Abstract
Presynaptic metabotropic glutamate receptors (mGluRs) are essential for the control of synaptic transmission. However, how the subsynaptic dynamics of these receptors is controlled and contributes to synaptic signaling remain poorly understood quantitatively. Particularly, since the affinity of individual mGluR subtypes for glutamate differs considerably, the activation of mGluR subtypes critically depends on their precise subsynaptic distribution. Here, using superresolution microscopy and single-molecule tracking, we unravel novel molecular mechanisms that control the nanoscale distribution and mobility of presynaptic mGluRs in hippocampal neurons. We demonstrate that the high-affinity group II receptor mGluR2 localizes diffusely along the axon, and is highly mobile, while the low-affinity group III receptor mGluR7 is stably anchored at the active zone. We demonstrate that intracellular interactions modulate surface diffusion of mGluR2, while immobilization of mGluR7 at the active zone relies on its extracellular domain. Receptor activation or increases in synaptic activity do not alter the surface mobility of presynaptic mGluRs. Finally, computational modeling of presynaptic mGluR activity revealed that this particular nanoscale arrangement directly impacts their ability to modulate neurotransmitter release. Altogether, this study demonstrates that distinct mechanisms control surface mobility of presynaptic mGluRs to contribute differentially to glutamatergic synaptic transmission.
INTRODUCTION
Activity-directed modulation of synaptic efficacy underlies the ability of neuronal networks to process and store information. Presynaptic mechanisms that impinge on the neurotransmitter release machinery are a critical factor in fine-tuning synaptic efficacy. In particular, presynaptic metabotropic glutamate receptors (mGluRs) are essential negative-feedback control elements that modulate transmission by dampening glutamate release (Pinheiro and Mulle, 2008; Reiner and Levitz, 2018). Disruptions in these receptor systems severely deregulate synaptic function and specific forms of synaptic plasticity, and aberrant mGluR function has been associated with several neurological disorders such as anxiety, epilepsy, and schizophrenia, further highlighting their physiological importance (Sansig et al., 2001; Muly et al., 2007; Woolley et al., 2008). Nevertheless, it remains poorly understood how these receptors are organized at presynaptic sites to efficiently modulate transmission.
The eight known mGluRs (mGluR1–mGluR8) belong to the class C G-protein-coupled receptors (GPCRs). These GPCRs exist as constitutive dimers and have distinctive large extracellular domains (ECD) that contain the ligand-binding domain connected to the prototypical 7-helix transmembrane domain (TMD) via a cysteine-rich domain. mGluRs are further divided into three groups based on their sequence homology, downstream signaling partners, and agonist selectivity (Niswender and Conn, 2010). These functionally diverse groups are expressed throughout the central nervous system but are generally targeted to specific subcellular locations. Group I mGluRs (mGluR1/5) are primarily expressed at postsynaptic sites, group II mGluRs (mGluR2/3) are present at both pre- and postsynaptic sites, and group III mGluRs (mGluR4, mGluR6-8) are located almost exclusively at presynaptic sites, except mGluR6, which is located at the postsynaptic site in retina bipolar cells (Petralia et al., 1996; Shigemoto et al., 1996). The presynaptic group II and III mGluRs mGluR2 and mGluR7 are both abundantly expressed in the hippocampus (Kinoshita et al., 1998), share substantial homology (∼60%), and both couple to inhibitory G-proteins (Gαi/o) that repress adenylyl cyclase activity. Nevertheless, these receptors differ significantly in their pharmacological characteristics and interactomes, conferring functionally distinct roles on them in synaptic transmission and plasticity.
Generally, activation of presynaptic mGluRs depresses synaptic transmission via inhibition of voltage-gated Ca2+-channels (VGCC), via activation of K+ channels, or by directly modulating components of the release machinery such as Munc13, Munc18, and RIM-1 (Pinheiro and Mulle, 2008; de Jong and Verhage, 2009). Thus, these receptors have been implicated in the regulation of both short-term plasticity and long-term depression of synaptic responses (Okamoto et al., 1994; Kamiya and Ozawa, 1999; Millán et al., 2002; Robbe et al., 2002; Pelkey et al., 2005, 2008; Martín et al., 2007). However, signaling events downstream of presynaptic mGluRs can also potentiate release, and mGluR7 particularly has been postulated to regulate synaptic transmission bidirectionally (Martín et al., 2010, 2018; Klar et al., 2015; Dasgupta et al., 2020). Thus, presynaptic mGluRs modulate synaptic transmission through a variety of downstream effectors, and the functional outcome of mGluR activation is probably determined by the frequency and duration of synaptic signals. Additionally, the subsynaptic distribution and dynamics of presynaptic mGluRs are likely to influence their ability to become activated and engage local downstream signaling partners. In particular, since these receptors have different affinities for glutamate, their subsynaptic position relative to the point of glutamate release ultimately determines their probability of activation. mGluR2 has a moderate to high affinity for glutamate (in the micromolar range), and its positioning relative to the release site might thus only modestly affect its contribution to regulating release probability. In contrast, when measured in nonneuronal cells, the affinity of mGluR7 for glutamate is exceptionally low, in the millimolar range (0.5–2.5 mM) (Schoepp et al., 1999). In addition, mGluRs are obligatory dimers, and activation of single subunits in an mGluR dimer produces only low-efficacy activation. Given that release events produce only brief peaks in glutamate concentration in the synaptic cleft (1–3 mM; Diamond and Jahr, 1997; Lisman et al., 2007), it has thus been questioned whether mGluR7 at neuronal synapses, even when placed immediately adjacent to release sites, will ever be exposed to sufficient levels of glutamate to become activated (Pinheiro and Mulle, 2008). However, this is in contrast to the wealth of physiological evidence from different model systems that show that mGluR7 is a key modulator of synaptic transmission (Sansig et al., 2001; Bushell et al., 2002; Millán et al., 2002; Pelkey et al., 2005, 2008; Klar et al., 2015; Martín et al., 2018). Thus, the precise localization of presynaptic mGluRs determines their activation probability and greatly impacts their ability to modulate synaptic transmission through local downstream effectors. Nevertheless, quantitative insight into the dynamic distribution of presynaptic mGluRs in live neurons and the mechanisms that control their dynamic positioning is lacking.
To understand how mGluR2 and mGluR7 contribute to synaptic transmission in rat hippocampal neurons, we studied how the dynamic positioning of the subsynaptic distribution of these receptors is mechanistically controlled. Using complementary superresolution imaging approaches, we found that mGluR2 is highly dynamic and localized throughout the axon, while mGluR7 is immobilized at presynaptic active zones. We found that the mobility of mGluR2 is mainly mediated by its intercellular domain. Surprisingly, we found that the specific positioning of mGluR7 at the active zone is not controlled by intracellular interactions but relies on extracellular interactions. Furthermore, a computational model of mGluR activation at presynaptic sites indicates that mGluR2 activation is only loosely coupled to release site location, while activation of mGluR7 is inefficient, even when localized within a few nanometers of the release site or during high-frequency stimulation patterns. Based on our findings, we propose that the different mechanisms that control presynaptic mGluR positioning ensure the differential contribution of these receptors to transmission.
RESULTS
Distinct differences in the subsynaptic distribution of presynaptic mGluR subtypes
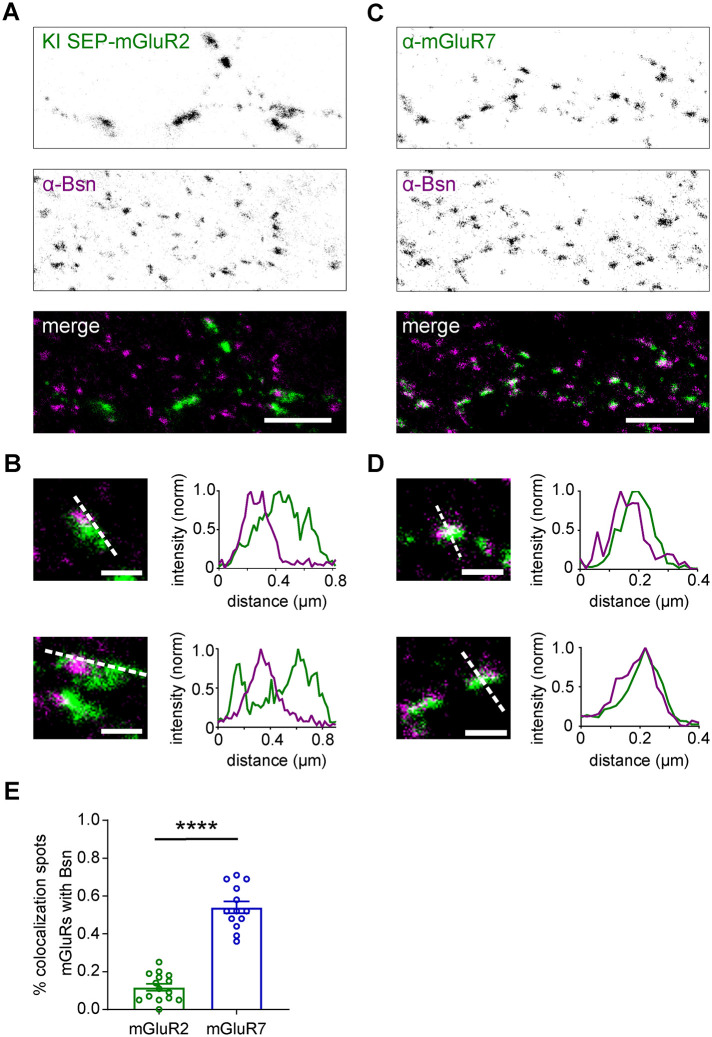
The precise spatial distribution of mGluR subtypes at presynaptic sites likely determines their functional contributions to the modulation of synaptic transmission. To compare the subsynaptic distribution of presynaptic group II and III mGluRs in dissociated hippocampal neurons isolated from embryonic day 18 (E18) Wistar rats, we determined the localization of mGluR2 (group II) and mGluR7 (group III) relative to the active zone marker Bassoon (Bsn) using two-color gated stimulated emission depletion (gSTED) superresolution microscopy. To visualize mGluR2, we tagged endogenous mGluR2 with superecliptic pHluorin (SEP), a pH-sensitive variant of GFP, using a recently developed CRISPR/Cas9-mediated knock-in approach (Willems et al., 2020). Because the level of endogenous mGluR2 expression was low, we enhanced the SEP signal using anti-GFP staining to reliably measure mGluR2 distribution. We found that mGluR2 was localized both in axons and dendrites (Supplemental Figure S1A), as reported previously (Ohishi et al., 1994), but even though an earlier study suggested that mGluR2 is located in the preterminal region of the axon, and not in presynaptic boutons (Shigemoto et al., 1997), we detected mGluR2 both in the axon shaft and within synaptic boutons (Figure 1A). However, as is apparent from line profiles of the fluorescence intensity of mGluR2 signal along Bsn-labeled puncta, the mGluR2 signal was largely excluded from presynaptic active zones (Figure 1B). Confirming this finding, a similar distribution pattern was observed using antibody labeling for mGluR2/3 (Figure S1B, C), further indicating that presynaptic group II mGluRs are distributed throughout the axon but excluded from active zones. Immunostaining for the group III mGluR, mGluR7 labeled a subset of neurons in our cultures (Figure 1C and S1D), consistent with previous studies (Shigemoto et al., 1996; Tomioka et al., 2014). In contrast to mGluR2, line profiles indicated that the maximum intensity of mGluR7 labeling coincided with the Bsn-marked active zone (Figure 1D). Colocalization analysis further confirmed this, showing that the majority of mGluR7-positive puncta overlap with Bsn-positive puncta, while mGluR2 labeling showed a striking lack of overlap with Bsn (co-localization with Bsn-positive puncta, mGluR2: 0.12 ± 0.02, mGluR7: 0.54 ± 0.03; Figure 1E). Together, these results indicate that two presynaptic mGluR subtypes that are both implicated in the regulation of presynaptic release properties, have distinct subsynaptic distribution patterns.
FIGURE 1:
Subsynaptic distribution of presynaptic mGluRs. (A) gSTED image of SEP-mGluR2 CRISPR/Cas9 knock-in neuron costained with anti-Bassoon (STAR635P; Bsn). Note that due to the low endogenous expression level of mGluR2, SEP signal was enhanced with anti-GFP (STAR580) staining. Scale bar, 2 µm. (B) Example images and intensity profiles of individual mGluR2 positive synapses from A. Scale bar, 500 nm. (C) gSTED image of neuron costained with anti-mGluR7 (STAR580) and anti-Bsn (STAR635P). Scale bar, 2 µm. (D) Example images and intensity profiles of individual mGluR7-positive synapses from C. Scale bar: 500 nm. (E) Quantification of colocalization between presynaptic mGluRs and Bsn. Unpaired t test, *** P < 0.001. See also Supplemental Figure S1.
