Abstract
Osteoclasts are highly specialized, multinucleated cells responsible for the selective resorption of the dense, calcified bone matrix. Microtubules (MTs) contribute to the polarization and trafficking events involved in bone resorption by osteoclasts; however, the origin of these elaborate arrays is less clear. Osteoclasts arise through cell fusion of precursor cells. Previous studies have suggested that centrosome MT nucleation is lost during this process, with the nuclear membrane and its surrounding Golgi serving as the major MT organizing centers (MTOCs) in these cells. Here we reveal that precursor cell centrosomes are maintained and functional in the multinucleated osteoclast and interestingly form large MTOC clusters, with the clusters organizing significantly more MTs compared with individual centrosomes. MTOC cluster formation requires dynamic MTs and minus-end directed MT motor activity. Inhibition of these centrosome clustering elements had a marked impact on both F-actin ring formation and bone resorption. Together these findings show that multinucleated osteoclasts employ unique centrosomal clusters to organize the extensive MTs during bone attachment and resorption.
INTRODUCTION
The microtubule (MT) network is the largest component of the cytoskeleton, which supports cellular shape and maintains intracellular organization (Goodson and Jonasson, 2018). MTs have important roles in protein trafficking, organelle transport, and mitosis (Goodson and Jonasson, 2018). Structurally, MTs are assembled by the GTP-driven polymerization of α- and β-tubulin subunits in a polarized manner. MTs have a fast-growing plus-end, a stable/slow-growing minus-end, and continuously undergo assembly and disassembly within the cell in a property termed dynamic instability (Voter and Erickson, 1984; Akhmanova and Steinmetz, 2015). Dynamic instability is crucial to many cellular processes and is regulated by a host of MT-associated proteins (MAPs) that stabilize, grow, or depolymerize MT filaments (Goodson and Jonasson, 2018). The nucleation of MTs generally occurs at the minus-end of the filament and is facilitated by γ-tubulin ring complexes (γ-TuRCs) (Kollman et al., 2011).
Nucleation and organization of MTs predominantly occur at MT organizing centers (MTOCs) (Akhmanova and Steinmetz, 2015; Goodson and Jonasson, 2018). In proliferative cells, the centrosome serves as the main MTOC. The centrosome consists of two centrioles, a mother and a daughter, surrounded by a matrix of proteins known as the pericentriolar material (PCM) (Bornens, 2002, 2012; Woodruff et al., 2014). The presence of distal appendages like CEP164 and cenexin, as well as subdistal appendages like ninein that aid in cilia development and MT anchoring, distinguishes the mother centriole from the daughter centriole (Bornens, 2002). While the PCM may be comprised of hundreds of proteins, only a core subset is involved with promoting centrosomal MT nucleation; pericentrin, CEP192 (centrosomal protein 192), and CDK5RAP2 are three such proteins (Bornens, 2002; Mennella et al., 2014).
It is largely believed that centrosome-based MTOCs are nonfunctional in most terminally differentiated cell types (Muroyama and Lechler, 2017; Martin and Akhmanova, 2018). Noncentrosomal (nc)MTOCs have been a recent topic of exploration in differentiated cell types and orchestrate unique MT arrays and cellular architectures to perform specialized functions. In differentiated muscle cells, MTs are nucleated at the nuclear envelope in parallel arrays to facilitate nuclear anchoring and positioning (Tassin et al., 1985; Bugnard et al., 2005; Starr, 2017). Neurons employ Golgi reorganization and MT nucleation at Golgi outposts that run along axons and dendrites for enhanced transport (Ori-McKenney et al., 2012). Functional nc-MTOCs require the coordinated accumulation of PCM components and the transport of existing MTs to new sites of MT organization and nucleation (Bartolini and Gundersen, 2006; Muroyama and Lechler, 2017; Martin and Akhmanova, 2018). The general mechanisms behind site designation, assembly, and function of nc-MTOCs still remain poorly understood.
Few studies (Lucht, 1973; Turksen et al., 1988; Mulari et al., 2003) have examined MTOCs in osteoclasts, and little consensus exists regarding their presence or function. Osteoclasts are multinucleated giant cells (Bar-Shavit, 2007; Yavropoulou and Yovos, 2008; Soysa et al., 2012) that are responsible for the selective degradation of bone during the bone remodeling process (Jurdic et al., 2006). The commitment and development of a mature osteoclast depend on multiple stages of differentiation (Mizoguchi et al., 2009; Oursler, 2010; Miyamoto, 2011; Motiur Rahman et al., 2015; Fiorino and Harrison, 2016a) including exit from the cell cycle (Okahashi et al., 2001; Mizoguchi et al., 2009; Motiur Rahman et al., 2015) followed by the migration (Wang et al., 2008; Brazier et al., 2009) and fusion (Kim et al., 2008; Helming and Gordon, 2009; Hobolt-Pedersen et al., 2014) of multiple osteoclast precursor cells. Activation and polarization of mature osteoclasts occur in response to attachment to a mineralized bone matrix (Saltel et al., 2004). Osteoclast polarization is characterized by major cytoskeletal reorganization and the formation of distinct domains that allow for bone resorption and subsequent secretion of digested elements (Bar-Shavit, 2007; Yavropoulou and Yovos, 2008).
The unique combination of cell fusion events, cytoskeletal changes, specialized cellular morphology, and the migratory capacity of multinucleated osteoclasts presents an interesting cell model to investigate centrosomal MTOCs. Utilizing high resolution microscopy and live imaging, we found that functional centrosomes are present throughout osteoclastogenesis and form aggregates/clusters early in the differentiation process. The MTOC clusters have potent MT nucleating activity compared with lone centrosomes. Nocodazole MT regrowth experiments identified centrosomes as a predominant location for MT nucleation and demonstrated that centrosome clustering requires an established MT network. MT dynamicity, KIFC1, and dynein were identified as regulators of centrosome clustering. Inhibitor assays demonstrated the importance of centrosome clustering during F-actin ring formation and bone resorption. Indeed, we have shown that active centrosomes persist throughout the lifespan of murine and human osteoclasts in vitro and have provided evidence, for first time, that centrosome clustering is functionally advantageous in multinucleated osteoclasts.
RESULTS
Structurally intact centrosomes are present and clustered in osteoclasts
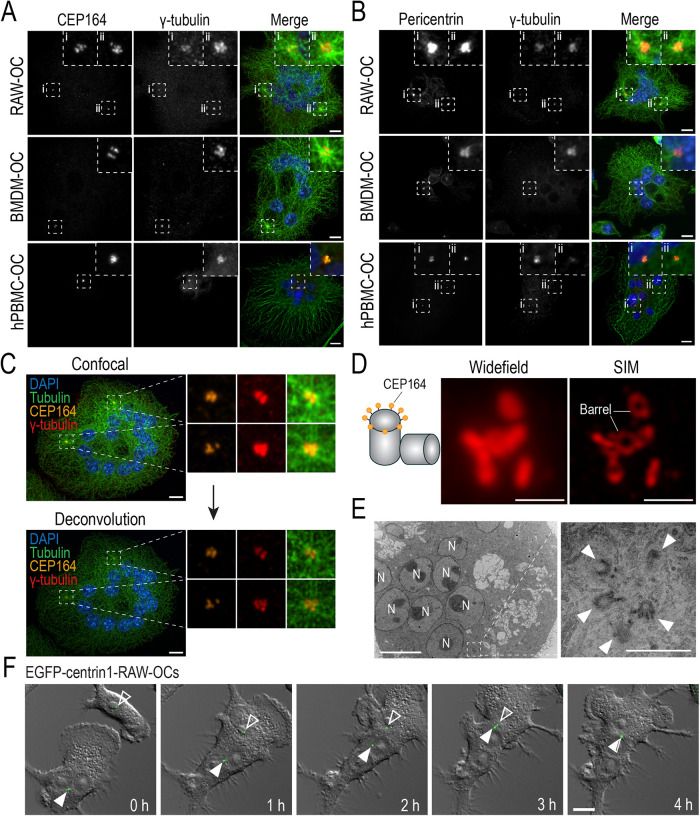
Early fluorescent analysis of osteoclasts suggested that mature mammalian osteoclasts possess few, if any, centrosomal MTOCs (Mulari et al., 2003). To perform our own evaluation of centrosomes in osteoclasts, immunofluorescence analysis and confocal imaging of the mother centriole (CEP164), pericentriolar matrix (pericentrin), and MT nucleation structures (γ-tubulin) were performed in osteoclasts derived from RAW 264.7 (RAW) cells, murine bone marrow-derived macrophages (BMDMs), and human peripheral blood monocytes (PBMCs) (Figure 1, A and B). Centrosomal components strongly colocalized in osteoclasts and were observed in every cell within the sampled populations (Figure 1, A and B). In addition, CEP164, pericentrin, and γ-tubulin signals were frequently observed clustered within aggregates from which extensive MT radial arrays emanated in murine osteoclasts. Centrosomal clusters were similarly observed over the 27-d differentiation period of human PBMC-derived osteoclasts (Figure 1, A and B).
FIGURE 1:
Structural components of functional centrosomes are present and organized into clusters in RAW-, BMDM-, and hPBMC-derived osteoclasts. (A and B) Representative immunofluorescent images of fixed end-point RAW-, BMDM-, and hPBMC-derived osteoclasts immunostained for centriolar and pericentriolar components. (A) Osteoclasts immunostained with DAPI (blue), tubulin (green), CEP164 (orange), and γ-tubulin (red) to label nuclei, MTs, the mother centriole, and γ-TuRCs, respectively. (B) Osteoclasts immunostained with DAPI (blue), tubulin (green), pericentrin (orange), and γ-tubulin (red) to label nuclei, MTs, the PCM, and γ-TuRCs, respectively. Dashed box insets show centrosomal signal clusters. (C) Iterative deconvolution of coalesced centrosomal signals in osteoclasts. Osteoclasts were immunostained with DAPI (blue), tubulin (green), CEP164 (orange), and γ-tubulin (red) to label nuclei, MTs, the mother centriole, and γ-TuRCs, respectively. (D) Widefield and SIM imaging of a CEP164-immunostained centrosome cluster in a RAW-derived osteoclast. (E) TEM imaging of centrosomes in RAW-derived osteoclasts. N represents nuclei and arrowheads indicate individual centrioles. Scale bars = 10 μm for panels A–C, F. Scale bars = 1 μm for D and E. (F) Representative epifluorescent live-cell imaging of EGFP-centrin1-transduced RAW cells cultured with 100 ng/ml RANKL for 2.5 d (see Supplemental Movie S1). Images display time-lapse frames of early osteoclast fusion and centrosome clustering. White arrowheads indicate clustered centrosomes in the osteoclast and open arrowheads indicate the lone centrosome in the fusion partner.
Movie S1.
Centrosome clustering occurs shortly after osteoclast fusion. Epifluorescent live-cell imaging of EGFP-centrin1-transduced osteoclasts after 2.5 days of differentiation. Osteoclasts were imaged every 5 minutes for 24 hours. Displayed is a 5-minute interval between frames; 9 hours total elapsed time.
To determine if the clusters were actually several centrosomes in close proximity, iterative deconvolution to a confidence level of 90% was employed. Indeed, clusters contained numerous discrete CEP164 and γ-tubulin foci, suggesting the presence of multiple bona fide centrosomes (Figure 1C). As a complement to deconvolution, structured illumination microscopy (SIM) was utilized to identify the individual barrels of multiple mother centrioles within the clustered signal (Figure 1D). Further high-resolution confirmation through transmission electron microscopy (TEM) confirmed centrioles in different spatial orientations that were organized into clusters in osteoclasts (Figure 1E). These observations indicate that functional centrosomes are present in differentiated murine- and human-derived osteoclasts.
