Abstract
Wheat NAC (TaNAC) transcription factors are important regulators of stress responses and developmental processes. This study proposes a new TaNAC nomenclature and identified defense-associated TaNACs based on the analysis of RNA-sequencing datasets of wheat tissue infected with major fungal pathogens. A total of 146 TaNACs were pathogen-responsive, of which 52 were orthologous with functionally characterized defense-associated NACs from barley, rice, and Arabidopsis, as deduced via phylogenetic analysis. Next, we focused on the phylogenetic relationship of the pathogen-responsive TaNACs and their expression profiles in healthy and diseased tissues. Three subfamilies (“a,” “e,” and “f”) were significantly enriched in pathogen-responsive TaNACs, of which the majority were responsive to at least 2 pathogens (universal pathogen response). Uncharacterized TaNACs from subfamily “a” enriched with defense-associated NACs are promising candidates for functional characterization in pathogen defense. In general, pathogen-responsive TaNACs were expressed in at least 2 healthy organs. Lastly, we showed that the wheat NAM domain is significantly divergent in sequence in subfamilies “f,” “g,” and “h” based on HMMER and motif analysis. New protein motifs were identified in both the N- and C-terminal parts of TaNACs. Three of those identified in the C-terminal part were linked to pathogen responsiveness of the TaNACs and 2 were linked to expression in grain tissue. Future studies should benefit from this comprehensive in silico analysis of pathogen-responsive TaNACs as a basis for selecting the most promising candidates for functional validation and crop improvement.
Keywords: wheat, NAC, transcription factor, phylogeny, pathogen
Introduction
An estimated 21.5% of global wheat yield is lost due to pests and pathogens (Savary et al. 2019). Breeding for resistance is an environmentally friendly and sustainable method to control diseases and an important component of an integrated disease management strategy. Recent advances in sequencing, bioinformatics, and data analytics have delivered the genomic and transcriptomic tools to expedite breeding via the identification of genes and genetic loci underpinning critical agronomic traits in wheat, including disease resistance [International Wheat Genome Sequencing Consortium (IWGSC) et al. 2018; Ramírez-González et al. 2018]. Transcription factor (TF) families regulate gene expression and are thus master regulators of cellular events. Many plant TF families involved in biotic stress responses have been identified. The NAC [no apical meristem (NAM), Arabidopsis thaliana transcription activation factor (ATAF1/2), and cup-shaped cotyledon (CUC2)] TFs represent one of the largest plant families of transcriptional regulators. They are delineated by their conserved NAC domains containing 5 subdomains A–E, of which A–D form the NAM domain (Ooka et al. 2003). The NAC domains are usually situated in the N-terminal (NT) part of the protein and are associated with DNA binding, transcriptional control, and homo- and heterodimerization (Welner et al. 2016). NAC proteins also encode a highly divergent transcriptional activation region (TAR) in the C-terminal (CT) part of the protein, associated with transcriptional activation and protein–protein interactions (Welner et al. 2016). NACs have been known to regulate developmental processes and stress response traits, including disease resistance (Xie et al. 2000; Jensen et al. 2007; Wu et al. 2009). NACs have been shown to play important roles in plant defense against pathogens with diverse lifestyles, via the regulation of plant immunity, modulation of the hypersensitive response, stomatal immunity, involvement in hormonal signaling pathways, and as targets of pathogen effectors (Yuan et al. 2019). Cross-talk between NAC proteins, phytohormones, and other signaling molecules, such as reactive oxygen species were shown to enhance or suppress the host response to pathogens (Yuan et al. 2019). However, the signaling pathways regulating NAC responses to pathogens remain elusive. The overexpression or suppression of NACs often leads to significantly improved resistance or tolerance to plant pathogens, making them promising candidates for improvement of disease resistance in crops (Yuan et al. 2019).
Four studies identified high confidence Triticum aestivum (wheat) NACs (TaNACs) in the cultivar Chinese Spring; Borrill et al. (2017) identified 453 TaNAC genes in the TGACv1 genome assembly, while Guérin et al. (2019) and Ma et al. (2021) identified, respectively, 460 and 462 TaNACs using a more recent assembly (RefSeq v1.0) and Lv et al. (2020) identified 460 TaNACs in the RefSeq v1.1 genome annotation. Studies from Borrill et al. (2017) and Guérin et al. (2019) mined RNA-sequencing (RNA-seq) datasets to determine the expression profiles of TaNACs in different tissues, and in response to biotic and/or abiotic stresses. Borrill et al. (2017) delineated TaNACs responsive to biotic stress, while other studies (Lv et al. 2020; Ma et al. 2021) delineated TaNAC genes responsive to specific pathogens, i.e. races of the causal agents of stripe rust disease (Puccinia striiformis f. sp. tritici) and powdery mildew disease (Blumeria graminis f. sp. tritici) in the rust and mildew resistant wheat line N9134.Lv et al. (2020) highlighted the significantly higher proportion of transcript variants of fungus-responsive TaNACs compared to nonresponsive TaNACs. Ma et al. (2021) concluded that the neofunctionalization of TaNACs plays a role in wheat adaptation to biotic stresses.
The aim of the current study was to go further than previous studies to (1) explore the phylogenetic relationship between all wheat TaNACs and well-characterized NACs from other plants and (2) delineate all high confidence TaNACs responsive to pathogens with diverse lifestyles and to explore their phylogenetic relationship. This study determined whether there were any associations between the pathogen responsiveness of TaNACs (within the wheat genome version RefSeqv1.1) and either the lifestyle of the pathogen, phylogeny, or divergence in their NAC subdomain structures or motif composition. A joined phylogenetic tree of NACs from monocots wheat, barley, rice, and the model dicot Arabidopsis was constructed to identify TaNAC orthologs closely related to characterized NAC genes involved in disease resistance. This study identified TaNACs responsive to a diverse set of diseases; 5 publicly available RNA-seq datasets were mined to identify TaNACs associated with Fusarium head blight (FHB), Fusarium crown rot, Septoria tritici blotch, powdery mildew, and stripe rust diseases. Focusing on the pathogen-responsive TaNACs, we studied their phylogenetic relationship, and their expression profiles in healthy and diseased tissue. In addition, in-depth motif analysis of full-length TaNAC amino acid sequences was done to investigate the divergence and structural diversity of TaNAC proteins and the potential association between the pathogen-responsiveness of TaNACs and their protein motif composition.
Materials and methods
Identification of NAC genes in T. aestivum, Hordeum vulgare, Oryza sativa subsp. japonica, and A. thaliana
High confidence wheat proteome sequences were downloaded from the IWGSC sequence repository (IWGSC RefSeq v1.1, https://wheat-urgi.versailles.inra.fr/Seq-Repository/Annotations, July 2017). Proteomes for Arabidopsis, rice IRGSP 1.0, and barley proteomes (splice variants from longest coding DNA sequences) were downloaded from the TAIR10 sequence repository (https://www.arabidopsis.org, June 2018) and Ensembl Plants release 37 (http://plants.ensembl.org, June 2018). The full alignment of the NAM domain (PF02365) was downloaded from the Pfam database (http://pfam.xfam.org/, August 2017) and used to build Hidden Markov Model (HMM) profile with hmmbuild in HMMER v.3.1 b2 (http://hmmer.org/). The HMM profile of the NAM family was then used as a query against wheat, barley, rice, and Arabidopsis proteomes with hmmsearch (default parameters; HMMER v.3.1 b2, http://hmmer.org/) and all nonredundant hits (E < 0.01) were retrieved (Supplementary Table 1).
Phylogenetic analysis
Multiple sequence alignment (MSA) of the full-length protein sequences was performed using MAFFT v7.407 (Katoh and Standley 2013) and the default parameters. The completeness of MSA was evaluated using AliStat v1.3 (Wong et al. 2020). Sequences that did not align with any of the sequences in the MSA and whose completeness score was Cij < 0.5 (Wong et al. 2020) were removed from the MSA used for joined phylogenetic tree (Supplementary Table 2). The MSA was masked in AliStat v1.3 (Wong et al. 2020) with a cutoff Cc = 0.6 (Wong et al. 2020), and manually inspected and corrected in Jalview v2.10.5 (Waterhouse et al. 2009). Identical masked sequences were removed to lower the sample size. Homo v1.3 (Rouse et al. 2013) confirmed that pairs of sequences in the MSA were consistent with evolution under stationary, reversible, and globally homogeneous conditions. ModelFinder (Kalyaanamoorthy et al. 2017) identified the best substitution model according to the lowest Bayesian information criterion. The best fitting substitution model used to infer joined phylogeny was Jones–Taylor–Thornton (JTT; Jones et al. 1992) with 7 categories of probability distribution free (PDF) model of rate heterogeneity across sites (RHAS; Kalyaanamoorthy et al. 2017), and JTT with 6 categories of PDF model of RHAS for phylogeny of TaNAC sequences. IQ-TREE (Nguyen et al. 2015) was used to estimate maximum likelihood phylogeny with an ascertainment bias correction used to correct for likelihood conditioned solely on variable sites (Lewis 2001). The goodness of fit of branch nodes with phylogenetic trees was estimated by ultrafast bootstrap approximation (UFBoot; Hoang et al. 2018) with 1,000 bootstrap alignments. After the tree inference, protein identifiers (IDs) of surplus sequences were added alongside the ID of its identical pair in the joined phylogenetic tree. The resulting trees were unrooted, although outgroup taxon TaNAC121-A1 (TraesCS1A02G190100.1) was randomly selected as a root in IQ-TREE. The joined phylogenetic tree was rerooted and subfamily “b” was selected as a root. A consensus tree was used for further analysis due to a higher maximum likelihood than the true tree. Tree figures were created in iTOL (https://itol.embl.de/; Letunic and Bork 2019).
Wheat NAC naming system
A naming system of TaNAC was defined, taking into account NAC rice nomenclature and their phylogenetic relationship, paralogy, gene tree and subgenome location, as described in detail in Schilling et al. (2020), with a few modifications. TaNAC homoeologs, paralogs, and rice orthologs were downloaded from Ensembl Plants (Biomart, release 45 of Ensembl Plants). The 145 rice NAC MSU IDs identified in Shen et al. (2009) were converted into RAP-DB IDs with ID converter on https://rapdb.dna.affrc.go.jp/tools/converter/run (September 2020). From these 145 O. sativa NAC (OsNAC) genes with MSU IDs, 4 were converted into 8 RAP-DB OsNACs and 8 were missing in RAP-DB. The RAP-DB OsNACs Os02g0745250, Os04g0508400, and Os12g0135850 not present in MSU DB did not have a name in any of the databases (RAP-DB, CGSNL, Oryzabase), so they were assigned the names ONAC150, ONAC151, and ONAC152, respectively, using the same nomenclature as described in Shen et al. (2009) (i.e. using the prefix O instead of the more widely used Os abbreviation for rice), resulting in a total of 144 OsNAC genes.
TaNAC homoeologs on subgenomes A, B, and D were primarily identified based on their phylogenetic relationship. In the case of TaNAC homoeologs orthologous to the rice ONAC007 and ONAC038 (Os06g0131700 and Os01g0946200, respectively), this approach was impossible due to either clade complexity and/or where the bootstrap value of the cluster’s nodes was <70%, and thus, the homoeologs were determined according to the Ensembl Plants. TaNAC paralogs were determined based on phylogeny and confirmed according to Ensembl Plants. In cases when orthologous clusters comprised multiple TaNAC paralogs located on the same chromosome with at least 80% protein identity, the paralogs were considered as duplicate genes (inparalogs) belonging to the same homoeologous group.
TaNAC genes were primarily named based on phylogeny to the closest rice ortholog. In cases where TaNACs were orthologous to more than 1 OsNAC paralog located on the same chromosome (e.g. ONAC020 [Os01g0104500] and ONAC026 [Os01g0393100]), they were named after the first OsNAC identified on the chromosome. TaNACs without a rice ortholog were named consecutively according to their position in a tree starting with subfamily “b” and starting with number 153. Genes from an “unknown” chromosome U that clustered with 2 putative homoeologs located on other subgenomes were given a common name (e.g. this was the case for TaNAC158-U1, TaNAC158-B1, and TaNAC158-D1).
NAC subfamily classification
NAC subfamilies a-h in the joined phylogenetic tree of wheat, barley, rice, and Arabidopsis were defined based on previous studies of NAC subfamilies from barley (Christiansen et al. 2011), rice, and Arabidopsis (Shen et al. 2009). Unclassified clades were those that either did not cluster with the subfamilies (with BS ≥ 70%) or were unsupported by bootstrap analysis. TaNAC sequences were assigned to the same subfamily as their characterized rice, barley, and Arabidopsis orthologs in the joined tree, and for those not included in the joined tree, the subfamily was assigned based on their location in the wheat NAC tree (e.g. TraesCS6D02G266000 and TraesCS7A02G152400). The lists of NAC sequences and their corresponding subfamilies in wheat, barley, rice, and Arabidopsis are given in Supplementary Table 3.
