Abstract
The adaptor protein complex-4 or AP-4 is known to mediate autophagosome maturation through regulating sorting of transmembrane cargo such as ATG9A at the Golgi. There is a need to understand AP-4 function in neurons, as mutations in any of its four subunits cause a complex form of hereditary spastic paraplegia (HSP) with intellectual disability. While AP-4 has been implicated in regulating trafficking and distribution of cargo such as ATG9A and APP, little is known about its effect on neuronal lysosomal protein traffic, lysosome biogenesis, and function. In this study, we demonstrate that in human iPSC-derived neurons AP-4 regulates lysosome composition, function, and transport via regulating the export of critical lysosomal receptors, including Sortilin 1, from the trans-Golgi network to endo-lysosomes. Additionally, loss of AP-4 causes endo-lysosomes to stall and build up in axonal swellings potentially through reduced recruitment of retrograde transport machinery to the organelle. These findings of axonal lysosome buildup are highly reminiscent of those observed in Alzheimer’s disease as well as in neurons modeling the most common form of HSP, caused by spastin mutations. Our findings implicate AP-4 as a critical regulator of neuronal lysosome biogenesis and altered lysosome function and axonal endo-lysosome transport as an underlying defect in AP-4-deficient HSP. Additionally, our results also demonstrate the utility of the human i3Neuronal model system in investigating neuronal phenotypes observed in AP-4-deficient mice and/or the human AP-4 deficiency syndrome.
INTRODUCTION
The adaptor protein complex-4 (AP-4) is a low abundance, ubiquitously expressed complex that belongs to the family of closely related heterotetrameric complexes (Dell’Angelica et al., 1999; Hirst et al., 1999) that are involved in sorting and trafficking of cargo in cells (Hirst et al., 2013). The AP-4 complex consists of four subunits encoded by different genes: ε (AP4E1), µ (AP4M1), β (AP4B1), and σ (AP4S1). Mutations in any of these four genes result in a complex form of hereditary spastic paraplegia (HSP) with intellectual disability, referred to as AP-4 deficiency syndrome (Verkerk et al., 2009; Abou Jamra et al., 2011; Moreno-De-Luca et al., 2011; Tüysüz et al., 2014; Abdollahpour et al., 2015; Hardies et al., 2015; Ebrahimi-Fakhari et al., 2018).
Early in vitro and yeast two-hybrid studies that examined AP-4 interactions indicated that it binds to lysosomal proteins such as LAMP1 and LAMP2a as well as CD63 through interactions with YXXø motif (Bonifacino and Dell’Angelica, 1999). However, these interactions are weak, and loss of AP-4 function does not alter the distribution of these lysosomal proteins. Loss of AP-4 function does, however, strongly affect the intracellular traffic and distribution of the autophagy protein ATG9A (Mattera et al., 2017; De Pace et al., 2018; Davies et al., 2018). ATG9A is a multispanning transmembrane protein in the core autophagy machinery (Rubinsztein et al., 2012) that cycles between the trans-Golgi network (TGN) and peripheral organelles including endosomes and preautophagosomal structures, providing membrane for the growth of the latter (Reggiori et al., 2004; Young et al., 2006; Orsi et al., 2012; Imai et al., 2016). Loss of AP-4 in several cell types, including HeLa cells, MEFs, and primary neurons, results in strong retention of ATG9A in the TGN and reduced amounts of peripheral ATG9A leading to defects in autophagosomal maturation (De Pace et al., 2018; Davies et al., 2018; Mattera et al., 2017; Ivankovic et al., 2020).
While the involvement of AP-4 in regulating the autophagic pathway has received considerable attention, very little is known about how it affects lysosomal composition, trafficking, and function. Studies have demonstrated that AP-4 ε KO mice exhibit axonal swellings in various regions of the brain and spinal cord (De Pace et al., 2018). Intriguingly, axonal swellings in the hippocampus and white matter tracts of the midbrain in AP-4 ε KO mice were enriched in the late endosomal and lysosomal protein, LAMP1 (Edmison et al., 2021; De Pace et al., 2018). This suggests that axonal endo-lysosome homeostasis is altered upon loss of AP-4.
Here we examine the composition, distribution, and function of lysosomes in AP-4-depleted human iPSC-derived neurons. We find that their neuronal lysosomes contain lower levels of certain lysosomal enzymes and exhibit compromised function. We show that loss of AP-4 dramatically alters distribution of Sortilin 1 (referred to as Sortilin here), a critical receptor involved in cargo transport between the TGN and the endo-lysosomes (Nielsen et al., 2001). Last, we show that loss of AP-4 causes LAMP1-positive organelles as well as MAPK8IP3/JIP3, a neuronally enriched putative adaptor protein that regulates retrograde axonal transport to build up in axonal swellings. Given AP-4 regulation of cargo sorting at the TGN, this could result from AP-4 loss altering TGN export of a regulator of axonal lysosome transport (and a potential interactor of MAPK8IP3) to the organelles. In addition to these new findings, we also validated the model by examining the distribution of a well-established AP-4 cargo, namely, ATG9A. We demonstrate that the AP-4-depleted human i3Neurons phenocopy the strong TGN-retention of ATG9A observed in AP-4-deficient mouse neurons and cultured cell lines. Thus using the human i3Neuron culture model, we demonstrate that the AP-4 complex is a key regulator of neuronal lysosome composition, function, and transport. Our studies indicate that altered lysosome function and movement likely contribute to the development of this form of complex HSP. Our studies also highlight the value of the i3Neuron model, in conjunction with the CRISPRi system in elucidating neuronal phenotypes linked with HSPs and other neurodegenerative conditions arising from loss/deficiency of specific proteins.
RESULTS
To determine the role of AP-4 complex in regulating neuronal lysosome function and traffic in human neurons, we made use of the CRISPRi-iPSC system (Wang et al., 2017; Fernandopulle et al., 2018; Tian et al., 2019; Wu et al., 2021). In iPSCs, the neurogenic transcription factor NGN2 is integrated under a doxycycline-responsive promoter at a safe harbor locus in the WTC11 iPSC line (Wang et al., 2017; Fernandopulle et al., 2018). This system allows simple and rapid generation of glutamatergic cortical neurons (i3Neurons) upon culture of the iPSCs in the presence of doxycycline for a brief period. These i3Neurons exhibit morphological and biochemical properties of neurons 14 d postinduction (Wang et al., 2017; Gowrishankar et al., 2021) and are electrically active after 21 d (Wang et al., 2017). We have employed a modified iPSC system, in which CRISPR-inhibition (CRISPRi) machinery is integrated into a safe harbor locus (Tian et al., 2019; Wu et al., 2021). In CRISPRi, an enzymatically dead Cas9 fused to a KRAB transcriptional repressor is targeted close to the transcriptional start site of the target gene by a single guide RNA (sgRNA), thereby inhibiting expression of the gene. This system has advantages over standard CRISPR-based knockout systems, which include high specificity with low toxicity and strikingly few off-targets (Tian et al., 2019; Wu et al., 2021).
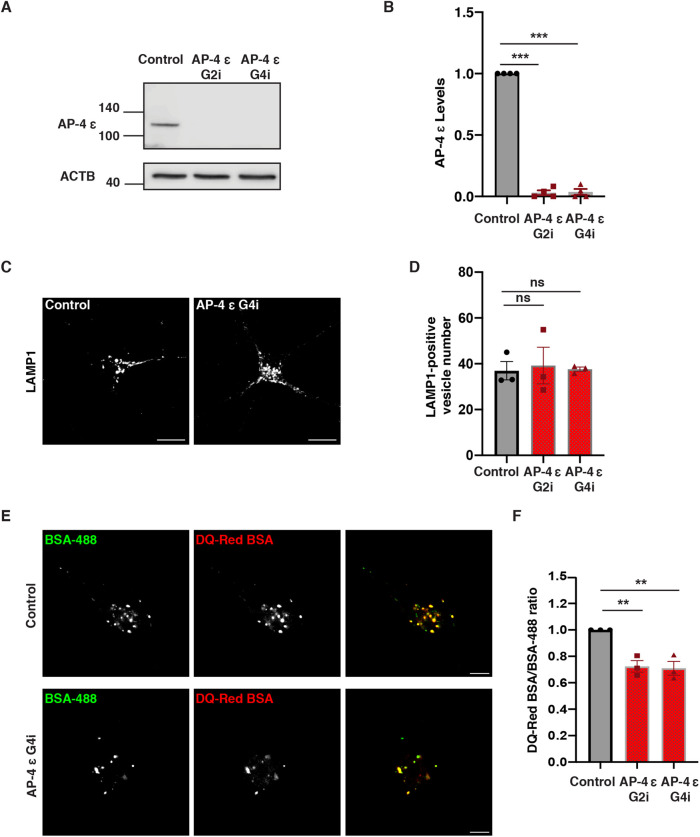
Depletion or loss of the ε subunit of AP-4 caused destabilization of the complex in cultured cells and in a mouse model (Mattera et al., 2017; Ivankovic et al., 2020) and should thus be effective in causing loss of AP-4 loss of function. We, therefore, transduced CRISPRi-iPSCs with either of two separate sgRNAs referred to as G2i or G4i (specific sequences of G2i and G4i in Supplemental Table S2), targeting the AP-4 ε transcriptional start site or a scrambled sequence (SC), then differentiated the resulting iPSC lines to neurons (henceforth AP-4 and Control i3Neurons). After 21 d in culture, efficient depletion of AP-4 ε was observed in AP-4 CRISPRi i3Neurons relative to scrambled sgRNA control (Figure 1, A and B).
