Abstract
Structural diversity of complex sphingolipids is important for maintenance of various cellular functions; however, the overall picture of the significance of this structural diversity remains largely unknown. To investigate the physiological importance of the structural diversity of complex sphingolipids, we here constructed a complex sphingolipid structural diversity disruption library in budding yeast, which comprises 11 mutants including with combinations of deletions of sphingolipid-metabolizing enzyme genes. The sensitivity of the mutants to various environmental stresses revealed that the more the structural variation of complex sphingolipids is limited, the more stress sensitivity tends to increase. Moreover, it was found that in mutant cells with only one subtype of complex sphingolipid, Slt2 MAP kinase and Msn2/4 transcriptional factors are essential for maintenance of a normal growth and compensation for reduced tolerance of multiple stresses caused by loss of complex sphingolipid diversity. Slt2 and Msn2/4 are involved in compensation for impaired integrity of cell walls and plasma membranes caused by loss of complex sphingolipid diversity, respectively. From these findings, it was suggested that loss of structural diversity of complex sphingolipids affects the environment of the cell surface, including both plasma membranes and cell walls, which could cause multiple environmental stress hypersensitivity.
INTRODUCTION
Biological membranes are based on a lipid bilayer composed of a wide variety of membrane lipids, and the structural diversity of these membrane lipids is considered to be an important factor that supports the multifunctions of biological membranes, such as signal transduction, transport of various molecules, and acquirement of stress tolerance. Complex sphingolipids, one of the classes in eukaryotic biomembranes, exhibit particularly complex structural diversity among membrane lipids. In mammals, complex sphingolipids have hundreds of hydrophilic head groups, and structural variation in long-chain bases (LCBs) and fatty acids in the ceramide (Cer) moiety of complex sphingolipids further complicates their structural diversity (Merrill, 2011). Although the overall picture of the significance of the structural diversity of complex sphingolipids in a single organism remains largely unknown, several lines of evidence have suggested the physiological significance of the structural diversity of complex sphingolipids in mammals; for example, the knockout mouse as to the lactosylCer α-2,3-sialyltransferase gene knockout mouse, which is responsible for the biosynthesis of glycosphingolipid GM3, exhibits altered sensitivity to insulin and complete hearing loss (Yamashita et al., 2003; Yoshikawa et al., 2009). However, establishment of mammalian cells having only one subtype of complex sphingolipids, which will be an effective means to understand the significance of the structural diversity, has not been reported, because the limitation of the diversity requires very complicated genetic manipulations.
The structural diversity of complex sphingolipids in budding yeast Saccharomyces cerevisiae is relatively simple as compared with that in mammals. This is mainly because the fatty acid chain length in the Cer moiety is mainly limited to C26 and complex sphingolipids in budding yeast have only three types of hydrophilic head group consisting of inositol phosphate and mannose (Dickson and Lester, 2002; Dickson et al., 2006; Tani, 2016). Owing to differences in hydrophilic structure, yeast complex sphingolipids are divided into inositol phosphorylceramide (IPC), mannosylinositol phosphorylceramide (MIPC), and mannosyldiinositol phosphorylceramide (M(IP)2C) (Figure 1, A and B) (Dickson and Lester, 2002; Dickson et al., 2006; Tani, 2016). In addition, the Cer moiety in complex sphingolipids can be classified into five types (types A, B, B′, C, and D) according to the hydroxylation state (Figure 1, A and B) (Dickson and Lester, 2002; Dickson et al., 2006; Tani, 2016). Thus, 15 subtypes of complex sphingolipids can be synthesized in budding yeast (Figure 1, A and B) (Tani, 2016). The structural diversity of complex sphingolipids in budding yeast is determined by five sphingolipid-metabolizing enzyme proteins, which are involved in extension of the hydrophilic head group (MIPC synthases Sur1 [Csg1] and Csh1 and M(IP)2C synthase Ipt1) and hydroxylation of the Cer moiety (sphingolipid C4-hydroxylase Sur2 and sphingolipid α-hydroxylase Scs7) (Figure 1, A and B) (Beeler et al., 1997; Dickson et al., 1997; Haak et al., 1997; Uemura et al., 2003). So far, it has been reported that deletion of any of these genes causes various abnormal phenotypes. Deletion of Csg1 and Csh1, which causes complete loss of MIPC biosynthesis, causes hypersensitivity to Ca2+ and low pH conditions (Uemura et al., 2003; Otsu et al., 2020), impairment of cell wall integrity and endosomal trafficking (Tani and Kuge, 2012b; Morimoto and Tani, 2015; Tanaka and Tani, 2018), and reduction of cell viability under nitrogen starvation (Yamagata et al., 2013; Knupp et al., 2017). The detrimental effect of loss of MIPC biosynthesis on sensitivity to Ca2+, low pH, or nitrogen starvation is not due to loss of MIPCs itself, but to the abnormal accumulation of IPCs, the precursor of MIPCs (Zhao et al., 1994; Yamagata et al., 2013; Knupp et al., 2017; Otsu et al., 2020). Several lines of evidence also suggest the importance of SUR2, SCS7, or IPT1. For instance, hydroxylation of the LCB moiety by SUR2 is required for formation of a lateral diffusion barrier in the endoplasmic reticulum (ER) between mother and daughter cells (Clay et al., 2014). IPT1 and SCS7 are essential for maintenance of normal cell growth when expression of phosphatidylserine (PS) synthase is repressed (Tani and Kuge, 2010b; Toda et al., 2020), indicating that the detailed structural features of complex sphingolipids determined by these genes are important when PS biosynthesis is repressed. In addition, it was also reported that deletion of IPT1 or SCS7 affects the sensitivities to some drugs (Hallstrom et al., 2001; Herrero et al., 2008).
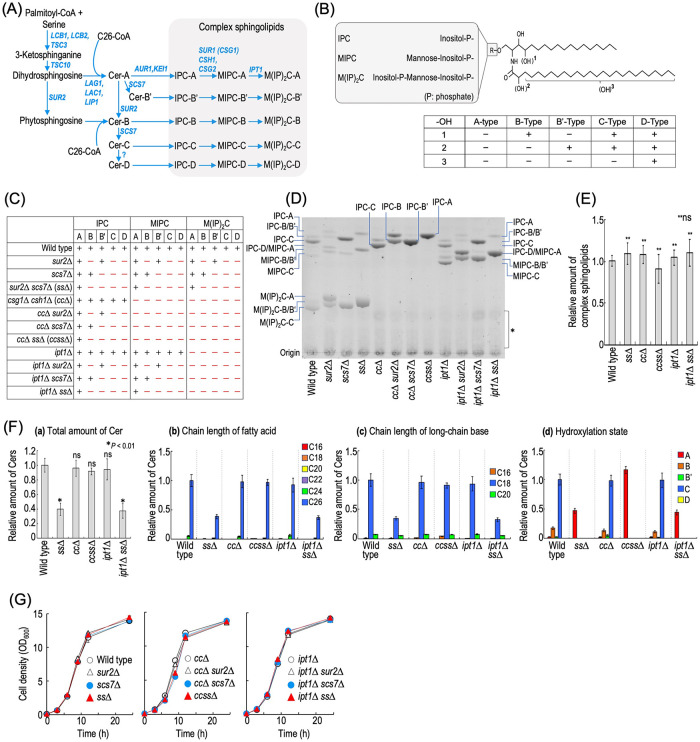
FIGURE 1:
Construction of a complex sphingolipid structural diversity disruption library. (A) Complex sphingolipid biosynthesis pathway in budding yeast S. cerevisiae. The pathway and genes responsible for the synthesis of complex sphingolipids in S. cerevisiae are shown. Owing to the different hydrophilic head groups and hydroxylation states of the ceramide (Cer) moiety, 15 subtypes of complex sphingolipid can be synthesized. For convenience, SUR1 encoding MIPC synthase is called CSG1 in this study. (B) Structure of S. cerevisiae complex sphingolipids. S. cerevisiae complex sphingolipids have three types of hydrophilic head group. Hydroxylation sites in the Cer moiety are labeled 1, 2, and 3. Sites 1 (the C-4 position of LCBs) and 2 (the C-2 position in fatty acids) are hydroxylated by Sur2 and Scs7, respectively. Site 3 is at an unknown position on the fatty acids, and the enzyme involved in the hydroxylation has not yet been identified. Owing to the difference in hydroxylation state, the Cer moiety of complex sphingolipids is classified into the A, B, B′, C, and D types. (C) The structural diversity of complex sphingolipid-disrupted mutants used in this study. A plus symbol indicates a subtype of complex sphingolipids that can be synthesized in each type of mutant cells. (D) TLC analysis of the mutant cells. Cells were cultured overnight in YPD medium, diluted (0.3 OD600 units/ml) in fresh YPD medium, and then incubated for 5 h at 30°C. Lipids were extracted, subjected to mild alkaline treatment, separated by TLC, and then visualized with a copper sulfate and orthophosphoric acid reagent. The asterisk indicates unidentified bands. (E) Amounts of complex sphingolipids in wild-type (MTY174), sur2∆ scs7∆ (ss∆), csg1∆ csh1∆ (cc∆), csg1∆ csh1∆ sur2∆ scs7∆ (ccss∆), ipt1∆, and ipt1∆ sur2∆ scs7∆ (ipt1∆ ss∆) cells. TLC analysis was performed as described in D. The amount of complex sphingolipids (IPCs, MIPCs, and M(IP)2Cs) in wild-type cells was taken as 1. (F) Analysis of free Cers by LC-ESI MS/MS. Cells were cultured, and lipids were extracted as described in D. Cers were measured by the MRM mode constructed by the combination of LCBs and fatty acids with different chain lengths or hydroxylation states. The total amount of Cers (panel a), carbon chain lengths of the fatty acid moieties (panel b), carbon chain lengths of the LCB moieties (panel c), and hydroxylation state of Cers (panel d) are shown. (G) Time course of cell growth. Cells were cultured overnight in YPD medium at 30°C and diluted (0.1 OD600 units/ml) in fresh YPD medium, and then aliquots of the cell suspensions were subjected to cell density measurements (OD600) at the indicated times. Data represent means ± SD for one experiment (triplicate) representative of three independent experiments. ns: no significant difference. The details are given in Materials and Methods.
The information as to which structural deficiency of complex sphingolipids causes what kind of abnormality is somewhat fragmentary even in budding yeast. For example, in high-throughput analysis, abnormal phenotypes caused by limitation of the structural diversity of complex sphingolipids are the result of a single deletion of a sphingolipid-metabolizing enzyme including MIPC synthases, M(IP)2C synthase, and sphingolipid hydroxylases (Dudley et al., 2005; Mira et al., 2010). Thus, there is not much information on the phenotypes caused by more severe disruption of the structural diversity due to the combination of deletion of multiple enzymes, for example, simultaneous defects of structural diversity of both the hydrophilic head group and the Cer moiety. In addition, budding yeast has two MIPC synthases, Csg1 and Csh1, and MIPC biosynthesis is not abolished completely with only one of the deletions (Uemura et al., 2003). Also, upon single deletion of CSG2 encoding the regulatory subunit of Csg1 and Csh1, a trace biosynthesis of MIPC still occur (Uemura et al., 2003; Tani and Kuge, 2012b). Furthermore, it was also shown that the abnormal phenotype caused by the single deletion of the gene involved in MIPC biosynthesis was weaker than that on double deletion of CSG1 and CSH1, which causes complete loss of MIPC biosynthesis (Morimoto and Tani, 2015; Otsu et al., 2020). Here, to systematically investigate the physiological importance of the structural diversity of complex sphingolipids, we created a complex sphingolipid structural diversity disruption library in budding yeast, which comprises mutants with various combinations of deletions of CSG1, CSH1, SUR2, SCS7, and IPT1 (Figure 1C). Analyses of this library revealed that, under various environmental stresses, the more the structural variation of both the hydrophilic group and the Cer moiety is limited, the more stress sensitivity tends to increase. Furthermore, it was found that mutant cells with only one subtype of complex sphingolipid exhibit impairment of both cell walls and plasma membranes, and these cell surface environment defects could be one of the causes of hypersensitivity to multiple environmental stresses due to the defect of structural diversity of complex sphingolipids.
RESULTS
Construction of a complex sphingolipid structural diversity disruption library
In S. cerevisiae, 15 subtypes of complex sphingolipid can be synthesized in total, the structural diversity being created by five sphingolipid-metabolizing enzymes, Csg1, Csh1, Sur2, Scs7, and Ipt1 (Figure 1, A and B). To systematically investigate the physiological importance of the structural diversity of complex sphingolipids, we constructed a complex sphingolipid diversity disruption library in budding yeast that comprises 11 mutants with various combinations of deletions of CSG1, CSH1, SUR2, SCS7, and IPT1 (Figure 1C). As shown in Figure 1D, the expected disappearance of specific subtypes of complex sphingolipid was observed in each library mutant; for example, in sur2∆ scs7∆ (ss∆) or csg1∆ csh1∆ (cc∆) cells, hydroxylated complex sphingolipids (B, B′, C, and D types) or MIPCs and M(IP)2Cs disappeared, respectively. Moreover, in csg1∆ csh1∆ sur2∆ scs7∆ (ccss∆) cells, only one subtype (IPC-A) was detected (Figure 1D). The total amount of complex sphingolipids was not significantly different among the wild-type, cc∆, ss∆, ccss∆, ipt1∆, and ipt1∆ sur2∆ scs7∆ (ipt1∆ ss∆) cells (Figure 1E). The results for ss∆, cc∆, and ccss∆ cells were consistent with those of a previous report (Uemura et al., 2014). Liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI MS/MS) analysis of free Cers revealed that the carbon chain lengths of fatty acid and LCB in Cers in wild-type, ss∆, cc∆, ccss∆, ipt1∆, and ipt1∆ ss∆ cells are mainly C26 and C18, respectively (Figure 1F), which is likely to reflect the composition of the Cer moiety of complex sphingolipids. Thus, it was suggested that the loss of biosynthesis of MIPCs and M(IP)2Cs and/or hydroxylation of the Cer moiety do not dramatically affect the carbon chain length of the Cer moiety. Although significant differences in the complex sphingolipid levels were not observed between these mutant cells (Figure 1E), a decrease in the total amount of Cers was observed in ss∆ and ipt1∆ ss∆ cells, but not in cc∆ and ccss∆ cells (Figure 1F). The levels of phosphatidylcholine (PC), phosphatidylserine/phosphatidylinositol (PS/PI), and phosphatidylethanolamine (PE) in ss∆, cc∆, ccss∆, ipt1∆, and ipt1∆ ss∆ cells did not change as compared with wild-type cells (Supplemental Figure S1), indicating that the alteration of the structural diversity of complex sphingolipids does not affect cellular levels of major glycerophospholipids. Although the growth rates of ccss∆ and cc∆ scs7∆ cells in YPD medium were slightly lower than that of wild-type cells, there was no significant difference for other mutants (Figure 1G), indicating that defects of the structural diversity of complex sphingolipids do not have a serious effect on cell growth under normal conditions. However, when the carbon source was changed from glucose to glycerol/ethanol, the growth of cc∆, cc∆ sur2∆, cc∆ scs7∆, and ccss∆ cells was decreased (Supplemental Figure S2). Furthermore, on changing to lactic acid, a decrease in the growth of the ccss∆ strain was also clearly observed (Supplemental Figure S2). In contrast, such a difference of growth among mutants was not observed under anaerobic conditions (Supplemental Figure S2). These results suggested that these mutations have a marked effect on respiratory growth.
