Abstract
Trisomy 21, the source of Down syndrome, causes a 0.5-fold protein increase of the chromosome 21-resident gene Pericentrin (PCNT) and reduces primary cilia formation and signaling. We investigate how PCNT imbalances disrupt cilia. Using isogenic RPE-1 cells with increased chromosome 21 dosage, we find PCNT accumulates around the centrosome as a cluster of enlarged cytoplasmic puncta that localize along microtubules (MTs) and at MT ends. Cytoplasmic PCNT puncta impact the density, stability, and localization of the MT trafficking network required for primary cilia. The PCNT puncta appear to sequester cargo peripheral to centrosomes in what we call pericentrosomal crowding. The centriolar satellite proteins PCM1, CEP131, and CEP290, important for ciliogenesis, accumulate at enlarged PCNT puncta in trisomy 21 cells. Reducing PCNT when chromosome 21 ploidy is elevated is sufficient to decrease PCNT puncta and pericentrosomal crowding, reestablish a normal density of MTs around the centrosome, and restore ciliogenesis to wild-type levels. A transient reduction in MTs also decreases pericentrosomal crowding and partially rescues ciliogenesis in trisomy 21 cells, indicating that increased PCNT leads to defects in the MT network deleterious to normal centriolar satellite distribution. We propose that chromosome 21 aneuploidy disrupts MT-dependent intracellular trafficking required for primary cilia.
INTRODUCTION
Primary cilia are signaling platforms required for development and homeostasis, and ciliopathies are genetic disorders affecting genes that impact ciliary function (Nigg and Raff, 2009; Reiter and Leroux, 2017; Breslow and Holland, 2019). Down syndrome (DS), or trisomy 21, is a distinct genetic condition caused by an extra copy of human somatic autosome 21 (HSA21) that also negatively impacts development (Hattori et al., 2000). Consistent with the phenotypic overlap between pathologies associated with DS and ciliopathies, trisomy 21 produces primary cilia and cilia associated signaling defects (Roper et al., 2006; Ripoll et al., 2012; Currier et al., 2012; Galati et al., 2018).
Primary cilia assemble from the centrosome comprised of two centrioles surrounded by a dense protein matrix called the pericentriolar material (PCM; Sorokin, 1962; Gould and Borisy, 1977; Bornens and Azimzadeh, 2007; Bettencourt-Dias et al., 2011; Nigg and Stearns, 2011). The mother centriole nucleates and organizes the primary cilium (Breslow and Holland, 2019). The centrosome PCM and associated centriolar appendage structures anchor and organize cytoplasmic microtubules (MTs) required for trafficking cargoes to and from centrosomes, including cargoes required for cilia (Sorokin, 1962; Kubo et al., 1999; Bornens, 2002; Kim et al., 2004; Sung and Leroux, 2013; Woodruff et al., 2014). In addition to the centrosome, the closely associated Golgi apparatus nucleates MTs responsible for cell migration in interphase cells (Efimov et al., 2007; Gavilan et al., 2018). Vesicles moving from the Golgi apparatus to the centrosome are critical for early events in cilia formation (Follit et al., 2006; Shakya and Westlake, 2021). In the case of primary cilia formation during interphase, MT minus ends emanate from the centrosome PCM and mother centriole appendages, establishing a distribution hub for MT motor-driven cargoes to and from the cilium (Kubo et al., 1999; Bornens, 2002; Dammermann and Merdes, 2002). To ensure accurate cargo translocations, the number and stability of MTs are tightly controlled (Doxsey, 2001; Woodruff et al., 2014; Vertii et al., 2016).
Within the trafficking milieu required for cilia formation and function are centriolar satellites. Centriolar satellites are 70–100 nm nonmembrane bound granule structures that localize around the centrosome and move along MTs (Bernhard and de Harven, 1960; Kubo et al., 1999; Dammermann and Merdes, 2002; Hori and Toda, 2017; Odabasi et al., 2019). Centriolar satellites positively and negatively modulate primary cilia through protein transport, degradation, and sequestration (Tollenaere et al., 2015; Odabasi et al., 2019; Prosser and Pelletier, 2020). The pericentriolar material-1 (PCM1) centriolar satellite scaffold protein ensures centriolar satellite integrity and is required for ciliogenesis (Gupta et al., 2015b; Wang et al., 2016). PCM1 loss disassembles centriolar satellites, reduces the PCM, and disrupts primary cilia (Dammermann and Merdes, 2002; Keryer et al., 2011; Odabasi et al., 2019; Quarantotti et al., 2019; Prosser and Pelletier, 2020). The dynamic interplay between centrosomal PCM and centriolar satellites is instrumental for primary cilia, yet how it is controlled and how it is disrupted in disease remains poorly understood.
Pericentrin (PCNT) is a centrosome PCM scaffold protein essential for ciliary protein recruitment and MT nucleation (Delaval and Doxsey, 2010; Sung and Leroux, 2013; Woodruff et al., 2014). Along with CDK5RAP2/CEP215, PCNT recruits γ-tubulin to the centrosome and promotes MT nucleation and organization (Dictenberg et al., 1998; Fong et al., 2008). PCNT also interacts with PCM1 and supports MT motor-dependent trafficking to and from centrosomes in particles that variably associate with centriolar satellites (Doxsey et al., 1994; Young et al., 2000; Dammermann and Merdes, 2002; Galati et al., 2018). PCNT is not necessary for interphase centrosome MT organization but is essential for mitotic spindle assembly (Dammermann and Merdes, 2002; Zimmerman et al., 2004; Gavilan et al., 2018). However, PCNT forms a complex with intraflagellar transport (IFT) proteins, and PCNT loss or overexpression disrupts IFT20 localization to the centrosome and primary cilia formation (Jurczyk et al., 2004; Galati et al., 2018). The precise roles for PCNT in promoting cilia formation are unknown. Moreover, PCM1 and PCNT interactions promote centrosome PCM assembly (Li et al., 2001; Dammermann and Merdes, 2002). The PCNT gene is on HSA21 and protein levels are elevated in DS (Salemi et al., 2013; Galati et al., 2018). Using heterologous and mosaic DS cell lines, elevated PCNT was found to disrupt primary cilia (Galati et al., 2018). Elevated PCNT in trisomy 21 produces enlarged centrosomes and cytoplasmic PCNT puncta that behave as phase dense particles (Galati et al., 2018; Jiang et al., 2021). As puncta enlarge, the movement of PCNT along MTs is suppressed, as is the localization of factors required for ciliogenesis (IFT20) and ciliary-dependent signaling (Smoothened; Galati et al., 2018). A mechanistic understanding for how elevated and enlarged PCNT puncta control MTs, centriolar satellite localization, and primary cilia formation is still required.
RESULTS AND DISCUSSION
Disrupted primary cilia in increased HSA21 isogenic lines is PCNT dependent
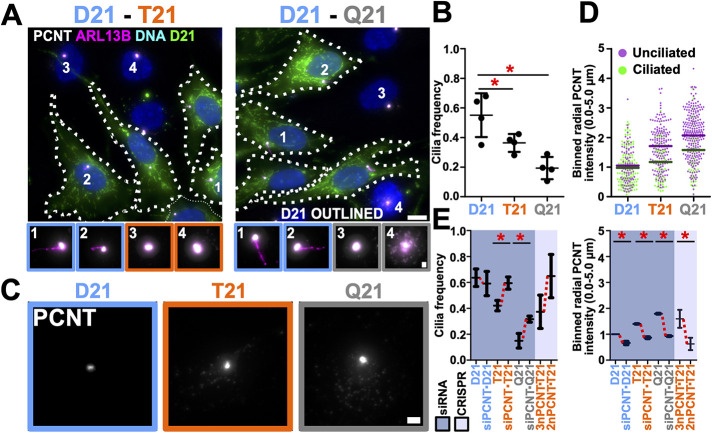
To study the controlled effects of HSA21 dosage on primary cilia, we utilized isogenic RPE-1 cell lines (disomy 21; D21) engineered with either one (trisomy 21; T21) or two (tetrasomy 21; Q21) extra copies of HSA21 (Lane et al., 2014). Primary cilia frequency was quantified in cells incubated for 24 h in reduced serum medium where T21 or Q21 cells were cocultured with D21 cells to control for confluency, cell cycle, and non-cell autonomous effects (Figure 1, A and B, and Supplemental Figure S1, A and B). Cilia frequency was reduced in an HSA21 dose-dependent manner (32% and 56% reduction for T21 and Q21; Figure 1B). Primary cilia length was not changed and Q21 cells were slightly larger than D21 and T21 cells (Supplemental Figure S1, E and F). Total cellular protein levels of the HSA21 resident gene, PCNT, in T21 and Q21 cells were increased as measured by both Western blot band intensity and immunofluorescence intensity quantification (Supplemental Figure S1, C and D). Importantly, PCNT levels at the centrosome are elevated, enlarging centrosomes (Galati et al., 2018), and levels within a 5 μm radius surrounding the centrosome in unciliated T21 and Q21 cells were 42% and 31% greater than their ciliated cell counterparts, respectively (Figure 1, C and D). Thus, elevated PCNT is associated with decreased cilia formation. However, there is no strict PCNT threshold across the three cell lines above which cilia do not form. This suggests that cells compensate for elevated PCNT and/or that increased HSA21 dosage suppresses some of the negative impacts elevated PCNT has on cilia. To confirm that elevated PCNT is sufficient to impact cilia frequency, PCNT was reduced in T21 and Q21 cells. The reduction of PCNT in T21 and Q21 cells to approximately D21 levels using siRNA increased cilia frequencies to 92% and 50% of D21 levels, respectively (Figure 1E). One possibility for the incomplete recovery in the siRNA experiments is because cells were coincidentally depleted for PCNT and induced for ciliogenesis such that ciliating cells do not have the complete 24 h of serum depletion with normal PCNT levels. Consistent with this, genetic ablation of a single PCNT allele in T21 cells using CRISPR-Cas9 (leaving cells with 2n PCNT) also increased the mean cilia frequency to 101% of D21 levels (Figure 1E, and Supplemental Figure S1, G–J). In summary, a dose-dependent increase in PCNT from HSA21 inhibits primary cilia formation in isogenic RPE-1 cells.
