Abstract
Prior work has identified signal sequences and motifs that are necessary and sufficient to target proteins to specific subcellular regions and organelles such as the plasma membrane, nucleus, endoplasmic reticulum, and mitochondria. In contrast, minimal sequence motifs that are sufficient for Golgi localization remain largely elusive. In this work, we identified a 37–amino acid alternative open reading frame (altORF) within the mRNA of the centromere protein CENP-R. This altORF peptide localizes specifically to the cytoplasmic surface of the Golgi apparatus. Through mutational analysis, we identify a minimal 10–amino acid sequence and a critical cysteine residue that are necessary and sufficient for Golgi localization. Pharmacological perturbations suggest that this peptide undergoes lipid modification to promote its localization. Together, our work defines a minimal sequence that is sufficient for Golgi targeting and provide a valuable Golgi marker for live cell imaging.
INTRODUCTION
The Golgi apparatus contributes to multiple critical cellular functions, including protein modification and sorting, vesicle transport, and lipid biosynthesis (Donaldson and Lippincott-Schwartz, 2000; Wilson et al., 2011; Bankaitis et al., 2012). The Golgi is also a highly dynamic membranous structure. Membrane trafficking through the secretory pathway results in frequent turnover of the Golgi membrane as proteins are delivered to and exported from the Golgi by vesicles (Lujan and Campelo, 2021). In addition, Golgi morphology can vary in response to cell state, such as during mitosis or apoptosis, where the Golgi disassembles through tightly regulated processes (Liu et al., 2021). For the Golgi to perform its critical functions, resident proteins must be targeted to this organelle in a way that allows them to persist despite this reorganization. One mechanism of targeting proteins to the Golgi involves trafficking through the secretory pathway, where proteins are first targeted to the endoplasmic reticulum (ER) before they are transported to the Golgi (Lujan and Campelo, 2021). Once at the Golgi, proteins are retained due to their association with unique features of the Golgi such as membrane thickness or their interaction with other resident Golgi proteins (Banfield, 2011). Alternatively, cytoplasmic–facing peripheral membrane proteins, such as a class of proteins called golgins, can be directly targeted to the Golgi. The association and retention of these peripheral membrane proteins at the Golgi is facilitated by lipid modifications such as myristoylation, palmitoylation, and prenylation, or indirectly through protein–protein interactions (Banfield, 2011).
For other organelles, such as nuclei or mitochondria, protein targeting requires the presence of organelle-specific signal sequences. For example, one class of nuclear localization sequences (NLS), termed “monopartitie,” is characterized by a cluster of four to eight basic amino acids in which four or more are arginine or lysine residues (Lu et al., 2021). Similarly, protein targeting to the mitochondria is mediated by a signal sequence found at a protein’s N-terminus and is characterized by the presence of positively charged and hydrophobic amino acids (Chacinska et al., 2009). In contrast, although prior work has identified protein domains that are required for the localization of resident or viral-derived Golgi proteins (Kjer-Nielsen et al., 1999a,b; Munro and Nichols, 1999; Nozawa et al., 2003; Liu and Zheng, 2007), a minimal consensus signal sequence for the directed targeting of proteins to the Golgi has not been identified.
In this work, we identify a novel 37 amino acid altORF peptide that localizes robustly to the Golgi apparatus. This altORF is expressed from the transcript of the CENP-R gene, a protein that canonically localizes to kinetochores (Hori et al., 2008). Given the unique localization behavior of this short peptide sequence, we dissected the mechanisms that underlie this localization behavior, including the identification of a critical cysteine residue and the definition of a minimal eight–amino acid consensus sequence. The robust localization of the altORF to the Golgi also provides a valuable imaging tool that can contribute to the analysis of dynamic Golgi behavior. This work provides valuable insight into sequences that can mediate protein targeting to the Golgi.
RESULTS AND DISCUSSION
A CENPR altORF peptide localizes to the Golgi compartment
Alternative open reading frames (altORF) are sequences present in transcribed mRNAs that are distinct from the canonically defined open reading frame. Such sequences have alternative translation start sites that result in the production of a novel protein sequence. AltORFs exist in a variety of proteins, and many have unknown functional roles and behavior (Orr et al., 2020). In our ongoing work, we identified a potential altORF that initiates upstream of the defined CENP-R open reading frame and has a unique reading frame (Figure 1A). Although hypothetical, this altORF was identified based on ribosome profiling data and was included in a proteomic database of small peptides produced by altORFs with a cutoff of less than 100 amino acids (Samandi et al., 2017). This CENPR altORF corresponds to a peptide sequence of 37 amino acids with a predicted molecular weight of 4.5 kDa and a pI of 9.84. This altORF sequence is evolutionarily conserved in primates (Figure 1B; Supplemental Figure 1A), but is highly divergent or absent from other vertebrates, including mice (Supplemental Figure 1B).
FIGURE 1:
A CENP-R altORF robustly localizes to the Golgi. (A) Visual representation of altORF translation from the canonical CENP-R transcript. The altORF begins upstream of the canonical ATG and has a different reading from the canonical protein. (B) Multiple sequence alignment of the altORF peptide sequence across New and Old World primates. (C) Localization of CENP-R altORF during interphase and mitosis as compared with the canonical CENP-R after transfection of GFP tagged constructs in HeLa cells. The CENP-R altORF shows localization distinct from that of canonical CENP-R, which localizes to centromeres. GFP boost was used to amplify GFP signal and kinetochores are stained with ACA. Images in this figure are deconvolved and max projected. Scale bars, 10 μm. (D) The CENP-R altORF colocalizes with the Golgi markers GM130 (cis-Golgi) and GGA2 (trans-Golgi), but not with the ER marker anti-KDEL. Representative images of fixed cells stably expressing altORF peptide are shown. Cells were imaged using high-resolution Airyscan confocal microscopy. Images represent a single Z position. Inset represents a 5 μm × 5 μm area. (E) The CENP-R altORF localizes to the Golgi in a variety of cell lines, including mouse fibroblast 3T3 cells. The CENP-R altORF was transiently transfected into each cell lines and assessed for colocalization with Golgi marker GM130. The GM130 antibody does not recognize the mouse protein so it was not included in the 3T3 panel. Images are deconvolved and max projected. Scale bar, 10 μm.
