Abstract
Previous studies suggesting a link between Escherichia coli phylogenetic groups and extraintestinal virulence have been hampered by the difficulty in establishing the intrinsic virulence of a bacterial strain. Indeed, unidentified virulence factors do exist, and the susceptibility of the host to infection is highly variable. To overcome these difficulties, we have developed a mouse model of extraintestinal virulence to test the virulence of the strains under normalized conditions. We then assessed the phylogenetic relationships compared to the E. coli reference (ECOR) collection, the presence of several known virulence determinants, and the lethality to mice of 82 human adult E. coli strains isolated from normal feces and during the course of extraintestinal infections. Commensal strains belong mainly to phylogenetic groups A and B1, are devoid of virulence determinants, and do not kill the mice. Strains exhibiting the same characteristics as the commensal strains can be isolated under pathogenic conditions, thus indicating the role of host-dependent factors, such as susceptibility linked to underlying disease, in the development of infection. Some strains of phylogenetic groups A, B1, and D are able to kill the mice, their virulence being most often correlated with the presence of virulence determinants. Lastly, strains of the B2 phylogenetic group represent a divergent lineage of highly virulent strains which kill the mice at high frequency and possess the highest level of virulence determinants. The observed link between virulence and phylogeny could correspond to the necessity of virulence determinants in a genetic background that is adequate for the emergence of a virulent clone, an expression of the interdependency of pathogenicity and metabolic activities in pathogenic bacteria.
In humans, strains of the species Escherichia coli can be commensal (since they are part of the normal intestinal microbial flora) and/or the cause of various infectious diseases (intestinal and extraintestinal infections) (2, 10). The barrier between commensalism and virulence results from a complex balance between the status of the host and the presence and expression of virulence factors in the bacteria. The genetic structure of E. coli can be considered clonal (37), and, indeed, various intestinal (41) or extraintestinal (1, 26, 38) E. coli infections have been linked to specific clones or groups of related clones. In several cases, pathogenicity has been correlated with the presence of genes encoding virulence factors organized on large blocks, called pathogenicity islands. It has been shown, further, that pathogenicity islands can disseminate horizontally between distinct E. coli strains whether they are located on plasmids, bacteriophages, or even the bacterial chromosome (16). Horizontal transfer in E. coli has also been reported for other systems, including the O antigen, the hsd restriction-modification system, and some housekeeping genes (28). It has been shown, however, that horizontal transfer, in general, does not disrupt the clonal structure of the species (9, 15, 27, 28). An attractive hypothesis to reconcile horizontal dissemination of virulence factors and oligoclonality of the virulent strains would be that of an interaction between virulence determinants and the genetic background of the bacteria. Recent attempts at establishing the phylogenetic position of virulent clones within the diversity of the species as a whole seem to support this hypothesis (5, 36). However, the conclusions derived from these studies are weakened by the difficulty in appreciating the intrinsic virulence of a given bacterial strain. Indeed, the direct search for virulence determinants is obviously limited to the known determinants, and unidentified virulence factors do exist. Second, it has been shown that the status of the host can be critical to the development of an infection, independent of the presence or absence of virulence factors and/or of chromosomal genetic markers (5, 20, 25, 33, 40). Finally, various other environmental parameters such as the amount of the inoculum can influence the course of an infection. Thus, the “virulent” or “nonvirulent” character of a strain cannot be systematically inferred from the circumstances in which the strain was isolated (commensal or pathogenic).
To clarify this issue, we have developed a mouse model to evaluate the intrinsic extraintestinal virulence of bacterial strains in normalized conditions. Using this model, we have determined the lethality to mice of a series of 82 human E. coli strains isolated from normal feces and during the course of extraintestinal infections. These strains were further characterized by studying the distribution of known virulence factors and their phylogenetic relationships with respect to the E. coli reference (ECOR) collection (30). Our data, taken together, establish the link between phylogeny and extraintestinal virulence in the E. coli species.
MATERIALS AND METHODS
Bacterial strains.
A total of 82 E. coli strains isolated from adult humans were studied that belonged to a previously published collection of 85 commensal isolates and 191 extraintestinal pathogenic isolates (isolated from patients with urinary tract infections, septicemia, and miscellaneous infections including cholecystitis, ascites, and lung infections) (12, 13). A study of the carboxylesterase B polymorphism distinguished two electrophoretic types among these strains: B1 (MF ≈ 68 to 72) and B2 (MF ≈ 57 to 63). Strains of carboxylesterase B type B2 were more frequently pathogenic and often produced alpha-hemolysin and mannose-resistant hemagglutinin (MRHA) whereas strains of type B1 were prevalent in commensal isolates and devoid of these virulent factors. The 82 E. coli strains used in this study were selected for their commensal (12) or pathogenic (13) origin, their carboxylesterase B electrophoretic types B1 and B2 (13), and their production of α-hemolysin and MRHA. Six groups were constituted according to these criteria: group 1 (15 strains), commensal, carboxylesterase B1 type, nonhemolytic, MRHA negative; group 2 (18 strains), pathogen, carboxylesterase B1 type, nonhemolytic, MRHA negative; group 3 (12 strains), pathogen, carboxylesterase B1 type, nonhemolytic, MRHA positive; group 4 (10 strains), pathogen, carboxylesterase B2 type, nonhemolytic, MRHA negative; group 5 (15 strains), pathogen, carboxylesterase B2 type, nonhemolytic, MRHA positive; group 6 (12 strains), pathogen, carboxylesterase B2 type, hemolytic, MRHA positive (Table 1).
TABLE 1.
