The quinoxaline portion of the title molecule is not quite planar and the conformation of the entire molecule is ‘U-shaped’, which is consolidated by an intramolecular antiparallel carbonyl electrostatic interaction. In the crystal, the molecules form corrugated layers through C—H⋯O and C—H⋯N hydrogen bonds and C—H⋯π(ring) and π-stacking interactions.
Keywords: crystal structure, quinoxaline, triazole, hydrogen bond, π-stacking
Abstract
The quinoxaline portion of the title molecule, C21H19N5O3, is not quite planar as indicated by a dihedral angle of 3.38 (7)° between the constituent rings. The molecule is ‘U-shaped’, which is consolidated by an intramolecular antiparallel carbonyl electrostatic interaction with C··O distances of 2.8905 (16) and 3.0221 (15) Å, in the crystal forms corrugated layers through C—H⋯O and C—H⋯N hydrogen bonds and C—H⋯π(ring) and π-stacking interactions.
Structure description
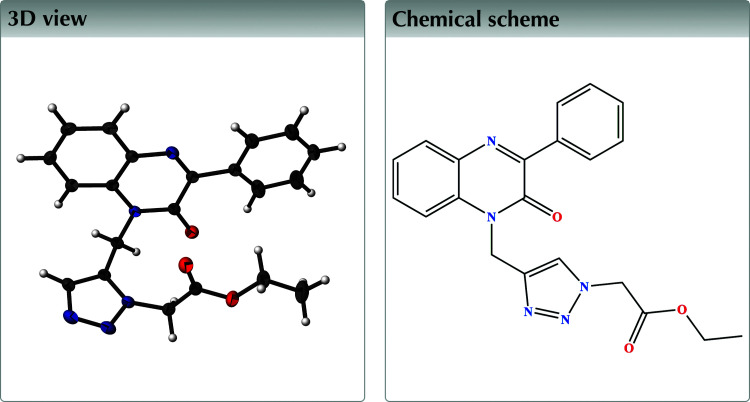
Quinoxaline derivatives exhibit a wide range of biological applications including antimicrobial (Teja et al., 2016 ▸), anti-inflammatory (Guirado et al., 2012 ▸), anticancer (Abbas et al., 2015 ▸), antidiabetic (Kulkarni et al., 2012 ▸) and antihistaminic (Sridevi et al., 2010 ▸) effects. As a continuation of our research on the synthesis and biological properties of quinoxaline derivatives (Missioui et al., 2022a ▸,b ▸,c ▸), the title compound (Fig. 1 ▸) was prepared and its crystal structure is reported here.
Figure 1.
The title molecule with the labeling scheme and 50% probability ellipsoids. The π interaction between the C19=O2 carbonyl group and the C1/C6/N1/C7/C8/N2 ring is shown by an orange dashed line.
The quinoxaline portion is not quite planar as indicated by a dihedral angle of 3.38 (7)° between the constituent rings. The dihedral angle between the C9–C14 and C1/C6/N1/C7/C8/N2 rings is 9.05 (8)° while that between the latter ring and the triazole ring is 78.47 (3)°. The molecule adopts a ‘U-shaped’ conformation, which is consolidated by an intramolecular antiparallel carbonyl electrostatic interaction (Allen et al., 1998 ▸) between the C8=O1 and C19=O2 groups with C19⋯O1 = 2.890 Å and C8⋯O2 = 3.022 Å. In the crystal, C12—H12⋯N3 hydrogen bonds (Table 1 ▸) lead to the formation of chains extending along the c-axis direction, which are linked into corrugated layers by C5—H5⋯N4 and C15—H15B⋯O2 hydrogen bonds and by C15—15A⋯Cg1 interactions (Table 1 ▸ and Fig. 2 ▸). These are accompanied by weak π-stacking interactions between C1/C6/N1/C7/C8/N2 and C1–C6 rings related by the symmetry operation x −
 , y, −z −
, y, −z −
 [centroid–centroid distance = 3.8105 (7) Å, dihedral angle = 6.13 (6)°].
[centroid–centroid distance = 3.8105 (7) Å, dihedral angle = 6.13 (6)°].
Table 1. Hydrogen-bond geometry (Å, °).