Gene expression analysis
Differential expression data from 5 independent RNA-seq studies (Table 1) on fungal diseases of wheat carried out under controlled environmental conditions were obtained from Benbow et al. (2019). Genes differentially expressed upon wheat infection with bacterial pathogen Xanthomonas translucens were extracted from Expression Atlas release 36 (log2-fold change >1; P < 0.05) (Table 1). Baseline RNA-seq expression data with transcript per million (tpm) above 0 were downloaded from wheat expression browser (http://wheat-expression.com/, October 2017; Choulet et al. 2014), gene-wise normalized, and expression values of developmental stages in 5 organs were averaged (cutoff = 0.1 tpm). IBM SPSS Statistics for Windows v24 (IBM Corp., Armonk, NY, USA) was used to define the expression range as either low (1st quartile, below 0.3 tpm), moderate (2nd and 3rd quartiles, between 0.3 and 3.7 tpm) or high (4th quartile, above 3.7 tpm). Baseline and differential expression data are presented alongside the tree in a heatmap generated using iTOL. Hierarchical clustering with Euclidian distance of the pathogen-responsive TaNACs was done in Genesis v1.8.1 (Sturn et al. 2002). Sets of differentially expressed genes were visualized using the IntercatiVenn (http://www.interactivenn.net/, March 2021) web-based tool (Heberle et al. 2015). Differential expression data of HvSNAC1 (HORVU5Hr1G111590) and ONAC060 (Os12g0610600) during pathogen infection were extracted from Expression Atlas release 36 (https://www.ebi.ac.uk/gxa/home, June 2021).
Table 1.
RNA-seq experiments used for the analysis of Triticum aestivum (bread wheat) NAC (TaNAC) genes in this study.
| Experiment accession | Organ | Treatment | Wheat line/cultivara | Timepoint | Reference |
|---|---|---|---|---|---|
| E-MTAB-4289b | Leaf | Puccinia striiformis | N9134 (R) | 1, 2, and 3 dpi | Zhang et al. (2014) |
| E-MTAB-4289b | Leaf | Blumeria graminis | N9134 (R) | 1, 2, and 3 dpi | Zhang et al. (2014) |
| E-MTAB-4222b | Spikelet | Fusarium graminearum | NIL38 (R), NIL54 (S) | 3, 6, 12, 24, 36, and 48 hpi | Schweiger et al. (2016) |
| E-MTAB-4308b | Shoot | F. pseudograminearum | NIL (R), NIL1S (S) | 3 and 5 dpi | Ma et al. (2014) |
| E-MTAB-4470b | Leaf | Zymoseptoria tritici | cv. Sevin (S) | 4, 10, and 13 dpi | Yang et al. (2013) |
| E-MTAB-4116b | Leaf | Z. tritici | cv. Riband (S) | 4, 9, 14, and 21 dpi | Rudd et al. (2015) |
| E-MTAB-5891b | Leaf | Xanthomonas translucens | cv. Chinese Spring (S) | 1 dpi | Garcia-Seco et al. (2017) |
| PRJEB12497c | Leaf | P. striiformis | cv. Vuka (S) | 1 dpi | Dobon et al. (2016) |
| E-MTAB-1729c | Spikelet | F. graminearum | cv. CM-82036 (R) | 50 hpi | Kugler et al. (2013) |
| SRP078208c | Coleoptile | F. pseudograminearum | cv. Chara (S) | 3 dpi | Powell et al. (2017) |
dpi, days postinoculation; hpi, hours postinoculation.
R, resistant; S, susceptible; NIL, near isogenic line; cv., cultivar; N9134 is a resistant wheat line carrying PmAS846 introgressed from wild emmer accession As846 (Triticum dicoccoides); NIL38 and NIL54 originate from a cross between cv. CM-82036 and cv. Remus, where NIL38 is homozygous for FHB-resistant alleles at Fhb1 and Qhfs.ifa-5A QTL, and NIL51 is homozygous for susceptible alleles at both QTL; NIL1R and NIL1S originate from the population of “Janz”*2/“CSCR6” using the heterogeneous inbred family method for the Fusarium crown rot QTL on chromosome arm 3BL (Qcrs-3B), where NIL1R carries the resistant allele and NIL1S carries the susceptible allele at the Qcrs-3B locus.
Used in the analysis of the response of all wheat NAC genes to the pathogen.
Used to validate the pathogen-responsiveness of specific NAC genes.
Publicly available tpm data from 3 additional RNA-seq studies on wheat infected with Fusarium graminearum, Fusarium pseudograminearum, and P. striiformis (Kugler et al. 2013; Dobon et al. 2016; Powell et al. 2017) (Table 1) were mined for select genes using the Wheat expression browser (http://wheat-expression.com/) and tpm data were analyzed to validate the pathogen-responsiveness of select genes for validation purposes.
Protein motif analysis
The HMM profile of NAM family proteins (PF02365) was aligned with the TaNAC amino acid sequences using hmmalign in HMMER. NAM subdomains (A–D) defined the NT part of the protein, and the remainder was trimmed (manually in Jalview v2.10.5) to extract CT sequences. When sequences had 2 NAM domains, the output given by hmmsearch was used to identify the coordinates of the second domain (output gives conditional E-values for each domain and their position in a sequence). Identification of motifs in the NT and CT of TaNACs was done separately in MEME v5.0.5 (http://meme-suite.org/, May 2019; Bailey et al. 2009). To search motifs in the NT, the parameters were: motif discovery mode—classic, motif site distribution—any number of repetitions, maximum number of motifs—20, and motif width—between 5 and 20 residues. For the CT, the parameters were: motif discovery mode—classic, motif site distribution—zero or 1 site per sequence, maximum number of motifs—40, and motif width—between 5 and 20 residues. Motifs were presented alongside the phylogenetic tree in iTOL. The position of the discovered motifs was compared with positions of subdomains A–E of the NAC domain defined by Ooka et al. (2003).
Statistical analysis
Statistical analysis was performed using IBM SPSS Statistics for Windows v24. The Shapiro–Wilk test was used to evaluate whether the data were normally distributed. The Kruskal–Wallis test with step-down comparison was used to analyze significant differences among distributions of sequence bit scores of subfamilies a-h, and a significance level was P < 0.05. In addition, Fisher’s exact test (conducted in Microsoft Excel 2011) was used to analyze differences in the proportion of pathogen-responsive TaNACs among subfamilies. The χ2 test (conducted in Microsoft Excel 2011) was used to analyze difference in proportions of pathogen-responsive TaNACs within the TaNAC family as compared to the proportion of a total number of pathogen-responsive high confidence genes within a wheat genome. The Moses test was used to analyze difference between tpm values in mock sample vs. pathogen-treated samples.
Results
Wheat NAC nomenclature
We propose a new T. aestivum (bread wheat) TaNAC nomenclature to simplify and streamline the naming of this gene family within this complex genome. Genes were named based on the relationship with their O. sativa (rice) OsNAC orthologs and conventional wheat gene nomenclature (McIntosh et al. 2013; Schilling et al. 2020) (Supplementary Tables 4 and 5). Thus, TaNACs were named according to the name of the phylogenetically closest OsNAC ortholog (TaNACXXX), their wheat subgenome (A, B, or D), the number of consecutive TaNAC singletons or homoeologous genes (1–8) that are orthologs of the OsNAC and, if relevant, the number of the consecutive TaNAC paralogs (2–5) within the gene cluster (TaNACXXX-A/B/D (1-8)-(2-5)). For example, TaNAC048-A1 (TraesCS3A02G406000), TaNAC048-B1 (TraesCS3B02G439600), and TaNAC048-D1 (TraesCS3D02G401200) are the only triad of homoeologs from A, B, and D wheat subgenomes named after the rice ortholog ONAC048. One of the more complex clusters consists of 10 TaNACs that are orthologs of ONAC066 (Os03g0777000); this includes the subgenome B singleton TaNAC066-B1 (TraesCS4B02G384700) orthologous to ONAC066 and 9 TaNACs from subgenomes A, B, and D that form a second homeologous cluster within the ONAC066 clade and that are inparalogs with >80% ID [e.g. TaNAC066-A2-1 (TraesCS5A02G411700), TaNAC066-A2-2 (TraesCS5A02G411800), and TaNAC066-A2-3 (TraesCS5A02G411900) are 3 consecutive inparalogs residing on chromosome A].
Phylogenetic relationship of defense-associated wheat, rice, barley, and Arabidopsis NACs
A total of 460, 138, 101, and 113 NAC proteins were respectively identified in the genome assemblies of wheat [IWGSC RefSeq v1.1; ∼17 Gb (2n = 6x = 42)], barley [IBSC v2; ∼5.3 Gb (2n = 14)], rice [IRGSP 1.0; ∼500 Mb (2n = 24)], and Arabidopsis [TAIR 10; ∼135 Mb (2n = 10)] (Supplementary Table 6). Although not further analyzed herein, a total of 766 low confidence wheat NAC proteins encoded by a total of 765 low confidence genes were also identified in this study (Supplementary Table 6). Of the 460 wheat high confidence TaNAC genes, 413–460 were also identified in previously mentioned studies (Borrill et al. 2017; Guérin et al. 2019; Lv et al. 2020; Ma et al. 2021). Notable differences between our results and other studies were (1) the wheat gene TraesCS5B02G271800 identified in Lv et al. (2020) was not identified in this study, (2) genes TraesCS5B01G550800 and TraesCS7B01G481500 identified in Ma et al. (2021) and Guérin et al. (2019) were not identified in this study, and (3) gene TaNAC118-D4 (TraesCS5D02G537600) from this study was not identified in Lv et al. (2020) and Guérin et al. (2019) (Supplementary Table 7).
From these 812 wheat, barley, rice, and Arabidopsis NACs, the evolutionary relationships of 751 unambiguous nonidentical NAC protein sequences were estimated using a joined maximum-likelihood phylogenetic tree. NACs excluded during tree construction (see Supplementary Table 2) were 21 that had insufficient overlap (Cij score <0.5) and 40 identical/surplus masked sequences (these were later positioned alongside their identical sequence in the tree). The final high-quality MSA (Supplementary File 1) used for the tree inference had 179 sites/columns and a completeness score (Ca) of 0.88.
As illustrated in Fig. 1a, there are 734 NACs, including 451 TaNACs, distributed in 8 subfamilies (a–h) and 17 unclassified NACs positioned outside of these subfamilies (7 of those were TaNACs). Similarly, 453 TaNACs were grouped in 8 subfamilies in the phylogenetic tree constructed by Borrill et al. (2017), and the majority of TaNACs delineated in Borrill et al. (2017) and the current study (i.e. 411 TaNACs) were placed in the same subfamily (Supplementary Table 3). Subfamilies “b” and “g” partitioned into 3 individual subclades based on the bootstrap analysis. All studied plants had NACs in all subfamilies (Fig. 1a and Table 2), with 1 subclade within subfamily “g” being Arabidopsis specific. Subfamily “d” was the most abundant subfamily in barley and rice with 23 and 20 NACs, respectively, while “b” was the subfamily most expanded in Arabidopsis with 33 NACs. Subfamily “h” was the most abundant for wheat and was almost exclusively restricted to monocots (including just 1 Arabidopsis NAC) and it is the largest subfamily in the plants we studied. It contained 23% (106), 16% (21), 12% (11), and 1% (1) of the wheat, barley, rice, and Arabidopsis NACs, respectively.
Fig. 1.
