E-26/211.128/2021, E-26/201.173/2021, E-26/211.134/2021, E-26/210.784/2021]; Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Brazil [grant number 312650/2021–3; 310361/2019–2]; National Institute of Science and Technology in Vaccines (INCTV), Brazil [grant number 465293/2014–0].
Ethics approval
This study was performed according to the principles of the Declaration of Helsinki under approval by the local ethics committee CONEP/CEP HUCFF/UFRJ #35303120.5.0000.5257 and IOC/Fiocruz CAAE #56246022.1.0000.5248.
Consent to participate
Informed consent was obtained from each participant included in this study.
CRediT author statement
Lorena O. Fernandes-Siqueira: conceptualization; design; methodology: ELISA; data curation; formal analysis; investigation; writing: original draft preparation, review and editing. Bruna G. Sousa: methodology: antigen expression and purification. Carlos E. Cleto: data curation; formal analysis. Luciana S. Wermelinger: conceptualization; design; sample preparation; writing: review and editing. Beatriz L. L. Caetano: methodology: Nabs quantification; formal analysis. Agatha R. Pacheco: methodology: Nabs quantification; formal analysis. Simone M. Costa: methodology: Nabs quantification; formal analysis. Fabio C. L. Almeida: conceptualization; design; resources. Gustavo C. Ferreira: conceptualization; design; writing: review and editing. Didier Salmon: conceptualization; design; writing: review and editing. Ada M. B. Alves: design; formal analysis; writing: review and editing; resources; funding acquisition. Andrea T. Da Poian: conceptualization; design; writing: review and editing; resources; funding acquisition; supervision.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to thank Dr. Marcos Fleury and all the members of Laboratório de Análises Clínicas, Faculdade de Farmácia (LACFar, UFRJ, Brazil), and Thiago Rodrigues Machado from the Laboratório de Biotecnologia e Fisiologia das Infecções Virais (IOC/Fiocruz), for performing the blood collections and sample preparation. We also thank Dr. Leda Castilho (COPPE, UFRJ, Brazil) for kindly providing the recombinant S protein.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jcvp.2022.100121.
Appendix. Supplementary materials
References
- 1.Lurie N., Saville M., Hatchett R., Halton J. Developing Covid-19 vaccines at pandemic speed. N. Engl. J. Med. 2020;382:1969–1973. doi: 10.1056/NEJMp2005630. [DOI] [PubMed] [Google Scholar]
- 2.WHO, https://www.who.int/emergencies/diseases/novel-coronavirus-2019/covid-19-vaccines/advice, 2022 (Accessed 26 September 2022).
- 3.WHO, https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines, 2022 (Accessed 26 September 2022).
- 4.H. Ritchie, E. Mathieu, L. Rodés-Guirao, C. Appel, C. Giattino, E. Ortiz-Ospina, J. Hasell, B. Macdonald, D. Beltekian, M. Roser, "Coronavirus Pandemic (COVID-19)", https://ourworldindata.org/coronavirus, 2020 (Accessed 26 September 2022).
- 5.Tregoning J.S., Flight K.E., Higham S.L., Wang Z., Pierce B.F. Progress of the COVID-19 vaccine effort: viruses, vaccines and variants versus efficacy, effectiveness and escape. Nat. Rev. Immunol. 2021;21(10):626–636. doi: 10.1038/s41577-021-00592-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung J.Y., Thone M.N., Kwon Y.J. COVID-19 vaccines: the status and perspectives in delivery points of view. Adv. Drug Deliv. Rev. 2021;170:1–25. doi: 10.1016/j.addr.2020.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaebler C., Wang Z., Lorenzi J.C.C., et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591(7851):639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lozano-Rodríguez R., Valentín-Quiroga J., Avendaño-Ortiz J., et al. Cellular and humoral functional responses after BNT162b2 mRNA vaccination differ longitudinally between naive and subjects recovered from COVID-19. Cell Rep. 2022;38(2) doi: 10.1016/j.celrep.2021.110235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lucas C., Klein J., Sundaram M.E., et al. Delayed production of neutralizing antibodies correlates with fatal COVID-19. Nat. Med. 2021;27:1178–1186. doi: 10.1038/s41591-021-01355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeyanathan M., Afkhami S., Smaill F., et al. Immunological considerations for COVID-19 vaccine strategies. Nat. Rev. Immunol. 2020;20:615–632. doi: 10.1038/s41577-020-00434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandes-Siqueira L.O., Ferreira F.A.P.…Da Poian A., et al. On the caveats of a multiplex test for SARS-CoV-2 to detect seroconversion after infection or vaccination. Sci. Rep. 2022;12:10366. doi: 10.1038/s41598-022-14294-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wheatley A.K., Juno J.A., Wang J.J., et al. Evolution of immune responses to SARS-CoV-2 in mild-moderate COVID-19. Nat. Commun. 2021;12(1):1162. doi: 10.1038/s41467-021-21444-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lumley S.F., Wei J., O'Donnell D., et al. The duration, dynamics, and determinants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibody responses in individual healthcare workers. Clin. Infect. Dis. 2021;73(3):e699–e709. doi: 10.1093/cid/ciab004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Greef J., Scohy A., Zech F., et al. Determinants of IgG antibodies kinetics after severe and critical COVID-19. J. Med. Virol. 