Abstract
Two fundamental elements of pre-implantation embryogenesis are cells’ intrinsic self-organization program and their developmental plasticity, which allows embryos to compensate for alterations in cell position and number; yet, these elements are still poorly understood. To be able to decipher these features, we established culture conditions that enable the two fates of blastocysts’ extraembryonic lineages—the primitive endoderm and the trophectoderm—to coexist. This plasticity emerges following the mechanisms of the first lineage segregation in the mouse embryo, and it manifests as an extended potential for extraembryonic chimerism during the pre-implantation embryogenesis. Moreover, this shared state enables robust assembly into higher-order blastocyst-like structures, thus combining both the cell fate plasticity and self-organization features of the early extraembryonic lineages.
Induction of an intermediate extraembryonic cell state reveals the cell fate plasticity of the early developmental program.
INTRODUCTION
The dividing zygote gives rise to a self-organizing entity of early embryonic and extraembryonic tissues that form the blastocyst. In mice, by embryonic day 4.5 (E4.5), the blastocyst consists of an epithelial layer of trophectoderm (TE) cells and an inner cell mass (ICM) positioned on one side of a fluid-filled cavity. The ICM contains pluripotent epiblast cells, which later give rise to all tissues of the fetus, and a surface layer of primitive endoderm (PE) cells, which will form the yolk sac.
Structurally, the TE defines the hollow-shaped architecture of the blastocyst and is subdivided into two compartments: the mural TE, which surrounds the blastocoel cavity, and the polar TE, in direct contact with the ICM. The mural TE mediates the process of implantation by differentiating into invasive trophoblast cells that penetrate the uterine wall. In contrast, the polar TE maintains its stemness and forms the extraembryonic ectoderm (ExE) of the nascent egg cylinder. The ExE is a pool of multipotent trophoblast stem cells (TSC) that later gives rise to the placental tissues supporting the fetal development to term (1).
Although the first TE cells are specified during the transition from the 8-cell to the 16-cell stage, their fate is not restricted exclusively to the trophoblast lineage until the 32-cell to the 64-cell stage. These cells can divide asymmetrically and give rise to ICM cells, thus exhibiting developmental plasticity within a limited time frame before implantation (2, 3).
Upon TE removal, the ICM cells of the 32-cell stage blastocyst can also activate the TE program, which depends on the cell responsiveness to modulation of Hippo signaling activity (4). This suggests that a poorly understood fail-safe mechanism enables the ICM cells to compensate TE cell deficiency. To decipher the developmental plasticity of the early lineages, here, we established culture conditions that enable the two fates of blastocysts’ extraembryonic tissues—the PE and the TE—to partially coexist. We found that the PE cells of the ICM, as well as extraembryonic endoderm (XEN) and XEN-like cells, acquire TE properties following the principles of the first lineage segregation. In addition, mimicking the tissue-scale organization of the TE, these cells efficiently cavitate and incorporate pluripotent cells to assemble hollowed-shaped, blastocyst-like embryoids.
RESULTS
Derivation of cells with TE-like properties from mouse blastocysts
Several stem cell lines have been established from the embryonic and the extraembryonic lineages of blastocyst and egg cylinder stage embryos, which can as an accessible model system of developmental plasticity. For instance, embryonic stem cells (ESC) and epiblast stem cells (EpiSC) capture features of the pre-implantation (naïve) and the postimplantation (primed) epiblast, respectively (5). Similarly, stem cell lines have also been derived from the trophoblast lineage of pre-implantation (E3.5) and postimplantation (E6.5) embryos. However, in contrast to ESC and EpiSC, which represent distinct pluripotent states and require different signaling cues for their self-renewal, both E3.5 and E6.5 TSC are established using an identical Fgf4/Heparin-supplemented medium (6). As a result of the culture environment, the TSC express postimplantation markers such as Sox2 and Elf5 regardless of the embryonic stage of origin and, overall, resemble the multipotent stem cell pool of the ExE (7, 8). Thus, the conventional TSC capture and represent an ExE-like state but not a pre-implantation TE-like state.
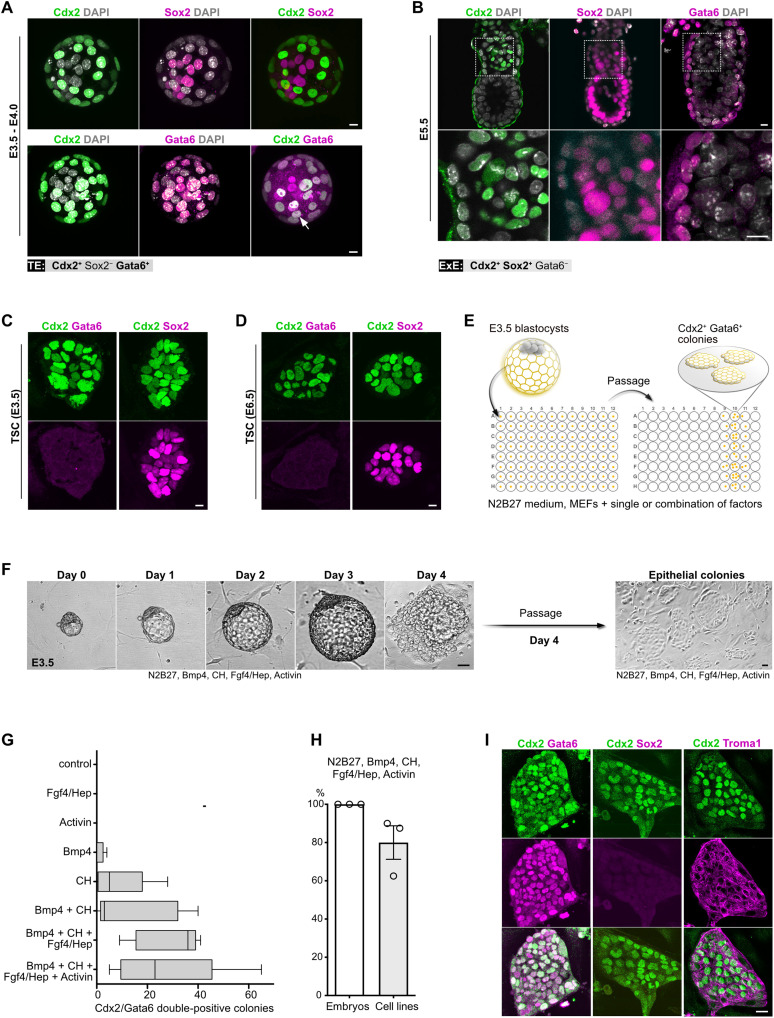
To examine whether cells with TE-like properties can be derived from early embryos, first, we defined a set of markers to discriminate between the pre-implantation (TE) and the postimplantation (ExE) trophoblast. Blastocyst-stage embryos showed mutually exclusive expression of Cdx2 in the TE and Sox2 in the epiblast. In addition to Cdx2, the TE was also positive for Gata6 (Fig. 1A), in agreement with previous reports (9–11). After implantation, Sox2 was detectable in both epiblast and ExE, whereas Gata6 expression was maintained only in the visceral endoderm (Fig. 1B). Thus, we defined the TE as Cdx2+/Sox2−/Gata6+ and the ExE as Cdx2+/Sox2+/Gata6− cell populations.
Fig. 1. Derivation of TE-like cells from mouse blastocysts.
(A) Blastocyst-stage embryos stained for Cdx2, Sox2, and Gata6. Arrow indicates Gata6 and Cdx2 colocalization. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). (B) E5.5 egg cylinders stained for Cdx2, Sox2, and Gata6. Nuclei were counterstained with DAPI. (C) TSC (E3.5) stained for Cdx2, Sox2, and Gata6. (D) TSC (E6.5) stained for Cdx2, Sox2, and Gata6. (E) Schematic representation of the experimental workflow used for TE-like cell derivation. (F) Snapshot images of mouse blastocyst cultured in iXTE medium and the subsequent establishment of epithelial colonies following the dissociation of the blastocyst outgrowth. (G) Number of Cdx2/Gata6 double-positive colonies formed in medium supplemented with different combinations of factors. Error bars represent SEM. (H) Efficiency of TE-like cell derivation from blastocyst-stage embryos using iXTE medium, n embryos = 26; three independent experiments, error bars represent SEM. (I) TE-like cells stained for Cdx2, Gata6, cytokeratin-8 (Troma1), and Sox2. Scale bars, 10 μm (A to D), 40 μm (F), and 20 μm (I).
Similar to the ExE, both blastocyst-derived (E3.5) and egg cylinder–derived (E6.5) TSC coexpressed Cdx2 and Sox2 but were Gata6 negative (Fig. 1, C and D), confirming that TSC capture features of the postimplantation trophoblast. To identify factors enabling derivation of TE-like cells directly from blastocyst stage embryos, we tested an array of ligands and small molecules that stimulate key developmental signaling pathways. We used N2B27 as a basal, chemically defined medium that was supplemented with either single or a combination of soluble factors. For each culture condition, we used 8 wells of a 96-well plate containing individual zona pellucida–free E3.5 blastocysts. The embryos were cultured for 4 days on mitotically inactivated mouse embryonic fibroblasts (MEFs), and after that, the outgrowths were dissociated and the cells were replated into fresh wells. Following an additional 4 days of culture, the emerging colonies were fixed and stained for Cdx2 and Gata6 (Fig. 1E).
We found that blastocysts cultured in medium supplemented with Bmp4, CH (Gsk3 inhibitor, CHIR99021), Fgf4, heparin, and activin (iXTE medium) gave rise to flat epithelial colonies that coexpress Cdx2 and Gata6 (Fig. 1, F and G). The efficiency of derivation of TE-like cells was approximately 80%, with 26 embryos giving rise to 21 individual cell lines (Fig. 1H). These cells were positive for the trophoblast-specific intermediate filament cytokeratin-8 (Troma1) and negative for Sox2 (Fig. 1I). Thus, in contrast to the E3.5 and E6.5 TSC, the blastocyst-derived TE-like cells expressed a set of markers akin to the pre-implantation trophoblast.
