Summary
Here we interrogate the factors responsible for SARS-CoV-2 breakthrough infections in a K18-hACE2 transgenic mouse model. We show that Delta and the closely related Kappa variant cause viral pneumonia and severe lung lesions in K18-hACE2 mice. Human COVID-19 mRNA post-vaccination sera after the 2nd dose are significantly less efficient in neutralizing Delta/Kappa than early 614G virus in vitro and in vivo. By 5 months post-vaccination, ≥50% of donors lack detectable neutralizing antibodies against Delta and Kappa and all mice receiving 5-month post-vaccination sera die after the lethal challenges. Although a 3rd vaccine dose can boost antibody neutralization against Delta in vitro and in vivo, the mean log neutralization titers against the latest Omicron subvariants are 1/3-1/2 of those against the original 614D virus. Our results suggest that enhanced virulence, greater immune evasion, and waning of vaccine-elicited protection account for SARS-CoV-2 variants caused breakthrough infections.
Subject areas: Immunology, Immune response, Virology
Graphical abstract
Highlights
-
•
COVID-19 vaccine-induced protection against SARS-CoV-2 variants wane after the 2nd dose
-
•
Delta and Kappa variants exhibit enhanced virulence in K18-hACE2 mice
-
•
Omicron subvariants show greater immune evasion than prior variants
Immunology; Immune response; Virology
Introduction
The COVID-19 pandemic caused by novel severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is one of the worst pandemics in history, with >585 million confirmed cases worldwide and >6 million deaths to date (https://covid19.who.int/). Since the first human infections were detected at the end of 2019, SARS-CoV-2 has acquired mutations in structural and nonstructural proteins via adaptation1,2,3 and has evolved into numerous different lineages (https://www.gisaid.org/). The World Health Organization (WHO) has classified emerging variants that cause global transmission and severe disease with enhanced resistance to neutralizing antibodies as a variant of concern (VOC), e.g. Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2) and the latest Omicron (B.1.1.529); while other variants that possess increased risks to regional public health are classified as a variant of interest (VOI), e.g. Kappa (B.1.617.1), Lambda (C.37) and Mu (B.1.621.1) (https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/)4
SARS-CoV-2 enters host cells via the surface glycoprotein spike which contains a receptor binding domain (RBD) with high affinity for human angiotensin converting enzyme 2 (hACE2).5,6,7 Amino acid substitutions in spike, especially those in RBD, may change virus infectivity and alter pathogenicity or viral antigenicity leading to immune escape.4,8,9,10 The signature substitution(s) in RBD as identified in early VOCs Alpha (N501Y), Beta (K417N/E484K/N501Y) and Gamma (K417T/E484K/N501Y), alone or in combination with other changes in spike, can reduce antibody neutralization and enable escape from convalescent or post-vaccination sera.3,4,11,12,13,14,15,16,17 Delta and the closely related Kappa variants contain different sets of mutations in the spike, including the signature substitutions of L452R/T478K (Delta) and L452R/E484Q (Kappa) in RBD.4,18 Positions 452, 478 and 484 are all located in the immunodominant antigenic site I.18,19 The L452R substitution not only increases virus infectivity but also confers neutralization resistance to monoclonal antibodies and convalescent/vaccine-elicited sera.10,18,20 The T478K substitution is an antibody escape mutation21 although it alone remains sensitive to neutralization by most monoclonal and polyclonal plasma antibodies.4,8,13,18,22,23,24 The Q or K substitution at E484 is a strong class 2 antibody escape mutation and particularly renders the sensitivity to polyclonal plasma antibodies.18,22,25
As of December 30, 2021, >9.3 billion doses of COVID-19 vaccines have been administered globally with nearly 50% of the world population being fully vaccinated (https://ourworldindata.org/covid-vaccinations?country=OWID_WRL), marking the biggest vaccination campaign in history. The two-dose mRNA vaccines developed by Pfizer and Moderna have each reportedly achieved ∼95% efficacy against symptomatic COVID-19 infections in persons of ≥16 years during the initial multinational placebo-controlled Phase III trials.26,27 Although having lower efficacy than mRNA vaccines, the one-dose Janssen adenoviral vector-based COVID-19 vaccine has also been reported >66% effective against severe to critical COVID-19 in the pivotal clinical trial.28 These vaccines are estimated to avert hundreds of thousands of severe COVID-19 cases including hospitalizations and deaths29,30,31 and their deployment has significantly curbed the surges in new infections in the US and other high-income countries.32,33,34 As Delta variant replaced Alpha, Beta and Gamma as the prevalent strain, reported vaccine breakthrough infections have increased.35,36,37 COVID-19 vaccine effectiveness (VE) against asymptomatic and symptomatic infections was reduced for Delta as compared to Alpha, though the protection against severe COVID-19 remained.38,39 Upon the emergence and circulation of the Omicron and its subvariants with greater immune evasion,40,41,42,43 the breakthrough infections are reportedly tripled compared to those caused by Delta at the peak in South Africa.44
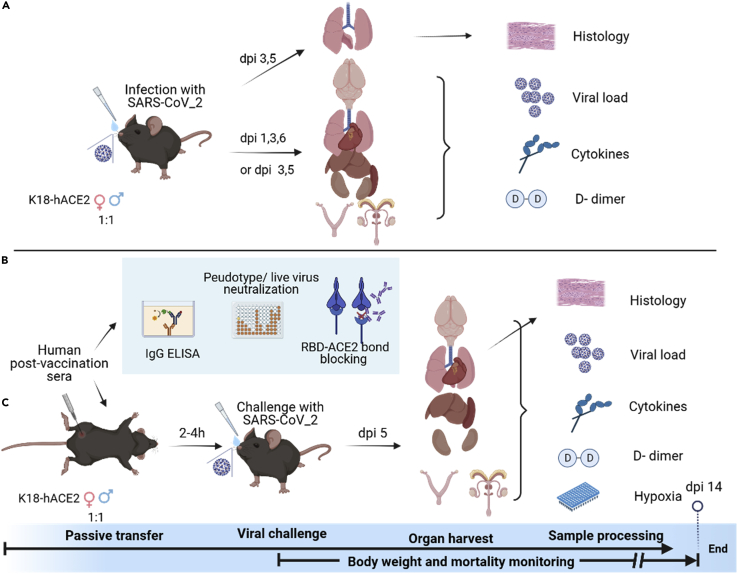
The K18-hACE2 transgenic mouse strain was originally developed by Paul B. McCray Jr. and Stanley Perlman at the University of Iowa for SARS-CoV-1 pathogenesis investigations.45 We and others have reported that K18-hACE2 mice are also highly susceptible to SARS-CoV-2 and produce many disease manifestations (e.g. acute lung injury, hypoxia, anosmia, abnormal blood clotting, severe systemic inflammation, morbidity, mortality, and so forth) that mimic severe COVID-19 in humans.46,47,48,49 Using the K18-hACE2 mouse strain as the passive transfer and challenge model for SARS-CoV-2 breakthrough infection, we interrogated the pathogenicity of Delta/Kappa and the protection of human COVID-19 post-vaccination (post-vac) sera as compared to New York-PV09158/2020 (NY (614G)) (a representative 614G clinical isolate from the early COVID-19 pandemic) in the present study. The schematic overview of the experimental design is shown in Figure 1. Our results suggest increased virulence, immune evasion, and waning of vaccine-induced protection likely account for breakthrough infections in humans.
Figure 1.
Schematic overview of in vivo and in vitro experiments
(A) Schematic diagram of experiments conducted on K18-hACE2 transgenic mice (female vs male, approximately 1:1 ratio) infected with SARS-CoV-2 New York-PV09158/2020 (NY (614G)) strain or Kappa or Delta variant. Lungs were collected on various days post-infection (dpi) for histopathology evaluation. Organs from separate sets of infected mice were harvested for tissue-specific viral loads, cytokine responses, and D-dimer deposition.
(B) Schematic drawing of in vitro characterization of human COVID-19 post-vaccination sera, including IgG titer by ELISA, neutralization capacity using pseudovirus or live infectious viruses, and blocking RBD-ACE2 binding using bio-layer interferometry (BLI).
(C) Schematic diagram of passive transfer experiments. Naive K18-hACE2 transgenic mice (female vs male, approximately 1:1 ratio) were injected intraperitonially with human COVID-19 post-vaccination sera followed by the lethal challenge of with SARS-CoV-2 614G strain or Kappa or Delta variant. Various organs were harvested on dpi 5 for tissue-specific viral loads, cytokines, and D-dimer measurements. Lungs from separate sets of mice were collected for histopathology and hypoxia PCR array.
Results
Heightened replication of Delta and Kappa variants and severe viral pneumonia in infected K18-hACE2 mice
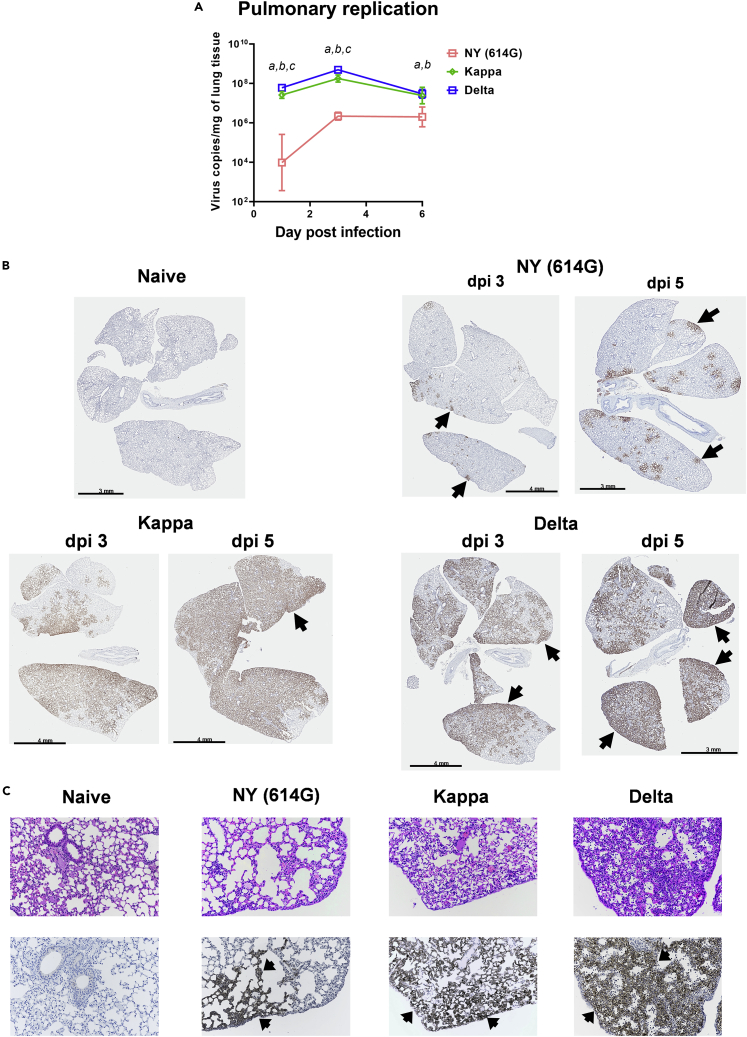
In K18-hACE2 transgenic mice of both sex (approximately 1:1 ratio), both Delta and Kappa variants given at 102 50% tissue culture infectious dose (TCID50) per mouse showed highly efficient pulmonary replication which peaked on 3 days post-infection (dpi) and maintained through dpi 6 (Figure 2A). In contrast, NY (614G) given at the same dose replicated much more slowly in mouse lungs (>2-log lower on dpi 1, p < 0.01) and did not reach the peak until dpi 6 which was still significantly lower than Delta or Kappa infection (Figure 2A). K18-hACE2 mice infected with NY (614G), Kappa or Delta variant also showed positive staining of viral antigen in lungs with the immunohistochemical (IHC) staining signal accumulating over time (Figure 2B). Consistent with RT-qPCR results (Figure 2A), NY (614G)-infected mice had considerably less antigen staining in lungs on both dpi 3 and 5 than those infected with the same dose of Kappa and Delta variants (Figure 2B). Kappa- and especially Delta-infected mouse lungs had widespread more diffuse alveolar viral antigen staining, primarily in alveolar Type I cells, in contrast to sporadic multiple alveolar foci of viral antigen staining in the mouse lungs infected with the same dose of NY (614G) (Figure 2C). Infection of Kappa and especially Delta variant also resulted in more extensive pulmonary infiltration of inflammatory cells (macrophages, lymphocytes, some neutrophils) than NY (614G) exposure at the same dose (Figure 2C).
Figure 2.
Lung histopathology staining after SARS-CoV-2 infection
K18-hACE2 mice were infected intranasally with live New York-PV09158/2020 (NY (614G)), Kappa or Delta variant at 102 TCID50/mouse.
(A) Pulmonary virus replication kinetics (geometric mean ± geometric SD of 4 mice/time point/group, female vs male at 1:1 ratio). Viral loads on 1, 3, and 6 days post-infection (dpi) were log transformed before two-way mixed ANOVA. a (Kappa vs NY (614G)), b (Delta vs NY (614G)), and c (Delta vs Kappa) indicate p < 0.05 at the same time point. Separate sets of infected mice were euthanized on dpi 3 and 5 and lungs were harvested for immunohistochemical (IHC) staining using rabbit polyclonal antibody specific for SARS-CoV-2 N/M/E. The same lung blocks were also sectioned for hematoxylin and eosin (H&E) staining. Representative images of lung pathology slides processed from 5 mice/virus/time point are shown.
(B). IHC of naïve mouse lung, or mouse lungs infected with NY (614G), Kappa or Delta variant on dpi 3 and 5.