2021;93(9):5416–5424. doi: 10.1002/jmv.27059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Efrati S., Catalogna M., Hamed R.A., et al. Early and long term antibody kinetics of asymptomatic and mild disease COVID-19 patients. Sci. Rep. 2021;11(1):13780. doi: 10.1038/s41598-021-93175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plūme J., Galvanovskis A., Šmite S., et al. Early and strong antibody responses to SARS-CoV-2 predict disease severity in COVID-19 patients. J. Transl. Med. 2022;20:176. doi: 10.1186/s12967-022-03382-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rangel-Ramírez V.V., Macías-Piña K.A., Garrido R.R.S., de Alba-Aguayo D.R., Moreno-Fierros L., Rubio-Infante N. A systematic review and meta-analysis of the IgA seroprevalence in COVID-19 patients; is there a role for IgA in COVID-19 diagnosis or severity? Microbiol. Res. 2022 doi: 10.1016/j.micres.2022.127105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zurac S., Vladan C., Dinca O., Constantin C., Neagu M. Immunogenicity evaluation after BNT162b2 booster vaccination in healthcare workers. Sci. Rep. 2022;12:12716. doi: 10.1038/s41598-022-16759-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takeuchi J.S., Fukunaga A., Yamamoto S., et al. SARS-CoV-2 specific T cell and humoral immune responses upon vaccination with BNT162b2: a 9 months longitudinal study. Sci. Rep. 2022;12:15447. doi: 10.1038/s41598-022-19581-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rammauro F., Carrión F., Olivero-Deibe N., Fló M., Ferreira A., Pritsch O., Bianchi S. Humoral immune response characterization of heterologous prime-boost vaccination with CoronaVac and BNT162b2. Vaccine. 2022;40:5189–5196. doi: 10.1016/j.vaccine.2022.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zervou F.N., Louie P., Stachel A., Zacharioudakis I.M., Ortiz-Mendez Y., Thomas K., Aguero-Rosenfeld M.E. SARS-CoV-2 antibodies: IgA correlates with severity of disease in early COVID-19 infection. J. Med. Virol. 2021;93(9):5409–5415. doi: 10.1002/jmv.27058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Planchais C., Fernández I., Bruel T., de Melo G.D., et al. Potent human broadly SARS-CoV-2-neutralizing IgA and IgG antibodies effective against Omicron BA.1 and BA.2. J. Exp. Med. 2022;4(7):219. doi: 10.1084/jem.20220638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khoury D.S., Cromer D., Reynaldi A., et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 24.Piccoli L., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peterhoff D., Glück V., Vogel M., et al. A highly specific and sensitive serological assay detects SARS-CoV-2 antibody levels in COVID-19 patients that correlate with neutralization. Infection. 2021;49(1):75–82. doi: 10.1007/s15010-020-01503-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis S.K., Selva K.J., Lopez E., et al. Heterologous SARS-CoV-2 IgA neutralising antibody responses in convalescent plasma (preprint) MedRxiv. 2022 doi: 10.1101/2022.02.06.22270359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sadarangani M., Marchant A., Kollmann T.R. Immunological mechanisms of vaccine-induced protection against COVID-19 in humans. Nat. Rev. Immunol. 2021;21(8):475–484. doi: 10.1038/s41577-021-00578-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Escobar A., Reyes-López F.E., Acevedo M.L., Alonso-Palomares L., Valiente-Echeverría F., et al. Evaluation of the immune response induced by coronaVac 28-day schedule vaccination in a healthy population group. Front. Immunol. 2022;12 doi: 10.3389/fimmu.2021.766278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanriover M.D., Doğanay H.L., Akova M., et al. Efficacy and safety of an inactivated whole-virion SARS-CoV-2 vaccine (CoronaVac): interim results of a double-blind, randomised, placebo-controlled, phase 3 trial in Turkey. Lancet. 2021;398(10296):213–222. doi: 10.1016/S0140-6736(21)01429-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ranzani O.T., Hitchings M.D.T., Dorion M., Agostini T.L., de Paula R.C., O.de Paula P.F., et al. Effectiveness of the CoronaVac vaccine in older adults during a gamma variant associated epidemic of covid-19 in Brazil: test negative case-control study. BMJ. 2021;374 doi: 10.1136/bmj.n2015. n2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Villela D.A., de Noronha T.G., Bastos L.S., Pacheco A.G., Cruz O.G., Carvalho L.M.…Struchiner C.J., et al. Effectiveness of mass vaccination in Brazil against severe COVID-19 cases. MedRxiv. 2021 doi: 10.1101/2021.09.10.21263084. (preprint) [DOI] [Google Scholar]
- 32.Madhi S.A., Baillie V., Cutland C.L., et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 Vaccine against the B.1.351 Variant. N. Engl. J. Med. 2021;384(20):1885–1898. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Favresse J., Gillot C., Di Chiaro L., et al. Neutralizing antibodies in COVID-19 patients and vaccine recipients after two doses of BNT162b2. Viruses. 2021;13(7):1364. doi: 10.3390/v13071364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sterlin D., Mathian A., Miyara M., Mohr A., Anna F., Gorochov G. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci. Transl. Med. 2021;13:eabd2223. doi: 10.1126/scitranslmed.abd2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z., Lorenzi J.C.C., Muecksch F., et al. Enhanced SARS-CoV-2 neutralization by dimeric IgA. Sci. Transl. Med. 2021;13:eabf1555. doi: 10.1126/scitranslmed.abf1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.