FIGURE 1:
Elevated chromosome 21 dosage and PCNT disrupt primary cilia formation. (A) Left panel, cocultured disomy 21 (D21) and trisomy 21 (T21) RPE-1 cells stained for centrosomes (Pericentrin [PCNT]; grayscale), cilia (ARL13B; magenta), and DNA (Hoescht33258; blue). Right panel, analogously stained cocultured D21 and Q21 cells. D21 cells were CFSE stained (green) and outlined. Cell number labels are referenced in bottom panels. Scale bars, 10 μm and 1 μm for insets. (B) Cilia frequency decreases with HSA21 ploidy. Mean ± SD. *, p < 0.05 (Supplemental Table S1). (C) PCNT florescence at the centrosome (PCNT; grayscale) in D21, T21, and Q21 RPE-1 cells. Scale bar, 2 μm. (D) PCNT level increase with HSA21 ploidy and in unciliated relative to ciliated cells. Intensity values are normalized to D21 average. Data points represent PCNT fluorescence intensity within a 5-μm radius surrounding the centrosome. (E) Reducing PCNT via siRNA or CRISPR-Cas9 knockout of one allele of PCNT increases cilia frequencies in T21 and Q21 RPE-1 cells. Changes in mean values indicated with red lines. Intensity values normalized to D21 average. Data points represent PCNT fluorescence intensity within a 5-μm radius of the centrosome. Mean ± SD. *, p < 0.05 (Supplemental Table S1).
Elevated PCNT forms enlarged puncta around the pericentrosomal region that is associated with reduced cilia
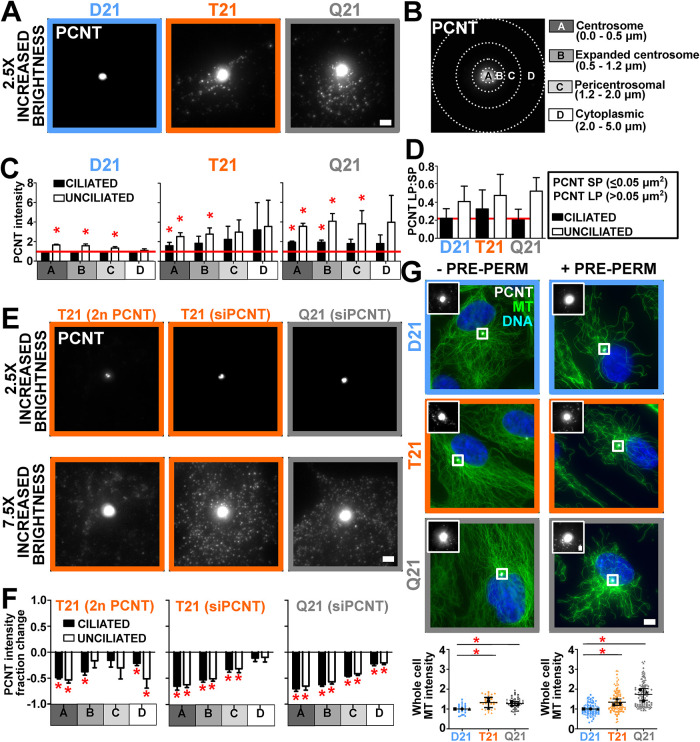
To understand how elevated PCNT inhibits cilia formation, we investigated differences in PCNT protein distribution at and around the centrosome relative to total cellular levels. In addition to increased PCNT at the centrosome (Figure 1C), enlarged PCNT puncta surround the centrosome (Figure 2A; Galati et al., 2018). The radial distribution of total PCNT fluorescence intensity around the centrosome was quantified and normalized to ciliated D21 PCNT fluorescence intensity levels by quantifying centrosomal (radius = 0.0–0.5 μm), expanded centrosome (radius = 0.5–1.2 μm), pericentrosomal (radius = 1.2–2.0 μm), and surrounding (radius = 2.0–5.0 μm) regions (Figure 2, B and C, and Supplemental Figure S2A). These regions were selected based on regions where differential localization was observed for PCNT. Because the distribution of PCNT is not always symmetric around the centrosome, PCNT asymmetries generate variance that is averaged across the data. The peripheral noncentrosomal PCNT surrounding the centrosome (radius = 1.2–5.0 μm) represents 44–53% of total cellular PCNT intensity (Supplemental Figure S2B). Although T21 and Q21 PCNT levels were elevated in all regions, PCNT is increased by 1.6- and 1.9-fold at the centrosome and 1.5- and 2.3-fold in regions surrounding the centrosome in unciliated T21 and Q21 cells as compared with their ciliated cell counterparts (Figure 2C, and Supplemental Figure S2A). The population of PCNT around the centrosome (radius = 1.2–5.0 μm) is comprised of both large and small puncta (Figure 2, A and D). Enlarged puncta (LP; greater than 0.05 μm2) commonly associate with MTs (see below; Supplemental Figure S2I). Unciliated cells tended to have more enlarged PCNT puncta than ciliated cells even in D21 cells (Figure 2D), consistent with the hypothesis that their accumulation inhibits primary cilia formation. Moreover, decreasing PCNT levels with siRNA or CRISPR-Cas9 reduces PCNT fluorescence intensity both at and around centrosomes (Figure 2, E and F). This suggests increases to PCNT surrounding centrosomes cause fewer cilia.
FIGURE 2:
Elevated PCNT forms large puncta peripheral to the centrosome that disrupt cilia formation. (A) PCNT fluorescence (PCNT; grayscale) in D21, T21, and Q21 RPE-1 cells. Brightness increased by a factor of 2.5. Scale bar, 2 μm. (B) Centrosome (A), expanded centrosome (B), pericentrosomal (C), and cytoplasmic (D) regions for binned radial fluorescence intensity analysis from the centroid of the centrosome. (C) D21, T21, and Q21 regional binned PCNT fluorescence intensities. Intensity values normalized to ciliated D21 average (indicated with red line). Statistical comparisons made to ciliated D21 averages. Mean ± SD. *, p < 0.05 (Supplemental Table S1). (D) PCNT large puncta (LP) to small puncta (SP) ratios (LP:SP) in ciliated and unciliated D21, T21, and Q21 cells. Mean ± SD. (E) Regional PCNT fluorescence (PCNT; grayscale) in PCNT CRISPR-Cas9 knockout line (T21 [2n PCNT]) and cells treated with PCNT siRNA (T21 [siPCNT], Q21 [siPCNT]). Scale bar, 2 μm. Top panels, brightness increased by a factor of 2.5. Bottom panels, brightness increased by a factor of 7.5. (F) Reducing PCNT via CRISPR-Cas9 knockout of one allele in T21 and siRNA in T21 and Q21 RPE-1 cells reduces PCNT intensity at and peripheral to the centrosome. Graphical and statistical comparisons made to control intensities. Mean ± SD. *, p < 0.05 (Supplemental Table S1). (G) Left panels, D21, T21, and Q21 RPE-1 cells stained for MTs (DM1A; green), PCNT (PCNT; grayscale), and DNA (Hoescht 33258; blue). PCNT insets are shown for centrosomes. Brightness was increased for insets by a factor of 4. MTs were fixed without prepermeabilization (−PRE-PERM). Scale bar, 5.5 μm and 1 μm for insets. Right panels, D21, T21, and Q21 RPE-1 cells stained for MTs (DM1A; green), PCNT (PCNT; grayscale), and DNA (Hoescht 33258; blue). PCNT insets are shown for boxed centrosomes. Brightness was increased for insets by a factor of 4. MTs in these images were fixed with prepermeabilization (+PRE-PERM). Scale bar, 5.5 μm and 1 μm for insets. Left graph, D21, T21, and Q21 whole-cell MT fluorescence intensities without prepermeabilization. Right graph, D21, T21, and Q21 whole-cell MT fluorescence intensities with prepermeabilization. Intensity values are normalized to D21 average. Statistical comparisons made to D21 averages. Mean ± SD. *, p < 0.05 (Supplemental Table S1).
