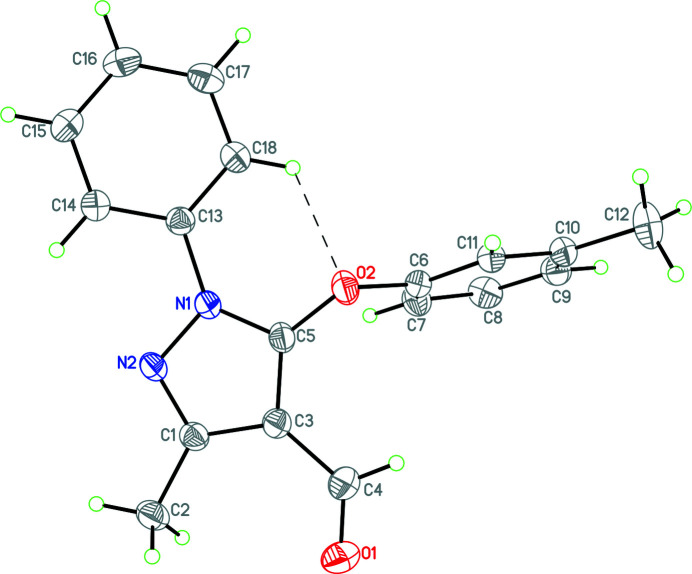
In the title compound, the phenyl and pyrazole rings subtend a dihedral angle of 22.68 (8)°.
Keywords: crystal structure, pyrazole, phenyl, aldehyde
Abstract
In the title compound, C18H16N2O2, the phenyl and pyrazole rings subtend a dihedral angle of 22.68 (8)°. The packing of the title compound features aromatic π–π stacking and weak C—H⋯π interactions.
Structure description
Pyrazoles possess many pharmacological activities such as the inhibition of protein glycation, antibacterial, antifungal, anticancer, antidepressant, anti-inflammatory, anti-tuberculosis and antioxidant activity as well as being used as antiviral agents (Fustero et al., 2011 ▸; Steinbach et al., 2000 ▸; García-Lozano et al., 1997 ▸). The crystal structures of (E)-1,3-dimethyl-5-p-tolyloxy-1H-pyrazole-4-carbaldehyde o-(6-chloropyridazin-3-yl)oxime (Hu et al., 2006 ▸), 1-(5-bromopyrimidin-2-yl)-3-phenyl-1H-pyrazole-4-carbaldehyde (Thiruvalluvar et al., 2007 ▸), 5-(2,4-dichlorophenoxy)-3-methyl-1-phenyl-1H-pyrazole-4-carbaldehyde (Kumar et al., 2016 ▸), four 1-aryl-1H-pyrazole-3,4-dicarboxylate derivatives (Asma et al., 2018 ▸), functionalized 3-(5-aryloxy-3-methyl-1-phenyl-1H-pyrazol-4-yl)-1-(4-substituted-phenyl)prop-2-en-1-ones (Kiran Kumar et al., 2020 ▸) and two isostructural 3-(5-aryloxy-3-methyl-1-phenyl-1H-pyrazol-4-yl)-1-(thiophen-2-yl)prop-2-en-1-ones (Shaibah et al., 2020 ▸) have been reported.
As part our studies in this area, we now report the synthesis and crystal structure of the title compound, C18H16N2O2, (1, Fig. 1 ▸) . Compound 1 crystallizes in the monoclinic space group P21/c with one molecule in the asymmetric unit. It consists of a C1/C3/C5/N1/N2 pyrazole ring linked to a C13–C18 phenyl ring by a carbon–nitrogen single bond [C13—N1 = 1.4285 (17) Å]. As a result of the single bond, the pyrazol and phenyl rings are twisted with a dihedral angle of 22.68 (8)°, perhaps due to the steric interaction between H18 and O2 and between H14 and N2. In the pyrazole ring, the aldehyde group (C3/C4/O1) is slightly twisted with a dihedral angle of 6.43 (10)° as a result of the steric interaction of this group with the C2 methyl substituent. The C6–C12 toluyl substituent makes dihedral angles of 79.44 (5) and 82.40 (5)° with the pyrazol and phenyl rings, respectively. A short intramolecular C18—H18⋯O2 contact closes an S(6) ring.