Developmental origin of the cells with TE-like properties
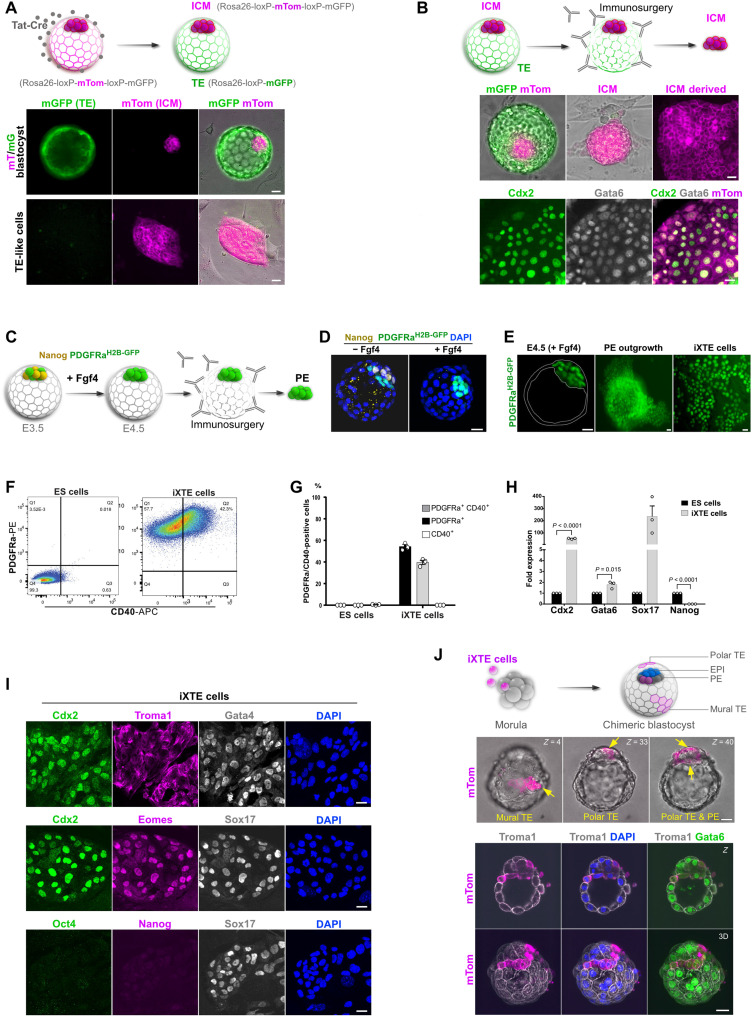
Although the marker signature of the TE-like cells indicated that they most likely originate from the pre-implantation trophoblast, all cell lineages of the early embryo were exposed to the iXTE medium. As the TE-like cells’ derivation procedure did not allow for tracing back the lineage of origin, there were two feasible options: either the TE-like cells derived directly from the TE or they arose via reprogramming of the ICM lineages (PE or epiblast). To tackle this distinction, we used a lineage-tracing approach using double-fluorescent reporter embryos expressing membrane-targeted tandem dimer Tomato (mTom) flanked by LoxP sites, followed by membrane-targeted green fluorescent protein (mGFP) (12). Intact embryos express only the mTom cassette, which can be excised via Cre-mediated recombination resulting in de novo expression of mGFP and loss of mTom fluorescence. As the blastocoel cavity is sealed by tight junctions (13), treatment with cell-permeant Cre protein (Tat-Cre) results in Tat-Cre uptake only by the directly exposed TE, whereas the ICM remains out of reach (Fig. 2A) (14). We removed zona pellucida of E3.5 embryos, and after 2 hours of incubation with Tat-Cre, the blastocysts were transferred into individual wells containing MEFs and iXTE medium. The next day, we observed that the TE gained mGFP expression, indicating successful excision of the mTom cassette, whereas the ICM remained mTom+ (Fig. 2A). Thus, this strategy allowed for permanent genetic labeling of the TE (mGFP+) and the ICM (mTom+) that was also inherited by the cells derived from the respective compartments. Unexpectedly, the TE-like cell colonies established after dissociation of the labeled blastocysts were mTom+, indicating that they originated from the ICM (Fig. 2A).
Fig. 2. Origin and developmental potential of the TE-like cells.
(A) Schematic representation of the Tat-Cre/loxP recombination of the mT/mG locus (top); mT/mG embryos after Tat-Cre treatment (middle) and cells derived from the Tat-Cre–treated embryos (bottom). (B) Schematic representation of TE immunosurgery using mT/mG embryos subjected to Tat-Cre recombination (top); embryo before and after removal of TE (middle); mTom+ ICM–derived TE-like cells stained for Cdx2 and Gata6 (bottom). (C) Schematic representation of the conversion of the ICM into PE and subsequent isolation of the PE via immunosurgery. (D) Control and Fgf4-treated embryos stained for Nanog, PDGFRaH2B-GFP, and DAPI. (E) E4.5 PDGFRaH2B-GFP blastocyst treated with Fgf4, PE outgrowth, and iXTE cells. (F) FACS analysis of iXTE cells and ESC using PDGFRα and CD40. (G) Quantification of PDGFRα- and CD40-expressing cells; three independent experiments, error bars represent SEM. (H) Quantitative polymerase chain reaction (qPCR) analysis in iXTE cells and ESC. β-Actin was used for normalization, and error bars represent SEM, unpaired Student’s t test, n = 3. (I) iXTE cells stained for TE markers—Cdx2, Troma1, and eomes; PE markers—Gata4 and Sox17; and epiblast markers—Oct4 and Nanog; nuclei were counterstained with DAPI. (J) Schematic representation of chimeric blastocyst formation (top); chimeric blastocyst with incorporated iXTE cells, arrows indicate the mTom-expressing iXTE cells (middle); chimeric embryos (n = 13) stained for mTom, Gata6, Troma1, and DAPI (bottom). Scale bars, 20 μm (A, B, D, E, and G) and 40 μm (H). See also fig. S1.
As an additional approach to verify the ICM as the source of TE-like cells, we eliminated the TE of Tat-Cre–labeled mTom/mGFP blastocysts via immunosurgery (Fig. 2B). We selected ICM clumps without any residual mGFP+ trophoblast cells for further analysis. Following culture in the iXTE medium, each of these clumps gave rise to colonies that coexpressed mTom, Cdx2, and Gata6, confirming the ICM origin of the TE-like cells (Fig. 2B). In contrast, dissected TE cultured in iXTE medium were not able to give rise to TE-like cells (fig. S1A).
Although we could clearly distinguish the ICM and TE in mTom/mGFP blastocysts, the Tat-Cre–mediated labeling strategy did not allow for direct discrimination between epiblast and PE, as the whole ICM was mTom+. Therefore, we used PDGFRaH2B-GFP reporter embryos to mark the PE (15) and converted all cells of the ICM into PE via treatment with Fgf4, as previously established (3). After that, we eliminated the TE via immunosurgery and plated the PE-only ICMs in iXTE medium, which resulted in the formation of TE-like cell colonies (Fig. 2, C to E). Conversely, pretreatment with Mek inhibitor (PD0325901) to suppress PE specification (3) diminished the formation of TE-like cell colonies (fig. S1, B and C).
Marker gene analysis revealed that the TE-like cells were positive for the endogenous PE surface marker platelet-derived growth factor receptor–α (PDGFRα; in contrast to the epiblast-derived ESC), and almost half of these cells coexpressed the trophoblast marker CD40 (Fig. 2, F and G). On both the transcriptional and protein levels, the TE-like cells expressed core PE genes, such as Gata4, Gata6, and Sox17, alongside trophoblast genes such as Cdx2, eomes, and cytokeratin-8 (Troma1). At the same time, these cells were negative for epiblast markers such as Oct4 and Nanog (Fig. 2, H and I). Thus, together, the lineage tracing and marker gene analysis show that the TE-like cells are derived from the PE. Therefore, to properly acknowledge the coexpression of TE and PE markers, these cells will be referred to as intermediate extraembryonic (iXTE) cells.
As the iXTE cells simultaneously express core PE and TE factors, the two extraembryonic developmental programs may coexist, allowing for reintegration into both extraembryonic lineages of the pre-implantation embryo. To test this, we aggregated the iXTE cells (mTom+) with 8-cell stage morulae and generated chimeric embryos. After 2 days of culture, we found that mTom-expressing cells incorporated in both the TE (predominantly mural and also polar regions) and the PE (Fig. 2J and fig. S1, D and E). In contrast, PE cells directly isolated from E4.5 embryos and aggregated with 8-cell stage morulae, contributed only to the PE (fig. S1, F to H). Together, this shows that the iXTE cells have an extended integration capacity in the context of the pre-implantation embryo.
XEN and XEN-like cells reprograming to iXTE cells
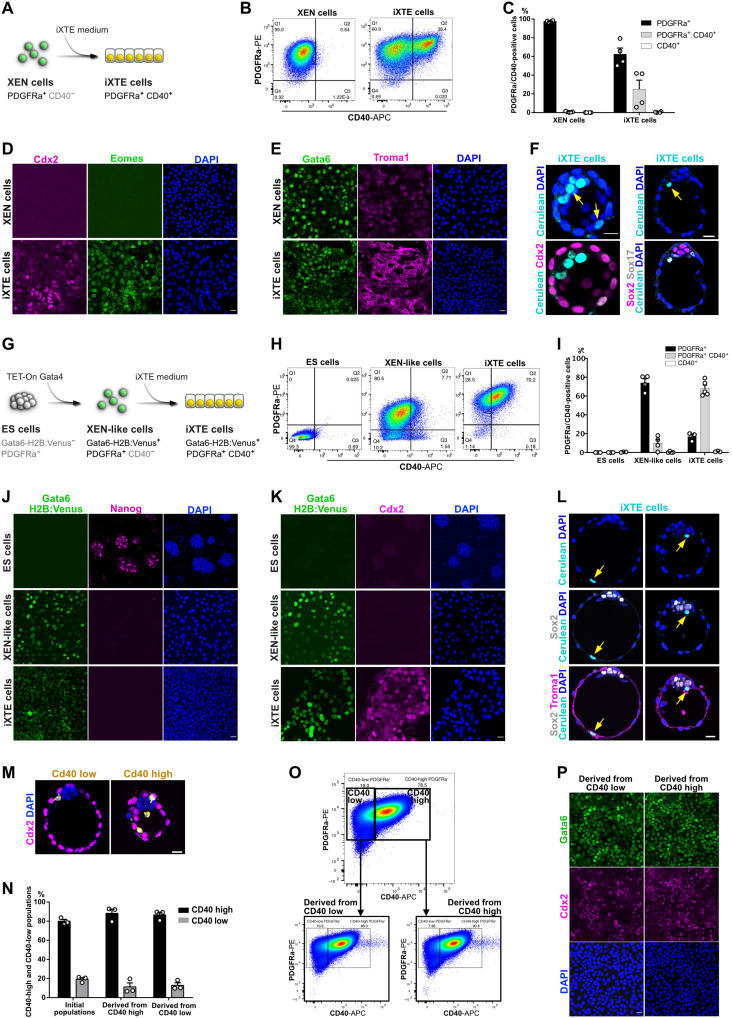
As the three-cell lineages that build the blastocyst exist only transiently during embryonic development, we asked whether the iXTE cells emerge from the PE only during direct derivation from pre-implantation embryos or whether they can emerge from the reprogramming of already established XEN cell lines. To address this question, we cultured XEN cells in iXTE medium and used the cell surface makers PDGFRα and CD40 as a readout for the establishment of iXTE cells. The XEN cells were PDGFRα+/CD40−, but within three to four passages in iXTE medium, they gradually gained CD40 expression, and approximately 30 to 60% of them became PDGFRα+/CD40+ iXTE cells (Fig. 3, A to C, and fig. S2A). At the same time, they up-regulated additional TE markers such as Cdx2, eomes, and Troma1 while maintaining the expression of XEN-specific genes such as Gata6 and Sox17 on both the transcriptional and protein levels (Fig. 3, D and E, and fig. S2B). Treatment with Bmp4 only, which was previously shown to promote XEN cells differentiation to visceral endoderm (16), did not result in up-regulation of CD40 and Cdx2 expression (fig. S2, C to E). Similar to the embryo-derived iXTE cells, the XEN-derived PDGFRα+/CD40+ iXTE cells showed extended integration capacity to populate both the TE and PE of pre-implantation embryos (Fig. 3F and fig. S2F). In contrast, conventional TSC and XEN cells were incorporated only in the TE or PE, respectively (fig. S2, G to J).
Fig. 3. Establishment of XEN state is required for the activation of core TE genes.
