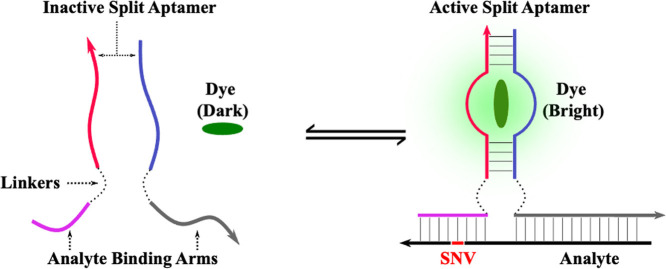
Abstract
Multiplex assays often rely on expensive sensors incorporating covalently linked fluorescent dyes. Herein, we developed a self-assembling aptamer-based multiplex assay. This multiplex approach utilizes a previously established split aptamer sensor in conjugation with a novel split aptamer sensor based upon a malachite green DNA aptamer. This system was capable of simultaneous fluorescent detection of two SARS COVID-19-related sequences in one sample with individual sensors that possesses a limit of detection (LOD) in the low nM range. Optimization of the Split Malachite Green (SMG) sensor yielded a minimized aptamer construct, Mini-MG, capable of inducing fluorescence of malachite green in both a DNA hairpin and sensor format.
Keywords: Aptamer, Biosensors, Split probe, Miniaturization, Malachite green, DNA
Graphical abstract
1. Introduction
With the potential threat of new or resurgent diseases, the need for both economical and rapidly deployable diagnostics are paramount. Commonly, nucleic acid analysis includes the use of fluorescently labelled oligonucleotides, such as TaqMan [1] or molecular beacons probes [2]. While these detection methods can achieve low limits of detection [3], the synthesis of fluorescently modified oligonucleotides incurs a significant economic cost. Several economical alternatives to these techniques have been proposed: including the use of functional nucleic acids such as deoxyribozymes [4], or aptamers [5], [6], [7], [8], [9].
Aptamers are functional nucleic acids generated through a series of iterative cycles of partitioning non-binding members from binding members of a randomized oligonucleotide library - a technique pioneered concurrently by the Szostak [10] and Gold [11] groups in the early 90s and termed SELEX (Sequential Evolution by Exponential Enrichment). The selected aptamers are generally short, less than 100 nucleotides, thus they are suitable for automated solid phase synthesis which decreases cost and increased production potential in comparison to antibodies. The relative ease in which aptamers can be selected has enabled the production of aptamers with a wide variety of target analytes including small molecules, toxins, [12] and whole cells [13]. A variety of detection schemes are possible with aptamers but one of the most widely utilized schemes is fluorophore and quencher conjugated oligonucleotides as their signaling elements [[14], [15]], analogous to molecular beacon technology. Some aptamers are capable of inducing a detectable signal upon binding their cognate target often through restriction of torsional rotation of the compound [16], [17], [18]. These aptamers, designated as Light-Up Aptamers, are enticing for the development of label-free and inexpensive biosensors as unmodified oligonucleotides can be utilized.
Light up aptamers have been successfully selected utilizing both RNA and DNA. In particular, RNA-based Light-Up aptamers have found use in intracellular imaging such as the spinach [19], mango [20], and broccoli [21] aptamers. The use of RNA allows intracellular imaging through the use of expression vectors but presents both economic and workflow challenges as the synthesis of RNA is more costly than DNA while also being susceptible to degradation. Thus, the use of DNA based Light-Up aptamers presents an alternative that is suitable to long term storage and provides a simpler workflow while being more amenable to in vitro diagnostic approaches.
Conventional hybridization probes can demonstrate a lack of selectivity towards mismatches in its target sequence. This lack of selectivity, particular at ambient temperatures, is a by-product of the length oligonucleotide probe that are commonly utilized in these hybridization-based assays [22]. The splitting paradigm relies on destabilization of the aptamer-core by scission of the intact strand into two smaller halves. These split aptamers can be converted to sensor by appending complimentary oligonucleotide sequences specific for the analyte of interest, named analyte-binding arms, to the ends of the split aptamer [23]. In the absence of its target, the two halves are incapable of hybridizing resulting in low signal. In the target's presence hybridization occurs between the analyte binding arms and analyte bringing the halves in close proximity, facilitating hybridization via a proximity effect with subsequent refolding of the aptamer core for signal generation. The advantage of this approach is that by utilizing shorter binding arms, selectivity for mismatches is increased as they are less tolerated by the shorter helices while affinity is maintained via cooperative binding of the small fragments [23]. The inverse relationship between affinity and selectivity central to hybridization-based technologies is circumvented by the splitting paradigm, as the affinity is imparted by cooperation between shorter fragments that retain selectivity as each individual piece is sensitive to mismatches. Split aptamer sensors have been produced for a variety of targets with a recent review highlighting some of the detection schemes and designs [24,25]. While many single target interrogating aptamer sensors have been developed, the literature is much less developed for multiplex aptamer-based assays. Addressing this gap could lead to substantial development of inexpensive multiplex approaches.
Multiplex assays allow the interrogations of one or more analytes of interest simultaneously and is commonly applied in PCR analysis of nucleic acid targets. Multiplexing reduces both cost and time necessary to determine if a sample contains an analyte of interest, while providing an avenue to generate more actionable data and unambiguous identification for potentially serious pathogens. While multiplex analysis is commonly accomplished with the use of expensive fluorescently modified primers, a gap in the literature exists for the application of split aptamer sensors in a multiplex format. With only a few examples of multiplex analysis utilizing aptamers in the literature and no examples of a split-aptamer based multiplex assays in solution. Our work here will lay the foundation for the development and optimization of other split aptamer based multiplex assays, describing some of the pitfalls and design approaches for this new direction in multiplex analysis.
Here in, we describe our efforts in the development of a self-assembling multiplex aptamer assay utilizing all-DNA split aptameric sensors: malachite green (MG) DNA aptamer [16] and Dapoxyl DNA aptamer [26]. This is the first demonstration of multiplex analysis utilizing self-assembling aptamers in solution. In this work we outline principles for the conversion and optimization of a new split MG DNA aptamer-based biosensor. First, we determine if the MG aptamer is capable of producing fluorescence after conversion to a split-aptamer sensor format, Split Malachite Green Sensor (SMG). Central to the split-sensor paradigm is a decoupling of the sensing and signaling portion of the aptamer sensor. This is accomplished via structural optimization, producing a platform which should allow for quick exchange of target specificity by altering the analyte binding arms while maintaining similar performance, an approach analogous to lateral flow-based assays. Following this paradigm, iterative structural optimization of the analyte binding arms, stem elements, and linkages between the sensor's sensing and signaling elements was performed to produce a sensor with detection limits in the low nM range. To demonstrate the ability to alter target specificity without time-consuming and costly optimization, a previously optimized split aptamer sensor, the split-dapoxyl aptamer (SDA) [27], was altered to detect new target analytes via exchange of its analyte binding arm with little loss in performance. Next, the individual aptamer sensors were then used in combination to interrogate two SARS-COVID19 DNA analytes simultaneously. Further work focused on quantifying the impact on the limit of detection (LOD) for the individual sensors in conditions that differ from their native buffer conditions and can provide guidelines for the selection of future aptamer pairs for multiplex detection. Lastly, we demonstrate that the MG aptamer can be further reduced to an active miniaturized construct, Mini-MG, which retains the ability to enhance MG fluorescence in a switch-sensor format.
2. Methods
General Fluorescent Assay for Split Malachite Green (SMG) Aptamer: Split malachite green aptamer (SMG#.A/B) strands were added to 5 × SMG buffer (100 mM Tris–HCl, pH 7.4) and diluted to a final volume of 99 µL by addition of water and the appropriate amount of analyte (NC-45) strand obtaining a final concentration of 500 nM for the aptamer strands. All samples were incubated at room temperature (22.5 °C) for 90 min before addition of 1.25 µL of 100 µM malachite green solution (final concentration of 1.25 µM) followed by additional incubation of 30 min at room temperature. Eighty µL fractions of these samples were transferred to a Thermo Fisher Scientific-Nunclon 96 Flat Bottom Transparent Polystyrene plate and the fluorescence recorded on a Tecan Infinite 200Pro at 658 nm with an excitation of 617 nm. The data of three independent trials was processed with Microsoft Excel.
General Fluorescent Assay for Split Dapoxyl (SDA) Aptamer: Split Dapoxyl aptamer (SDA#.A/B) strands were added to 5 × SDA buffer (100 mM Tris–HCl, 1 M KCl, 50 mM MgCl2,pH 7.4) and diluted to a final volume of 99 µL by addition of water and the appropriate amount of analyte (NC-14) strand obtaining a final concentration of 500 nM for the aptamer strands. All samples were incubated at room temperature (22.5 °C) for 90 min before addition of 1.0 µL of 200 µM Auramine O solution (final concentration of 2 µM) followed by additional incubation of 30 min at room temperature. Eighty µL fractions of these samples were transferred to a Thermo Fisher Scientific-Nunclon 96 Flat Bottom Transparent Polystyrene plate and the fluorescence recorded on a Tecan Infinite 200Pro at 540 nm with an excitation of 475 nm. The data of three independent trials was processed with Microsoft Excel.
Discrimination Assays: All discrimination assays followed the same procedure but utilized 150 nM of the match or mismatched analyte and 250 nM of the sensor strands.