Elevated PCNT puncta increase cytoplasmic MTs
Cytoplasmic PCNT puncta arise either from self-assembly or from expansion of the centrosome population into the pericentrosomal space. We suggest that these puncta are analogous to the cytoplasmic MT organizing centers found when centrioles, centrosomes, and the Golgi apparatus are disrupted (Gavilan et al., 2018). To test this, we measured the effects of increased HSA21 dosage on MTs. Total cell MT intensities were elevated with increased HSA21 dosage (Figure 2G, and Supplemental Figure S2, C and D). Because centrosomes and the Golgi apparatus are in close proximity and because MTs of interphase RPE-1 cells nucleate from both the centrosome and Golgi apparatus (Efimov et al., 2007; Sutterlin and Colanzi, 2010; Gavilan et al., 2018), we tested whether elevated MT intensity was associated with the Golgi apparatus (Supplemental Figure S2E). In contrast to the centrosome and pericentrosomal regions, only a minor increase in MT intensity overlapping with the Golgi apparatus was observed in Q21 but not T21 cells (Supplemental Figure S2E). In addition, the contribution of Golgi apparatus MTs relative to the increase in whole-cell MTs was not significantly elevated across D21, T21, and Q21 cells (Supplemental Figure S2E). Moreover, we did not observe significant changes to the Golgi apparatus size or proximity to centrosomes relative to HSA21 dosage (Jewett et al., 2021). Furthermore, the proximity of the centrosome to the Golgi apparatus is not functionally necessary for protein transport in RPE-1 cells (Tormanen et al., 2019). Trafficking from the Golgi apparatus to the centrosome may be disrupted in trisomy 21 (Galati et al., 2018) and the likeliest explanation is the elevated MTs at the centrosome and pericentrosomal regions, either due to more MT nucleation or enhanced MT stability, and not to changes in Golgi apparatus MT nucleation or Golgi apparatus localization.
To test whether MTs are more stable in T21 and Q21 cells, cells were exposed to either a 5-min cell prepermeabilization or an acute 10-min cold treatment (cold MT depolymerization [CD]), both of which disassemble dynamic MTs (Figure 2G, and Supplemental Figure S2, F and G). The 5-min prepermeabilization before fixation resulted in increased MT densities in T21 and Q21 cells when compared with D21 cells receiving the same treatment (Figure 2G). Moreover, consistent with T21 and Q21 having greater MT stability, increasing MT densities were found in T21 and Q21 cells as compared with D21 cells after a 10-min CD (Supplemental Figure S2, F and G). Together, these results suggest that MT stability is increased in T21 and Q21 cells.
To address whether MT posttranslational modifications (PTMs) contribute to increased MT stability in T21 and Q21 cells, tubulin and tubulin PTM densities were quantified (Supplemental Figure S2H). Tubulin staining density was slightly elevated in Q21 cells; however, this increase was not enough to account for the differences in the whole-cell MT intensities that were observed in these cells (Supplemental Figure S2H). Moreover, detyrosinated tubulin was slightly increased in Q21 cells and acetylation and polyglutamylation remained unchanged under the conditions tested (Supplemental Figure S2H). In summary, increased MT stability in T21 and Q21 cells is likely independent of MT PTMs.
Enlarged PCNT puncta increase MTs and are associated with reduced ciliation
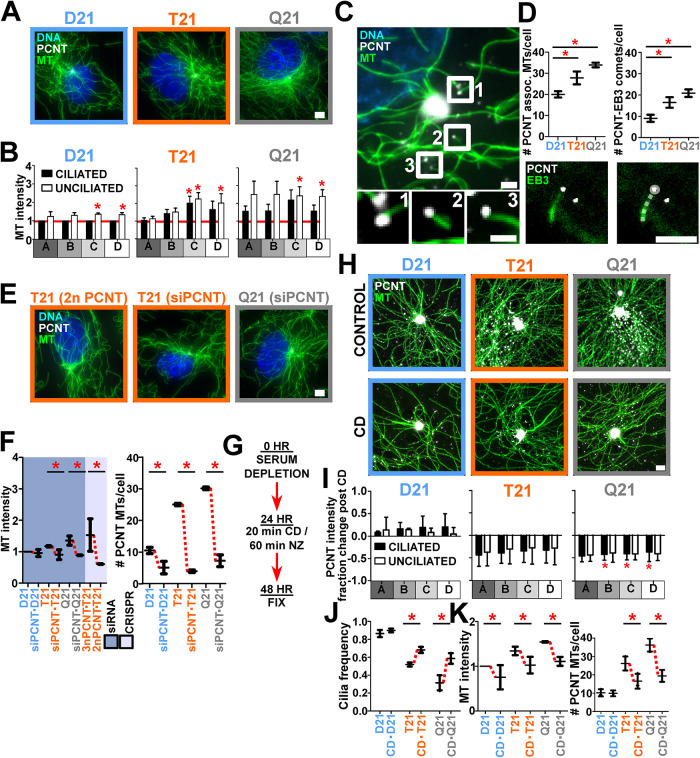
We next investigated whether elevated PCNT and PCNT puncta in T21 and Q21 cells contribute to the increased MT density at and around the centrosome. Indeed, both regional and whole-cell MT densities using preextraction fixation methods were increased in both unciliated cells and cells with elevated HSA21 dosage (Figure 3, A and B, and Supplemental Figure S3, A and B). D21 and Q21 cells exhibited significant positive correlation coefficients when comparing whole-cell PCNT and MT intensities (Supplemental Figure S3B). Coincident with increased PCNT intensities (Figure 2C, and Supplemental Figure S2A), MTs in T21 and Q21 cells were elevated in all regions surrounding centrosomes (Figure 3B, and Supplemental Figure S3A). The levels of PCNT interacting partners, CEP215 and γ-tubulin, were also increased at and surrounding centrosomes (Supplemental Figure S3, C–E). In T21 and Q21 cells, an increased number of free MT ends colocalize with γ-tubulin and PCNT (Supplemental Figure S3E). This suggests that the increase in PCNT associated with HSA21 dosage and unciliated cells increases MT-nucleation factors and MTs. Because a larger increase in MTs is observed in the pericentrosomal region in unciliated cells, we hypothesize this PCNT and MT population is responsible for disrupting cilia formation.
FIGURE 3:
Large PCNT puncta nucleate free MTs that are associated with decreased ciliation. (A) D21, T21, and Q21 RPE-1 cells stained for MTs (DM1A; green), PCNT (PCNT; grayscale), and DNA (Hoescht 33258; blue). Prepermeabilization was performed before fixation. Scale bar, 4 μm. (B) D21, T21, and Q21 regional binned MT fluorescence intensities. Intensity values normalized to ciliated D21 average (indicated with red line). Statistical comparisons made to ciliated D21 averages. Mean ± SD. *, p < 0.05 (Supplemental Table S1). (C) PCNT localizes to MT ends. Unciliated D21 RPE-1 cell stained for MTs (DM1A; green) and PCNT (PCNT; grayscale). Labels are referenced in bottom panels. Scale bars, 2 μm and 0.75 μm for insets. (D) Top, HSA21 dosage increases PCNT colocalization at MT ends and PCNT-associated growing MTs. Bottom, RPE-1 cell labeled with EB3-mNeon and PCNT (PCNT; grayscale). Right panel includes a line trace of the EB3 comet. Scale bars, 2.5 μm. Mean ± SD. *, p < 0.05 (Supplemental Table S1). (E) CRISPR-Cas9 knockout line (T21 [2n PCNT]) and cells treated with PCNT siRNA (T21 [siPCNT], Q21 [siPCNT]) stained for MTs (DM1A; green), PCNT (PCNT; grayscale), and DNA (Hoescht33258; blue). Scale bar, 4 μm. (F) Left, reducing PCNT via siRNA in T21 and Q21 RPE-1or CRISPR-Cas9 knockout of one allele in T21 cells reduces MT intensities. Intensity values normalized to D21 average. Data points represent 5-μm binned radial MT fluorescence intensities. Right, reducing PCNT via siRNA reduces PCNT-associated MTs in D21, T21, and Q21 RPE-1 cells. Mean ± SD. *, p < 0.05 (Supplemental Table S1). (G) Timeline for cold-depolymerization (CD) and nocodazole (NZ) treatment experiments. (H) Top panels, D21, T21, and Q21 RPE-1 cells stained for MTs (DM1A; green) and PCNT (PCNT; grayscale). Bottom panels, D21, T21, and Q21 RPE-1 CD-treated cells stained for MTs (DM1A; green) and PCNT (PCNT; grayscale). Scale bar, 1.5 μm. (I) CD reduces PCNT intensities at and peripheral to the centrosome in T21 and Q21 RPE-1 cells. Statistical comparisons made to relative control intensities. Mean ± SD. *, p < 0.05 (Supplemental Table S1). (J) CD increases cilia frequencies in T21 and Q21 RPE-1 cells. Mean ± SD. *, p < 0.05 (Supplemental Table S1). (K) Left, CD reduces MT intensities in T21 and Q21 RPE-1 cells. Intensity values normalized to D21 average. Data points represent 5-μm binned radial MT fluorescence intensities. Right, CD reduces PCNT-associated MTs in D21, T21, and Q21 RPE-1 cells. Mean ± SD. *, p < 0.05 (Supplemental Table S1).