(C) H&E and IHC staining (20X magnification) of the same lung sections from naïve mice or mice infected with NY (614G), Kappa or Delta variant on dpi 3. Brownish staining indicates positive SARS CoV2 antigen staining (arrows).
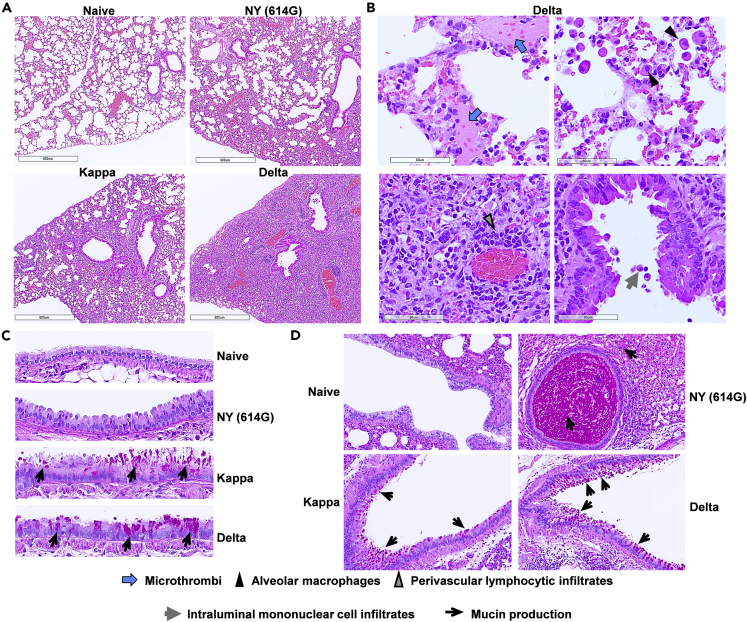
Previously we have determined that the Delta variant at the infectious dose of 102 TCID50/mouse is lethal to K18-hACE2 mice,50 while it requires NY (614G) at a 10-fold higher infectious dose (≥103 TCID50/mouse) to achieve 100% lethality.49,50 Thus, we used the lethal dose of 103 TCID50/mouse for NY (614G) and 102 TCID50/mouse for Kappa and Delta variants in the subsequent infection and passive transfer experiments, unless otherwise specified. K18-hACE2 mice inoculated with the lethal dose of NY (614G) exhibited gross pathological lesions in multiple organs (brain, lung, stomach, and intestines) (Figure S1), which was consistent with our early report.49 In contrast, mice infected with the lethal dose of Delta or Kappa variant showed more severe lung damage but lacked gastrointestinal lesions (Figure S1). Kappa and especially Delta variant caused more severe and extensive histological changes in lung parenchyma than NY (614G), including interstitial pneumonia (characterized by interstitial inflammatory cell infiltrates, interstitial edema, and perivascular lymphocytic infiltrates), vasculitis, alveolar accumulation of proteinaceous exudate (alveolar edema), bronchiolar cytomegaly, atypical alveolar Type II hyperplasia, and micro-thrombi-like lesions (Figures 3A, 3B and S2). Half of Kappa- or Delta-infected mice had micro-thrombi-like lesions present in lungs and a few had alveolar hemorrhage associated with the micro-thrombi (Figures S2C and S2D). Of note, Kappa and Delta-infected mice had mucus in bronchiolar and bronchial epithelial cells as revealed by PAS staining (Figures 3C and 3D). This was unlikely caused by the allergic reaction since no or minimum eosinophil infiltration was observed in these mouse lungs (Figure 3B). In contrast, NY (614G)-infected mouse lungs had intense PAS staining extracellularly in the lumen of larger bronchioles with the presence of neutrophils (Figure 3D) but lacked PAS staining within bronchiolar epithelial cells (Figure 3C), suggesting mucus was inhaled from the upper respiratory tract probably by aspiration or inhalation pneumonia as previously described in SARS-CoV-1 infected K18-hACE2 mice.51
Figure 3.
Histological changes in lung parenchyma of K18-hACE2 mice infected with SARS-CoV-2 and variants
K18-hACE2 mice were infected intranasally with 103 TCID50/mouse of New York-PV09158/2020 (NY (614G)), or 102 TCID50/mouse of Kappa and Delta variants. Whole lungs were harvested on 6 days post-infection. Lung tissues embedded in paraffin were sectioned for hematoxylin and eosin (H&E) staining and for Periodic acid-Schiff (PAS) staining for mucin. Uninfected mouse lungs were stained in parallel as negative controls. Representative lung H&E images (A and B) and PAS staining (c & d) of 6 mice/virus processed are shown.
(A) Low magnification (bar = 600 μm) of H&E stained sections show increased cellular infiltrates in NY (614G), Kappa and Delta-infected lung tissues compared to naïve lung.
(B) Higher magnification (bar = 60 μm) of H&E stained sections show micro-thrombi-like lesions (blue arrows), infiltrating alveolar macrophages (black triangles, upper right), perivascular lymphocytic infiltrates (gray triangles, lower left) and intraluminal mononuclear cell infiltrates in the bronchiole tube lined predominately with club cells (gray arrows, lower right).
(C) PAS staining (40X magnification) shows increased mucin (black arrows) in Kappa- and Delta-infected lungs as compared to naïve and NY (614G)-infected lungs.
(D) PAS staining (10X magnification) shows mucin secretion by goblet cells in Kappa- and Delta-infected mouse lungs vs extracellular mucin staining in the bronchiole lumen of NY (614G)-infected mice (black arrows).
Distinct tissue-specific replication patterns and comparable inflammatory profiles elicited by Delta and Kappa variants compared to NY (614G) strain
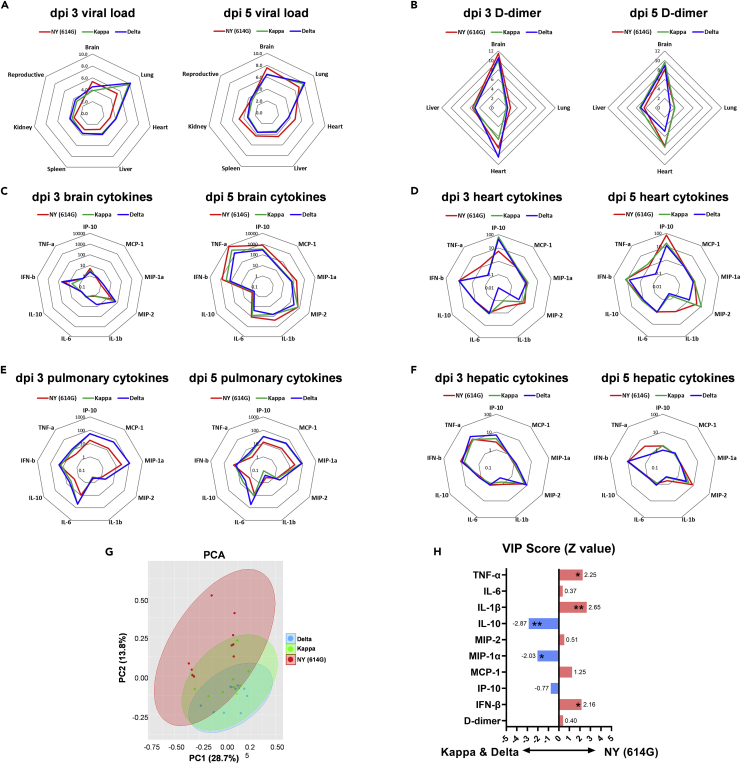
Delta and Kappa variants exhibited similar tissue-specific virus replication patterns in K18-hACE2 mice on both dpi 3 and 5, which lungs had the highest mean viral loads (8-log viral RNA copies/mg of tissue) and other organs including brain, liver, kidney, spleen, and reproductive tissues had lower viral loads (4-6 log viral RNA copies/mg of tissues on average) (Figure 4A and individual viral titers shown in Figure S3). In contrast, mice infected with a 10-fold higher dose of NY (614G) had higher mean viral loads in brains and lungs (approximately 5-log viral RNA copies/mg of tissue) than other organs on dpi 3 (Figure 4A). As NY (614G) infection progressed, the viral RNA copies detected in most vital organs increased with the highest mean viral loads detected in brains (approximately 8-log viral RNA copies/mg of tissue), followed by lungs (7-log viral RNA) and then hearts/livers/kidneys/spleens (approximately 5-log) on dpi 5 (Figure 4A and individual viral titers shown in Figure S3). Elevated D-dimer is a diagnostic marker for pulmonary embolism and venous/arterial thromboembolism in severe COVID-19.52 Interestingly, Delta and Kappa variant-infected K18-hACE2 mice exhibited tissue-specific D-dimer deposition patterns similar to those infected with NY (614G) that only differed in the magnitude (Figure 4B and individual D-dimer levels shown in Figure S4A). All three viruses induced D-dimer deposition in vital organs on dpi 3 with the highest D-dimer levels detected in brains (12-fold on average vs naive mice), followed by hearts (4-10-folds) and livers (approximately 6-fold) and the least D-dimer deposition in lungs (<2-fold) (Figure 4B). K18-hACE2 mice infected with a lethal dose of NY (614G), Kappa or Delta variant also exhibited overall similar cytokine profiles in brain, heart, lung, and liver homogenates (determined by ELISA) that differed in magnitude and limiting to a few cytokines (Figures 4C-4F and individual cytokine levels shown in Figures S4B-S4J). In general, both Kappa and Delta variants caused significantly higher tissue-specific inflammation than NY (614G) on dpi 3, including IP10 in hearts (Figure S4B), MCP-1 in brains (Figure S4C), MCP-1α in brains (Figure S4D) and IL-10 in lungs and livers (Figure S4H). This was consistent with that both Kappa and Delta had significantly higher tissue-specific replications than NY (614G) during early infection (Figure S3A). By dpi 5 when NY (614G) replication reached a similar or even higher level than those of Delta and Kappa in individual organs (Figure S3B), the differences in most tissue-specific cytokines diminished (Figures S4B-S4J).
Figure 4.
Tissue-specific viral loads, D-dimer and cytokine profiles after infections of SARS-CoV-2 and variants
K18-hACE2 mice of both sexes (1:1 ratio, n = 5 mice/group) were infected intranasally with 103 TCID50/mouse of New York-PV09158/2020 (NY (614G)), or 102 TCID50/mouse of Kappa and Delta variants. Viral RNA copies in various organs including reproductive organs (ovary or testis) on 3 and 5 days post-infection (dpi) were measured by RT-qPCR (Individual viral titers shown in Figure S4) and were log transformed. Tissue-specific D-dimer and cytokines were determined by ELISA (Individual D-dimer and cytokine levels shown in Figure S5) and were normalized over uninfected naïve mice.
(A) Mean viral loads in various organs (log).
(B) Tissue-specific D-dimer (fold vs naïve).
(C) Brain-specific cytokines (fold vs naïve).
(D) Heart-specific cytokines (fold vs naïve).
(E) Pulmonary cytokines (fold vs naïve).
(F) Hepatic cytokines (fold vs naïve).
(G) Multivariate principal component analysis (PCA) of tissue-specific D-dimer and cytokines after NY (614G), Kappa or Delta infection. Two most significant principal components (PC1 and PC2) are shown.
(H) The variable importance in projection (VIP) scores are shown after the logistic regression accounting for different tissues. ∗p < 0.05, ∗∗p < 0.01 by the Wald test.
We then performed Principal Component Analysis (PCA) on tissue-specific D-dimer and cytokines measured on both dpi 3 and 5 (Figure 4G). Through PCA dimensionality-reduction, the most significant two principal components PC1+PC2 that grouped infected mice according to the virus exposure revealed that the pathogenic patterns induced by NY (614G) did not overlap with those induced by Kappa or Delta especially (Figure 4G). The Variable Importance in Projection (VIP) scores, after adjusted for all four tissues analyzed, indicated that mice infected with Kappa and Delta variants tended to have overall significantly higher MIP-1α and IL-10 secretions than NY (614G)-infected mice (Figure 4H). In contrast, NY (614G) infection tended to induce overall significantly higher IL-1β, TNF-α, and IFN-β secretions in K18-hACE2 mice than Kappa or Delta infection (Figure 4H).