FIGURE 1:
Loss of AP-4 affects lysosome function. (A and B) Immunoblotting reveals decreased levels of AP-4 protein in AP-4 i3Neurons compared with Control i3Neurons (21 DIV; ACTB used as loading Control; mean ± SEM from four independent experiments; ***P < 0.001). (C) Control and AP-4 G4i i3Neurons (14 DIV) stained for LAMP1. (D) Quantification of number of LAMP1-positive vesicles in soma of AP-4 i3Neurons compared with Control i3Neurons (14 DIV; mean ± SEM from three independent experiments; >40 neurons per genotype; ns, not significant). (E) Images of DQ-Red BSA and A488 BSA fluorescence in Control and AP-4 G4i i3Neurons (14 DIV). Bar, 10 μm. (F) Quantification of lysosomal degradation (mean normalized DQ-Red BSA/BSA-488 ratio) in AP-4 i3Neurons compared with Control i3Neurons (14 DIV; mean ± SEM from three independent experiments; >40 neurons per genotype; **P < 0.01).
We next examined the distribution of different lysosomal proteins in these i3Neurons at 14 d differentiation. Staining with LAMP1, a marker of late endosomes as well as degradative lysosomes (Cheng et al., 2018; Yap et al., 2018; Gowrishankar et al., 2021; Lie et al., 2021), revealed that there was not a significant difference in the number or distribution of LAMP1-positive vesicles in the cell bodies of AP-4 i3Neurons (Figure 1, C and D; Supplemental Figure S1, A and B). AP-4 i3Neurons exhibited strong accumulation of ATG9A in the TGN (Supplemental Figure S2, A, B, and D) when compared with Control i3Neurons, recapitulating the phenotype observed in other systems upon loss of AP-4 function (De Pace et al., 2018; Davies et al., 2018). In addition to this strong TGN-retention phenotype in the neuronal soma, ATG9A vesicles accumulated in Tau-positive swellings in AP-4 i3Neurons (Supplemental Figure S2C), while relatively few ATG9A puncta are observed in neurites of Control i3Neurons. This is consistent with the previous observation of ATG9 vesicle buildup in axonal swellings in AP-4 ε KO mice (De Pace et al., 2018; Edmison et al., 2021).
Since LAMP1 labels heterogenous organelles that include nondegradative vesicles as well as protease-rich degradative lysosomes (Gowrishankar et al., 2015; Cheng et al., 2018; Yap et al., 2018), we next examined lysosome function more directly in these i3Neurons using the DQ-Red bovine serum albumin (BSA) trafficking assay (Marwaha and Sharma, 2017) with some modifications for use in i3Neurons. This assay employs BSA that is heavily labeled with a BODIPY TR-X dye (DQ-Red BSA), that is self-quenched until the BSA is degraded in lysosomes to smaller protein fragments resulting is dequenching of the isolated dye molecules and increased fluorescence, pulsed along with A488 BSA whose fluorescence serves as a control for endocytic uptake. Lysosomal degradative efficiency, as read out by fluorescence of DQ-Red BSA normalized to fluorescence of A488 BSA for each cell was found to decrease by 25–30% in AP-4 i3Neurons compared with the Control i3Neurons (Figure 1, E and F; Supplemental Figure S1, C and D).
In view of the reduced efficiency of lysosome proteolytic function in AP-4 i3Neurons, we next examined the maturation and distribution of lysosomal proteases in these cells. We found that the maturation of the cysteine protease, Cathepsin L, was severely affected in AP-4 i3Neurons, with the amount of mature processed Cathepsin L in AP-4 i3Neurons about 50% of that in Control i3Neurons (Figure 2, A and B). We also found that the number of Cathepsin L-positive organelles in the soma of AP-4 i3Neurons was about 50% of that in Control i3Neurons (Figure 2, C and D; Supplemental Figure S3, A and B). Likewise, there was a strong reduction in the number of PPT-1 enzyme-containing lysosomes (Figure 2, E and F; Supplemental Figure S3, C and D), as well as some reduction in total cellular PPT-1 enzyme levels (Supplemental Figure S3, E and F). While the number of PPT-1 and Cathepsin L-positive vesicles are reduced, they each colocalize extensively with LAMP1 indicating they are lysosomal (Supplemental Figures S4 and S5). Given AP-4′s previously established role in regulating cargo sorting and export out the Golgi, we hypothesized that the reduction in levels of proteases at lysosomes likely resulted from perturbed Golgi to endo-lysosome transport of these cargo. While Cathepsin L and PPT-1 were affected, another protease, Cathepsin B, was not affected (Supplemental Figure S6). AP-4 i3Neurons have a similar number of Cathepsin B-positive lysosomes when compared with Control i3Neurons (Supplemental Figure S6, A–C), suggesting that AP-4 specifically controls trafficking of a subset of lysosomal proteases.
FIGURE 2:
Loss of AP-4 affects neuronal lysosomal composition. (A and B) Immunoblotting reveals decreased levels of Cleaved/Pro-Cathepsin L protein in AP-4 i3Neurons compared with Control i3Neurons (21 DIV; α-Tubulin used as loading Control; mean ± SEM from three independent experiments; *P < 0.05; **P < 0.01). (C) Control and AP-4 G4i i3Neurons (14 DIV) stained for Cathepsin L (green) and neurofilament (blue) showing a reduced number of Cathepsin L-enriched lysosomes in cell bodies of AP-4 i3Neurons. Bar, 10 μm. (D) Quantification of Cathepsin L-positive vesicles in AP-4 i3Neurons compared with Control i3Neurons (14 DIV; mean ± SEM from three independent experiments; 25–30 neurons per genotype; *P < 0.05). (E) Control and AP-4 G4i i3Neurons (14 DIV) stained for PPT-1 (green) and Tau (red). Bar, 10 μm. (F) Quantification of PPT-1 positive vesicles in AP-4 i3Neurons compared with Control i3Neurons (14 DIV; mean ± SEM from three independent experiments; >30 neurons per genotype; **P < 0.01).
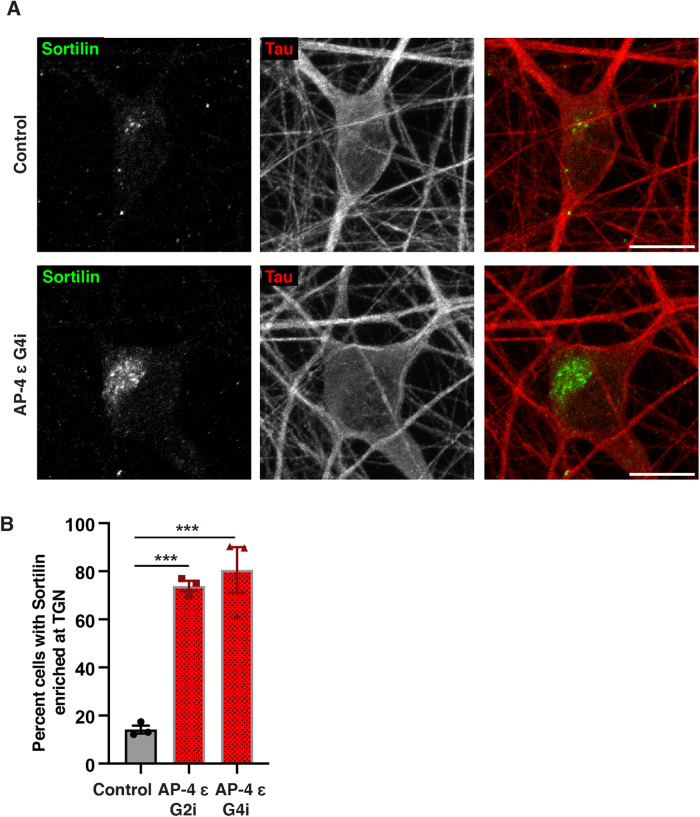
The trafficking of lysosomal proteases from the Golgi to endo-lysosomes is dependent on their interaction with receptors such as Mannose-6-phosphate receptor (M6PR) and Sortilin (Koster and Yoshii, 2019; Qian et al., 2008; Braulke and Bonifacino, 2009). Examination of Sortilin revealed a dramatic change in its intracellular distribution. While there was a strong buildup of Sortilin at the TGN area in AP-4 i3Neurons (Figure 3, A and B; Supplemental Figures S7, A and B, and S8), Sortilin signal in Control i3Neurons appears much dimmer with relatively few discrete and bright puncta visible (Figure 3, A and B; Supplemental Figure S7A). We confirmed that the Sortilin accumulation in the soma of AP-4 i3Neurons is in the TGN as it colocalized with TGN46 (Supplemental Figure S8). This retention of Sortilin at TGN in AP-4 i3Neurons suggests that Sortilin export from TGN is AP-4 dependent. Reduction in PPT-1 and Cathepsin L- containing lysosomes would also suggest that the delivery of these lysosomal enzymes may be Sortilin dependent in these neurons.