Defect of tolerance of various environmental stresses due to defects of the structural diversity of complex sphingolipids
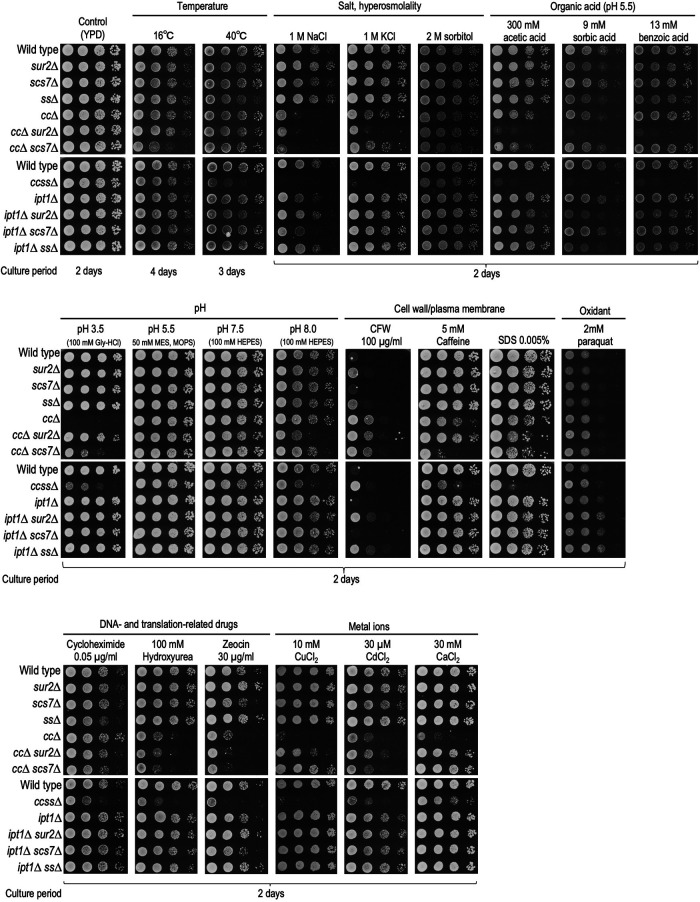
It has been reported that yeast cells with impaired structural diversity of complex sphingolipids exhibit increased sensitivity to cell wall–related stresses, low pH, metal ions, and some drugs (Zhao et al., 1994; Hallstrom et al., 2001; Herrero et al., 2008; Morimoto and Tani, 2015; Tanaka and Tani, 2018; Otsu et al., 2020). Here, by using the complex sphingolipid disruption library, we investigated which structural deficiencies of complex sphingolipids affect what kind of environmental stresses, including those previously reported (Figure 2). MIPC biosynthesis–deficient cells (cc∆ cells in this study), which have been most frequently reported as mutants showing various abnormal phenotypes among mutants included in the library, exhibited increased sensitivity to NaCl, KCl, acetic acid (pH 5.5), pH 3.5, caffeine, SDS, hydroxyurea, zeocin, CuCl2, CdCl2, and CaCl2. In contrast, deletion of SUR2, SCS7, or both (ss∆) did not have marked effects as compared with cc∆; however, when these mutations were combined with cc∆ (ccss∆, cc∆ sur2∆, and cc∆ scs7∆), they had synergistic effects on stress sensitivity. That is, for 40°C, NaCl, KCl, organic acids (acetic acid, sorbic acid, and benzoic acid), pH 8.0, SDS, cycloheximide, and zeocin, ccss∆ cells were more sensitive than cc∆ or ss∆ cells. Conversely, it was also observed that the deletion of SUR2 and/or SCS7 suppressed the increased sensitivity due to cc∆; that is, the hypersensitivity to pH 3.5, CaCl2, or CuCl2 due to cc∆ was partly recovered on deletion of SUR2 or SCS7 (cc∆ vs. cc∆ sur2∆ or cc∆ scs7∆ cells). The recovery of sensitivity to CdCl2 was observed only in cc∆ sur2∆ cells. The recovery of sensitivity to pH 3.5 or CaCl2 was also observed in ccss∆ cells. The suppressive effect of sur2∆ or scs7∆ on the hypersensitivity to pH 3.5 or CaCl2 due to cc∆ was consistent with previous reports (Tani and Toume, 2015; Otsu et al., 2020). As compared with wild-type cells, cc∆ cells exhibited resistance to calcofluor white (CFW), which binds to chitin at the cell wall and subsequently perturbs the function of the cell wall (Ram and Klis, 2006); however, the effect of this resistance was reduced in ccss∆ cells (this will be discussed later). For M(IP)2C biosynthesis–deficient mutants (ipt1∆, ipt1∆ sur2∆, ipt1∆ scs7∆, and ipt1∆ ss∆ cells), there was very little change in stress sensitivities as compared with the MIPC biosynthesis–deficient mutants (Figure 2). Overall, these results indicated that most of the stress hypersensitivity due to limitation of structural diversity of complex sphingolipids is mainly caused by the loss of biosynthesis of MIPCs, but not M(IP)2Cs, and the loss of hydroxylation in the Cer moiety promotes or suppresses the detrimental effects due to loss of MIPC biosynthesis. Notably, it was also indicated that ccss∆ cells having only one subtype of complex sphingolipid are the most severely sensitive to the majority of stresses.
FIGURE 2:
Sensitivities of the complex sphingolipid structural diversity disruption library to various stress conditions and different carbon sources. Cells cultured overnight in YPD medium at 30°C were spotted onto agar plates containing the indicated compounds: YPD containing 1 M NaCl, 1 M KCl, 2 M sorbitol, 100 µg/ml CFW, 5 mM caffeine, 0.005% SDS, 2 mM paraquat, 0.05 µg/ml cycloheximide, 100 mM hydroxyurea, 30 µg/ml zeocin, 10 mM CuCl2, 30 µM CdCl2, or 30 mM CaCl2; YPD buffered with 100 mM Gly-HCl (for pH 3.5), 50 mM MES and MOPS (for pH 5.5), or 100 mM HEPES (for pH 7.5 and 8.0); YPD buffered at pH 5.5 by the addition of 50 mM MES and MOPS, and containing 300 mM acetic acid, 9 mM sorbic acid, or 13 mM benzoic acid (unless otherwise stated, pH of the medium was performed at 6.0). Plates were incubated at 30°C for the indicated numbers of days. When indicated, plates were also incubated at 16 or 40°C.
Abnormal phenotypes due to accumulation of IPCs or disappearance of MIPC and M(IP) 2C
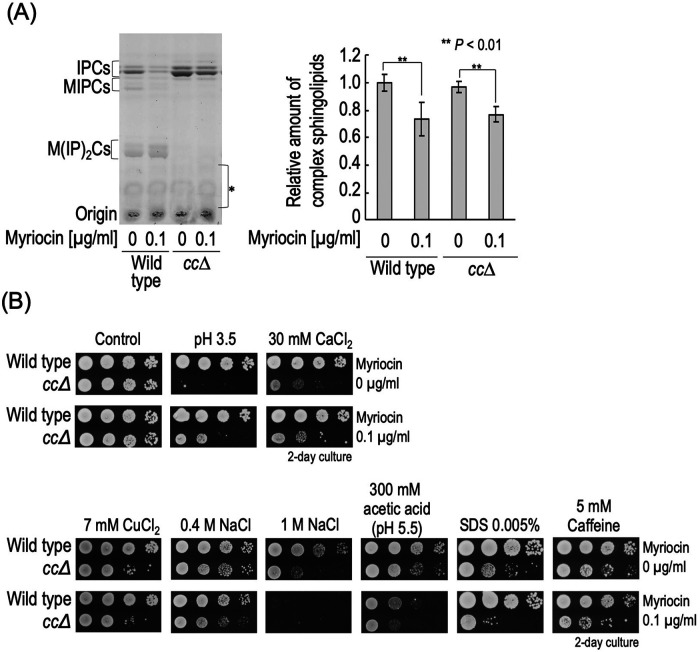
It was reported that hypersensitivity to Ca2+ or low pH due to a defect of MIPC biosynthesis is suppressed not only by the deletion of SUR2 or SCS7 but also by suppression of de novo sphingolipid biosynthesis, which includes inhibition of serine palmitoyltransferase (SPT) (Zhao et al., 1994; Otsu et al., 2020) (Figure 1A). This implies that excess accumulation of IPCs, precursors of MIPCs, triggers these abnormal phenotypes in MIPC biosynthesis–deficient cells. Next, we examined how the various stress sensitivities seen in cc∆ cells were altered by inhibition of the de novo sphingolipid biosynthesis pathway. To repress de novo biosynthesis of all sphingolipids, myriocin, an inhibitor of SPT, was used. As shown in Figure 3A, treatment with 0.1 µg/ml myriocin significantly reduced the complex sphingolipid levels in both wild-type and cc∆ cells. We also used a mutant strain that carries the LIP1 gene encoding the regulatory subunit of Cer synthases under the control of a tetracycline-regulatable (Tet) promoter (tet-LIP1) for repression of biosynthesis of Cers (Supplemental Figure S3) (Tani and Kuge, 2010a; Toume and Tani, 2016). tet-LIP1 and tet-LIP1 cc∆ cells also exhibited a decrease in complex sphingolipid levels when they were treated with doxycycline (Dox), which represses expression of the gene under the Tet promoter (Supplemental Figure S3A). As reported previously, under these de novo sphingolipid biosynthesis–repressive conditions, hypersensitivity to pH 3.5 or CaCl2 due to cc∆ was suppressed (Figure 3B and Supplemental Figure S3B). However, the sensitivities to CuCl2, NaCl, acetic acid, SDS, and caffeine in cc∆ cells were promoted by both myriocin treatment and LIP1 repression, or either, suggesting that hypersensitivity to these stresses is not caused by the accumulation of IPCs, but probably by loss of MIPCs (and also M(IP)2Cs) (Figure 3B and Supplemental Figure S3B). It should be noted that CuCl2 hypersensitivity due to cc∆ was suppressed by scs7∆ or sur2∆ (Figure 2), suggesting that the rescue mechanism through a defect of hydroxylation in the Cer moiety is not necessarily associated with the rescue mechanism through avoidance from accumulation of IPCs. Collectively, these results indicated that the abnormal phenotypes due to the loss of biosynthesis of MIPC are caused by multiple factors. In addition, the hypersensitivities to CuCl2, NaCl, acetic acid, SDS, and caffeine due to ccss∆ were also promoted or unaffected by myriocin treatment or LIP1 repression (Supplemental Figure S4), suggesting that, in ccss∆ cells, the hypersensitivities to these stresses are not also caused by accumulation of IPC-A.
FIGURE 3:
Stress sensitivity caused by loss of MIPCs or accumulation of IPCs. (A) Effect of myriocin on complex sphingolipid levels. Cells were cultured overnight in YPD medium at 30°C, diluted (0.3 OD600 units/ml) in fresh YPD medium with or without 0.1 µg/ml myriocin, and then incubated for 3 h at 30°C. TLC analysis were performed as described in Figure 1D. The amount of complex sphingolipids (IPCs, MIPCs, and M(IP)2Cs) in myriocin-untreated wild-type cells was taken as 1. (B) Effect of myriocin on sensitivity to stresses. Cells cultured overnight in YPD medium were spotted onto YPD plates containing 30 mM CaCl2, 7 mM CuCl2, 0.4 or 1 M NaCl, 300 mM acetic acid (adjusted to pH 5.5 by the addition of 50 mM MES and MOPS), SDS 0.005%, 5 mM caffeine, or 0.1 µg/ml myriocin, and then incubated for 2 d. YPD plates (pH 3.5) were prepared by the addition of 100 mM Gly-HCl.