Reduced neutralization capacity by human COVID-19 post-vac sera against Delta and Kappa variants
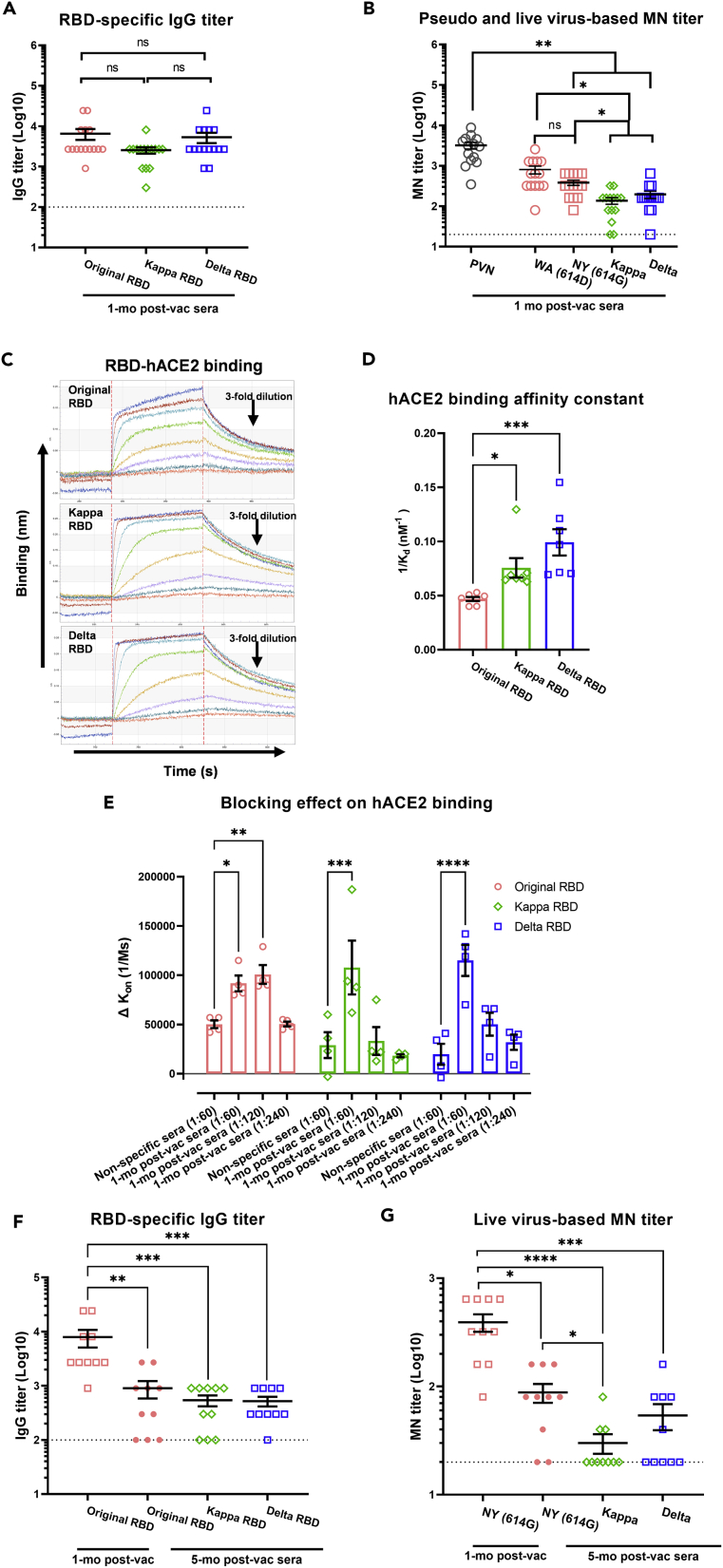
Human COVID-19 post-vac sera collected approximately 1 month (mo) after the 2nd dose Pfizer or Moderna mRNA vaccines exhibited similar ELISA IgG binding titers and IgG avidity in the presence of a chaotropic agent (urea) toward RBD derived from the original Wuhan-Hu-1 strain, Kappa and Delta variants (Figures 5A and S5). Compared to pseudovirus-based neutralization (PVN) titers using the spike of Wuhan-Hu-1 strain (bearing 614D), the 1-mo human post-vac sera exhibited >25% reduction in microneutralization (MN) titers against live wild type NY (614G), Kappa or Delta (p < 0.001 vs PVN, Figure 5B). The 1-mo human post-vac sera also showed significantly reduced MN titers toward live Kappa and Delta variants than toward NY (614G) (p < 0.05, Figure 5B). To test if COVID-19 vaccine-elicited antibodies could block RBD-hACE2 interaction, we first measured RBD binding to hACE2 immobilized to streptavidin biosensors interaction using biolayer interferometry (BLI) (Figure 5C). Both Delta RBD and Kappa RBD showed strong binding to immobilized hACE2 (Figure 5D). We then pre-incubated recombinant RBD with pooled 1-mo human post-vac sera before applying it to hACE2-loaded BLI biosensors (Figure 5E). Compared to the inhibition on original RBD, the capacity of 1-mo human post-vac sera to block Delta RBD-hACE2 or Kappa RBD-hACE2 interaction was quickly diluted out at 1:120 dilution (Figure 5E). This suggests that vaccine-elicited antibodies have reduced inhibition of Delta/Kappa RBD. Of the initial 14 donors that had provided 1-mo post-vac sera, ten had returned to provide 5-month post-vac sera which had significantly reduced RBD-specific IgG binding titers and live virus-based MN titers against NY (614G), Kappa or Delta variant, respectively (Figures 5F and 5G). Moreover, ≥50% of returned donors had undetectable MN titers against live Kappa or Delta variant 5 months after the receipt of the 2nd vaccine dose (Figure 5G). Due to a small sample size, no analysis of biological variables such as biological sex was performed.
Figure 5.
Human COVID-19 post-vaccination antibody responses to Kappa and Delta variants
Human post-vaccination (post-vac) sera were collected from 14 adult volunteers (6 with Pfizer and 8 with Moderna) at approximately 1 month (mo) after the receipt of 2nd dose mRNA vaccines. Ten of the original donors (4 with Pfizer and 6 with Moderna) have provided additional sera at approximately 5-month after the 2nd dose. The receptor binding domain (RBD)-specific IgG titers were determined by ELISA. Pseudovirus-based neutralization (PVN) titers against original Wuhan-Hu-1 strain, or microneutralization (MN) titers against live USA-WA1/2020 (WA (614D)), New York-PV09158/2020 (NY (614G)), Kappa and Delta variants were also determined. The 1-mo post-vaccination sera were also pooled and tested for blocking human ACE2 (hACE2)-RBD binding by bio-layer interferometry (BLI).
(A) RBD-specific IgG and (B) neutralizing antibody titers of 1-mo human post-vac sera (mean ± s.e.m., n = 14 donors/group).
(C) Representative BLI sensorgrams of RBD derived from original Wuhan-Hu-1 strain, Kappa and Delta variants binding to immobilized monomeric hACE2.
(D) Monomeric hACE2-RBD binding affinity constant (mean ± s.e.m., n = 7 replicates/group).
(E) The blocking effect of pooled 1-mo human post-vac sera on hACE2-RBD binding (mean ± s.e.m., n = 4 replicates/group)
(F) RBD-specific IgG and (G) live virus-based MN titers of the same donors at 1-mo and 5-month after the 2nd dose (mean ± s.e.m., n = 10 donors/group). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 by One-way ANOVA with nonparametric test. IgG or MN titers were log transformed for statistical analysis. Dashed lines indicate the lowest serum dilutions tested. ns: not significant.
Reduced protection against Delta and Kappa variants in the mouse passive transfer model
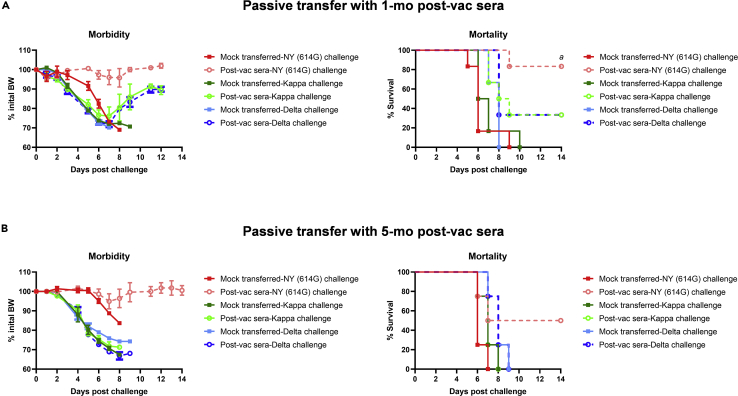
We next conducted passive transfer experiments in K18-hACE2 mice with pooled human post-vac sera. Following a lethal challenge with NY (614G) at 103 TCID50/mouse,49,50 mock-transferred K18-hACE2 mice all died within 9 dpi after losing substantial body weight (BW) (Figure 6A). In contrast, K18-hACE2 mice receiving 1-mo pooled human post-vac sera showed no obvious BW loss with 83% survival after the lethal NY (614G) challenge (Figure 6A). The post-vac sera recipient mice surviving the lethal NY (614G) challenge showed minimum lung lesions on dpi 15 (Table 1). When mock-transferred K18-hACE2 mice were challenged with a lethal dose of Kappa or Delta at 102 TCID50/mouse, they suffered not only significant morbidity but also 100% mortality (Figure 6A). Mice receiving 1-mo human post-vac sera also dropped significant BW with 33% survival after the lethal Kappa or Delta challenge (Figure 6A). When K18-hACE2 mice were given 5-month pooled human post-vac sera, protection against NY (614G), Kappa or Delta variant all decreased, resulting in 50% mortality for NY (614G)-challenged mice and 100% mortality for Kappa- and Delta-challenged mice, respectively (Figure 6B). These results suggest that both Kappa and Delta variants are more resistant to human COVID-19 post-vac sera than NY (614G), and the neutralization and protective capacity of human COVID-19 mRNA post-vac sera waned substantially in recipient mice by 5 months after the 2nd dose.
Figure 6.
Morbidity and mortality of K18-hACE2 mice receiving human post-vaccination sera after lethal challenges of SARS-CoV-2 and variants
K18-hACE2 mice of both sexes (1:1 ratio) were injected intraperitoneally with 200 μL/mouse of PBS (mock-transferred) or pooled human post-vaccination (post-vac) sera collected at 1 month (mo) or 5 months after the 2nd dose of COVID-19 mRNA vaccines (post-vac sera transferred). Recipient mice were then challenged intranasally with 103 TCID50/mouse of New York-PV09158/2020 (NY (614G)), or 102 TCID50/mouse of Kappa and Delta variants. Morbidity and mortality of infected mice were monitored for up to 14 days post-challenge. % body weight drops (mean ± s.e.m.) and % cumulative survivals were determined.
(A) Morbidity and mortality of mice receiving 1-mo post-vac sera (n = 6 mice/group).
(B) Morbidity and mortality of mice receiving 5-month post-vac sera (n = 4 mice/group). a indicates p < 0.05 vs mock-transferred group with NY (614G) challenge by Log rank (Mantel-Cox) survival test.
Table 1.
Lung histopathology of K18-hACE2 mice infected with SARS-CoV2 Delta or Kappa variant∗
| Score category# | Uninfected naive (n = 2 mice) | NY (614G) |
Kappa |
Delta |
|||
|---|---|---|---|---|---|---|---|
| Mock (n = 5 mice) | Post-vac sera (n = 6 mice) | Mock (n = 6 mice) | Post-vac sera (n = 4 mice) | Mock (n = 6 mice) | Post-vac sera (n = 4 mice) | ||
| H&E staining | |||||||
| Total lung score | 0 ± 0 | 2.2 ± 0.2 | 0.8 ± 0.2a | 3.5 ± 0.2b | 3.3 ± 0.3 | 3.3 ± 0.3b | 3.0 ± 0.0 |
| Interstitial pneumonia | 0 ± 0 | 1.2 ± 0.5 | 0.3 ± 0.2 | 3.3 ± 0.2b | 3.3 ± 0.3 | 3.2 ± 0.4b | 3.0 ± 0.0 |
| Alveolar type II cell hyperplasia | 0 ± 0 | 0.2 ± 0.2 | 0 ± 0 | 1.0 ± 0.4 | 1.3 ± 0.3 | 1.2 ± 0.4 | 2.3 ± 0.3 |
| Bronchiolar cytomegaly | 0 ± 0 | 0.4 ± 0.4 | 0.3 ± 0.2 | 2.7 ± 0.2b | 2.3 ± 0.3 | 1.7 ± 0.2b | 2.0 ± 0.0 |
| Vasculitis/perivascular inflammation | 0 ± 0 | 0.8 ± 0.4 | 0.7 ± 0.2 | 2.0 ± 0.0b | 2.0 ± 0.0 | 2.2 ± 0.2b | 2.3 ± 0.3 |
| Alveolar edema | 0 ± 0 | 0.4 ± 0.4 | 0 ± 0 | 0.5 ± 0.3 | 0.5 ± 0.3 | 0.2 ± 0.2 | 0.3 ± 0.3 |
| Micro-thrombi like lesions | 0 ± 0 | 0.6 ± 0.2 | 0.3 ± 0.2 | 0.7 ± 0.3 | 0.3 ± 0.3 | 0.8 ± 0.4 | 0.8 ± 0.3 |
| Eosinophils | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0.2 ± 0.2 | 0 ± 0 | 0 ± 0 | 0.3 ± 0.3 |
| PAS staining | |||||||
| Mucin in large bronchioles | 0 ± 0 | 0.8 ± 0.5 | 1.0 ± 0.3 | 2.0 ± 0.3b | 2.0 ± 0.4 | 1.5 ± 0.2 | 1.8 ± 0.6 |
| Mucin in small bronchioles | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0.3 ± 0.2 | 0.3 ± 0.3 | 0 ± 0 | 0.5 ± 0.6 |
| IHC staining | |||||||
| SARS CoV2 Viral antigen | 0 ± 0 | 2.0 ± 0.4 | 0 ± 0a | 3.8 ± 0.2b | 3.8 ± 0.3b | 3.3 ± 0.3b | 3.3 ± 0.5 |
| Sum of scores | 0 | 7.8 | 2.5 | 19.3 | 18.8 | 17.8 | 21 |
∗K18-hACE2 mice of both sexes (1:1 ratio) were injected intraperitoneally with 200 μL/mouse of PBS (mock-transferred) or pooled human post-vaccination (post-vac) sera collected at 1-month after the 2nd dose of COVID-19 mRNA vaccines. Recipient mice were then challenged intranasally with 103 TCID50/mouse of New York-PV09158/2020 (NY (614G)), or 102 TCID50/mouse of Kappa and Delta variants. Most mice reached the moribund stage on 6-7 days post-challenge and were humanely euthanized for lung harvest. Post-vac sera transferred mice inoculated with NY (614G) survived the challenged and were euthanized 15 days post-challenge. Whole lungs were embedded in paraffin and sectioned for hematoxylin and eosin (H&E) staining and immunohistochemistry (IHC) staining using in-house raised rabbit polyclonal antibody specific for SARS-CoV-2 N/M/E. Sectioned lung tissues were also stained with Periodic acid-Schiff (PAS) for mucin. Uninfected mouse lungs were stained in parallel as negative controls.
