Abstract
During the meiotic divisions in oocytes, microtubules are sorted and organized by motor proteins to generate a bipolar spindle in the absence of centrosomes. In most organisms, kinesin-5 family members crosslink and slide microtubules to generate outward force that promotes acentrosomal spindle bipolarity. However, the mechanistic basis for how other kinesin families act on acentrosomal spindles has not been explored. We investigated this question in Caenorhabditis elegans oocytes, where kinesin-5 is not required to generate outward force and the kinesin-12 family motor KLP-18 instead performs this function. Here we use a combination of in vitro biochemical assays and in vivo mutant analysis to provide insight into the mechanism by which KLP-18 promotes acentrosomal spindle assembly. We identify a microtubule binding site on the C-terminal stalk of KLP-18 and demonstrate that a direct interaction between the KLP-18 stalk and its adaptor protein MESP-1 activates nonmotor microtubule binding. We also provide evidence that this C-terminal domain is required for KLP-18 activity during spindle assembly and show that KLP-18 is continuously required to maintain spindle bipolarity. This study thus provides new insight into the construction and maintenance of the oocyte acentrosomal spindle as well as into kinesin-12 mechanism and regulation.
INTRODUCTION
When a cell divides, a bipolar microtubule-based spindle forms to mediate chromosome segregation. In most cell types, structures called centrosomes act to nucleate microtubules and define and organize the spindle poles. However, oocytes of most species lack centrosomes and therefore utilize a unique pathway to form acentrosomal spindles during the meiotic divisions. In this process, microtubules nucleate and then are sorted by motor proteins to generate a bipolar spindle (Mullen et al., 2019). How these motors generate force to sort and organize microtubules into bipolar structures in the absence of centrosomes is important to understand, because errors in spindle construction can lead to chromosome missegregation that results in aneuploid gametes.
One major class of force-generating motors is the kinesin-5 family of plus end–directed microtubule motors. These tetrameric motors can cross-link and slide antiparallel microtubules (Sawin et al., 1992; Kashina et al., 1996; Kapitein et al., 2005; Scholey et al., 2014); this ability enables them to generate outward force that promotes spindle bipolarity in both mitosis (Sawin and Mitchison, 1991; Sawin et al., 1992; Kapoor et al., 2000) (reviewed in Mann and Wadsworth, 2019) and oocyte meiosis (Walczak et al., 1998; Mailhes et al., 2004; Schuh and Ellenberg, 2007; Yang et al., 2008; Radford et al., 2017; Wan et al., 2018; Roeles and Tsiavaliaris, 2019) in many species. However, in mammalian mitosis, overexpression of the kinesin-12 motor Kif15 is sufficient to promote spindle bipolarity when kinesin-5 is inhibited, suggesting that kinesin-12 family motors can also provide outward force to separate spindle poles. Therefore, kinesin-12s may provide a backup to the predominant kinesin-5–driven mechanism (Tanenbaum et al., 2009; Vanneste et al., 2009; Sturgill and Ohi, 2013).
Interestingly, the roles of these two families of motors appear to be reversed in Caenorhabditis elegans oocytes. In these cells, the kinesin-12 family motor KLP-18 is required to establish spindle bipolarity (Segbert et al., 2003; Wignall and Villeneuve, 2009; Connolly et al., 2014; Wolff et al., 2016), whereas kinesin-5 (BMK-1) is not essential (Bishop et al., 2005); a role for BMK-1 in spindle assembly can be seen only under conditions where KLP-18 is depleted (Cavin-Meza et al., 2022). Therefore, KLP-18 is the primary force-generating motor in this system. KLP-18 acts with adaptor protein MESP-1 (meiotic spindle 1) to sort microtubule minus ends to the periphery of a microtubule array, where they coalesce into spindle poles (Wolff et al., 2016). If either of these proteins is depleted, outward sorting of microtubules is lost and minus ends converge to form a monopole. However, the mechanistic basis for how these proteins promote bipolarity is not understood.
Here we use a combination of in vitro biochemical assays and in vivo mutant analysis to provide insight into the mechanism by which these proteins collaborate to promote acentrosomal spindle assembly. Moreover, we show that KLP-18 is continuously required to maintain spindle bipolarity, demonstrating the importance of this motor in maintaining force balance in the acentrosomal spindle. Altogether, this work provides initial biochemical characterization of the essential KLP-18/MESP-1 complex during C. elegans oocyte meiosis, providing insight into how kinesin motors construct acentrosomal spindles.
RESULTS
The KLP-18 coiled-coil stalk domain contains a regulated microtubule binding site
Kinesin-12 family motors contain a globular N-terminal motor domain that can walk along microtubules in a plus end–directed manner. However, in order to cross-link and sort microtubules within the spindle, these motors must bind a second microtubule. Multiple microtubule cross-linking strategies have been proposed for Kif15: 1) Kif15 forms a homotetramer with antipolar motor domains, allowing both ends of the complex to bind to microtubules (Drechsler et al., 2014; Drechsler and McAinsh, 2016; Mann et al., 2017; McHugh et al., 2018), and 2) a nonmotor microtubule binding site in the middle of the stalk domain mediates binding to a second microtubule (Sturgill et al., 2014; Reinemann et al., 2017). Because the kinesin-12 family motor KLP-18 is the major microtubule sorting kinesin in the C. elegans oocyte spindle, we reasoned that this would be an ideal system to investigate the mechanisms by which kinesin-12 motors promote spindle bipolarity, starting with the question of whether this motor contains multiple microtubule binding sites.
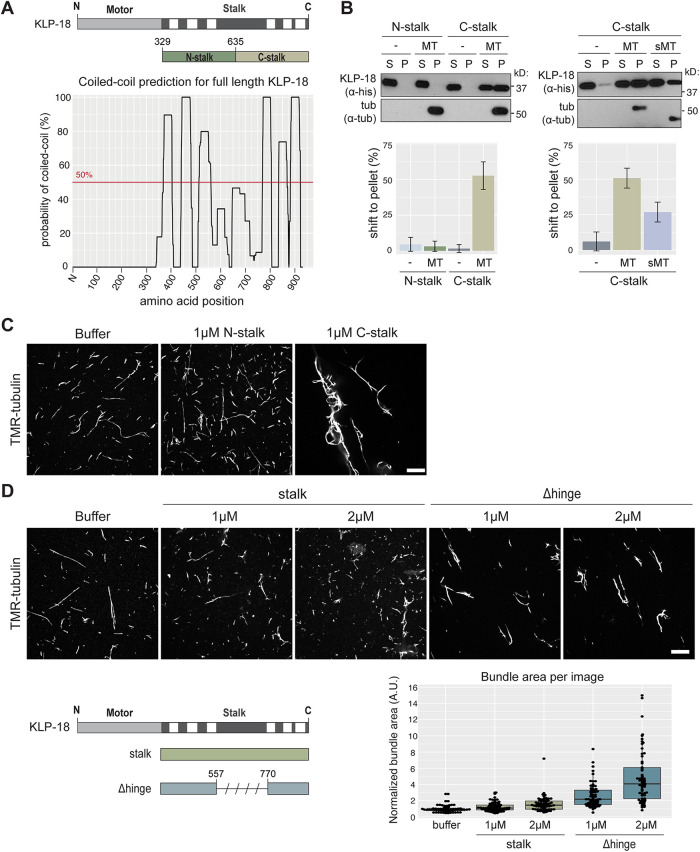
Like other kinesin-12 motors, KLP-18 contains a C-terminal stalk domain (Segbert et al., 2003), so we first examined whether this domain could mediate cross-linking by directly binding to microtubules. Structural prediction of the KLP-18 stalk revealed that it contains discrete coiled-coil domains (Figure 1A), similar to Kif15 (Segbert et al., 2003; Sturgill et al., 2014). To test whether the stalk contains a microtubule binding site, we expressed and purified the N- and C-terminal halves of the stalk (“N-stalk” and “C-stalk”; Figure 1A and Supplemental Figure S1A) and performed a microtubule cosedimentation assay (Alfaro-Aco et al., 2017) (Figure 1B). After microtubules were pelleted, N-stalk remained in the supernatant, while a sizable fraction of C-stalk pelleted, reflecting its ability to bind microtubules in vitro (Figure 1B, left). To investigate the nature of this interaction, we treated microtubules with subtilisin to cleave E-hooks, which are negatively charged regions at the tubulin C-terminus that can promote microtubule association of proteins through an electrostatic interaction (Serrano et al., 1984; Rodionov et al., 1990). We found that C-stalk bound to microtubules lacking E-hooks, but with decreased affinity (Figure 1B, right). These results are consistent with previous studies of the nonmotor microtubule binding sites in Kif15 and kinesin-1 (Seeger and Rice, 2010; Reinemann et al., 2017), and suggest that an electrostatic interaction increases the affinity of the KLP-18 stalk to microtubules. To further assess the microtubule binding activity of C-stalk and N-stalk, we incubated these proteins with fluorescently labeled microtubules in vitro and visualized microtubule organization; during purification, both N-stalk and C-stalk elute from size exclusion chromatography as oligomeric complexes, potentially providing multiple binding sites that could facilitate microtubule bundling. Addition of C-stalk caused dramatic microtubule bundling compared with buffer alone or N-stalk (Figure 1C), confirming that C-stalk is able to bind microtubules. Together, these data show that the stalk domain of KLP-18 contains a C-terminal microtubule binding site.
FIGURE 1:
The kinesin-12/KLP-18 coiled-coil stalk domain contains a regulated microtubule binding site. (A) Coiled-coil prediction software paircoil2 (McDonnell et al., 2006) shows discrete coiled-coil domains in the KLP-18 stalk (bottom) marked on a KLP-18 domain diagram (top, coiled-coil domains denoted in white). N- and C-stalk constructs shown relative to full stalk. (B) N-stalk and C-stalk microtubule binding activity tested by microtubule cosedimentation assay with no microtubules added (–), undigested (MT), and subtilisin-digested (sMT) microtubules. Blots show supernatant (S) and pellet (P) samples; quantification is of average shift ± SD. N = 3 experiments for each set. (C, D) Microtubule binding activity assessed as microtubule bundling ability. Representative images of TMR-microtubules incubated with buffer alone, N-stalk, and C-stalk (C) or with buffer alone, full-length stalk, and Δhinge (D). Quantification of bundling below. Boxplot represents first quartile to third quartile, and the median is indicated by a horizontal line. Quantified images were acquired over two independent experiments. Scale bars = 10 μm.
Next, we purified the full KLP-18 stalk (termed “stalk”; Supplemental Figure S1A) and paradoxically found that it showed very little microtubule bundling activity (Figure 1D), suggesting that it cannot bind microtubules. Because an autoinhibitory mechanism has been proposed for mammalian Kif15, where the motor folds its stalk domain to block the nonmotor microtubule binding site (Sturgill et al., 2014), we tested whether KLP-18 could employ a similar regulatory mechanism. The coiled-coil prediction for the KLP-18 stalk contains a region of low probability in the center of the stalk domain (Figure 1A), and we hypothesized that this region may be a flexible hinge that could fold, thus preventing the C-terminal region from binding microtubules. To test this mechanism, we purified a version of the stalk with this putative hinge deleted (termed “Δhinge”), which is predicted to be completely coiled-coil (Supplemental Figures S1A and S2, A and B). Consistent with an autoinhibitory mechanism, deletion of the hinge region restored microtubule bundling activity, reflecting an ability to bind microtubules (Figure 1D). In addition, we assessed the flexibility of the KLP-18 stalk by running an MBP-stalk fusion protein (Supplemental Figure S2A) through a size exclusion column in high salt (300 mM) and low salt (20 mM) buffers. If the KLP-18 stalk was able to fold via an electrostatic interaction, high salt would disrupt folding and lead to an extended conformation; this conformational change would be apparent in the elution volume. Indeed, in high salt, a small population of MBP-stalk eluted at a lower elution volume, indicating that this subpopulation contains extended molecules (Supplemental Figure S2C). Although this is a subtle shift and therefore is not definitive proof of a conformational change, these results are consistent with the possibility that the KLP-18 stalk can exist in either an inactive folded state or an unfolded state that is capable of binding to microtubules.