Relationship between multiplestress hypersensitivity and stress response systems in ccss∆ cells
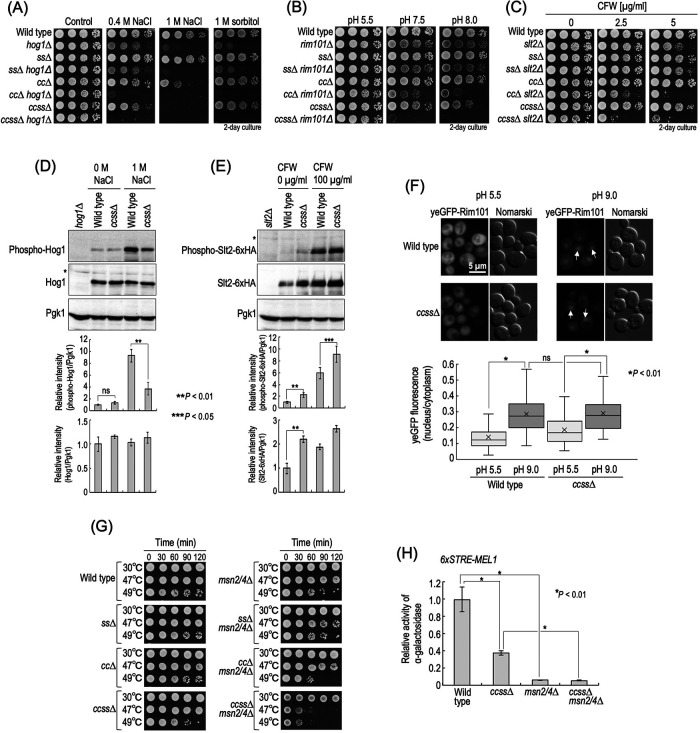
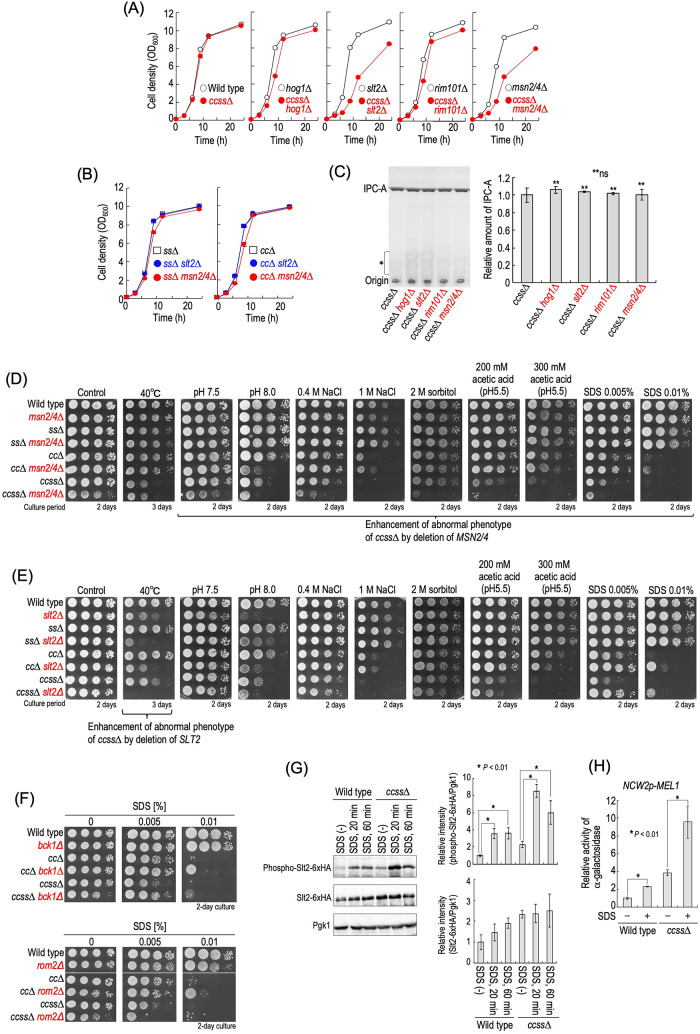
Yeast cells have a variety of stress responsive signal transduction systems to compensate for their growth under stress, and thus it was assumed that the multiplestress hypersensitivity of complex sphingolipid diversity–deficient cells is caused by impairment of stress response system(s). Therefore, we examined the stress sensitivity of ss∆, cc∆, and ccss∆ cells in the absence of the typical stress response systems. Because ccss∆ cells exhibited hypersensitivity to NaCl and alkaline pH, Hog1 MAP kinase in the high-osmolarity glycerol (HOG) pathway (Brewster and Gustin, 2014) or alkaline pH–responsive transcription factor Rim101 in the Rim pathway (Futai et al., 1999) was deleted. In addition, because altered sensitivity to CFW, SDS, and caffeine may imply an abnormality in cell wall integrity (Levin, 2005; Ram and Klis, 2006), Slt2 MAP kinase in the cell wall integrity (CWI) pathway was also deleted. As shown in Figure 4, A and B, ccss∆ caused hypersensitivity to NaCl and sorbitol or alkaline pH in the absence of Hog1 or Rim101, respectively (hog1∆ vs. ccss∆ hog1∆ cells or rim101∆ vs. ccss∆ rim101∆ cells). In many cases, hypersensitivity to CFW suggests impairment of cell wall integrity (Ram and Klis, 2006); however, ccss∆ cells exhibited weak resistance to CFW (Figure 2). In contrast, the sensitivity of ccss∆ slt2∆ cells to CFW was much greater than that of slt2∆ cells (Figure 4C), implying that ccss∆ cells have a stronger defect of cell wall integrity in the absence of SLT2. Similar results were obtained for cc∆ cells (Figure 4C) (Morimoto and Tani, 2015). Notably, in the absence of Hog1, Slt2, or Rim101, the enhancing effects of stress sensitivities caused by ccss∆ were much more robust than that in the case of cc∆ or ss∆ (Figure 4, A–C). Next, we examined activation of these signaling pathways in wild-type and ccss∆ cells. As shown in Figure 4D, enhancement of phosphorylation of Hog1 in the presence of NaCl was observed in ccss∆ cells, although the degree of the increase was lower than that in wild-type cells. However, it is unlikely that the decrease in activation of the HOG pathway causes the hypersensitivity to NaCl in ccss∆ cells because salt/hyperosmotic stress hypersensitivity due to ccss∆ is observed even in the absence of HOG1 (Figure 4A). In ccss∆ cells, the level of phosphorylated Slt2-6xHA in both the presence and absence of CFW was higher than that in wild-type cells (Figure 4E), suggesting weak constitutive activation of Slt2 due to ccss∆ even in the absence of stress. Similar results were obtained when Slt2 was not tagged with 6xHA (unpublished data). It should be noted that the total expression level of Slt2-6xHA in ccss∆ cells was also higher than that in wild-type cells regardless of the presence or absence of CFW (Figure 4E). These increases were consistent with the previous finding for cc∆ cells (Morimoto and Tani, 2015; Tanaka and Tani, 2018). The increased activity of Slt2 may be the reason why ccss∆ cells exhibited CFW resistance as compared with wild-type cells (Figure 2) because csss∆ slt2∆ cells were more sensitive to CFW than wild-type and slt2∆ cells (Figure 4C). A significant difference in nuclear translocation of yeGFP-Rim101 due to alkalinization of the medium was not observed between wild-type and ccss∆ cells (Figure 4F). Collectively, these results suggested that the hypersensitivity of ccss∆ cells to these stresses is not due to impairment of these stress response systems.
FIGURE 4:
Relationship between stress sensitivity of complex sphingolipid diversity–disrupted mutants and stress response systems. (A–C) Cells cultured overnight in YPD medium at 30°C were spotted onto YPD plates containing 0.4 or 1 M NaCl, 2 M sorbitol (A) or 2.5 or 5 µg/ml CFW (C) and then incubated for 2 d. Buffered YPD plates were prepared by the addition of 50 mM MES and MOPS (for pH 5.5) or 100 mM HEPES (for pH 7.5 and 8.0) (B). (D) Western blotting analysis of phosphorylation of Hog1. Cells were cultured overnight in YPD medium, diluted (0.1 OD600 units/ml) in fresh YPD medium, and then incubated for 5 h at 30°C. Then, cells were treated with 1 M NaCl (final concentration) for 20 min by the addition of fresh YPD containing 5 M NaCl to the culture medium. For the control experiment (0 M NaCl), an equal volume of fresh YPD medium was added to the culture medium. Yeast cell extracts were immunoblotted using anti–phospho-p38 MAPK (recognizes phospho-Hog1), anti-Hog1, or anti-Pgk1. The relative amount of phospho-Hog1/Pgk1 or phospho-Hog1/Hog1 in wild-type cells without NaCl was taken as 1. (E) Western blotting analysis of phosphorylation of Slt2-6xHA. Cells expressing Slt2-6xHA were precultured as described in D. Then, cells were treated with 100 µg/ml CFW for 30 min. Yeast cell extracts were immunoblotted using anti–phospho-p44/42 MAPK (recognizes phospho-Slt2-6xHA), anti-HA, or anti-Pgk1. The relative amount of phospho-Slt2-6xHA/Pgk1 or Slt2-6xHA/Pgk1 in wild-type cells without CFW was taken as 1. Data represent means ± SD for one experiment (triplicate) representative of three independent experiments. The asterisk indicates unidentified bands. (F) Translocation of yeGFP-Rim101. Cells expressing yeGFP-Rim101 were cultured overnight in YPD medium, diluted (0.1 OD600 units/ml) in fresh YPD medium, and then incubated for 4 h at 30°C. For alkaline treatment to activate the Rim101 pathway, Tris-HCl (pH 9.0) was added to the culture medium at a final concentration of 100 mM. For the control experiment, MES and MOPS (pH 5.5) were added at a final concentration of 50 mM. Cells were incubated for 60 min at 30°C, fixed, and then viewed under a fluorescence microscope. The arrows indicate nuclear localization of yeGFP-Rim101. The ratio of yeGFP fluorescence in cytoplasm and the nucleus in individual cells is expressed as a boxplot. Data represent the values for 100 cells for individual strains. (G) Heat stability. Cells (0.7 OD600 units) grown in YPD medium were collected by centrifugation, washed with water, and then suspended in 1 ml of water. The cell suspensions were incubated for the indicated times at the indicated temperatures, and samples (3.5 µl) were spotted onto YPD plates and then incubated for 2 d. (H) STRE-mediated transcriptional activity. Cells harboring pRS416-6xSTRE-MEL1 were cultured overnight in SC medium lacking uracil (SC–Ura), diluted (0.3 OD600 units/ml) in fresh SC–Ura medium, and then incubated for 6 h at 30°C. Cells were harvested, and α-galactosidase activity was measured. Data represent means ± SD for one experiment (triplicate) representative of three independent experiments. ns: no significant difference. The details are given in Materials and Methods.
Transcriptional regulation via Msn2 and Msn4 is also involved in the adaptation of cells to various environmental changes, which is known as a “general stress response” (Schmitt and McEntee, 1996; Moskvina et al., 1998; Gasch et al., 2000). Thus, we also examined the effect of deletion of MSN2/4 in ccss∆ cells. Deletion of MSN2/4 caused reduced heat stability (wild-type vs. msn2/4∆ cells) (Figure 4G), a typical phenotype of msn2/4∆ cells (Wei et al., 2008). ccss∆ cells exhibited a slight decrease in heat stability as compared with wild-type cells, and this decrease was more pronounced in the absence of MSN2/4 (msn2/4∆ vs. ccss∆ msn2/4∆ cells) (Figure 4G). In ccss∆ cells, basal transcriptional activity mediated by Msn2/4 was lower than that in wild-type cells but much higher than that in msn2/4∆ cells, when estimated using a reporter gene plasmid that contained the α-galactosidase gene and six tandem repeats of the stress response element (STRE), a binding site of Msn2/4 (Martinez-Arias et al., 1984; Schmitt and McEntee, 1996) (Figure 4H). Similar results were obtained when cells were exposed to heat shock (47 or 48°C for 60 min) (Supplemental Figure S5). These results suggested that Msn2/4 is still functional even with mutation of ccss∆.
Slt2 and Msn2/4 are important for maintenance of the growth rate and stress tolerance in ccss∆ cells
During evaluation of the stress sensitivities of stress response systems–deleted cells, we noticed that the deletion of SLT2 or MSN2/4 dramatically reduced the growth rate of ccss∆ cells under normal growth conditions (YPD medium) (Figure 5A and Supplemental Figure S6A), while the deletion of HOG1 or RIM101 had only a mild effect (Figure 5A). Although Msn2 and Msn4 are largely, that is, not completely, functionally redundant (Schmitt and McEntee, 1996; Yamaguchi et al., 2018), a single deletion of MSN2 or MSN4 does not cause reduction of the growth rate of ccss∆ cells (Supplemental Figure S6B), indicating that both Msn2 and Msn4 are required for maintenance of the growth rate of ccss∆ cells. The effect of deletion of SLT2 or MSN2/4 on the cell growth rate was not severe for ss∆ and cc∆ cells (Figure 5B). Significant differences in the complex sphingolipid levels between ccss∆ and ccss∆ slt2∆ or ccss∆ msn2/4∆ cells were not observed (Figure 5C), indicating that the slow growth phenotype due to the deletion of SLT2 or MSN2/4 is not caused by reduction of the complex sphingolipid levels. The deletion of MSN2/4 enhanced the sensitivity of ccss∆ cells to pH 8.0, NaCl, sorbitol, acetic acid, and SDS (ccss∆ vs. ccss∆ msn2/4∆ cells) (Figure 5D). Notably, a marked difference was not observed between wild-type and msn2/4∆ cells under the experimental conditions used in this study (Figure 5D), indicating that Msn2/4 are involved in compensation for reduced tolerance to these stress conditions due to ccss∆. In contrast, the deletion of SLT2 did not enhance the hypersensitivity to NaCl, sorbitol, or acetic acid due to ccss∆ (Figure 5E). As reported previously (Imazu and Sakurai, 2005), slt2∆ cells exhibited hypersensitivity to 40°C, probably because of a defect of the response to cell wall damage due to elevated temperature. Notably, ccss∆ slt2∆ cells showed very strong sensitivity to 40°C compared with ccss∆ and slt2∆ cells (Figure 5E); however, ccss∆ msn2/4∆ cells exhibited sensitivity similar to that of ccss∆ cells (Figure 5D). Although ccss∆ slt2∆ cells were more sensitive to pH 8.0 than ccss∆ cells, the sensitivity of slt2∆ cells was more severe than that of ccss∆ and ccss∆ slt2∆ cells (Figure 5E). Thus, it was suggested that the deletion of SLT2 does not contribute to compensation for the pH 8.0 hypersensitivity due to ccss∆. Collectively, these results suggested that both Msn2/4 and Slt2 contribute to compensation for the multiplestress hypersensitivity due to ccss∆; however, their roles are distinctly different.
FIGURE 5:
Slt2- and Msn2/4-dependent cell growth and stress tolerance of ccss∆ cells. (A) Effect of deletion of HOG1, SLT2, RIM101, or MSN2/4 on cell growth of ccss∆ cells. Cells were cultured overnight in YPD medium at 30°C and diluted (0.1 OD600 units/ml) in fresh YPD medium, and then aliquots of cell suspensions were subjected to cell density measurements (OD600) at the indicated times. (B) Effect of deletion of SLT2 or MSN2/4 on the cell growth of cc∆ and ss∆ cells. (C) Effect of deletion of HOG1, SLT2, RIM101, or MSN2/4 on the IPC-A level in ccss∆ cells. TLC analysis was performed as described in Figure 1, D and E. Data represent means ± SD for one experiment (triplicate) representative of three independent experiments. ns: no significant difference. (D, E) Effects of deletion of MSN2/4 (D) or SLT2 (E) on stress sensitivities. Cells cultured overnight in YPD medium were spotted onto YPD plates (the details of the composition of the medium are given in Figure 2). Plates were incubated at 30 or 40°C for the indicated numbers of days. (F) Effects of deletion of BCK1 or ROM2 on SDS sensitivity. (G) Phosphorylation of Slt2-6xHA in SDS-treated cells. Cells expressing Slt2-6xHA were cultured overnight in YPD medium, diluted (0.3 OD600 units/ml) in fresh YPD medium, incubated for 5 h at 30°C, and then treated with 0.01% SDS for 20 or 60 min. Yeast cell extracts were immunoblotted using anti–phospho-p44/42 MAPK, anti-HA, or anti-Pgk1. The relative amount of phospho-Slt2-6xHA/Pgk1 or Slt2-6xHA/Pgk1 in SDS-untreated wild-type cells was taken as 1. (H) Promoter activity of NCW1 in SDS-treated cells. Cells harboring pRS415-NCW2p-MEL1 were cultured overnight in SC–Leu medium, diluted (0.3 OD600 units/ml) in fresh SC–Leu medium, and then incubated for 5 h at 30°C. Cells were treated with or without 0.01% SDS for 60 min and harvested, and then α-galactosidase activity was measured. The activity in wild-type cells without SDS was taken as 1. Data represent means ± SD for one experiment (triplicate) representative of three independent experiments. The details are given in Materials and Methods.
Notably, deletion of MSN2/4 increased the hypersensitivity to SDS due to ccss∆, whereas that of SLT2 suppressed the hypersensitivity (Figure 5, D and E). Deletion of BCK1 encoding MAPKKK that functions upstream of Slt2 also suppressed the SDS hypersensitivity of ccss∆ and cc∆ cells (ccss∆ vs. ccss∆ bck1∆ cells or cc∆ vs. cc∆ bck1∆ cells) (Figure 5F). Deletion of ROM2 encoding a guanine nucleotide exchange factor for Rho1, an upstream factor of the CWI pathway, suppressed the SDS hypersensitivity of cc∆ cells (cc∆ vs. cc∆ rom2∆ cells), but did not have such an effect on ccss∆ cells (ccss∆ vs. ccss∆ rom2∆ cells) (Figure 5F). This is probably because Rho1 is an essential GTP-binding protein, and thus the deletion of ROM2 may affect the growth of ccss∆ cells by factor(s) other than the defect of the CWI pathway. SDS treatment enhances phosphorylation of Slt2 (Sakata et al., 2022). As shown in Figure 5G, an increase in the level of phosphorylated Slt2-6xHA by the SDS treatment was observed in both wild-type and ccss∆ cells, suggesting that the activation of the CWI pathway by SDS also occurs in ccss∆ cells. In addition, in both wild-type and ccss∆ cells, promoter activity of NCW2, one of the downstream targets of Slt2 (this will be described later), was increased by the SDS treatment (Figure 5H). SDS is recognized as a reagent for evaluation of the integrities of both plasma membranes and cell walls; that is, the detrimental effects of SDS are caused by perturbation of the integrity of the cell membrane (Kono et al., 2016; Zhao et al., 2020), and defects of cell wall also lead to increased sensitivity to SDS because a weakened cell wall allows an increase in the accessibility of SDS on plasma membranes (Levin, 2005; Kono et al., 2016; Zhao et al., 2020). Thus, the fact that the defect of the CWI pathway confers resistance to SDS in ccss∆ cells probably reflects the impaired integrity of plasma membranes but not cell walls (this will be discussed later).