We first sought to assess the cellular behavior of this altORF peptide by ectopically expressing this altORF sequence with an N-terminal GFP tag in human HeLa cells. Strikingly, we found that this peptide displayed clear localization to what appeared to be membrane-bound structures during interphase (Figure 1C). These membranous structures localized peripherally to the nucleus and were typically compact, clustering adjacent to the nucleus. In mitosis, the altORF localized to distinct puncta throughout the cytoplasm of the dividing cell (Figure 1C). This localization behavior contrasts with that of the canonical CENP-R protein, which localizes to centromeres in both interphase and mitosis (Figure 1C). We also observed similar localization when the altORF was tagged at its C-terminus (altORF-GFP; Supplemental Figure 1C) or when expressed with a HaloTag (Supplemental Figure 1D).
Based on this observed localization to a membrane compartment adjacent to the nuclear periphery, we hypothesized that the altORF localizes to either the ER or the Golgi apparatus. To test this, we compared the altORF localization with established markers for the ER—using an anti-KDEL antibody—and the Golgi—using an anti-GM130 antibody. We found that the altORF peptide did not colocalize with ER markers but overlapped closely with GM130 in fixed cells (Figure 1D; Supplemental Figure 2A). The Golgi network is further divided into cis, medial, and trans compartments. To define the localization behavior of this altORF peptide, we additionally compared its localization with that of GM130, a marker of the cis-Golgi, and GGA2, a marker of the trans-Golgi (Nakamura et al., 1995; Mardones et al., 2007). We observed partial colocalization between the altORF peptide and GM130 (Figure 1D; Supplemental Figure 2A) in both interphase and mitosis. In contrast, the altORF peptide did not overlap with the trans-Golgi marker GGA2 in either cell cycle phase (Figure 1D; Supplemental Figure 2A). We conclude that the altORF peptide localizes to the Golgi in HeLa cells, although we cannot definitively specify in which subcompartment(s) of the Golgi the altORF resides.
We next sought to test whether the observed localization behavior for the CENPR altORF also occurred in other cell lines. To test this, we transfected the N-terminal GFP construct into a panel of human cell lines, including the lung carcinoma A549 cell line, the osteosarcoma U2OS cell line, and the human embryonic kidney 293T cell line. In each case, we observed localization to the nuclear periphery similar to that in HeLa cells, as well as colocalization with GM130 (Figure 1E). In addition to these human cell lines, we tested the localization of the human altORF in mouse fibroblast NIH3T3 cells. Although this altORF sequence is absent from mice, we observed a similar localization of the altORF to the Golgi in NIH3T3 cells (Figure 1E). We therefore conclude that the altORF robustly localizes to the Golgi across a variety of mammalian cell lines.
The CENPR altORF remains associated during Golgi remodeling and dynamics
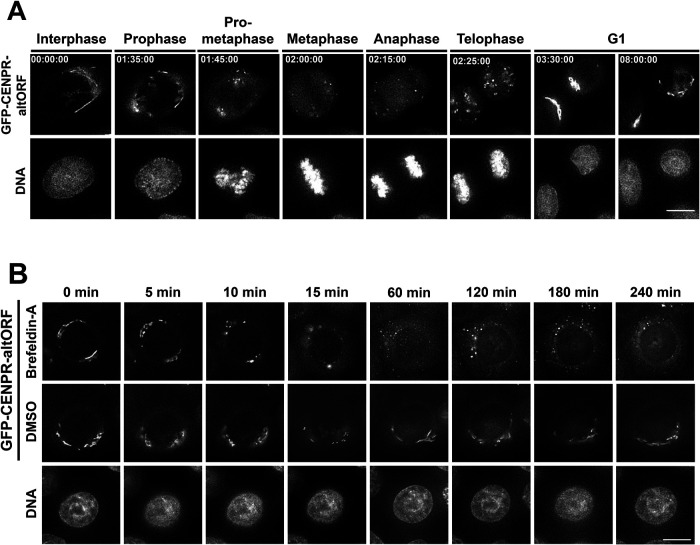
To assess the dynamic localization of the altORF to the Golgi, we used the altORF construct to generate a cell line stably expressing the N-terminally tagged GFP-altORF using retroviral integration. Stable expression of the altORF had no apparent effect on cell growth or Golgi morphology (Supplemental Figure 2, B and C). Using this cell line, we analyzed the association of the altORF with the Golgi under conditions where Golgi morphology is dynamic. First, we visualized the localization of the altORF peptide throughout the cell cycle. The Golgi undergoes a cycle of assembly and disassembly as cells progress through cell division (Tang and Wang, 2013). Using time-lapse imaging, we observed the rapid fragmentation of the Golgi upon mitotic entry, with the altORF appearing as small puncta throughout the cytoplasm of the dividing cell, consistent with prior fixed-cell analysis. As cells exited mitosis in telophase, we observed the reformation of larger stacks and ultimately the full reformation of the Golgi into ribbonlike structures in G1 (Figure 2A).
FIGURE 2:
The CENP-R altORF marks the Golgi throughout dynamic processes. (A) Images from a time-lapse sequence in a cell line stably expressing the CENP-R altORF. DNA was stained using SiR-DNA. Cells were imaged for 12 h at 5-min intervals at 37°C in CO2-independent media. The time-lapse video was deconvolved. Scale bar, 10 μm. (B) Images from time-lapse sequence of a cell line stably expressing the CENP-R altORF following treatment with 0.2 μM Brefeldin A. DNA was stained with SiR-DNA. Cells were imaged every 5 min for 4 h immediately after the addition of Brefeldin A. Selected frames show the breakdown of the Golgi and the association of the altORF to the Golgi throughout this process. Scale bar, 10 μm.