The KLP-18 stalk microtubule interaction domain is essential for spindle assembly
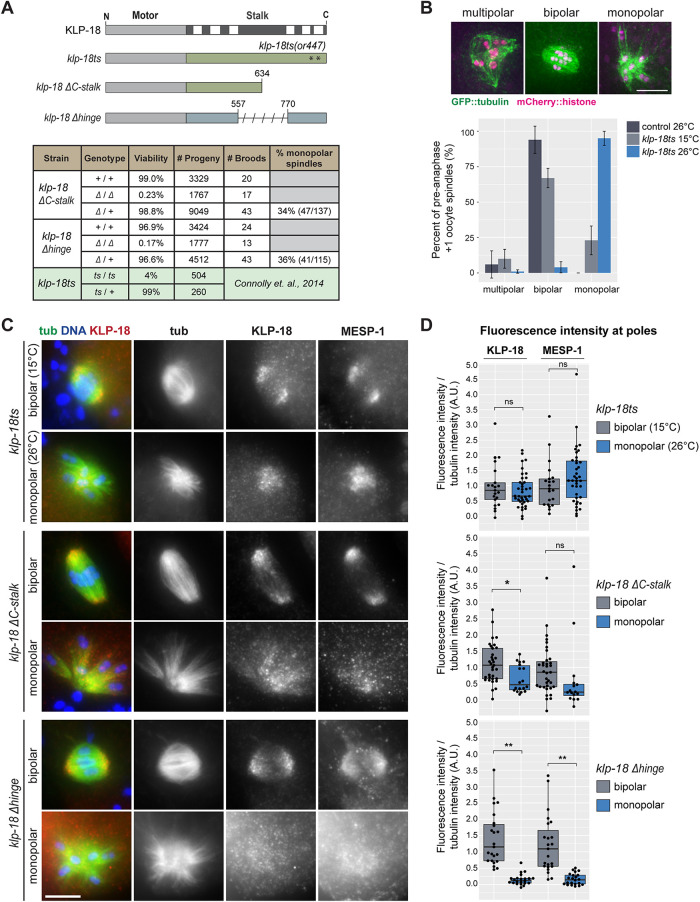
During acentrosomal spindle assembly in C. elegans oocytes, KLP-18 sorts microtubule minus ends to the outside of a microtubule array, enabling the formation of multiple poles that then coalesce to form a bipolar spindle (Segbert et al., 2003; Wignall and Villeneuve, 2009; Connolly et al., 2014; Wolff et al., 2016). Given our identification of the C-terminal microtubule binding site, we hypothesized that KLP-18 might sort microtubules by binding a microtubule with its stalk and walking on a second microtubule with its motor domain. To test this hypothesis, we analyzed a set of klp-18 mutants in vivo. We previously demonstrated that a mutant lacking most of the N-terminal motor domain, klp-18(ok2519), formed monopolar oocyte spindles, confirming that this domain is required for KLP-18 function (Wolff et al., 2016). To assess requirements for other domains, we made use of the previously described klp-18(or447) temperature-sensitive mutant, which contains two substitutions (V854M and G876S) in the C-terminal microtubule binding domain (Figure 2A, mutant hereafter referred to as “klp-18ts”) (Connolly et al., 2014). At the restrictive temperature of 26°C, klp-18ts has high embryonic lethality, defects in polar body extrusion, and aberrant chromosome dynamics, suggesting that bipolar spindles do not form (Connolly et al., 2014), but this has not been directly demonstrated. In addition, we generated two new klp-18 mutants using CRISPR: klp-18ΔC-stalk, a deletion of the C-terminal microtubule binding site, and klp-18Δhinge, a deletion of the putative hinge (Figure 2A), and we assessed spindle assembly in all three mutants.
FIGURE 2:
KLP-18 hinge and stalk microtubule interaction domain are essential for spindle assembly. (A) Domain diagram of klp-18 mutants used: klp-18ts, klp-18ΔC-stalk, and klp-18Δhinge. Asterisks indicate approximate location of or447 mutations in the klp-18ts mutant. Characterization of progeny from heterozygous (Δ /+) or homozygous (Δ/Δ or +/+) klp-18ΔC-stalk and klp-18Δhinge parents. Data for klp-18ts were adapted from Connolly et al. (2014) and are shaded in green. (B) Morphology of preanaphase oocyte spindles in the most recently fertilized (+1) embryo of control (dark gray) and klp-18ts worms expressing GFP::tubulin and mCherry::histone. During normal spindle assembly, minus ends are sorted away from chromosomes and form multiple poles (these forming spindles are denoted “multipolar”) that coalesce to form a bipolar spindle (“bipolar”) (Wolff et al., 2016). If minus ends are not sorted outward, they converge to form a monopolar spindle (“monopolar”) (Segbert et al., 2003; Wignall and Villeneuve, 2009; Connolly et al., 2014; Wolff et al., 2016). Spindle morphology in klp-18ts was quantified at permissive (15°C, light gray) and restrictive (26°C, blue) temperatures. Note that spindles that had entered anaphase were not included in this graph; quantification of all +1 oocytes is shown in Supplemental Figure S3A. Bars represent mean percentage ± SD. For klp-18ts conditions, n = 4 experiments; for control, n = 3 experiments. Scale bar = 5 μm. (B) DNA (blue), tubulin (green), KLP-18 (red), and MESP-1 (not shown in merge) localization in mutant strains described in A. Representative images of bipolar and mutant monopolar spindles are shown for each strain. Scale bar = 5 μm. (D) Quantification of KLP-18 and MESP-1 fluorescence intensity on the poles of bipolar (gray) and monopolar (blue) spindles in each mutant. KLP-18/MESP-1 intensity was normalized to tubulin intensity. Box represents first quartile to third quartile, and the median is indicated by a horizontal line. Welch two-sample t test, ns = p > 0.05, * = p < 0.05, ** = p < 0.0005.
Eggs laid by homozygous parents of both the klp-18ΔC-stalk and klp-18Δhinge mutants are largely inviable (Figure 2A, 0.23 and 0.17% viability, respectively), so we were unable to propagate homozygous mutant strains. Instead, we analyzed progeny of heterozygous worms; 25% of their progeny should be homozygous mutants. Homozygous mutants reach adulthood because they have a maternal supply of wild-type KLP-18 that enables proper development but produce oocytes containing only the mutant form of KLP-18. Notably, about 35% of preanaphase oocyte spindles from the adult progeny of heterozygous parents were monopolar in both strains (Figure 2A), demonstrating that deleting either the putative hinge or the C-terminal domain affects KLP-18 function. To similarly quantify spindle defects in the klp-18ts strain, we shifted worms to the restrictive temperature for 1 h, thus disrupting KLP-18 before the initiation of spindle assembly. We then quantified spindle morphology in the most recently fertilized oocyte; although by this stage some control spindles had progressed to anaphase (Supplemental Figure S3A), many were either forming or bipolar (Figure 2B and Supplemental Figure S3A), enabling us to assess effects on spindle formation. In klp-18ts oocytes at the permissive temperature (15°C), we found that 22% of preanaphase spindles (23/105) were monopolar. This indicates that KLP-18 function is partially compromised, consistent with previously reported embryonic lethality at this temperature (Connolly et al., 2014). Upon shift to the restrictive temperature, the percentage of preanaphase spindles that were monopolar increased to 94% (124/132) (Figure 2B). As expected, the poles of monopolar spindles in klp-18ts, klp-18ΔC-stalk, and klp-18Δhinge oocytes are marked by the minus-end marker ASPM-1 (Jiang et al., 2017), phenocopying klp-18(RNAi) (Wignall and Villeneuve, 2009) and confirming that microtubule sorting was aberrant (Supplemental Figure S3B). However, ASPM-1 also localized to some microtubule ends on the outside of the aster in 24/30 klp-18ts monopolar spindles analyzed (Supplemental Figure S3B, arrowheads), suggesting that klp-18ts may allow weak sorting activity that enables some minus ends to be pushed outward, even at the restrictive temperature. Supporting this idea, ∼5% of preanaphase spindles (7/132) are bipolar at 26°C (Figure 2B).
Although our results suggest that the C-terminal microtubule binding and hinge domains are required for KLP-18 function, it is also possible that these mutations merely destabilize the KLP-18 protein. However, we did not detect a decrease in KLP-18 abundance in klp-18ts worms at 15°C or 26°C by Western blot compared with control worms (Supplemental Figure S3C), and we found that the KLP-18ΔC-stalk and KLP-18Δhinge mutant proteins are expressed (Supplemental Figure S3D), suggesting that the phenotypes in the klp-18 mutants are not due to loss of KLP-18 protein.
In both Xenopus and mammals, the adaptor protein TPX2 is required for targeting kinesin-12 to the spindle (Wittmann et al., 1998, 2000; Drechsler et al., 2014; Mann et al., 2017; McHugh et al., 2018; Malaby et al., 2019), and our previous work indicates that MESP-1 performs this kinesin-12 targeting function in C. elegans: KLP-18 and MESP-1 colocalize on spindle microtubules, are found in a complex in worm extract, have identical depletion phenotypes, and are interdependent for localization (Wolff et al., 2016). Therefore, to better understand the phenotype of the klp-18 mutants, we investigated whether targeting of the KLP-18/MESP-1 complex to microtubules was affected. We quantified the fluorescence intensity of each protein at spindle poles (either an average of the two bipolar poles or the one monopolar pole) and normalized this value to the average fluorescence intensity of tubulin. In klp-18ts oocytes there was no significant difference between KLP-18 and MESP-1 localization to the monopole at the restrictive temperature compared with localization on the bipolar poles at the permissive temperature (Figure 2, C and D, top), suggesting that the KLP-18/MESP-1 complex is able to localize to spindle microtubules but is inactivated by the temperature shift, possibly through disruption of the C-terminal microtubule binding domain that contains the two klp-18ts mutations. In klp-18ΔC-stalk oocytes there was persistent but decreased localization of KLP-18 and MESP-1 to monopoles in most oocytes, indicating that these proteins are able to target to spindle poles with decreased efficiency (Figure 2, C and D, middle). Finally, we saw a striking decrease of KLP-18 and MESP-1 localization in klp-18Δhinge oocytes, suggesting that the “hinge” region is essential for spindle localization of either of these proteins (Figure 2, C and D, bottom). Altogether, these results show that the C-terminal microtubule binding site and the putative hinge region that we identified in vitro are essential for KLP-18 activity and spindle assembly in vivo.
KLP-18 is targeted to microtubules through a direct interaction with microtubule-associated protein MESP-1
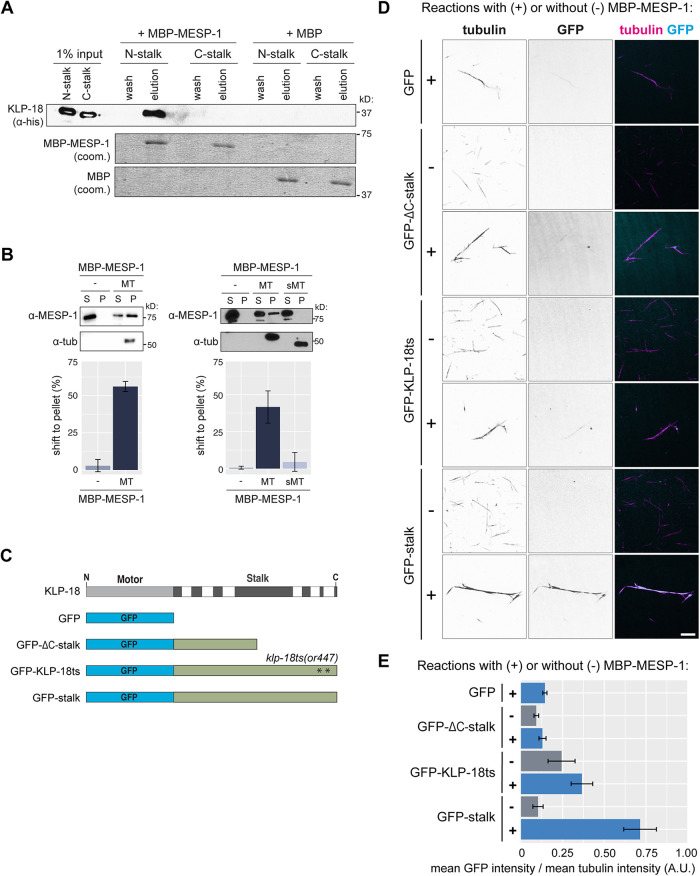
Next, we aimed to better understand the contribution of MESP-1 to KLP-18 function; MESP-1 is required for KLP-18 spindle localization, but how MESP-1 regulates KLP-18 is not known. Considering that MESP-1 can localize to monopolar spindle microtubules in the klp-18ΔC-stalk and klp-18ts strains, in which C-terminal microtubule binding is disrupted, but does not localize in the klp-18Δhinge mutant, we hypothesized that MESP-1 may bind to a region overlapping with the hinge. To test this in vitro, we first attempted to express GST-MESP-1, but this protein degraded significantly during purification (Supplemental Figure S1B). This suggests that MESP-1 is unstable, consistent with the prediction that portions of MESP-1 are disordered (Supplemental Figure S1C) similar to TPX2 (Alfaro-Aco et al., 2017; Zhang et al., 2017; King and Petry, 2020). Therefore, we switched to an MBP tag (Wang et al., 1999; Esposito and Chatterjee, 2006; Reuten et al., 2016), which increased MESP-1 stability (Supplemental Figure S1B). We incubated purified MBP-MESP-1 with the N- and C-terminal KLP-18 stalk truncations individually and added amylose resin to retrieve MBP-MESP-1. We found that N-stalk was present in the eluted fraction but C-stalk was not, indicating that MESP-1 binds to the N-terminal half of the KLP-18 stalk (Figure 3A). However, this interaction appears to be weak, because only a small fraction of N-stalk was pulled out by MBP-MESP-1; this is similar to what has been reported for the interaction between mammalian TPX2 and Kif15 (Drechsler et al., 2014). Importantly, this N-terminal MESP-1 binding region overlaps significantly with the hinge region (see Figure 5A later in this article), perhaps explaining why MESP-1 localization is decreased in klp-18Δhinge spindles.