(A) Conversion of XEN to iXTE cells. (B) FACS analysis of XEN reprogramming to iXTE cells. (C) Quantification of PDGFRα- and CD40-expressing cells based on FACS analysis; error bars represent SEM, n = 4. (D) XEN and XEN-derived iXTE cells stained for Cdx2, eomes, and DAPI. (E) XEN and XEN-derived iXTE cells stained for Gata6, Troma1, and DAPI. (F) Chimeric blastocysts stained for Cdx2 and DAPI (n = 18) or Sox2, Sox17, and DAPI (n = 11). Arrows indicate the Histone H2B:Cerulean-positive iXTE cells. (G) ESC reprogramming to iXTE cells via an intermediate XEN-like state. (H) FACS analysis for PDGFRα and CD40 expression in ESC, XEN-like cells, and iXTE cells. (I) Quantification of PDGFRα- and CD40-expressing cells based on the FACS analysis; error bars represent SEM, n = 3. (J) ESC, XEN-like, and iXTE cells stained for Gata6 H2B:Venus, Nanog, and DAPI. (K) ESC, XEN-like, and iXTE cells stained for Gata6 H2B:Venus, Cdx2, and DAPI. (L) Chimeric blastocyst (n = 10) stained for Troma1, Sox2, and DAPI. Arrows indicate the Histone H2B:Cerulean-positive iXTE cells. (M) Incorporation of CD40-low or CD40-high cells in chimeric embryos (n = 9) stained from Cdx2, mTom, and DAPI. (N) Quantification of CD40-low and CD40-high expressing cells based on FACS analysis, three independent experiments. (O) FACS analysis of the initial CD40-high and CD40-low expressing populations and after one passage. (P) Cells derived from CD40-high and CD40-low populations and stained for Cdx2, Gata6, and DAPI. Scale bars, 20 μm (D to F, J to M, and P); see also figs. S2 and S3.
Next, we asked whether the activation of PE gene expression is a prerequisite for reprogramming to iXTE cells. To activate the PE program de novo, we used ESC that constitutively express Histone H2B:Cerulean construct and carry a doxycycline (Dox)–inducible Gata4 transgene and a Histone H2B:Venus knock-in cassette in the Gata6 locus (Gata6-H2B:Venus) (17). Following a 24-hour pulse of Dox driving transient Gata4 expression, the ESC were converted into XEN-like cells, marked by the up-regulation of the Gata6-H2B:Venus reporter and gain of PDGFRα expression (Fig. 3, G to J). In addition, the XEN-like cells down-regulated Nanog and up-regulated Gata6 and Sox17 expression (Fig. 3J and fig. S3A).
Using this system, we examined whether an intermediate XEN-like state is required for the conversion of ESC into iXTE cells. We found that directly exposing ESC (as well as TSC) to the reprogramming cocktail failed to generate iXTE cells (fig. S3, B to E). However, culturing the Gata6-H2B:Venus+ (XEN-like) cells in the iXTE medium resulted in the establishment of PDGFRα+/CD40+ cells (within 1 passage), which also activated Cdx2 expression (Fig. 3, G to K, and fig. S3, A and F). Extending the duration of the Dox pulse from 24 to 48 and 72 hours increased the amount of Gata6-H2B:Venus+/PDGFRα+ XEN-like cells, without affecting the overall efficiency of the reprogramming to PDGFRα+/CD40+ iXTE cells (fig. S3G).
Upon engraftment into pre-implantation embryos, the iXTE cells showed similar integration capacity as the PDGFRα+/CD40+ iXTE cells derived from embryos or generated from XEN cells (Fig. 3L and fig. S3, H and I).
Together, these analyses confirm the PE origin of the iXTE cells and show that the activation of the PE program, either induced de novo in ESC/XEN-like cells or already established in vivo (PE) or in vitro (XEN cells), is a prerequisite for the subsequent activation of TE genes.
As we used CD40 as a marker indicating the acquisition of TE features in PDGFRα+ cells, we asked whether the level of CD40 expression is also indicative for the integration capacity of the cells in vivo. To test this, we sorted embryo-derived iXTE cells into PDGFRα+/CD40-low and PDGFRα+/CD40-high fractions and used these cells directly in morula aggregation experiments. Examining the TE/PE integration capacity, the CD40-high cells showed a more pronounced tendency for TE incorporation than the Cd40-low cells (Fig. 3M and fig. S3J). In vitro, within one passage, the CD40-low cells gave rise to both CD40-low and CD40-high populations. The CD40-high fraction also gave rise to CD40-low and CD40-high expressing cells in a similar ratio (Fig. 3, N to P). This indicates that in vitro, the culture conditions promote dynamic fluctuation of CD40-high and CD40-low expressing populations in a metastable iXTE state.
Differentiation potential of the iXTE cells
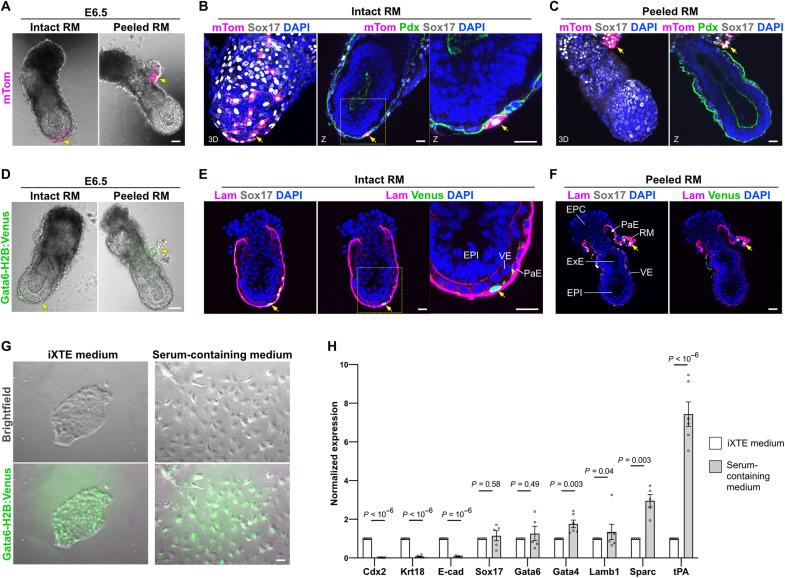
To examine the differentiation potential of the iXTE cells, we transferred chimeric blastocysts to foster mothers and analyzed the contribution of the donor cells progeny to E5.5 and E6.5 postimplantation embryos. We used the mTom+ iXTE cells (embryo derived) or Gata6-H2B:Venus iXTE cells (derived from ESC/XEN-like cells) as donor cells. We found that the iXTE cells gave rise to parietal endoderm (PaE) cells, positioned in the inner side of the Reichert’s membrane (RM). These cells were positive for Sox17 and Gata6 (Gata6-H2B:Venus) (Fig. 4, B, C, E, and F). Peeling the RM revealed that there is no contribution of donor cells to other extraembryonic compartments (ExE or visceral endoderm) of the egg cylinder (Fig. 4, A to F, and fig. S4, A and B).
Fig. 4. Differentiation potential of iXTE cells.
(A) E6.5 chimeric embryo with intact (left) and peeled RM (right), containing mTom-positive cells. (B) E6.5 chimeric embryo with intact RM, containing mTom-positive cells (n chimeric E6.5 embryos = 23), stained for Sox17, mTom, Pdx (podocalyxin), and DAPI. All chimeric embryos showed iXTE contribution to the PaE but not to VE or ExE. (C) E6.5 chimeric embryo with peeled RM, containing mTom-positive cells, Sox17, mTom, Pdx, and DAPI. (D) E6.5 chimeric embryo with intact (left) and peeled RM (right), containing Gata6-H2B:Venus–positive cells. (E) E6.5 chimeric embryo with intact RM, containing Gata6-H2B:Venus–positive cells (n chimeric E6.5 embryos = 4), stained for Sox17, Laminin B1, Venus, and DAPI. (F) E6.5 chimeric embryo with peeled RM, containing Gata6-H2B:Venus–positive cells (n chimeric E6.5 embryos = 4), stained for Sox17, Laminin B1, Venus, and DAPI. (G) Gata6-H2B:Venus iXTE cells cultured for 2 days on fibronectin-coated plates in the presence of iXTE medium (left) or serum-containing medium (right). (H) qPCR analysis of marker gene expression in undifferentiated and differentiated iXTE cells; error bars represent SEM, n > 4; unpaired Student’s t test. Scale bars, 40 μm (A, D, and G) and 20 μm (B, C, E, and F). See also fig. S4.
In agreement with the in vivo analysis, culturing iXTE cells in a serum-containing medium [Dulbecco’s modified Eagle’s medium (DMEM) and 15% fetal bovine serum (FBS)] resulted in the differentiation of the iXTE cells into migratory cells (Fig. 4G). Marker gene analysis revealed that these cells down-regulate TE factors (Cdx2 and Krt18), as well as E-cadherin (E-cad), but maintain the expression of Gata6, Gata4, and Sox17 and up-regulate PaE markers (Lamb1, Sparc, and tPA) (Fig. 4H). The observed E-cad suppression is also in line with the previously reported down-regulation of this adhesion molecule in the PaE cells in vivo (18). Thus, although the iXTE cells have an extended integration capacity in the pre-implantation embryo, these cells show restricted differentiation potential to PaE cells, both in vitro and in the context of the postimplantation conceptus.
Transcriptional state of the iXTE cells
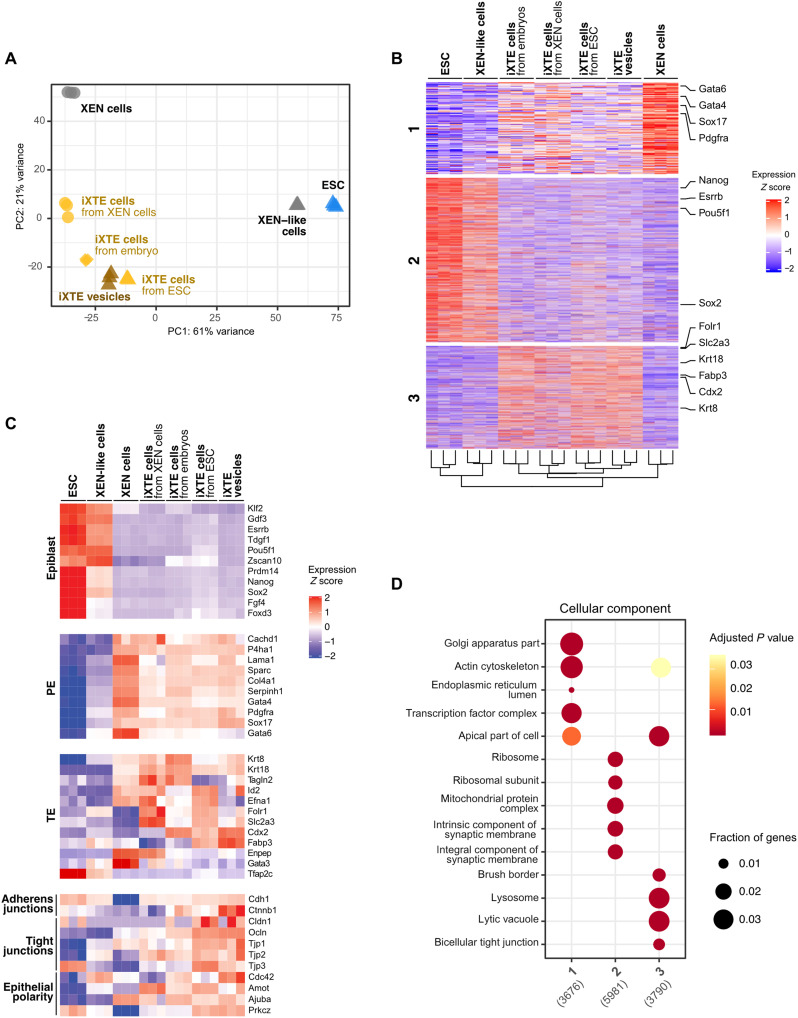
To further characterize the iXTE cells, we collected the PDGFRα+/CD40+ cell populations derived from embryos, XEN, or ESC/XEN-like cells and analyzed their transcriptome using bulk RNA sequencing (RNA-seq). This approach “masks” cellular heterogeneity, providing information about the average gene expression in the examined cell populations. Principal components analysis (PCA) showed that all iXTE cells clustered together, aside from the parental cell lines, namely, XEN, ESC, and XEN-like cells, indicating that a common transcriptional state is established regardless of the source of iXTE cells (Fig. 5A and fig. S5A).