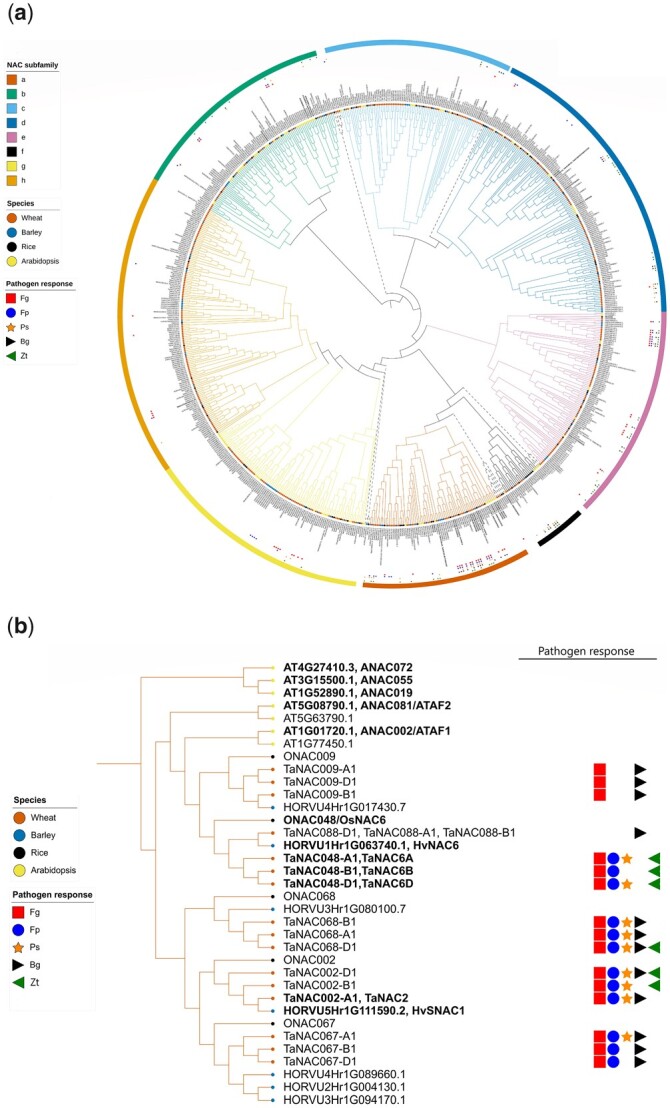
Phylogenetic relationship of NACs in wheat, rice, barley, and Arabidopsis. a) Unrooted consensus maximum likelihood joined phylogenetic tree of wheat, barley, rice and Arabidopsis NACs (https://itol.embl.de/tree/372282299412371613249495) and b) a close-up of the “TaNAC2 subclade” of subfamily “a.” Bootstrap scores >50% are indicated on each node in the online version. Branch lines are colored according to NAC subfamily and terminate in a colored circle, which denotes the plant species; alongside the tree, the colored symbols denote the TaNACs that are responsive (at the transcriptome level) to pathogens. All NACs genetically characterized for their role in diseases are highlighted in bold. Identical sequences positioned on the same branch were assigned the pathogen symbol only if all of them were responsive to the specific fungus. Pathogen abbreviations: Fg, Fusarium graminearum; Fp, Fusarium pseudograminearum; Ps, Puccinia striiformis; Bg, Blumeria graminis; Zt, Zymoseptoria tritici.
Table 2.
Number of NAC genes in subfamilies a-h in wheat, barley, rice, and Arabidopsis.
| Subfamily | Number of NACs per subfamily (% of total NACsa) |
||||
|---|---|---|---|---|---|
| Wheat b | Barley | Rice | Arabidopsis | Total | |
| a | 41 (9) | 17 (13) | 12 (13) | 12 (11) | 82 (10) |
| b | 43 (9) | 18 (14) | 10 (10) | 33 (30) | 104 (13) |
| c | 53 (12) | 12 (9) | 10 (10) | 13 (12) | 88 (11) |
| d | 78 (17) | 23 (18) | 20 (20) | 14 (13) | 135 (17) |
| e | 64 (14) | 17 (13) | 11 (11) | 8 (7) | 100 (13) |
| f | 12 (3) | 3 (2) | 8 (8) | 3 (3) | 26 (3) |
| g | 56 (12) | 16 (12) | 9 (9) | 21 (19) | 102 (13) |
| h | 106 (23) | 21 (16) | 11 (12) | 1 (1) | 139 (18) |
| Unclassified | 5 (1) | 1 (1) | 5 (5) | 4 (4) | 15 (2) |
| Total numberc | 458 | 128 | 96 | 109 | 791 |
Number expressed as a percentage of the total number of NACs in the given species is given in parenthesis.
The final classification of wheat NACs was based on the phylogenetic tree constructed for TaNACs and described below rather than the combined tree constructed for all plants studied. There were differences in wheat NAC classification based on which tree was used; for example, the joined phylogenetic tree had 7 unclassified TaNACs but 2 of those were classified into subfamilies based on the TaNAC tree below.
Note that the total number of classified NAC genes differs from the total number of identified NAC genes since some of the NAC genes were excluded from the phylogenetic analysis as explained in the main text.
The 29 wheat, barley, rice, and Arabidopsis NACs functionally characterized (with genetic evidence) for their role in defense against pathogens (Table 3) are highlighted in bold within the NAC phylogenetic tree (Fig. 1a). These were distributed across each subfamily, except “f” and “g,” with “a” containing the highest number (15) of them (Fig. 1a and Table 3). Ten of them were from wheat and were distributed in subfamilies “a,” “c,” “d,” “e,” and “h.” Five uncharacterized defense-associated TaNACs were positioned directly next to their functionally characterized ortholog (TaNAC002-A1 and TaNAC048 homoeologs in subfamily “a” next to HvSNAC1 and HvNAC6, respectively, and TaNAC060-B1 in subfamily “d” next to rice ONAC060) (Fig. 1b). Orthologs TaNAC002-A1 and HvSNAC1 were differentially expressed in response to B. graminis (Supplementary Files 2 and 3, respectively). Expression of orthologs TaNAC060-B1 and ONAC060 was, respectively, shown to regulate defense against the obligate biotroph P. striiformis and the hemibiotroph Magnaporthe oryzae (Supplementary File 4 and Table 3). TaNAC048 homoeologs and their characterized ortholog HvNAC6 were involved in positive defense against B. graminis (Table 3) and thus are likely isofunctional orthologs.
Table 3.
Functionally characterized NACs in Triticum aestivum, Hordeum vulgare, Oryza sativa and Arabidopsis thaliana.
| Subfamily | Species | Cultivar/ecotype | Gene ID | Gene namea | mRNA/protein AC | Gene function | Genetic evidence | Reference |
|---|---|---|---|---|---|---|---|---|
| a | T. aestivum | Suwon11 | TraesCS5A02G468300 | TaNAC2, TaNAC002-A1 | AAU08786.1b | Suppressed resistance to Puccinia striiformis f. sp. tritici | VIGS | Zhang et al. (2018) |
| NAU9918, Yangmai158, OEStpk-V | TraesCS3A02G406000 | TaNAC6A, TaNAC048-A1 | FN396829.1b | Enhanced resistance to Blumeria graminis | Overexpression and VIGS | Zhou et al. (2018) | ||
| TraesCS3B02G439600 | TaNAC6B, TaNAC048-B1 | FN396830.1b | Enhanced resistance to B. graminis | VIGS | Zhou et al. (2018) | |||
| TraesCS3D02G401200 | TaNAC6D, TaNAC048-D1 | FN396831.1b | Enhanced resistance to B. graminis | VIGS | Zhou et al. (2018) | |||
| NIL-R from Sumai 3 | TraesCS3B02G194000 | TaNAC032, TaNAC160-B1 | MT512636b | Enhanced resistance to Fusarium graminearum | VIGS | Soni et al, (2021) | ||
| H. vulgare | P-01 | HORVU1Hr1G063740 | HvNAC6 | CAM57978.1b | Enhanced resistance to B. graminis | Overexpression, VIGS, RNA interference | Chen et al. (2013); Jensen et al. (2007) | |
| Haruna Nijo | HORVU5Hr1G111590 | HvSNAC1 | AK249102.1b | Enhanced resistance to Ramularia collo-cygni | Overexpression | McGrann et al. (2015) | ||
| O. sativa | Nipponbare | Os11g0126900 | ONAC122 | LOC_Os11g03300.1c | Enhanced resistance to Magnaporthe oryzae | VIGS | Sun et al. (2013) | |
| Os12g0123700 | ONAC131 | LOC_Os12g03040.1c | Enhanced resistance to M. oryzae | VIGS | Sun et al. (2013) | |||
| Os01g0884300 | OsNAC6, ONAC048 | BAG90892.1b | Enhanced resistance to M. oryzae | Overexpression | Nakashima et al. (2007) | |||
| A. thaliana | Col-0 | At1g01720 | ATAF1, ANAC002 | BT024513.1b | Enhanced resistance to B. graminis | T-DNA insertion | Jensen et al. (2007) | |
| Conferred susceptibility to Botrytis cinerea | Overexpression | Wu et al. (2009) | ||||||
| Suppressed resistance to B. cinerea, Pseudomonas syringae pv. Tomato (PsPto) strain DC3000 or Alternaria brassicicola | T-DNA insertion, overexpression | Wang et al. (2009) | ||||||
| Age‐related resistance to Hyaloperonospora parasitica | T-DNA insertion | Carviel et al. (2009) | ||||||
| At5g08790 | ATAF2, ANAC081 | BAF00699.1b | Suppressed resistance to B. cinerea | Overexpression | Delessert et al. (2005) | |||
| At1g52890 | ANAC019 | AAM51299.1b | Suppressed resistance to Fusarium oxysporum | T-DNA insertion, overexpression | Bu et al. (2008) | |||
| Suppressed resistance to P. syringae pv. maculicola (Psm) strain ES4326 | T-DNA insertion | Zheng et al. (2012) | ||||||
| At3g15500 | ATNAC3, ANAC055 | ABF74720.1b | Suppressed resistance to F. oxysporum | T-DNA insertion, overexpression | Bu et al. (2008) | |||
| Suppressed resistance to Psm strain ES4326 | T-DNA insertion | Zheng et al. (2012) | ||||||
| Age‐related resistance to H. parasitica | T-DNA insertion | Carviel et al. (2009) | ||||||
| At4G27410 | ANAC072 | NM_001084983.1b | Suppressed resistance to Psm strain ES4326 | T-DNA insertion | Zheng et al. (2012) | |||
| b | O. sativa | Nipponbare | Os03g0119966 | RIM1, ONAC054 | BAH03170.1b | Conferred susceptibility to Rice Dwarf Virus (RDV) | Tos17 insertion, overexpression | Yoshii et al. (2009) |
| A. thaliana | Col-0 | At5g24590 | TIP, ANAC091 | AAF87300.1b | Conferred resistance to Turnip Crinkle Virus (TCV) | Resistant Di-17 inbred line | Ren et al. (2000) | |
| At3g49530 | NTL6, ANAC062 | AAL24091.1b | Enhanced resistance to PsPto strain DC3000 | Overexpression of truncated NTL6-6ΔC, RNA interference | Seo et al. (2010) | |||
| At4g35580 | NTL9, CBNAC | NM_001125650.1b | Suppressed resistance to PsPto strain DC3000 | T-DNA insertion | Kim et al. (2012) | |||
| c | T. aestivum | Suwon11 | TraesCS2D02G576400 | TaNAC30, TaNAC031-D3-2 | Tae062585c | Suppressed resistance to P. striiformis f. sp. tritici | VIGS | Wang et al. (2018) |
| d | TraesCS5B02G054200 | TaNAC21/22, TaNAC060-B1 | AGV08300.1b | Suppressed resistance to P. striiformis f. sp. tritici | VIGS | Feng et al. (2014) | ||
| Taichung 29 | TraesCS7A02G305200 | TaNAC1, TaNAC104-A2 | ADG85703.1b | Suppressed resistance to P. striiformis f. sp. tritici, and to PsPto strain DC3000 | VIGS (Wheat), ectopic overexpression (Arabidopsis) | Wang et al. (2015) | ||
| O. sativa | Nipponbare | Os12g0610600 | OMTN3, ONAC060 | XM_015765089.2b | Enhanced resistance to M. oryzae | RNA interference | Wang et al. (2018) | |
| A. thaliana | Col-0 | At5g39610 | ANAC092 | AY091191.1b | Age‐related resistance to H. parasitica | T-DNA insertion | Carviel et al. (2009) | |
| e | T. aestivum | Thatcher + Lr14b | TraesCS3D02G398200 | TaNAC35, TaNAC075-D1 | KY461080.1b | Suppressed resistance to Puccinia triticina | VIGS | Zhang et al. (2021) |
| O. sativa | Nipponbare | Os11g0154500 | OsNAC111, ONAC017 | XM_015760375.2b | Enhanced resistance to M. oryzae | Overexpression | Yokotani et al. (2014) | |
| Os03g0777000 | ONAC066 | LOC_Os03g56580.1c | Enhanced resistance to M. oryzae and Xanthomonas oryzae pv. Oryzae [Xoo] | Overexpression | Liu et al. (2018) | |||
| H | T. aestivum | CM82036 | TraesCS5D02G111300 | TaNACL-D1, TaNAC175-D1 | MG701911.1b | Enhanced resistance to F. graminearum | Overexpression | Perochon et al. (2019) |
| Unknown | A. thaliana | Col-0, Bur-0 | At5g64530 | XND1, ANAC104 | NM_001345642.1b | Enhanced resistance to Ralstonia solanacearum | T-DNA insertion, complementation | Tang et al. (2018) |
AC, accession number; VIGS, virus-induced gene silencing.
T. aestivum NAC names include the ones that are established based on the nomenclature proposed in this study.
GeneBank AC.
TFDB (Transcription factor database) AC.
