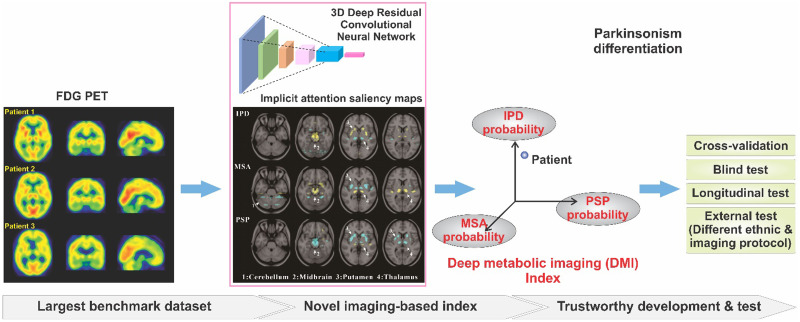
Visual Abstract
Keywords: Parkinson disease, atypical parkinsonian syndrome, differential diagnosis, deep learning, deep metabolic imaging indices
Abstract
The clinical presentations of early idiopathic Parkinson disease (IPD) substantially overlap with those of atypical parkinsonian syndromes such as multiple system atrophy (MSA) and progressive supranuclear palsy (PSP). This study aimed to develop metabolic imaging indices based on deep learning to support the differential diagnosis of these conditions. Methods: A benchmark Huashan parkinsonian PET imaging (HPPI, China) database including 1,275 parkinsonian patients and 863 nonparkinsonian subjects with 18F-FDG PET images was established to support artificial intelligence development. A 3-dimensional deep convolutional neural network was developed to extract deep metabolic imaging (DMI) indices and blindly evaluated in an independent cohort with longitudinal follow-up from the HPPI and an external German cohort of 90 parkinsonian patients with different imaging acquisition protocols. Results: The proposed DMI indices had less ambiguity space in the differential diagnosis. They achieved sensitivities of 98.1%, 88.5%, and 84.5%, and specificities of 90.0%, 99.2%, and 97.8%, respectively, for the diagnosis of IPD, MSA, and PSP in the blind-test cohort. In the German cohort, they resulted in sensitivities of 94.1%, 82.4%, and 82.1%, and specificities of 84.0%, 99.9%, and 94.1%, respectively. Using the PET scans independently achieved a performance comparable to the integration of demographic and clinical information into the DMI indices. Conclusion: The DMI indices developed on the HPPI database show the potential to provide an early and accurate differential diagnosis for parkinsonism and are robust when dealing with discrepancies between populations and imaging acquisitions.
Idiopathic Parkinson disease (IPD) is one of the most common neurodegenerative disorders. Although extensively studied, its accurate diagnosis remains clinically challenging, particularly in early stage patients, because their symptoms overlap largely with atypical parkinsonian syndromes such as multiple system atrophy (MSA) and progressive supranuclear palsy (PSP) (1). Approximately 20%–30% of patients with initial diagnoses of IPD were subsequently demonstrated to be either MSA or PSP at pathologic examination (1). The development of accurate indices for parkinsonism’s differential diagnosis is of importance and potential utility when determining therapeutic strategies.
18F-FDG PET detects a wide spectrum of neurobiologic abnormalities and has been reported of advantage in the differential diagnosis of parkinsonism in advance of structural damage to brain (2). Metabolic patterns of IPD, MSA, and PSP identified by principal component analysis (PCA) (3,4), which were used as features for a machine learning method of logistic regression, have been found as effective surrogates for the early and accurate differential diagnosis (5). However, the PCA decomposition takes the 3-dimensional (3D) image volume of a subject as a squeezed 1-dimensional vector without considering the high-level spatial interrelation during the pattern extraction.
The differences among parkinsonism are reflected in the complex interaction of interrelated brain regions. The differential indices may be obscured by complexity within the metabolic imaging signal. We hypothesized that deep learning may reveal characteristic imaging indices from complex metabolic alterations and provide accurate classifications (6). Therefore, a 3D deep residual convolutional neural network termed PD Diagnosis Network (PDD-Net) was built for the automatic identification of imaging-related indices to support parkinsonism’s differential diagnosis.
MATERIALS AND METHODS
Subjects and Study Protocol
Huashan Parkinsonian PET Imaging (HPPI) Database
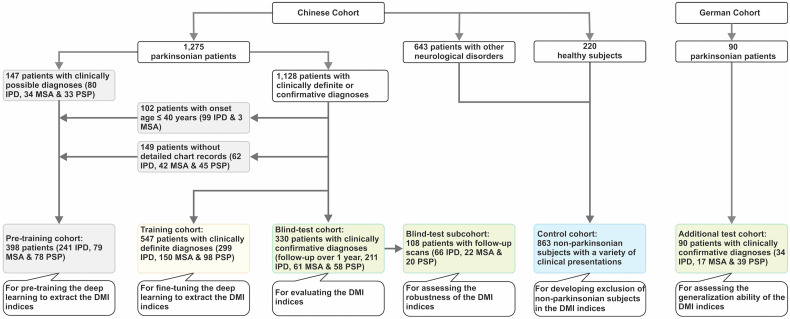
A unique HPPI database, the largest to our knowledge, has been established to benchmark the imaging-based artificial intelligence development for parkinsonism. This database includes 3 cohorts with a total of 1,275 parkinsonian patients (subset of PD Database and Samples Bank of Huashan Hospital) (Fig. 1; Supplemental Tables 1 and 2 [supplemental materials are available at http://jnm.snmjournals.org]) (7–11). Among the cohorts, 85.7% of patients underwent dopaminergic imaging at the same time as 18F-FDG to assist the diagnosis, and the remaining patients were followed for 3–8 y (5.6 ± 2.1 y) to determine the diagnosis. A control cohort of 643 patients with various neurologic disorders and 220 healthy subjects was also enrolled (Fig.1; Supplemental Tables 3 and 4; Supplemental Fig. 1).