# Histopathology lesion score was determined in a blind manner by a board-certified veterinary pathologist based on the following categories: (1) Total lung H&E and IHC scoring: 0, no lesion; 1, 1-25%; 2, 26-50%; 3, 51-75%; 4, 76-100% of the lung involved with lesions; (2) Individual lesion and PAS scoring: 0, no lesions or normal; 1, minimal; 2, mild; 3, moderate; 4, severe. Accumulative scores of each group are reported as mean ± SEM.
Indicate p < 0.05 vs mock-transferred mice challenged with NY (614G) by two-way ANOVA test.
Indicate p < 0.05 vs mock-transferred mice challenged with NY (614G) by two-way ANOVA test.
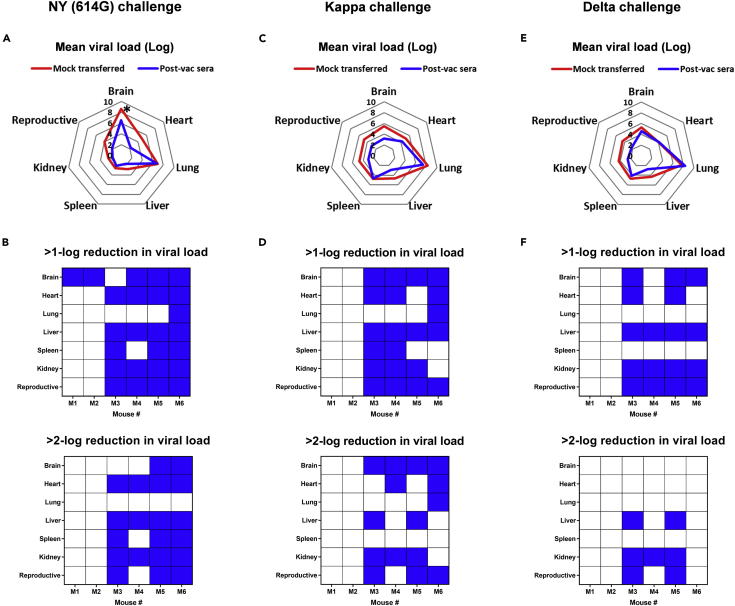
Tissue-specific viral loads reduced by human COVID-19 post-vac sera following SARS-CoV-2 variant challenges
K18-hACE2 mice receiving 1-mo pooled human COVID-19 post-vac sera exhibited reduced mean viral loads (log) in the brain (p < 0.05 vs mock-transferred), heart, liver, kidney, and reproductive organs on dpi 5 following the NY (614G) challenge (Figures 7A and S6A). Compared to mock-transferred mice, all 6 mice receiving post-vac sera had at least 1 organ showing >1-log lower viral titers, including 3 recipient mice with 5-6 organs showing >2-log reduction in viral loads after the NY (614G) challenge (Figure 7B). The receipt of post-vac sera also reduced mean viral loads in brain, liver, kidney, and reproductive organs of 4 out of 6 mice after Kappa or Delta challenge (Figures 7C-7F, S6B and S6C). However, lung was one of the two organs (the other was spleen) in Kappa- or Delta-challenged mice that had the least viral load reductions after post-vac sera transfer (Figures 7C-7F). Many lung lesions (i.e., alveolar type II cell hyperplasia and mucin secretion in large or small bronchioles) were not alleviated by post-vac sera transfer after Kappa or Delta challenge (Table 1). These results are consistent with the live virus-based MN titers, suggesting that human COVID-19 post-vac sera are less efficient at blocking tissue-specific virus replication in K18-hACE2 mice challenged with Kappa or Delta variant than NY (614G).
Figure 7.
Effects of human COVID-19 post-vaccination sera on tissue-specific viral burden of K18-hACE2 mice challenged with SARS-CoV-2 and variants
K18-hACE2 mice of both sexes (1:1 ratio, n = 6 mice/group) were injected intraperitoneally with 200 μL/mouse of PBS (mock-transferred) or pooled human post-vaccination (post-vac) sera collected at 1-month after the 2nd dose of COVID-19 mRNA vaccines (post-vac sera transferred). Recipient mice were then challenged intranasally with 103 TCID50/mouse of New York-PV09158/2020 (NY (614G)), or 102 TCID50/mouse of Kappa and Delta variants. Viral RNA copies in brain, heart, lung, liver, spleen, kidney, and reproductive organs (ovary or testis) on 5 days post-infection (dpi) were measured by RT-qPCR (Individual viral titers shown in Figure S6).
(A, C, and E) Mean viral loads (log) in various organs.
(B, D, and F) Post-vac sera transferred mice that showed >1-log or >2-log reduction in tissue-specific viral loads as compared to mock-transferred are highlighted in blue. ∗p < 0.05 vs mock-transferred by Mann-Whitney test.
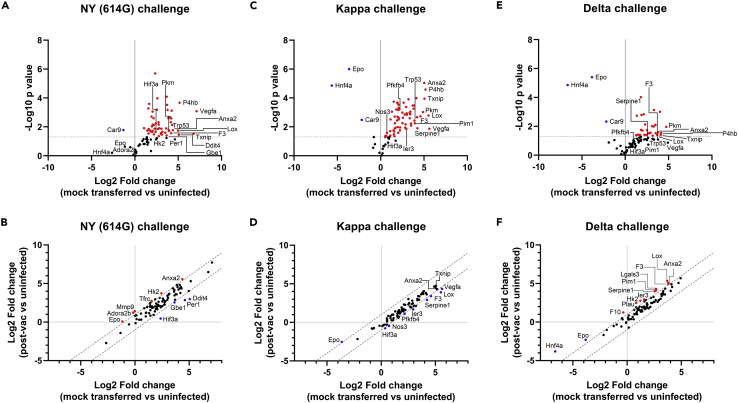
Downregulation of pulmonary hypoxia signaling by human COVID-19 post-vac sera following SARS-CoV-2 variant challenges
Exposure to NY (614G), Kappa, or Delta variant resulted in extensive activation of the pulmonary hypoxia signaling pathway in K18-hACE2 mice on dpi 5 (Figures 8A, 8C, and 8E). Many activated hypoxic genes (e.g. Anxa2, Lox, Pkm, P4hb, Txnip, and Car9) were shared between NY (614G)-infected mice and those infected with Kappa or Delta variant and only differed in the fold changes relative to uninfected naive controls (Figures 8A, 8C, and 8E). Some hypoxic genes activated by NY (614G) or Kappa variant were significantly downregulated in mice transferred with the 1-mo pooled human COVID-19 post-vac sera (Figures 8B and 8D), while most hypoxic genes especially those active by Delta remained unchanged compared to those without post-vac sera transferred (Figure 8F). Nevertheless, infection of NY (614G) or Delta/Kappa variants resulted in similar hypoxic gene activation in the lungs of K18-hACE2 mice and the receipt of 1-mo post-vac sera had limited effects in suppressing pulmonary hypoxia signaling pathway.
Figure 8.
The hypoxia pathway transcriptional gene expression in K18-hACE2 mice challenged with SARS-CoV-2 and variants
K18-hACE2 mice of both sexes (1:1 ratio) were injected intraperitoneally with 200 μL/mouse of PBS (mock-transferred) or pooled human post-vaccination sera collected at 1-month after the 2nd dose of COVID-19 mRNA vaccines (post-vac sera transferred). Recipient mice were then challenged intranasally with 103 TCID50/mouse of New York-PV09158/2020 (NY (614G)), or 102 TCID50/mouse of Kappa and Delta variants. Hypoxia signaling pathway PCR array was performed using RNA extracted from lung homogenates of infected mice (n = 6 mice/group, female vs male at 1:1 ratio) 5 days post-infection (dpi). Gene regulation as fold changes over uninfected naïve mice (n = 3 mice/group including 2 females and 1 male) was used to construct the volcano plots (A, C, and E) or correlation plots (B, D, and F). The dashed lines indicate the thresholds of p value < 0.05. Selected genes that show a significant rise (red) or fall (blue) are highlighted.
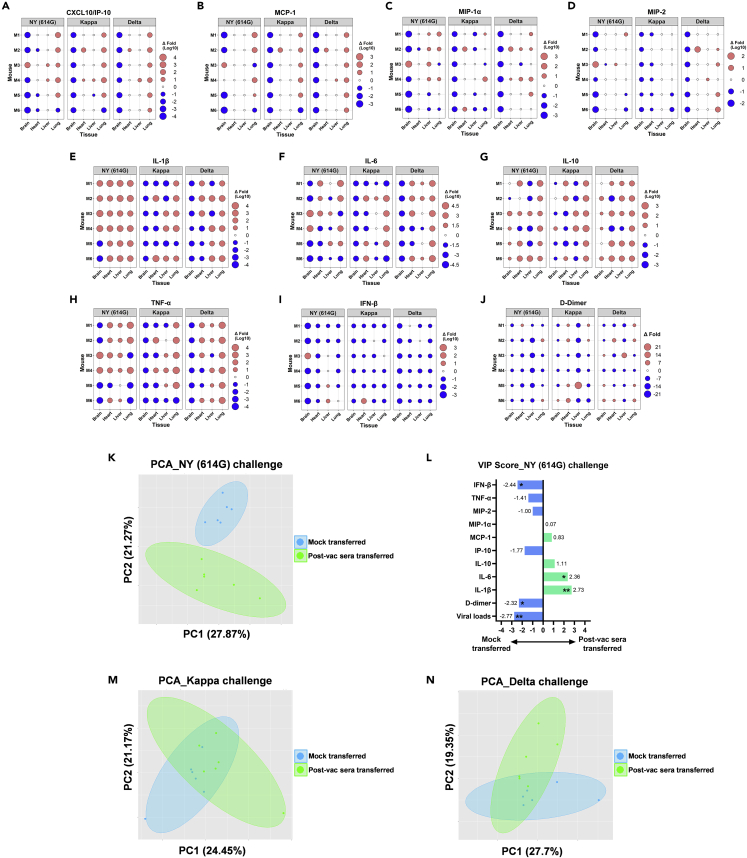
Effects of human COVID-19 post-vac sera on tissue-specific inflammation and in vivo protection against SARS-CoV-2 variant challenges
Compared to mock-transferred mice, K18-hACE2 mice receiving 1-mo pooled human COVID-19 post-vac sera exhibited notably reduced chemokine secretions in brain tissues (e.g., CXCL10/IP-10, MCP-1, MIP-1α, and MIP2) on dpi 5 following the challenge of NY (614G), Kappa or Delta variant (Figures 9A-9D and S7A-S7L). In other organs (heart, liver, and lung), virus-induced chemokine secretions remained largely unchanged after the post-vac sera transfer (Figures 9A-9D and S7A-S7L). Most post-vac sera transferred mice also had reduced IFN-β secretion (Figures 9I and S8J-S8L) and D-dimer deposition (Figure 9J and S7M-S7O) in vital organs, regardless of challenge virus. Compared to mock-transferred mice, post-vac sera transferred mice after NY (614G) challenge had significantly elevated tissue-specific IL-1β secretion, especially in heart, liver, and lung (Figure 9E and S8A). In Kappa- or Delta-challenged mice, pulmonary IL-1β levels were also significantly increased (p < 0.05 or 0.01 vs mock-transferred) (Figures 9E, S8B and S8C). Other cytokines (e.g. IL-6, TNF-α or IL-10) in post-vac sera transferred mice trended low or high depending on specific tissue or challenge virus (Figures 9F-9H, S8E-S8I, S8N, and S8O).
Figure 9.
Effects of human COVID-19 post-vaccination sera on tissue-specific cytokines and D-dimer deposition of K18-hACE2 mice challenged with SARS-CoV-2 and variants
K18-hACE2 mice of both sexes (1:1 ratio, n = 6 mice/group) were injected intraperitoneally with 200 μL/mouse of PBS (mock-transferred) or pooled human post-vaccination (post-vac) sera collected at 1-month after the 2nd dose of COVID-19 mRNA vaccines (post-vac sera transferred). Recipient mice were then challenged intranasally with 103 TCID50/mouse of New York-PV09158/2020 (NY (614G)), or 102 TCID50/mouse of Kappa and Delta variants. D-dimer and cytokines in brain, heart, lung, and liver homogenates harvested on 5 days post-infection were measured (Individual D-dimer and cytokine levels shown in Figures S7 and S8). Fold changes in tissue-specific cytokine and D-dimer levels of post-vac sera transferred mice vs mock-transferred mice are shown, including (A) CXCL10/IP-10, (B) MCP-1, (C)CMIP-1α, (D) MIP-2, (E) IL-1β, (F) IL-6, (G) IL-10, (H) TNF-α, (I) IFN-β and (J) D-dimer. Tissue-specific viral loads, cytokines, and D-dimer of post-vac sera transferred mice and mock-transferred mice were also subjected to multivariate principal component analysis (PCA).
(K, M, and N) The most significant two principal components (PC1 and PC2) are shown for NY (614G) (K), Kappa (M) or Delta challenge (N), respectively. Only in NY (614G) challenged mice, those receiving post-vac sera were fully separated from mock-transferred (K).
(L) The variable importance in projection (VIP) scores for NY (614G) challenge is shown after the logistic regression accounting for different tissues. ∗p < 0.05, ∗∗p < 0.01 by the Wald test.
