Abstract
RNA interference is sequence-specific gene silencing triggered by double-stranded RNA. Systemic RNA interference is where double-stranded RNA, expressed or introduced into 1 cell, is transported to and initiates RNA interference in other cells. Systemic RNA interference is very efficient in Caenorhabditis elegans and genetic screens for systemic RNA interference-defective mutants have identified RNA transporters (SID-1, SID-2, and SID-5) and a signaling protein (SID-3). Here, we report that SID-4 is nck-1, a C. elegans NCK-like adaptor protein. sid-4 null mutations cause a weak, dose-sensitive, systemic RNA interference defect and can be effectively rescued by SID-4 expression in target tissues only, implying a role in double-stranded RNA import. SID-4 and SID-3 (ACK-1 kinase) homologs interact in mammals and insects, suggesting that they may function in a common signaling pathway; however, a sid-3; sid-4 double mutants showed additive resistance to RNA interference, suggesting that these proteins likely interact with other signaling pathways as well. A bioinformatic screen coupled to RNA interference sensitivity tests identified 23 additional signaling components with weak RNA interference-defective phenotypes. These observations suggest that environmental conditions may modulate systemic RNA interference efficacy, and indeed, sid-3 and sid-4 are required for growth temperature effects on systemic RNA interference silencing efficiency.
Keywords: NCK-1, C. elegans, RNAi, dsRNA
Introduction
RNA interference (RNAi) is sequence-specific gene silencing triggered by natural or experimentally introduced double-stranded RNA (dsRNA) (Fire et al. 1998). In many animals, including Caenorhabditis elegans, experimentally introduced dsRNA is mobile (Jose and Hunter 2007). This dsRNA mobility allows dsRNA introduced by localized injection and transgenic expression and in some animals by ingestion, to produce a whole-animal systemic silencing response. We previously reported the results of a visual screen for systemic RNAi-defective (Sid) mutants that has led to the identification of dsRNA transport and signal transduction proteins (Winston et al. 2002, 2007; Hinas et al. 2012; Jose et al. 2012).
Characterization of the sid genes has so far identified 4 SID proteins. SID-1 is a dsRNA channel protein that selectively transports long dsRNA into cells (Feinberg and Hunter 2003; Shih and Hunter 2011). In sid-1 mutants, systemic RNAi is undetectable, but expressed or injected dsRNA can cause robust autonomous RNAi (Winston et al. 2002). Genetic mosaic and tissue-specific rescue experiments demonstrate that SID-1 is required for import but not export of silencing information (presumably dsRNA) (Winston et al. 2002; Jose et al. 2009; Whangbo et al. 2017). SID-2 is an intestinally expressed transmembrane protein, present at the intestinal lumen, that selectively endocytoses ingested dsRNA (Winston et al. 2007; McEwan et al. 2012). While SID-2 is required only for feeding RNAi, SID-1 is also required for feeding RNAi; thus, it is presumed that SID-1 releases endocytosed dsRNA into the cytoplasm to initiate RNAi. SID-3 is a broadly expressed ACK1 tyrosine kinase homolog (Jose et al. 2012). sid-3 mutants are partially defective for the import of silencing signals. The requirement of a signal transduction protein suggests that environmental or physiological conditions may regulate systemic RNAi. SID-5 is a small novel protein associated with late endosomes (Hinas et al. 2012). Like sid-3 mutants, sid-5 mutants are partially defective for RNAi, but sid-5 is also essential for initiation of parental RNAi (transgenerational silencing) (Wang and Hunter 2017). Here, we report the identification and characterization of sid-4.
Several other activities are also required for systemic RNAi. RME-2 is an endocytosis receptor that, in the absence of SID-1, is required to transport dsRNA into oocytes to support parental RNAi (Wang and Hunter 2017). Similar to feeding RNAi, SID-1 activity is subsequently required in the embryo for RNAi silencing, likely to release endosome trapped dsRNA into the cytoplasm to initiate RNAi. In addition, mutations in rde-10, -11, and -12 and rrf-1, which fail to produce abundant secondary siRNAs resulting in dose-dependent RNAi-silencing defects (Yang et al. 2014), are also required for effective systemic RNAi. dsRNA introduced locally (injection or expression) in rde-12 and rrf-1 mutants results in RNAi silencing locally, but not systemically, indicating that limiting amounts of dsRNA are transported. Thus, systemic RNAi is likely a dose-sensitive process. Our analysis of sid-4 provides additional evidence for this hypothesis.
Materials and methods
Strains
Unless otherwise indicated, all strains were grown at 20°C on NGM plates seeded with OP50 as a food source (Brenner 1974). Strains used in this analysis are listed in Table 1 and for the candidate SID-3–SID-4-interacting screen in Supplementary Table 1.
Table 1.
Strains used in this study (see also Supplementary Table 1 for candidate RNAi-defective strains).