To test whether PCNT puncta are associated with free MT ends, the number of centrosome-free MTs associated with PCNT puncta was quantified (Figure 3, C and D). An increase of 39% and 70% of centrosome-free MTs associated with PCNT puncta was observed in T21 and Q21 compared with D21 cells, respectively (Figure 3D). To determine whether these are sites of active MT growth, the number of growing MT plus ends (EB3 comets) associated with the enlarged PCNT puncta was quantified in D21, T21, and Q21 cells (Figure 3D). PCNT-associated EB3 comets increased with elevated HSA21 dosage (Figure 3D). This suggests that enlarged PCNT puncta in the pericentrosomal region act to organize MTs peripheral to the centrosome. Radial analysis of MT distribution using structured illumination microscopy (SIM) imaging further supports the model that MTs are elevated in the expanded centrosome and pericentrosomal regions (Supplemental Figure S2K). Moreover, depletion of PCNT using siRNA or CRISPR-Cas9 reduced T21 and Q21 MT intensities and PCNT-associated free MTs to or below D21 levels (Figure 3, E and F). Thus, enlarged cytoplasmic PCNT puncta promote increased MT growth in both unciliated cells and cells with elevated HSA21 dosage. We suggest that these MT populations repress cilia formation.
To investigate whether increased MTs caused by elevated PCNT contribute to primary cilia defects in T21 and Q21 cells, we reduced MTs using CD (Figure 3, G–K). Following a 20-min CD treatment and 24-h recovery, MTs were reduced when compared with cells that did not receive the CD treatment (Figure 3, H and K). Importantly, whole-cell PCNT and PCNT at and surrounding T21 and Q21 centrosomes was reduced (Figure 3, H and I, and Supplemental Figure S3I). The relative mean primary cilia frequency increased by 31% and 86% in T21 and Q21 cells after CD treatment, respectively (Figure 3J). To address the model that PCNT puncta nucleate free MTs contribute to primary cilia defects in T21 and Q21 cells, the number of PCNT-associated free MTs post CD was quantified (Figure 3K). CD reduced the number of PCNT-associated free MTs in T21 and Q21 cells (Figure 3K). Thus, PCNT-associated free MTs likely contribute to primary cilia defects.
Enlarged PCNT puncta form along and at the ends of MTs
Because MT-dependent intracellular trafficking to and from the centrosome is disrupted by T21 (Galati et al., 2018), we propose two nonmutually exclusive models by which enlarged PCNT puncta that associate with MTs inhibit intracellular trafficking. First, PCNT puncta localized along MTs act as trafficking roadblocks. In this model, enlarged PCNT puncta either sterically block the movement of other cargoes or bind and sequester cargoes as they encounter PCNT puncta. While cargo can translocate around objects on MTs (Conway et al., 2012; Can et al., 2014), our prior work suggests that the trafficking of enlarged PCNT puncta is reduced along MTs (Galati et al., 2018), and PCNT-associated cargoes no longer traffic efficiently. This would suggest that, despite the ability for motor-driven cargoes to move around objects, the enlarged PCNT puncta inhibit continuous cargo movement along MTs. Second, given its MT nucleation capacity with CEP215 and γ-tubulin (Doxsey et al., 1994; Gavilan et al., 2018), PCNT puncta promote promiscuous free-MT nucleation at cytoplasmic puncta peripheral to the centrosome that generate trafficking dead ends.
To visualize single MTs and PCNT puncta, cells were prepared using a 5-min prepermeabilization to focus on stable MTs and to facilitate the discernment of PCNT puncta associated with MTs (Figure 2G). Consistent with both models, PCNT puncta localize along MTs and at MT ends (Figure 4D). The average size of PCNT puncta along MTs increased in T21 and Q21 cells, while the average size of PCNT puncta at MT ends was unchanged in T21 and Q21 cells (Supplemental Figure S2I). On average, 78% were associated with MTs (Supplemental Figure S2J). Decreasing PCNT levels with siRNA or CRISPR-Cas9 reduces the number and size of enlarged PCNT puncta around the centrosome (Figure 2, E and F). Because PCNT reduction in T21 and Q21 also increases ciliation, we suggest that elevated and enlarged MT-associated PCNT puncta disrupt primary cilia formation.
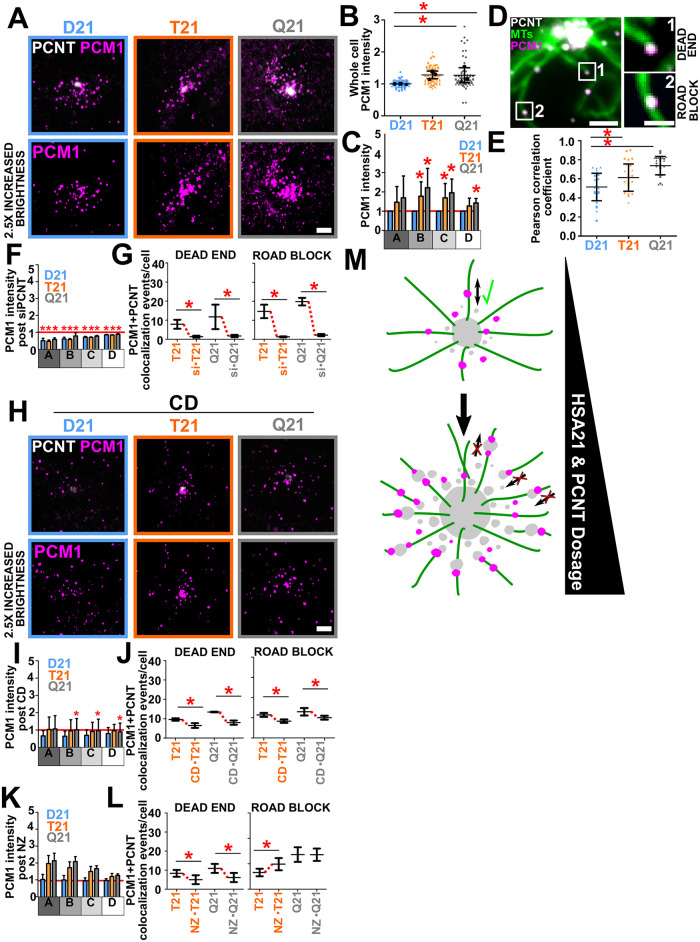
FIGURE 4:
PCNT-associated free MTs and trafficking puncta inhibit proper localization of centriolar satellites. (A) D21, T21, and Q21 RPE-1 cells stained for PCNT (PCNT; grayscale) and PCM1 (PCM1; magenta). Brightness increased by a factor of 2.5. Scale bar, 3 μm. (B) D21, T21, and Q21 whole-cell PCM1 fluorescence intensity. Intensity values normalized to D21 average. Statistical comparisons made to D21 averages. Mean ± SD. *, p < 0.05 (Supplemental Table S1). (C) D21, T21, and Q21 regional binned PCM1 fluorescence intensities. Intensity values normalized to D21 average (indicated with red line). Statistical comparisons made to D21 averages. Mean ± SD. *, p < 0.05 (Supplemental Table S1). (D) PCM1 colocalizes with PCNT along MTs and at MT ends. Unciliated D21 RPE-1 cell stained for PCNT (PCNT; grayscale), MTs (DM1A; green), and PCM1 (PCM1; magenta). Labels are referenced in right panels. Scale bar, 2 μm and 1 μm for insets. (E) PCM1 and PCNT colocalization increases with HSA21 ploidy. Mean ± SD. *, p < 0.05 (Supplemental Table S1). (F) Reducing PCNT via siRNA reduces regional PCM1 intensities in D21, T21, and Q21 RPE-1 cells. Red line denotes D21 siControl intensity. Statistical comparisons made to control intensities for each cell type. Mean ± SD. *, p < 0.05 (Supplemental Table S1). (G) Reducing PCNT via siRNA reduces PCM1 colocalization with PCNT at MT ends (dead end) and along MTs (roadblock) in T21 and Q21 RPE-1 cells. Mean ± SD. *, p < 0.05 (Supplemental Table S1). (H) D21, T21, and Q21 RPE-1 CD-treated cells stained for PCNT (PCNT; grayscale) and PCM1 (PCM1; magenta). Fluorescence intensity brightness increased by a factor of 2.5. Scale bar, 3 μm. (I) CD reduces regional PCM1 intensities in D21, T21, and Q21 RPE-1 cells. Red line denotes D21 control intensity. Statistical comparisons made to relative control intensities for each cell type. Mean ± SD. *, p < 0.05 (Supplemental Table S1). (J) CD reduces PCM1 colocalization with PCNT at MT ends and along MTs in T21 and Q21 RPE-1 cells. Mean ± SD. *, p < 0.05 (Supplemental Table S1). (K) NZ does not reduce regional PCM1 intensity. Red line indicates D21 control intensity. (L) NZ reduces PCM1 colocalization with PCNT at MT ends and increases PCM1 colocalization with PCNT along MTs in T21 cells. Mean ± SD. *, p < 0.05 (Supplemental Table S1). (M) Elevated PCNT caused by HSA21 dosage increases PCNT at the centrosome (Figure 1; Galati et al., 2018), along MTs, and at MT ends. PCNT associated with MT ends are more distant from the centrosome and we propose that this population creates trafficking dead ends, inhibiting the ability for PCM1 to traffic to the centrosome. Enlarged PCNT puncta along MTs might increase trafficking roadblocks that inhibit the ability for PCM1 to traffic to and from the centrosome. The inability for proteins to efficiently traffic to and from the centrosome likely inhibits primary cilia formation. PCM1; magenta. PCNT; gray. MTs; green.
