Abstract
Cancer-associated fibroblasts (CAFs) are the predominant components of the tumor microenvironment (TME) and influence cancer hallmarks, but without systematic investigation on their ubiquitous characteristics across different cancer types. Here, we perform pan-cancer analysis on 226 samples across 10 solid cancer types to profile the TME at single-cell resolution, illustrating the commonalities/plasticity of heterogenous CAFs. Activation trajectory of the major CAF types is divided into three states, exhibiting distinct interactions with other cell components, and relating to prognosis of immunotherapy. Moreover, minor CAF components represent the alternative origin from other TME components (e.g., endothelia and macrophages). Particularly, the ubiquitous presentation of endothelial-to-mesenchymal transition CAF, which may interact with proximal SPP1+ tumor-associated macrophages, is implicated in endothelial-to-mesenchymal transition and survival stratifications. Our study comprehensively profiles the shared characteristics and dynamics of CAFs, and highlight their heterogeneity and plasticity across different cancer types. Browser of integrated pan-cancer single-cell information is available at https://gist-fgl.github.io/sc-caf-atlas/.
Subject terms: Cancer microenvironment, Cancer genomics, Cancer genomics, Tumour heterogeneity
Cancer-associated fibroblasts (CAFs) are a predominant and critical component of the tumour microenvironment. Here, the authors integrate and analyse single-cell RNA-seq data of CAFs across 10 common solid cancer types, identifying their plasticity and interactions with other cell types.
Introduction
Along with malignant cells, heterogeneous tumor microenvironment (TME) composites are important parts of tumors. They interact with malignant cells and contribute to the hallmarks of cancer1,2. Different types of nonmalignant cells are present in the TME, mainly including fibroblasts, immune cells (e.g., myeloid cells, lymphocytes, and macrophages), and endothelial cells. Previous studies have highlighted the indispensable role of the TME in the biological capabilities of cancer, such as tumor progression, treatment resistance, angiogenesis induction, and metastasis2–4. Mechanistically, the TME influences cancer cells via complicated and dynamic pathways to regulate cancer-related signaling5, involving ligand–receptor interactions (e.g., PD-L1 of cancer cells binding to PD1 of T cells), cytokine/metabolite responses, and deposition of extracellular matrix (ECM)6–10.
Of all types of stromal cells, fibroblasts are the predominant component in the TME, and cancer-associated fibroblasts (CAFs) play prominent and diverse tumor-supporting roles in cancer10,11. Besides directly interacting with malignant epithelial cells, CAFs create a tumor-permissive TME, including inducing the activation of normal fibroblasts into CAFs, promoting angiogenesis with endothelial cells12, recruiting myeloid cells13, and immunosuppression of T cells14. Therefore, CAFs play a key role in sculpting the TME by interacting with other TME components7,10,11, exhibiting their potential value as a prognostic factor and therapeutic target15. On the other hand, CAFs are primarily derived from normal fibroblasts (NFs) via several cancer type-specific mechanisms, and the modulation of CAFs to the TME is diverse and vague due to their heterogeneity and plasticity7. Recent studies have specified various subtypes of CAFs7, but the definitive origin of CAFs is controversial, and the lack of generalized characterization hinders CAF-targeted therapy in clinical practice. Meanwhile, it is difficult to define small subtypes of CAFs and investigate the plasticity of CAFs with limited cell populations, thus hindering the investigation on the evolutionary trajectory of CAFs and possible switches between CAFs and other TME components during cancer development and progression.
In recent years, the development of single-cell RNA sequencing (scRNA-seq) technology has provided the opportunity to investigate fluctuations in cell status and the strength of cell plasticity15–17. The characteristics of cancer cells and the TME have been profiled in multiple types of cancer, revealing the heterogeneity of cancer samples with different components at single-cell resolution and suggesting their possible involvement in the biological capabilities of cancer11. Additionally, increasing scRNA-seq-related software and strategies have been developed to improve the accuracy of bioinformatics analysis and upgrade the analytical dimensions, including batch effect correction18, cell–cell interaction evaluation19, and evolutionary trajectory20. However, recent scRNA-seq studies are still limited by the sample size, mainly due to fresh sample availability and expenditure. On the other hand, a large sample size would facilitate characterizing the complexity of cancer by excluding individual variants and by enriching the distinct components with a small cell population, particularly those shared across different cancer types. Due to the similarity of TME cells but not cancer cells among patients with different cancer types21,22, a few scRNA-seq-based pan-cancer studies have recently been conducted to integrate the increasing accessibility of scRNA-seq profiles, thus maximizing resolution and analyzing cell quantity within controlled bias, and exhibiting the ubiquitous characteristics of TME cells21–24. However, these pan-cancer studies have only focused on the characteristics of immune cells, omitting the interactions between different cell components.
In this study, we combine public and inhouse scRNA-seq data to profile the TME across 10 common solid cancer types, characterizing the interconvertibility and interaction among different types of stromal cells, with a particular focus on the ubiquitous characteristics of CAFs among diverse cancer types and the plasticity of CAF subtypes. Our systematic investigation on CAFs and their subtypes across cancers at single-cell resolution highlights the possible heterogeneity and plasticity of CAFs in cancer biology.
Results
Landscape of the TME in pan-cancer illustrated using scRNA-seq analysis
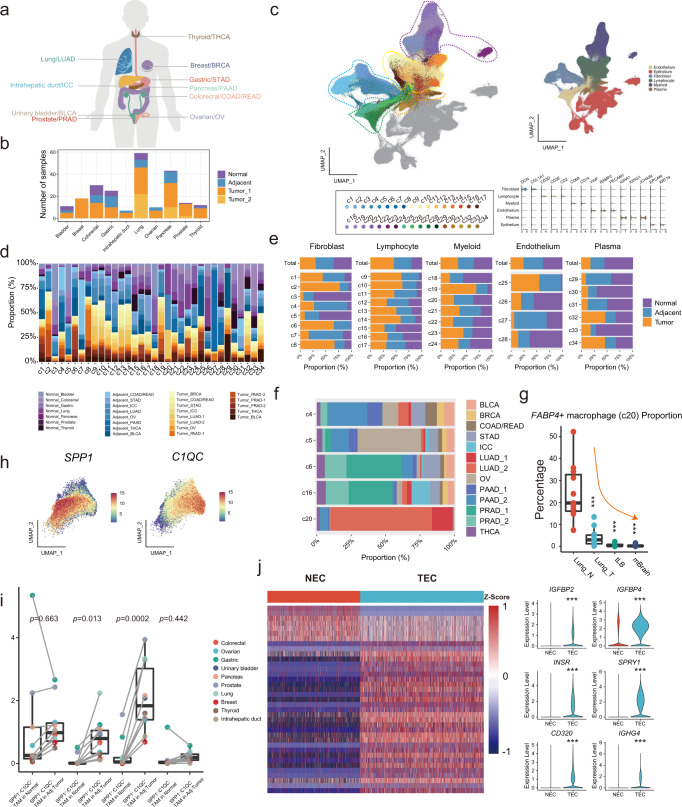
To profile the TME landscape of solid cancers, we compiled a single-cell transcriptional atlas in 10 common solid cancer types (Fig. 1a), with 148 primary tumor, 53 adjacent, and 25 normal samples from 164 donors enrolled in 12 studies (Fig. 1b and Supplementary Data 1). Available independent cohorts of patients with the same cancer type (i.e., pancreatic, lung and prostate cancers) were included to establish internal validation (Fig. 1b). Additionally, scRNA-seq data of the counterpart normal tissues from public resources were included to match the selected cancer type (Fig. 1b)25. To exclude possible technology-induced bias, all scRNA-seq data were generated using the same platform (i.e., 10× Genomics) without specific sorting. After strict quality control and filtering, a total of 855,271 cells from 226 samples were included and integrated based on a batch effect correction algorithm (Supplementary Fig. S1a). Next, unsupervised clustering generated a total of 34 TME-related clusters (c1-c34, 569,759 cells), which were separated from cancerous/normal epithelial cells (Fig. 1c and d). These clusters were divided into five major cell components based on canonical markers of different cell types, including fibroblasts (c1-c8, DCN, COL1A1), lymphocytes (c9-c17, CD3D, CD3E), myeloid cells (c18-c24, CD68, CD14), endothelial cells (c25-c28, VWF, PECAM1), and plasma cells (c29-c34, IGHG1, JCHAIN) (Fig. 1c, e and Supplementary Data 2). The remaining cells were clustered as epithelial cells (EPCAM, KRT19) (Fig. 1c). No obvious bias of the TME was observed regardless of malignant state and tissue type (Supplementary Fig. S1b and c), compared to the tissue type-specific distribution of epithelial cancer cells (Supplementary Fig. S1d). The normalized proportion of different clusters varied substantially among the different malignant states and cancer types (Fig. 1d and Supplementary Data 3). Although the composition of the five major components was relatively homogenously distributed among normal, adjacent, and tumor tissues, cells from cancer/adjacent tissues were predominantly enriched in specific clusters (e.g., c2 and c9), and vice versa (e.g., c3) (Fig. 1e). Additionally, some clusters, particularly c20 (FABP4+ macrophages), exhibited cancer type enrichment, which was consistent with an independent study on lung cancer26 (Fig. 1f). Intriguingly, a significant stepwise decrease in the proportion of FABP4+ macrophages (c20) but not other macrophage clusters (i.e., IL1B+, SPP1+, and APOE+) was observed along with adjacent controls, early- and late-stage primary tumors, and brain metastasis of lung cancer26 (all p < 0.001 to normal tissue) (Fig. 1g and Supplementary Fig. S1e), suggesting that FABP4+ macrophages may negatively correlate with lung cancer progression.
Fig. 1. Landscape of the TME in pan-cancer illustrated using scRNA-seq analysis.
a The cancer types included in this pan-cancer study. b The sample size histography of the selected normal/adjacent/tumor tissues. The sample size of replicates is shown when applicable. c Uniform Manifold Approximation and Projection (UMAP) plots of pan-cancer with 34 TME clusters, which are grouped into 4 main parts (i.e., endothelial cells, fibroblasts, lymphocytes/plasma cells, and myeloid cells). d Histography of the composition proportion of different tissue types in each TME cluster. e Clustering of TME components and their composition proportions in normal, adjacent, and tumor tissues. The proportion was normalized to the total cell number in each cancer. f Cancer type composition histography of tissue-enriched clusters according to cellular origin (c4-6, c16 and c20). g Significantly decreased proportions of FABP4+ macrophages along adjacent normal lung (Lung_N, n = 11), lung tumor (Lung_T, n = 11), advanced stage of tumor (tLB, n = 4), and brain metastasized (mBrain, n = 10) tissues, The box is bounded by the first and third quartile with a horizontal line at the median and whiskers extend to the maximum and minimum value. Dunnett-t two-sided test is used to test the significance of FABP4 + macrophages proportion between different tumor and normal tissue categories, Lung_N vs Lung_T p-value is 5.87 × 10−7, Lung_N vs tLB, p-value is 1.26 × 10−5, Lung_N vs mBrain, p-value is 2.97 × 10−8; ***: p < 0.001. h Feature plot of SPP1 and C1QC expression in the tumor-associated macrophage cluster. i Comparison of C1QC+ TAMs, SPP1+ TAMs, C1QC+/SPP1+ TAMs and C1QC-/SPP1- TAMs in normal and adjacent/tumor tissues, normal tissue n = 43, adjacent/tumor tissue n = 159. The box is bounded by the first and third quartile with a horizontal line at the median and whiskers extend to the maximum and minimum value. Mann–Whitney two-sided test is used to test the significance of proportion between different cell types. ***p < 0.001. j Differentially expressed genes clustering and specifically altered genes between tumor endothelial cells (TECs) and normal endothelial cells (NECs). Mann–Whitney two-sided test is used to test the significance of gene expression level between NEC and TEC categories. Source data are provided as a Source Data file.