FIGURE 3:
KLP-18 C-terminal domain is essential for microtubule binding in vitro and requires a direct interaction with microtubule-associated protein MESP-1. (A) MBP-MESP-1 direct interaction with N-stalk or C-stalk was tested by MBP pull down. Reactions using MBP-MESP-1 or MBP as bait are shown. KLP-18 truncations visualized on Western blot with anti-His antibody; MBP and MBP-MESP-1 visualized by Coomassie stain. (B) Microtubule cosedimentation assay for MBP-MESP-1 with no microtubules added (–), undigested (MT), and subtilisin-digested (sMT) microtubules. Representative blots show supernatant (S) and pellet (P) samples; quantification is of average shift ± SD. N = 2 experiments for nonsubtilisin experiment; n = 3 experiments for subtilisin experiment. (C) Schematic of GFP-tagged proteins used in D and E. (D) KLP-18 recruitment to microtubules was tested by visualization of GFP-tagged stalk proteins on TMR-microtubules. Representative images with (+) and without (–) MBP-MESP-1 added to the indicated GFP-tagged proteins are shown for each condition. Black and white images are inverted to better visualize the signals. Scale bar = 5 μm. (E) Quantification of mean GFP intensity overlaid on the microtubule normalized to mean tubulin intensity. Bars represent mean percentage ± SD for each experiment over n = 3 experiments.
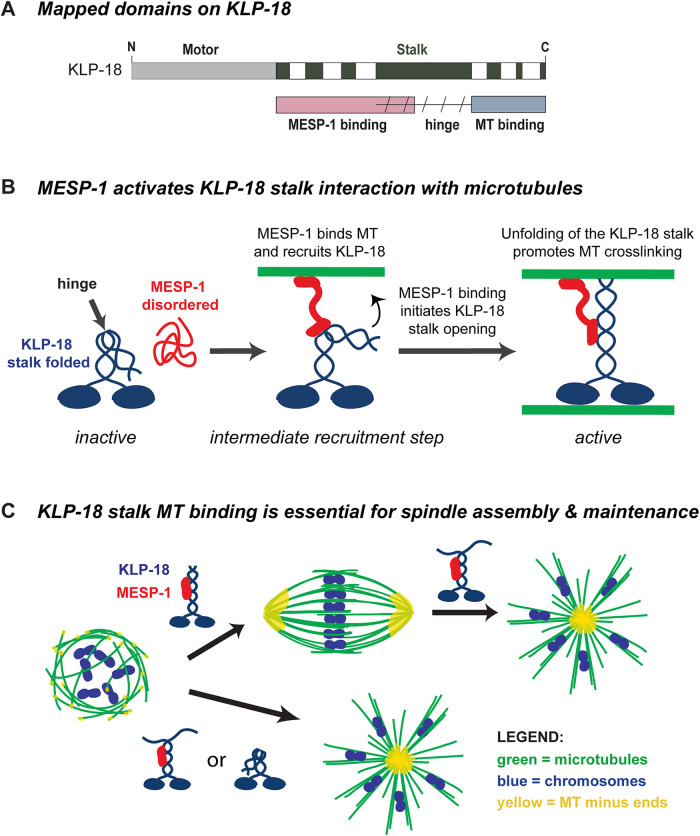
FIGURE 5:
Model. (A) KLP-18 stalk contains a MESP-1 binding site in its N-terminal half (pink), a microtubule binding site in its C-terminal half (blue), and likely a flexible hinge region (slanted dashed lines) that functions to self-inhibit the motor. The MESP-1 binding site and the hinge region overlap to some extent, as shown. (B) Before complex formation, KLP-18 is in an autoinhibited inactive state and MESP-1 is disordered and nonfunctional. Upon complex formation, MESP-1 properly folds and targets KLP-18 to spindle microtubules and promotes KLP-18 stalk opening. Once KLP-18 is activated through unfolding, the C-terminal microtubule binding site can bind to microtubules, allowing the KLP-18/MESP-1 complex to cross-link microtubules and promote spindle bipolarity. After the initial targeting of the complex, MESP-1 may remain bound to one of the two cross-linked microtubules. In this simplified model, we show MESP-1 bound to the microtubule associating with the KLP-18 C-terminal binding site, but it is also possible that MESP-1 could bind the microtubule associated with the KLP-18 motor domain. Note that some studies have proposed that KLP-18 forms a homotetramer (Drechsler et al., 2014; Drechsler and McAinsh, 2016; Mann et al., 2017; McHugh et al., 2018). While we depict KLP-18 as a dimer in this model, our data do not exclude the possibility that KLP-18 forms a tetramer and that MESP-1 regulates this tetrameric form. (C) Disruption of the KLP-18 C-terminal microtubule binding site impairs microtubule sorting during spindle assembly and leads to the collapse of preformed bipolar spindles. Both the C-terminal microtubule binding site and interaction with MESP-1 are essential for KLP-18 function; MESP-1 is likely essential for initial targeting to spindle microtubules, and the C-terminal microtubule binding site is essential for motor function. KLP-18 is shown as blue and MESP-1 is shown as red in the cartoon. On the spindle diagrams, microtubules are green, chromosomes are blue, and microtubule minus ends are yellow.
Next, we tested the microtubule binding ability of MESP-1 to begin to clarify the role of MESP-1 in activating KLP-18 and targeting it to microtubules. MBP-MESP-1 pelleted with microtubules in a cosedimentation assay (Figure 3B) and was able to bundle microtubules (Supplemental Figure S1D), confirming that MESP-1 is a microtubule binding protein. In addition, we found that MESP-1’s ability to bind microtubules was greatly diminished when we used subtilisin-digested microtubules (Figure 3B, right). Thus, MESP-1 is a microtubule-associated protein, similar to TPX2. However, TPX2 binds along the microtubule lattice and does not require tubulin E-hooks (Balchand et al., 2015; Zhang et al., 2017), suggesting that MESP-1 employs a different microtubule binding mechanism.
Considering that MESP-1 can bind to both KLP-18 and microtubules, we hypothesized that MESP-1 may recruit KLP-18 to microtubules directly. To test this model, we purified GFP-tagged versions of the full-length KLP-18 stalk (termed “GFP-stalk”) and the KLP-18ΔC-stalk (“GFP-KLP-18ΔC-stalk”) and a version of the stalk containing the two mutations present in the klp-18ts strain (“GFP-KLP-18ts”) and then quantified GFP localization to microtubules in the presence and absence of MESP-1 (Figure 3, C–E). We attempted to purify a GFP-KLP-18Δhinge protein but it was unstable and did not yield consistent results. In these experiments, GFP-stalk did not localize to microtubules when incubated alone (Figure 3, D and E), consistent with our model of KLP-18 autoinhibition. However, we observed strong localization of GFP-stalk to microtubules in the presence of MBP-MESP-1 (Figure 3, D and E), showing that MESP-1 is sufficient to enable KLP-18 microtubule binding. Next, we analyzed GFP-ΔC-stalk, which contains the N-terminal MESP-1 binding region (Figure 3A) but lacks the C-terminal microtubule binding domain (Figure 1B). We did not see any enrichment of GFP-ΔC-stalk onto microtubules with or without MBP-MESP-1, showing that MESP-1 alone is not sufficient to target KLP-18 to microtubules and that both the C-terminal microtubule binding domain and MESP-1 are required. Finally, we tested whether the substitution mutations present in klp-18ts affect the microtubule binding ability of the stalk. We quantified an approximately twofold decrease in microtubule-associated GFP-KLP-18ts with MBP-MESP-1 compared with wild-type GFP-stalk with MBP-MESP-1, suggesting that the in vivo phenotype that we observe in the klp-18ts mutant is a result of decreased microtubule binding ability of the C-terminal domain.
KLP-18 activity is essential to maintain spindle bipolarity
Given the importance of KLP-18 in spindle assembly, we next asked whether KLP-18 is also necessary to maintain spindle bipolarity. In human oocytes, the acentrosomal meiotic divisions proceed through an extended bipolar stage that can become unstable, and this instability is correlated with errors in chromosome segregation that result in aneuploidy (Holubcová et al., 2015). Therefore, it is important to understand the molecular mechanisms that maintain acentrosomal spindle stability.
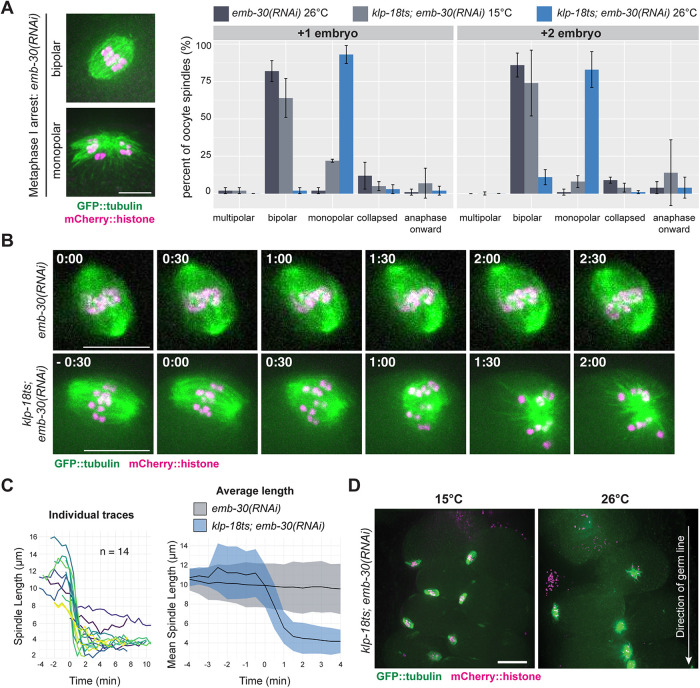
Kinesin-5 is essential for acentrosomal spindle maintenance in mouse and Drosophila oocytes (Mailhes et al., 2004; Duncan et al., 2012; Costa and Ohkura, 2019), but whether kinesin-12 can similarly maintain spindle bipolarity has not been tested in oocytes of any system. The rapid klp-18ts temperature-sensitive mutant allowed us to address this question by inactivating KLP-18 function after spindles have already formed. To this end, we induced metaphase I arrest by depleting anaphase-promoting complex (APC) component EMB-30 using RNA interference (RNAi) (Furuta et al., 2000). Because the C. elegans germ line is organized in a production-line manner, oocytes continue to be fertilized despite this depletion, and each forms a spindle that arrests at metaphase I; this leads to a buildup of bipolar spindles in the germ line (with the most recently fertilized oocyte containing the spindle that was most recently formed). In klp-18ts; emb-30(RNAi) worms at the permissive temperature (15°C) and control emb-30(RNAi) worms at 26°C, at least 60% of the two most recently arrested oocyte spindles were bipolar (these embryos are denoted +1 and +2; Figure 4A). In contrast, when klp-18ts; emb-30(RNAi) worms were shifted to the restrictive temperature (26°C), the majority of oocyte spindles were monopolar (93% [±6%] in +1 and 83% [±12%] in +2 embryos). Moreover, we also observed monopolar spindles at positions beyond the +2 position in the germ line (Figure 4D). Under the conditions of our temperature shift, spindles in the +1 position may have formed after KLP-18 was inactivated, but spindles in oocytes that had been arrested longer should have established bipolarity before KLP-18 inactivation. These results suggest that KLP-18 not only is required to sort microtubule minus ends outward during spindle assembly but is also required to maintain spindle bipolarity.