FIGURE 1.
Study profile, showing a Chinese cohort (Huashan parkinsonian PET imaging dataset) and a German cohort. Clinically definite diagnoses = diagnoses by clinical experts after return visit but without a formal clinical follow-up; clinically confirmative diagnoses = diagnoses resulting from at least 1 formal clinical follow-up over 1 y after PET imaging.
The HPPI database includes the following cohorts: pretraining (398 subjects with possible diagnoses), training (547 subjects with definite diagnoses), and blind test (330 subjects with confirmative diagnoses with follow-up) (Fig.1; Table 1). These patients were routinely assessed by movement disorder specialists in Huashan Hospital before PET examination between June 2011 and April 2019. Routine MRI examinations were performed before PET scans and those patients with structural brain abnormalities were excluded. After PET examination, patients had at least one return visit and the movement disorders specialists made a clinical diagnosis according to the latest clinical criteria (9–11).
TABLE 1.
Demographic and Clinical Data
| Clinical parameters | Huashan parkinsonian PET imaging dataset (Chinese cohort) | German cohort | ||||||
|---|---|---|---|---|---|---|---|---|
| Pretraining cohort | Training cohort | Blind-test cohort | ||||||
| Overall | Short symptom duration (≤2 y) | Long symptom duration (>2 y) | Overall | Baseline | Follow-up | |||
| IPD* | ||||||||
| Patient (n) | 241 | 299 | 136 | 163 | 211 | 66 | 66 | 34 |
| Sex (male/female) | 154/87 | 166/133 | 73/63 | 93/70 | 130/81 | 43/23 | 43/23 | 21/13 (34/34) |
| Age at PET (y) | 50.0 ± 15.5 | 60.2 ± 8.5 | 59.1 ± 9.0 | 61.0 ± 8.0 | 60.0 ± 7.6 | 60.0 ± 7.9 | 62.1 ± 7.9 | 72.9 ± 9.5 (34/34) |
| Symptom duration at PET (mo) | – | 45.3 ± 46.0 | 13.0 ± 5.9 | 72.3 ± 47.4 | 39.0 ± 41.3 | 26.0 ± 24.1 | 53.4 ± 24.2 | 44.5 ± 32.9 (18/34) |
| Hoehn and Yahr stage† | – | 2.2 ± 1.0 | 1.7 ± 0.6 | 2.7 ± 1.0 | 1.9 ± 0.9 | 1.6 ± 0.7 | 1.9 ± 0.6 | 1.6 ± 0.8 (22/34) |
| UPDRS III | – | 27.0 ± 14.3 | 18.9 ± 8.9 | 33.8 ± 14.5 | 22.8 ± 12.1 | 19.6 ± 9.1 | 24.2 ± 10.1 | 12.0 ± 3.6 (3/34) |
| Clinical follow-up (mo) | – | – | – | – | 46.8 ± 30.4 | – | 64.5 ± 25.3 | 19.1 ± 21.8 (14/34) |
| MSA* | ||||||||
| Patient (n) (MSA-C/MSA-P) | 79 | 150 (57/93) | 90 (39/51) | 60 (18/42) | 61 (21/40) | 22 (8/14) | 22 (8/14) | 17 (8/8/1) |
| Sex (male/female) | 42/37 | 78/72 | 47/43 | 31/29 | 32/29 | 14/8 | 14/8 | 10/7 (17/17) |
| Age at PET (y) | 57.5 ± 10.6 | 57.8 ± 8.0 | 56.5 ± 8.1 | 59.6 ± 7.4 | 58.5 ± 6.3 | 58.3 ± 7.4 | 60.3 ± 7.3 | 61.3 ± 8.3 (17/17) |
| Symptom duration at PET (mo) | – | 24.3 ± 17.1 | 13.9 ± 6.0 | 39.9 ± 16.5 | 27.0 ± 20.1 | 22.1 ± 11.8 | 45.6 ± 12.5 | 30.0 ± 22.2 (17/17) |
| Hoehn and Yahr stage† | – | 3.1 ± 0.8 | 3.0 ± 0.8 | 3.5 ± 0.7 | 2.9 ± 0.8 | 2.6 ± 0.6 | 3.4 ± 0.8 | 2.4 ± 1.1 (15/17) |
| UPDRS III | – | 30.6 ± 14.5 | 25.9 ± 12.4 | 37.6 ± 14.7 | 29.3 ± 14.4 | 23.5 ± 8.2 | 36.4 ± 11.1 | 34.6 ± 12.8 (11/17) |
| Clinical follow-up (mo) | – | – | – | – | 30.7 ± 18.2 | – | 41.7 ± 16.4 | 22.6 ± 22.4 (17/17) |
| PSP* | ||||||||
| Patient (n) | 78 | 98 | 34 | 64 | 58 | 20 | 20 | 39 |
| Sex (male/female) | 45/33 | 60/38 | 23/11 | 37/27 | 39/19 | 17/3 | 17/3 | 21/18 (39/39) |
| Age at PET (y) | 64.6 ± 8.6 | 67.2 ± 8.0 | 65.0 ± 9.3 | 68.5 ± 6.9 | 65.1 ± 6.6 | 64.8 ± 7.5 | 67.0 ± 7.2 | 70.0 ± 7.1 (39/39) |
| Symptom duration at PET (mo) | – | 35.0 ± 20.7 | 15.3 ± 5.4 | 45.5 ± 18.0 | 34.1 ± 22.7 | 32.4 ± 22.0 | 58.8 ± 22.8 | 22.4 ± 15.7 (37/39) |
| Hoehn and Yahr stage† | – | 3.2 ± 0.8 | 2.9 ± 0.6 | 3.4 ± 0.8 | 3.0 ± 0.8 | 2.7 ± 1.0 | 3.6 ± 0.8 | 2.6 ± 1.1 (37/39) |
| UPDRS III | – | 30.1 ± 13.5 | 28.0 ± 11.0 | 31.2 ± 14.6 | 26.8 ± 11.0 | 23.0 ± 10.4 | 34.6 ± 15.9 | 37.0 ± 15.9 (20/39) |