Characteristics of the 82 E. coli clinical strainsa
| Strain | Origin | Carboxylesterase B (MF) | Expression of:
|
Group | Detection of
|
Phylogenetic group | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hemolysin | MRHA | K1 antigen | sfa/foc operon | pap operon | afa operon | hly operon | aer operon | ibe10 gene | Lethality in mice | |||||
| 1 | Feces | 70 | − | 0 | 1 | − | − | − | − | − | − | − | 0 | B1 |
| 2 | Feces | 70 | − | 0 | 1 | − | − | − | − | − | − | − | 0 | B1 |
| 3 | Feces | 70 | − | 0 | 1 | − | − | − | − | − | − | − | 0 | B1 |
| 4 | Feces | 70 | − | 0 | 1 | − | − | − | − | − | − | − | 0 | B1 |
| 5 | Feces | 70 | − | 0 | 1 | − | − | − | − | − | + | − | 0 | B1 |
| 6 | Feces | 70 | − | 0 | 1 | + | − | − | − | − | − | − | 0 | A |
| 7 | Feces | 70 | − | 0 | 1 | − | − | − | − | − | − | − | 0 | B1 |
| 8 | Feces | 70 | − | 0 | 1 | − | − | − | − | − | − | − | 0 | A |
| 9 | Feces | 70 | − | 0 | 1 | − | − | − | − | − | − | − | 0 | B1 |
| 10 | Feces | 70 | − | 0 | 1 | − | − | − | − | − | − | − | 0 | A |
| 11 | Feces | 70 | − | 0 | 1 | − | − | − | − | − | + | − | 0 | A |
| 12 | Feces | 70 | − | 0 | 1 | − | − | − | − | − | − | − | 0 | B1 |
| 13 | Feces | 70 | − | 0 | 1 | − | − | − | − | − | − | − | 0 | A |
| 14 | Feces | 70 | − | 0 | 1 | − | − | − | − | − | + | − | 0 | A |
| 15 | Feces | 70 | − | 0 | 1 | − | − | − | − | − | − | − | 0 | A |
| 16 | Blood | 72 | − | 0 | 2 | − | − | − | − | − | − | − | 0 | A |
| 17 | Urine | 72 | − | 0 | 2 | − | − | − | − | − | − | − | 0 | A |
| 18 | Urine | 70 | − | 0 | 2 | − | − | + | − | + | − | − | 9 | B1 |
| 19 | Urine | 70 | − | 0 | 2 | − | − | − | − | − | − | − | 0 | A |
| 20 | Miscellaneous | 68 | − | 0 | 2 | − | − | − | − | − | − | − | 0 | A |
| 21 | Urine | 72 | − | 0 | 2 | − | − | − | − | − | − | − | 0 | B1 |
| 22 | Urine | 70 | − | 0 | 2 | − | − | − | − | − | − | − | 0 | A |
| 23 | Urine | 70 | − | 0 | 2 | − | − | − | − | − | − | − | 0 | A |
| 24 | Urine | 70 | − | 0 | 2 | − | − | − | − | − | − | − | 0 | B1 |
| 25 | Urine | 70 | − | 0 | 2 | + | − | − | − | − | − | − | 0 | A |
| 26 | Urine | 70 | − | 0 | 2 | − | − | − | − | − | − | − | 0 | B1 |
| 27 | Urine | 70 | − | 0 | 2 | − | − | − | − | − | − | − | 0 | B1 |
| 28 | Urine | 70 | − | 0 | 2 | − | − | − | − | − | − | − | 0 | A |
| 29 | Urine | 70 | − | 0 | 2 | − | − | − | − | − | − | − | 0 | A |
| 30 | Urine | 70 | − | 0 | 2 | − | − | − | − | − | − | − | 0 | B1 |
| 31 | Urine | 70 | − | 0 | 2 | − | − | − | − | − | − | − | 0 | B1 |
| 32 | Urine | 70 | − | 0 | 2 | − | − | − | − | − | − | − | 0 | B1 |
| 33 | Urine | 68 | − | 0 | 2 | − | − | − | − | − | − | − | 0 | B1 |
| 34 | Blood | 73 | − | 4 | 3 | − | − | − | − | − | − | − | 0 | A |
| 35 | Blood | 72 | − | 2 | 3 | − | − | + | + | − | − | − | 10+ | D |
| 36 | Miscellaneous | 70 | − | 3 | 3 | − | − | + | − | − | + | − | 10+ | B1 |
| 37 | Miscellaneous | 70 | − | 2 | 3 | − | − | − | + | − | + | − | 0 | A |
| 38 | Miscellaneous | 68 | − | 2 | 3 | − | − | + | − | − | + | − | 10+ | D |
| 39 | Urine | 72 | − | 2 | 3 | + | − | − | − | − | + | − | 10+ | B1 |
| 40 | Urine | 70 | − | 3 | 3 | − | − | − | + | − | + | − | 0 | A |
| 41 | Urine | 72 | − | 3 | 3 | − | − | + | − | − | − | − | 0 | B1 |
| 42 | Urine | 70 | − | 3 | 3 | − | − | + | − | − | − | − | 5 | B1 |
| 43 | Urine | 70 | − | 4 | 3 | − | − | + | − | − | + | − | 0 | B1 |
| 44 | Urine | 70 | − | 2 | 3 | − | − | − | − | − | − | − | 7 | A |
| 45 | Urine | 70 | − | 3 | 3 | − | − | − | − | − | − | − | 0 | B1 |
| 46 | Urine | 62 | − | 0 | 4 | + | − | + | − | − | + | − | 9 | B2 |
| 47 | Urine | 62 | − | 0 | 4 | − | − | − | − | − | − | − | 0 | B2 |
| 48 | Blood | 60 | − | 0 | 4 | − | − | − | − | − | − | − | 0 | B2 |
| 49 | Urine | 57 | − | 0 | 4 | − | − | − | − | − | − | + | 10+ | B2 |
| 50 | Urine | 57 | − | 0 | 4 | − | − | − | − | − | + | − | 0 | B2 |
| 51 | Urine | 57 | − | 0 | 4 | + | + | − | − | − | − | − | 10+ | B2 |
| 52 | Urine | 57 | − | 0 | 4 | + | + | − | − | − | − | − | 10+ | B2 |
| 53 | Urine | 57 | − | 0 | 4 | + | − | − | − | − | + | + | 0 | B2 |
| 54 | Urine | 57 | − | 0 | 4 | − | − | − | − | + | − | − | 0 | B2 |
| 55 | Urine | 57 | − | 0 | 4 | − | − | + | − | − | − | + | 10 | B2 |
| 56 | Urine | 57 | − | 3 | 5 | + | − | + | − | − | + | − | 9 | B2 |
| 57 | Urine | 57 | − | 1 | 5 | − | + | − | − | + | − | − | 9+ | B2 |
| 58 | Urine | 63 | − | 2 | 5 | + | − | − | − | − | − | − | 9+ | B2 |
| 59 | Urine | 62 | − | 2 | 5 | + | − | + | − | − | + | − | 10+ | B2 |
| 60 | Blood | 62 | − | 2 | 5 | + | − | + | − | − | + | − | 1 | B2 |
| 61 | Miscellaneous | 62 | − | 2 | 5 | + | − | + | − | − | + | − | 9 | B2 |
| 62 | Urine | 57 | − | 3 | 5 | − | + | + | − | − | − | − | 10+ | B2 |
| 63 | Urine | 57 | − | 2 | 5 | − | + | + | − | − | − | − | 10+ | B2 |
| 64 | Urine | 57 | − | 1 | 5 | − | + | + | − | − | − | + | 10+ | B2 |
| 65 | Miscellaneous | 62 | − | 2 | 5 | + | − | + | − | − | − | − | 10+ | B2 |
| 66 | Urine | 60 | − | 3 | 5 | + | − | + | − | − | + | − | 9+ | B2 |
| 67 | Urine | 57 | − | 3 | 5 | − | − | + | − | − | + | − | 0 | B2 |