To better comprehend these data, we then performed PCA on tissue-specific viral loads, D-dimer, and cytokines with and without the post-vac transfer (Figures 9K, 9M, and 9N). In NY (614G)-challenged mice, the most dominant PC1+PC2 completely separated those receiving post-vac sera from mock-transferred mice (Figure 9K). After adjusting for different tissues, mock-transferred mice tended to have significantly higher viral loads, D-dimer deposition, and IFN-β secretion than the post-vac sera transferred mice after the NY (614G) challenge (Figure 9L). In contrast, the post-vac sera transferred mice after the NY (614G) challenge more likely developed significantly higher IL-1β and IL-6 levels (Figure 9L). Neither Kappa- nor Delta-challenged mice were completely separated according to their receipt of the post-vac sera transfer (Figures 9M and 9N), suggesting no distinct patterns extracted by PCA via dimensionality-reduction.
Neutralization of human COVID-19 post-vac sera after the 3rd dose of mRNA vaccines
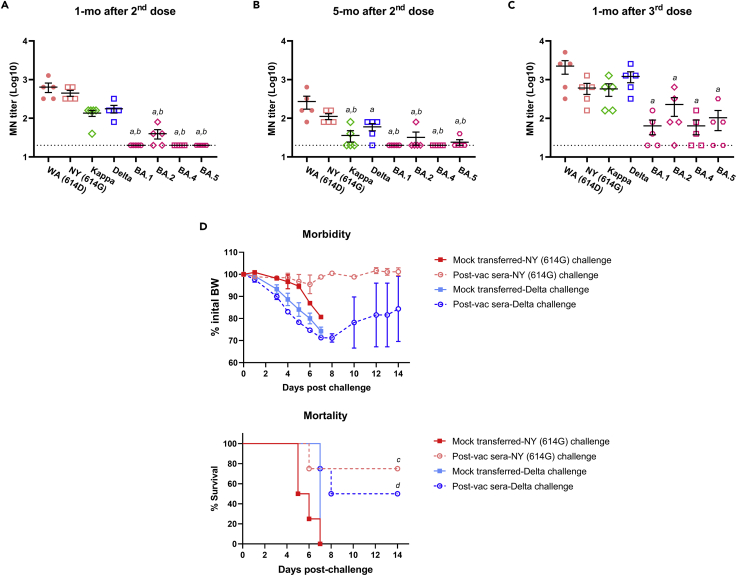
During the composition of this article, the 3rd dose of COVID-19 mRNA vaccines was approved for persons of ≥12-17 years (Pfizer) or ≥18 years (Moderna) at least 5 mo after completing the initial two-dose vaccination regimen. Five of the original donors provided post-vac sera at 1-mo after receiving the 3rd dose of mRNA vaccines (2 Pfizer and 3 Moderna). Regardless testing virus, all five donors showed markedly enhanced neutralizing antibody titers after receiving the booster dose of mRNA vaccines compared to 1-mo and 5-mo after finishing the 2nd dose (Figures 10A-10C). Of note, post-vac sera from 1-mo and 5-mo after the 2nd dose of mRNA vaccines had barely detectable MN titers against emerging Omicron subvariants (BA.1, BA.2, BA.4 and BA.5) (Figures 10A and 10B). Although a 3rd mRNA vaccine dose boosted neutralizing antibodies against Omicron subvariants, the mean log MN titers were still 1/3-1/2 of those against the original WA (614D) virus (Figure 10C). Similarly, 1-mo convalescent sera from the three donors that suffered the Delta breakthrough infection after being fully vaccinated also had lower MN titers against Omicron subvariants as compared to the titers against the original WA (614D) virus (Figure S9). These results indicate that Omicron subvariants exhibit great immune escape from vaccination or infection elicited pre-existing immunity.
Figure 10.
In vitro and in vivo neutralization of human COVID-19 post-vaccination sera after the booster dose
Five healthy donors (3 with Moderna and 2 with Pfizer) provided post-vaccination (post-vac) sera at approximately 1 month (mo) and 5-mo after the receipt of the 2nd dose and 1-mo after the 3rd dose (booster) of mRNA vaccines. The microneutralization (MN) titers against live USA-WA1/2020 (WA (614D)), New York-PV09158/2020 (NY (614G)), Kappa, Delta, and emerging Omicron subvariants (BA.1, BA.2, BA.4 and BA.5) were determined.
(A-C) MN titers of the same donors at 1-mo and 5-mo after 2nd dose and 1-mo after 3rd dose of mRNA vaccines (mean ± s.e.m., n = 5 donors/group). MN titers were log transformed before One-way ANOVA with nonparametric test. (A and B) indicate p < 0.05 vs WA (614D) and NY (614G), respectively. Dashed lines indicate the lowest serum dilutions tested. ns: not significant. The 1-mo post-vac sera after the booster dose were also pooled and were injected intraperitoneally at 200 μL/mouse into naïve K18-hACE2 mice of both sexes (1:1 ratio, n = 4 mice/group). Mice receiving 200 μL/mouse of PBS (mock-transferred) served as the controls. Recipient mice were then challenged intranasally with 103 TCID50/mouse of NY (614G), or 102 TCID50/mouse of Delta variant. Morbidity and mortality of infected mice were monitored for up to 14 days post-challenge. % body weight drops (mean ± s.e.m.) and % cumulative survivals were determined.
(D) Morbidity and mortality of mice receiving post-vac sera from 1-mo booster. (C and D) indicate p < 0.05 vs mock-transferred group with the same challenge by Log rank (Mantel-Cox) survival test.
In the K18-hACE2 passive transfer model, mice receiving 1-mo booster post-vac sera showed no obvious BW loss and had 75% survival after the lethal NY (614G) challenge (Figure 10D). Fifty percent of recipient mice transferred with 1-mo booster post-vac sera also survived the lethal Delta challenge despite initial substantial BW drop (Figure 10D). In contrast, all mock-transferred mice died within 7 dpi following the NY (614G) or Delta challenge (Figure 10D). These results suggest that a 3rd mRNA vaccine dose can boost antibody neutralization and protection against NY (614G) and Delta variant.
In K18-hACE2 mice infected with 103 TCID50/mouse of Omicron BA.1, the highest viral loads were detected in lungs (∼7-log viral RNA copies/mg of tissues) on dpi 3 and 5, while extrapulmonary organs had low viral titers (1-3 log viral RNA copies/mg of tissues on average) that were almost undetectable by dpi 5 (Figure S10A). In general, Omicron infection caused mild lung lesions and infected mice developed no morbidity nor mortality (Figures S10B and S10C), which is consistent with the literature report.53 Thus, we did not perform the passive and challenge experiment for Omicron because of attenuated lung infection and a lack of lethality in K18-hACE2 mice.
Discussion
In this study, we showed that Delta/Kappa-induced pathogenic patterns in K18-hACE2 mice are distinct from those caused by NY (614G) representing the strains widespread in the early COVID-19 pandemic. Mice infected with Delta and Kappa variants had higher pulmonary viral loads but developed no gastrointestinal lesions compared to those infected with NY (614G). This also contrasts with early VOC Alpha and Beta that cause severe blood clot formation in vital organs and severe systemic pathology.49 The different viral pathogenicity is attributed to the unique cumulative mutations acquired in the surface spike and internal viral proteins of Delta/Kappa.18,54,55 For instance, the L452R mutation harbored by Delta/Kappa enhances RBD-ACE2 binding and spike stability and promotes viral infectivity and fusogenicity resulting in increased in vitro virus replication.10,56 In K18-hACE2 transgenic mice, Delta/Kappa replicate more efficiently in the lung than NY (614G) resulting in extensive lung lesions, including severe interstitial pneumonia, massive immune cell infiltration, vasculitis, edema, type 2 pneumocyte hyperplasia, alveolar capillary micro-thrombi, and so forth. A few groups have also observed micro-thrombi-like lesions in SARS-CoV-2 infected K18-hACE2 mice.46,57 Similar lesions have been identified in autopsied patients with COVID-19.58 Of note, we observed that Delta- or Kappa-infected mice had increased mucus accumulation in the bronchiolar and bronchial epithelial cells with abundant viral antigens present in the lung, consistent with viral pneumonia. Large mucus plugs were also found in the upper lung airways of NY (614G)-infected mice, indicating aspiration or inhalation pneumonia. Aspiration pneumonia has been observed in SARS-CoV-1-infected K18-hACE2 mice.51 Delta- or Kappa-infected mice had mucus clogged larger airways with probably reduced airflow in the lung, which can lead to shortness of breath. Similarly, mucus plugs in smaller airways could also cause collapsed alveoli resulting in reduced pulmonary oxygen levels. Hypoxia is one of the early clinical signs of severe COVID-19 and many hospitalized patients require supplemental oxygen and/or ventilation.59 Following SARS-CoV-2 exposures, K18-hACE2 mice exhibited extensive activation of pulmonary hypoxic genes before succumbing to death.49 In view of overall disease pathology developed, our results suggest that viral pneumonia may be the root of cause for Delta- and Kappa-induced lethality in K18-hACE2 mice.
The current study also confirms that human post-vac sera collected at 1-mo after the 2nd mRNA COVID-19 vaccine dose are effective against NY (614G) strain in vitro and in vivo. Recipient K18-hACE2 mice had overall reduced viral loads in the vital organs and decreased inflammation and tissue damage after the lethal NY (614G) challenge. However, we were surprised to observe elevated systemic IL-1β in mice treated with post-vac sera transfer following NY (614G) challenge. Unlike IL-6 and TNF-α levels that correlate with COVID severity and death,49,60 IL-1β is not a signature proinflammatory cytokine aggravated in SARS-CoV-2 infections. Given that most recipient mice survived the lethal NY (614G) challenge without severe lung lesions, further investigations are warranted to elucidate the role of IL-1β in resolving COVID-19.
Under the same passive transfer condition, the 1-mo human post-vac sera showed substantially reduced in vivo efficacy against Delta and Kappa variants, which is consistent with in vitro MN data that Delta and Kappa variants are more resistant to current mRNA vaccines-elicited sera than NY (614G). Both L452R and E484Q substitutions alone confer great resistance to neutralization by monoclonal antibodies and polyclonal plasma antibodies elicited by infection and vaccination. Additionally, the L452R/T478K (Delta) or L452R/E484Q (Kappa) double substitution is reported to strengthen the RBD-hACE2 binding,13,61 which also makes Delta and Kappa less susceptible to antibody neutralization.3,13 Other mutations in the N-terminal domain (NTD) of Delta or Kappa spike S1 subunit, such as T19R, G142D, E156G and Δ157-158 are known to reshape antigenic sites in NTD and to provide additional glycan shield from neutralizing antibodies, resulting in enhanced resistance to convalescent and post-vac sera than previous VOCs.18,61,62,63 Substantially reduced VE against Delta variant has been reported in adolescents and adults ≥3 mo after the receipt of the 2nd COVID-19 vaccine dose.64,65,66,67 An Israeli study has also reported that the viral loads in adults with Delta breakthrough infections increase as the effectiveness of mRNA vaccine gradually declines.68 The results of our passive transfer study corroborate these epidemiological reports that vaccine-elicited protection wane over time leading to breakthrough infections during the Delta-predominant period.
In January 2022, the US CDC recommended a 3rd dose of Pfizer or Moderna mRNA vaccine for all adults 5 mo after completing the initial two-dose vaccination regimen.67 Our results show that the five donors that took the 3rd dose of COVID-19 mRNA vaccines had boosted neutralizing antibodies toward not only early WA (614D) and NY (614G) viruses but also Delta and Kappa variants compared to the titers after the 2nd dose vaccination. K18-hACE2 mice receiving the 1-mo booster post-vac sera showed improved survival rates from the lethal NY (614G) or Delta challenge compared to those transferred with the 5-mo post-vac sera from the 2nd dose vaccination. These results are consistent with the epidemiology reports that VE against laboratory-confirmed COVID-19 infections is higher among the populations who had received the 3rd mRNA vaccine dose than those without during the Delta wave.66,67 Despite the overall boosted antibody responses after the 3rd mRNA vaccine dose, those donors still exhibited lower neutralizing antibody titers against the latest Omicron subvariants compared to the titers against Delta and early SARS-CoV-2 strains. This is consistent with the report that the COVID-19 VE during the Omicron-predominant period was lower than during the Delta-predominant period regardless of the receipt of 2 or 3 doses of mRNA vaccines.66 The Omicron and its subvariants acquire >30 mutations in the spike including 15 mutations located in RBD, the total substitutions almost triple of those identified in Delta RBD.18,40,55 The cumulative mutations in RBD and NTD of Omicron spike cause significant reduction in antibody neutralization leading to greater immune escape than all previous VOCs.40,41,42,43
Taken together, our current study confirmed the epidemiological studies that mRNA vaccines-elicited antibodies and protection wane quickly within 5 mo after the 2nd dose. Our results suggest enhanced pathogenicity, greater immune evasion and waning immunity after vaccination are responsible for breakthrough infections during the Delta wave. Although a 3rd dose of COVID-19 mRNA vaccines can boost the overall antibody response, it may not confer sufficient protection against the Omicron subvariants with superior immune escape.