FIGURE 3:
Sortilin distribution is dramatically altered in AP-4 i3Neurons. (A) Representative images of Control and AP-4 G4i i3Neurons stained for Sortilin (green) and Tau (red), showing Sortilin is strongly accumulated in the TGN area in soma of AP-4 i3Neurons. Bar, 10 μm. (B) Quantification of the percentage of AP-4 i3Neurons with Sortilin accumulation in the TGN area compared with Control i3Neurons (42 DIV; mean ± SEM from three independent experiments; >100 neurons per genotype; ***P < 0.001).
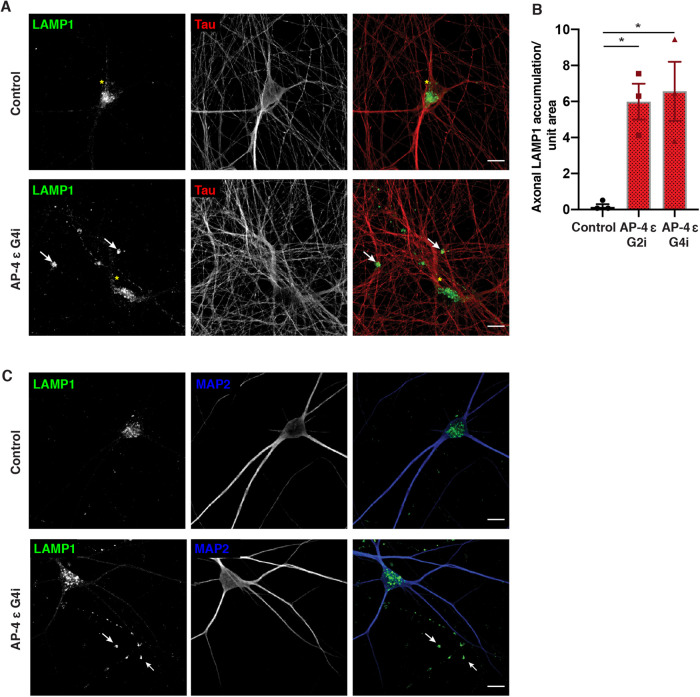
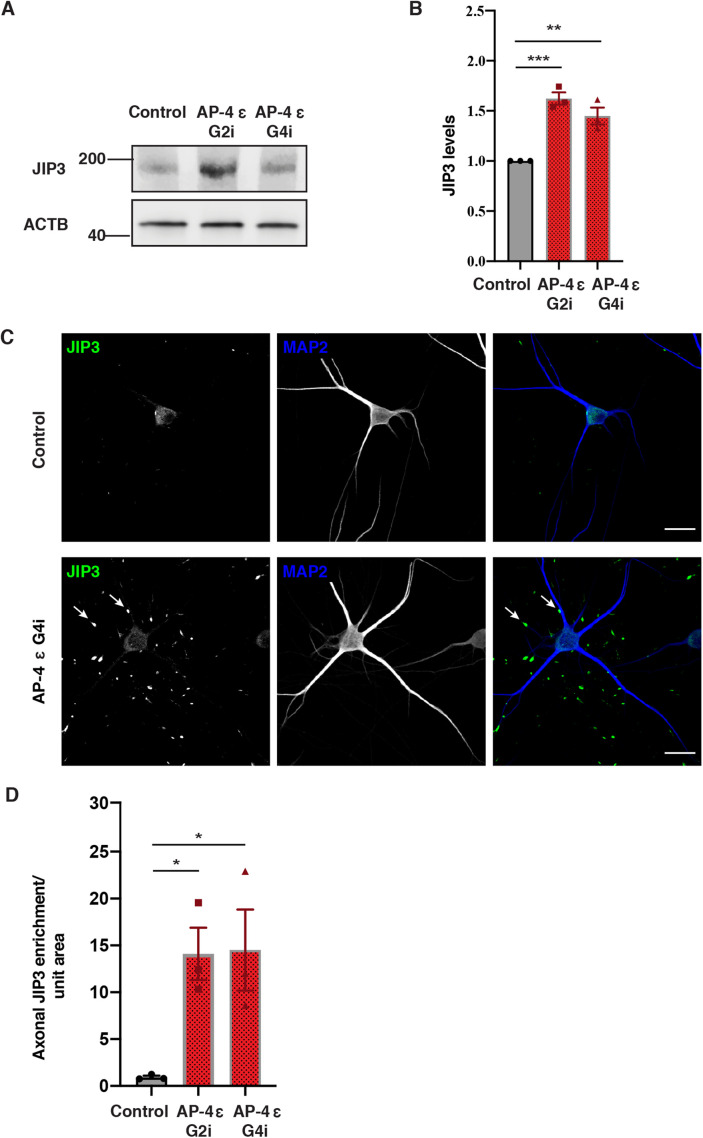
Having observed changes in lysosome function and composition in the soma, we next focused on LAMP1 distribution in axons. In Control i3Neurons, consistent with previous reports (Gowrishankar et al., 2021), most LAMP1 vesicles are largely concentrated in the soma with relatively fewer vesicles observed in neurites (Figure 4, A and B). In contrast, we found that AP-4 i3Neurons robustly developed Tau-positive axonal dystrophies with age (at DIV 42) and these axonal swellings were filled with LAMP1-positive vesicles (Figure 4, A and B; Supplemental Figure S9, A and D, white arrows). We confirmed that these LAMP1 accumulations were axonal as they were negative for the dendritic marker, MAP2B (Figure 4C; Supplemental Figure S9B). Airyscan imaging of such axonal swellings revealed the presence of numerous ringlike and punctate LAMP1-positive vesicles in them (Supplemental Video S1). Since axonal LAMP1-vesicles can be heterogenous, composed of both biosynthetic LAMP1- carriers as well as maturing endo-lysosomes (Gowrishankar et al., 2021; Lie et al., 2021), we used LysoTracker staining to determine if the vesicles that built up were acidic. Indeed, large LysoTracker-positive (acidic) vesicles moved retrogradely toward or were stalled in axonal swellings in AP-4 i3Neurons (Supplemental Video S2; Supplemental Figure S9C). Since LAMP1-positive acidic vesicles exhibit a predominantly retrograde movement in axons (Gowrishankar et al., 2021; Lie et al., 2021), we examined the distribution and levels of JIP3/MAPK8IP3, a neuronally enriched scaffolding protein that potentially links axonal endo-lysosomes and motors and regulates their retrograde axonal transport (Gowrishankar et al., 2021; Rafiq et al., 2020). Interestingly, JIP3 protein level is significantly increased in AP-4 i3Neurons compared with Control i3Neurons (Figure 5, A and B). When we examined distribution of endogenous JIP3 in the i3Neurons, we found that the AP-4 i3Neurons exhibited multiple JIP3-positive axonal swellings, while there was almost no JIP3 buildup in Control i3Neurons (Figure 5, C and D; Supplemental Figure S10, A and B). Thus loss of AP-4 in neurons causes LAMP1-positive vesicles as well JIP3/MAPK8IP3 to build up in axonal dystrophies.