| Strain | Genotype reference |
|---|---|
| N2 | Wild type |
| HC57 | ccIs4251 [pSAK2 (myo-3::NGFP-LacZ), pSAK4 (myo-3::mtGFP); dpy-20] I; |
| qtIs3(myo-2::GFP dsRNA) III; | |
| mIs11[myo-2p::GFP + pes-10p::GFP + F22B7.9::GFP] IV | |
| HC259 | ccIs4251 I; mIs11 IV; sid-4(qt15)X |
| HC119 | ccIs4251 I; mIs11IV; sid-4(qt17)X |
| HC158 | ccIs4251 I; qtIs3 III; mIs11 IV; sid-4(qt33)X |
| HC160 | ccIs4251 I; qtIs3 III; mIs11 IV; sid-4(qt35)X |
| HC114 | ccIs4251 I; qtIs3 III; mIs11 IV; sid-1(qt9)V |
| HC122 | ccIs4251 I; sid-2(qt13), qtIs3 III; mIs11 IV; |
| HC770 | sid-3(tm342) X |
| HC1088 | sid-4(qt33) X |
| HC1093 | sid-4(ok694) X [out crossed 8X to N2] |
| HC1098 | ccIs4251 I; qtIs3 III; mIs11 IV; sid-4 (ok694) X |
| HC1133 | nrIs20 [sur-5::NLS-GFP] IV; sid-4 (ok694) X |
| HC1139 | sid-4(ok694) X; qtEx159 [sid-4::GFP] |
| HC1140 | nrIs20 IV; sid-4 (ok694) X; qtEx214 [ZK470.5a, PCFJ90(Pmyo-2::mCherry)]; line 1 |
| HC1141 | nrIs20 IV; sid-4 (ok694) X; qtEx215 [ZK470.5a, PCFJ90(Pmyo-2::mCherry)]; line 2 |
| HC1142 | nrIs20 IV; sid-4 (ok694) X; qtEx216 [ZK470.5a, PCFJ90(Pmyo-2::mCherry)]; line 3 |
| HC1143 | nrIs20 IV; sid-4 (ok694) X; qtEx217 [ZK470.5b, PCFJ90(Pmyo-2::mCherry)]; line 1 |
| HC1144 | nrIs20 IV; sid-4 (ok694) X; qtEx218 [ZK470.5b, PCFJ90(Pmyo-2::mCherry)]; line 2 |
| HC1145 | nrIs20 IV; sid-4 (ok694) X; qtEx219 [ZK470.5b, PCFJ90(Pmyo-2::mCherry)]; line 3 |
| HC1146 | sid-4(ok694), sid-3 (tm342) X |
| HC1147 | ver-1(ok1738)III; sid-4(ok694)X |
| HC1148 | sid-3(tm342), ver-1(ok1738) X |
Whole genome sequencing
Genomic DNA was extracted from each of the 4 sid-4 strains: HC259 sid-4(qt15); HC119 sid-4(qt17); HC158 sid-4(qt33); and HC160 sid-4(qt35). Samples were sheared to 250 bp and prepared according to NEB Ultra DNA library kit E7370S. Sample concentrations were quantified via qPCR (Kapa Kit KK4824) and sequenced (Illumina), recovering a total of 250 million reads with 80× coverage. Reads were aligned using bowtie2 and exon variants on linkage group X were called using Samtools. Coding sequence variants in only a single gene, nck-1, were identified in all 4 strains.
SID-4 isoform expression constructs
Full-length sid-4A (ZK470.5A) and sid-4B (ZK470.5B) cDNA constructs (Open Biosystems) were verified and individually injected (4.5 ng/μl) with a Pmyo-2::mCherry (10 ng/μl) coinjection marker (PCFJ90) and 1 kb DNA ladder (10 ng/μl) (New England Biolabs) into sur-5::NLS-GFP; sid-4(ok694) (HC1133). For each isoform plasmid, 3 recovered independent complex extrachromosomal array lines (ZK470.5a; HC1140, HC1141, HC1142; ZK470.5b; HC1143, HC1144 HC1145) were analyzed.
Feeding RNAi assays
L4-young adults were placed on RNAi food (Kamath and Ahringer 2003) at room temperature (unless otherwise specified) and in every case, F1 adult progeny were scored for sensitivity to RNAi. Bacteria containing the empty-vector L4440 was used as the control for all feeding RNAi experiments. The following phenotypes were scored for each food: gfp, GFP silencing; dpy-11, body length (qualitative); bli-1, full-body (hyp7) blisters; unc-45, paralysis; unc-22, twitching; act-5, F1 larval growth; fkh-6, F1 fertility (presence of internal eggs); pos-1, F2 embryonic viability.
sid-4 mosaic analysis
HC1139 sid-4(ok694); qtEX159 sid-4::gfp young adults were cultured on bli-1 RNAi plates for 3 days. RNAi-sensitive blistered animals were counted and RNAi “resistant” nonblistered animals were collected and scored for GFP expression. All resistant worms expressing detectable GFP were then mounted for widefield microscopy and scored for GFP expression in the hypodermis.
dsRNA synthesis and injection
pal-1 dsRNA was made by amplifying a 1.2-kb region from the pal-1 plasmid (Ahringer library, Kamath and Ahringer 2003) using forward and reverse T7 primers (pal-1-F-T7 TAATACGACTCACTATAGGTCCCATTTTAGGCAGTGAGTTA; pal-1-R GTTGCCAGCTCGTTATTTTATTG; pal-1-F TCCCATTTTAGGCAGTGAGTTTA; pal-1-R-T7 TAATACGACTCACTATAGGCTCGAGAAGAAAAAGAACGACAA). A T7-flash Ampliscribe kit was used to make single-stranded sense and anti-sense RNA strands, which were annealed, quantified, diluted as necessary, and injected into either 1 or both gonad arms of sid-4(ok694) and N2 animals. Six hours after injection, each recovered animal was singled to an OP50 plate. To score embryonic lethality, the adult was removed after 24 h and laid eggs counted. Three days later, the number of hatched progeny were counted.
SID-3–SID-4 interactor screen
All known mammalian NCK and ACK physical interactors were obtained from BioGrid (build 3.4.140) and Human protein reference databases (release 9) and all mammalian network information for both was obtained from PathCards (version 4.4) (Keshava Prasad et al. 2009; Belinky et al. 2015; Chatr-Aryamontri et al. 2017). Cross referencing all NCK interactors against both ACK interactors and the network data and ACK interactors against the NCK network data identified 113 unique mammalian proteins. We used wormbase (WS262) to identify 116 C. elegans orthologs of these proteins as well as to identify viable mutations. Fifty-three mutant strains were ordered from the C. elegans Genetic stock Center (cgc.umn.edu). To test each strain for RNAi defects, young adult hermaphrodites were placed on fkh-6 RNAi plates and dpy-11 RNAi plates. Three days later, the fkh-6 RNAi plates were scored for presence of laid eggs and the dpy-11 RNAi plates were scored for Dpy progeny.
Microscopy
Figure 6 images were obtained with a Zeiss Axiovert 200m spinning disk confocal microscope (63× and 100× objectives) equipped with a Hamamatsu Orca-ER digital camera. All the other images were obtained with an Olympus SZX2-TR30PT fluorescent microscope [filter (GFP-470)] equipped with a Hamamatsu digital camera. ImageJ was used to adjust contrast and brightness and Adobe illustrator was used to crop images.