Cell wall–related genes are abundant among genes whose expression is changed by ccss∆
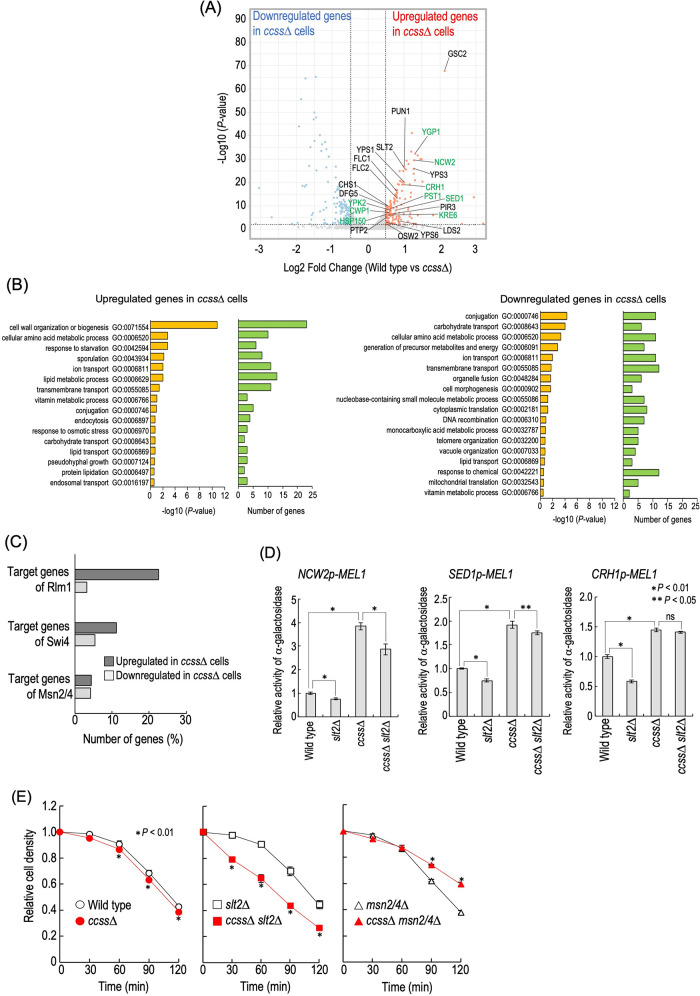
To investigate how loss of structural diversity of complex sphingolipids affects the gene expression profile, we performed transcriptome analysis by RNA-sequencing for wild-type and ccss∆ cells (Figure 6A). It was found that expression of 161 and 160 genes was significantly up-regulated and down-regulated in ccss∆ cells, respectively (Figure 6A and Supplemental Table S1). Gene ontology (GO) enrichment analysis of biological processes revealed that genes annotated as “cell wall organization or biogenesis (GO:0071554)” were the most abundant among the genes up-regulated in the ccss∆ cells, indicating that cell wall–related genes were predominantly up-regulated in ccss∆ cells (Figure 6B). It should be noted that nine genes (YGP1, NCW2, CRH1, PST1, SED1, KRE6, YPK2, CWP1, and HSP150) annotated as “cell wall organization or biogenesis” were listed in the Saccharomyces Genome Database (SGD) (https://www.yeastgenome.org/) as putative downstream targets of Rlm1 and/or Swi4, transcriptional factors of the CWI pathway (Figure 6A), which may be related to the constitutive activation of Slt2 MAP kinase in ccss∆ cells (Figure 4E). Thus, we next examined the promoter activity of NCW2 (target of Swi4), SED1 (target of Rlm1), and CRH1 (target of Rlm1 and Swi4) in wild-type, slt2∆, ccss∆, and ccss∆ slt2∆ cells by using reporter gene plasmids that contain the α-galactosidase gene and a potential promoter region of each gene. As shown in Figure 6D, an increase in these promoter activities was observed in ccss∆ cells as compared with wild-type cells. In both wild-type and ccss∆ cells, the deletion of SLT2 slightly but significantly decreased the promoter activity of NCW2 and SED1 (wild-type vs. slt2∆ cells or ccss∆ vs. ccss∆ slt2∆ cells); however, the deletion did not have such an effect on the promoter activity of CRH1 in ccss∆ cells. Moreover, these promoter activities in ccss∆ slt2∆ cells were greater than that in slt2∆ cells (Figure 6D). These results suggested that the up-regulation of these genes in ccss∆ cells at least partly depends on the CWI pathway, but another factor(s) is also involved. In contrast, most of the genes that are listed in the SGD as putative downstream targets of Msn2/4 were not up-regulated in ccss∆ cells (Figure 6C), which coincided with the results of reporter gene assays (Figure 4H). These results suggested that the expression profile of cell wall–related genes was greatly affected by loss of structural diversity of complex sphingolipids.
FIGURE 6:
Cell wall–related phenotypes of ccss∆ cells. (A) Volcano plot for comparison of RNA-sequencing data between wild-type and ccss∆ cells. Up-regulated and down-regulated genes due to ccss∆ (P value ≤ 0.01 and fold change ≥1.4) are denoted in red and blue, respectively. Among up-regulated genes in ccss∆ cells, genes annotated as “cell wall organization or biogenesis (GO:0071554)” are marked (23 genes). In addition, of these genes, the genes listed as putative downstream targets of Rlm1 and/or Swi4 (transcriptional factors of the CWI pathway) in the Saccharomyces Genome Database (SGD) (https://www.yeastgenome.org/) are in green. (B) Gene ontology (GO) enrichment analysis. GO (biological process) enrichment analysis of up-regulated or down-regulated genes due to ccss∆ was performed using the Gene Ontology Slim Term Mapper in the SGD (https://www.yeastgenome.org/goSlimMapper). GO terms significantly enriched in the up-regulated or down-regulated genes (–log10[P value] ≥ 0.5) are shown. The enrichment P values were calculated using Fisher’s exact test. (C) Percentages of numbers of genes up-regulated or down-regulated in ccss∆ cells among the putative target genes of Rlm1, Swi4, or Msn2/4 listed in the SGD. (D) Promoter activities of NCW1, SED1, and CRH1. Cells harboring pRS415-NCW2p-MEL1, pRS415-SED1p-MEL1, or pRS415-CRH1p-MEL1 were cultured overnight in SC–Leu medium, diluted (0.5 OD600 units/ml) in fresh SC–Leu medium, and then incubated for 5 h at 30°C. Cells were harvested, and then α-galactosidase activity was measured. The activity in wild-type cells was taken as 1. Data represent means ± SD for one experiment (triplicate) representative of three independent experiments. ns: no significant difference. (E) Zymolyase sensitivity. Cells were cultured overnight in YPD medium at 30°C, diluted (0.3 OD600 units/ml) in fresh YPD medium, and then incubated for 5 h at 30°C. They were then washed with 20 mM HEPES buffer (pH 7.5), resuspended (1.5 OD600 units/ml) in the same buffer containing 15 µg/ml zymolyase-20T (Nacalai Tesque), and incubated at 30°C. At the indicated times, the cell density (OD600) in the cell suspensions was measured. Data represent means ± SD for one experiment (triplicate) representative of three independent experiments. The details are given in Materials and Methods.
Defect of cell wall integrity in ccss∆ cells
Transcriptome analysis (Figure 6A) and the effect of deletion of SLT2 on the sensitivities to CFW and 40°C (Figures 4C and 5E) prompted us to investigate cell wall integrity in ccss∆ cells. Thus, we examined the sensitivity to zymolyase, a cell wall–digesting enzyme, for evaluation of the cell wall integrity. As shown in Figure 6E, ccss∆ cells were slightly but significantly more sensitive to zymolyase than wild-type cells, suggesting decreased cell wall integrity due to ccss∆. Moreover, deletion of SLT2 further promoted the increased sensitivity to zymolyase due to ccss∆ (Figure 6E), whereas a notable difference was not observed between wild-type and slt2∆ cells (Figure 6E and Supplemental Figure S7). Thus, it was indicated that, unlike wild-type cells, ccss∆ cells are strongly dependent on maintenance of cell wall integrity by the CWI pathway even in the absence of stresses. This notion is supported by the fact that Slt2 is constitutively activated in ccss∆ cells (Figure 4E). Previously, we reported that cc∆ cells also exhibit increased sensitivity to zymolyase (Morimoto and Tani, 2015); however, the zymolyase sensitivity of ccss∆ slt2∆ cells was greater than that of cc∆ slt2∆ cells (Supplemental Figure S7). In addition, ccss∆ slt2∆ cells were more sensitive to CFW than cc∆ slt2∆ and ss∆ slt2∆ cells (Figure 4C). Thus, it was indicated that the additional defect of hydroxylation of the Cer moiety causes a more serious defect of cell wall integrity in MIPC biosynthesis–deficient cells. Interestingly, deletion of MSN2/4 suppressed the zymolyase hypersensitivity due to ccss∆ (Figure 6E). The exact reason for this is unclear, but at least this suggests that Msn2/4 do not contribute to maintenance of cell wall integrity in ccss∆ cells.
Maintenance of plasma membrane integrity in ccss∆ cells by Msn2/4
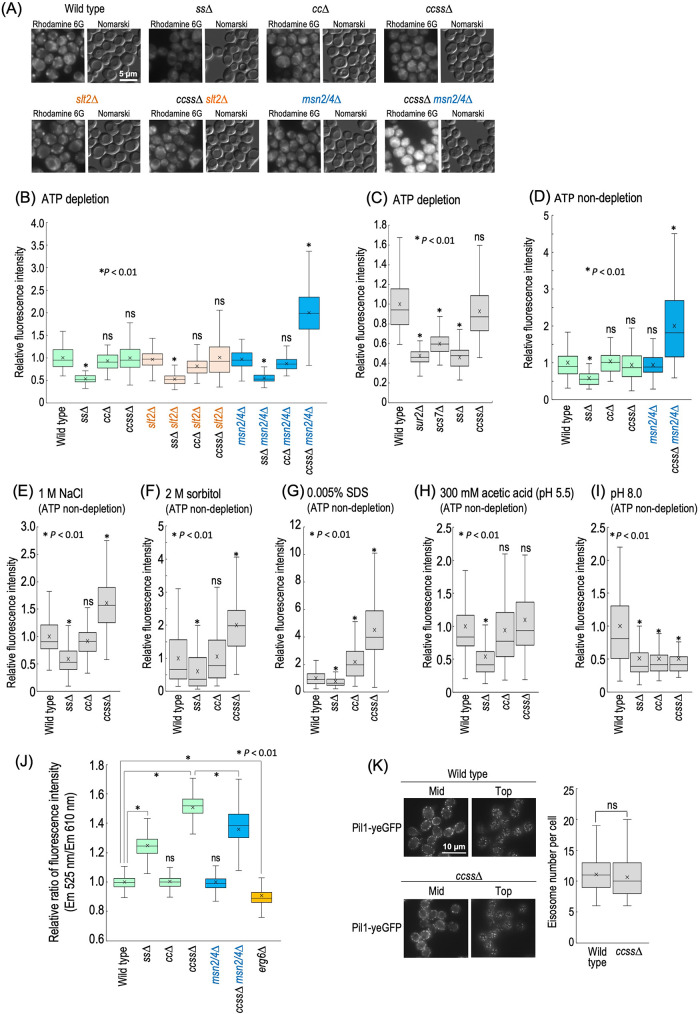
Maintenance of integrity of plasma membranes is important for acquirement of tolerance to various stresses (Mioka et al., 2018; Kishimoto et al., 2021). ccss∆ cells exhibit abnormal lateral diffusion of membrane proteins at plasma membranes (Uemura et al., 2014), suggesting a change in the properties of plasma membranes due to loss of structural diversity of complex sphingolipids. Thus, it became important to investigate whether or not ccss∆ affects the integrity of plasma membranes. To examine this, we evaluated the permeability of plasma membranes by observing the efficiency of incorporation of a lipofilic fluorescent dye, rhodamine 6G, into cells (Mioka et al., 2018). Because incorporated rhodamine 6G is extruded in an ATP-dependent manner (Mioka et al., 2018), the examination of incorporation of the dye was performed under ATP-depleted conditions. Intracellular accumulation of the dye was significantly reduced in ss∆ cells (Figure 7, A and B), and a reduction was also observed in sur2∆ or scs7∆ cells (Figure 7C), indicating that loss of either SUR2 or SCS7 causes decreased permeability of plasma membranes. In contrast, notable differences were not observed between wild-type, cc∆, and ccss∆ cells; however, ccss∆ msn2/4∆ cells exhibited a marked increase in accumulation of the dye (Figure 7, A and B). Such an increase was not observed in msn2/4∆, cc∆ msn2/4∆, and ss∆ msn2/4∆ cells (Figure 7B), suggesting that Msn2/4 are involved in suppression of the increase in plasma membrane permeability due to ccss∆. In contrast, the deletion of SLT2 did not affect the plasma membrane permeability in ccss∆ cells (Figure 7, A and B). Deletion of MSN2/4 further promotes hypersensitivity to NaCl, sorbitol, SDS, acetic acid, and pH 8.0 due to ccss∆ (Figure 5D), and thus it is possible that, even in the presence of MSN2/4, the permeability of plasma membranes of ccss∆ cells changes under these stresses. Therefore, we next investigated the accumulation of rhodamine 6G in wild-type, ss∆, cc∆, and ccss∆ cells under these stresses. To eliminate the possibility that depletion of ATP has detrimental effects on cells in the presence of each stress, these experiments were performed under the condition that ATP is not depleted. As show in Figure 7D, similar to the ATP-depleted conditions, a decrease or increase in plasma membrane permeability was observed in ss∆ or ccss∆ msn2/4∆ cells, respectively, when ATP was not depleted. In the presence of 1 M NaCl, 2 M sorbitol, or 0.005% SDS, the accumulation of rhodamine 6G in ccss∆ cells was increased as compared with that in wild-type cells (Figure 7, E–G), whereas the accumulation of the dye in ccss∆ cells was not increased in the presence of acetic acid or under pH 8.0 conditions (Figure 7, H and I). Because hyperosmotic stresses cause a change in the tension of plasma membranes due to shrinkage of cells (Dupont et al., 2010; Petelenz-Kurdziel et al., 2011), and SDS directly damages plasma membranes (Kono et al., 2016; Zhao et al., 2020), these results support the notion that the properties of plasma membranes are altered due to ccss∆. These results may also suggest that multiplestress hypersensitivity due to loss of structural diversity of complex sphingolipids is caused, at least partly, by the abnormal plasma membrane property. To gain further insight into the physical properties of plasma membranes, we used di-4-ANEPPDHQ, a probe for evaluating membrane lipid order (Owen et al., 2011). When cells were stained with di-4-ANEPPDHQ for 1 min, most of fluorescence signal was observed at the cell surface (Supplemental Figure S8). di-4-ANEPPDHQ has green fluorescence when residing in the ordered phase of membranes and red fluorescence in the disordered phase (Owen et al., 2011). Figure 7J shows the ratio of green (525 nm) and red (610 nm) fluorescences in each cell, measured by a flow cytometer. ERG6-deleted cells, which exhibit increases in both plasma membrane permeability and fluidity (Abe and Hiraki, 2009; Khmelinskaia et al., 2020), showed a decrease in the green/red fluorescence ratio as compared with wild-type cells, suggesting lower lipid order in plasma membranes (Figure 7J). In contrast, ss∆ and ccss∆ cells exhibited higher lipid order in plasma membranes as compared with wild-type cells (Figure 7J). These results are probably consistent with the fact that speed of lateral diffusion of plasma membrane–localized hexose transporter 1 is decreased due to ccss∆ (Uemura et al., 2014). The deletion of MSN2/4 caused a decrease in the green/red fluorescence ratio in ccss∆ cells (ccss∆ vs. ccss∆ msn2/4∆ cells), whereas no significant difference was observed between wild-type and msn2/4∆ cells (Figure 7J). These results suggested that a defect of structural diversity of complex sphingolipids alters physical properties of plasma membranes. Eisosomes, which are furrow-like structures at plasma membranes, have important roles in acquirement of stress tolerance (Dupont et al., 2010; Sakata et al., 2022), and several reports suggest functional relationship between eisosomes and sphingolipids (Walther et al., 2007; Luo et al., 2008; Frohlich et al., 2009). However, difference of distribution pattern of eisosomes was not observed between wild-type and ccss∆ cells when eisosomes were detected by Pil1-yeGFP, an eisosome marker protein (Walther et al., 2007) (Figure 7K).