| 68 | Urine | 57 | − | 3 | 5 | − | + | + | − | − | + | − | 3 | B2 |
| 69 | Miscellaneous | 62 | − | 2 | 5 | − | − | + | − | − | + | − | 10+ | B2 |
| 70 | Urine | 57 | − | 3 | 5 | − | + | + | − | − | − | + | 10+ | B2 |
| 71 | Urine | 62 | + | 3 | 6 | − | + | + | − | + | + | − | 10+ | B2 |
| 72 | Urine | 57 | + | 2 | 6 | − | − | − | − | + | − | − | 10+ | B2 |
| 73 | Blood | 57 | + | 2 | 6 | − | + | + | − | + | − | − | 10+ | B2 |
| 74 | Urine | 57 | + | 3 | 6 | + | + | + | − | + | − | − | 10+ | B2 |
| 75 | Miscellaneous | 57 | + | 3 | 6 | − | + | + | − | + | − | − | 10+ | B2 |
| 76 | Urine | 57 | + | 2 | 6 | − | + | + | − | + | − | − | 3 | B2 |
| 77 | Urine | 60 | + | 2 | 6 | − | + | + | − | + | − | − | 10+ | B2 |
| 78 | Urine | 57 | + | 4 | 6 | − | − | − | − | + | + | − | 5 | B2 |
| 79 | Urine | 57 | + | 3 | 6 | − | − | + | − | + | + | − | 10+ | B2 |
| 80 | Urine | 57 | + | 3 | 6 | − | − | + | − | + | + | − | 10+ | B2 |
| 81 | Urine | 57 | + | 4 | 6 | − | − | + | − | + | + | − | 9+ | B2 |
| 82 | Urine | 57 | + | 3 | 6 | − | + | + | − | + | − | − | 10+ | B2 |
Strains originating from feces are commensal (group 1), while the remainder were isolated from patients with pathological conditions (groups 2 to 6). Strains exhibiting a B1-type carboxylesterase B (MF ≈ 68 to 72) correspond to groups 1 to 3, while strains exhibiting a B2-type carboxylesterase B (MF ≈ 57 to 63) correspond to groups 4 to 6. MHRA was quantified from 0 to 4 according to the intensity of the reaction. The six groups (see Materials and Methods) are listed in order. The lethality in mice is indicated as the number of mice killed by the bacterial strain (0 to 10). A plus sign indicates that the bacterial strain killed at least six mice in less than 18 h. Phylogenetic groups A, B1, B2, and D are indicated as in reference 17.
Animal model.
Pathogen-free female of the outbred white Swiss mice lineage (Rj: Swiss-IOPS Orl) (6 to 8 weeks old, 25 to 30 g) were purchased from the Centre d’Elevage R. Janvier (Le Genest, Saint Isle, France). E. coli strains were stored in glycerol solution at −70°C. The strains were isolated in Trypticase soy agar (AES Laboratories, Combourg, France) and then grown in Trypticase soy broth medium (pH 7.3, osmolarity = 271 mOsm/l) (AES Laboratories) and incubated with shaking at 37°C to a density of 109 bacteria per ml (approximately 4 h). The bacterial count was determined by measuring the optical density at 530 nm. A 2-ml sample of the culture was then centrifuged at 2,500 × g for 10 min, washed twice with Ringer solution, and resuspended in Ringer solution. These suspensions were used to inoculate the mice subcutaneously in the abdomen or intraperitoneally. Ten mice were inoculated with each strain. After inoculation, the mice were observed every hour for the first 6 h, at 18 h and daily for up to 1 week. Death and its delay were noted for each mouse. Dead animals were dissected, and the liver, spleen, and kidneys were inspected macroscopically and sampled. Each organ was washed with Ringer solution, weighed, and crushed with 5 ml (spleen and kidneys) or 10 ml (liver) of sterile Fontainebleau sand. Successive dilutions of the crushed organs were obtained (from 10−1 to 10−7) with Ringer solution and inoculated on Trypticase soy plates. One drop of heart blood was also inoculated on the same medium. The plates were incubated at 37°C for 24 h, and the colonies were counted.
rrn restriction fragment length polymorphism (RFLP) analysis (ribotyping).
Total E. coli DNA was prepared as previously described (3). It was digested with EcoRI and HindIII and subjected to Southern blotting analysis with ribosomal 16S+23S RNA from E. coli as a probe. The probe was labeled by random oligopriming with a mixture of hexanucleotides (Pharmacia, Uppasala, Sweden) and cloned Moloney murine leukemia virus reverse transcriptase (Bethesda Research Laboratories, Inc., Gaithersburg, Md.) in the presence of 0.35 mM DIG-11-dUTP (digoxigenin-11–deoxyuridine-5′ triphosphate; Boehringer, Mannheim, Germany). The procedure for chemiluminescence detection was as already reported (4).
Detection of virulence determinants.
The virulence determinants surveyed in this work for the 82 E. coli strains were chosen as they were relevant to extraintestinal infections (29). The capsular type K1 common among pathogenic E. coli strains was detected phenotypically with an antiserum to Neisseria meningitidis group B (8).
The genes encoding extraintestinal E. coli attachment factors (pap, sfa, and afa), an E. coli transmembrane protein involved in neonatal meningitis (ibe10), the alpha-hemolysin, a toxin often produced by urinary tract infection strains leading to cell lysis (hly), and the aerobactin determinant (aer) which competes with serum transferrin for iron uptake were detected by PCR. The pap, sfa/foc, and afa adhesin-encoding operons were detected by the procedure described by Le Bouguenec et al. (22), while the ibe10 gene and the hly and aer operons were detected as described by Bingen et al. (5).
