Abstract
Background:
Perfluorooctane sulfonate (PFOS) is a persistent environmental pollutant that has become a significant concern around the world. Exposure to PFOS may alter gut microbiota and liver metabolic homeostasis in mammals, thereby increasing the risk of cardiometabolic diseases. Diets high in soluble fibers can ameliorate metabolic disease risks.
Objectives:
We aimed to test the hypothesis that soluble fibers (inulin or pectin) could modulate the adverse metabolic effects of PFOS by affecting microbe-liver metabolism and interactions.
Methods:
Male C57BL/6J mice were fed an isocaloric diet containing different fibers: a) inulin (soluble), b) pectin (soluble), or c) cellulose (control, insoluble). The mice were exposed to PFOS in drinking water () for 7 wk. Multi-omics was used to analyze mouse liver and cecum contents.
Results:
In PFOS-exposed mice, the number of differentially expressed genes associated with atherogenesis and hepatic hyperlipidemia were lower in those that were fed soluble fiber than those fed insoluble fiber. Shotgun metagenomics showed that inulin and pectin protected against differences in microbiome community in PFOS-exposed vs. control mice. It was found that the plasma PFOS levels were lower in inulin-fed mice, and there was a trend of lower liver accumulation of PFOS in soluble fiber-fed mice compared with the control group. Soluble fiber intake ameliorated the effects of PFOS on host hepatic metabolism gene expression and cecal content microbiome structure.
Discussions:
Results from metabolomic, lipidomic, and transcriptomic studies suggest that inulin- and pectin-fed mice were less susceptible to PFOS-induced liver metabolic disturbance, hepatic lipid accumulation, and transcriptional changes compared with control diet-fed mice. Our study advances the understanding of interaction between microbes and host under the influences of environmental pollutants and nutrients. The results provide new insights into the microbe-liver metabolic network and the protection against environmental pollutant-induced metabolic diseases by high-fiber diets. https://doi.org/10.1289/EHP11360
Introduction
Environmental exposures impact the human body throughout life. The gut and liver mostly serve as the first line of insult if exposure occurs through the oral route. They are also major players in the defense against environmental exposure-induced toxicity. The gut, along with its harbored microbiome, is not only the primary organ for digestion and absorption, but it also plays an important role in regulating the host’s metabolism.1 The liver is pivotal for xenobiotic and endobiotic biotransformation, maintenance of energy homeostasis in the body, and regulation of physiology and disease pathologies.2 Evidence from animal3,4 and epidemiologic5 studies has suggested that gut–liver crosstalk plays extremely vital roles in the occurrence and development of diseases related with environmental exposures (e.g., as reviewed by Di Ciaula et al.6) The accumulating evidence that prebiotics, probiotics, and antibiotics might improve severe liver disorders supports the crucial function of the gut microbiome in hepatic diseases.7 Modulation of the gut microbiota may be an intervention strategy to lessen the harmful consequences of environmental exposure, given the gut microbiome’s growing importance to human health and susceptibility to environmental insults.8,9
Perfluoroalkyl substances (PFAS) are synthetic chemicals widely used in water and oil repellents, coating agents, surfactant additives, and firefighting foams. PFAS are resistant to degradation and thus have been detected in environmental biota, including drinking water, groundwater, soils, and air.10,11 One of the most widely used PFASs is perfluorooctane sulfonate (PFOS). People are mainly exposed to PFOS through drinking water and contaminated food.10 Biomonitoring data from the U.S. Centers for Disease Control and Prevention (years 2011–2018) have indicated that PFOS can be universally detected in the general U.S. population’s serum.12 The half-life of PFOS has been estimated to be 3.4 y13 and 1–2 months14 in humans and rodents, respectively. Enterohepatic circulation was reported to contribute to the high bioaccumulation and long biological half-lives of PFOS.15 After PFOS administration to rodents, 6% of the dose could be recovered in urine and feces at 48 h post dose, and 79% of the dose was recovered in the carcass, indicating that PFOS persists and accumulates in tissues.14 Because of its broad exposure and documented toxicity, concern regarding PFOS’s health effects is high. In a study about PFAS distribution in human tissues, it was found that the liver is a major organ of PFOS bioaccumulation.16 Epidemiologic studies have reported the elevation of serum hepatic enzymes after PFOS exposure, which is suggestive of liver damage.17–19 Alterations in cholesterol and circulating lipid levels resulting from PFOS exposure have also been demonstrated.20 Rodents exposed to PFOS have exhibited hepatocellular hypertrophy,21 hepatic steatosis,22,23 and microscopic hepatic lesions associated with greater liver weight20; transcriptome analysis in zebrafish exposed to PFOS has revealed differences in genes related to hepatic hyperlipidemia.24 In addition, PFOS administration has been shown to disrupt hepatic lipid and glucose homeostasis in mouse models.25–27 In vitro studies have suggested that PFOS exposure can up-regulate the expression of genes involved in lipogenesis in primary hepatocytes28,29 and that the activation of peroxisome proliferator-activated receptor () can be partially responsible for these effects caused by PFOS.30 Furthermore, PFOS has been shown to activate other nuclear receptors that regulate liver functions, such as the constitutive androstane receptor, farnesoid X receptor, and pregnane X receptor.20,29 According to recent studies in mice, the gut microbiota can modify host metabolome and gene expression, mediating some of the molecular and biochemical alterations caused by PFOS.31,32
Healthful nutrition, such as diets rich in plant-derived polyphenols and fiber, and nutritional intervention in general, has been described as a sensible and successful means of developing prevention strategies against diseases associated with environmental exposures.33,34 Dietary fibers are carbohydrate polymers consisting of more than three monosaccharide units that withstand the host digestive enzymes. Dietary fibers include water soluble and insoluble forms.35 Insoluble forms, such as cellulose, are resistant to fermentation by the gut microbiome. Soluble dietary fibers, such as inulin and pectin, are fermented by different gut bacteria through complex mechanisms.35 Mounting evidence has suggested that soluble fibers could reduce systemic inflammation, promote the growth of beneficial bacteria, and enhance intestinal barrier function.36 In addition, inulin and pectin administration have been shown to protect against liver diseases through multiple mechanisms, including antioxidant effects in hepatic stellate cells,37 improvement of lipid profiles in patients with nonalcoholic fatty liver disease (NAFLD)38 and in animal models,39,40 and affecting hepatic drug-metabolizing enzymes in rodents.41,42 According to our previous studies, exposure to dioxin-like polychlorinated biphenyls (PCBs) led to gut microbiota dysbiosis and hepatic metabolism disorder in mice.40,43,44 The soluble dietary fiber inulin was protective against these toxicities.40 In addition to differences in solubility, dietary fibers have distinct chemical structures; for example, pectin is poly d-galacturonate, whereas inulin is poly fructose. Such differences could affect their biological effects.45 Therefore, the aims of this study were to investigate the role of different dietary fiber interventions (inulin and pectin) on PFOS-induced disruption of hepatic and gut microbiota health. Understanding the mechanisms might lead to improved dietary intervention strategies to reduce environmental exposure-related disease risk.
Methods
Chemicals and Reagents
PFOS (heptadecafluorooctanesulfonic acid potassium salt; 77282; Sigma-Aldrich; purity ). (300616; Sigma-Aldrich) was used as the internal standard for metabolomic analysis. The SPLASH LIPIDOMIX Mass Spec Standard (330707) used for lipidomic assay was purchased from Avanti Polar Lipids. , sodium salt (98% chemical purity), obtained from Cambridge Isotopes Laboratories, was used as the internal standard for PFOS quantification. All solvents used were of high-performance liquid chromatography (HPLC) grade.
Diets
Inulin, pectin, and cellulose (control) were incorporated directly into the formulation of the OpenStandard diet (Research Diets Inc.) at 8% to produce isocaloric diets ( calories). All diets were gamma-irradiated. Inulin, a polyfructosan (Orafti HP; source: chicory root) was procured from Beneo. Pectin, a polygalacturonate (source: apple peel) was purchased from Spectrum Chemical MFG Corp. The level of fiber in this study was chosen based on our previous studies,39,40 in which 8% of inulin was demonstrated to improve plasma lipid profiles and protect against dioxin-like PCB126-induced hepatotoxicity in a hyperlipidemic mouse model. The established level of fiber in purified rodent diets is 5%, equating to per 1,000 calories. The recommended intake of fiber in humans is per 1,000 calories,46 and the 8% level of fiber in the OpenStandard diet equates to per 1,000 calories, which can be classified as a high-fiber diet. Detailed diet compositions can be found in Table S1.
Study Design
Wild-type male C57BL/6J mice (7 wk old) were purchased from the Jackson Laboratory. The animals were allowed access to chow and water ad libitum and maintained at 23°C with a 14:10-h light/dark cycle. The mice were randomly divided into six groups. Random numbers were generated using an online random order generator (https://www.random.org/lists/). Eight mice were allocated to each experimental group. There were two animal losses at the beginning of the experiment (one in the group and one in the group), and one loss (in the group) in the middle of the study for unknown reasons. The mice were fed with the diet supplemented with one of the three fibers: cellulose as control, inulin, or pectin. PFOS was mixed with drinking water to get a final concentration at . The exposure dose corresponded to (calculated based on an average mouse body weight of and mean volume of drinking water consumption of at 10 wk of age). Mouse drinking water was prepared from NERL high purity water (23-249-589; Thermo Fisher Scientific), which is produced using the most efficacious methods to remove PFAS contaminates (i.e., reverse osmosis and carbon filtering).47 The animals in each group received drinking water with or without PFOS for 7 wk (Figure 1A). The selection of the dosage schedule was based on a previous study that suggested that mice exposed to PFOS at for 7 wk exhibited metabolic function changes.32 All mice received food and water ad libitum and measures of food and water intake and body weight were recorded weekly. At the end of the study, the mice were fasted for 16 h, anesthetized using isoflurane, and blood was collected via cardiac puncture. Liver and cecum content (feces) samples were collected (weight of the liver was recorded), snap frozen in liquid nitrogen, and stored at until analysis. All experimental procedures were approved by the institutional animal care and use committee of the University of Kentucky.
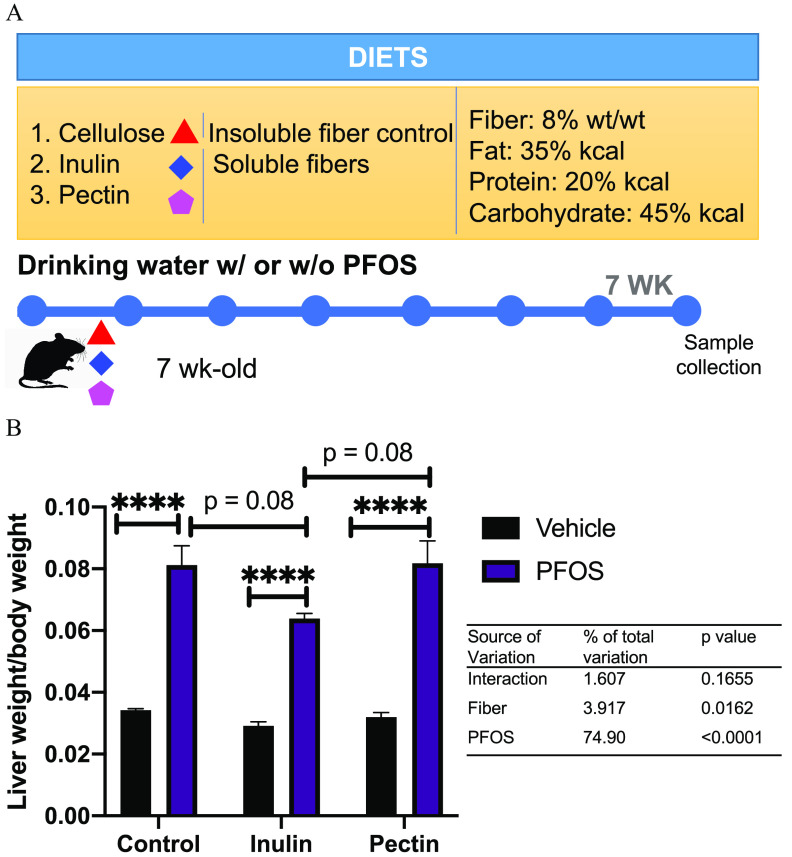
Figure 1.
Effects of PFOS exposure and dietary interventions on liver/body weight ratio. (A) Study design. Seven-week-old C57BL/6 male mice () were placed on an irradiated diet supplemented with one of three fibers: cellulose (control), inulin, or pectin. The mice were exposed to PFOS at BW per day by drinking water and maintained on the diet for 7 wk. Body weight, food, and water intake were measured weekly. The mice were fasted 16 h before euthanasia, and tissues and plasma were collected at the end of the study. (B) The effects of PFOS and dietary fibers on liver/body weight ratio. Data were analyzed by two-way ANOVA followed by Tukey’s post hoc test for multiple comparisons. Bars represent of 6–8 mice in each group. ***. No significant interactions between PFOS exposure and dietary fiber on liver/body weight ratio were observed. Detailed data of liver/body weight ratio are listed in Excel Table S11. Note: ANOVA, analysis of variance; BW, body weight; PFOS, perfluorooctane sulfonate; SEM, standard error of the mean.
Transcriptomic Analysis of Mouse Liver
RNA was extracted from mouse liver samples () with the Qiagen RNeasy Plus Mini Kit. Nucleic acid concentration and purity were determined using the NanoDrop 2000 spectrophotometer (Thermo Scientific), and the quality of RNA was assessed using the Agilent 2100 Bioanalyzer system (Agilent Technologies). Samples were sequenced by Novogene Corporation (https://en.novogene.com). The complementary DNA library was constructed using the RNA-NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs). NovaSeq 6000 (Illumina) was used for the sequencing. Raw reads of FASTQ format were processed through fastp, and clean data were obtained by filtering raw data with reads containing adapter and poly-N sequences and reads with low quality. For data analysis, paired-end clean reads were aligned to the reference genome using STAR48 (version 2.5), and the maximal Mappable Prefix method, which can generate a precise mapping result for junction reads, was used. HTSeq49 (version 0.6.1) was used to count the read numbers mapped of each gene, and then fragments per kilobase of transcript per million mapped reads of each gene were calculated based on the length of the gene and reads count mapped to that gene. Alignments were parsed using the TopHat2 program50 (http://ccb.jhu.edu/software/tophat) and differential expressions were determined through DESeq251 R package (version 2_1.6.3; R Development Core Team). -Values were adjusted using the Benjamini and Hochberg method.52 Significantly differentially expressed genes with and an absolute are listed in Excel Tables S2–S4. ClusterProfiler53 R package (version 3.8.1; R Development Core Team) was used to test the statistical enrichment of differential expression genes in the Kyoto Encyclopedia of Genes and Genomes (KEGG)54 pathways (http://www.genome.jp/kegg/). KEGG terms with an adjusted were considered significantly enriched.
Sample Extraction for Metabolome and Lipidome Analyses
Liver and cecum content samples were extracted and analyzed using a modified method of that reported previously.39 Briefly, samples were homogenized in of 0.1% ammonium formate. The homogenate was spiked with an internal standard solution, then mixed with methanol and methyl tertiary-butyl ether (MTBE). After shaking for 30 min at room temperature, water was added to induce phase separation. After centrifugation, the upper lipid phase was collected and dried under nitrogen flow, and the lower aqueous (polar) phase was pooled and lyophilized. For liquid chromatography–mass spectrometry (LC-MS) analysis, the lipid residue was dissolved in of a chloroform and methanol mixture (2:1, vol/vol), and the polar residue was dissolved in of a mixture of methanol and water (4:1, vol/vol). Quality control (QC) samples were used to monitor the overall quality of the MS analyses, which were prepared from pooled mouse liver and cecum content extracts.
Untargeted Metabolomics and Lipidomics
Metabolomic and lipidomic analyses were performed using a Q Exactive Orbitrap MS coupled to a Dionex UltiMate 3000 ultra HPLC system (Thermo Fisher Scientific). The MS was equipped with an Ion Max API source and a HESI II probe. External mass calibration under both negative and positive modes was performed using the standard calibration mixture every 7 d.