Next, we sought to visualize altORF localization and Golgi behavior by inducing morphological changes to the Golgi using Brefeldin A treatment. Brefeldin A is an inhibitor of the secretory pathway that causes the Golgi to disassemble and redistribute into the ER (Fujiwara et al., 1988). Upon treatment with Brefeldin A, we observed the breakdown of the ribbonlike structures of the Golgi into small puncta—behavior that has been observed previously for Golgi-associated proteins such as GM130 (Figure 2B; Mardones et al., 2006). Indeed, we observed similar localization of GFP-altORF and GM130 staining following BFA treatment (unpublished data). Thus, the altORF peptide remains robustly associated with the Golgi membrane under conditions where the organelle is disassembled, providing a valuable tool for the live cell imaging of Golgi dynamics.
The CENPR altORF localizes to the cytoplasmic surface of the Golgi
The robust localization of this 37-amino acid altORF peptide to the Golgi raises questions as to how this peptide associates with and is retained at the Golgi. We first sought to determine whether this altORF peptide is associated with the cytoplasmic or luminal side of the Golgi membrane. To test this, we utilized a linker that contains a TEV protease cleavage site between GFP and the start of the altORF sequence (Cheeseman and Desai, 2005). We then expressed mCherry-tagged TEV protease in cells stably expressing this GFP-TEV-altORF peptide fusion. If the altORF peptide faces the cytoplasm, then the TEV cleavage site should be exposed and susceptible to cleavage by the cytoplasmic TEV protease, thereby resulting in the loss of Golgi-localized GFP signals. Indeed, upon TEV expression, we observed a substantial increase in cytoplasmic GFP fluorescence and a loss of Golgi-specific GFP localization (Figure 3A), suggesting that the altORF localizes to the cytoplasmic surface of the Golgi.
FIGURE 3:
CENP-R altORF associates to the cytoplasmic surface of the Golgi. (A)Localization of the GFP-CENPR altORF peptide stably expressed in HeLa cells in the presence of transfected mCherry-TEV protease construct. Localization to the Golgi was determined based on colocalization with Golgi marker GM130. Images are deconvolved and maximally projected. Scale bar, 10 μm. (B) Hydrophobicity plot of the CENPR altORF peptide sequence. Sequence was run through the Phobius program (https://phobius.sbc.su.se/poly.html). Probability values assigned to amino acids to determine transmembrane topology are plotted. (C) Western showing the presence of GFP-altORF or GM130 following lysis with buffers of various amounts of the detergent NP40. Asynchronous HeLa cells stably expressing the altORF peptide were used in this experiment. Following cell lysis, lysates were centrifuged to pellet the cellular debris. From this the supernatant was taken and labeled as the “Sup.” sample and the pellet was resuspended in loading sample buffer and used as the “Pellet” sample.
We next sought to elucidate the sequence requirements for targeting the altORF peptide to the Golgi. Resident Golgi proteins can associate with the Golgi membrane as transmembrane or peripheral membrane proteins. Golgi-associated transmembrane domains typically span ∼20 aa in length and are enriched in aromatic and hydrophobic amino acids (Sharpe et al., 2010). To determine whether this altORF peptide could act as a transmembrane protein at the Golgi, we utilized the Phobius online transmembrane topology tool to test for the presence of predicted transmembrane domains (Kall et al., 2004; Kall et al., 2007). Analysis of the altORF sequence alone found a low probability for the presence of a transmembrane domain, suggesting that the altORF likely associates with the Golgi as a peripheral membrane protein (Figure 3B). To further assess the association of the altORF with the Golgi, we used cell lysis in the presence of NP40 to assess the conditions required to solubilize the altORF peptide. We compared this solubilization behavior with that of GM130, a well-characterized Golgi protein known to act as a strongly associated peripheral membrane protein (Nakamura et al., 1995). We found that the altORF peptide was solubilized in the presence of 0.05% NP40 detergent, whereas GM130 required 1% NP40 to be liberated to the supernatant fraction (Figure 3C). This indicates that the altORF is weakly associated with the Golgi relative to GM130.
Some proteins that are peripherally associated with the Golgi are associated through protein–protein interactions with Golgi-resident proteins. To determine whether altORF association with the Golgi was mediated through its interaction with another Golgi-associated protein, we sought to identify altORF-interacting proteins by immunoprecipitation–mass spectrometry (IP-MS) analysis. Although we were able to isolate the altORF peptide as a GFP fusion, we did not detect any additional Golgi-associated proteins in our mass spectrometry analysis relative to controls (unpublished data). Thus, although our IP experiments do not exclude the possibility that protein–protein interactions help to promote altORF localization, robust and persistent protein–protein interactions are unlikely to be responsible for retaining the altORF at the Golgi.
Cysteine residues are important for localization of the altORF peptide to the Golgi
We next sought to determine the amino acid residues that are required for Golgi localization. The altORF peptide has regions with hydrophobic amino acid residues and aromatic residues, and contains two cysteine residues (Supplemental Figure 2D), which may play roles in Golgi localization (Banfield, 2011). To test the contributions of these residues, we generated truncation mutants to determine the minimal sequence required for Golgi localization (Figure 4A). We found that truncating the last 23 amino acids did not affect Golgi localization, as the GFP-altORF21aa and the GFP-altORF14aa mutants colocalized with GM130 (Figure 4B). However, truncating the altORF peptide sequence further to its first nine amino acids resulted in the loss of Golgi localization and an increase in the cytoplasmic GFP signal (Figure 4B). Next, we truncated an additional four amino acids from the N-terminus of the altORF14aa mutant, resulting in a 10–amino acid sequence (amino acids 5–14). This 10–amino acid sequence was sufficient to direct Golgi localization (Figure 4B). Of the residues eliminated through the truncation of the altORF sequence from 14 to 9 amino acids, the loss of the Cys11 residue was particularly interesting, given its potential to be modified (Figure 4A). To test the role of cysteine residues in altORF localization, we generated an altORF mutant in which both cysteine residues were mutated to alanine (altORFC11A, C32A). The altORFC11A, C32A mutant failed to localize robustly to the Golgi (Figure 4C), suggesting that the cysteine residues play a critical role in targeting and retaining this peptide to the Golgi.