Furthermore, the presence of the eaeA gene, belonging to the locus of enterocyte effacement, which is involved in intestinal adherence (29), was assessed. Indeed, many E. coli isolates from urinary tract infections derive from a fecal source and fecal isolates are known to carry locus of enterocyte effacement determinants. The assay was performed by PCR under the same conditions as described by Bingen et al. (5) with the eaeA5′ GACCCGGCACAAGCATAAGCT and eaeA3′ ACCTAARASWGGTAAGGCACT primers.
Statistical analysis.
The 82 clinical strains were compared to the 72 strains of the ECOR collection by a statistical treatment of the rrn RFLP data to assign these strains to the phylogenetic groups A, B1, B2, and D previously delineated among the ECOR strains (17). The rrn RFLP data were summarized as two-way tables of 72 rows for the 72 ECOR strains (9) and of 82 rows for the 82 E. coli clinical strains, with the number of columns corresponding to the number of rrn-containing DNA fragments detected by HindIII and EcoRI endonucleases. For each column, the rrn-containing fragment was coded as a binary code, present = 1 or absent = 0, according to the strains. A factorial analysis of correspondence (FAC) (14, 21, 39) was first conducted on the ECOR strain data. FAC is an eigenvector method of ordination similar to a principal-component analysis. However, it differs from the latter by the use of a covariance matrix based on χ2 distances rather than a covariance matrix based on Euclidean distances. Both techniques describe the dispersion and shape of a cloud of n objects (here, the ECOR strains) or p variables (here, p was the number of rrn containing DNA fragments detected by HindIII and EcoRI) in a multidimensional space, by replacing the original data set by a new set of orthogonal linear coordinates in a space of significantly lower dimension. The explained variances of the elements of the data set (the strains and the rrn DNA fragments) are in decreasing order of magnitude with respect to these new coordinates. Furthermore, the variables used for FAC are categories, whereas those used for principal-component analysis are quantitative or continuous variables. The computation determines a plane defined by the first two principal axes of the analysis; the first axis, F1, accounts for most of the variance, and the second axis F2, orthogonal to F1, accounts for the largest part of the variance that is not accounted for by F1. Each of the 72 ECOR strains is thus projected on the plane F1/F2, and their respective projections on this plane allows the subclassification of E. coli into four main phylogenetic groups. In a second step, the 82 clinical strain data were considered to be supplementary observations and projected on the principal plane F1/F2 obtained with the ECOR strains. Their projections in the different areas corresponding to each phylogenetic group allows their classification into these groups.
In a third step, the whole data were summarized as a third table of 82 rows for the E. coli clinical strains with the number of columns corresponding to the following characters (variables): pathologic or commensal origin; carboxylesterase B types B1 or B2; phylogenetic groups A, B1, B2, or D; detection of the K1 antigen, the sfa/foc, pap, afa, hly, and aer operons, and the ibe 10 gene; and lethality to mice (0 or 1 mouse, 2 to 8 mice, or 9 or 10 mice killed). A FAC was performed from this table. First, the projections of the variables (the characters of the strains) on the plane F1/F2 were analyzed and possible links between those characters were searched through their closeness on the plane. Then the projections of the strains on the plane F1/F2 or F1/F3 were analyzed in the same manner. In all cases, computations were performed with SPAD.N software (Centre International de Statistiques et Informatique Appliquées, St Mandé, France). Distribution of virulence determinants was analyzed among the phylogenetic groups, and the statistical significance between groups was tested by the χ2 test.
RESULTS
Lethality in mice.
To obtain reproducible results and a clear-cut interpretation of the data, it was important first to standardize the model. To do so, the size of the inoculum from 103 to 109 CFU was tested for one strain of group 1 with no virulence determinants (strain 1) and for one strain of group 6 with numerous virulent determinants (strain 71). For these two strains, two routes of inoculation (subcutaneous and intraperitoneal) were assayed. The mice were observed continuously during the first 6 h following inoculation to detect whether rapid death was linked to endotoxinic shock. From these preliminary steps, we retained the less invasive subcutaneous route of inoculation with 108 CFU in 0.2 ml of Ringer solution. Under these conditions, no mouse died within the first 6 h and the animals could be clearly classified as “killed” from infection or “survivors.” Bacterial counts in the liver, spleen, and kidneys of the killed mice were greater than or equal to 108 CFU per ml. Animals surviving for more than 7 days were considered permanent survivors, since tissue and blood cultures from these mice, sacrificed on this date after the inoculation, were sterile.
Of the 82 E. coli strains we studied, 38 killed at least one mouse (Table 1). Of these 38 strains, 32 (84.2%) killed 9 or all 10 of the 10 inoculated mice. Thus, “lethality” is a rather clear-cut parameter since when it is noted for a given strain, it is so most of the time for all or almost the inoculated mice. In addition, of the 32 strains which killed 9 or 10 mice, 27 (84.4%) did so in a short period, since more than 6 of the 10 mice inoculated with each strain died in less than 18 h.
A clear difference in lethality was observed among the six groups of strains. For the strains of groups 1 and 2, no strains, except one (group 2), killed the mice. Thus, a discrepancy is observed between the pathogenic origin of the strains of group 2 and their lack of virulence in the animal model. In contrast, all the strains of groups 5 and 6, except one, did kill the mice. Strains of groups 3 and 4 exhibited an intermediate behavior, with approximately 50% of them killing the mice.
Considering only the strains which were lethal for the mice, no significant difference was observed between the groups in terms of the number of mice killed by a given strain or the rapidity with which the strains killed the mice.
Phylogenetic distribution of the E. coli strains.
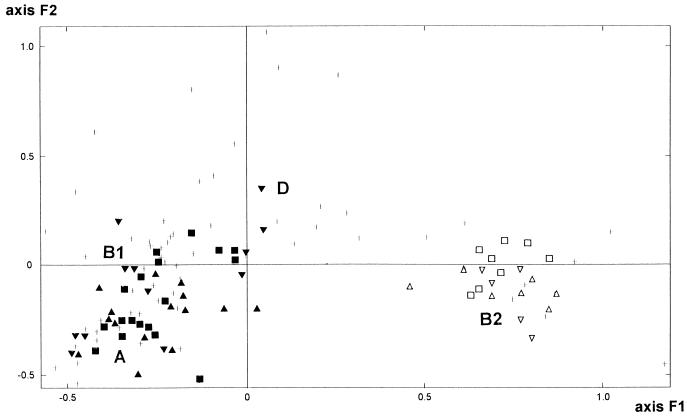
To assess the phylogenetic relationships among strains, statistical analysis by FAC was performed with the rrn RFLP data obtained for the clinical and ECOR (9) strains. The results are expressed as the projections of each strain on a plane defined by two axes (F1 and F2) which account for most of the total variance (Fig. 1). As previously described (5, 9), this analysis divides the ECOR strains into four main phylogenetic groups (A, B1, B2, and D) and these groups correspond to the classification initially reported by Hertzer et al. (17) from their analysis of 38 metabolic enzymes.