A total of 146 out of the total 460 TaNACs have been shown to be pathogen responsive based on 5 publicly available RNA-seq studies (Supplementary File 2) and their phylogenetic relationship with the 29 NACs with a validated role in defense was investigated (Fig. 1a). A total of 52 of these pathogen-responsive TaNACs were positioned within the same subclade as a functionally characterized defense-associated NAC, and 35 of them were positioned within the same subclade as a defense-associated TaNAC. Interestingly, 14 of them were within the “TaNAC2 subclade,” which includes the functionally characterized TaNAC048 homoeologs and TaNAC002-A1 (Fig. 1b).
The expression pattern and phylogenetic relationship of pathogen-responsive TaNACs
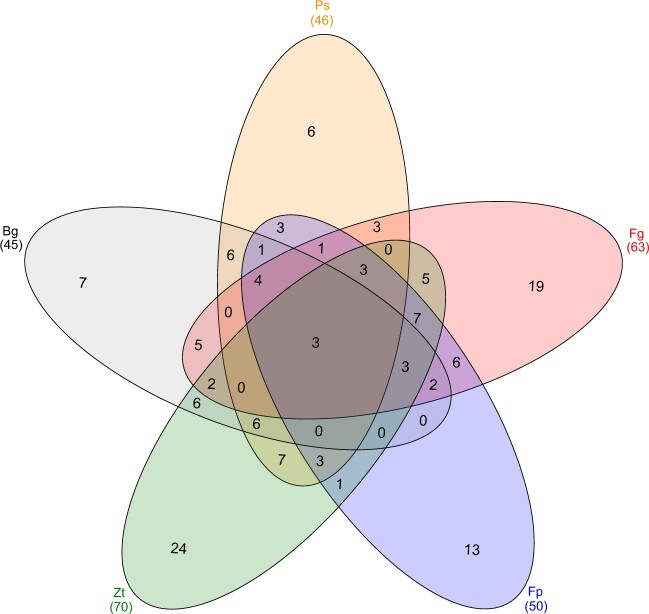
We investigated the expression profile of the 146 pathogen-responsive TaNACs (Supplementary File 2) and examined if there was any association with the lifestyle (hemibiotrophic or biotrophic) of the activating fungus. Some were specific for a disease (as illustrated in Fig. 2). Fifty-one percentage of all pathogen-responsive TaNACs were specific to hemibiotrophs (F. graminearum, F. pseudograminearum, Z. tritici), and 13% to biotrophs (B. graminis and P. striiformis) (Figs. 2 and 3 and Supplementary Table 8). Within the 127 TaNACs that were responsive to hemibiotrophic pathogens, a larger proportion were responsive to only hemibiotrophs (53–59% for the different pathogens) as compared to both hemibiotrophs and biotrophs (41–47%). In contrast, a larger proportion of the 71 TaNACs responsive to biotrophic pathogens were responsive to both biotrophs and hemibiotrophs (71–74%) as compared to only biotrophs (26–29%). It is clear from the heatmap (Fig. 3), and time point gene expression data (Supplementary Table 9) that many more TaNACs were upregulated by hemibiotrophs as compared to biotrophs, while many more TaNACs were downregulated by biotrophs as compared to hemibiotrophs. The number of upregulated TaNACs generally increased with time postinfection with hemibiotrophic pathogens (except in cv. Riband infected with Z. tritici) and decreased with time postinfection for biotrophic pathogens (Fig. 3 and Supplementary Table 9). The TaNAC family had more Fusarium (F. graminearum and F. pseudograminearum)- responsive genes than expected (14% and 11%, respectively), as compared to the whole genome response to these fungi (10% and 6%, respectively; χ2 test, P-value <0.001); the same was not observed with the other 3 diseases (Supplementary Table 10). Out of curiosity, we also investigated whether fungal-responsive TaNACs were responsive to a bacterial pathogen; of the 146 TaNACs responsive to fungal pathogens, 39 (27%) were also responsive to a bacterial hemibiotroph (X. translucens) and 92% of those clustered in subclades enriched with pathogen-responsive TaNACs (see Supplemental Results). For validation purposes, we also conducted analysis to determine if the TaNACs that were most induced by F. graminearum, F. pseudograminearum, and P. striiformis (up to 5 per pathogen) were induced by the same pathogens in other distinct RNA-seq studies (Kugler et al. 2013; Dobon et al. 2016; Powell et al. 2017). All such TaNACs were significantly induced by pathogens in these studies (see Supplemental Results and Supplementary Table 11). In addition, we compared TaNACs responsive to B. graminis and/or P. striiformis with those delineated in studies by Lv et al. (2020) and Ma et al. (2021) who analyzed the same RNA-seq study (E-MTAB-4289). There were likely differences in analysis and cross-comparison was not directly possible for all RNA-seq results from Lv et al. (2020), which may explain why many of our pathogen-responsive TaNACs were not detected as being so in their studies and vice versa. But 18 and 11 TaNACs, respectively, responsive to B. graminis and P. striiformis herein were also responsive to the same pathogens in Lv et al. (2020) and/or Ma et al. (2021) (see Supplemental Results and Supplementary Tables 12 and 13).
Fig. 2.
Venn diagram showing the numbers of TaNAC genes that are positively and/or negatively transcriptionally responsive to pathogens. Each colored oval represents a specific pathogen response. Pathogen abbreviations: Fg, Fusarium graminearum; Fp, Fusarium pseudograminearum; Ps, Puccinia striiformis; Bg, Blumeria graminis; Zt, Zymoseptoria tritici. The total number of TaNACs responsive to each pathogen is indicated between brackets.
Fig. 3.
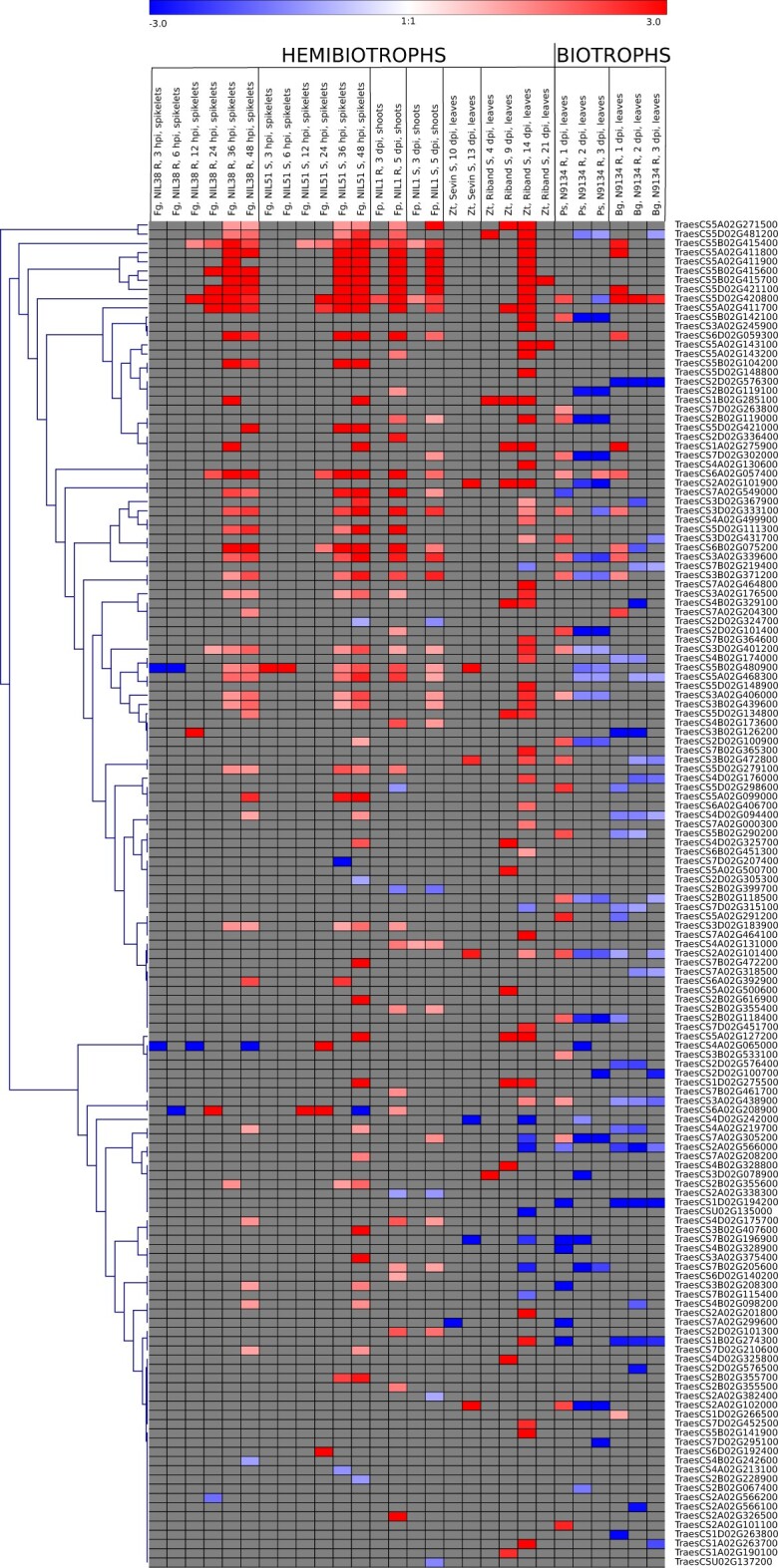
A heatmap of hierarchically clustered TaNAC genes indicating the temporal responsiveness to hemibiotrophic and biotrophic fungal pathogens. Color scale at the top of the heatmap represents log2-fold change of gene expression: blue represents downregulated expression and red upregulated gene expression, relative to mock treatment. “R” and “S” next to the cultivar or NIL (near-isogenic line) names indicate resistance and susceptibility to a pathogen, respectively. Pathogen abbreviations: Fg, Fusarium graminearum; Fp, Fusarium pseudograminearum; Ps, Puccinia striiformis; Bg, Blumeria graminis; Zt, Zymoseptoria tritici.
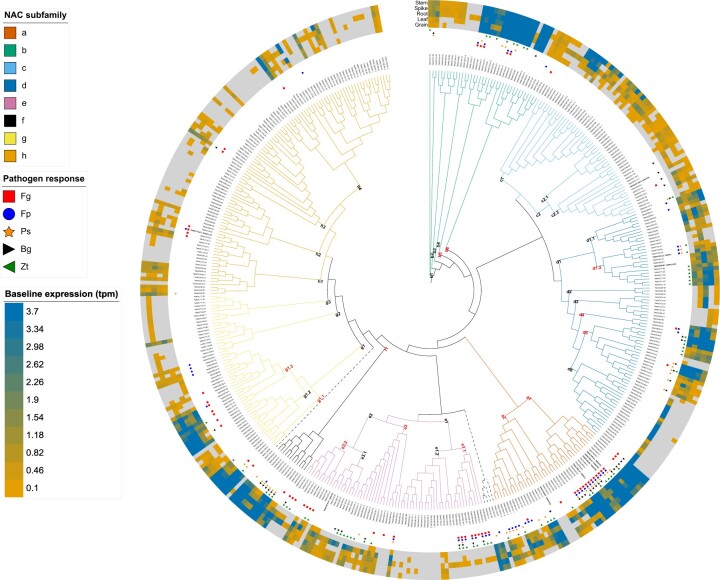
A wheat-specific phylogenetic tree was constructed to examine the distribution of pathogen-responsive TaNACs within NAC subfamilies (Fig. 4). The tree was inferred based on the MSA of 446 TaNAC sequences and 218 sites/columns with completeness score (Ca) of 0.9 (Supplementary File 6); 14 surplus identical sequences were removed from the phylogenetic analysis (Supplementary Table 2). Eighty-six percentage (119) of the pathogen-responsive TaNACs were densely clustered into 13 pathogen-responsive-enriched subclades within subfamilies “a,” “b,” “d,” “e,” “f,” and “g” (delineated by red labels at the nodes of the tree; ≥60% of TaNACs within these subclades being pathogen responsive). All pathogen-responsive TaNACs in subclade “g1.3” were only responsive to hemibiotrophs. At the subfamily level, “a,” “e,” and “f” were significantly enriched in pathogen-responsive TaNACs, as compared to “c” and “h” (representing 80, 45, 67, 11, and 8% of the total number of TaNACs in the subfamily, respectively; P-value <0.05, Fisher’s exact test) (Supplementary Table 14). The majority (>66%) of the TaNACs in subfamilies “a,” “e,” and “f” were responsive to more than 1 pathogen, while most (>78%) of the pathogen-responsive TaNACs in subfamilies “c” and “h” were responsive to only 1 pathogen (Supplementary Table 14). Subfamilies “a” and “e” had the highest number (10-22) of TaNACs responsive to F. graminearum, B. graminis, and Z. tritici, as compared to other subfamilies, and subfamily “a” had the highest number of TaNACs responsive to F. pseudograminearum and P. striiformis (20 and 17, respectively) (Fig. 4 and Supplementary Fig. 1).