FIGURE 4:
Loss of AP-4 in i3Neurons results in axonal lysosome buildup. Control and AP-4 G4i i3Neurons (42 DIV) stained for LAMP1 (green) and Tau (red). White arrows highlight LAMP1 accumulation in the neurites of AP-4 i3Neurons. Yellow asterisks indicate the LAMP1-positive vesicles in the cell body. Bar, 20 µm. (B) Quantification of axonal LAMP-positive swellings per unit area in AP-4 i3Neurons and Control i3Neurons (42 DIV; mean ± SEM from three independent experiments; >20 areas imaged at random per genotype; *P < 0.05). (C) Representative images of Control and AP-4 G4i i3Neurons (42 DIV) stained for LAMP1 (green) and MAP2 (blue). Arrowheads highlight LAMP1 accumulation in the MAP2-negative neurites (axons) of AP-4 i3Neurons. Bar, 20 µm.
FIGURE 5:
Loss of AP-4 in i3Neurons results in accumulation of JIP3 in axonal swellings. (A and B) Immunoblotting reveals increased levels of JIP3 in AP-4 i3Neurons compared with Control i3Neurons (21 DIV; ACTB used as loading Control; mean ± SEM from three independent experiments; **P < 0.01; ***P < 0.001). (C) Representative images of Control and AP-4 G4i i3Neurons (28 DIV) stained for JIP3 (green) and MAP2 (blue). Arrowheads highlight JIP3 accumulation in the axons of AP-4 G4i i3Neurons. Bar, 20 µm. (D) Quantification of axonal JIP3-positive swellings per unit area in AP-4 i3Neurons compared with Control i3Neurons (28 DIV; mean ± SEM from three independent experiments; > 20 areas chosen at random per genotype; *P < 0.05).
Movie S1.
3D reconstruction of Airyscan image of an axonal swelling filled with LAMP1-positive vesicles in AP-4 i3Neurons.
Movie S2.
Large number of LysoTracker-positive vesicles (acidic) in both the swelling and rest of the axon in an AP-4 i3Neuron. Images were acquired at 1 frame/s, playback rate 7 frames/s. Scale bar, 5 μm.
DISCUSSION
Loss of AP-4 complex function leads to “AP-4 deficiency syndrome” which is also a form of complex HSP with intellectual disability. While AP-4 is ubiquitously expressed and its loss affects different systems, the nervous system is particularly adversely impacted, consistent with the idea that neurons exhibit greater vulnerability to impaired protein trafficking and lysosome dysfunction. A key aspect to understanding disease pathology arising from loss of function of this coat protein complex is to identify the different cargoes whose transport it regulates in neurons. While studies have demonstrated that AP-4 regulates traffic of proteins linked to the autophagy pathway, relatively little is known about how it may affect the closely related lysosomal pathway, a central player in maintaining protein and organelle homeostasis in neurons. In this study, we present evidence that neuronal lysosome composition and function as well as axonal movement and distribution of these organelles are affected by loss of AP-4.
AP-4 CRISPRi i3Neurons as a model for examining pathology
The AP-4-depleted i3Neurons based on CRISPRi technology (Tian et al., 2019) are a good model system for studying alterations in cargo traffic in human neurons upon AP-4 loss. Apart from a robust depletion of the AP-4 ε subunit, we demonstrate that these i3Neurons phenocopy the strong TGN retention of ATG9A as well as an increased abundance of ATG9A (Supplemental Figure S2) observed in other neuronal and non-neuronal model systems (Davies et al., 2018; De Pace et al., 2018). We have also identified changes to neuronal lysosome composition, function, as well as changes in the neuron-enriched protein JIP3/MAPK8IP3, alterations to which have been linked to a neurodevelopmental disorder (Platzer et al., 2019; Iwasawa et al., 2019). This reiterates the importance of exploring cellular changes in response to loss of AP-4 function in a neuronal model.
AP-4 and neuronal lysosome composition and function
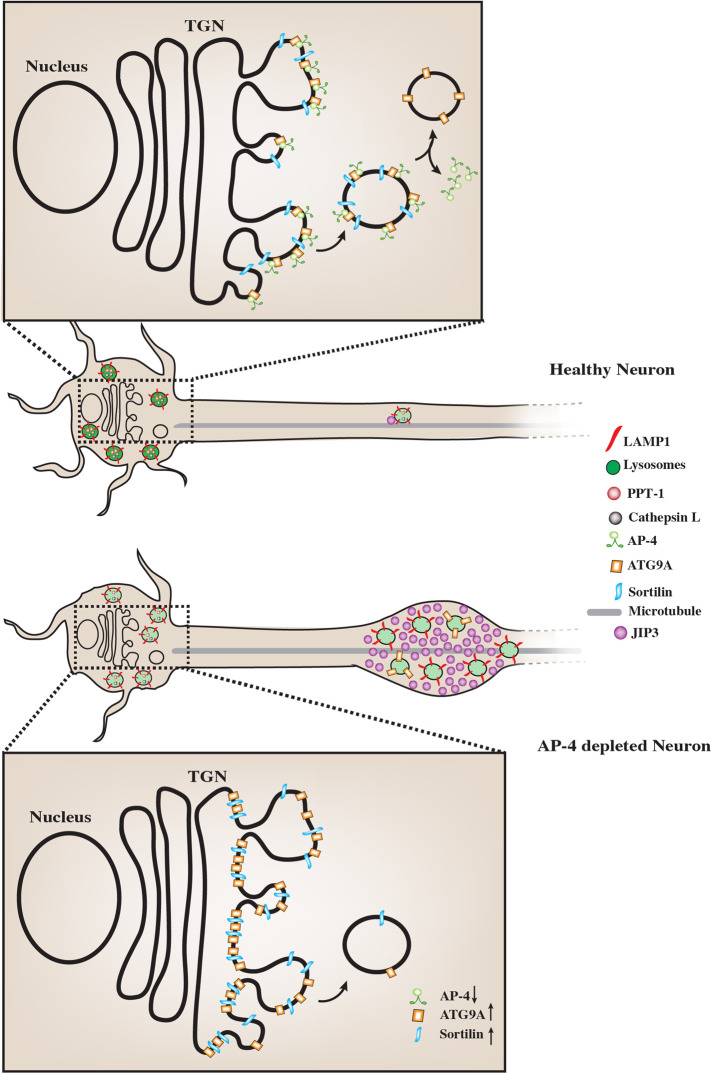
Studies in non-neuronal cells indicate that AP-4 can interact with sorting motifs in tails of receptor proteins such as M6PR, Furin, and LDL receptor (Jacobsen et al., 2001; Nielsen et al., 2001; Dennes et al., 2002; Nilsson et al., 2008; Braulke and Bonifacino, 2009). Here we have demonstrated that loss of AP-4 causes reduced abundance of certain lysosomal enzymes in LAMP1-positive organelles. Considering the role of AP-4 in sorting and export of other cargo from the TGN, this probably results from reduced biosynthetic delivery of these enzymes from the TGN to endo-lysosomes. Since many lysosomal enzymes are dependent on M6PR and/or Sortilin to get sorted at the TGN and trafficked to endo-lysosomes (Braulke and Bonifacino, 2009; Saftig and Klumperman, 2009), we examined the distribution of these receptors in AP-4 i3Neurons. We found that Sortilin is strongly retained in the TGN in AP-4 i3Neurons (Figure 6, magnified portion of schematic of soma in AP-4 i3Neurons), in contrast to the more vesicular distribution in Control i3Neurons. Interestingly, we found that while loss of AP-4 resulted in reduced Cathepsin L delivery to neuronal lysosomes, Cathepsin B delivery to lysosomes was not severely perturbed. Our finding that AP-4 loss specifically affects some lysosomal enzymes while others may be unaffected is consistent with several previous reports that these different lysosomal enzymes can exhibit differential dependence on enzyme receptors such as M6PR or Sortilin, often in a cell-type-dependent manner (van Meel et al., 2011; Coutinho et al., 2015; Boonen et al., 2016; Markmann et al., 2017). Indeed, there is growing evidence that Sortilin can aid in the sorting transport of different cathepsins that exhibit varying degrees of M6PR independence (Canuel et al., 2009). For instance, sorting and traffic of Cathepsin D and H in COS-7 cells has been shown to depend mainly on Sortilin (Canuel et al., 2008). Sortilin is also implicated in the traffic of a luminal lysosomal protein progranulin (PGRN) which is linked to frontotemporal dementia (Hu et al., 2010) and interestingly, Cathepsin L has been linked to the intracellular cleavage of PGRN (Lee et al., 2017). Studies on distribution and levels of PGRN in AP-4 i3Neurons will thus be interesting and may shed more light on neuronal traffic of this lysosome-localized protein. Sortilin has also been implicated in the sorting and traffic of other enzymes including prosaposin and acid sphingomyelinase to phagosomes in macrophages (Wähe et al., 2010). Further dissection of sorting and localization of these as well as other lysosomal enzymes such as legumain in the AP-4 i3Neurons will shed light on how these different enzymes are trafficked in human neurons and their preference/dependence on the enzyme receptors in this cell type.