FIGURE 4:
Cysteine residues are required for altORF localization to the Golgi. (A) Figures representing the truncation mutants generated and tested for localization. (B) Localization of the truncated altORF constructs transfected in HeLa cells. Localization to the Golgi was determined based on colocalization with Golgi marker GM130. Cells were fixed and imaged 48 h post transfection. Images are deconvolved and max projected. Scale bar, 10 μm. (C) Localization of the dual cysteine mutant altORF constructs transfected in HeLa cells. Localization to the Golgi was determined based on colocalization with Golgi marker GM130. Cells were fixed and imaged 48 h post transfection. Images are deconvolved and max projected. Scale bar is 10 μm. (D) Localization of the altORF peptide following treatment with 2-bromopalmitate. Cells were treated with either DMSO or 10 μM 2-bromopalmitate for 3 h before being fixed. Images are deconvolved. Scale bar, 10 μm. GFP signal is graphed as a ratio of GM130 fluorescence per cell. Quantification of GFP signal in DMSO and 2-BP treated cells with N = 34 and N = 22, respectively. The Golgi area was determined blindly using GM130 as the guide; the GFP fluorescence intensity was then recorded. Background signal was subtracted from fluorescence intensity. Background signal was determined by measuring the fluorescence intensity of a region of the same area as the Golgi selected from an area of the image neighboring the selection. Error bars represent the mean and SD. Statistical significance **** p < 0.0001 as determined by Mann–Whitney test.
We next took an evolution-guided approach to define the requirements for altORF Golgi localization. The altORF sequence present in New World primates differs from that in hominids and Old World primates due to an insertion and deletion event that disturbs and restores the reading frame, resulting in a peptide sequence that differs from the human altORF peptide sequence (Supplemental Figure 1, A and B; Harmit Malik, personal communication). Although both the C11 and C32 cysteine residues are present in the Old World primate altORF peptide sequences, only the C32 residue is conserved in New World primates (Figure 1B). To evaluate the consequences of these changes in Golgi localization, we tested the localization of the Capuchin altORF sequence, which lacks the C11 residue. Using transfection of a GFP-tagged construct into HeLa cells, we found that the Capuchin altORF sequence did not localize to the Golgi (Figure 4C).
Based on both our observations from the mutational analysis and the evolutionary conservation of the altORF sequence, we hypothesized that the Cys11 residue would be required for Golgi localization. To test this, we generated altORF mutants in which individual cysteine residues were mutated to alanine (altORFC11A and altORFC32A). We found that the altORFC11A failed to localize to the Golgi and was instead localized diffusely throughout the cytoplasm (Figure 4C). In contrast, the altORFC32A mutant behaved similarly to the wild-type sequence, localizing to the Golgi (Figure 4C). We therefore conclude that the Cys11 residue is necessary for the altORF to localize to the Golgi.
Lipid modification of the altORF peptide contributes to Golgi localization
Our results suggest that the altORF peptide associates with the Golgi as a peripheral membrane protein and that the cysteine residues are required for this localization. Therefore, we hypothesized that the localization of the altORF to the Golgi is facilitated by a posttranslational modification that targets the cysteine residues in this peptide. Potential posttranslational modifications that occur at cysteine residues and are known to contribute to Golgi localization include myristoylation, prenylation, and palmitoylation (Aicart-Ramos et al., 2011; Banfield, 2011; Palsuledesai and Distefano, 2015; Udenwobele et al., 2017). Due to the absence of clear myristoylation or prenylation consensus motifs within the altORF sequence (Palsuledesai and Distefano, 2015; Udenwobele et al., 2017), we hypothesized that palmitoylation may be involved in facilitating the localization of the altORF to the Golgi. Palmitoylation is a reversible posttranslational lipid modification that typically functions to enhance protein hydrophobicity and promote the membrane association of modified proteins (Guan and Fierke, 2011). The process of palmitoylation is catalyzed by a large family of enzymes termed palmitoyl acyltransferases (PATs), consisting of 23 distinct proteins that all share a core DHHC motif (Philippe and Jenkins, 2019). Most of these enzymes are enriched at the Golgi, although they additionally localize to the ER and plasma membrane (Ohno et al., 2006; Rocks et al., 2010; Ernst et al., 2018). The function of these PAT enzymes is largely redundant, as multiple PAT enzymes have been shown to have overlapping substrates. Currently, no generalized consensus motifs have been identified for palmitoylated proteins (Salaun et al., 2010).
To test whether palmitoylation is involved in altORF Golgi localization, we first sought to identify PAT enzymes that are required for altORF localization. Utilizing, a CRISPR/Cas9 inducible knockout strategy (McKinley and Cheeseman, 2017), we specifically targeted the PAT enzymes that localize to the Golgi, including ZDHHC3, ZDHHC7, ZDHHC9, ZDHHC11, ZDHHC13, ZDHHC15, ZDHHC17, ZDHHC21, and ZDHHC22 (Rocks et al., 2010; Ernst et al., 2018). However, likely due to the functional redundancy of these enzymes, knocking out individual enzymes or two enzymes combinations (specifically ZDHHC 3/7; 3/9, 7/9) did not result in the mislocalization of the altORF (data not shown) (Rocks et al., 2010; Greaves et al., 2017; Philippe and Jenkins, 2019). Thus, as an alternative approach, we sought to pharmacologically perturb cellular palmitoylation utilizing the inhibitor 2-bromopalmitate (2-BP), a palmitate analog that inhibits this process (Jennings et al., 2009; Salaun et al., 2010). Following treatment with 2-BP for 4 h, we observed the mislocalization of the altORF from the Golgi and an increase in cytoplasmic GFP signal (Figure 4D). Based on this observation, together with the fact that the peptide lacks a transmembrane domain and requires a cysteine residue for localization, our results suggest that the peptide likely requires a posttranslational modification, such as palmitoylation, to promote its localization to the Golgi.