Differential stability of mGluR2 and mGluR7 at presynaptic boutons
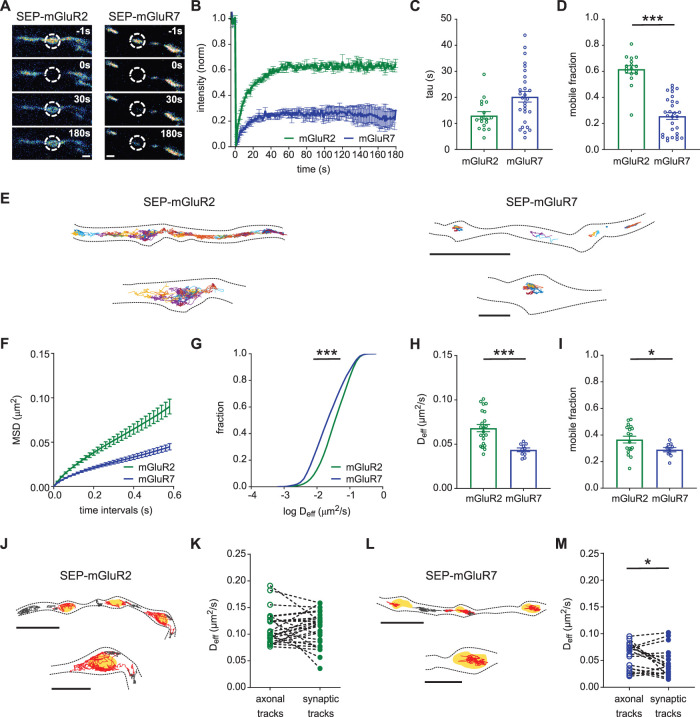
To test if the observed receptor distributions reflect differences in surface mobility in the axonal membrane, we expressed SEP-tagged mGluR2 and mGluR7 to visualize surface-expressed receptors in live cells and performed fluorescence recovery after photobleaching (FRAP) experiments. Importantly, we found that expressed receptors were efficiently targeted to axons and their localization was consistent with the observed endogenous distributions. SEP-mGluR7 was enriched in presynaptic boutons, while SEP-mGluR2 expression was more diffuse throughout the axon (Supplemental Figure S2, A–C). Additionally, we verified that expression levels of SEP-tagged mGluRs are comparable (on average ∼50% increase) with endogenous levels of mGluR2 and mGluR7 (Supplemental Figure S2, D and E). We photobleached the fluorescence in small regions overlapping with presynaptic boutons and monitored the recovery of fluorescence over time. Strikingly, the recovery of fluorescence was much more rapid and pronounced for SEP-mGluR2 than for SEP-mGluR7 (Figure 2, A and B). Indeed, quantification of the fluorescence recovery curves showed that the mobile fraction (SEP-mGluR2: 0.61 ± 0.03, SEP-mGluR7: 0.25 ± 0.03, P < 0.0001, one-way ANOVA; Figure 2D) of SEP-mGluR2 was significantly higher than observed for SEP-mGluR7. Thus, these results indicate that mGluR2 is highly mobile in axons, while mGluR7 is immobilized at presynaptic sites and displays minor exchange between synapses.
FIGURE 2:
Distinct surface diffusion behavior of mGluR2 and mGluR7. (A) Example images from a FRAP time series in neurons overexpressing SEP-mGluR2 and SEP-mGluR7. The dotted circles indicate the bleached boutons. Scale bar, 1 µm. (B) Normalized fluorescence recovery of SEP-mGluR2 and SEP-mGluR7 (n = 17 boutons for SEP-mGluR2, 28 boutons for SEP-mGluR7 from three independent experiments). (C, D) Quantification of tau of fluorescence recovery, C, and the mobile fraction, D, of SEP-tagged mGluRs. One-way ANOVA followed by Tukey’s multiple comparisons test (see Supplemental Figure S3 for full dataset); ***P < 0.001. Error bars represent SEM. (E) Example trajectories of SEP-mGluR2 and SEP-mGluR7. Trajectories are displayed with random colors. Outlines of cells are based on the TIRF image of SEP signal. Scale bar, 5 µm; zooms, 1 µm. (F) Average mean squared displacement (MSD) plot of SEP-mGluR2 and SEP-mGluR7 (n = 22 fields of view for mGluR2, 10 fields of view for mGluR7 from three independent experiments). (G) Frequency distribution of instantaneous diffusion coefficient (Deff) of SEP-mGluR2 and SEP-mGluR7 (n = 22,821 trajectories for SEP-mGluR2, 5161 trajectories for SEP-mGluR7). Kolmogorov–Smirnov test; ***P < 0.0001. (H, I) Quantification of the average instantaneous diffusion coefficient (Deff), H, and the mobile fraction, I, of SEP-tagged mGluRs (n = 22 fields of view for mGluR2, 10 fields of view for mGluR7 from three independent experiments). Unpaired t test; *P < 0.05, ***P < 0.0005. Error bars represent SEM. (J) Trajectories of SEP-mGluR2 plotted on top of the mask marking the presynaptic bouton. Red track—synaptic tracks, gray tracks—axonal tracks, yellow areas—bouton mask based on Syp-mCherry signal. Scale bar, 2 µm; zooms, 1 µm. (K) Quantification of the instantaneous diffusion coefficient (Deff) of axonal and synaptic tracks of SEP-mGluR2 (n = 27 fields of view from 2 independent experiments). (L) Trajectories of SEP-mGluR7 plotted on top of the mask marking the presynaptic bouton, as in J. (M) Quantification of instantaneous diffusion coefficient (Deff) of axonal and synaptic tracks of SEP-mGluR7 (n = 18 fields of view from five independent experiments). Paired t test, *P < 0.05. See also Supplemental Figure S2.
Single-molecule tracking reveals differences in diffusional behavior of mGluR2 and mGluR7
To resolve the dynamics of mGluR2 and mGluR7 at high spatial resolution and to investigate whether the diffusional behavior of these receptors is heterogeneous within axons, we next performed live-cell single-molecule tracking experiments using universal point accumulation in nanoscale topography (uPAINT) (Giannone et al., 2010). SEP-tagged receptors were labeled with anti-GFP nanobodies conjugated to the photostable dye ATTO-647N at low concentrations, which made it possible to reliably detect, localize, and track single receptors over time for up to several seconds. The acquired receptor tracks were then compiled into trajectory maps revealing the spatial distribution of receptor motion. These maps were consistent with the receptor distribution patterns as resolved with gSTED imaging. SEP-mGluR2 seemed to diffuse rapidly throughout the axon and synaptic boutons, while SEP-mGluR7 motion was limited and highly confined within synaptic boutons, with only a few molecules occasionally diffusing along the axon shaft (Figure 2E). The mean squared displacement (MSD) versus elapsed time curves (Figure 2F) display a sublinear relationship for both receptor types, indicating that the majority of these receptors undergo anomalous diffusion. The instantaneous diffusion coefficients (Deff) for both receptors were estimated by fitting the slope through the four initial points of the MSD curves. Histograms of Deff estimated from individual trajectories (Figure 2G) and the average Deff per field of view (Figure 2H) revealed a significantly higher diffusion coefficient for SEP-mGluR2 than for SEP-mGluR7 (Deff for SEP-mGluR2: 0.068 ± 0.004 µm2/s, for SEP-mGluR7: 0.044 ± 0.002 µm2/s, P < 0.0005, unpaired, two-tailed t test), further indicating that mGluR2 diffuses much more rapidly in the axonal membrane than mGluR7. In addition, we classified the receptors’ diffusional states as either mobile or immobile in a manner independent of MSD-based diffusion coefficient estimation, by determining the ratio between the radius of gyration and the mean displacement per time step of individual trajectories (Supplemental Figure S2, F and G) (Golan and Sherman, 2017). Using this approach, we found that SEP-mGluR2 showed a larger fraction of mobile tracks than SEP-mGluR7 (mobile fraction SEP-mGluR2: 0.37 ± 0.03, SEP-mGluR7: 0.29 ± 0.02, P < 0.05, unpaired, two-tailed t test; Figure 2I and Supplemental Figure S2H) further confirming that in axons, mGluR2 is overall more mobile than mGluR7.
To determine whether the surface mobility of these receptors is differentially regulated at synaptic sites, we coexpressed SEP-tagged mGluRs together with a marker of presynaptic boutons, Synaptophysin1 (Syp1) fused to mCherry. Based on epifluorescence images of Syp1-mCherry, we created a mask of presynaptic boutons and compared the Deff of receptors diffusing inside or outside synapses (Figure 2, J and L). The diffusion coefficients of SEP-mGluR2 within presynaptic boutons and along axons did not differ significantly (Deff axonal tracks: 0.113 ± 0.006 µm2/s, Deff synaptic tracks: 0.110 ± 0.006 µm2/s, P > 0.05, paired, two-tailed t test; Figure 2, J and K), suggesting that mGluR2 diffusion is not hindered at synaptic sites. Comparing the diffusion coefficients of axonal SEP-mGluR7 tracks with synaptic tracks showed that on average the mobility of SEP-mGluR7 is considerably lower inside boutons (Deff axonal tracks: 0.058 ± 0.006 µm2/s, Deff synaptic tracks: 0.045 ± 0.006 µm2/s, P < 0.05, paired, two-tailed t test; Figure 2, L and M). However, the reduction in mobility was not observed at all boutons, suggesting that mobility of mGluR7 is regulated at most, but not all presynaptic sites. Taken together, the FRAP and single-molecule tracking data indicate a striking difference in the dynamic behavior of presynaptic mGluRs. mGluR2 diffuses seemingly unhindered throughout the axon, while mGluR7 is largely immobilized, preferentially at presynaptic active zones.
The intracellular domain of mGluR2 regulates receptor mobility
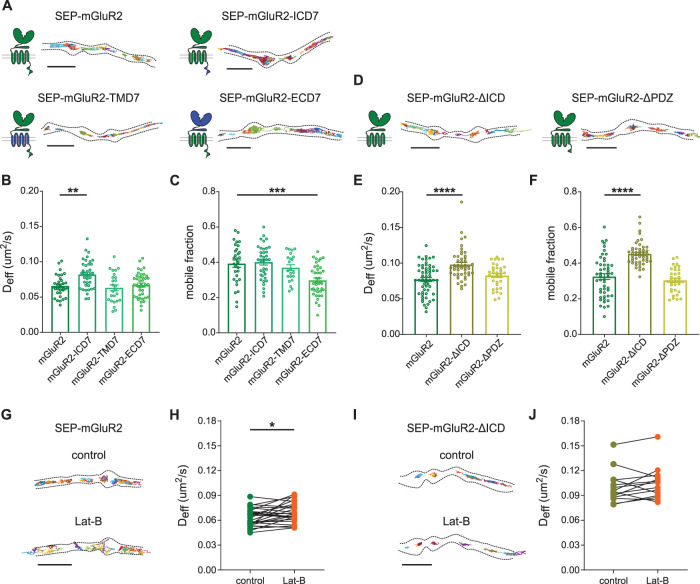
To gain insight into the structural mechanisms that control the dynamics of presynaptic mGluRs and to explain the distinct diffusional properties of mGluR2 and mGluR7, we next sought to identify the receptor domains that are involved in controlling mGluR mobility. mGluRs consist of three regions: the intracellular domain (ICD) containing a PDZ binding motif, the prototypical seven-helix transmembrane domain (TMD) involved in G-protein coupling, and the large extracellular domain (ECD) that includes the ligand-binding site (Niswender and Conn, 2010). First, to unravel which segment of mGluR2 regulates its mobility, we created three chimeric receptors of mGluR2 by exchanging the ICD, TMD, or ECD domains of mGluR2 with the corresponding domains of mGluR7 to maintain the overall structure of the receptor. All SEP-tagged chimeric mGluR2 variants were targeted to the axon and dendrites, similarly to wild-type mGluR2, indicating that axonal targeting and surface expression were not altered by replacing these domains (Supplemental Figure S3A). Moreover, single-molecule tracking showed that all chimeric mGluR2 variants diffused throughout the axon and presynaptic boutons, similarly to wild-type mGluR2 (Figure 3A). Interestingly, though, the mGluR2 chimera containing the ICD of mGluR7 revealed a significantly higher diffusion coefficient compared with wild-type mGluR2 (Deff SEP-mGluR2-ICD7: 0.082 ± 0.003 µm2/s, SEP-mGluR2: 0.065 ± 0.003 µm2/s, P < 0.005, one-way ANOVA), while exchanging the TMD or ECD did not affect the diffusion kinetics of mGluR2 (Deff for SEP-mGluR2-TMD7: 0.063 ± 0.004 µm2/s; for SEP-mGluR2-ECD7: 0.067 ± 0.003 µm2/s, P > 0.05, one-way ANOVA; Figure 3B). Thus, comparing the diffusional behavior of this set of chimeric mGluR2 variants indicates that intracellular interactions mediate mGluR2 mobility in axons.
FIGURE 3:
Intracellular interactions regulate the mobility of presynaptic mGluR2. (A) Schematic diagrams and example trajectories of wild-type and chimeric variants of mGluR2 (green) with the ICD, TMD, and ECD exchanged with the corresponding mGluR7 domains (blue). Scale bar, 2 µm. (B, C) Quantification of average diffusion coefficient (Deff), B, and the mobile fraction, C, of SEP-tagged chimeric mGluR2 variants (n = 30 fields of view for mGluR2, 40 fields of view for mGluR2-ECD7 from five independent experiments, 36 fields of view for mGluR2-ICD7 and 22 fields of view for mGluR2-TMD7 from four independent experiments). (D) Schematic diagrams and example trajectories of C-terminal deletion variants of mGluR2. Scale bar, 2 µm. (E, F) Quantification of average diffusion coefficient (Deff), E, and the mobile fraction, F, of SEP-tagged C-terminal deletion variants of mGluR2 (n = 50 fields of view for mGluR2, 49 fields of view form mGluR2-ΔICD, and 34 fields of view for mGluR2-ΔPDZ from three independent experiments). One-way ANOVA followed by Dunnett’s multiple comparison test; **P < 0.005, ***P < 0.0005. (G) Example trajectories of SEP-mGluR2 before and after incubation with 5 µM latrunculin B (Lat-B). Scale bar, 2 µm. (H) Quantification of diffusion coefficient (Deff) of SEP-mGluR2 before and after incubation with Lat-B (n = 27 fields of view, from three independent experiments). Paired t test, * P < 0.05. (I) Example trajectories of SEP-mGluR2-ΔICD before and after incubation with Lat-B. Scale bar, 2 µm. (J) Quantification of diffusion coefficient (Deff) of SEP-mGluR2-ΔICD before and after incubation with Lat-B (n = 14, from two independent experiments). Error bars represent SEM. All trajectories are displayed with random colors. Outlines of cells are based on the TIRF image of SEP signal. See also Supplemental Figure S3.