Fig. 5. Transcriptomic profiling of iXTE cells.
(A) PCA of gene expression in embryo-, ESC-, and XEN cell–derived iXTE cells and ESC/iXTE vesicles, including the parental cell lines, namely, XEN cells, ESC, and XEN-like cells. (B) K-means clustering of gene expression reveals three major clusters corresponding to gene expression enriched in XEN cells (cluster 1), ESC (cluster 2), and iXTE cells (cluster 3). (C) Expression of epiblast, PE, and TE markers and factors involved in the formation of adherens junctions, tight junctions, and epithelial polarity in iXTE cells, ESC, XEN-like, and XEN cells. (D) GO “cellular component” enrichment analysis for each gene cluster defined in (B). See also fig. S5.
Differential expression analysis delineated three large groups of actively transcribed genes enriched in XEN cells (cluster 1), ESC (cluster 2), and iXTE cells (cluster 3), respectively (Fig. 5B). In contrast to ESC and in accord with their XEN state of origin, the iXTE cells were negative for epiblast markers but expressed an array of PE-specific genes. Although the PE-specific gene activity was maintained in the iXTE cells, the expression level of the PE genes was toned down in comparison to XEN cells (Fig. 5C). This fine-tuning of the PE transcriptional circuit may allow the TE program to coexist, thus combining features of both extraembryonic lineages in a shared iXTE state. In addition, the expression cluster enriched in iXTE cells contained trophoblast genes, including the master TE factor Cdx2 (Fig. 5, B and C). However, we found that some TE genes, such as Gata3, Enpep, and Tfap2c, were not activated in the iXTE cells, indicating that the iXTE cells do not have the complete transcriptional wiring inherent to the TE. This can provide one explanation for the inability of the iXTE cells to give rise to postimplantation trophoblast, despite the engraftment into the TE at blastocyst stage.
Gene ontology (GO) analysis revealed that the iXTE cells up-regulate sets of genes that are associated with intercellular junctions and apical cellular compartments (Fig. 5D and fig. S5, B and C). More specifically, we found that expression of key adherens junction components, such as E-cad (Cdh1) and β-catenin (Ctnnb1), tight junction proteins, and apical polarity determinants such as Cdc42 (Cdc42) and the aPKC zeta isoform (Prkcz) was enriched in iXTE cells (Fig. 5C). These are bona fide epithelial factors, which suggest that reprogramming to iXTE cells is associated with a concomitant process of epithelialization.
Reprogramming to iXTE cells through the in vivo TE specification program
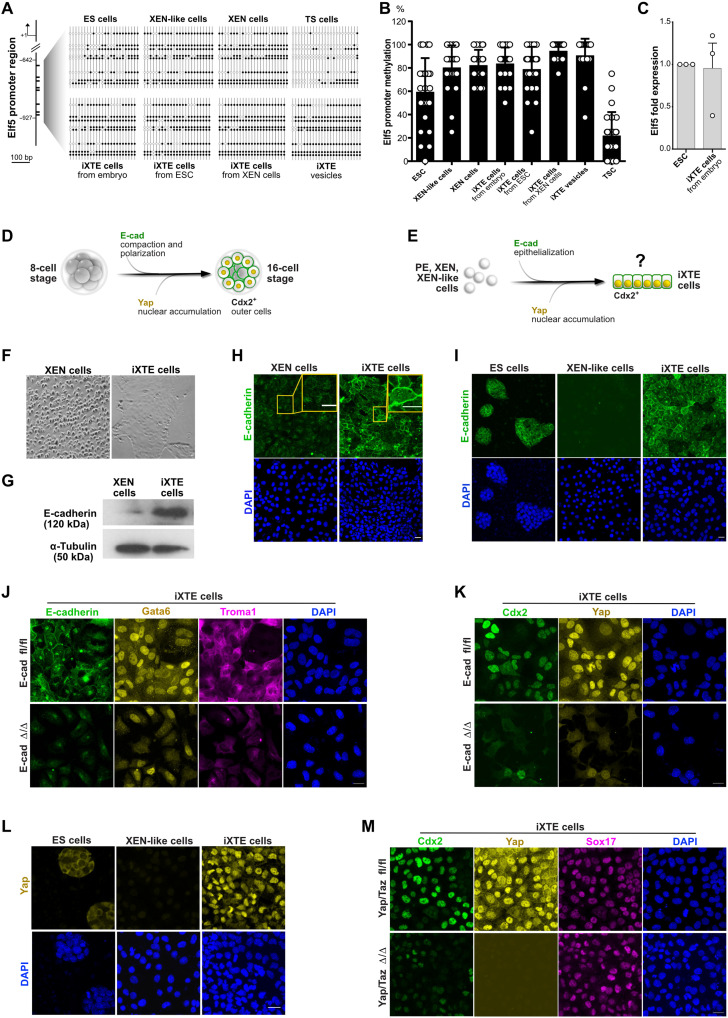
During reprogramming to iXTE cells, the XEN state acquires TE-specific gene expression manifested by the up-regulation of trophoblast markers, particularly Cdx2. Cdx2 is the master regulator of the TE programme in the early embryo and is both indispensable and sufficient to drive the TE fate (19). Thus, we next aimed to determine the mechanism of Cdx2 activation in iXTE cells. A previously reported genome-wide screen for promoter methylation identified Elf5 as a gatekeeper of Cdx2 expression (20). In wild-type ESC, the Elf5 promoter is methylated and Elf5 expression is low. However, in ESC with a DNA methyltransferase 1 knockout (Dnmt1 ko ESC), the Elf5 promoter is hypomethylated, resulting in Elf5 up-regulation. In turn, Elf5 directly binds and activates Cdx2, which enables trophoblast and pluripotent fates to coexist in Dnmt1 ko ESC (20). To understand whether Elf5 drives the TE program in the iXTE cells, we analyzed and compared the methylation status of the Elf5 promoter in TSC, ESC, XEN-like, XEN, and iXTE cells. We found that in contrast to TSC, where the Elf5 promoter is hypomethylated, the Elf5 regulatory sequences in iXTE cells were hypermethylated, and the levels of methylation were comparable to levels in XEN, XEN-like cells, and ESC (Fig. 6, A and B). Also, similar to ESC, the iXTE cells had low Elf5 transcript levels (Fig. 6C and fig. S5B), indicating that the Cdx2 expression in these cells is activated via an alternative mechanism.
Fig. 6. Mechanism of XEN-state reprogramming into iXTE cells.
(A) Methylation status of the Elf5 promoter in ESC, XEN-like cells, XEN cells, iXTE cells, and TSC determined by bisulfite sequencing. White and black circles represent unmethylated and methylated CpGs, respectively. (B) Quantification of Elf5 promoter methylation. (C) qPCR analysis of Elf5 expression in ESC and embryo-derived iXTE normalized to β-actin expression; error bars represent SEM, n = 4. (D) Schematic representation of the 8- to 16-cell-stage transition during mouse pre-implantation embryogenesis including the process of E-cad–mediated compaction and Yap-mediated Cdx2 activation in the outer cells. (E) Schematic representation of the potential mechanism of XEN state reprogramming to iXTE cells, following the principles of the TE specification in mouse embryos. (F) Bright-field images of XEN cells and XEN-derived iXTE cells. (G) Western blot analysis for E-cad expression in XEN and iXTE cells. α-Tubulin was used an internal control. (H) XEN and XEN-derived iXTE cells stained for E-cad and DAPI. (I) Expression of E-cad in ESC, XEN-like cells, and ESC-derived iXTE cells. (J) Expression of E-cad, Gata6, Troma1, and DAPI in E-cad fl/fl iXTE cells and E-cad Δ/Δ iXTE cells. (K) E-cad fl/fl iXTE cells and E-cad Δ/Δ iXTE cells stained for Cdx2, Yap, and DAPI. (L) Expression of Yap in ESC, XEN-like cells, and iXTE cells. (M) Yap/Taz fl/fl iXTE cells and Yap/Taz Δ/Δ iXTE cells stained for Cdx2, Yap, Sox17, and DAPI. Scale bars, 20 μm (H to M). See also fig. S6.
During the pre-implantation development, the blastomeres of the late 8-cell stage embryo undergo a process of compaction, dependent on E-cad–mediated intercellular adhesion (21). At the 16- to 32-cell stage, the epithelial polarity of the outer cells suppresses the activity of the Hippo signaling pathway, which results in the transcriptional coactivator Yap translocating to the nucleus; there, together with Tead4, Yap triggers Cdx2 expression and subsequent TE fate specification (Fig. 6D) (22). As the RNA-seq analysis revealed that reprogramming to iXTE cells is associated with an elevated expression of epithelial factors, we hypothesized that Cdx2 activation in iXTE cells follows similar principles as the TE specification program in vivo. Therefore, we examined whether E-cad–mediated epithelialization of iXTE cells is associated with nuclear accumulation of Yap that would, in turn, drive Cdx2 up-regulation (Fig. 6E).
To test this hypothesis, first we analyzed E-cad expression dynamics during reprogramming of XEN cells to iXTE cells. The XEN cells typically grow as single loosely connected cells, but conversion to iXTE cells resulted in E-cad up-regulation and formation of an epithelial monolayer (Fig. 6, F to H). Similarly, the embryo-derived iXTE cells also expressed E-cad and formed epithelial colonies (fig. S6A).
In addition, we examined E-cad expression patterns during ESC differentiation into XEN-like cells and subsequent reprogramming into iXTE cells. E-cad was down-regulated during the transition from ESC to XEN-like cells, and then it was up-regulated during the conversion from XEN-like to iXTE cells (Fig. 6I). β-Catenin, which is another key element of the adherens junctions complex, also followed a similar expression pattern (fig. S6, B and C). Moreover, E-cad expression was associated with the establishment of Ezrin/Par6-positive apical domain (fig. S6E). To understand whether E-cad–mediated adhesion is required for activating Cdx2 expression, we used E-cad floxed ESC (23) expressing 4-hydroxy tamoxifen (4OHT)–inducible Cre-ERT2 and a Dox-inducible Gata6 transgene. Following a pulse of Dox, the E-cad floxed CreERT2 ESC were converted into PDGFRα+ XEN-like cells. After that, the XEN-like cells were cultured in iXTE medium in the presence of 4OHT to induce E-cad deletion (Fig. 6J and fig. S6D). As a result, the E-cad–deficient cells (+4OHT) were Ezrin/Par6 negative, failed to efficiently accumulate Yap in the nucleus, and failed to up-regulate Cdx2 expression (Fig. 6K and fig. S6E).
Next, we analyzed the dynamics of Yap localization during the conversion of XEN to iXTE cells, as well as during the reprogramming of ESC/XEN-like to iXTE cells. We found that Yap accumulates in the nucleus of the resulting iXTE cells in both reprogramming approaches (Fig. 6L and fig. S6F). To analyze whether Yap signaling promotes Cdx2 expression, we used Yap/Taz double-floxed ESC (24) that express Cre-ERT2 and Dox-inducible Gata6. After conversion to XEN-like cells and subsequent culture in iXTE medium, Yap/Taz were depleted, resulting in loss of Cdx2 expression (Fig. 6M, and fig. S6, G and H). As an alternative approach, we treated iXTE cells with peptide 17, an inhibitor of Yap-Tead interactions, and that also diminished Cdx2 expression (fig. S6I).