A Sequant ZIC-pHILIC column (, ) (Merck) was used for the chromatographic separation of polar metabolites. The analytical conditions were similar to our previously published method.55 The mobile phase was composed of ammonium carbonate with 0.1% ammonium hydroxide (buffer A) and acetonitrile (buffer B). The chromatographic gradient was delivered at a flow rate of as follows: 0–20 min, linear gradient from 80% to 20% B; 20–21 min, maintained at 20% B min; 21–22 min, linear gradient to 80% B; and 22–28 min, re-equilibration at 80% B. The sample injection volume was . The MS was operated under both positive and negative modes with the heated capillary held at 275°C and the HESI probe held at 350°C. The spray voltage was set to 3.0 kV. The sheath gas flow was 40 units, the auxiliary gas flow was 15 units, and the sweep gas flow was 1 unit. The MS data acquisition was performed in a range of m/z 59–850. The full scan and MS2 spectra were collected at a resolution of 70,000 and 17,500, respectively; the AGC target was set at ; and the maximum injection time was 100 ms.
A Waters ACQUITY BEH C8 column (, ) was used for the separation of lipids. The analytical conditions were similar to our previously published method.39 The mobile phases consisted of 60:40 water/acetonitrile (A), and 90:10 isopropanol/acetonitrile (B), both containing ammonium formate and 0.1% formic acid. Gradient elution started from 32% B and increased to 97% B over 25 min, the mobile phase was maintained at 97% B for 4 min, followed by re-equilibration with the initial mobile phase (32% B) for 6 min. The flow rate was . The column oven was maintained at 40°C, and the sample injection volume was . The MS was operated under both positive and negative ionization modes. The full scan and fragment spectra were collected at a resolution of 70,000 and 17,500, respectively. Experimental samples were randomized across the run and QC samples were spaced evenly among the injections.
Metabolites were assigned and the LC-MS peaks were integrated using Compound Discoverer version 3.0; (Thermo Fisher Scientific), and a customized workflow was established based on our previous report55 (Figure S1). Multidimensional peak tables that included molecular weight (or m/z), retention time, compound formula, and internal standard normalized peak area were generated (peak areas were listed in Excel Table S5). Raw data were log-transformed to correct for heteroscedasticity and to balance distributions, then the data were normalized by median centering.56 Metabolic pathway analysis was employed using the MetaboAnalyst (version 5.0)57 to identify the most relevant metabolic pathways involved in PFOS exposure. Metabolites showing significant change were mapped to the KEGG54 pathways (http://www.genome.jp/kegg/).
Lipidsearch (version 4.1; Thermo Fisher) was used for lipid data analysis and identification. Within a lipid class, MainArea values output by LipidSearch were assigned to each fatty acid moiety of a given lipid species and Microsoft Excel was used to sum the data. Positive and negative electrospray ionization data were processed separately, and peak area tables were combined into an Excel file. The peak areas’ metabolites and lipids were normalized by internal standards and sample weight [liver: peak area of metabolite or lipid/(peak area of ), plasma: peak area of metabolite or lipid/peak area of internal standard] before statistical analysis (Figure S1).
Quantification of Linear and Branched PFOS in Mouse Plasma and Liver
Plasma and liver samples were analyzed at the Laboratory of Exposure Assessment and Development for Environmental Health Research (LEADER) at Emory University. Liver samples were homogenized with a tissue grinder with of methanol. Plasma () and homogenized liver () samples were spiked with an isotopically labeled standard solution (, ) and diluted with a formic acid/methanol solution ( for plasma; for liver) prior to sample preparation so that their concentrations were within the calibration range of the method and to deproteinate the sample. After centrifugation, of the supernatant of the samples were introduced onto a Strata RP on-line extraction column (). The on-line extraction column was washed with formic acid/acetonitrile (90/10, vol/vol), then the target analytes were transferred from the on-line extraction column to a Betasil C18 analytical column (, ) for chromatographic separation and detection using HPLC interfaced with tandem MS (6460 Triple Quadrupole LC/MS; Agilent Technologies). In each analytical batch, a matrix-based calibration curve, a blank sample, National Institute Standards Technology (NIST) standard reference materials (SRMs) and QC samples were prepared and injected alongside unknown plasma or liver samples. Quantification was performed using isotope dilution calibration with the limits of detection of for branched PFOS and for linear PFOS. The limit of detection was defined as the lowest concentrations in the calibration curve, where the signal/noise ratio of the observed signal was and the accuracy derived from NIST SRMs samples was . The method precision, calculated as the relative standard deviation (RSD) of QC materials, was . The LEADER successfully participates in and is certified semiannually for PFAS measurements by the German External Quality Assessment Scheme (https://app.g-equas.de/web/).
Shotgun Metagenomics of Cecum Contents
Genomic DNA was isolated from murine cecal content () using the QIAamp DNA Stool Mini Kit (Qiagen). The quality and integrity of DNA samples were checked by 1% agarose gel electrophoresis (loading of DNA). Meanwhile, DNA yield and purity were measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher) and a Qubit 2.0 fluorometer (Thermo Fisher), and the A260/280 values were or higher. Shotgun metagenomics was performed under contract by Psomagen, Inc. DNA integrity was checked by Psomagen using E Gel 48 Agarose Gels with SYBR Safe DNA Gel Stain 1% Kit (Thermo Fisher) on an E-base Electrophoresis Device (Thermo Fisher). Two nanograms of genomic DNA were used as the input material for the shotgun metagenome library. The samples mixed with the bead-linked transposomes from the Nextera DNA Flex Library Prep Kit (Illumina) were incubated at 55°C for 15 min to fragment and tag the DNA with adapter sequences. Tagmented samples bound on the beads were resuspended with polymerase chain reaction (PCR) master mix and index from IDT for Illumina Nextera DNA Unique Dual Indexes kit. Then they were amplified using a limited-cycle PCR program to add the index and sequences required for sequencing cluster generation on an Axygen MaxyGene II thermal cycler. The PCR product was purified using magnetic beads and validated using the Agilent D5000 ScreenTape assay and Picogreen assay (G820841; Thermo Fisher). The validated libraries were then normalized to and loaded onto an Illumina Hiseq X platform with PE150 (pair-end sequencing, 150 bp reads) after diluting to the desired loading concentration ().
Shotgun metagenomics data sets were analyzed by the Bioinformatics group at the University of Kentucky. Raw shotgun sequencing reads were preprocessed using KneadData (version 0.10.0) QC pipeline.58 Briefly, the paired-end reads were first trimmed to remove low-quality positions, adapter bases, and repetitive sequences. The resulting reads were then queried against the Bowtie2 Mus musculus reference database mouse_C57BL_6NJ to remove contaminating host reads from the sequencing files.59 After preprocessing, a total of paired-end reads remained.
Taxonomy assignment of shotgun sequencing reads was performed using Kraken2.60 In brief, the kraken2 function was used to run the kneadphyla data-filtered shotgun sequencing reads against the Kranen2 Refseq database (data accessed 17 May 2021) containing the “archaea,” “bacteria,” and “viral” reference databases. The taxonomy labels assigned by Kraken2 for each sample were passed to Bayesian Reestimation of Abundance with Kraken (Bracken2) to estimate the relative abundance at the phylum, genus, and species levels.61 The relative abundance outputs from Braken2 can be found in Excel Tables S6–S8.
Alpha- and beta-diversity analyses were performed using Phyloseq (version 1.38.0)51 and Vegan (version 2.5-7)53 in R (version 4.1.2; R Development Core Team). Kraken2 outputs were combined and imported into a phyloseq object. The Shannon Diversity Index was used to measure alpha-diversity (within-sample diversity). Bray–Curtis distance matrix and principal coordinate analysis (PCoA) values were obtained using the ordinate function to measure beta-diversity. Permutational multivariate analysis of variance (PERMANOVA) was performed to determine significance in the distance metrices across samples by dietary fibers or PFOS exposure using the adonis3 function in the GUniFrac package. Microbiota changes focusing on relative abundances of taxa were compared between groups using linear discriminant analysis (LDA) effect size (LEfSe) with the default -value () and the LDA score of 2.0.62
Statistical Analyses
Partial least-squares discriminant analysis (PLS-DA) was used to identify initial trends and clusters in data sets. Variable importance in projection (VIP) scores, which indicated the importance of the variable to the whole model, were calculated using the Metaboanalyst (version 5.0; www.metaboanalyst.ca). GraphPad Prism (version 7.04 for Windows; GraphPad Software Inc.) was used for the statistical analyses of metabolomic and lipidomic data. Data are expressed as . Comparisons between groups were made by one-way (comparison of PFOS levels in exposed mice) or two-way ANOVA with post hoc Tukey’s test. Statistical significance was set at a determined .
Results
Effects of PFOS Exposure and Dietary Fiber Intervention on the Mouse Liver/Body Weight Ratio
No significant differences were observed in water consumption, food intake, and body weight (Figure S2, Excel Tables S9 and S10), suggesting that the mice from different groups had equivalent dietary fiber intake and PFOS exposure via drinking water and that the dose of the PFOS used did not cause hypophagia. We observed that mice exposed to PFOS had a higher liver/body weight ratio regardless of diet (Figure 1B; Excel Table S11). There was a trend in PFOS-exposed mice that those fed with the inulin-supplemented diet demonstrated an lower liver/body weight ratio than control- and pectin-diet-fed mice (; Figure 1B). Although significant effects of PFOS exposure and fiber on liver/body weight ratio were observed, there were no interactions between PFOS and fiber intake (Figure 1B).
Mouse Liver Lipidomic Changes in Response to PFOS Exposure and Dietary Fiber Intervention
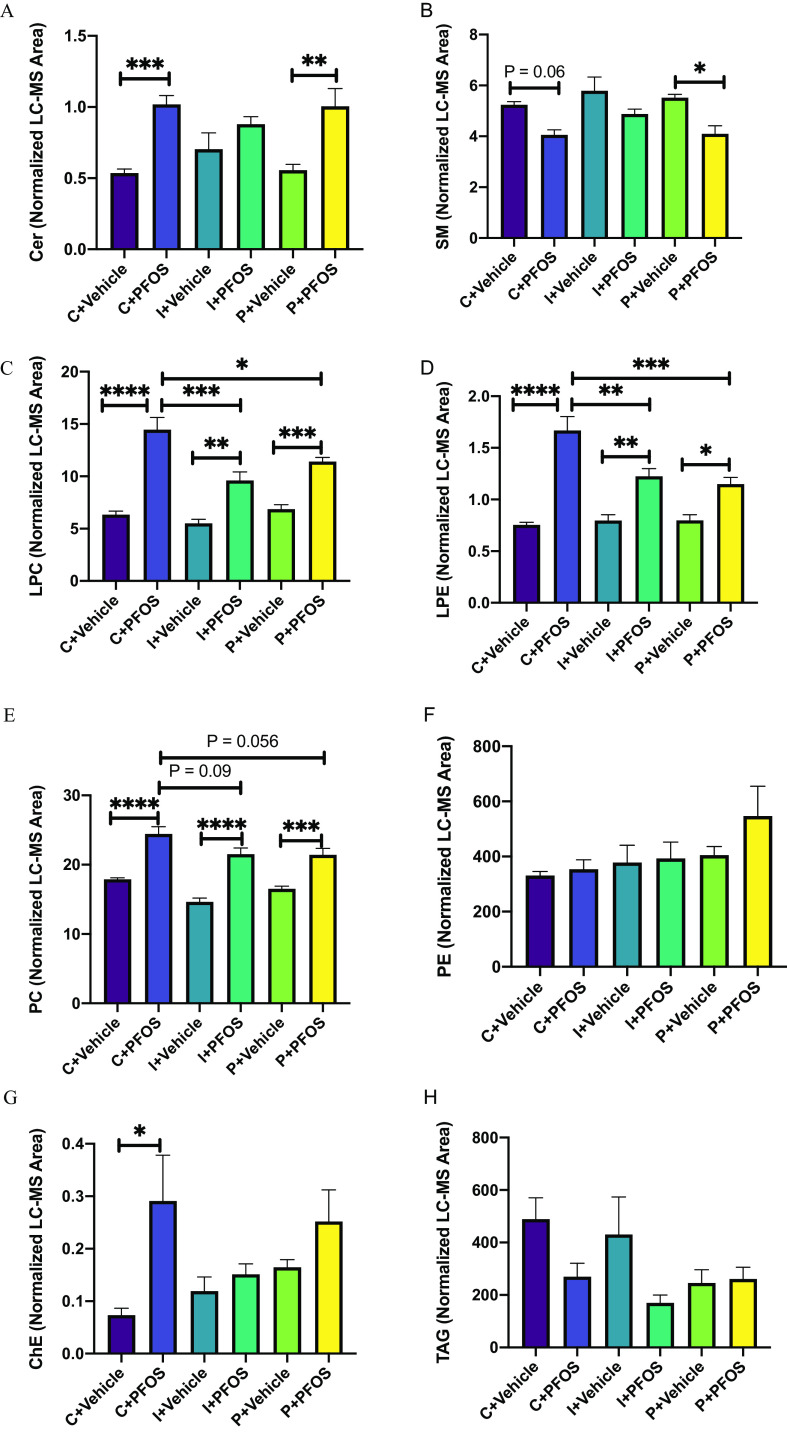
The LC-MS–based lipidomic analysis identified a total of 1,651 lipid species in mouse liver. Analysis of lipidomic data (Excel Table S12) by two-way ANOVA revealed a higher abundance of particular classes of hepatic lipids in PFOS-exposed mice regardless of diet (Figure 2A–H), including lysophosphatidylcholine (LPC), lysophosphatidylethanolamine (LPE), and phosphatidylcholine (PC). Comparing PFOS-exposed mice and vehicle-treated mice, significantly higher levels of ceramide (Cer) were observed in control- and pectin-fed groups, but not in the inulin-fed group (Figure 2A). The level of sphingomyelin (SM) was significantly lower after PFOS exposure in control- and pectin-fed mice (Figure 2B). In addition, the PFOS-induced accumulation of hepatic LPC and LPE was ameliorated by both inulin and pectin feeding compared with control diet (Figure 2C,D). Furthermore, the significantly higher cholesterol ester seen in mice on the control diet exposed to PFOS was not observed with PFOS-exposed inulin- and pectin-fed mice (Figure 2G). There were no significant differences in hepatic triacylglycerol (TAG) and phosphatidylethanolamine (PE) after PFOS exposure and different dietary fiber treatments, although there was a trend of lower TAG levels in PFOS-exposed mice in the control- and inulin-fed groups.
Figure 2.
Liver lipid profiles in mice exposed to PFOS and fed with diets supplemented with different fibers (C: cellulose as control, I: inulin, P: pectin). Lipids including sphingolipids (Cer, A; SM, B), lysophospholipids (LPC, C; LPE, D), phospholipids (PC, E; PE, F), cholesterol ester (ChE, G), and a neutral lipid (TAG, H) were analyzed using UHPLC-Q exactive MS. The normalized peak areas of lipid species in each lipid class are summarized. Bars represent of 6–8 mice in each group. Data were compared using two-way ANOVA and Tukey’s post hoc test for multiple comparisons, *; **; ***; and ****. Detailed lipidomic data are listed in Excel Table S12. Note: ANOVA, analysis of variance; Cer, ceramide; LC-MS, liquid chromatography-mass spectrometry; LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PFOS, perfluorooctane sulfonate; SEM, standard error of the mean; SM, sphingomyelin; UHPLC-Q exactive MS, ultra-high-performance liquid chromatography coupled with quadrupole-exactive mass spectrometer; TAG, triacylglycerol.