To further investigate whether intracellular interactions regulate mGluR2 dynamic, we created two deletion variants of mGluR2 by removing the entire ICD or only the distal PDZ binding motif. SEP-tagged C-terminal deletion mGluR2 variants were targeted to the axon, similar to the wild-type receptor (Supplemental Figure S3B). Also, trajectory maps revealed their diffusion throughout the axon (Figure 3D). Interestingly, deletion of the PDZ binding motif did not alter the diffusion behavior of mGluR2; however, removal of the entire ICD increased the diffusion coefficient (Deff for SEP-mGluR2-ΔICD: 0.098 ± 0.003 µm2/s, for SEP-mGluR2-ΔPDZ: 0.082 ± 0.003 µm2/s, for SEP SEP-mGluR2: 0.077 ± 0.003 µm2/s, P < 0.0001, one-way ANOVA; Figure 3E) and the mobile fraction of mGluR2 (mobile fraction SEP-mGluR2-ΔICD: 0.45 ± 0.01, SEP-mGluR2-ΔPDZ: 0.30 ± 0.01, SEP-mGluR7: 0.32 ± 0.02, P < 0.0001, one-way ANOVA; Figure 3F) confirming that the ICD of mGluR2 is involved in controlling receptor mobility but this regulation is not mediated by PDZ-mediated interactions.
Little is known about mGluR2 C-tail-mediated interactions, but we reasoned that direct or indirect interactions with the actin cytoskeleton, which have an important role in controlling membrane organization in axons (He et al., 2016; Zhou et al., 2019), could influence mGluR2 diffusion. Disruption of the actin cytoskeleton with latrunculin-B (Lat-B) increased the diffusion coefficient of wild-type mGluR2 (Deff control: 0.063 ± 0.002 µm2/s, Lat-B: 0.069 ± 0.002 µm2/s, P < 0.05, paired, two-tailed t test; Figure 3, G and H) but did not cause a further increase in the diffusion coefficient of mGluR2 lacking the ICD (Deff control: 0.100 ± 0.005 µm2/s, Lat-B: 0.104 ± 0.005 µm2/s, P > 0.05, paired, two-tailed t test; Figure 3, I and J). These results suggest that intracellular interactions with the actin cytoskeleton may regulate mGluR2 mobility.
mGluR7 immobilization at presynaptic active zones is controlled by extracellular domain
While mGluR2 diffuses rapidly through the axon, we found that mGluR7 is stably anchored and concentrated at active zones. Therefore, we decided to focus further on the mechanisms that could underlie the immobilization of mGluR7 at presynaptic sites. To test which region of mGluR7 is involved in the immobilization of mGluR7 at the active zone, we generated five chimeric variants of mGluR7 to exchange the ICD, TMD, or ECD of mGluR7 with the corresponding domains of mGluR2 or mGluR1. Because the C-terminal domain of mGluR1 is involved in targeting the receptor to the dendritic compartment we decided to not substitute the ICD of mGluR7 for the ICD of mGluR1 (Francesconi and Duvoisin, 2002). All SEP-tagged chimeric variants of mGluR7 were readily detected in axons, similarly to wild-type mGluR7 (Supplemental Figure S3C), indicating that these receptors are correctly targeted to the axonal membrane.
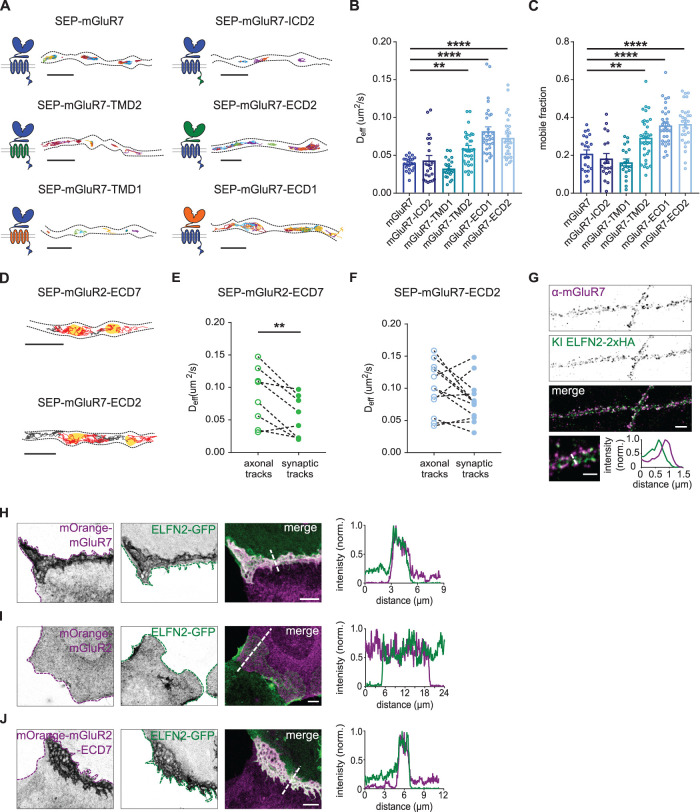
In contrast to mGluR2, the exchange of the ICD of mGluR7 did not change the diffusional behavior of the receptor. Trajectory maps obtained from single-molecule tracking showed that diffusion of the SEP-tagged mGluR7 chimera containing the ICD of mGluR2 was still restricted to presynaptic boutons (Figure 4A) and the diffusion coefficient (Deff for SEP-mGluR7-ICD2: 0.043 ± 0.004 µm2/s, for SEP-mGluR7: 0.039 ± 0.002 µm2/s, P > 0.05, one-way ANOVA; Figure 4B) and mobile fraction were similar to those for wild-type SEP-mGluR7 (mobile fraction SEP-mGluR7-ICD2: 0.18 ± 0.03, SEP-mGluR7: 0.21 ± 0.02, P > 0.05, one-way ANOVA; Figure 4C), suggesting that intracellular interactions do not contribute to mGluR7 immobilization. Diffusion of SEP-tagged TMD chimeric variants of mGluR7 was also mostly restricted to presynaptic boutons (Figure 4A), although we found that replacing the mGluR7 TMD with the TMD of mGluR2 slightly increased the diffusion coefficient (Deff for SEP-mGluR7-TMD2 0.059 ± 0.004 µm2/s, P < 0.05, one-way ANOVA; Figure 4B) and the mobile fraction (SEP-mGluR7-TMD2 0.29 ± 0.02, P < 0.05, one-way ANOVA; Figure 4C). However, the substitution of the mGluR7 TMD with the mGluR1 TMD did not alter its diffusional behavior (Deff for SEP-mGluR7-TMD1: 0.033 ± 0.003 µm2/s, mobile fraction: 0.16 ± 0.02, P > 0.05, one-way ANOVA; Figure 4, B and C), suggesting that the faster diffusion of the mGluR7 variant containing the TMD of mGluR2 is most likely due to specific properties of the mGluR2 TMD and cannot be attributed to a mGluR7-specific mechanism. Indeed, a previous study reported stronger interactions between transmembrane regions in mGluR2 homodimers compared with other mGluR subtypes (Gutzeit et al., 2019).
FIGURE 4:
Extracellular domain regulates the mobility of mGluR7. (A) Schematic diagrams and example trajectories of wild-type and chimeric variants of mGluR7 (blue) with the ICD, TMD, and ECD exchanged with the corresponding domains from mGluR2 (green) or mGluR1 (orange). Scale bar, 2 µm. (B, C) Quantification of average diffusion coefficient (Deff), B, and the mobile fraction, C, of SEP-tagged chimeras of mGluR7 (n = 22 fields of view for mGluR7, 31 fields of view for mGluR7-ECD1, 29 fields of view for mGluR7-ECD2 from five independent experiments; 19 fields of view for mGluR7-ICD2, 20 fields of view for mGluR7-TMD1, and 32 fields of view for mGluR7-TMD2 from four independent experiments). One-way ANOVA followed by Dunnett’s multiple comparison test; **P < 0.05, ****P < 0.0005. (D) Trajectories of extracellular chimeras SEP-mGluR2-ECD7 and SEP-mGluR7-ECD2 plotted on top of the mask of the presynaptic bouton. Red tracks—synaptic tracks, gray tracks—axonal tracks, yellow areas—bouton mask based on Syp-mCherry signal. Scale bar, 2 µm. (E, F) Quantification of diffusion coefficient (Deff) of axonal and synaptic tracks of SEP-mGluR2-ECD7, E, and SEP-mGluR7-ECD2, F (n = 8 fields of view for SEP-mGluR2-ECD7, 12 fields of view for SEP-mGluR7-ECD2 from two independent experiments). Paired t test, ** P < 0.005. Error bars represent SEM. All trajectories are displayed with random colors. Outlines of cells are based on the TIRF image of the SEP signal. (G) Confocal images of ELFN2-2xHA CRISPR/Cas9 knock-in neuron costained with anti-mGluR7 (STAR635P). HA-tag was visualized with anti-HA (Alexa Fluor 594) antibodies. Scale bar, 5 µm. Example images and intensity profiles of individual ELFN2 and mGluR7-positive synapses. Scale bar, 2 µm. (H–J) Example images of mixed cocultures of U2OS cells expressing ELFN2-GFP and mOrange-mGluR7, H; mOrange-mGluR2, I; and mOrange-mGluR2-ECD7, J and normalized intensity profiles along interface between cells expressing different proteins indicated with dashed lines. Dotted line—outline of cell. Scale bar, 10 µm. See also Supplemental Figure S3.
Interestingly, replacing the ECD of mGluR7 drastically altered its diffusional behavior. In contrast to the wild-type receptor, SEP-tagged chimeric mGluR7 variants containing the ECD of mGluR2 or mGluR1 diffused throughout the axon (Figure 4A) and displayed almost a twofold increase in diffusion coefficient (Deff for SEP-mGluR7-ECD1: 0.082 ± 0.006 µm2/s, for SEP-mGluR7-ECD2: 0.073 ± 0.005 µm2/s, P < 0.0005, one-way ANOVA; Figure 4B) and larger mobile fraction compared with wild-type SEP-mGluR7 (SEP-mGluR7-ECD1: 0.36 ± 0.02, SEP-mGluR7-ECD2: 0.37 ± 0.02, SEP-mGluR7: 0.21 ± 0.02, P < 0.0005, one-way ANOVA; Figure 4C). Thus, the immobilization of mGluR7 at presynaptic sites likely relies on extracellular interactions with its ECD. To assess if the ECD of mGluR7 is sufficient to immobilize receptors, we replaced the ECD of mGluR2 with the ECD of mGluR7. Indeed, we found a significant decrease in the mobile fraction of the SEP-tagged chimeric mGluR2 variant containing the mGluR7 ECD (SEP-mGluR2-ECD7: 0.30 ± 0.02, SEP-mGluR2: 0.39 ± 0.02, P < 0.0005, one-way ANOVA; Figure 3C) supporting the role of the mGluR7 ECD in immobilizing the receptor. To further substantiate these results, we performed FRAP experiments and found a significant increase in fluorescence recovery of SEP-tagged mGluR7 variants with substituted ECDs (Supplemental Figure S3E) and slower recovery kinetics of SEP-tagged chimeric mGluR2 with the ECD of mGluR7 (Supplemental Figure S3D). These results are in striking agreement with the single-molecule tracking data and confirm the dominant role of the mGluR7 ECD in regulating receptor mobility.
Based on our findings that the localization of mGluR7 is restricted to the active zone and that mGluR7 diffusion is hindered at presynaptic boutons, we hypothesized that the ECD of mGluR7 mediates receptor immobilization specifically at presynaptic sites. To test this, we resolved receptor mobility at synapses by coexpressing ECD chimeric variants of mGluR2 and mGluR7 with Syp1-mCherry (Figure 4D). Although the mGluR2 chimera containing the ECD of mGluR7 displayed rather high diffusion coefficients in the axonal shaft, the pool of chimeric receptors inside presynaptic boutons showed a significantly lower diffusion coefficient (Deff for synaptic tracks: 0.054 ± 0.011 µm2/s, for axonal tracks: 0.087 ± 0.015 µm2/s, P < 0.005, paired, two-tailed t test; Figure 4E). Replacing the ECD of mGluR7 with the ECD of mGluR2 resulted in similar diffusion coefficients of axonal and synaptic tracks (Deff for synaptic tracks: 0.081 ± 0.01 µm2/s, for axonal tracks: 0.1 ± 0.01 µm2/s, P > 0.05, paired, two-tailed t test; Figure 4F) suggesting that the ECD of mGluR7 is indeed sufficient to immobilize receptors at presynaptic sites. Altogether, these results indicate that mGluR7 immobilization at synaptic sites is in large part mediated by extracellular domain.