General Fluorescent Multiplex Assay: Split malachite green aptamer strands (SMG42AT and SMG42B) and split Dapoxyl aptamer (SDA) strands (SDA4A and SDA4B) were added to 5 × Multiplex buffer (100 mM Tris–HCl, 50 mM KCl and 2 mM MgCl2, pH 7.4) and diluted to a final volume of 98 µL by addition of water and the appropriate matched analyte (NC-45G and NC-1450 for SMG and SDA, respectively) (with a final composition of 500nM for SMG4.2T, 150nM SDA4, and 150 nM of each matched analyte. All samples were incubated at room temperature (22.5 °C) for 90 min before addition of 1.0 µL of 100 µM of malachite green (final concentration of 1 µM) and 1.0 µL of 200 µM Auramine O solution (final concentration of 2 µM) followed by an additional incubation of 30 min at room temperature. Eighty µL fractions of these samples were transferred to a Thermo Fisher Scientific-Nunclon 96 Flat Bottom Transparent Polystyrene plate and the fluorescence recorded at 658 nm with an excitation of 617 nm to detect malachite green fluorescence and at 540 nm with an excitation of 475 nm on a Tecan Infinite 200Pro. The data of three independent trials was processed with Microsoft Excel.
Limit of Detection (LOD) Calculations: The LOD of split aptamer assays (SMG and SDA) were calculated by determining the linear trendline of the curves (EXCEL) and calculating the concentration in which the fluorescence surpassed the threshold value (defined as 3 SD of the blank sample).
Structure Prediction and Sequence Schematic Generation: All structural predictions utilized NUPACK which were exported as .svg files and edited in InkScape.
3. Results and discussion
3.1. Analyte model systems for the development of a multiplex aptamer sensor
For our model system we selected two fragments of the SARS-CoV-2 genome as pulled from the NCBI reference sequence (NC_045512.2). Here we chose a region of the coronavirus genome that contains single nucleotide variation, SNV, that results in a glutamate to glycine mutation in the spike protein [28]. This mutation is associated with higher infectivity and more severe diseases and is present in several variants of concern such as B1.617.2 (Delta) [28]. In this study, we used synthetic DNA sequences, titled NC-45 and NC-14, and corresponding mutant sequences containing single or double mismatches, NC-45A containing a guanine to adenine mutation and NC-14C containing a thymine to cytosine mutation. Derivatives of the NC-45 analyte consisting of the same mismatch (G>A) were designed that encompasses more of the reference genome (NC-45L or NC-45AL).
3.2. Conversion of an intact MG aptamer into a split aptamer sensor
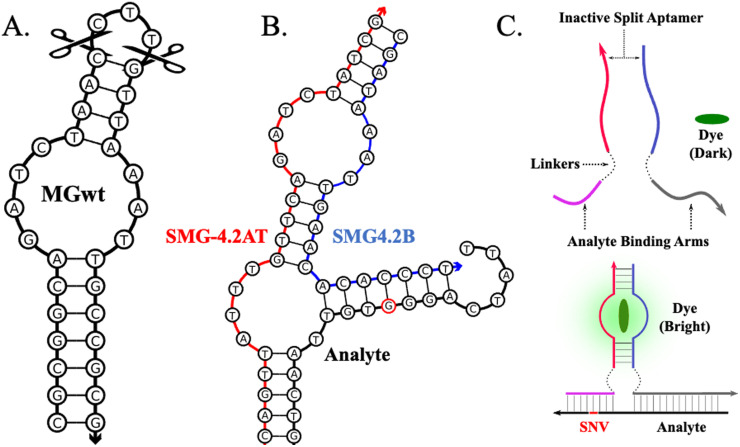
Fig. 1 contains the sequence diagram of the optimized MG aptamer as reported by Hyung et al. [16]. The MG aptamer is a relatively small aptamer of 32 nucleotides with a predicted secondary structure consisting of a centralized asymmetric bulge flanked by two stem elements. Alterations to aptamer's structure, such as sequence minimization or splitting, is a laborious process that relies on combination of experience and intuition as most aptamers lack structural detail such as crystal structures or in depth nucleotide contributions to the binding pocket [29,30]. This editing can be guided by utilizing general observations related to aptamer-target interactions. Two such relevant observations are that target-aptamer interaction are mostly mediated by free bases rather than those that are base-paired, such as the found in stem elements. Secondly, nucleotide contribution to binding tends to decrease as the nucleotide's distance from the purported aptamer core increases [31]. This allows for exchange or elimination of nucleotides with only small changes in aptamer affinity to the ligand [31]. A structural analysis of the folded MG aptamer via NUPACK [32] shows a trinucleotide loop, consisting of CTT, that is removed from the proposed core by 4 base-paired nucleotides. This decreases the likelihood that the loop is intrinsic to MG aptamer's function and represented the first attempted structural modification to achieve a split MG DNA aptamer. This trithymine loop was excised, severing the strands thus generating a split MG aptamer (SMG). The functioning of this split aptamer was verified by fluorescence spectroscopy with no significant impact on fluorescent enhancement of MG or its affinity toward MG (data not shown). The split aptameric sensor paradigm relies on the inability of the two aptamer strands to reform an active binding pocket in the absence of target; instead relying on a proximity effect induced by binding to the target analyte to stabilize hybridization of the stem elements to reform the active pocket as shown via the diagram in Fig. 1C [[23], [27], [33]] Once split, analyte binding arms specific for our model analytes were appended onto the 5′ and 3′ ends of the new MG aptamer strand halves, SMG-#A and SMG-#B, as shown in Fig. 1. The two strands are unable to stably hybridize in the absence of the target analyte as the melting temperature of the resulting stem elements are below ambient temperatures, resulting in low fluorescence output. Once hybridized to the target analyte, the aptamer halves hybridize via a proximity-induced effect with an enhancement in the fluorescence of MG as the core is reformed. The process of developing the SMG sensor utilized an iterative approach. Thus, the convention for naming our sensor designs follow the format of SMG-#A/B where # refers to the iteration of the sensor, A indicates strong analyte binding arm, and B for the weak analyte binding arm. Within each iteration, designs are further classified (SMG-A/B-#.#L/T) to denote different pairs within the same generation. The usage of the letters L or T specifies the inclusion of a hexaethylene glycol or tri-thymidine linkers, respectively.
Fig. 1.
Sequence Diagrams of the MG aptamer, its optimized split sensor (SMG4.2T), and the split aptamer sensor paradigm. Panel A illustrates the MG aptamer as selected by Hyuang et al. [16]. Conversion to a split aptamer was accomplished by excision of the tri-thymidine loop. Panel B depicts the sequence diagram of the optimized split malachite green aptamer sensor (SMG4.2T) in complex with its target analyte (NC-45, shown with black backbone). The single nucleotide variation (SNV) sensitive and strong analyte binding arm are indicated with a blue and red backbone, respectively. Panel C: split aptamer sensor paradigm. In the absence of analyte, the strands are incapable of association into a functional aptamer thus resulting in low malachite green (MG, show in between red and blue strands) fluorescence. Hybridization of the analyte binding arms to the target brings each half into close proximity, allowing the core to reform enhancing MG fluorescence.
Our initial work focused on establishing the lowest limit of detection (LOD) of the SMG sensor platform with no regards to the specificity. To this end, a sensor was developed utilizing the split aptamer sequence with long near symmetric analyte binding arms of 20 and 19 nucleotides. At these lengths, the binding between the aptamer sensor and its target analyte will be strong (–65.0 kcal/mol as predicted via NUPACK [32]) but will lack SNV selectivity particularly at ambient temperatures. The resulting complex between the sensor and its analyte is shown in Fig. 2 (top) with its accompanying calibration curve (bottom). With this design, an LOD was obtained in the low nM range (1.4 nM) which is suitable for post-PCR amplification analysis of nucleic acid targets.
Fig. 2.
Sequence Diagram SMG-2 in complex with its target analyte (NC-45A) and its calibration curve. The complex between SMG-2 and its target analyte (NC-45A) is shown as a sequence diagram (top) and its resulting calibration curve is shown bottom. This sensor design demonstrated an LOD of 1.4 nM. The threshold signal, defined as the is indicated by the horizontal solid line. The above curve is the average of 3 independent trials and was processed on Microsoft Excel. Fluorescence was recorded at 658 nm with an excitation at 617 nm.