Fig. 4.
Consensus maximum likelihood tree of TaNAC proteins with their corresponding healthy tissue and disease-responsive gene expression profiles indicated (https://itol.embl.de/tree/372282299294211613865063). Branch lines are colored according to NAC subfamily (unclassified branches and clades are indicated with a dashed line). Within a subfamily, TaNAC subclades are coded at the node based on the subfamily letter and a specific number. For example, “e1” is the first subclade of subfamily “e,” and “e1.1” and “e1.2” are two distinct subclades within “e1” subclade. Red font indicates subclades enriched with pathogen-responsive TaNACs. TaNACs highlighted in bold are functionally characterized in pathogen defense (Table 3). The colored symbols denote the TaNACs that are responsive (at the transcriptome level) to pathogens. Pathogen abbreviations: Fg, Fusarium graminearum; Fp, Fusarium pseudograminearum; Ps, Puccinia striiformis; Bg, Blumeria graminis; Zt, Zymoseptoria tritici. TaNAC expression within healthy tissues (grain, leaf, root, spike, and stem) is represented by a color scale that represents baseline expression in tpm values, and the color gradient maximum is set to 3.7 tpm.
In general, pathogen-responsive TaNACs within subclades did not follow similar expression patterns for different pathogens (Supplementary Fig. 2). However, all TaNACs in the “a1” subclade (the “TaNAC2 subclade”) were pathogen responsive, with 83% and 61% of the TaNACs upregulated by F. graminearum and F. pseudograminearum, respectively, and 56% up- and/or downregulated to B. graminis, in at least 1 timepoint (Fig. 1b and Supplementary Fig. 2). Similarly, >80% of the genes in the “e1.1” subclade were upregulated by F. graminearum, F. pseudograminearum, and Z. tritici.
The expression profile of TaNACs in healthy wheat organs (grain, leaf, root, spike, and stem) was assessed to determine if this differed for pathogen vs. nonpathogen responsive genes (Fig. 4 and Supplementary File 7; Choulet et al. 2014). The majority (92%) of pathogen-responsive TaNACs were expressed in healthy tissue and in at least 2 organs, while 6% were expressed in 1 organ and 2% were not expressed in any organ. The baseline expression of pathogen-responsive TaNACs was high compared to nonpathogen responsive TaNACs (mean tpm in all tissues >4.7 as compared to <1.2 for all other TaNACs).
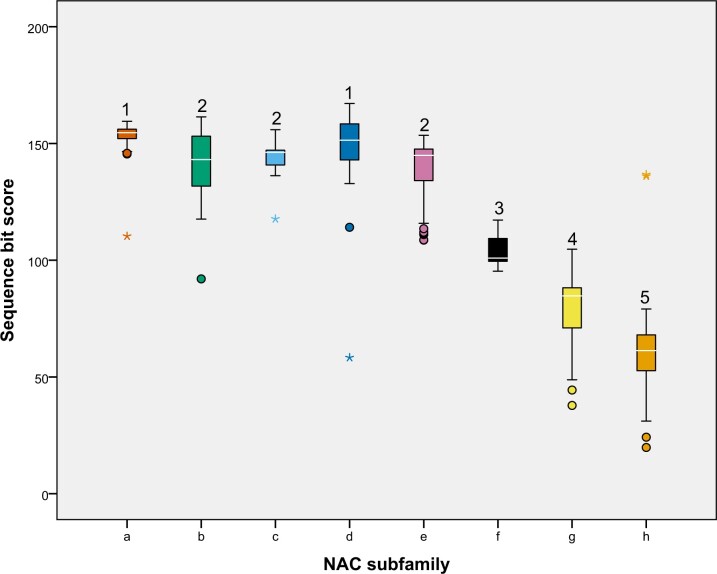
TaNACs in subfamilies “f,” “g,” and “h” encode a diverged NAM domain
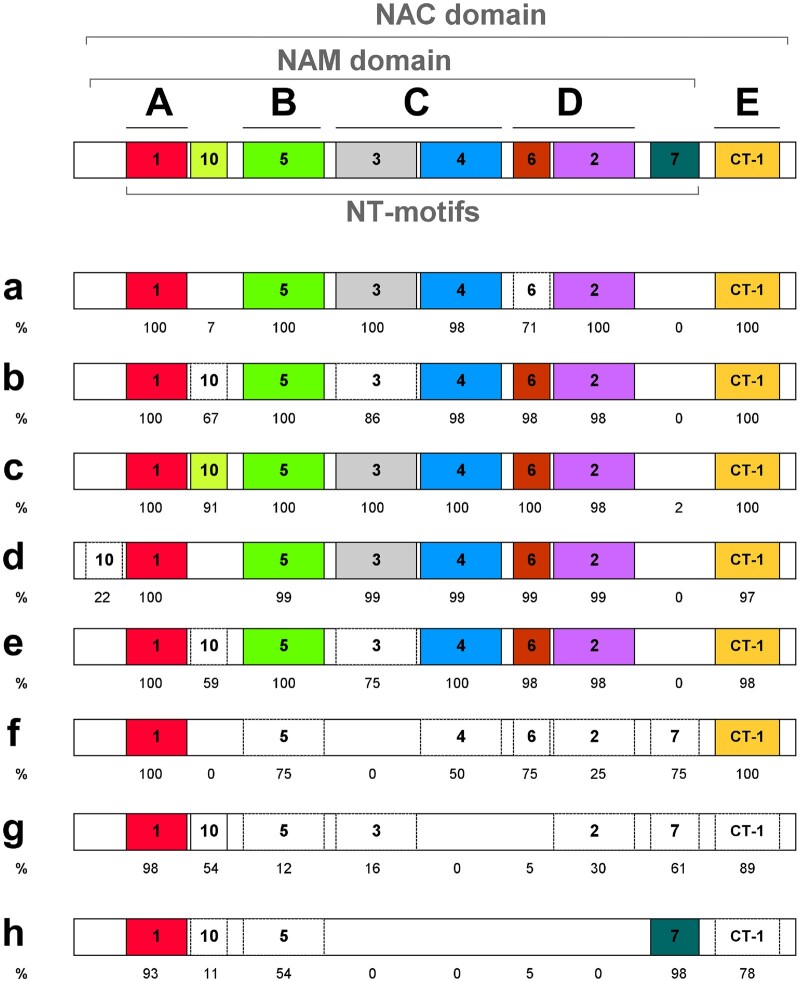
The highly conserved NAM domain has not previously been analyzed in wheat and hence was investigated herein based on HMM (the higher the score the better the match with the profile of the NAM domain), motif and domain analysis (Figs. 5 and 6). Bit scores within subfamilies a–e were high (116–167), with some small but significant differences between these subfamilies. In comparison, there was greater NAM domain divergence within subfamilies f–h as indicated by the lower scores (31–117) (Fig. 5). We examined the distribution of a total of 21 motifs within the NT NAC domain and 39 motifs within the putative CT TAR of all wheat TaNACs (Supplementary Fig. 3, a and b and Supplementary Tables 15 and 16). Eight NT motifs were specific for a subfamily and were present in 8–61% of subfamily members. The 7 most conserved motifs (NT-1 to NT-6, and CT-1) were all present in more than 50% of TaNACs (Supplementary Fig. 3a); all others were present in less than 40% of the TaNACs. Figure 6 schematically illustrates how the motifs map to the conserved NAC subdomains A–E within each TaNAC subfamily. MEME analysis deduced that the 7 conserved motifs mapped to subdomains A–E. An additional 2 NT motifs (NT-7 and NT-10) were found to be positioned between the NAC subdomains.
Fig. 5.
The sequence bit scores from HMMER output across the TaNAC subfamilies. The sequence bit scores indicate the probability of a TaNAC sequence being homologous to the hmm profile of the NAM domain (Finn et al. 2011). Within the boxplots, the white line denotes the median. Outliers and extreme values, below or above whiskers, are marked with the circle or the star, respectively. Boxplots that do not share the same number above whiskers are significantly different (P-value <0.05).
Fig. 6.
Schematic representation of a conserved NAC domain (on the top) within TaNAC proteins and the distribution of NAC motifs within the protein subfamilies. Only highly conserved motifs present in at least 90% of the TaNACs in at least 1 subfamily are presented in the scheme. Based on Ooka et al. (2003), a canonical NT NAC domain comprises the NAM domain (encompassing subdomains A–D) and a subdomain E. Motif distribution within subdomains is illustrated; a motif is colored if it considered a signature of a subfamily, being present in more than 90% of TaNACs, while a motif is uncolored if present in 10–90% of TaNACs in the subfamily; a motif is absent when less than 10% of TaNACs in the subfamily contain it. The values below each motif represent the percentage of TaNACs in the subfamily that contain it.
All subfamilies had well-conserved subdomains A and E (motifs NT-1 and CT-1, respectively; Fig. 6). In addition, subdomains B, C, and D (motifs NT-2, -3, -4, -5, and -6) were well conserved within subfamilies a–e, while in subfamilies f–h, these subdomains were partially present or absent. Thus, this confirms the significant divergence of NAM domain in subfamilies f–h. Interestingly, motif NT-7 is specific to subfamilies f–h and found nearly in all TaNACs from family “h” (98%). It may have diverged from NT-2 because they both have conserved amino acid residues T[x]W[x]MHE when comparing motif logos (Supplementary Table 15). None of the 39 motifs within TAR were conserved within the TaNAC family, all being present in <19% of the total number of TaNACs. But 28 of them were conserved within subfamilies, being specific to a subfamily (Supplementary Fig. 3b) and present in 10–34% of the total number of TaNACs in the subfamily (Supplementary Fig. 4). Twenty-six of these 28 CT motifs were specific for a subclade (all except CT-7 and CT-17) (Supplementary Fig. 4).
CT motifs of TaNACs specific for the subclade may be linked to a pathogen responsiveness or an expression in grain
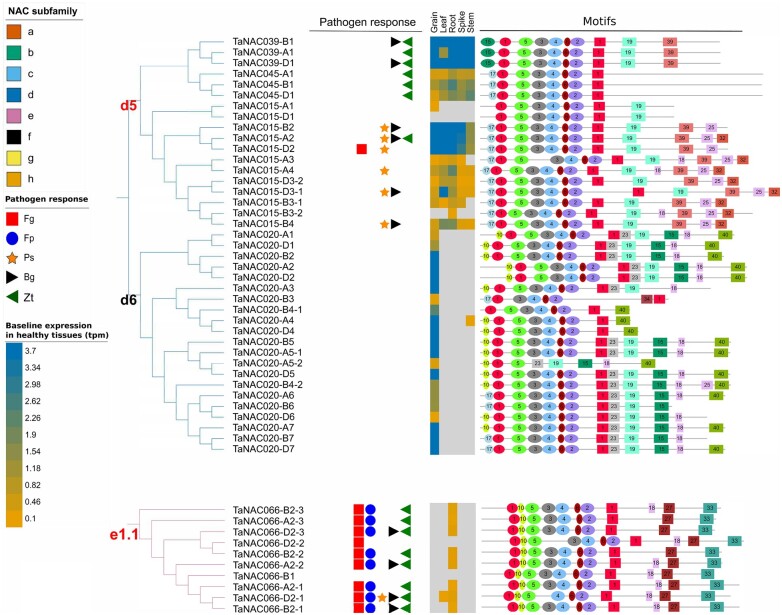
The association, if any, between NT or CT motif composition and expression pattern of TaNACs in either healthy tissues, or in response to pathogens, was investigated (detailed motif composition is illustrated in Supplementary Fig. 4). In general, the developmental and pathogen response of TaNACs was not associated with any specific NT or CT motif. The exceptions are illustrated in Fig. 7. Motifs CT-27 and CT-33 specific for the “e1” subclade were found in all pathogen-responsive TaNACs within that subclade. In addition, CT-39 specific for the “d5” subclade that is enriched with pathogen-responsive TaNACs was present in 72% of TaNACs of the subclade. In terms of tissue-specific expression patterns, all TaNACs in the subclade “d6” were expressed only in grain, except 1 gene that was expressed also in stem tissue at a relatively low level. Motifs CT-23 and CT-15 were specific for the subclade “d6” and present in 81% and 76% of the TaNACs within the subclade, respectively.
Figure 7.