Given the large contribution of the ECD of mGluR7 to surface mobility, we sought to gain further insights into the ECD-mediated interactions that could underlie the anchoring of mGluR7 at presynaptic boutons. It was recently shown that the postsynaptic adhesion molecules ELFN1 and ELFN2 can interact transsynaptically with mGluR7 and modulate its activity (Tomioka et al., 2014; Dunn et al., 2019b). To investigate the possibility that ELFN2 could anchor mGluR7 at presynaptic sites, we assessed whether ELFN2 is expressed in hippocampal neurons. To visualize endogenous ELFN2, we created an ELFN2-2xHA CRISPR/Cas9 knock-in construct. We found that endogenous ELFN2-positive clusters colocalized with endogenous mGluR7-positive puncta (Figure 4G), suggesting that ELFN2 could interact in trans with mGluR7 localized at the active zone and play a role in positioning mGluR7.
To further test whether mGluR7 can be recruited and clustered by ELFN2, we cocultured a population of U2OS cells transfected with mOrange-tagged mGluR7 with a population of cells expressing ELFN2-GFP to detect possible interactions in trans between these proteins at the junctions between the two populations of transfected cells. We observed a strong accumulation of both mGluR7 and ELFN2 at the interfaces between cells expressing mOrange-mGluR7 and ELFN2-GFP (Figure 4H). In contrast, we did not find recruitment of mOrange-mGluR2 to junctions with ELFN2-expressing cells (Figure 4I), suggesting that trans interactions with ELFN2 can indeed specifically recruit mGluR7, in line with recent findings (Dunn et al., 2019b). To further investigate if this interaction is mediated by the extracellular domain of mGluR7, we tested whether replacing the mGluR2 ECD with the mGluR7 ECD would be sufficient to recruit mGluR2 to the junctions with ELFN2 expressing cells. Indeed, mGluR2 harboring the ECD of mGluR7 was strongly recruited to the junctions with ELFN2 expressing cells (Figure 4J). These results indicate that ELFN2 can potently recruit mGluR7 to cellular junctions and that the ECD of mGluR7 is both required and sufficient for receptor recruitment by ELFN2. Thus, ELFN2 could be a potential candidate to mediate immobilization of mGluR7 at the presynaptic active zone.
Surface mobility of presynaptic mGluRs is altered by decreased but not increased synaptic activity
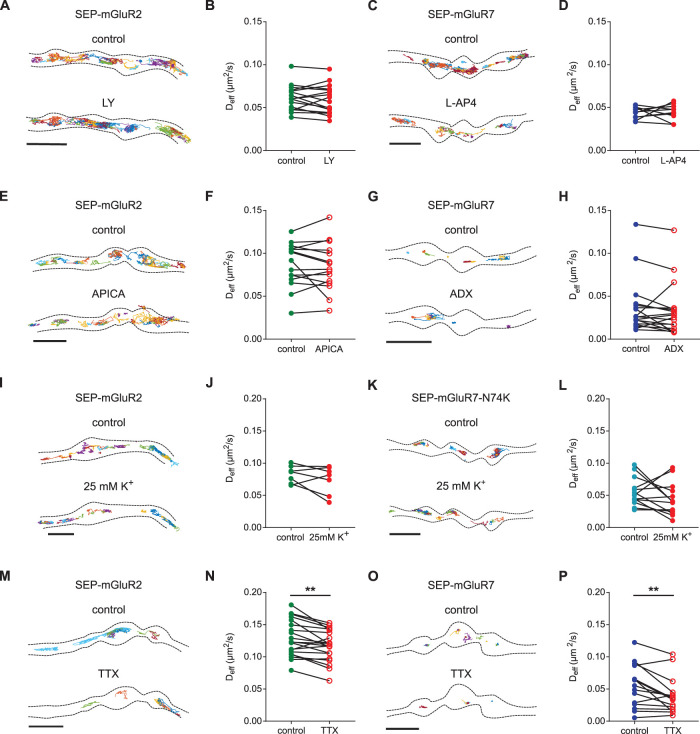
Our results so far suggest that, under resting conditions, the diffusional properties of presynaptic mGluRs are largely controlled by distinct intra- and extracellular interactions. However, ligand-induced activation of GPCRs involves a dramatic change in receptor conformation and has been shown to change the oligomerization and diffusion behavior of various GPCRs, including mGluRs, in nonneuronal cells (Calebiro et al., 2013; Kasai and Kusumi, 2014; Sungkaworn et al., 2017; Yanagawa et al., 2018). To test whether receptor activation alters the diffusion of presynaptic mGluRs in neurons, we performed single-molecule tracking of mGluR2 and mGluR7 before and after stimulation with their specific agonists. We found that the selective group II mGluR agonist LY379268 (LY) did not change the diffusion of SEP-mGluR2 (Deff for control: 0.06 ± 0.003 µm2/s, for LY: 0.058 ± 0.004 µm2/s, P > 0.05, paired, two-tailed t test; Figure 5, A and B). Similarly, direct activation of mGluR7 with the potent group III mGluR agonist L-AP4 did not change the diffusional behavior of SEP-mGluR7 (Deff for control: 0.044 ± 0.002 µm2/s, for L-AP4: 0.045 ± 0.003 µm2/s; P > 0.05, paired, two-tailed t test; Figure 5, C and D). Thus, these experiments indicate that in neurons, the dynamics of presynaptic mGluRs are not modulated by agonist-stimulated receptor activation.
FIGURE 5:
Lateral diffusion of presynaptic mGluRs is not regulated by activity. (A) Example trajectories of SEP-mGluR2 before and after incubation with 100 µM LY. Scale bar, 2 µm. (B) Quantification of diffusion coefficient (Deff) of SEP-mGluR2 before and after incubation with LY (n = 17 fields of view from two independent experiments). (C) Example trajectories of SEP-mGluR7 before and after incubation with 500 µM L-AP4. Scale bar, 2 µm. (D) Quantification of diffusion coefficient (Deff) of SEP-mGluR7 before and after incubation with L-AP4 (n = 10 fields of view from two independent experiments). (E) Example trajectories of SEP-mGluR2 before and after incubation with 100 µM APICA. Scale bar, 2 µm. (F) Quantification of diffusion coefficient (Deff) of SEP-mGluR2 before and after incubation with APICA (n = 15 fields of view from three independent experiments). (G) Example trajectories of SEP-mGluR7 before and after incubation with 10 µM ADX. Scale bar, 2 µm. (H) Quantification of diffusion coefficient (Deff) of SEP-mGluR7 before and after incubation with ADX (n = 18, from four independent experiments). (I) Example tracks of SEP-mGluR2 before and after incubation with 25 mM K+. Scale bar, 2 µm. (J) Quantification of diffusion coefficient (Deff) of SEP-mGluR2 before and after incubation with 25 mM K+ (n = 7 fields of view from two independent experiments). (K) Example tracks of SEP-mGluR7-N74K before and after incubation with 25 mM K+. Scale bar, 2 µm. (L) Quantification of diffusion coefficient (Deff) of SEP-mGluR7-N74K before and after incubation and with 25 mM K+ (n = 13 fields of view from two independent experiments). (M) Example trajectories of SEP-mGluR2 before and after incubation with 1 µM TTX. Scale bar, 2 µm. (N) Quantification of diffusion coefficient (Deff) of SEP-mGluR2 before and after incubation with TTX (n = 20 from three independent experiments). (O) Example trajectories of SEP-mGluR7 before and after incubation with 1 µM TTX. Scale bar, 2 µm. (P) Quantification of diffusion coefficient (Deff) of SEP-mGluR7 before and after incubation with TTX (n = 15 from three independent experiments). All trajectories are displayed with random colors. Outlines of cells are based on the TIRF image of SEP signal. Paired t test, **P < 0.005. See also Supplemental Figure S4.
Next, we tested whether inhibition of presynaptic mGluRs influences their mobility. We found that inhibition of SEP-mGluR2 with the group II mGluR antagonist APICA did not alter the diffusion rate of mGluR2 (Deff for control: 0.087 ± 0.007 µm2/s, for APICA: 0.084 ± 0.007 µm2/s, P > 0.05, paired, two-tailed t test; Figure 5, E and F). Similarly, inhibition of mGluR7 activity with the negative allosteric modulator ADX71743 (ADX) did not change the distribution of trajectories (Figure 5G) or the diffusion coefficient of the receptor (Deff for control: 0.036 ± 0.007 µm2/s, for ADX: 0.033 ± 0.007 µm2/s, P > 0.05, paired, two-tailed t test; Figure 5H). Altogether, these results suggest that the diffusion of presynaptic mGluRs is not impacted by changes in the activation state of the receptor.
Global changes in neuronal activity could alter receptor mobility, either directly by receptor stimulation by their endogenous ligand glutamate, or perhaps indirectly through structural changes in synapse organization. To test this, we next determined whether strong synaptic stimulation by application of the potassium channel blocker 4-AP together with the glutamate reuptake blocker TBOA, to acutely increase synaptic glutamate levels, changed receptor diffusion. However, we did not find a significant effect of synaptic stimulation on the diffusion coefficient of SEP-mGluR2 (Deff for control: 0.085 ± 0.011 µm2/s, for 4-AP + TBOA: 0.069 ± 0.009 µm2/s, P > 0.05, paired, two-tailed t test; Supplemental Figure S4, A and B). Additionally, even under strong depolarizing conditions (25 mM K+, 5–10 min), the diffusion coefficient of SEP-mGluR2 remained unaltered (Deff for control: 0.082 ± 0.005 µm2/s, for 25 mM K+: 0.074 ± 0.008 µm2/s, P > 0.05, paired, two-tailed t test; Figure 5, I and J). We found similar results for SEP-mGluR7 (unpublished data). However, since the affinity of mGluR7 for glutamate is very low, in the range of 0.5–1 mM (Schoepp et al., 1999), we reasoned that the unaltered diffusion of mGluR7 during synaptic stimulation could be due to the incomplete activation of the receptor. Therefore, we analyzed the mobility of an mGluR7 mutant with a two-fold increased affinity for glutamate (mGluR7-N74K; Kang et al., 2015) during strong depolarization. Importantly, we found that the diffusion rate of SEP-mGluR7-N74K was not significantly different from that of wild-type SEP-mGluR7 under control conditions (Deff for SEP-mGluR7-N74K: 0.049 ± 0.005 µm2/s, for SEP-mGluR7: 0.039 ± 0.002 µm2/s, P > 0.05, unpaired, two-tailed t test; Supplemental Figure S4, C–E). However, despite having a twofold higher affinity for glutamate, the diffusion kinetics of SEP-mGluR7-N74K remained unaltered under strong depolarizing conditions (Deff for control: 0.056 ± 0.006 µm2/s, for 25 mM K+: 0.044 ± 0.007 µm2/s, P > 0.05, paired, two-tailed t test; Figure 5, K and L). Thus, these single-molecule tracking experiments demonstrate that the lateral diffusion of presynaptic mGluRs on the axonal membrane is not modulated by direct activation with ligands, or an acute increase in neuronal activity.
Furthermore, we determined whether a reduction in neuronal activity modulates the mobility of presynaptic mGluRs. We analyzed the diffusion of mGluRs after acute inhibition of spontaneous action potential by blocking sodium channels with TTX. Interestingly, we found a reduction of SEP-mGluR2 mobility after treatment with TTX (Deff for control: 0.129 ± 0.006 µm2/s, for TTX: 0.117 ± 0.005 µm2/s, P < 0.005, paired, two-tailed t test; Figure 5, M and N). Moreover, inhibition of spontaneous activity also decreased the diffusion coefficient of SEP-mGluR7 (Deff for control: 0.055 ± 0.009 µm2/s, for TTX: 0.040 ± 0.007 µm2/s, P < 0.05, paired, two-tailed t test; Figure 5, M and N). Thus, our data revealed that the mobility of presynaptic mGluRs is reduced under conditions of reduced neuronal activity.
A computational model of presynaptic mGluR activation reveals that different levels of receptor activation depend on subsynaptic localization
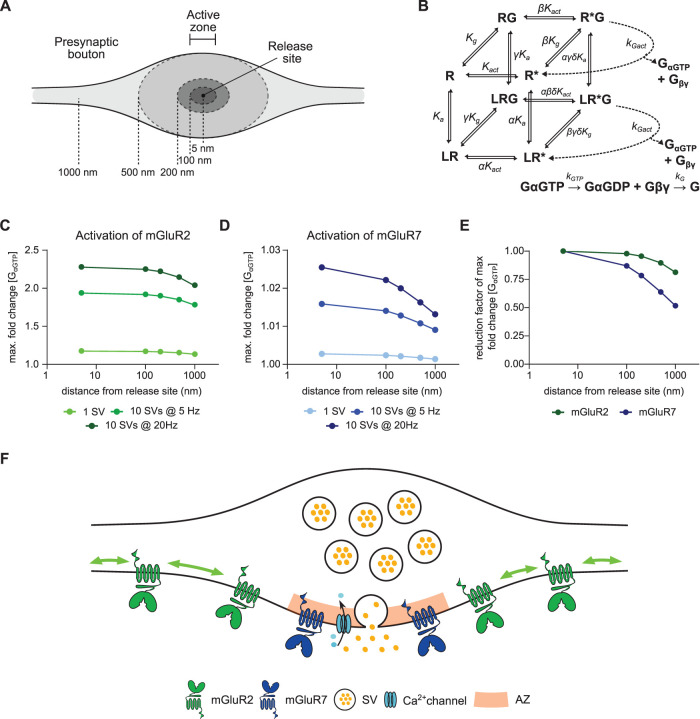
Our data show that mGluR7 is immobilized at the active zone, close to the release site, while mGluR2 is distributed along the axon and synaptic boutons, seemingly excluded from the active zone. Moreover, their localization and dynamics did not change during increased synaptic activity. We hypothesized that these distinct distribution patterns differentially influence the contribution of presynaptic mGluRs to the modulation of synaptic transmission. To test this hypothesis, we investigated a computational model of presynaptic mGluR activation combining the cubic ternary complex activation model (cTCAM) of GPCRs signaling (Figure 6B; Kinzer-Ursem and Linderman, 2007) with a model of time-dependent diffusion of glutamate release after a single synaptic vesicle (SV) fusion or multivesicle release at different frequencies. To determine the effect of mGluR localization, we compared receptor activation at varying distances (5 nm to 1 µm) from the release site (Figure 6A). We calibrated the activation model of mGluR2 and mGluR7 by solving cTCAM with different values of the association constant (Ka), keeping other parameters constant (Supplemental Table S1), to match the model outputs: the relative number of receptor–ligand complexes (Supplemental Figure S5A) and the GαGTP concentration (Supplemental Figure S5B) with previously published EC50 values for mGluR2 and mGluR7 (Schoepp et al., 1999). Because two out of four liganded receptor states in the cTCAM represent inactive receptors, we used the GαGTP concentration as a readout of receptor activation to compare responses of mGluRs to different synaptic activity patterns.