FIGURE 6:
AP-4 regulates neuronal lysosome composition, function, and transport. Schematic representation of the effect of AP-4 loss on neuronal lysosome trafficking and function. In AP-4 i3Neurons there are lysosomes in the soma that contain reduced levels of certain lysosomal enzymes, such as PPT-1 and Cathepsin L. Magnified insets of the TGN area in the soma of the neurons show molecular detail of cargo potentially regulated by AP-4 action. Upon loss of AP-4 complex function, concentration and transport of Sortilin out of the TGN is affected when compared with healthy Control i3Neurons. Thus strong Sortilin accumulation in the TGN area is observed in the AP-4 i3Neurons. ATG9A, previously well-established cargo whose sorting out of the TGN is dependent on AP-4 complex function, shows a similar strong accumulation in TGN area, validating these CRISPRi-based AP-4 i3Neurons as a model for studying neuronal membrane trafficking events regulated by AP-4 complex. Also, unlike healthy Control i3Neurons, where few LAMP1-positive vesicles are observed in axons, AP-4 loss causes axonal lysosomes (LAMP1-positive vesicles) to build up in dystrophies/swellings. Additionally, there is increased accumulation of JIP3/MAPK8IP3, a regulator of retrograde axonal lysosome transport, in these swellings. We propose that the accumulation of cytosolic JIP3 in the axonal swellings in AP-4 i3Neurons could also be due to absence/reduced levels of these interacting partners on lysosomes, whose sorting from TGN to the organelle is AP-4 dependent, as with ATG9A and potentially Sortilin.
AP-4 function and links to APP processing and Alzheimer’s disease pathology
The TGN-retention of Sortilin is likely to have deleterious effects on many other cargoes in addition to the lysosomal enzymes we have examined here. Sortilin is a proneurotrophin receptor with strong links to Alzheimer’s disease (AD) (Carlo et al., 2013), having been demonstrated to bind APP (Gustafsen et al., 2013; Yang et al., 2013), BACE1 (Finan et al., 2011), as well as APOE and is also implicated in the clearance of APOE/Aβ complex in neurons (Carlo, 2013). Interestingly, Sortilin is particularly enriched in neurites and thought to promote non-amyloidogenic cleavage of APP in neurites (Gustafsen et al., 2013; Yang et al., 2013). Thus reduced traffic of Sortilin to neurites could promote amyloidogenic APP processing in AP-4 i3Neurons. In addition to the possible links to AD through Sortilin, previous studies in non-neuronal cells suggest a robust and specific interaction between AP-4 and APP (Burgos et al., 2010). Depletion of AP-4 from cultured cell lines or disrupting its interaction with exogenously expressed APP has been demonstrated to increase amyloidogenic processing of APP. While this suggests a neuroprotective role for AP-4 in AD, the physiological relevance of AP-4-APP interaction in neurons has not been established. This CRISPRi-based i3Neuron culture system could serve as an excellent model system in the future, for exploring these interactions and determining the contribution of AP-4-dependent trafficking on neuroprotection from amyloidogenic APP processing.
AP-4 function and axonal endo-lysosome transport
In addition to changes in the composition and function of lysosomes in the soma, LAMP1-positive organelles build up in axonal swellings in AP-4 i3Neurons with age. Since alterations to retrograde axonal transport of LAMP1-positive organelles (a mixture of late endosomes and amphisomes arising from the fusion of endo-lysosomes with autophagosomes) have been found to cause such focal lysosomal accumulation in axonal swellings (Gowrishankar et al., 2017, 2021), we examined distribution of JIP3/MAPK8IP3, a potential adaptor that links axonal lysosomes to motors (Gowrishankar et al., 2017). We found that there were large accumulations of JIP3 in axonal swellings. While the buildup of axonal LAMP1-positive vesicles could trigger a compensatory increase in JIP3 levels, the accumulation of JIP3 we observed in these swellings could also result from a failure of recruitment of cytosolic JIP3 to axonal lysosomes in AP-4 i3Neurons (Figure 6). De novo variants in JIP3/MAPK8IP3 have been causally implicated in a neurodevelopmental disease characterized by intellectual disability, spasticity, cerebral atrophy, and thin corpus callosum (Iwasawa et al., 2019; Platzer et al., 2019). Notably, these features overlap with the AP-4 deficiency syndrome, suggesting that the pathogenesis of these conditions may be linked, and the proteins may function in the same pathway.
Thus upon loss of AP-4, we observe changes in the composition of the lysosomes in the soma of neurons as well as altered distribution of endo-lysosomes in the axon. While the alteration in traffic of Sortilin, a known lysosomal enzyme receptor, likely contributes to the altered lysosomal enzyme composition, its role in regulating axonal endo-lysosomal distribution is unclear. It remains to be determined if Sortilin or one of its potential cargo is necessary for normal retrograde transport of axonal endo-lysosomes.
In addition to the strong TGN retention of ATG9A, we observed some accumulation of ATG9A-positive vesicles in axonal swellings of older AP-4 i3Neurons, as has been previously observed in AP-4-deficient mice (Edmison et al., 2021). We speculate that this arises from gradual accumulation of immature axonal autophagosomes which depend on the fusion with and retrograde transport of axonal LAMP1-positive organelles for their clearance (Maday et al., 2012; Cheng et al., 2015).
AP-4 complex has been linked to retrograde trafficking machinery recently. The AP-4 complex has recently been demonstrated to interact with Hook proteins (Mattera et al., 2020) which act as dynein-dynactin activators and thus aid in the retrograde transport of organelles. Indeed, through interaction with FTS (fused toes homologue) Hook and FHIP (FTS and Hook-interacting protein) proteins (FHF complex), AP-4 complex has been suggested to effect perinuclear clustering of ATG9A containing vesicles (Mattera et al., 2020). Hook proteins have been implicated in retrograde transport of BDNF-signaling endosomes in neurons (Olenick et al., 2019). The accumulation of cytosolic JIP3 in the axonal swellings in AP-4 i3Neurons could also be due to absence/reduced levels of interacting partners on lysosomes, whose sorting from TGN to the organelle is AP-4 dependent, as with ATG9A and potentially Sortilin. Indeed, a recent study has highlighted how TGN-derived transport carriers deliver lysosomal components to the maturing axonal organelles (Lie et al., 2021). Thus in the absence of optimal AP-4 complex function, sorting of this putative JIP3-binding lysosomal protein (which may or may not depend on Sortilin) into TGN-derived transport carriers would be compromised, resulting in stalled LAMP1-positive organelles in axons. Identifying AP-4 interactors in this i3Neuron system will shed more light on the how AP-4 regulates neuronal lysosome composition, function, and transport and the mechanism underlying pathology of AP-4 deficiency syndrome.
MATERIAL AND METHODS
Generation of AP-4 CRISPRi iPSC lines
Constructs: Two different sgRNA sequences targeting the transcription start site of AP4E1 (G2i and G4i) were selected from the Weissman CRISPRi v2 library (Horlbeck et al., 2016). Sense and antisense sgRNA oligonucleotides were designed with 5′CACC and 3′CAAA overhangs, respectively, and cloned into pKLV-U6gRNA-EF(BbsI)-PGKpuro2ABFP for lentivirus production. Sequences of sgRNA used are detailed in Supplemental Table S2. pKLV-U6gRNA-EF(BbsI)-PGKpuro2ABFP was a gift from Kosuke Yusa (Addgene plasmid # 50946; RRID: Addgene_50946).