Cg1 is the centroid of the triazole ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C5—H5⋯N4i | 0.973 (16) | 2.462 (16) | 3.2183 (17) | 134.3 (12) |
| C12—H12⋯N3ii | 0.988 (19) | 2.572 (19) | 3.4094 (18) | 142.5 (15) |
| C15—H15A⋯Cg1iii | 0.997 (16) | 2.657 (15) | 3.3580 (14) | 127.5 (10) |
| C15—H15B⋯O2iv | 0.986 (15) | 2.464 (15) | 3.2459 (16) | 135.9 (11) |
Symmetry codes: (i)
 ; (ii)
; (ii)
 ; (iii)
; (iii)
 ; (iv)
; (iv)
 .
.
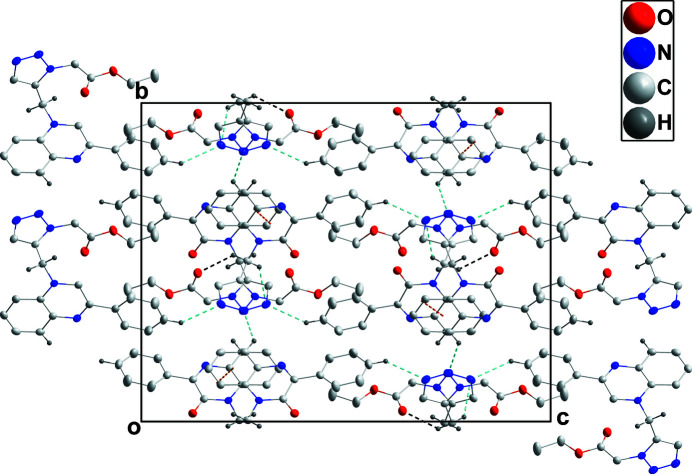
Figure 2.
Packing viewed along the a-axis direction. C—H⋯O and C—H⋯N hydrogen bonds are shown, respectively, by black and light-blue dashed lines while the π-stacking interactions are shown by orange dashed lines.
Synthesis and crystallization
To a solution of 3-phenyl-1-(prop-2-yn-1-yl)quinoxalin-2(1H)-one (0.68 mmol) in ethanol (15 ml) was added ethyl 2-azidoacetate (1.03 mmol). The reaction mixture was stirred under reflux for 72 h. After completion of the reaction (monitored by TLC), the solution was concentrated and the residue was purified by column chromatography on silica gel by using a hexane/ethyl acetate mixture (9:1) as eluent. The solid product obtained was crystallized from ethanol solution to afford colorless crystals. Yield 80%, m.p. = 408–410 K. 1H MNR (300 MHz, CDCl3) δ (p.p.m.):1.22–1.26 (t, 3H, CH3, J = 6 Hz); 4.12-4.19 (q, 2H, O—CH2, J = 6 Hz); 5.57 (s, 2H, N—CH2CO2); 5.60 (s, 2H, N—CH2); 7.72 (s, H,CHtriazole); 7.44–8.31 (m, 9Harom); 13C MNR (75 MHz,CDCl3) δ (p.p.m.):13.95 (CH3); 34.99 (O—CH2); 50.01(N—CH2C=O); 62.48 (N—CH2); 113.48, 124.61, 128.19 (traizole), 129.52, 130.70, 130.85, 131.16, 131.79, (CHarom); 132.79, 133.53, 134.34, 135.52, 153.69 (Cq); 154.32 (C=Oarom);166.80 (C=Oacetate)
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C21H19N5O3 |
| M r | 389.41 |
| Crystal system, space group | Orthorhombic, P b c a |
| Temperature (K) | 150 |
| a, b, c (Å) | 8.8585 (3), 18.0405 (5), 23.1961 (7) |
| V (Å3) | 3707.0 (2) |
| Z | 8 |
| Radiation type | Cu Kα |
| μ (mm−1) | 0.79 |
| Crystal size (mm) | 0.21 × 0.10 × 0.02 |
| Data collection | |
| Diffractometer | Bruker D8 VENTURE PHOTON 100 CMOS |
| Absorption correction | Multi-scan (SADABS; Krause et al., 2015 ▸) |
| T min, T max | 0.89, 0.98 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 26694, 3662, 3086 |
| R int | 0.047 |
| (sin θ/λ)max (Å−1) | 0.618 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.035, 0.087, 1.05 |
| No. of reflections | 3662 |
| No. of parameters | 339 |
| H-atom treatment | All H-atom parameters refined |
| Δρmax, Δρmin (e Å−3) | 0.24, −0.21 |
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2414314622006939/bx4022sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2414314622006939/bx4022Isup2.hkl
Supporting information file. DOI: 10.1107/S2414314622006939/bx4022Isup3.cml
CCDC reference: 2184531
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
Author contributions are as follows. Conceptualization, YR and NA; methodology, MM and AS; investigation, NA and MM; writing (original draft), JTM and YR; writing (review and editing of the manuscript), YR; formal analysis, AA and YR; supervision, YR and EME; crystal-structure determination and validation, JTM; synthesis, NA.