Effects of PFOS Exposure and Dietary Fiber Intervention on Mouse Liver Sphingolipids
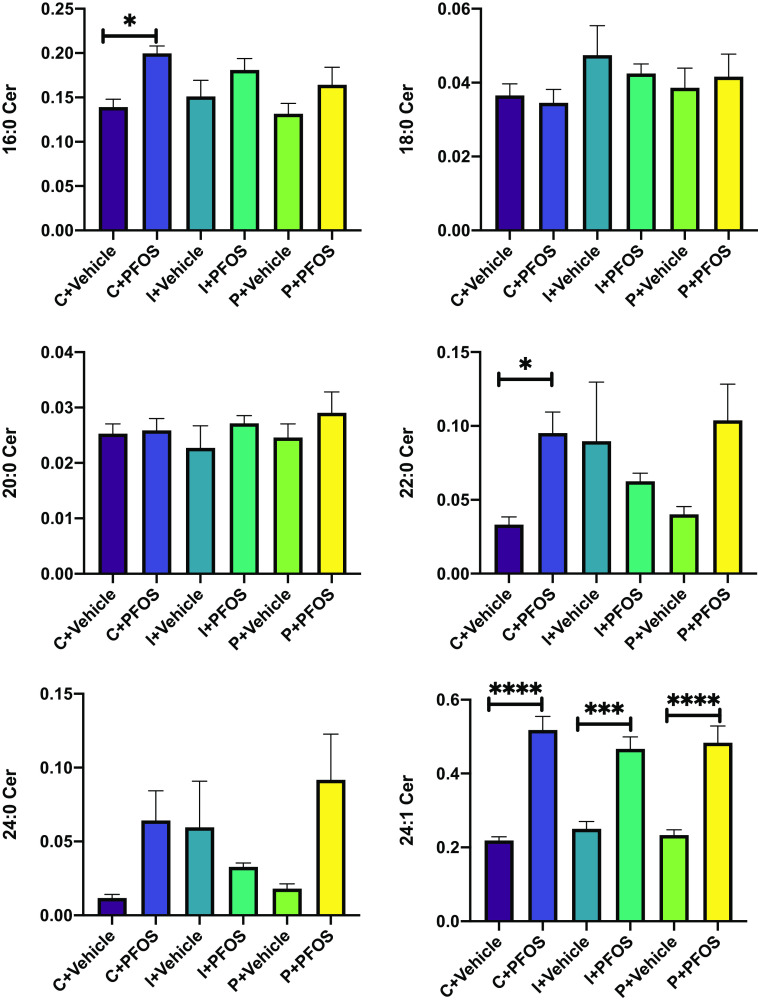
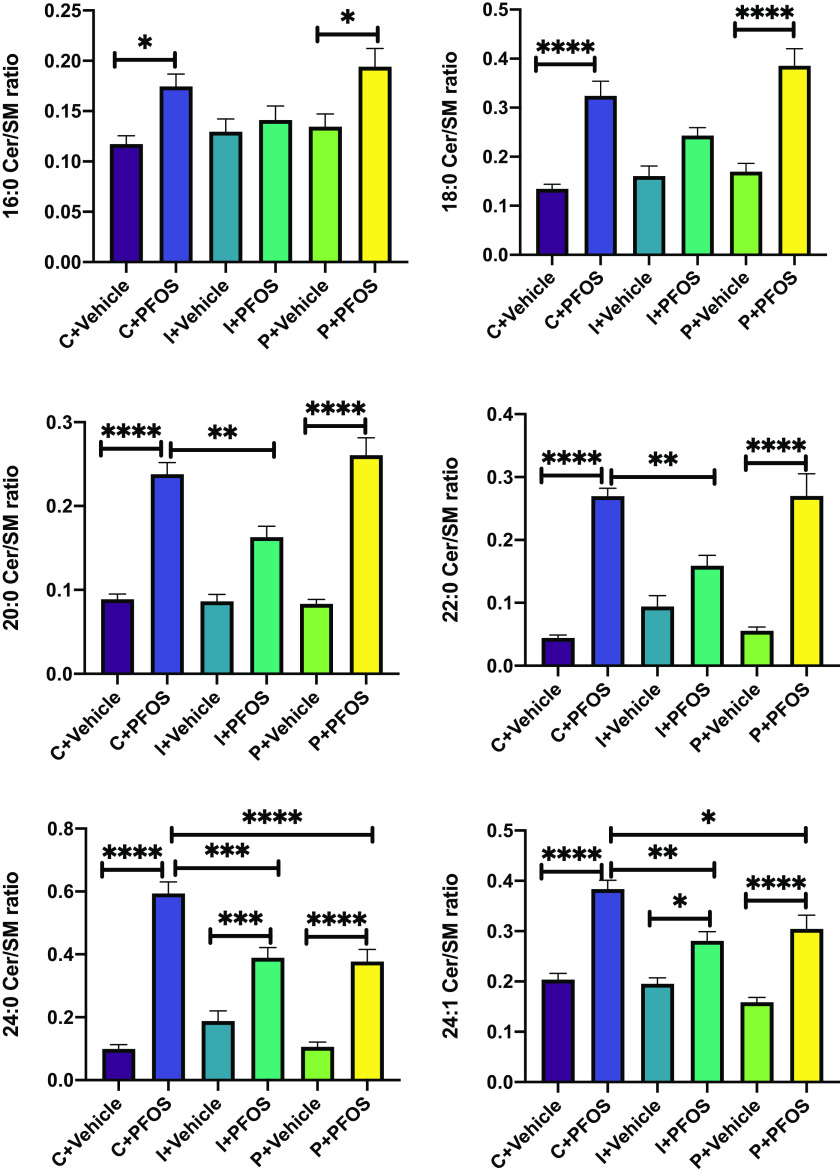
Ceramide and sphingomyelin are two major sphingolipids that form a complex class of bioactive lipids. Certain species with distinct acyl chains are closely associated with the development of metabolic syndrome.63 The analysis of individual ceramides indicated that Cer 24:1 was significantly higher in PFOS-exposed mice regardless of diet (Figure 3; Excel Table S12). Cer 16:0 and 22:0 were both significantly higher in PFOS-exposed mice than vehicle-treated mice, but only in the cellulose control diet group. There was a trend for higher Cer 16:0 after PFOS exposure in the inulin and pectin groups, but it did not reach statistical significance (Figure 3). Similarly, there was a trend for higher Cer 22:0 in the pectin group that did not reach statistical significance (Figure 3). Sphingolipid is a major precursor for ceramide, and hydrolysis of sphingolipid by sphingomylinase is one of the major biosynthesis pathways for ceramide.64 The ratio between ceramide and sphingomyelin (Cer/SM), an indicator of sphingomyelinase activity,65 was significantly higher in PFOS-exposed mice than in vehicle-treated mice. This effect was partially attenuated in inulin-fed mice (Figure 4; Excel Table S13), especially for long-chain species (C18:0, C20:0, C22:0), whose Cer/SM ratios were lower compared with control diet-fed mice ( vs. ). In addition, a higher C16:0 Cer/SM ratio in response to PFOS exposure was observed in control and pectin-fed mice, but not in the inulin-fed group. For very long-chain species (C24:0 and C24:1), although the Cer/SM ratio was higher in all three dietary groups after PFOS exposure, significantly lower values were observed in both inulin- and pectin-fed mice compared with the control group (Figure 4).
Figure 3.
Liver ceramide levels in mice exposed to PFOS and fed diets supplemented with different fibers (C: cellulose as control, I: inulin, P: pectin). Ceramides were analyzed using UHPLC-Q Exactive MS. Bars represent of 6–8 mice in each group. Data were compared using two-way ANOVA and Tukey test for multiple comparisons, *; **; ***; and ****. Detailed ceramide data are listed in Excel Table S12. Note: ANOVA, analysis of variance; PFOS, perfluorooctane sulfonate; SEM, standard error of the mean; UHPLC-Q exactive MS, ultra-high-performance liquid chromatography coupled with quadrupole-exactive mass spectrometer.
Figure 4.
Liver Cer/SM ratio in mice exposed to PFOS and fed diets supplemented with different fibers (C: cellulose as control, I: inulin, P: pectin). Bars represent of 6–8 mice in each group. Data were compared using two-way ANOVA and Tukey test for multiple comparisons, *; **; ***; and ****. Detailed data of liver Cer/SM ratio are listed in Excel Table S13. Note: ANOVA, analysis of variance; Cer, ceramide; PFOS, perfluorooctane sulfonate; SEM, standard error of the mean; SM, sphingomyelin.
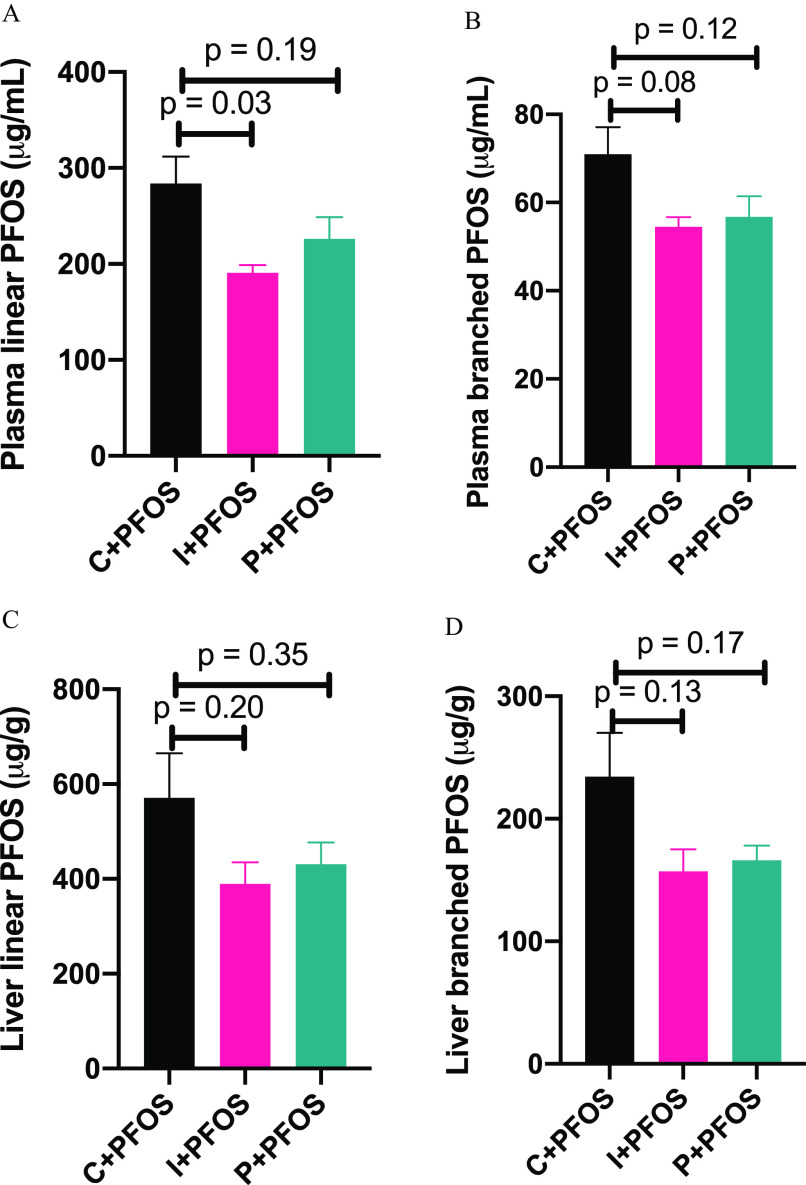
Effects of Dietary Fiber Intervention on Plasma and Liver PFOS Levels
PFOS levels in plasma and liver were analyzed using the LC-MS method (Figure 5; Excel Table S14). It was found that in control diet-fed mice, the levels of linear and branched PFOS in plasma were and , respectively, and the levels were at and in liver tissue. The plasma level of linear PFOS was lower by 33% in inulin-fed mice compared with control diet-fed mice, and the level was lower by 20% in pectin-fed mice, although not statistically significant (; Figure 5A). There were trends that plasma levels of branched PFOS were lower in the inulin- and pectin-diet-fed mice, with , respectively (Figure 5B). Similarly, trends of lower hepatic linear and branched PFOS were observed in inulin- and pectin-diet-fed mice (Figure 5C,D).
Figure 5.
Plasma and liver levels of (A,C) linear and (B,D) branched PFOS in mice fed with cellulose- (control), inulin-, and pectin-supplemented diets. Bars represent of 6–8 mice in each group and -values are labeled on the chart. Data were compared using one-way ANOVA and Tukey’s post hoc test for multiple comparisons. Detailed data of PFOS levels are listed in Excel Table S14. Note: ANOVA, analysis of variance; PFOS, perfluorooctane sulfonate; SEM, standard error of the mean.
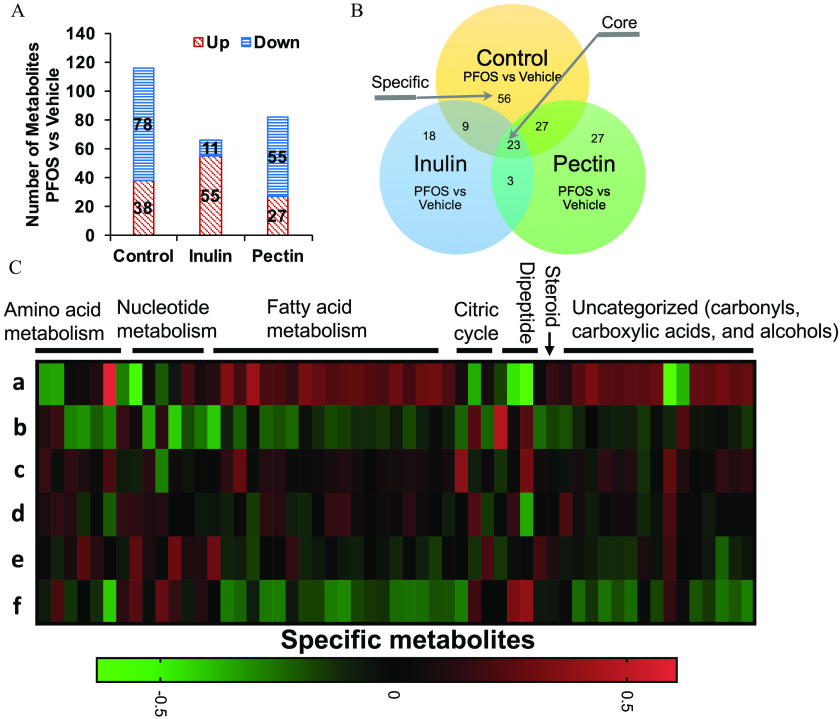
Effects of PFOS Exposure and Dietary Fiber Intervention on the Mouse Liver Metabolome
The LC-MS–based metabolomic analysis identified a total of 516 metabolites in mouse liver (Excel Table S5). Metabolites found in QC samples had a median RSD of 8%, indicating the analysis was highly reproducible. To identify potential metabolites that contribute to the divergent effects of diet and PFOS exposure on the mouse liver metabolome, a PLS-DA was carried out. In all three diet-treated groups, there were clear differences between the PFOS- and vehicle-treated mice (Figure 6A), indicating considerable variations in the hepatic metabolite composition after PFOS exposure regardless of diet. Patterns of metabolite abundance in the heatmap of liver metabolites indicated apparent impacts of the PFOS exposure and of the diet composition (Figure S3, full metabolomic data is listed in Table S5). Two different approaches were used to analyze these data.
Figure 6.