FIGURE 4:
KLP-18 activity is essential to maintain spindle bipolarity. (A) Spindle morphology in the +1 and +2 embryos of metaphase I–arrested control emb-30(RNAi) (dark gray) or klp-18ts; emb-30(RNAi) worms expressing GFP::tubulin and mCherry::histone. Spindle morphology in the klp-18ts strain was quantified at permissive (15°C, light gray) or restrictive (26°C, blue) temperature; example images of the categories are shown in Supplemental Figure S3A. Bars represent mean percentage ± SD; example images shown on the left. For all conditions n = 3 experiments. Scale bar = 5 μm. (B) Metaphase I–arrested spindles filmed in oocytes dissected from control emb-30(RNAi) (top) and klp-18ts; emb-30(RNAi) (bottom) worms expressing GFP::tubulin and mCherry::histone; the klp-18ts spindle shortens and then forms a monopole while the control spindle maintains bipolarity. Timestamps represent minutes; for klp-18ts t = 0 was set as first frame after shortening began; for the control t = 0 was the first frame acquired because there was no observable shortening throughout the time lapse (see Supplemental Video S1). Scale bars = 10 μm. (C) Spindle length measurements for individual spindles are shown on left. Average ± SD for control emb-30(RNAi) (n = 7, gray) and klp-18ts; emb-30(RNAi) (n = 14, blue) are shown on the right; average is the black line, and SD is shown as shading. For klp-18ts; emb-30(RNAi), time = 0 min was set as first frame after shortening began; for control emb-30(RNAi), time = 0 was set as first frame acquired. Scale bar = 10 μm. (D) Representative images of klp-18(or447ts); emb-30(RNAi) germ lines at 15° and 26°C. Arrow indicates direction of germ line; the most recently arrested spindle is at the top of each image. Scale bar = 15 μm.
Next, we used live imaging to confirm these results and to visualize the dynamics of spindle reorganization. Upon dissecting and mounting klp-18ts; emb-30(RNAi) embryos at room temperature (23–25°C), previously assembled bipolar spindles shortened and then formed a monopole within 2 min (Figure 4, B and C, and Supplemental Video S1). In contrast, bipolar spindles in control emb-30(RNAi) embryos did not show any shortening or collapse (Figure 4, B and C, and Supplemental Video S2). This result confirms that KLP-18 activity is essential to stabilize spindles after bipolarity is achieved and also demonstrates the existence of an inward force that drives spindle collapse when KLP-18 is inactivated. Thus, KLP-18 could maintain spindle bipolarity either by providing outward force through its own motor activity or by antagonizing this inward force in another way. We again confirmed persistent KLP-18 and MESP-1 localization to the monopolar spindle in temperature-shifted, metaphase-arrested oocytes (Supplemental Figure S3E), suggesting that KLP-18 remains spindle associated but is inactive. Taken together, these results show that KLP-18 activity is essential to maintain a bipolar spindle and that this activity relies on the KLP-18 C-terminal microtubule binding domain.
Movie S1.
Metaphase I-arrested oocyte spindle shortens and then forms a monopole upon temperature shift in the klp-18(or447ts) mutant. Representative movie of spindle reorganization in the klp-18 (or447ts) mutant upon temperature shift; corresponds to Figure 4B. Chromosomes (mCherry::histone, magenta) and microtubules (GFP::tubulin, green) visualized in an emb-30(RNAi) klp-18(or447ts) oocyte dissected then shifted to the restrictive temperature. Scale bar = 10μm.
Movie S2.
Control Metaphase I-arrested oocyte spindle maintains bipolarity. Representative movie of a Metaphase I-arrested control spindle; corresponds to Figure 4B. Chromosomes (mCherry::histone, magenta) and microtubules (GFP::tubulin, green) visualized in an emb-30(RNAi) oocyte dissected then shifted to the restrictive temperature. Scale bar = 10μm.
DISCUSSION
This work is the first biochemical characterization of kinesin-12/KLP-18 and MESP-1 and provides the first mechanistic dissection of motor activity in a system in which kinesin-12 is naturally the dominant microtubule sorting motor. We propose a model wherein KLP-18 exists in a folded autoinhibited state that is alleviated through MESP-1 binding; MESP-1 binds to a region overlapping with the hinge and disrupts the folding of the stalk to activate C-terminal microtubule binding (Figure 5B). This could allow KLP-18 to bind one microtubule with its motor domain and another at its C-terminus, potentially enabling it to cross-link and slide microtubules within the spindle. In addition to unhinging KLP-18, we hypothesize that MESP-1 also promotes the initial targeting of the complex to spindle microtubules, as evidenced by its ability to bind microtubules in vitro and its localization to spindles in vivo in the klp-18ts and klp-18ΔC-stalk mutants (conditions in which the C-terminal binding site is inactivated or deleted). However, MESP-1 was not sufficient to localize GFP-KLP-18ΔC-stalk to microtubules in vitro, suggesting that multiple associations between the KLP-18/MESP-1 complex and microtubules may be required for stable spindle targeting in vivo. Consistent with this view, our previous studies showed that a KLP-18 mutant with the motor domain deleted, klp-18(ok2519), abolished both KLP-18 and MESP-1 localization to the monopolar spindle (Wolff et al., 2016), showing that the motor domain is essential for spindle localization.
After the spindle forms, KLP-18 is unable to maintain spindle bipolarity if the C-terminal microtubule binding domain is disrupted, even if the complex is still associated with microtubules (Figure 5C). This is consistent with the possibility that KLP-18 generates an outward force on spindle microtubules that is required to maintain bipolarity. Although we were not able to directly measure the force generated by this motor in vitro, the lack of microtubule sorting in vivo, evidenced by the monopolar spindle phenotype observed in three klp-18 mutants, suggests that a direct stalk–microtubule interaction facilitates force generation during acentrosomal spindle assembly and during metaphase to maintain spindle length. However, an alternative possibility is that KLP-18 may not itself provide an outward force and instead may antagonize an inward force. Our experiments demonstrated that an inward force exists, because the metaphase spindle rapidly shortened and then collapsed upon KLP-18 inhibition. Moreover, we recently found that dynein provides an inward force on the oocyte spindle, suggesting that it could antagonize KLP-18 (Cavin-Meza et al., 2022). Future biophysical studies will be valuable to distinguish between these models.
MESP-1 is predicted to be a heavily disordered protein and was difficult to purify without the stabilizing MBP tag (Supplemental Figure S1, B and C). This suggests that MESP-1 may be disordered and inactive before it associates with KLP-18 in vivo (as depicted in our model; Figure 5B), which may explain why MESP-1 cannot localize to spindle microtubules when KLP-18 is depleted (Wolff et al., 2016). Our previous work also implicated MESP-1 as a functional orthologue of TPX2 in C. elegans (Wolff et al., 2016), which in combination with our current findings stimulates additional hypotheses that could be tested in the future. First, it is possible that once MESP-1 targets KLP-18 to microtubules, MESP-1 may regulate KLP-18 motor activity through persistent association to both the microtubule and the motor (Figure 5B), similar to TPX2 regulation of kinesin-12/Kif15 (Drechsler and McAinsh, 2016; Mann et al., 2017) and kinesin-5/Eg5 (Ma et al., 2011) in mammalian systems. In addition, TPX2 has been shown to inhibit Kif15 motility and help Kif15 associate more tightly to microtubules in vitro (Drechsler et al., 2014; Mann et al., 2017; McHugh et al., 2018). It remains to be seen whether MESP-1 regulates KLP-18 in a similar way, though this type of regulation could enable KLP-18 to counteract inward forces in the spindle, should this be the mechanism by which it acts to maintain spindle bipolarity. Finally, the relatively weak interaction between KLP-18 N-stalk and MESP-1 (Figure 3A) that appears to be stimulated by the addition of microtubules (Figure 3, D and E) suggests that the interaction between MESP-1 and KLP-18 may require microtubules as has been shown for Xenopus and mammalian TPX2 and kinesin-12 (Wittmann et al., 1998; Drechsler et al., 2014). Although it is still not clear what regulates the association between KLP-18 and MESP-1 in vivo, if this interaction requires microtubules, it is possible that microtubule nucleation after nuclear envelope breakdown may trigger the formation of the active KLP-18/MESP-1 complex to temporally synchronize microtubule sorting by KLP-18 with other activities essential for proper spindle assembly (Mullen et al., 2019).
In mammalian somatic cells, kinesin-5 generates an outward force on microtubules to achieve spindle bipolarity (Sawin et al., 1992; Blangy et al., 1995; Kapoor et al., 2000), and when kinesin-5 function is altered, kinesin-12 can take over this role (Tanenbaum et al., 2009; Vanneste et al., 2009; Raaijmakers et al., 2012; Sturgill and Ohi, 2013; Sturgill et al., 2016). The molecular basis for this activity has been characterized in vitro and in cell culture (Drechsler et al., 2014; Drechsler and McAinsh, 2016; Sturgill et al., 2014; Mann et al., 2017; Reinemann et al., 2017; McHugh et al., 2018; Dumas et al., 2019). Our work confirms that certain properties of kinesin-12s are shared across species: the presence of discrete coiled-coil domains in the C-terminal stalk, the requirement of a C-terminal nonmotor microtubule binding site, self-inhibition of nonmotor microtubule binding, and regulation by a TPX2-like adaptor protein. Further characterization of the mechanism of KLP-18 self-inhibition, activation by MESP-1, and force generation on microtubules will be essential for fully understanding the role of KLP-18 in meiosis and to put KLP-18 in proper context with other essential motors, particularly mammalian kinesin-12 and kinesin-5.
Previously, we and others have proposed that KLP-18 acts to organize microtubules and provide outward force on the spindle during anaphase (Laband et al., 2017; Mullen and Wignall, 2017); because KLP-18 sorts microtubule minus ends outward during spindle assembly and is required for maintaining spindle length in metaphase, it is possible that it also exerts outward force to drive microtubule sliding during anaphase B. We recently found that when KLP-18 and dynein are both depleted, anaphase B can still occur (Cavin-Meza et al., 2022), demonstrating that this motor is not essential for outward sliding. However, it is possible that KLP-18 does play a role during normal anaphase in combination with other mechanisms; this remains to be directly tested. Understanding the role of KLP-18 may help to resolve different models of chromosome segregation proposed in C. elegans oocytes (reviewed in Severson et al., 2016; Mullen et al., 2019) (Dumont et al., 2010; Muscat et al., 2015; McNally et al., 2016; Davis-Roca et al., 2017, 2018; Laband et al., 2017; Mullen and Wignall, 2017; Redemann et al., 2018; Danlasky et al., 2020). Further characterization of KLP-18’s role in spindle assembly, spindle maintenance, and chromosome segregation will be essential to fully understand C. elegans oocyte meiosis.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
C. elegans strains
Strains and their sources were as follows: OD868: ltSi220[pOD1249/pSW077; Pmex-5::GFP::tbb-2::operon_linker::mCherry::his-11; cb-unc-119(+)] I (Wang et al., 2015);
HR1160: klp-18(or447ts), dpy-20 IV (gift from Bruce Bowerman, University of Oregon);
SMW29 (Δhinge): klp-18(wig2) IV; unc-119(ed3) III; ltIs37[pAA64; pie-1::mCherry::his-58; unc-119(+)] IV; ltIs25[pAZ132; pie-1::GFP::tba-2; unc-119 (+)] IV (transgenes were from strain OD57; gift from Arshad Desai, University of California, San Diego);
SMW34 (ΔC-stalk): klp-18(wig3), ieSi38 [sun-1p::TIR1::mRuby::sun-1 3′UTR + Cbr-unc-119(+)] IV; unc-119(ed3) III; mMaple3::tba-1 I; and
SMW36: HR1160 x OD868 (klp-18(or447ts), dpy-20 IV; ItSi220[pOD1249/pSW077; Pmex-5::GFP::tbb-2::operon_linker::mCherry::his-11; cb-unc-119(+)] I).