| Clinical follow-up (mo) | – | – | – | – | 25.1 ± 15.7 | – | 37.5 ± 12.9 | 22.2 ± 13.8 (17/39) |
Diagnosis information: Supplemental Table 1.
Detailed Hoehn and Yahr stage information: Supplemental Table 2.
Data are shown as mean ± SD. In German cohort, associated numbers of subjects with these items are provided together with statistics information (subject number with certain item/total subject number).
UPDRS = Unified Parkinson’s Disease Rating Scale; MSA-C/MSA-P = MSA-cerebellar/MSA-parkinsonian.
After a low-dose CT for attenuation correction, the emission data were acquired at 60 min (lasting 10 min) after injection of approximately 185 MBq of 18F-FDG using the Biograph 64 HD PET/CT (Siemens). After corrections for attenuation, scatter, dead time, and random coincidences, PET images were reconstructed using the ordered-subset expectation maximization method.
German Parkinsonian Cohort
A German cohort with 34 IPD, 17 MSA, and 39 PSP patients from the University Hospital of Munich was included for external validation. These patients were scanned on 3 different PET/CT systems (ECAT Exact HR+ [Siemens], Discovery 690 [GE Healthcare], and Biograph 64) according to the European Association of Nuclear Medicine protocol (12) using a slow bolus injection of approximately 150 MBq of 18F-FDG (Supplemental Table 5). The uptake differences between cohorts are presented in Supplemental Figure 2.
The institutional review boards (IRB or equivalent from Huashan Hospital and University of Munich) approved this study, and all subjects signed a written informed consent form.
Image Preprocessing
PET images were spatially normalized into Montreal Neurologic Institute brain space and smoothed by a 3D gaussian filter of 10 mm in full width at half maximum by SPM5 software (Institute of Neurology). Before inputting the PET image into the deep neural network, z score normalization was applied to convert PET image values into a certain range for facilitating the network training. In addition, the performances of z score normalization and global mean normalization were also compared (Supplemental Table 6).
PDD-Net and DMI Indices
The deep learning method contains 2 PDD-Nets (Supplemental Fig. 3). The PDD-Net-1 sought to exclude patients without parkinsonism. The PDD-Net-2 performed computation of DMI indices and classification of IPD, MSA, or PSP. Both PDD-Nets were based on a 3D residual convolutional neural network. The PDD-Net 2 was trained preliminarily in the pretraining cohort and then fine-tuned in the training cohort. The performance of the DMI indices was evaluated with cross-validation (6-fold) in the training cohort and then an independent test in the blind-test cohort and the external German cohort.
At the end of the PDD-Net computation, the extracted features were mapped to 3 classification probabilities of IPD, MSA, and PSP correspondingly, which were proposed as the DMI indices. The highest probability among the DMI indices was considered for the prediction of IPD, MSA, or PSP. An additional option of confidence inspection was provided to warn the predictions without sufficiently high probability. A confidence threshold can be customized. By default, a set of confidence thresholds were derived in the cross-validation stage based on the generalized Youden’s index. Predictions lying below these thresholds were flagged as uncertain cases (Supplemental Table 7). We generated saliency maps using the full-gradient method (13) to assist the interpretation of the DMI indices. The saliency maps assign importance scores to both the input features and the individual neurons in a network, reflecting the contribution of groups of pixels to the DMI probabilities.