Stable cell lines: The sgRNA lentivectors described earlier were packaged into lentivirus for transduction of iPSCs as previously described (Rodger et al., 2020). In brief, HEK293T cells were cotransfected with a lentiviral sgRNA expression plasmid and the packaging vectors pCMVΔ8.91 and pMD VSV-G (1:0.7:0.3 mix) using TransIT-293 (Mirus Bio). HEK293T media were collected 48 h post-transfection, filtered using a 0.45-μm filter, and applied to target iPSCs in the presence of 10 μg/ml polybrene (Sigma-Aldrich). Cells were transduced for 16 h and selected using 1 μg/ml puromycin 24 h later.
iPSC culture and neuronal differentiation
iPSCs were cultured and i3Neurons were differentiated from them as previously described (Fernandopulle et al., 2018; Gowrishankar et al., 2021), with slight modifications. Briefly, on day 0, iPSCs were dissociated into single cells using Accutase (Thermo Fisher Scientific) and seeded at a density of 100,000 cells/well on a Matrigel-coated 6-well plate in Induction Medium composed of KO DMEM, 1× N-2 Supplement, 1× MEM nonessential amino acids solution, 1× GlutaMAX Supplement (Thermo Fisher Scientific), 10 μM Y-27632 (Tocris Biosciences), and 2 μg/ml doxycycline hydrochloride (Sigma-Aldrich). Predifferentiated cells were maintained in imaging media (IM) for 3 d with daily changes of media. After the 3-day differentiation period, cells were dissociated with Accutase and seeded at 30,000 cells per 35-mm glass bottom dish (MatTek Life Sciences) or 12-mm glass coverslip (Deckglaser) coated with 0.1 mg/ml poly-L-ornithine (Sigma-Aldrich) and 10 μg/ml mouse laminin (Life Technologies). Cells were maintained in Cortical Neuron Culture Medium composed of KO DMEM (Life Technologies), 1× B-27 Supplement (Thermo Fisher Scientific), 10 ng/ml BDNF (PeproTech), 10 ng/ml NT-3 (PeproTech), and 1 μg/ml mouse laminin (Thermo Fisher Scientific) with half media changes carried out every 3–4 d.
When required, as an alternative to Matrigel, vitronectin (VTN-N; Life Technologies) was used to coat the surface prior to iPSC culture as described previously (Chen et al., 2011). Briefly, VTN-N aliquots from –80°C were thawed and resuspended in calcium- and magnesium-free phosphate-buffered saline (PBS) to a final working concentration of 5 μg/ml and used to coat the surface of 6-well plates (1 ml/well). The plates were incubated at room temperature for 1 h and then aspirated prior to passaging the cells onto VTN-N-coated plates. i3Neurons were plated at 35% higher density for experiments when the induction and passaging of iPSCs were on VTN-N-coated plates.
Immunofluorescence analysis of i3Neurons
i3Neurons were differentiated for 1–6 wk on 35-mm glass bottom dishes (MatTek Life Sciences) and processed for immunostaining as described previously (Gowrishankar et al., 2021). See Supplemental Table S1 for antibody information.
Immunoblotting experiments
i3Neurons were grown on PLO/laminin-coated 6-well plates (500,000 cells/well). After 21 d of differentiation, i3Neurons were washed with ice-cold PBS and lysed in lysis buffer (Tris-buffered saline with 1% Triton, protease inhibitor cocktail and phosphatase inhibitor) and then spun at 13,000 × g for 5 min. The supernatant was collected and incubated at 95°C for 5 min in SDS sample buffer before SDS–PAGE, transfer to nitrocellulose membranes, and immunoblotting. See Supplemental Table S1 for antibody information.
Microscopy
Standard confocal images were acquired using a Zeiss 880 Airyscan confocal microscope via a 100× plan-apochromatic objective (1.46 NA) with 2× optical zoom. Live imaging of lysosome dynamics in i3Neurons was carried out using Airyscan imaging mode using 100× objective with 1.5× or 2× optical zoom and scan speeds of 1–2 frames per second. Zeiss Zen software was used for processing of the Airyscan images. Further image analysis was performed using FIJI/ImageJ software (Schindelin et al., 2012).
DQ-BSA assay
DQ-BSA assay was carried out to examine degradative/functional lysosomes in DIV 14 i3Neurons grown on 35-mm glass bottom dishes (MatTek) as previously described (Marwaha and Sharma, 2017; Kulkarni et al., 2021), with minor modifications. DIV 14 i3Neurons were pulsed with equal concentrations of the DQ-Red BSA and Alexa 488-BSA (BSA-488) probes# (25 µg/ml, Thermo Fisher) for a period of 5 h, followed by gentle washes (two times with warm IM*). They were then imaged live in IM at 37°C using a Zeiss 880 confocal microscope in Airyscan mode with a 60× objective (NA 1.4). Healthy i3Neurons were identified using brightfield mode and imaged using the 561 and 488 lasers to capture fluorescence of DQ-Red BSA and BSA-488. Fluorescence intensity for each of the two channels was measured following outlining of individual cells using Image J software and the normalized ratio of DQ-Red BSA to BSA-488 intensity was computed for each cell. The mean of these per cell ratios was then computed and normalized to the value of Control i3Neurons to compare across multiple experiments.
#DQ-Red BSA and BSA-488 probe mixture: 10% Cortical Neuron Culture Medium (Gowrishankar et al., 2021) in KO DMEM F12 was equilibrated at 37°C and 5% CO2. Probes were added to the 10% Cortical Neuronal Culture Medium and incubated at 37°C and 5% CO2 for 5 min for equilibration.
*Composition of IM: 20 mM HEPES, 5 mM KCL, 1 mM CaCl2, 150 mM NaCl, 1 mM MgCl2, 1.9 mg/ml glucose and BSA, pH 7.4. The probe mixture pulses were staggered by 45 min between dishes to enable live imaging.
Analysis of vesicle count in i3Neurons
Confocal (LAMP1, Cathepsin B, and L, PPT-1) and Airy scan images (DQ-BSA) were used for analysis of vesicle size and number in i3Neurons. Vesicles were identified using “threshold” function and then their number and size determined using “Analyze particles” function in FIJI/ImageJ software. Detailed statistics are included in the figure legend (Figures 1 and 2; Supplemental Figure S1, S3 and S6).
Analysis of axonal accumulation of LAMP1 and JIP3
Maximum intensity projections of confocal images obtained of LAMP1 or JIP3 staining in i3Neurons were used for analyzing the extent of axonal accumulation of these proteins. Images from Control and AP-4 i3Neurons were thresholded to the same intensity values and accumulations in the axons identified as those larger than 2 μm and above the threshold. All such “accumulations” were counted irrespective of relative size and intensity. The axonal accumulations were reported as the number per 18,211 μm2 (area of each confocal image).
Colocalization analysis of lysosomal enzymes with LAMP1
Maximum intensity projections of confocal images obtained of LAMP1 costained with Cathepsin L or PPT-1 in i3Neurons were used for analyzing the fraction of each of those enzymes that overlaps with LAMP1. The images are thresholded and the extent/fraction of PPT-1/Cathepsin L that overlapped with LAMP1 (i.e., overlap of PPT-1 or Cathepsin L channel with the LAMP1 channel) was computed for each soma using JACOP plugin of Image J.
Statistical analysis
Data are represented as mean ± SEM unless otherwise specified. Statistical analysis was performed using Prism 8 software. Groups were compared using one-way ANOVA with Dunnett’s multiple comparison test. Detailed statistical information (number of independent experiments, and p values) is described in the respective figure legends.
Supplementary Material
Acknowledgments
We thank Daniel McBride and Eduardo Pallares for technical assistance. We thank Dr. Hofmann (UT Southwestern) for generous gift of the PPT-1 antibody. We thank Michael Ward (NIH) for the generous gift of CRISPRi – i3Neurons. S.G. is funded by the Wolverine foundation, the Dr. Ralph and Marian Falk Medical Research Trust, and the NIH (1 RF1 AG076653-01). E.R. was funded by the NIHR Cambridge Biomedical Research Centre (BRC-1215-20014), the UK Medical Research Council (Project Grant MR/R026440/1), and a generous donation from Hazel and Keith Satchell. The views expressed are those of the authors and not necessarily those of the NIHR or the UK Department of Health and Social Care. The authors declare no competing financial interests.
Abbreviations used:
- AD
Alzheimer’s disease
- AP-4
adaptor protein complex-4
- APOE
apolipoprotein E
- APP
amyloid precursor protein
- ATG9A
autophagy-related protein 9A
- BACE1
beta-site APP cleaving enzyme 1
- BDNF
brain-derived neurotrophic factor
- BSA
bovine serum albumin
- CD63
cluster of differentiation 63 (Tetraspanin/LAMP3)
- CRISPRi
CRISPR-inhibition
- DIV
days in vitro
- FHIP Protein
FTS and hook-interacting protein
- FTS Hook
fused toes homolog
- HSP
hereditary spastic paraplegia
- i3Neurons
iPSC-derived glutamatergic cortical neurons
- iPSC
induced pluripotent stem cells
- KRAB
krüppel-associated box
- LAMP1
lysosome associated membrane protein1
- LAMP2
lysosome associated membrane protein2
- M6PR
mannose-6-phosphate receptor
- MAP2B
microtubule-associated protein 2B
- MAPK8IP3/JIP3
mitogen-activated protein kinase 8 interacting protein 3/JNK-interacting protein 3
- MEF
mouse embryonic fibroblast
- NGN2
neurogenin 2
- PGRN
progranulin
- PPT-1
palmitoyl-protein thioesterase 1
- sgRNA
single guide RNA
- TGN
trans-Golgi network.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-09-0473) on August 17, 2022.