To understand the kinetics of centrosome clustering, we next performed live-cell imaging of 2-d differentiated RAW-derived osteoclasts transduced with EGFP-centrin1. Centrin is primarily localized on the luminal side of centrioles (Paoletti et al., 1996; Ruiz-Binder et al., 2002), which allowed us to track both mother and daughter centrioles of the centrosome throughout osteoclast differentiation. Cells were imaged over a 24-h period at 5-min intervals in order to track the behaviour of the centrosomes prior and during cell fusion (Figure 1F; Supplemental Movie S1). Using DIC and epifluorescent imaging, initially both mononuclear cells and small osteoclasts were observed, with the latter containing clustered EGFP-centrin1 signals (Figure 1F; Supplemental Movie S1). Postcell fusion, the EGFP-centrin1 signal in the fusion partner (originally mononuclear) began migrating toward the centrosome cluster in the pre-existing small osteoclast (Figure 1F; Supplemental Movie S1). Complete coalescence of centrin1 signal originating from the fusion partner and osteoclast could be observed just 4 h postfusion (Figure 1F; Supplemental Movie S1).
Multinucleated osteoclasts retain a full complement of centrosomes and display different clustering phenotypes relative to cell size
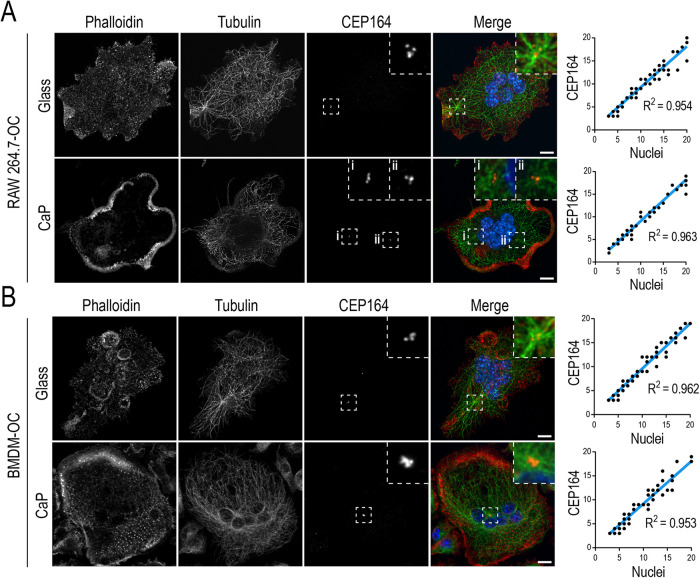
Osteoclast precursor cell differentiation requires cell cycle withdrawal prior to initiation of cell–cell fusion (Okahashi et al., 2001; Mizoguchi et al., 2009; Motiur Rahman et al., 2015). Each postmitotic mononuclear osteoclast precursor cell could theoretically donate a single centrosome to its fusion partner, producing a 1:1 ratio of centrosomes to nuclei in each resultant osteoclast. However, some differentiated cells inactivate their centrosomes and move to ncMT nucleation for their specialized functions (Yang and Feldman, 2015; Zebrowski et al., 2015; Muroyama and Lechler, 2017; Magescas et al., 2019). To determine the fate of centrosomes in osteoclasts, we compared centrosome number to nuclei number in RAW- and BMDM-derived osteoclasts cultured on either glass (nonpolarized) or bone mimetic calcium phosphate (CaP; polarized) substrates (Figure 2, A and B). Osteoclasts imaged via spinning disk confocal were subjected to iterative deconvolution to distinguish between individual CEP164 signals within clusters. Each CEP164 signal was representative of a mother centriole and served as a marker of a single centrosome (Graser et al., 2007). Osteoclasts cultured on glass contained characteristic F–actin-rich individual podosomes or podosome belts. F–actin-rich sealing zones were observed in osteoclasts cultured on CaP, indicative of their polarized states (Saltel et al., 2004). Most CEP164 signals were found in clusters colocalizing with asters of MTs; however, lone CEP164 signals were still present in RAW- and BMDM-derived osteoclasts in both resorbing and nonresorbing environments. Regardless of the osteoclast polarization status, the number of centrosomes was similar to the number of nuclei irrespective of osteoclast size. Linear regression analyses revealed an R2 of 0.95 and 0.96 for RAW-derived osteoclasts cultured on glass and CaP, respectively (Figure 2A). Similarly, BMDM-derived osteoclasts showed R2 values of 0.96 and 0.95 for glass and bone mimetic, respectively (Figure 2B). These data suggest that centrosomes inherited from osteoclast precursor cells persist in mature osteoclasts regardless of activation status.
FIGURE 2:
Centrosome number correlates with nuclei number in nonresorbing and resorbing RAW- and BMDM-derived osteoclasts. (A) Representative fixed RAW-derived osteoclasts cultured on glass (n = 70) and biomimetic CaP-coated (n = 59) glass coverslips. (B) Representative fixed BMDM-derived osteoclasts cultured on glass (n = 69) and biomimetic CaP-coated (n = 71) glass coverslips. Osteoclasts were labeled for nuclei, F-actin, MTs, and centrosomes with DAPI (blue), phalloidin (red), tubulin (green), and CEP164 (orange), respectively. F-actin labeling was performed to observe bone resorptive structures. Linear regression curves of centrosomes in a range of small (3–5 nuclei) to large (11+ nuclei) osteoclasts were quantified by scoring the number of CEP164 signals against the number of nuclei. Dashed box insets show centrosome clusters. Scale bar = 10 μm.
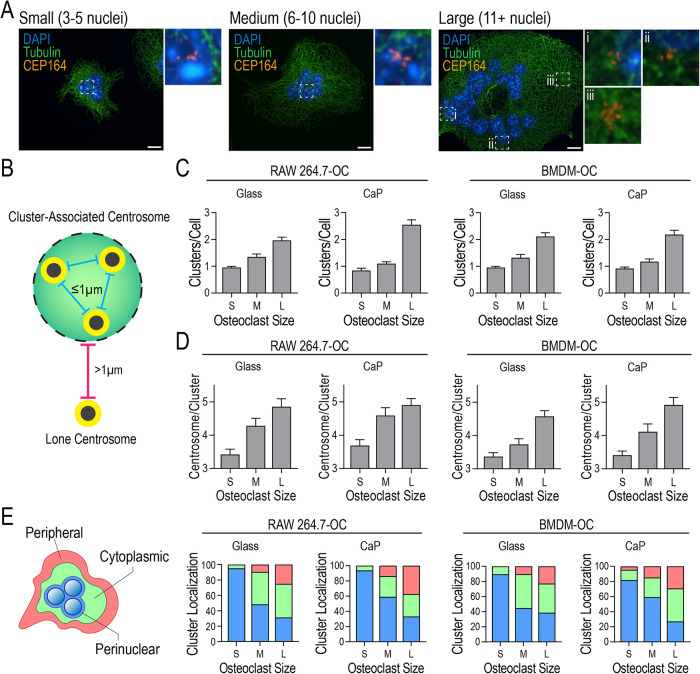
In addition to centrosome number, cluster frequency, size, and localization in both nonresorbing and resorbing environments were analyzed in RAW- and BMDM-derived osteoclasts (Figure 3, A–E). To identify size-dependent differences, the heterogeneous multinucleated osteoclast population was subdivided into groups of small (3–5 nuclei), medium (6–10 nuclei), and large (11+ nuclei) osteoclasts (Figure 3A). Centrosome clusters were defined as having 3 or more adjacent CEP164 signals within 1 µm of each other (Figure 3B). Cluster frequency was similar across culturing surfaces, with small and medium osteoclasts typically possessing a single centrosome cluster, while more than one cluster was often visible in large osteoclasts (Figure 3C). Additionally, smaller clusters of 3 CEP164 signals were observed in small osteoclasts whereas medium and large osteoclasts often contained larger centrosome clusters consisting of 4+ CEP164 signals (Figure 3D). Cluster localization was scored using the spatial designations outlined in Figure 3E, panel 1. Perinuclear localization was assigned when CEP164 signal colocalized directly with nuclei. Peripheral localization was defined as a cluster located within 2 μm of the periphery of the cell or found within pseudopodlike structures. Cytoplasmic localization was centrosomes in between these 2 regions. Spatial distributions of clusters in each osteoclast subpopulation were similar across culture conditions (Figure 3E). Small osteoclasts predominantly contained cytoplasmic and perinuclear-localized centrosome clusters. Medium osteoclasts contained centrosome clusters in all three zones with perinuclear and cytoplasmic localizations being the most prevalent. Large osteoclasts possessed the most varied distribution of centrosome cluster localizations with perinuclear, cytoplasmic, and peripheral positions observed in nearly equal proportions (Figure 3E). Overall, centrosome persistence and clustering are features of both resorbing and nonresorbing osteoclasts, regardless of cell size.
FIGURE 3:
Centrosome clustering and localization in nonresorbing and resorbing RAW- and BMDM-derived osteoclasts varies with cell size. (A) Representative images of centrosome clusters in small (S; 3–5 nuclei/cell), medium (M; 6–10 nuclei/cell), and large (L; 11+ nuclei/cell) osteoclasts stained for DAPI (blue), tubulin (green), and CEP164 (orange). Dashed box insets show centrosome clusters. Scale bars = 10 μm. (B) Illustration of lone and cluster-associated centrosomes. A cluster is defined as at least three centrosomes with less than 1 μm distance from adjacent centrosomes. (C) Distribution of the number of centrosome clusters per osteoclast for RAW-derived osteoclasts cultured on glass or CaP and BMDM-derived osteoclasts on glass and CaP (n ≥ 60). (D) Distribution of the number of centrosomes within each cluster in RAW-derived osteoclasts plated on glass or CaP and BMDM-derived osteoclasts plated on glass and CaP (n ≥ 60). (E) Schematic of three cellular localization zones and the corresponding distributions of cluster locations in osteoclasts plated on glass or CaP.
Clustered centrosomes are potent MTOCs with each centrosome contributing to MT nucleation
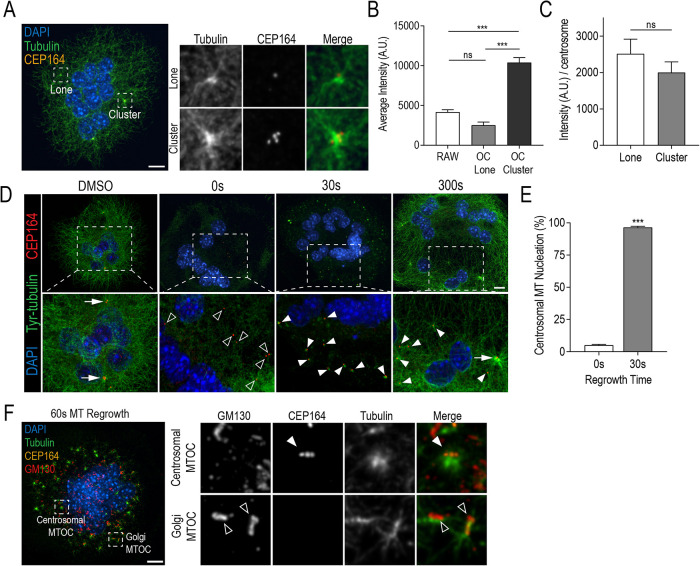
To determine if the unique clustering of centrosomes augmented the MT network in osteoclasts, we measured MT nucleation relative to centrosome cluster size. Specifically, we assessed whether larger clusters were capable of organizing more MTs. We approached this through the comparison of mean MT fluorescence signal between lone and clustered centrosomes (Figure 4, A and B). Background-corrected tubulin fluorescence was quantified within regions of interest (ROI) segmented from centrosomal signals from osteoclasts differentiated for 4 d and unstimulated RAWs. On average, centrosome clusters were associated with significantly more tubulin fluorescence than their lone counterparts in osteoclasts. No difference in MT signal intensity was observed between lone centrosomes in unstimulated RAW cells compared with lone centrosomes in osteoclasts (Figure 4B). Interestingly, when MT intensity values were normalized by cluster size, no significant difference was observed relative to lone centrosomes in osteoclasts (Figure 4C). This suggests that centrosomes within clusters do not individually nucleate more (or less) MTs.