3.3. Linker position and composition influence SMG fluorescence
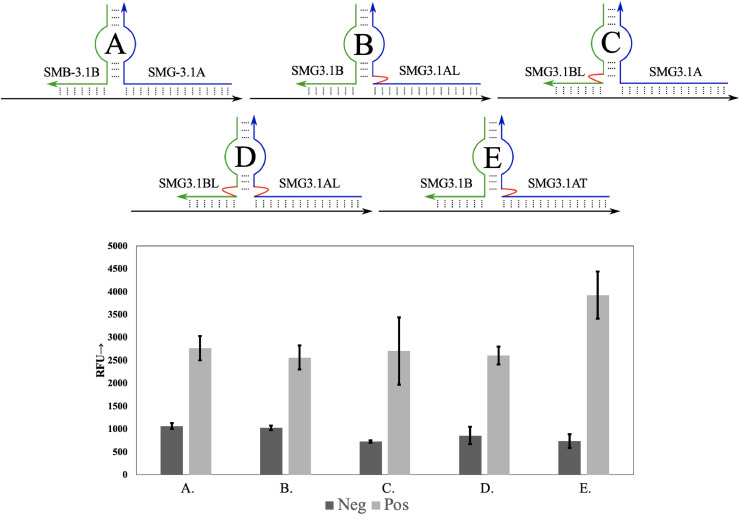
From this initial design, a series of SMG sensor variants (SMG-3.#A/B) were designed to determine the influence of linkers between the sensing elements (analyte binding arms) and reporter unit (the aptamer core) on sensor functioning. The analyte binding arms from SMG-2 were edited to produce two asymmetric arms consisting of 17 nucleotides (SMG-3A) and 8 nucleotides (SMG-3B) for the next generation of sensor, SMG-3. The longer arm, designated SMG-3A, provides a stable and high affinity interaction with the target while the shorter arm, SMG-3B, provides selectivity by being intolerant of mismatches within its shortened helix. This intolerance is attributed to the short length, 8 nucleotides, which results in a melting temperature below ambient temperatures and requires stabilization from SMG-3.1A to facilitate binding to its target site. Thus, binding of SMG-3.1A to the target is required to facilitate the binding of SMG-3.1B which is sensitive to the single nucleotide variation found in the sequence, increasing the selectivity of the sensor with little reduction in affinity via cooperative binding. However, addition of the analyte binding arms to the aptamer can result in alterations to the binding pocket as the geometry of the complex differs from that of the native aptamer lowering performance. This tension between binding pocket formation and tight analyte binding can be circumvented by use of linkers, such as hexaethylene glycol or trithymine linkers [27], which separates the sensing element of the SMG sensor (the analyte binding arms) from the signaling core. Similar investigations focused on linker length optimization to improve affinity for its target or its performance when functionalized onto a solid support, such as gold nanoparticles [34,35]. Presumably, these linkers provide the aptameric sensor flexibility to facilitate binding between the sensor and target while preserving its aptameric core's fold. To this end, derivatives of the split SMG-3 sensor were designed that incorporated trithymine or hexaethylene glycol linkers in SMG3.1A, SMG3.1B or both (Fig. 3 ). These variations were screened for their ability to induce MG fluorescence with emphasis being placed on determining which architecture achieves lower background signal and a high turn on fluorescence in the presence of target. As shown in Fig. 3, linker position and composition had effect on both the background and turn on fluorescence signal for the SMG sensor. The inclusion of hexaethylene glycol linkers did not provide a fluorescence increase but did result in reduction of the background fluorescence when compared to the split aptamer without the linkers. This effect was present when the linker was present on either or both analyte binding arms (compare columns A-D, with background RFU values of 1063, 1026, 725 and 858 respectively). The lowest background fluorescence was obtained when the hexaethylene glycol linker was placed on both analyte binding arm (SMG3.1AL and SMG3.1BL), reducing the background fluorescence by 32% from a RFU of 1063 (for SMG-3, column A) to 725 (For SMG-3.1AL/BL, column C) for the unmodified and modified sensor respectively. The use of a trithymidine linker on the strong analyte binding arm (SMG-3.1AT, where T signifies the trithymidine linker) produced a simultaneous increase in turn on signal and a reduced background fluorescence (compare columns A, B, and E). This reduction was similar to the value when utilizing ethylene glycol linkers on the weak analyte binding arm or both arms (SMG-3.1AL and SMG3.1BL, column C), with RFUs of 725 and 736 for column C (dark bar) and column E (dark bar), respectively. In contrast, the inclusion of the trithymidine linker on the strong analyte binding arm) resulted in a significant enhancement of ON fluorescence when compared to the unmodified sensor (Columns A and E, light bars with values of 2777 and 3925 respectively). While the structural impact behind this improvement has not been studied in great depth, previous work with aptamers have shown similar preferences [27,33]. The exact mechanism behind how these modifications influence aptamer function is not well understood and may require individualized study due to heterogenous folding patterns across aptamers. Qualitatively, the inclusion of linkers results in separation of the target analyte-analyte binding arm helix from the aptamer core. This increased space may allow for better accommodation of the folded core, thus improving the ON signal in presence of the analyte. The difference in activity between the use of hexaethylene glycol linkers and trithymidine linkers may be attributed to a difference in flexibility or the separation distance between the core and binding arm-target helix. The internal hexaethylene glycol linker is roughly equivalent to two base pairs and provides more flexibility than the naturally occurring phosphodiester backbone of the oligonucleotide. When the trithymidine are utilized as linkers, the linkage is longer and more rigid due to base stacking interactions between the thymines. This increase in flexibility imparted by the linkers may lower background fluorescence by stabilizing the secondary structures of the un-complexed aptamer sensor strands, reducing the proportion of strands folded into an active confirmation in the absence of target. When complexed to its target analyte, the trithymidine linker being slightly longer than its hexaethylene glycol counterpart may allow for the adoption of a more natural fold. This more natural fold could increase the activity of the aptamer core, which is reflected in higher fluorescence signal by the tri-thymidine modified analyte binding arm.
Fig. 3.
Linker Composition and Position Influence SMG-3 Performance. Top a schematic of SMG-3.1 complexed with its target analyte (NC-45) is shown. The strong analyte binding arm is depicted blue, with the SNV sensitive arm depicted green. Linker position is denoted via red line. Composition of the linker is shown by the use of L (hexaethylene glycol) and T (tri-thymidine). Bottom is MG fluorescence measured when 250 nM of sensor strands (SMG3.1A and SMG3.1B) was complexed with 250 nM of its target analyte (NC-45). Inclusion of a trithymidine linker on SMG3.1A resulted in simultaneous lowering of the background fluorescence (dark gray) and elevation of the fluorescence in the ON condition (gray). The above image is the average of 3 independent trials and was processed on Microsoft Excel. Fluorescence was recorded at 658 nm with an excitation at 617 nm.
3.4. Structural optimization of the SMG architecture
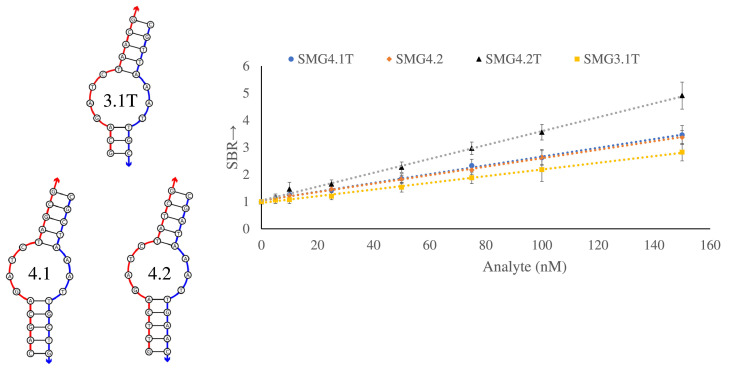
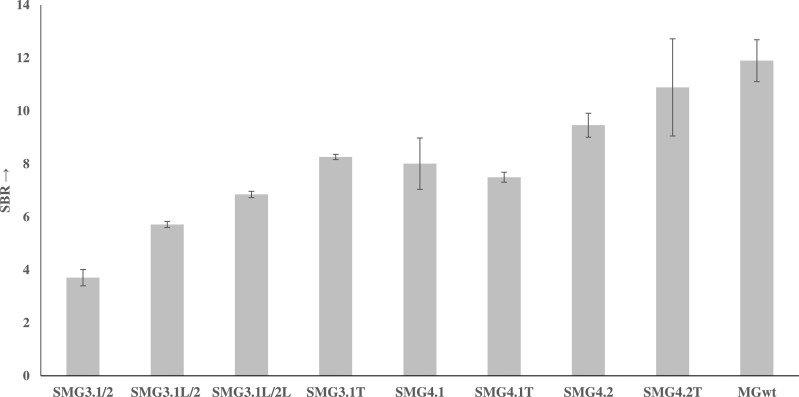
Having established that the presence of a trithymidine linker on the strong analyte binding arm, SMG-3.1AT, strand produces the most highly active complex, further optimization was performed to improve the signal-to-background ratio (SBR), which is defined as the ratio of ON signal (in presence of analyte) and its absence (OFF). Optimization focused on the influence of the upper and lower stem on the development of MG fluorescence with emphasis given to the stem length and GC content. Both of these factors will influence the stability of the aptamer core with longer stems or higher GC content expected to give a more stable structure to aid the core folding. The respective free energies, as calculated by NUPACK, for each of the core-stem complexes (shown in Fig. 4 ) are –8.2, –12.7 and –9.9 kcal/mol for SMG-3, SMG-4.1 and SMG-4.2 respectively. While this increase in stability is intended to elevate the sensor's fluorescence when complexed to its target, unintended increases in background fluorescence can occur as a higher proportion of aptamer sensor strands are folded into an active confirmation. To study this effect, SMG4.1 and SMG4.2 were designed to have equivalent total binding arm lengths of 24 nucleotides, which are comparable to SMG-3′s total binding length of 25 nucleotides. Since these sensors all interrogate the same portion of their target sequence, there binding arms are nearly identical sequences (see SI Fig. 1). This design allows for a direct comparison of the stem's influence on the sensor's function. As can be seen in Fig. 4, shown below, the increase in stem stability resulted in higher SBR ratios between generation 3 (SMG-3.1T) and generation 4 (SMG-4.1 and SMG-4.2). Further SBR improvement was elicited by altering the GC-content the stems from 60% in SMG-4.1 to 40% in SMG-4.2, reducing the stem stability to an intermediate value between SMG3.1 and SMG4.1. Higher GC content is associated with higher melting temperatures, so the reduction in GC content lowers the stability of both the upper and lower stem between SMG-4.1 and SMG-4.2. This improvement at first appears counter intuitive, as an increase in stability demonstrated a higher SBR between SMG-3 and SMG-4. However, the noted improvement in SBR was a consequence of a reduction in background signal for 4.2T as opposed to a significant increase in fluorescence magnitude (see Fig. 9, compare the first bar and second bar between SMG-4.1T and SMG-4.2T). This reduction in background can be attributed to a smaller proportion of associated SMG-A and SMG-B strands in the absence of its target, which is influenced by the stems ability to form a stable complex in the absence of target. The results of these structural optimizations are summarized in Fig. 5 , shown below, which demonstrates that SMG-4.2T has activity comparable to the intact MG aptamer at equimolar concentrations of 1 µM (SBR equals 10.8 and 11.8 for SMG-4.2T and MGwt, respectively).