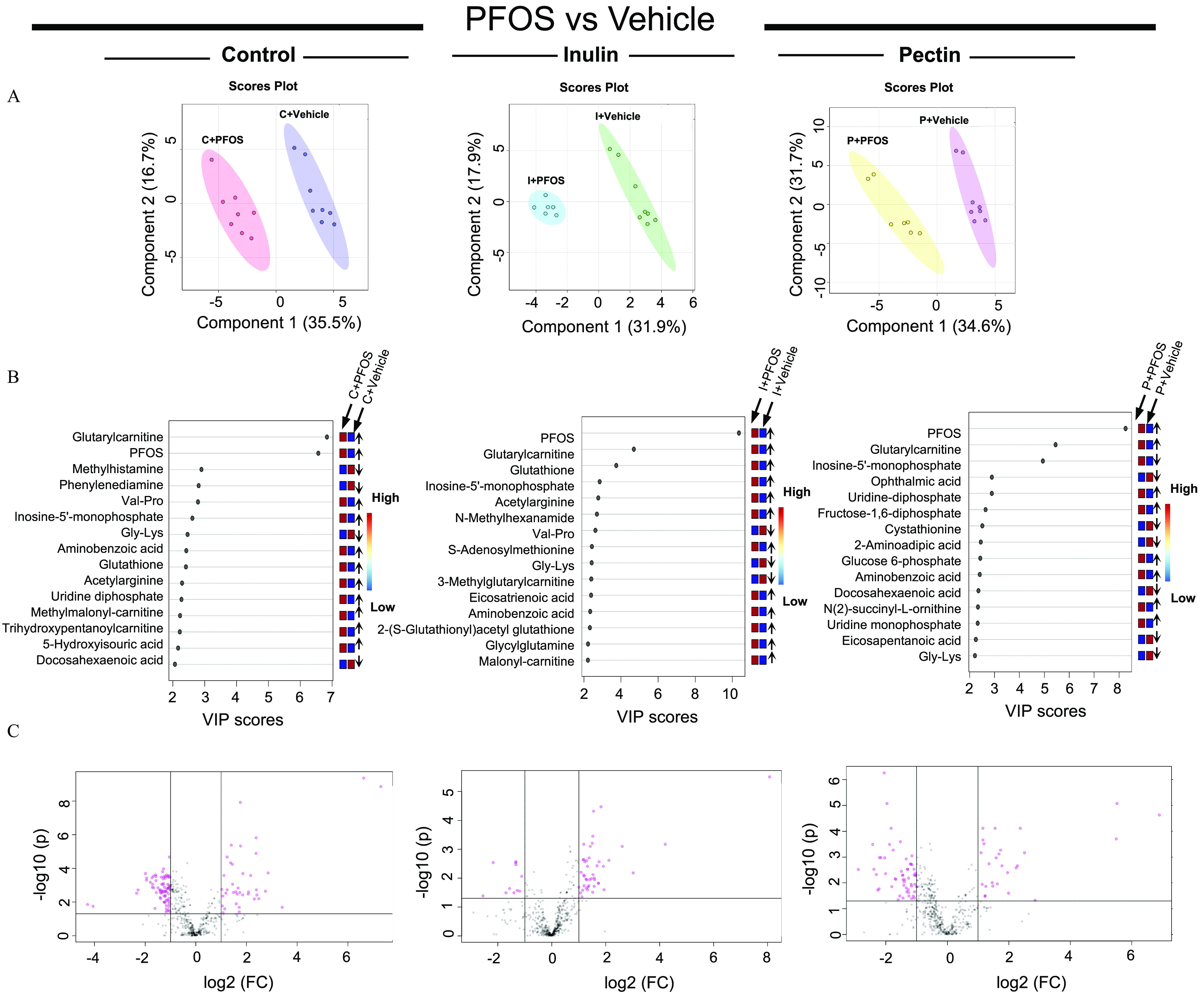
Metabolomic analysis of liver samples from mice exposed to PFOS and fed with diets supplemented with cellulose (control), inulin, or pectin. (A) Liver metabolite profiles were analyzed by PLS-DA and significant separations between PFOS and vehicle treatments were observed in all three dietary groups (). (B) Main metabolites responsible for separation between vehicle- and PFOS-treated groups by PLS-DA were analyzed by variable in projection (VIP). Up arrow indicates the level of the metabolite was higher in PFOS-exposed mice than that in the vehicle group, down arrow indicates the level of the metabolite was lower in PFOS-exposed mice than that in the vehicle group. Detailed data of VIP scores are listed in Excel Table S15. (C) Analysis of statistically significant metabolites were identified by volcano plot (, and ). Detailed metabolomic data are listed in Excel Table S5. Note: FC, fold change; Gly, glycine; Lys, lysine; PFOS, perfluorooctane sulfonate; PLS-DA, partial least-squares discriminant analysis; Pro, proline; Val, valine.
First, PLS-DA identified main metabolites responsible for separation between vehicle and PFOS treated groups, which were further analyzed by VIP, and the importance of individual metabolites that altered by PFOS treatment in different dietary groups were ranked by VIP scores (Figure 6B; Excel Table S15). In addition to PFOS, glutarylcarnitine was scored the highest across all three dietary groups, indicating that it was a main contributor to the separation of PFOS-vehicle groups. Other metabolites that contributed to the separation across the three dietary groups included inosine monophosphate (IMP), Gly-Lys, and aminobenzoic acid. Interestingly, the ranking (VIP score) of metabolite was not equivalent between diets although the overall pattern of metabolic profile change was similar, suggesting that metabolic responses to PFOS were specific to dietary fiber types.
Second, analysis of statistically significant metabolites identified by volcano plot ( and ; Figure 6C) revealed the overall differences in metabolite profiles. It was found that control diet-fed mice had the highest number of differentially expressed metabolites after PFOS exposure among all three dietary groups (Figure 7A,B). As shown in the Venn diagrams (Figure 7B), 23 core metabolites that were different between PFOS- and control diet-fed mice in all three diet groups were identified (Table 1). Those included metabolites from amino acid metabolism, nucleotide metabolism, and fatty acid biosynthesis (Figure S4, Excel Table S5). Among these, the 4 major differential metabolites identified in the VIP analysis were also detected. Importantly, 55 specific metabolites that were altered by PFOS exposure in the control diet group but not in the inulin- and pectin-fed groups were identified (Figure 7C; Excel Table S16) that were enriched into pathways of amino acid metabolism, nucleotide metabolism, fatty acid metabolism, and the citric cycle.
Figure 7.
Liver metabolome in mice exposed to PFOS and fed with diets supplemented with different fibers. (A) The total number of different hepatic metabolites in mice exposed to PFOS and treated with cellulose- (control), inulin-, and pectin-supplemented diets (). (B) Venn diagrams depicting the distribution of metabolites in the liver of mice on the cellulose, inulin, or pectin diet in response to PFOS exposure. Shared elements constitute “core” metabolites and are associated with PFOS exposure regardless of diets. “Specific” metabolites are associated with PFOS exposure in the cellulose diet group only but not in inulin- or pectin-fed groups. (C) Heatmap of liver metabolomic profiling of mice fed with diets supplemented with cellulose (b vs. a), inulin (d vs. c), and pectin (f vs. e) compared with mice treated with vehicle. Log fold changes of profiled metabolites are shown. Metabolites are ordered within each category: amino acid metabolism, nucleotide metabolism, fatty acid metabolism, citric cycle, dipeptide, steroid, and uncategorized (carbonyls, carboxylic acids, and alcohols). Detailed metabolomic data are listed in Excel Table S5. Heatmap data are listed in Excel Table S16. Note: a, ; b, ; c, ; d, ; e, ; f, ; PFOS, perfluorooctane sulfonate.
Table 1.
Metabolomic analysis of liver samples from mice exposed to PFOS and fed with diets supplemented with cellulose (control), inulin, or pectin.
| Metabolite | Category | MW | Rt (min) | Fold difference: PFOS vs. vehicle | ||
|---|---|---|---|---|---|---|
| Cellulose | Inulin | Pectin | ||||
| Glutarylcarnitine | Short-chain acylcarnitine | 275.136 | 5.7 | 158.0 | 18.4 | 45.9 |
| Malonylcarnitine | Short-chain acylcarnitine | 247.105 | 7.1 | 3.4 | 3.5 | 2.9 |
| Pimelylcarnitine | Short-chain acylcarnitine | 303.168 | 2.4 | 0.3 | 0.3 | 0.3 |
| Inosine-5′-monophosphate | Nucleotide metabolism | 348.046 | 9.7 | 7.2 | 6.1 | 45.1 |
| Inositol cyclic phosphate | Nucleotide metabolism | 242.019 | 10.9 | 3.2 | 2.3 | 3.3 |
| Uridine monophosphate | Nucleotide metabolism | 324.035 | 9.2 | 2.2 | 2.9 | 5.2 |
| Uridine diphosphate | Nucleotide metabolism | 404.001 | 10.4 | 4.5 | 2.8 | 5.7 |
| UDP--acetylglucosamine | Nucleotide metabolism | 607.080 | 9.3 | 2.8 | 2.6 | 3.4 |
| Cytidine | Nucleotide metabolism | 243.085 | 6.4 | 0.3 | 0.4 | 0.4 |
| Cytosine | Nucleotide metabolism | 111.043 | 6.4 | 0.3 | 0.4 | 0.4 |
| Isocytosine | Nucleotide metabolism | 111.043 | 5.2 | 0.3 | 0.4 | 0.4 |
| Nicotinic acid | Nucleotide metabolism | 123.032 | 2.7 | 3.0 | 2.6 | 2.4 |
| Acetylarginine | Arginine metabolism | 216.122 | 9.0 | 5.2 | 4.3 | 2.2 |
| Aceglutamide | Glutamine metabolism | 188.079 | 4.7 | 2.4 | 2.9 | 2.3 |
| -methylglutathione | Glutathione metabolism | 321.099 | 7.0 | 3.6 | 2.9 | 2.7 |
| -acetyl-aminoadipic acid | Lysine metabolism | 203.079 | 9.3 | 3.0 | 2.4 | 3.0 |
| Aminobenzoic acid | Tryptophan metabolism | 137.048 | 9.3 | 3.6 | 3.0 | 3.9 |
| Aminobutyric acid | Alanine metabolism | 103.064 | 7.8 | 0.4 | 0.3 | 0.3 |
| Dihydroxyphenylacetic acid | Amine metabolism | 168.042 | 10.9 | 2.7 | 2.3 | 3.2 |
| Glycine hydroxyproline dipeptide (Gly-Hyp) | Dipeptide | 188.080 | 2.4 | 2.6 | 2.4 | 2.2 |
| Gly-Lys | Dipeptide | 203.127 | 6.3 | 0.2 | 0.2 | 0.2 |
| Hypotaurocyamine | Other | 151.041 | 9.7 | 0.5 | 0.4 | 0.2 |
| Benzoxazolone | Other | 135.032 | 3.8 | 0.4 | 0.5 | 0.4 |
Note: Core metabolite differences in response to PFOS exposure across all three dietary groups are listed (). The fold difference data were obtained from volcano plot results as determined by using MetaboAnalyst (version 5.0). Detailed metabolomic data are listed in Excel Table S5. Lys, lysine; MW, molecular weight; PFOS, perfluorooctane sulfonate; Rt, retention time; UDP, uridine diphosphate.
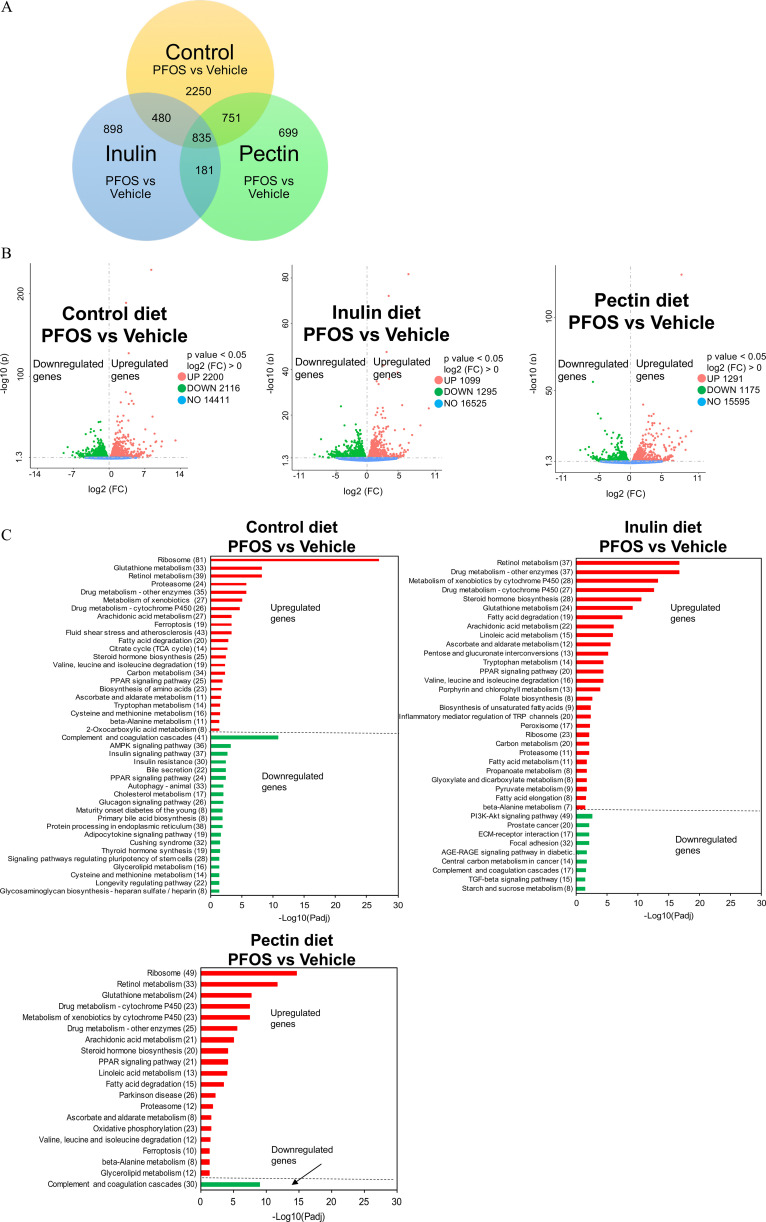
Effects of PFOS Exposure and Dietary Fiber Intervention on the Mouse Liver Transcriptome
Soluble fiber feeding paradigms were associated with effects on liver/body weight ratio (inulin only) and liver metabolism after PFOS exposure. To gain insight into the underlying mechanisms, we conducted RNA sequencing analysis of liver samples (Excel Tables S2–S4). As shown in the Venn diagram and volcano plots (Figure 8A,B), pairwise comparisons of PFOS-vehicle treatment identified 4,316, 2,394, and 2,466 differentially expressed gene transcripts in the control and the inulin- and pectin-fed groups, respectively. In addition, a total of 835 shared genes that were altered by PFOS exposure but independent of diet were identified in the liver transcriptome (Figure 8A).
Figure 8.
Liver transcriptome in mice exposed to PFOS and fed with diets supplemented with different fibers (). (A) Venn diagrams depicting the distribution of transcriptions in the liver of mice on cellulose (control), inulin, or pectin diet in response to PFOS exposure. (B) Analysis of statistically significant transcripts identified by volcano plot. (C) Enrichment analysis of the differentially expressed genes to identify biological functions or pathways that differ between PFOS-exposed and control mice in three dietary groups. The horizontal axis is (Padj) of the significantly enriched pathway and the vertical axis is the enriched pathway, and the number of differentially expressed genes is shown in the parentheses. Detailed transcriptomic data are listed in Excel Tables S2–S4. Note: AGE, advanced glycation end products; Akt, protein kinase B; AMPK, adenosine monophosphate-activated protein kinase; ECM, extracellular matrix; FC, fold change; Padj, adjusted -value; PFOS, perfluorooctane sulfonate; PI3K, phosphatidylinositol-3-kinase; PPAR, peroxisome proliferator-activated receptor; RAGE, receptor for advance glycation end products; TCA, tricarboxylic acid; TGF, transforming growth factor; TRP, transient receptor potential.