Statistical Analysis
The CIs were calculated with DeLong’s method. The optimal cutoff points of the receiver-operating-characteristic curves were estimated using the generalized Youden’s index. For continuous variables, the Wilcoxon test was used to compare 2 paired groups and the Kolmogorov–Smirnov test was used to compare 2 unpaired groups; for categoric variables, the χ2 test was used. Four standard metrics, that is, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV), were used to illustrate the diagnostic performance of the DMI indices.
RESULTS
Performance of the DMI Indices in Cross-Validation
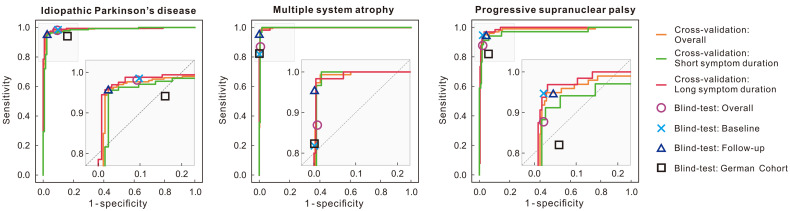
The performance of the DMI indices in the cross-validation is illustrated in Figure 2. The area under the curves was 0.986, 0.997, and 0.982 for IPD, MSA and PSP, respectively. The sensitivity, specificity, PPV, and NPV are summarized in Table 2, and all values were above 90% except for sensitivity and PPV for PSP with short symptom durations. Compared with those with short symptom durations, the specificity for those with long symptom durations slightly increased for IPD and MSA, whereas they remained the same for PSP.
FIGURE 2.
Accuracy of DMI indices in development phase in training cohort and blind-test phase on both Chinese and German test cohorts. Results in cross-validation were plotted using receiver-operating-characteristic curves. Results in Chinese blind-test cohort were illustrated as single points, where overall represents results of all tested 330 patients. One hundred eight patients in blind test underwent follow-up scanning, and performance of these at baseline and follow-up was plotted. Blind-test results in German cohort (90 patients) are also included and denoted with black rectangle for easy comparison.
TABLE 2.
Accuracy of DMI Indices in Cross-Validation on Training Cohort
| Diagnosis | Metrics | Overall | Short symptom duration (≤2 y) | Long symptom duration (>2 y) |
|---|---|---|---|---|
| IPD | AUC | 0.986 (0.977–0.996) | 0.981 (0.965–0.997) | 0.991 (0.981–1.000) |
| Sensitivity | 95.7% (92.7%–97.7%) | 94.9% (89.7%–97.9%) | 95.7% (91.4%–98.3%) | |
| Specificity | 97.6% (94.8%–99.1%) | 97.6% (93.1%–99.5%) | 98.4% (94.3%–99.8%) | |
| PPV | 97.9% (95.6%–98.9%) | 97.7% (93.5%–99.1%) | 98.7% (95.5%–99.5%) | |
| NPV | 94.9% (91.5%–98.1%) | 94.5% (89.1%–98.8%) | 94.6% (89.2%–99.3%) | |
| MSA | AUC | 0.997 (0.994–1.000) | 0.996 (0.988–1.000) | 0.998 (0.995–1.000) |
| Sensitivity | 97.3% (93.3%–99.3%) | 100% (96.0%–100%) | 98.3% (91.1%–100%) | |
| Specificity | 99.5% (98.2%–99.9%) | 98.2% (94.9%–99.6%) | 99.6% (97.6%–100%) | |
| PPV | 98.6% (95.3%–99.6%) | 96.8% (91.0%–100%) | 98.3% (91.3%–100%) | |
| NPV | 99.0% (97.4%–99.9%) | 100% (97.8%–100%) | 99.6% (97.5%–100%) | |
| PSP | AUC | 0.982 (0.965–0.998) | 0.968 (0.925–1.000) | 0.990 (0.980–1.000) |
| Sensitivity | 91.8% (84.5%–96.4%) | 88.2% (72.5%–96.7%) | 93.8% (84.8%–98.3%) | |
| Specificity | 98.2% (96.5%–99.2%) | 98.2% (95.5%–99.5%) | 98.2% (95.5%–99.5%) | |
| PPV | 91.8% (85.0%–96.4%) | 88.2% (74.3%–96.7%) | 93.7% (85.2%–98.3%) | |
| NPV | 98.2% (96.4%–99.2%) | 98.2% (95.1%–99.5%) | 98.2% (95.3%–99.5%) |
AUC = area under the curve.
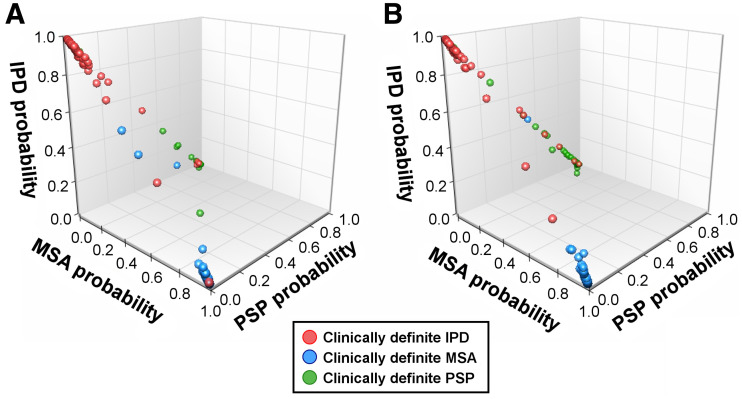
The probabilities of IPD, MSA, and PSP according to the DMI indices for individual subjects are plotted in 3D coordination space in Figure 3. These probabilities tended to distribute aggregately to their expected centers: IPD for [1,0,0], MSA for [0,1,0], and PSP for [0,0,1]. If the probability for a category was high, the probabilities for the other 2 categories were much smaller. The aggregation distance, which is the mean distance of the probabilities to the corresponding expected centers, illustrates the determinability of the DMI indices. The probabilities of those with long symptom durations (aggregative distance = 0.103) were more aggregated (P = 0.020) compared with the subjects with short symptom durations (aggregative distance = 0.114). Overall, the probabilities among the DMI indices had less ambiguity space for differential diagnosis.