FIGURE 7:
Plasma membrane properties in ccss∆ cells. (A) Detection of rhodamine 6G incorporated into cells under ATP-depleted conditions. Cells were cultured overnight in YPD medium at 30°C, diluted (0.3 OD600 units/ml) in fresh YPD medium, and then incubated for 5 h at 30°C. Rhodamine 6G (10 µM) was added to cells (1 OD600 units/300 µl) under ATP-depleted conditions, followed by incubation for 30 min at 30°C. Cells were viewed under a fluorescence microscope. (B, C) Efficiency of incorporation of rhodamine 6G into cells under ATP-depleted conditions. The fluorescence intensity of individual cells is expressed as a boxplot. Data represent the value for 100 cells for individual strains. The average (marked as x) of fluorescence intensity in wild-type cells was taken as 1. ns: no significant difference. (D) Efficiency of incorporation of rhodamine 6G into cells under ATP-nondepleted conditions. (E–I) Efficiency of incorporation of rhodamine 6G under stress conditions. Cells (1 OD600 units/300 µl) were incubated with 10 µM rhodamine 6G in YPD medium containing 1 M NaCl (E), 2 M sorbitol (F), 0.005% SDS (G), or 300 mM acetic acid (adjusted to pH 5.5 by the addition of 50 mM MES and MOPS) (H) for 30 min. YPD medium buffered at pH 8.0 was prepared by the addition of 100 mM HEPES (I). (J) Evaluation of membrane lipid order by using di-4-ANEPPDHQ. Cells were cultured overnight in YPD medium at 30°C, diluted (0.3 OD600 units/ml) in fresh YPD medium, and then incubated for 5 h at 30°C. di-4-ANEPPDHQ (5 µM) was added to cells (1 OD600 units/100 µl), followed by incubation for 1 min at 30°C. The ratio of green (525 nm) and red (610 nm) fluorescences in individual cells, which was measured by a flow cytometer, is expressed as a boxplot. Data represent the value for 1000 cells for individual strains. The average (marked as x) of the fluorescence ratio in wild-type cells was taken as 1. (K) Distribution of eisosomes in wild-type and ccss∆ cells. Cells expressing Pil1-yeGFP were cultured overnight in YPD medium at 30°C, diluted (0.3 OD600 units/ml) in fresh YPD medium, and then incubated for 5 h at 30°C. Cells were observed under a fluorescence microscope. Eisosomes in top section of images of individual cells (n = 30) were counted by using Fiji (Schindelin et al., 2012; Sakata et al., 2022).
Rescue of the integrity of cell walls, but not plasma membranes, in ccss∆ cells by sterols
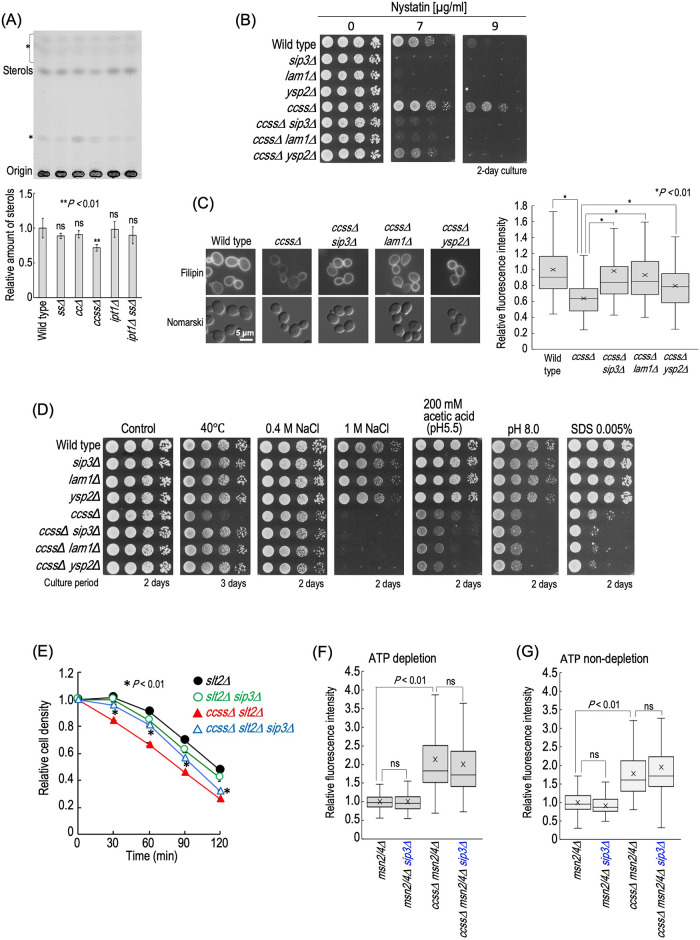
Complex sphingolipids and sterols are known to function coordinately (Guan et al., 2009; Tanaka and Tani, 2018; Otsu et al., 2020). Thus, we next investigated cellular sterol levels in wild-type, cc∆, ss∆, ccss∆, ipt1∆, and ipt1∆ ss∆ cells. As shown in Figure 8A, ccss∆ cells exhibited a significant decrease in the sterol levels as compared with wild-type cells. Moreover, ccss∆ cells exhibited decreased sensitivity to the polyene antifungal nystatin (Figure 8B), which exerts cytotoxicity through binding to ergosterol at plasma membranes and subsequently formation of channels (Serhan et al., 2014). When cells were stained with filipin, a fluorescent probe that stains sterols, the fluorescence intensity at plasma membranes in ccss∆ cells was decreased as compared with that in wild-type cells, also suggesting a decrease in sterol levels (Figure 8C). To investigate whether or not this decrease is related to hypersensitivity of ccss∆ cells to multiplestresses, SIP3, LAM1, or YSP2 encoding sterol-transfer protein involved in trafficking of sterols from plasma membranes to the ER (Gatta et al., 2015) was deleted in order to alter the distribution pattern of sterols at plasma membranes. Although ccss∆ cells, compared with wild-type cells, exhibited resistance to nystatin, the resistance was suppressed by the deletion of SIP3, LAM1, or YSP2 (Figure 8B). Furthermore, when cells were stained with filipin, the deletion of SIP3, LAM1, or YSP2 recovered the decrease in the fluorescence intensity due to ccss∆ (Figure 8C). These results suggested a change of the distribution pattern of sterols at plasma membranes in ccss∆ cells by the deletion of SIP3, LAM1, or YSP2. The deletion of SIP3, LAM1, or YSP2 did not suppress the hypersensitivities to NaCl, acetic acid, pH 8.0, and SDS due to ccss∆ (Figure 8D). In contrast, these deletions abolished the hypersensitivity to 40°C due to ccss∆ (Figure 8D). The hypersensitivities to NaCl, acetic acid, pH 8.0, and SDS due to ccss∆ were enhanced by the deletion of MSN2/4 (Figure 5D), whereas the hypersensitivity to 40°C was enhanced by that of SLT2 (Figure 5E), which may imply that the deletion of SIP3, LAM1, or YSP2 improves the defect of cell wall integrity due to ccss∆. Thus, we next examined the zymolyase sensitivity of SIP3-deleted cells. In this experiment, SLT2 was deleted in all strains in order to allow clear observation of the increase in zymolyase sensitivity due to ccss∆. As shown in Figure 8E, the hypersensitivity to zymolyase in ccss∆ slt2∆ cells was suppressed by the deletion of SIP3 (ccss∆ slt2∆ vs. ccss∆ slt2∆ sip3∆ cells); however, the deletion did not improve the zymolyase sensitivity of slt2∆ cells (slt2∆ vs. slt2∆ sip3∆ cells). Thus, it was suggested that alteration of the distribution pattern of sterols at plasma membranes compensates for the impaired cell wall integrity due to ccss∆. In contrast, the increase in accumulation of rhodamine 6G due to ccss∆ msn2/4∆ was not suppressed by deletion of SIP3 in either the absence or the presence of ATP (ccss∆ msn2/4∆ vs. ccss∆ msn2/4∆ sip3∆ cells) (Figure 8, F and G), suggesting that sterols do not improve the impairment of plasma membrane integrity due to ccss∆, which coincided with the fact that the deletion of SIP3 did not suppress the hypersensitivities to NaCl, acetic acid, pH 8.0, and SDS due to ccss∆ (Figure 8D). It should be noted that the deletion of MSN2/4 did not enhance the reduction in the sterol levels due to ccss∆ (Supplemental Figure S9), supporting the notion that sterols are not involved in compensation for the impaired integrity of plasma membranes due to ccss∆. Moreover, the deletion of SLT2 also did not affect the sterol levels in ccss∆ cells (Supplemental Figure S9), indicating that increased impairment of cell wall integrity in ccss∆ slt2∆ cells is not caused by enhancement of the decrease in the sterol levels.
FIGURE 8:
Alteration of distribution of sterols at plasma membranes restores impairment of cell wall but not plasma membrane integrity in ccss∆ cells. (A) TLC analysis of sterols in wild-type, ss∆, cc∆, ccss∆, ipt1∆, and ipt1∆ ss∆ cells. Cells were cultured overnight in YPD medium, diluted (0.3 OD600 units/ml) in fresh YPD medium, and then incubated for 5 h at 30°C. Lipids were extracted, separated by TLC, and then visualized with a copper sulfate and orthophosphoric acid reagent. The intensity of band of sterols in wild-type cells was taken as 1. Data represent means ± SD for one experiment (triplicate) representative of three independent experiments. ns: no significant difference. The asterisk indicates unidentified bands. (B) Nystatin sensitivity. Cells cultured overnight in YPD medium at 30°C were spotted onto YPD plates containing the indicated concentrations of nystatin and then incubated at 30°C for 2 d. (C) Filipin staining. Cells were cultured overnight in YPD medium, diluted (0.3 OD600 units/ml) in fresh YPD medium, and then incubated for 5 h at 30°C. Cells were stained with filipin and observed under a fluorescence microscope. The fluorescence intensity of individual cells is expressed as a boxplot. Data represent the value for 100 cells for individual strains. The average (marked as x) of fluorescence intensity in wild-type cells was taken as 1. (D) Effects of deletion of SIP3, LAM1, or YSP2 on stress sensitivities. Cells cultured overnight in YPD medium were spotted onto YPD plates (the details of the composition of the medium are given in Figure 2). Plates were incubated at 30 or 40°C for the indicated numbers of days. (E) The zymolyase sensitivities of slt2∆, slt2∆ sip3∆, ccss∆ slt2∆, and ccss∆ slt2∆ sip3∆ cells were examined as described in Figure 6E. (F, G) The efficiency of rhodamine 6G incorporation into msn2/4∆, msn2/4∆ sip3∆, ccss∆ msn2/4∆, and ccss∆ msn2/4∆ sip3∆ under ATP-depleted (F) or ATP-nondepleted (G) conditions was examined as described in Figure 7. The details are given in Materials and Methods.
DISCUSSION
In this study, we created a complex sphingolipid structural diversity disruption library and evaluated various environmental stress sensitivities of the library mutants. As a general tendency, it was found that the more the structural variation of complex sphingolipids is limited, the more stress sensitivity tends to increase (Figure 2). In particular, in many cases, loss of MIPC biosynthesis (cc∆) triggers the occurrence of stress hypersensitivity, and the detrimental effects were promoted by the loss of hydroxylation of the Cer moiety (sur2∆, scs7∆, or ss∆) (Figure 2). Moreover, it was suggested that these abnormal phenotypes, except for hypersensitivities to low pH and CaCl2, are caused by loss of production of MIPCs, but not by accumulation of IPCs (Figure 3 and Supplemental Figure S3). Thus, these results imply that, among complex sphingolipid subtypes, MIPCs are the most important factor for the acquirement of stress tolerance in budding yeast. Loss of MIPC biosynthesis causes loss of M(IP)2Cs as well as MIPCs; however, loss of M(IP)2C biosynthesis (ipt1∆) hardly affected the stress sensitivities, suggesting the importance of MIPCs but not M(IP)2Cs. However, it should be noted that it was not possible to establish a mutant in which M(IP)2Cs are produced but MIPCs are not produced (Figure 1D). Furthermore, MIPCs were accumulated, as compared with in wild-type cells, in IPT1-deleted cells (Figure 1D). Thus, it is not possible to conclude whether the occurrence of the stress hypersensitivities due to cc∆ is caused by the disappearance of MIPCs alone or by the disappearance of both MIPCs and M(IP)2Cs. ccss∆ cells, which were the most severely sensitive to many stresses, exhibited an increase in permeability of plasma membranes and enhancement of impairment of cell wall integrity upon deletion of MSN2/4 and SLT2, respectively, both of which may affect the acquirement of tolerance of multiple stresses in ccss∆ cells (Figures 6E, 7, and 5, D and E). In contrast, the detrimental effects of the deletion of MSN2/4 or SLT2 on wild-type cells were mild or hardly detectable (Figures 5, 6E, and 7B), indicating that ccss∆ cells are more dependent on Msn2/4 and Slt2 than wild-type cells. It should be noted that there are hundreds of target genes in Msn2/4, and thus it is unclear whether the detrimental effects of the deletion of MSN2/4 on ccss∆ cells can be explained only by the impairment of plasma membrane integrity.