After deeply investigating the components of the TME to evaluate its heterogeneity, potentially functional subclusters were identified. For instance, heterogeneity of APOE+ tumor-associated macrophages (TAM, c19) were exhibited by the complementary distribution of C1QC+ and SPP1+ cells (Fig. 1h), which is consistent with a previous report on pan-cancer scRNA-seq analysis21. The SPP1+ TAMs showed distinct transcriptomic profiles compared to C1QC+ TAMs, which are involved in several metabolism-related pathways (Supplementary Fig. S1f). The proportion of SPP1+C1QC-/SPP1+C1QC+ TAMs but not SPP1-C1QC+/SPP1-C1QC- TAMs significantly increased from normal to adjacent/tumor samples among different cancer types (Fig. 1i), suggesting the potential role of SPP1+ TAMs in tumorigenesis and tumor metabolism. On the other hand, tissue preferential distribution of endothelial cells promoted their classification into tumor endothelial cells (TECs) (i.e., c25) and normal endothelial cells (NECs) (i.e., c26 and c28) (Fig. 1e). The distinct transcriptional profiles of TECs and NECs were identified, including TEC significantly upregulated genes involved in the insulin response (e.g., INSR and IGFBPs), MAPK regulators (e.g., SPRY1), and immunoglobulins (e.g., CD320 and IGHG4)27,28 (Fig. 1j), suggesting the possible role of TECs in angiogenesis, tumor growth, and immune modulation. Moreover, pan-cancer analysis enhanced the possibility of delineating the potentially important but low-quantity subsets of immune cells in the TME. For instance, tumor/adjacent confined suppressive dendritic cells (i.e., DC subcluster 5) have been identified as a subset of dendritic cells (c21) with high expression of CD274, LAMP3, and CCL22, which may negatively regulate immune cells29,30 (Supplementary Fig. S1g). Intriguingly, small portions of both DCs and plasmacytoid DCs (pDCs, c24) expressed T-cell-specific markers (e.g., CD3D and CD3E) (Supplementary Figs. S1g and 1h). As validation, confocal images of multiplexed immunofluorescence (mIF) with co-staining CD3, CD86 and CD11c in stromal compartments of three cancer types (i.e., anaplastic thyroid cancer, gastric cancer and colorectal cancer), revealed the consistent existence of CD3+ DCs (CD3+CD11c+CD86+) (Supplementary Fig. S1i), ranging from 3 to 13% of myeloid cells (CD11c+) (Supplementary Fig. S1j), which has also been reported through flow cytometry31. On the other hand, heterogeneity of tumor-infiltrating B lymphocytes was found through subclustering. For instance, a subset of the B-cell cluster (i.e., c13) expressed markers consistent with suppression of B-cell differentiation (RGS13+, subclusters 7–8) (Supplementary Fig. S1k). Future studies should be conducted to investigate the origin and potential biofunction of these B-cell subpopulations in tumors.
Moreover, we also estimate the ubiquitous characteristics of epithelia among different cancer types. First, a total of 23 clusters (E1-E23) were divided with unsupervised clustering (Supplementary Fig. S2a and Supplementary Data 4). Unlike TME components, epithelia clusters exhibited bias in terms of both malignant status (e.g., predominant normal in E17 and tumor in E6) and cancer type (e.g., predominant thyroid cancer in E1 and prostate cancer in E6) (Supplementary Data 5 and Supplementary Fig. S2b). Not surprisingly, similarity among different cancer types was identified in only 6 out of 23 clusters, including E3 (IRS2, KRT6A), E5 (CD24, STMN1), E8 (GKN1, MUC5AC), E9 (PHGR1, TFF3), E10 (TFF3, TPO) and E13 (VTN and ITIH5) (Supplementary Fig. S2c, d). Interestingly, IRS2 and CD24 was widely present in epithelia and exhibited the strongest expression in E3 and E5 cluster respectively (Supplementary Fig. S2d), suggesting the ubiquitous activated insulin signaling32 and “don’t eat me” signal33 shared by different cancer types.
Generalized activation of CAFs in the TME
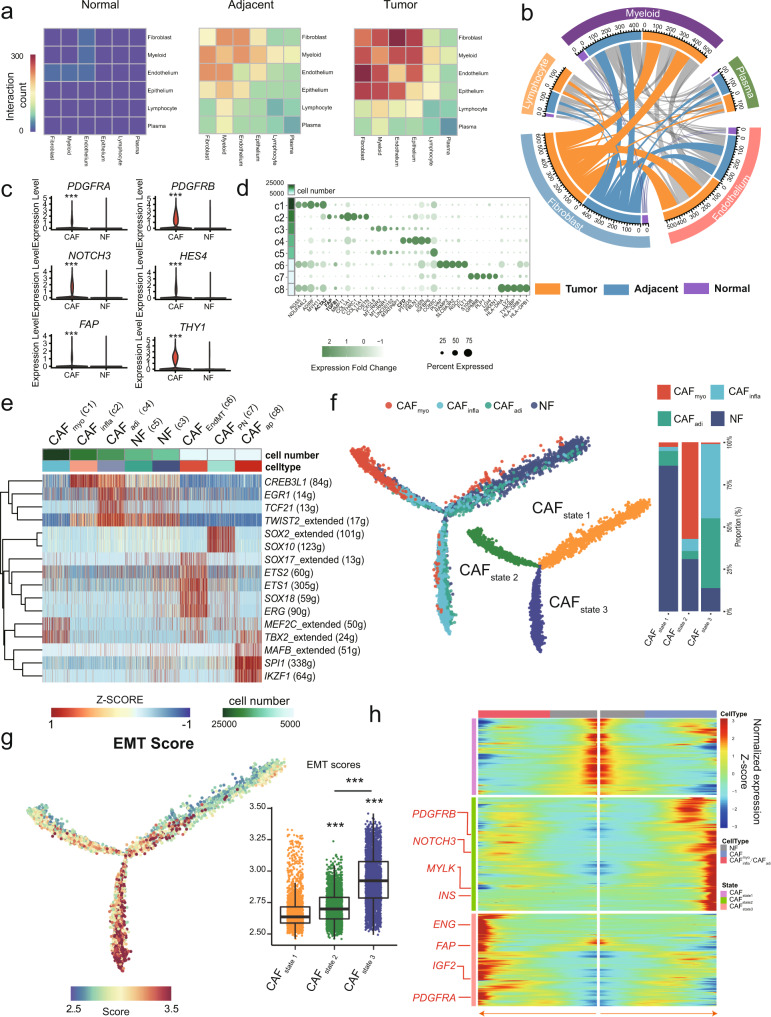
Compared to the biased distribution of epithelial cells (Supplementary Fig. S1d), TME components from different cancer types were clustered together by unsupervised dendrograms. This proved that different lineages of TME components shared similar transcriptomic profiles across different cancer types (Supplementary Fig. S3a). Given complicated intercommunications among TME components play a critical role in tumor progression and treatment response, CellphoneDB-based analysis was conducted to evaluate the interactions between each major TME component and epithelial cells. The overall interactions significantly increased in the order of normal, adjacent, and tumor regardless of the tissue type (p < 0.001) (Fig. 2a). Notably, crosstalk among fibroblasts, endothelial cells, and myeloid cells was dominant in the TME, and fibroblasts presented the most prolific interactions with other TME components in tumor/adjacent samples regardless of the tissue type (all p < 0.001) (Fig. 2a, b and Supplementary Fig. S3b), suggesting the possible important role of fibroblasts in cancer biology through communication with other TME components.
Fig. 2. Generalized activation of CAFs in the TME.
a The mutual interaction among the main TME components and epithelial cells in different tissue origins. b The interaction between fibroblasts and other TME components. The length of arcs represents the predicted interaction counts. c Violin plot of specific marker genes in cancer-associated fibroblasts (CAFs) and normal fibroblasts (NFs). Mann–Whitney two-sided test is used to test the significance of gene expression level between CAF and NF categories. ***p < 0.001. d Bubble plot showing the expression of tag genes between CAFs and NFs. e Regulons enriched in each fibroblast cluster detected via SCENIC analysis. f Left: activation trajectory of CAFs, which are divided into three states (CAFState1/2/3). Right: histography of the different CAF components in each CAF state. g Left: epithelial-mesenchymal transition (EMT) score enriched along the evolutionary trajectory of CAFs. Right: comparison of EMT scores among three CAF states, state1: n = 2130, state2: n = 4948, state3: n = 8667. The box is bounded by the first and third quartile with a horizontal line at the median and whiskers extend to the maximum and minimum value. Mann–Whitney two-sided test is used to test the significance of EMT scores among different state categories. ***p < 0.001. h Fluctuation of genes along different states. Source data are provided as a Source Data file.
A total of eight clusters (c1-c8) were annotated as fibroblasts (ACTA2 and ACTG2) and presented in all tissue types (Fig. 1e). According to the differences in the distribution of cell proportions, cells in c3 and c5 were dominantly derived from normal tissues and referred to as NFs, while the remaining six tumor-enriched clusters were considered as CAFs due to their predominant deviation from tumor/adjacent tissues (Fig. 1e). Compared to the transcriptional profile of NFs, collagen activation- and matrix metalloproteinase-related genes were highly expressed in CAFs, suggesting the activation of fibroblasts in CAFs (Supplementary Fig. S3c and Supplementary Data 6). Furthermore, angiogenesis- and immunomodulation-related genes (e.g., PDGFRA, PDGFRB, FAP, NOTCH3, HES4, and THY1) were significantly upregulated in CAFs (Fig. 2c). Specifically, according to canonical markers34–36 (Fig. 2d), the three quantity-predominant major components (i.e., c1, c2, and c4) were defined as cancer-associated myofibroblasts (CAFmyo), inflammatory CAFs (CAFinfla), and adipogenic CAFs (CAFadi) by over-presenting ACTA2, FAP/TGFB1, and CFD, respectively (Fig. 2d and Supplementary Fig. S3d). Given the consensus in the origin of CAFs7, the other three minor components (i.e., c6-c8) were identified as endothelial-to-mesenchymal transition CAF (CAFEndMT), peripheral nerve-like CAF (CAFPN), and antigen-presenting CAF (CAFap) by overpresenting specific marker genes (Fig. 2d and Supplementary Data 2). As expected, these CAF clusters are involved indistinct pathways (Supplementary Fig. S3e).