Figure 1.
Diagram of 1 showing displacement ellipsoids at the 30% probability level. The C18—H18⋯O2 intramolecular contact is shown by a dashed line.
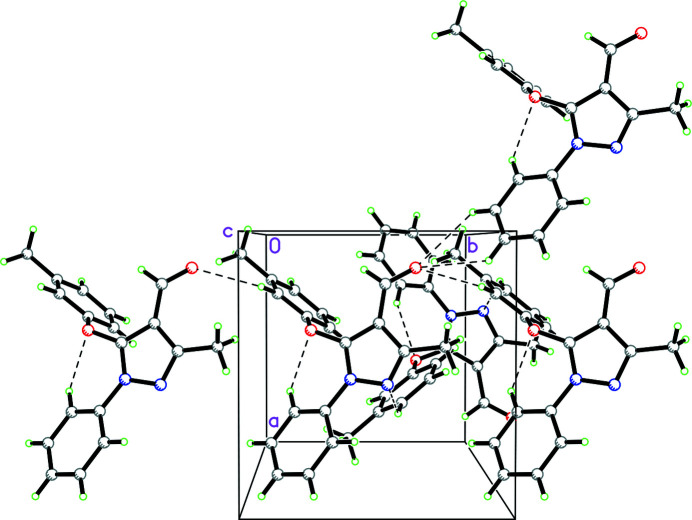
In the packing, a very weak C11—H11⋯O1 hydrogen bond generates [010] chains (Fig. 2 ▸, Table 1 ▸). In addition, aromatic π–π stacking involving the pyrazole rings in adjacent molecules [centroid-to-centroid distance = 3.8908 (9) Å, slippage = 1.233 Å, symmetry operation 1 − x, 1 − y, 1 − z] is observed. There is also a weak C—H⋯π interaction involving the toluene rings in adjacent molecules [distance between ring centroid and carbon atom = 3.8075 (17) Å, C—H⋯Cg = 148°, symmetry operation 1 − x,
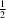 + y,
+ y,
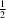 − z].
− z].
Figure 2.
Packing diagram for 1 (viewed along the c-axis direction) showing C—H⋯O and C—H⋯N contacts as dashed lines.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C18—H18⋯O2 | 0.95 | 2.42 | 2.9593 (19) | 116 |
| C11—H11⋯O1i | 0.95 | 2.75 | 3.626 (2) | 153 |
Symmetry code: (i)
 .
.
Synthesis and crystallization
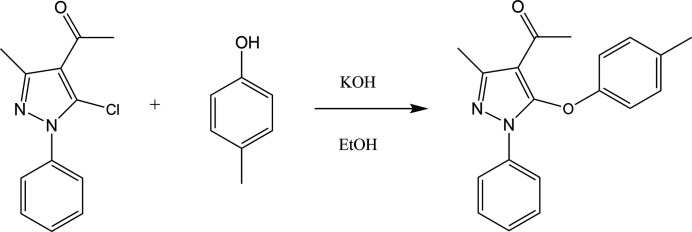
To a solution of p-cresol (0.1 mol) dissolved in 10 ml of dimethylsulfoxide,1-phenyl-5-chloro-3-methyl-1H-pyrazol-4-carbaldehyde (3.22 g, 0.1 mol) and potassium hydroxide (0.8 g, 0.1 mol) were added and the resulting solution was heated on a water bath for 5 h. The reaction mixture was cooled to room temperature and poured onto crushed ice. The solid that separated was filtered off and washed with water and the dried product was recrystallized from ethanol solution. The reaction scheme is shown in Fig. 3 ▸.
Figure 3.
Reaction scheme.
Yield: 82%; m.p. 320–322 K; MS (m/z) 293.1 (M + + 1). 1H NMR (400 MHz, CDCl3, δ p.p.m.), 2.22 (s, 3H, pyrazole methyl), 2.34 (s, 3H, o-tolyloxy methyl), 6.93 (d, 2H, J = 8.3 Hz, Ar—H), 7.04 (d, 2H, J = 8.3 Hz, Ar—H), 7.23 (d, 1H, J = 7.3 Hz), 7.46 (d, 2H, J = 8.1 Hz, Ar—H), 7.81 (d, 2H, J = 8.1 Hz, Ar—H), 8.61 (s, 1H, aldehyde-H).