FIGURE 3.
3D plot of probabilities of IPD, MSA, and PSP of DMI indices in training cohort. (A) Patients with short symptom duration (≤2 y). (B) Patients with long symptom duration (>2 y).
The saliency maps are showed in Supplemental Figure 4–6 (13). Regions with relatively higher contribution to the DMI indices were putamen and midbrain for IPD, MSA, and PSP as well as cerebellum for MSA.
Performance of the DMI Indices in the Blind Test
Table 3 illustrates the predictive accuracy of the DMI indices in the blind-test cohort. The image-based classification resulted in 98.1% sensitivity, 90.0% specificity, 94.5% PPV, and 96.4% NPV for PD and also accurate for MSA (88.5% sensitivity, 99.2% specificity, 96.4% PPV, and 97.4% NPV) and PSP (84.5% sensitivity, 97.8% specificity, 89.1% PPV, and 97.0% NPV). For the 108 patients in the blind-test cohort with follow-up PET scans, the DMI indices had slightly better performance comparing follow-up to baseline (P = 0.017).
TABLE 3.
Accuracy of DMI Indices on Blind-Test Cohort from Huashan Parkinsonian PET Imaging Dataset (Chinese Cohort) and German Cohort
| Diagnosis | Metrics | Huashan parkinsonian PET imaging dataset (Chinese cohort) | German cohort | ||
|---|---|---|---|---|---|
| Overall | Baseline | Follow-up | |||
| IPD | Sensitivity | 98.1% | 98.5% | 95.5% | 94.1% |
| Specificity | 90.0% | 88.1% | 97.6% | 84.0% | |
| PPV | 94.5% | 92.9% | 98.4% | 78.0% | |
| NPV | 96.4% | 97.4% | 93.2% | 95.9% | |
| MSA | Sensitivity | 88.5% | 81.8% | 95.4% | 82.4% |
| Specificity | 99.2% | 99.9% | 98.8% | 99.9% | |
| PPV | 96.4% | 99.9% | 95.5% | 99.9% | |
| NPV | 97.4% | 95.6% | 98.8% | 96.1% | |
| PSP | Sensitivity | 84.5% | 90.0% | 95.0% | 82.1% |
| Specificity | 97.8% | 97.7% | 96.6% | 94.1% | |
| PPV | 89.1% | 90.0% | 86.4% | 91.4% | |
| NPV | 97.0% | 97.7% | 98.8% | 87.3% | |
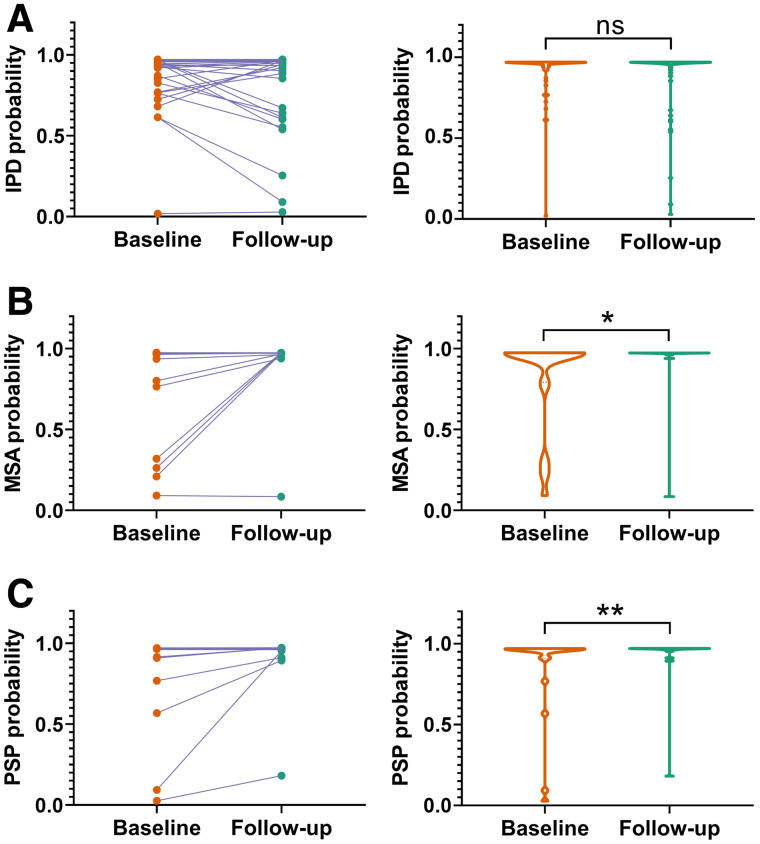
The probabilities among the DMI indices for subjects with follow-up imaging in the blind-test cohort are plotted in Figure 4. The probabilities of MSA and PSP increased at follow-up imaging (MSA: P = 0.028, PSP: P = 0.002). The probabilities of IPD between at follow-up and baseline imaging were comparable (P = 0.894), but the median and most of the IPD probabilities (38/66) increased. Nine cases presented relative significant lower probabilities of IPD at follow-up (over 0.1) compared with the baseline.