Limitations of study
Our current study has limitations. No paired human pre-vac sera were collected which could be better mock-transferred controls than PBS-injected mice for the mouse transfer study. The main reason is that the study was initiated in the middle of the COVID-19 pandemic and there was a shortage of healthcare workers to manage donors and collect samples. The other limitation is that our current passive transfer model only addressed in vivo protective efficiency of human COVID-19 mRNA vaccines-elicited sera and does not consider vaccines-induced cellular immunity (i.e. B cell memory and T cells). mRNA COVID-19 vaccines are found to induce high frequencies of spike-binding germinal center B cells and plasmablasts that last for at least 3 mo after the 2nd dose.69 A recently published longitudinal study also shows that mRNA vaccines may prevent Delta variant hospitalizations by eliciting robust functional memory T cell immunity.70
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Peroxidase-conjugated goat anti-human IgG (H + L) | Seracare | 5220-0330 |
| Peroxidase-conjugated goat anti-rabbit IgG (H + L) | Seracare | 5220-0336 RRID: AB_2857917 |
| Peroxidase-conjugated goat anti-mouse IgG (H + L) | Seracare | 5450-0011 RRID: AB_2687537 |
| Mouse monoclonal antibody specific for SARS-CoV-2 N | Gene Universal | UDA9001-6 |
| Rabbit polyclonal antibody specific for SARS-CoV-2 N/M/E | Radvak et al.49 | N/A |
| Bacterial and virus strains | ||
| SARS-CoV-2 Kappa (B.1.617.1) strain | CDC | N/A |
| SARS-CoV-2 Delta (B.1.617.2) strain | CDC | N/A |
| SARS-CoV-2 USA-WA1/2020 (WA (614D)) | BEI Resources | ATCC# NR-52281 |
| SARS-CoV-2 New York-PV09158/2020 (NY (614G)) | BEI Resources | ATCC# NR-53516 |
| SARS-CoV-2 USA/MD-HP20874/2021 (Omicron BA.1) | BEI Resources | ATCC# NR-56462 |
| SARS-CoV-2 USA/CO-CDPHE-2102544747/2021 (Omicron BA.2) | BEI Resources | ATCC# NR-56522 |
| SARS-CoV-2 USA/MD-HP30386/2022 (BA.4) | BEI Resources | ATCC# NR-56806 |
| SARS-CoV-2 USA/COR-22-063113/2022 (BA.5) | BEI Resources | ATCC# NR-58620 |
| Pseudotyped virus VSVdG-EGFP-SARS2-Sgp expressing the original Wuhan-Hu-1 spike | Sagara et al.71 | N/A |
| Biological samples | ||
| Healthy adult volunteer post-vaccination sera | University of Maryland | N/A |
| SARS-CoV-2 Infected mouse tissues | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Hygromycin B Gold | InvivoGen | 31282-04-9 |
| Streptavidin biosensor | Sartorius | 18-5019 |
| 1X Kinetics Buffer | Sartorius | 18-1105 |
| Biotinylated human ACE2/ACEH recombinant protein | Acrobiosystems | AC2-H82E6 |
| His-tagged recombinant original RBD protein | Acrobiosystems | SPD-C52H3 |
| His-tagged recombinant Kappa RBD bearing L452R and E484Q proteins | Acrobiosystems | SPD-C52Hv |
| His-tagged recombinant Delta RBD bearing L452R and T478K protein | Acrobiosystems | SPD-C52Hh |
| 1-Step Ultra TMB-ELISA substrate solution | ThermoFisher Scientific | 34028 |
| Critical commercial assays | ||
| Custom MSD V-Plex kits | Meso Scale Diagnostic | K152A0H-2 |
| Mouse D-Dimer ELISA Kit | MyBioSource | MBS764560 |
| Mouse IFN-b ELISA kit | PBL Assay Science | 42410-1 |
| RNeasy Plus Mini Kit | Qiagen | 74136 |
| High-Capacity cDNA Reverse Transcription Kit | Thermo Fisher Scientific | 4368813 |
| QuantiNova SYBR Green PCR Kit | Qiagen | 208052 |
| Deposited data | ||
| Raw and analyzed PCR array data | This paper | GEO: GSE193839 |
| Experimental models: Cell lines | ||
| Vero E6 | ATCC | CRL-1586 RRID:CVCL_0574 |
| TMPRSS2-E6 | BPS Bioscience | 78081 |
| BHK-21-ACE2 | Sagara et al.71 | N/A |
| Experimental models: Organisms/strains | ||
| Hemizygous B6. Cg-Tg(K18-ACE2)2Prlmn/J (K18-hACE2) transgenic mice/Strain #:034860 | The Jackson Laboratories | RRID:IMSR_JAX:034860 |
| Oligonucleotides | ||
| 2019-nCoV RUO Kit | IDT (Integrated DNA Technologies) | 10006713 |
| RT2 Profiler™ PCR Array Mouse Hypoxia Signaling Pathway | Qiagen | PAMM-032ZA-24/330231 |
| Recombinant DNA | ||
| pCC1-CoV2-F7 | Xie et al.72 | N/A |
| Software and algorithms | ||
| Qiagen web-based GeneGlobe data analysis tool | Qiagen | https://geneglobe.qiagen.com/us/analyze |
| GraphPad Prism version 9.3.1 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| FlowJo version 10.8.0 | TreeStar | https://www.flowjo.com/ |
| Wallac 1420 Workstation version 3 revision 4 | PerkinElmer | https://wallac-1420-d-workstation.software.informer.com/download/ |
| R version 4.0.3 | R Core Team | https://www.r-project.org/ |
| ForteBio data acquisition software version 10.0 and data analysis software version 10.0 | Sartorius | https://www.sartorius.com/en/products/protein-analysis/octet-bli-detection/octet-systems-software |
| MSD Discovery Workbench version 4.0.12 | Meso Scale Diagnostic | https://www.mesoscale.com/en/products_and_services/software |
| MxPro qPCR software version 3.0 | Agilent | https://www.agilent.com/en/product/real-time-pcr-%28qpcr%29/real-time-pcr-%28qpcr%29-instruments/mx3000-mx3005p-real-time-pcr-system-software/mxpro-qpcr-software-232751 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Hang Xie (hang.xie@fda.hhs.gov).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Human subjects
A total of 14 healthy, consenting, adult volunteers provided post-vaccination (post-vac) sera at approximately 1 month (mo) after completion of the 2nd dose of mRNA vaccines (6 Pfizer and 8 Moderna). Ten of the original donors (4 Pfizer and 6 Moderna) also returned to provide sera at approximately 5-mo after the 2nd dose. Among these donors, five (2 Pfizer and 3 Moderna) also provided sera at 1-mo after receiving the 3rd dose of mRNA vaccines. Another 3 fully vaccinated donors that had the Delta breakthrough infection also provided convalescent sera at 1 mo after recovery (Table S1). All donors were HIV/HBV/HCV negative and were not infected with SARS-CoV-2 before and during vaccination and during blood donations. Written informed consent was provided before enrollment. This study was approved by the University of Maryland, Baltimore Institutional Review Board.
Mouse model
Hemizygous B6.Cg-Tg(K18-ACE2)2Prlmn/J (K18-hACE2) transgenic mice (JAX Stock No. 034860)45 were bred at FDA White Oak Vivarium. All K18-hACE2 mice were ear tagged and were individually genotyping-confirmed (Transnetyx) before experiments. All procedures were performed according to the animal study protocols approved by the FDA White Oak Animal Program Animal Care and Use Committee.
Cell lines
Vero E6 (CRL-1586; American Type Culture Collection) was grown in Gibco high-glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin and 10 mM HEPES pH 7.3 (the base growth medium) at 37°C, 5% CO2. Recombinant Vero E6 constitutively expressing the full-length human TMPRSS2 (TMPRSS2-E6) (BPS Bioscience #78081) was maintained in the base growth medium plus 3 μg/mL of Puromycin and 1% Na pyruvate. hACE2 overexpression cell line – BHK-21-ACE2 was generated at LVD/NIAID, Bethesda71 and was maintained in the base growth medium plus 250 μg/mL hygromycin.
Viruses
All research on live SARS-CoV-2 viruses were performed in an animal biosafety level (ABSL) 3 laboratory equipped with advanced access control devices and by personnel equipped with powered air-purifying respirators. The seeds of Kappa (B.1.617.1) and Delta (B.1.617.2) variants were isolated at CDC. The SARS-CoV-2 clinical isolates including (1) USA-WA1/2020 (WA (614D)) (ATCC# NR-52281), (2) New York-PV09158/2020 (NY (614G)) (ATCC# NR-53516), (3) USA/MD-HP20874/2021 (Omicron BA.1) (ATCC# NR-56462), (4) USA/CO-CDPHE-2102544747/2021 (Omicron BA.2) (ATCC# NR-56522), (5) USA/MD-HP30386/2022 (BA.4) (ATCC# NR-56806), and (6) USA/COR-22-063,113/2022 (BA.5) (ATCC# NR-58620) were obtained through BEI Resources (Manassas, VA). All seed viruses were amplified once by inoculating 90% confluent TMPRSS2-E6 cells in DMEM supplemented with 3% FBS and were incubated at 37°C, 5% CO2 for 3 days or until observation of cytopathic effect. Supernatants were clarified and aliquots were stored in a secured −80°C freezer until use. Virus titers were determined using an ELISA-based 50% tissue culture infectious dose (TCID50) method.49
Method details
Bio-layer interferometry
RBD-ACE2 binding measurements were performed as described before73 using an Octet® RED96 system (ForteBio) with integrated data acquisition software 10.0. Streptavidin (SA) biosensors (Sartorius #18-5019) were pre-hydrated in 1X Kinetics Buffer (Sartorius #18-1105) and were then loaded with biotinylated human ACE2/ACEH recombinant protein (Acrobiosystems #AC2-H82E6) for up to 0.5-1 nm thickness. The ACE2 loaded SA biosensors were then dipped in RBD or its mutants in the concentration of 0.07-50 μg/mL in 1X Kinetics Buffer for 600 s for association, followed by being immersed in 1X Kinetics Buffer alone for another 600 s for dissociation. All steps were conducted at 26°C with constant shaking at 1,000 RPM. Collected data were analyzed using ForteBio Data Analysis software 10.0. The Kon and Koff values were determined using a built-in 1:1 global curve-fitting model and the Kd values were calculated. In antibody blocking experiments, diluted human sera were pre-incubated with 10 μg/mL of RBD or its mutants at 37°C for 60 min before being applied to ACE2-loaded SA biosensors as described above.
RBD-specific IgG ELISA
The following His-tagged recombinant RBD proteins (SEC-MALS verified) were purchased from Acrobiosystems, including (1) original RBD (#SPD-C52H3), (2) Kappa RBD bearing L452R and E484Q (#SPD-C52Hv), and (3) Delta RBD bearing L452R and T478K (#SPD-C52Hh). Serially diluted human sera were added to 96-well microtiter plates pre-coated with 1 μg/mL of recombinant RBD protein and were incubated at room temperature for 1h. For avidity assays, the plates were washed and overlaid with 100 μL/well of 4 M urea for 15 min.74 The plates were washed and re-blocked in blocking buffer for another hour. Bound IgG was detected using peroxidase-conjugated goat anti-human IgG (H + L) (Seracare #5220-0330, 1:2000) followed by 1-Step Ultra TMB-ELISA substrate (ThermoFisher # 34028). Optical density (OD) at 450 nm was measured using a Victor V multilabel reader (PerkinElmer) equipped with Wallac 1420 Workstation (Version 3 Revision 4). Endpoint ELISA titers were calculated based on OD values > 2-fold of blank. Avidity index was calculated as the area under curve (AUC) using Prism 9.3.1 (GraphPad).
Live virus-based microneutralization (MN) assay
Human sera were heated inactivated at 56°C for 30 min before use. Heat-inactivated human sera were serially diluted and then incubated with 100 TCID50/well of live virus (1:1, v/v) at room temperature for 1 h. The virus-serum mixtures were added to Vero E6 cells pre-seeded in 96-well tissue culture plates and were incubated at 37°C, 5% CO2 for 2 days.49 Virus-infected cells were detected using in-house rabbit polyclonal antibody specific for SARS-CoV-2 N/M/E (1:1000)49 paired with peroxidase-conjugated goat anti-rabbit IgG (H + L) secondary antibody (Seracare # 5220-0336, 1:2000) or mouse monoclonal antibody specific for SARS-CoV-2 N (Gene Universal #UDA9001-6, 1:1000) paired with peroxidase-conjugated goat anti-mouse IgG (H + L) secondary antibody (Seracare # 5450-0011, 1:2000). MN titer represented the reciprocal of the highest serum dilution that resulted in >50% reduction in OD value as compared to wells containing virus only. An MN titer of 20 was assigned if no neutralization was observed at the initial serum dilution of 1:40.
Pseudovirus neutralization (PVN) assay
The flow cytometry-based pseudovirus neutralization assay was performed using non-replicative self-reporting VSVdG pseudotyped virus expressing the spike glycoprotein (Sgp) of the original Wuhan-Hu-1 strain – VSVdG-EGFP-SARS2-Sgp.71 Human sera were diluted in 1/2 log10 in DMEM supplemented with 8% FBS, 10 mM HEPES and 50 μg/mL of gentamicin (Sigma #G1397). Serially diluted sera were incubated with VSVdG-EGFP-SARS2-Sgp virus in non-tissue culture treated polypropylene 96 round-bottom well plates at 37°C, 5% CO2 for 1 h. The pseudovirus-serum mixtures were then added to BHK-21-ACE2 cells (MOI = 0.05) and incubated at 37°C, 5% CO2 overnight.71 The BHK-21-ACE2 cells infected with VSVdG-EGFP-SARS2-Sgp virus were detected using a BD FACSCelesta™ Cell Analyzer (BD Biosciences). The frequencies of EGFP-positive cells were analyzed using FlowJo version 10.8.0 (TreeStar). PVN titers were determined based on 50% maximal effective concentration (EC50) by nonlinear regression curve fitting dose-response inhibition model after the data were normalized over the controls without antibody treatment.