Fig. 6.

sid-4 is required in the importing tissue. Progeny of sid-4(ok694); [Ex: sid-4::gfp] animals were scored for bli-1 RNAi sensitivity and SID-4::GFP expression. a) A SID-4::GFP positive bli-1 RNAi-sensitive animal. Scale bar: 0.1 mm. b, b′, and b″) Confocal image of a bli-1 RNAi-sensitive animal showing SID-4 GFP expression in hyp7. In (b), the bright gut GFP obscures dim diffuse GFP in syncytial Hyp7, which (b′) is outlined in the enlarged gray-scale image that excludes the bright intestinal signal. b″) The same image without outlining. Scale bars: 25 and 5 µm. c) Intestine and d) pharyngeal SID-4::GFP expression in bli-1 RNAi-resistant animals.
Results
sid-4/nck-1 encodes the C. elegans ortholog of the mammalian noncatalytic region of tyrosine kinase adaptor protein
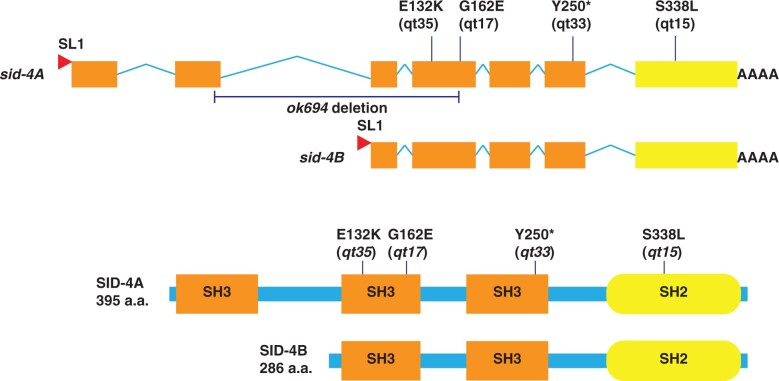
The Sid screen recovered 4 X-linked, noncomplementing alleles that define the sid-4 locus (Winston et al. 2002). Whole genome sequencing of these 4 strains identified protein-altering nucleotide variants in ZK470.5 (nck-1) in each isolate: sid-4(qt15) [S338L], sid-4(qt17) [G162E], sid-4(qt33) [Y250-stop], and sid-4(qt35) [E132K] (Fig. 1). For subsequent characterization, we used the 1,814 base pair (bp) deletion allele, sid-4/nck-1 (ok694) (Consortium 2012) (Fig. 1). SID-4/NCK-1 is homologous to adaptor proteins that promote the formation of signaling protein complexes either at the plasma membrane or in the cytoplasm (Buday 1999). Interestingly, SID-3 is homologous to ACK1 (activated CDC42 kinase) (Jose et al. 2012) and mammalian and Drosophila ACK1 proteins physically interact with NCK proteins (Teo et al. 2001; Worby et al. 2002). A translational nck-1 genomic::gfp reporter is broadly expressed in embryos and adults in a manner similar to SID-3::GFP reporters, supporting the possibility that sid-3 and sid-4 may interact to support systemic RNAi (Mohamed and Chin-Sang 2011; Jose et al. 2012).
Fig. 1.
sid-4 encodes NCK-1, a noncatalytic region of tyrosine kinase adaptor protein. Schematic representation of protein-altering SNPs identified in ZK470.5 and their location in SID-4A and SID-4B (adapted from Mohamed and Chin-Sang 2011). sid-4A and sid-4B are produced by independent promoters and trans-spliced to SL1.
Both sid-4 isoforms function to support systemic RNAi
The mammalian NCK homologs, nck-1/nckα and nck-2/nckβ, may function redundantly as well as have distinct roles in signal transduction (Chen et al. 1998; Li et al. 2001). C. elegans sid-4/nck-1, which is equally similar to both mammalian NCK homologs, encodes 2 isoforms: NCK-1A and NCK-1B (Fig. 1). NCK-1A is the larger isoform and contains 3 SH3 domains followed by a single SH2 domain, while NCK-1B lacks the first SH3 domain (Fig. 1). The screen identified missense mutations in the SH2 domain and each of the SH3 domains common to both isoforms (Fig. 1). The lack of a recovered mutation in the first SH3 domain fails to provide the evidence of function for one or the other isoform. The 2 isoforms SID-4A and SID-4B are produced by 2 different sid-4 promoters and transcription initiation sites (Fig. 1) (Mohamed and Chin-Sang 2011). To test each isoform independently, we rescued the sid-4(ok694) with isoform-specific cDNA constructs driven by their endogenous promoters.
To score RNAi efficacy in multiple tissues, we monitored the expression of a sur-5::gfp transgene that expresses nuclear localized GFP in all somatic cells. The progeny of sur-5::gfp hermaphrodites grown on bacteria expressing gfp dsRNA are fully silenced, while roughly three-quarters of the progeny of sid-4(ok694); sur-5::gfp hermaphrodites grown on bacteria expressing gfp dsRNA are resistant or partially resistant to silencing (Fig. 2). We then tested 3 independent SID-4A or SID-4B expressing extrachromosomal array lines for rescue of the silencing defect. GFP expression in nonintestinal cells was blind scored in the progeny of animals placed on gfp RNAi food (Fig. 2); the sur-5::gfp transgene line is not reliably expressed in the intestine in the presence of other extrachromosomal arrays (Jose et al. 2012). We found that both SID-4A and SID-4B similarly and partially rescued silencing in sid-4; sur-5::gfp strains (Fig. 2). This result shows that expression of either SID-4 isoform is sufficient to produce SID-4 activity and, therefore, that the first SH3 domain is not required for SID-4 to support systemic RNAi. However, the partial rescue suggest that the isoforms may function together for full activity. To test this, we injected both SID-4A and SID-4B cDNA constructs into the sid-4(ok694); sur-5::gfp strain; however, all identified transgenic progeny were developmentally arrested. This phenotype is unlikely to be related to dsRNA transport; thus, we did not investigate further.
Fig. 2.