Furthermore, we determined the possible involvement of regulons in CAFmyo, CAFinfla, and CAFadi by SCENIC analysis. Tumorigenesis- (e.g., TBX237) and myogenesis-related regulons (e.g., MEF2C38) were highly enriched in CAFmyo. In addition, dedifferentiation-related (e.g., CREB3L139) and epithelial-mesenchymal transition (EMT)-related regulons (e.g., TWIST225) were enriched in CAFinfla and CAFadi, respectively (Fig. 2e). Therefore, we speculated that the activation of CAFmyo was different from CAFadi and CAFinfla. Through similarity analysis as described previously21, the same major lineages of CAFs from different cancer types were clustered together, illustrating their shared characteristics among diverse cancer types (Supplementary Fig. S4a). Therefore, we pooled the CAFs from all cancer types together to explore the possible general activation process of CAFs. Two distinct activation paths from NFs to CAFs were revealed via evolutionary trajectory, enhancing the definition of three different states, CAFstate1 (NFs dominant), CAFstate2 (CAFmyo dominant), and CAFstate3 (CAFadi/CAFinfla dominant) (Fig. 2f), all of which were not biased in terms of the constitution of tissue type (Supplementary Fig. S4b). Both the EMT score (Supplementary Data 7) and CREB3L1 expression gradually increased along the activation trajectory of CAFs and were significantly higher in CAFstate3 than in CAFstate1/2 (p < 0.001) (Fig. 2g and Supplementary Fig. S4c) regardless of the tissue type (Supplementary Fig. S4d), suggesting a general dedifferentiated process along CAF activation. Consistently, the regulon activity of CREB3L1 (Supplementary Data 7) significantly increased along with the expression of CREB3L1 and peaked in CAFstate3 (p < 0.001) (Supplementary Fig. S4c–e). Finally, tracing the gene fluctuation along biforked trajectories, CAFstate2 tended to act in angiogenesis represented by overexpressing INS and PDGFRB, while CAFstate3 had a high expression of both pro-angiogenic (e.g., PDGFRA)40 and immunomodulation-related genes (e.g., ENG and FAP)36 (Fig. 2h and Supplementary Data 8). Collectively, the major CAFs were derived from NFs and evolved into different differentiated states that may exhibit distinct effects on the TME.
Interactions of CAFs with TME and epithelia
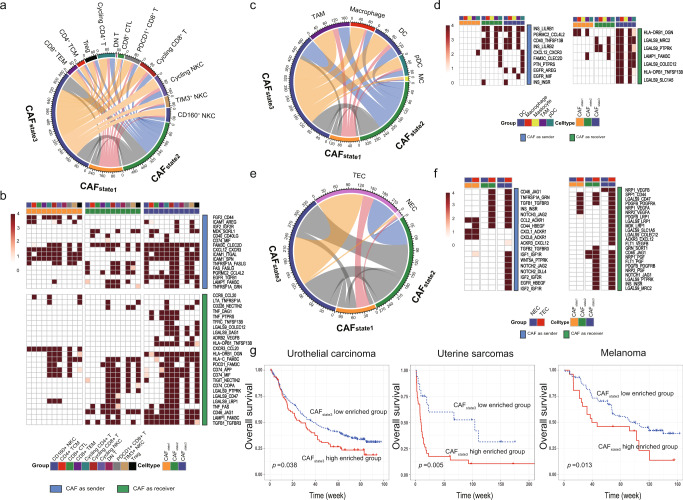
Immunosurveillance escape is one of the major hallmarks in cancer, and CAFs can facilitate this process not only by providing a physical barrier but also by impacting on the immune TME. Based on canonical gene expression, tumor-infiltrating- natural killer cell (NKC)/T-cells were divided into three NKC clusters (e.g., TIM3+ NKCs) and eight T cell clusters (e.g., CD8+ cytotoxic T cells and regulatory T cells (Tregs)) (Supplementary Fig. S5a). The interaction counts between CAFstate3 and NK/T subclusters were superior to CAFstate1 and CAFstate2 (Fig. 3a), highlighting its potential immunoregulatory role. Shared and specific reciprocal communication was observed between different CAF states and NK/T cells. In particular, CD44, an important index of T cell activation and navigation in antitumor immunity41, exclusively interacts with FGF2 in CAFstate1 among different NK/T subclusters, while TGF-β (TGFB1)42, progranulin (GRN)43, galectin-9 (LGALS9)44, and tumor necrosis factor (TNF)45, which regulate adaptive immune cells, tend to interact with their partner receptors in CAFstate2 and CAFstate3 (Fig. 3b). Subcluster-specific interactions were also observed, such as the absence of most interactions in double-negative T cells (DN T) and specific LTA_TNFRSF1A binding in Treg/cycling CD4+ cells for all CAF states (Fig. 3b). In addition to interact with NK and T cells, CAFs may also regulate tumor-infiltrating B-cell by overexpressing the TNF ligand superfamily. For instance, CAFstate3 interacted with the majority of B-cell subclusters via TNFSF13B_TNFRSF13B/C (B-cell survival/maturation-related genes)46 (Supplementary Figs. S1k and S5b). In addition to adaptive immunity, CAFs may also modulate innate immune cells, illustrating predominant interaction counts between CAFstate3 and myeloid components (Fig. 3c). TAM was the most prolific partner, whereas mastocytes hardly interacted with CAFs (Fig. 3d). Through the CXCL12_CXCR3 interaction, CAF state1/2/3 specifically may recruit pDCs (Fig. 3d), which can secrete granzyme B to constrict the expansion of T cells47 (Supplementary Fig. S1h). In addition to the suppressive function of CAFs in priming DCs, CAFstate3 may also stimulate DC activation and maturation by interacting with galectin-9 (LGALS9)48 and CD40 [ref. 49] on DC subtypes (Supplementary Fig. S5c). Therefore, CAFs, particularly CAFstate3, may present immunomodulatory capability.
Fig. 3. CAFs orchestrate the immune TME and angiogenesis.
a and b Predicted and detailed interactions between different CAF states and NK/T cells. c and d Predicted and detailed interactions between different CAF states and subgroups of dendritic cells. e and f Predicted and detailed interactions between different CAF states and endothelial cells (TECs and NECs). g Estimation of the prognostic value of the CAFstate3 signature score in three immunotherapy cohorts (urothelial carcinoma, uterine sarcoma, and melanoma) Kaplan–Meier curves for overall survival in all patients according to the number of positive ligands. p-values for all survival analyses have been calculated using the log-rank test.ue.
Next, we examined prime activity on endothelial cells, which are essential for angiogenesis. More dramatic interactions were observed between TECs and CAFstate2/3 than CAFstate1 (Fig. 3e), including NOTCH1/3_JAG1/JAG2, TGFB1_TGFBR3 and INS_INSR (Fig. 3f), which are involved in angiogenesis-related VEGF and NOTCH signaling pathways. In particular, interactions of NOTCH2_JAG2/DLL4, ACKR3_CXCL12 and IGF1/2_IGFR1R/2 R were specifically present between CAFstate3 and TECs.
Moreover, communications between three CAF states and each epithelia cluster were estimated. Interestingly, CAFstate3 also exhibited more interactions with epithelia than CAFstate1/2, especially with E3/E13 (clusters shared by all cancer types), E4 (dominant by digestive system tumor), and E18 (dominant by breast and ovarian cancer) (Supplementary Figs. S2b and S5d). Moreover, we focused on the crosstalk between each CAF state with the epithelia clusters exhibited similarities across cancer types (i.e., E3, E5, E8, E9, E10, and E13) (Supplementary Fig. S5e), a series of ligand-receptor pairs were identified, which are involved in cancer related pathways, including EGFR (e.g., EGFR_TGFB1), NOTCH (e.g., JAG1_NOTCH2/3), WNT pathways (e.g., FZD6_WNT5A) (Supplementary Fig. S5f).
Given the prolific communication between CAFs and immune cells, particularly CAFstate3 (e.g., PDCD1_FAM3C interaction in CAFstate3) (Fig. 3b), it is possible that CAF states may play an important role in checkpoint blockade immunotherapy. For instance, galectin-9 (encoded by LGALS9) related interactions have been linked to tolerogenic macrophage programming and adaptive immune suppression50, and regulate T-cell death for cancer immunotherapy51. Interestingly, a series of interactions between LGALS9 and its partners were enriched in crosstalk between T-cell/myeloid/endothelial/epithelial and CAFstate3 but not CAFstate1/state2 (Fig. 3b, d, f, Supplementary Fig. S5c and f). Therefore, we conducted ssGSEA-based deconvolution analysis to estimate the proportion of CAFs in each state using bulk transcriptome profiles of patients from three independent immunotherapy cohorts with open accessible sequencing data and follow-up information across different cancer types (i.e., melanoma52, urothelial cancer53, and uterine sarcoma54). Intriguingly, compared to the insignificant associations observed for CAFstate1 and CAFstate2 (Supplementary Fig. S6a), a high proportion of CAFstate3 was significantly associated with poor overall survival in these cohorts (p = 0.038, p = 0.005, and p = 0.013, respectively) (Fig. 3g), even after adjusting available covariates in each cohort study by multivariate analysis (p = 0.049, p < 0.001, and p < 0.001, respectively) (Supplementary Fig. S6b), suggesting the potential independent prognostic value of CAFstate3 for immunotherapy
Characterization of the plasticity of fibroblasts via pan-cancer analysis
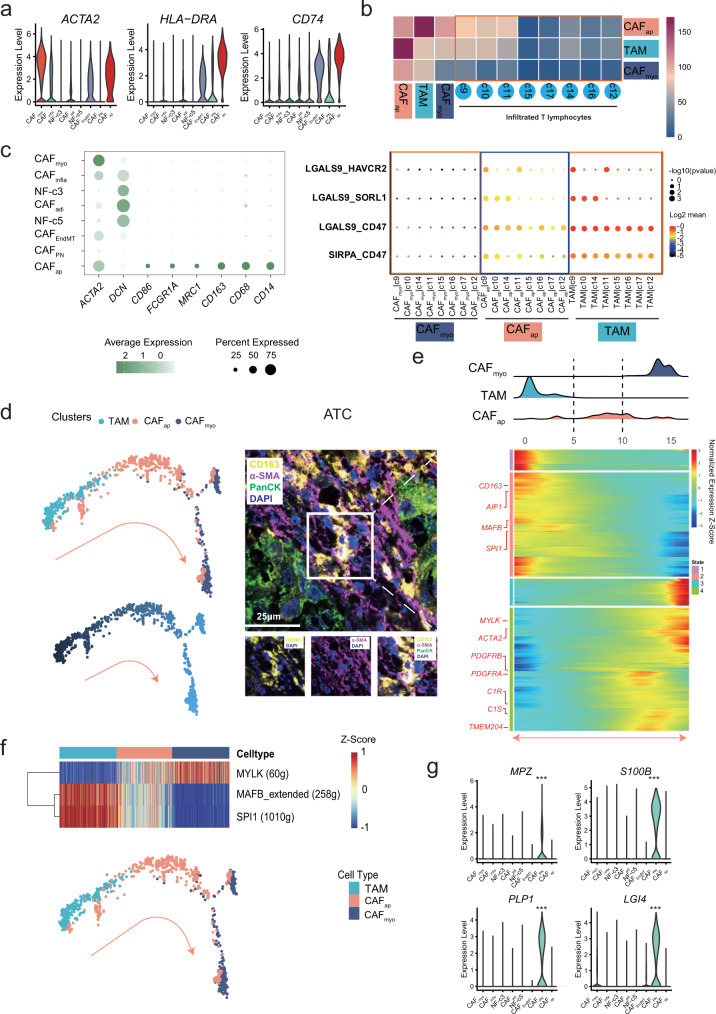
Fibroblast activation is an important source of mesenchyme-derived stromal components7. Except for the main origin of CAFs from NFs (Fig. 2f), CAFs may have alternative origins with various biofunctions in the TME7. As described above, three distinct clusters of CAFs with a small number of cells (i.e., c6-c8) were shared by all cancer types (Fig. 1d and Supplementary Data 3). As reported, c8 was defined as antigen-presenting CAFs (CAFap) that overexpressed ACTA2, HLA-DRA, and CD74 (Fig. 4a)55 and was enriched in pancreatic cancer (Supplementary Fig. S7a). Intriguingly, CAFap presented significantly more interactions with tumor-infiltrating T-cell clusters than CAFmyo (p = 0.002), with a similar interactive pattern as that of TAM (e.g., LGALS9_HAVCR2/SORL1/CD47) (Fig. 4b). Moreover, CAFap exhibited a higher transcriptional similarity with mono-macrophage-related clusters (c18-c22) than with other fibroblast clusters (c1-c7) (Supplementary Fig. S7b) and expressed mono-macrophage-specific markers (e.g., CD68, CD163, and CD14) (Fig. 4c). Using the SCENIC analysis described above, several regulons were highly enriched in CAFap (e.g., MAFB, SPI1, and IKZF1) (Fig. 2e), which are well-known mediators of polarization and function in TAMs56–58. This evidence implied a possible association between CAFap and TAM.