To address the contribution of the elevated and enlarged PCNT puncta to the primary cilia defect, we treated cells with nocodazole (NZ), which disassembles MTs without severely disrupting large PCNT puncta (Figure 3G, and Supplemental Figure S3, G, I, and J). Under these conditions, enlarged PCNT puncta remain while MTs are transiently depolymerized allowing us to test whether puncta are detrimental to cilia. Both NZ treatment and CD dissembled 60–85% of MTs in D21, T21, and Q21 cells (Supplemental Figure S3, J and K). MTs partially recover by 10 min and do not completely recover to their predepolymerization levels even 24 h after CD or NZ treatment (Supplemental Figure S3K). After transient NZ treatment and a 24-h incubation in low serum conditions, the relative mean cilia frequency increased by 15% and 144% in T21 and Q21 cells, respectively (Supplemental Figure S3F). Though mean increases in cilia post NZ are comparable to CD, large variability in cilia frequency post NZ, but not CD, suggests NZ rescues cilia less robustly than CD (% coefficients of variation [%CV] for NZ and CD treatment are 10% and 2% for D21, 13% and 4% for T21, and 18% and 8% for Q21; Figure 3J, and Supplemental Figure S3F). We attribute the less robust cilia rescue in NZ relative to CD to the negative effects of PCNT puncta on cilia formation that remain after NZ treatment but are dispersed after CD (Supplemental Figure S3I). We suggest that these residual PCNT puncta following NZ treatment are sufficient to repress trafficking to the centrosome and ciliogenesis.
Like PCNT reduction and CD treatment, MT intensities and PCNT-associated free MTs were reduced in T21 and Q21 cells post-NZ treatment (Supplemental Figure S3G). Following either PCNT siRNA, CD, or NZ treatment, the remaining PCNT-associated, free MTs were closer to the centrosome (Supplemental Figure S3H). This suggests that a dynamic reorganization of PCNT puncta and MTs reestablishes the MT network required for primary cilia. We propose that MT minus ends are now closer to the centrosome PCM, allowing for normal trafficking and ciliogenesis. However, unlike PCNT reduction or CD, NZ did not significantly reduce the elevated pericentrosomal PCNT in T21 and Q21 cells (Supplemental Figure S3K). NZ treatment alters cellular MTs without disturbing enlarged PCNT puncta. Our results are consistent with the hypothesis that PCNT-associated centrosome-free MTs lead to MT-dependent trafficking dead ends that are disrupted with NZ, and these dead ends may contribute, in part, to primary cilia defects in T21 and Q21 cells. The remaining PCNT puncta may create trafficking roadblocks that also reduce primary cilia. Further studies are required to determine whether trafficking events toward the centrosome are obstructed at these sites.
Elevated PCNT puncta and associated MTs mislocalize centriolar satellites
To investigate the impact that enlarged PCNT puncta have on intracellular trafficking, we tested whether centriolar satellites that traffic along MTs are mislocalized with elevated HSA21 dosage. PCM1 is a major centriolar scaffold protein that interacts with PCNT (Li et al., 2001; Gupta et al., 2015a; Hein et al., 2015). The fluorescence intensity of PCM1 was quantified in D21, T21, and Q21 cells (Figure 4, A–C). Consistent with elevated PCNT localization (Figure 2, A–C), PCM1 increases around centrosomes in T21 and Q21 cells (Figure 4C). Though whole-cell PCM1 intensity is elevated 1.3-fold in both T21 and Q21 cells, PCM1 intensity in regions surrounding centrosomes are elevated to an even greater extent (1.6- and 1.8-fold) in T21 and Q21 cells, respectively (Figure 4, B and C). To determine whether colocalization of PCNT puncta and centriolar satellites is dependent on HSA21 ploidy, we calculated Pearson correlation coefficients denoting colocalization of PCNT and PCM1 (Figure 4, D and E). Colocalization is increased in T21 and Q21 cells compared with D21 cells (Figure 4E). Thus, PCM1 and PCNT increasingly colocalize when PCNT is elevated. Importantly, PCM1 colocalizes with PCNT at both PCNT-associated free MT ends (dead ends) and along MTs (roadblocks) suggesting that proper trafficking of PCM1 to and from centrosomes is disrupted with increases to either of these PCNT populations (Figure 4D).
To test whether increased accumulation of PCM1 peripheral to the centrosome is caused by elevated PCNT in these regions, PCM1 intensities were measured after PCNT reduction (Figure 4F). Indeed, reducing PCNT by siRNA also reduces PCM1 accumulation surrounding the centrosome. To determine whether colocalization of PCM1 and PCNT at MT ends (dead ends) or along MTs (roadblock) changed after PCNT siRNA treatment, the number of colocalization events for each population was quantified (Figure 4G). Importantly, colocalization of PCNT and PCM1 at both populations drastically decreases after PCNT reduction. Conversely, PCM1 knockdown by siRNA does not have a significant effect on PCNT puncta morphology or distribution (Supplemental Figure S4, A and B). CD, which reduces PCNT puncta and MTs, also reduces PCM1 accumulation peripheral to the centrosome in T21 and Q21 cells (Figure 4, H and I). Moreover, colocalization of PCNT and PCM1 at both dead ends and roadblocks was reduced after CD (Figure 4J). These data indicate that when PCNT is elevated, PCM1 accumulates at PCNT puncta both along MTs and at free MT ends within the pericentrosomal crowd.
Consistent with NZ affecting the cell’s MT population without affecting enlarged PCNT puncta accumulation (Supplemental Figure S3, H–K), PCM1 intensity around the centrosome remained elevated in T21 and Q21 cells post NZ treatment (Figure 4K). Moreover, only colocalization of PCNT and PCM1 at MT ends (dead ends) was reduced after NZ treatment while PCNT and PCM1 remained colocalized along MTs (Figure 4L), suggesting that the centrosome is a more efficient MT nucleator than enlarged PCNT puncta. PCNT and PCM1 colocalization along MTs (roadblock) was even increased in T21 cells and unchanged in Q21 cells after NZ treatment (Figure 4L). For T21 cells, this suggests a redistribution of enlarged PCNT puncta with associated PCM1 from MT ends to along MTs. For Q21 cells, this could suggest a saturation of enlarged PCNT puncta along MTs, limiting the redistribution of PCNT puncta along MTs (Figure 4L). That there is more variability in the NZ rescue of cilia frequency than PCNT knockdown or CD suggests the remaining enlarged cytoplasmic PCNT puncta along MTs inhibit efficient ciliogenesis (Supplemental Figure S3, H, I, and J). This suggests that MT dead end and trafficking roadblock populations of enlarged PCNT puncta contribute to primary cilia formation defects.
To test whether satellite mislocalization caused by elevated PCNT was unique to PCM1, we stained cells for CEP131 and CEP290. The frequency of colocalization of PCNT, PCM1, CEP290, and CEP131 at cytoplasmic puncta revealed there was strong, yet variable, overlap between these molecules (Supplemental Figure S4, C and D). Interestingly, PCNT puncta containing both CEP131 and PCM1 or CEP131 and CEP290 were most common in D21, T21, and Q21 cells (Supplemental Figure S4D). To determine whether CEP131 and CEP290 were elevated in T21 and Q21 cells, we quantified CEP131 and CEP290 fluorescence intensities in regions at and surrounding the centrosome (Supplemental Figure S4, E and G). Consistent with the notion that the increased PCM1 puncta associated with PCNT puncta represent centriolar satellites, CEP290 centriolar satellite proteins are also found in the pericentrosomal crowd of T21 and Q21 cells (Supplemental Figure S4, E and G). Though CEP290 fluorescence intensities were consistently elevated in T21 and Q21 cells on average, CEP131 intensities were only elevated in Q21 cells, and to a lesser extent than either PCM1 or CEP290 fluorescence intensities (Supplemental Figure S4, E and G). To determine whether CD or NZ altered CEP131 and CEP290 protein levels at and/or around the centrosome, we quantified CEP131 and CEP290 fluorescence intensities post CD and NZ treatments (Supplemental Figure S4, F and G). Consistent with PCM1 results, NZ did not affect CEP131 or CEP290 intensities compared with control intensities (Supplemental Figure S4, F and G). To our surprise, neither CEP131 nor CEP290 intensities were reduced by CD (Supplemental Figure S4, F and G). Instead, CD subtly elevates CEP131 and CEP290 intensities both at and around centrosomes (Supplemental Figure S4, F and G). These results suggest unique responses of satellite components PCM1, CEP131, and CEP290 to CD, and illuminate differences between components of centriolar satellites. While the PCNT siRNA and CD in T21 and Q21 cells both rescue mislocalization of PCM1 but not CEP131 and CEP290, among other trafficking events, the mechanism by which centriolar satellites might modulate ciliation remains to be discovered.