full crystallographic data
Crystal data
| C21H19N5O3 | Dx = 1.395 Mg m−3 |
| Mr = 389.41 | Cu Kα radiation, λ = 1.54178 Å |
| Orthorhombic, Pbca | Cell parameters from 9932 reflections |
| a = 8.8585 (3) Å | θ = 3.8–72.3° |
| b = 18.0405 (5) Å | µ = 0.79 mm−1 |
| c = 23.1961 (7) Å | T = 150 K |
| V = 3707.0 (2) Å3 | Plate, colourless |
| Z = 8 | 0.21 × 0.10 × 0.02 mm |
| F(000) = 1632 |
Data collection
| Bruker D8 VENTURE PHOTON 100 CMOS diffractometer | 3662 independent reflections |
| Radiation source: INCOATEC IµS micro–focus source | 3086 reflections with I > 2σ(I) |
| Mirror monochromator | Rint = 0.047 |
| Detector resolution: 10.4167 pixels mm-1 | θmax = 72.5°, θmin = 3.8° |
| ω scans | h = −10→10 |
| Absorption correction: multi-scan (SADABS; Krause et al., 2015) | k = −22→22 |
| Tmin = 0.89, Tmax = 0.98 | l = −28→27 |
| 26694 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.035 | All H-atom parameters refined |
| wR(F2) = 0.087 | w = 1/[σ2(Fo2) + (0.0384P)2 + 1.2273P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.05 | (Δ/σ)max < 0.001 |
| 3662 reflections | Δρmax = 0.24 e Å−3 |
| 339 parameters | Δρmin = −0.21 e Å−3 |
| 0 restraints | Extinction correction: SHELXL 2018/1 (Sheldrick, 2015b), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.00064 (5) |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger.All H atom positional and Uiso values were freely refined. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.43111 (11) | 0.52576 (5) | 0.37440 (4) | 0.0275 (2) | |
| O2 | 0.09359 (10) | 0.46632 (5) | 0.36206 (4) | 0.0270 (2) | |
| O3 | 0.20937 (11) | 0.40033 (5) | 0.43166 (4) | 0.0303 (2) | |
| N1 | 0.18289 (12) | 0.67590 (6) | 0.34317 (5) | 0.0226 (2) | |
| N2 | 0.34767 (11) | 0.56638 (5) | 0.28730 (4) | 0.0193 (2) | |
| N3 | 0.16535 (13) | 0.36820 (6) | 0.19501 (5) | 0.0252 (2) | |
| N4 | 0.16515 (13) | 0.34789 (6) | 0.24943 (5) | 0.0250 (2) | |
| N5 | 0.26117 (12) | 0.39291 (6) | 0.27772 (5) | 0.0208 (2) | |
| C1 | 0.26318 (14) | 0.61609 (6) | 0.25427 (5) | 0.0195 (3) | |
| C2 | 0.26183 (16) | 0.61555 (7) | 0.19390 (6) | 0.0238 (3) | |
| H2 | 0.3228 (19) | 0.5802 (9) | 0.1724 (7) | 0.034 (4)* | |
| C3 | 0.17264 (16) | 0.66602 (7) | 0.16465 (6) | 0.0264 (3) | |
| H3 | 0.1743 (18) | 0.6650 (9) | 0.1222 (7) | 0.033 (4)* | |
| C4 | 0.08259 (16) | 0.71695 (7) | 0.19408 (6) | 0.0260 (3) | |
| H4 | 0.0164 (19) | 0.7523 (9) | 0.1731 (7) | 0.034 (4)* | |
| C5 | 0.08467 (15) | 0.71842 (7) | 0.25326 (6) | 0.0236 (3) | |