Fig. 4. Characterization of fibroblast plasticity.
a Violin plot of specific gene expression in antigen-presenting CAFs (CAFap). b Upper: peer comparison of the interactions of CAFap, TAM, and CAFmyo with T-cell clusters. Lower: the instance of interaction pattern presentation in CAFmyo, TAM, and CAFap with T-cell clusters. c Bubble plot of mono-macrophage-specific markers in each fibroblast cluster, one-sided Wilcoxon rank-sum test is used to assess the statistical significance of each interaction score. d The evolutionary trajectory along the TAM-CAFap-CAFmyo path. Confocal image of multiplexed immunofluorescence staining of PanCK, α-SMA, and CD163 in anaplastic thyroid cancer tissues. Multiplexed immunofluorescence assays are performed twice on tumor samples following assay optimization. e Gene expression alteration and ridgeline plot along the reciprocal trajectory. f Regulon enrichment along the evolutionary trajectory in different cell types. g Violin plot of peripheral nerve-specific genes (MPZ, S100B, PLP1, and LGI4) in CAFPN. Kruskal-Wallis two-sided test is used to test the significance of gene expression level among different fibroblast clusters. ***p < 0.001.
The transition between macrophages and myofibroblasts has recently been discovered as a pathogenic process that plays a regulatory role in renal fibrosis59, renal allograft injury60 and myocardial infarction healing61. However, the dynamic alteration between myofibroblasts and macrophages remains unclear. We speculated that CAFap might be a transitional position between CAFs and TAMs based on previous reports and our findings. With pseudotime trajectory analysis (Fig. 4d), a possible evolutionary TAM-CAFap-CAFmyo path was implied. As validation, we found a small population of cells co-expressing α-SMA and CD163 in the stromal compartment of anaplastic thyroid cancer, colorectal cancer and stomach cancer through confocal mIF imaging (Fig. 4d and Supplementary Fig. S7c). Cells with double positivity (α-SMA+ CD163+) ranged from 10.2% to 13.6% of all α-SMA+ cells in these cancer types, which is consistent with our pan-cancer single-cell analysis and a previous report in pancreatic cancer55. Given that the regulatory role of CAFap in Tregs has been experimentally determined in pancreatic cancer62, the similar functions of this CAF subtype may be shared by different cancer types.
Expression of a series of fibroblast-specific (e.g., ACTA2 and MYLK) and macrophage-specific genes (e.g., CD163, MAFB, and SPI1) gradually changed from TAM to CAFmyo, with CAFap located at the intermediate position (Fig. 4e). Moreover, CAF- (e.g., MYLK) and TAM-specific regulons (e.g., MAFB and SPI1) were gradually enriched along the TAM-CAFap-CAFmyo trajectory, and CAFap was present in both regulons (Fig. 4f), suggesting a the possible transitional position of CAFap between CAFs and TAMs, which may be an alternative origin of CAFs.
Similarly, c7 was identified as fibroblast-like peripheral nerve cells (CAFPN), specifically expressing peripheral nerve-related genes (e.g., MPZ, S100B, LGI4, and PLP1) (Fig. 4g). CAFPN was abundant in STAD, COAD/READ, PAAD, PRAD and OV (Supplementary Fig. S7d), in which perineural invasion was considered as an indication of poor prognosis63. Additionally, CAFPN enriched unique transcriptional regulons (i.e., SOX2 and SOX10) in regulating myelination of peripheral nerves64,65 with distinct metabolic hallmarks (e.g., peroxisome, bile acid, and cholesterol homeostasis) (Fig. 2e and Supplementary Fig. S3e).
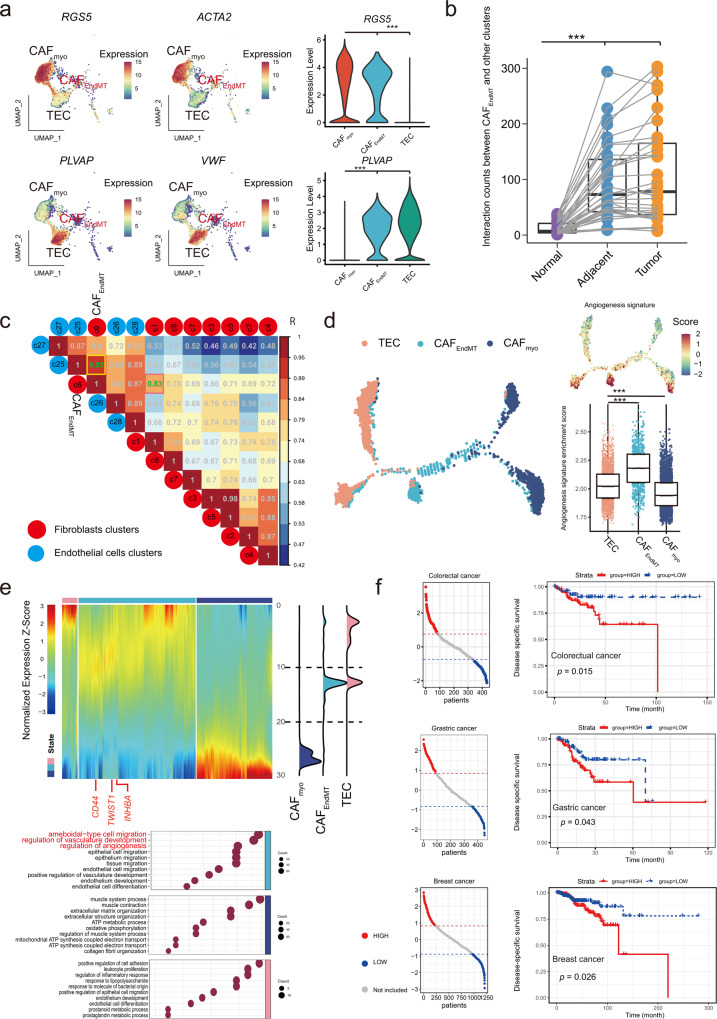
Besides macrophages and peripheral nerve-derived CAFs, CAFs also exhibited potential plasticity from endothelial cells by endothelial-mesenchymal transition (EndMT), defining c6 as CAFEndMT. Intriguingly, CAFEndMT exhibited dual expression of canonical lineage markers of fibroblasts (e.g., RGS5 and ACTA2) and endothelial cells (e.g., PLVAP and VWF), highlighting the possibility of its involvement in EndMT, which has recently been described in gastric cancer66 (Fig. 5a). It was confined to all types of tumor/adjacent tissues (Supplementary Fig. S8a) and had more prolific communications with other TME components in tumor/adjacent tissue than normal tissue (Fig. 5b). Moreover, CAFEndMT exhibited dramatic transcriptional similarity with both CAF (e.g., correlation coefficient R = 0.83 with CAFmyo) and TEC (R = 0.92) (Fig. 5c). Besides, compared to all the other CAFs and endothelial cells, a unique signature (e.g., ESM1) of CAFEndMT (Supplementary Data 9 and Supplementary Fig. S8b) was enriched in ameboidal-type cell/epithelial cell migration signaling (Supplementary Fig. S8c), which was associated with angiogenesis, and consistent with previous hallmark enrichment67 (Supplementary Fig. S3e). Moreover, CAFEndMT distinctly presented endothelial differentiation- and conversion-related regulons (e.g., SOX17/18 and ETS1/2)68,69 (Fig. 2e). Consistently, CAFEndMT was located at the transitional position from TEC to CAFmyo in the evolutionary trajectory (Fig. 5d). As expected, the angiogenesis hallmark signature (Supplementary Data 7) was highly enriched in CAFEndMT, which was significantly higher than both TEC and CAFmyo (both p < 0.001) (Fig. 5d), suggesting its possible role in angiogenesis. A series of genes were distinctly enriched in each type of cells, and CAFEndMT tended to be involved in several pathways, including ameboidal-type cell migration and vasculature development (Fig. 5e). Moreover, we estimated the CAFEndMT signature in each patient from The Cancer Genome Atlas (TCGA) cohort using ssGSEA-based deconvolution analysis. Due to the small proportion of CAFEndMT in tumors, we compared patients with the top and bottom 20% percentile of the CAFEndMT score. A higher proportion of CAFEndMT is associated with poor prognosis in several types of cancers with large sample sizes, including breast, gastric, and colorectal cancers (Fig. 5f and Supplementary Fig. S9a). Based on the evidence above, CAFEndMT reflects the plasticity of TECs to CAFs and is linked to the initial step of angiogenesis among multiple cancer types, which is associated with metastasis and cancer progression70.