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C18H16N2O2 |
| M r | 292.33 |
| Crystal system, space group | Monoclinic, P21/c |
| Temperature (K) | 100 |
| a, b, c (Å) | 8.2745 (5), 7.9167 (6), 23.0663 (17) |
| β (°) | 93.225 (4) |
| V (Å3) | 1508.60 (18) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.09 |
| Crystal size (mm) | 0.33 × 0.29 × 0.21 |
| Data collection | |
| Diffractometer | Bruker APEXII CCD |
| Absorption correction | Multi-scan (SADABS; Krause et al., 2015 ▸) |
| T min, T max | 0.560, 0.746 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 22729, 4612, 3061 |
| R int | 0.075 |
| (sin θ/λ)max (Å−1) | 0.717 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.051, 0.150, 1.08 |
| No. of reflections | 4612 |
| No. of parameters | 201 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.24, −0.22 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2414314622009245/hb4413sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2414314622009245/hb4413Isup2.hkl
Supporting information file. DOI: 10.1107/S2414314622009245/hb4413Isup3.cml
CCDC reference: 2207948
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
SDA and HAN are grateful to Mysore & Mangalore Universities, respectively for the provision of research facilities.
full crystallographic data
Crystal data
| C18H16N2O2 | F(000) = 616 |
| Mr = 292.33 | Dx = 1.287 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 8.2745 (5) Å | Cell parameters from 6530 reflections |
| b = 7.9167 (6) Å | θ = 3.1–29.6° |
| c = 23.0663 (17) Å | µ = 0.09 mm−1 |
| β = 93.225 (4)° | T = 100 K |
| V = 1508.60 (18) Å3 | Block, pale yellow |
| Z = 4 | 0.33 × 0.29 × 0.21 mm |
Data collection
| Bruker APEXII CCD diffractometer | 3061 reflections with I > 2σ(I) |
| φ and ω scans | Rint = 0.075 |
| Absorption correction: multi-scan (SADABS; Krause et al., 2015) | θmax = 30.6°, θmin = 2.5° |
| Tmin = 0.560, Tmax = 0.746 | h = −11→11 |
| 22729 measured reflections | k = −11→9 |
| 4612 independent reflections | l = −32→32 |
Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.051 | H-atom parameters constrained |
| wR(F2) = 0.150 | w = 1/[σ2(Fo2) + (0.0554P)2 + 0.2283P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.08 | (Δ/σ)max < 0.001 |
| 4612 reflections | Δρmax = 0.24 e Å−3 |
| 201 parameters | Δρmin = −0.21 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. All hydrogen atoms were placed geometrically and refined as riding atoms with their Uiso values 1.2 × (1.5 × for CH3) that of their attached atoms. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.14639 (15) | 0.70581 (16) | 0.38374 (6) | 0.0644 (3) | |