| H5 | 0.0237 (18) | 0.7530 (9) | 0.2755 (7) | 0.031 (4)* | |
| C6 | 0.17717 (14) | 0.66902 (7) | 0.28405 (5) | 0.0207 (3) | |
| C7 | 0.26837 (14) | 0.63234 (7) | 0.37345 (5) | 0.0211 (3) | |
| C8 | 0.35681 (14) | 0.57104 (7) | 0.34670 (5) | 0.0208 (3) | |
| C9 | 0.27185 (16) | 0.64586 (7) | 0.43697 (6) | 0.0248 (3) | |
| C10 | 0.1715 (2) | 0.69806 (8) | 0.45951 (6) | 0.0356 (3) | |
| H10 | 0.101 (2) | 0.7214 (11) | 0.4333 (8) | 0.053 (6)* | |
| C11 | 0.1707 (2) | 0.71453 (9) | 0.51787 (7) | 0.0435 (4) | |
| H11 | 0.098 (2) | 0.7522 (12) | 0.5330 (9) | 0.062 (6)* | |
| C12 | 0.2704 (2) | 0.67988 (9) | 0.55500 (6) | 0.0408 (4) | |
| H12 | 0.268 (2) | 0.6896 (11) | 0.5969 (8) | 0.050 (5)* | |
| C13 | 0.3689 (2) | 0.62824 (10) | 0.53366 (7) | 0.0434 (4) | |
| H13 | 0.442 (2) | 0.6034 (12) | 0.5596 (9) | 0.063 (6)* | |
| C14 | 0.37072 (19) | 0.61077 (9) | 0.47502 (7) | 0.0369 (4) | |
| H14 | 0.443 (2) | 0.5717 (11) | 0.4606 (8) | 0.051 (5)* | |
| C15 | 0.42864 (14) | 0.50430 (7) | 0.25935 (6) | 0.0206 (3) | |
| H15A | 0.5028 (17) | 0.4856 (8) | 0.2882 (6) | 0.025 (4)* | |
| H15B | 0.4831 (17) | 0.5225 (8) | 0.2251 (6) | 0.022 (4)* | |
| C16 | 0.32363 (14) | 0.44361 (6) | 0.24135 (5) | 0.0191 (3) | |
| H16 | 0.2784 (19) | 0.4473 (9) | 0.1516 (7) | 0.036 (4)* | |
| C17 | 0.26194 (15) | 0.42622 (7) | 0.18895 (6) | 0.0226 (3) | |
| C18 | 0.28965 (15) | 0.37793 (7) | 0.33815 (6) | 0.0232 (3) | |
| H18A | 0.3932 (19) | 0.3894 (9) | 0.3470 (7) | 0.030 (4)* | |
| H18B | 0.2764 (17) | 0.3237 (9) | 0.3441 (6) | 0.026 (4)* | |
| C19 | 0.18500 (14) | 0.42063 (7) | 0.37736 (5) | 0.0221 (3) | |
| C20 | 0.11923 (18) | 0.43913 (9) | 0.47469 (6) | 0.0329 (3) | |
| H20A | 0.1346 (19) | 0.4932 (10) | 0.4678 (7) | 0.036 (4)* | |
| H20B | 0.0111 (19) | 0.4247 (9) | 0.4681 (7) | 0.032 (4)* | |
| C21 | 0.1730 (2) | 0.41458 (12) | 0.53292 (7) | 0.0456 (4) | |
| H21A | 0.165 (2) | 0.3587 (12) | 0.5382 (9) | 0.062 (6)* | |
| H21B | 0.111 (2) | 0.4381 (11) | 0.5639 (9) | 0.057 (6)* | |
| H21C | 0.277 (2) | 0.4295 (11) | 0.5392 (8) | 0.052 (6)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0328 (5) | 0.0238 (5) | 0.0258 (5) | 0.0042 (4) | −0.0065 (4) | 0.0000 (4) |
| O2 | 0.0241 (5) | 0.0317 (5) | 0.0251 (5) | 0.0058 (4) | −0.0013 (4) | 0.0000 (4) |
| O3 | 0.0350 (5) | 0.0339 (5) | 0.0219 (5) | 0.0116 (4) | 0.0037 (4) | 0.0036 (4) |
| N1 | 0.0247 (5) | 0.0210 (5) | 0.0222 (5) | 0.0002 (4) | −0.0005 (5) | −0.0029 (4) |
| N2 | 0.0204 (5) | 0.0169 (5) | 0.0204 (5) | −0.0001 (4) | −0.0007 (4) | −0.0021 (4) |