Determination of a minimal Golgi-targeting sequence
Given the critical role the Cys11 residue plays in the localization of this peptide, we next sought to determine what other amino acid residues within the minimal 10–amino acid sequence, identified in the altORFaa5-14 truncation mutant, are required for Golgi localization (Figure 4A). Within this 10–amino acid sequence, there is an “FCF” motif that is conserved across Old World primates, with varying amino acids surrounding this motif (Figure 1B). Therefore, we sought to assess the functional importance of the amino acids directly surrounding the cysteine residue. Mutating the phenylalanine residues to tyrosine to retain their aromatic character resulted in a loss of localization. In contrast, mutating these residues to leucine to preserve the hydrophobicity did not affect Golgi localization (Figure 5A). This suggests that the presence of hydrophobic amino acids on either side of the cysteine is important for Golgi localization.
FIGURE 5:
FCF motif not sufficient to confer Golgi localization. (A) Representative images of the FCF mutant localization in HeLa cells. Cells were fixed and imaged 48 h posttransfection. Images are deconvolved and max projected. Scale bar is 10 μm. (B) Localization of GFP-tagged altORF mutant testing sufficiency of FCF motif. Constructs were transiently transfected in HeLa cells and expressed for 48 h before fixation. Images are deconvolved and max projected. Scale bar, 10 μm. (C) Localization of GFP-tagged Sabaeus altORF and motif mutant testing sufficiency of FCF motif. Constructs were transiently transfected in HeLa cells and expressed for 48 h before fixation. Images are deconvolved and max projected. Scale bar, 10 μm. (D) Representative images of the alanine mutants of the 10–amino acid peptide sequence. Constructs were transiently expressed in HeLa cells for 48 h before fixation. Images were deconvolved and max projected. Scale bar, 10 μm. (E) Table summarizing the mutants that failed to localize and the proposed consensus sequence required for Golgi localization. In the consensus sequence the () brackets represent the amino acids that can be substituted into that position and the {} brackets represent amino acids that cannot be included within a given position. The φ represents amino acids that are hydrophobic.
Next, we examined whether the FCF motif is sufficient to direct Golgi localization. To test this, we took the altORFC11A mutant, which does not localize to the Golgi (Figure 4B), and mutated the two amino acid residues adjacent to the C32 residue to phenylalanine (Supplemental Figure 2E). This mutant failed to localize to the Golgi, indicating that the FCF motif is not sufficient to confer Golgi localization (Figure 5B). Finally, we tested the functional importance of the amino acid residues surrounding the FCF motif. First, we utilized the evolutionarily conserved altORF sequences to probe whether the surrounding amino acids are needed to promote the Golgi localization. Based on this analysis, we selected the altORF sequences from two distinct primates—the Gorilla sequence, which contains one amino acid substitution, and the Sabaeus sequence, which contains two amino acid substitutions. Despite these limited changes, neither sequence localized to the Golgi (Figure 5C), suggesting stringent sequence requirements for Golgi localization. Thus, we took an unbiased approach to dissect the amino acid requirements at each position within the altORF sequence. To do this, we systematically performed site directed mutagenesis in which we targeted each amino acid in the peptide sequence (Figure 5D; Supplemental Figure 3A). We generated mutant constructs in which we individually mutated each amino acid within the peptide sequence to alanine or an amino that alters the characteristics of a given residue to determine which amino acids can be substituted at each position (Supplemental Figure 3A). From this analysis, we established the following generalized motif required for Golgi localization of this peptide: X-[ILVM]-{DN}-X-{RKDN}-[FILVM]-C-[IFLVM]-X-[RK]-X (Figure 5E).
A minimal Golgi-targeting peptide
In conclusion, our work identifies a small, 37–amino acid peptide derived from the CENPR transcript that localizes robustly to the Golgi apparatus. This alternative peptide sequence is present across primates but is absent in other vertebrates, including mice. Moreover, our analysis indicates that only a subset of the altORF peptide sequences that are generated in primates are able to localize to the Golgi. This indicates that the localization behavior of the human altORF sequence is a newly acquired behavior of this peptide, representing an example of evolution in action where minor sequence changes can enable new capabilities for a protein. Although this peptide could play roles in selected tissues or cell types for a unique feature of primate biology, due to its limited conservation it is unlikely to play an essential role in facilitating a core biological function at the level of individual cells. Additionally, the levels at which this peptide is expressed endogenously in human cells remain an open question. Analyses of ribosome profiling data suggests that the peptide is expressed at lower levels than the full-length CENP-R protein. Additionally, mass spectrometry analysis to identify this peptide in proteomic samples is challenging due to the short length and specific amino acid composition of the peptide sequence. Although this does not preclude the possibility that this peptide is endogenously expressed, our work demonstrates that the localization behavior of this peptide sequence can be harnessed to use as a Golgi labeling tool.