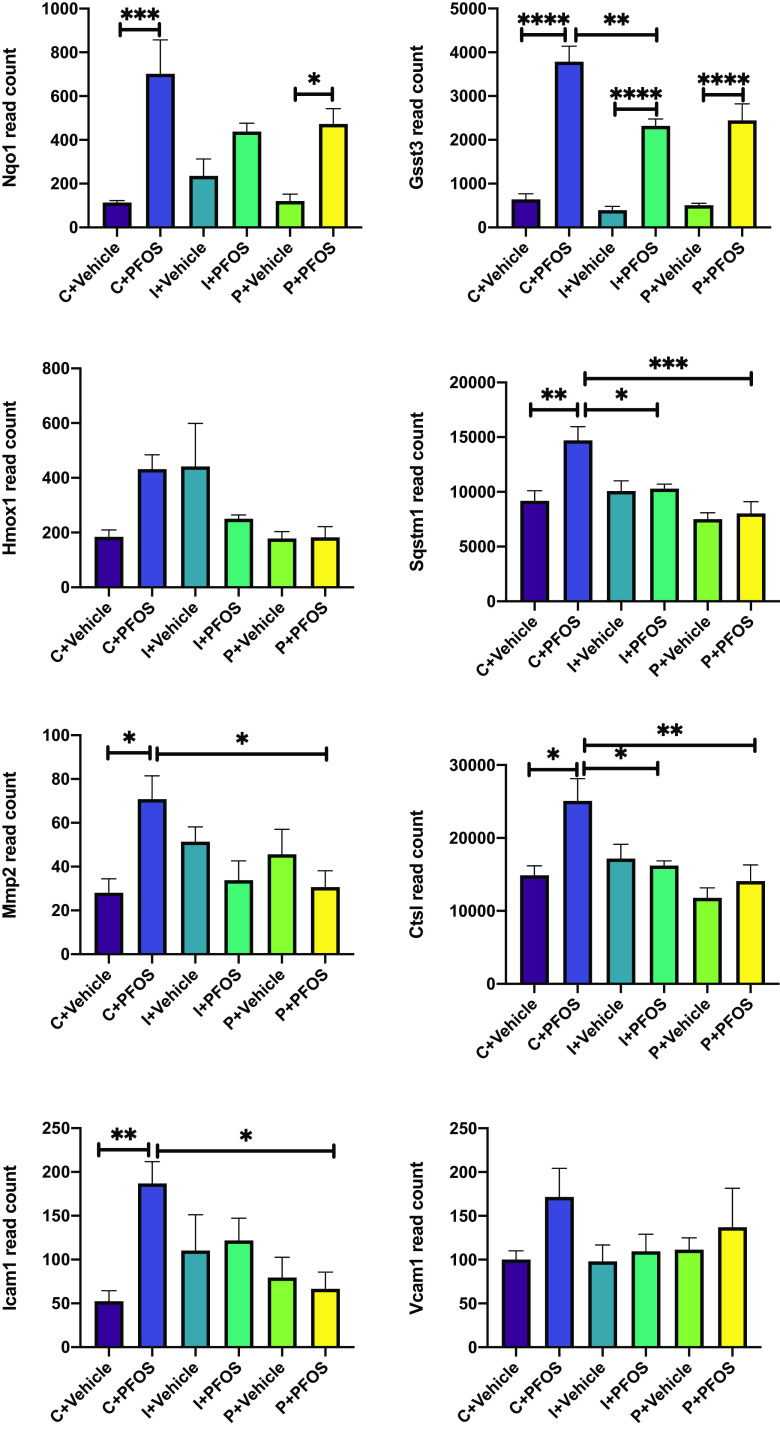
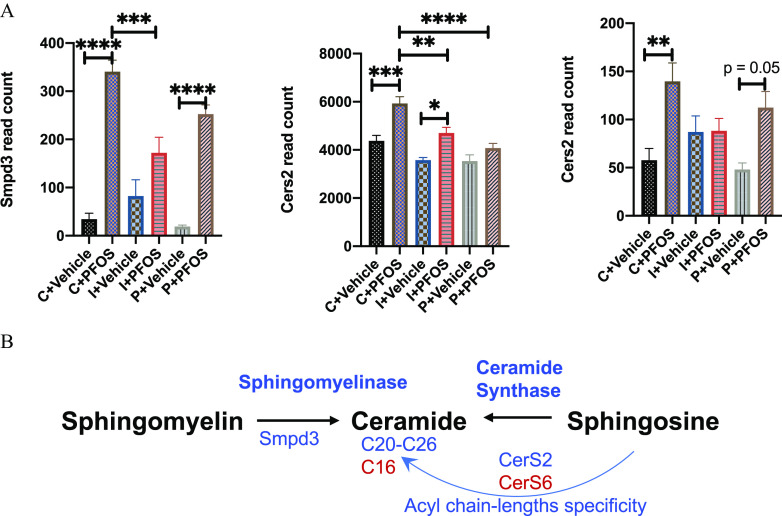
Enrichment analysis showed that control diet-fed mice exposed to PFOS demonstrated a higher expression of genes associated with ribosome, amino acid metabolism, PPAR pathway, and fatty acid and lipid metabolism (Figure 8C). The main genes with lower expression were associated with pathways of complement and coagulation cascades, adenosine monophosphate–activated protein kinase (AMPK) signaling, insulin pathway, and bile acids biosynthesis and excretion. Although mice consuming inulin- and pectin-supplemented diets showed some similarities in the affected pathways as those in the control diet, the number of differentially expressed genes in each pathway were less (Figure 8C), namely, those associated with the ribosome [control/inulin/pectin (C/I/P): 81/37/49 genes], glutathione metabolism (C/I/P: 33/24/24 genes), retinol metabolism (C/I/P: 39/37/33 genes), protostome (C/I/P: 24/11/12 genes), arachidonic acid metabolism (C/I/P: 27/22/21 genes), fatty acid degradation (C/I/P: 20/19/15 genes), branched chain amino acid metabolism (C/I/P: 19/16/12 genes), and PPAR signaling pathway (C/I/P: 49/20/21 genes). Genes with lower expression after PFOS exposure in mice fed with the three diets showed the overlapping KEGG pathway of complement and coagulation cascades. Notably, the number of enriched genes in this pathway were dramatically less in inulin- and pectin-fed mice compared with the control group (C/I/P: 41/17/30 genes). Genes associated with the ferroptosis pathway was up-regulated after PFOS exposure in control- and pectin-fed mice but not in inulin-fed mice. Genes associated with cysteine and methionine metabolism were significantly different in control diet-fed mice exposed to PFOS, with 16 genes up-regulated and 14 genes down-regulated, but these genes were not significantly different in inulin- and pectin-fed mice. Furthermore, 43 up-regulated genes were enriched into the pathway of fluid shear stress and atherosclerosis in control mice after PFOS exposure, whereas this pathway was not observed as significantly different in the inulin- and pectin-fed groups. Results of the two-way ANOVA of differentially expressed atherosclerosis-related genes in response to PFOS exposure in the three diets fed mice are shown in Figure 9, including anti-atherogenesis (Nqo1, Gstt3, Hmox1, and Sqstm1) and pro-atherogenesis genes (Mmp2, Ctsl, Icam1, and Vcam1). The AMPK signaling pathway, related genes of which were significantly down-regulated in PFOS-exposed mice, was protected in the inulin- and pectin-fed groups. The major genes involved in ceramide metabolism that were higher after PFOS exposure included Smpd3, CerS2, and CerS6 (Figure 10). Hepatic gene expression of Smpd3 was significant lower in the inulin-fed mice. The effects of PFOS on CerS2 and CerS6 were either diminished or protected in the inulin- and pectin-fed mice (Figure 10).
Figure 9.
Hepatic expression of anti-atherogenesis (Nqo1, Gstt3, Hmox1, and Sqstm1) and pro-atherogenesis genes (Mmp2, Ctsl, Icam1, and Vcam1) determined by transcriptomics. Bars represent of gene read count for 5 mice in each group. Data were compared using two-way ANOVA and Tukey’s post hoc test for multiple comparisons, *; **; ***; and ****. Detailed transcriptomic data are listed in Excel Tables S2–S4. Note: ANOVA, analysis of variance; PFOS, perfluorooctane sulfonate; SEM, standard error of the mean.
Figure 10.
Hepatic expression of ceramide biosynthesis-related genes determined by transcriptomics. (A) Gene expression of Smpd3, CerS2, and CerS6 determined by transcriptomics. Bars represent of gene read count for 5 mice in each group. (B) Biosynthetic pathways of ceramide that were modulated by PFOS exposure and dietary fibers. Data were compared using two-way ANOVA and Tukey’s post hoc test for multiple comparisons, *; **; ***; and ****. Detailed transcriptomic data are listed in Excel Tables S2–S4. Note: ANOVA, analysis of variance; PFOS, perfluorooctane sulfonate; SEM, standard error of the mean; Smpd3, sphingomyelin phosphodiesterase 3.
Effects of PFOS Exposure and Dietary Fiber Intervention on the Cecal Content Microbiome
Relative abundance analysis demonstrated that Bacteroidetes was the dominant phylum in mice from the six groups (Excel Table S6). Actinobacteria was the major phylum whose abundance was significantly higher in inulin-fed mice than the control diet group treated with the vehicle (Figure 11A), and there were no significant differences among other groups. The cecal content microbial alpha-diversity at genus level was evaluated using the Shannon Diversity Index. Pairwise comparisons indicate that alpha-diversity of the inulin group was remarkably higher than the pectin group () in the vehicle-treated mice. In contrast, no significant shifts were observed among the other experimental groups (Figure 11B; Excel Table S17). The PCoA using Bray–Curtis distance (beta-diversity) showed a separation of the inulin treatment from other dietary groups at the genus level (Figure 11C). PERMANOVA showed that the beta-diversity derived from the inulin differed from other dietary interventions (), supporting the notion that feeding inulin led to significant changes in the microbiome community compared with feeding pectin or cellulose.
Figure 11.
![Figure 11A is a set of six stacked bar graph, plotting Relative Abundance, ranging from 0 to 100 in increments of 50 (y-axis) across uppercase c plus vehicle, ranging as lowercase a 1, lowercase a 2, lowercase a 3, lowercase a 4, and lowercase a 5; uppercase c plus P F O S, ranging as lowercase b 1, lowercase b 2, lowercase b 3, lowercase b 4, and lowercase b 5; uppercase i plus vehicle, ranging as lowercase c 1, lowercase c 2, lowercase c 3, lowercase c 4, and lowercase c 5; uppercase i plus P F O S, ranging as lowercase d 1, lowercase d 2, lowercase d 3, lowercase d 4, and lowercase d 5; uppercase p plus vehicle, ranging as lowercase e 1, lowercase e 2, lowercase e 3, lowercase e 4, and lowercase e 5; and uppercase p plus P F O S, ranging as lowercase f 1, lowercase f 2, lowercase f 3, lowercase f 4, and lowercase f 5 (x-axis) for Cyanobacteria, Chordata, Actinobacteria, Proteobacteria, Firmicutes, Verrucomicrobia, and Bacteroidetes. Figure 11B is a box and whisker plot, plotting alpha diversity, ranging from 0 to 5 in unit increments (y-axis) across uppercase c plus vehicle, uppercase c plus P F O S, uppercase i plus vehicle, uppercase i plus P F O S, uppercase p plus vehicle, and uppercase p plus P F O S (x-axis). Figure 11C is an dot graph titled P Co A, plotting Axis 2 [23 percent], ranging from negative 30 to 20 in increments of 10 (y-axis) across Axis 1 [74.4 percent], ranging from negative 60 to 40 in increments of 20 (x-axis) for uppercase a 1 to uppercase a 5: uppercase c plus vehicle; uppercase b 1 to uppercase b 5: uppercase c plus P F O S; uppercase c 1 to uppercase c 5: uppercase i plus vehicle; uppercase d 1 to uppercase d 5: uppercase p plus vehicle; and uppercase f 1 to uppercase f 5: uppercase p plus P F O S.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/b6ef/9635512/32a796509685/ehp11360_f11.jpg)
Cecal content microbial communities in mice exposed to PFOS and fed with diets supplemented with different fibers. Shotgun metagenomic analysis of cecal contents of mice fed with different diets containing cellulose (control), inulin (I), or pectin (P), and exposed to PFOS by drinking water (). (A) The major bacterial phyla presented in mice cecum content samples, shown as relative abundance in each sample (Excel Table S6). (B) Alpha-diversity index of microbial communities at the genus level. Data were analyzed by two-way ANOVA and presented as a box and whisker plot. Detailed data of alpha-diversity index are listed in Excel Table S17. * (midline, median; box limits, upper and lower quartiles; whiskers, 10th and 90th percentiles). (C) Principal coordinates analysis (PCoA) at the genus level comparing microbial beta-diversity (Bray–Curtis distance) among different groups. Note: ANOVA, analysis of variance; PFOS, perfluorooctane sulfonate.
To identify the differences in specific bacterial taxa in response to PFOS exposure and dietary interventions, LEfSe was used to compare the cecal microbiota composition (Figure 12), and the LDA score was used to discriminate specific taxa between different groups at the genus level (Figure 13; Excel Table S18). It was found that fewer bacterial taxa were different after PFOS exposure (Figures 12 and 13) in inulin- and pectin-fed mice compared with the control diet group. For control diet groups, compared with the vehicle-treated mice, PFOS-exposed mice had a lower abundance of Alistipes, Oscillibacter, Flavonifractor, Flintibacter, Dysosmobacter, Paenibacillus, Citrobacter, Intestinimonas, Acutalibacter, and Streptomyces (), which mostly belonged to the phylum of Firmucutes and Actinobacteria. Higher abundances of Duncaniella, Muribaculum, Prevotella, and Sodaliphilus were observed in control diet-fed mice after PFOS exposure (Figure 13A). On the contrary, in inulin-fed mice, abundances of Flavonifractor, Dysosmobacter, Streptomyces, Ruminococcus, and Faecalitalea were higher in PFOS-exposed mice (Figure 13B). In pectin-fed mice, abundances of genus Paraprevotella and Staphylococcus were found to be lower in PFOS-exposed mice than in the vehicle control group (Figure 13C).
Figure 12.
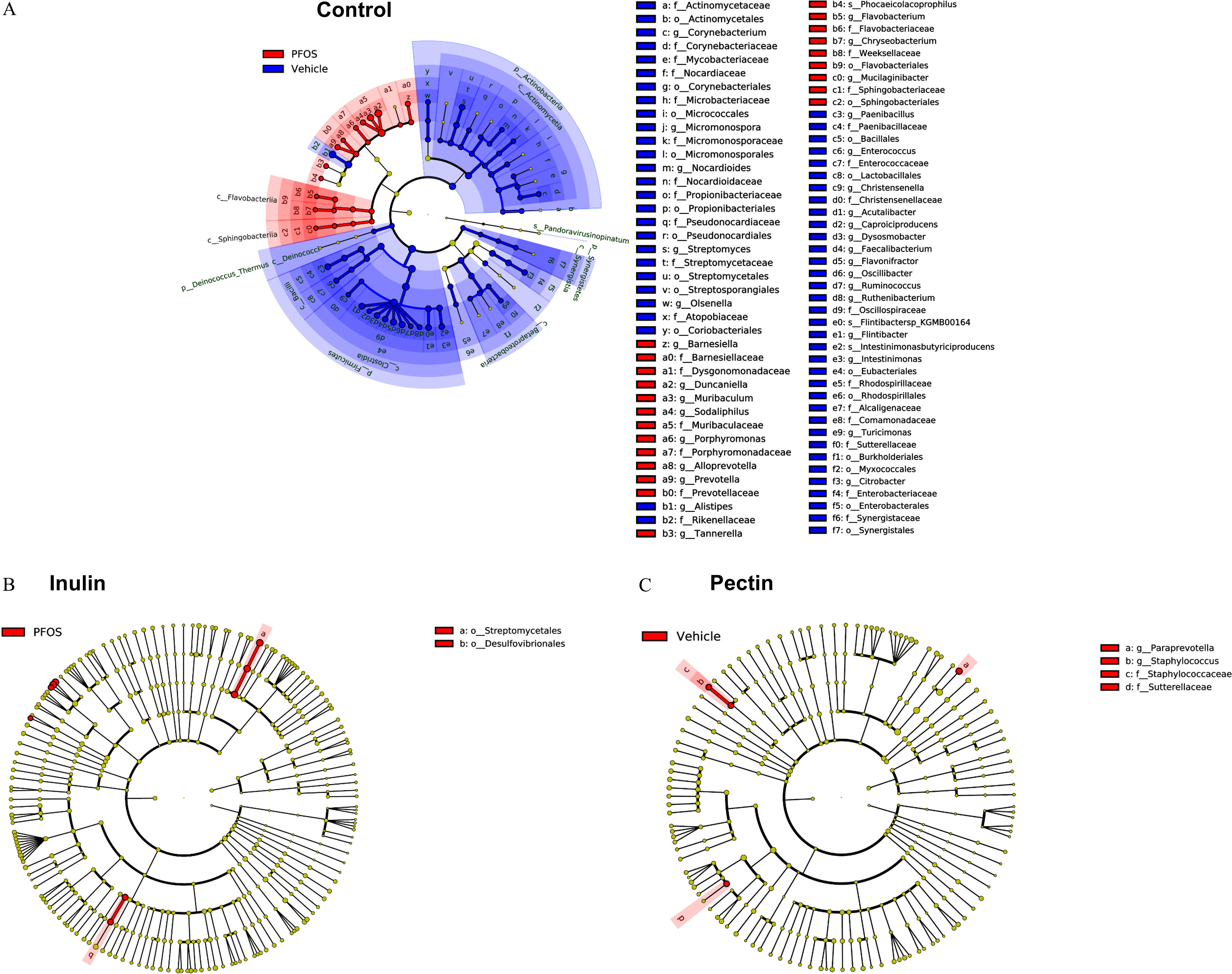
Cladograms of cecal content microbiome generated using linear discriminant analysis (LDA) effect size (LEfSe) analysis. Relative abundances of taxa were compared between groups using LEfSe with the default -value () and the LDA score of 2.0. Comparison results are presented for PFOS vs. vehicle in each dietary group. Colors distinguish taxa differences between PFOS and vehicle treatments. (A) control diet; (B) inulin diet; and (C) pectin diet. Note: PFOS, perfluorooctane sulfonate.
Figure 13.