Fig. 5. Characterization of CAFs in endothelial-mesenchymal transition (EndMT).
a Feature and violin plots of specific genes in CAFEndMT and TECs. b The interaction counts between CAFEndMT and other components in different tissue origins. Normal: n = 34, Adjacent: n = 34, Tumor: n = 34. The box is bounded by the first and third quartile with a horizontal line at the median and whiskers extend to the maximum and minimum value. Mann–Whitney two-sided test is used to test the significance of interaction counts between CAFEndoMT and other clusters. Normal vs Adjacent p-value is 2.52 × 10−6, Normal vs Tumor p-value is 5.87 × 10−8, ***p < 0.001. c Genetic similarity between clusters of CAFs and endothelial cells. d The evolutionary trajectory along the TECs-CAFEndMT-CAFmyo path with the angiogenesis hallmark signature enriched along the trajectory and CAFEndMT. State1: n = 3550, state2: n = 754, state3: n = 3062. The box is bounded by the first and third quartile with a horizontal line at the median and whiskers extend to the maximum and minimum value. Mann–Whitney two-sided test is used to test the significance of Angiogenesis signatures enrichment scores among TECs-CAFEndMT-CAFmyo clusters. TEC vs CAFEndoMT p-value is 4.67 × 10−102. TEC vs CAFmyo p-value is 5.53 × 10−27, ***p < 0.001. e Gene expression alteration with gene ontology and ridgeline plot along the reciprocal trajectory. f Estimation on the prognostic value of the CAFEndMT signature in colorectal, gastric and breast cancer in terms of disease-specific survival. Kaplan–Meier curves for overall survival in all patients according to the number of positive ligands. p-values for all survival analyses have been calculated using the log-rank test. Source data are provided as a Source Data file.
Collectively, we characterized the shared plasticity of fibroblasts across different cancer types and suggested alternative cell origins of CAFs from TECs at single-cell profiles across cancers.
The triple interplay between CAFs, TECs, and TAMs in the TME
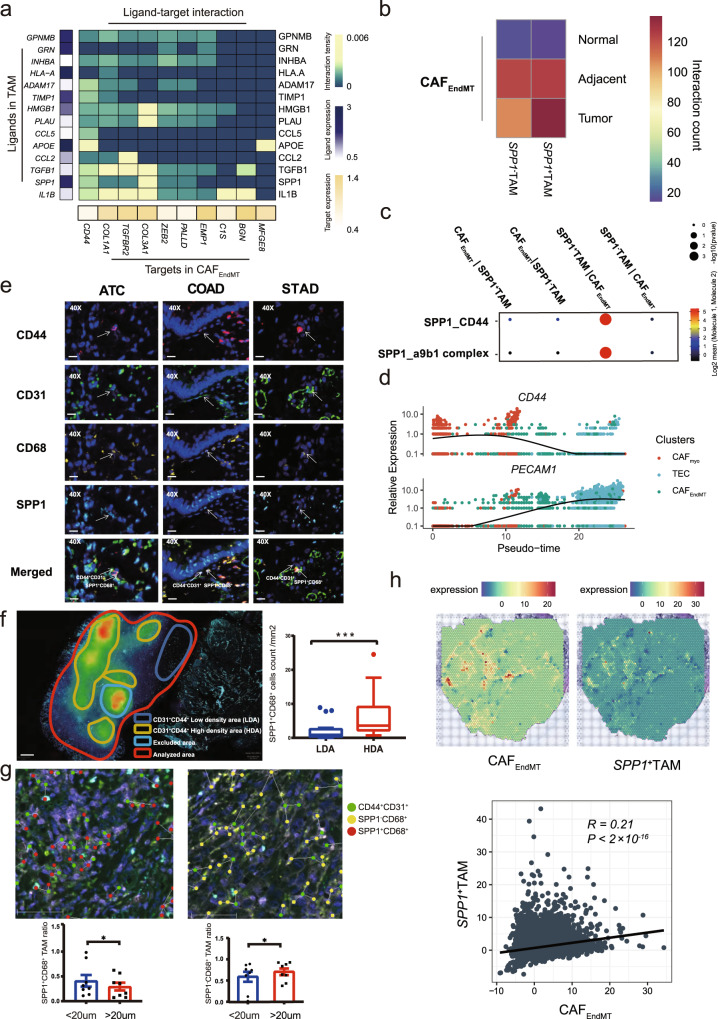
The triple interplay among fibroblasts, endothelial cells, and myeloid cells was predominant in the TME among all tissue types (Fig. 2b). As stated, increasing interactions between each TME component were observed along normal, adjacent, and tumor tissues (p < 0.001). CAFEndMT had the most prolific communications with other TME components, particularly TAM (Supplementary Fig. S9b). Therefore, we speculated that TAMs may play a role in the EndMT process and subsequent tumor angiogenesis. Using a linear regression model, we screened out top 30 genes that were gradually expressed along the EndMT process from CAFmyo to TECs via CAFEndMT (Supplementary Data 10). Intriguingly, CD44 ranked the top in terms of the interaction counts between CAFEndMT and TAM estimated by NicheNet analysis71, with SPP1 and APOE exhibited the highest expression in TAM among all its partners (Fig. 6a). Since APOE was expressed in nearly 90% of TAMs, we focused on SPP1+ TAMs and found more interactions of CAFEndMT with SPP1+ TAMs than that with SPP1- TAMs in tumor samples (Fig. 6b). Particularly, the SPP1_a9b1 complex and SPP1_CD44 axes were exclusively enriched in the reciprocal interaction between SPP1+ TAMs and CAFEndMT (Fig. 6c). However, only CD44 expression increased in a stepwise manner along the EndMT trajectory (Fig. 6d), while no trend was observed for ITGA9 and ITGB1 (Supplementary Fig. S9c), whose protein products formed the a9b1 complex. This result suggested the possible involvement of the SPP1_CD44 interaction in the EndMT process. We next validated this hypothesis by mIF to assess the spatial distribution of SPP1+ TAMs (SPP1 and CD68) and CAFEndMT (CD44 and CD31, canonical endothelial markers encoded by PECAM1, which decline in a stepwise manner during the EndMT process (Fig. 6d)). In three cancer types (i.e., anaplastic thyroid, colorectal, and gastric cancers), mIF consistently illustrated the proximity of some SPP1+ TAMs (SPP1+CD68+) to CAFEndMT (CD44+CD31+) (Fig. 6e). Phenotypic images and density maps were used to quantify marker expression and spatial distribution, respectively (Fig. 6f and Supplementary Fig. S9d). After excluding the defective regions (Supplementary Fig. S9e), the spatial density distribution of CAFEndMT cells was quantified and classified into high-density (HDA) and low-density areas (LDA), and SPP1+ TAMs were significantly enriched in the HDA compared to the LDA (Fig. 6f). Furthermore, when quantifying the spatial distribution, we found that the SPP1+ TAM ratio normalized by the total number of TAMs was significantly higher within 20 μm of CAFEndMT than that outside 20 μm (Fig. 6g). In contrast, the SPP1- TAM ratio was significantly lower within 20 μm (Fig. 6g). Moreover, a significantly high correlation (R = 0.23, p < 0.001) was identified between the signature enrichment of CAFEndMT and SPP1+ TAMs in the spatial transcriptomic profile from seven colorectal tumor samples (Fig. 6h). Additionally, the significantly positive associations of enrichment scores of CAFEndMT and SPP1+ TAMs were also validated in 25 of 28 cancer types in the TCGA dataset (Supplementary Fig. S10), further supporting the possible proximity of SPP1+ TAMs and CAFEndMT. The evidence described above suggested that pro-angiogenic SPP1+ TAMs may play a role in the EndMT process to facilitate intratumoral angiogenesis through the SPP1_CD44 interaction between SPP1+ TAMs and CAFEndMT and thus implied poor prognosis in cancer patients. However, mechanistic verifications are largely needed in the future.
Fig. 6. Triple interplay between CAFEndMT and SPP1+ TAMs.
a NicheNet analysis screening potential ligands of CD44. The left bar presents the expression scale of the potential ligand in TAM. b Predicted interaction counts between CAFEndMT and SPP1+ TAM/SPP1- TAM using CellphoneDB analysis. c SPP1-involved specific ligand–receptor interaction between CAFEndMT and SPP1+/SPP1- TAMs, one-sided Wilcoxon rank-sum test is used to assess the statistical significance of each interaction score. d Dynamic alterations in CD44 and PECAM1 during EndMT. e Multiplexed immunofluorescence staining of CD44, CD31, CD68, and SPP1 in anaplastic thyroid cancer, gastric cancer, and colorectal cancer tissues, Scale bar: 20 μm. Multiplexed immunofluorescence assays are performed twice on tumor samples following assay optimization. f Illustration of CD44+CD31+ high- and low-density areas (HDA and LDA, respectively) and the quantified results (LDA: n = 23, HDA: n = 17, The box is bounded by the first and third quartile with a horizontal line at the median and whiskers extend to the maximum and minimum value. Mann–Whitney two-sided test is used to test the significance of proportion between LDA and HDA categories. ***p < 0.001, p-value is 0.0008), Scale bar: 500 μm Multiplexed immunofluorescence assays are performed twice on tumor samples following assay optimization. g The spatial distance quantification. The left panel compares the SPP1+CD68+ and the right panel compares SPP1-CD68+ macrophage ratios (normalized by the total number of macrophage) between within 20 μm and outside 20 μm of CAFEndMT (<20 μm: n = 8, >20 μm: n = 8, Wilcoxon two-sided test is used to test the significance of ratio between within 20 μm and outside 20 μm of CAFEndMT. *p < 0.05, left panel: p-value is 0.0391, right panel: p-value is 0.0391). Scale bar: 50 μm. h Upper: Illustration of the spatial transcriptomic spot of colorectal cancer tissues with CAFEndMT and SPP1+TAM signature enrichment. Lower: The scatter plot and correlation between the CAFEndMT enrichment score and SPP1+TAM enrichment score (R represents Pearson’s correlation and its coefficient of determination, p-value is 2.26 × 10−10), suggesting the co-localization of these two cell types. Source data are provided as a Source Data file.
Discussion
Single-cell profiles were investigated in multiple cancer types to reveal the heterogeneity and cancer biology of both cancer and stromal cells. A few studies have conducted pan-cancer analysis to systematically illustrate shared and cancer-type specific characteristics of different cell components, particularly for myeloid and T cells21,22,24. Overall, this is a systematic investigation of a single-cell transcriptional atlas of fibroblasts across cancer types.