SID-4 isoform rescue of systemic RNAi. Representative images (top) of variable sur-5::GFP silencing. Quantification (bottom) of rescue of silencing in 3 independent lines for each SID-4 isoform. Scale bar: 0.1 mm
sid-4 alleles have weak systemic RNAi-defective phenotypes
The above analysis showed that sid-4 mutants partially disable RNAi silencing. To directly compare sid-4 systemic silencing defects to that of other Sid mutants, we crossed the deletion allele sid-4(ok694) into the transgenic HC57 background used for the screen. In this transgenic strain, pharyngeal expressed gfp dsRNA autonomously silences pharyngeal GFP and systemically silences GFP in anterior body wall muscle cells. When cultured on gfp dsRNA expressing bacteria, the ingested dsRNA silences GFP in all body wall muscle cells. A sid-1 mutant in this transgenic background disrupts the silencing of the body wall muscle GFP from both pharyngeal expressed gfp dsRNA and ingested bacterial gfp dsRNA (Fig. 3). In contrast, sid-2 mutants disrupt only silencing in response to ingested dsRNA. Thus, sid-2 mutants in the transgenic background have the same silencing phenotype whether grown on normal bacteria or bacteria expressing gfp dsRNA (Fig. 3). In contrast to sid-1 and sid-2 mutants, sid-4(qt33) and sid-4(ok694) show incomplete systemic RNAi defects (Fig. 2a and Supplementary Fig. 1). Like sid-1 and sid-2, neither allele disrupts pharyngeal silencing, confirming that the mutations do not noticeably disrupt RNAi. However, in contrast to sid-1 and sid-2, many anterior body wall muscle cells are silenced when grown on bacteria expressing gfp dsRNA. While this indicates that sid-4 is not required for the uptake of ingested dsRNA, the lack of complete silencing may indicate a dose-dependent response to exported pharyngeal expressed or ingested gfp dsRNA. Detailed comparison of the extent of body-wall muscle GFP silencing shows no discernable difference between the 2 sid-4 alleles (Supplementary Fig. 1).
Fig. 3.
sid-4 mutants are partially defective for RNAi. Adult progeny of indicated sid mutant L4 hermaphrodites in the HC57 background (pharyngeal GFP; body wall muscle GFP; pharynx expressed gfp dsRNA) placed on bacteria expressing gfp dsRNA (GFP RNAi) or L440 empty-vector bacteria (control RNAi). All animals approximately 1 mm. sid-4 (ok694) and sid-4 (qt33) progeny showed indistinguishable penetrance and expressivity of systemic RNAi-silencing defects (Supplementary Fig. 1).
Weak sid-1 alleles show gene-specific patterns of silencing that correlate with strong and weak RNAi foods (Whangbo et al. 2017). We tested both sid-4 alleles on that same set of strong and weak RNAi foods. Wild-type worms were completely sensitive (100% silencing) to all these RNAi foods (Fig. 4), while strong sid-1 and sid-2 mutant worms were completely resistant (0% silencing, not shown). sid-4(qt33) and sid-4(ok694) worms were completely or nearly completely resistant to fkh-6 (gonad), unc-45 (muscle), and bli-1 (hypodermis); completely or mostly sensitive to act-5 (intestine) and unc-22 (muscle); and partially resistant to pos-1 (germline) and dpy-11 (hypodermis). The discrepancy in sensitivity to dpy-11 between qt33 (nonsense) and ok694 (deletion) indicates that the nonsense mutant qt33 may have residual activity, perhaps reflecting tissue-specific read-through translation of the stop codon (Fig. 4). The similarity in the pattern of RNAi sensitivity and resistance between strong sid-4 alleles and weak sid-1 alleles is consistent with sid-4 mutations compromising systemic RNAi generally, rather than reflecting a tissue or dsRNA delivery-specific effect.
Fig. 4.
sid-4 and sid-3 have similar RNAi phenotypes. The adult progeny (F1) of L4 animals (F0) placed on RNAi foods were scored for RNAi sensitivity (fraction sensitive on each plate). Each circle represents the plate mean sensitivity for each F0 (n = 10) with ≥150 F1’s for each F0. 100% sensitive (left) to 100% resistant (right).
Because sid-3 is also a weak Sid mutant and mammalian and Drosophila SID-3 and SID-4 homologs have been shown to interact, we tested sid-3(tm342) on the same panel of RNAi foods (Fig. 4). We observed a similar, but not identical pattern of resistance and sensitivity. These results are consistent with sid-3 and sid-4 acting in concert to promote systemic RNAi. In summary, these results indicate that the sid-4 null phenotype is not a tissue-specific defect but an incomplete RNAi defect.
sid-4 is a dose-dependent Sid mutant
sid-4 mutant worms retain RNAi-silencing activity in the pharynx and anterior body wall muscle cells (Fig. 2a) indicating that sid-4 mutations do not compromise RNAi-silencing activity. However, weak RNAi-defective (Rde) mutants that are suppressed by high levels of pharyngeal expressed dsRNA can produce wild-type silencing in the pharynx (Yang et al. 2014). To explicitly test sid-4 mutants for a weak Rde phenotype, we injected pal-1 dsRNA directly into the syncytial germline of wild-type and sid-4 mutant hermaphrodites and scored the frequency of the pal-1(RNAi) phenotype, embryonic lethality. Because the germline is syncytial, injections into the anterior or posterior gonad do not require dsRNA transport for effective silencing. If sid-4 mutants are weak RNAi defective, then wild-type and sid-4 mutant worms should show distinct embryonic lethality frequencies in response to decreasing injected dsRNA dose. We injected both gonad arms to eliminate the effect of gonad to gonad transfer.
Furthermore, for each dsRNA concentration, we used a single needle and identical injection times to limit variability and enable direct comparison between strains. We found that both wild-type and sid-4(ok694) mutants showed proportionate changes in embryonic lethality when injected with changing concentrations of pal-1 dsRNA (Fig. 5). This indicates that sid-4 mutants have a wild-type level of RNAi activity in the germline and are not weak Rde mutants.
Fig. 5.