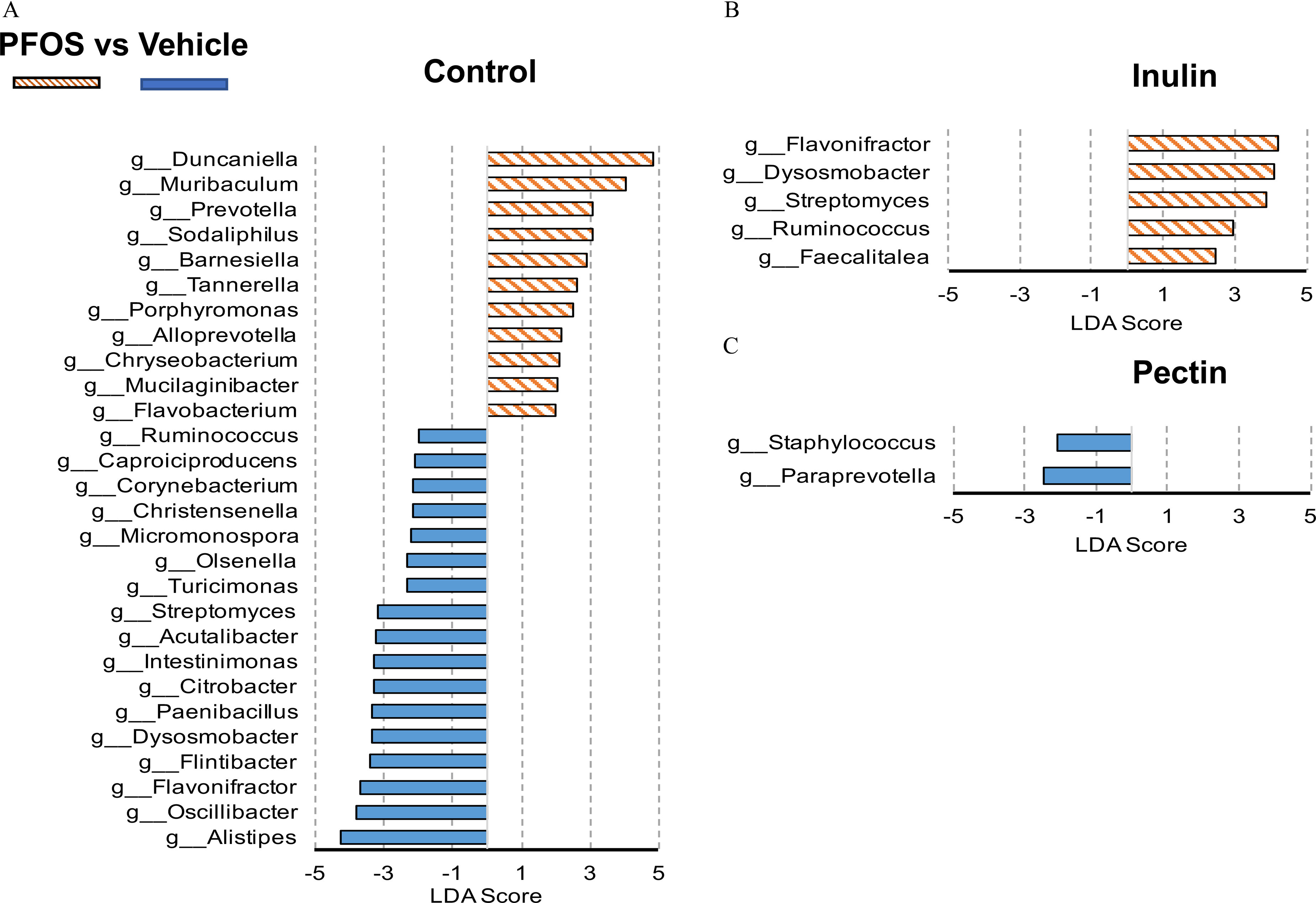
Linear discriminant analysis (LDA) scores of differentially abundant taxa in cecal content between the PFOS- and vehicle-treated mice fed the (A) control diet, (B) inulin diet, or (C) pectin diet using the linear discriminant analysis effect size method. Only taxa meeting an LDA significant threshold of 2 and are shown. The mice were fed with a diet supplemented with dietary fibers. Detailed LDA score data are listed in Excel Table S18. Note: PFOS, perfluorooctane sulfonate.
The effects of dietary fiber on bacterial taxa were investigated by comparing vehicle-treated mice fed with different diets (without PFOS exposure). LEfSe suggested that Bifidobacterium (), Duncaniella (), and Muribaculum () were the top three genera that were up-regulated by inulin feeding compared with the control diet group. The two-way ANOVA revealed a significant effect of PFOS-inulin fiber interaction on the relative abundance of Muribaculum (Figure S5). Duncaniella (), Muribaculum (), and Bifidobacterium () are the major genera that were up-regulated in the pectin-fed mice. The two-way ANOVA revealed a significant interaction effect between PFOS and pectin fiber on the relative abundance of Duncaniella and Muribaculum (Figure S5).
Discussion
PFOS has been considered to disturb lipid metabolism owing to its structural similarity to fatty acids.66 Some in vitro studies have demonstrated that PFOS elicited its hepatotoxicity by activation of the PPAR pathway.29,67 Other studies have suggested that PFOS toxicity may not be relevant to the transcription of PPAR; for example, Wan et al.22 reported that PFOS can inhibit of fatty acids in mice by up-regulating the hepatic gene expression of lipoprotein lipase and fatty acid translocase, as well as the reduction of apolipoprotein B, leading to the hepatic accumulation of fatty acids and triglycerides. Another mouse study suggested that PFOS inhibited the secretion and function of low-density lipoproteins, which may block the exportation of lipids from the liver to peripheral tissues.68 Recently, Sen et al. reported that interactions between PFOS and bile acids led to lipid-related changes in human liver.69 In the present study, we uncovered hepatic lipidomic, metabolomic, and transcriptional differences in PFOS-exposed mice compared with control mice. Liver lipids of Cer, LPC, LPE, PC, and ChE were higher after PFOS exposure (Figure 2), which was consistent with the recently reported results from female BALB/c mice exposed to PFOS that higher levels of hepatic Cer, LPC, LPE, and PC were observed.70 Metabolomic analysis revealed 23 core metabolites that were differentially expressed after PFOS exposure across all dietary groups, with short-chain carnitines being the major affected metabolites (Figure S4), and this may be attributed to the activated PPAR pathway after PFOS exposure. Transcriptomics revealed that PFOS-exposed mice had higher expression of hepatic genes enriched into pathways associated with lipid metabolism (Figure 6C), including PPAR signaling, fatty acid degradation, primary bile acid synthesis, and cholesterol metabolism, which contributed to the hepatic lipid accumulation after PFOS exposure. Taken together, the present study suggests that both PPAR and non-PPAR pathways are involved in the effects of PFOS on liver lipid homeostasis. Interestingly, the number of disturbed genes were lower in inulin- and pectin-fed mice (Figure 8A,B), suggesting that lipids and metabolites that were disturbed during PFOS exposure were rescued by dietary fiber diets.
Ceramides are fatty acid amide derivatives of sphingosine and precursors of complex sphingolipids. Previous in vitro and in vivo studies have shown that the accumulation of ceramides can lead to the activation of signaling pathways that induce hepatic steatosis, insulin resistance, and cardiometabolic disorders,71–74 suggesting a causative role of ceramides in the metabolic disorders. It has been reported that liver ceramide levels were increased in BALB/c mice exposed to PFOS.70 In the liver of NAFLD patients, the levels of hepatic ceramides were positively associated with PFAS exposure.69 Furthermore, PFAS-associated perturbations in the sphingolipid pathway were observed in human and rodent serum/plasma metabolomes.43,75 Our recent study in mice reported that inulin feeding down-regulated ceramide biosynthesis in a hyperlipidemia mouse model through reductions in neutral sphingomyelinase expression and activity.39 In the present study, we found that PFOS exposure resulted in higher ceramide levels in liver tissue (Figure 3), particularly Cer 16:0, 22:0, and 24:1 species, among which Cer 16:0 was reported as the major contributor to metabolic diseases76; moreover, Cer 16:0 is more involved than other ceramide species in liver disease onset and progression.77 Inulin- and pectin-supplemented diets protected against the elevation of ceramide in mouse liver (Figure 3). In addition, the ratio of Cer/SM, an indicator of sphingomyelinase activity, was higher after PFOS exposure, and this effect was ameliorated greatly by the inulin diet and, at a modest extent, by the pectin diet (Figure 4). Compared with the cellulose-fed mice exposed to PFOS, hepatic gene expression of Smpd3 was significantly lower in inulin-fed mice (Figure 10), which was consistent with our previous findings.39 Ceramide synthases (CerS) catalyze the acylation of sphinganine, and the specificity of each CerS toward the acyl CoA chain length used for N-acylation is high.78 CerS6 is responsible for the biosynthesis of C16-ceramide, and CerS2 synthesizes very long acyl chain ceramides (C20–C26).78 PFOS-exposed mice had higher CerS6 and CerS2 gene expression (Figure 10), corresponding to the observed elevation of Cer 16:0, 22:0, and 24:1, respectively (Figure 3). On the other hand, the effects of PFOS on CerS6 and CerS2 were either diminished or protected in soluble fiber-fed mice (Figure 10), which explained the unchanged Cer 16:0 and Cer 22:0 levels in those mice after PFOS challenge (Figure 3).
Previous studies have reported that PFOS exposure was associated with increased cardiovascular disease risk.43,79 In the present study, we found that pathologies associated with fluid shear stress and atherosclerosis were higher after PFOS exposure in control diet-fed mice but not in inulin- and pectin-fed mice (Figure 8). In the three diets fed mice, the atheroprotection genes, including Nqo1, Gstt3, and Sqstm1, were more highly expressed in response to PFOS exposure, which might be related to PFOS-induced redox stress.80 In addition, PFOS exposure was related to a higher expression of Mmp2, Ctsl, and Icam1, whereas inulin and pectin diets protected against PFOS-induced pro-atherogenesis effects as demonstrated by the unaffected Mmp2, Ctsl, and Icam1 genes after PFOS exposure (Figure 9). Two-way ANOVA suggested significant interactions between diet and PFOS exposure for Hox1, Sqstm, Mmp2, Ctsl, and Icam1, indicating that dietary fiber could modulate PFOS-induced changes in atherosclerosis-related genes. These results, together with the lipid and metabolite profiles, suggest that high-fiber diets could potentially decrease the cardiometabolic disease risk associated with PFOS exposure.
Laboratory mice are usually fed a standardized chow diet that typically contains 5% fiber; for example, AIN-93G and AIN-93M contain 5% cellulose as the fiber source. Therefore, the 8% fiber-supplemented diet used in the present study represents an increase in the fiber contents compared with the standard chow. For humans, it has been estimated that dietary fiber intake for adults was 16–24 g/d81 and 82 in European countries and the United States, respectively. Current recommendations by the European Food Safety Authority83 and the U.S. Institute of Medicine84 for dietary fiber intake for adults are between per day, and it has been suggested that dietary fiber intake should be increased by compared with the current intake.85 Therefore, the diets used in the present animal study represent the increase in dietary fiber intake recommended for humans. Inulin is a polyfructosan that contains linear chains of fructosyl-groups linked by glycosydic bonds,86 and pectin mainly consists of linear 1,4-d-galacturonan (homogalacturonan) segments.87 Considering that inulin and pectin are structurally distinct and require specific carbohydrate-active enzymes for their fermentation in the gut,88 it seems conceivable that these fibers may differentially cater to diverse groups of bacteria, resulting in disparate physiological effects. Metagenomic results suggested that both soluble fibers protected against PFOS-induced changes in the microbiome community (Figure 12 and 13). However, the effects of inulin and pectin on the cecal content microbiome by themselves were different. LEfSe analysis indicated that the most significant genus difference after inulin feeding was in the higher numbers of Bifidobacterium, which is consistent with the effects of inulin supplementation on the human gut microbiome.89 It is well established that Bifidobacterium confer positive health benefits to the human host, and they have been incorporated into many functional foods as active ingredients.90 The primary up-regulated genus induced by pectin feeding is Duncaniella, which is particularly abundant in the mouse intestinal tract, although the function is yet to be studied.91 Significant interactions between dietary fibers and PFOS exposure on the abundance of Duncaniella and Muribaculum were observed (Figure S5), suggesting that PFOS interacts with a dietary component in modulating the microbial structure, and such interactions should be considered when evaluating the effects of PFOS exposure on microbial ecology.
Short-chain fatty acids (SCFAs) were the major end products of dietary fiber fermentation. It was reported that acetate, propionate, and butyrate were the major SCFAs derived from dietary fibers.35 The mole percentage ratio of these three SCFAs in the rodent cecum after inulin intervention was 63:18:19, whereas it was 84:11:5 for pectin,92 suggesting that inulin was preferably used by the gut microbiome to produce higher levels of propionate and butyrate than pectin. The SCFAs are thought to be absorbed from the intestinal mucosa into the portal vein. In inulin-fed mice the most significantly elevated SCFA in the portal vein was propionate,93 which could regulate host metabolism by down-regulating lipogenic enzymes94 and inhibit fatty acid synthesis.95 The molecular mechanisms by which dietary fiber alter the physiology of the liver is likely through gut-localized events (i.e., microbe-microbe and microbe-intestinal cell interactions, microbial metabolism), coupled with metabolites that emanate from the gut microbe in response to soluble fibers, which signal to or affect the physiology of the host liver. Therefore, we reasoned that the beneficial liver profile conferred by inulin and pectin during PFOS exposure, was in part strongly associated with their ability to modulate, as well as to be metabolized by, the microbiome community, which is known to produce bioactive metabolites, such as SCFAs, that could affect host metabolism. In our previous study, using stable isotope-resolved metabolomics, we found that the fecal microbiome degraded inulin and produced metabolites other than SCFAs, including amino acids, neurotransmitters, vitamin B5, and other coenzymes.55 The interactions between the gut microbiome and the host mediated by these bioactive metabolites warrant further investigation. Our ongoing research is investigating the in vivo metabolites of inulin by using stable isotope-labeled metabolomics, and further analysis on portal vein blood will identify bioactive mediators that link dietary fiber fermentation to the regulations of host metabolism.
In our previous animal study, inulin feeding protected against dioxin-like PCB126-induced hepatotoxicity and gut dysbiosis in a hyperlipidemic mouse model.40 Fiber-rich food intake has been associated with lower serum PFAS concentration in humans.96,97 In a recent study, the relation of serum PFAS concentrations to intake of dietary fiber was evaluated using data from 6,482 National Health and Nutrition Examination Survey participants. Dietary fiber intake was found to be negatively associated with serum PFAS levels; the authors suggested that fiber may have a role in the gastrointestinal excretion of PFOA, PFOS, and PFNA.98 The present mouse study supports this hypothesis and found that a higher intake of soluble fiber (inulin or pectin) was associated with lower plasma levels of PFOS compared with a higher intake of the insoluble fiber (cellulose) in treated mice (Figure 5). In addition, the effect was dependent on fiber type, with inulin exerting more pronounced effects on PFOS accumulation in plasma than pectin, suggesting that the low PFOS accumulation resulting from inulin and pectin intake could partially contribute to the protective effects associated with these soluble fibers. Previous in vitro studies have demonstrated that PFOS could be transported by apical sodium-dependent bile acid transporter (ASBT), taurocholate cotransport polypeptide (NTCP), and organic anion transporting polypeptide (OATP).99,100 Molecular docking calculations have suggested that PFOS has similar binding poses to those of taurocholic acid, an endogenous substrate of ASBT and NTCP.15 Therefore, PFOS could be taken up from the intestinal lumen into the blood, resulting in reduced excretion into the feces. It was reported that inulin treatment down-regulated the gene expressions of ASBT, NTCP, and OATP in mouse liver and increased fecal excretion of bile acids.101 Taken together, soluble fiber could potentially reduce the reabsorption of PFOS by regulating ASBT, NTCP, and OATP transporters, and thus increasing PFOS excretion in the feces. Further studies are needed to investigate fecal excretion of PFOS in mice treated with dietary fiber-supplemented diets. Inulin is a heterogeneous collection of fructose polymers with a degree of polymerization ranging from 3 to 60. Pectin is a complex molecule with many structural characteristics. Most studies, including the present one, investigating the health effects of pectin and inulin have used fibers with a combination of several structural characteristics. Additional studies with inulin and pectin molecules with well-defined structures are needed to determine which specific structural patterns of fibers induce the health-promoting effects observed in the present study.