FIGURE 4:
Clustered centrosomes have strong MTOC capacity with individual centrosomes retaining MT nucleation ability. (A) Representative RAW-derived osteoclasts stained with DAPI (blue), tubulin (green), and CEP164 (orange). (B) Quantification of total tubulin fluorescence at lone centrosomes and centrosome clusters in unstimulated RAWs and osteoclasts. Graph displays total mean intensity ± SEM from three independent experiments (n = 30). Significance was determined via a one-way ANOVA followed by Tukey’s multiple comparison (***P < 0.001). (C) Quantification of total tubulin fluorescence per centrosome for lone and clustered centrosomes in osteoclasts. Significance was determined by Student’s t test (*P < 0.05). (D) Representative images of osteoclasts after MT regrowth assays. Cells were fixed and stained for DAPI (blue), tyrosinated (Tyr) tubulin (green), and CEP164 (orange) at indicated recovery time points in order to visualize MT regrowth. Dashed box insets show detailed centrosomal and MT nucleation signals. White arrows indicate clustered CEP164 signals, open arrowheads indicate lone CEP164 signal, and closed arrowheads indicate centrosomal MT nucleation sites. (E) Quantification of % MT nucleation at 0 and 30 s of MT regrowth. Nucleating centrosomes were defined as CEP164 signal colocalized with Tyr-tubulin signal. Graph displays mean ± SEM from three independent samples (n = 30) where significance was determined by Student’s t test (***P < 0.001). (F) Representative osteoclast after 60 s of MT regrowth. Cells were stained with DAPI (blue), tubulin (green), GM130 (red), and CEP164 (orange). Dashed boxes show location of expanded panels for centrosomal and Golgi MTOCs. White arrowhead indicates a radial MT aster colocalizing with centrosomes and open arrowheads indicate MTs colocalizing with Golgi. Scale bars = 10 μm.
Centrosomes are the main MTOC in proliferating cells and are often inactivated in terminally differentiated cells (Muroyama and Lechler, 2017). To examine the individual centrosome functionality in osteoclasts, we performed MT regrowth assays (Nguyen et al., 1999). MT depolymerization was achieved by treating RAW-derived osteoclasts with 10 µM of nocodazole for 1 h followed by washout for MT regrowth. Osteoclasts were fixed at regular time intervals and stained for tyrosinated tubulin (Tyr-tubulin) and CEP164 to visualize newly polymerized MTs and centrosomes, respectively (Figure 4D). At 0 s in recovery medium, the MT network was completely depolymerized and lone centrosomes were observed. After 30 s, small MT asters were observed colocalizing with or directly adjacent to CEP164 signals, indicating MT nucleation. Recovery of the MT network was observed after 300 s of incubation in medium. The percentage of MT nucleation in medium-sized (6–10 nuclei) osteoclasts was quantified at 0 and 30 s of MT regrowth, with a nucleating centrosome defined as having a CEP164 signal colocalizing with or directly adjacent to a Tyr-tubulin signal (Figure 4E). Nearly all the centrosomes (96.4 ± 0.9%) in medium-sized osteoclasts were capable of MT nucleation after nocodazole-induced depolymerization. Immunostaining for Golgi with a GM-130 antibody also revealed asymmetric MT arrays colocalizing with GM130 in addition to radial centrosomal MT arrays (Figure 4F). These data confirmed the presence of these nc-MTOCs in osteoclasts previously identified by Mulari et al. (2003) and Vergarajauregui et al. (2020). Taken together, we find that individual centrosomes retain their MTOC ability and cluster to organize more MTs.
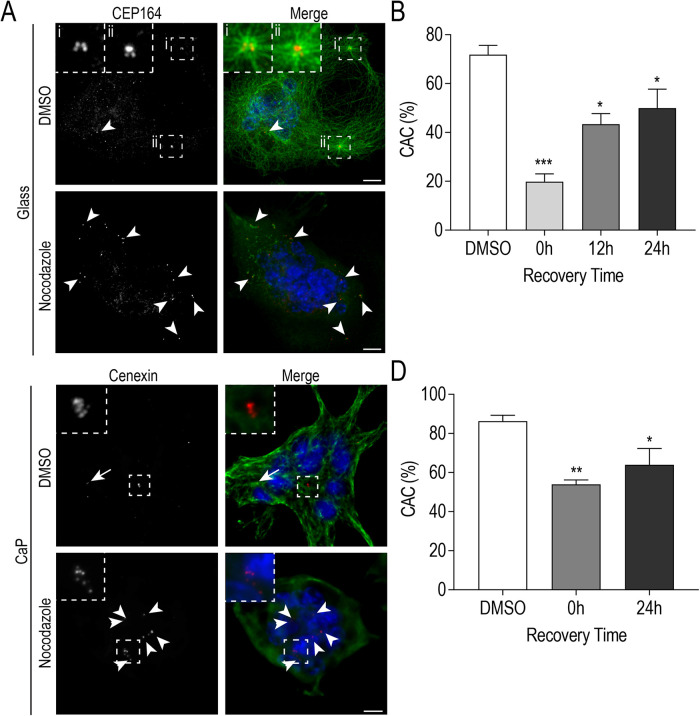
Interestingly, we frequently observed that nocodazole-induced MT depolymerization produced centrosome declustering that was not rescued after 5 min in recovery culture medium (Figure 5A). We expanded our time points postnocodazole treatment to examine whether normal levels of clustering could be recovered in medium-sized (6–10 nuclei) osteoclasts (Figure 5B). Utilizing the same definition of clustering as in Figure 3B, we determined that MT depolymerization significantly reduced the proportion of cluster-associated centrosomes, with osteoclasts immediately following nocodazole treatment (0 h) containing 19.8 ± 3.3% cluster-associated centrosomes compared with controls (71.9 ± 3.8%). Only partial reclustering values were observed after MT polymerization, with 43.4 ± 4.4% cluster-associated centrosomes at 12 h and 49.9 ± 7.8% cluster-associated centrosomes at 24 h (Figure 5B). The extended MT regrowth assay in Figure 5B was repeated with osteoclasts replated onto CaP substrate to determine whether centrosome clustering was similarly affected in polarized, activated osteoclasts. RAW-derived osteoclasts were transferred onto CaP-coated coverslips prior to a 1-h nocodazole treatment (Figure 5, C and D), followed by 24 h of recovery. Similarly, only partial centrosome reclustering (64.0 ± 8.3%) after 24 h of recovery was observed when compared with control conditions (86.3 ± 3.0%) (Figure 5D). This suggests that an established MT network is important in the development and maintenance of centrosome clusters in both nonresorbing and polarized osteoclasts.
FIGURE 5:
Centrosome clustering is maintained by an established MT network. (A) Representative images of fixed RAW-derived osteoclasts after nocodazole-induced MT depolymerization (5 min recovery) cultured on glass (C) Representative images of fixed RAW-derived osteoclasts after nocodazole-induced MT depolymerization (0 min recovery) replated on CaP substrate. Cells were stained with DAPI (blue), tubulin (green), and CEP164 or cenexin (red). White arrows indicate centrosome pairs and arrowheads indicate lone centrosomes. Centrosome clusters are shown in dashed box insets. Scale bars = 10 μm. (B and D) Quantification of centrosome clustering (%) for osteoclasts cultured on glass (B; n = 60) or CaP substrate (D; n = 34) at indicated time points post-MT regrowth from three independent experiments. Significance relative to DMSO was determined via a one-way ANOVA followed by Dunnett’s multiple comparison (*P < 0.05; **P < 0.01; ***P < 0.001).
MT dynamicity, KIFC1, and dynein contribute to centrosome clustering.
After establishing the prevalence of centrosome clustering within osteoclasts and the requirement for intact MTs for centrosome clustering, additional regulators of this process were explored. Centrosome clustering, as a result of unregulated centrosomal amplification, is associated with many human cancers (Fukasawa, 2005). In cancer cells, extra centrosomes are clustered during mitosis to avoid multipolar spindle formation and eventual cell death (Marthiens et al., 2012). Using cancer studies as a model, we next examined the role of the minus-end directed motors, dynein and KIFC1 (kinesin motor), and MT dynamicity as potential mediators of centrosome clustering in osteoclasts. The role of dynein has been implicated in cells with supernumerary centrosomes, where the displacement of dynein from the spindle resulted in centrosome declustering and subsequent multipolarity (Quintyne et al., 2005). KIFC1 is capable of cross-linking MTs (Mountain et al., 1999; Cai et al., 2009) and has been identified in RNAi screens as a requirement for centrosome clustering in cancer cells (Kwon et al., 2008). Finally, disruption of MT dynamic instability has recently been shown to decluster centrosomes in cancer cells (Rebacz et al., 2007; Pannu et al., 2014).
Centrosome declustering agents were administered for 24 h at different stages of osteoclast differentiation in order to test their effects on centrosome clustering (Figure 6 and Supplemental Figure S1). Dynarrestin (175 μM DYN) is a cell-permeable reversible and specific blocker of cytoplasmic dynein (Höing et al., 2018). CW069 (80 μM CW) is a specific small molecule inhibitor of KIFC1 and induces multipolar spindles in cancer cells (Watts et al., 2013; Sekino et al., 2019). Griseofulvin (40 μM GF) is an antimitotic agent that reduces MT dynamicity. GF prevents centrosome coalescence not only during mitosis but also in interphase (Rebacz et al., 2007; Pannu et al., 2014).