Summary
Here, we compare ciliated and unciliated RPE-1 cells with modified HSA21 dosage to show elevated levels of PCNT, an HSA21 resident gene up-regulated in Down syndrome, increases the number of enlarged ectopic PCNT foci peripheral to the centrosome (Figure 4M). These foci disrupt the formation of primary cilia by associating with MTs away from the centrosome, by generating MT dead ends, and by preventing the normal distribution of the centriolar satellites required for efficient ciliogenesis by acting as roadblocks along MTs. We suggest that this disrupts the movement of trafficking events from the cytoplasm to the centrosome by blocking cargo recruitment to mother centriole appendages and the cilium. Further studies will uncover the trafficking dynamics and molecules responsible for ciliary formation and signaling defects. Resetting MT distributions via depolymerization or reducing the number of enlarged PCNT foci allows for normal trafficking and for primary cilia formation. Future work will explore which components are waylaid in the pericentrosomal crowd and their dynamics during trafficking. Moreover, we envision that dysregulated MTs and trafficking could also impact cell types like pancreatic β cells that rely on massive changes to the interphase MT array for signaling and secretion (Zhu et al., 2015).
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Cell culture
Disomy 21, trisomy 21, and tetrasomy 21 hTERT-immortalized retinal pigment epithelial (RPE-1) cells were generated by Andrew Lane and David Pellman (Lane et al., 2014). Cells were grown in DMEM:F12 (SH30023; Cytiva) supplemented with 10% fetal bovine serum (FBS; Peak Serum; PS-FB2) and 1% penicillin/streptomycin at 37°C and 5% CO2. Cells were passaged 1:5 at ∼80–90% confluency with 0.25% trypsin (15090-046; Life Technologies). Coculture assays to label one strain and compare to an unlabeled strain were performed by incubating cells in 2.5 μM CFSE (65-0850-84; eBiosciences) in phosphate-buffered saline (PBS; 1 mM KH2PO4, 155 mM NaCl, 3 mM Na2HPO4-7H2O, pH 7.4) for 8 min at room temperature before washing three times in complete media. Cells were then mixed and plated with unlabeled cells on 12-mm glass coverslips. Cells were screened for mycoplasma every 6–12 mo.
Immunofluorescence
Glass coverslips (12 mm) were washed in 1 M HCL heated to 50°C for 16 h. Coverslips were then washed with water, 50%, 70%, and 95% ethanol in a sonicating water bath for 30 min. Coverslips were coated with collagen (C9791; Sigma) and left to air dry for 20 min before crosslinking under UV light for 30 min. Coverslips were then washed three consecutive times in PBS before the addition of cells. All cells were serum starved in DMEM:F12 (SH30023; Cytiva) supplemented with 0.5% FBS (PS-FB2; Peak serum) for 24 or 48 h before fixation. All cells were fixed at ∼90% confluency. Prepermeabilized MT fixation was performed according to Waterman-Storer and Salmon (1997). Briefly, cells were prepermeabilized in 0.5% Triton X-100 in PHEM (60 mM Pipes, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, 6.9 pH) for 5 min. Cells were then fixed with 4% paraformaldehyde/0.5% glutaraldehyde diluted in PHEM for 20 min. Coverslips were quenched three times, for 5 min each, in 0.1% sodium borohydride freshly diluted in PHEM. Coverslips were then washed three times, for 5 min each, in 0.1% Triton X-100 in PHEM (PHEM-T) and stored in PHEM at 4°C until immunostaining. For non-prepermeabilized cells, MT fixation was performed identically, but the 5-min prepermeabilization step was eliminated. For methanol MT fixation and all other fixations, cells were fixed in 100% methanol at −20°C for 2 min. Cells were then washed once in PBS for 5 min before a 10-min permeabilization step in 0.5% Triton X-100 in PBS. Cells were then washed three times, for 5 min each, in PBS and stored in PBS at 4°C. For immunostaining, fixed cells were blocked with 0.5% bovine serum albumin (BSA; A3912-50G; Sigma) in PBS with 0.25% Triton X-100 for 1 h at room temperature. Cells were incubated at 4°C overnight with primary antibodies diluted in blocking buffer. For MT immunostaining, coverslips were washed three times, for 5 min each, in PHEM-T before incubation with secondary antibodies. For all other immunostaining, coverslips were washed three times, for 5 min each, in PBS before incubation with secondary antibodies. Secondary antibodies and Hoechst33258 (1 μg/ml; Invitrogen) were diluted in blocking buffer and incubated for 1 h at room temperature. Coverslips were washed four times, for 5 min each, in PHEM (MT immunostaining) or PBS (all other immunostaining) before mounting to slides using Citifluor (17970-25; Electron Microscopy Sciences). Coverslips were sealed to slides using clear nail polish.
RNAi
Human PCNT siRNA (Smart Pool; M-012172-01-0005; Dharmacon) was transfected into cells with lipofectamine RNAi MAX (13778100; ThermoFisher Scientific) according to the manufacturer’s protocol. Mission siRNA universal negative control #1 was used for all negative controls (SIC001-1NMOL; Sigma). All siRNAs were used at a final concentration of 25 nM. Cells were treated with siRNA in starvation media for 24 h before fixation and subsequent immunostaining steps. For PCM1 knockdown, ON-TARGET siRNAs (J-005165-07: GCGCCUUACUCAUCUAAUA, and J-005165-09: AGAAUAAUGUUCAGAGGUU; Dharmacon) were used at 20 nM for 48 h in starvation medium before fixation.
DNA FISH
Fluorescence in situ hybridization (FISH) was performed by first washing cells in PBS and treating them with 0.59% KCl for 15 min at 37°C. Cells were then fixed and washed three times in methanol:acetic acid 3:1 at −20°C for 5 min each. Cells were then hybridized using a 21qter subtelomere-specific probe (LPT21QG-A; Cytocell) according to the manufacturer’s protocol.
CRISPR-Cas9
PCNT-specific crRNA (Hs.Cas9.PCNT.1.AD and Hs.Cas9.PCNT.1.AG) were designed using Integrated DNA Technologies (IDT’s) online platform. Editing efficiencies of both gRNAs were confirmed by in vitro Cas9 digestion assay. For editing PCNT in T21 RPE-1 cells, approximately 450,000 mycoplasma-free cells were seeded into two wells of a six-well plate in DMEM (11995-065; ThermoFisher Scientific) containing 10% FBS (PS-FB3; Peak serum) and 1% Anti-Anti (100X; 15240-02; Life Technologies). Cells were then cultured for 24 h at 37°C and 5% CO2. Cells were cotransfected with PCNT-specific ribonucleoproteins that were created by complexing 1 μM Alt-R S.p. Cas9 nuclease V3 (1081059; IDT) with 1 μM PCNT gRNAs AD and AG, which were generated by annealing the PCNT crRNA with Alt-R CRISPR-Cas9 tracerRNA ATTO550 (1075927; IDT). Lipofectamine RNAi MAX (13778100; ThermoFisher Scientific) was used for transfection. Media was changed after 24 h, and cells were maintained as usual (37°C and 5% CO2). The genomic DNA (gDNA) from control (nontransfected) and transfected pooled cell line was extracted and used for PCR to evaluate for the presence of a subpopulation with PCNT edits. Single-cell clones were plated from the edited pooled populations by manually seeding approximately one cell into each well of 96-well plates. CloneSelect Imager (Molecular Devices; CSI8086) was used to visually select wells with a single colony. These single colonies were expanded and evaluated for edit in PCNT using PCR (Supplemental Figure S1, G–J). With increasing passage number, CRISPR-Cas9 cells changed in PCNT levels resulting in ∼0.1X compared with D21 PCNT levels. We included analysis on cells that exhibited approximately 1X D21 PCNT levels.
Qiagen dPCR
Genetic CRISPR/Cas9 correction of PCNT copy number from 3n to 2n in trisomy 21 RPE-1 cells was confirmed using the Qiagen digital PCR system (dPCR).
Nocodazole and CD
CD experiments were performed by first placing cells in starvation media for 24 h. At 24 h, cells were placed on ice for 10 or 20 min at 4°C. For CD MT stability assays, cells were treated with a 10-min CD, immediately followed by fixation. For 20-min CD rescue experiments, cells were treated with a 20-min CD and returned to incubation at 37°C for an additional 24 h before fixation. All control cells were left at 37°C. Nocodazole treatments were performed by first placing cells in starvation media for 24 h. At 24 h, cells were treated with 1 μM nocodazole for 60 min at 37°C. After 60 min, coverslips were washed five times, for 5 min each, in starvation media. After washes, cells were returned to incubation at 37°C for an additional 24 h before fixation. Control cells were treated with the same volume of dimethyl sulfoxide (D2650-100ML; Sigma) for 60 min at 37°C.