Fig. 4.
Sequence diagrams of stem alterations between generation 3 and 4 and their perfomance in a MG flourescecne assays. Sequence diagrams of the aptamer core region of SMG-3, SMG-4.1, and SMG-4.2 depicting alterations to the stem elements. To increase the stability of the upper and lower stem between SMG-3.1T and SMG-4.1 an A-T base pair in the upper stem was replaced with a G-C pair with extension of the lower stem by 2 base pairs. 4.2 destabilizes these stem elements by replacement of a pair of A-T with G-C base pairs in the central base pair of the stems. The resulting SBR calibration curve is shown right, with the SMG4.2T showing improved performance at all analyte concentrations. The above curve is the average of 3 independent trials and was processed on Microsoft Excel. Fluorescence was recorded at 658 nm with an excitation at 617 nm.
Fig. 9.
Discrimination Results for both SDA4 and SMG4.2T (top) and various earlier iterations of SMG sensor (bottom). As can be seen in the top left of the image, 250 nM SDA4 exhibited excellent discrimination of single or double mismatch analytes (150 nM) with the double mismatch producing no signal above background. In contrast, SMG4.2T exhibits much greater difficult in distinguishing G>A mutation (top right) a trend that is consistent throughout all generations of SMG sensors (bottom). The above curve is the average of 3 independent trials and was processed on Microsoft Excel. Fluorescence was recorded at 658 nm with an excitation at 617 nm.
Fig. 5.
Overall optimization of iterative SMG sensors (SMG-3 – SMG-4.2T) as demonstrated by improvements by SBR. The overall improvement in performance, a 3-fold increase in SBR, across generations of sensors. The final iteration of the SMG sensor (SMG-4.2T) has performance comparable to the original intact aptamer at equimolar (1 µM) concentrations. The above curve is the average of 3 independent trials and was processed on Microsoft Excel. Fluorescence was recorded at 658 nm with an excitation at 617 nm.
3.5. The split dapoxyl aptamer (SDA) sensor represents a general sensing platform
The original dapoxyl aptamer was highly active, capable of increasing the intrinsic fluorescence of its cognate dye over 700-fold [26]. Work by Kikuchi resulted in generation of an optimized SDA sensor which relied on a combination of both hexaethylene glycol linker and dithymidine linkers to produce the most active complex with a reported LOD of 0.44 nM [27]. In this work, we create a series of derivatives of this sensor quickly and efficiently by exchange of the analyte binding arms from the original publication with two sets of COVID-19 analyte specific arms. The results of these exchanges for an SDA sensor for NC-45 and NC-14 are found in Table 1 and Fig. 6 . The original dapoxyl aptamer was selected against a non-commercially available dye, dapoxyl sulfonyl ethylenediamine. Later work with the dapoxyl aptamer found that it exhibits promiscuous binding to a range of dyes, including commercially available Auramine O which was utilized for all experiments with the SDA sensor [36].
Table 1.
LOD of constructs in native or multiplex buffer conditions.
| Construct | Target | LOD Native | LOD MultiplexC |
|---|---|---|---|
| SMG4.2TA | NC-45 | 5.3 nM | 6.3 nM |
| Mini-MG 1.2 | NC-45 | —— | 3.9 nM |
| Mini-MG 1.3 | NC-45 | —— | 3.2 nM |
| SDAB | —— | 0.44 nM | —— |
| SDA3 | NC-45 | 1.8 nM | —— |
| SDA4 | NC-14 | 1.3 nM | 2.2 nM |
Native buffer consist of 20 mM Tris-HCL (pH 7.4).
Native Buffer consist of 20 mM Tris-HCL (pH 7.4), 200 mM KCL and 10 mM MgCl2.
Multiplex Buffer consist of 20 mM Tris-HCL (pH 7.4), 10 mM KCL and 0.4 mM MgCl2.
Fig. 6.
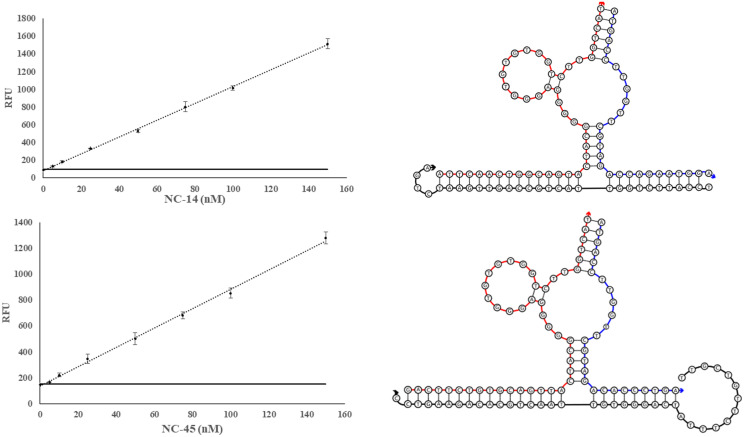
Calibration Curves of SDA4 and SDA3 sensors with sequence diagrams. Left depicts calibration curves for SDA4 (top) and SDA3 (bottom). The sequence diagrams of each sensor in complex with its target analyte (SDA4:NC-14 and SDA3:NC-45, top and bottom, respectively). The obtained LOD for SDA3 and SDA4 were 1.8 and 1.3 nM, respectively. The threshold signal, defined as the , is indicated by the horizontal solid line. The above curve is the average of 3 independent trials and was processed on Microsoft Excel. Fluorescence was recorded at 525 nm with an excitation at 475 nm.
These results strengthen the argument that SDA sensing platform represents an optimal design, which can facilitate its development for other targets quickly with minimal reduction in performance. In an analogous method to sandwich based lateral flow assays, in which specificity can be altered by exchange of an antibody specific to the desired analyte in question. While not exhaustively examined, the narrow range of LODs achieved with different binding arms, instrumentation and experimenters provides evidence that the SDA platform represents an optimal and reliable design which could be quickly developed into sensors for different nucleic acid targets.
3.6. The SBR and LOD difference between SDA3 and SMG4.2T
When comparing SBRs, all derivates of the SDA sensor far outstrip any SMG sensors when challenged by a common analyte (NC-45) in our assay conditions with SBRs of 10 and 4 (SDA3 and SMG4.2T), respectively with 500 nM of sensor strands and 150 nM of analyte. SDA3 and the SMG-4.2T sensors utilized common analyte binding arms in their designs, eliminating a difference in hybridization efficiency as an explanation for this discrepancy in performance. Instead, this performance gap may be attributable to the difference in aptamer affinity and mode of interaction with their cognate dyes. The dapoxyl aptamer exhibits a higher affinity for auramine O (Kd = 7–25 nM) [36] in comparison to the MG aptamer for MG (2.1 µM) [16]. This difference in affinity may contribute to the difference in the limits of detection between the two aptamers, 5.3 nM for SMG4.2T and 1.2 nM SDA3, as the dye complex between MG and its aptamer may not be as stable reducing its ability to induce torsional fluorescence. Another potential contributing factor could be the mode of recognition differs between the aptamers. Aptamer interactions generally progress through two modes, shape complementarity between the target and aptamer surface [37] or via incorporation of the molecule into the folded structure of the aptamer (encapsulation) [38]. Given the size difference between the aptamers and structural differences between their cognate dyes, it is plausible that the interaction mode differs with one strategy conferring a lower detection limit floor. Confirmation of this could be a fruitful endeavor for further development of aptamer-based biosensors.
3.7. Multiplex detection
Multiplex detection assays are a workflow that allows simultaneous detection of several targets within a single sample. Since the detection is simultaneous, spectral overlap of dye pairs could lead to reduced performance. The fluorescence spectra of MG and auramine O have very little overlap making them suitable for simultaneous detection. In comparison to traditional hybridization approaches, the aptamer-based approach is more complex to develop. As sequence interactions between the aptamer pairs can occur unexpectedly and the aptamer pairs could possess different environmental requirements which are determined in the aptamer's initial selection.
Selection of the dapoxyl and MG aptamers utilized different buffer conditions with the dapoxyl aptamer selection incorporating Mg2+ and K+ in comparison to the salt free selection of the MG aptamer. The SDA sensor platform demonstrates a K+ dependence for signaling (data not shown) which in combination with its G-rich sequence implies that the aptamer's folded confirmation incorporates a G-quadruplex (a planar arrangement of guanine tetrads) structural element, as stabilization of G-quadruplex structures requires the presence of mono-valent cations to fold correctly [39]. Initial results when performing SMG assays with SDA buffer resulted in low fluorescence enhancement even at high target concentrations (data not shown). The SDA aptamer similarly experienced a significant decrease in fluorescence when utilized in a Tris only buffer, producing no appreciable increase in fluorescence above the background. This salt dependence is well established for aptamers, as aptamer cores often incorporate cations for stabilization of binding pockets [40]. As such, aptamers demonstrate an intrinsic sensitivity to environmental factors, in particular the ionic strength of the solution [[41], [42], [43]], and function best at the buffer composition similar to the one used during their selection. A screening was performed in which the buffer composition was varied between the two . Selection of the final buffer composition focused on achieving SBRs > 2 for both aptamer pairs. This resulted in a final buffer composition (Multiplex Buffer) of 20 mM Tris–HCl (pH 7.4), 10 mM KCl and 0.4 mM MgCl2.