Mouse infections and challenges
Age-matched male and female K18-hACE2 adult mice (8–12 weeks old) were randomly assigned at 1:1 ratio per experimental group. Under isoflurane anesthesia, mice were inoculated intranasally with NY (614G), Kappa or Delta variant at an infectious dose specified in individual experiments. Separate sets of infected mice were euthanized on dpi 1–6 and various organs were harvested for viral loads and whole lungs were collected for pathology, respectively.
Human sera collected at approximately 1-mo or 5-mo after the 2nd dose Pfizer or Moderna mRNA vaccines were pooled after heat-inactivation. Pooled human post-vaccination sera were then injected intraperitoneally into naïve K18-hACE2 mice at 0.2 mL/mouse (post-vac transferred). K18-hACE2 mice receiving 0.2 mL/mouse of PBS intraperitoneally served as controls (mock-transferred). Within 2–4 h after passive transfer, recipient mice were challenged intranasally with 103 TCID50/mouse of NY (614G) or 102 TCID50/mouse of Kappa or Delta variant. Infected mice were monitored daily for body weight (BW) and mortality for up to 2 weeks. Mice becoming moribund or reaching humane endpoints (e.g., severe hypothermia and/or 30% BW loss) were immediately euthanized and necropsied for pathology evaluation. Separate sets of mock-transferred and post-vac transferred mice were euthanized on dpi 5 and tissues were harvested for viral loads, cytokine and D-dimer determination and pulmonary hypoxia PCR array, respectively. All infections and challenged were performed in a secured ABSL3 biocontainment laboratory.
Viral burden
Equal numbers of female and male mice after infection or with passive transfer were euthanized on various dpi and various tissues were harvested for viral load determination, including brain, heart, whole lung, liver, spleen, kidneys and reproductive organs (ovary or testis). Collected tissues were homogenized in sterile PBS (pH7.2) (1:1, v/w) using a Fisherbrand Bead Mill 24 Homogenizer (FisherScientific). Total RNA was extracted from tissue homogenates using the RNeasy Plus Mini Kit (Qiagen #74136) and cDNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific #4368813) following the manufacturer’s protocol. QuantiNova SYBR Green PCR kit (Qiagen #208052) in combination of 500 nM of 2019-nCoV RUO Kit (Integrated DNA Technologies #10006713) was used to amplify nucleocapsid (N) gene in homogenized tissues in Stratagene MX3000p qPCR system (Agilent) according to the following program: 95°C for 120 s, 95°C for 5 s and 60°C for 18 s (50 cycles).49 Threshold cycle (Ct) values were calculated using MxPro qPCR software (Agilent) and the N gene copies in individual tissues were interpolated from a standard curve constructed by serial dilutions of a pCC1-CoV2-F7 plasmid expressing SARS-CoV-2 N74. A value of 1 was assigned if gene copies were below the detection limits.
Hypoxia signaling pathway PCR array
Total RNA was extracted from lung homogenates and was reverse transcribed as described above. cDNA was mixed with RT2 SYBR Green ROX qPCR Mastermix (Qiagen #330523) to perform RT2 Profiler™ PCR Array Mouse Hypoxia Signaling Pathway (Qiagen #PAMM-032ZA-24/330231) real-time PCR in Stratagene MX3000p qPCR system (Agilent). Ct values were determined using MxPro qPCR software (Version 3.0) after the following cycling program: hold for 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 60 s at 60°C. Individual gene expressions based on Ct values were normalized using Gapdh as the internal housekeeping gene and were calculated for fold changes using Qiagen web-based GeneGlobe data analysis tool. Correlation plot and volcano plot were generated using Prism 9.3.1(GraphPad).
Cytokine and chemokine detection
Multiplex proinflammatory cytokines and chemokines in tissue homogenates were determined using custom MSD V-Plex kits (Meso Scale Diagnostic #K152A0H-2) and a MESO QuickPlex SQ 120 imager equipped with MSD Discovery Workbench 4.0.12 (LSR_4_0_12). Tissue-specific IFN-β was quantitated using Verikine High Sensitivity mouse IFN-β ELISA kit (PBL Assay Science #42410-1).
D-dimer ELISA
Tissue-specific D-dimer was determined using mouse D-Dimer ELISA kit (MyBioSource #MBS764560) according to the manufacturer’s instructions.
Histopathology
Whole mouse lungs were fixed in 10% neutral buffered formaldehyde, embedded in paraffin, sections prepared and stained with hematoxylin and eosin (H&E) by Histoserv Inc. (Germantown, MD) or by Toxicologic Pathology Associates (Jefferson, AR) according to the standard histology protocols. Immunohistochemical (IHC) staining was performed using in-house raised rabbit polyclonal antibody specific for SARS-CoV-2 N/M/E (1:1000)49 (Toxicologic Pathology Associates and Histoserv). Sectioned lung tissues were also stained with Periodic acid–Schiff (PAS) for mucin (Histoserv). Uninfected mouse tissues were used as negative controls and stained in parallel. Tissue sections were visualized using a Leica Aperio AT2 slide scanner at Histoserv or Leica Aperio Scanscope System at Toxicologic Pathology Associates. High magnification images were obtained with a Nikon Eclipse Ni-E Upright Motorized Microscope or an Olympus BX-41 microscope. Histopathology lesions were scored in a blind fashion by a board-certified veterinary pathologist based on the following categories: (1) total lung H&E and IHC scoring: 0, no lesion; 1, 1–25%; 2, 26–50%; 3, 51–75%; 4, 76–100% of the lung involved with lesions; (2) individual lesion and PAS scoring: 0, no lesions or normal; 1, minimal; 2, mild; 3, moderate; 4, severe.
Quantification and statistical analysis
Data of biological replicates were reported for all experiments, except for the BLI binding experiments which technical replicates were performed and reported. Prism 9.3.1 (GraphPad) was used to conduct one-way nonparametric ANOVA or two-way mixed ANOVA, or Mann-Whitney test, or Log rank (Mantel-Cox) survival test as specified in individual figure legends. Data were log transformed before statistical analysis. A p value of <0.05 was considered statistically significant. Multivariate Principal Components Analysis (PCA) were used to analyze viral loads, D-dimer and cytokine profile among different virus infections and between mock-transferred vs post-vac transferred groups for NY (614G) challenge. The variables of interest for the logistic regression model consisted of viral load, D-dimer, and cytokines. The cytokines that resulted in unstable regression model were excluded. The variable importance in projection (VIP) scores, which measures the variable’s importance, were reported as the z values from the Wald test in the logistic regression models accounting for different tissues. R version 4.0.3 (https://www.r-project.org/) was used to perform PCA, logistic regression and Wald test.
Acknowledgments
This work was supported by FDA/CBER intramural SARS-CoV-2 pandemic fund to H.X. I. Kang was on the ORISE Research Fellowship supported by the FDA Office of Women's Health grant to H.X. The SARS-CoV-2 clinical isolates USA-WA1/2020 (ATCC# NR-52281), New York-PV09158/2020 (ATCC# NR-53516), USA/MD-HP20874/2021 (Omicron BA.1) (ATCC# NR-56462), USA/CO-CDPHE-2102544747/2021 (Omicron BA.2) (ATCC# NR-56522), USA/MD-HP30386/2022 (BA.4) (ATCC# NR-56806), and USA/COR-22-063113/2022 (BA.5) (ATCC# NR-58620) were obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2. The authors sincerely appreciate the support of the FDA/CBER Biosafety team and White Oak Vivarium staff in this study.
Author contributions
Conceptualization, H. X.; Investigation, H-J.K., M.K., W. T., U.O-R., P.R. I. Kang, H.X., K.E.M., L.M., K.D. and J.M.W.; Statistical analysis, R.X. and H.X.; Methodology, I. Kosik, J.H. and J.W.Y.; Clinical samples, W.H.C., M.C.S., R.S.B. and M.F.P.; Resources, H.X., E.P.P., B.Z. and D.E.W.; Writing-Original draft, H.X.; Writing-review and editing, H.X., R.X., J.W.Y, M.F.P., J.M.W., K.E.M., E.P.P., W.H.C., M.C.S., M.K., K.D., I.K, J.H., H-J.K., W-C.T., U.R.O., P.R., B.Z. and D.E.W.; Supervision, H.X.
Declaration of interests
The authors have no competing interests to declare.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: December 22, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105507.
Supplemental information
Data and code availability
This study did not report original code. PCR array data that support the findings of this study have been deposited at GEO and are publicly available as of the date of publication. The accession number is listed in the key resources table. All other data reported in this manuscript will be shared by the lead contact upon reasonable request.
References
- 1.Gupta R., Charron J., Stenger C.L., Painter J., Steward H., Cook T.W., Faber W., Frisch A., Lind E., Bauss J., et al. SARS-CoV-2 (COVID-19) structural and evolutionary dynamicome: insights into functional evolution and human genomics. J. Biol. Chem. 2020;295:11742–11753. doi: 10.1074/jbc.RA120.014873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCarthy K.R., Rennick L.J., Nambulli S., Robinson-McCarthy L.R., Bain W.G., Haidar G., Duprex W.P. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science. 2021;371:1139–1142. doi: 10.1126/science.abf6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gobeil S.M.C., Janowska K., McDowell S., Mansouri K., Parks R., Stalls V., Kopp M.F., Manne K., Li D., Wiehe K., et al. Effect of natural mutations of SARS-CoV-2 on spike structure, conformation, and antigenicity. Science. 2021;373:eabi6226. doi: 10.1126/science.abi6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tao K., Tzou P.L., Nouhin J., Gupta R.K., de Oliveira T., Kosakovsky Pond S.L., Fera D., Shafer R.W. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 2021;22:757–773. doi: 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 8.Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182:1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., COVID-19 Genomics UK COG-UK Consortium, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Motozono C., Toyoda M., Zahradnik J., Saito A., Nasser H., Tan T.S., Ngare I., Kimura I., Uriu K., Kosugi Y., et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe. 2021;29:1124–1136.e11. doi: 10.1016/j.chom.2021.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cherian S., Potdar V., Jadhav S., Yadav P., Gupta N., Das M., Rakshit P., Singh S., Abraham P., Panda S., Team N. SARS-CoV-2 spike mutations, L452R, T478K, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. Microorganisms. 2021;9:1542. doi: 10.3390/microorganisms9071542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuzmina A., Khalaila Y., Voloshin O., Keren-Naus A., Boehm-Cohen L., Raviv Y., Shemer-Avni Y., Rosenberg E., Taube R. SARS-CoV-2 spike variants exhibit differential infectivity and neutralization resistance to convalescent or post-vaccination sera. Cell Host Microbe. 2021;29:522–528.e2. doi: 10.1016/j.chom.2021.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saunders K.O., Lee E., Parks R., Martinez D.R., Li D., Chen H., Edwards R.J., Gobeil S., Barr M., Mansouri K., et al. Neutralizing antibody vaccine for pandemic and pre-emergent coronaviruses. Nature. 2021;594:553–559. doi: 10.1038/s41586-021-03594-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 17.Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C., Muecksch F., Rutkowska M., Hoffmann H.H., Michailidis E., et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020;9:e61312. doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCallum M., Walls A.C., Sprouse K.R., Bowen J.E., Rosen L.E., Dang H.V., De Marco A., Franko N., Tilles S.W., Logue J., et al. Molecular basis of immune evasion by the Delta and Kappa SARS-CoV-2 variants. Science. 2021;374:1621–1626. doi: 10.1126/science.abl8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng X., Garcia-Knight M.A., Khalid M.M., Servellita V., Wang C., Morris M.K., Sotomayor-González A., Glasner D.R., Reyes K.R., Gliwa A.S., et al. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell. 2021;184:3426–3437.e8. doi: 10.1016/j.cell.2021.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muecksch F., Weisblum Y., Barnes C.O., Schmidt F., Schaefer-Babajew D., Wang Z., C Lorenzi J.C., Flyak A.I., DeLaitsch A.T., Huey-Tubman K.E., et al. Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations. Immunity. 2021;54:1853–1868.e7. doi: 10.1016/j.immuni.2021.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29:463–476.e6. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29:44–57.e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Starr T.N., Greaney A.J., Addetia A., Hannon W.W., Choudhary M.C., Dingens A.S., Li J.Z., Bloom J.D. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371:850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greaney A.J., Starr T.N., Barnes C.O., Weisblum Y., Schmidt F., Caskey M., Gaebler C., Cho A., Agudelo M., Finkin S., et al. Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies. Nat. Commun. 2021;12:4196. doi: 10.1038/s41467-021-24435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. Overseas. Ed. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sadoff J., Gray G., Vandebosch A., Cárdenas V., Shukarev G., Grinsztejn B., Goepfert P.A., Truyers C., Fennema H., Spiessens B., et al. Safety and efficacy of single-dose Ad26.COV2.S vaccine against covid-19. N. Engl. J. Med. 2021;384:2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vilches T.N., Sah P., Moghadas S.M., Shoukat A., Fitzpatrick M.C., Hotez P.J., Schneider E.C., Galvani A.P. COVID-19 hospitalizations and deaths averted under an accelerated vaccination program in northeastern and southern regions of the USA. Lancet Reg. Health. Am. 2022;6 doi: 10.1016/j.lana.2021.100147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mesle M.M., Brown J., Mook P., Hagan J., Pastore R., Bundle N., Spiteri G., Ravasi G., Nicolay N., Andrews N., et al. Estimated number of deaths directly averted in people 60 years and older as a result of COVID-19 vaccination in the WHO European Region, December 2020 to November 2021. Euro Surveill. 2021;26:2101021. doi: 10.2807/1560-7917.ES.2021.26.47.2101021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta S., Cantor J., Simon K.I., Bento A.I., Wing C., Whaley C.M. Vaccinations against COVID-19 may have averted up to 140, 000 deaths in the United States. Health Aff. 2021;40:1465–1472. doi: 10.1377/hlthaff.2021.00619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossman H., Shilo S., Meir T., Gorfine M., Shalit U., Segal E. COVID-19 dynamics after a national immunization program in Israel. Nat. Med. 2021;27:1055–1061. doi: 10.1038/s41591-021-01337-2. [DOI] [PubMed] [Google Scholar]