sid-4 is a dose-dependent Sid. Wild-type and sid-4(ok694) young adults were injected with varying pal-1 dsRNA concentrations into 1 (open symbols) or both gonad arms (filled symbols) and then 6 h postinjection singled and allowed to lay eggs for 24 h. The fraction of hatched eggs for each injected adult (n ≥ 10) was scored 3 days later.
We used the same assay to determine whether sid-4 is required to transfer silencing information between gonad arms. In wild-type animals, injecting a single gonad arm can result in 100% embryonic lethality while in strong sid-1 mutants only progeny from the injected gonad are affected (Winston et al. 2002; Wang and Hunter 2017). To test sid-4(ok694) mutants for systemic RNAi defects (Sid), we injected single gonad arms with either a high or low pal-1 dsRNA dose. Consistent with effective systemic RNAi, there was no detectable difference in embryonic lethality among the progeny of wild-type worms injected with pal-1 dsRNA into either a single or both gonad arms at either high or low dsRNA dose (Fig. 5). Similarly, the frequency of embryonic lethality among the progeny of sid-4(ok694) animals injected with the high concentration of pal-1 dsRNA into either a single or both gonad arms was nearly identical (Fig. 5). However, the frequency of embryonic lethality among the progeny of sid-4(ok694) animals injected with the low concentration of pal-1 dsRNA into single gonad arm was half that of animals injected into both gonad arms (Fig. 5). This dose-dependent systemic RNAi result is consistent with inefficient transport of dsRNA between gonad arms, indicating that sid-4 is a weak Sid mutant.
sid-4 is import defective
We used genetic mosaic analysis to determine whether sid-4 is required in importing cells for efficient feeding RNAi. Specifically, we rescued sid-4(ok694) animals with a sid-4::gfp construct maintained as a mitotically unstable extrachromosomal array. About 82% of the progeny inherited the array and were GFP positive and formed full-body blisters when cultured on bli-1 RNAi food. The remaining 18% of the progeny did not inherit the array and were mostly GFP negative and fully resistant to bli-1 RNAi. bli-1 is expressed in the large syncytial cell hyp7 (139 nuclei, 29 from embryonic cells that fuse to form hyp7 and 110 from postembryonic cells that fuse with hyp7), which produces the full-body blister in bli-1 RNAi-treated animals (Chisholm and Hsiao 2012). Among 976-resistant worms, we identified 74 with detectable GFP expression. If sid-4 is required for import, then in these resistant animals, we would expect all or most of the hyp7 nuclei to lack the sid-4::gfp array and fail to express detectable GFP in hyp7. Indeed, in none of these animals, did we detect GFP expression in hyp7. It is notable that all of the hyp7 nuclei arise from descendants of 3 distinct early embryonic cells (ABarp, ABp, and C; Supplementary Fig. 2); thus, to produce a sid-4::gfp mosaic lacking sid-4::GFP expression in most hyp7 nuclei would require 2 or 3 losses. For example (see Supplementary Fig. 1b), the array may fail to segregate to AB at the first division and fail to segregate to P2 at the second division producing a mosaic animal that lacks sid-4::gfp in all hyp7 nuclei but expresses sid-4::gfp in the posterior pharynx, the gut, and anterior body wall muscle cells. Twenty-two bli-1 RNAi-resistant animals were identified that had pharynx and gut GFP expression and lacked detectable GFP in hyp7 (Fig. 6c). As a second example (Supplementary Fig. 1c), the array could be lost in P1, ABp, and ABar(pp), producing an animal that lacks sid-4::gfp in all hyp7 nuclei, but that expresses GFP in the anterior pharynx only. Fifty-two animals were recovered that are consistent with this scenario (Fig. 6d). It is important to note that by selecting for resistant animals that maintained some GFP, we greatly enriched for mosaic animals that failed to segregate the array at multiple cell divisions. In summary, the results of the sid-4 genetic mosaic analysis indicate that sid-4 is required in the target cell for effective RNAi.
sid-3 and sid-4 genetically interact but do not function in a simple linear pathway
Mammalian and Drosophila SID-3/ACK1 and SID-4/NCK-1 homologs interact (Teo et al. 2001; Worby et al. 2002); thus, it is likely that they do so in C. elegans as well. Both sid-4/nck-1 and sid-3 fluorescent protein fusion constructs are broadly expressed cytoplasmic proteins that localize to intracellular membranes (Mohamed and Chin-Sang 2011; Jose et al. 2012). Furthermore, both sid-3 and sid-4 are weak Sid mutants required in importing cells for effective systemic RNAi (Jose et al. 2012; Figs. 3–6). If these genes function together as a kinase and exclusive adapter, then we expect the double mutant to resemble both single mutants. To test this, we constructed and tested a sid-3(tm342); sid-4(ok694) double mutant on a broad panel of RNAi foods (Supplementary Fig. 3). For many of the foods, the sensitivity of either single mutant was maximal or minimal, precluding interpretation of the double mutant sensitivity. But for 2 foods, unc-22 (muscle) and pos-1 (germline), the sensitivity of both single mutants was intermediate, and in both cases, the double mutant was less sensitive (Fig. 7). This result shows that the effect of sid-3 and sid-4 are at least partially additive, indicating that in these 2 tissues, these 2 signaling proteins do not function in a simple linear pathway but likely interact independently with other signaling proteins to modulate systemic RNAi silencing.
Fig. 7.

Double Sid weak mutants show enhanced RNAi resistance. The F1 adult progeny of F0 L4 animals placed on RNAi foods were scored for RNAi sensitivity (fraction sensitive on each plate). Each circle represents the plate mean sensitivity for each F0 (n = 10) with ≥40 F1’s per F0. 100% sensitive (left) to 100% resistant (right). t-test P-values: *<0.01, **<0.0005.