Previously, it was found that the HOG pathway is activated on repression of IPC synthase or SPT, which causes reduction in total levels of complex sphingolipids, and the activation is required for maintenance of cell growth under complex sphingolipid biosynthesis–defective conditions (Tanigawa et al., 2012; Yamaguchi et al., 2018). In contrast, the phosphorylation of Hog1 was not enhanced by ccss∆ (Figure 4D), and the deletion of HOG1 had only a mild effect on the growth of ccss∆ cells as compared with that of IPC synthase- or SPT-repressed cells (Figure 5A) (Yamaguchi et al., 2018). Because the total amount of complex sphingolipids was not decreased in ccss∆ cells (Figure 1E), it is suggested that the HOG pathway is activated through reduction of the total amount of complex sphingolipids, but not through a defect of structural diversity of complex sphingolipids. These results also suggest that the activation of the HOG pathway is not necessarily required for compensation for the defect of structural diversity of complex sphingolipid due to ccss∆. Msn2/4 are activated by Hog1 and involved in the rescue effect of the HOG pathway under complex sphingolipid biosynthesis–defective conditions (Yamaguchi et al., 2018; Urita et al., 2022). Msn2/4 are involved in the maintenance of plasma membrane integrity and acquirement of tolerance of several stresses in ccss∆ cells, whereas the Msn2/4-mediated transcriptional activity was decreased rather than increased in ccss∆ cells (Figure 4H). Moreover, in ccss∆ cells, overexpression of MSN2, which suppresses the growth defect due to reduction of the total amount of complex sphingolipids (Yamaguchi et al., 2018) (Supplemental Figure S10A), did not confer marked resistance to stresses whose sensitivity was enhanced by the deletion of MSN2/4 (Supplemental Figure S10B and Figure 5D). Thus, it was suggested that, in ccss∆ cells, activation of Msn2/4 is not necessarily required as well as the HOG pathway; however, the basal activity of Msn2/4 is necessary and sufficient for the protective effects.
How does ccss∆ cause abnormality in plasma membrane properties? Through in silico analysis, Lindahl et al. (2016) suggested that a decrease in the ratio of complex sphingolipids in membranes causes a decrease in thickness and density of the lipid bilayer. In addition, it was also proposed that the formation of hydrogen bonds between complex sphingolipid molecules via the hydroxy group of the Cer moiety contributes to stabilization of membranes (Slotte, 2016). These results suggest that complex sphingolipids and their hydroxylation state are important for the maintenance of membrane integrity. However, in ss∆ cells, the permeability of plasma membranes was decreased rather than increased (Figure 7, A–C). Furthermore, lateral diffusion of plasma membrane–localized hexose transporter 1 is decreased due to ccss∆, which may imply decreased fluidity of plasma membranes (Uemura et al., 2014). In addition, lipid order in plasma membranes of ss∆ and ccss∆ cells, which was evaluated by di-4-ANEPPDHQ, was higher than that of wild-type cells (Figure 7J). Thus, it is assumed that the abnormal plasma membrane properties due to the defect of structural diversity of complex sphingolipids is caused by complex factors. One possible explanation is that the defect of the structural diversity may result in alteration of the complex sphingolipid subcellular distribution, including in plasma membranes. It was reported that, in wild-type cells, the distribution pattern of complex sphingolipids depends on their structure; that is, MIPCs and M(IP)2Cs are relatively abundant in plasma membranes, whereas IPCs are also abundantly detected in the Golgi apparatus and vacuoles, other than plasma membranes (Hechtberger et al., 1994). In addition, the deletion of SUR2 causes loss of production of phytosphingosine from dihydrosphingosine, both of which function as molecules regulating various signal transduction systems (Chung et al., 2001; Pina et al., 2018; Yabuki et al., 2019; Arita et al., 2020). Furthermore, deletion of either or both SCS7 and SUR2 also results in structural changes in free Cer, a precursor of complex sphingolipids (Haak et al., 1997). In most cases, the deletion of SUR2, SCS7, or both did not cause stress hypersensitivity unless there was an additional deletion of CSG1 and CSH1 (Figure 2), and the intracellular amounts of free LCBs and Cers are much lower than that of complex sphingolipids in the steady state (Ejsing et al., 2009); however, it may also be necessary to consider the effect of structural alteration of free LCBs and Cers on plasma membrane properties.
When plasma membrane stress occurs due to an increase in the tension of membranes, the target of rapamycin complex 2(TORC2)/Ypk1 pathway is activated and subsequently increases the sphingolipid levels for a stress response (Roelants et al., 2017). Furthermore, the phosphorylation of Ypk1 by TORC2 is also increased on deletion of CSG2 or CSG1 (Berchtold et al., 2012), which may suggest that abnormalities in plasma membrane properties caused by these mutations are alleviated by the TORC2/Ypk1 pathway. Thus, it is possible that the slow growth phenotype and increases in stress sensitivities of ccss∆ msn2/4∆ cells are caused by disturbance of the TORC2/Ypk1 pathway due to the deletion of MSN2/4. However, the level of IPC-A in ccss∆ msn2/4∆ cells was not decreased as compared with that in ccss∆ cells (Figure 5C), suggesting that the TORC2/Ypk1 pathway is not involved in the abnormal phenotypes due to the deletion of MSN2/4. This also suggests that Msn2/4 are involved in the rescue of abnormal phenotypes due to ccss∆ by affecting other functions rather than regulation of complex sphingolipid biosynthesis. This notion also applies to Slt2, because the deletion of SLT2 also did not cause a decrease in the IPC-A level in ccss∆ cells (Figure 5C). However, it should also be considered the fact that the activation of Slt2 downregulates the activity of TORC2, which may suggest the connection between Slt2 and the TORC2/Ypk1 pathway in ccss∆ cells (Leskoske et al., 2018).
In summary, the present study indicated a relationship between the structural diversity of complex sphingolipids and tolerance to various environmental stresses and compensation mechanisms against the defect of structural diversity. To further investigate the relationship between complex sphingolipids and stress tolerance, it will be necessary to pursue mechanisms that protect themselves by actively altering the composition of complex sphingolipids in the presence of various stresses. For example, it was reported that spontaneous alteration of the composition of complex sphingolipids is required for adaptation of yeast cells to extracellular low pH conditions and impairment of intracellular H+ homeostasis due to a defect of vacuolar H+-ATPase (Tani and Toume, 2015; Otsu et al., 2020). Elucidation of such mechanisms will provide deeper insights into the physiological significance of the structural diversity of complex sphingolipids.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Yeast strains and media
The S. cerevisiae strains used are listed in Table 1, and the methods used for genetic modification of yeast strains and construction of plasmids are described in the Supplemental Materials and Methods. The cells were cultured in YPD medium (1% yeast extract, 2% peptone, and 2% glucose [pH 6.0]) or SC (synthetic complete) medium (0.67% yeast nitrogen base without amino acids [BD Difco, Heidelberg, Germany] and 2% glucose [pH 6.0]) containing nutritional supplements. Buffered medium was prepared by the addition of 100 mM glycine-HCl (for pH 3.5), 50 mM MES, and 50 mM MOPS (for pH 5.5), or 100 mM HEPES (for pH 7.5 and 8.0).
TABLE 1:
Strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| BY4741 | MATa his3∆1 leu2∆0 met15∆0 ura3∆0 | Brachmann et al., 1998 |
| MTY174 | BY4741, URA3 | Arita et al., 2020 |
| MTY128 | BY4741, sur2∆::URA3 | Tani and Kuge, 2012a |
| MTY211 | BY4741, scs7∆::URA3 | Tani and Kuge, 2012a |
| AKY55 | BY4741, sur2∆::kanMX4 scs7∆::URA3 | This study |
| AKY56 | BY4741, csg1∆::natMX4 csh1∆::URA3 | This study |
| AKY57 | BY4741, csg1∆::natMX4 csh1∆::hphNT1 sur2∆::URA3 | This study |
| AKY58 | BY4741, csg1∆::natMX4 csh1∆::hphNT1 scs7∆::URA3 | This study |
| AKY59 | BY4741, csg1∆::natMX4 csh1∆::hphNT1 sur2∆:URA3 scs7∆::kanMX4 | This study |
| AKY61 | BY4741, ipt1∆::URA3 | This study |
| AKY62 | BY4741, ipt1∆::URA3 sur2∆::kanMX4 | This study |
| AKY63 | BY4741, ipt1∆::URA3 scs7∆::hphNT1 | This study |
| AKY64 | BY4741, ipt1∆::URA3 sur2∆::kanMX4 scs7∆::hphNT1 | This study |
| AKY75 | BY4741, hog1∆::LEU2 URA3 | This study |
| AKY80 | BY4741, sur2Δ::kanMX4 scs7Δ::URA3 hog1Δ::LEU2 | This study |
| AKY83 | BY4741, csg1∆::natMX4 csh1∆::URA3 hog1∆::LEU2 | This study |
| AKY86 | BY4741, csg1∆::natMX4 csh1∆::hphNT1 sur2∆:URA3 scs7∆::kanMX4 hog1∆::LEU2 | This study |
| AKY76 | BY4741, slt2∆::LEU2 URA3 | This study |
| AKY78 | BY4741, sur2∆::kanMX4 scs7∆::URA3 slt2∆::LEU2 | This study |
| AKY85 | BY4741, csg1∆::natMX4 csh1∆::URA3 slt2∆::LEU2 | This study |
| MTY2884 | BY4741, csg1∆::natMX4 csh1∆::hphNT1 sur2∆:URA3 scs7∆::kanMX4 slt2∆::LEU2 | This study |
| MTY2886 | BY4741, rim101∆::LEU2 URA3 | This study |
| AKY109 | BY4741, sur2∆::URA3 scs7∆::LEU2 rim101∆::natNT2 | This study |
| AKY111 | BY4741, csg1∆::URA3 csh1∆::LEU2 rim101∆::natNT2 | This study |
| MTY2885 | BY4741, csg1∆::natMX4 csh1∆::hphNT1 sur2∆:URA3 scs7∆::kanMX4 rim101∆::LEU2 | This study |
| MTY2941 | BY4741, SLT2-6xHA::hphNT1 URA3 LEU2 | This study |
| AKY275 | BY4741, csg1∆::URA3 csh1∆::LEU2 sur2∆::kanMX4 scs7∆::natNT2 SLT2-6HA::hphNT1 | This study |
| AKY280 | BY4741, TEFp-yeGFP-RIM101::natNT2 URA3 LEU2 | This study |
| AKY283 | BY4741, csg1∆::URA3 csh1∆::LEU2 sur2∆::kanMX4 scs7∆::hphNT1 TEFp-yeGFP-RIM101::natNT2 | This study |
| AKY312 | BY4741, msn2∆::HIS3 msn4∆::LEU2 URA3 | This study |
| AKY127 | BY4741, sur2∆::kanMX4 scs7∆::URA3 msn2∆::HIS3 msn4∆::LEU2 | This study |
| AKY316 | BY4741, csg1∆::natMX4 csh1∆::hphNT1 msn2∆::HIS3 msn4∆::LEU2 URA3 | This study |
| MTY2957 | BY4741, csg1∆::natMX4 csh1∆::hphNT1 sur2∆::URA3 scs7∆::kanMX4 msn2∆::HIS3 msn4∆::LEU2 | This study |
| AKY244 | BY4741, LEU2 harboring pRS416-6xSTRE-MEL1 | This study |
| AKY247 | BY4741, csg1∆::natMX4 csh1∆::hphNT1 sur2∆::kanMX4 scs7∆::LEU2 harboring pRS416-6xSTRE-MEL1 | This study |
| AKY248 | BY4741, msn2∆::natNT2 msn4∆::hphMX4 harboring pRS416-6xSTRE-MEL1 | This study |
| AKY249 | BY4741, csg1∆::natMX4 csh1∆::hphNT1 sur2∆::LEU2 scs7∆::kanMX4 msn2∆::HIS3 msn4∆::MET15 harboring pRS416-6xSTRE-MEL1 | This study |
| MTY3090 | BY4741, bck1∆::hphNT1 URA3 | This study |
| MTY3091 | BY4741, csg1∆::natMX4 csh1∆::URA3 bck1∆::hphNT1 | This study |
| MTY3092 | BY4741, csg1∆::URA3 csh1∆::LEU2 sur2∆::kanMX4 scs7∆::natNT2 bck1∆::hphNT1 | This study |
| MTY3093 | BY4741, rom2∆::hphNT1 URA3 | This study |
| MTY3094 | BY4741, csg1∆::natMX4 csh1∆::URA3 rom2∆::hphNT1 | This study |
| MTY3089 | BY4741, csg1∆::URA3 csh1∆::LEU2 sur2∆::kanMX4 scs7∆::natNT2 rom2∆::hphNT1 | This study |
| MTY3097 | BY4741 harboring pRS415-NCW2p-MEL1 | This study |
| MTY3098 | BY4741, slt2∆::hphMX4 harboring pRS415-NCW2p-MEL1 | This study |
| MTY3099 | BY4741, csg1∆::natMX4 csh1∆::hphNT1 sur2∆:URA3 scs7∆::kanMX4 harboring pRS415-NCW2p-MEL1 | This study |
| MTY3100 | BY4741, csg1∆::natMX4 csh1∆::hphNT1 sur2∆:URA3 scs7∆::kanMX4 slt2∆::MET15 harboring pRS415-NCW2p-MEL1 | This study |
| MTY3101 | BY4741 harboring pRS415-SED1p-MEL1 | This study |
| MTY3102 | BY4741, slt2∆::hphMX4 harboring pRS415-SED1p-MEL1 | This study |
| MTY3103 | BY4741, csg1∆::natMX4 csh1∆::hphNT1 sur2∆:URA3 scs7∆::kanMX4 harboring pRS415-SED1p-MEL1 | This study |
| MTY3104 | BY4741, csg1∆::natMX4 csh1∆::hphNT1 sur2∆:URA3 scs7∆::kanMX4 slt2∆::MET15 harboring pRS415-SED1p-MEL1 | This study |
| MTY3105 | BY4741 harboring pRS415-CRH1p-MEL1 | This study |
| MTY3106 | BY4741, slt2∆::hphMX4 harboring pRS415-CRH1p-MEL1 | This study |
| MTY3107 | BY4741, csg1∆::natMX4 csh1∆::hphNT1 sur2∆:URA3 scs7∆::kanMX4 harboring pRS415-CRH1p-MEL1 | This study |
| MTY3108 | BY4741, csg1∆::natMX4 csh1∆::hphNT1 sur2∆:URA3 scs7∆::kanMX4 slt2∆::MET15 harboring pRS415-CRH1p-MEL1 | This study |
| STY89 | BY4741, erg6∆::kanMX4 | Tanaka and Tani, 2018 |
| MTY3095 | BY4741, PIL1-yeGFP::hphNT1 URA3 | This study |
| MTY3096 | BY4741, csg1∆::URA3 csh1∆::LEU2 sur2∆::kanMX4 scs7∆::natNT2 PIL1-yeGFP::hphNT1 | This study |
| MTY2297 | BY4741, sip3∆::kanMX4 | Otsu et al., 2020 |
| MTY2300 | BY4741, lam1∆::kanMX4 | Otsu et al., 2020 |
| MTY2338 | BY4741, ysp2∆::kanMX4 | Otsu et al., 2020 |
| AKY260 | BY4741, csg1∆::URA3 csh1∆::HIS3 sur2∆::kanMX4 scs7∆::hphNT1 sip3∆::natNT2 | This study |
| AKY261 | BY4741, csg1∆::URA3 csh1∆::HIS3 sur2∆::kanMX4 scs7∆::hphNT1 lam1∆::natNT2 | This study |
| AKY262 | BY4741, csg1∆::URA3 csh1∆::HIS3 sur2∆::kanMX4 scs7∆::hphNT1 ysp2∆::natNT2 | This study |
| AKY318 | BY4741, slt2∆::LEU2 sip3∆::kanMX4 | This study |
| MTY3003 | BY4741, csg1∆::URA3 csh1∆::HIS3 sur2∆::kanMX4 scs7∆::hphNT1 slt2∆::LEU2 sip3∆::natNT2 | This study |
| MTY3000 | BY4741, msn2∆::HIS3 msn4∆::LEU2 sip3∆::kanMX4 URA3 | This study |
| MTY3007 | BY4741, csg1∆::URA3 csh1∆::HIS3 sur2∆::kanMX4 scs7∆::hphNT1 msn2∆::LEU2 msn4∆::MET15 sip3∆::natNT2 | This study |
Spot assays
Cells were cultured overnight in YPD medium at 30°C and then spotted onto YPD plates in 10-fold serial dilutions starting with a density of 0.7 OD600 units/ml. All plates were incubated at 30°C and photographed after 1–3 d. When indicated, plates were also incubated at 16 or 40°C.