Once an operable buffer was found, the next stage of optimization focused on the relative concentrations of each aptamer pair. This optimization was a necessity as the dapoxyl aptamer demonstrates promiscuous dye binding, and could enhance the fluorescence of a variety of tri-phenyl or aromatic compounds, including MG [36]. At equimolar concentrations of 500 nM for both sensors, SDA4 produced a high background fluorescence for MG (crosstalk), which obscured the enhancement from the SMG aptamer. However, this difficulty was overcome by exploiting the SDA sensor more efficient generation of fluorescence by lowering the concentration of the SDA4 strands (from 500 nM to 150 nM). Since the dapoxyl aptamer binds MG less efficiently than auramine O (Kds of 6.9 µM and 7–25 nM, respectively [36]). This allowed a reliable increase in auramine O fluorescence while reducing the crosstalk between the SDA and SMG-4.2T sensors. Ultimately, an SBR of greater than 2 was maintained for each aptamer sensor. As seen in Fig. 7 , the multiplex assay was able to successfully detect the presence of one or both analytes. First giving a positive signal only in the presence of their respective analytes or in the presence of both unrelated analytes with very little increase in signal above the background in the absence of their appropriate target. This, to the author's knowledge, represents the first reported solution based multiplex assay utilizing self-assembling aptamers.
Fig. 7.
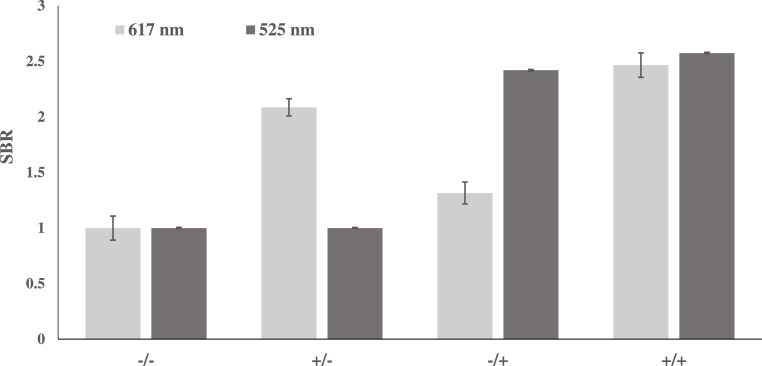
Multiplex Assay Utilizing the SDA4 and SMG4.2T sensors. SMG4.2T (500 nM) and SDA4 (150 nM) were incubated for 2 h at room temperature (22 °C) in a buffer consisting of 20 mM Tris–HCl (pH 7.4), 10 mM KCl and 0.4 mM MgCl2. The light gray bars represent fluorescence at 617 nm specific for SMG-4.2T, while dark gray bars signify fluorescence at 525 nm for SDA4. Sample +/- contained 150 nM of NC-45 (SMG4.2T's target analyte), while sample -/+ contained 150 nM of NC-14 (SDA4’s target analyte). Some cross talk between the SDA4 and MG was observed (compare light gray columns in the -/- condition and -/+ condition) but both achieved an SBR > 2 for detection of 150 nM of their respective analytes. The above image is the average of 3 independent trials and was processed on Microsoft Excel. Fluorescence was recorded at 658 nm with an excitation at 617 nm.
3.8. Split aptamer sensitivity
As discussed in section 1.2.7, the influence of non-native buffer conditions on aptamer function was observed but not unexpected. To quantify the impact of these differing buffer conditions on the split aptamer sensors performances, a comparison between the achievable LOD with our sensor constructs in their native versus multiplex buffer conditions was performed. As can be seen in Fig. 8 and Table 1, the LODs of both sensors were negatively impacted by use of the multiplex buffer. However, this impact was not evenly distributed between the aptamer pairs. The SMG-4.2T sensor experienced an increase in the LOD of roughly 19%, from 5.3 to 6.3 nM. For comparison, LOD for SDA4 increased by 70%, from 1.3 nM to 2.2 nM. This degradation in performance by SDA4 can be attributed to the role of the cations in folding and signaling of the dapoxyl aptamer as opposed to the MG aptamer which demonstrates no such salt requirement.
Fig. 8.
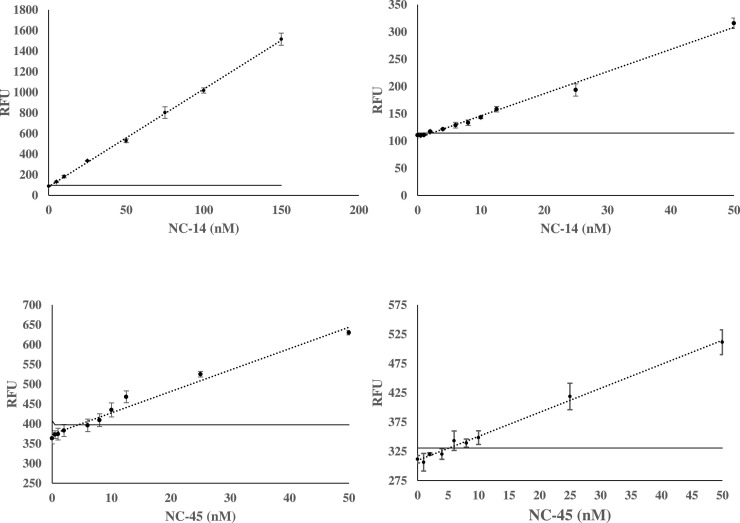
Comparison of SDA4 and SMG4.2T performance in native and multiplex buffer. The top two curves represent the fluorescence response of SDA4 in native (left) and multiplex (right) buffer. Calibration curves from SMG4.2T are shown bottom and follow the same convention. The sensitivity of both sensors was impacted in multiplex buffer but to different degrees with the impact on SDA4 more severe in comparison to SMG4.2T. The threshold signal, defined as the , is indicated by the horizontal solid line. The above curves are the average of 3 independent trials and was processed on Microsoft Excel. Fluorescence was recorded at 658 nm with an excitation at 617 nm.
3.9. Selectivity
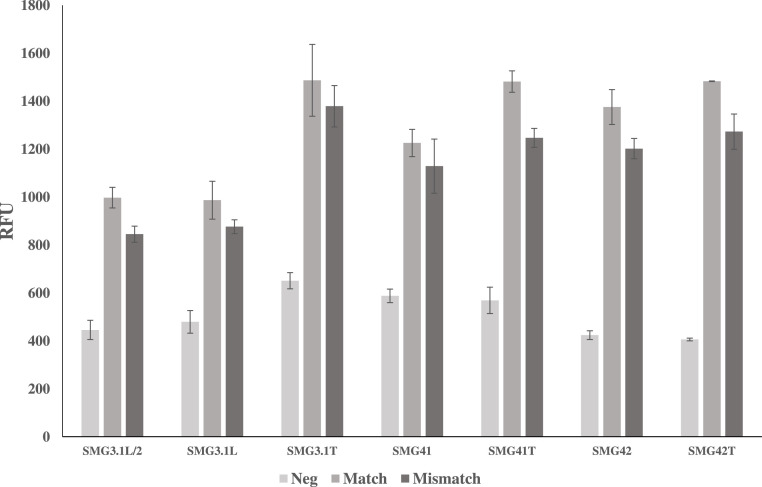
With established LODs in both native and multiplex conditions, the selectivity of each sensor was examined by comparing the signal between analyte sets containing single or double nucleotide mismatches between the analyte binding arms and analyte. As can be seen in Fig. 9 (top left), SDA4 exhibited selectivity for its target when was challenged to differentiate both the single and double mismatch analytes. At 30 min, the SBRs obtained for the match (13.5), single mismatch (1.74) and double mismatch (0.97) demonstrate a highly selective sensor which is in line with previous reports and is suitable for qualitative determinations about the presence or absence of the target SNV. In contrast, SMG4.2T demonstrated very little difference between matched or mismatched analyte with a signal difference of only 20% between matched and mismatched analytes (top right). Given this lack of selectivity, earlier iterations of the SMG sensor were challenged to the same discrimination assay and demonstrated consistent inability to discriminate between the analytes NC-45 and NC-45A. This lack of selectivity was consistent throughout all generations of SMG sensors tested. This result was unexpected as discrimination was expected given the small size of the SNV sensitive analyte binding arm (8 nucleotides).
In an effort to improve the selectivity performance of SMG4.2T, the concentration of the SNV sensitive arm (SMG4.2B) was varied from 100 to 400 nM and challenged to discriminate between its target analyte, NC-45, and mismatched analyte, NC-45A. This is a common optimization approach utilized by our group, as the formation of mismatched complex is reduced in tandem with lower strand concentrations [23]. As can be seen in Fig. 10 , all concentrations of SMG4.2B demonstrated a lack of selectivity towards it target. Upon closer analysis of the results, the magnitude of the fluorescence and background signal was independent of the concentration the SNV sensitive arm (SMG4.2B). This was unexpected as the anticipated results would demonstrate improved discrimination (a decrease in the dark grey bar) at the cost of fluorescence magnitude. This anticipation relied on the assumption that both halves of the aptamer sequence encompassing the central region of free bases contribute to the development of fluorescence. These unexpected results led to the hypothesis that the contribution of each half of the aptamer sensor (SMG4.2AT and SMG4.2B) to developing fluorescence is not equal and seems to rely more on the presence of the SMG4.2AT. To verify this, the ability of each sensor component to induce fluorescence was checked in all possible permutations. The results of this line of questioning are shown in Fig. 11 and present a different image of the true MG aptamer sequence.
Fig. 10.