Generation of Δhinge and ΔC-stalk klp-18 mutant strains
The endogenous klp-18 gene was edited using a CRISPR-Cas9–based approach similar to what has previously been described (Arribere et al., 2014; Paix et al., 2015; Mullen and Wignall, 2017; Zdraljevic et al., 2019). Genomic DNA was cut through CRISPR RNA (crRNA) targeting to two cut sites: one at the 5′ end of the desired mutation and one at the 3′ end. A single-strand DNA (ssDNA) repair template homologous to sequences upstream and downstream of the two cut sites was used to bridge the excised DNA. For ΔC-stalk (SMW34), residues 635–932 of KLP-18 were deleted. For Δhinge, residues 557–770 were deleted. The dpy-10 coinjection marker was used to screen successful genome editing events. See Table 1 for sequence and concentration of all oligonucleotides used for CRISPR-Cas9–directed cutting and homology-directed repair. First, 13.6 µM Alt-R CRISPR-Cas9 tracrRNA (IDT), dpy-10 crRNA, and both gene-specific crRNA guides were incubated at 95°C for 5 min and then 10°C for 5 min in a thermocycler. Half of this mixture was removed and placed into a cold PCR tube on ice and then 27 µM recombinant Cas9 (IDT) was added and incubated at room temperature for 5 min. The dpy-10 ssDNA oligo (IDT) and gene-specific ssDNA ultramer (IDT) repair templates were then added and nuclease free water was added to a final injection mix volume of 5 µl. The injection mix was loaded into a capillary tube through a mouth pipette and then injected into the germ line of young adult worms. Injected parental worms (POs) were recovered in M9 (22 mM KH2PO4, 22 mM Na2HPO4, 85 mM NaCl, 1 mM MgSO4) and then pooled onto a single plate and allowed to recover at 15°C overnight. The genetic background of each mutant can be found above in C. elegans strains. The next day, injected POs were singled out to individual plates, grown at 20°C, and allowed to lay eggs. The resulting F1 progeny were screened for rollers or dumpys indicating dpy-10/+ or dpy-10/dpy-10, respectively (Arribere et al., 2014). Individual animals from “jackpot” broods (plates with many dumpys and rollers) were isolated, allowed to produce progeny, and then screened with single-worm PCR. Mutations in both strains were confirmed through Sanger sequencing. The primers used to screen and sequence mutants are shown in Table 1. Homozygous Δhinge and ΔC-stalk mutants have high embryonic lethality (Figure 2A); therefore these strains were propagated by picking individual worms to plates, letting them lay eggs, and then PCR screening each adult to verify the genotype. Adult progeny of animals heterozygous for the mutations were used for experiments, with the assumption that ¼ will be wild type (+/+), ½ will be heterozygous (Δ/+), and ¼ will be homozygous mutant (Δ/Δ). Although homozygous mutant worms lack the ability to produce wild-type KLP-18, they are able to successfully complete the embryonic divisions and reach adulthood, presumably due to a maternal load of functional KLP-18 from their heterozygous mothers. However, they then produce embryos that contain only mutant KLP-18, and a high percentage of these eggs do not hatch (Figure 2A).
TABLE 1:
Editing oligomers and genotyping primers.
| Sequence (5′ to 3′) | Working concentration (µM) | |
|---|---|---|
| CRISPR-Cas9 editing DNA oligomers | ||
| dpy-10 crRNA | GCTACCATAGGCACCACGAG | 4 |
| klp-18(Δhinge) crRNA 5′ | AACGTCAAAGTCAATCGAGA | 4.8 |
| klp-18(Δhinge) crRNA 3′ | AAAAGGATATTGACCAATGC | 4.8 |
| klp-18(ΔC-stalk) crRNA 5′ | CCATGATTGAAGATCTCGAA | 4.8 |
| klp-18(ΔC-stalk) crRNA 3′ | CAGTCAGACCACAAAGCCAT | 4.8 |
| dpy-10 ssDNA repair template | CACTTGAACTTCAATACGGCAAGATGAGAATGACTGGAAACCGTACCGCATGCGGTGCCTATGGTAGCGGAGCTTCACATGGCTTCAGACCAACAGCCTAT | 1.34 |
| klp-18(Δhinge) ssDNA ultramer repair template | TTGAAAGACAAGCACGAAGAGGCAACGTCAAAGTCAATCGAGACGAAACGAAAGAAGAAATTGGAAATTCAATCAATGCAACTCTCGCTGGACAATGCGAAC | 4 |
| klp-18(ΔC-stalk) ssDNA ultramer repair template | TTTAATTTTTAATATTACTCAGTTTTTACGGTTCTTGTTTCAGAACAACGTAAGACAAATCGTCTTAATTTATTTCTGTAAATATTAGCCGAATACTCACT | 4 |
| Genotyping primers | ||
| klp-18 FWD | GAAGCTCAGTTGTGAACTGAGC | 1 |
| klp-18(Δhinge) REV | CGCCATTTCATTGAGCATCG | 1 |
Embryonic lethality assay
Embryonic lethality scoring was performed as previously described (Mullen and Wignall, 2017) with slight modifications: L4 progeny of unbalanced klp-18 mutant strains (SMW29 or SMW34) were isolated onto individual nematode growth medium (NGM) plates and allowed to lay eggs at 20°C for 24 h before being transferred to a fresh NGM plate. The eggs were allowed to hatch for 24 h at 20°C, and then the total progeny, unhatched eggs and hatched worms, were scored. This process was repeated two times per isolated worm for a total of three scored plates per worm. Following the third day of egg laying, parent worms were screened for genotype (homozygote background, klp-18 heterozygote mutant, or klp-18 homozygote mutant) by PCR.
Fixed imaging
For quantification of spindle morphology, intact worms were fixed in EtOH as previously described (Wolff et al., 2022): ∼30–45 worms were picked into a 15°C drop of M9, the drop was dried with Whatman paper, and 10 µl of 100% EtOH was added directly to worms. The EtOH was allowed to dry completely, and another drop of 100% EtOH was added, and this was repeated for a total of three times. A 1:1 mixture of Vectashield:M9 was added to the worms, and then a coverslip was added and sealed with nail polish. Slides were stored at 4°C until imaging. Slides were visualized on a DeltaVision Core microscope (GE Biosciences), and spindle morphology was quantified by eye with a 40× objective or by taking a snapshot of the spindle with a 100× objective. Oocyte spindles in embryos in the +1 or +2 positions were quantified as indicated on the figure.
Immunofluorescence was performed as previously described (Wolff et al., 2022). Oocytes were dissected into drops of M9, snap-frozen in liquid N2, freeze cracked, and fixed in –20°C MeOH for 35 min. Slides were washed twice with phosphate-buffered saline (PBS), blocked with Abdil (1× PBS, 4% bovine serum albumin, 0.1% Triton X-100, 0.02% sodium azide) for 30 min at room temperature, and then primary antibody was applied overnight at 4°C. The next day slides were washed three times with PBS + 0.1% Triton X-100 (PBST) at room temperature, incubated with secondary antibody for 2 h at room temperature, washed three times with PBST, incubated with Hoechst for 15 min at room temperature, washed twice with PBST, then mounted in mounting media (90% glycerol, 20 mM Tris, pH 8.8, 0.5% p-phenylenediamine), and sealed with nail polish. Primary antibodies and concentrations used: rat anti–KLP-18 (1:500; gift of Olaf Bossinger, RWTH Aachen University [Segbert et al., 2003]), rabbit anti–MESP-1 (1:3000 [Wolff et al., 2016]), rabbit ASPM-1 (1:5000 [Wignall and Villeneuve, 2009]). Secondary antibodies and concentrations used: goat anti-rat (1:500; Invitrogen), goat anti-rabbit (1:500; Invitrogen). Slides were imaged using a DeltaVision Core microscope with a 100× objective (NA = 1.40). All image acquisition and processing was performed using softWoRx software (GE Biosciences). Image stacks were acquired with 0.2 µm z-steps. All immunofluorescence images are full maximum-intensity projections of the entire spindle structure and are not deconvolved unless indicated in the figure legend.
Microscopy was performed at the Biological Imaging Facility at Northwestern University.
Protein domain analysis
KLP-18 coiled-coil domains were identified using Paircoil2 coiled-coil prediction software (http://cb.csail.mit.edu/cb/paircoil2/) (McDonnell et al., 2006). Probability scores were calculated with a cutoff of 0.5. MESP-1 disordered regions predicted using PONDR (http://www.pondr.com/) (Xue et al., 2010).
Protein expression and purification
Purification results, affinity tag, and expression system for each protein can be found in Supplemental Figure S1A. C. elegans cDNA was amplified from extracted mRNA from wild-type worms using the iScript Select cDNA Synthesis kit (Bio-Rad). KLP-18 cDNA was amplified from whole-worm cDNA with gene-specific primers and Q5 DNA polymerase and then used for cloning all KLP-18 pET expression constructs. Δhinge was created via site-directed mutagenesis (NEB) from the stalk construct. MESP-1 cDNA was amplified from a GST-MESP-1 construct (Wolff et al., 2016) and inserted into an MBP vector (gift of Laura Lackner, Northwestern University, and Marijn Ford, University of Pittsburgh). pET His6 GFP TEV LIC cloning vector (1GFP [Pédelacq et al., 2006]) was a gift from Scott Gradia, University of California, Berkeley (Addgene plasmid #29663; http://n2t.net/addgene:29663; RRID: Addgene_29663). KLP-18 cDNA was inserted into the GFP construct via Gibson Assembly to create the GFP-stalk construct, and the GFP-KLP-18ts and GFP-ΔC-stalk were created via site-directed mutagenesis (NEB) from this GFP-stalk construct. KLP-18 cDNA was inserted into the MBP construct via restriction digest to make MBP-stalk.
All proteins were purified using the same protocol and the same buffers. All expression vectors were transformed into BL21 DE3 Escherichia coli cells and grown at 37°C to an OD of ∼0.6. Cells were induced with 0.1 mM Isopropyl-β-d-thiogalactoside (IPTG) and grown for various expression times and at various temperatures; see Supplemental Figure S1A for growth conditions for individual proteins. Growth cultures were spun at 4700 rpm and resuspended in lysis buffer (80 mM PIPES, pH 6.8, 2 mM MgCl2, 1 mM ethyleneglycol-bis(β-aminoethyl)-N,N,N′,N′-tetraacetic acid (EGTA), 250 mM NaCl, 5–10% glycerol, 0.02% Tween, leupeptin, aprotinin, pepstatin, 2 mM imidazole), and then cells were lysed with 1 mg/ml lysozyme (incubated for 20 min at 4°C) and with sonication. Lysate was cleared by centrifugation for 45 min at 11,900 rpm in a Ti50.2 rotor. Ni-NTA resin was equilibrated with lysis buffer and then added to cleared lysate and incubated at 4°C for 1–2 h. Slurry was applied to a plastic column and washed with 30 ml of lysis buffer + 20 mM imidazole. Bound protein was eluted with 5–10 ml of lysis buffer + 500 mM imidazole. Ni-NTA elution was either kept at 4°C overnight before application or applied directly to a HiLoad 26/600 Superdex 200 gel filtration column run on the Aktaprime FPLC system (GE Biosciences) and equilibrated with lysis buffer. Eluted fractions from individual peaks were tested by SDS–PAGE, and fractions containing pure protein of interest were combined, concentrated, frozen by dripping into liquid N2, and stored at –80°C.
Microtubule cosedimentation assay
Purified porcine brain tubulin (100 µM; self-purified, gift of Sarah Rice, Northwestern University) was incubated with 1 mM dithiothreitol (DTT) and 1 mM GTP in BRB80 (80 mM PIPES, pH 6.8, 2 mM MgCl2, 1 mM EGTA) on ice for 5 min and then spun at 80,000 rpm for 10 min at 4°C. The supernatant was removed and incubated at 37°C for 1 h. Taxol was added stepwise to the following final concentrations at 37°C: 1 µM Taxol, then incubated for 10 min; 10 µM Taxol, then incubated for 10 min; and 100 µM Taxol, then incubated for 15 min. Polymerized microtubules were diluted 1:1 in BRB80 + 50 µM Taxol and kept at room temperature until use. For subtilisin treatment, 1 mg/ml subtilisin was added to 20 µM polymerized tubulin and incubated overnight at 37°C. The reaction was quenched with 2 mM final concentration phenylmethylsulfonyl fluoride (PMSF) followed by incubation at 37°C for 1 h and then spun over a cushion (200 µl BRB80, 60% glycerol, 20 µM Taxol) at 80,000 rpm for 20 min. The cushion and pellet were washed in 50 µl of BRB80 + 20 µM Taxol and then resuspended in 10 µl of BRB80 + 50 µM Taxol.