It was previously shown that Notch signaling regulates Cdx2 expression in the pre-implantation embryo, in parallel with the Hippo pathway (8, 25). Accordingly, using embryos expressing CBF:H2B-Venus reporter, which contains Recombination Signal Binding Protein For Immunoglobulin Kappa J Region (RBPJ) binding sites driving nuclear H2B-Venus expression (26), we detected Notch signaling activity in the TE of the E3.5 embryo. After implantation, at E5.5, the reporter activity was up-regulated in the postimplantation epiblast and down-regulated in the ExE (fig. S6J). iXTE cells derived from CBF:H2B-Venus blastocyst also showed Notch activity, exhibited by nuclear H2B-Venus fluorescence, similar to the pre-implantation trophoblast (fig. S6K).
Together, these results show that the establishment of an epithelial phenotype during reprogramming to iXTE cells enables Yap/Taz and potentially Notch signaling–mediated up-regulation of Cdx2, similar to the process of TE lineage specification during the first cell fate decision in vivo.
Assembly of blastocyst-like embryoids
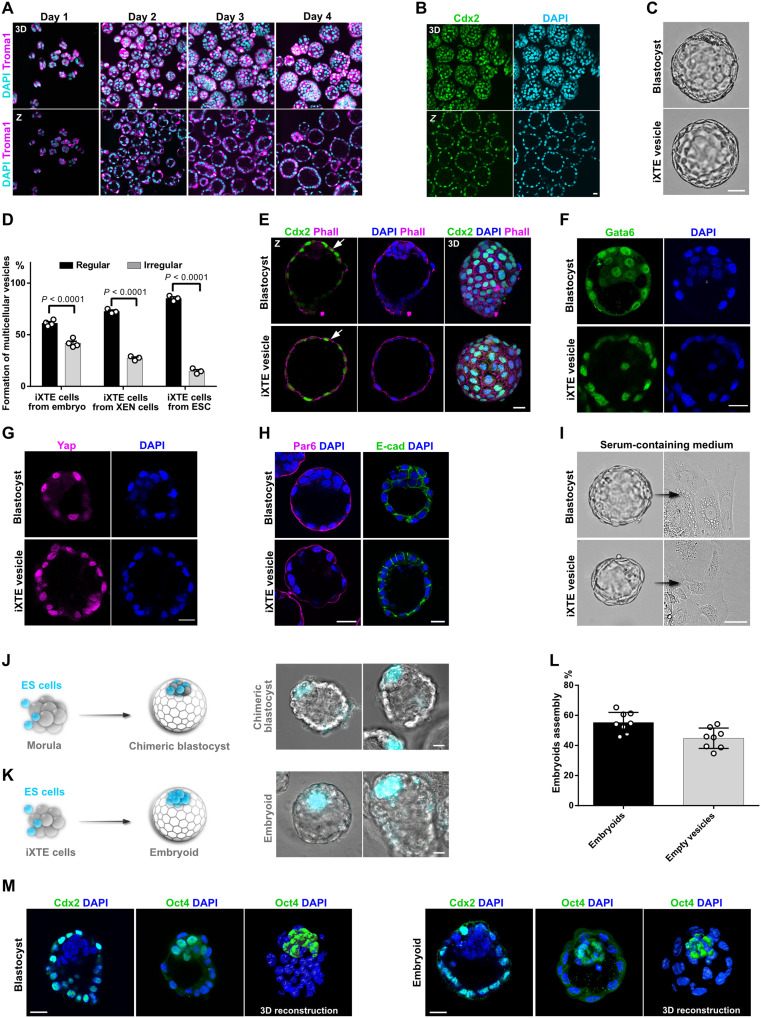
The TE encloses the ICM and the blastocoel cavity, which defines the hollow-shaped architecture of the blastocyst. To determine whether the iXTE cells capture these features, in the following experiments, we examined their tissue-scale organization properties. We cultured the iXTE cells on cell-repellent plates and found that within 24 hours, small clumps were formed instead of the typical flat colonies. In the next days, a central cavity emerged and expanded, transforming the clumps into multicellular vesicles, closely resembling the TE of E3.5 to E4.5 blastocyst-stage embryos (Fig. 7, A to C). Independent of the source of origin, all iXTE cells were able to form blastocoel-like cavities. We classified the vesicles into two groups: regular, spherical structures containing a cavity surrounded by a continuous single cell layer and irregular and deformed oval structures with one or more smaller cavities containing few cells or cell debris (fig. S7A). Regular vesicles were established with up to 85% efficiency, showing that the iXTE cells have a very robust tissue-scale organization capacity (Fig. 7D). The iXTE vesicles were Cdx2 and Gata6 positive and exhibited nuclear accumulation of Yap similar to the TE of pre-implantation embryos (Fig. 7, E to G). The transcriptome and the Elf5 promoter methylation status of the iXTE vesicles were similar to the iXTE cells grown as epithelial colonies (Figs. 5, A and B, and 6, A and B). The vesicles showed proper orientation of the apical-basal polarity axis, with Par6 and cortical actin localized on the outer membrane and E-cad localized on the basolateral membrane, closely mimicking the general architecture of the TE of blastocyst-stage embryos (Fig. 7, E and H).
Fig. 7. Tissue-scale organization properties of iXTE cells and assembly of blastocyst-like embryoids.
(A) iXTE cells were grown on cell-repellent plates for a period of four days in iXTE medium to form multicellular vesicles. The vesicles were fixed at 24-hour intervals and stained for Troma1 and DAPI. (B) Day 4 iXTE vesicles stained for Cdx2 and DAPI. (C) Bright-field images of mouse blastocyst and an iXTE vesicle. (D) Percentage of cavitation representing the efficiency of vesicle formation of iXTE cells derived from embryo, XEN cells, and ESC. The regular vesicles are represented in black and irregular ones in gray. Error bars represent SEM, three independent experiments. (E) Blastocyst and iXTE vesicles stained for Cdx2, Phall, and DAPI. (F) Blastocyst and iXTE vesicles stained for Gata6 and DAPI. (G) Blastocyst and iXTE vesicles stained for Yap and DAPI. (H) Blastocyst and iXTE vesicles stained for Par6, E-cad, and DAPI. (I) Bright-field images of a blastocyst and iXTE vesicle placed in serum-containing medium. (J) Schematic representation of ESC/morula aggregation and formation of chimeric blastocyst (left); chimeric blastocyst with incorporated ESC (right). (K) Schematic representation of ESC/iXTE cell aggregation and formation of blastocyst-like embryoids (left); iXTE cell–based embryoids with incorporated ESC (right). (L) Efficiency of embryoids assembly, eight independent experiments. (M) Blastocysts and embryoids stained for Cdx2 or Oct4 and DAPI. 3D reconstruction of blastocyst and embryoid stained for Oct4 and DAPI is represented in the right. Scale bars, 20 μm. See also fig. S7 and movies S1 and S2.
Similar to the flat iXTE colonies, the iXTE vesicles also responded to serum-containing medium and differentiated into migratory PaE cells (Fig. 7I, fig. S7B, and movie S1). Although on a first look the PaE cells appeared morphologically similar to trophoblast giant cells (TGCs), which emerge in blastocyst outgrowths (Fig. 7I, fig. S7B, and movie S2), we confirmed that these cells are not polyploid TGCs (fig. S7, C and D).
Next, we analyzed whether the iXTE cells can incorporate ESC for complementing the vesicles with an “artificial ICM.” As a control, we used 8-cell stage embryos aggregated with fluorescently labeled (Histone H2B-Cerulean) ESC that integrated into the ICM and established chimeric blastocysts (Fig. 7J). We used the same approach to generate artificial blastocyst-like embryoids. After 24 hours of culture in cell-repellent plates, small clumps of iXTE cells were placed into individual microwells containing iXTE medium. Histone H2B-Cerulean+ ESC were added to the iXTE clumps and subsequently internalized and positioned at one side of the emerging cavity, assembling blastocyst-like embryoids (Fig. 7, K to M, and fig. S7E).
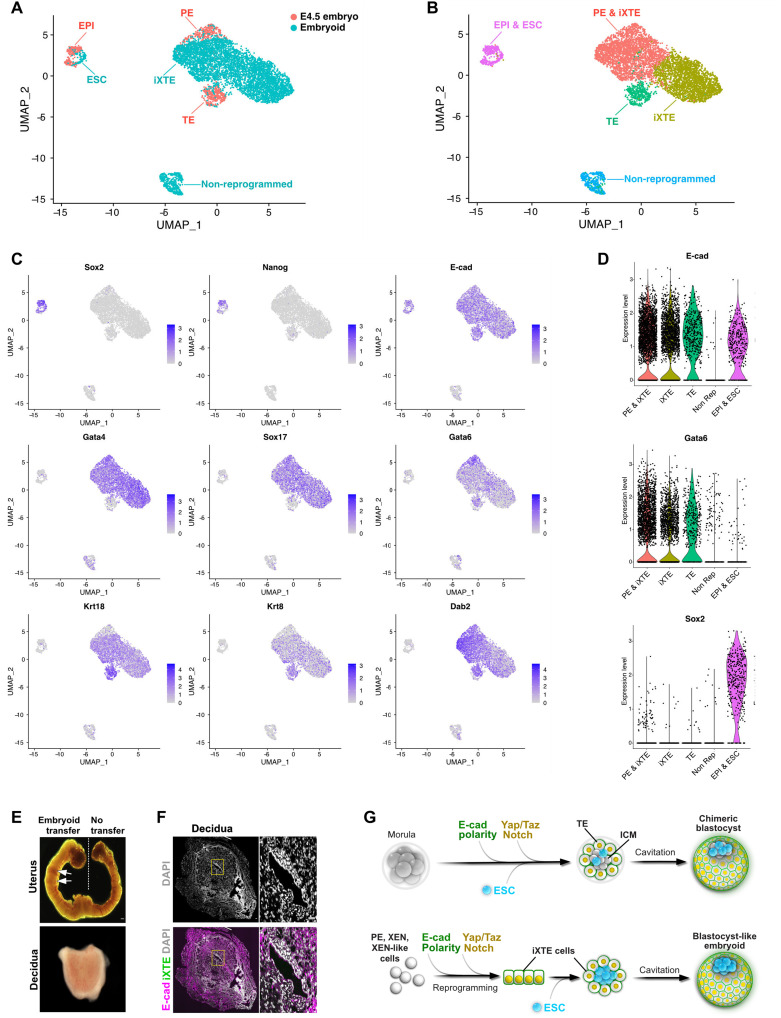
To further characterize the iXTE-based embryo-like structures, we performed single-cell RNA sequencing (scRNA-seq). To get a fair overview of the in vitro system, we used approximately 200 structures of cocultured ESC and iXTE cells, without preselecting individual embryoids. Thus, empty and irregular vesicles were also randomly included in the analysis. As a reference, we used 220 E4.5 blastocysts for scRNA-seq analysis. The cells of the E4.5 embryos distributed into three distinct groups: epiblast (Sox2 and Nanog positive), TE (Krt8 and Krt18 positive), and PE (Gata4 and Sox17 positive) (Fig. 8, A and C). The iXTE cells positioned between the TE and PE lineages, whereas the ESC clustered together with the epiblast and nonreprogrammed cells (E-cad negative) were positioned separately away from all other clusters (Fig. 8, A to D). In addition, we incorporated our scRNA-seq datasets with previously published datasets of E6.5, E7.0, E7.25, E7.5, E8.0, E8.25, and E8.5 embryos (27) and found that the iXTE cells cluster closely to the early extraembryonic tissues rather than the later derivatives of the PE and definitive endodermal lineages (fig. S8A). Essentially, the iXTE cells expressed both TE- and PE-specific genes, as well as shared markers, such as Dab2 and Gata6 (Fig. 8C), together showing that the iXTE cells transcriptionally capture features of both PE and TE lineages of the pre-implantation embryo. In line with the bulk RNA-seq analysis, the iXTE cells did not express Gata3, Enpep, and Tfap2c (fig. S8B), potentially reflecting their restricted differentiation potential.