- 33.Milman O., Yelin I., Aharony N., Katz R., Herzel E., Ben-Tov A., Kuint J., Gazit S., Chodick G., Patalon T., Kishony R. Community-level evidence for SARS-CoV-2 vaccine protection of unvaccinated individuals. Nat. Med. 2021;27:1367–1369. doi: 10.1038/s41591-021-01407-5. [DOI] [PubMed] [Google Scholar]
- 34.CDC COVID-19 Vaccine Breakthrough Case Investigations Team COVID-19 vaccine breakthrough infections reported to CDC - United States, January 1-April 30, 2021. MMWR Morb. Mortal. Wkly. Rep. 2021;70:792–793. doi: 10.15585/mmwr.mm7021e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown C.M., Vostok J., Johnson H., Burns M., Gharpure R., Sami S., Sabo R.T., Hall N., Foreman A., Schubert P.L., et al. Outbreak of SARS-CoV-2 infections, including COVID-19 vaccine breakthrough infections, associated with large public gatherings - barnstable county, Massachusetts, July 2021. MMWR Morb. Mortal. Wkly. Rep. 2021;70:1059–1062. doi: 10.15585/mmwr.mm7031e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chia P.Y., Ong S.W.X., Chiew C.J., Ang L.W., Chavatte J.M., Mak T.M., Cui L., Kalimuddin S., Chia W.N., Tan C.W., et al. Virological and serological kinetics of SARS-CoV-2 Delta variant vaccine breakthrough infections: a multicentre cohort study. Clin. Microbiol. Infect. 2022;28:612.e1–612.e7. doi: 10.1016/j.cmi.2021.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Christensen P.A., Olsen R.J., Long S.W., Subedi S., Davis J.J., Hodjat P., Walley D.R., Kinskey J.C., Ojeda Saavedra M., Pruitt L., et al. Delta variants of SARS-CoV-2 cause significantly increased vaccine breakthrough COVID-19 cases in Houston, Texas. Am. J. Pathol. 2022;192:320–331. doi: 10.1016/j.ajpath.2021.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harder T., Kulper-Schiek W., Reda S., Treskova-Schwarzbach M., Koch J., Vygen-Bonnet S., Wichmann O. Effectiveness of COVID-19 vaccines against SARS-CoV-2 infection with the Delta (B.1.617.2) variant: second interim results of a living systematic review and meta-analysis, 1 January to 25 August 2021. Euro Surveill. 2021;26:2100920. doi: 10.2807/1560-7917.ES.2021.26.41.2100920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopez Bernal J., Andrews N., Gower C., Gallagher E., Simmons R., Thelwall S., Stowe J., Tessier E., Groves N., Dabrera G., et al. Effectiveness of covid-19 vaccines against the B.1.617.2 (delta) variant. N. Engl. J. Med. 2021;385:585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCallum M., Czudnochowski N., Rosen L.E., Zepeda S.K., Bowen J.E., Walls A.C., Hauser K., Joshi A., Stewart C., Dillen J.R., et al. Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement. Science. 2022;375:864–868. doi: 10.1126/science.abn8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Q., Guo Y., Iketani S., Nair M.S., Li Z., Mohri H., Wang M., Yu J., Bowen A.D., Chang J.Y., et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4, & BA.5. Nature. 2022;608:603–608. doi: 10.1038/s41586-022-05053-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mannar D., Saville J.W., Zhu X., Srivastava S.S., Berezuk A.M., Tuttle K.S., Marquez A.C., Sekirov I., Subramaniam S. SARS-CoV-2 Omicron variant: antibody evasion and cryo-EM structure of spike protein-ACE2 complex. Science. 2022;375:760–764. doi: 10.1126/science.abn7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cui Z., Liu P., Wang N., Wang L., Fan K., Zhu Q., Wang K., Chen R., Feng R., Jia Z., et al. Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Cell. 2022;185:860–871.e13. doi: 10.1016/j.cell.2022.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goga A., Bekker L.G., Garrett N., Reddy T., Yende-Zuma N., Fairall L., Moultrie H., Takalani A., Trivella V., Faesen M., et al. Breakthrough SARS-CoV-2 infections during periods of delta and omicron predominance, South Africa. Lancet. 2022;400:269–271. doi: 10.1016/S0140-6736(22)01190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCray P.B., Jr., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H.P., Halabi C., Sigmund C.D., et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng J., Wong L.Y.R., Li K., Verma A.K., Ortiz M.E., Wohlford-Lenane C., Leidinger M.R., Knudson C.M., Meyerholz D.K., McCray P.B., Jr., Perlman S. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature. 2021;589:603–607. doi: 10.1038/s41586-020-2943-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yinda C.K., Port J.R., Bushmaker T., Offei Owusu I., Purushotham J.N., Avanzato V.A., Fischer R.J., Schulz J.E., Holbrook M.G., Hebner M.J., et al. K18-hACE2 mice develop respiratory disease resembling severe COVID-19. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., Fox J.M., Chen R.E., Earnest J.T., Keeler S.P., et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020;21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Radvak P., Kwon H.J., Kosikova M., Ortega-Rodriguez U., Xiang R., Phue J.N., Shen R.F., Rozzelle J., Kapoor N., Rabara T., et al. SARS-CoV-2 B.1.1.7 (alpha) and B.1.351 (beta) variants induce pathogenic patterns in K18-hACE2 transgenic mice distinct from early strains. Nat. Commun. 2021;12:6559. doi: 10.1038/s41467-021-26803-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hong J., Kwon H.J., Cachau R., Chen C.Z., Butay K.J., Duan Z., Li D., Ren H., Liang T., Zhu J., et al. Dromedary camel nanobodies broadly neutralize SARS-CoV-2 variants. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2201433119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Netland J., Meyerholz D.K., Moore S., Cassell M., Perlman S. Severe acute respiratory syndrome coronavirus infection causes neuronal death in the absence of encephalitis in mice transgenic for human ACE2. J. Virol. 2008;82:7264–7275. doi: 10.1128/JVI.00737-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ackermann M., Verleden S.E., Kuehnel M., Haverich A., Welte T., Laenger F., Vanstapel A., Werlein C., Stark H., Tzankov A., et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in covid-19. N. Engl. J. Med. 2020;383:120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Halfmann P.J., Iida S., Iwatsuki-Horimoto K., Maemura T., Kiso M., Scheaffer S.M., Darling T.L., Joshi A., Loeber S., Singh G., et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature. 2022;603:687–692. doi: 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thorne L.G., Bouhaddou M., Reuschl A.K., Zuliani-Alvarez L., Polacco B., Pelin A., Batra J., Whelan M.V.X., Hosmillo M., Fossati A., et al. Evolution of enhanced innate immune evasion by SARS-CoV-2. Nature. 2022;602:487–495. doi: 10.1038/s41586-021-04352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mittal A., Khattri A., Verma V. Structural and antigenic variations in the spike protein of emerging SARS-CoV-2 variants. PLoS Pathog. 2022;18 doi: 10.1371/journal.ppat.1010260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Golden J.W., Cline C.R., Zeng X., Garrison A.R., Carey B.D., Mucker E.M., White L.E., Shamblin J.D., Brocato R.L., Liu J., et al. Human angiotensin-converting enzyme 2 transgenic mice infected with SARS-CoV-2 develop severe and fatal respiratory disease. JCI insight. 2020;5:142032. doi: 10.1172/jci.insight.142032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Borczuk A.C., Salvatore S.P., Seshan S.V., Patel S.S., Bussel J.B., Mostyka M., Elsoukkary S., He B., Del Vecchio C., Fortarezza F., et al. COVID-19 pulmonary pathology: a multi-institutional autopsy cohort from Italy and New York City. Mod. Pathol. 2020;33:2156–2168. doi: 10.1038/s41379-020-00661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xie J., Covassin N., Fan Z., Singh P., Gao W., Li G., Kara T., Somers V.K. Association between hypoxemia and mortality in patients with COVID-19. Mayo Clin. Proc. 2020;95:1138–1147. doi: 10.1016/j.mayocp.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Del Valle D.M., Kim-Schulze S., Huang H.H., Beckmann N.D., Nirenberg S., Wang B., Lavin Y., Swartz T.H., Madduri D., Stock A., et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J., Xiao T., Cai Y., Lavine C.L., Peng H., Zhu H., Anand K., Tong P., Gautam A., Mayer M.L., et al. Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant. Science. 2021;374:1353–1360. doi: 10.1126/science.abl9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang T.-J., Yu P.-Y., Chang Y.-C., Chang N.-E., Tsai Y.-X., Liang K.-H., Draczkowski P., Lin B., Wang Y.-S., Chien Y.-C., et al. Structure-activity relationships of B.1.617 and other SARS-CoV-2 spike variants. bioRxiv. 2021 doi: 10.1101/2021.09.12.459978. Preprint at. [DOI] [Google Scholar]
- 63.Lipsitch M., Krammer F., Regev-Yochay G., Lustig Y., Balicer R.D. SARS-CoV-2 breakthrough infections in vaccinated individuals: measurement, causes and impact. Nat. Rev. Immunol. 2022;22:57–65. doi: 10.1038/s41577-021-00662-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prunas O., Weinberger D.M., Pitzer V.E., Gazit S., Patalon T. Waning effectiveness of the BNT162b2 vaccine against infection in adolescents. medRxiv. 2022 doi: 10.1101/2022.01.04.22268776. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eyre D.W., Taylor D., Purver M., Chapman D., Fowler T., Pouwels K.B., Walker A.S., Peto T.E. Effect of covid-19 vaccination on transmission of alpha and delta variants. N. Engl. J. Med. Overseas. Ed. 2022;386:744–756. doi: 10.1056/NEJMoa2116597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ferdinands J.M., Rao S., Dixon B.E., Mitchell P.K., DeSilva M.B., Irving S.A., Lewis N., Natarajan K., Stenehjem E., Grannis S.J., et al. Waning 2-dose and 3-dose effectiveness of mRNA vaccines against COVID-19-associated emergency department and urgent care encounters and hospitalizations among adults during periods of delta and omicron variant predominance - VISION network, 10 states, August 2021-January 2022. MMWR Morb. Mortal. Wkly. Rep. 2022;71:255–263. doi: 10.15585/mmwr.mm7107e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thompson M.G., Natarajan K., Irving S.A., Rowley E.A., Griggs E.P., Gaglani M., Klein N.P., Grannis S.J., DeSilva M.B., Stenehjem E., et al. Effectiveness of a third dose of mRNA vaccines against COVID-19-associated emergency department and urgent care encounters and hospitalizations among adults during periods of delta and omicron variant predominance - VISION network, 10 states, August 2021-January 2022. MMWR Morb. Mortal. Wkly. Rep. 2022;71:139–145. doi: 10.15585/mmwr.mm7104e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Levine-Tiefenbrun M., Yelin I., Alapi H., Katz R., Herzel E., Kuint J., Chodick G., Gazit S., Patalon T., Kishony R. Viral loads of Delta-variant SARS-CoV-2 breakthrough infections after vaccination and booster with BNT162b2. Nat. Med. 2021;27:2108–2110. doi: 10.1038/s41591-021-01575-4. [DOI] [PubMed] [Google Scholar]
- 69.Turner J.S., O'Halloran J.A., Kalaidina E., Kim W., Schmitz A.J., Zhou J.Q., Lei T., Thapa M., Chen R.E., Case J.B., et al. SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature. 2021;596:109–113. doi: 10.1038/s41586-021-03738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goel R.R., Painter M.M., Apostolidis S.A., Mathew D., Meng W., Rosenfeld A.M., Lundgreen K.A., Reynaldi A., Khoury D.S., Pattekar A., et al. mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science. 2021;374:abm0829. doi: 10.1126/science.abm0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sagara I., Woodford J., Dicko A., Zeguime A., Doucoure M., Kwan J., Zaidi I., Doritchamou J., Snow-Smith M., Alani N., et al. SARS-CoV-2 seroassay optimization and performance in a population with high background reactivity in Mali. medRxiv. 2021 doi: 10.1101/2021.03.08.21252784. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xie X., Muruato A., Lokugamage K.G., Narayanan K., Zhang X., Zou J., Liu J., Schindewolf C., Bopp N.E., Aguilar P.V., et al. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe. 2020;27:841–848.e3. doi: 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vogel M., Augusto G., Chang X., Liu X., Speiser D., Mohsen M.O., Bachmann M.F. Molecular definition of severe acute respiratory syndrome coronavirus 2 receptor-binding domain mutations: receptor affinity versus neutralization of receptor interaction. Allergy. 2022;77:143–149. doi: 10.1111/all.15002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kosikova M., Li L., Radvak P., Ye Z., Wan X.F., Xie H. Imprinting of repeated influenza A/H3 exposures on antibody quantity and antibody quality: implications for seasonal vaccine strain selection and vaccine performance. Clin. Infect. Dis. 2018;67:1523–1532. doi: 10.1093/cid/ciy327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not report original code. PCR array data that support the findings of this study have been deposited at GEO and are publicly available as of the date of publication. The accession number is listed in the key resources table. All other data reported in this manuscript will be shared by the lead contact upon reasonable request.