Motif composition of TaNACs in subclades “d5” and “e1.1” enriched with pathogen-responsive TaNACs, and the grain-specific subclade “d6” as part of consensus maximum likelihood tree of TaNAC amino acid sequences (https://itol.embl.de/tree/372282299294211613865063). “D5” and “d6” are 2 of the 6 subclades within subfamily “d,” and “e1.1” is one of the 2 distinct subclades within “e1” subclade within subfamily “e.” Pathogen responsiveness is indicated with colored symbols (see legend). Pathogen abbreviations: Fg, Fusarium graminearum; Fp, Fusarium pseudograminearum; Ps, Puccinia striiformis; Bg, Blumeria graminis; Zt, Zymoseptoria tritici. For tissue-specific expression, the colored scale represents tpm baseline expression values for the genes in healthy tissues (grain, leaf, root, spike, and stem) (see baseline expression legend). Motifs: NT (circle) and CT (rectangle) motifs are represented with a different colors and numbers. Gray lines flanking the motifs represent nonconserved regions of TaNACs.
Discussion
This study represents the first in-depth in silico analysis of TaNACs responsive to diverse fungal diseases, which included assessment of their phylogenetic relationship with defence-associated NACs from Arabidopsis, rice, and barley. Subfamily “a” had the highest number of functionally characterized defense-associated NACs from wheat, barley, rice, and Arabidopsis. Therefore, the function in pathogen defense seems to be conserved and characteristic for subfamily “a,” as first suggested by Shen et al. (2009). Subfamily “a” has the lowest evolutionary rate, being the most conserved NAC subfamily that originated in mosses (Zhu et al. 2012; Jin et al. 2017), and this is supported by our findings. Hence, subfamily “a” NACs may be a crucial component of plants’ immune systems since the origin of basal land plants.
Within subfamily “a,” 14 pathogen-responsive TaNACs orthologous to defense-associated NACs were positioned in the “TaNAC2 subclade.” Several heterospecies and isospecies homologs within the “TaNAC2 subclade” share the same proven function, e.g. HvNAC6, ATAF1, and TaNAC048 homoeologs were positive regulators of penetration resistance against B. graminis (Jensen et al. 2007; Chen et al. 2013; Zhou et al. 2018), and ATAF1 and ATAF2 were both negative regulators of resistance to Botrytis cinerea (Delessert et al. 2005; Wang et al. 2009). Therefore, one could speculate regarding the function of uncharacterized NACs in the “TaNAC2 subclade.” For example, TaNAC067-A1, TaNAC002-B1 and -D1 homoeologs, and TaNAC068 homoeologs were all responsive to P. striiformis and were the closest homologs of the TaNAC2 involved in P. striiformis defense based on their phylogenetic relationship. Thus, these NACs are very likely involved in defense against P. striiformis. Eighty-three percentage of TaNACs within the “TaNAC2 subclade” were responsive to F. graminearum and are likely involved in defense against that fungus. In addition, the “TaNAC2 subclade” contains orthologs (1) from all 4 species included in this study that are pathogen responsive (based on the data in the public Expression Atlas database; results not shown) and (2) from rice and Arabidopsis that are functionally characterized as playing a role in pathogen defense. Thus, the “TaNAC2 subclade” contains NACs that are important for immunity in both monocot and dicot plants.
Many of the disease-responsive TaNACs were within phylogenetic subfamilies “a,” “e,” and “f,” and many were universally responsive to pathogens. Interestingly, the Arabidopsis ortholog of defense-associated rice ONAC017 and TaNAC017 from pathogen-enriched tree subclade “e3.2” (AT3G44350) was identified as a “core immunity response gene” upregulated by 6 different elicitors of pattern-triggered immunity (Bjornson et al. 2021). Universal response TaNACs warrant further investigation for their potential as broad-spectrum defense-associated signaling hub genes, as do subclade “g1.3” for their potential as hemibiotroph-specific TaNACs. TaNACs in defense-associated subfamilies “a,” “e,” and “f” were also responsive to abiotic stressors (Borrill et al. 2017) or have a proven role in abiotic stress resistance (see Supplemental Results), suggesting that these subfamilies evolved to regulate biotic and abiotic responses in wheat. Several characterized NACs within defense-associated subfamily “a” were proven to be multifunctional and 6 uncharacterized TaNACs shared a subfamily with orthologs functionally characterized as playing a role in either abiotic stress responses, senescence, and/or starch synthesis (results not shown). Genes providing resistance against multiple pathogens or stressors are highly valued in breeding programs. The rapid advances in wheat genome sequencing will give us insights into the level of allelic diversity within TaNACs and follow-on studies will determine whether allelic divergence affects function and the potential of TaNACs as target genes in wheat breeding programs.
This study demonstrated that response of TaNACs to pathogens is conserved in certain subfamilies and subclades. Similarly, Borrill et al. (2017) previously demonstrated that more phylogenetically related TaNACs shared more similar expression patterns compared to the more distant TaNACs. Thus, this study supports the previous finding. Our study also highlighted TaNACs previously identified as being pathogen-responsive. By comparing TaNACs responsive to B. graminis and P. striiformis delineated herein with the ones determined in studies by Lv et al. (2020) and Ma et al. (2021), we highlighted pathogen-responsive genes common to all studies. But many could not be compared (due to data available) or were not commonly pathogen-responsive across studies. We attempted to determine if the latter was due to differences in methodology used for differential expression analysis, but the available information was insufficient to enable such a comparison. For the TaNACs, commonly responsive across studies to B. graminis and P. striiformis were placed in the subclades enriched with pathogen-responsive TaNACs and the majority of them exhibited a universal pathogen response, and as such should be further explored for their function in biotic stress responses. Within the subclades enriched with pathogen-responsive TaNACs, the ones with the universal pathogen response and candidates in the “TaNAC2 subclade” could be the strongest candidates for further functional validation and breeding purposes. Of course, since this study mined pathogen-responsive TaNACs by analyzing a limited number of RNA-seq studies, it is expected that other candidates responsive to certain pathogens in specific cultivars and at specific timepoints and the ones that are not regulated at the transcriptional level were missed herein.
TaNAC2 subclade genes identified herein exhibit allelic variation and locate within disease resistance quantitative trait loci (QTL). Based on the current data in the EnsemblPlants website (https://plants.ensembl.org/index.html), all except 3 TaNACs (TaNAC088-B1, TaNAC002-B1, and TaNAC068-A1) within the “TaNAC2 subclade” had at least 1 gene variant that would result in a truncated protein or a protein with a missense mutation (results not shown). Three genes from the “TaNAC2 subclade” (TaNAC088-A1, TaNAC088-B1, TaNAC009-A1, and TaNAC009-A1) are, respectively, located within QTL MQTL1A.2, MQTL1B.4, MQTL4A.1, and MQTL4D.1 (Saini et al. 2022), which were associated with resistance to multiple diseases. Thus, future studies should use new available resources to explore natural allelic and copy number variation for TaNACs within the wheat pangenome. These genetic resources could provide the tools for further functional exploration of the gene family and for breeding purposes as explained in detail in a previous review (Borrill et al. 2019). These and other genomic tools can be used to determine the functional importance of TaNACs located within QTL associated with disease resistance.
The observation that the majority of TaNACs were specific to the hemibiotrophs (at least those tested herein) is interesting, and further studies should determine whether they are involved in 1 or both phases (biotrophic and necrotrophic) of these diseases. The observation that the number of upregulated TaNACs increased with time for hemibiotrophs suggests their implication in the later necrotrophic stage of the infection, but this remains to be determined. The majority of TaNACs responsive to the hemibiotrophic bacteria X. translucens (76%) were also responsive to hemibiotrophic fungi, further supporting the hypothesis that many TaNACs might had preferably evolved to regulate defense against hemibiotrophic pathogens. This hypothesis can be tested as more wheat disease transcriptional data becomes available in the future. Our data showed that TaNAC2 was downregulated upon infection with the biotrophs and upregulated upon infection with Fusarium fungi at the later stage of infection (36 hpi), most likely being the necrotrophic phase. Silencing of TaNAC2 and TaNAC30, respectively, from subfamilies “a” and “c,” enhanced resistance against biotrophic P. striiformis via accumulation of hydrogen peroxide and decreased hyphal growth at the early stage of infection (Wang et al. 2018; Zhang et al. 2018). Thus, it is very likely that these 2 TaNACs negatively regulate programmed cell death at the early stage of biotrophic infection.
In the joined phylogenetic tree of wheat, barley, rice, and Arabidopsis NACs, the proteins were distributed in 8 distinct subfamilies a-h, which agrees with the classification first proposed by Shen et al. (2009). Herein, it was shown that subfamily “h” expanded in grasses, particularly wheat; wheat contained 5 and 10 times more NAC genes than in barley and rice, respectively, while Arabidopsis had only 1 NAC in subfamily “h.” Shen et al. (2009) reported a higher proportion of subfamily “h” NAC sequences in rice (24%) than in this study (12%), which can be explained by the lower number of rice sequences used to infer phylogeny herein. It should also be noted, compared to this study, respectively, 29, 50, and 4 more NACs were identified in extensive research of barley, rice, and Arabidopsis (Nuruzzaman et al. 2010; Murozuka et al. 2018), which can be explained by differences in analyses. Expansion of the TaNAC family has been explained through small-scale duplication and retroposition events (Guérin et al. 2019), but the ecological drivers remain to be determined.
To our knowledge, this is the first study to quantitatively assess the divergence of the NAM domain of TaNAC proteins. It deduced that the NAM domain significantly diverged in sequence in subfamilies f–h. These subfamilies partially or completely lost subdomains C and D, which are implicated in DNA binding. Motif NT-4 in subdomain C contained the DNA-recognition motif (WKATGTDK) that binds DNA (Welner et al. 2012). Also, the NT-2 motif in subdomain D contained conserved amino acids of the β4–β5 loop, which is also a possible DNA-binding site (Duval et al. 2002; Ernst et al. 2004). The NT-4 motif was absent in NACs in subfamilies “g” and “h,” and NT-2 was partially lost in subfamilies “f” and “g” and absent in “h.” Whether or not this affects their DNA-binding activity remains to be determined. Only subdomains A and E were conserved in all subfamilies suggesting that these subdomains could be the most important for the NAC domain function. Indeed, it has been proven that the disruption of the NAC dimer interface located in subdomain A reduced the affinity for DNA-binding (Ernst et al. 2004; Olsen et al. 2005). Similarly, subdomain A was proven to be the dimerization domain in wheat NAM-A1 and mutation within the subdomain impaired the physiological function of NAM-A1 (Harrington et al. 2019). However, in vitro site-directed mutagenesis studies of NAC subdomains in TaNAC69 showed that all 5 subdomains were important for the DNA-binding function, and subdomains A, D and E were necessary for dimerization (Xue et al. 2006). In total, we found 14 newly identified putative NT motifs that were not associated with conserved subdomains A–E and the majority were present in divergent subfamilies “f,” “g,” and “h.” These features may contribute to the functional diversity of wheat NACs in these divergent and as yet uninvestigated subfamilies.
Within the TAR, we identified 39 motifs, 5 of where were previously identified in Borrill et al. (2017) who annotated TaNAC genes from the previous wheat genome version. The CT-7 motif matched with a transcriptional activation domain called the “WQ-box” (Ko et al. 2007). The “WQ-box” was found in NST (NAC secondary wall-thickening promoting factor) and VND (vascular-related NAC domain) proteins in subfamily “c” and is specific for vascular plants (Shen et al. 2009). The motif CT-24 was identified in subfamily “h” (Borrill et al. 2017), and in this study also in subfamilies “a,” “e,” “f,” and “g.” The motif was part of the nuclear localization signal recently functionally characterized in the FHB resistance gene TaNACL-D1 described by Perochon et al. (2019), which belonged to subfamily “h.” TaNACL-D1 did not have subdomain C and beyond that, this study differed from Perochon et al. (2019) in that our analysis indicates it also lacked the subdomains D and E. The differences between the 2 studies are most likely reflective of different parameters and numbers of sequences used for analysis, the larger number of sequences used in this study increasing the chances of detecting intraspecies motif differences.
Variations in NT motifs were not associated with TaNACs expressed in healthy or diseased wheat organs. However, within the TAR, CT-27 and CT-33 were specific for the subclade “e1.1,” and CT-39 was specific for the subclade “d5,” and both subclades were enriched in pathogen-responsive TaNACs. But to date, no NAC motifs have been experimentally proven to directly influence their function in pathogen defense. Similarly, CT-15 and CT-23 in the TAR were specific for the subclade “d6” wherein TaNACs were specifically expressed in grain. The “grain NACs” in maize (ZmNAC128 and ZmNAC130), and rice (ONAC020 and ONAC026) positively regulated starch and storage protein synthesis in grain (Zhang et al. 2019; Wang et al. 2020) and had a CT motif that matched CT-23 described herein (Murozuka et al. 2018). The 2 rice “grain NACs,” ONAC020 and ONAC026, were orthologs of all grain-specific TaNACs in subclade “d6” described herein. Of those grain-specific TaNACs, TaNAC020-A1 had a very short CT compared to other “grain NACs” from the subclade “d6” and it lacked the CT-15 and CT-23 motifs. TaNAC020-A1 was proven to be transcriptional repressor that negatively affected grain weight and starch synthesis (Liu et al. 2020). However, whether the presence or absence of CT-15 and CT-23 in the “grain-NACs” is associated with positive or negative effect on grain development remains to be investigated.