C. elegans homologs of mammalian NCK/ACK-interacting proteins have weak RNAi phenotypes
The above double mutant analysis suggests that SID-3/4-interacting proteins may modulate systemic RNAi. To identify candidate C. elegans sid-3 and sid-4-interacting proteins, we compiled a list 113 mammalian proteins that likely interact (directly or indirectly) with both an NCK and an ACK ortholog (see Materials and Methods). Among 116 C. elegans orthologs of these proteins, 80 viable mutants have been described and we tested 52 by feeding RNAi for reduced sensitivity to fkh-6 and dpy-11 RNAi food (Supplementary Table 1). Twenty-two candidates were detectably resistant to fkh-6 RNAi, 7 of which were also partially resistant to dpy-11 RNAi, and 1 strain was only partially resistant to dpy-11 RNAi (Table 2). In addition, 5 of the 29 strains that were fully sensitive produced enhanced Dpy-11 phenotypes, possibly signifying enhanced RNAi.
Table 2.
RNAi-defective SID-3–SID-4 candidate-interacting proteins.
| Strain | Genotypea | fkh-6 (RNAi) eggs on plate? | dpy-11 (RNAi) % non-Dpy adults | Homology | Lethal null? |
|---|---|---|---|---|---|
| (n = 2) | (n = 2) | ||||
| N2 | WT | None | 0 | – | – |
| CB96 | vab-2(e96) IV | Many | 44 | Ephrin ligand | Y |
| CZ375 | vab-1(e856) II | Many | 0 | Ephrin receptor | Y |
| EM305 | efn-4(bx80) IV | Many | 15 | Ephrin ligand | Y |
| RB942 | cdc-42(ok825) II | Many/few | 19 | GTPase | Y |
| MT12615 | mys-1(n3681) V | Many | 0 | Histone acetylase | Y |
| VC1263 | ver-1(ok1738) III | Many | 0 | Receptor tyrosine kinase | |
| RB2513 | C26C6.6(ok3481) I | Many | 29 | Lim domain | |
| VC664 | ras-1(ok977) II | Few | 18 | GTPase | |
| MT4434 | ced-5(n1812) IV | Few | 15 | DOCK | |
| CB3257 | ced-2(e1752) IV | Few | 0 | SH2/SH3 adapter | |
| CX51 | dyn-1(ky51) X | Few | 0 | Dynactin | Y |
| MT5267 | soc-1(n1789) V | Few | 0 | Pleckstrin homology domain | |
| RB1591 | ddr-1(ok1956) X | Few | 0 | Tyr kinase | |
| RB689 | pak-1(ok488) X | Few | 0 | Ser/Thr kinase | Y |
| NW1549 | efn-2(ev658) IV; efn-3(ev696) X | Few | 0 (n = 1) | Ephrin ligand | Sterile |
| CZ414 | vab-1(e699) II | Few | 0 | Ephrin receptor | Y |
| RB1267 | ensh-1 (ok1349) X | Few | 0 | Fibrogenin like | |
| RB1751 | rga-5(ok2241) IV | Few | 0 | RhoGAP | |
| RB759 | akt-1(ok525) V | Few | 0 | Ser/Thr kinase | |
| ZD500 | hecw-1(ok1347) III | Few | 0 | E3 ubiquitin ligase | |
| RB776 | kin-32(ok166) I | Few | 0 | Tyr kinase |
Some strains include nonrelevant mutations, listed in full in Supplementary Table 1.
Among the top candidates was the receptor tyrosine kinase VER-1 (Table 2), which is orthologous to the mammalian VEGF Receptor 1, an interactor of NCK1 (Igarashi et al. 1998). In C. elegans, VER-1 is expressed in the intestine and neuronal sheath cells (Procko et al. 2012). To explore the RNAi sensitivity of the ver-1(ok1783) deletion mutant more thoroughly, we scored the RNAi phenotypes of the progeny of adult animals placed on a panel of RNAi foods targeting a variety of tissues (Supplementary Fig. 4). The progeny of wild-type animals grown on fkh-6 RNAi food fail to lay any eggs and while ver-1(ok1783) animals lay eggs (Table 2); however, this analysis showed that the proportion of egg-laying adults is small (Supplementary Fig. 3). Similarly, the penetrance of dpy-11 RNAi resistance was low. These results suggest that ver-1 is a very weak RNAi-defective mutant and would most likely not be recovered in a forward genetic screen. Because sid-3, sid-4 double mutants show a stronger RNAi resistance phenotype than either single mutant, we constructed and tested ver-1(ok1783); sid-3(tm342) double mutant on the same panel of RNAi foods (Supplementary Fig. 3). Consistent with VER-1 acting in the SID-4 pathway, we found that the double mutant was more resistant to feeding RNAi targeting unc-22 and bli-1 than either single mutant (Fig. 7).
In summary, 23 of 52 candidate SID-3/4-interacting protein mutants had detectable RNAi defects, supporting the hypothesis that additional signaling components and likely pathways modulate systemic RNAi. Null mutants for many of these genes are lethal, thus perhaps explaining the low penetrance and expressivity of the observed RNAi defects. This, combined with the high level of noise in the experimental assays, makes it challenging to verify the significance of these interactions.
SID-3/SID-4 is required for temperature-dependent RNAi silencing
Efficient systemic RNAi requires signaling proteins, indicating that physiological or environmental conditions may modulate systemic RNAi. We previously reported that temperature affects systemic RNAi; we noted more efficient silencing of body wall muscle GFP in response to exported pharyngeal gfp dsRNA at 20°C than at 25°C (Winston et al. 2002). To extend this analysis, we determined whether temperature can affect the silencing of the endogenous gene unc-22 in response to ingested unc-22 dsRNA. The progeny of 10 singled wild-type L4’s placed on unc-22 RNAi food at 15, 20, and 25°C were scored for unc-22 twitching as adults. Consistent with the previous findings, 2 independent trials showed more efficient silencing at 15°C and less efficient silencing at 25°C (Fig. 8). Thus, the temperature effect on systemic RNAi silencing is independent of the method of dsRNA delivery (expression vs. ingestion) and target gene (transgene vs. endogenous). We then repeated the experiment with sid-3 and sid-4 mutants (Fig. 8). We found that the progeny of both sid-3 and sid-4 worms showed less efficient silencing at all 3 growth temperatures. This experiment was then repeated in a second large trial (Fig. 8), which confirmed that sid-3 and sid-4 mutants are indistinguishable from each other and wild type at 25°C and that the penetrance of both mutants were most different from wild type at 15°C (P < 10−8; t-test). These results indicate that sid-3 and sid-4 do not contribute to systemic RNAi at 25°C and are required primarily for the low temperature enhanced penetrance of systemic RNAi.