| O2 | 0.41488 (13) | 0.23801 (12) | 0.37711 (4) | 0.0434 (3) | |
| N1 | 0.61932 (13) | 0.41501 (13) | 0.41767 (5) | 0.0370 (3) | |
| N2 | 0.63851 (14) | 0.57702 (14) | 0.43941 (5) | 0.0395 (3) | |
| C1 | 0.49777 (17) | 0.65200 (17) | 0.42911 (6) | 0.0388 (3) | |
| C2 | 0.4761 (2) | 0.83049 (19) | 0.44777 (7) | 0.0502 (4) | |
| H2A | 0.576401 | 0.870910 | 0.467780 | 0.075* | |
| H2B | 0.387621 | 0.836723 | 0.474185 | 0.075* | |
| H2C | 0.450214 | 0.901154 | 0.413627 | 0.075* | |
| C3 | 0.38270 (17) | 0.54160 (18) | 0.40121 (6) | 0.0398 (3) | |
| C4 | 0.21536 (19) | 0.5708 (2) | 0.38257 (7) | 0.0492 (4) | |
| H4 | 0.154397 | 0.476467 | 0.368230 | 0.059* | |
| C5 | 0.46660 (16) | 0.39239 (16) | 0.39555 (6) | 0.0373 (3) | |
| C6 | 0.36301 (16) | 0.21805 (16) | 0.31822 (6) | 0.0358 (3) | |
| C7 | 0.41496 (19) | 0.32023 (19) | 0.27473 (7) | 0.0453 (3) | |
| H7 | 0.486871 | 0.411651 | 0.283142 | 0.054* | |
| C8 | 0.3587 (2) | 0.2851 (2) | 0.21804 (7) | 0.0510 (4) | |
| H8 | 0.393472 | 0.353203 | 0.187199 | 0.061* | |
| C9 | 0.25337 (19) | 0.1535 (2) | 0.20582 (6) | 0.0473 (4) | |
| H9 | 0.214581 | 0.133416 | 0.166890 | 0.057* | |
| C10 | 0.20347 (17) | 0.05002 (18) | 0.25005 (6) | 0.0424 (3) | |
| C11 | 0.26116 (15) | 0.08271 (17) | 0.30678 (6) | 0.0372 (3) | |
| H11 | 0.230471 | 0.011963 | 0.337554 | 0.045* | |
| C12 | 0.0879 (2) | −0.0940 (3) | 0.23791 (8) | 0.0687 (5) | |
| H12A | 0.086719 | −0.123627 | 0.196639 | 0.103* | |
| H12B | −0.021126 | −0.060187 | 0.247824 | 0.103* | |
| H12C | 0.122673 | −0.192035 | 0.261368 | 0.103* | |
| C13 | 0.75428 (16) | 0.30235 (16) | 0.42240 (6) | 0.0371 (3) | |
| C14 | 0.87596 (18) | 0.3355 (2) | 0.46428 (7) | 0.0470 (3) | |
| H14 | 0.867780 | 0.429849 | 0.489387 | 0.056* | |
| C15 | 1.0098 (2) | 0.2308 (2) | 0.46956 (8) | 0.0554 (4) | |
| H15 | 1.093906 | 0.254084 | 0.498186 | 0.067* | |
| C16 | 1.0220 (2) | 0.0931 (2) | 0.43365 (8) | 0.0540 (4) | |
| H16 | 1.113977 | 0.021373 | 0.437420 | 0.065* | |
| C17 | 0.8997 (2) | 0.0602 (2) | 0.39220 (8) | 0.0566 (4) | |
| H17 | 0.907241 | −0.035666 | 0.367730 | 0.068* | |
| C18 | 0.7662 (2) | 0.1650 (2) | 0.38573 (7) | 0.0505 (4) | |
| H18 | 0.683509 | 0.143019 | 0.356447 | 0.061* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0580 (7) | 0.0630 (8) | 0.0711 (8) | 0.0205 (6) | −0.0064 (6) | −0.0004 (6) |
| O2 | 0.0538 (6) | 0.0350 (5) | 0.0400 (5) | −0.0067 (4) | −0.0104 (4) | −0.0001 (4) |
| N1 | 0.0410 (6) | 0.0327 (5) | 0.0367 (6) | 0.0002 (4) | −0.0036 (4) | −0.0047 (4) |
| N2 | 0.0464 (6) | 0.0325 (5) | 0.0391 (6) | −0.0002 (5) | −0.0018 (5) | −0.0069 (5) |