| N3 | 0.0228 (6) | 0.0248 (6) | 0.0280 (6) | 0.0015 (4) | −0.0014 (5) | −0.0069 (4) |
| N4 | 0.0232 (5) | 0.0218 (5) | 0.0301 (6) | −0.0013 (4) | −0.0003 (5) | −0.0062 (4) |
| N5 | 0.0212 (5) | 0.0189 (5) | 0.0224 (5) | −0.0007 (4) | 0.0010 (4) | −0.0026 (4) |
| C1 | 0.0197 (6) | 0.0167 (5) | 0.0222 (6) | −0.0030 (5) | −0.0013 (5) | 0.0005 (5) |
| C2 | 0.0270 (7) | 0.0221 (6) | 0.0223 (6) | −0.0021 (5) | 0.0022 (5) | −0.0002 (5) |
| C3 | 0.0328 (7) | 0.0249 (6) | 0.0215 (7) | −0.0052 (6) | −0.0021 (6) | 0.0031 (5) |
| C4 | 0.0280 (7) | 0.0198 (6) | 0.0302 (7) | −0.0029 (5) | −0.0068 (6) | 0.0041 (5) |
| C5 | 0.0230 (6) | 0.0184 (6) | 0.0295 (7) | −0.0002 (5) | −0.0029 (5) | −0.0013 (5) |
| C6 | 0.0219 (6) | 0.0185 (6) | 0.0216 (6) | −0.0028 (5) | −0.0010 (5) | −0.0014 (5) |
| C7 | 0.0231 (6) | 0.0189 (6) | 0.0213 (6) | −0.0031 (5) | −0.0003 (5) | −0.0013 (5) |
| C8 | 0.0217 (6) | 0.0195 (6) | 0.0213 (6) | −0.0032 (5) | −0.0016 (5) | −0.0012 (5) |
| C9 | 0.0306 (7) | 0.0215 (6) | 0.0225 (7) | −0.0046 (5) | −0.0001 (6) | −0.0022 (5) |
| C10 | 0.0537 (10) | 0.0285 (7) | 0.0246 (7) | 0.0063 (7) | 0.0022 (7) | −0.0013 (6) |
| C11 | 0.0717 (12) | 0.0329 (8) | 0.0260 (8) | 0.0098 (8) | 0.0073 (8) | −0.0043 (6) |
| C12 | 0.0684 (12) | 0.0340 (8) | 0.0201 (7) | −0.0059 (8) | 0.0021 (7) | −0.0038 (6) |
| C13 | 0.0540 (10) | 0.0499 (10) | 0.0263 (8) | 0.0030 (8) | −0.0102 (7) | −0.0036 (7) |
| C14 | 0.0395 (8) | 0.0450 (9) | 0.0263 (8) | 0.0067 (7) | −0.0072 (6) | −0.0068 (6) |
| C15 | 0.0179 (6) | 0.0198 (6) | 0.0241 (6) | 0.0009 (5) | 0.0017 (5) | −0.0031 (5) |
| C16 | 0.0173 (5) | 0.0178 (5) | 0.0222 (6) | 0.0026 (5) | 0.0025 (5) | −0.0024 (5) |
| C17 | 0.0211 (6) | 0.0240 (6) | 0.0226 (6) | 0.0029 (5) | 0.0008 (5) | −0.0044 (5) |
| C18 | 0.0251 (7) | 0.0219 (6) | 0.0226 (7) | 0.0024 (5) | 0.0014 (5) | 0.0008 (5) |
| C19 | 0.0219 (6) | 0.0232 (6) | 0.0212 (6) | −0.0020 (5) | 0.0006 (5) | 0.0004 (5) |
| C20 | 0.0366 (8) | 0.0404 (8) | 0.0218 (7) | 0.0107 (7) | 0.0051 (6) | 0.0006 (6) |
| C21 | 0.0474 (10) | 0.0652 (12) | 0.0241 (8) | 0.0154 (9) | 0.0001 (7) | 0.0041 (7) |
Geometric parameters (Å, º)
| O1—C8 | 1.2301 (16) | C9—C14 | 1.395 (2) |
| O2—C19 | 1.2088 (16) | C9—C10 | 1.397 (2) |
| O3—C19 | 1.3293 (16) | C10—C11 | 1.386 (2) |
| O3—C20 | 1.4574 (17) | C10—H10 | 0.97 (2) |
| N1—C7 | 1.2978 (17) | C11—C12 | 1.383 (2) |
| N1—C6 | 1.3780 (16) | C11—H11 | 1.00 (2) |
| N2—C8 | 1.3827 (16) | C12—C13 | 1.369 (2) |