Proteins of interest were thawed from –80°C storage and precleared by spinning at 80,000 rpm for 10 min at 25°C. Soluble protein in the supernatant was removed and added to 5 µM microtubules quickly to maintain protein solubility. The reactions were assembled in BRB80 + 20 µM Taxol with equal salt, detergent, and glycerol concentrations based on the final purification buffers (see the Protein expression and purification section for details) to a final volume of 25 µl. The exact protein concentrations for each experiment can be found in the Figure quantification section. Reactions were incubated at room temperature for 30 min and then spun through a 100 µl BRB80 + 40% glycerol + 20 µM Taxol cushion at 90,000 rpm for 15 min at 25°C. From the very top of the solution 25 µl was removed and added to 2× SDS Laemmli sample buffer (4% SDS [Sodium dodecyl sulfate], 20% glycerol, 120 mM Tris-HCl pH 6.8, 0.02% bromophenol blue) to make the “supernatant” sample. The cushion was washed with BRB80 + 20 µM Taxol and removed, and then the pellet was washed with BRB80 + 20 µM Taxol. The pellet was resuspended with 25 µl of cold BRB80 + 10 mM CaCl2 and then added to 2× SDS Laemmli sample buffer to make the “pellet” sample. All spins were performed in a TLA120.2 rotor (Beckman).
Supernatant and pellet samples were probed by Western blot using the following antibody concentrations: 1:5000 anti-6XHis-HRP (horseradish peroxidase) (Abcam), 1:5000 mouse anti-tubulin (Invitrogen), 1:5000 anti-mouse HRP (Invitrogen). See Figure quantification section for Western blot quantification details.
Microtubule bundling assay
Fluorescent microtubules were polymerized by incubating 20 µM porcine brain tubulin and 2 µM TMR-tubulin (Cytoskeleton) with 1 mM DTT and 1 mM guanosine triphosphate (GTP) in BRB80 (80 mM PIPES, pH 6.8, 2 mM MgCl2, 1 mM EGTA) on ice for 5 min and incubated at 37°C for 2 min. Taxol was added stepwise to the following final concentrations at 37°C: 0.2 µM Taxol, then incubated for 10 min; 2 µM Taxol, then incubated for 10 min; and 20 µM Taxol, then incubated for 10 min. Polymerized microtubules were kept at room temperature until use.
Proteins of interest were thawed from –80°C storage and precleared by spinning at 80,000 rpm for 10 min at 25°C. Protein concentrations were measured by Bradford assay and then added to 25 µl reactions with 200 nM TMR-microtubules in BRB80 + 20 µM Taxol with equal salt, detergent, and glycerol concentrations based on the final purification buffers (see the Protein expression and purification section for details). Reactions were incubated at room temperature for 30 min and then fixed with 1% glutaraldehyde and incubated for an additional 3 min. Reactions were then squashed onto a poly-l-lysine slide, sealed with nail polish, and imaged on a spinning-disk confocal microscope (see Time-lapse imaging for microscope details). For exact protein concentrations and quantification details, see the Figure quantification section.
GFP-tagged protein recruitment to microtubules
Fluorescent microtubules were polymerized as previously described (Reinemann et al., 2017). First, GMPCPP seeds were nucleated by incubating 13 µl of PEM104 (104 mM PIPES, pH 6.9, 1.3 mM EGTA, 6.3 mM MgCl2), 2.2 µl of 10 mM GMPCPP, 2.2 µl of dimethyl sulfoxide (DMSO), 4 µl of 13 mg/ml homemade porcine brain tubulin (gift of Sarah Rice), and 4 µl of 10 mg/ml TMR-tubulin (Cytoskeleton) at 37°C for 40 min. A microtubule elongation mixture of 13 µl of PEM104, 2.2 µl of 10 mM GTP, 2.2 µl of DMSO, 2 µl of 13 mg/ml homemade porcine brain tubulin, and 1.5 µl of 1 mg/ml TMR-tubulin (Cytoskeleton) was incubated at 37°C for 1 min, 1.5 µl of the GMPCPP seed mixture was added, and then the reaction was incubated at 37°C for 40 min. Microtubules were stabilized by adding 2 µl of stabilization buffer (38.6 µl of BRB80, 0.5 µl of 100 mM GTP, and 6 µl of 2 mM Taxol dissolved in DMSO) and then kept at room temperature until use. Microtubules were used within 1 wk of polymerization. Proteins of interest were thawed from –80°C storage then added to 25 µl reactions with 5 µl of 1:20 diluted TMR-microtubules in BRB80 + 20 µM Taxol with equal salt, detergent, and glycerol concentrations based on the final purification buffers (see the Protein expression and purification section for details). Reactions were incubated, fixed, and imaged exactly as described above in the Microtubule bundling assay section.
MBP pull down
N-stalk/C-stalk (1µM) and MBP/MBP-MESP-1 (2 µM) were added to a 100 µl reaction in BRB80 (80 mM PIPES, pH 6.8, 2 mM MgCl2, 1 mM EGTA) with equal salt, detergent, and glycerol concentrations based on the final purification buffers (see the Protein expression and purification section for details). An equal volume of protein was added to 200 µl of 1× Laemmli sample buffer for the “input” gel samples. The reactions were incubated at 4°C overnight and then equilibrated amylose resin (New England Biolabs) was added to pull out MBP-tagged proteins. Amylose resin was incubated with reactions for 1 h at 4°C and then the resin was washed four times with 900 µl of BRB80. Final wash (300 µl) was saved for acetone precipitation to make “wash” samples. To elute bound proteins from amylose resin, 900 µl of BRB80 + 50 mM NaCl + 10 mM maltose was added to beads and incubated for 20 min at 4°C. Eluate (300 µl) was removed for acetone precipitation to make “elution” gel samples.
Wash and elution samples were concentrated via acetone precipitation as follows: 1.2 ml of –20°C acetone was added to 300 µl of wash or elution samples, vortexed, and incubated at –20°C overnight. The next day samples were spun at 16,000 × g for 10 min at room temperature and incubated at 50–55°C for ∼20 min until the acetone was completely evaporated and the pellets were dry. Pellet was resuspended in 40 µl of 1× Laemmli sample buffer and boiled for 10 min at 95°C to make gel samples. Samples were run on a Western blot and probed with 1:5000 anti–6XHis-HRP to visualize His-tagged KLP-18 truncations. To visualize MBP or MBP-MESP-1 samples were run on an SDS–PAGE gel and stained with Coomassie. The input lane represents 1% of protein added to the reaction, and wash/elution lanes represent 12.5% of all protein in those samples.
RNAi
RNAi was performed as previously described (Wolff et al., 2022). Briefly, individual RNAi clones picked from an RNAi feeding library (Fraser et al., 2000; Kamath et al., 2003) were used to inoculate Luria broth (LB) plus ampicillin (100 μg/ml) and grown overnight at 37°C. These cultures were used to seed NGM/ampicillin (100 μg/ml)/1 mM IPTG plates, and the plates were left overnight at room temperature. Synchronized L1 worms were plated on induced plates and grown at 15°C for 5–6 d until they became gravid adults.
Time-lapse imaging
Worms were prepared for two-color live imaging as previously described (Laband et al., 2018). klp-18ts; emb-30(RNAi) and control emb-30(RNAi) adult worms were kept at 15°C and room temperature before dissection, respectively. Oocytes were dissected out of worms in a drop of room temperature Meiosis Medium (final concentrations in ddH2O: 60% Leibovitz L-15, 20% heat-inactivated FBS, 25 mM HEPES, pH 7.5, 0.5 mg/ml inulin), and then a coverslip was added. Oocytes that settled onto the coverslip were filmed. All live imaging was performed at ambient temperature (∼23–25°C).
Imaging was performed using a spinning-disk confocal microscope with a 63× HC PL APO 1.40 NA objective lens. A spinning-disk confocal unit (CSU-X1; Yokogawa Electric Corporation) attached to an inverted microscope (Leica DMI6000 SD) and a Spectral Applied Imaging laser merge ILE3030 and a back-thinned electron-multiplying charge-coupled device camera (Evolve 521 Delta) were used for image acquisition. The microscope and attached devices were controlled using MetaMorph Image Series Environment software (Molecular Devices). Frames were acquired every 30 s at 2 µm z-stack intervals.
Hydrodynamic analysis
A purification of MBP-stalk was split and diluted into BRB80 + 300 mM NaCl (high salt) or BRB80 + 20 mM NaCl (low salt). The high-salt and low-salt dilutions were then applied to a HiLoad 26/600 Superdex 200 column equilibrated with appropriate buffer. Identical fractions were collected for each dilution and probed by Western blot using rabbit anti–KLP-18 at 1:5000 and anti–rabbit-HRP at 1:5000. See Figure quantification section for further details.
Western blots
For klp-18(or447ts) whole-worm Western blots, control (OD868) or klp-18(or447ts) (SMW36) plates were shifted to 26°C for 1 h. Fifty to one hundred worms were picked from a shifted plate to a prewarmed unseeded plate for 5 min to avoid transfer of bacteria and then washed off the plate with room temperature M9. Worms were pelleted by spinning at 800 × g for 1 min, supernatant was removed, and an equal volume of 2× SDS Laemmli sample buffer was added. Gel samples were boiled for 10 min at 95°C, briefly vortexed, and then boiled for an additional 10 min at 95°C. A volume of sample corresponding to 50 worms was loaded onto the gel. Antibodies used: rat anti–KLP-18 1:1000, anti–rat-HRP 1:5000 (Invitrogen), mouse anti-tubulin 1:5000, and anti–mouse-HRP 1:5000 (Invitrogen).
Samples were run on 8–12% SDS–PAGE gel and transferred to a nitrocellulose membrane using a Trans-Blot Turbo Transfer System (BioRad). Membrane was blocked in 5% milk in TBST (TBS [20 mM Tris Base pH 7.6, 150 mM NaCl] + 0.1% Tween) blocking solution, incubated with primary antibody in blocking solution at room temperature for 1 h or at 4°C overnight, washed in TBST, incubated in secondary antibody at room temperature for 1 h, washed in TBST, incubated with Clarity Western ECL substrate (BioRad) for 2 min, and then imaged.
Figure quantification
Figure 1B. Western blots were developed on film and then scanned, and the ImageJ “analyze gels” function was used to quantify band intensity in each lane (Schindelin et al., 2012; Rueden et al., 2017). For each reaction, the total band intensity was calculated by adding the “supernatant” and “pellet” band intensities, and “shift to pellet (%)” was calculated by dividing the “pellet” band intensity by the total band intensity for the reaction. The mean ± SD for n = 3 independent experiments is shown.
For each experiment quantified, [tubulin] = 5 µM, [N-stalk] = 1.0–1.26 µM, [C-stalk] = 0.14–1 µM. In the representative blots shown for Figure 1B, left: [N-stalk] = 1.26 µM and [C-stalk] = 0.53 µM. For the subtilisin experiment shown in Figure 1B, right: [C-stalk] = 0.14 µM for all reactions. For the nonsubtilisin experiment: Shift to pellet (%) ± SD for N-stalk (– MT) = 4.5 ± 4.9%, N-stalk (+ MT) = 3.2 ± 3.6%, C-stalk (– MT) = 1.6 ± 2.8%, C-stalk (+ MT) = 52.7 ± 9.7%. For the subtilisin experiment: Shift to pellet (%) ± SD for C-stalk (– MT) = 6.0 ± 6.7, C-stalk (+ MT) = 50.8 ± 7.0%, C-stalk (+ sMT) = 26.7 ± 7.0%.
Figure 1D. Microtubule bundling assays were imaged on a spinning-disk microscope with consistent exposure times across the experiment. Images were quantified in ImageJ: raw images were made into a grayscale composite, and then an automatic threshold using the “Threshold” command was applied. Across an experiment, a threshold was applied to each image individually but with the same automatic thresholding algorithm. After the thresholding was applied, fluorescent particles were selected and the “mean size” was calculated. This measurement is the mean size of all fluorescent particles in an image and represents the degree of microtubule bundling (large bundles will have a larger mean size than individual microtubules). To normalize mean bundle area, the area from experimental images was divided by the mean area of all of the buffer-only control images, making the mean size of unbundled microtubules equal to 1. Each plotted data point is one image’s normalized bundle area.