NAFLD has become a burgeoning health problem worldwide, and it has been estimated to affect of the general population.102 Recent studies have recognized that PFAS exposure could contribute to the progression of NAFLD under preexisting liver disease conditions,69 and it has been associated with more advanced disease in pediatric NAFLD patients.103 Consumption of soluble fibers markedly ameliorated the phenotype of NAFLD/nonalcoholic steatohepatitis in murine models.41,42 Given the substantial prevalence of liver disease in the general population, and reports of liver toxicity with PFOS exposures, the potential of dietary fibers to protect individuals with exacerbated hepatic dysfunction as a result of PFOS exposure warrants further investigation.
In the present study, wild-type C57BL/6 mice were used to investigate the effects of dietary fiber on PFOS-induced toxicity. We found that genes associated with PPAR signaling and lipid metabolism pathways were enriched in PFOS-exposed mice by using transcriptomic analysis. Previous in vitro studies have shown that PFOS could differentially activate mouse and human .29,30 Recently, Su et al. found that human was not as responsive to PFOS as mouse by comparing wild-type, Ppara-null, and PPARA-humanized mice.104 In addition, mice, which have a lipoprotein metabolism that is similar to humans, have been used to investigate perfluorinated compound-induced effects on lipid metabolism.68 Therefore, PPARA-humanized mice and mice would be useful models for investigating the mechanisms that mediate species differences in PFOS exposure. In addition, recent toxicology research has suggested that females may be more susceptible to PFAS exposure-associated liver diseases69; therefore, future studies are needed to investigate the sex differences in dietary fiber-modulated PFOS toxicity.
It has been reported that the geometric mean concentration of total PFOS in the serum of workers in a fluorochemical manufacturing plant was 105 and the median serum concentration was in Chinese citizens,106 which are higher than those reported in U.S.12 or European studies.107 Therefore, the mouse plasma level of total PFOS in the present study was -fold higher than observed during human occupational exposures. Lower doses of PFOS exposure should be evaluated to produce more environmental relevance, providing valuable insight for human risk assessment when comparing data from rodents.
In conclusion, the present study applied an integrated multi-omics approach based on lipidomics, metabolomics, transcriptomics, and metagenomics, which provided pathway enrichment and connectivity between cecal content microbiome and liver, underscoring their overall relevance in PFOS exposure and dietary interventions. Our results support the potential of enriching diets with soluble fiber as a means to ameliorate metabolic perturbance of liver metabolism and the gut microbiota induced by PFOS. The distinct effects mediated by dietary fiber studied herein also serve to emphasize the fact that soluble fibers do not equally modulate PFOS-induced metabolic dysfunctions. A deeper understanding on the various aspects of soluble fibers and the mechanism of their health beneficial effects to the microbiome and host may pave the way forward for the development of personalized fiber-based interventions for individuals who are vulnerable to environmental disease risks.
Supplementary Material
Acknowledgments
We thank D. Barr, Emory University, and her research team, P. Esilda, P. Panuwet, and V. Yakimavets, for the perfluorooctane sulfonate analysis.
This work was supported by the National Institute of Environmental Health Sciences, National Institutes of Health (NIH; P42 ES007380, to B.H.), the Markey Cancer Center Biostatistics and Bioinformatics Core Facility (P30 CA177558, to C.W.), the Emory University Human Exposome Research Center (HERCULES; P30 ES019776), the University of Kentucky Agricultural Experiment Station, and the Priority Academic Program Development of Jiangsu Higher Education Institutes (PAPD). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
References
- 1.Tu P, Chi L, Bodnar W, Zhang Z, Gao B, Bian X, et al. 2020. Gut microbiome toxicity: connecting the environment and gut microbiome-associated diseases. Toxics 8(1):19, PMID: , 10.3390/toxics8010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parthasarathy G, Revelo X, Malhi H. 2020. Pathogenesis of nonalcoholic steatohepatitis: an overview. Hepatol Commun 4(4):478–492, PMID: , 10.1002/hep4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang C, Zhang Y, Deng M, Wang X, Tu W, Fu Z, et al. 2019. Bioaccumulation in the gut and liver causes gut barrier dysfunction and hepatic metabolism disorder in mice after exposure to low doses of OBS. Environ Int 129:279–290, PMID: , 10.1016/j.envint.2019.05.056. [DOI] [PubMed] [Google Scholar]
- 4.Liao J, Liu Y, Yi J, Li Y, Li Q, Li Y, et al. 2022. Gut microbiota disturbance exaggerates battery wastewater-induced hepatotoxicity through a gut-liver axis. Sci Total Environ 809:152188, PMID: , 10.1016/j.scitotenv.2021.152188. [DOI] [PubMed] [Google Scholar]
- 5.Peters A, Nawrot TS, Baccarelli AA. 2021. Hallmarks of environmental insults. Cell 184(6):1455–1468, PMID: , 10.1016/j.cell.2021.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Ciaula A, Baj J, Garruti G, Celano G, De Angelis M, Wang HH, et al. 2020. Liver steatosis, gut-liver axis, microbiome and environmental factors. A never-ending bidirectional cross-talk. J Clin Med 9(8):2648, PMID: , 10.3390/jcm9082648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tilg H, Cani PD, Mayer EA. 2016. Gut microbiome and liver diseases. Gut 65(12):2035–2044, PMID: , 10.1136/gutjnl-2016-312729. [DOI] [PubMed] [Google Scholar]
- 8.Chi L, Lai Y, Tu P, Liu CW, Xue J, Ru H, et al. 2019. Lipid and cholesterol homeostasis after arsenic exposure and antibiotic treatment in mice: potential role of the microbiota. Environ Health Perspect 127(9):97002, PMID: , 10.1289/EHP4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hennig B, Deng P. 2020. Healthful nutrition as a prevention and intervention paradigm to decrease the vulnerability to environmental toxicity or stressors and associated inflammatory disease risks. Food Front 1(1):13–14, PMID: , 10.1002/fft2.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sunderland EM, Hu XC, Dassuncao C, Tokranov AK, Wagner CC, Allen JG. 2019. A review of the pathways of human exposure to poly- and perfluoroalkyl substances (PFASs) and present understanding of health effects. J Expo Sci Environ Epidemiol 29(2):131–147, PMID: , 10.1038/s41370-018-0094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Susmann HP, Schaider LA, Rodgers KM, Rudel RA. 2019. Dietary habits related to food packaging and population exposure to PFASs. Environ Health Perspect 127(10):107003, PMID: , 10.1289/EHP4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.CDC (Centers for Disease Control and Prevention). 2019. Fourth National Report on Human Exposure to Environmental Chemicals. Updated Tables, January 2019. http://www.cdc.gov/exposurereport [accessed 9 February 2022].
- 13.Li Y, Fletcher T, Mucs D, Scott K, Lindh CH, Tallving P, et al. 2018. Half-lives of PFOS, PFHxS and PFOA after end of exposure to contaminated drinking water. Occup Environ Med 75(1):46–51, PMID: , 10.1136/oemed-2017-104651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang SC, Noker PE, Gorman GS, Gibson SJ, Hart JA, Ehresman DJ, et al. 2012. Comparative pharmacokinetics of perfluorooctanesulfonate (PFOS) in rats, mice, and monkeys. Reprod Toxicol 33(4):428–440, PMID: , 10.1016/j.reprotox.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Cao H, Zhou Z, Hu Z, Wei C, Li J, Wang L, et al. 2022. Effect of enterohepatic circulation on the accumulation of per- and polyfluoroalkyl substances: evidence from experimental and computational studies. Environ Sci Technol 56(5):3214–3224, PMID: , 10.1021/acs.est.1c07176. [DOI] [PubMed] [Google Scholar]
- 16.Pérez F, Nadal M, Navarro-Ortega A, Fàbrega F, Domingo JL, Barceló D, et al. 2013. Accumulation of perfluoroalkyl substances in human tissues. Environ Int 59:354–362, PMID: , 10.1016/j.envint.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Gallo V, Leonardi G, Genser B, Lopez-Espinosa MJ, Frisbee SJ, Karlsson L, et al. 2012. Serum perfluorooctanoate (PFOA) and perfluorooctane sulfonate (PFOS) concentrations and liver function biomarkers in a population with elevated PFOA exposure. Environ Health Perspect 120(5):655–660, PMID: , 10.1289/ehp.1104436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.ATSDR (Agency for Toxic Substances and Disease Registry). 2021. Toxicological Profile for Perfluoroalkyls. Last updated March 2020. Atlanta, GA: ATSDR. https://www.atsdr.cdc.gov/ToxProfiles/tp200-p.pdf [accessed 1 October 2022]. [PubMed] [Google Scholar]
- 19.EFSA CONTAM Panel (EFSA Panel on Contaminants in the Food Chain), Knutsen HK, Alexander J, Barregård L, Bignami M, Brüschweiler B, et al. 2018. Risk to human health related to the presence of perfluorooctane sulfonic acid and perfluorooctanoic acid in food. EFSA J 16(12):e05194, PMID: , 10.2903/j.efsa.2018.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.U.S. EPA (U.S. Environmental Protection Agency). 2016. Health Effects Support Document for Perfluorooctane Sulfonate (PFOS). EPA 822-R-16-002. Washington, DC: U.S. EPA, Office of Water. https://www.epa.gov/sites/default/files/2016-05/documents/pfos_hesd_final_508.pdf [accessed 1 October 2022]. [Google Scholar]
- 21.Qazi MR, Abedi MR, Nelson BD, DePierre JW, Abedi-Valugerdi M. 2010. Dietary exposure to perfluorooctanoate or perfluorooctane sulfonate induces hypertrophy in centrilobular hepatocytes and alters the hepatic immune status in mice. Int Immunopharmacol 10(11):1420–1427, PMID: , 10.1016/j.intimp.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 22.Wan HT, Zhao YG, Wei X, Hui KY, Giesy JP, Wong CKC. 2012. PFOS-induced hepatic steatosis, the mechanistic actions on β-oxidation and lipid transport. Biochim Biophys Acta 1820(7):1092–1101, PMID: , 10.1016/j.bbagen.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 23.Fai Tse WK, Li JW, Kwan Tse AC, Chan TF, Hin Ho JC, Sun Wu RS, et al. 2016. Fatty liver disease induced by perfluorooctane sulfonate: novel insight from transcriptome analysis. Chemosphere 159:166–177, PMID: , 10.1016/j.chemosphere.2016.05.060. [DOI] [PubMed] [Google Scholar]
- 24.Wang Q, Huang J, Liu S, Wang C, Jin Y, Lai H, et al. 2022. Aberrant hepatic lipid metabolism associated with gut microbiota dysbiosis triggers hepatotoxicity of novel PFOS alternatives in adult zebrafish. Environ Int 166:107351, PMID: , 10.1016/j.envint.2022.107351. [DOI] [PubMed] [Google Scholar]
- 25.Pfohl M, Ingram L, Marques E, Auclair A, Barlock B, Jamwal R, et al. 2020. Perfluorooctanesulfonic acid and perfluorohexanesulfonic acid alter the blood lipidome and the hepatic proteome in a murine model of diet-induced obesity. Toxicol Sci 178(2):311–324, PMID: , 10.1093/toxsci/kfaa148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salter DM, Wei W, Nahar PP, Marques E, Slitt AL. 2021. Perfluorooctanesulfonic acid (PFOS) thwarts the beneficial effects of calorie restriction and metformin. Toxicol Sci 182(1):82–95, PMID: , 10.1093/toxsci/kfab043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marques E, Pfohl M, Auclair A, Jamwal R, Barlock BJ, Sammoura FM, et al. 2020. Perfluorooctanesulfonic acid (PFOS) administration shifts the hepatic proteome and augments dietary outcomes related to hepatic steatosis in mice. Toxicol Appl Pharmacol 408:115250, PMID: , 10.1016/j.taap.2020.115250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marques E, Pfohl M, Wei W, Tarantola G, Ford L, Amaeze O, et al. 2022. Replacement per- and polyfluoroalkyl substances (PFAS) are potent modulators of lipogenic and drug metabolizing gene expression signatures in primary human hepatocytes. Toxicol Appl Pharmacol 442:115991, PMID: , 10.1016/j.taap.2022.115991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bjork JA, Butenhoff JL, Wallace KB. 2011. Multiplicity of nuclear receptor activation by PFOA and PFOS in primary human and rodent hepatocytes. Toxicology 288(1–3):8–17, PMID: , 10.1016/j.tox.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 30.Takacs ML, Abbott BD. 2007. Activation of mouse and human peroxisome proliferator-activated receptors (α, β/δ, γ) by perfluorooctanoic acid and perfluorooctane sulfonate. Toxicol Sci 95(1):108–117, PMID: , 10.1093/toxsci/kfl135. [DOI] [PubMed] [Google Scholar]
- 31.Zhang L, Rimal B, Nichols RG, Tian Y, Smith PB, Hatzakis E, et al. 2020. Perfluorooctane sulfonate alters gut microbiota-host metabolic homeostasis in mice. Toxicology 431:152365, PMID: , 10.1016/j.tox.2020.152365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai KP, Ng AHM, Wan HT, Wong AYM, Leung CCT, Li R, et al. 2018. Dietary exposure to the environmental chemical, PFOS on the diversity of gut microbiota, associated with the development of metabolic syndrome. Front Microbiol 9:2552, PMID: , 10.3389/fmicb.2018.02552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hennig B, Ettinger AS, Jandacek RJ, Koo S, McClain C, Seifried H, et al. 2007. Using nutrition for intervention and prevention against environmental chemical toxicity and associated diseases. Environ Health Perspect 115(4):493–495, PMID: , 10.1289/ehp.9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hennig B, Ormsbee L, McClain CJ, Watkins BA, Blumberg B, Bachas LG, et al. 2012. Nutrition can modulate the toxicity of environmental pollutants: implications in risk assessment and human health. Environ Health Perspect 120(6):771–774, PMID: , 10.1289/ehp.1104712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cronin P, Joyce SA, O’Toole PW, O’Connor EM. 2021. Dietary fibre modulates the gut microbiota. Nutrients 13(5):1655, PMID: , 10.3390/nu13051655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Makki K, Deehan EC, Walter J, Bäckhed F. 2018. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe 23(6):705–715, PMID: , 10.1016/j.chom.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 37.Abu-Elsaad NM, Elkashef WF. 2016. Modified citrus pectin stops progression of liver fibrosis by inhibiting galectin-3 and inducing apoptosis of stellate cells. Can J Physiol Pharmacol 94(5):554–562, PMID: , 10.1139/cjpp-2015-0284. [DOI] [PubMed] [Google Scholar]
- 38.Chambers ES, Byrne CS, Rugyendo A, Morrison DJ, Preston T, Tedford C, et al. 2019. The effects of dietary supplementation with inulin and inulin-propionate ester on hepatic steatosis in adults with non-alcoholic fatty liver disease. Diabetes Obes Metab 21(2):372–376, PMID: , 10.1111/dom.13500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng P, Hoffman JB, Petriello MC, Wang CY, Li XS, Kraemer MP, et al. 2020. Dietary inulin decreases circulating ceramides by suppressing neutral sphingomyelinase expression and activity in mice. J Lipid Res 61(1):45–53, PMID: , 10.1194/jlr.RA119000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoffman JB, Petriello MC, Morris AJ, Mottaleb MA, Sui Y, Zhou C, et al. 2020. Prebiotic inulin consumption reduces dioxin-like PCB 126-mediated hepatotoxicity and gut dysbiosis in hyperlipidemic Ldlr deficient mice. Environ Pollut 261:114183, PMID: , 10.1016/j.envpol.2020.114183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugatani J, Sadamitsu S, Wada T, Yamazaki Y, Ikari A, Miwa M. 2012. Effects of dietary inulin, statin, and their co-treatment on hyperlipidemia, hepatic steatosis and changes in drug-metabolizing enzymes in rats fed a high-fat and high-sucrose diet. Nutr Metab (Lond) 9(1):23, PMID: , 10.1186/1743-7075-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sugatani J, Wada T, Osabe M, Yamakawa K, Yoshinari K, Miwa M. 2006. Dietary inulin alleviates hepatic steatosis and xenobiotics-induced liver injury in rats fed a high-fat and high-sucrose diet: association with the suppression of hepatic cytochrome P450 and hepatocyte nuclear factor 4α expression. Drug Metab Dispos 34(10):1677–1687, PMID: , 10.1124/dmd.106.010645. [DOI] [PubMed] [Google Scholar]
- 43.Deng P, Wang C, Wahlang B, Sexton T, Morris AJ, Hennig B. 2020. Co-exposure to PCB126 and PFOS increases biomarkers associated with cardiovascular disease risk and liver injury in mice. Toxicol Appl Pharmacol 409:115301, PMID: , 10.1016/j.taap.2020.115301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng P, Barney J, Petriello MC, Morris AJ, Wahlang B, Hennig B. 2019. Hepatic metabolomics reveals that liver injury increases PCB 126-induced oxidative stress and metabolic dysfunction. Chemosphere 217:140–149, PMID: , 10.1016/j.chemosphere.2018.10.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh V, Yeoh BS, Walker RE, Xiao X, Saha P, Golonka RM, et al. 2019. Microbiota fermentation-NLRP3 axis shapes the impact of dietary fibres on intestinal inflammation. Gut 68(10):1801–1812, PMID: , 10.1136/gutjnl-2018-316250. [DOI] [PubMed] [Google Scholar]
- 46.U.S. Department of Health and Human Services, U.S. Department of Agriculture. 2015–2020 Dietary Guidelines for Americans. 8th ed. December 2015. https://health.gov/sites/default/files/2019-09/2015-2020_Dietary_Guidelines.pdf [accessed 1 October 2022].