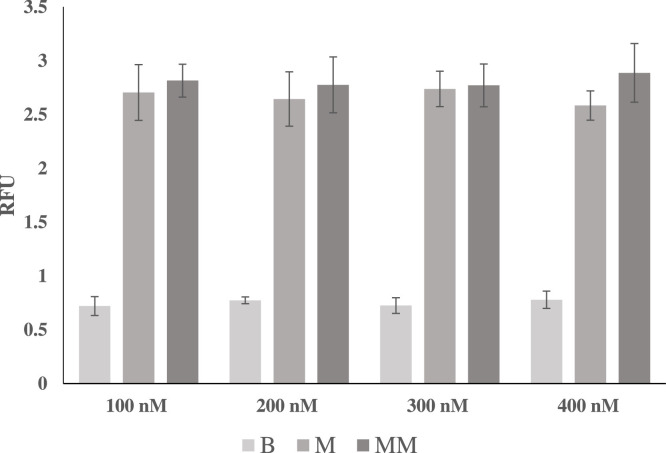
Discrimination Assay with SMG4.2T and a variable concentration of SMG4.2B The concentration of the SNV sensitive arm (SMG4.2B) was varied between 100 and 400 nM with 250 nM of SMG-4.2AT and challenged to discriminate 250 nM of NC-45 (matched analyte, M) and 250 nM of NC-45A (mismatched analyte, MM) while monitoring the signal in the absence of target (blank sample, B). As can be seen, the magnitude of fluorescence in all conditions did not very substantially with the concentration of SMG4.2B The above curve is the average of 3 independent trials and was processed on Microsoft Excel. Fluorescence was recorded at 658 nm with an excitation at 617 nm.
Fig. 11.
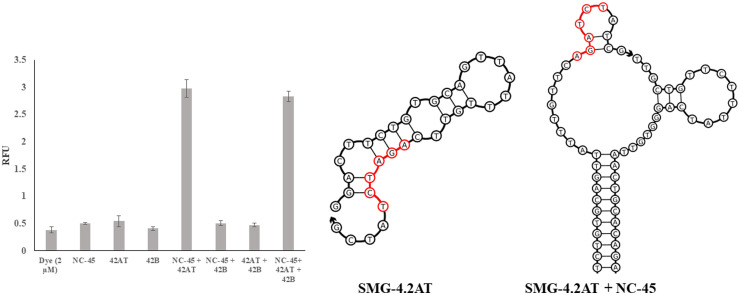
Fluorescence response of SMG4.2T components Assembled Piecewise. The fluorescence results of SMG4.2T (at 1 µM) encompassing the MG in the absence of any sensor strands (Dye), each component singular (NC-45 through 42B) or in combination. As can be seen, no component of the sensor induced fluorescence of MG above the background elevation that occurs via non-specific interactions with DNA. The presence of SMG4.2AT and NC-45 is sufficient to increase the fluorescence of MG by 5-fold with no observable increase when both halves of the SMG4.2T are present. Structural analysis via NUPACK of the SMG4.2AT in the absence (center) and presence of NC-45 (right) are shown on right side of the image. In the absence of NC-45, SMG4.2AT exhibits a tightly folded secondary structure with the core sequence derived from the MG aptamer (highlighted red) occluded by unintended hybridization with the analyte binding arm. In the presence of NC-45, hybridization of the analyte binding arm to the NC-45 liberates this sequence which is sufficient to produce detectable fluorescence. The above graph is the average of 3 independent trials and was processed on Microsoft Excel. Fluorescence was recorded at 658 nm with an excitation at 617 nm.
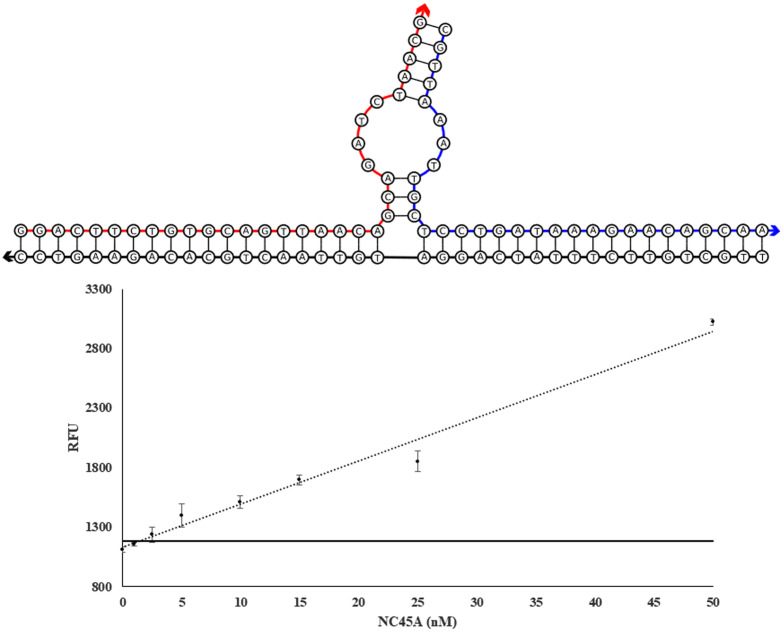
As can be seen Fig. 11, the combination of SMG4.2AT and NC-45 is sufficient to generate fluorescence, with only minor differences between the partial complex (SMG4.2AT+NC-45) and the full complex (SMG4.2AT+SMG4.2B+NC-45). Structural analysis of the strong binding arm, shown in Fig. 11, detailed that the hexanucleotide fragment (highlighted red) derived from the original MG aptamer is occluded in the absence of analyte. Upon addition of the target analyte, this occlusion is removed by the more complete hybridization between the analyte and analyte binding arm. When combined with the analysis presented in Fig. 10, implies that the hexanucleotide sequence, 5′-AGATCT-3′, represents the functional aptamer core. While convincing, it is possible that a portion of the analyte sequence, NC-45, could contribute nucleotides that allow for the core to fold correctly. To determine if this was the case, a series of minimized constructs, Mini-MG, were designed which contained solely the hexanucleotide sequence either in a free state or confined into a hairpin of various length, as seen in Fig. 12 , to eliminate this possible contribution from the analyte sequence.
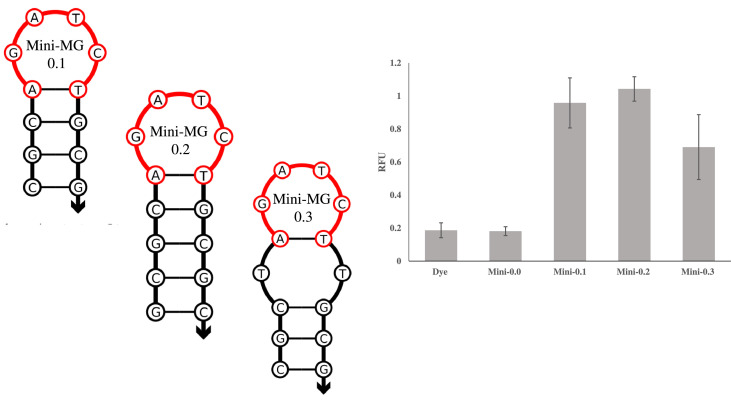
Fig. 12.
Sequence diagrams of Mini-MG constructs and their associated fluorescence results. The sequence diagrams of Mini-MG 0.1–0.3 are shown left with their corresponding fluorescence results shown right. Excluded from the image is Mini-MG 0.0, which consisted solely of the hexanucleotide sequence, 5′-AGATCT-3′. Mini-MG 0.2 was able to elevate the fluorescence of MG 5-fold. The image was produced via the average of 3 independent trials and was processed on Microsoft Excel. Fluorescence was recorded at 658 nm with an excitation at 617 nm.
The sequence diagrams and fluorescence results are shown in Fig. 12. The hexanucleotide sequence along with a stabilizing stem is sufficient to induce fluorescence of MG, resulting in a fluorescence increase of 5-fold. In contrast the free sequence, Mini-MG 0.0, was unable to induce fluorescence of MG. When comparing the stems across each construct, the fluorescence signal is correlated with stem stability with the most stable stem (Mini-MG 0.2) producing the highest fluorescence output. This small size puts the MG aptamer as one of the smaller functional aptamers and investigations continue into determining structural requirements for MG fluorescence and its exploitation as a bioanalytical tool. In the context of this work, the lack of selectivity across all generations of SMG sensors is attributed to the fact that this short sequence was present on the strong binding arm resulting in little differentiation between matched or mismatched analyte.
3.1.1. MG aptamer switch-based sensor
With this new information, a redesign of the MG aptamer-based sensor was required. The small size of the active aptamer sequence along with its efficient inhibition via partial hybridization allowed for the generation of a single stranded switch sensor, which is fluorescent only in the presence of matched analyte, whose design is illustrated in Fig. 13 .
Fig. 13.
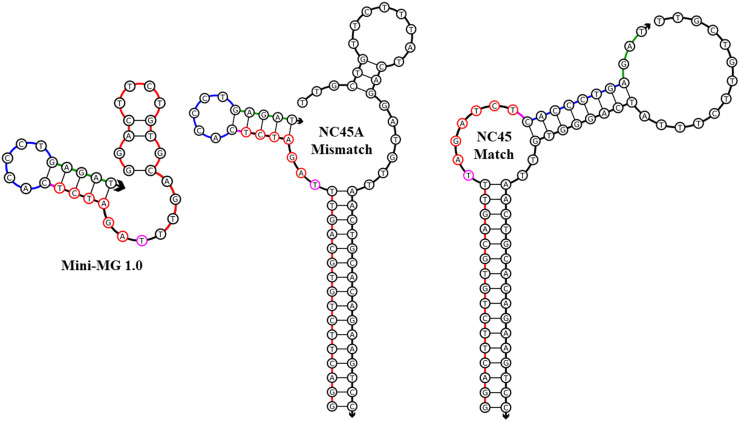
Design of Mini-MG 1.0 in complex with its mismatched analyte (NC-45A) and matched analyte (NC-45). The design of Mini-MG 1.0, show left, consist of a strong analyte binding portion (shown red), the hexanucleotide active sequence (highlighted red nucleotides), the SNV sensitive arm (shown blow) and blocking sequence (shown green). When hybridized with its target analyte, the blocking sequence is removed from the active sequence allowing MG fluorescence. Prospective linker site is highlighted magenta.