Fig. 8. scRNA-seq analysis.
(A) Uniform manifold approximation and projection (UMAP) plot of the single-cell transcriptomes of E4.5 blastocysts (orange) and iXTE/ESC-based embryoids (blue). (B) UMAP plot displaying cells clustering. (C) UMAP plots of lineage markers expression. (D) Violin plot of E-cad, Gata6, and Sox2 expression in the different cell clusters. (E) Embryoids transferred into the uterus of pseudopregnant females induce decidualization, arrows indicate deciduae. (F) Sections of decidua induced by embryoid transfer and stained for E-cad and iXTE cells expressing Venus. (G) TE lineage specification and incorporation of ESC (top). Reprogramming of XEN state cells to iXTE cells, subsequent cavitation and embryoids’ self-assembly (bottom). Scale bars, 40 μm (E and F). See also fig. S7.
Last, we also transferred the embryoids into the uterus of pseudopregnant females and found that the embryoids induced uterine decidualization (Fig. 8E), but as expected, egg cylinder–like structures were not formed (Fig. 8F). This is in accord with the limited developmental capacity of the iXTE cells, which were not able to generate functional trophoblast, therefore setting a time frame of embryoid “development” from morula-like up to blastocyst-like stages (Fig. 8G).
DISCUSSION
During the 8- to 16-cell stage transition, two cell populations are established in the early mouse embryo: outside cells that form the TE and the inside ICM cells. The outside cells are not restricted to the TE fate until the 32-cell stage, as they can divide asymmetrically and give rise to ICM cells. Even an additional minor wave of asymmetric division has been found as late as the 64-cell stage (2, 3). Thus, the 16- to 64-cell stage TE has the capacity to generate both epiblast and PE progenitors, and this potential rapidly decreases after E3.5. In addition to the master TE factor Cdx2, the pre-implantation TE expresses the PE determinant Gata6. Therefore, the early TE cells may occupy a state where more than one developmental programme coexists, enabling developmental plasticity that allows the early embryo to adapt and compensate for alterations in cell position and overall cell number.
Gata6 is also expressed in the ICM, which, at E3.5, is a heterogeneous population of PE (Gata6-positive) and epiblast (Nanog-positive) progenitors that segregate into two layers by E4.5 (28). A recent study by Posfai et al. (4) showed that the ICM loses its ability to form TE, as it becomes refractory to Hippo signaling beyond 32-cell stage. Since the ICM of the 32-cell stage blastocyst is a mix of Gata6- and Nanog-expressing cells, it will be of interest to examine whether any of these subpopulations are more prone to activate Cdx2 expression upon TE removal. Our results suggest that this subpopulation might be the Gata6-expressing cells, and the putative activation of Cdx2 may act as a fail-safe mechanism for compensating for the loss of TE cells. We found that cells with PE gene expression up-regulate Cdx2 in response to the accumulation of nuclear Yap and Notch intracellular domain (ICD), which requires the establishment of epithelial polarity. The potential activation of a compensatory TE program in the ICM in response to the loss of TE cells may follow the same principle, namely, de novo polarization of ICM cells, enabling Hippo signaling inactivation, concurrent activation of the Notch pathway, and up-regulation of Cdx2. Thus, the process of reprogramming to iXTE cells may capture certain features of a putative fail-safe mechanism that enables Gata6-positive ICM cells to compensate TE cell deficiency.
Previous studies showed that the XEN cells can reprogram also into chemically induced pluripotent stem cells (ciPSC) (29, 30), acquiring characteristics akin to the pre-implantation epiblast. The generation of both ciPSC and iXTE cells does not require a forced expression of transgenes but depends on the establishment of the XEN state, which enables reprogramming by small molecules and ligands. Moreover, the XEN-like cells can be directly converted into functional neurons and hepatocyte-like cells, bypassing the establishment of pluripotency (31). We found that such cells can up-regulate Cdx2, establishing Gata6+/Cdx2+ iXTE cells that combine features of both TE and PE lineages. Together, this shows that the XEN/XEN-like state has a remarkable potential to respond to external signaling cues and adopt embryonic and extraembryonic fates.
The XEN-like cells, which were generated by a pulse of ectopic Gata4 expression in ESC, reprogrammed faster and more efficiently, in comparison to the conventional XEN cells. Moreover, the PCA indicated that the XEN-like cells are overall transcriptionally closer to the ESC than to the conventional XEN cells. We found that the down-regulation of pluripotency genes and up-regulation of core PE factors were sufficient to enable the reprogramming of the XEN-like cells to iXTE cells. Thus, the partial (transient) XEN-like state is rewired faster and reprogrammed more efficiently, in comparison to the fully settled (stable) conventional XEN state.
We determined that cells derived from the PE lineage of the ICM and XEN and XEN-like cells acquire TE properties following similar principles as the TE specification in vivo. Activation of Cdx2 in iXTE cells requires establishment of epithelial polarity and accumulation of Yap/Taz and Notch in the nucleus (Fig. 7G). At the same time, these cells maintain the PE transcriptional circuit, enabling extended integration capacity of the iXTE cells in both extraembryonic lineages of the blastocyst. However, after implantation, the iXTE cells give rise only to PaE cells and do not contribute to the ExE and visceral endoderm. Thus, the iXTE cells are not bona fide stem cells of either trophoblast or PE linages but instead exhibit certain PE and TE characteristics and terminal developmental capacity. These cells exhibit a “hybrid” identity, which most likely does not represent a physiological state during normal embryogenesis and this identity does not progress into a “real” TE or PE state. This also sets the margin of the iXTE-based embryoid development up to a blastocyst-like stage but not beyond. Nevertheless, further adjustment of the cocktail may enable the complete conversion of the XEN state cells into a functional trophoblast.
The iXTE cells showed robust tissue-scale organization potential, enabling highly efficient formation of multicellular vesicles that resemble the overall three-dimensional (3D) architecture of the TE. Previous reports used conventional TSC or extended pluripotent stem (EPS) cells to form similar vesicles with approximately 10 to 15% efficiency (32–34). This low efficiency indicates that the TSC and EPS cells may contain a subpopulation of vesicle-initiating cells that manifest cavitation properties under specific culture conditions. The culture medium used for cavity induction in these pioneering studies contains factors such as Bmp4, CHIR99021, Fgf4, and heparin, a composition that is notably similar to our iXTE medium, which induces TE-like properties in PE, XEN, and XEN-like cells. Moreover, it was previously reported that EPS cells can give rise to XEN cells (32), and, in addition, TSC are often coderived together with XEN cells (35, 36). Thus, it is plausible that a vesicle-initiating subpopulation with XEN cell identity may exist or emerge in TSC or EPS cell cultures. Although such a subpopulation can gain TE properties and trigger decidual response, these cells will subsequently fail to generate functional placental tissues. This can provide one explanation why despite uterine decidualization, none of the artificial blastocyst-like structures reported to date can properly develop further, following transfer into a foster mother.
In summary, we showed that the PE of the pre-implantation embryo, as well as conventional XEN and ESC-derived XEN-like cells, can acquire TE properties. These cells activate the pre-implantation TE program following the trophoblast specification cascade of the first lineage segregation. The resulting iXTE cells combine characteristics of the two extraembryonic lineages, enabling integration into both the TE and PE domains, but exhibit restricted differentiation potential. Mimicking the TE of morula/blastocyst-stage embryos, the iXTE cells can establish a blastocoel-like cavity and integrate pluripotent cells, forming blastocyst-like structures. The iXTE cells can provide a platform for deciphering the hidden compensatory mechanisms of cell fate specification, as well as the processes of polarization, cavitation, cell-cell recognition, and sorting in the pre-implantation embryo.
METHODS
Mice
The mice used in the study were at age from 6 weeks to 5 months. The animals were maintained under a 14-hour light/10-hour dark cycle with free access to food and water. Male mice were kept individually, whereas the female mice were housed in groups of up to four per cage. Embryos for experiments were obtained from wild-type and transgenic strains from matings using females with natural ovulation cycle or after superovulation in case of morula aggregations. The mouse strains used in the study are B6C3F1, CD1, mT/mG (12), Yapfl/fl/Tazfl/fl (24), CBF:H2B-Venus (26), and PDGFRaH2B-GFP (37). Animal experiments and husbandry were performed according to the German Animal Welfare guidelines and approved by the Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen (State Agency for Nature, Environment and Consumer Protection of North Rhine-Westphalia).
Cell lines
XEN cells were maintained in DMEM supplemented with 15% FBS, 2 mM l-glutamine, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, penicillin-streptomycin (50 U/ml), and 0.1 mM 2-mercaptoethanol. For reprogramming experiments, the XEN cells were passaged and maintained in N2B27 medium. TSC were grown on mitotically inactivated MEFs in the presence of Fgf4 (25 ng/ml) and heparin (1 μg/ml). ESC were maintained in DMEM medium supplemented with 15% FBS, 2 mM l-glutamine, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, penicillin-streptomycin (50 U/ml), 0.1 mM 2-mercaptoethanol, 0.4 μM PD0325901, 3 μM CHIR99021, and Lif (4 ng/ml) on gelatin-coated culture plates. All cell types were grown at 37°C/5% CO2 and passaged using 0.05% trypsin-EDTA. Yap/Taz double-floxed ESC were derived from Yap/Taz double-floxed mouse strain (24). E3.5 embryos were plated into individual wells of a 96-well plate with inactivated MEFs and grown in ESC medium. After 4 days of culture, the blastocyst outgrowths were trypsinized, transferred into fresh wells, and expanded once ESC colonies emerged. Yap/Taz double-floxed ESC were stably transfected with Cre-ERT2 and Dox-inducible Gata6 cloned into a pIB12-PB-hCMV1-cHA-IRES-Venus plasmid, with lipofectamine 2000 (Invitrogen) using the manufacturer’s protocol. Expression of Tet-on Gata6 transgene was stimulated using Dox (1 μg/ml), and Cre/loxP recombination was induced using 500 nM 4OHT. E-cad fl/fl ESC were stably transfected with Cre-ERT2 and transiently transfected with Dox-inducible Gata6 cloned into a pIB12-PB-hCMV1-cHA-IRES-Venus plasmid using lipofectamine 2000 (Invitrogen) following the manufacturer’s protocol.
Establishment of iXTE cells
iXTE were derived from E3.5 blastocysts by plating embryos into individual wells of a 96-well plate coated with mitotically inactivated MEFs in iXTE medium [N2B27 supplemented with 3 μM CHIR99021, BMP4 (50 ng/ml), Fgf4 (25 ng/ml), heparin (1 μg/ml), and activin (20 ng/ml)]. The blastocyst outgrowths were dissociated after 4 days using 0.05% trypsin-EDTA and seeded into fresh wells to allow the emergence of iXTE colonies. The iXTE cells were then sorted by fluorescence-activated cell sorting (FACS) to select for PDGFRα/CD40 double-positive population and maintained in iXTE medium on fibronectin-coated wells.
Conversion of ESC to iXTE cells is as follows: ESC that carries Dox-inducible Gata4 transgene and Histone H2B:Venus knock-in cassette in the Gata6 locus (Gata6-H2B:Venus) (17) or Dox-inducible Gata6 was converted to XEN-like cells by a 24-hour pulse of Dox. After that, the XEN-like cells were sorted for expression of Venus and PDGFRα and seeded on fibronectin-coated plates either in N2B27 medium (for experiments using XEN-like cells) or in iXTE medium (for conversion to iXTE cells). The XEN-like cells were cultured in iXTE medium for 5 days allowing conversion to iXTE cells that were subsequently sorted by FACS for PDGFRα/CD40 expression.