In conclusion, this study provided TaNAC nomenclature, which will facilitate consistent naming of all such wheat genes and insights into the phylogeny and pathogen responsive of this gene family in wheat. We determined that subfamilies “a,” “e,” and “f” and subclades enriched with pathogen-responsive TaNACs are rich in defense-associated TaNACs. For the first time, this study demonstrated that the NAC domain diverged in wheat subfamilies “f,” “g,” and “h.” Diverged motifs determined herein may be implicated in yet uninvestigated diverged function of the NAC domain in these subfamilies. Lastly, we identified 3 CT motifs associated with pathogen responsiveness and 2 associated with specific expression in grain. Their functional validation will provide a “fingerprint” to identify defense- or grain-related function in otherwise noncharacterized NACs in wheat and related crops.
Supplementary Material
Acknowledgments
We thank Prof. Lars Jermiin for the shared knowledge and advice on the data analysis and construction of phylogenetic trees.
Funding
This work received funding from Science Foundation Ireland Project No. 14/1A/2508, the European Union’s Horizon 2020 research and innovation program under Grant Agreement No. 818144.
Author contributions
MV, AP, and FMD designed the research; AP and FMD supervised the experiments; MV performed the experiments and analyzed the data, except differential expression, which was conducted by HB; MV wrote the article, edited, and amended by AP, HB, and FMD; FMD won the funding to conduct the research.
Conflicts of interest
None declared.
Contributor Information
Monika Vranic, UCD School of Biology and Environmental Science and Earth Institute, College of Science, University College Dublin, Dublin 4, Ireland.
Alexandre Perochon, UCD School of Biology and Environmental Science and Earth Institute, College of Science, University College Dublin, Dublin 4, Ireland.
Harriet Benbow, UCD School of Biology and Environmental Science and Earth Institute, College of Science, University College Dublin, Dublin 4, Ireland.
Fiona M Doohan, UCD School of Biology and Environmental Science and Earth Institute, College of Science, University College Dublin, Dublin 4, Ireland.
Data Availability
The data generated/analyzed during the study are available in the supplementary files, supplementary figures, and supplementary tables.
The supplemental files contain the Supplemental Results (Supplemental_Results.docx) and the following data: Supplementary Files 1–7, Supplementary Tables 1–16, and Supplementary Figs. 1–4.
Online versions of Figs. 1a and 4 are available at https://itol.embl.de/tree/372282299412371613249495 and https://itol.embl.de/tree/372282299294211613865063, respectively.
DE_dataset_Vranic_et_al.xlsx contains the DESeq2 output used to extract differential expression profiles of pathogen-responsive TaNACs and was submitted on Figshare: https://doi.org/10.25387/g3.19326374.
Supplemental material is available at G3 online.
Literature cited
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS.. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202–208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benbow HR, Jermiin LS, Doohan FM.. Serpins: genome-wide characterisation and expression analysis of the serine protease inhibitor family in Triticum aestivum. G3 (Bethesda). 2019;9(8):2709–2722. 10.1534/g3.119.400444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornson M, Pimprikar P, Nürnberger T, Zipfel C.. The transcriptional landscape of Arabidopsis thaliana pattern-triggered immunity. Nat Plants. 2021;7(5):579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrill P, , HarringtonSA, , Uauy C.. Applying the latest advances in genomics and phenomics for trait discovery in polyploid wheat. Plant J. 2019;97(1):56–72. 10.1111/tpj.14150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrill P, Harrington SA, Uauy C.. Genome-wide sequence and expression analysis of the NAC transcription factor family in polyploid wheat. G3 (Bethesda). 2017;7(9):3019–3029. 10.1534/g3.117.043679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q, Jiang H, Li C-B, Zhai Q, Zhang J, Wu X, Sun J, Xie Q, Li C.. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res. 2008;18(7):756–767. 10.1038/cr.2008.53 [DOI] [PubMed] [Google Scholar]
- Carviel JL, Al-Daoud F, Neumann M, Mohammad A, Provart NJ, Moeder W, Yoshioka K, Cameron RK.. Forward and reverse genetics to identify genes involved in the age-related resistance response in Arabidopsis thaliana. Mol Plant Pathol. 2009;10(5):621–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-J, Perera V, Christiansen MW, Holme IB, Gregersen PL, Grant MR, Collinge DB, Lyngkjær MF.. The barley HvNAC6 transcription factor affects ABA accumulation and promotes basal resistance against powdery mildew. Plant Mol Biol. 2013;83(6):577–590. [DOI] [PubMed] [Google Scholar]
- Choulet F, Alberti A, Theil S, Glover N, Barbe V, Daron J, Pingault L, Sourdille P, Couloux A, Paux E, et al. Structural and functional partitioning of bread wheat chromosome 3B. Science. 2014;345(6194):1249721. 10.1126/science.1249721 [DOI] [PubMed] [Google Scholar]
- Christiansen MW, Holm PB, Gregersen PL.. Characterization of barley (Hordeum vulgare L.) NAC transcription factors suggests conserved functions compared to both monocots and dicots. BMC Res Notes. 2011;4:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delessert C, Kazan K, Wilson IW, Van Der Straeten D, Manners J, Dennis ES, Dolferus R.. The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. Plant J. 2005;43(5):745–757. 10.1111/j.1365-313X.2005.02488.x [DOI] [PubMed] [Google Scholar]
- Dobon A, Bunting DCE, Cabrera-Quio LE, Uauy C, Saunders DGO.. The host-pathogen interaction between wheat and yellow rust induces temporally coordinated waves of gene expression. BMC Genomics. 2016;17:380. 10.1186/s12864-016-2684-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval M, Hsieh T-F, Kim SY, Thomas TL.. Molecular characterization of AtNAM: a member of the Arabidopsis NAC domain superfamily. Plant Mol Biol. 2002;50(2):237–248. 10.1023/a:1016028530943 [DOI] [PubMed] [Google Scholar]
- Ernst HA, Olsen AN, Larsen S, Lo Leggio L.. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 2004;5(3):297–303. 10.1038/sj.embor.7400093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Duan X, Zhang Q, Li X, Wang B, Huang L, Wang X, Kang Z.. The target gene of tae‐miR164, a novel NAC transcription factor from the NAM subfamily, negatively regulates resistance of wheat to stripe rust. Mol Plant Pathol. 2014;15(3):284–296. 10.1111/mpp.12089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR.. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39(Web Server issue):W29–W37. 10.1093/nar/gkr367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Seco D, Chiapello M, Bracale M, Pesce C, Bagnaresi P, Dubois E, Moulin L, Vannini C, Koebnik R.. Transcriptome and proteome analysis reveal new insight into proximal and distal responses of wheat to foliar infection by Xanthomonas translucens. Sci Rep. 2017;7(1):10157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guérin C, Roche J, Allard V, Ravel C, Mouzeyar S, Bouzidi MF.. Genome-wide analysis, expansion and expression of the NAC family under drought and heat stresses in bread wheat (T. aestivum L.). PLoS One. 2019;14(3):e0213390. 10.1371/journal.pone.0213390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington SA, Overend LE, Cobo N, Borrill P, Uauy C.. Conserved residues in the wheat (Triticum aestivum) NAM-A1 NAC domain are required for protein binding and when mutated lead to delayed peduncle and flag leaf senescence. BMC Plant Biol. 2019;19(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R.. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. 2015;16(1): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS.. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35(2):518–522. 10.1093/molbev/msx281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium (IWGSC); Appels R, Eversole K, Stein N, Feuillet C, Keller B, Rogers J, Pozniak CJ, Choulet F, Distelfeld A, Poland, et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. 2018;361(6403): 10.1126/science.aar7191 [DOI] [PubMed] [Google Scholar]
- Jensen MK, Rung JH, Gregersen PL, Gjetting T, Fuglsang AT, Hansen M, Joehnk N, Lyngkjaer MF, Collinge DB.. The HvNAC6 transcription factor: a positive regulator of penetration resistance in barley and Arabidopsis. Plant Mol Biol. 2007;65(1–2):137–150. [DOI] [PubMed] [Google Scholar]
- Jin X, Ren J, Nevo E, Yin X, Sun D, Peng J.. Divergent evolutionary patterns of NAC transcription factors are associated with diversification and gene duplications in angiosperm. Front Plant Sci. 2017;8:1156. 10.3389/fpls.2017.01156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM.. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8(3):275–282. 10.1093/bioinformatics/8.3.275 [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14(6):587–589. 10.1038/nmeth.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Park HC, Kim KE, Jung MS, Han HJ, Kim SH, Kwon YS, Bahk S, An J, Bae DW, et al. A NAC transcription factor and SNI1 cooperatively suppress basal pathogen resistance in Arabidopsis thaliana. Nucleic Acids Res. 2012;40(18):9182–9192. 10.1093/nar/gks683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J-H, Yang SH, Park AH, Lerouxel O, Han K-H.. ANAC012, a member of the plant-specific NAC transcription factor family, negatively regulates xylary fiber development in Arabidopsis thaliana. Plant J. 2007;50(6):1035–1048. 10.1111/j.1365-313X.2007.03109.x [DOI] [PubMed] [Google Scholar]
- Kugler KG, , SiegwartG, , NussbaumerT, , AmetzC, , SpannaglM, , SteinerB, , LemmensM, , MayerKFX, , BuerstmayrH, , Schweiger W.. Quantitative trait loci-dependent analysis of a gene co-expression network associated with Fusarium head blight resistance in bread wheat (Triticum aestivum L.). BMC Genomics. 2013;14:728. 10.1186/1471-2164-14-728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P.. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47(W1):W256–W259. 10.1093/nar/gkz239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PO. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst Biol. 2001;50(6):913–925. 10.1080/106351501753462876 [DOI] [PubMed] [Google Scholar]
- Liu Q, Yan S, Huang W, Yang J, Dong J, Zhang S, Zhao J, Yang T, Mao X, Zhu X, et al. NAC transcription factor ONAC066 positively regulates disease resistance by suppressing the ABA signaling pathway in rice. Plant Mol Biol. 2018;98(4–5):289–302. [DOI] [PubMed] [Google Scholar]
- Liu Y, Hou J, Wang X, Li T, Majeed U, Hao C, Zhang X.. The NAC transcription factor NAC019-A1 is a negative regulator of starch synthesis in wheat developing endosperm. J Exp Bot. 2020;71(19):5794–5807. 10.1093/jxb/eraa333 [DOI] [PubMed] [Google Scholar]
- Lv S, Guo H, Zhang M, Wang Q, Zhang H, Ji W.. Large-scale cloning and comparative analysis of TaNAC genes in response to stripe rust and powdery mildew in wheat (Triticum aestivum L.). Genes. 2020;11(9):1073. 10.3390/genes11091073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Stiller J, Zhao Q, Feng Q, Cavanagh C, Wang P, Gardiner D, Choulet F, Feuillet C, Zheng Y-L, et al. Transcriptome and allele specificity associated with a 3BL locus for Fusarium crown rot resistance in bread wheat. PLoS One. 2014;9(11):e113309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Yuan M, Sun B, Zhang D, Zhang J, Li C, Shao Y, Liu W, Jiang L.. Evolutionary divergence and biased expression of NAC transcription factors in hexaploid bread wheat (Triticum aestivum L.). Plants. 2021;10(2):382. 10.3390/plants10020382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrann GRD, Steed A, Burt C, Goddard R, Lachaux C, Bansal A, Corbitt M, Gorniak K, Nicholson P, Brown JKM.. Contribution of the drought tolerance-related stress-responsive NAC1 transcription factor to resistance of barley to Ramularia leaf spot. Mol Plant Pathol. 2015;16(2):201–209. 10.1111/mpp.12173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh R, Yamazaki Y, Dubcovsky J, Rogers J, Morris C, Xia X. Catalogue of gene symbols for wheat. In: Proceedings of the 12th International Wheat Genetics Symposium, Yokohama, Japan. 2013; pp. 9–10.