Lipid extraction and TLC analysis
Lipids were extracted from S. cerevisiae as described previously (Hanson and Lester, 1980) with minor modifications. Briefly, cells (3 OD600 units [for detection of complex sphingolipids] or 1.5 OD600 units [for detection of sterols]) were suspended in 350 µl of ethanol/water/diethyl ether/pyridine/15 M ammonia (15:15:5:1:0.018, vol/vol) and then incubated at 65°C for 15 min. The lipid extract was centrifuged at 10,000 × g for 1 min and then extracted once more in the same manner. The resulting supernatants were dried. For analysis of complex sphingolipids but not sterols, the lipid extracts were dissolved in 130 µl of monomethylamine (40% methanol solution)/water (10:3, vol/vol), incubated for 1 h at 53°C (mild alkaline treatment), and then dried. The lipids were suspended in 60 µl of chloroform/methanol/water (5:4:1, vol/vol), and then separated on Silica Gel 60 TLC (thin-layer chromatography) plates (Merck, Whitehouse Station, NJ) with chloroform/methanol/4.2 M ammonia (9:7:2, vol/vol) (for detection of complex sphingolipids) or hexane/diethyl ether/acetic acid (30:70:1, vol/vol) (for detection of sterols) as the solvent system. The TLC plates were sprayed with 10% copper sulfate in 8% orthophosphoric acid and then heated at 180°C to visualize lipids. The relative intensity of each lipid band was determined with ImageJ software (National Institutes of Health, Bethesda, MD). Identification of the band of each complex sphingolipid subtype and sterols was performed as described in previous papers (Uemura et al., 2014; Tani and Toume, 2015; Yamaguchi et al., 2018).
LC-ESI MS/MS analysis of Cers
Lipids were extracted from cells (3 OD600 units), subjected to mild alkaline treatment, and then dried as described above. The lipids mainly containing sphingolipids were dissolved in 100 µl of chloroform/methanol/water (5:4:1, vol/vol) with sonication and then mixed with 500 µl of 2-propanol. After centrifugation at 18,000 × g for 3 min, 550 µl of the supernatant was transferred to autoinjector vials, and then Cers were measured using LC-ESI MS/MS (3200 QTRAP; SCIEX, USA) equipped with an InertSustain C18 reverse-phase column (2.1 × 150 mm, 5 μm; GL Sciences, Tokyo, Japan). The gradient was started with 40% B (2-propanol with 0.1% formic acid and 0.028% ammonium) in buffer A (acetonitrile/methanol/distilled water, 19:19:2, vol/vol/vol containing 0.1% formic acid and 0.028% ammonium), reached 70% B for 10 min, and maintained at 70% B for 5 min. The gradient was returned to the starting conditions and the column was equilibrated for 5 min before the next run. For the measurement of molecular species of Cers, MRM (multiple reaction monitoring) analysis with positive ion mode was constructed with the combinations LCBs phytosphingosine [t16:0, t18:0, t20:0] or dihydrosphingosine [d16:0, d18:0, d20:0]), fatty acids with different chain lengths (C16:0, C18:0, C20:0, C22:0, C24:0, or C26:0), and fatty acids with different hydroxylation states (0 [A and B type], 1 [B′ and C type], or 2 [D type]). Collision energy for generating product ion was 35. The peak intensity of each Cer molecular species was analyzed using MultiQuant 3.0.1 (SCIEX).
Yeast protein extraction, SDS–PAGE, and Western blotting
Protein extraction, SDS–PAGE, and Western blotting were performed as described elsewhere (Tani and Kuge, 2010b) with some modifications. For protein extraction, yeast cells (2 OD600 units) grown in YPD medium were collected by centrifugation, washed with water, and then resuspended in 100 µl of 0.2 N NaOH containing 0.5% 2-mercaptoethanol. The suspension was incubated on ice for 15 min. One milliliter of ice-cold acetone was added to the suspension, followed by incubation for 30 min at –25°C, and then the proteins were precipitated by centrifugation for 10 min at 10,000 × g. The pellet was resuspended in 100 µl of SDS sample buffer (156 mM Tris-HCl, pH 6.8, containing 5% SDS, 25% glycerol, 5% 2-mercaptoethanol and 0.001% bromophenol blue). The suspension was mixed well, heated for 3 min at 95°C, and then centrifuged for 2 min at 10,000 × g. Then the supernatant was separated by SDS–PAGE according to the method of Laemmli (1970). For Western blotting, anti-HA (HA-7; Sigma-Aldrich), anti–phospho-p38 MAPK, anti–phospho-p44/42 MAPK (Cell Signaling Technology, Danvers, MA), anti-Hog1 (Santa Cruz Biotechnology, Santa Cruz, CA), and anti-Pgk1 (Thermo Fisher Scientific, Waltham, MA) were used as primary antibodies. Horseradish peroxidase–conjugated anti-mouse or anti-rabbit immunoglobulin G (Thermo Fisher Scientific) was used as the secondary antibody. The relative intensity of each protein band was determined with ImageJ software.
Enzyme assay for α-galactosidase
α-Galactosidase assays were carried out as described previously (Yamaguchi et al., 2018) with minor modifications. Briefly, cells (0.7 or 1 OD 600 units for measurement of 6xSTRE or NCW2, SED1, and CRH1 promoters, respectively) were suspended in 200 µl of 20 mM Tris-HCl buffer, pH 7.5, containing 0.35% 2-mercaptoethanol and 0.002% SDS (buffer A), 5 µl of chloroform was added, and then the samples were vortexed for 10 s. For measurement of the activity of α-galactosidase expressed by NCW2, SED1, and CRH1 promoters, the samples were diluted 10-fold with buffer A. After 5 min of preincubation at 30°C, the samples (200 µl) were mixed with 80 µl of 7 mM p-nitrophenyl-α-galactoside (Sigma-Aldrich) in 100 mM citric acid buffer, pH 4.0, and then incubated at 30°C for 10 min (for 6xSTRE) or 15 min (for NCW2, SED1, and CRH1 promoters). Following quenching of the reaction with 900 µl of 0.1 M NaOH, samples were centrifuged at 10,000 × g for 1 min, and the A410 of the supernatants was measured.
RNA-sequencing
Wild-type (n = 3) and ccss∆ (n = 3) cells were cultured overnight in YPD medium, diluted (0.3 OD600 units/ml) in fresh YPD medium, and then incubated for 5 h at 30°C. The total RNA was extracted from cells (10 OD600 units) by the hot-phenol methods (Collart and Oliviero, 2001) and then purified using the TRIzol LS Reagent (Thermo Fisher Scientific) and an SV Total RNA Isolation System (Promega, Madison, WI) according to the manufacturers’ instructions. RNA samples were quantified with an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE), and the quality was confirmed with a Tapestation (Agilent Technologies, Santa Clara, CA). The sequencing libraries were prepared from 200 ng of total RNA with the NEBNext Magnesium RNA Fragmentation Module (New England Biolab, USA) and the MGIEasy RNA Directional Library Prep Set (MGI, Shenzhen, China) according to the manufacturers’ instructions. The libraries were sequenced with a DNBSEQ-G400 FAST Sequencer (MGI) using the paired-end 150 nt strategy. Quality trimming and adapter clipping of the read data were performed using Trimmomatic version 0.38 (Bolger et al., 2014). Trimmed reads were mapped to the transcript in the reference S288C (https://www.yeastgenome.org/strain/s288c) using the Bowtie2 aligner within RSEM (RNA-Seq by expectation-maximization) (Li and Dewey, 2011). The abundance estimation for genes and isoforms with RSEM generated basic counts data (expected counts). The edgeR (Robinson et al., 2010) program was used to detect the differentially expressed genes (DEGs). Normalized counts per million (CPM) values, log fold changes (logFC), and P values were obtained from the normalized CPM values. Then criteria for DEGs were established: P value ≤ 0.01 and fold change ≥1.4. The data sets of RNA-sequencing are listed in Supplemental Table S1.
Translocation of yeGFP-Rim101 into nuclei
Cells were collected by centrifugation, fixed with 500 μl of 2% paraformaldehyde in phosphate-buffered saline (PBS) (–), and then incubated at room temperature for 30 min. Cells were washed twice with PBS (–) and then viewed under a fluorescence microscope. For quantification of the nuclear localization of yeGFP-Rim101, the fixed cells were stained with 10 µg/ml 4’,6-diamidino-2-phenylindole (Dojindo Chemical Laboratories, Kumamoto, Japan) for 30 min. The fluorescence intensity of yeGFP in cytoplasm and the nucleus in individual cells was quantified with ImageJ software.
Rhodamine 6G incorporation into cells
Cells were cultured overnight in YPD medium, diluted (0.3 OD600 units/ml) in fresh YPD medium, and then incubated for 5 h at 30°C. For measurement of the incorporation rate of rhodamine 6G under ATP-depleted conditions, cells (1 OD600 units) were preincubated in 300 µl of YP (1% yeast extract and 2% peptone) containing 50 mM 2-deoxy-d-glucose (Nacalai Tesque) and 10 mM NaN3 and NaF for 30 min at 30°C and then incubated with 10 µM rhodamine 6G (Tokyo Chemical Industry, Tokyo, Japan) for 30 min at 30°C. When ATP was not depleted, cells (1 OD600 units) were resuspended in 300 µl of YPD medium containing 10 µM rhodamine 6G and then incubated for 30 min at 30°C. The cells were collected by centrifugation, washed with 10 mM NaN3 and NaF, and then viewed under a fluorescence microscope. The fluorescence intensity of rhodamine 6G in individual cells was quantified with ImageJ software.
di-4-ANEPPDHQ staining
di-4-ANEPPDHQ staining of yeast cells was performed as described in Owen et al. (2011) with several modifications. Briefly, cells (1 OD600 units) were suspended in 100 µl of YPD medium containing 5 µM di-4-ANEPPDHQ and incubated for 1 min at 30°C, and then measurement of the intensity of green fluorescence (emission wavelength of 525 ± 20 nm) and red fluorescence (emission wavelength of 610 ± 10 nm), which were excited at 488 nm, was performed by flow cytometry.
Filipin staining
Filipin staining of yeast cells was performed as described in Nakase et al. (2010). Briefly, filipin was added to the medium at a final concentration of 15 µg/ml, and cells were observed immediately using a fluorescence microscope.
Statistical analysis
Statistical analysis was performed using Student’s t test, and the P values obtained are indicated.
Supplementary Material
Acknowledgments
We thank O. Kuge, T. Ogishima, and N. Miyata (Kyushu University) for valuable suggestions regarding this study. This study was supported by KAKENHI (21H02118) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan, the Noda Institute for Scientific Research, Japan, and the Ohsumi Frontier Science Foundation, Japan.
Abbreviations used:
- Cer
ceramide
- CFW
calcofluor white
- CWI
cell wall integrity
- Dox
doxycycline
- GO
gene ontology
- HOG
high osmolarity glycerol
- IPC
inositol phosphorylceramide
- LCB
long chain base
- MIPC
mannosylinositol phosphorylceramide
- M(IP) 2C
mannosyldiinositol phosphorylceramide
- SPT
serine palmitoyltransferase.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E22-04-0117) on July 27, 2022.