| C1 | 0.0459 (7) | 0.0362 (7) | 0.0343 (7) | 0.0031 (6) | 0.0017 (5) | −0.0008 (5) |
| C2 | 0.0614 (9) | 0.0379 (7) | 0.0512 (9) | 0.0061 (7) | 0.0014 (7) | −0.0067 (6) |
| C3 | 0.0430 (7) | 0.0388 (7) | 0.0371 (7) | 0.0020 (6) | −0.0023 (5) | −0.0002 (5) |
| C4 | 0.0478 (8) | 0.0526 (9) | 0.0463 (8) | 0.0051 (7) | −0.0060 (6) | 0.0003 (7) |
| C5 | 0.0431 (7) | 0.0343 (6) | 0.0337 (6) | −0.0028 (5) | −0.0051 (5) | −0.0013 (5) |
| C6 | 0.0355 (6) | 0.0352 (6) | 0.0361 (6) | 0.0020 (5) | −0.0026 (5) | −0.0026 (5) |
| C7 | 0.0492 (8) | 0.0409 (7) | 0.0458 (8) | −0.0069 (6) | 0.0023 (6) | 0.0007 (6) |
| C8 | 0.0635 (10) | 0.0480 (8) | 0.0422 (8) | 0.0023 (7) | 0.0080 (7) | 0.0061 (7) |
| C9 | 0.0554 (9) | 0.0496 (8) | 0.0364 (7) | 0.0078 (7) | −0.0025 (6) | −0.0052 (6) |
| C10 | 0.0400 (7) | 0.0435 (7) | 0.0431 (7) | 0.0011 (6) | −0.0022 (5) | −0.0084 (6) |
| C11 | 0.0362 (6) | 0.0374 (7) | 0.0378 (7) | −0.0011 (5) | 0.0006 (5) | −0.0018 (5) |
| C12 | 0.0738 (12) | 0.0718 (12) | 0.0589 (11) | −0.0261 (10) | −0.0100 (9) | −0.0122 (9) |
| C13 | 0.0395 (7) | 0.0351 (6) | 0.0366 (7) | 0.0014 (5) | 0.0020 (5) | 0.0008 (5) |
| C14 | 0.0492 (8) | 0.0456 (8) | 0.0453 (8) | 0.0040 (6) | −0.0061 (6) | −0.0054 (6) |
| C15 | 0.0495 (9) | 0.0597 (10) | 0.0557 (9) | 0.0102 (7) | −0.0088 (7) | −0.0006 (8) |
| C16 | 0.0510 (9) | 0.0535 (9) | 0.0581 (10) | 0.0158 (7) | 0.0073 (7) | 0.0061 (7) |
| C17 | 0.0617 (10) | 0.0485 (9) | 0.0601 (10) | 0.0101 (7) | 0.0085 (8) | −0.0112 (7) |
| C18 | 0.0515 (9) | 0.0487 (8) | 0.0507 (9) | 0.0049 (7) | −0.0030 (7) | −0.0125 (7) |
Geometric parameters (Å, º)
| O1—C4 | 1.2129 (19) | C8—C9 | 1.377 (2) |
| O2—C5 | 1.3555 (15) | C9—H9 | 0.9500 |
| O2—C6 | 1.4103 (16) | C9—C10 | 1.389 (2) |
| N1—N2 | 1.3830 (15) | C10—C11 | 1.3917 (19) |
| N1—C5 | 1.3478 (17) | C10—C12 | 1.505 (2) |
| N1—C13 | 1.4285 (17) | C11—H11 | 0.9500 |
| N2—C1 | 1.3167 (18) | C12—H12A | 0.9800 |
| C1—C2 | 1.4909 (19) | C12—H12B | 0.9800 |
| C1—C3 | 1.4197 (19) | C12—H12C | 0.9800 |
| C2—H2A | 0.9800 | C13—C14 | 1.381 (2) |
| C2—H2B | 0.9800 | C13—C18 | 1.384 (2) |
| C2—H2C | 0.9800 | C14—H14 | 0.9500 |
| C3—C4 | 1.445 (2) | C14—C15 | 1.383 (2) |
| C3—C5 | 1.3800 (19) | C15—H15 | 0.9500 |
| C4—H4 | 0.9500 | C15—C16 | 1.376 (2) |
| C6—C7 | 1.376 (2) | C16—H16 | 0.9500 |
| C6—C11 | 1.3794 (18) | C16—C17 | 1.377 (2) |
| C7—H7 | 0.9500 | C17—H17 | 0.9500 |
| C7—C8 | 1.391 (2) | C17—C18 | 1.383 (2) |
| C8—H8 | 0.9500 | C18—H18 | 0.9500 |
| C5—O2—C6 | 118.48 (10) | C8—C9—C10 | 120.45 (14) |
| N2—N1—C13 | 118.56 (11) | C10—C9—H9 | 119.8 |