As an important mesenchyme-derived stromal component, fibroblasts are plastic in phenotype7. Pan-cancer analysis offered us an opportunity to characterize the cells in a fluctuating state across various tissues, which was difficult to detect in the original research due to its low population. In the present analysis, the state fluctuation or transition of fibroblasts was associated with multiple biological functions (e.g., angiogenesis, immune modulation, and EMT) and clinical outcomes, particularly after immunotherapy. Although it is crucial to investigate the biological function of CAF subtypes, such as FAP+ CAFs72, ENG+ CAFs, and THY1+ CAFs, it is more important to explore the generalized characteristics of CAFs in the TME. Here, we found that CAFs had divergent differential states with specific biofunctions. Of note, CAFstate3, which was at the most dedifferential state, predicted a worse outcome of immunotherapy. Thus, CAF differentiation may promote the stratification of patients with immunotherapy. In addition, the origin of CAFs was delineated in our study. Consistent with consensus7, the trajectory of CAFs from state1 to state2/state3 exemplified that most CAFs were likely derived from the activation of local normal fibroblasts. Moreover, the three minor clusters of CAFs (CAFEndMT, CAFpn and CAFap) imply alternative origins from endothelial cells, peripheral nerves, and macrophages, respectively.
The SPP1 gene encodes an integrin-binding glyco-phosphoprotein, named as osteopontin. It is secreted by various tumors and is associated with tumor progression, invasion, and metastasis73,74. Although it could be chemokines that recruit macrophages75, SPP1 expression was significantly related to TAMs in multiple tumors in the TIMER study76. Thus SPP1+TAM should be abounded in tumors. Furthermore, a previous atlas of tumor-infiltrating myeloid cells identified angiogenesis-associated macrophages in 8 cancer types, which was marked by the expression of SPP121. However, the potential mechanism was not postulated. In the present study, we found that SPP1+ TAMs may be involved in tumor angiogenesis by interacting with adjacent CAFEndMT, which is regarded as the initial step of angiogenesis70.
Limitations of the study should be noted to avoid overinterpretation. First, some canonical markers may not exhibit restrictive expression in different cell types, thus necessitating expanding evidence to support the findings and avoid possible misleading. Although the expression of RGS5 and ACTA2 defined that CAFEndMT was also reported in a recent study77, the possibility of pericytes could not be excluded. It is well established that pericytes are an important resource of CAFs in tumors78. Pericytes recruited by EndMT78 or pericyte-fibroblast transition79 lead to tumor vasculature. Overall, pericytes are an important origin of CAFs dependent on EndMT (mainly) or pericyte-fibroblast transition that play roles in angiogenesis80. It would be interesting to demarcate or subcluster pericytes from CAFs and determine the difference between them in pan-cancer level in future studies. Second, mechanistic analysis has determined the potential functions of some CAF populations in specific cancer types (e.g., key role of CAFap on Treg in pancreatic cancer58). Although we profiled the ubiquitous characteristics of different CAF subpopulations at single-cell resolution, experimental validation is still warranted to determine whether these functional CAF cells can play a similar role across diverse cancer types. Third, we only demonstrated the possible alternative origin of CAFs mainly through bioinformatic approaches for evolutionary trajectory and illustrated the possible cells at an intermediate state through mIF with specific markers. However, further verification on the dynamics of CAFs and the underlying mechanisms/crucial regulatory factors is lacking, which should be explored in the future.
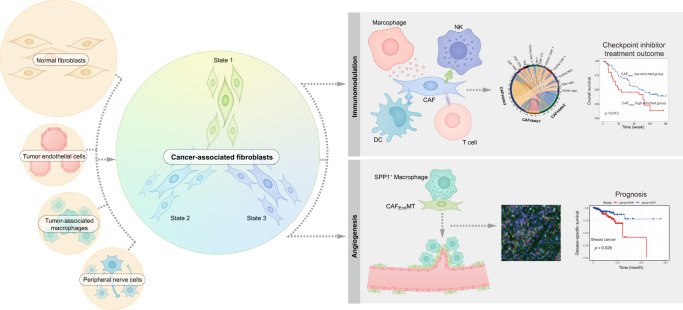
In conclusion, we systematically characterized CAFs across cancers, not only providing the possible origins of CAFs but also highlighting the different states of CAFs in the outcome of immunotherapy and prognosis. To accelerate the related data mining and application in wide research, we established an interactive website-based tool (https://gist-fgl.github.io/sc-caf-atlas/). Collectively, although further experimental verification is warranted to establish the functional role of each CAF cluster and diverse origins of CAFs, our pan-cancer study on CAFs may facilitate CAF-targeted therapy development and application in the future (Fig. 7).
Fig. 7. Summary illustration of the study.
Left part: four circles stand for the origins of cancer associated fibroblast (CAF). The big circle indicates the main origin of CAF-derived from normal fibroblasts activation, whereas the three small circles indicate the alternative origin of CAF. In general, the activation trajectory is divided into three states (state 1–3). Right part: The state of CAF is associated with immunomodulation, thus may predicting the prognosis of checkpoint-inhibitor-based treatment for specific cancer types, and it is also associated with angiogenesis by interacting with proximal SPP1+ macrophages and prognosis of cancer patients.
Methods
Single-cell RNA-seq data processing
Publicly available and inhouse FASTQ files generated from 10× Genomics were aligned and quantified against the GRCh38 human reference genome using Cell Ranger software (Version 6.1.2) with default settings. The output of the cellranger and count matrix were read using the Read10X function from the Seurat package (Version 4.0.4) and read.table function, respectively, and the latter was further converted to dgCMatrix format. Potential doublets predicted by Scrublet81 were removed to avoid interference with the analysis. The merge function was used to integrate all individual objects into an aggregate object, and the RenameCells function was used to ensure that all cell labels were unique. In total, 990,990 cells from different studies were pooled. Furthermore, quality control was applied to the cells based on several criteria. Briefly, cells with <200 detected genes as well as those with >20% mitochondrial content were removed. Cells having over 6000 detected genes were eliminated to further exclude the possible doublets. After filtering, 855,271 high-quality cells were preserved for subsequent analyses. A global-scaling normalization method (“LogNormalize”) was employed to ensure that the total gene expression in each cell was equal, and the scale factor was set to 10,000. The top 2000 variably expressed genes were returned for downstream analysis using the FindVariableFeatures function. The ScaleData function, “vars.to.regress” option UMI, and percent mitochondrial content were used to regress out unwanted sources of variation. Principal component analysis (PCA) incorporating highly variable features reduced the dimensionality of this dataset, and the first 30 PCs were identified for analysis. To remove batch effects, the RunFastMNN function in SeuratWrappers package (Version 0.3.0) was selected to perform sample batch correction. Clustering analysis was performed based on the edge weights between any two cells, and a shared nearest-neighbor graph was produced using the Louvain algorithm, which was implanted in the FindNeighbors and FindClusters functions. The identified clusters were visualized using the UMAP method. For subclustering analysis, a similar procedure was applied, including normalization, variably expressed feature selection, dimension reduction, batch correction with RunFastMNN, and clustering identification. To annotate the cell clusters, differentially expressed markers of the resulting clusters were identified with the FindAllMarkers function using the default nonparametric Wilcoxon rank sum test with Bonferroni correction.
Comparison dendrograms
To demonstrate that the subpopulations of CAFs or myeloid cells were not heterogeneous across tumor types, an unsupervised comparison dendrogram was performed. We selected the top 2000 highly variable genes across different subclusters. The mean expression of these genes in each cluster was used to calculate the Pearson correlation coefficient with the psych package (Version 2.2.5). The distance defined as (1-Pearson correlation coefficient)/2 was adopted for hierarchical clustering. For visualization, the factoextra package (Version 1.0.7) was applied.
Cell–cell interaction analysis
CellphonedDB19 was used to analyze cell–cell interactions among all TME components. Input files for the statistical analysis function comprised a raw count matrix extracted from the Seurat object and an annotation file of cell types. The heatmap_plot function from CellphoneDB and the Circlize package (Version 0.4.14) were used to display the frequency of interactions between two cell subsets. Visualization of the potential interaction strength between ligand and receptor, which was predicted based on their average expression, was performed using the dot_plot function and pheatmap package (Version 1.0.12). Significant ligand–receptor pairs (p < 0.01) were extracted for illustration.
Trajectory analysis
To investigate dynamic biological processes, such as interconversion and evolutionary trajectories of different cell types, we applied the Monocle (Version 2.22.0) algorithm82. The NewCellDataSet function was used to create a new object for the monocle using transcript count data of the included cell populations. The results generated from estimateSizeFactors and estimateDispersions function assisted us in normalizing for differences in mRNA recovered across cells and performing differential expression analysis later. Signature genes expressed in at least 10% cells of the dataset and with a p < 0.01 calculated using the differentialGeneTest function were included to define the trajectory progress. The ReduceDimension function reduced the space down to two dimensions, and the orderCells function ordered the cells according to gene expression. Pseudotime-dependent genes were calculated using differentialGeneTest and the “fullModelFormulaStr” option “~sm.ns(Pseudotime)”, and smooth expression curves were generated with the plot_pseudotime_heatmap function. The Ggridges package (Version 0.5.3) was used to analyze the frequency of distributed cells in different groups on the pseudotime axis.
Enrichment analysis
Pseudotime-dependent genes were further subjected to GO and KEGG enrichment analysis using the clusterProfiler package (Version 3.0.4) with default settings. Fifty hallmark gene sets in the MSigDB database (https://www.gsea-msigdb.org/gsea/msigdb) were used for GSEA of clusters c6/c7/c8 with the escape package (Version 1.4.0). A nonparametric and unsupervised algorithm from the gene set variation analysis (GSVA) package (Version 1.14.1) was selected to assess the EMT and ANGIOGENESIS scoring of different states generated with Monocle. The signature genes of EMT and ANGIOGENESIS were obtained from fifty hallmark gene sets.
Single-cell regulatory network inference and clustering (SCENIC) analysis
Cells in different states confirmed using Monocle were further included in the SCENIC package (Version 1.2.4)83, and they were then sorted based on clusters and states. To remove noise, genes with low expression levels or low positive rates were filtered using the geneFiltering function with default settings. Additionally, only the genes that matched the Rcis target databases were retained for downstream analysis. After reconstruction of the gene regulatory network, GENIE3 detected the relationship between transcription factors and potential targets. A total of 24,453 motifs from the cisTarget Human motif database v9 were used for enrichment of gene signatures, which were pruned for targets according to cis-regulatory cues using default settings. The enrichment of regulons across single cells was identified using the “aucell” positional argument, and the results were visualized using the pheatmap package (Version 1.0.12).
NicheNet analysis
NicheNet (Version 1.0.0)71 was used to identify potential ligands that drive the phenotype of cluster c6. The top 20 differentially expressed genes were ordered using log2FC between cluster c6, and the remaining clusters were treated as potential targets. Genes with a positive rate of >10% in clusters c6 and c19 were considered background and potential ligands, respectively. Sender cells from c19 and target cells from c6 were included to construct the expressed ligand–receptor interactions and calculate the ligand activity. We used the active_ligand_target_links function to compute the potential intensity of regulation between the ligand and target.