Fig. 8.

sid-3 and sid-3 temperature-dependent systemic RNAi. F1 progeny of F0 animals placed on unc-22 food at the indicated temperature were scored in 3 mM Levamisole for paralysis/twitching (sensitivity). Mean (large filled circle), 95% confidence interval (vertical line), and each F0 plate mean (small unfilled circle) are shown for each genotype temperature. Distribution of 10 plate means for 2 independent trials is shown to the left and right of each mean.
Discussion
The identity of SID-4 as a NCK homolog complements the identity of SID-3 as an ACK1 homolog, one of the presumed SID-4/NCK-1-binding partners. Establishing that these proteins interact to regulate systemic RNAi in C. elegans and understanding what conditions regulate their activity will lead to further insight into the mechanisms and functions of systemic RNAi. Unlike the dsRNA transporters SID-1 and SID-2, these 2 proteins have been implicated in a variety of presumably systemic RNAi-independent functions in C. elegans, including cell migration (Martynovsky et al. 2012; Mohamed et al. 2012), virus entry (Jiang et al. 2017; Tanguy et al. 2017), and endocytic trafficking (Lazetic et al. 2018) and cell polarity (Goh et al. 2012). All these functions impinge on endocytosis.
Endocytosis has been implicated in feeding RNAi as well as SID-1-independent parental RNAi. In parental RNAi, the endocytosis receptor RME-2 is required for SID-1-independent transfer of extracellular dsRNA into oocytes (Wang and Hunter 2017). SID-1 is then subsequently required in the embryo for RNAi silencing, presumably to release the endocytosed dsRNA into the cytoplasm to initiate RNAi. In feeding RNAi, the SID-2 transmembrane protein, which is expressed in the intestine and localizes to the apical (lumenal) membrane, appears to act as a dsRNA specific endocytosis receptor (Winston et al. 2007; McEwan et al. 2012). SID-1 is also required for feeding RNAi induced silencing in intestinal cells, again, presumably to release endocytosed dsRNA into the cytoplasm to initiate RNAi. Furthermore, homologs of several of the candidate SID-3/SID-4-interacting proteins that had detectable Rde phenotypes have roles in or are implicated in endocytosis, including dyn-1 (dynactin), pak-1(p21 activated kinase), and cdc-42. It should be emphasized that only viable alleles of candidate SID-3–SID-4-interacting proteins were tested and for many of these genes null alleles are inviable (Table 2). Thus, these weak RNAi-defective phenotypes are associated with, in many cases, weak loss-of-function alleles of essential genes. Thus, even if endocytosis is critical for dsRNA transport, we are unlikely to recover strong viable Sid mutants disrupted for endocytosis.
The RNAi potency that makes systemic RNAi possible also makes it difficult to identify and analyze weak RNAi-defective mutants. The vanishing small amounts of dsRNA that are transported into recipient cells are processed and then amplified by RNA-directed RNA polymerases to produce abundant secondary siRNAs. Thus, to observe a Sid phenotype, dsRNA transport must be nearly blocked, not merely disrupted. This is aptly illustrated by the dose sensitivity we documented for sid-4 (Fig. 5). This dose sensitivity may also explain our results with fkh-6(RNAi). Among the tested RNAi foods, sid-4 mutants were most resistant to fkh-6 silencing (Fig. 4). A reasonable interpretation for this high penetrance and expressivity is that fkh-6 is a poor RNAi target, such that even a modest reduction in dsRNA transport is sufficient to cause a silencing defect. These observations indicate that it will be difficult to directly observe disrupted transport of dsRNA in sid-4 mutants.
The original large visual screen identified many alleles in sid-1 and sid-2 with a strong Sid phenotype and relatively few alleles in sid-3, sid-4, and sid-5, which produce a weak Sid phenotype. This bias likely reflects the relative ease of identifying worms with bright GFP in many cells in the strong mutants (sid-1 and sid-2 mutants) vs. the variable and relatively sparse GFP expression in the weak mutants (sid-3, sid-4, and sid-5, as well as a partial loss-of-function sid-1 alleles; Whangbo et al. 2017). Our screen and analysis of candidate SID-3/4-interacting proteins extends this pattern, suggesting that most or all strong Sid mutants have been identified. Our analyses also showed that sid-3 and sid-4 are dose-dependent SIDs, that is, increasing the dose of dsRNA bypasses the dsRNA import defect.
Furthermore, and unexpectedly, we discovered that combining 2 weak mutants resulted in a stronger Sid phenotype, suggesting that multiple independent processes contribute to dsRNA import. Some of these processes may involve putative NCK/ACK1-interacting proteins, several of which have weak RNAi-defective phenotypes. Finally, the sid-3- and sid-4-dependent process is most apparent at lower growth temperatures, confirming that the temperature effect on silencing is due to more efficient systemic RNAi at lower growth temperature.
Supplementary Material
Acknowledgments
We thank members of the Hunter lab, Alexander Weisman, Andrey Shubin, and Nicole Bush for comments of the article.
Funding
This work was supported by National Institutes of Health (NIH) grant GM089795. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Conflicts of interest
None declared.
Contributor Information
Sonya Bhatia, Department of Molecular and Cellular Biology, Harvard University, Cambridge MA 02138, USA.
Craig P Hunter, Department of Molecular and Cellular Biology, Harvard University, Cambridge MA 02138, USA.
Data Availability
Strains not available at the CGC (cgc.umn.edu) are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplemental material is available at G3 online.