FIGURE 6:
A computational model of mGluRs activation shows that subsynaptic localization of presynaptic mGluRs tunes receptor activation. (A) Schematic of presynaptic bouton highlighting subsynaptic localizations used in modeling. (B) Kinetics and rate equations described in the cubic ternary complex activation model of presynaptic mGluRs signaling. All parameters used in the model are summarized in Supplemental Table S1. (C, D) Receptor response to glutamate release during different release patterns (1 SV, 10 SVs at 5 Hz, and 10 SVs at 20 Hz) at different distances from release site (5 nm to 1 µm) for mGluR2, C, and mGluR7, D. Note that the x-axes are on a logarithmic scale. (E) The normalized reduction factor of mGluR2 and mGluR7 activation at different distances from the release site after release of 10 SVs at 20Hz. Note that the x-axis is on a logarithmic scale. (F) A model of subsynaptic distribution and mobility of presynaptic mGluRs. mGluR2 is distributed along the axon and displays high mobility that is modulated by its intracellular interactions. mGluR7 is enriched and immobilized at the active zone. Immobilization of mGluR7 is regulated by its extracellular domain. SV—synaptic vesicle, AZ—active zone. See also Supplemental Figure S5, Supplemental Tables S1 and S2.
The release of glutamate from a single SV, representing release during spontaneous synaptic activity, caused only a slight increase in the activation of mGluR2 when located close to the release site (r = 5 nm) and outside the active zone (r ≥ 100 nm, Figure 6C and Supplemental Figure S5C). Release of 10 SVs, corresponding to the size of the readily releasable pool, at low frequency (5 Hz) increased the activity of mGluR2 almost twofold inside presynaptic boutons (r ≤ 500 nm; Figure 6C and Supplemental Figure S5D). Elevation of the fusion frequency to 20 Hz further increased receptor activation to ∼2.3-fold of basal activity (Figure 6 and Supplemental Figure S5E). Together, these data suggest that mGluR2 is activated during moderate synaptic stimulation patterns, in line with an earlier study suggesting use-dependent activation of group II mGluRs (Scanziani et al., 1997). Surprisingly, for all patterns of synaptic activity, levels of mGluR2 activation were almost identical next to the release site (r = 5 nm) and at the edge of the active zone (r = 100 nm) and decreased only slowly with increasing distance from the active zone (r > 100 nm, Figure 6, C and E). These results suggest that mGluR2 is efficiently activated, even at further distances from the release site, and its activation is only loosely coupled to release site location. This finding is in line with the localization of mGluR2 along the axon and inside the presynaptic bouton but not inside the active zone.
In contrast, mGluR7, having a distinctively low affinity for glutamate, was not efficiently activated by the release of a single SV, even when positioned close to the release site. At r = 5 nm, we found less than 0.3% change in activation compared with basal receptor activity (Figure 6D and Supplemental Figure S5G). The release of 10 SVs at 5 Hz caused a relatively small increase (∼1.5%) in mGluR7 activity (Figure 6D and Supplemental Figure S5H). However, the fusion of the same number of SVs at higher frequency (20 Hz) almost doubled mGluR7 response to glutamate (∼2.6% increase of GαGTP concentration at r = 5 nm, Figure 6D and Supplemental Figure S5I), suggesting that the level of mGluR7 activation depends strongly on the frequency of release and the peak of maximal glutamate concentration in the cleft. Additionally, the activity profiles of mGluR7 further away from the release site showed a reduction in mGluR7 response indicating that mGluR7 activation is most likely to occur in locations close to release sites (Figure 6, D and E). Altogether, these data indicate that mGluR7 is involved in the modulation of synaptic transmission only during repetitive, high-frequency release and its localization at the active zone close to the release site is curtail for its function.
A subset of parameters in the cTCAM model describing the interaction between GPCR and G-protein could be different between cell types and between the different receptors. However, there is only limited experimental data available. Therefore, we tested the influence of parameter changes on the cTCAM model output and our conclusions. We solved the model with different numerical values for the receptor activation rate, the receptor–G protein association rate, the receptor–G protein collision efficiency, and the receptor–G protein binding affinity (Supplemental Table S2). All parameter values were in the ranges previously reported (Kinzer-Ursem and Linderman, 2007). For all tested parameters, we found a similar dependency of mGluRs activation on the distance from the release site (Supplemental Figure S5, F and J). However, we found that receptor–G protein collision efficiency has a greater impact on the mGluR2 and mGluR7 activation profiles.
DISCUSSION
Despite the functional importance of presynaptic mGluRs in modulating the efficacy of synaptic transmission, the mechanisms that control their dynamic distribution at excitatory synapses remain poorly understood. Here, we provide new insights into the molecular mechanisms that determine the spatial distribution and mobility of presynaptic mGluRs in hippocampal neurons (Figure 6F). We observed that presynaptic mGluR subtypes display striking differences in their subsynaptic localization and dynamics that are controlled by distinct structural mechanisms. We found that the intracellular domain of mGluR2 controls its mobility and that the actin cytoskeleton contributes to the regulation of mGluR2 diffusion behavior. In contrast, we found that the extracellular domain of mGluR7 is critical for immobilization of the receptor at presynaptic sites which could be mediated by transsynaptic interactions with the postsynaptic adhesion molecule ELFN2. Finally, a computational model of receptor activation showed that mGluR2 activation is only loosely coupled to release site location. In contrast, even when placed immediately next to the release site, there is only modest activation of mGluR7 by physiologically relevant synaptic stimulation patterns.
We found that mGluR2 was distributed along the axon and was largely excluded from the active zone. In contrast, we found that mGluR7 was highly enriched at the presynaptic active zone, close to the release site of synaptic vesicles. This is in line with earlier immuno-EM studies that showed that mGluR2 is present in the preterminal part of axons, but rarely found in boutons (Shigemoto et al., 1997), and that group III mGluRs, including mGluR7, is almost exclusively localized in the presynaptic active zone (Shigemoto et al., 1996, 1997; Siddig et al., 2020). Interestingly, these differences in localization were reflected in the surface diffusion behavior of these receptors. mGluR2 was highly mobile throughout the axon and within boutons, similarly to other presynaptic receptors such as the cannabinoid type 1 receptor (CB1R; Mikasova et al., 2008) and the mu-type opioid receptor (MOR; Jullié et al., 2020). In contrast to these mobile receptors, diffusion of mGluR7 was severely hindered at presynaptic boutons. Such differences in the distribution of presynaptic receptors are likely associated with their function and may provide a means for synapses to spatially and temporally compartmentalize receptor signaling.
The differences in the distance of these mGluR2 and mGluR7 to the release site imply that these receptors respond differentially to synaptic activity. Indeed, our computational modeling studies indicate that mGluR2 activation is only loosely coupled to release site location, while mGluR7 activation is limited, even when placed in immediate proximity to the release site. These two receptor types might thus encode different modes of synaptic activity patterns: mGluR2 responding to lower-frequency stimulation patterns, and mGluR7 being activated only during intense, high-frequency synaptic stimulation. It has been suggested that group III mGluRs act as autoreceptors during repetitive stimulations and modulate release probability (Billups et al., 2005; Pinheiro and Mulle, 2008). On the other hand, it has been described that mGluR7 is constitutively active (Kammermeier, 2015; Dunn et al., 2018; Stachniak et al., 2019) and that activity of group III mGluRs is regulated by the transsynaptic interaction with ELFN1/2 at excitatory synapses (Dunn et al., 2019a; Stachniak et al., 2019). Allosteric modulation of mGluR7 by ELFN1/2 could thus alter the threshold for receptor activation or increase its basal activity. Nevertheless, the results from our computational model indicate that mGluR7 positioning relative to the release site is a critical factor increasing the probability of receptor activation.
Both mGluR2 and mGluR7 couple to inhibitory Gαi proteins that repress adenylyl cyclase activity, decreasing cAMP production, and both receptors are described to modulate VGCC activity (Robbe et al., 2002; Martín et al., 2007; de Jong and Verhage, 2009; Ferrero et al., 2013), in part through inhibition by membrane-anchored βγ subunits of the G-protein (Niswender and Conn, 2010; Kammermeier, 2015). An important rate-limiting factor in this mechanism is probably the distance between the Gβγ subunits and VGCCs. The effect of mGluR2 activation on synaptic transmission could thus be delayed by the diffusion time of βγ subunits to VGCCs enriched at the active zone. For mGluR7, being immobilized in close proximity to release sites, the inhibition of VGCCs might occur much more instantaneously. Altogether, our data indicate that the specific modulatory effects of presynaptic mGluRs on synaptic transmission could be determined by their differential localization relative to the release site and their distinct surface diffusion properties.
Given the distinct distribution and diffusion properties of mGluR2 and mGluR7, we speculated that distinct mechanisms control the surface mobility of these receptors. Our data indeed suggest that intracellular interactions mediated by the C-terminal region of mGluR2 regulate receptor diffusion but are not dependent on PDZ binding motifs. Instead, we found that the mobility of mGluR2 is increased by depolymerization of the actin cytoskeleton. The striking periodic organization of the actin cytoskeleton is proposed to control the distribution of receptors in the axons (Zhou et al., 2019), but the protocol of actin depolymerization used in our single-molecule tracking experiments should not cause loss of the ring structures (Zhong et al., 2014). Therefore, we propose that the ICD of mGluR2 introduces steric hindrance for receptor diffusion. However, little is known about the molecular interaction of the C-terminal region of mGluR2 and we cannot exclude a possible direct or indirect molecular link between mGluR2 and actin. Thus, molecular mechanisms that control mGluR2 diffusion remains to be elucidated.
Also, for mGluR7, it has been suggested that stable surface expression and clustering in presynaptic boutons are controlled by the intracellular interaction with the PDZ-domain, containing scaffold protein PICK1 (Boudin et al., 2000; Suh et al., 2008). In contrast, another study showed that the synaptic distribution of a mGluR7 mutant lacking the PDZ binding motif was unaltered (Zhang et al., 2008). Our findings suggest that the ICD of mGluR7 does not contribute to receptor clustering and immobilization at presynaptic boutons are consistent with this. Rather, we found an unexpected role of the extracellular domain of mGluR7 in its immobilization at the presynaptic plasma membrane. Chimeric mGluR7 variants with substituted ECDs displayed higher diffusion coefficients than wild-type mGluR7 and surface diffusion was no longer restricted to the presynaptic bouton but was virtually unrestricted along the axon. Our data thus suggest that extracellular interactions can efficiently cluster the receptor and that the extracellular domain of mGluR7 is essential for immobilizing and concentrating the receptor at active zones.
The dramatic effect of replacing the ECD of mGluR7 on localization and diffusion suggests that transsynaptic interactions might effectively concentrate mGluR7 at synaptic sites. This is consistent with the emerging notion that transcellular interactions greatly impact GPCR biology (Dunn et al., 2019a). Specifically for group III mGluRs, interactions with the adhesion molecules ELFN1 and ELFN2 have been found to modulate the functional properties of these receptors and potently impact synaptic function (Tomioka et al., 2014; Dunn et al., 2018, 2019b; Stachniak et al., 2019). Thus, the transsynaptic interaction with ELFN1/2 could also play a role in anchoring mGluR7 at specific synaptic sites while simultaneously regulating receptor activity via allosteric modulation. However, contribution of the transsynaptic interactions of mGluR7 to its immobilization at the active zone remains to be further investigated.
Previous studies have suggested that ligand-induced GPCR activation, alters their surface diffusion and oligomerization properties (Calebiro et al., 2013; Kasai and Kusumi, 2014; Sungkaworn et al., 2017; Yanagawa et al., 2018). Surprisingly, our data in neurons indicate that the surface mobility of mGluRs is not altered by agonist-induced receptor activation or acute increase in neuronal activity, but was only reduced by acute silencing of neuronal activity. Our data thus indicate that for presynaptic mGluRs, structural factors, such as interactions with intra- and extracellular components predominantly instruct receptor localization, and that these mechanisms act independently of the receptor activation status. This has potentially important implications for the contribution of these receptors to the regulation of synaptic transmission. mGluR7 is likely to exert its effects very locally, restricted to individual synapses. For mGluR2 on the other hand, it could be speculated that the unchanged, high surface mobility of mGluR2 after activation allows the receptor to activate downstream effectors over larger areas, as has been suggested for the opioid receptor (Jullié et al., 2020). This would imply that, once activated, mGluR2 could spread its effects to neighboring synapses and dampen transmission much more globally than mGluR7 does. This could also imply that the activity of mGluR2 not only modulates synaptic transmission, but perhaps also controls other axonal processes such as protein synthesis, cargo trafficking, or cytoskeleton reorganization.