- Murozuka E, Massange-Sánchez JA, Nielsen K, Gregersen PL, Braumann I.. Genome wide characterization of barley NAC transcription factors enables the identification of grain-specific transcription factors exclusive for the Poaceae family of monocotyledonous plants. PLoS One. 2018;13(12):e0209769. 10.1371/journal.pone.0209769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Tran L-SP, Nguyen DV, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi Shinozaki K, et al. Functional analysis of a NAC‐type transcription factor OsNAC6 involved in abiotic and biotic stress‐responsive gene expression in rice. Plant J. 2007;51(4):617–630. 10.1111/j.1365-313X.2007.03168.x [DOI] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32(1):268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, Kikuchi S.. Genome-wide analysis of NAC transcription factor family in rice. Gene. 2010;465(1–2):30–44. 10.1016/j.gene.2010.06.008 [DOI] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K.. DNA-binding specificity and molecular functions of NAC transcription factors. Plant Sci. 2005;169(4):785–797. 10.1016/j.plantsci.2005.05.035 [DOI] [PubMed] [Google Scholar]
- Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Research. Int J Rapid Pub Rep Genes Genomes. 2003;10(6):239–247. [DOI] [PubMed] [Google Scholar]
- Perochon A, Kahla A, Vranić M, Jia J, Malla KB, Craze M, Wallington E, Doohan FM.. A wheat NAC interacts with an orphan protein and enhances resistance to Fusarium head blight disease. Plant Biotechnol J. 2019;17(10):1892–1904. 10.1111/pbi.13105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JJ, , CarereJ, , FitzgeraldTL, , StillerJ, , CovarelliL, , XuQ, , GublerF, , ColgraveML, , GardinerDM, , Manners JM,. et al. The Fusarium crown rot pathogen Fusarium pseudograminearum triggers a suite of transcriptional and metabolic changes in bread wheat (Triticum aestivum L.). Ann Bot. 2017;119(5):853–867. 10.1093/aob/mcw207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-González RH, Borrill P, Lang D, Harrington SA, Brinton J, Venturini L, Davey M, Jacobs J, Ex F, van Pasha A, et al. ; International Wheat Genome Sequencing Consortium. The transcriptional landscape of polyploid wheat. Science. 2018;361(6403):1–12. 10.1126/science.aar6089 [DOI] [PubMed] [Google Scholar]
- Ren T, Qu F, Morris TJ.. HRT gene function requires interaction between a NAC protein and viral capsid protein to confer resistance to turnip crinkle virus. Plant Cell. 2000;12(10):1917–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse GW, Jermiin LS, Wilson NG, Eeckhaut I, Lanterbecq D, Oji T, Young CM, Browning T, Cisternas P, Helgen LE, et al. Fixed, free, and fixed: the fickle phylogeny of extant Crinoidea (Echinodermata) and their Permian-Triassic origin. Mol Phylogenet Evol. 2013;66(1):161–181. 10.1016/j.ympev.2012.09.018 [DOI] [PubMed] [Google Scholar]
- Rudd JJ, Kanyuka K, Hassani-Pak K, Derbyshire M, Andongabo A, Devonshire J, Lysenko A, Saqi M, Desai NM, Powers SJ, et al. Transcriptome and metabolite profiling of the infection cycle of Zymoseptoria tritici on wheat reveals a biphasic interaction with plant immunity involving differential pathogen chromosomal contributions and a variation on the hemibiotrophic lifestyle definition. Plant Physiol. 2015;167(3):1158–1185. 10.1104/pp.114.255927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini DK, , ChahalA, , PalN, , SrivastavaP, , Gupta PK.. Meta-analysis reveals consensus genomic regions associated with multiple disease resistance in wheat (Triticum aestivum L.). Mol Breeding. 2022;42(3),11. 10.21203/rs.3.rs-773587/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savary S, Willocquet L, Pethybridge SJ, Esker P, McRoberts N, Nelson A.. The global burden of pathogens and pests on major food crops. Nat Ecol Evol. 2019;3(3):430–439. [DOI] [PubMed] [Google Scholar]
- Schilling S, Kennedy A, Pan S, Jermiin LS, Melzer R.. Genome-wide analysis of MIKC-type MADS-box genes in wheat: ervasive duplications, functional conservation and putative neofunctionalization. New Phytol. 2020;225(1):511–529. 10.1111/nph.16122 [DOI] [PubMed] [Google Scholar]
- Schweiger W, Steiner B, Vautrin S, Nussbaumer T, Siegwart G, Zamini M, Jungreithmeier F, Gratl V, Lemmens M, Mayer KFX, et al Suppressed recombination and unique candidate genes in the divergent haplotype encoding Fhb1, a major Fusarium head blight resistance locus in wheat. Theor Appl Genet. 2016;129(8):1607–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Kim MJ, Park J-Y, Kim S-Y, Jeon J, Lee Y-H, Kim J, Park C-M.. Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 2010;61(4):661–671. 10.1111/j.1365-313X.2009.04091.x [DOI] [PubMed] [Google Scholar]
- Shen H, Yin Y, Chen F, Xu Y, Dixon RA.. A Bioinformatic analysis of NAC genes for plant cell wall development in relation to lignocellulosic bioenergy production. Bioenerg Res. 2009;2(4):217–232. [DOI] [Google Scholar]
- Soni N, Altartouri B, Hegde N, Duggavathi R, Nazarian-Firouzabadi F, Kushalappa A.. TaNAC032 transcription factor regulates lignin-biosynthetic genes to combat Fusarium head blight in wheat. Plant Sci. 2021;304:110820. 10.1016/j.plantsci.2021.110820 [DOI] [PubMed] [Google Scholar]
- Sturn A, Quackenbush J, Trajanoski Z.. Genesis: cluster analysis of microarray data. Bioinformatics (Oxford, England). 2002;18(1):207–208. 10.1093/bioinformatics/18.1.207 [DOI] [PubMed] [Google Scholar]
- Sun L, Zhang H, Li D, Huang L, Hong Y, Ding XS, Nelson RS, Zhou X, Song F.. Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea. Plant Mol Biol. 2013;81(1–2):41–56. [DOI] [PubMed] [Google Scholar]
- Tang N, Shahzad Z, Lonjon F, Loudet O, Vailleau F, Maurel C.. Natural variation at XND1 impacts root hydraulics and trade-off for stress responses in Arabidopsis. Nat Commun. 2018;9(1):3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Wei J, Song N, Wang N, Zhao J, Kang Z.. A novel wheat NAC transcription factor, TaNAC30, negatively regulates resistance of wheat to stripe rust. J Integr Plant Biol. 2018;60(5):432–443. 10.1111/jipb.12627 [DOI] [PubMed] [Google Scholar]
- Wang F, Lin R, Feng J, Chen W, Qiu D, Xu S.. TaNAC1 acts as a negative regulator of stripe rust resistance in wheat, enhances susceptibility to Pseudomonas syringae, and promotes lateral root development in transgenic Arabidopsis thaliana. Front Plant Sci. 2015;6:108. 10.3389/fpls.2015.00108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Chen Z, Zhang Q, Meng S, Wei C.. The NAC transcription factors OsNAC20 and OsNAC26 regulate starch and storage protein synthesis. Plant Physiol. 2020;184(4):1775–1791. 10.1104/pp.20.00984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Basnayake BMVS, Zhang H, Li G, Li W, Virk N, Mengiste T, Song F.. The Arabidopsis ATAF1, a NAC transcription factor, is a negative regulator of defense responses against necrotrophic fungal and bacterial pathogens. Mol Plant Microbe Interact. 2009;22(10):1227–1238. 10.1094/MPMI-22-10-1227 [DOI] [PubMed] [Google Scholar]
- Wang Z, Xia Y, Lin S, Wang Y, Guo B, Song X, Ding S, Zheng L, Feng R, Chen S, et al. Osa-miR164a targets OsNAC60 and negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Plant J. 2018;95(4):584–597. 10.1111/tpj.13972 [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ.. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welner DH, Deeba F, Lo Leggio L, Skriver K.. NAC transcription factors: from structure to function in stress-associated networks. In Gonzalez DH, editor. Plant Transcription Factors. Boston: Academic Press; 2016. p. 199–212(Chapter 13). [DOI] [Google Scholar]
- Welner DH, Lindemose S, Grossmann JG, Møllegaard NE, Olsen AN, Helgstrand C, Skriver K, Lo Leggio L.. DNA binding by the plant-specific NAC transcription factors in crystal and solution: a firm link to WRKY and GCM transcription factors. Biochem J. 2012;444(3):395–404. 10.1042/BJ20111742 [DOI] [PubMed] [Google Scholar]
- Wong TKF, Kalyaanamoorthy S, Meusemann K, Yeates DK, Misof B, Jermiin LS.. A minimum reporting standard for multiple sequence alignments. NAR Genom Bioinform. 2020;2(2):lqaa024. 10.1093/nargab/lqaa024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Deng Z, Lai J, Zhang Y, Yang C, Yin B, Zhao Q, Zhang L, Li Y, Yang C, et al. Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 2009;19(11):1279–1290. 10.1038/cr.2009.108 [DOI] [PubMed] [Google Scholar]
- Xie Q, Frugis G, Colgan D, Chua N-H.. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000;14(23):3024–3036. 10.1101/gad.852200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue G-P, Bower NI, McIntyre CL, Riding GA, Kazan K, Shorter R, Xue G-P, Bower NI, McIntyre CL, Riding GA, et al. TaNAC69 from the NAC superfamily of transcription factors is up-regulated by abiotic stresses in wheat and recognises two consensus DNA-binding sequences. Funct Plant Biol. 2006;33(1):43–57. 10.1071/FP05161 [DOI] [PubMed] [Google Scholar]
- Yang F, Li W, Jørgensen HJL.. Transcriptional reprogramming of wheat and the hemibiotrophic pathogen Septoria tritici during two phases of the compatible interaction. PLoS One. 2013;8(11):e81606. 10.1371/journal.pone.0081606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokotani N, Tsuchida-Mayama T, Ichikawa H, Mitsuda N, Ohme-Takagi M, Kaku H, Minami E, Nishizawa Y.. OsNAC111, a blast disease-responsive transcription factor in rice, positively regulates the expression of defense-related genes. Mol Plant Microbe Interact. 2014;27(10):1027–1034. [DOI] [PubMed] [Google Scholar]
- Yoshii M, Shimizu T, Yamazaki M, Higashi T, Miyao A, Hirochika H, Omura T.. Disruption of a novel gene for a NAC-domain protein in rice confers resistance to Rice dwarf virus. Plant J. 2009;57(4):615–625. 10.1111/j.1365-313X.2008.03712.x [DOI] [PubMed] [Google Scholar]
- Yuan X, Wang H, Cai J, Li D, Song F.. NAC transcription factors in plant immunity. Phytopathol Res. 2019;1(1):3. 10.1186/s42483-018-0008-0 [DOI] [Google Scholar]
- Zhang H, Yang Y, Wang C, Liu M, Li H, Fu Y, Wang Y, Nie Y, Liu X, Ji W.. Large-scale transcriptome comparison reveals distinct gene activations in wheat responding to stripe rust and powdery mildew. BMC Genomics. 2014;15(1):898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Yuan S, Zhao C, Park RF, Wen X, Yang W, Zhang N, Liu D.. TaNAC35 acts as a negative regulator for leaf rust resistance in a compatible interaction between common wheat and Puccinia triticina. Mol Genet Genomics. 2021;296(2):279–287. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhang Q, Pei C, Li X, Huang X, Chang C, Wang X, Huang L, Kang Z.. TaNAC2 is a negative regulator in the wheat-stripe rust fungus interaction at the early stage. Physiol Mol Plant Pathol. 2018;102:144–153. 10.1016/j.pmpp.2018.02.002 [DOI] [Google Scholar]
- Zhang Z, Dong J, Ji C, Wu Y, Messing J.. NAC-type transcription factors regulate accumulation of starch and protein in maize seeds. Proc Natl Acad Sci U S A. 2019;116(23):11223–11228. 10.1073/pnas.1904995116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X-Y, Spivey NW, Zeng W, Liu P-P, Fu ZQ, Klessig DF, He SY, Dong X.. Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe. 2012;11(6):587–596. 10.1016/j.chom.2012.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Qian C, Li R, Zhou S, Zhang R, Xiao J, Wang X, Zhang S, Xing L, Cao A.. TaNAC6s are involved in the basal and broad-spectrum resistance to powdery mildew in wheat. Plant Sci. 2018;277:218–228. 10.1016/j.plantsci.2018.09.014 [DOI] [PubMed] [Google Scholar]
- Zhu T, Nevo E, Sun D, Peng J.. Phylogenetic analyses unravel the evolutionary history of Nac proteins in plants. Evolution. 2012;66(6):1833–1848. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated/analyzed during the study are available in the supplementary files, supplementary figures, and supplementary tables.
The supplemental files contain the Supplemental Results (Supplemental_Results.docx) and the following data: Supplementary Files 1–7, Supplementary Tables 1–16, and Supplementary Figs. 1–4.
Online versions of Figs. 1a and 4 are available at https://itol.embl.de/tree/372282299412371613249495 and https://itol.embl.de/tree/372282299294211613865063, respectively.
DE_dataset_Vranic_et_al.xlsx contains the DESeq2 output used to extract differential expression profiles of pathogen-responsive TaNACs and was submitted on Figshare: https://doi.org/10.25387/g3.19326374.
Supplemental material is available at G3 online.