FIGURE 4.
Comparison of probabilities of IPD (A), MSA (B), and PSP (C) of DMI indices on the 108 patients in blind-test cohort with repeated PET scans. (Left column) Comparison of the probability of DMI indices extracted from baseline and follow-up PET for individuals. (Right column) Violin plots demonstrate statistical distribution of probabilities of DMI indices. *P ≤ 0.05. **P ≤ 0.01. ns = no significance.
Besides, differential diagnosis performance of using the DMI indices only and using the combination of the DMI indices with demographic and clinical features were compared, and no difference was found (P = 0.999) (Supplemental Table 8) (14). Besides, DMI indices made predictions inconsistent with the clinical diagnosis in 6 cases obvious probability decrease during follow-up (Supplemental Table 9).
Test on the External German Cohort
The DMI indices achieved 94.1% sensitivity, 84.0% specificity, 78.0% PPV, and 95.9% NPV for the diagnosis of the IPD on the German cohort (Table 3). The diagnoses were also accurate for MSA (82.4% sensitivity, 99.9% specificity, 99.9% PPV, and 96.1% NPV) and PSP (82.1% sensitivity, 94.1% specificity, 91.4% PPV, and 87.3% NPV). Although the performance metrics were slightly lower than those for the Chinese cohort, no significant difference has been observed in the performance of the diagnosis of IPD (P = 0.14), MSA (P = 0.25) and PSP (P = 0.50).
DISCUSSION
An effective imaging-based tool may contribute to earlier and more precise diagnosis in parkinsonian conditions and may help with the development and monitoring of individualized disease-modifying treatments (15,16). This study confirms that deep learning can identify accurate imaging-based indices from 18F-FDG PET.
Similar to pattern expression scores of PCA analysis (5), the DMI indices herein identified 3 probability scores from 18F-FDG PET for each individual and a prediction was generated by comparing these 3 probabilities. The conventional pattern related scores are derived from linear weightings of imaging intensities. In contrast, the DMI indices can reveal hyper-level interrelations such as textures, which may better describe the complex heterogeneous pathogenesis of parkinsonian disorders. Compared with previously reported studies (5), in our study the extensive test in relatively large cohorts found that the DMI indices can achieve competitive or possibly better performance in the differential diagnosis of parkinsonism.
The probabilities among the DMI indices have low ambiguity and a dominant maximal probability is definable for resulting in a robust diagnosis prediction. Nevertheless, we also support confidence inspection to differentiate predictions with different confidence levels. The confidence thresholds can be customized (Supplemental Table 7). For a default setting according to the optimization of generalized Youden’s index, the confidence threshold for MSA was higher than for IPD or PSP. In this study, the MSA patients were mixed with MSA-parkinsonian (MSA-P) and MSA-cerebellar (MSA-C) types and had greater heterogeneity in metabolic pathologic phenotype. Therefore, it could be posited that a higher confidence threshold is required to obtain a robust prediction.
The DMI indices can be combined with demographic and clinical information as well as other indices, such as impairment of olfactory function (for IPD vs. MSA) or skin biopsy positivity for phospho-α-synuclein aggregates (for IPD and MSA vs PSP) (17), to comprehensively generate diagnostic classifications. In our study, using the PET scans independently achieved a performance comparable to the integration of demographic and clinical information into the DMI indices, indicating that the most discriminative information for the parkinsonism diagnosis was included in the PET scan modality and could be extracted by the proposed method into the DMI indices. In addition, the 2-stage design (Supplemental Fig. 3) (13,18,19) of our work allows the DMI indices to reduce the risk of erroneous predictions through exclusion of nonparkinsonian subjects in the control stage, which aims at further improving the robustness of diagnostic classifications.
In general, the DMI indices developed from the Chinese HPPI database achieved comparable performance in a German cohort. However, there were substantial differences between the 2 cohorts: in contrast to the Chinese cohort, the German cohort used different scanners. The imaging protocols (i.e., acquisition time, reconstruction method, tracer dose) and patient preparation (i.e., eye patch and noise-cancelling differences) (Supplemental Table 5) varied. Significantly different metabolic uptake was observed in the cerebellum, midbrain, and caudate between these 2 cohorts (Supplemental Fig. 2), for which population-based differences (3,20) may exist. The domain difference between data can present an obstacle to the wider clinical translation of conventional methods. A prerequisite for spatial covariance analysis in the established population-based patterns for IPD, MSA, and PSP is to bridge the difference between various populations (5). In contrast to pattern analysis, the hierarchical feature representation of deep learning is more flexible and affords migration of domain differences during the learning phase (21). Similar to previous studies (22), our test confirmed that deep learning can be robust to the discrepancies inherent in molecular imaging acquisitions. This finding suggests the DMI marker extracted using deep learning in this study may be more generalizable and better suited for clinical translation.