FIG. 1.
Factorial analysis of correspondence of the 72 strains of the ECOR collection (+) based on the rrn RFLP data (9). The 82 E. coli clinical isolates belonging to groups 1 (▴), 2 (■), 3 (▾), 4 (▵), 5 (□), and 6 (▿), considered supplementary observations, are projected in the F1/F2 plane. This plane, obtained by computation, is defined by the two principal axes of the analysis; the first axis, F1, explains most of the variance, and the second axis, F2 (orthogonal to F1), explains most of the variance which is not explained by F1. Phylogenetic groups A, B1, B2, and D (17) of E. coli are indicated. For clarity, when several strains are projected on the same point, only one is represented.
Projecting the 82 clinical E. coli strains as supplementary individuals on the plane F1/F2, which in our analysis accounted for 34.85% of the total variance, allows the sorting of the studied strains among the four phylogenetic groups mentioned above (Fig. 1). Phylogenetic group B2 was clearly distinguished from the others by the positive values on the F1 axis (which accounts for 23.67% of the total variance). Phylogenetic group D was distinguished from groups B1 and A by the positive values on the F2 axis (which accounts for 11.18% of the total variance). Finally, phylogenetic group A was distinguished from group B1 by negative values on both the F1 and F2 axes. This analysis shows that the group 1 of commensal carboxylesterase B1 strains without alpha-hemolysin and MRHA belong to phylogenetic groups A (seven strains) and B1 (eight strains). A comparable result was obtained for group 2 of pathogenic carboxylesterase B1 strains without alpha-hemolysin and MRHA and no lethality in the mice, since nine strains belonged to phylogenetic group A and nine strains belonged to phylogenetic group B1. Group 3 of carboxylesterase B1 pathogenic strains producing MRHA without alpha-hemolysin appeared genetically more heterogeneous, since six strains belonged to phylogenetic group B1, four belonged to phylogenetic group A, and two belonged to phylogenetic group D. Interestingly, these last two strains exhibited the more rarely evidenced MF ≈ 72 and MF ≈ 68 electrophoretic variants of carboxylesterase B (13). As expected from previous studies (34, 35), pathogenic strains of the three remaining groups, 4, 5, and 6, which all belonged to the carboxylesterase B type B2, were classified in phylogenetic group B2.
Detection of virulence determinants.
Overall, a progressive enrichment of virulence determinants was observed, from the strains of groups 1 and 2, which lack virulence factors, to the strains of group 6, which possess the highest level of virulence determinants. The K1 antigen was detected in 15 isolates. Of these, 13 belong to phylogenetic group B2. There was a good phenotype-genotype correlation for the adhesin and hemolysin determinants. The rare discrepancies observed between phenotype and genotype were not judged significant, since previous reports indicate variable in vitro phenotypes of virulence factors due to the in vitro culture conditions (11, 25, 31). The afa operon was rarely detected (three strains in group 3). The pap operon was found in strains of phylogenetic groups D, B1, and B2 but was significantly more frequent in the B2 group (χ2 = 25.27; P < 0.0005), whereas the sfa/foc operon is strictly restricted to strains of the B2 phylogenetic group. Thus, the strains of the B2 phylogenetic group more often possess an adhesin-encoding operon than do strains of the other phylogenetic groups, whatever the level of MRHA expression. As expected, the ibe10 gene, which is involved in the pathogenesis of meningitis in newborns (18), was rarely found (five strains) and was strictly restricted to strains belonging to the B2 phylogenetic group. The hly determinant was significantly more frequent in strains of phylogenetic group B2 (χ2 = 12.29; P < 0.005). The aer determinant was widespread in strains of all the phylogenetic groups. This can be explained by the fact that we did not differentiate the episomal from the chromosomal aer operon. It has been shown that carboxylesterase B type B1 strains carry a plasmid-encoded aerobactin system whereas carboxylesterase B type B2 strains carry chromosomal aerobactin determinants (19). The eaeA determinant was not detected in any of the 82 studied clinical strains.
Whole-data analysis.
A FAC analysis was conducted on the whole data, taking as variables the origin of the strains, carboxylesterase B type, phylogenetic group, virulence determinants, and lethality to mice. When the distribution of these variables was studied, the first two principal axes, F1 and F2, accounted for 47.41% of the total variance and projections of the variables on the F1/F2 plane showed very striking features (Fig. 2A). Indeed, the F1 axis, which accounted for 37.25% of the total variance, clearly showed two opposite classes of determinants. Its negative values were lethality to mice, phylogenetic group B2, carboxylesterase B type B2, detection of sfa/foc, hly, and pap operons and of the ibe10 gene, detection of K1 antigen, and, to a lesser degree, detection of aer and pathogenic origin. Its positive values were commensal origin, absence of lethality to mice, carboxylesterase B type B1, and phylogenetic groups B1 and A. Phylogenetic group D and the afa detection were clearly singled out from their highly negative values on the F2 axis.
FIG. 2.
Graphical representation of the results of the FAC carried out with the whole data from the 82 E. coli clinical strains. (A) Projections of the variables on the F1/F2 plane (defined as in Fig. 1): hly, detection of the hly operon; aer, detection of the aer operon; sfa, detection of the sfa/foc operon; ibe, detection of the ibe10 gene; pap, detection of the pap operon, afa, detection of the afa operon; K1, detection of the K1 antigen; b1, carboxylesterase B type B1; b2, carboxylesterase B type B2; B2, phylogenetic group B2; B1, phylogenetic group B1; A, phylogenetic group A; D, phylogenetic group D; pat, pathologic origin; co, commensal origin; 0/1, low level of lethality to mice; 2/8, medium level of lethality to mice; 9/10, high level of lethality to mice. The + and − signs after the variables indicate the presence or absence of the variable, respectively. (B) Projections of the 82 E. coli strains on the F1/F3 plane. The third axis F3, orthogonal to the F1/F2 plane, explains the largest part of the variance which is not explained by the F1/F2 plane. The strains (▴) are numbered as in Table 1, and for clarity, when several strains are projected on the same point, only one is represented.