| C5—N1—N2 | 110.27 (11) | C9—C10—C11 | 118.70 (13) |
| C5—N1—C13 | 131.16 (11) | C9—C10—C12 | 121.51 (14) |
| C1—N2—N1 | 105.68 (11) | C11—C10—C12 | 119.79 (14) |
| N2—C1—C2 | 119.63 (13) | C6—C11—C10 | 119.87 (13) |
| N2—C1—C3 | 111.60 (12) | C6—C11—H11 | 120.1 |
| C3—C1—C2 | 128.76 (13) | C10—C11—H11 | 120.1 |
| C1—C2—H2A | 109.5 | C10—C12—H12A | 109.5 |
| C1—C2—H2B | 109.5 | C10—C12—H12B | 109.5 |
| C1—C2—H2C | 109.5 | C10—C12—H12C | 109.5 |
| H2A—C2—H2B | 109.5 | H12A—C12—H12B | 109.5 |
| H2A—C2—H2C | 109.5 | H12A—C12—H12C | 109.5 |
| H2B—C2—H2C | 109.5 | H12B—C12—H12C | 109.5 |
| C1—C3—C4 | 130.04 (13) | C14—C13—N1 | 118.10 (12) |
| C5—C3—C1 | 103.98 (12) | C14—C13—C18 | 120.17 (13) |
| C5—C3—C4 | 125.97 (13) | C18—C13—N1 | 121.72 (13) |
| O1—C4—C3 | 125.34 (15) | C13—C14—H14 | 120.1 |
| O1—C4—H4 | 117.3 | C13—C14—C15 | 119.77 (14) |
| C3—C4—H4 | 117.3 | C15—C14—H14 | 120.1 |
| O2—C5—C3 | 130.53 (13) | C14—C15—H15 | 119.8 |
| N1—C5—O2 | 120.75 (12) | C16—C15—C14 | 120.46 (15) |
| N1—C5—C3 | 108.45 (11) | C16—C15—H15 | 119.8 |
| C7—C6—O2 | 123.07 (12) | C15—C16—H16 | 120.3 |
| C7—C6—C11 | 121.98 (13) | C15—C16—C17 | 119.48 (15) |
| C11—C6—O2 | 114.90 (12) | C17—C16—H16 | 120.3 |
| C6—C7—H7 | 121.1 | C16—C17—H17 | 119.6 |
| C6—C7—C8 | 117.77 (14) | C16—C17—C18 | 120.83 (15) |
| C8—C7—H7 | 121.1 | C18—C17—H17 | 119.6 |
| C7—C8—H8 | 119.4 | C13—C18—H18 | 120.4 |
| C9—C8—C7 | 121.20 (14) | C17—C18—C13 | 119.28 (15) |
| C9—C8—H8 | 119.4 | C17—C18—H18 | 120.4 |
| C8—C9—H9 | 119.8 | ||
| O2—C6—C7—C8 | 178.25 (13) | C5—N1—C13—C14 | −157.00 (15) |
| O2—C6—C11—C10 | −179.47 (12) | C5—N1—C13—C18 | 24.1 (2) |
| N1—N2—C1—C2 | 179.50 (12) | C5—C3—C4—O1 | −174.07 (16) |
| N1—N2—C1—C3 | 0.71 (15) | C6—O2—C5—N1 | −118.79 (14) |
| N1—C13—C14—C15 | −178.97 (14) | C6—O2—C5—C3 | 67.98 (19) |
| N1—C13—C18—C17 | 179.89 (14) | C6—C7—C8—C9 | 0.5 (2) |
| N2—N1—C5—O2 | −173.26 (12) | C7—C6—C11—C10 | −2.3 (2) |
| N2—N1—C5—C3 | 1.32 (16) | C7—C8—C9—C10 | −1.4 (2) |
| N2—N1—C13—C14 | 21.84 (19) | C8—C9—C10—C11 | 0.4 (2) |
| N2—N1—C13—C18 | −157.01 (13) | C8—C9—C10—C12 | 179.67 (16) |
| N2—C1—C3—C4 | 179.55 (14) | C9—C10—C11—C6 | 1.4 (2) |
| N2—C1—C3—C5 | 0.05 (16) | C11—C6—C7—C8 | 1.3 (2) |
| C1—C3—C4—O1 | 6.5 (3) | C12—C10—C11—C6 | −177.89 (15) |
| C1—C3—C5—O2 | 173.04 (14) | C13—N1—N2—C1 | 179.67 (11) |
| C1—C3—C5—N1 | −0.83 (15) | C13—N1—C5—O2 | 5.7 (2) |
| C2—C1—C3—C4 | 0.9 (3) | C13—N1—C5—C3 | −179.76 (13) |
| C2—C1—C3—C5 | −178.59 (14) | C13—C14—C15—C16 | −0.5 (3) |
| C4—C3—C5—O2 | −6.5 (2) | C14—C13—C18—C17 | 1.1 (2) |