In conclusion, we identified novel regulatory mechanisms that differentially control the spatial distribution and dynamics of presynaptic glutamate receptors, that have important implications for how these receptors can contribute to the modulation of synaptic transmission. The coexistence of various other and distinct receptor types at presynaptic sites likely provides flexibility and allows synapses to differentially respond to incoming stimulation patterns. Defining the molecular mechanisms that control the dynamic spatial distribution of these receptors will be important to further our understanding of synaptic modulation.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Animals
All experiments that required animals were approved by the Dutch Animal Experiments Committee (Dier Experimenten Commissie [DEC]). All animals were treated in accordance with the regulations and guidelines of Utrecht University, and conducted in agreement with Dutch law (Wet op de Dierproeven, 1996) and European regulations (Directive 2010/63/EU).
Primary rat neuronal culture and transfection
Dissociated hippocampal cultures from embryonic day 18 (E18) Wistar rat (Janvier Labs) brains of both genders were prepared as described previously (Cunha-Ferreira et al., 2018). Neurons were plated on 18-mm glass coverslips coated with poly-l-lysine (37.5 mg/ml, Sigma-Aldrich) and laminin (1.25 mg/ml, Roche Diagnostics) at a density of 100,000 neurons per well in a 12-well plate. Neurons were growing in Neurobasal Medium (NB; Life Technologies) supplemented with 2% B27 (Life Technologies), 0.5 mM l-glutamine (Life Technologies), 15.6 µM l-glutamic acid (Sigma), and 1% penicillin/streptomycin (Life Technologies). Once per week, starting from DIV1, half of the medium was refreshed with BrainPhys neuronal medium (BP, STEMCELL Technologies) supplemented with 2% NeuroCult SM1 supplement (STEMCELL Technologies) and 1% penicillin/streptomycin (Life Technologies). Neurons were transfected at DIV3-4 (knock-in constructs) or DIV10-11 (overexpression constructs) using Lipofectamine 2000 reagent (Invitrogen). Shortly before transfection, neurons were transferred to a plate with fresh NB medium with supplements. Next, a mixture of 2 µg of DNA and 3.3 µl of Lipofectamine in 200 µl of NB medium was incubated for 15–30 min and added to each well. After 1–2 h, neurons were briefly washed with NB medium and transferred back to the plate with conditioned medium. All experiments were performed using neurons at DIV21-24.
Antibodies and reagents
In this study the following primary antibodies were used: mouse anti-Bassoon (1:500 dilution, Enzo, #ADI-VAM-PS003-F, RRID AB_10618753); rabbit anti-GFP (1:2000 dilution, MBL Sanbio, #598, RRID AB_591819); rabbit anti-mGluR2/3 (1:50 dilution, EMD Millipore, #AB1553, RRID AB_90767); rabbit anti-mGluR7 (1:100 dilution, Merck Millipore, #07-239, RRID AB_310459); rat anti-HA (dilution 1:400, Sigma Aldrich, #11867423001; RRID AB_390919); and anti-GFP nanobodies conjugated with ATTO647N (1:15000 dilution, GFPBooster-ATTO647N, Chromotek, #gba647n). The following secondary antibodies were used: goat Alexa Fluor594-conjugated anti-rat (1:200 dilution, Life Technologies, #A-11007); goat Abberior STAR580-conjugated anti-rabbit (1:200 dilution, Abberior GmbH, #2-0012-005-8); and goat Abberior STAR635P-conjugated anti-mouse (1:200 dilution, Abberior GmBH, #2-0002-007-5). The following chemical reagents were used: 4-aminopyridine (4-AP, TOCRIS, #940), ADX 71743 (TOCRIS, #5715), DL-TBOA (TOCRIS, #1223), L-AP4 (TOCRIS, #0103), Latrunculin B (Bio-Connect, #SC-203318), LY379268 (TOCRIS, #2453), (RS)-APICA (TOCRIS, #1073) and Tetrodotoxin citrate (TTX, TOCRIS, #1069).
DNA plasmids
The SEP-mGluR2 and ELFN2-2xHA CRISPR/Cas9 knock-in constructs were designed as described in (Willems et al., 2020). SEP tag was inserted into exon 2 of the Grm2 gene using the following target sequence: 5′-AGGGTCAGCACCTTCTTGGC-3′. 2xHA tag was inserted into exon 2 of Elfn2 gene using following target sequence: 5′-AGACCCCCTTCCAGTAATCA-3′. Plasmids pRK5-mGluR2-GFP and pRK5-myc-mGluR7a (gift from J. Perroy) were used as PCR template to generate pRK5-SEP-mGluR2 and pRK5-SEP-mGluR7. pRK5-mOrange-mGluR2 and pRK5-mOrange-mGluR7 were created by exchanging SEP with mOrange in pRK5-SEP-mGluR2 and pRK5-SEP-mGluR7. pRK5-SEP-mGluR7-N74K was cloned using site-directed mutagenesis with the following primers: forward: 5′-GGCGACATCAAGAGGGAGAAAGGGATCCACAGGCTGGAAGC-3′ and reverse: 5′-GCTTCCAGCCTGTGGATCCCTTTCTCCCTCTTGATGTCGCC-3′. To create SEP-tagged chimeric variants of mGluR2 and mGluR7, sequences of wild-type receptors in pRK5-SEP-mGluR2 and pRK5-SEP-mGluR7 were replaced by the sequence of the chimeric receptor. Chimeric receptors were cloned by fusing sequences encoding different domains of mGluR2, mGluR7, and mGluR1 as follows:
mGluR2-ICD7: 1-819 aa mGluR2 + 849-913 aa mGluR7;
mGluR2-TMD7: 1-556 aa mGluR2 + 578-848 aa mGluR7 + 820-872 mGluR2;
mGluR2-ECD7: 1-583 aa mGluR7 + 562-872 aa mGluR2;
mGluR7-ICD2: 1-848 aa mGluR7 + 820-872 aa mGluR2;
mGluR7-TMD1: 1-588 aa mGluR7 + 591-839 aa mGluR1 + 849-914 aa mGluR7;
mGluR7-TMD2: 1-588 aa mGluR7 + 568-819 aa mGluR2 + 849-914 aa mGluR7;
mGluR7-ECD1: 1-585 aa mGluR1 + 584-913 aa mGluR7;
mGluR7-ECD2: 1-556 aa mGluR2 + 584-913 aa mGluR7.
Amino acid numbering is based on sequences in the UniPortKB database (mGluR1-Q13255-1, mGluR2-P31421-1, mGluR7-P35400-1) and starts with the first amino acid of the signal peptide. pRK5-SEP-mGluR1 (Scheefhals et al., 2019) was used as a PCR template for the transmembrane and extracellular domain of mGluR1. pRK5-SEP-mGluR2-ΔICD (lacing ICD, 823–872 aa) and pRK5-SEP-mGluR2-ΔPDZ (lacing PDZ binding motif, 869–872 aa) were cloned using primers containing the desired mutation. All chimeric and deletion mGluR variants were cloned using Gibson assembly (NEBuilder HiFi DNA assembly cloning kit). pRK5-mOrange-mGluR2-ECD7 was generated by replacing SEP tag in pRK5-SEP-mGluR2-ECD7. Synaptophysin1-mCherry plasmid was generated by replacing the pHluorin-tag in Synaptophysin1-pHluorin (gift from L. Lagnado, Addgene plasmid #24478, Granseth et al., 2006) with mCherry from pmCherry-N1 (Invitrogen). ELFN2-GFP plasmid was a gift from E. Sylwestrak (Sylwestrak and Ghosh, 2012). All sequences were verified by DNA sequencing.
U2OS cells coculture assays
U2OS cells (ATCC HTB-96) were cultured in DMEM (Lonza) supplemented with 10% fetal calf serum (Sigma), 2 mM glutamine, and 1% penicillin/streptomycin (Life Technologies). The day before transfection U2OS cells were seeded in a six-well plate. Next, cells were transfected using 6 µg of polyethylenimine (PEI, Polysciences) and 4 µg of DNA per well. Cells were transfected either with ELFN2-GFP or with mOrange-tagged mGluR2/7. At 24 h after transfection, cells were trypsinized, and ELFN2-GFP transfected cells were mixed with mOrange-mGluR2/7 transfected cells in a 1:1 ratio and seeded on 18-mm glass coverslips. 48 h after trypsinization, U2OS cells were fixed with 4% PFA for 10 min at RT, washed three times with phosphate-buffered saline (PBS), and mounted in Mowiol mounting medium (Sigma). Imaging of U2OS cells was performed with a Zeiss LSM 700 confocal microscope using a 63 × NA 1.40 oil objective.
Immunostaining and gSTED imaging
Neurons at DIV21 were fixed with 4% PFA and 4% sucrose in PBS for 10 min at RT and washed three times with PBS supplemented with 100 mM glycine. Next, cells were permeabilized and blocked with 0.1% Triton-X (Sigma), 10% normal goat serum (Abcam), and 100 mM glycine in PBS for 1 h at 37°C. Neurons were incubated with primary antibodies diluted in PBS supplemented with 0.1% Triton-X, 5% normal goat serum, and 100 mM glycine for 3–4 h at RT. After three times washing cells with PBS with 100 mM glycine, neurons were incubated with secondary antibodies diluted in PBS supplements with 0.1% Triton-X, 5% normal goat serum, and 100 mM glycine for 1 h at RT. Cells were washed two times with PBS with 100 mM glycine and two times with PBS. Neurons were mounted in Mowiol mounting medium (Sigma). Due to low endogenous level of mGluR2, the signal of endogenously tagged protein was enhanced by immunostaining with rabbit anti-GFP antibodies (1:2000 dilution, MBL Sanbio). Dual-color gated STED imaging was performed with a Leica TCS SP8 STED 3 microscope using an HC PL APO 100/1.4 oil-immersion STED WHITE objective. Abberior STAR 580 and 635P were excited with 561-nm and 633-nm pulsed laser light (white light laser, 80 MHz), respectively. Both Abberior STAR 580 and 635P were depleted with a 775-nm pulsed depletion laser. Fluorescence emission was detected using a Leica HyD hybrid detector with a gating time from 0.5 to 6 ns.
Live-cell imaging and FRAP experiments
For all live-cell imaging experiments, cells were kept in a modified Tyrode’s solution (pH 7.4) containing 25 mM HEPES, 119 mM NaCl, 2.4 mM KCl, 2 mM CaCl2, 2 mM MgCl2, and 30 mM glucose. FRAP experiments were carried out in an environmental chamber at 37°C (TokaHit) on an inverted Nikon Ti Eclipse microscope equipped with a confocal spinning disk unit (Yokogawa), an ILas FRAP unit (Roper Scientific France/ PICT-IBiSA, Institut Curie), and a 491-nm laser (Cobolt Calypso). Fluorescence emission was detected using a 100× oil-immersion objective (Nikon Apo, NA 1.4) together with an EM-CCD camera (Photometirc Evolve 512) controlled by MetaMorph 7.7 software (Molecular Devices). Images were acquired at 1 Hz with an exposure time between 100 and 200 ms. Three to five ROIs covering single boutons were bleached per field of view.
Single-molecule tracking with uPAINT
Single-molecule tracking was carried out in modified Tyrode’s solution supplement with 0.8% BSA and ATTO647N-conjugated anti-GFP nanobodies (1:15,000 dilution, GFPBooster-ATTO647N, Chromotek, #gba647n) on a Nanoimager microscope (Oxford Nanoimaging; ONI) equipped with a 100× oil-immersion objective (Olympus Plans Apo, NA 1.4), an XYZ closed-loop piezo stage, and 471-, 561-, and 640-nm lasers used for excitation of SEP, mCherry, and ATTO647N, respectively. Fluorescence emission was detected using an sCMOS camera (ORCA Flash 4, Hamamatsu). Three thousand images were acquired in stream mode at 50 Hz in TIRF. Before every tracking acquisition, 30 frames of SEP and mCherry signal were taken to visualize cell morphology or boutons. To determined how the activity of receptors influences their diffusion, first control acquisitions (2–3 fields of view per coverslip) were taken, and then chemical reagents (with final concentrations 200 µM 4-aminopyridine [4-AP] + 10 µM DL-TBOA; 30 µM ADX; 500 µM L-AP4; 100 µM APICA; 100 µM LY379268; 5 µM latrunculin-B; 1 µM TTX) or high K+ solution (2×) were added to the imaging chamber and incubated for 3–5 min, and final acquisitions of previously imaged fields of views were performed. A high K+ solution was prepared by replacing 45 mM NaCl with KCl. Total incubation times with chemical reagents or high K+ solution did not exceed 15 min.
Computational modeling of mGluR activity
Receptor model.