Recently, concerns have been raised regarding the reproducibility or stability of deep learning methods: methods optimized in one cohort may have limited performance in other cohorts or in other applications (23). We subjected our DMI indices to a blind test as a means of independent in-depth validation (24). The performance of the DMI indices under conditions of a blind test was consistent with the cross-validation test. Thus, the DMI indices are reproducible. Deep learning is impeded by the black-box nature of the derived model, which precludes the drawing of any links to the underlying pathophysiology. To address this concern, we used saliency maps to understand the decision mechanism behind the neural networks. The saliency maps indicated that the DMI indices derived probabilities largely based on parkinsonism-related brain regions, which are consistent with the critical regions of IPD-, MSA-, and PSP-related covariance pattern (5,25).
Dopaminergic imaging is critical for diagnosing parkinsonian disorders, although it has not been confirmed to be suitable for the reliable differential diagnosis. Most patients with parkinsonism in our study underwent contemporary dopaminergic imaging as 18F-FDG. Therefore, this study can be regarded as performed based on dopaminergic imaging. Whether 18F-FDG imaging and deep learning can be used to diagnose parkinsonian disorders with blinded dopaminergic imaging results is an interesting future direction to explore.
One limitation of this study is that we did not use MRI for partial-volume correction and spatial normalization. Although MRI is generally included in the neurologic work-up of these patients, many of them were scanned at external centers with a variety of protocols and the 3D images were not always retrievable. We conceded that the cortical thickness derived from MR images might also assist the differentiation of parkinsonism (26). The integration of these morphometries in any future study may further enhance the imaging-based indices. In addition, although performance on the training cohort, blind-test cohort, and German cohort—which have different data distributions (IPD:MSA:PSP)—has indicated that the DMI indices have a certain level of ability to handle the distribution-different problems, different distributions may still be a factor influencing performance on another future cohort. It is worthy to conduct multicenter studies to further validate our method. Meanwhile, we evaluated only one possible multimodality fusion method in this work. In the future, to further improve the diagnosis performance, other fusion methods such as gating- and attention-mechanism–based late fusion will be evaluated.
CONCLUSION
We developed a 3D deep residual convolutional neural network to extract DMI indices for the automated differential diagnosis of parkinsonism. The indices were evaluated with the cross-validation experiment and blind tests on both Chinese and German cohorts, demonstrating that the proposed method was both robust and accurate, which may complement diagnoses made by expert clinicians.
DISCLOSURE
This work was supported by the National Natural Science Foundation of China (81771483, 81671239, 81361120393, 81401135, 81971641, 81902282, 91949118, 81771372), the Ministry of Science and Technology of China (2016YFC1306504), Shanghai Municipal Science and Technology Major Project (no. 2017SHZDZX01, 2018SHZDZX03) and ZJ Lab, Youth Medical Talents–Medical Imaging Practitioner Program by Shanghai Municipal Health Commission and Shanghai Medical and Health Development Foundation (SHWRS(2020)_087), Shanghai Sailing Program by Shanghai Science and Technology Committee (18YF1403100), the Swiss National Science Foundation (188350), Jacques & Gloria Gossweiler Foundation and Siemens Healthineers. Wolfgang H. Oertel is Hertie Senior Research Professor, supported by the Charitable Hertie Foundation, Frankfurt/Main, Germany. Axel Rominger and Kuangyu Shi received research support from Novartis and Siemens Healthineers. No other potential conflict of interest relevant to this article was reported.
KEY POINTS
QUESTION: Can deep learning effectively extract indices from brain glucose metabolic imaging (18F-FDG PET) to improve the differential diagnosis of Parkinson disease and atypical parkinsonian syndromes?
PERTINENT FINDINGS: The developed DMI indices prediction using deep learning provides an early and accurate method for differential diagnosis that may complement diagnoses made by expert clinicians. The reliable artificial intelligence development was achieved by training on large-scale benchmark data on 18F-FDG PET and extensive testing on longitudinal data and independent external data with different ethnicity or examination protocols.
IMPLICATIONS FOR PATIENT CARE: These developed DMI indices may assist early differential diagnosis of parkinsonism and the development of disease-modifying treatment strategies.