| C4—C3—C5—N1 | 179.64 (14) | C14—C15—C16—C17 | 0.1 (3) |
| C5—O2—C6—C7 | 25.02 (19) | C15—C16—C17—C18 | 0.9 (3) |
| C5—O2—C6—C11 | −157.83 (12) | C16—C17—C18—C13 | −1.5 (3) |
| C5—N1—N2—C1 | −1.26 (15) | C18—C13—C14—C15 | −0.1 (2) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C18—H18···O2 | 0.95 | 2.42 | 2.9593 (19) | 116 |
| C11—H11···O1i | 0.95 | 2.75 | 3.626 (2) | 153 |
Symmetry code: (i) x, y−1, z.
Funding Statement
HSY and BK are grateful to UGC, New Delhi, for the award of BSR Faculty Fellowships.
References
- Asma, Kalluraya, B., Yathirajan, H. S., Rathore, R. S. & Glidewell, C. (2018). Acta Cryst. E74, 1783–1789. [DOI] [PMC free article] [PubMed]
- Bruker (2002). SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2005). APEX2. Bruker AXS Inc., Madison, Wisconsin, USA.
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Fustero, S., Sánchez-Roselló, M., Barrio, P. & Simón-Fuentes, A. (2011). Chem. Rev. 111, 6984–7034. [DOI] [PubMed]
- García-Lozano, J., Server-Carrió, J., Escrivà, E., Folgado, J.-V., Molla, C. & Lezama, L. (1997). Polyhedron, 16, 939–944.
- Hu, F.-Z., Chang, Y.-Q., Zhu, Y.-Q., Zou, X.-M. & Yang, H.-Z. (2006). Acta Cryst. E62, o3676–o3677.
- Kiran Kumar, H., Yathirajan, H. S., Harish Chinthal, C., Foro, S. & Glidewell, C. (2020). Acta Cryst. E76, 488–495. [DOI] [PMC free article] [PubMed]
- Krause, L., Herbst-Irmer, R., Sheldrick, G. M. & Stalke, D. (2015). J. Appl. Cryst. 48, 3–10. [DOI] [PMC free article] [PubMed]
- Kumar, S. M., Manju, N., Asma, Kalluraya, B., Byrappa, K. & Warad, I. (2016). IUCrData, 1, x161111.
- Shaibah, M. A. E., Yathirajan, H. S., Manju, N., Kalluraya, B., Rathore, R. S. & Glidewell, C. (2020). Acta Cryst. E76, 48–52. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Steinbach, G., Lynch, P. M., Phillips, R. K. S., Wallace, M. H., Hawk, E., Gordon, G. B., Wakabayashi, N., Saunders, B., Shen, Y., Fujimura, T., Su, L.-K., Levin, B., Godio, L., Patterson, S., Rodriguez-Bigas, M. A., Jester, S. L., King, K. L., Schumacher, M., Abbruzzese, J., DuBois, R. N., Hittelman, W. N., Zimmerman, S., Sherman, J. W. & Kelloff, G. (2000). N. Engl. J. Med. 342, 1946–1952. [DOI] [PubMed]
- Thiruvalluvar, A., Subramanyam, M., Kalluraya, B. & Lingappa, B. (2007). Acta Cryst. E63, o3362. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2414314622009245/hb4413sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2414314622009245/hb4413Isup2.hkl
Supporting information file. DOI: 10.1107/S2414314622009245/hb4413Isup3.cml
CCDC reference: 2207948
Additional supporting information: crystallographic information; 3D view; checkCIF report