The localization behavior of this altORF peptide is dependent on the Cys11 residue present within the peptide. Chemical perturbation of cellular palmitoylation disrupted Golgi localization of the altORF suggesting that palmitoylation is a posttranslational modification that is important for its Golgi localization. We further identified a minimal 10–amino acid sequence that is sufficient to target GFP to the Golgi, suggesting that this sequence could function as a Golgi-targeting sequence. In addition to revealing the requirements for Golgi localization, this altORF peptide provides a valuable marker for visualizing the Golgi apparatus in intact cells. This small peptide, when tagged with a fluorescent protein, can be used to image the Golgi by either transient transfection or stable expression, and remains associated with the Golgi throughout its dynamic rearrangement. In addition, this minimal peptide sequence can effectively target both GFP and Halotag to the Golgi, two proteins that are distinct in structure and size. This supports the effectiveness of this peptide as a potential strategy that can be used to target fusion proteins to the outer surface of the Golgi. Additionally, the altORF sequence can also be used in a variety of cell lines, including mouse fibroblasts, for both fixed and live imaging of the Golgi. We hope that this easy-to-use Golgi labeling construct will provide a valuable tool for researchers conducting studies of Golgi localization and dynamics in addition to the existing Golgi labeling proteins (Kall et al., 2004; Aicart-Ramos et al., 2011; Udenwobele et al., 2017).
METHODS
Request a protocol through Bio-protocol.
Cell culture, cell transfection, and cell line generation
HeLa, U2Os, A549, and 3T3 cells were cultured in DMEM supplemented with 10% fetal bovine serum (GE Healthcare), 100 U/ml penicillin and streptomycin, and 2 mM L-glutamine at 37°C with 5% CO2. Cell lines were tested for mycoplasma monthly. All GFP-tagged altORF constructs were cloned into a pBABEblast vector with an N terminal GFP-Lap tag (pIC242) as described previously (Cheeseman and Desai, 2005). A cell line stably expressing the GFP-altORF was generated by retroviral transduction in HeLa parental cell lines. Following retroviral infection, cells were selected with 2 μg/ml blasticidin (Life Technologies) for two weeks, changing media every 4 d. Cells were then FACS sorted for GFP expression into single clones. After two weeks of growth, cells were screened for proper GFP expression. Transient transfections were performed with Xtremegene-9 and Optimem per manufacturer’s instructions. Cells were transfected 24 h after seeding on poly-L-lysine–coated coverslips. Media were changed on transfected cells 16 h posttransfection; cells were then fixed and processed for immunofluorescence 48 h posttransfection. A sample of 500 ng of each plasmid was transfected. Plasmids used to express the GFP-altORF: (pAN65). Plasmids used for altORF mutants: GFP-CENPR-uORF1-STOP-1-21aa (pAN329); GFP-CENPR-uORF1-STOP-1-14aa (pAN330); GFP-CENPR-uORF1-STOP-1-9aa (pAN331); GFP-altORF_5_14aa (pAN368); GFP-altORF-C11A-C32A (pAN340); GFP-altORF-C11A (pAN345); GFP_altORF_C32A (pAN346); GFP-Capuchin-altORF (pAN323); GFP-Gorilla-altORF (pAN369); GFP-Sabaeus-altORF (pAN370); GFP-altORF-YCY (pAN372); GFP-altORF-LCL (pAN378); GFP-altORF-C11A-FC32F (pAN381); HALO-altORF (pAN385). Plasmids used in point mutation screen, in order from top to bottom as listed in the table in Supplemental Figure 3: pAN372; pAN378; pAN393; pAN406; pAN407; pAN394; pAN411; pAN412; pAN395; pAN415; pAN416; pAN396; pAN418; pAN420; pAN397; pAN423; pAN424; pAN401; pAN403; pAN402; pAN398; pAN426; pAN428; pAN399; pAN429; pAN430. Plasmid for expression of mCherry-MammalianTEVprotease: pAN327.
Immunoblotting
Cells were harvested from cell lines stably expressing the GFP-altORF (cell line: cAN34). Cells were harvested by trypsinization with 0.05% trypsin/EDTA and resuspended in DMEM. Cells were counted and pellets with 2 million cells were collected. Cells were then lysed in lysis buffer (50 mM HEPES, pH 7.4; 1 mM EGTA; 1 mM MgCl2; 100 mM KCl; 10% glycerol; 1X Complete EDTA-free protease inhibitor cocktail (Roche); 1 mM PMSF; 20 mM beta-glycerophosphate; 1 mM sodium fluoride; and 0.4 mM sodium orthovanadate, pH 7.4) with the corresponding percentage of NP40. Lysates were then sonicated. Cellular debris was removed by centrifugation. Following centrifugation, the supernatant was taken and used as the “Sup.” sample while the pellet was resuspended in loading sample buffer and used as the “Pellet.” Samples were then run on a 12% SDS–PAGE gel and transferred to PVDF membrane using 1 h semidry transfer (BioRad). Membranes were blocked for 1 h in Blocking buffer (2% nonfat dry milk + 0.1% Tween-20 in TBS). Primary antibodies (Mouse anti-GFP from Roche Prod No. 1181446001; anti-GM130 from Cell Signaling D6B1 #12480) were diluted in 0.2% nonfat dry milk in TBS +0.1% Tween and incubated on membrane overnight at 4°C. HRP-conjugated secondary antibodies (antiMouse IgG-HRP from Biorad #1705047; antiRabbit IgG-HRP from Kindle Biosciences #R1006) were diluted 1:10,000 in 0.2% nonfat dry milk in TBS +0.1% Tween for 1 h at room temperature. Clarity-enhanced chemiluminescence substrate (Bio-Rad; 1705061) was added to the membrane according to the manufacturer’s instructions. Membranes were imaged with a KwikQuant Imager (Kindle Biosciences).