When the distribution of the E. coli strains was studied, taking into account the above variables, the largest part of the total variance (45.8%) was accounted for by the F1/F3 plane, where the third axis, F3, is orthogonal to the F1/F2 plane. Figure 2B shows the projections of the 82 clinical E. coli strains on the F1/F3 plane. Strains of the phylogenetic group B2 all were projected on the negative values of F1, and they segregated in two clusters, one of 31 strains, with the lowest negative values on F1, and one of 6 strains, with higher negative values on F1 (strains 47, 48, 50, 53, 54, and 60). Interestingly, these six strains were characterized by the absence of lethality to mice (only one strain killed a single mouse) and by the presence of few (one to three) virulence factors. Strains of phylogenetic groups B1 and A were projected on the positive values of F1, and each segregated in two major clusters. The two clusters with the highest positive values on the F1 axis were composed of eight phylogenetic group B1 strains and seven phylogenetic group A strains, respectively, and in both cases they corresponded to strains of commensal origin. Two clusters of intermediate positive values on the F1 axis were composed of 12 phylogenetic group B1 strains and 12 phylogenetic group A strains, respectively, and in both cases they corresponded to strains of pathologic origin. Three phylogenetic group B1 strains (strains 18, 36, and 39), which were characterized by a high level of lethality to mice (9 to 10 mice) and the detection of two virulence determinants by strain (pap operon in four strains, aer operon in three strains, afa operon in one strain, K1 antigen in two strains), were projected near the zero value of F1 close to two phylogenetic group D strains (strains 35 and 38). The two clusters of phylogenetic group A strains, on the one hand, and those of phylogenetic group B1 strains, on the other, were clearly distinguished by their opposite values on the F3 axis. One phylogenetic group A strain (strain 44) was distinguished by its strongly negative value on the F3 axis and its low positive value on the F1 axis; it corresponded to a strain characterized by a medium level of lethality to mice (seven mice).
DISCUSSION
To our knowledge, this is the first report to provide an overview of extraintestinal virulence in E. coli by combining data on commensal and pathogenic strains tested for global virulence in a standardized mouse model, for the presence or absence of several known virulence determinants, and for their classification into the different phylogenetic groups of the E. coli species. Our mouse model is able to account for a large potential of virulence and is not hampered by the variability of the susceptibility to infection of the host.
From the whole-data analysis (Fig. 2A), we clearly show that there is a link between phylogenetic group, detection of virulence determinants, and lethality in mice. Carboxylesterase B type B2 strains, which correspond to B2 phylogenetic group strains (34, 35), are rare in the feces in commensal situations. They represent 7% (in Western countries) to 3% (in developing countries) of the commensal E. coli population (12). Similarly, phylogenetic group D strains are underrepresented in the human commensal strains; in our series, the commensal strains, which all belong to carboxylesterase B type B1 (group 1), are exclusively composed in equal proportion of strains of phylogenetic groups A and B1, devoid of pathogenic determinants and not lethal for the mouse. The pathogenic strains of group 2 exhibit exactly the same characteristics as the commensal strains since all but one of them do not kill the mice, they are devoid of the tested virulence genes, and they belong to the phylogenetic groups A and B1 (Table 1). Indeed, if the “pathogenic” and “commensal” parameters are not taken into account for the whole-data analysis, the two clearly separated clusters of 7 and 12 strains for the A phylogenetic group and of 8 and 12 strains for the B1 phylogenetic group in Fig. 2B, respectively, merge into a single cluster for the A group and a single cluster for the B1 group (data not shown) without affecting the position of the rare virulent strains in these two groups (strains 44, 18, 36, and 39). This suggests that the pathogenic origin of the strains of group 2 could be related to various host-dependent factors, such as susceptibility linked to an underlying disease or site whereby the organisms gain entry. This is in agreement with the previous epidemiological studies, which have shown that strains with no virulence determinants can infect a host that has been made susceptible (5, 20, 25, 33, 40). This also explains that the character “pathogenic origin” (pat+) in Fig. 2A is at an intermediate position on the first axis as opposed to the parameter “lethality.” Still, strains of all phylogenetic groups are able to be lethal for mice. Even though phylogenetic D group strains are clearly underrepresented in our panel, it is clear that strains of this phylogenetic group as well as strains of the B1 phylogenetic group can be highly virulent for the mice, since they killed more than six mice in less than 18 h, in agreement with the presence of virulence determinants. Lastly, B2 phylogenetic group strains are clearly the most virulent strains for the mice, again in agreement with the presence of virulence determinants. Their wide distribution in the B2 area of the F1/F2 plane of the FAC analysis (Fig. 1) illustrates that they are genetically heterogeneous in accordance with the carboxylesterase B2 polymorphism (Table 1). It should be noted that in the rrn RFLP FAC analysis (Fig. 1), a clear-cut distribution between phylogenetic groups A, B1, and D and phylogenetic group B2 is observed, reinforcing the deep genetic divergence of this particular group of strains among the E. coli species (23, 32).
Our set of adult extraintestinal E. coli strains can be linked to other extraintestinal E. coli strain collections, strengthening the prominent role of the B2 phylogenetic group strains in extraintestinal infection. The EcoRI rrn RFLP pattern presented in Fig. 2 of the paper by Maslow et al. (26) which describes 187 bloodstream E. coli isolates from patients in Boston and Nairobi, Kenya, can be compared to our data. Indeed, it is clear that their ribotypes A, B2, and E, which are found only in their cluster III containing 65% of the isolates (26), correspond to ribotypes that we have observed only within phylogenetic B2 group strains (Fig. 3 in the paper by Picard et al. [34] and data not shown). Furthermore, sfa-positive strains of Maslow et al. belong only to cluster III, and we have shown in this work and also previously (5) that sfa/foc operons are found only in the phylogenetic B2 group. Similarly, EcoRI rrn RFLPs and hly operon data presented by Arthur et al. (1), when studying uropathogenic isolates, allows the conclusion that their group IV, which encompasses the majority of the strains, corresponds to phylogenetic B2 group strains. According to the common O18:K1:H7 strains studied by Selander et al. (38) and Bingen et al. (5) and to the virulence determinant distribution, most of the neonatal meningitis strains of group 1 of Selander et al. correspond to phylogenetic B2 strains. This is in accordance with the data of Bingen et al., who found that 68% of the neonatal meningitis isolates belong to the B2 phylogenetic group (5). This high proportion of B2 phylogenetic group strains in human extraintestinal infections is not specific to humans and is also observed in nonhuman mammalian extraintestinal infections (7). Finally, although the ECOR collection is not representative of human isolates, the phylogenetic B2 group and, to a lesser extent, the phylogenetic D group strains have been shown to exhibit numerous virulence determinants compared to phylogenetic A and B1 strains (5, 6).