FIGURE 6:
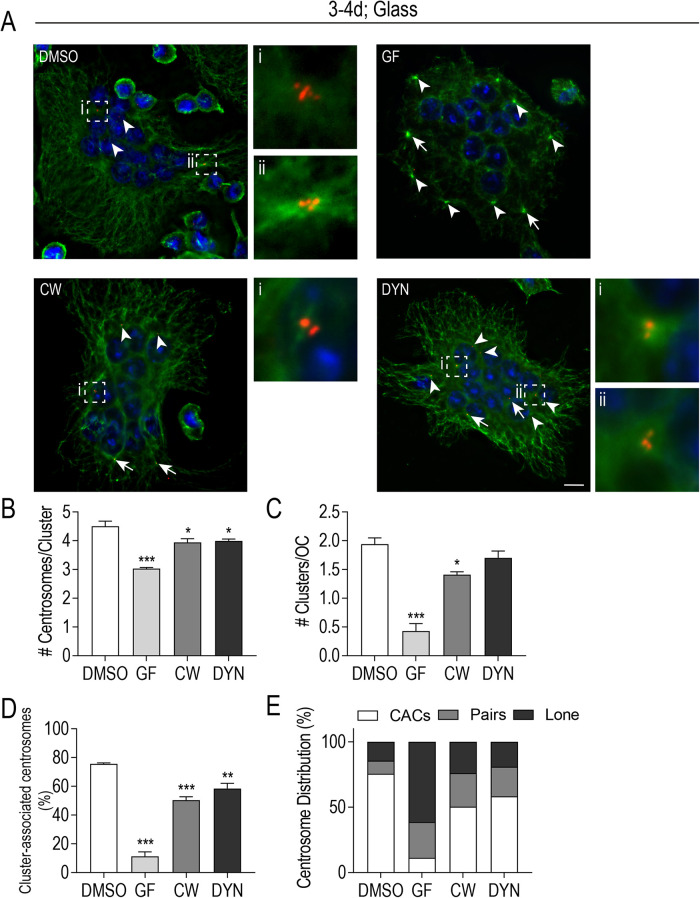
MT dynamicity, KIFC1, and dynein activity contribute to centrosome clustering in osteoclasts. (A) Representative images of fixed, glass-plated RAW-derived osteoclasts after treatment with centrosome declustering agents: 40 μM GF, 80 μM CW069 (CW), and 175 μM DYN. Cells were fixed 24 h posttreatment and stained with DAPI (blue), tubulin (green), and cenexin (red). White arrows indicate centrosome pairs and arrowheads indicate lone centrosomes. Centrosome clusters are shown in dashed box insets. Scale bars = 10 μm. (B and C) The average number of centrosomes per cluster (B) and the average number of clusters/cell (C) after treatment with declustering drugs was calculated from four independent experiments (n = 60). Significance relative to DMSO was determined through a one-way ANOVA followed by Dunnett’s multiple comparison (*P < 0.05; ***P < 0.001) (D) The % cluster-associated centrosomes after treatment with individual declustering drugs from four independent experiments. Significance relative to DMSO was determined through a one-way ANOVA followed by Dunnett’s multiple comparison (**P < 0.01; ***P < 0.001). Each graph displays mean ± SEM (B–D). (E) Centrosome distribution (%) in clusters, pairs, or individually after treatment with individual declustering drugs from four independent experiments.
First, early stage osteoclasts (days 3 and 4 of differentiation), when fusion activities predominate and centrosome clusters are being formed, were treated with centrosome declustering drugs (Figure 6). We observed a decrease in centrosome clustering in all three treatment groups in medium-large-sized osteoclasts (7–20 nuclei) (Figure 6A). Under control conditions, osteoclasts with two large clusters/cell (1.9 ± 0.1) of at least 4 centrosomes/cluster (4.5 ± 0.2) were frequently observed (Figure 6, B and C). In CW and DYN- treated cells, it was more common to see a single, smaller centrosome cluster (CW 1.4 ± 0.1; DYN 1.7 ± 0.1) of at most 4 centrosomes/cluster (CW 3.4 ± 0.1; DYN 4.0 ± 0.1). The GF-treated osteoclasts rarely contained centrosome clusters (0.4 ± 0.1) and individual centrosomes were often localized to the periphery of osteoclasts (Figure 6, A–C). Further analysis revealed significant differences in the percentage of cluster-associated centrosomes, with control conditions having 75.6 ± 0.6% cluster-associated centrosomes compared with the decreases observed in GF (11.3 ± 3.1%), CW (50.4 ± 2.3%), and DYN (58.4 ± 3.7%) conditions (Figure 6D). As a complement to the percentage of cluster-associated centrosome data, the relative proportions of centrosomes located in clusters, pairs, or as individuals are represented in Figure 6E. Of note, nearly 61% of centrosomes were individually distributed in GF-treated osteoclasts, whereas CW and DYN exposure resulted in approximately equal distributions (20% each) of lone or paired centrosomes in treated osteoclasts (Figure 6E). In addition, CaP-transferred mature osteoclasts (days 4 and 5 of differentiation), which had established cluster-associated centrosomes, were challenged with centrosome declustering drugs (Supplemental Figure S1). In contrast with osteoclasts differentiated on glass (Figure 6, B and C), larger centrosome clusters (8–11 centrosomes/cluster) in control osteoclasts and (5 and 6 centrosomes/cluster) in CW- and DYN-treated osteoclasts (Supplemental Figure S1) were observed. Overall, trends in the percentage of cluster-associated centrosomes remained similar when osteoclasts were replated onto CaP, which indicate MT dynamicity, KIFC1, and dynein are important for centrosome clustering in both differentiating and polarized osteoclasts.
Centrosome declustering drugs impact both F-actin ring formation and bone resorption by osteoclasts
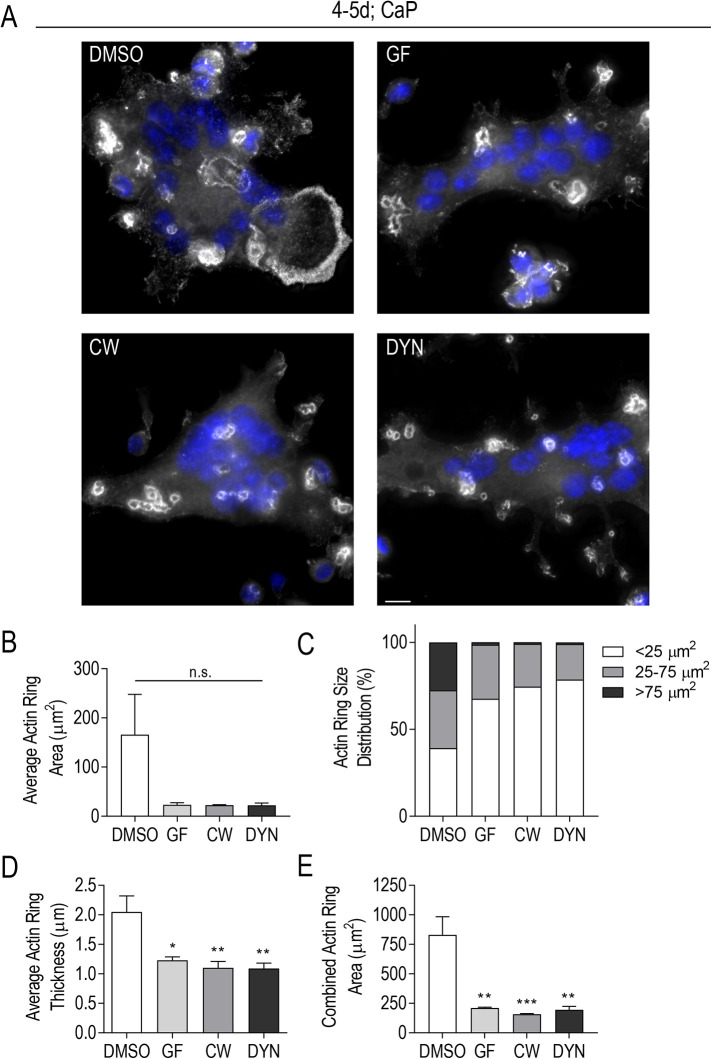
We speculated that centrosome persistence and clustering provided a robust, well-orchestrated MT array for osteoclast activation and bone degradation (Bar-Shavit, 2007; Yavropoulou and Yovos, 2008). Osteoclast polarization and activation is achieved through the formation of a sealing zone at the substrate interface (Batsir et al., 2017). The sealing zone is characterized by a dense F-actin ring adhesion structure that isolates the resorption area from the external environment. MTs enriched along the sealing zone region in osteoclasts are implicated in regulating sealing zone stability (Jurdic et al., 2006; Batsir et al., 2017). To determine whether centrosome clustering played a role in osteoclast activation, we examined characteristics of F-actin ring size and thickness (Figure 7). A continuous actin ring was defined as an F-actin ring with no visible break in fluorescence signal. We continued to utilize the inhibitors from Figure 6 and Supplemental Figure S1, as each was shown to significantly impact centrosome clustering in RAW-derived osteoclasts. A large range of F-actin ring sizes was observed in all conditions, including control osteoclasts (Figure 7, A–C). Control cells had the greatest variation, with 25% of all F-actin rings > 75 μm2. In contrast, the GF-treated osteoclasts had 68% of all F-actin rings < 25 μm2, while both CW- and DYN-treated osteoclast populations had at least 75% of their F-actin rings represented in the small group (<25 μm2) (Figure 7C). In addition, we examined the F-actin ring thickness by taking three randomly selected width measurements of three different rings/osteoclast (Figure 7D). A significant difference in F-actin ring thickness was observed in each treatment condition relative to control. The average F-actin ring thickness for control osteoclasts was 2.1 ± 0.3 μm compared with <1.5 μm for GF-, CW-, and DYN-treated osteoclasts (Figure 7D). Finally, we considered the combined total actin ring area as an indirect measure of the resorptive potential of each osteoclast (Figure 7E). When F-actin ring areas were combined per osteoclast, a dramatic and significant difference was observed. DMSO-treated osteoclasts had an average total F-actin ring area of 830.1 ± 153.7 μm2 compared with 210.9 ± 6.0 μm2, 157.0 ± 5.9 μm2, and 195.34 ± 28.3 μm2 for GF-, CW-, and DYN-treated osteoclasts, respectively. Taken together, centrosome clustering impacts F-actin ring formation in osteoclasts.
FIGURE 7:
Centrosome declustering drugs impact F-actin ring size and thickness. RAW-derived osteoclasts were lifted and replated on CaP prior to 24-h treatment with centrosome declustering agents: 40 μM GF, 80 μM CW069 (CW), and 175 μM DYN. (A) Representative immunofluorescent images of CaP-plated osteoclasts were stained for actin (gray) and DAPI (blue). Scale bars = 10 μm. (B, D, E) Average Actin Ring Area (μm2) (B), Average Actin Ring Thickness (μm) (D), and Combined Actin Ring Area (μm2) (E) was determined from three independent experiments (n = 40). Significance relative to DMSO was determined through a one-way ANOVA followed by Dunnett’s multiple comparison (*P < 0.05; **P < 0.01; ***P < 0.001). Each graph displays mean ± SEM. (C) Actin Ring Size Distribution (%) is presented as small (<25 μm2), midsize (25-75 μm2), and large rings (>75 μm2) from three independent experiments.