Conversion of XEN cells to iXTE cells is as follows: XEN cells were grown in iXTE medium on fibronectin-coated plates for three to four passages followed by sorting for PDGFRα/CD40 double-positive population of iXTE cells. As the iXTE cells are not homogeneous population, but metastable, we used cells up to passages five to six for downstream analysis.
Lineage tracing
mT/mG reporter mouse strain (12) was used for lineage-tracing experiment. This strain harbors membrane-targeted tandem dimer Tomato (mT) cassette flanked by loxP sites, followed by membrane-targeted GFP (mG), integrated in the Rosa26 locus. Zona-free mT/mG blastocysts were incubated with 1.5 μM recombinant cell-permeant Cre protein (Tat-Cre, Millipore) in prewarmed KSOM medium (Millipore) for 2 hours at 37°C in 5% CO2 atmosphere in air, according to a previously published protocol (14). After that, the embryos were washed and cultured in KSOM overnight at 37°C in 5% CO2. The expression of mTom and mGFP was examined on the next day.
Immunosurgery
ICMs of E3.5 mT/mG embryos subjected to Tat-Cre treatment were isolated by immunosurgery (38), and the ICM was then placed on inactivated MEFs in iXTE medium for derivation of iXTE cells.
Morula aggregation
E2.5 embryos were collected from oviducts by flushing with M2 medium. Zona pellucida was removed by brief exposure to Tyrode’s solution. Individual embryos were places into microwells filled with KSOM medium, and approximately 10 cells were added in close proximity to the embryo.
Vesicle formation
For generation of multicellular vesicles, 10,000 to 20,000 PDGFRα/CD40 double-positive iXTE cells were seeded per well of a 96-well plate (CELLSTAR cell repellent plates, Greiner Bio-One) containing 100 μl of iXTE medium. The cells were allowed to self-organize for 4 days.
Embryoid generation
Embryoids were grown by seeding ES cells (10,000 cells) and XTEs (50,000 to 80,000) in an AggreWell400 plates (StemCell Technologies) in 24-well format for 48 hours, and then the aggregates were transferred to cell repellent plates for 48 hours to facilitate cavitation and formation of embryoids.
Quantification of polyploidy
TSC and iXTE cells were grown in serum-containing medium for 5 days. After that, the cells were dissociated using 0.25% trypsin and processed using a CyStain UV Precise T kit according to the manufacturer’s protocol (Sysmex Deutschland GmbH). The DNA content was analyzed using a FACSAria IIu sorter (BD Biosciences).
Bulk RNA-seq and bioinformatic analysis
RNA was isolated from cells using a NucleoSpin RNA isolation kit following the manufacturer’s protocol (Macherery and Nagel) with a purity ratio of 2 for absorbances at 260 and 280 nm, measured using Nanodrop 2000c. One microgram of total RNA was then used as the starting material for mRNA enrichment, which was done using the NEBNext Poly(A) mRNA Magnetic Isolation Module, and complementary DNA library was prepared using the NEBNext Ultra II Directional RNA Library Prep Kit. Sequencing was performed on the NextSeq 500 system (75 cycles, high output, v2.5) in the Core Facility Genomics of the Medical Faculty, University of Münster.
Transcript expression from RNA-seq was quantified using Salmon (39) (version 0.13.0) with parameters “--seqBias -l A” using a transcriptome index created from Mus musculus genome version GRCm38 (Ensembl release 93). Transcript quantifications were imported into R and associated with genes using the tximport package [version 1.13.12 (40)]. Differential expression analysis was performed using DESeq2 [version 1.25.10 (41)]. Genes were considered significantly differentially expressed between two conditions if the comparison had an absolute log2 fold change of at least 2 and an adjusted P value less than 0.05.
The variance-stabilizing transformation was applied to data for the purposes of visualization using PCA or heatmaps. PCA was performed using the “prcomp” R function on the top 500 most variable genes across all samples. For gene expression clustering, we considered genes that were significantly differentially expressed in at least one comparison. Gene expression matrices were transformed to row Z scores and clustered using k-means clustering using the “kmeans” R function. The appropriate number of clusters was assessed using the elbow method (fig. S2A).
GO enrichment analysis was carried out using the “compareCluster” function from the clusterProfiler package [version 3.13.0 (42)] with a q value threshold of 0.1. Results were simplified to remove highly similar GO terms based on semantic similarity using the “simplify” function with the Wang method and a similarity threshold of 0.7 [GOSemSim version 2.11.0 (43)].
RNA-seq data analysis and visualization were carried out using R [version 3.6.1 (44)]. The ggplot2 [version 3.2.1 (45)] and ComplexHeatmap [version 2.1.0 (46)] R packages were used for data visualization, and the dplyr [version 0.8.3 (47)] and purrr [version 0.3.2 (48)] packages were used for general data analysis.
scRNA-seq analysis
Around 200 E4.5 embryos or embryoids were collected and dissociated into single cells by incubating with TrypLE for 15 min at 37°C. The single cells were then counted using a Luna-II automated cell counter (Logos Biosystems), and a total 1500 cells for E4.5 embryos or 25,000 cells for embryoids were loaded on to a microwell cartridge of BD Rhapsody Express system (BD Biosciences). BD Rhapsody cell capture beads of embryoids were subsampled by half aiming to recover around 6000 cells. Single-cell whole transcriptome analysis libraries were prepared according to the manufacturer’s instructions using a BD Rhapsody WTA Reagent kit (BD Biosciences) and sequenced on the Illumina NextSeq 500 using the High Output Kit v2.5 (150 cycles, Illumina). Sequencing data were processed with UMI tools (version 1.0.1) (49), aligned to the mouse reference genome (mm10) with STAR (version 2.7.1a) (50), and quantified with Subread featureCounts (version 1.6.4) (51). Data normalization and further analysis were performed using Seurat (version 3.1.3) (52). For initial quality control of the extracted gene-cell matrices, we filtered cells with parameters nFeature_RNA > 500 & nFeature_RNA < 6000 for number of genes per cell and percent.mito <25 for percentage of mitochondrial genes and genes with parameter min.cell = 3. Filtered matrices were normalized by LogNormalize method with scale factor = 10,000. Variable genes were found with parameters of mean.function = ExpMean, dispersion.function = LogVMR, x.low.cutoff = 0.0125, x.high.cutoff = 3, and y.cutoff = 0.5, trimmed for the genes related to cell cycle (GO:0007049) and then used for PCA. FindIntegrationAnchors and IntegrateData with default options were used for the data integration. Statistically significant principal components were determined by JackStraw method, and the first eight principle components were used for nonlinear dimensional reduction and unsupervised hierarchical clustering analysis with a resolution = 0.3. Cluter-specific markers were identified using a function, FindAllMarkers with parameters only.pos = TRUE, min.pct = 0.25, and logfc.threshold = 0.25.
Quantitative PCR
Total RNA was isolated using a NucleoSpin RNA isolation kit following the manufacturer’s protocol (Macherery and Nagel) and reverse-transcribed using MMLV-Reverse transcriptase (Applied Biosystems). Transcript levels were quantified using iTaq SYBR Green Supermix (Bio-Rad), and the gene expression was normalized to the housekeeping gene β-actin. The calculations were done using the delta Ct algorithm. The primer sequences are listed in table S1.
Genomic DNA isolation and PCR
The PureLink Genomic DNA Mini Kit was used to isolate genomic DNA according to the manufacturer’s protocol. Polymerase chain reaction (PCR) was performed using the primers listed in table S2.
Bisulfite sequencing
To determine the DNA methylation status at regulatory regions of Elf5 gene, genomic DNA was isolated using the PureLink Genomic DNA Mini Kit, and bisulfite conversion was carried out using 2 μg of isolated genomic DNA with an EZ DNA methylation kit (Zymo Research) according to the manufacturer’s protocol. The bisulfite-converted DNA was amplified by PCR using the previously described primers (20). The PCR products were cloned using the TOPO-TA kit (Invitrogen) according to the manufacturer’s protocol. Individual clones were then sequenced, and sequences were analyzed using the Quantification Tool for Methylation Analysis (http://quma.cdb.riken.jp).
Morula aggregation
The zona pellucida of E2.5 embryos was removed via brief exposure to Tyrode’s solution (Sigma-Aldrich). After that, ESC or iXTE cells were aggregated with the zona-free morulae in individual microwells filled with KSOM medium and covered with mineral oil. The embryos were cultured to blastocyst stage in vitro at 37°C in a humidified atmosphere of 5% CO2.
Immunofluorescence labeling and microscopy
The samples were fixed using 4% paraformaldehyde for 20 min, washed twice with 1% fetal calf serum (FCS) in phosphate-buffered saline (PBS; wash buffer) and permeabilized with 0.1 M glycin/0.3% Triton X-100 (3 to 5 min for cells and pre-implantation embryos and 10 min for postimplantation embryos). After washing, the samples were incubated with primary antibodies diluted in blocking buffer (2% FCS in PBS) for 24 hours at 4°C. The specimens were then washed twice and incubated with secondary antibody and 4′,6-diamidino-2-phenylindole (DAPI) overnight at 4°C. On the following day, the samples were washed again twice and kept in the wash buffer for imaging. The embryos were mounted in drops of 1% FCS in PBS on glass-bottom plates and covered with mineral oil, whereas cells were directly grown on the ibidi μ-plates coated with fibronectin and processed further for imaging. The antibodies used in the study are listed in table S3. Conventional epifluorescence microscopy was performed using Leica AF600, and confocal microscopy was performed using Leica SP5 and Zeiss LSM780 systems. Images were processed using Fiji, Icy, and Imaris (Bitplane) software.
Fluorescence-activated cell sorting
Cells were dissociated using 0.05% trypsin and resuspended in PBS supplemented with 3% FCS (dilution buffer). For sorting cells based on PDGFRα and CD40 expression, the cells were incubated with the respective antibodies (table S2) for 1 hour on ice and washed three times with dilution buffer to remove traces of unbound antibodies. After that, the cells were resuspended in the dilution buffer and subjected to sorting in FACSAria IIu sorter (BD Biosciences). Single viable cells were first selected on the basis of FSC and SSC gating, and either PDGFRα+ and Venus+ cells or PDGFRα+ and CD40+ cells were collected depending on the experimental setup. FlowJo software was used for data analysis and plotting.
Western blot
Cells were lysed using buffer containing 10 mM tris-HCl (pH 7.6), 150 mM NaCl, 2 mM MgCl2, 2 mM EDTA, 0.1% Triton X-100, 10% glycerol, and 1× protease inhibitors cocktail (cOmplete ULTRA). The lysis buffer was added directly to the culture dish, and the cells were scraped off the surface. The lysate was kept on ice for 20 min and then sonicated for 5 min. The total protein concentration was measured using the Pierce BCA Protein Assay Kit. Twenty-five micrograms of the total protein per sample was run on SDS–polyacrylamide gel alongside with the Precision Plus Protein Kaleidoscope Standard marker and subsequently transferred onto a polyvinylidene difluoride membrane. Next, the membrane was incubated in 5% dry milk in PBS with Tween 20 (PBST) for 30 min at room temperature for blocking and then incubated with primary antibodies at 4°C overnight. On the next day, the membrane was washed tree times with PBST and incubated with secondary antibodies conjugated to horseradish peroxidase for 2 hours at room temperature. The proteins were detected using the ECL Prime kit (GE Healthcare) by exposing the membranes to ECL Hyperfilms (GE Healthcare).