In conclusion, within the E. coli species, there is a phylogenetic group of strains, the B2 phylogenetic group, which is deeply divergent from the other strains and which represents the vast majority of strains involved in various extraintestinal infections. Such a distribution of virulence within phylogenetic groups in a species which remains clonal despite horizontal gene transfer (9, 15, 27, 28) allows the evolutionary hypothesis of an early genetic divergence within the E. coli species which led to phylogenetic group A and B1 commensal strains, on the one hand, and to the B2 phylogenetic group of pathogenic strains, on the other. This latter group of strains evolved toward extraintestinal virulence by acquisition of numerous pathogenic determinants (23). The observed stable link between virulence and phylogeny could correspond to the necessity of having virulence determinants into the right genetic background for the emergence of a virulent clone, an expression of the interdependence of microbial pathogenicity and metabolic activities in pathogenic bacteria (10). Selection could have favored strains of intermediate or high levels of virulence, since they compensate for the loss of transmission opportunities that result from killing or debilitating their hosts by increased fitness in the presence of host defenses or other bacterial strains (24).
ACKNOWLEDGMENTS
We are grateful to Xavier Nassif for helpful discussions, to Juliette Chapron for her advice on statistical analysis, and to Francine Grimont for providing us with E. coli E2348/69.
This work was supported in part by the Programme Hospitalier de Recherche Clinique (grant AOM 96069).
Footnotes
This paper is dedicated to the memory of Philippe Goullet, a pioneer in the analysis of the genetic structure of E. coli.
REFERENCES
- 1.Arthur M, Arbeit R D, Kim C, Beltran P, Crowe H, Steinbach S, Campanelli C, Wilson R A, Selander R K, Goldstein R. Restriction fragment length polymorphisms among uropathogenic Escherichia coli isolates: pap-related sequences compared with rrn operons. Infect Immun. 1990;58:471–479. doi: 10.1128/iai.58.2.471-479.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg R D. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4:430–435. doi: 10.1016/0966-842x(96)10057-3. [DOI] [PubMed] [Google Scholar]
- 3.Bingen E H, Denamur E, Lambert-Zechovsky N Y, Bourdois A, Mariani-Kurkdjian P, Cezard J P, Navarro J, Elion J. DNA restriction-fragment length polymorphism differentiates crossed from independent infections in nosocomial Xanthomonas maltophilia bacteremia. J Clin Microbiol. 1991;29:1348–1350. doi: 10.1128/jcm.29.7.1348-1350.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bingen E, Denamur E, Lambert-Zechovsky N, Aujard Y, Brahimi N, Geslin P, Elion J. Analysis of DNA restriction-fragment length polymorphism extends the evidence for breast milk transmission in Streptococcus agalactiae late-onset neonatal infection. J Infect Dis. 1992;165:569–573. doi: 10.1093/infdis/165.3.569. [DOI] [PubMed] [Google Scholar]
- 5.Bingen E, Picard B, Brahimi N, Mathy S, Desjardins P, Elion J, Denamur E. Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent B2 group strains. J Infect Dis. 1998;177:642–650. doi: 10.1086/514217. [DOI] [PubMed] [Google Scholar]
- 6.Boyd E F, Hartl D L. Chromosomal regions specific to pathogenic isolates of Escherichia coli have a phylogenetically clustered distribution. J Bacteriol. 1998;180:1159–1165. doi: 10.1128/jb.180.5.1159-1165.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherifi A, Contrepois M, Picard B, Goullet P, Orskov I, Orskov F, De Rycke J. Clonal relationship among Escherichia coli serogroup O6 isolates from human and animal infections. FEMS Microbiol Lett. 1991;80:225–230. doi: 10.1016/0378-1097(91)90600-f. [DOI] [PubMed] [Google Scholar]
- 8.Cross A, Orskov I, Orskov F, Sadoff J, Gemski P. Identification of K1 Escherichia coli antigen. J Clin Microbiol. 1984;20:302–304. doi: 10.1128/jcm.20.2.302-304.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desjardins P, Picard B, Kaltenböck B, Elion J, Denamur E. Sex in Escherichia coli does not disrupt the clonal structure of the population: evidence from random amplified polymorphic DNA and restriction-fragment length polymorphism. J Mol Evol. 1995;41:440–448. doi: 10.1007/BF00160315. [DOI] [PubMed] [Google Scholar]
- 10.Falkow S. The evolution of pathogenicity in Escherichia coli, Shigella, and Salmonella. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 2723–2729. [Google Scholar]
- 11.Göransson M, Forsman K, Uhlin B E. Regulatory genes in the thermoregulation of Escherichia coli pili gene transcription. Genes Dev. 1989;3:123–130. doi: 10.1101/gad.3.1.123. [DOI] [PubMed] [Google Scholar]
- 12.Goullet P, Picard B. Comparative esterase electrophoretic polymorphism of Escherichia coli isolates obtained from animal and human sources. J Gen Microbiol. 1986;132:1843–1851. doi: 10.1099/00221287-132-7-1843. [DOI] [PubMed] [Google Scholar]
- 13.Goullet P, Picard B. Highly pathogenic strains of Escherichia coli revealed by the electrophoretic patterns of carboxylesterase B. J Gen Microbiol. 1986;132:1853–1858. doi: 10.1099/00221287-132-7-1853. [DOI] [PubMed] [Google Scholar]
- 14.Greenacre M J. Theory and applications of correspondence analysis. London, United Kingdom: Academic Press, Ltd.; 1984. [Google Scholar]
- 15.Guttman D S. Recombination and clonality in natural populations of Escherichia coli. Trends Ecol Evol. 1997;12:16–22. doi: 10.1016/s0169-5347(96)10057-4. [DOI] [PubMed] [Google Scholar]