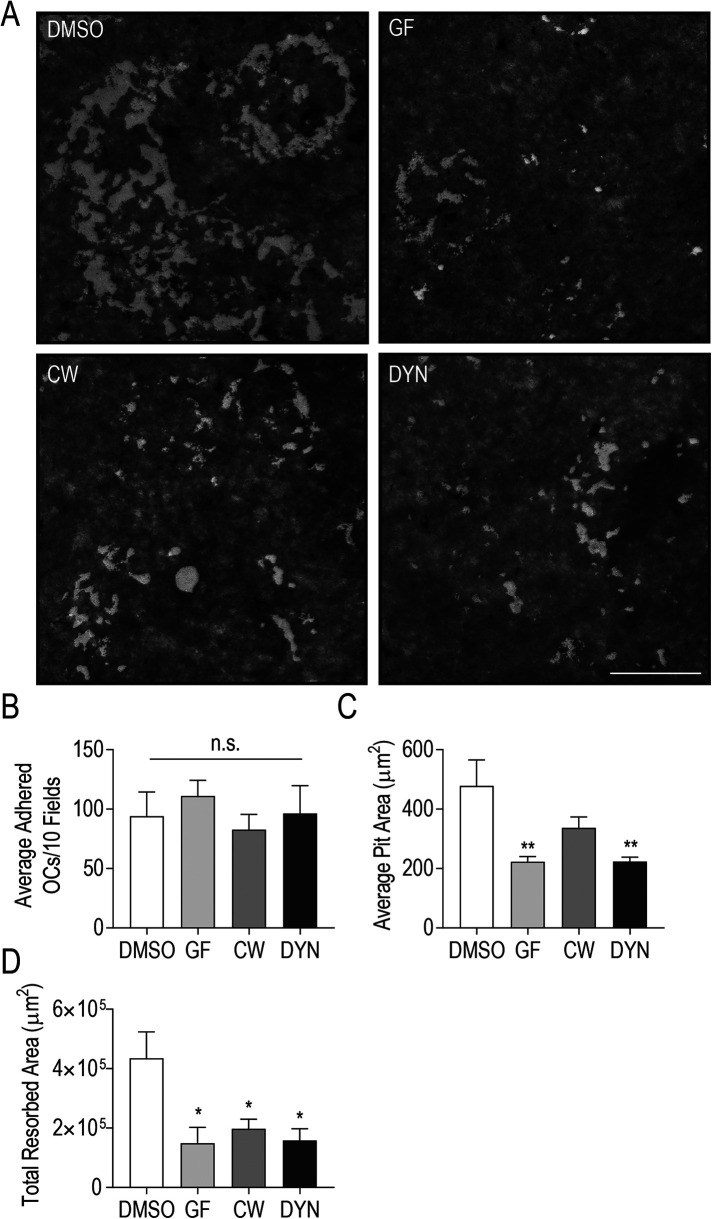
The striking differences in osteoclast resorptive potential determined in Figure 7E prompted us to measure bone resorption more directly. To examine osteoclast resorptive capacity, we plated osteoclasts on bone mimetic surfaces and measured resorption pits in control cells and those treated with centrosome declustering agents (Figure 8). Multinucleated osteoclasts were transferred onto Corning Osteo Assay multiwell plates and were allowed to degrade the bone mimetic substrate for 24 h in the presence of centrosome declustering drugs. We observed large regions with resorption pits in control wells, but only small resorption pits were generated by GF- and DYN-treated osteoclasts (Figure 8A). Occasionally, some larger pits were observed in bone surfaces exposed to CW-treated osteoclasts (Figure 8A). Prior to quantification of population-level resorption areas, we confirmed that total osteoclast adherence to the substrate was not significantly impacted by lifting and replating procedures or by the addition of declustering drugs (Figure 8B). In agreement with Figure 8A, the average pit area generated by control osteoclasts was 478.3 ± 87.1 μm2, compared with resorption pit areas of 222.9 ± 17.7 μm2, 336.9 ± 37.1 μm2, and 223.7 ± 14.5 μm2 for wells containing GF-, CW-, and DYN-treated osteoclasts (Figure 8C). Significant differences in average resorption pit areas were observed in GF- and DYN-treated osteoclast populations relative to control. However, when the total resorbed area was compared across treatments, a significant difference was observed in each drug-treated osteoclast population, relative to control cells (Figure 8D). Indeed, the total resorbed area by control osteoclasts was 2.9-fold greater than by osteoclasts treated with GF and 2.2-fold and 2.7-fold greater for CW- and DYN-treated osteoclasts, respectively. Centrosome clustering in osteoclasts is strongly correlated with bone degradation by osteoclasts.
FIGURE 8:
Centrosome declustering drugs markedly reduce osteoclast bone resorption. RAW-derived osteoclasts differentiated on Corning Osteo Assay plates prior to 24-h treatment with centrosome declustering agents: 40 μM GF, 80 μM CW069 (CW), and 175 μM DYN. (A) Representative images of bone mimetic stained with 2.5% AgNO3. Resorption pits are indicated by an absence of stain. Scale bar = 10 μm. (B) Osteoclast adherence was assessed as the total number of adhered osteoclasts in 16 randomly chosen fields in four independent experiments. (C and D) Average Pit Area (μm2) (C) and Total Resorbed Area (μm2) (D) was determined from 16 fields of view and four independent experiments. Significance relative to DMSO was determined through a one-way ANOVA followed by Dunnett’s multiple comparison (*P < 0.05; **P < 0.01).
DISCUSSION
In this study, we describe the presence and role of clustered centrosomes in terminally differentiated murine and human osteoclasts. Our findings show the persistence of centrosomes on osteoclast differentiation and confirm that centrosomes retain their ability to nucleate MTs. Once donated to the osteoclast cell body on cellular fusion, we show that centrosomes adopt a clustered conformation that is important for actin ring formation and subsequent bone resorption.
Early research suggested that precursor cell centrosomal MTOCs were lost during osteoclastogenesis (Mulari et al., 2003). To conclusively identify functional centrosomes in osteoclasts, we probed for structural proteins including CEP164 as well as γ-tubulin to clearly denote the location and number of centrosomes in osteoclasts. We observed a near one-to-one ratio of centrosomes with osteoclast nuclei, indicating that centrosomes donated by cell fusion were retained within multinucleated osteoclasts. Interestingly, osteoclast centrosomes often aggregated together, forming clusters that were consistently linked to large radial MT arrays and high-resolution imaging enabled the discernment of individual centrioles within these clusters. Using MT depolymerization and regrowth assays, we further demonstrated that centrosomes in osteoclasts individually retained their ability to nucleate MTs. Centrosome aggregates were previously observed and acknowledged as “centrospheres,” a former term used to describe large centrosomal signals within osteoclasts (Lucht, 1973; Lewis and Albrecht-Buehler, 1987; Turksen et al., 1988; Mulari et al., 2003). With the limited probing of centrosomal components as well as imaging resolution constraints, centrospheres were often considered as a single centrosomal MTOC within these studies. This misnomer could explain discrepancies in the literature where the quantification of centrospheres underestimated contributing centrosomes in osteoclasts.
Terminal cell differentiation is typically associated with major cytoskeletal restructuring to promote specialized cell shapes and functions. This often occurs in concert with the attenuation of the centrosome’s MTOC ability by PCM relocalization to other cell regions via site-specific adaptor proteins to form nc-MTOCs (Yang and Feldman, 2015; Zebrowski et al., 2015; Muroyama and Lechler, 2017; Magescas et al., 2019). At first glance, multinucleated myotubes are the most directly comparable differentiated cell type to osteoclasts as they also undergo multiple fusion events and share differentiation mechanisms before specialization (Bugnard et al., 2005; Rochlin et al., 2010). To achieve specialization, myotubes transfer MT nucleating and organization responsibilities from the centrosome to the nuclear envelope and Golgi (Bugnard et al., 2005). In both osteoclasts and myotubes, the Golgi structure adopts a circumnuclear positioning with a coordinated redistribution of pericentrin, γ-tubulin, and ninein to the nuclear membrane and Golgi via AKAP6 and AKAP9 (Mulari et al., 2003; Bugnard et al., 2005; Vergarajauregui et al., 2020). Using MT regrowth experiments, we also identified the Golgi as a site of MT nucleation, and shared contributions from centrosomal and nc-MTOCs likely enable the dynamic, polarized function of multinucleated osteoclasts. Both subsets of MTOCs may also contribute to bone resorption as we do not see complete abolishment of resorptive activity when anticlustering drugs are added to osteoclasts. An important distinction between myotubes and osteoclasts is that myotubes are nonmotile cells and require parallel MT arrays for the maintenance of nuclear positioning (Bugnard et al., 2005). In contrast, osteoclasts are highly motile and are capable of moving across the bone surface as different regions are resorbed (Hu et al., 2011; Søe and Delaissé, 2017). We suspect that the unique MT network adopted by osteoclasts is in large part due to differences in the apicobasal polarization and migratory requirements of osteoclasts, which may necessitate both centrosomal and ncMT arrays.
The clustering of centrosomes in osteoclasts is a unique phenotype only described thus far in cancer cells (Pannu et al., 2014). The presence of supernumerary centrosomes in cancer cells is not normally tolerated during cell division, leading to mitotic arrest and cell death. However, to facilitate their survival, cancer cells cluster their extra centrosomes toward the spindle poles during mitosis, thereby allowing for viable cell division (Ganem et al., 2009; Milunovic´-Jevtic´ et al., 2016). To date, many positive and negative regulators of centrosome clustering in cancer cells have been reported (Leber et al., 2010; Sabat-Pospiech et al., 2019). These molecular regulators span kinetochore, chromosome, centrosome, motor, spindle, and cortical proteins that all affect efficient centrosome clustering in cancer cells. Many of these regulators are specific to cycling cells. However, MT motors were immediately attractive to us given how quickly centrosomes clustered in osteoclasts during live-cell imaging. We found that inhibition of either KIFC1 or cytoplasmic dynein attenuated centrosome clustering in osteoclasts. KIFC1 is a unique MT minus-end-directed motor within the kinesin-14 family and is able to bind, cross-link, and slide MTs (Mountain et al., 1999; Cai et al., 2009). KIFC1 was identified to be critical in the coalescence of supernumerary centrosomes in cancer cells as knockdown or inhibition of KIFC1 resulted in the formation of multipolar spindles (Kwon et al., 2008; Watts et al., 2013; Wu et al., 2013; Xiao et al., 2016). Cytoplasmic dynein in cancer centrosome clustering is typically cortex-anchored to the spindle poles as well as associated with the spindle and facilitates centrosome clustering toward the poles by pulling on astral MTs (Dujardin and Vallee, 2002; Quintyne et al., 2005; Sommi et al., 2011).
Given the role of these motors in centrosome clustering in osteoclasts, it followed that intact MTs were required as a platform for centrosomal aggregation. Likewise, MT depolymerization with colchicine had a centrosome declustering effect in triple negative breast cancer cells (Thomopoulou et al., 2016). While these molecular motors in cancer cells exploit dynamic astral MTs in the mitotic spindle and the spindle itself, it is still unclear where and how MT motors facilitate centrosome clustering in postmitotic osteoclasts. What is clear is that MT dynamics drive these events, as inhibition of MT dynamics using GF resulted in significant dispersal of centrosomes in osteoclasts. Dynamic instability is also a key requirement of cancer centrosome clustering as treatment of cancer cells with drugs that mildly attenuate MT dynamics results in multipolar spindles (Rebacz et al., 2007; Karna et al., 2011). Furthermore, MT dynamic instability was shown for interphase-specific maintenance of centrosome clusters in cancer cells (Pannu et al., 2014). How MT dynamics facilitate centrosome clustering in cancer cells remains unknown. GF and MT motor inhibitors are being explored as promising chemotherapeutics but off-target effects particularly in bone could be given special consideration in light of our findings in osteoclasts.
Why might a terminally differentiated osteoclast cluster its centrosomes? We speculate that the clustering of centrosomes in osteoclasts may be necessary for the formation of concerted central hubs in which centrosomes functionally aggregate their PCM and MT nucleation sites to act as a supercentrosomal MTOCs. Indeed, clustered MTOCs organized more MTs compared with lone centrosomes and these clusters were readily observed across osteoclasts of various sizes. The increased prevalence of peripheral centrosome clusters in large osteoclasts may allow pooled MT nucleation to facilitate trafficking within the exceptionally large cytoplasm of these cells.