| N2—C1 | 1.3970 (16) | C12—H12 | 0.988 (19) |
| N2—C15 | 1.4797 (15) | C13—C14 | 1.396 (2) |
| N3—N4 | 1.3144 (16) | C13—H13 | 0.99 (2) |
| N3—C17 | 1.3592 (18) | C14—H14 | 1.01 (2) |
| N4—N5 | 1.3468 (15) | C15—C16 | 1.4962 (17) |
| N5—C16 | 1.3618 (16) | C15—H15A | 0.997 (16) |
| N5—C18 | 1.4497 (17) | C15—H15B | 0.986 (15) |
| C1—C2 | 1.4005 (18) | C16—C17 | 1.3691 (18) |
| C1—C6 | 1.4034 (17) | C17—H16 | 0.957 (17) |
| C2—C3 | 1.3832 (19) | C18—C19 | 1.5100 (18) |
| C2—H2 | 0.974 (17) | C18—H18A | 0.963 (17) |
| C3—C4 | 1.395 (2) | C18—H18B | 0.995 (16) |
| C3—H3 | 0.984 (16) | C20—C21 | 1.499 (2) |
| C4—C5 | 1.3732 (19) | C20—H20A | 0.997 (18) |
| C4—H4 | 0.994 (17) | C20—H20B | 1.005 (17) |
| C5—C6 | 1.4055 (18) | C21—H21A | 1.02 (2) |
| C5—H5 | 0.973 (16) | C21—H21B | 1.00 (2) |
| C7—C8 | 1.4905 (17) | C21—H21C | 0.97 (2) |
| C7—C9 | 1.4937 (17) | ||
| C19—O3—C20 | 115.32 (11) | C13—C12—C11 | 119.30 (14) |
| C7—N1—C6 | 120.37 (11) | C13—C12—H12 | 119.4 (11) |
| C8—N2—C1 | 122.62 (10) | C11—C12—H12 | 121.2 (11) |
| C8—N2—C15 | 117.02 (10) | C12—C13—C14 | 120.87 (16) |
| C1—N2—C15 | 120.34 (10) | C12—C13—H13 | 120.3 (12) |
| N4—N3—C17 | 108.35 (11) | C14—C13—H13 | 118.8 (12) |
| N3—N4—N5 | 107.40 (10) | C9—C14—C13 | 120.44 (15) |
| N4—N5—C16 | 111.08 (10) | C9—C14—H14 | 120.3 (11) |
| N4—N5—C18 | 117.95 (10) | C13—C14—H14 | 119.2 (11) |
| C16—N5—C18 | 130.82 (11) | N2—C15—C16 | 112.02 (10) |
| N2—C1—C2 | 123.27 (11) | N2—C15—H15A | 106.4 (9) |
| N2—C1—C6 | 117.23 (11) | C16—C15—H15A | 110.4 (9) |
| C2—C1—C6 | 119.49 (12) | N2—C15—H15B | 109.8 (8) |
| C3—C2—C1 | 119.38 (12) | C16—C15—H15B | 108.9 (8) |
| C3—C2—H2 | 119.8 (10) | H15A—C15—H15B | 109.3 (12) |
| C1—C2—H2 | 120.8 (10) | N5—C16—C17 | 103.54 (11) |
| C2—C3—C4 | 121.35 (12) | N5—C16—C15 | 124.84 (11) |
| C2—C3—H3 | 118.0 (10) | C17—C16—C15 | 131.57 (12) |
| C4—C3—H3 | 120.7 (10) | N3—C17—C16 | 109.62 (12) |
| C5—C4—C3 | 119.61 (12) | N3—C17—H16 | 119.7 (10) |
| C5—C4—H4 | 119.1 (10) | C16—C17—H16 | 130.7 (10) |
| C3—C4—H4 | 121.3 (10) | N5—C18—C19 | 112.36 (11) |
| C4—C5—C6 | 120.24 (12) | N5—C18—H18A | 109.3 (10) |
| C4—C5—H5 | 122.3 (9) | C19—C18—H18A | 110.3 (10) |
| C6—C5—H5 | 117.4 (9) | N5—C18—H18B | 107.3 (9) |
| N1—C6—C1 | 122.08 (11) | C19—C18—H18B | 110.2 (9) |
| N1—C6—C5 | 118.04 (11) | H18A—C18—H18B | 107.1 (13) |
| C1—C6—C5 | 119.86 (12) | O2—C19—O3 | 125.10 (12) |
| N1—C7—C8 | 122.05 (11) | O2—C19—C18 | 125.61 (12) |
| N1—C7—C9 | 116.55 (11) | O3—C19—C18 | 109.29 (11) |