Quantification and statistical analysis
Values represented in bar graphs are shown as mean and error bars represent SEM. Graphs were generated using GraphPad Prism. All significances were calculated using two-tailed unpaired Student’s t test and are represented on the figures.
Acknowledgments
We thank C. Schröter (MPI-MP) for providing Gata6-H2B:Venus ESC, H. Schorle (University of Bonn) for sharing the TSC, K. Adachi for providing the XEN cells, N. K. Rink and L. Kremer (Transgenic facility, MPI-MB) for the embryo transfers, and all laboratory members for suggestions and constructive criticism.
Funding: This work was supported by the German Research Foundation (DFG) Emmy Noether grant (BE 5800/1-1 and BE 5800/1-2) and the Collaborative Research Center 1348 “Dynamic Cellular Interfaces” grant (1348/2, B09) to I.B. PCT patent application is filed.
Author contributions: I.B. and A.S. conceived the study, designed and executed the experiments, and interpreted results; I.B. and A.S. wrote the manuscript; E.I.-S. performed bioinformatic analysis; R.C. standardized and carried out the scRNA-seq for E4.5 embryos; H.-W.J. performed the scRNA-seq analysis; H.O.O. standardized the lineage-tracing experiments; K.-P.K. performed bisulfite sequencing analysis; Y.S.K. generated the E-cad floxed/CreERT2 ESC; B.D. helped with qPCR and microscopy experiments; M.S., R.F., and A.S. performed FACS analysis; H.B. provided general technical support; R.H.A., J.M.V., and H.R.S. provided support of key infrastructure, reagents, and discussion of the results.
Competing interest: The authors declare that they have no competing interests.
Data and materials availability: Bulk RNA-seq data are deposited to ArrayExpress and can be accessed via www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-8788/. scRNA-seq data are deposited to NCBI GEO and can be accessed via GEO accession GSE159883: Go to https://ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE159883. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Tables S1 to S3
Other Supplementary Material for this manuscript includes the following:
Movies S1 and S2
REFERENCES AND NOTES
- 1.Govindasamy N., Duethorn B., Oezgueldez H. O., Kim Y. S., Bedzhov I., Test-tube embryos–Mouse and human development in vitro to blastocyst stage and beyond. Int. J. Dev. Biol. 63, 203–215 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Morris S. A., Teo R. T. Y., Li H., Robson P., Glover D. M., Zernicka-Goetz M., Origin and formation of the first two distinct cell types of the inner cell mass in the mouse embryo. Proc. Natl. Acad. Sci. U.S.A. 107, 6364–6369 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamanaka Y., Lanner F., Rossant J., FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development 137, 715–724 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Posfai E., Petropoulos S., de Barros F. R. O., Schell J. P., Jurisica I., Sandberg R., Lanner F., Rossant J., Position- and Hippo signaling-dependent plasticity during lineage segregation in the early mouse embryo. eLife 6, e22906 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nichols J., Smith A., Pluripotency in the embryo and in culture. Cold Spring Harb. Perspect. Biol. 4, a008128 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka S., Kunath T., Hadjantonakis A. K., Nagy A., Rossant J., Promotion of trophoblast stem cell proliferation by FGF4. Science 282, 2072–2075 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Adachi K., Nikaido I., Ohta H., Ohtsuka S., Ura H., Kadota M., Wakayama T., Ueda H. R., Niwa H., Context-dependent wiring of Sox2 regulatory networks for self-renewal of embryonic and trophoblast stem cells. Mol. Cell 52, 380–392 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Rayon T., Menchero S., Rollán I., Ors I., Helness A., Crespo M., Nieto A., Azuara V., Rossant J., Manzanares M., Distinct mechanisms regulate Cdx2 expression in the blastocyst and in trophoblast stem cells. Sci. Rep. 6, 27139 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freyer L., Schröter C., Saiz N., Schrode N., Nowotschin S., Martinez-Arias A., Hadjantonakis A.-K., A loss-of-function and H2B-Venus transcriptional reporter allele for Gata6 in mice. BMC Dev. Biol. 15, 38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng Y., Moore R., Tao W., Smith E. R., Tse J. D., Caslini C., Xu X.-X., GATA6 phosphorylation by Erk1/2 propels exit from pluripotency and commitment to primitive endoderm. Dev. Biol. 436, 55–65 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koutsourakis M., Langeveld A., Patient R., Beddington R., Grosveld F., The transcription factor GATA6 is essential for early extraembryonic development. Development 126, 723–732 (1999). [PubMed] [Google Scholar]
- 12.Muzumdar M. D., Tasic B., Miyamichi K., Li L., Luo L., A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Kan N. G., Stemmler M. P., Junghans D., Kanzler B., de Vries W. N., Dominis M., Kemler R., Gene replacement reveals a specific role for E-cadherin in the formation of a functional trophectoderm. Development 134, 31–41 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Ozguldez H. O., Fan R., Bedzhov I., Placental gene editing via trophectoderm-specific Tat-Cre/loxP recombination. Development 147, dev190371 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Plusa B., Piliszek A., Frankenberg S., Artus J., Hadjantonakis A.-K., Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development 135, 3081–3091 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paca A., Séguin C. A., Clements M., Ryczko M., Rossant J., Rodriguez T. A., Kunath T., BMP signaling induces visceral endoderm differentiation of XEN cells and parietal endoderm. Dev. Biol. 361, 90–102 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Schroter C., Rue P., Mackenzie J. P., Martinez Arias A., FGF/MAPK signaling sets the switching threshold of a bistable circuit controlling cell fate decisions in embryonic stem cells. Development 142, 4205–4216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filimonow K., Saiz N., Suwińska A., Wyszomirski T., Grabarek J. B., Ferretti E., Piliszek A., Plusa B., Maleszewski M., No evidence of involvement of E-cadherin in cell fate specification or the segregation of Epi and PrE in mouse blastocysts. PLOS ONE 14, e0212109 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strumpf D., Mao C.-A., Yamanaka Y., Ralston A., Chawengsaksophak K., Beck F., Rossant J., Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132, 2093–2102 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Ng R. K., Dean W., Dawson C., Lucifero D., Madeja Z., Reik W., Hemberger M., Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat. Cell Biol. 10, 1280–1290 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vestweber D., Kemler R., Identification of a putative cell adhesion domain of uvomorulin. EMBO J. 4, 3393–3398 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishioka N., Inoue K. I., Adachi K., Kiyonari H., Ota M., Ralston A., Yabuta N., Hirahara S., Stephenson R. O., Ogonuki N., Makita R., Kurihara H., Morin-Kensicki E. M., Nojima H., Rossant J., Nakao K., Niwa H., Sasaki H., The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell 16, 398–410 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Boussadia O., Kutsch S., Hierholzer A., Delmas V., Kemler R., E-cadherin is a survival factor for the lactating mouse mammary gland. Mech. Dev. 115, 53–62 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Reginensi A., Scott R. P., Gregorieff A., Bagherie-Lachidan M., Chung C., Lim D.-S., Pawson T., Wrana J., McNeill H., Yap- and Cdc42-dependent nephrogenesis and morphogenesis during mouse kidney development. PLOS Genet. 9, e1003380 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rayon T., Menchero S., Nieto A., Xenopoulos P., Crespo M., Cockburn K., Cañon S., Sasaki H., Hadjantonakis A.-K., de la Pompa J. L., Rossant J., Manzanares M., Notch and Hippo converge on Cdx2 to specify the trophectoderm lineage in the mouse blastocyst. Dev. Cell 30, 410–422 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nowotschin S., Xenopoulos P., Schrode N., Hadjantonakis A. K., A bright single-cell resolution live imaging reporter of Notch signaling in the mouse. BMC Dev. Biol. 13, 15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pijuan-Sala B., Griffiths J. A., Guibentif C., Hiscock T. W., Jawaid W., Calero-Nieto F. J., Mulas C., Ibarra-Soria X., Tyser R. C. V., Ho D. L. L., Reik W., Srinivas S., Simons B. D., Nichols J., Marioni J. C., Göttgens B., A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566, 490–495 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chazaud C., Yamanaka Y., Pawson T., Rossant J., Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev. Cell 10, 615–624 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Hou P., Li Y., Zhang X., Liu C., Guan J., Li H., Zhao T., Ye J., Yang W., Liu K., Ge J., Xu J., Zhang Q., Zhao Y., Deng H., Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science 341, 651–654 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Zhao Y., Zhao T., Guan J., Zhang X., Fu Y., Ye J., Zhu J., Meng G., Ge J., Yang S., Cheng L., du Y., Zhao C., Wang T., Su L., Yang W., Deng H., A XEN-like state bridges somatic cells to pluripotency during chemical reprogramming. Cell 163, 1678–1691 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Li X., Liu D., Ma Y., Du X., Jing J., Wang L., Xie B., Sun D., Sun S., Jin X., Zhang X., Zhao T., Guan J., Yi Z., Lai W., Zheng P., Huang Z., Chang Y., Chai Z., Xu J., Deng H., Direct reprogramming of fibroblasts via a chemically induced XEN-like state. Cell Stem Cell 21, 264–273.e7 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Li R., Zhong C., Yu Y., Liu H., Sakurai M., Yu L., Min Z., Shi L., Wei Y., Takahashi Y., Liao H.-K., Qiao J., Deng H., Nuñez-Delicado E., Esteban C. R., Wu J., Izpisua Belmonte J. C., Generation of blastocyst-like structures from mouse embryonic and adult cell cultures. Cell 179, 687–702.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rivron N. C., Frias-Aldeguer J., Vrij E. J., Boisset J.-C., Korving J., Vivié J., Truckenmüller R. K., van Oudenaarden A., van Blitterswijk C. A., Geijsen N., Blastocyst-like structures generated solely from stem cells. Nature 557, 106–111 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Sozen B., Cox A. L., De Jonghe J., Bao M., Hollfelder F., Glover D. M., Zernicka-Goetz M., Self-organization of mouse stem cells into an extended potential blastoid. Dev. Cell 51, 698–712.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Golding M. C., Generation of trophoblast stem cells. Methods Mol. Biol. 925, 49–59 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Himeno E., Tanaka S., Kunath T., Isolation and manipulation of mouse trophoblast stem cells. Curr. Protoc. Stem Cell Biol. Chapter 1, Unit 1E.4 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Hamilton T. G., Klinghoffer R. A., Corrin P. D., Soriano P., Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Mol. Cell. Biol. 23, 4013–4025 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Solter D., Knowles B. B., Immunosurgery of mouse blastocyst. Proc. Natl. Acad. Sci. U.S.A. 72, 5099–5102 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soneson C., Love M. I., Robinson M. D., Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Res 4, 1521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu G. C., Wang L. G., Han Y. Y., He Q. Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu G. C., Li F., Qin Y., Bo X., Wu Y., Wang S., GOSemSim: An R package for measuring semantic similarity among GO terms and gene products. Bioinformatics 26, 976–978 (2010). [DOI] [PubMed] [Google Scholar]
- 44.R. C. Team (R Foundation for Statistical Computing, 2019).
- 45.H. Wickham, ggplot2: Elegant Graphics for Data Analysis. (Springer-Verlag New York, 2016). [Google Scholar]
- 46.Gu Z., Eils R., Schlesner M., Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
- 47.H. Wickham, R. François, L. Henry, K. Müller, dplyr: A Grammar of Data Manipulation (2018); https://dplyr.tidyverse.org, https://github.com/tidyverse/dplyr.
- 48.L. Henry, H. Wickham, purrr: Functional Programming Tools (2022); http://purrr.tidyverse.org, https://github.com/tidyverse/purrr.
- 49.Smith T., Heger A., Sudbery I., UMI-tools: Modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 27, 491–499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Tables S1 to S3
Movies S1 and S2