Similarity analysis
To quantify the similarity among different subclusters of fibroblasts and endothelial cells, the top 5000 variably expressed genes were included in the corr.test function from the psych package (Version 2.2.5), and the corrplot package (Version 0.92) was used for visualization.
Survival analysis
mRNA expression counts and clinical information from TCGA84 were downloaded from Firebrowse (http://firebrowse.org/). Based on the enrichment scores of CAFEnoMT signature gene sets (SPP1, CD44, CD68, PECAM1, VCAN, CD14, and MARCO) calculated with the GSVA package, patients with top and bottom 20% scores were selected for subsequent survival analysis. CIBERSORT85 was used to infer the abundance of each CAF state from available bulk RNA-seq data in four immunotherapy cohorts with detailed follow-up information. TPM or TMM values were used because their results show small root-mean-square error (RMSE) and high Pearson correlation values. B-mode was used to remove the batch effect. The algorithm was run with a web portal autogenerated signature matrix and 1000 permutations. Survival (Version 2.42–3) and Survminer (Version 0.4.9) packages were used for analysis and visualization.
Spatial transcriptomics
Raw base call files were converted to FASTQ reads using bcl2fastq. Reads were mapped to the human reference genome GRCh38-2020 using Space Ranger (Version 1.3.1) software86, and 332 million high-quality uniquely mapped reads were obtained. The median numbers of reads and genes detected per spot were 98,830 and 1,803, respectively. Read10X_h5 and the CreateSeuratObject function from the Seurat package were used to create an object with the output of Space Ranger. With the help of the Read10X_Image function, we loaded the H&E image data and used a standard logNormalize function to normalize the dataset. The SpatialFeaturePlot function showed the expression level of a single gene at a spatial location. Spots where the expression of the four genes (CD44, SPP1, PECAM1, and CD68) was simultaneously nonzero were recolored, and the color intensity reflected the average expression level of the signature set. We selected the corr.test function from the psych package (Version 2.2.5) to calculate the correlation of genes belonging to the signature set.
Multiplexed immunofluorescence (mIF) analysis
All involved surgical tissue samples were processed into paraffin blocks and cut into 5-μm-thick FFPE sections. Multiplex IHC staining was performed using an Opal 7-color kit (Akoya Bioscience, NEL801001KT). The relative markers CD68 (ab213363, Abcam, 1:1000, Opal 620), SPP1 (ab214050, Abcam, 1:1000, Opal 520), CD31 (ab182981, Abcam, 1:2000, Opal 480), and CD44 (ab213363, Abcam, 1:2000, Opal 690) were evaluated via IHC. Briefly, the sections were dewaxed with xylene for 20 min, and ethanol was used for rehydration. Microwave treatment was performed for antigen retrieval with buffer (pH 9.0). Next, all sections were cooled for 30 min to room temperature. Endogenous peroxidase activity was blocked using an antibody diluent/block (72424205; Akoya Bioscience) at room temperature for 10 min. Slides were incubated with a primary antibody at room temperature for 1 h, followed by secondary reagents at 37 °C for 20 min and tyramide signal amplification reagents at room temperature for 10 min (Opal 480, Opal 520, Opal 620, and Opal 690, Akoya Bioscience, 1:100). MWT antigen retrieval was performed until all markers were stained. Nuclear staining was performed using DAPI (Akoya Bioscience, 1:5) at room temperature for 5 min. Slides were mounted using anti-Fade fluorescence mounting medium (ab104135, Abcam), and they were stored at 4 °C until image acquisition. Slides were scanned using a PerkinElmer Vectra Polaris (PerkinElmer) and a confocal microscope (TCS SP8, Leica). The percentage of positively stained cells among all nucleated cells was determined. Multispectral image unmixing was performed using QuPath software (version 3.0)87 and ImageJ (version 1.53i). Briefly, DAPI-positive cells were identified using the “cell detection” command, and each single channel threshold was selected. Following this, all detected cells were divided into different subgroups for further analysis, and defective samples or areas with staining artifacts were reanalyzed or excluded. The in-house data were obtained from Novogene Co., Ltd.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
Appreciates the assistance of Mis. Xinyue Hu in illustration (Fig. 7) design and drawing. This study was supported by the National Key R&D Program of China (No. 2021YFA1301203(H.X.), 2018YFC2000305(L.Z.D.)), National Natural Science Foundation of China (82103031(H.L.), 81973408(H.X.), 82272933 (H.L.), 82273445(Y.S.)), International Cooperation Project of Chengdu Municipal Science and Technology Bureau (2020-GH02-00017-HZ(H.L.)), 1.3.5 Project for Disciplines of Excellence, West China Hospital, Sichuan University (No. ZYYC20003(H.X.), ZYJC18035(H.L.), ZYJC18025(Z.H.L.), ZYJC18003(Y.L.), ZYJC18004(H.X.), and ZYYC20007(L.D.)), and the National Research Foundation of Korea funded by the Korean government (2019R1C1C1005403(J.P), 2019R1A4A1028802(J.P), 2021M3H9A2097520(J.P)), Clinical Research Incubation Project, West China Hospital, Sichuan University (22HXFH019(H.L)).
Source data
Author contributions
H.L., J.P., and H.X. designed and supervised the project. X.Y.X., M.Y.C., Y.S., Z.X.R., K.D and P.-H.L. analyzed the data. H.L., H.R.T., B.B., W.G.J., Yi.L., W.Z., L.Y., Y.P., L.Z.D., H.B.H., Y.G.H., J.Q.Z., J.P., C.C., and H.X., interpreted the data. H.L., H.-N.C., W.-H.Z., Yang.L., R.H.S., C.L., and Z.H.L. collected the clinical samples and information. L.-B.H., X.Y.K., and H.J. performed multiplexed immunofluorescence staining and conducted related analysis. H.A. and J.P. built the website. Y.J. conducted pathological evaluation. X.Y.K. draw the diagrams. H.L., J.P., and H.X. wrote and revised the manuscript.
Peer review
Peer review information
Nature Communications thanks the other anonymous reviewer(s) for their contribution to the peer review of this work. Peer review reports are available.
Data availability
All the expression data can be obtained from the Gene Expression Omnibus, and the selected studies are listed in Supplementary Data 1. Analysis and visualization of the scRNA-seq datasets in this study can also be performed at https://gist-fgl.github.io/sc-caf-atlas/. Additionally, the integrated single-cell RNA sequencing matrix data that support the findings of this study are deposited in Gene expression Omnibus (accession No. GSE210347). Previously published scRNA-seq data reanalyzed here are available under accession codes GSE134355 (Normal data by Han et al.25), GSE141445 (Prostate cancer data by Chen et al.88), E-MTAB-8107 [https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-8107/sdrf] (Ovary/Breast/Colorectum cancer data by Qian et al.24), GSE157703 (Prostate cancer data by Ma et al.89), GSE131907 (Lung cancer data by Kim et al.26), GSE138709 (Intrahepatic cholangiocarcinoma data by Zhang et al.90), E-MTAB-6149 [https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-6149/sdrf], E-MTAB-6653 [https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-6653/sdrf] (Lung carcinomas data by Qian et al.24), CRA001160 (PDAC data by Peng et al.91), GSE154778 (PDAC data by Lin et al.92), HRA000212 (Bladder cancer data by Chen et al.93), HRA000686 (Thyroid cancer data by Luo et al.39). The gastric cancer data by Sathe et al.94 were downloaded from [http://dna-discovery.stanford.edu/download/1401/]. The raw sequencing data for the spatial transcriptome from this study have been deposited in the Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, under accession numbers (HRA003299 and HRA003300) that can be accessed at https://ngdc.cncb.ac.cn/gsa-human/. Source data are provided with this paper.
Code availability
The bioinformatic analysis code has been uploaded into the github (https://github.com/Xiaxy-XuLab/PanCAF, 10.5281/zenodo.7095147)95.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Han Luo, Xuyang Xia, Li-Bin Huang.
Contributor Information
Han Luo, Email: luohan-hx@scu.edu.cn.
Jihwan Park, Email: jihwan.park@gist.ac.kr.
Heng Xu, Email: xuheng81916@scu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-022-34395-2.
References
- 1.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Pitt JM, et al. Targeting the tumor microenvironment: removing obstruction to anticancer immune responses and immunotherapy. Ann. Oncol. 2016;27:1482–1492. doi: 10.1093/annonc/mdw168. [DOI] [PubMed] [Google Scholar]
- 4.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meurette O, Mehlen P. Notch signaling in the tumor microenvironment. Cancer Cell. 2018;34:536–548. doi: 10.1016/j.ccell.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 6.Kaymak I, Williams KS, Cantor JR, Jones RG. Immunometabolic interplay in the tumor microenvironment. Cancer cell. 2021;39:28–37. doi: 10.1016/j.ccell.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sahai E, et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer. 2020;20:174–186. doi: 10.1038/s41568-019-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L, Han X. Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J. Clin. Investig. 2015;125:3384–3391. doi: 10.1172/JCI80011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ai L, Xu A, Xu J. Roles of PD-1/PD-L1 pathway: signaling, cancer, and beyond. Adv. Exp. Med. Biol. 2020;1248:33–59. doi: 10.1007/978-981-15-3266-5_3. [DOI] [PubMed] [Google Scholar]
- 10.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat. Rev. Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 11.Kalluri R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer. 2016;16:582–598. doi: 10.1038/nrc.2016.73. [DOI] [PubMed] [Google Scholar]
- 12.Fukumura D, et al. Tumor induction of VEGF promoter activity in stromal cells. Cell. 1998;94:715–725. doi: 10.1016/s0092-8674(00)81731-6. [DOI] [PubMed] [Google Scholar]
- 13.Kumar V, et al. Cancer-associated fibroblasts neutralize the anti-tumor effect of CSF1 receptor blockade by inducing PMN-MDSC infiltration of tumors. Cancer Cell. 2017;32:654–668.e655. doi: 10.1016/j.ccell.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fearon DT. The carcinoma-associated fibroblast expressing fibroblast activation protein and escape from immune surveillance. Cancer Immunol. Res. 2014;2:187–193. doi: 10.1158/2326-6066.CIR-14-0002. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y, McAndrews KM, Kalluri R. Clinical and therapeutic relevance of cancer-associated fibroblasts. Nat. Rev. Clin. Oncol. 2021;18:792–804. doi: 10.1038/s41571-021-00546-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rauch A, Mandrup S. Transcriptional networks controlling stromal cell differentiation. Nat. Rev. Mol. cell Biol. 2021;22:465–482. doi: 10.1038/s41580-021-00357-7. [DOI] [PubMed] [Google Scholar]
- 17.Li PH, et al. Recent developments in application of single-cell RNA sequencing in the tumour immune microenvironment and cancer therapy. Mil. Med. Res. 2022;9:52. doi: 10.1186/s40779-022-00414-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oller-Moreno, S., Kloiber, K., Machart, P. & Bonn, S. J. C. O. I. S. B. Algorithmic advances in Machine Learning for single cell expression analysis. Curr. Opin. Syst. Biol.25, 27–33 (2021).