- 16.Hacker J, Blum-Oehler G, Mühldorfer I, Tschäpe H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 17.Herzer P J, Inouye S, Inouye M, Whittman T S. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J Bacteriol. 1990;172:6175–6181. doi: 10.1128/jb.172.11.6175-6181.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang S H, Wass C, Fu Q, Prasadarao N V, Stins M, Kim K S. Escherichia coli invasion of brain microvascular endothelial cells in vitro and in vivo: molecular cloning and characterization of invasion gene ibe10. Infect Immun. 1995;63:4470–4475. doi: 10.1128/iai.63.11.4470-4475.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson J R, Goullet P, Picard B, Moseley S L, Roberts P L, Stamm W E. Association of carboxylase B electrophoretic pattern with presence and expression of urovirulence factor determinants and antimicrobial resistance among strains of Escherichia coli that cause urosepsis. Infect Immun. 1991;59:2311–2315. doi: 10.1128/iai.59.7.2311-2315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson J R, Orskov I, Orskov F, Goullet P, Picard B, Moseley S L, Roberts P L, Stamm W E. O, K, and H antigens predict virulence factors, carboxylesterase B pattern, antimicrobial resistance, and host compromise among Escherichia coli strains causing urosepsis. J Infect Dis. 1994;169:119–126. doi: 10.1093/infdis/169.1.119. [DOI] [PubMed] [Google Scholar]
- 21.Lebart L, Morineau A, Warwick K M. Multivariate descriptive analysis: correspondence analysis and related technique for large matrices. New York, N.Y: Wiley-Interscience; 1984. [Google Scholar]
- 22.Le Bouguenec C, Archambaud M, Labigne A. Rapid and specific detection of the pap, afa, and sfa adhesin-encoding operons in uropathogenic Escherichia coli strains by polymerase chain reaction. J Clin Microbiol. 1992;30:1189–1193. doi: 10.1128/jcm.30.5.1189-1193.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lecointre G, Rachdi L, Darlu P, Denamur E. Escherichia coli molecular phylogeny using the incongruence length difference test. Mol Biol Evol. 1998;15:1685–1695. doi: 10.1093/oxfordjournals.molbev.a025895. [DOI] [PubMed] [Google Scholar]
- 24.Lipsitch M, Moxon E R. Virulence and transmissibility of pathogens: what is the relationship? Trends Microbiol. 1997;5:31–37. doi: 10.1016/S0966-842X(97)81772-6. [DOI] [PubMed] [Google Scholar]
- 25.Maslow J N, Mulligan M E, Adams K S, Justis J C, Arbeit R D. Bacterial adhesins and host factors: role in the development and outcome of Escherichia coli bacteremia. Clin Infect Dis. 1993;17:89–97. doi: 10.1093/clinids/17.1.89. [DOI] [PubMed] [Google Scholar]
- 26.Maslow J N, Whittam T S, Gilks C F, Wilson R A, Mulligan M E, Adams K S, Arbeit R D. Clonal relationships among bloodstream isolates of Escherichia coli. Infect Immun. 1995;63:2409–2427. doi: 10.1128/iai.63.7.2409-2417.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maynard Smith J, Smith N H, O’Rourke M, Spratt B G. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milkman R. Recombination and population structure in Escherichia coli. Genetics. 1997;146:745–750. doi: 10.1093/genetics/146.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mühldorfer I, Hacker J. Genetic aspects of Escherichia coli virulence. Microb Pathog. 1994;16:171–181. doi: 10.1006/mpat.1994.1018. [DOI] [PubMed] [Google Scholar]
- 30.Ochman H, Selander R K. Standard reference strains of Escherichia coli from natural population. J Bacteriol. 1984;157:690–692. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ott M, Hacker J. Analysis of the variability of S-fimbriae expression in an Escherichia coli pathogen. FEMS Microbiol Lett. 1991;63:233–238. doi: 10.1016/0378-1097(91)90091-n. [DOI] [PubMed] [Google Scholar]
- 32.Picard B, Goullet P, Krishnamoorthy R. A novel approach to study of the structural basis of enzyme polymorphism. Analysis of carboxylesterase B of Escherichia coli as a model. Biochem J. 1987;241:877–881. doi: 10.1042/bj2410877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Picard B, Goullet P. Correlation between electrophoresis types B1 and B2 of carboxylesterase B and host-dependent factors in Escherichia coli septicaemia. Epidemiol Infect. 1988;100:51–61. doi: 10.1017/s0950268800065559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Picard B, Picard-Pasquier N, Krishnamoorthy R, Goullet P. Characterization of highly virulent Escherichia coli strains by ribosomal DNA restriction fragment length polymorphism. FEMS Microbiol Lett. 1991;82:183–188. doi: 10.1016/0378-1097(91)90330-d. [DOI] [PubMed] [Google Scholar]
- 35.Picard B, Journet-Mancy C, Picard-Pasquier N, Goullet P. Genetic structures of the B2 and B1Escherichia coli strains responsible for extra-intestinal infections. J Gen Microbiol. 1993;139:3079–3088. doi: 10.1099/00221287-139-12-3079. [DOI] [PubMed] [Google Scholar]
- 36.Pupo G M, Karaolis D R K, Lan R, Reeves P S. Evolutionary relationship among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect Immun. 1997;65:2685–2692. doi: 10.1128/iai.65.7.2685-2692.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Selander R K, Lewin B R. Genetic diversity and structure in Escherichia coli populations. Science. 1980;210:545–547. doi: 10.1126/science.6999623. [DOI] [PubMed] [Google Scholar]
- 38.Selander R K, Korhonen T K, Väisanen-Rhen V, Williams P H, Pattison P E, Caugant D A. Genetic relationships and clonal structure of strains of Escherichia coli causing neonatal septicemia and meningitis. Infect Immun. 1986;52:213–222. doi: 10.1128/iai.52.1.213-222.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tenehaus M, Young F W. An analysis and synthesis of multiple correspondence analysis, optimal scaling, dual scaling, homogeneity analysis and other methods for quantifying categorical multivariate data. Psychometrika. 1985;50:91–119. [Google Scholar]
- 40.Tullus K, Brauner A, Fryklund B, Munkhammar T, Rabsch W, Reissbrodt R, Burman L G. Host factors versus virulence-associated bacterial characteristics in neonatal and infantile bacteraemia and meningitis caused by Escherichia coli. J Med Microbiol. 1992;36:203–208. doi: 10.1099/00222615-36-3-203. [DOI] [PubMed] [Google Scholar]
- 41.Whittam T S, Wolfe M L, Wachsmuth I K, Orskov F, Orskov I, Wilson R A. Clonal relationship among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect Immun. 1993;61:1619–1629. doi: 10.1128/iai.61.5.1619-1629.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]