Two experiments are shown on the graph, and the same result (Δhinge bundles MTs while full-length stalk does not) was shown in six total experiments. In each reaction, [tubulin] = 200 nM. Mean “Normalized bundle area (A.U.)” and n for each condition are buffer alone: 1, n = 62; 1 µM stalk: 1.22, n = 67; 2 µM stalk: 1.59, n = 62; 1 µM Δhinge: 2.63, n = 63; 2 µM Δhinge: 5.27, n = 62. Using a two-sample equal variance Student’s t test, all conditions compared with buffer and Δhinge compared with equal concentrations of stalk showed statistically significant (p < 0.01) increases in normalized bundle area.
Figure 2B. Spindle morphology in control or klp-18(or447ts) worms fixed in EtOH was quantified by eye using a 40× objective or by taking snapshots at 100×. The percentages shown in Figure 2B are calculated by considering only preanaphase oocyte spindles (multipolar, bipolar, monopolar); representative images are shown. However, we did assess all +1 spindles (including anaphase and MII spindles), and that full data set is graphed in Supplemental Figure S3A. For klp-18(or447ts), four experiments were quantified (for 15°C n = 28, 16, 32, 29 preanaphase spindles; for 26°C n = 27, 21, 42, 42 preanaphase spindles); for control 26°C, three experiments were quantified (n = 12, 11, 14 preanaphase spindles). Mean “percent of preanaphase +1 oocytes (%)” ± SD for control 26°C: multipolar = 6 ± 10%, bipolar = 94 ± 10%, monopolar = 0 ± 0%. Mean “percent of preanaphase +1 oocytes (%)” ± SD for klp-18(or447ts) 15°C: multipolar = 10 ± 7%, bipolar = 67 ± 7%, monopolar = 23 ± 10%. Mean “percent of preanaphase +1 oocytes (%)” ± SD for klp-18(or447ts) 26°C: multipolar = 1 ± 1%, bipolar = 4 ± 4%, monopolar = 95% ± 5.
Figure 2, C and D. Fluorescence intensity of KLP-18 or MESP-1 immunofluorescence staining at poles of fixed oocyte spindles. MI or MII bipolar or monopolar spindles were quantified for each condition. Raw undeconvolved images were used for quantification, and each image was acquired at similar exposures and laser power. For the klp-18ΔC-stalk and klp-18Δhinge strains, each slide contained a mix of bipolar and multipolar spindles allowing for an internal control. Every image of a spindle on the same slide was acquired identically. All image analysis was performed in ImageJ (Schindelin et al., 2012; Rueden et al., 2017). First, each pole of the bipolar spindle or single monopole was qualitatively identified in the tubulin channel. A five-slice sum projection was then made containing two z-stacks above and below the center z-slice at the center of the pole for each channel; then a region of interest (ROI) was drawn around the pole in the tubulin channel of the sum projection. The exact ROI was applied to the KLP-18 and MESP-1 channels, and the integrated density (mean pixel intensity × area) was recorded for each channel. To calculate background, the same ROI was moved to an area of the cytoplasm adjacent to each pole and the same intensity measurements were recorded. The background measurements were then subtracted from the pole measurements. For bipolar spindles, the two background-subtracted poles were averaged together. To calculate final fluorescence intensity on spindle poles, the background-subtracted KLP-18 or MESP-1 intensity was divided by the background-subtracted tubulin intensity to normalize for variability in staining or the number of microtubules present on mutant spindles. Each data point represents one spindle. For klp-18ts bipolar n = 21 spindles; klp-18ts monopolar n = 38 spindles; klp-18ΔC-stalk bipolar n = 38 spindles; klp-18ΔC-stalk monopolar n = 18 spindles; klp-18Δhinge bipolar n = 23 spindles; klp-18ΔC-stalk monopolar n = 22 spindles.
Figure 3B. Quantified exactly as described in Figure 1B, except some blots were imaged with an Azure Biosystems digital imager. Concentrations for each experiment quantified: [tubulin] = 5 µM, [MBP-MESP-1] = 1.0–5.77 µM. In the representative blots shown, [MBP-MESP-1] = ∼1.0 µM (nonsubtilisin experiment) and 5.77 µM (subtilisin experiment). Two nonsubtilisin experiments were quantified, and three subtilisin experiments were quantified. Shift to pellet (%) ± SD for MBP-MESP-1: (– MT) = 2.9 ± 4.1%, (+ MT) = 56.1 ± 3.4%. For subtilisin experiments, shift to pellet (%) ± SD for MBP-MESP-1: (– MT) = 0.6 ± 0.1%, (+ MT) = 41.6 ± 10.9%, (+ sMT) = 4.5 ± 6.4%.
Figure 3, D and E. Images were acquired and threshold applied as described for Figure 1D. Exposure times were held constant across the experiment. To quantify GFP localization, fluorescent particles were selected in the microtubule channel and then the mean intensity in the GFP channel was measured within the selection. Therefore, only GFP signal that was overlaid on microtubule bundles was measured. Mean intensity in the tubulin channel was also measured. To normalize to background, the selection was inverted to select the entire image that was not previously selected as microtubules and the mean intensity in both the GFP and red tubulin channel was measured. The mean background intensity was subtracted from mean GFP intensity for each image. “Normalized GFP enrichment” was calculated by dividing background-subtracted GFP signal by background-subtracted tubulin signal to normalize for variability in the amount and size of microtubule bundles. The mean “Normalized GFP enrichment” value for an experiment was calculated and averaged over n = 3 experiments. The mean ± SD for each condition are as follows: GFP + MBP-MESP-1 = 0.14 ± 0.1; GFP-ΔC-stalk = 0.09 ± 0.01; GFP-ΔC-stalk + MBP-MESP-1 = 0.13 ± 0.02; GFP-klp-18ts = 0.24 ± 0.08; GFP-klp-18ts + MBP-MESP-1 = 0.37 ± 0.06; GFP-stalk = 0.10 ± 0.03; GFP-stalk + MBP-MESP-1 = 0.71 ± 0.10. Each reaction contained 250 nM GFP-tagged proteins and 1 µM MBP-MESP-1 (if added as indicated).
Figure 4A. Quantified as in Figure 2B and Supplemental Figure S3A. For all strains and conditions, three experiments were quantified (control 26°C n = 52, 26, 28 +1 spindles, n = 50, 26, 27 +2 spindles; klp-18(or447ts) 15°C n = 42, 81, 52 +1 spindles, n = 38, 77, 55 +2 spindles; klp-18(or447ts) 26°C n = 39, 50, 56 +1 spindles, n = 36, 42, 56 +2 spindles). The mean percent of oocytes (%) ± SD for each condition was calculated. For control emb-30(RNAi) 26°C +1 oocytes: multipolar = 2 ± 2%; bipolar = 82 ± 7%; monopolar = 2 ± 2%; collapsed = 12 ± 9%; anaphase onward = 1 ± 2%. For control emb-30(RNAi) 26°C +2 oocytes: multipolar = 0 ± 0%; bipolar = 86 ± 8%; monopolar = 1 ± 2%; collapsed = 9 ± 2%; anaphase onward = 4 ± 4%. For klp-18(or447ts); emb-30(RNAi) 15°C +1 oocytes: multipolar = 2 ± 2%; bipolar = 64 ± 13%; monopolar = 22 ± 1%; collapsed = 5 ±3%; anaphase onward = 7 ± 1%. For klp-18(or447ts); emb-30(RNAi) 15°C +2 oocytes: multipolar = 0%; bipolar = 74 ± 22%, monopolar = 8 ± 4%; collapsed = 4 ± 3%; anaphase onward = 4 ± 7%. For klp-18(or447ts); emb-30(RNAi) 26°C +1 oocytes: multipolar = 0 ± 0%; bipolar = 2 ± 2%; monopolar = 93 ± 6%; collapsed = 3 ± 3%; anaphase onward = 2 ± 3%. For klp-18(or447ts); emb-30(RNAi) 26°C +2 oocytes: multipolar = 0 ± 0%; bipolar = 11 ± 5%; monopolar = 83 ± 12%; collapsed = 1 ± 1%; anaphase onward = 4 ± 7%.
Figure 4C. To quantify spindle length in time-lapse videos, raw data were loaded in ImageJ and the distance between poles was measured with the line tool: spindle poles were identified in the tubulin channel by a bright circular area of tubulin, and a straight line was drawn between the outer edges of pole tubulin signal to measure the spindle length. This was done for each frame of the video. Because spindle collapse in klp-18ts; emb-30(RNAi) oocytes happened at slightly different times after beginning filming, the timescale was normalized such that the first frame showing shortening was set as time = 0. Each trace for 14 spindles is shown on the left, and the mean spindle length (± SD) over time for the same data set is shown on the right (blue shading). For control emb-30(RNAi) spindles, because there was no obvious spindle collapse, the beginning of the video was set at time = –4 and the time was not readjusted.
Supplementary Material
Acknowledgments
We thank members of the Wignall lab and the WiLa ICB for support and thoughtful discussions; Gabriel Cavin-Meza, Hannah Horton, Rachel Kadzik, and Tim Mullen for critical reading of the manuscript; Sarah Rice for reagents and technical advice; Olaf Bossinger, Bruce Bowerman, Arshad Desai, Marijn Ford, Scott Gradia, and Laura Lackner for reagents; and Vladimir Gelfand, Laura Lackner, and John Marko for helpful suggestions. Microscopy was performed at the Biological Imaging Facility at Northwestern University, supported by the Chemistry for Life Processes Institute, the NU Office for Research, and the Rice Foundation. This work was supported by National Institutes of Health (NIH) Grant R01GM124354 (to S.M.W.), American Heart Association Predoctoral Fellowship 17PRE33440016 (to I.D.W.), and NIH/National Institute of General Medical Sciences Molecular Biophysics Training Grant T32GM008382 (to I.D.W.).
Abbreviations used:
- ASPM
abnormal spindles and primary microcephaly
- CRISPR
clustered regularly interspaced short palindromic repeats
- crRNA
CRISPR RNA
- EMB
abnormal embryogenesis
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- KLP
kinesin-like protein
- MBP
maltose-binding protein
- MESP
meiotic spindle
- PCR
polymerase chain reaction
- RNAi
RNA interference
- TPX2
targeting protein for Xklp2.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E22-05-0153).