Generation of tetracycline-inducible mNeon-EB3 RPE-1 D21, T21, and Q21 cell lines
Tetracycline-inducible lentiviral stable RPE-1 cell lines were generated according to methods outlined in Sankaran et al. (2020). Briefly, HEK293T cells were transfected with the tetracycline-inducible mNeon-EB3 construct and lentivirus packaging plasmids using lipofectamine 2000 (11668-027; Invitrogen). HEK293T media containing virus was collected and added to target cells (RPE-1 D21, T21, and Q21 cells) in the presence of 2 μg/ml polybrene. Target cells were incubated with viral media for 24 h. After 24 h, new viral media was added to target cells and target cells were left to incubate for an additional 24 h. Transduced cells were selected using 10 μg/ml puromycin for 3 days. Cells were flow-sorted to isolate EB3-mNeon–positive clones. Before fixation, cells were induced overnight with 0.125 μg/ml doxycycline.
Fluorescence microscopy
Wide-field images were acquired with a Nikon Eclipse Ti-E microscope (Nikon) equipped with a 100× Plan Apochromat objective (NA 1.40) and an Andor Xyla 4.2 scientific CMOS camera (Andor). Nikon NIS Elements imaging software was used for wide-field image acquisition. Confocal images were acquired with a Nikon Eclipse Ti inverted microscope stand equipped with a 100× Plan Apochromat objective (NA 1.45), Andor iXon X3 camera, and CSU-X1 (Yokogawa) spinning disk. SIM images were acquired using a Nikon SIM (N-SIM) with a Nikon Ti2 (Nikon Instruments; LU-N3-SIM) microscope equipped with a 100× SR Apo TIRF, NA 1.49 objective. Images were captured using a Hamamatsu ORCA-Flash 4.0 Digital CMOS camera (C13440) with 0.1-μm Z step sizes. Raw SIM images were reconstructed using the image slice reconstruction algorithm (NIS Elements). Slidebook 6 digital microscopy software was used for confocal image acquisition. Image acquisition times were kept constant within a given experiment and ranged from 30 to 500 msec. All images were acquired at room temperature. Identical acquisition settings were used for quantitatively compared images. Confocal imaging was used to assess colocalization of two proteins. All other quantification was performed using wide-field acquired images. Images in Supplemental Figures S2C, S2F, 4A, 4D, 4H, S4A, S4C, S4E, and S4F were acquired using spinning-disk confocal microscopy. Images in Supplemental Figures S2K and 3H are SIM images. All other figure images were acquired using wide-field microscopy. All images presented in figures are brightest pixel maximum projections. Images of MTs fixed without a prepermeabilization step to show MT colocalization with PCNT and PCM1 were selected single z-plane images for clarity.
Fluorescence quantification
To quantify the presence of cilia, cilia structures were labeled with ARL13B, 6-11-B1 (acetylated tubulin) or DM1A (α-tubulin) primary antibodies. Radial fluorescence intensity analysis was completed using the Radial Profile Extended ImageJ plugin. Briefly, this analysis plots average fluorescence intensity as a function of distance from the user-identified centroid. Whole-cell MT density analysis was performed by outlining cell boundaries (defined by MTs) in ImageJ. Free MTs are defined as being traceable and having two clear ends (identified by moving through slices of the z-stack). To evaluate colocalization of two proteins, regions of interest (ROIs) with radius 10 μm were centered over the centrosome. Pearson correlation coefficients were recorded for two proteins of interest within the ROI using ImageJ plugin Coloc2. To quantify PCNT puncta ratios, ImageJ particle analysis was performed on 10-μm radius ROIs centered over the centrosome of cells. To identify puncta, a threshold minimum intensity fluorescence value cutoff was the greatest 5% of the total fluorescence intensity for D21 samples. Only pixel values above the established minimum fluorescence intensity value were included in the particle identification. This intensity value was applied to all cells within each replicate. Puncta ≤ 0.05 μm2 were defined as “small puncta” (SP) and those >0.05 μm2 were defined as “large puncta” (LP). For reference, a 0.05 μm2 puncta is 3.44 pixels (0.2236 μm) × 3.44 pixels (0.2236 μm) in size. All other fluorescence intensity analyses were performed by recording the integrated density (intensity) of defined ROIs in ImageJ. All intensity analysis was performed on maximum projected images. Excluding quantification performed on CEP290 fluorescence intensity, all other fluorescence intensity background subtraction was performed by subtracting the minimum pixel intensity per field of view for each channel quantified. Due to variability in CEP290 cell background intensity, an in-cell background subtraction was performed for all CEP290 fluorescence intensity quantification. To determine the region of MT nucleation and distribution, cells were serum starved for 24 h and fixed using paraformaldehyde and glutaraldehyde as described previously with the exception that an initial 5-min simultaneous permeabilization and fixation step was employed in PHEM with 0.5% Triton X-100 and six drops of 4% paraformaldehyde, 0.5% glutaraldehyde per 500 µl. Cells were stained for MTs and either PCNT or CEP192 and images were collected by SIM. A Gaussian blur was applied to the PCNT or CEP192 channel and the centroid of the centrosome was calculated from this blurred image. Circular line scans were conducted on circles centered around the centroid in maximum projections of the seven planes (0.1 µm step size) encompassing the centrosome in the MT channel with radii of 0.5, 1.2, 2.0, and 5.0 µm. Fluorescence intensities were normalized for each cell to the average value of the 0.5-µm radius circle within that cell. The average relative intensity for each sized circle across each cell line was plotted.
Statistics
Data were analyzed and organized with Microsoft Excel (Microsoft) and GraphPad Prism (GraphPad). Data center values represent averages while error bars represent SD. All experiments utilized at least three biological replicates. The D’Agostino and Pearson omnibus normality test was used to test for normality of data (Supplemental Table S1). A Student’s two-tailed unpaired t test was used to test the significance between two normal unpaired distributions. The Mann-Whitney U test was used to test the significance between two nonnormal unpaired distributions. Differences between distributions were considered statistically significant if p values were less than 0.05. Only values that were statistically significant were denoted. p values, sample sizes, and statistical tests used for samples are denoted in Supplemental Table S1. %CV was calculated by dividing the SD of a distribution by the mean of the distribution and multiplying by 100.
Supplementary Material
Acknowledgments
We thank Andrew Lane and David Pellman for generously providing the HSA21 RPE-1 cell lines; A. Soh for ImageJ code contributions; and J. Black and L. Porritt for assistance with FISH experiments. We acknowledge the Pearson lab and Nick Galati (Western Washington University) for helpful discussions. This research was funded by Grants no. NIH-NIGMS R01 GM-138415 and no. NIH-NIGMS R35 GM-140813 to C.G.P., the Linda Crnic Institute for Down Syndrome, and the Global Down Syndrome Foundation.
Abbreviations used:
- DS
Down syndrome
- HSA21
Human somatic autosome 21
- PCNT
Pericentrin
- MT
microtubule
- SIM
structured illumination microscopy.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-10-0517-T) on April 27, 2022.