This design utilizes the same analyte binding arms as SMG4.2T but incorporates an inhibitory fragment (color-coded green in the image). Tight binding regardless of presence or absence of a mismatch occurs along the 5′ side of the sensor. When the matching analyte (NC-45) is present, the inhibitory fragment is separated from the active sequence via hybridization of the SNV sensitive arm to its target analyte. This allows for the active sequence to fold, producing detectable fluorescence.
Similar to our initial work in Section 3.3, a structural optimization focused on incorporation of linkers, hexaethylene glycol or trithymidine linkers, and their impact on the background and fluorescent output of the aptamer sensor. Given the reduced complexity of the construct, these linkers were incorporated into either at the 5′ or 3′ end of the hexanucleotide sequence. As can be seen in Fig. 14 , the inclusion of hexaethylene glycol linkers on the 3′ side of the active sequence resulted in a lower background and elevated fluorescence in the presence of matching analyte. This increase can be recapitulated in part by use of trithymidine linkers (data not shown) in place of the hexaethylene glycol linker significantly lowering the cost of the sensor. These dual effects, lowering background and raising the magnitude of fluorescence in the ON state, could be related to the increase in length of the sensor. Incorporation of hexaethylene glycol linkers add roughly the equivalent of 2 nucleotides in length. This additional length could relieve the tension caused by the tight turn on the inhibitory fragment in the off state and relax some tension formed by the matched complex. This sensor exhibited a lower LOD in comparison to the SMG4.2T with an LOD of 3.6 nM (compared to 5.3 nM for SMG4.2T). This improvement likely stems from a higher equilibrium concentration of productive signaling complex in comparison to the split aptamer approach at any given analyte concentration.
Fig. 14.
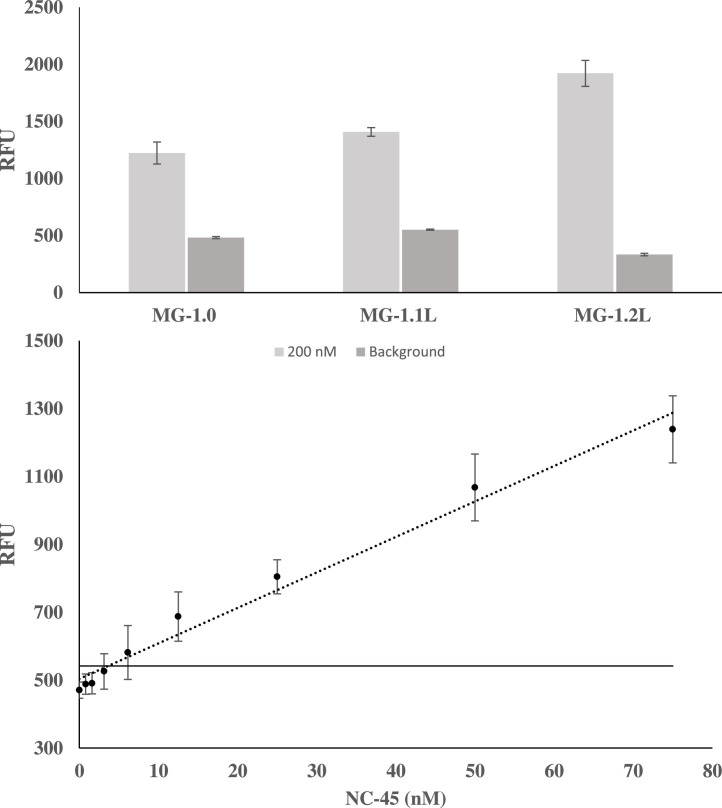
Fluorescence data for MG-1.0 derivatives with accompanying calibration curve Mini-MG 1.2 L. The fluorescence response at 658 nM of Mini-MG 1.0, 1.1 L and 1.2 L is shown on top. The inclusion of a hexaethylene glycol linker on the 3′ side of the active sequence simultaneously lowered background signal (dark gray) with an accompanying elevation of the fluorescence at 200 nM. Given the superior performance of Mini-MG 1.2 L, a calibration curve was created with a calculated LOD of 3.6 nM. The threshold signal, defined as the , is indicated by the horizontal solid line. The above curve is the average of 3 independent trials and was processed on Microsoft Excel. Fluorescence was recorded at 658 nm with an excitation at 617 nm.
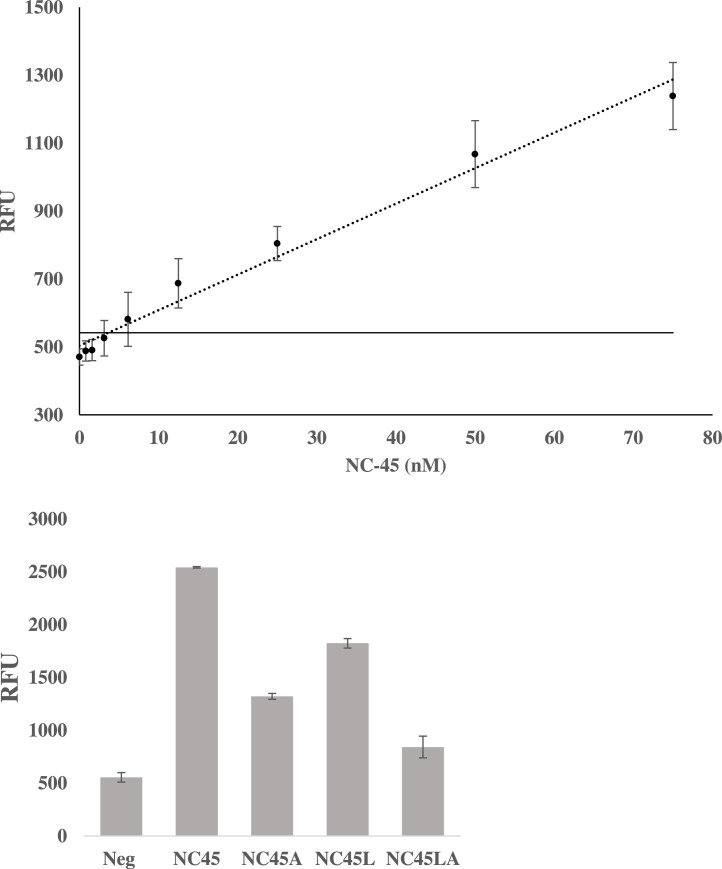
Even with improvements of LOD conferred by this new design, selectivity still remained an issue. To this end bases were removed from the strong analyte binding portion of the sensor, producing a construct that contains 15 nucleotides as the strong analyte binding arm, a sensor designated Mini-MG 1.3. This reduction did not significantly impact the LOD of the construct but improved the discrimination between matched and mismatched analytes (NC-45 and NC-45A, respectively). The resulting calibration curve and the results of its discrimination assay are shown in Fig. 15 .
Fig. 15.
Calibration Curve and Discrimination Assay with Mini-MG 1.3. Top, the calibration curve of Mini-MG1.3 L is shown with a calculated LOD of 3.2 nM. The ability of Mini-MG 1.3 to discriminate between its target analyte NC-45 and mismatched analyte NC-45A and their corresponding long derivatives (NC-45 L and NC-45LA) is shown on the bottom. Mini-MG 1.3 was able to differentiate a G>A SNV with roughly a two-fold difference in signal which was further improved upon when the long-mismatched analyte, NC-45A, was used. The threshold signal, defined as the , is indicated by the horizontal solid line. The above curve is the average of 3 independent trials and was processed on Microsoft Excel. Fluorescence was recorded at 658 nm with an excitation at 617 nm.
4. Conclusions
In this work, a DNA aptamer selected for increasing the intrinsic fluorescence of MG was successfully split and utilized to develop a sensor that achieved limits of detection in the low nM range. A previously reported split dapoxyl aptamer (SDA) was utilized for a new analyte by exchange of its analyte binding arms with very little change in sensitivity and exhibiting the expected high selectivity towards a T to C SNV. These aptamers were then combined into a multiplex assay utilizing self-assembling aptamer sensors for the simultaneous detection of two SARS-COVID19 gene fragments. This represents the first demonstration of such an approach and helps develop guidance for further development of such multiplex aptamer pairs. Further development of aptameric multiplex assays should consider the following points in selection of their aptamer partners, minimization of crosstalk between the aptamer partners and selection of compatible aptamers. Minimization of crosstalk between components of the multiplex assay can be achieved by selection of aptamer pairs that bind disparate dyes. The SDA sensor is promiscuous in its binding, resulting in additional optimization steps as it was able to induce fluorescence of MG, while the SMG sensor showed no interaction with Auramine O. Additional consideration should be given to the composition requirements for each utilized aptamer. Ideally these aptamers would be selected under the same environmental conditions, thus ensuring full activity in the multiplex detection assay conditions. Further development of multiplex aptameric assays may include the selection scheme for several dye-binding DNA under the same buffer conditions, preferably those utilized in PCR and/or isothermal amplification (LAMP, NASBA, RPA) buffers. Such set of DNA aptamers will facility the multiplex assay development and may contribute the widespread use of stable and label-free inexpensive fluorescent assays for nucleic acid analysis.