To study the time-dependent response of mGluRs upon glutamate release, a G-protein-coupled receptor model was combined with the time-dependent concentration profile of glutamate released from synaptic vesicles. The cubic ternary complex activation model (cTCAM) of GPCR signaling describes the interaction of the receptors R, ligands L, and G-proteins G (Kinzer-Ursem and Linderman, 2007). The receptors can complex with G-proteins to form RG and furthermore can be in an active state R* denoted by the asterisk. G-proteins are produced by a cascade of GαGTP hydrolysis and Gβγ binding. The reactions are described by the following differential equations:
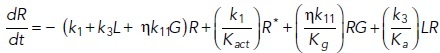 |
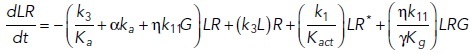 |
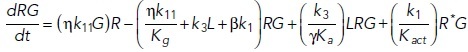 |
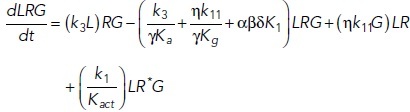 |
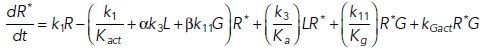 |
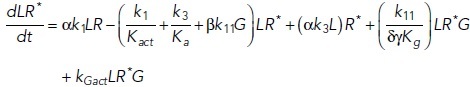 |
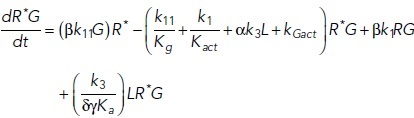 |
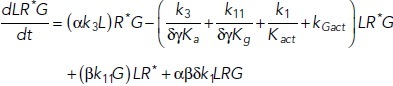 |
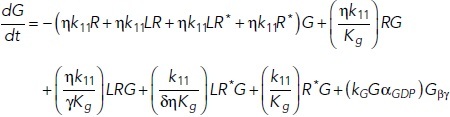 |
To find the steady-state solution without ligand (L = 0), these equations were solved with initial conditions R = 100, G = 1000, and the remaining variables set to zero using the NDSolve function of Mathematica (version 12.0, Wolfram Research Inc.). The numerical values for the used parameters have been described previously (Kinzer-Ursem and Linderman, 2007) and are summarized in Supplemental Table S1. The number of receptors and G-proteins in presynaptic bouton are estimated based on quantitative mass-spectrometry data published in (Wilhelm et al., 2014). To describe the different behavior of mGluR2 and mGluR7, only the association constant Ka was adjusted to match previously published EC50 values: 10 µM for mGluR2 and 1 mM for mGluR7 (Schoepp et al., 1999). The EC50 value is the concentration of the ligand that gives the half-maximum response. The response was estimated by the number of GαGTP. The steady-state solution without ligand was used as the initial state of the system and the new steady-state values for different amounts of the ligand were numerically determined. The relative normalized change of GαGTP gives the response:
To obtain the EC50 value, the following function was fitted to the data points from the numerical solution (Supplemental Figure S5B):
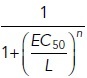 |
In this way, a parameterization of mGluR2 with Ka = 0.7 × 104 M–1 and respective EC50 = 10 µM, and mGluR7 with Ka = 60 M–1 and respective EC50 = 1.15 mM was obtained. To investigate the ligand–receptor affinity, the normalized response of the sum of all formed receptor-ligand complexes was determined as (Supplemental Figure S5A)
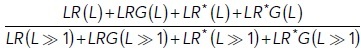 |
Diffusion model.
The time-dependent concentration of glutamate released from a synaptic vesicle was described as a point source on an infinite plane. The solution of the diffusion equation gives the surface density:
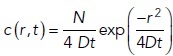 |
in which
r: the distance from the source,
N = 3000: the total amount of glutamate released,
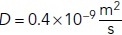 : the diffusion constant of glutamate (Kessler, 2013).
: the diffusion constant of glutamate (Kessler, 2013).
To transform the surface density into a concentration the following formula was used:
in which
rv = 25 nm: the radius of a vesicle,
C0: the glutamate concentration inside the vesicle.
Next, the surface density was divided by the width d = 20 nm of the synaptic cleft to obtain
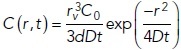 |
Hence, the initial concentration is given by
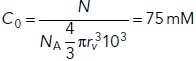 |
in which NA is Avogadro’s constant.
To describe the glutamate concentration from a sequence of vesicles release events, superposition was used as follows:
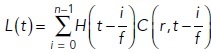 |
in which
n: the number of vesicles released,
f: the release frequency,
 : a step function.
: a step function.
The diffusion profile was combined with the receptor model and the differential equations were solved numerically for a given distance r from the release site. For the initial conditions, the steady-state solution without ligand was used. Because of the nonlinearities in the equations and the possible large values of the concentration profile for small times, to solve the equations numerically, we reduced the accuracy and precision of the numerical integration method in Mathematica’s NDSolve function. This adjustment potentially introduced an error of less than 5%, which is small enough to be neglected in our analysis and conclusions.
Testing the model with different sets of parameters.
To test the model’s output sensitivity to the numerical values of the model parameters, the receptor activation rate, the G-protein association rate, the G-protein collision efficiency, and the G-protein binding affinity were changed in the ranges reported previously (Kinzer-Ursem and Linderman, 2007). After the value of one of the parameters was changed, the Ka was always adjusted to match the EC50 values of the specific receptors. Note, the nonlinearity and the multiple pathways of the model also cause changes in Ka values in some cases (Supplemental Table S2). Next, the diffusion profile of glutamate released from 10 SV at 20 Hz was combined with the receptor models described by different sets of parameters. A change of the numerical values for the parameter changed the absolute value of the model output, but the overall trend of receptor activation remained the same. For better comparison of how changes in parameter values influence receptor activation at different distances from the release site, the maximum fold changes of GαGTP concentration for each receptor at different distances were normalized to the maximum of this concentration at 5 nm away from the release site to obtain the reduction factor of the maximum fold change of GαGTP concentration (Supplemental Figure S5, F and J).
Quantification of colocalization
Analysis of colocalization between Bsn and mGluRs was done using Spot Detector and Colocalization Studio plug-ins built in in Icy software (De Chaumont et al., 2012). Objects detected with Spot Detector (size of detected spots: ∼7 pixels with sensitivity 100 and ∼13 pixels with sensitivity 80) were loaded into Colocalization Studio and statistical object distance analysis (SODA; Lagache et al., 2018) was performed to obtain the fraction of mGluR spots colocalized with Bsn spots.
Quantification of SEP-mGluRs bouton enrichment
Neurons coexpressing cytosolic mCherry and SEP-mGluR2 or SEP-mGluR7 were fixed at DIV21 with 4% PFA and 4% sucrose from 10 min in RT. Next, cells were washed three times with PBS and mounted in Mowiol mounting medium (Sigma). Imaging was performed with a Zeiss LSM 700 confocal microscope using a 63 × NA 1.40 oil objective. To analyze the enrichment of mGluRs in presynaptic boutons, line profiles along boutons and neighboring axonal region were drawn in ImageJ (linewidth 3 pixels). Next, intensity profiles were fitted with a Gaussian function in GraphPad Prism. To calculate the ratio of intensity in bouton over axon, the amplitude of the Gaussian fit was divided by the minimum value of the fit.
Quantification of the expression level of SEP-mGluRs
Neurons transfected with SEP-mGluR2 or SEP-mGluR7 at DIV 10 were fixed and immunostained at DIV21. To compare expression levels of SEP-tagged mGluRs with the endogenous levels, neurons expressing SEP-mGluR2 were costained with anti-mGluR2/3, and cells expressing SEP-mGluR7 were costained with anit-mGluR7. Imaging was performing on a Leica TCS SP8 STED 3 microscope using an HC PL APO 100/1.4 oil-immersion STED WHITE objective keeping all imaging parameters constant. In ImageJ, circular ROIs with the size of boutons were drawn around boutons in neurons expressing SEP-tagged mGluRs and untransfected boutons positive for mGluR2 or mGluR7. Fluorescent intensity values of control boutons (untransfected) and expressing SEP-tagged mGluRs were normalized by dividing by the average intensity values of all control boutons.
Quantification of FRAP experiments
Time series obtained during FRAP experiments were corrected for drift when needed using Template Matching plug-in in ImageJ. Circular ROIs with the size of the bleached area were drawn in ImageJ. Fluorescent intensity transients were normalized by subtracting the intensity values of the first frame after bleaching and dividing by the average intensity value of the baseline (five frames before bleaching). The mobile fraction was calculated by averaging the values of the last five points of fluorescent transients. τ of recovery was determined by fitting a single exponential function to the recovery traces.
Single-molecule tracking analysis
NimOS software (Oxford Nanoimager; ONI) was used to detect the localization of single molecules in uPAINT experiments. Molecules with a localization precision <50 nm and photon count >200 photons were used for analysis. To filter out unspecific background localizations from outside neurons, a cell mask based on the SEP image was created using an adaptive image threshold in Matlab (sensitivity 40–55). Only localizations inside the mask were selected for further analysis. Tracking and calculation of the diffusion coefficient were performed in custom-written Matlab (MathWorks) scripts described previously (Willems et al., 2020). Only trajectories longer than 30 frames were used to estimate the instantaneous diffusion coefficient. Classification of molecule state as mobile or immobile was based on the ratio between the radius of gyration and mean step size of individual trajectories using formula
(Golan and Sherman, 2017; Supplemental Figure S2, F–H). Molecules with a ratio < 2.11 were considered immobile (Golan and Sherman, 2017). A mask of presynaptic boutons was created based on the TIRF image of Synaptophysin1-mCherry as previously described (Li and Blanpied, 2016). Synaptic trajectories were defined as trajectories that had at least one localization inside the bouton mask. In all graphs representing Deff or mobile fraction, a single data point is an average Deff or mobile fraction from one complete field of view. Only experiments with at least three fields of view with a minimum of 30 trajectories were included in analyses.
Statistical analysis
All used in this study statistical tests are described in figure legends and the main text. All average values presented in the main text are means ± SEM. All statistical analyses and graphs were prepared in GraphPad Prism 9. Figures were created in Inkscape.
Resource availability
Further information and requests for resources, reagents, and code should be directed to and will be fulfilled by Harold MacGillavry (h.d.macgillavry@uu.nl).
Supplementary Material
Acknowledgments
We would like to thank Arthur de Jong for critical reading of the manuscript, Manon Westra for help with Matlab scripts, and all members of the MacGillavry lab for helpful discussions. This work was supported by the European Research Council (ERC-StG 716011) to H.D.M.
Abbreviations used:
- Bsn
Bassoon
- ECD
extracellular domain
- mGluR
metabotropic glutamate receptor
- ICD
intracellular domain
- TMD
transmembrane domain.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-10-0484) on May 5, 2022.