Immunofluorescence and microscopy
Cells for immunofluorescence were seeded on glass coverslips coated with poly-L-lysine (Sigma-Aldrich). For all experiments, except those determining kinetochore levels throughout the cell cycle, cells were fixed in 4% formaldehyde (Sigma-Aldrich) diluted in PBS for 10 min at room temperature. Coverslips were washed with PBS plus 0.1% Triton X-100 (PBS-TX). Blocking and primary antibody dilutions were performed in Abdil (20 mM Tris, 150 mM NaCl, 0.1% Triton X-100, 3% BSA and 0.1% NaN3, pH 7.5). GFP booster (Chromotek gba-488-100; 1:500 dilution) was used to amplify GFP fluorescence for all experiments involving visualization of GFP-tagged transgenes. Microtubules were stained with DM1a antibody (Sigma-Aldrich T9026; 1:3000 dilution). For staining of Golgi for colocalization experiments GM130 antibody (Cell Signaling Technologies #12480S; 1:3200 dilution) and GGA2 antibody (BD Transduction Laboratories #612612; 1:500) were used. For staining of the ER KDEL antibody (abcam; ab176333; 1:250), Cy3- and Cy5-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories; 1:200 dilution) were diluted in PBS plus 0.1% Triton X-100. DNA was visualized by incubating cells in 1 μg/ml Hoechst-33342 (Invitrogen; H1399) in PBS plus 0.1% Triton X-100 for 10 min. Coverslips were mounted using PPDM (0.5% p-phenylenediamine and 20 mM Tris-Cl, pH 8.8, in 90% glycerol) and sealed with nail polish. Assessment of altORF localization experiment were performed blindly, where cells were randomly chosen based on visualization of the reference organelle markers (commonly GM130). For each experiment, we assessed the behavior of at least 50 cells throughout the coverslip. For live-cell imaging, cells were seeded into eight-well glass-bottomed chambers (Ibidi; Fisher Scientific cat no. 50-305-795) and moved into CO2-independent media (Life Technologies; Thermo Fisher; 18045088) with 10% fetal bovine serum (GE Healthcare), 100 U/ml penicillin and streptomycin, and 2 mM L-glutamine before imaging at 37°C. For time-lapse imaging, DNA was stained with 1 μM SiR-DNA (Chromotek; SC007).
Immunofluorescence imaging was performed using a DeltaVision Core deconvolution microscope (Applied Precision) equipped with a CoolSnap HQ2 charge-coupled device camera (Photometrics). Fixed cells were imaged at 100× magnification and live imaging was performed using 60× magnification. For fixed images, 40 Z-sections were acquired at 0.2-μm steps; for live imaging, images were acquired with three Z-sections at 5-μm steps. Images were maximally projected and deconvolved where noted. For altORF co-localization imaging, cells were imaged using Airyscan microscopy conducted on a Zeiss LSM980 confocal microscope equipped with a Plan-Apochromat 63×/1.4 oil objective. Raw Airyscan images were processed using a Zen v3.5 program. Statistical analyses were performed using Prism (Graphpad software). Details of statistical tests and sample sizes are provided in the figure legends.
Small molecule treatments
Cells were treated with 0.2 μM Brefeldin A (Sigma Aldrich B5936; resuspended in DMSO) diluted in media. Brefeldin A was diluted in CO2 independent media before the start of time-lapse imaging. 2-Bromopalmitate (Sigma Aldrich 21604; resuspended in DMSO) was used at 10 μM. For all small molecule treatments, control cells were treated with the same volume of DMSO alone. Cells were treated for 4 h before fixation.
Movie S1.
Supplementary Material
Acknowledgments
The authors thank the members of the Cheeseman lab for their support, and Lawrence Welch, Isheir Raote, Yamuna Krishnan, Sam Campos, and John Ngo for their suggestions and comments (via Twitter). Additional thanks to Harmit Malik for his help in analyzing the evolutionary conservation of this altORF sequence. This work was supported by grants from the National Science Foundation (2029868) and the NIH/National Institute of General Medical Sciences (R35GM126930) to I.M.C. and a National Science Foundation Graduate Research Fellowship to A.P.N.
Abbreviations used:
- Aa
amino acid
- altORF
alternative open reading frame
- BFA
Brefeldin A
- 2-BP
2-bromopalmitate
- CENP
centromere protein
- Cys
cysteine
- EGTA
ethylene glycol-bis(β-aminoethyl ether)- N, N, N′, N′-tetraacetic acid
- ER
endoplasmic reticulum
- FACS
fluorescence-activated cell sorting
- GFP
green fluorescent protein
- HEPES
N-2-hydroxyethylpiperazine- N′-2-ethanesulfonic Acid
- HRP
horseradish peroxidase
- IP-MS
immunoprecipitation-mass spectrometry
- PAT
palmitoylacyl transferase
- PMSF
phenylmethanesulfonyl fluoride.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E22-03-0091) on August 3, 2022.