| C8—C7—C9 | 121.40 (11) | O3—C20—C21 | 107.51 (13) |
| O1—C8—N2 | 120.76 (12) | O3—C20—H20A | 106.6 (10) |
| O1—C8—C7 | 123.81 (12) | C21—C20—H20A | 112.9 (10) |
| N2—C8—C7 | 115.41 (11) | O3—C20—H20B | 107.1 (9) |
| C14—C9—C10 | 117.95 (13) | C21—C20—H20B | 111.4 (9) |
| C14—C9—C7 | 124.25 (13) | H20A—C20—H20B | 111.0 (14) |
| C10—C9—C7 | 117.79 (12) | C20—C21—H21A | 112.3 (12) |
| C11—C10—C9 | 120.89 (15) | C20—C21—H21B | 110.4 (12) |
| C11—C10—H10 | 121.1 (12) | H21A—C21—H21B | 107.3 (16) |
| C9—C10—H10 | 118.0 (12) | C20—C21—H21C | 110.8 (12) |
| C12—C11—C10 | 120.55 (16) | H21A—C21—H21C | 108.7 (17) |
| C12—C11—H11 | 120.0 (12) | H21B—C21—H21C | 107.2 (16) |
| C10—C11—H11 | 119.4 (12) | ||
| C17—N3—N4—N5 | 0.03 (14) | N1—C7—C9—C14 | −171.75 (14) |
| N3—N4—N5—C16 | 0.56 (13) | C8—C7—C9—C14 | 9.3 (2) |
| N3—N4—N5—C18 | −175.36 (10) | N1—C7—C9—C10 | 6.99 (18) |
| C8—N2—C1—C2 | 175.04 (12) | C8—C7—C9—C10 | −172.00 (13) |
| C15—N2—C1—C2 | −6.76 (18) | C14—C9—C10—C11 | 0.2 (2) |
| C8—N2—C1—C6 | −4.48 (17) | C7—C9—C10—C11 | −178.63 (15) |
| C15—N2—C1—C6 | 173.72 (11) | C9—C10—C11—C12 | 0.5 (3) |
| N2—C1—C2—C3 | 178.84 (12) | C10—C11—C12—C13 | −0.9 (3) |
| C6—C1—C2—C3 | −1.65 (19) | C11—C12—C13—C14 | 0.6 (3) |
| C1—C2—C3—C4 | −0.7 (2) | C10—C9—C14—C13 | −0.5 (2) |
| C2—C3—C4—C5 | 1.5 (2) | C7—C9—C14—C13 | 178.27 (15) |
| C3—C4—C5—C6 | 0.1 (2) | C12—C13—C14—C9 | 0.1 (3) |
| C7—N1—C6—C1 | −0.69 (19) | C8—N2—C15—C16 | 103.20 (13) |
| C7—N1—C6—C5 | −178.96 (12) | C1—N2—C15—C16 | −75.10 (14) |
| N2—C1—C6—N1 | 4.56 (18) | N4—N5—C16—C17 | −0.90 (13) |
| C2—C1—C6—N1 | −174.98 (12) | C18—N5—C16—C17 | 174.34 (12) |
| N2—C1—C6—C5 | −177.20 (11) | N4—N5—C16—C15 | 176.59 (11) |
| C2—C1—C6—C5 | 3.26 (18) | C18—N5—C16—C15 | −8.2 (2) |
| C4—C5—C6—N1 | 175.80 (12) | N2—C15—C16—N5 | −76.76 (15) |
| C4—C5—C6—C1 | −2.51 (19) | N2—C15—C16—C17 | 99.98 (15) |
| C6—N1—C7—C8 | −3.34 (18) | N4—N3—C17—C16 | −0.61 (14) |
| C6—N1—C7—C9 | 177.67 (11) | N5—C16—C17—N3 | 0.91 (13) |
| C1—N2—C8—O1 | 179.23 (11) | C15—C16—C17—N3 | −176.34 (12) |
| C15—N2—C8—O1 | 0.97 (17) | N4—N5—C18—C19 | −92.47 (13) |
| C1—N2—C8—C7 | 0.80 (17) | C16—N5—C18—C19 | 92.57 (15) |
| C15—N2—C8—C7 | −177.45 (10) | C20—O3—C19—O2 | −1.8 (2) |
| N1—C7—C8—O1 | −175.08 (12) | C20—O3—C19—C18 | 178.01 (12) |
| C9—C7—C8—O1 | 3.85 (19) | N5—C18—C19—O2 | −4.81 (19) |
| N1—C7—C8—N2 | 3.29 (18) | N5—C18—C19—O3 | 175.42 (10) |
| C9—C7—C8—N2 | −177.78 (11) | C19—O3—C20—C21 | −174.39 (14) |
Hydrogen-bond geometry (Å, º)