REFERENCES
- Abdollahpour H, Alawi M, Kortüm F, Beckstette M, Seemanova E, Komárek V, Rosenberger G, Kutsche K (2015). An AP4B1 frameshift mutation in siblings with intellectual disability and spastic tetraplegia further delineates the AP-4 deficiency syndrome. Eur J Hum Genet EJHG 23, 256–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abou Jamra R, Philippe O, Raas-Rothschild A, Eck SH, Graf E, Buchert R, Borck G, Ekici A, Brockschmidt FF, Nöthen MM, et al. (2011). Adaptor protein Complex 4 deficiency causes severe autosomal-recessive intellectual disability, progressive spastic paraplegia, shy character, and short stature. Am J Hum Genet 88, 788–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Dell’Angelica EC (1999). Molecular bases for the recognition of tyrosine-based sorting signals. J Cell Biol 145, 923–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonen M, Staudt C, Gilis F, Oorschot V, Klumperman J, Jadot M (2016). Cathepsin D and its newly identified transport receptor SEZ6L2 can modulate neurite outgrowth. J Cell Sci 129, 557–568. [DOI] [PubMed] [Google Scholar]
- Braulke T, Bonifacino JS (2009). Sorting of lysosomal proteins. Biochim Biophys Acta BBA - Mol Cell Res 1793, 605–614. [DOI] [PubMed] [Google Scholar]
- Burgos PV, Mardones GA, Rojas AL, daSilva LLP, Prabhu Y, Hurley JH, Bonifacino JS (2010). Sorting of the Alzheimer’s disease amyloid precursor protein mediated by the AP-4 Complex. Dev Cell 18, 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canuel M, Korkidakis A, Konnyu K, Morales CR (2008). Sortilin mediates the lysosomal targeting of cathepsins D and H. Biochem Biophys Res Commun 373, 292–297. [DOI] [PubMed] [Google Scholar]
- Canuel M, Libin Y, Morales CR (2009). The interactomics of sortilin: an ancient lysosomal receptor evolving new functions. Histol Histopathol 24, 481–492. [DOI] [PubMed] [Google Scholar]
- Carlo A-S (2013). Sortilin, a novel APOE receptor implicated in Alzheimer disease. Prion 7, 378–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlo A-S, Gustafsen C, Mastrobuoni G, Nielsen MS, Burgert T, Hartl D, Rohe M, Nykjaer A, Herz J, Heeren J, et al. (2013). The Pro-Neurotrophin Receptor Sortilin Is a major neuronal apolipoprotein E receptor for catabolism of amyloid-β peptide in the brain. J Neurosci 33, 358–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, Propson NE, et al. (2011). Chemically defined conditions for human iPSC derivation and culture. Nat Methods 8, 424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X-T, Xie Y-X, Zhou B, Huang N, Farfel-Becker T, Sheng Z-H (2018). Characterization of LAMP1-labeled nondegradative lysosomal and endocytic compartments in neurons. J Cell Biol 217, 3127–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X-T, Zhou B, Lin M-Y, Cai Q, Sheng Z-H (2015). Axonal autophagosomes recruit dynein for retrograde transport through fusion with late endosomes. J Cell Biol 209, 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho MF, Lacerda L, Pinto E, Ribeiro H, Macedo-Ribeiro S, Castro L, Prata MJ, Alves S (2015). Molecular and computational analyses of genes involved in mannose 6-phosphate independent trafficking. Clin Genet 88, 190–194. [DOI] [PubMed] [Google Scholar]
- Davies AK, Itzhak DN, Edgar JR, Archuleta TL, Hirst J, Jackson LP, Robinson MS, Borner GHH (2018). AP-4 vesicles contribute to spatial control of autophagy via RUSC-dependent peripheral delivery of ATG9A. Nat Commun 9, 3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Angelica EC, Mullins C, Bonifacino JS (1999). AP-4, a novel protein complex related to clathrin adaptors. J Biol Chem 274, 7278–7285. [DOI] [PubMed] [Google Scholar]
- Dennes A, Madsen P, Nielsen MS, Petersen CM, Pohlmann R (2002). The yeast Vps10p cytoplasmic tail mediates lysosomal sorting in mammalian cells and interacts with human GGAs*. J Biol Chem 277, 12288–12293. [DOI] [PubMed] [Google Scholar]
- De Pace RD, Skirzewski M, Damme M, Mattera R, Mercurio J, Foster AM, Cuitino L, Jarnik M, Hoffmann V, Morris HD, et al. (2018). Altered distribution of ATG9A and accumulation of axonal aggregates in neurons from a mouse model of AP-4 deficiency syndrome. PLOS Genet 14, e1007363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi-Fakhari D, Cheng C, Dies K, Diplock A, Pier DB, Ryan CS, Lanpher BC, Hirst J, Chung WK, Sahin M, et al. CureSPG47 (2018). Clinical and genetic characterization of AP4B1-associated SPG47. Am J Med Genet A 176, 311–318. [DOI] [PubMed] [Google Scholar]
- Edmison D, Wang L, Gowrishankar S (2021). Lysosome Function and Dysfunction in Hereditary Spastic Paraplegias. Brain Sci 11, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandopulle MS, Prestil R, Grunseich C, Wang C, Gan L, Ward ME (2018). Transcription-factor mediated differentiation of human iPSCs into neurons. Curr Protoc Cell Biol 79, e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan GM, Okada H, Kim T-W (2011). BACE1 retrograde trafficking is uniquely regulated by the cytoplasmic domain of sortilin. J Biol Chem 286, 12602–12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowrishankar S, Lyons L, Rafiq NM, Roczniak-Ferguson A, De Camilli P, Ferguson SM (2021). Overlapping roles of JIP3 and JIP4 in promoting axonal transport of lysosomes in human iPSC-derived neurons. Mol Biol Cell 32, 1094–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowrishankar S, Wu Y, Ferguson SM (2017). Impaired JIP3-dependent axonal lysosome transport promotes amyloid plaque pathology. J Cell Biol 216, 3291–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowrishankar S, Yuan P, Wu Y, Schrag M, Paradise S, Grutzendler J, De Camilli P, Ferguson SM (2015). Massive accumulation of luminal protease-deficient axonal lysosomes at Alzheimer’s disease amyloid plaques. Proc Natl Acad Sci 112, E3699–E3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsen C, Glerup S, Pallesen LT, Olsen D, Andersen OM, Nykjær A, Madsen P, Petersen CM (2013). Sortilin and SorLA display distinct roles in processing and trafficking of amyloid precursor protein. J Neurosci Off J Soc Neurosci 33, 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardies K, May P, Djémié T, Tarta-Arsene O, Deconinck T, Craiu D, AR working group of the EuroEPINOMICS RES Consortium, Helbig I, Suls A, Balling R, Weckhuysen S, et al. (2015). Recessive loss-of-function mutations in AP4S1 cause mild fever-sensitive seizures, developmental delay and spastic paraplegia through loss of AP-4 complex assembly. Hum Mol Genet 24, 2218–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Bright NA, Rous B, Robinson MS (1999). Characterization of a Fourth Adaptor-related Protein Complex. Mol Biol Cell 10, 2787–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Irving C, Borner GHH (2013). Adaptor protein complexes AP-4 and AP-5: New players in endosomal trafficking and progressive spastic paraplegia. Traffic. 14, 153–164. [DOI] [PubMed] [Google Scholar]
- Horlbeck MA, Gilbert LA, Villalta JE, Adamson B, Pak RA, Chen Y, Fields AP, Park CY, Corn JE, Kampmann M, Weissman JS (2016). Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife. 5. doi:10.7554/eLife.19760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F, Padukkavidana T, Vægter CB, Brady OA, Zheng Y, Mackenzie IR, Feldman HH, Nykjaer A, Strittmatter SM (2010). Sortilin-mediated endocytosis determines levels of the frontotemporal dementia protein. Progranulin Neuron 68, 654–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K, Hao F, Fujita N, Tsuji Y, Oe Y, Araki Y, Hamasaki M, Noda T, Yoshimori T (2016). Atg9A trafficking through the recycling endosomes is required for autophagosome formation. J Cell Sci 129, 3781–3791. [DOI] [PubMed] [Google Scholar]
- Ivankovic D, Drew J, Lesept F, White IJ, Doménech GL, Tooze SA, Kittler JT (2020). Axonal autophagosome maturation defect through failure of ATG9A sorting underpins pathology in AP-4 deficiency syndrome. Autophagy 16, 391–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasawa S, Yanagi K, Kikuchi A, Kobayashi Y, Haginoya K, Matsumoto H, Kurosawa K, Ochiai M, Sakai Y, Fujita A, et al. (2019). Recurrent de novo MAPK8IP3 variants cause neurological phenotypes. Ann Neurol 85, 927–933. [DOI] [PubMed] [Google Scholar]