REFERENCES
- Bernhard W, de Harven E (1960). L’ultrastructure du centriole et d’autres éléments de l’appareil achromatique. In Vierter Internationaler Kongress für Elektronenmikroskopie/Fourth International Conference on Electron Microscopy/Quatrième Congrès International de Microscopie Électronique: Berlin, September 10–17, 1958, Bargmann W., Möllenstedt G., Niehrs H., Peters D., Ruska E., Wolpers C. (eds.) (Springer, Berlin Heidelberg; ), pp. 217–227. [Google Scholar]
- Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G, Godinho SA (2011). Centrosomes and cilia in human disease. Trends Genet 27, 307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M (2002). Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol 14, 25–34. [DOI] [PubMed] [Google Scholar]
- Bornens M, Azimzadeh J (2007). Origin and evolution of the centrosome. Adv Exp Med Biol 607, 119–129. [DOI] [PubMed] [Google Scholar]
- Breslow DK, Holland AJ (2019). Mechanism and regulation of centriole and cilium biogenesis. Annu Rev Biochem 88, 691–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Can S, Dewitt MA, Yildiz A (2014). Bidirectional helical motility of cytoplasmic dynein around microtubules. eLife 3, e03205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway L, Wood D, Tüzel E, Ross JL (2012). Motor transport of self-assembled cargos in crowded environments. Proc Natl Acad Sci USA 109, 20814–20819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currier DG, Polk RC, Reeves RH (2012). A Sonic hedgehog (Shh) response deficit in trisomic cells may be a common denominator for multiple features of Down syndrome. Prog Brain Res 197, 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A, Merdes A (2002). Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol 159, 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaval B, Doxsey SJ (2010). Pericentrin in cellular function and disease. J Cell Biol 188, 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dictenberg JB, Zimmerman W, Sparks CA, Young A, Vidair C, Zheng Y, Carrington W, Fay FS, Doxsey SJ (1998). Pericentrin and γ-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J Cell Biol 141, 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey S (2001). Re-evaluating centrosome function. Nat Rev Mol Cell Biol 2, 688–698. [DOI] [PubMed] [Google Scholar]
- Doxsey SJ, Stein P, Evans L, Calarco PD, Kirschner M (1994). Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell 76, 639–650. [DOI] [PubMed] [Google Scholar]
- Efimov A, Kharitonov A, Efimova N, Loncarek J, Miller PM, Andreyeva N, Gleeson P, Galjart N, Maia AR, McLeod IX, et al. (2007). Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev Cell 12, 917–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit JA, Tuft RA, Fogarty KE, Pazour GJ (2006). The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol Biol Cell 17, 3781–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong KW, Choi YK, Rattner JB, Qi RZ (2008). CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the γ-tubulin ring complex. Mol Biol Cell 19, 115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galati DF, Sullivan KD, Pham AT, Espinosa JM, Pearson CG (2018). Trisomy 21 represses cilia formation and function. Dev Cell 46, 641–650.e646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavilan MP, Gandolfo P, Balestra FR, Arias F, Bornens M, Rios RM (2018). The dual role of the centrosome in organizing the microtubule network in interphase. EMBO Rep 19, e45942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould RR, Borisy GG (1977). The pericentriolar material in Chinese hamster ovary cells nucleates microtubule formation. J Cell Biol 73, 601–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta GD, Coyaud E, Goncalves J, Mojarad BA, Liu Y, Wu Q, Gheiratmand L, Comartin D, Tkach JM, Cheung SW, et al. (2015a). A dynamic protein interaction landscape of the human centrosome-cilium interface. Cell 163, 1484–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta GD, Coyaud É, Gonçalves J, Mojarad BA, Liu Y, Wu Q, Gheiratmand L, Comartin D, Tkach JM, Cheung SWT, et al. (2015b). A dynamic protein interaction landscape of the human centrosome-cilium interface. Cell 163, 1484–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M, Fujiyama A, Taylor TD, Watanabe H, Yada T, Park HS, Toyoda A, Ishii K, Totoki Y, Choi DK, et al. (2000). The DNA sequence of human chromosome 21. Nature 405, 311–319. [DOI] [PubMed] [Google Scholar]
- Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, Gak IA, Weisswange I, Mansfeld J, Buchholz F, et al. (2015). A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163, 712–723. [DOI] [PubMed] [Google Scholar]
- Hori A, Toda T (2017). Regulation of centriolar satellite integrity and its physiology. Cell Mol Life Sci 74, 213–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett CE, McCurdy BL, O’Toole ET, Given KS, Lin CH, Olsen V, Martin W, Reinholdt LG, Espinosa JM, Sullivan KD, et al. (2021). Trisomy 21 induces pericentrosomal crowding disrupting early stages of primary ciliogenesis and mouse cerebellar development. BioRxiv 11, 468107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Ho DBT, Mahe K, Mia J, Sepulveda G, Antkowiak M, Jiang L, Yamada S, Jao LE (2021). Condensation of pericentrin proteins in human cells illuminates phase separation in centrosome assembly. J Cell Sci 134, jcs258897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurczyk A, Gromley A, Redick S, San Agustin J, Witman G, Pazour GJ, Peters DJ, Doxsey S (2004). Pericentrin forms a complex with intraflagellar transport proteins and polycystin-2 and is required for primary cilia assembly. J Cell Biol 166, 637–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keryer G, Pineda JR, Liot G, Kim J, Dietrich P, Benstaali C, Smith K, Cordelières FP, Spassky N, Ferrante RJ, et al. (2011). Ciliogenesis is regulated by a huntingtin-HAP1-PCM1 pathway and is altered in Huntington disease. J Clin Investigat 121, 4372–4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JC, Badano JL, Sibold S, Esmail MA, Hill J, Hoskins BE, Leitch CC, Venner K, Ansley SJ, Ross AJ, et al. (2004). The Bardet-Biedl protein BBS4 targets cargo to the pericentriolar region and is required for microtubule anchoring and cell cycle progression. Nat Genet 36, 462–470. [DOI] [PubMed] [Google Scholar]
- Kubo A, Sasaki H, Yuba-Kubo A, Tsukita S, Shiina N (1999). Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J Cell Biol 147, 969–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane AA, Chapuy B, Lin CY, Tivey T, Li H, Townsend EC, van Bodegom D, Day TA, Wu SC, Liu H, et al. (2014). Triplication of a 21q22 region contributes to B cell transformation through HMGN1 overexpression and loss of histone H3 Lys27 trimethylation. Nat Genet 46, 618–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Hansen D, Killilea A, Joshi HC, Palazzo RE, Balczon R (2001). Kendrin/pericentrin-B, a centrosome protein with homology to pericentrin that complexes with PCM-1. J Cell Sci 114, 797–809. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Raff JW (2009). Centrioles, centrosomes, and cilia in health and disease. Cell 139, 663–678. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Stearns T (2011). The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol 13, 1154–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odabasi E, Gul S, Kavakli IH, Firat-Karalar EN (2019). Centriolar satellites are required for efficient ciliogenesis and ciliary content regulation. EMBO Rep 20, e47723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser SL, Pelletier L (2020). Centriolar satellite biogenesis and function in vertebrate cells. J Cell Sci 133, jcs239566. [DOI] [PubMed] [Google Scholar]
- Quarantotti V, Chen JX, Tischer J, Gonzalez Tejedo C, Papachristou EK, D’Santos CS, Kilmartin JV, Miller ML, Gergely F (2019). Centriolar satellites are acentriolar assemblies of centrosomal proteins. EMBO J 38, e101082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JF, Leroux MR (2017). Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol 18, 533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoll C, Rivals I, Ait Yahya-Graison E, Dauphinot L, Paly E, Mircher C, Ravel A, Grattau Y, Bléhaut H, Mégarbane A, et al. (2012). Molecular signatures of cardiac defects in Down syndrome lymphoblastoid cell lines suggest altered ciliome and Hedgehog pathways. PLoS One 7, e41616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper RJ, Baxter LL, Saran NG, Klinedinst DK, Beachy PA, Reeves RH (2006). Defective cerebellar response to mitogenic Hedgehog signaling in Down [corrected] syndrome mice. Proc Natl Acad Sci USA 103, 1452–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salemi M, Barone C, Romano C, Salluzzo R, Caraci F, Cantarella RA, Salluzzo MG, Drago F, Romano C, Bosco P (2013). Pericentrin expression in Down’s syndrome. Neurol Sci 34, 2023–2025. [DOI] [PubMed] [Google Scholar]
- Sankaran DG, Stemm-Wolf AJ, McCurdy BL, Hariharan B, Pearson CG (2020). A semi-automated machine learning-aided approach to quantitative analysis of centrosomes and microtubule organization. J Cell Sci 133, jcs243543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakya S, Westlake CJ (2021). Recent advances in understanding assembly of the primary cilium membrane. Fac Rev 10, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin S (1962). Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J Cell Biol 15, 363–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung CH, Leroux MR (2013). The roles of evolutionarily conserved functional modules in cilia-related trafficking. Nat Cell Biol 15, 1387–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterlin C, Colanzi A (2010). The Golgi and the centrosome: building a functional partnership. J Cell Biol 188, 621–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollenaere MA, Mailand N, Bekker-Jensen S (2015). Centriolar satellites: key mediators of centrosome functions. Cell Mol Life Sci 72, 11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tormanen K, Ton C, Waring BM, Wang K, Sutterlin C (2019). Function of Golgi-centrosome proximity in RPE-1 cells. PloS One 14, e0215215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertii A, Hehnly H, Doxsey S (2016). The centrosome, a multitalented Renaissance organelle. Cold Spring Harb Perspect Biol 8, a025049. 10.1101/cshperspect.a025049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lee K, Malonis R, Sanchez I, Dynlacht BD (2016). Tethering of an E3 ligase by PCM1 regulates the abundance of centrosomal KIAA0586/Talpid3 and promotes ciliogenesis. eLife 5, e12950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer CM, Salmon ED (1997). Actomyosin-based retrograde flow of microtubules in the lamella of migrating epithelial cells influences microtubule dynamic instability and turnover and is associated with microtubule breakage and treadmilling. J Cell Biol 139, 417–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff JB, Wueseke O, Hyman AA (2014). Pericentriolar material structure and dynamics. Philos Trans R Soc Lond B Biol Sci 369, 20130459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A, Dictenberg JB, Purohit A, Tuft R, Doxsey SJ (2000). Cytoplasmic dynein-mediated assembly of pericentrin and γ tubulin onto centrosomes. Mol Biol cell 11, 2047–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Hu R, Brissova M, Stein RW, Powers AC, Gu G, Kaverina I (2015). Microtubules negatively regulate insulin secretion in pancreatic β cells. Dev Cell 34, 656–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman WC, Sillibourne J, Rosa J, Doxsey SJ (2004). Mitosis-specific anchoring of γ tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol Biol Cell 15, 3642–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.