Centrosome declustering drugs had a marked effect on F-actin ring formation and bone resorption in osteoclasts. F-actin rings form the sealing zone in resorbing osteoclasts, which is associated with a large radial array of MTs that are important for the maturation and maintenance of these specialized adhesion structures (Mulari et al., 2003; Matsumoto et al., 2013; Biosse Duplan et al., 2014; Ti et al., 2015; Batsir et al., 2017). Treatment of osteoclasts with centrosome declustering drugs resulted in smaller actin rings which may not have had sufficient MT penetration and MT-mediated delivery to elaborate the massive circular sealing zone structures. We cannot rule out that disrupting retrograde motors and MT dynamicity may have additional effects on osteoclast function beyond centrosome declustering (Biosse Duplan et al., 2014); however, the strong attenuation of actin rings and bone resorption in the presence of all the inhibitors favors a necessity for centrosome clustering in osteoclast activation. Most compelling is the role of KIFC1, which has not otherwise been implicated in osteoclast sealing zone formation and function. KIFC1 is a compelling candidate, however, as its levels are known to increase in osteoclasts in an inflammatory disease mouse model (Chen et al., 2020). Future work will investigate the role of other candidates, including adhesion proteins, in centrosome clustering in osteoclasts. Our lab has shown that E-cadherin levels change during osteoclastogenesis (Fiorino and Harrison, 2016b) and a landmark paper revealed that E-cadherin knockdown increases cellular contractility, leading to more centrosome movement and clustering in cancer cells (Rhys et al., 2018).
Together, this work demonstrates that centrosomes coalesce in multinucleated cells and play an active role in terminally differentiated osteoclasts. It will be of interest to identify other molecular regulators of this process in osteoclasts and determine how clustered centrosomes coordinate their MT nucleating activities with noncentrosomal MTOCs like the Golgi complex.
MATERIALS AND METHODS
Reagents and antibodies
DMEM, fetal bovine serum (FBS), phosphate-buffered saline (PBS), and 0.25% Trypsin-EDTA were purchased from Wisent Inc. (St-Bruno, QC). Alpha Modified Eagle Medium (AMEM) and RPMI 1640 GlutaMAX was purchased from ThermoFisher Scientific (Waltham, MA). TrypLE Select Enzyme was purchased from ThermoFisher Scientific (Waltham, MA). Recombinant murine and human macrophage-colony stimulating factor (M-CSF) were purchased from Peprotech (Rocky Hill, NJ) while recombinant receptor activator of nuclear factor kappa-B ligand (RANKL) was produced from BL21 Escherichia coli transformed with a pGEX-GST-hRANKL vector (a gift from Morris Manolson, Faculty of Dentistry, University of Toronto). Mouse monoclonal anti-γ-tubulin, anticentrin (clone 20H5) and anti-Tyr-tubulin antibodies were purchased from MilliporeSigma Canada Co. (Oakville, ON). Rat monoclonal antitubulin and rabbit polyclonal antipericentrin antibodies were purchased from Abcam Plc. (Cambridge, UK). Rabbit polyclonal anti-CEP164 antibody and anti-ODF2 (cenexin) were purchased from Proteintech Group (Rosemont, IL). Mouse monoclonal anti-GM130 antibody was purchased from BD Biosciences Canada (Mississauga, ON). Mouse monoclonal pan-actin antibody (clone 7A8.2.1) was purchased from Cytoskeleten (Denver, CO). AffiniPure donkey polyclonal antirabbit, donkey antimouse, and donkey antirat cyanine Cy2, Cy3, Cy5 secondary antibodies were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA); 4′,6-diamidine-2′-phenylindole dihydrochloride (DAPI) was purchased from MilliporeSigma Canada Co. (Oakville, ON). Alexa Fluor 488 and 647 phalloidin was purchased from ThermoFisher Scientific (Waltham, MA). DAKO Fluorescence mounting media was purchased from Agilent Technologies Canada (Mississauga, ON). Kinesin KIFC1 inhibitor CW069 was purchased from Selleck Chemicals LLC (Houston, TX). Antimitotic agents Nocodazole and GF were purchased from MilliporeSigma Canada Co. (Oakville, ON). DYN inhibitor was purchased from Tocris Bioscience (Toronto, ON).
Cell culture and osteoclast formation
The RAW 264.7 (RAW) murine macrophage cell line was purchased from the American Type Culture Collection (ATCC; Manassas, VA). RAW cells were cultured and maintained in complete DMEM containing 10% heat-inactivated FBS at 37°C, 5% CO2, and 90% relative humidity in T75 flasks. To generate osteoclasts, confluent RAW cells were subcultured at a density of 10,000 cells/cm2 into 12-well or 24-well tissue culture plates in AMEM supplemented with 10% FBS and 25–100 ng/ml RANKL. The medium was replaced every 2 d with cell fusion typically occurring within 2 d from initial seeding and large multinucleated osteoclasts forming on days 3 and 4.
BMDM isolation and differentiation
Primary BMDMs were obtained by bone marrow extraction from the femurs and tibias of 6- to 12-wk-old C57BL/6 mice as previously described (Boraschi-Diaz and Komarova, 2016). Briefly, mice were killed after which their femurs and tibias were dissected. After clearing the bones of tissue, the bones were rinsed with PBS, snipped at both ends, and placed into Eppendorf tubes. The bones were spun at 13,000 rpm, 3× for 30 s to isolate the bone marrow. The bone marrow was collected and treated with red blood cell lysis buffer (MilliporeSigma Canada Co., Oakville, ON) before being plated in a flask overnight in AMEM containing 10% heat-inactivated FBS, 1% penicillin/streptomycin, and 25 ng/ml M-CSF. Following a 24-h incubation, the nonadherent precursor cells were collected, counted, and seeded into 12-well plates or 10-cm dishes at 150,000 cells/cm2 in AMEM containing 10% FBS, 50 ng/ml M-CSF, and 100 ng/ml RANKL. The medium was replaced every 2 d, with mature osteoclasts observed on days 4 and 5.
hPBMC differentiation
Isolated human PBMC cells were purchased from the American Type Culture Collection (ATCC; Manassas, VA). The cells were seeded onto 24-well plates at 1.7 × 106 cells/well in RPMI 1640 GlutaMAX containing 10% heat-inactivated FBS, 1% penicillin/streptomycin, and 20 ng/ml M-CSF. Following a 24-h incubation, the nonadherent cells were removed and osteoclast differentiation was initiated by adding RPMI along with 20 ng/ml M-CSF and 60 ng/ml RANKL. The medium was replaced every 3 d, with mature osteoclasts observed on days 10–27, depending on donor age.
Biomimetic CaP substrate production and osteoclast culture
The procedure for precipitating and coating surfaces was carried out as previously described (Patntirapong et al., 2009; Maria et al., 2014). Briefly, three stock solutions were initially made which included a Tris buffer solution, calcium stock solution (CSS), and a phosphate stock solution (PSS). The Tris buffer solution contained 50 mM Tris base, pH = 7.4. The CSS contained 25 mM CaCl2 · 2H2O, 1.37 M NaCl, 15 mM MgCl2 · 6H2O in Tris buffer, pH 7.4. The PSS contained 11.1 mM Na2HPO4 · H2O, 42 mM NaHCO3 in Tris buffer, pH 7.4. The three solutions were then combined in a 2:1:1 ratio of Tris buffer:CSS:PSS in order to prepare a 2.5× concentrated simulated body fluid (SBF) solution. A final supersaturated CaP solution (CPS) was made by adding 41 ml of 1 M HCl to 800 ml of MilliQ water prior to dissolving 2.25 mM Na2HPO4 · H2O, 4 mM CaCl2 · 2H2O, 0.14 M NaCl, and 50 mM Tris, then bringing the pH to 7.4 and the volume to 1 l. Solutions were sterilized by filtration with a 0.22-μm filter. For the coating process, glass coverslips were incubated with freshly mixed 2.5× SBF (1 ml in each well/12-well plate) for 24 h at room temperature for 3 d. The precipitated amorphous CaP acted as the nucleation layer for the CPS solution to produce CaP crystal. After 3 d, the final CPS solution was added for 1 d at room temperature. CaP-coated coverslips were washed with 70% ethanol to sterilize the surface, washed twice with distilled water, and allowed to dry overnight.
The procedure for lifting and replating RAW-derived osteoclasts onto CaP or Corning Osteo Assay 16-well plates (Tewksbury, MA) was adapted from Maria et al. (2014). RAW-derived osteoclasts were first generated in 10-cm dishes for 4 d. Once large multinucleated osteoclasts were observed, the culture media was aspirated and cells were washed twice in PBS. To begin, 1 ml of TrypLE Select was added and incubated for 3–5 min at 37°C to lift and remove mononuclear cells. To lift osteoclasts, another 1 ml of TrypLE Select was added for 4–5 min at 37°C. Osteoclasts were dislodged using tapping and TrypLE Select was inactivated by adding 4 ml of AMEM. Resuspended osteoclasts were spun at 100 × g for 5 min before careful resuspension in 7–10 ml of AMEM and 25 ng/ml RANKL. An identical volume of suspended osteoclasts was added to each CaP-coated coverslip and osteoclasts were allowed to adhere for a minimum of 2.5 h before any further treatment. CaP-coated coverslips were preincubated with media and declustering drugs for 24 h prior to osteoclast plating.
Live-cell imaging
For live-cell imaging, stably transduced RAW EGFP-centrin1 cells were seeded 2 d prior to imaging into 35-mm glass-bottom single well dishes (MatTek, Ashland, MA) at 10,000 cells/cm2 in supplemented AMEM containing 100 ng/ml RANKL. After 2 d, the cell population was largely mononucleate. To capture early fusion events, cells were refed with 100 ng/ml RANKL prior to imaging. Live-cell imaging was conducted using a Zeiss AxioObserver Z1 inverted epifluorescence microscope with a 40× 1.4 NA oil immersion lens (Carl Zeiss Canada, Toronto, ON) using AxioVision 4.8 software for image capture. Time-lapse imaging was acquired over a 24-h period at 5-min intervals after replacing the medium containing RANKL. Cells were kept at 37°C and 5% CO2 throughout imaging.
Immunofluorescence
Osteoclasts generated on coverslips were initially washed with PBS three times prior to fixation with 4% paraformaldehyde (PFA) in PBS for 20 min at room temperature. After fixation and subsequent washes in PBS, cells were permeabilized with 0.25% Triton X-100 for 20 min. Blocking was done with a 5% fetal calf serum (FCS) (GE Healthcare Bio-Sciences, Pittsburg, PA) in PBS solution for 1 h, followed by labeling with primary antibodies in a 1% FCS in PBS blocking solution for 1 h. Alternatively, some osteoclasts were fixed with 100% methanol at –20°C for 10 min. Primary antibodies included mouse monoclonal anti-γ-tubulin (1:500), mouse monoclonal anti-Tyr-tubulin (1:500), rat monoclonal antitubulin (1:1000), rabbit polyclonal anti-CEP164 (1:500), rabbit polyclonal antipericentrin (1:500), and mouse monoclonal anti-GM130 (1:200). Following labeling with primary antibodies, cells were washed three times with PBS and labeled with secondary antibodies for 1 h. All Cy2–, Cy3–, and Cy5–conjugated secondary antibodies were used at a 1:500 dilution in 1% FCS in PBS. Nuclei were stained with DAPI for 10 min prior to mounting.