- Jacobsen L, Madsen P, Jacobsen C, Nielsen MS, Gliemann J, Petersen CM (2001). Activation and functional characterization of the mosaic receptor SorLA/LR11. J Biol Chem 276, 22788–22796. [DOI] [PubMed] [Google Scholar]
- Koster KP, Yoshii A (2019). Depalmitoylation by palmitoyl-protein thioesterase 1 in neuronal health and degeneration. Front Synaptic Neurosci 11. doi:10.3389/fnsyn.2019.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni VV, Anand A, Herr JB, Miranda C, Vogel MC, Maday S (2021). Synaptic activity controls autophagic vacuole motility and function in dendrites. J Cell Biol 220, e202002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CW, Stankowski JN, Chew J, Cook CN, Lam Y-W, Almeida S, Carlomagno Y, Lau K-F, Prudencio M, Gao F-B, et al. (2017). The lysosomal protein cathepsin L is a progranulin protease. Mol Neurodegener 12, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie PPY, Yang D-S, Stavrides P, Goulbourne CN, Zheng P, Mohan PS, Cataldo AM, Nixon RA (2021). Post-Golgi carriers, not lysosomes, confer lysosomal properties to predegradative organelles in normal and dystrophic axons. Cell Rep 35, 109034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S, Wallace KE, Holzbaur ELF (2012). Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol 196, 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmann S, Krambeck S, Hughes CJ, Mirzaian M, Aerts JMFG, Saftig P, Schweizer M, Vissers JPC, Braulke T, Damme M (2017). Quantitative proteome analysis of mouse liver lysosomes provides evidence for mannose 6-phosphate-independent targeting mechanisms of acid hydrolases in mucolipidosis II. Mol Cell Proteomics MCP 16, 438–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marwaha R, Sharma M (2017). DQ-Red BSA trafficking assay in cultured cells to assess cargo delivery to lysosomes. Bio-Protoc 7. doi:10.21769/BioProtoc.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattera R, Park SY, De Pace R, Guardia CM, Bonifacino JS (2017). AP-4 mediates export of ATG9A from the trans-Golgi network to promote autophagosome formation. Proc Natl Acad Sci USA 114, E10697–E10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattera R, Williamson CD, Ren X, Bonifacino JS (2020). The FTS-Hook-FHIP (FHF) complex interacts with AP-4 to mediate perinuclear distribution of AP-4 and its cargo ATG9A. Mol Biol Cell 31, 963–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meel E, Boonen M, Zhao H, Oorschot V, Ross FP, Kornfeld S, Klumperman J (2011). Disruption of the Man-6-P Targeting Pathway in Mice Impairs Osteoclast Secretory Lysosome Biogenesis. Traffic Cph Den 12, 912–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-De-Luca A, Helmers SL, Mao H, Burns TG, Melton AMA, Schmidt KR, Fernhoff PM, Ledbetter DH, Martin CL (2011). Adaptor protein complex-4 (AP-4) deficiency causes a novel autosomal recessive cerebral palsy syndrome with microcephaly and intellectual disability. J Med Genet 48, 141–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen MS, Madsen P, Christensen EI, Nykjær A, Gliemann J, Kasper D, Pohlmann R, Petersen CM (2001). The sortilin cytoplasmic tail conveys Golgi–endosome transport and binds the VHS domain of the GGA2 sorting protein. EMBO J 20, 2180–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson SK, Christensen S, Raarup MK, Ryan RO, Nielsen MS, Olivecrona G (2008). Endocytosis of Apolipoprotein A-V by members of the low density lipoprotein receptor and the Vps10p domain receptor families. J Biol Chem 283, 25920–25927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olenick MA, Dominguez R, Holzbaur ELF (2019). Dynein activator Hook1 is required for trafficking of BDNF-signaling endosomes in neurons. J Cell Biol 218, 220–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi A, Razi M, Dooley HC, Robinson D, Weston AE, Collinson LM, Tooze SA (2012). Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol Biol Cell 23, 1860–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platzer K, Sticht H, Edwards SL, Allen W, Angione KM, Bonati MT, Brasington C, Cho MT, Demmer LA, Falik-Zaccai T, et al. (2019). De novo variants in MAPK8IP3 cause intellectual disability with variable brain anomalies. Am J Hum Genet 104, 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian M, Sleat DE, Zheng H, Moore D, Lobel P (2008). Proteomics analysis of serum from mutant mice reveals lysosomal proteins selectively transported by each of the two mannose 6-phosphate receptors. Mol Cell Proteomics MCP 7, 58–70. [DOI] [PubMed] [Google Scholar]
- Rafiq NM, Lyons LL, Gowrishankar S, De Camilli PD, Ferguson SM (2020). JIP3 links lysosome transport to regulation of multiple components of the axonal cytoskeleton. Commun Biol 5, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ (2004). The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell 6, 79–90. [DOI] [PubMed] [Google Scholar]
- Rodger C, Flex E, Allison RJ, Sanchis-Juan A, Hasenahuer MA, Cecchetti S, French CE, Edgar JR, Carpentieri G, Ciolfi A, et al. (2020). De novo VPS4A mutations cause multisystem disease with abnormal neurodevelopment. Am J Hum Genet 107, 1129–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Shpilka T, Elazar Z (2012). Mechanisms of autophagosome biogenesis. Curr Biol CB 22, R29–R34. [DOI] [PubMed] [Google Scholar]
- Saftig P, Klumperman J (2009). Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol 10, 623–635. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian R, Gachechiladze MA, Ludwig CH, Laurie MT, Hong JY, Nathaniel D, Prabhu AV, Fernandopulle MS, Patel R, Abshari M, et al. (2019). CRISPR interference-based platform for multimodal genetic screens in human iPSC-derived neurons. Neuron. 104, 239–255.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tüysüz B, Bilguvar K, Koçer N, Yalçınkaya C, Çağlayan O, Gül E, Şahin S, Çomu S, Günel M (2014). Autosomal recessive spastic tetraplegia caused by AP4M1 and AP4B1 gene mutation: Expansion of the facial and neuroimaging features. Am J Med Genet A 164, 1677–1685. [DOI] [PubMed] [Google Scholar]
- Verkerk AJMH, Schot R, Dumee B, Schellekens K, Swagemakers S, Bertoli-Avella AM, Lequin MH, Dudink J, Govaert P, van Zwol AL, et al. (2009). Mutation in the AP4M1 gene provides a model for neuroaxonal injury in cerebral palsy. Am J Hum Genet 85, 40–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wähe A, Kasmapour B, Schmaderer C, Liebl D, Sandhoff K, Nykjaer A, Griffiths G, Gutierrez MG (2010). Golgi-to-phagosome transport of acid sphingomyelinase and prosaposin is mediated by sortilin. J Cell Sci 123, 2502–2511. [DOI] [PubMed] [Google Scholar]
- Wang C, Ward ME, Chen R, Liu K, Tracy TE, Chen X, Xie M, Sohn PD, Ludwig C, Meyer-Franke A, et al. (2017). Scalable production of iPSC-derived human neurons to identify tau-lowering compounds by high-content screening. Stem Cell Rep. 9, 1221–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Hill SE, Nathan WJ, Paiano J, Callen E, Wang D, Shinoda K, van Wietmarschen N, Colón-Mercado JM, Zong D, et al. (2021). Neuronal enhancers are hotspots for DNA single-strand break repair. Nature 593, 440–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Virassamy B, Vijayaraj SL, Lim Y, Saadipour K, Wang Y-J, Han Y-C, Zhong J-H, Morales CR, Zhou X-F (2013). The intracellular domain of sortilin interacts with amyloid precursor protein and regulates its lysosomal and lipid raft trafficking. PLoS One 8, e63049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap CC, Digilio L, McMahon LP, Garcia ADR, Winckler B (2018). Degradation of dendritic cargos requires Rab7-dependent transport to somatic lysosomes. J Cell Biol 217, 3141–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ARJ, Chan EYW, Hu XW, Köchl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA (2006). Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci 119, 3888–3900. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.