REFERENCES
- 1. Hughes AJ, Daniel SE, Ben‐Shlomo Y, Lees AJ. The accuracy of diagnosis of parkinsonian syndromes in a specialist movement disorder service. Brain. 2002;125:861–870. [DOI] [PubMed] [Google Scholar]
- 2. Stoessl AJ, Lehericy S, Strafella AP. Imaging insights into basal ganglia function, Parkinson’s disease, and dystonia. Lancet. 2014;384:532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ge J, Wu J, Peng S, et al. Reproducible network and regional topographies of abnormal glucose metabolism associated with progressive supranuclear palsy: multivariate and univariate analyses in American and Chinese patient cohorts. Hum Brain Mapp. 2018;39:2842–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eckert T, Tang C, Ma Y, et al. Abnormal metabolic networks in atypical parkinsonism. Mov Disord. 2008;23:727–733. [DOI] [PubMed] [Google Scholar]
- 5. Tang CC, Poston KL, Eckert T, et al. Differential diagnosis of parkinsonism: a metabolic imaging study using pattern analysis. Lancet Neurol. 2010;9:149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Choi BW, Kang S, Kim HW, Kwon OD, Vu HD, Youn SW. Faster region-based convolutional neural network in the classification of different parkinsonism patterns of the striatum on maximum intensity projection images of [18F] FP-CIT positron emission tomography. Diagnostics (Basel). 2021;11:1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Litvan I, Agid Y, Calne D, et al. Clinical research criteria for the diagnosis of progressive supranuclear palsy (Steele-Richardson-Olszewski syndrome) report of the NINDS-SPSP international workshop. Neurology. 1996;47:1–9. [DOI] [PubMed] [Google Scholar]
- 8. Hughes AJ, Daniel SE, Kilford L, Lees AJ. Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: a clinico-pathological study of 100 cases. J Neurol Neurosurg Psychiatry. 1992;55:181–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gilman S, Wenning GK, Low PA, et al. Second consensus statement on the diagnosis of multiple system atrophy. Neurology. 2008;71:670–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Höglinger GU, Respondek G, Stamelou M, et al. Clinical diagnosis of progressive supranuclear palsy: the movement disorder society criteria. Mov Disord. 2017;32:853–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Postuma RB, Berg D, Stern M, et al. MDS clinical diagnostic criteria for Parkinson’s disease. Mov Disord. 2015;30:1591–1601. [DOI] [PubMed] [Google Scholar]
- 12. Varrone A, Asenbaum S, Vander Borght T, et al. EANM procedure guidelines for PET brain imaging using [18F]FDG, version 2. Eur J Nucl Med Mol Imaging. 2009;36:2103–2110. [DOI] [PubMed] [Google Scholar]
- 13. Srinivas S, Fleuret F. Full-gradient representation for neural network visualization. Cornell University, arxiv website. https://arxiv.org/pdf/1905.00780v4.pdf. Last revised December 3, 2019. Accessed August 17, 2022.
- 14. Chen T, Guestrin C. Xgboost: A scalable tree boosting system. Cornell University, arxiv website. https://arxiv.org/pdf/1603.02754v3.pdf. Last revised June 10, 2016. Accessed August 17, 2022.
- 15. Strafella AP, Bohnen NI, Perlmutter JS, et al. Molecular imaging to track Parkinson’s disease and atypical parkinsonisms: new imaging frontiers. Mov Disord. 2017;32:181–192. [DOI] [PubMed] [Google Scholar]
- 16. Meles SK, Teune LK, de Jong BM, Dierckx RA, Leenders KL. Metabolic imaging in Parkinson disease. J Nucl Med. 2017;58:23–28. [DOI] [PubMed] [Google Scholar]
- 17. Devine MJ, Gwinn K, Singleton A, Hardy J. Parkinson’s disease and α-synuclein expression. Mov Disord. 2011;26:2160–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. Cornell University, arxiv website. https://arxiv.org/pdf/1512.03385v1.pdf. Submitted December 10, 2015. Accessed August 17, 2022.
- 19. Skrede O-J, De Raedt S, Kleppe A, et al. Deep learning for prediction of colorectal cancer outcome: a discovery and validation study. Lancet. 2020;395:350–360. [DOI] [PubMed] [Google Scholar]
- 20. Shi L, Liang P, Luo Y, et al. Using large-scale statistical Chinese brain template (Chinese2020) in popular neuroimage analysis toolkits. Front Hum Neurosci. 2017;11:414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ghafoorian M, Mehrtash A, Kapur T, et al.Transfer learning for domain adaptation in MRI: application in brain lesion segmentation. Cornell University, arxiv website. https://arxiv.org/pdf/1702.07841.pdf. Submitted on February 25, 2017. Accessed on August 17, 2022.
- 22. Wenzel M, Milletari F, Krüger J, et al. Automatic classification of dopamine transporter SPECT: deep convolutional neural networks can be trained to be robust with respect to variable image characteristics. Eur J Nucl Med Mol Imaging. 2019;46:2800–2811. [DOI] [PubMed] [Google Scholar]
- 23. Maier-Hein L, Eisenmann M, Reinke A, et al. Why rankings of biomedical image analysis competitions should be interpreted with care. Nat Commun. 2018;9:5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Segler MH, Preuss M, Waller MP. Planning chemical syntheses with deep neural networks and symbolic AI. Nature. 2018;555:604–610. [DOI] [PubMed] [Google Scholar]
- 25. Matthews DC, Lerman H, Lukic A, et al. FDG PET Parkinson’s disease-related pattern as a biomarker for clinical trials in early stage disease. Neuroimage Clin. 2018;20:572–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Möller L, Kassubek J, Südmeyer M, et al. Manual MRI morphometry in Parkinsonian syndromes. Mov Disord. 2017;32:778–782. [DOI] [PubMed] [Google Scholar]