Alterations to the above method were performed for methanol-fixed cells: Cells were blocked in 3% bovine serum albumin (BSA) and 1% FCS buffer O/N. Primary rabbit polyclonal anti-ODF2 (1:900) and secondary antibody dilutions were prepared in 3% BSA, 1% FCS buffer. Fixed images of osteoclasts were obtained using a Quorum WaveFX-X1 spinning disk confocal microscope (Quorum Technologies Inc., Guelph, ON) configured with MetaMorph image acquisition software (Molecular Devices LLC, Sunnyvale, CA). Optical slices were taken every 0.2 μm (z-axis) to reconstruct whole osteoclasts. SIM was carried out on a Zeiss Elyra PS1 (Carl Zeiss Canada, Toronto, ON) configured with Zeiss Zen 2012 software for image capture and SIM processing. Additional images were taken with a Zeiss AxioObserver Z1 inverted epifluorescence microscope with a 63× 1.4 NA oil immersion lens (Carl Zeiss Canada, Toronto, ON) using Zen 3.1 (blue edition) software for image capture. Optical slices of 0.4 μm (z-axis) were taken to observe the entire osteoclast.
TEM
Mature RAW-derived osteoclasts were fixed in 2% glutaraldehyde in a 0.1 M sodium cacodylate buffer at pH 7.2 for 2 h at room temperature. Osteoclasts were postfixed in 1% osmium tetroxide and 1.25% potassium ferrocyanide in sodium cacodylate buffer at room temperature for 45 min and stained for 30 min with 4% uranyl acetate in water. Dehydration was carried out by incubating in a graded series of ethanol ranging from 70 to 100%. To embed the cells, samples were infiltrated with a 1:1 mixture of SPI-812 epoxy (SPI Supplies, West Chester, PA) and ethanol for 45 min, followed by two incubations with 100% epoxy for 120 min and overnight subsequently. After hardening of the resin, ultrathin sections (80 nm) were cut and collected onto copper grids prior to being poststained with uranyl acetate and lead citrate. Sections were imaged using a Hitachi H7500 transmission electron microscope with images being captured with an Olympus SIS Megaview II digital camera.
MT regrowth assay
MT regrowth assays in osteoclasts first involved depolymerizing the MT network. End-point osteoclasts would be treated with culture media containing 10 μM nocodazole for 1 h at 37°C. To ensure precise regrowth with no premature MT nucleation, osteoclasts were placed on ice for a further 10 min. After depolymerization, cells were washed 5–7 times with ice-cold PBS in order to remove the nocodazole. Osteoclasts were then allowed to re-equilibrate in prewarmed culture medium to induce MT nucleation and repolymerization. Osteoclasts were fixed in 4% PFA for 20 min at room temperature or in methanol at –20°C for 10 min at multiple time points of MT regrowth to visualize the MTOCs using immunofluorescence.
Centrosome declustering assays
Centrosome declustering assays in osteoclasts were performed with the use of minus-end motor inhibitors, DYN and CW069, in addition to the antimitotic agent GF. RAW cells were initially seeded as described above on glass coverslips. After 3 d of differentiation, culture medium and RANKL was replenished. Cells were treated with declustering agents 4 h postrefeed in order to test their effects early in the fusion process. After 24 h of treatment, cells were fixed in methanol at –20°C for 10 min prior to immunostaining. An average osteoclast size of 13 nuclei/cell was maintained across treatment conditions for quantification purposes.
Additional declustering assays were performed on CaP-coated surfaces with some modifications. Mature osteoclasts were initially formed in 10-cm dishes as described above. Lifted osteoclasts were replated on CaP coverslips and were allowed to adhere to the substrate for 2.5 h prior to drug addition. After 24 h of treatment, cells were fixed in methanol at –20°C for 10 min prior to immunostaining for centrosomes, MTs, or actin ring structures. An average osteoclast size of 13 nuclei/cell was maintained across treatment conditions for quantification purposes. For resorption pit analysis, cells were removed with 1 M sodium chloride in 0.2% Triton X-100 for 5 min. The CaP coverslips were treated with 2.5% AgNO3 for at least 30 min before imaging with a Zeiss AxioObserver Z1 inverted microscope with a 20× air lens.
Image processing
Images acquired via epifluorescence microscopy were deconvolved using Fiji version 2.3.0 (National Institutes of Health, Bethesda, MD) and DeconvolutionLab2 and Extended Depth of Field plugins. Resorption pit areas were determined using the Analyze Particles standard feature in Fiji. Linear adjustments to contrast and brightness, iterative deconvolution (90% confidence interval), and generation of image projections acquired by spinning disk confocal microscopy were performed with Volocity version 6.5.1 (Quorum Technologies Inc., Puslinch, ON). Images acquired by TEM were acquired, processed, and exported using iTEM version 5.2 (Olympus Soft Imaging Solutions GmbH, Muenster, DEU). To measure fluorescence intensity, a routine was created in CellProfiler 4.2.1 (Broad Institute, Cambridge, MA) where segmentation was initially carried out based on the colocalized intensity of CEP164 and γ-tubulin channels of fluorescent osteoclast images. The segmentations were then converted into ROIs to measure total tubulin intensity and subsequently subtracted from the background.
Statistical analyses
Linear regression analyses were used to determine the linearity of datasets. An unpaired two-tailed Student’s t test was used to determine statistical significance between two groups (P < 0.05 was considered statistically significant). When comparing three groups or more, a nonparametric one-way ANOVA was used, followed by Tukey’s test for multiple comparisons. When comparing three groups or more to a control group, a nonparametric one-way ANOVA followed by Dunnett’s test for multiple comparisons was used. The mean ± SEM of each group was displayed in all cases. Prism (version 9.3.1) was used to conduct the statistical analyses (GraphPad Software Inc., La Jolla, CA).
Supplementary Material
Acknowledgments
We thank Vaishali Sridhar and Laurence Pelletier for the EGFP-centrin1 construct and viral transduction that enabled the tracking of centrosome clustering live in osteoclasts. This project was funded by a Natural Sciences and Engineering Research Council grant (RGPIN 2017-06087) and a grant from the Canadian Space Agency (15FASTB03) to R.E.H.
Abbreviations used:
- BMDM
bone marrow-derived macrophage
- CaP
calcium phosphate
- CPS
calcium phosphate solution
- CSS
calcium stock solution
- CW
CW069
- DAPI
4′,6-diamidine-2′-phenylindole dihydrochloride
- DMSO
dimethyl sulfoxide
- GF
griseofulvin
- MTOC
microtubule organizing centre
- nc-MTOC
non-centrosomal microtubule organizing centre
- PCM
pericentriolar material
- PSS
phosphate stock solution
- RANKL
receptor activator of nuclear factor kappa-B ligand
- SIM
structured illumination microscopy
- TEM
transmission electron microscopy
- γ-TuRC
γ-tubulin ring complex
- Tyr-tubulin
tyrosinated tubulin.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E22-03-0098) on May 5, 2022.
REFERENCES
- Akhmanova A, Steinmetz MO (2015). Control of microtubule organization and dynamics: Two ends in the limelight. Nat Rev Mol Cell Biol 16, 711–726. [DOI] [PubMed] [Google Scholar]
- Bar-Shavit Z (2007). The osteoclast: a multinucleated, hematopoietic-origin, bone-resorbing osteoimmune cell. J Cell Biochem 102, 1130–1139. [DOI] [PubMed] [Google Scholar]
- Bartolini F, Gundersen GG (2006). Generation of noncentrosomal microtubule arrays. J Cell Sci 119, 4155–4163. [DOI] [PubMed] [Google Scholar]
- Batsir S, Geiger B, Kam Z (2017). Dynamics of the sealing zone in cultured osteoclasts. Cytoskeleton 74, 72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biosse Duplan M, Zalli D, Stephens S, Zenger S, Neff L, Oelkers JM, Lai FPL, Horne W, Rottner K, Baron R (2014). Microtubule dynamic instability controls podosome patterning in osteoclasts through EB1, cortactin, and Src. Mol Cell Biol 34, 16–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boraschi-Diaz I, Komarova SV (2016). The protocol for the isolation and cryopreservation of osteoclast precursors from mouse bone marrow and spleen. Cytotechnology 68, 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M (2002). Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol 14, 25–34. [DOI] [PubMed] [Google Scholar]
- Bornens M (2012). The centrosome in cells and organisms. Science 335, 422–426. [DOI] [PubMed] [Google Scholar]
- Brazier H, Pawlak G, Vives V, Blangy A (2009). The Rho GTPase Wrch1 regulates osteoclast precursor adhesion and migration. Int J Biochem Cell Biol 41, 1391–1401. [DOI] [PubMed] [Google Scholar]
- Bugnard E, Zaal KJM, Ralston E (2005). Reorganization of microtubule nucleation during muscle differentiation. Cell Motil Cytoskeleton 60, 1–13. [DOI] [PubMed] [Google Scholar]
- Cai S, Weaver LN, Ems-Mcclung SC, Walczak CE (2009). Kinesin-14 family proteins HSET/XCTK2 control spindle length by cross-linking and sliding microtubules. Mol Biol Cell 20, 1348–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Pang DD, Dai SM (2020). Expression profile of osteoclasts following the stimulation with interleukin-23 in mice. Arch Rheumatol 35, 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin DL, Vallee RB (2002). Dynein at the cortex. Curr Opin Cell Biol 14, 44–49. [DOI] [PubMed] [Google Scholar]
- Fiorino C, Harrison RE (2016a). E-cadherin is important for cell differentiation during osteoclastogenesis. Bone 86, 106–118. [DOI] [PubMed] [Google Scholar]
- Fiorino C, Harrison RE (2016b). E-cadherin is important for cell differentiation during osteoclastogenesis. Bone 86, 106–118. [DOI] [PubMed] [Google Scholar]
- Fukasawa K (2005). Centrosome amplification, chromosome instability and cancer development. Cancer Lett 230, 6–19. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D (2009). A mechanism linking extra centrosomes to chromosomal instability. Nature 460, 278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson HV, Jonasson EM (2018). Microtubules and microtubule-associated proteins. Cold Spring Harb Perspect Biol 10, a022608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, le Clech M, Nigg EA (2007). Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol 179, 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helming L, Gordon S (2009). Molecular mediators of macrophage fusion. Trends Cell Biol 19, 514–522. [DOI] [PubMed] [Google Scholar]
- Hobolt-Pedersen AS, Delaissé JM, Søe K (2014). Osteoclast fusion is based on heterogeneity between fusion partners. Calcif Tissue Int 95, 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höing S, Yeh TY, Baumann M, Martinez NE, Habenberger P, Kremer L, Drexler HCA, Küchler P, Reinhardt P, Choidas A, et al. (2018). Dynarrestin, a novel inhibitor of cytoplasmic dynein. Cell Chem Biol 25, 357–369.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Planus E, Georgess D, Place C, Wang X, Albiges-Rizo C, Jurdic P, Géminard JC (2011). Podosome rings generate forces that drive saltatory osteoclast migration. Mol Biol Cell 22, 3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurdic P, Saltel F, Chabadel A, Destaing O (2006). Podosome and sealing zone: Specificity of the osteoclast model. Eur J Cell Biol 85, 195–202. [DOI] [PubMed] [Google Scholar]