During the course of developing the multiplex assay we have demonstrated the creation of a new aptamer construct, Mini-MG, which is a minimalist aptamer sequence consisting of just 6 nucleotides. Even at this reduced size and without much optimization, the sequence was able to induce a 5-fold increase in MG fluorescence. This hexanucleotide sequence was then utilized in a switch sensor format, which should better activity than the parent split aptamer. The small size of this aptamer is enticing for the development of future bioanalytical tools and should be further explored to determine the structural requirements necessary to induce high fluorescence of MG.
Author contributions
MO, designed experiments, collected, analyzed the data, wrote the paper; DMK analyzed data, corrected the manuscript.
Declaration of Competing Interest
The authors declare no conflict of interests.
Acknowledgments
This work was supported by the National Science Foundation through the CCF: Software and Hardware Foundations under cooperative agreement SHF-1907824.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.snr.2022.100125.
Appendix. Supplementary materials
Data availability
Data will be made available on request.
References
- 1.Holland P.M., et al. Detection of specific polymerase chain reaction product by utilizing the 5′—-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 1991;16:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tyagi S., Kramer F.R. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;3:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 3.Kolpashchikov D.M. An elegant biosensor molecular beacon probe: challenges and recent solutions. Scientifica (Cairo) 2012 doi: 10.6064/2012/928783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith A.L., Kolpashchikov D.M. Divide and control: comparison of split and switch hybridization sensors. ChemistrySelect. 2017;19:5427–5431. doi: 10.1002/slct.201701179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Y.-.W., et al. Application of aptamer-based biosensor for rapid detection of pathogenic escherichia coli. Sensors. 2018;8:2518. doi: 10.3390/s18082518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamula C.L.A., et al. Selection and analytical applications of aptamers binding microbial pathogens. TrAC Trends Anal. Chem. 2011;10:1587–1597. doi: 10.1016/j.trac.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang Y.-.C., et al. Rapid single cell detection of Staphylococcus aureus by aptamer-conjugated gold nanoparticles. Sci. Rep. 2013;1:1863. doi: 10.1038/srep01863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berlina A.N., et al. Rapid visual detection of lead and mercury via enhanced crosslinking aggregation of aptamer-labeled gold nanoparticles. J. Nanosci. Nanotechnol. 2019;9:5489–5495. doi: 10.1166/jnn.2019.16575. [DOI] [PubMed] [Google Scholar]
- 9.Bala R., et al. A supersensitive silver nanoprobe based aptasensor for low cost detection of malathion residues in water and food samples. Spectrochim. Acta Part A: Mol. Biomol. Spectroscopy. 2018:268–273. doi: 10.1016/j.saa.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 10.Ellington A.D., Szostak J.W. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature. 1992;6363:850–852. doi: 10.1038/355850a0. [DOI] [PubMed] [Google Scholar]
- 11.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;4968:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 12.Hong K.L., Sooter L.J. Single-stranded DNA aptamers against pathogens and toxins: identification and biosensing applications. Biomed. Res. Int. 2015 doi: 10.1155/2015/419318. -419318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi H., et al. Whole cell-SELEX aptamers for highly specific fluorescence molecular imaging of carcinomas in vivo. PLoS ONE. 2013;8:e70476. doi: 10.1371/journal.pone.0070476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stojanovic M.N., de Prada P., Landry D.W. Aptamer-based folding fluorescent sensor for cocaine. J. Am. Chem. Soc. 2001;21:4928–4931. doi: 10.1021/ja0038171. [DOI] [PubMed] [Google Scholar]
- 15.Zhou H., Zhang S. Recent development of fluorescent light-up RNA aptamers. Crit. Rev. Anal. Chem. 2021:1–18. doi: 10.1080/10408347.2021.1907735. [DOI] [PubMed] [Google Scholar]
- 16.Wang H., et al. Selection and characterization of malachite green aptamers for the development of light–up probes. ChemistrySelect. 2016;8:1571–1574. doi: 10.1002/slct.201600154. [DOI] [Google Scholar]
- 17.Wang H., et al. Selection and characterization of thioflavin T aptamers for the development of light-up probes. Anal. Methods. 2016;48:8461–8465. doi: 10.1039/C6AY02890J. [DOI] [Google Scholar]
- 18.Wang J., et al. Selection and analysis of DNA aptamers to berberine to develop a label-free light-up fluorescent probe. New Journal of Chemistry. 2016;11:9768–9773. doi: 10.1039/C6NJ02290A. [DOI] [Google Scholar]
- 19.Pothoulakis G., et al. The spinach RNA aptamer as a characterization tool for synthetic biology. ACS Synth. Biol. 2014;3:182–187. doi: 10.1021/sb400089c. [DOI] [PubMed] [Google Scholar]
- 20.Dolgosheina E.V., et al. RNA mango aptamer-fluorophore: a bright, high-affinity complex for RNA labeling and tracking. ACS Chem. Biol. 2014;10:2412–2420. doi: 10.1021/cb500499x. [DOI] [PubMed] [Google Scholar]
- 21.Filonov G.S., et al. Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J. Am. Chem. Soc. 2014;46:16299–16308. doi: 10.1021/ja508478x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsourkas A., et al. Hybridization kinetics and thermodynamics of molecular beacons. Nucleic Acids Res. 2003;4:1319–1330. doi: 10.1093/nar/gkg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolpashchikov D.M. Binary malachite green aptamer for fluorescent detection of nucleic acids. J. Am. Chem. Soc. 2005;36:12442–12443. doi: 10.1021/ja0529788. [DOI] [PubMed] [Google Scholar]
- 24.Luo Y., et al. Label-free, visual detection of small molecules using highly target-responsive multimodule split aptamer constructs. Anal. Chem. 2019;11:7199–7207. doi: 10.1021/acs.analchem.9b00507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolpashchikov D.M., Spelkov A.A. Binary (Split) light-up aptameric sensors. Angew. Chem. Int. Ed. 2021;10:4988–4999. doi: 10.1002/anie.201914919. [DOI] [PubMed] [Google Scholar]
- 26.Kato T., et al. Light-up fluorophore–DNA aptamer pair for label-free turn-on aptamer sensors. Chem. Commun. 2016;21:4041–4044. doi: 10.1039/C5CC08816J. [DOI] [PubMed] [Google Scholar]
- 27.Kikuchi N., et al. Split dapoxyl aptamer for sequence-selective analysis of nucleic acid sequence based amplification amplicons. Anal. Chem. 2019;4:2667–2671. doi: 10.1021/acs.analchem.8b03964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plante J.A., et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;7852:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu H., et al. Structure-guided development of Pb2+-binding DNA aptamers. Sci. Rep. 2022;1:460. doi: 10.1038/s41598-021-04243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.VarnBuhler B.S., et al. Detection of SARS-CoV-2 RNA using a DNA aptamer mimic of green fluorescent protein. ACS Chem. Biol. 2022;4:840–853. doi: 10.1021/acschembio.1c00893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katilius E., Flores C., Woodbury N.W. Exploring the sequence space of a DNA aptamer using microarrays. Nucleic Acids Res. 2007;22:7626–7635. doi: 10.1093/nar/gkm922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zadeh J.N., et al. NUPACK: analysis and design of nucleic acid systems. J. Comput. Chem. 2011;1:170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- 33.Kikuchi N., Kolpashchikov D.M. A universal split spinach aptamer (USSA) for nucleic acid analysis and DNA computation. Chem. Commun. (Camb) 2017;36:4977–4980. doi: 10.1039/c7cc01540b. [DOI] [PubMed] [Google Scholar]
- 34.Gribas A.V., et al. Structure–activity relationship study for design of highly active covalent peroxidase-mimicking DNAzyme. RSC Adv. 2015;64:51672–51677. doi: 10.1039/C5RA03167B. [DOI] [Google Scholar]
- 35.Merkle T., Holder I.T., Hartig J.S. The dual aptamer approach: rational design of a high-affinity FAD aptamer. Org. Biomol. Chem. 2016;2:447–450. doi: 10.1039/C5OB02026C. [DOI] [PubMed] [Google Scholar]
- 36.Connelly R.P., et al. Promiscuous dye binding by a light-up aptamer: application for label-free multi-wavelength biosensing. Chem. Commun. 2021;30:3672–3675. doi: 10.1039/D1CC00594D. [DOI] [PubMed] [Google Scholar]
- 37.Dolot R., et al. Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity. Nucleic Acids Res. 2018;9:4819–4830. doi: 10.1093/nar/gky268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernandez-Millan P., et al. Crystal structure and fluorescence properties of the iSpinach aptamer in complex with DFHBI. RNA. 2017;12:1788–1795. doi: 10.1261/rna.063008.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhattacharyya D., Mirihana Arachchilage G., Basu S. Metal cations in G-quadruplex folding and stability. Front. Chem. 2016;38 doi: 10.3389/fchem.2016.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zimmermann G.R., et al. Molecular interactions and metal binding in the theophylline-binding core of an RNA aptamer. RNA. 2000;5:659–667. doi: 10.1017/S1355838200000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bruno J.G., et al. Aptamer Beacon Assay for C-telopeptide and handheld fluorometer to monitor bone resorption. J. Fluoresc. 2011;5:2021. doi: 10.1007/s10895-011-0903-6. [DOI] [PubMed] [Google Scholar]
- 42.Jeong S., Paeng I.R. Sensitivity and selectivity on aptamer-based assay: the determination of tetracycline residue in bovine milk. Sci. World J. 2012 doi: 10.1100/2012/159456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bruno J.G. Effects of various additives on cancer biomarker aptamer-magnetic pull-down in human serum. J. Bionanosci. 2017;1:45–51. doi: 10.1166/jbns.2017.1409. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.