REFERENCES
- Alfaro-Aco R, Thawani A, Petry S (2017). Structural analysis of the role of TPX2 in branching microtubule nucleation. J Cell Biol 216, 983–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere JA, Bell RT, Fu BXH, Artiles KL, Hartman PS, Fire AZ (2014). Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balchand SK, Mann BJ, Titus J, Ross JL, Wadsworth P (2015). TPX2 inhibits Eg5 by interactions with both motor and microtubule. J Biol Chem 290, 17367–17379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop JD, Han Z, Schumacher JM (2005). The Caenorhabditis elegans Aurora B kinase AIR-2 phosphorylates and is required for the localization of a BimC kinesin to meiotic and mitotic spindles. Mol Biol Cell 16, 742–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangy A, Lane HA, d’Hérin P, Harper M, Kress M, Nigg EA (1995). Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell 83, 1159–1169. [DOI] [PubMed] [Google Scholar]
- Cavin-Meza G, Kwan MM, Wignall SM (2022). Multiple motors cooperate to establish and maintain acentrosomal spindle bipolarity in C. elegans oocyte meiosis. eLife 11, e72872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly AA, Osterberg V, Christensen S, Price M, Lu C, Chicas-Cruz K, Lockery S, Mains PE, Bowerman B (2014). Caenorhabditis elegans oocyte meiotic spindle pole assembly requires microtubule severing and the calponin homology domain protein ASPM-1. Mol Biol Cell 25, 1298–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa MFA, Ohkura H (2019). The molecular architecture of the meiotic spindle is remodeled during metaphase arrest in oocytes. J Cell Biol 218, 2854–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danlasky BM, Panzica MT, McNally KP, Vargas E, Bailey C, Li W, Gong T, Fishman ES, Jiang X, McNally FJ (2020). Evidence for anaphase pulling forces during C. elegans meiosis. J Cell Biol 219, e202005179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis-Roca AC, Divekar NS, Ng RK, Wignall SM (2018). Dynamic SUMO remodeling drives a series of critical events during the meiotic divisions in Caenorhabditis elegans. PLoS Genet 14, e1007626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis-Roca AC, Muscat CC, Wignall SM (2017). Caenorhabditis elegans oocytes detect meiotic errors in the absence of canonical end-on kinetochore attachments. J Cell Biol 216, 1243–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsler H, McAinsh AD (2016). Kinesin-12 motors cooperate to suppress microtubule catastrophes and drive the formation of parallel microtubule bundles. Proc Natl Acad Sci USA 113, E1635–E1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsler H, McHugh T, Singleton MR, Carter NJ, McAinsh AD (2014). The kinesin-12 Kif15 is a processive track-switching tetramer. eLife 3, e01724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas ME, Chen GY, Kendrick ND, Xu G, Larsen SD, Jana S, Waterson AG, Bauer JA, Hancock W, Sulikowski GA, et al. (2019). Dual inhibition of Kif15 by oxindole and quinazolinedione chemical probes. Bioorg Med Chem Lett 29, 148–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont J, Oegema K, Desai A (2010). A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat Cell Biol 12, 894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan FE, Hornick JE, Woodruff TK (2012). Bipolar-to-monopolar spindle collapse in human eggs. Mol Reprod Dev 79, 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito D, Chatterjee DK (2006). Enhancement of soluble protein expression through the use of fusion tags. Curr Opin Biotechnol 17, 353–358. [DOI] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J (2000). Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408, 325–330. [DOI] [PubMed] [Google Scholar]
- Furuta T, Tuck S, Kirchner J, Koch B, Auty R, Kitagawa R, Rose AM, Greenstein D (2000). EMB-30: an APC4 homologue required for metaphase-to-anaphase transitions during meiosis and mitosis in Caenorhabditis elegans. Mol Biol Cell 11, 1401–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holubcová Z, Blayney M, Elder K, Schuh M (2015). Human oocytes. Error-prone chromosome-mediated spindle assembly favors chromosome segregation defects in human oocytes. Science 348, 1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Rezabkova L, Hua S, Liu Q, Capitani G, Altelaar AFM, Heck AJR, Kammerer RA, Steinmetz MO, Akhmanova A (2017). Microtubule minus-end regulation at spindle poles by an ASPM-katanin complex. Nat Cell Biol 19, 480–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. (2003). Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Peterman EJG, Kwok BH, Kim JH, Kapoor TM, Schmidt CF (2005). The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature 435, 114–118. [DOI] [PubMed] [Google Scholar]
- Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ (2000). Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol 150, 975–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashina AS, Baskin RJ, Cole DG, Wedaman KP, Saxton WM, Scholey JM (1996). A bipolar kinesin. Nature 379, 270–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MR, Petry S (2020). Phase separation of TPX2 enhances and spatially coordinates microtubule nucleation. Nat Commun 11, 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laband K, Lacroix B, Edwards F, Canman JC, Dumont J (2018). Live imaging of C. elegans oocytes and early embryos. Methods Cell Biol 145, 217–236. [DOI] [PubMed] [Google Scholar]
- Laband K, Le Borgne R, Edwards F, Stefanutti M, Canman JC, Verbavatz J-M, Dumont J (2017). Chromosome segregation occurs by microtubule pushing in oocytes. Nat Commun 8, 1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, Titus J, Gable A, Ross JL, Wadsworth P (2011). TPX2 regulates the localization and activity of Eg5 in the mammalian mitotic spindle. J Cell Biol 195, 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailhes JB, Mastromatteo C, Fuseler JW (2004). Transient exposure to the Eg5 kinesin inhibitor monastrol leads to syntelic orientation of chromosomes and aneuploidy in mouse oocytes. Mutat Res 559, 153–167. [DOI] [PubMed] [Google Scholar]
- Malaby HLH, Dumas ME, Ohi R, Stumpff J (2019). Kinesin-binding protein ensures accurate chromosome segregation by buffering KIF18A and KIF15. J Cell Biol 218, 1218–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann BJ, Balchand SK, Wadsworth P (2017). Regulation of Kif15 localization and motility by the C-terminus of TPX2 and microtubule dynamics. Mol Biol Cell 28, 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann BJ, Wadsworth P (2019). Kinesin-5 regulation and function in mitosis. Trends Cell Biol 29, 66–79. [DOI] [PubMed] [Google Scholar]
- McDonnell AV, Jiang T, Keating AE, Berger B (2006). Paircoil2: improved prediction of coiled coils from sequence. Bioinformatics 22, 356–358. [DOI] [PubMed] [Google Scholar]
- McHugh T, Drechsler H, McAinsh AD, Carter NJ, Cross RA (2018). Kif15 functions as an active mechanical ratchet. Mol Biol Cell 29, 1743–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally KP, Panzica MT, Kim T, Cortes DB, McNally FJ (2016). A novel chromosome segregation mechanism during female meiosis. Mol Biol Cell 27, 2576–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TJ, Davis-Roca AC, Wignall SM (2019). Spindle assembly and chromosome dynamics during oocyte meiosis. Curr Opin Cell Biol 60, 53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TJ, Wignall SM (2017). Interplay between microtubule bundling and sorting factors ensures acentriolar spindle stability during C. elegans oocyte meiosis. PLoS Genet 13, e1006986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muscat CC, Torre-Santiago KM, Tran MV, Powers JA, Wignall SM (2015). Kinetochore-independent chromosome segregation driven by lateral microtubule bundles. eLife 4, e06462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A, Folkmann A, Rasoloson D, Seydoux G (2015). High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9 ribonucleoprotein complexes. Genetics 201, 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pédelacq J-D, Cabantous S, Tran T, Terwilliger TC, Waldo GS. (2006). Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol 24, 79–88. [DOI] [PubMed] [Google Scholar]
- Raaijmakers JA, van Heesbeen RGHP, Meaders JL, Geers EF, Fernandez-Garcia B, Medema RH, Tanenbaum ME (2012). Nuclear envelope-associated dynein drives prophase centrosome separation and enables Eg5-independent bipolar spindle formation. EMBO J 31, 4179–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford SJ, Go AMM, McKim KS (2017). Cooperation between kinesin motors promotes spindle symmetry and chromosome organization in oocytes. Genetics 205, 517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redemann S, Lantzsch I, Lindow N, Prohaska S, Srayko M, Müller-Reichert T (2018). A switch in microtubule orientation during C. elegans meiosis. Curr Biol 28, 2991–2997.e2. [DOI] [PubMed] [Google Scholar]
- Reinemann DN, Sturgill EG, Das DK, Degen MS, Vörös Z, Hwang W, Ohi R, Lang MJ (2017). Collective force regulation in anti-parallel microtubule gliding by dimeric Kif15 kinesin motors. Curr Biol 27, 2810–2820.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuten R, Nikodemus D, Oliveira MB, Patel TR, Brachvogel B, Breloy I, Stetefeld J, Koch M (2016). Maltose-binding protein (MBP), a secretion-enhancing tag for mammalian protein expression systems. PLoS One 11, e0152386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov VI, Gyoeva FK, Kashina AS, Kuznetsov SA, Gelfand VI (1990). Microtubule-associated proteins and microtubule-based translocators have different binding sites on tubulin molecule. J Biol Chem 265, 5702–5707. [PubMed] [Google Scholar]
- Roeles J, Tsiavaliaris G (2019). Actin-microtubule interplay coordinates spindle assembly in human oocytes. Nat Commun 10, 4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, Eliceiri KW (2017). ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin KE, LeGuellec K, Philippe M, Mitchison TJ (1992). Mitotic spindle organization by a plus-end-directed microtubule motor. Nature 359, 540–543. [DOI] [PubMed] [Google Scholar]
- Sawin KE, Mitchison TJ (1991). Mitotic spindle assembly by two different pathways in vitro. J Cell Biol 112, 925–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey JE, Nithianantham S, Scholey JM, Al-Bassam J (2014). Structural basis for the assembly of the mitotic motor kinesin-5 into bipolar tetramers. eLife 3, e02217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M, Ellenberg J (2007). Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell 130, 484–498. [DOI] [PubMed] [Google Scholar]
- Seeger MA, Rice SE (2010). Microtubule-associated protein-like binding of the kinesin-1 tail to microtubules. J Biol Chem 285, 8155–8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segbert C, Barkus R, Powers J, Strome S, Saxton WM, Bossinger O (2003). KLP-18, a Klp2 kinesin, is required for assembly of acentrosomal meiotic spindles in Caenorhabditis elegans. Mol Biol Cell 14, 4458–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano L, de la Torre J, Maccioni RB, Avila J (1984). Involvement of the carboxyl-terminal domain of tubulin in the regulation of its assembly. Proc Natl Acad Sci USA 81, 5989–5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson AF, von Dassow G, Bowerman B (2016). Oocyte Meiotic Spindle Assembly and Function. Curr Top Dev Biol 116, 65–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill EG, Das DK, Takizawa Y, Shin Y, Collier SE, Ohi MD, Hwang W, Lang MJ, Ohi R (2014). Kinesin-12 Kif15 targets kinetochore fibers through an intrinsic two-step mechanism. Curr Biol 24, 2307–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill EG, Norris SR, Guo Y, Ohi R (2016). Kinesin-5 inhibitor resistance is driven by kinesin-12. J Cell Biol 213, 213–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill EG, Ohi R (2013). Kinesin-12 differentially affects spindle assembly depending on its microtubule substrate. Curr Biol 23, 1280–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Macůrek L, Janssen A, Geers EF, Alvarez-Fernández M, Medema RH (2009). Kif15 cooperates with eg5 to promote bipolar spindle assembly. Curr Biol 19, 1703–1711. [DOI] [PubMed] [Google Scholar]
- Vanneste D, Takagi M, Imamoto N, Vernos I (2009). The role of Hklp2 in the stabilization and maintenance of spindle bipolarity. Curr Biol 19, 1712–1717. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Vernos I, Mitchison TJ, Karsenti E, Heald R (1998). A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr Biol 8, 903–913. [DOI] [PubMed] [Google Scholar]
- Wan X, Zhang Y, Lan M, Pan M-H, Tang F, Zhang H-L, Ou X-H, Sun S-C (2018). Meiotic arrest and spindle defects are associated with altered KIF11 expression in porcine oocytes. Environ Mol Mutagen 59, 805–812. [DOI] [PubMed] [Google Scholar]
- Wang C, Castro AF, Wilkes DM, Altenberg GA (1999). Expression and purification of the first nucleotide-binding domain and linker region of human multidrug resistance gene product: comparison of fusions to glutathione S-transferase, thioredoxin and maltose-binding protein. Biochem J 338 (Pt 1), 77–81. [PMC free article] [PubMed] [Google Scholar]
- Wang S, Wu D, Quintin S, Green RA, Cheerambathur DK, Ochoa SD, Desai A, Oegema K (2015). NOCA-1 functions with γ-tubulin and in parallel to patronin to assemble non-centrosomal microtubule arrays in C. elegans. eLife 4, e08649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wignall SM, Villeneuve AM (2009). Lateral microtubule bundles promote chromosome alignment during acentrosomal oocyte meiosis. Nat Cell Biol 11, 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T, Boleti H, Antony C, Karsenti E, Vernos I (1998). Localization of the kinesin-like protein Xklp2 to spindle poles requires a leucine zipper, a microtubule-associated protein, and dynein. J Cell Biol 143, 673–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T, Wilm M, Karsenti E, Vernos I (2000). TPX2, a novel xenopus MAP involved in spindle pole organization. J Cell Biol 149, 1405–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff ID, Divekar NS, Wignall SM (2022). Methods for investigating cell division mechanisms in C. elegans. Methods Mol Biol 2415, 19–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff ID, Tran MV, Mullen TJ, Villeneuve AM, Wignall SM (2016). Assembly of Caenorhabditis elegans acentrosomal spindles occurs without evident microtubule-organizing centers and requires microtubule sorting by KLP-18/kinesin-12 and MESP-1. Mol Biol Cell 27, 3122–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN (2010). PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim Biophys Acta 1804, 996–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Cameron LA, Maddox PS, Salmon ED, Danuser G (2008). Regional variation of microtubule flux reveals microtubule organization in the metaphase meiotic spindle. J Cell Biol 182, 631–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdraljevic S, Fox BW, Strand C, Panda O, Tenjo FJ, Brady SC, Crombie TA, Doench JG, Schroeder FC, Andersen EC (2019). Natural variation in C. elegans arsenic toxicity is explained by differences in branched chain amino acid metabolism. eLife 8, e40260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Roostalu J, Surrey T, Nogales E (2017). Structural insight into TPX2-stimulated microtubule assembly. eLife 6, e30959. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.