REFERENCES
- Aicart-Ramos C, Valero RA, Rodriguez-Crespo I (2011). Protein palmitoylation and subcellular trafficking. Biochim Biophys Acta 1808, 2981–2994. [DOI] [PubMed] [Google Scholar]
- Banfield DK (2011). Mechanisms of protein retention in the Golgi. Cold Spring Harb Perspect Biol 3, a005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis VA, Garcia-Mata R, Mousley CJ (2012). Golgi membrane dynamics and lipid metabolism. Curr Biol 22, R414–R424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N (2009). Importing mitochondrial proteins: machineries and mechanisms. Cell 138, 628–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A (2005). A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci STKE 2005, pl1. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Lippincott-Schwartz J (2000). Sorting and signaling at the Golgi complex. Cell 101, 693–696. [DOI] [PubMed] [Google Scholar]
- Ernst AM, Syed SA, Zaki O, Bottanelli F, Zheng H, Hacke M, Xi Z, Rivera-Molina F, Graham M, Rebane AA, et al. (2018). S-Palmitoylation sorts membrane cargo for anterograde transport in the Golgi. Dev Cell 47, 479–493.e477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Oda K, Yokota S, Takatsuki A, Ikehara Y (1988). Brefeldin A causes disassembly of the Golgi complex and accumulation of secretory proteins in the endoplasmic reticulum. J Biol Chem 263, 18545–18552. [PubMed] [Google Scholar]
- Greaves J, Munro KR, Davidson SC, Riviere M, Wojno J, Smith TK, Tomkinson NC, Chamberlain LH (2017). Molecular basis of fatty acid selectivity in the zDHHC family of S-acyltransferases revealed by click chemistry. Proc Natl Acad Sci USA 114, E1365–E1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X, Fierke CA (2011). Understanding protein palmitoylation: biological significance and enzymology. Sci China Chem 54, 1888–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Okada M, Maenaka K, Fukagawa T (2008). CENP-O class proteins form a stable complex and are required for proper kinetochore function. Mol Biol Cell 19, 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings BC, Nadolski MJ, Ling Y, Baker MB, Harrison ML, Deschenes RJ, Linder ME (2009). 2-Bromopalmitate and 2-(2-hydroxy-5-nitro-benzylidene)-benzo[b]thiophen-3-one inhibit DHHC-mediated palmitoylation in vitro. J Lipid Res 50, 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kall L, Krogh A, Sonnhammer EL (2004). A combined transmembrane topology and signal peptide prediction method. J Mol Biol 338, 1027–1036. [DOI] [PubMed] [Google Scholar]
- Kall L, Krogh A, Sonnhammer EL (2007). Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res 35, W429–W432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjer-Nielsen L, Teasdale RD, van Vliet C, Gleeson PA (1999a). A novel Golgi-localisation domain shared by a class of coiled-coil peripheral membrane proteins. Curr Biol 9, 385–388. [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen L, van Vliet C, Erlich R, Toh BH, Gleeson PA (1999b). The Golgi-targeting sequence of the peripheral membrane protein p230. J Cell Sci 112 (Pt 11), 1645–1654. [DOI] [PubMed] [Google Scholar]
- Liu J, Huang Y, Li T, Jiang Z, Zeng L, Hu Z (2021). The role of the Golgi apparatus in disease (Review). Int J Mol Med 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zheng XF (2007). Endoplasmic reticulum and Golgi localization sequences for mammalian target of rapamycin. Mol Biol Cell 18, 1073–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Wu T, Zhang B, Liu S, Song W, Qiao J, Ruan H (2021). Types of nuclear localization signals and mechanisms of protein import into the nucleus. Cell Commun Signal 19, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan P, Campelo F (2021). Should I stay or should I go? Golgi membrane spatial organization for protein sorting and retention. Arch Biochem Biophys 707, 108921. [DOI] [PubMed] [Google Scholar]
- Mardones GA, Burgos PV, Brooks DA, Parkinson-Lawrence E, Mattera R, Bonifacino JS (2007). The trans-Golgi network accessory protein p56 promotes long-range movement of GGA/clathrin-containing transport carriers and lysosomal enzyme sorting. Mol Biol Cell 18, 3486–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardones GA, Snyder CM, Howell KE (2006). Cis-Golgi matrix proteins move directly to endoplasmic reticulum exit sites by association with tubules. Mol Biol Cell 17, 525–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley KL, Cheeseman IM (2017). Large-scale analysis of CRISPR/Cas9 cell-cycle knockouts reveals the diversity of p53-dependent responses to cell-cycle defects. Dev Cell 40, 405–420.e402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S, Nichols BJ (1999). The GRIP domain—a novel Golgi-targeting domain found in several coiled-coil proteins. Curr Biol 9, 377–380. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Rabouille C, Watson R, Nilsson T, Hui N, Slusarewicz P, Kreis TE, Warren G (1995). Characterization of a cis-Golgi matrix protein, GM130. J Cell Biol 131, 1715–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa N, Daikoku T, Koshizuka T, Yamauchi Y, Yoshikawa T, Nishiyama Y (2003). Subcellular localization of herpes simplex virus type 1 UL51 protein and role of palmitoylation in Golgi apparatus targeting. J Virol 77, 3204–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno Y, Kihara A, Sano T, Igarashi Y (2006). Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. Biochim Biophys Acta 1761, 474–483. [DOI] [PubMed] [Google Scholar]
- Orr MW, Mao Y, Storz G, Qian SB (2020). Alternative ORFs and small ORFs: shedding light on the dark proteome. Nucleic Acids Res 48, 1029–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsuledesai CC, Distefano MD (2015). Protein prenylation: enzymes, therapeutics, and biotechnology applications. ACS Chem Biol 10, 51–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe JM, Jenkins PM (2019). Spatial organization of palmitoyl acyl transferases governs substrate localization and function. Mol Membr Biol 35, 60–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocks O, Gerauer M, Vartak N, Koch S, Huang ZP, Pechlivanis M, Kuhlmann J, Brunsveld L, Chandra A, Ellinger B, et al. (2010). The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell 141, 458–471. [DOI] [PubMed] [Google Scholar]
- Salaun C, Greaves J, Chamberlain LH (2010). The intracellular dynamic of protein palmitoylation. J Cell Biol 191, 1229–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samandi S, Roy AV, Delcourt V, Lucier JF, Gagnon J, Beaudoin MC, Vanderperre B, Breton MA, Motard J, Jacques JF, et al. (2017). Deep transcriptome annotation enables the discovery and functional characterization of cryptic small proteins. eLife 6, 27860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe HJ, Stevens TJ, Munro S (2010). A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell 142, 158–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Wang Y (2013). Cell cycle regulation of Golgi membrane dynamics. Trends Cell Biol 23, 296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udenwobele DI, Su RC, Good SV, Ball TB, Varma Shrivastav S, Shrivastav A (2017). Myristoylation: an important protein modification in the immune response. Front Immunol 8, 751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C, Venditti R, Rega LR, Colanzi A, D’Angelo G, De Matteis MA (2011). The Golgi apparatus: an organelle with multiple complex functions. Biochem J 433, 1–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.