- 47.Appleman TD, Higgins CP, Quiñones O, Vanderford BJ, Kolstad C, Zeigler-Holady JC, et al. 2014. Treatment of poly- and perfluoroalkyl substances in U.S. full-scale water treatment systems. Water Res 51:246–255, PMID: , 10.1016/j.watres.2013.10.067. [DOI] [PubMed] [Google Scholar]
- 48.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21, PMID: , 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anders S, Pyl PT, Huber W. 2015. HTSeq—a python framework to work with high-throughput sequencing data. Bioinformatics 31(2):166–169, PMID: , 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14(4):R36, PMID: , 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12):550, PMID: , 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57(1):289–300, 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 53.Yu G, Wang LG, Han Y, He QY. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16(5):284–287, PMID: , 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28(1):27–30, PMID: , 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deng P, Valentino T, Flythe MD, Moseley HNB, Leachman JR, Morris AJ, et al. 2021. Untargeted stable isotope probing of the gut microbiota metabolome using 13C-labeled dietary fibers. J Proteome Res 20(5):2904–2913, PMID: , 10.1021/acs.jproteome.1c00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van den Berg RA, Hoefsloot HCJ, Westerhuis JA, Smilde AK, van der Werf MJ. 2006. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics 7:142, PMID: , 10.1186/1471-2164-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pang Z, Chong J, Zhou G, de Lima Morais DA, Chang L, Barrette M, et al. 2021. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res 49(W1):W388–W396, PMID: , 10.1093/nar/gkab382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McIver LJ, Abu-Ali G, Franzosa EA, Schwager R, Morgan XC, Waldron L, et al. 2018. bioBakery: a meta’omic analysis environment. Bioinformatics 34(7):1235–1237, PMID: , 10.1093/bioinformatics/btx754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10(3):R25, PMID: , 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wood DE, Lu J, Langmead B. 2019. Improved metagenomic analysis with Kraken 2. Genome Biol 20(1):257, PMID: , 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu J, Breitwieser FP, Thielen P, Salzberg SL. 2017. Bracken: estimating species abundance in metagenomics data. Peerj Comput Sci 3(1):e104, 10.7717/peerj-cs.104. [DOI] [Google Scholar]
- 62.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12(6):R60, PMID: , 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Choi RH, Tatum SM, Symons JD, Summers SA, Holland WL. 2021. Ceramides and other sphingolipids as drivers of cardiovascular disease. Nat Rev Cardiol 18(10):701–711, PMID: , 10.1038/s41569-021-00536-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kitatani K, Idkowiak-Baldys J, Hannun YA. 2008. The sphingolipid salvage pathway in ceramide metabolism and signaling. Cell Signal 20(6):1010–1018, PMID: , 10.1016/j.cellsig.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Awojoodu AO, Keegan PM, Lane AR, Zhang Y, Lynch KR, Platt MO, et al. 2014. Acid sphingomyelinase is activated in sickle cell erythrocytes and contributes to inflammatory microparticle generation in SCD. Blood 124(12):1941–1950, PMID: , 10.1182/blood-2014-01-543652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang L, Wang Y, Liang Y, Li J, Liu Y, Zhang J, et al. 2014. PFOS induced lipid metabolism disturbances in BALB/c mice through inhibition of low density lipoproteins excretion. Sci Rep 4:4582, PMID: , 10.1038/srep04582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Louisse J, Rijkers D, Stoopen G, Janssen A, Staats M, Hoogenboom R, et al. 2020. Perfluorooctanoic acid (PFOA), perfluorooctane sulfonic acid (PFOS), and perfluorononanoic acid (PFNA) increase triglyceride levels and decrease cholesterogenic gene expression in human HepaRG liver cells. Arch Toxicol 94(9):3137–3155, PMID: , 10.1007/s00204-020-02808-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bijland S, Rensen PCN, Pieterman EJ, Maas ACE, van der Hoorn JW, van Erk MJ, et al. 2011. Perfluoroalkyl sulfonates cause alkyl chain length-dependent hepatic steatosis and hypolipidemia mainly by impairing lipoprotein production in APOE*3-Leiden CETP mice. Toxicol Sci 123(1):290–303, PMID: , 10.1093/toxsci/kfr142. [DOI] [PubMed] [Google Scholar]
- 69.Sen P, Qadri S, Luukkonen PK, Ragnarsdottir O, McGlinchey A, Jäntti S, et al. 2022. Exposure to environmental contaminants is associated with altered hepatic lipid metabolism in non-alcoholic fatty liver disease. J Hepatol 76(2):283–293, PMID: , 10.1016/j.jhep.2021.09.039. [DOI] [PubMed] [Google Scholar]
- 70.Li X, Li T, Wang Z, Wei J, Liu J, Zhang Y, et al. 2021. Distribution of perfluorooctane sulfonate in mice and its effect on liver lipidomic. Talanta 226:122150, PMID: , 10.1016/j.talanta.2021.122150. [DOI] [PubMed] [Google Scholar]
- 71.Summers SA. 2006. Ceramides in insulin resistance and lipotoxicity. Prog Lipid Res 45(1):42–72, PMID: , 10.1016/j.plipres.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 72.Holland WL, Summers SA. 2008. Sphingolipids, insulin resistance, and metabolic disease: new insights from in vivo manipulation of sphingolipid metabolism. Endocr Rev 29(4):381–402, PMID: , 10.1210/er.2007-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Summers SA. 2015. The ART of lowering ceramides. Cell Metab 22(2):195–196, PMID: , 10.1016/j.cmet.2015.07.019. [DOI] [PubMed] [Google Scholar]
- 74.Chaurasia B, Summers SA. 2015. Ceramides—lipotoxic inducers of metabolic disorders. Trends Endocrinol Metab 26(10):538–550, PMID: , 10.1016/j.tem.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 75.Stratakis N, V Conti D, Jin R, Margetaki K, Valvi D, Siskos AP, et al. 2020. Prenatal exposure to perfluoroalkyl substances associated with increased susceptibility to liver injury in children. Hepatology 72(5):1758–1770, PMID: , 10.1002/hep.31483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Summers SA. 2018. Could ceramides become the new cholesterol? Cell Metab 27(2):276–280, PMID: , 10.1016/j.cmet.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 77.Hajduch E, Lachkar F, Ferré P, Foufelle F. 2021. Roles of ceramides in non-alcoholic fatty liver disease. J Clin Med 10(4):792, PMID: , 10.3390/jcm10040792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Levy M, Futerman AH. 2010. Mammalian ceramide synthases. IUBMB Life 62(5):347–356, PMID: , 10.1002/iub.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Meneguzzi A, Fava C, Castelli M, Minuz P. 2021. Exposure to perfluoroalkyl chemicals and cardiovascular disease: experimental and epidemiological evidence. Front Endocrinol (Lausanne) 12:706352, PMID: , 10.3389/fendo.2021.706352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Qian Y, Ducatman A, Ward R, Leonard S, Bukowski V, Lan Guo N, et al. 2010. Perfluorooctane sulfonate (PFOS) induces reactive oxygen species (ROS) production in human microvascular endothelial cells: role in endothelial permeability. J Toxicol Environ Health A 73(12):819–836, PMID: , 10.1080/15287391003689317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stephen AM, Champ MMJ, Cloran SJ, Fleith M, van Lieshout L, Mejborn H, et al. 2017. Dietary fibre in Europe: current state of knowledge on definitions, sources, recommendations, intakes and relationships to health. Nutr Res Rev 30(2):149–190, PMID: , 10.1017/S095442241700004X. [DOI] [PubMed] [Google Scholar]
- 82.Quagliani D, Felt-Gunderson P. 2017. Closing America’s fiber intake gap: communication strategies from a food and fiber summit. Am J Lifestyle Med 11(1):80–85, PMID: , 10.1177/1559827615588079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.EFSA NDA Panel (EFSA Panel on Dietetic Products, Nutrition, and Allergies). 2010. Scientific opinion on dietary reference values for carbohydrates and dietary fibre. EFSA J 8(3):1462, 10.2903/j.efsa.2010.1462. [DOI] [Google Scholar]
- 84.U.S. Institute of Medicine. 2005. Dietary Reference Intakes for Energy, Carbohydrate, Fiber, Fat, Fatty Acids, Cholesterol, Protein, and Amino Acids (Macronutrients). Washington, DC: National Academies Press. [Google Scholar]
- 85.Barber TM, Kabisch S, Pfeiffer AFH, Weickert MO. 2020. The health benefits of dietary fibre. Nutrients 12(10):3209, PMID: , 10.3390/nu12103209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mensink MA, Frijlink HW, van der Voort Maarschalk K, Hinrichs WLJ. 2015. Inulin, a flexible oligosaccharide. II: review of its pharmaceutical applications. Carbohydr Polym 134:418–428, PMID: , 10.1016/j.carbpol.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 87.Voragen AGJ, Coenen GJ, Verhoef RP, Schols HA. 2009. Pectin, a versatile polysaccharide present in plant cell walls. Struct Chem 20:263–275, 10.1007/s11224-009-9442-z. [DOI] [Google Scholar]
- 88.Ndeh D, Gilbert HJ. 2018. Biochemistry of complex glycan depolymerisation by the human gut microbiota. FEMS Microbiol Rev 42(2):146–164, PMID: , 10.1093/femsre/fuy002. [DOI] [PubMed] [Google Scholar]
- 89.Le Bastard Q, Chapelet G, Javaudin F, Lepelletier D, Batard E, Montassier E. 2020. The effects of inulin on gut microbial composition: a systematic review of evidence from human studies. Eur J Clin Microbiol Infect Dis 39(3):403–413, PMID: , 10.1007/s10096-019-03721-w. [DOI] [PubMed] [Google Scholar]
- 90.O’Callaghan A, van Sinderen D. 2016. Bifidobacteria and their role as members of the human gut microbiota. Front Microbiol 7:925, PMID: , 10.3389/fmicb.2016.00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lagkouvardos I, Lesker TR, Hitch TCA, Gálvez EJC, Smit N, Neuhaus K, et al. 2019. Sequence and cultivation study of Muribaculaceae reveals novel species, host preference, and functional potential of this yet undescribed family. Microbiome 7(1):28, PMID: , 10.1186/s40168-019-0637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. 2013. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res 54(9):2325–2340, PMID: , 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kawasoe J, Uchida Y, Kawamoto H, Miyauchi T, Watanabe T, Saga K, et al. 2022. Propionic acid, induced in gut by an inulin diet, suppresses inflammation and ameliorates liver ischemia and reperfusion injury in mice. Front Immunol 13:862503, PMID: , 10.3389/fimmu.2022.862503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Weitkunat K, Schumann S, Nickel D, Kappo KA, Petzke KJ, Kipp AP, et al. 2016. Importance of propionate for the repression of hepatic lipogenesis and improvement of insulin sensitivity in high-fat diet-induced obesity. Mol Nutr Food Res 60(12):2611–2621, PMID: , 10.1002/mnfr.201600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nishina PM, Freedland RA. 1990. Effects of propionate on lipid biosynthesis in isolated rat hepatocytes. J Nutr 120(7):668–673, PMID: , 10.1093/jn/120.7.668. [DOI] [PubMed] [Google Scholar]
- 96.Lin PID, Cardenas A, Hauser R, Gold DR, Kleinman KP, Hivert MF, et al. 2020. Dietary characteristics associated with plasma concentrations of per- and polyfluoroalkyl substances among adults with pre-diabetes: cross-sectional results from the Diabetes Prevention Program Trial. Environ Int 137:105217, PMID: , 10.1016/j.envint.2019.105217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Skuladottir M, Ramel A, Rytter D, Haug LS, Sabaredzovic A, Bech BH, et al. 2015. Examining confounding by diet in the association between perfluoroalkyl acids and serum cholesterol in pregnancy. Environ Res 143(pt A):33–38, PMID: , 10.1016/j.envres.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 98.Dzierlenga MW, Keast DR, Longnecker MP. 2021. The concentration of several perfluoroalkyl acids in serum appears to be reduced by dietary fiber. Environ Int 146:106292, PMID: , 10.1016/j.envint.2020.106292. [DOI] [PubMed] [Google Scholar]
- 99.Zhao W, Zitzow JD, Weaver Y, Ehresman DJ, Chang SC, Butenhoff JL, et al. 2017. Organic anion transporting polypeptides contribute to the disposition of perfluoroalkyl acids in humans and rats. Toxicol Sci 156(1):84–95, PMID: , 10.1093/toxsci/kfw236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhao W, Zitzow JD, Ehresman DJ, Chang SC, Butenhoff JL, Forster J, et al. 2015. Na+/taurocholate cotransporting polypeptide and apical sodium-dependent bile acid transporter are involved in the disposition of perfluoroalkyl sulfonates in humans and rats. Toxicol Sci 146(2):363–373, PMID: , 10.1093/toxsci/kfv102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang R, Ren Y, Bao T, Wang T, Li Y, Liu Y, et al. 2022. Inulin activates FXR-FGF15 signaling and further increases bile acids excretion in non-alcoholic fatty liver disease mice. Biochem Biophys Res Commun 600:156–162, PMID: , 10.1016/j.bbrc.2022.02.033. [DOI] [PubMed] [Google Scholar]
- 102.Ge X, Zheng L, Wang M, Du Y, Jiang J. 2020. Prevalence trends in non-alcoholic fatty liver disease at the global, regional and national levels, 1990–2017: a population-based observational study. BMJ Open 10(8):e036663, PMID: , 10.1136/bmjopen-2019-036663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jin R, McConnell R, Catherine C, Xu S, Walker DI, Stratakis N, et al. 2020. Perfluoroalkyl substances and severity of nonalcoholic fatty liver in children: an untargeted metabolomics approach. Environ Int 134:105220, PMID: , 10.1016/j.envint.2019.105220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Su S, Billy LJ, Chang S, Gonzalez FJ, Patterson AD, Peters JM. 2022. The role of mouse and human peroxisome proliferator-activated receptor-α in modulating the hepatic effects of perfluorooctane sulfonate in mice. Toxicology 465:153056, PMID: , 10.1016/j.tox.2021.153056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gao Y, Fu J, Cao H, Wang Y, Zhang A, Liang Y, et al. 2015. Differential accumulation and elimination behavior of perfluoroalkyl acid isomers in occupational workers in a manufactory in China. Environ Sci Technol 49(11):6953–6962, PMID: , 10.1021/acs.est.5b00778. [DOI] [PubMed] [Google Scholar]
- 106.Duan Y, Sun H, Yao Y, Meng Y, Li Y. 2020. Distribution of novel and legacy per-/polyfluoroalkyl substances in serum and its associations with two glycemic biomarkers among Chinese adult men and women with normal blood glucose levels. Environ Int 134:105295, PMID: , 10.1016/j.envint.2019.105295. [DOI] [PubMed] [Google Scholar]
- 107.Göckener B, Weber T, Rüdel H, Bücking M, Kolossa-Gehring M. 2020. Human biomonitoring of per- and polyfluoroalkyl substances in German blood plasma samples from 1982 to 2019. Environ Int 145:106123, PMID: , 10.1016/j.envint.2020.106123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.