REFERENCES
- Abe F, Hiraki T (2009). Mechanistic role of ergosterol in membrane rigidity and cycloheximide resistance in Saccharomyces cerevisiae. Biochim Biophys Acta 1788, 743–752. [DOI] [PubMed] [Google Scholar]
- Arita N, Sakamoto R, Tani M (2020). Mitochondrial reactive oxygen species-mediated cytotoxicity of intracellularly accumulated dihydrosphingosine in the yeast Saccharomyces cerevisiae. FEBS J 287, 3427–3448. [DOI] [PubMed] [Google Scholar]
- Beeler TJ, Fu D, Rivera J, Monaghan E, Gable K, Dunn TM (1997). SUR1 (CSG1/BCL21), a gene necessary for growth of Saccharomyces cerevisiae in the presence of high Ca2+ concentrations at 37 degrees C, is required for mannosylation of inositolphosphorylceramide. Mol Gen Genet 255, 570–579. [DOI] [PubMed] [Google Scholar]
- Berchtold D, Piccolis M, Chiaruttini N, Riezman I, Riezman H, Roux A, Walther TC, Loewith R (2012). Plasma membrane stress induces relocalization of Slm proteins and activation of TORC2 to promote sphingolipid synthesis. Nat Cell Biol 14, 542–547. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132. [DOI] [PubMed] [Google Scholar]
- Brewster JL, Gustin MC (2014). Hog1: 20 years of discovery and impact. Sci Signal 7, re7. [DOI] [PubMed] [Google Scholar]
- Chung N, Mao C, Heitman J, Hannun YA, Obeid LM (2001). Phytosphingosine as a specific inhibitor of growth and nutrient import in Saccharomyces cerevisiae. J Biol Chem 276, 35614–35621. [DOI] [PubMed] [Google Scholar]
- Clay L, Caudron F, Denoth-Lippuner A, Boettcher B, Buvelot Frei S, Snapp EL, Barral Y (2014). A sphingolipid-dependent diffusion barrier confines ER stress to the yeast mother cell. eLife 3, e01883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart MA, Oliviero S (2001). Preparation of yeast RNA. Curr Protoc Mol Biol Chapter 13, Unit13.12. [DOI] [PubMed] [Google Scholar]
- Dickson RC, Lester RL (2002). Sphingolipid functions in Saccharomyces cerevisiae. Biochim Biophys Acta 1583, 13–25. [DOI] [PubMed] [Google Scholar]
- Dickson RC, Nagiec EE, Wells GB, Nagiec MM, Lester RL (1997). Synthesis of mannose-(inositol-P)2-ceramide, the major sphingolipid in Saccharomyces cerevisiae, requires the IPT1 (YDR072c) gene. J Biol Chem 272, 29620–29625. [DOI] [PubMed] [Google Scholar]
- Dickson RC, Sumanasekera C, Lester RL (2006). Functions and metabolism of sphingolipids in Saccharomyces cerevisiae. Prog Lipid Res 45, 447–465. [DOI] [PubMed] [Google Scholar]
- Dudley AM, Janse DM, Tanay A, Shamir R, Church GM (2005). A global view of pleiotropy and phenotypically derived gene function in yeast. Mol Syst Biol 1, 2005.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Beney L, Ritt JF, Lherminier J, Gervais P (2010). Lateral reorganization of plasma membrane is involved in the yeast resistance to severe dehydration. Biochim Biophys Acta 1798, 975–985. [DOI] [PubMed] [Google Scholar]
- Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, Simons K, Shevchenko A (2009). Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci USA 106, 2136–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohlich F, Moreira K, Aguilar PS, Hubner NC, Mann M, Walter P, Walther TC (2009). A genome-wide screen for genes affecting eisosomes reveals Nce102 function in sphingolipid signaling. J Cell Biol 185, 1227–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futai E, Maeda T, Sorimachi H, Kitamoto K, Ishiura S, Suzuki K (1999). The protease activity of a calpain-like cysteine protease in Saccharomyces cerevisiae is required for alkaline adaptation and sporulation. Mol Gen Genet 260, 559–568. [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000). Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11, 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatta AT, Wong LH, Sere YY, Calderon-Norena DM, Cockcroft S, Menon AK, Levine TP (2015). A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport. eLife 4, e07253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan XL, Souza CM, Pichler H, Dewhurst G, Schaad O, Kajiwara K, Wakabayashi H, Ivanova T, Castillon GA, Piccolis M, et al. (2009). Functional interactions between sphingolipids and sterols in biological membranes regulating cell physiology. Mol Biol Cell 20, 2083–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haak D, Gable K, Beeler T, Dunn T (1997). Hydroxylation of Saccharomyces cerevisiae ceramides requires Sur2p and Scs7p. J Biol Chem 272, 29704–29710. [DOI] [PubMed] [Google Scholar]
- Hallstrom TC, Lambert L, Schorling S, Balzi E, Goffeau A, Moye-Rowley WS (2001). Coordinate control of sphingolipid biosynthesis and multidrug resistance in Saccharomyces cerevisiae. J Biol Chem 276, 23674–23680. [DOI] [PubMed] [Google Scholar]
- Hanson BA, Lester RL (1980). The extraction of inositol-containing phospholipids and phosphatidylcholine from Saccharomyces cerevisiae and Neurospora crassa. J Lipid Res 21, 309–315. [PubMed] [Google Scholar]
- Hechtberger P, Zinser E, Saf R, Hummel K, Paltauf F, Daum G (1994). Characterization, quantification and subcellular localization of inositol-containing sphingolipids of the yeast, Saccharomyces cerevisiae. Eur J Biochem 225, 641–649. [DOI] [PubMed] [Google Scholar]
- Herrero AB, Astudillo AM, Balboa MA, Cuevas C, Balsinde J, Moreno S (2008). Levels of SCS7/FA2H-mediated fatty acid 2-hydroxylation determine the sensitivity of cells to antitumor PM02734 . Cancer Res 68, 9779–9787. [DOI] [PubMed] [Google Scholar]
- Imazu H, Sakurai H (2005). Saccharomyces cerevisiae heat shock transcription factor regulates cell wall remodeling in response to heat shock. Eukaryot Cell 4, 1050–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskaia A, Marques JMT, Bastos AEP, Antunes CAC, Bento-Oliveira A, Scolari S, Lobo G, Malho R, Herrmann A, Marinho HS, de Almeida RFM (2020). Liquid-ordered phase formation by mammalian and yeast sterols: a common feature with organizational differences. Front Cell Dev Biol 8, 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto T, Mioka T, Itoh E, Williams DE, Andersen RJ, Tanaka K (2021). Phospholipid flippases and Sfk1 are essential for the retention of ergosterol in the plasma membrane. Mol Biol Cell 32, 1374–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knupp J, Martinez-Montanes F, Van Den Bergh F, Cottier S, Schneiter R, Beard D, Chang A (2017). Sphingolipid accumulation causes mitochondrial dysregulation and cell death. Cell Death Differ 24, 2044–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono K, Al-Zain A, Schroeder L, Nakanishi M, Ikui AE (2016). Plasma membrane/cell wall perturbation activates a novel cell cycle checkpoint during G1 in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 113, 6910–6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Leskoske KL, Roelants FM, Emmerstorfer-Augustin A, Augustin CM, Si EP, Hill JM, Thorner J (2018). Phosphorylation by the stress-activated MAPK Slt2 down-regulates the yeast TOR complex 2. Genes Dev 32, 1576–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DE (2005). Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 69, 262–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl L, Genheden S, Eriksson LA, Olsson L, Bettiga M (2016). Sphingolipids contribute to acetic acid resistance in Zygosaccharomyces bailii. Biotechnol Bioeng 113, 744–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G, Gruhler A, Liu Y, Jensen ON, Dickson RC (2008). The sphingolipid long-chain base-Pkh1/2-Ypk1/2 signaling pathway regulates eisosome assembly and turnover. J Biol Chem 283, 10433–10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Arias A, Yost HJ, Casadaban MJ (1984). Role of an upstream regulatory element in leucine repression of the Saccharomyces cerevisiae leu2 gene. Nature 307, 740–742. [DOI] [PubMed] [Google Scholar]
- Merrill AH Jr (2011). Sphingolipid and glycosphingolipid metabolic pathways in the era of sphingolipidomics. Chem Rev 111, 6387–6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mioka T, Fujimura-Kamada K, Mizugaki N, Kishimoto T, Sano T, Nunome H, Williams DE, Andersen RJ, Tanaka K (2018). Phospholipid flippases and Sfk1p, a novel regulator of phospholipid asymmetry, contribute to low permeability of the plasma membrane. Mol Biol Cell 29, 1203–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira NP, Palma M, Guerreiro JF, Sa-Correia I (2010). Genome-wide identification of Saccharomyces cerevisiae genes required for tolerance to acetic acid. Microb Cell Fact 9, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto Y, Tani M (2015). Synthesis of mannosylinositol phosphorylceramides is involved in maintenance of cell integrity of yeast Saccharomyces cerevisiae. Mol Microbiol 95, 706–722. [DOI] [PubMed] [Google Scholar]
- Moskvina E, Schuller C, Maurer CT, Mager WH, Ruis H (1998). A search in the genome of Saccharomyces cerevisiae for genes regulated via stress response elements. Yeast 14, 1041–1050. [DOI] [PubMed] [Google Scholar]
- Nakase M, Tani M, Morita T, Kitamoto HK, Kashiwazaki J, Nakamura T, Hosomi A, Tanaka N, Takegawa K (2010). Mannosylinositol phosphorylceramide is a major sphingolipid component and is required for proper localization of plasma-membrane proteins in Schizosaccharomyces pombe. J Cell Sci 123, 1578–1587. [DOI] [PubMed] [Google Scholar]
- Otsu M, Toume M, Yamaguchi Y, Tani M (2020). Proper regulation of inositolphosphorylceramide levels is required for acquirement of low pH resistance in budding yeast. Sci Rep 10, 10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen DM, Rentero C, Magenau A, Abu-Siniyeh A, Gaus K (2011). Quantitative imaging of membrane lipid order in cells and organisms. Nat Protoc 7, 24–35. [DOI] [PubMed] [Google Scholar]
- Petelenz-Kurdziel E, Eriksson E, Smedh M, Beck C, Hohmann S, Goksor M (2011). Quantification of cell volume changes upon hyperosmotic stress in Saccharomyces cerevisiae. Integr Biol 3, 1120–1126. [DOI] [PubMed] [Google Scholar]
- Pina F, Yagisawa F, Obara K, Gregerson JD, Kihara A, Niwa M (2018). Sphingolipids activate the endoplasmic reticulum stress surveillance pathway. J Cell Biol 217, 495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram AF, Klis FM (2006). Identification of fungal cell wall mutants using susceptibility assays based on Calcofluor white and Congo red. Nat Protoc 1, 2253–2256. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants FM, Leskoske KL, Martinez Marshall MN, Locke MN, Thorner J (2017). The TORC2-dependent signaling network in the yeast Saccharomyces cerevisiae. Biomolecules 7, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata KT, Hashii K, Yoshizawa K, Tahara YO, Yae K, Tsuda R, Tanaka N, Maeda T, Miyata M, Tabuchi M (2022). Coordinated regulation of TORC2 signaling by MCC/eisosome-associated proteins, Pil1 and tetraspan membrane proteins during the stress response. Mol Microbiol 117, 1227–1244. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AP, McEntee K (1996). Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 93, 5777–5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serhan G, Stack CM, Perrone GG, Morton CO (2014). The polyene antifungals, amphotericin B and nystatin, cause cell death in Saccharomyces cerevisiae by a distinct mechanism to amphibian-derived antimicrobial peptides. Ann Clin Microbiol Antimicrob 13, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte JP (2016). The importance of hydrogen bonding in sphingomyelin’s membrane interactions with co-lipids. Biochim Biophys Acta 1858, 304–310. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Tani M (2018). Mannosylinositol phosphorylceramides and ergosterol coodinately maintain cell wall integrity in the yeast Saccharomyces cerevisiae. FEBS J 285, 2405–2427. [DOI] [PubMed] [Google Scholar]
- Tani M (2016). Structure-function relationship of complex sphingolipids in yeast. Trends Glycosci Glycotechnol 28, E109–E116. [Google Scholar]
- Tani M, Kuge O (2010a). Defect of synthesis of very long-chain fatty acids confers resistance to growth inhibition by inositol phosphorylceramide synthase repression in yeast Saccharomyces cerevisiae. J Biochem 148, 565–571. [DOI] [PubMed] [Google Scholar]
- Tani M, Kuge O (2010b). Requirement of a specific group of sphingolipid-metabolizing enzyme for growth of yeast Saccharomyces cerevisiae under impaired metabolism of glycerophospholipids. Mol Microbiol 78, 395–413. [DOI] [PubMed] [Google Scholar]
- Tani M, Kuge O (2012a). Hydroxylation state of fatty acid and long-chain base moieties of sphingolipid determine the sensitivity to growth inhibition due to AUR1 repression in Saccharomyces cerevisiae. Biochem Biophys Res Commun 417, 673–678. [DOI] [PubMed] [Google Scholar]
- Tani M, Kuge O (2012b). Involvement of complex sphingolipids and phosphatidylserine in endosomal trafficking in yeast Saccharomyces cerevisiae. Mol Microbiol 86, 1262–1280. [DOI] [PubMed] [Google Scholar]
- Tani M, Toume M (2015). Alteration of complex sphingolipid composition and its physiological significance in yeast Saccharomyces cerevisiae lacking vacuolar ATPase. Microbiology 161, 2369–2383. [DOI] [PubMed] [Google Scholar]
- Tanigawa M, Kihara A, Terashima M, Takahara T, Maeda T (2012). Sphingolipids regulate the yeast high-osmolarity glycerol response pathway. Mol Cell Biol 32, 2861–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Urita A, Koga A, Takayama C, Tani M (2020). ROS-mediated synthetic growth defect caused by impaired metabolism of sphingolipids and phosphatidylserine in budding yeast. Biosci Biotechnol Biochem 84, 2529–2532. [DOI] [PubMed] [Google Scholar]
- Toume M, Tani M (2016). Yeast lacking the amphiphysin-family protein Rvs167 are sensitive to disruptions in sphingolipid levels. FEBS J 283, 2911–2928. [DOI] [PubMed] [Google Scholar]
- Uemura S, Kihara A, Inokuchi J, Igarashi Y (2003). Csg1p and newly identified Csh1p function in mannosylinositol phosphorylceramide synthesis by interacting with Csg2p. J Biol Chem 278, 45049–45055. [DOI] [PubMed] [Google Scholar]
- Uemura S, Shishido F, Tani M, Mochizuki T, Abe F, Inokuchi JI (2014). Loss of hydroxyl groups from the ceramide moiety can modify the lateral diffusion of membrane proteins in S. cerevisiae. J Lipid Res 55, 1343–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urita A, Ishibashi Y, Kawaguchi R, Yanase Y, Tani M (2022). Crosstalk between protein kinase A and the HOG pathway under impaired biosynthesis of complex sphingolipids in budding yeast. FEBS J 289, 766–786. [DOI] [PubMed] [Google Scholar]
- Walther TC, Aguilar PS, Frohlich F, Chu F, Moreira K, Burlingame AL, Walter P (2007). Pkh-kinases control eisosome assembly and organization. EMBO J 26, 4946–4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M, Fabrizio P, Hu J, Ge H, Cheng C, Li L, Longo VD (2008). Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genet 4, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki Y, Ikeda A, Araki M, Kajiwara K, Mizuta K, Funato K (2019). Sphingolipid/Pkh1/2-TORC1/Sch9 signaling regulates ribosome biogenesis in tunicamycin-induced stress response in yeast. Genetics 212, 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata M, Obara K, Kihara A (2013). Unperverted synthesis of complex sphingolipids is essential for cell survival under nitrogen starvation. Genes Cells 18, 650–659. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Katsuki Y, Tanaka S, Kawaguchi R, Denda H, Ikeda T, Funato K, Tani M (2018). Protective role of the HOG pathway against the growth defect caused by impaired biosynthesis of complex sphingolipids in yeast Saccharomyces cerevisiae. Mol Microbiol 107, 363–386. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Hashiramoto A, Haluzik M, Mizukami H, Beck S, Norton A, Kono M, Tsuji S, Daniotti JL, Werth N, et al. (2003). Enhanced insulin sensitivity in mice lacking ganglioside GM3. Proc Natl Acad Sci USA 100, 3445–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Go S, Takasaki K, Kakazu Y, Ohashi M, Nagafuku M, Kabayama K, Sekimoto J, Suzuki S, Takaiwa K, et al. (2009). Mice lacking ganglioside GM3 synthase exhibit complete hearing loss due to selective degeneration of the organ of Corti. Proc Natl Acad Sci USA 106, 9483–9488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Beeler T, Dunn T (1994). Suppressors of the Ca(2+)-sensitive yeast mutant (csg2) identify genes involved in sphingolipid biosynthesis. Cloning and characterization of SCS1, a gene required for serine palmitoyltransferase activity. J Biol Chem 269, 21480–21488. [PubMed] [Google Scholar]
- Zhao F, Yang J, Li J, Li Z, Lin Y, Zheng S, Liang S, Han S (2020). Multiple cellular responses guarantee yeast survival in presence of the cell membrane/wall interfering agent sodium dodecyl sulfate. Biochem Biophys Res Commun 527, 276–282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.