Literature cited
- Belinky F, Nativ N, Stelzer G, Zimmerman S, Iny Stein T, Safran M, Lancet D.. PathCards: multi-source consolidation of human biological pathways. Database (Oxford). 2015;2015:bav006. 10.1093/database/bav006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-Aryamontri A, Oughtred R, Boucher L, Rust J, Chang C, Kolas NK, O'Donnell L, Oster S, Theesfeld C, Sellam A, et al. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 2017;45(D1):D369–D379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, She H, Davis EM, Spicer CM, Kim L, Ren R, Le Beau MM, Li W.. Identification of Nck family genes, chromosomal localization, expression, and signaling specificity. J Biol Chem. 1998;273(39):25171–25178. [DOI] [PubMed] [Google Scholar]
- Chisholm AD, Hsiao TI.. The Caenorhabditis elegans epidermis as a model skin. I: development, patterning, and growth. Wiley Interdiscip Rev Dev Biol. 2012;1(6):861–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium CeDM. large-scale screening for targeted knockouts in the Caenorhabditis elegans genome. G3 (Bethesda). 2012;2:1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg EH, Hunter CP.. Transport of dsRNA into cells by the transmembrane protein SID-1. Science. 2003;301(5639):1545–1547. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC.. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. [DOI] [PubMed] [Google Scholar]
- Goh KY, Ng NW, Hagen T, Inoue T.. p21-activated kinase interacts with Wnt signaling to regulate tissue polarity and gene expression. Proc Natl Acad Sci U S A. 2012;109(39):15853–15858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinas A, Wright AJ, Hunter CP.. SID-5 is an endosome-associated protein required for efficient systemic RNAi in C. elegans. Curr Biol. 2012;22(20):1938–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi K, , IsoharaT, , KatoT, , ShigetaK, , YamanoT, , Uno I.. Tyrosine 1213 of Flt-1 is a major binding site of Nck and SHP-2. Biochem Biophys Res Commun. 1998;246(1):95–99. [DOI] [PubMed] [Google Scholar]
- Jiang H, Chen K, Sandoval LE, Leung C, Wang D.. An evolutionarily conserved pathway essential for orsay virus infection of Caenorhabditis elegans. mBio. 2017;8(5):e00940-17. 10.1128/mBio.00940-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose AM, Hunter CP.. Transport of sequence-specific RNA interference information between cells. Annu Rev Genet. 2007;41(1):305–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose AM, Kim YA, Leal-Ekman S, Hunter CP.. Conserved tyrosine kinase promotes the import of silencing RNA into Caenorhabditis elegans cells. Proc Natl Acad Sci U S A. 2012;109(36):14520–14525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose AM, Smith JJ, Hunter CP.. Export of RNA silencing from C. elegans tissues does not require the RNA channel SID-1. Proc Natl Acad Sci U S A. 2009;106(7):2283–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J.. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30(4):313–321. [DOI] [PubMed] [Google Scholar]
- Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human protein reference database—2009 update. Nucleic Acids Res. 2009;37(Database issue):D767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazetic V, Joseph BB, Bernazzani SM, Fay DS.. Actin organization and endocytic trafficking are controlled by a network linking NIMA-related kinases to the CDC-42-SID-3/ACK1 pathway. PLoS Genet. 2018;14(4):e1007313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Fan J, Woodley DT.. Nck/Dock: an adapter between cell surface receptors and the actin cytoskeleton. Oncogene. 2001;20(44):6403–6417. [DOI] [PubMed] [Google Scholar]
- Martynovsky M, Wong MC, Byrd DT, Kimble J, Schwarzbauer JE.. mig-38, a novel gene that regulates distal tip cell turning during gonadogenesis in C. elegans hermaphrodites. Dev Biol. 2012;368(2):404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan DL, Weisman AS, Hunter CP.. Uptake of extracellular double-stranded RNA by SID-2. Mol Cell. 2012;47(5):746–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed AM, Boudreau JR, Yu FP, Liu J, Chin-Sang ID.. The Caenorhabditis elegans Eph receptor activates NCK and N-WASP, and inhibits Ena/VASP to regulate growth cone dynamics during axon guidance. PLoS Genet. 2012;8(2):e1002513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed AM, Chin-Sang ID.. The C. elegans nck-1 gene encodes two isoforms and is required for neuronal guidance. Dev Biol. 2011;354(1):55–66. [DOI] [PubMed] [Google Scholar]
- Procko C, , LuY, , Shaham S.. Sensory organ remodeling in Caenorhabditis elegans requires the zinc-finger protein ZTF-16. Genetics. 2012;190(4):1405–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih JD, Hunter CP.. SID-1 is a dsRNA-selective dsRNA-gated channel. RNA. 2011;17(6):1057–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanguy M, Véron L, Stempor P, Ahringer J, Sarkies P, Miska EA.. An alternative STAT signaling pathway acts in viral immunity in Caenorhabditis elegans. mBio. 2017;8(5):e00924-17. https://doi.org/10.1128/ mBio.00924-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo M, Tan L, Lim L, Manser E.. The tyrosine kinase ACK1 associates with clathrin-coated vesicles through a binding motif shared by arrestin and other adaptors. J Biol Chem. 2001;276(21):18392–18398. [DOI] [PubMed] [Google Scholar]
- Wang E, Hunter CP.. SID-1 functions in multiple roles to support parental RNAi in Caenorhabditis elegans. Genetics. 2017;207(2):547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whangbo JS, Weisman AS, Chae J, Hunter CP.. SID-1 domains important for dsRNA import in Caenorhabditis elegans. G3 (Bethesda). 2017;7(12):3887–3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston WM, Molodowitch C, Hunter CP.. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295(5564):2456–2459. [DOI] [PubMed] [Google Scholar]
- Winston WM, Sutherlin M, Wright AJ, Feinberg EH, Hunter CP.. Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proc Natl Acad Sci U S A. 2007;104(25):10565–10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worby CA, Simonson-Leff N, Clemens JC, Huddler D, Muda M, Dixon JE.. Drosophila Ack targets its substrate, the sorting nexin DSH3PX1, to a protein complex involved in axonal guidance. J Biol Chem. 2002;277(11):9422–9428. [DOI] [PubMed] [Google Scholar]
- Yang H, Vallandingham J, Shiu P, Li H, Hunter CP, Mak HY.. The DEAD box helicase RDE-12 promotes amplification of RNAi in cytoplasmic foci in C. elegans. Curr Biol. 2014;24(8):832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains not available at the CGC (cgc.umn.edu) are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplemental material is available at G3 online.