REFERENCES
- Billups B, Graham BP, Wong AYC, Forsythe ID (2005). Unmasking group III metabotropic glutamate autoreceptor function at excitatory synapses in the rat CNS. J Physiol 565, 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudin H, Doan A, Xia J, Shigemoto R, Huganir RL, Worley P, Craig AM (2000). Presynaptic clustering of mGluR7a requires the PICK1PDZ domain binding site. Neuron 28, 485–497. [DOI] [PubMed] [Google Scholar]
- Bushell TJ, Sansig G, Collett VJ, van der Putten H, Collingridge GL (2002). Altered short-term synaptic plasticity in mice lacking the metabotropic glutamate receptor mGlu7. ScientificWorldJournal 2, 730–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calebiro D, Rieken F, Wagner J, Sungkaworn T, Zabel U, Borzi A, Cocucci E, Zürn A, Lohse MJ (2013). Single-molecule analysis of fluorescently labeled G-protein-coupled receptors reveals complexes with distinct dynamics and organization. Proc Natl Acad Sci USA 110, 743–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha-Ferreira I, Chazeau A, Buijs RR, Stucchi R, Will L, Pan X, Adolfs Y, van der Meer C, Wolthuis JC, Kahn OI, et al. (2018). The HAUS complex is a key regulator of non-centrosomal microtubule organization during neuronal development. Cell Rep 24, 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta A, Lim YJ, Kumar K, Baby N, Pang KLK, Benoy A, Behnisch T, Sajikumar S (2020). Group III metabotropic glutamate receptors gate long-term potentiation and synaptic tagging/capture in rat hippocampal area CA2. Elife 9, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Chaumont F, Dallongeville S, Chenouard N, Hervé N, Pop S, Provoost T, Meas-Yedid V, Pankajakshan P, Lecomte T, Le Montagner Y, et al. (2012). Icy: an open bioimage informatics platform for extended reproducible research. Nat Methods 9, 690–696. [DOI] [PubMed] [Google Scholar]
- de Jong AP, Verhage M (2009). Presynaptic signal transduction pathways that modulate synaptic transmission. Curr Opin Neurobiol 19, 245–253. [DOI] [PubMed] [Google Scholar]
- Diamond JS, Jahr CE (1997). Transporters buffer synaptically released glutamate on a submillisecond time scale. J Neurosci 17, 4672–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn HA, Orlandi C, Martemyanov KA (2019a). Beyond the ligand: extracellular and transcellular G protein-coupled receptor complexes in physiology and pharmacology. Pharmacol Rev 71, 503–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn HA, Patil DN, Cao Y, Orlandi C, Martemyanov KA (2018). Synaptic adhesion protein ELFN1 is a selective allosteric modulator of group III metabotropic glutamate receptors in trans. Proc Natl Acad Sci 115, 5022–5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn HA, Zucca S, Dao M, Orlandi C, Martemyanov KA (2019b). ELFN2 is a postsynaptic cell adhesion molecule with essential roles in controlling group III mGluRs in the brain and neuropsychiatric behavior. Mol Psychiatry 24, 1902–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrero JJ, Bartolomé-Martín D, Torres M, Sánchez-Prieto J (2013). Potentiation of mGlu7 receptor-mediated glutamate release at nerve terminals containing N and P/Q type Ca2+channels. Neuropharmacology 67, 213–222. [DOI] [PubMed] [Google Scholar]
- Francesconi A, Duvoisin RM (2002). Alternative splicing unmasks dendritic and axonal targeting signals in metabotropic glutamate receptor 1. J Neurosci 22, 2196–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannone G, Hosy E, Levet F, Constals A, Schulze K, Sobolevsky AI, Rosconi MP, Gouaux E, Tampe R, Choquet D, et al. (2010). Dynamic superresolution imaging of endogenous proteins on living cells at ultra-high density. Biophys J 99, 1303–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golan Y, Sherman E (2017). Resolving mixed mechanisms of protein subdiffusion at the T cell plasma membrane. Nat Commun 8, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granseth B, Odermatt B, Royle S, Lagnado L (2006). Clathrin-mediated endocytosis is the dominant mechanism of vesicle retrieval at hippocampal synapses. Neuron 51, 773–786. [DOI] [PubMed] [Google Scholar]
- Gutzeit VA, Thibado J, Stor DS, Zhou Z, Blanchard SC, Andersen OS, Levitz J (2019). Conformational dynamics between transmembrane domains and allosteric modulation of a metabotropic glutamate receptor. Elife 8, 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Zhou R, Wu Z, Carrasco MA, Kurshan PT, Farley JE, Simon DJ, Wang G, Han B, Hao J, et al. (2016). Prevalent presence of periodic actin-spectrin-based membrane skeleton in a broad range of neuronal cell types and animal species. Proc Natl Acad Sci USA 113, 6029–6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullié D, Stoeber M, Sibarita JB, Zieger HL, Bartol TM, Arttamangkul S, Sejnowski TJ, Hosy E, von Zastrow M (2020). A discrete presynaptic vesicle cycle for neuromodulator receptors. Neuron 105, 663–677.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya H, Ozawa S (1999). Dual mechanism for presynaptic modulation by axonal metabotropic glutamate receptor at the mouse mossy fibre-CAS synapse. J Physiol 518, 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammermeier PJ (2015). Constitutive activity of metabotropic glutamate receptor 7. BMC Neurosci 16, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Wilkins AD, Lichtarge O, Wensel TG (2015). Determinants of endogenous ligand specificity divergence among metabotropic glutamate receptors. J Biol Chem 290, 2870–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai RS, Kusumi A (2014). Single-molecule imaging revealed dynamic GPCR dimerization. Curr Opin Cell Biol 27, 78–86. [DOI] [PubMed] [Google Scholar]
- Kessler JP (2013). Control of cleft glutamate concentration and glutamate spill-out by perisynaptic glia: uptake and diffusion barriers. PLoS One 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A, Shigemoto R, Ohishi H, Van Der Putten H, Mizuno N (1998). Immunohistochemical localization of metabotropic glutamate receptors, mGluR7a and mGluR7b, in the central nervous system of the adult rat and mouse: a light and electron microscopic study. J Comp Neurol 393, 332–352. [PubMed] [Google Scholar]
- Kinzer-Ursem TL, Linderman JJ (2007). Both ligand- and cell-specific parameters control ligand agonism in a kinetic model of G protein-coupled receptor signaling. PLoS Comput Biol 3, 0084–0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar R, Walker AG, Ghose D, Grueter Ba, Engers DW, Hopkins CR, Lindsley CW, Xiang Z, Conn PJ, Niswender CM (2015). Activation of metabotropic glutamate receptor 7 is required for induction of long-term potentiation at SC-CA1 synapses in the hippocampus. J Neurosci 35, 7600–7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagache T, Grassart A, Dallongeville S, Faklaris O, Sauvonnet N, Dufour A, Danglot L, Olivo-Marin JC (2018). Mapping molecular assemblies with fluorescence microscopy and object-based spatial statistics. Nat Commun 9, 102–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li TP, Blanpied TA (2016). Control of transmembrane protein diffusion within the postsynaptic density assessed by simultaneous single-molecule tracking and localization microscopy. Front Synaptic Neurosci 8, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman JE, Raghavachari S, Tsien RW (2007). The sequence of events that underlie quantal transmission at central glutamatergic synapses. Nat Rev Neurosci 8, 597–609. [DOI] [PubMed] [Google Scholar]
- Martín R, Durroux T, Ciruela F, Torres M, Pin JP, Sánchez-Prieto J (2010). The metabotropic glutamate receptor mGlu7 activates phospholipase C, translocates munc-13-1 protein, and potentiates glutamate release at cerebrocortical nerve terminals. J Biol Chem 285, 17907–17917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín R, Ferrero JJ, Collado-Alsina A, Aguado C, Luján R, Torres M, Sánchez-Prieto J (2018). Bidirectional modulation of glutamatergic synaptic transmission by metabotropic glutamate type 7 receptors at Schaffer collateral–CA1 hippocampal synapses. J Physiol 596, 921–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín R, Torres M, Sánchez-Prieto J (2007). mGluR7 inhibits glutamate release through a PKC-independent decrease in the activity of P/Q-type Ca2+ channels and by diminishing cAMP in hippocampal nerve terminals. Eur J Neurosci 26, 312–322. [DOI] [PubMed] [Google Scholar]
- Mikasova L, Groc L, Choquet D, Manzoni OJ (2008). Altered surface trafficking of presynaptic cannabinoid type 1 receptor in and out synaptic terminals parallels receptor desensitization. Proc Natl Acad Sci USA 105, 18596–18601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millán C, Luján R, Shigemoto R, Sánchez-Prieto J (2002). The inhibition of glutamate release by metabotropic glutamate receptor 7 affects both [Ca2+]c and cAMP: evidence for a strong reduction of Ca2+ entry in single nerve terminals. J Biol Chem 277, 14092–14101. [DOI] [PubMed] [Google Scholar]
- Muly EC, Mania I, Guo J-D, Rainnie DG (2007). Group II metabotropic glutamate receptors in anxiety circuitry: correspondence of physiological response and subcellular distribution. J Comp Neurol 505, 682–700. [DOI] [PubMed] [Google Scholar]
- Niswender CM, Conn PJ (2010). Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu Rev Pharmacol Toxicol 50, 295–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohishi H, Ogawa-Meguro R, Shigemoto R, Kaneko T, Nakanishi S, Mizuno N (1994). Immunohistochemical localization of metabotropic glutamate receptors, mGluR2 and mGluR3, in rat cerebellar cortex. Neuron 13, 55–66. [DOI] [PubMed] [Google Scholar]
- Okamoto N, Hori S, Akazawa C, Hayashi Y, Shigemoto R, Mizuno N, Nakanishi S (1994). Molecular characterization of a new metabotropic glutamate receptor mGluR7 coupled to inhibitory cyclic AMP signal transduction. J Biol Chem 269, 1231–1236. [PubMed] [Google Scholar]
- Pelkey KA, Lavezzari G, Racca C, Roche KW, McBain CJ (2005). mGluR7 is a metaplastic switch controlling bidirectional plasticity of feedforward inhibition. Neuron 46, 89–102. [DOI] [PubMed] [Google Scholar]
- Pelkey KA, Topolnik L, Yuan XQ, Lacaille JC, McBain CJ (2008). State-dependent cAMP sensitivity of presynaptic function underlies metaplasticity in a hippocampal feedforward inhibitory circuit. Neuron 60, 980–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia RS, Wang Y-X, Niedzielski AS, Wenthold RJ (1996). The metabotropic glutamate receptors, MGLUR2 and MGLUR3, show unique postsynaptic, presynaptic and glial localizations. Neuroscience 71, 949–976. [DOI] [PubMed] [Google Scholar]
- Pinheiro PS, Mulle C (2008). Presynaptic glutamate receptors: physiological functions and mechanisms of action. Nat Rev Neurosci 9, 423–436. [DOI] [PubMed] [Google Scholar]
- Reiner A, Levitz J (2018). Glutamatergic signaling in the central nervous system: ionotropic and metabotropic receptors in concert. Neuron 98, 1080–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbe D, Alonso G, Chaumont S, Bockaert J, Manzoni OJ (2002). Role of p/q-Ca2+ channels in metabotropic glutamate receptor 2/3-dependent presynaptic long-term depression at nucleus accumbens synapses. J Neurosci 22, 4346–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansig G, Bushell TJ, Clarke VR, Rozov A, Burnashev N, Portet C, Gasparini F, Schmutz M, Klebs K, Shigemoto R, et al. (2001). Increased seizure susceptibility in mice lacking metabotropic glutamate receptor 7. J Neurosci 21, 8734–8745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanziani M, Salin PA, Vogt KE, Malenka RC, Nicoll RA (1997). Use-dependent increases in glutamate concentration activate presynaptic metabotropic glutamate receptors. Nature 385, 630–634. [DOI] [PubMed] [Google Scholar]
- Scheefhals N, Catsburg LAE, Westerveld ML, Blanpied TA, Hoogenraad CC, MacGillavry HD (2019). Shank proteins couple the endocytic zone to the postsynaptic density to control trafficking and signaling of metabotropic glutamate receptor 5. Cell Rep 29, 258-269.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoepp DD, Jane DE, Monn JA (1999). Pharmacological agents acting at subtypes of metabotropic glutamate receptors. Neuropharmacology 38, 1431–1476. [DOI] [PubMed] [Google Scholar]
- Shigemoto R, Kinoshita A, Wada E, Nomura S, Ohishi H, Takada M, Flor PJ, Neki A, Abe T, Nakanishi S, et al. (1997). Differential presynaptic localization of metabotropic glutamate receptor subtypes in the rat hippocampus. J Neurosci 17, 7503–7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigemoto R, Kulik A, Roberts JDB, Ohishi H, Nusser Z, Kaneko T, Somogyi P (1996). Target-cell-specific concentration of a metabotropic glutamate receptor in the presynaptic active zone. Nature 381, 523–525. [DOI] [PubMed] [Google Scholar]
- Siddig S, Aufmkolk S, Doose S, Jobin M-L, Werner C, Sauer M, Calebiro D (2020). Super-resolution imaging reveals the nanoscale organization of metabotropic glutamate receptors at presynaptic active zones. Sci Adv 6, eaay7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachniak XTJ, Sylwestrak XEL, Scheiffele XP, Hall BJ, Ghosh A, Stachniak TJ, Sylwestrak EL, Scheiffele P, Hall BJ, Ghosh A (2019). Elfn1-induced constitutive activation of mGluR7 determines frequency-dependent recruitment of somatostatin interneurons. J Neurosci 39, 4461–4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh YH, Pelkey KA, Lavezzari G, Roche PA, Huganir RL, McBain CJ, Roche KW (2008). Corequirement of PICK1 binding and PKC phosphorylation for stable surface expression of the metabotropic glutamate receptor mGluR7. Neuron 58, 736–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungkaworn T, Jobin ML, Burnecki K, Weron A, Lohse MJ, Calebiro D (2017). Single-molecule imaging reveals receptor-G protein interactions at cell surface hot spots. Nature 550, 543–547. [DOI] [PubMed] [Google Scholar]
- Sylwestrak EL, Ghosh A (2012). Elfn1 regulates target-specific release probability at CA1-interneuron synapses. Science 338, 536–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomioka NH, Yasuda H, Miyamoto H, Hatayama M, Morimura N, Matsumoto Y, Suzuki T, Odagawa M, Odaka YS, Iwayama Y, et al. (2014). Elfn1 recruits presynaptic mGluR7 in trans and its loss results in seizures. Nat Commun 5, 1–16. [DOI] [PubMed] [Google Scholar]
- Wilhelm BG, Mandad S, Truckenbrodt S, Kröhnert K, Schäfer C, Rammner B, Koo SJ, Claßen Ga, Krauss M, Haucke V, et al. (2014). Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science 344, 1023–1028. [DOI] [PubMed] [Google Scholar]
- Willems J, de Jong APH, Scheefhals N, Mertens E, Catsburg LAE, Poorthuis RB, de Winter F, Verhaagen J, Meye FJ, MacGillavry HD (2020). ORANGE: A CRISPR/Cas9-based genome editing toolbox for epitope tagging of endogenous proteins in neurons. PLoS Biol 18, e3000665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley ML, Pemberton DJ, Bate S, Corti C, Jones DNC (2008). The mGlu2 but not the mGlu3 receptor mediates the actions of the mGluR2/3 agonist, LY379268, in mouse models predictive of antipsychotic activity. Psychopharmacology (Berl) 196, 431–440. [DOI] [PubMed] [Google Scholar]
- Yanagawa M, Hiroshima M, Togashi Y, Abe M, Yamashita T, Shichida Y, Murata M, Ueda M, Sako Y (2018). Single-molecule diffusion-based estimation of ligand effects on G protein-coupled receptors. Sci Signal 11, eaao1917. [DOI] [PubMed] [Google Scholar]
- Zhang C-S, Bertaso F, Eulenburg V, Lerner-Natoli M, Herin GA, Bauer L, Bockaert J, Fagni L, Betz H, Scheschonka A (2008). Knock-In mice lacking the PDZ-ligand motif of mGluR7a show impaired PKC-dependent autoinhibition of glutamate release, spatial working memory deficits, and increased susceptibility to pentylenetetrazol. J Neurosci 28, 8604–8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong G, He J, Zhou R, Lorenzo D, Babcock HP, Bennett V, Zhuang X (2014). Developmental mechanism of the periodic membrane skeleton in axons. Elife 3, e04581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Han B, Xia C, Zhuang X (2019). Membrane-associated periodic skeleton is a signaling platform for RTK transactivation in neurons. Science 365, 929–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.