- Karna P, Rida PCG, Pannu V, Gupta KK, Dalton WB, Joshi H, Yang VW, Zhou J, Aneja R (2011). A novel microtubule-modulating noscapinoid triggers apoptosis by inducing spindle multipolarity via centrosome amplification and declustering. Cell Death Differ 18, 632–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Lee SH, Jung HK, Choi Y, Kim N (2008). NFATc1 induces osteoclast fusion via up-regulation of Atp6v0d2 and the dendritic cell-specific transmembrane protein (DC-STAMP). Mol Endocrinol 22, 176–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Merdes A, Mourey L, Agard DA (2011). Microtubule nucleation by γ-tubulin complexes. Nat Rev Mol Cell Biol 12, 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M, Godinho SA, Chandhok NS, Ganem NJ, Azioune A, Thery M, Pellman D (2008). Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev 22, 2189–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber B, Maier B, Fuchs F, Chi J, Riffel P, Anderhub S, Wagner L, Ho AD, Salisbury JL, Boutros M, Krämer A (2010). Proteins required for centrosome clustering in cancer cells. Sci Transl Med 2, 1–11. [DOI] [PubMed] [Google Scholar]
- Lewis L, Albrecht-Buehler G (1987). Distribution of multiple centrospheres determines migration of BHK syncitia. Cell Motil Cytoskeleton 7, 282–290. [DOI] [PubMed] [Google Scholar]
- Lucht U (1973). Electron microscope observations of centrioles in osteoclasts. Z Anat Entwicklungsgesch 140, 143–152. [DOI] [PubMed] [Google Scholar]
- Magescas J, Zonka JC, Feldman JL (2019). A two-step mechanism for the inactivation of microtubule organizing center function at the centrosome. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maria SM, Prukner C, Sheikh Z, Mueller F, Barralet JE, Komarova Sv (2014). Reproducible quantification of osteoclastic activity: Characterization of a biomimetic calcium phosphate assay. J Biomed Mater Res B Appl Biomater 102, 903–912. [DOI] [PubMed] [Google Scholar]
- Marthiens V, Piel M, Basto R (2012). Never tear us apart-the importance of centrosome clustering. J Cell Sci 125, 3281–3292. [DOI] [PubMed] [Google Scholar]
- Martin M, Akhmanova A (2018). Coming into focus: Mechanisms of microtubule minus-end organization. Trends Cell Biol 28, 574–588. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Nagase Y, Hirose J, Tokuyama N, Yasui T, Kadono Y, Ueki K, Kadowaki T, Nakamura K, Tanaka S (2013). Regulation of bone resorption and sealing zone formation in osteoclasts occurs through protein kinase b-mediated microtubule stabilization. J Bone Miner Res 28, 1191–1202. [DOI] [PubMed] [Google Scholar]
- Mennella V, Agard DA, Huang B, Pelletier L (2014). Amorphous no more: Subdiffraction view of the pericentriolar material architecture. Trends Cell Biol 24, 188–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milunovic´-Jevtic´ A, Mooney P, Sulerud T, Bisht J, Gatlin JC (2016). Centrosomal clustering contributes to chromosomal instability and cancer. Curr Opin Biotechnol 40, 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T (2011). Regulators of osteoclast differentiation and cell-cell fusion. Keio J Med 60, 101–105. [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Muto A, Udagawa N, Arai A, Yamashita T, Hosoya A, Ninomiya T, Nakamura H, Yamamoto Y, Kinugawa S, et al. (2009). Identification of cell cycle-arrested quiescent osteoclast precursors in vivo. J Cell Biol 184, 541–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motiur Rahman M, Takeshita S, Matsuoka K, Kaneko K, Naoe Y, Sakaue-Sawano A, Miyawaki A, Ikeda K (2015). Proliferation-coupled osteoclast differentiation by RANKL: Cell density as a determinant of osteoclast formation. Bone 81, 392–399. [DOI] [PubMed] [Google Scholar]
- Mountain V, Simerly C, Howard L, Ando A, Schatten G, Compton DA (1999). The kinesin-related protein, hset, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J Cell Biol 147, 351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulari MTK, Patrikainen L, Kaisto T, Metsikkö K, Salo JJ, Väänänen HK (2003). The architecture of microtubular network and Golgi orientation in osteoclasts–-major differences between avian and mammalian species. Exp Cell Res 285, 221–235. [DOI] [PubMed] [Google Scholar]
- Muroyama A, Lechler T (2017). Microtubule organization, dynamics and functions in differentiated cells. Development (Cambridge) 144, 3012–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HL, Gruber D, Bulinski JC (1999). Microtubule-associated protein 4 (MAP4) regulates assembly, protomer-polymer partitioning and synthesis of tubulin in cultured cells. J Cell Sci 112, 1813–1824. [DOI] [PubMed] [Google Scholar]
- Okahashi N, Murase Y, Koseki T, Sato T, Yamato K, Nishihara T (2001). Osteoclast differentiation is associated with transient upregulation of cyclin-dependent kinase inhibitors p21(WAF1/CIP1) and p27(KIP1). J Cell Biochem 80, 339–345. [PubMed] [Google Scholar]
- Ori-McKenney KM, Jan LY, Jan YN (2012). Golgi outposts shape dendrite morphology by functioning as sites of acentrosomal microtubule nucleation in neurons. Neuron 76, 921–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oursler MJ (2010). Recent advances in understanding the mechanisms of osteoclast precursor fusion. J Cell Biochem 110, 1058–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannu V, Rida PCG, Celik B, Turaga RC, Ogden A, Cantuaria G, Gopalakrishnan J, Aneja R (2014). Centrosome-declustering drugs mediate a two-pronged attack on interphase and mitosis in supercentrosomal cancer cells. Cell Death Dis 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti A, Moudjou M, Bornens M, Paintrand M, Salisbury JL (1996). Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J Cell Sci 109, 3089–3102. [DOI] [PubMed] [Google Scholar]
- Patntirapong S, Habibovic P, Hauschka Pv (2009). Effects of soluble cobalt and cobalt incorporated into calcium phosphate layers on osteoclast differentiation and activation. Biomaterials 30, 548–555. [DOI] [PubMed] [Google Scholar]
- Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS (2005). Spindle multipolarity is prevented by centrosomal clustering. Science 307, 127–129. [DOI] [PubMed] [Google Scholar]
- Rebacz B, Larsen TO, Clausen MH, Rønnest MH, Löffler H, Ho AD, Krämer A (2007). Identification of griseofulvin as an inhibitor of centrosomal clustering in a phenotype-based screen. Cancer Res 67, 6342–6350. [DOI] [PubMed] [Google Scholar]
- Rhys AD, Monteiro P, Smith C, Vaghela M, Arnandis T, Kato T, Leitinger B, Sahai E, McAinsh A, Charras G, et al. (2018). Loss of E-cadherin provides tolerance to centrosome amplification in epithelial cancer cells. J Cell Biol 217, 195–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochlin K, Yu S, Roy S, Baylies MK (2010). Myoblast fusion: when it takes more to make one. Dev Biol 341, 66–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Binder NE, Geimer S, Melkonian M (2002). In vivo localization of centrin in the green alga Chlamydomonas reinhardtii. Cell Motil Cytoskeleton 52, 43–55. [DOI] [PubMed] [Google Scholar]
- Sabat-Pospiech D, Fabian-Kolpanowicz K, Prior IA, Coulson JM, Fielding AB (2019). Targeting centrosome amplification, an Achilles’ heel of cancer. Biochem Soc Trans 47, 1209–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltel F, Destaing O, Bard F, Eichert D, Jurdic P (2004). Apatite-mediated Actin Dynamics in Resorbing Osteoclasts. Mol Biol Cell 15, 5231–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekino Y, Oue N, Koike Y, Shigematsu Y, Sakamoto N, Sentani K, Teishima J, Shiota M, Matsubara A, Yasui W (2019). KIFC1 inhibitor CW069 induces apoptosis and reverses resistance to docetaxel in prostate cancer. J Clin Med 8, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Søe K, Delaissé JM (2017). Time-lapse reveals that osteoclasts can move across the bone surface while resorbing. J Cell Sci 130, 2026–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommi P, Cheerambathur D, Brust-Mascher I, Mogilner A (2011). Actomyosin-dependent cortical dynamics contributes to the prophase force-balance in the early drosophila embryo. PLoS One 6, e18366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soysa NS, Alles N, Aoki K, Ohya K (2012). Osteoclast formation and differentiation: an overview. J Med Dent Sci 59, 65–74. [PubMed] [Google Scholar]
- Starr DA (2017). Muscle development: nucleating microtubules at the nuclear envelope. Curr Biol 27, R1071–R1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin AM, Maro B, Bornens M (1985). Fate of microtubule-organizing centers during myogenesis in vitro. J Cell Biol 100, 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomopoulou P, Sachs J, Teusch N, Mariappan A, Gopalakrishnan J, Schmalz HG (2016). New colchicine-derived triazoles and their influence on cytotoxicity and microtubule morphology. ACS Med Chem Lett 7, 188–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ti Y, Zhou L, Wang R, Zhao J (2015). Inhibition of microtubule dynamics affects podosome belt formation during osteoclast induction. Cell Biochem Biophys 71, 741–747. [DOI] [PubMed] [Google Scholar]
- Turksen K, Kanehisa J, Opas M, Heersche JNM, Aubin JE (1988). Adhesion patterns and cytoskeleton of rabbit osteoclasts on bone slices and glass. J Bone Miner Res 3, 389–400. [DOI] [PubMed] [Google Scholar]
- Vergarajauregui S, Becker R, Steffen U, Sharkova M, Esser T, Petzold J, Billing F, Kapiloff MS, Schett G, Thievessen I, et al. (2020). Akap6 orchestrates the nuclear envelope microtubule-organizing center by linking golgi and nucleus via akap9. eLife 9, 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voter WA, Erickson HP (1984). The kinetics of microtubule assembly. Evidence for a two-stage nucleation mechanism. J Biol Chem 259, 10430–10438. [PubMed] [Google Scholar]
- Wang Y, Lebowitz D, Sun C, Thang H, Grynpas MD, Glogauer M (2008). Identifying the relative contributions of Rac1 and Rac2 to osteoclastogenesis. J Bone Miner Res 23, 260–270. [DOI] [PubMed] [Google Scholar]
- Watts CA, Richards FM, Bender A, Bond PJ, Korb O, Kern O, Riddick M, Owen P, Myers RM, Raff J, et al. (2013). Design, synthesis, and biological evaluation of an allosteric inhibitor of HSET that targets cancer cells with supernumerary centrosomes. Chem Biol 20, 1399–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff JB, Wueseke O, Hyman AA (2014). Pericentriolar material structure and dynamics. Philos Trans R Soc Lond B Biol Sci 369, 20130459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Mikule K, Wang W, Su N, Petteruti P, Gharahdaghi F, Code E, Zhu X, Jacques K, Lai Z, et al. (2013). Discovery and mechanistic study of a small molecule inhibitor for motor protein KIFC1. ACS Chem Biol 8, 2201–2208. [DOI] [PubMed] [Google Scholar]
- Xiao Y-X, Yang W-X, Xiao Y-X, Yang W-X (2016). KIFC1: a promising chemotherapy target for cancer treatment? Oncotarget 7, 48656–48670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Feldman JL (2015). SPD-2/CEP192 and CDK are limiting for microtubule-organizing center function at the centrosome. Curr Biol 25, 1924–1931. [DOI] [PubMed] [Google Scholar]
- Yavropoulou MP, Yovos JG (2008). Osteoclastogenesis–current knowledge and future perspectives - PubMed. J Musculoskelet Neuronal Interact 8, 204–216. [PubMed] [Google Scholar]
- Zebrowski DC, Vergarajauregui S, Wu CC, Piatkowski T, Becker R, Leone M, Hirth S, Ricciardi F, Falk N, Giessl A, et al. (2015). Developmental alterations in centrosome integrity contribute to the postmitotic state of mammalian cardiomyocytes. Elife 4, e05563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.