Cg1 is the centroid of the triazole ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C5—H5···N4i | 0.973 (16) | 2.462 (16) | 3.2183 (17) | 134.3 (12) |
| C12—H12···N3ii | 0.988 (19) | 2.572 (19) | 3.4094 (18) | 142.5 (15) |
| C15—H15A···Cg1iii | 0.997 (16) | 2.657 (15) | 3.3580 (14) | 127.5 (10) |
| C15—H15B···O2iv | 0.986 (15) | 2.464 (15) | 3.2459 (16) | 135.9 (11) |
Symmetry codes: (i) −x, y+1/2, −z+1/2; (ii) −x+1/2, −y+1, z+1/2; (iii) x−1/2, y, −z−1/2; (iv) x+1/2, y, −z+1/2.
Funding Statement
The support of NSF–MRI grant No. 1228232 for the purchase of the diffractometer and Tulane University for support of the Tulane Crystallography Laboratory are gratefully acknowledged.
References
- Abbas, H. S., Al-Marhabi, A. R., Eissa, S. I. & Ammar, Y. A. (2015). Bioorg. Med. Chem. 23, 6560–6572. [DOI] [PubMed]
- Allen, F. H., Baalham, C. A., Lommerse, J. P. M. & Raithby, P. R. (1998). Acta Cryst. B54, 320–329.
- Brandenburg, K. & Putz, H. (2012). DIAMOND, Crystal Impact GbR, Bonn, Germany.
- Bruker (2016). APEX3, SAINT and SADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
- Guirado, A., López Sánchez, J. I., Ruiz-Alcaraz, A. J., Bautista, D. & Gálvez, J. (2012). Eur. J. Med. Chem. 54, 87–94. [DOI] [PubMed]
- Krause, L., Herbst-Irmer, R., Sheldrick, G. M. & Stalke, D. (2015). J. Appl. Cryst. 48, 3–10. [DOI] [PMC free article] [PubMed]
- Kulkarni, N. V., Revankar, V. K., Kirasur, B. N. & Hugar, M. H. (2012). Med. Chem. Res. 21, 663–671.
- Missioui, M., Lgaz, H., Guerrab, W., Lee, H., Warad, I., Mague, J. T., Ali, I. H., Essassi, E. M. & Ramli, Y. (2022a). J. Mol. Struct. 1253, 132132–143.
- Missioui, M., Said, M. A., Demirtaş, G., Mague, J. T., Al-Sulami, A., Al-Kaff, N. S. & Ramli, Y. (2022b). Arab. J. Chem. 15, 103595–613. [DOI] [PMC free article] [PubMed]
- Missioui, M., Said, M. A., Demirtaş, G., Mague, J. T. & Ramli, Y. (2022c). J. Mol. Struct. 1247, 131420–433.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Sridevi, K. B. C. H., Naidu, A. & Sudhakaran, R. (2010). Eur. J. Chem. 7, 234–238.
- Teja, R., Kapu, S., Kadiyala, S., Dhanapal, V. & Raman, A. N. (2016). J. Saudi Chem. Soc. 20, S387–S392.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2414314622006939/bx4022sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2414314622006939/bx4022Isup2.hkl
Supporting information file. DOI: 10.1107/S2414314622006939/bx4022Isup3.cml
CCDC reference: 2184531
Additional supporting information: crystallographic information; 3D view; checkCIF report