- 19.Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 20.Qiu X, et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng S, et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell. 2021;184:792–809.e723. doi: 10.1016/j.cell.2021.01.010. [DOI] [PubMed] [Google Scholar]
- 22.Zheng L, et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science. 2021;374:abe6474. doi: 10.1126/science.abe6474. [DOI] [PubMed] [Google Scholar]
- 23.Hong F, et al. Single-cell analysis of the pan-cancer immune microenvironment and scTIME portal. Cancer Immunol. Res. 2021;9:939–951. doi: 10.1158/2326-6066.CIR-20-1026. [DOI] [PubMed] [Google Scholar]
- 24.Qian J, et al. A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res. 2020;30:745–762. doi: 10.1038/s41422-020-0355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han X, et al. Construction of a human cell landscape at single-cell level. Nature. 2020;581:303–309. doi: 10.1038/s41586-020-2157-4. [DOI] [PubMed] [Google Scholar]
- 26.Kim N, et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat. Commun. 2020;11:2285. doi: 10.1038/s41467-020-16164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nowak-Sliwinska P, et al. Oncofoetal insulin receptor isoform A marks the tumour endothelium; an underestimated pathway during tumour angiogenesis and angiostatic treatment. Br. J. Cancer. 2019;120:218–228. doi: 10.1038/s41416-018-0347-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montico B, et al. Loss of Spry1 reduces growth of BRAF(V600)-mutant cutaneous melanoma and improves response to targeted therapy. Cell Death Dis. 2020;11:392. doi: 10.1038/s41419-020-2585-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rapp M, et al. CCL22 controls immunity by promoting regulatory T cell communication with dendritic cells in lymph nodes. J. Exp. Med. 2019;216:1170–1181. doi: 10.1084/jem.20170277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oh SA, et al. PD-L1 expression by dendritic cells is a key regulator of T-cell immunity in cancer. Nat. Cancer. 2020;1:681–691. doi: 10.1038/s43018-020-0075-x. [DOI] [PubMed] [Google Scholar]
- 31.Romani N, et al. A small number of anti-CD3 molecules on dendritic cells stimulate DNA synthesis in mouse T lymphocytes. J. Exp. Med. 1989;169:1153–1168. doi: 10.1084/jem.169.3.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mao N, et al. Oncogenic ERG represses PI3K signaling through downregulation of IRS2. Cancer Res. 2020;80:1428–1437. doi: 10.1158/0008-5472.CAN-19-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barkal AA, et al. CD24 signalling through macrophage Siglec-10 is a target for cancer immunotherapy. Nature. 2019;572:392–396. doi: 10.1038/s41586-019-1456-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cremasco V, et al. FAP delineates heterogeneous and functionally divergent stromal cells in immune-excluded breast tumors. Cancer Immunol. Res. 2018;6:1472–1485. doi: 10.1158/2326-6066.CIR-18-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feig C, et al. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc. Natl Acad. Sci. USA. 2013;110:20212–20217. doi: 10.1073/pnas.1320318110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kraman M, et al. Suppression of antitumor immunity by stromal cells expressing fibroblast activation protein-alpha. Science. 2010;330:827–830. doi: 10.1126/science.1195300. [DOI] [PubMed] [Google Scholar]
- 37.Jacobs JJ, et al. Senescence bypass screen identifies TBX2, which represses Cdkn2a (p19(ARF)) and is amplified in a subset of human breast cancers. Nat. Genet. 2000;26:291–299. doi: 10.1038/81583. [DOI] [PubMed] [Google Scholar]
- 38.Dodou E, Xu SM, Black BL. mef2c is activated directly by myogenic basic helix-loop-helix proteins during skeletal muscle development in vivo. Mechanisms Dev. 2003;120:1021–1032. doi: 10.1016/s0925-4773(03)00178-3. [DOI] [PubMed] [Google Scholar]
- 39.Luo, H. et al. Characterizing dedifferentiation of thyroid cancer by integrated analysis. Sci. Adv.10.1126/sciadv.abf3657 (2021). [DOI] [PMC free article] [PubMed]
- 40.Crawford Y, et al. PDGF-C mediates the angiogenic and tumorigenic properties of fibroblasts associated with tumors refractory to anti-VEGF treatment. Cancer Cell. 2009;15:21–34. doi: 10.1016/j.ccr.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 41.Mrass P, et al. CD44 mediates successful interstitial navigation by killer T cells and enables efficient antitumor immunity. Immunity. 2008;29:971–985. doi: 10.1016/j.immuni.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li MO, Flavell RA. TGF-beta: a master of all T cell trades. Cell. 2008;134:392–404. doi: 10.1016/j.cell.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang, A. et al. Progranulin prevents regulatory NK cell cytotoxicity against antiviral T cells. JCI Insight10.1172/jci.insight.129856 (2019). [DOI] [PMC free article] [PubMed]
- 44.Cummings RD. T cells are Smad’ly in love with galectin-9. Immunity. 2014;41:171–173. doi: 10.1016/j.immuni.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 45.Calzascia T, et al. TNF-alpha is critical for antitumor but not antiviral T cell immunity in mice. J. Clin. Investig. 2007;117:3833–3845. doi: 10.1172/JCI32567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mackay F, Browning JL. BAFF: a fundamental survival factor for B cells. Nat. Rev. Immunol. 2002;2:465–475. doi: 10.1038/nri844. [DOI] [PubMed] [Google Scholar]
- 47.Jahrsdörfer B, et al. Granzyme B produced by human plasmacytoid dendritic cells suppresses T-cell expansion. Blood. 2010;115:1156–1165. doi: 10.1182/blood-2009-07-235382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.John S, Mishra R. Galectin-9: From cell biology to complex disease dynamics. J. Biosci. 2016;41:507–534. doi: 10.1007/s12038-016-9616-y. [DOI] [PubMed] [Google Scholar]
- 49.Morrison AH, Diamond MS, Hay CA, Byrne KT, Vonderheide RH. Sufficiency of CD40 activation and immune checkpoint blockade for T cell priming and tumor immunity. Proc. Natl Acad. Sci. USA. 2020;117:8022–8031. doi: 10.1073/pnas.1918971117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Daley D, et al. Dectin 1 activation on macrophages by galectin 9 promotes pancreatic carcinoma and peritumoral immune tolerance. Nat. Med. 2017;23:556–567. doi: 10.1038/nm.4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang R, et al. Galectin-9 interacts with PD-1 and TIM-3 to regulate T cell death and is a target for cancer immunotherapy. Nat. Commun. 2021;12:832. doi: 10.1038/s41467-021-21099-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Riaz N, et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017;171:934–949.e916. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mariathasan S, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554:544–548. doi: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gultekin O, et al. FOXP3+ T cells in uterine sarcomas are associated with favorable prognosis, low extracellular matrix expression and reduced YAP activation. NPJ Precis. Oncol. 2021;5:97. doi: 10.1038/s41698-021-00236-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elyada E, et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov. 2019;9:1102–1123. doi: 10.1158/2159-8290.CD-19-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zakrzewska A, et al. Macrophage-specific gene functions in Spi1-directed innate immunity. Blood. 2010;116:e1–e11. doi: 10.1182/blood-2010-01-262873. [DOI] [PubMed] [Google Scholar]
- 57.Cuevas VD, et al. MAFB determines human macrophage anti-inflammatory polarization: relevance for the pathogenic mechanisms operating in multicentric carpotarsal osteolysis. J. Immunol. 2017;198:2070–2081. doi: 10.4049/jimmunol.1601667. [DOI] [PubMed] [Google Scholar]
- 58.Mougiakakos D, et al. The IKZF1-IRF4/IRF5 axis controls polarization of myeloma-associated macrophages. Cancer Immunol. Res. 2021;9:265–278. doi: 10.1158/2326-6066.CIR-20-0555. [DOI] [PubMed] [Google Scholar]
- 59.Tang PM, et al. Neural transcription factor Pou4f1 promotes renal fibrosis via macrophage-myofibroblast transition. Proc. Natl Acad. Sci. USA. 2020;117:20741–20752. doi: 10.1073/pnas.1917663117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang YY, et al. Macrophage-to-myofibroblast transition contributes to interstitial fibrosis in chronic renal allograft injury. J. Am. Soc. Nephrology: JASN. 2017;28:2053–2067. doi: 10.1681/ASN.2016050573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haider N, et al. Transition of macrophages to fibroblast-like cells in healing myocardial infarction. J. Am. Coll. Cardiol. 2019;74:3124–3135. doi: 10.1016/j.jacc.2019.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang, H. et al. Mesothelial cell-derived antigen-presenting cancer-associated fibroblasts induce expansion of regulatory T cells in pancreatic cancer. Cancer Cell10.1016/j.ccell.2022.04.011 (2022). [DOI] [PMC free article] [PubMed]
- 63.Amit M, Na’ara S, Gil Z. Mechanisms of cancer dissemination along nerves. Nat. Rev. Cancer. 2016;16:399–408. doi: 10.1038/nrc.2016.38. [DOI] [PubMed] [Google Scholar]
- 64.Roberts SL, et al. Sox2 expression in Schwann cells inhibits myelination in vivo and induces influx of macrophages to the nerve. Dev. (Camb., Engl.) 2017;144:3114–3125. doi: 10.1242/dev.150656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bremer M, et al. Sox10 is required for Schwann-cell homeostasis and myelin maintenance in the adult peripheral nerve. Glia. 2011;59:1022–1032. doi: 10.1002/glia.21173. [DOI] [PubMed] [Google Scholar]
- 66.Kumar, V. et al. Single-cell atlas of lineage states, tumor microenvironment and subtype-specific expression programs in gastric cancer. Cancer Discov.10.1158/2159-8290.Cd-21-0683 (2021). [DOI] [PMC free article] [PubMed]
- 67.Rocha SF, et al. Esm1 modulates endothelial tip cell behavior and vascular permeability by enhancing VEGF bioavailability. Circulation Res. 2014;115:581–590. doi: 10.1161/CIRCRESAHA.115.304718. [DOI] [PubMed] [Google Scholar]
- 68.Lilly AJ, Lacaud G, Kouskoff V. SOXF transcription factors in cardiovascular development. Semin. Cell Develop. Biol. 2017;63:50–57. doi: 10.1016/j.semcdb.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 69.Wei G, et al. Ets1 and Ets2 are required for endothelial cell survival during embryonic angiogenesis. Blood. 2009;114:1123–1130. doi: 10.1182/blood-2009-03-211391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Welch-Reardon KM, Wu N, Hughes CC. A role for partial endothelial-mesenchymal transitions in angiogenesis? Arterioscler. Thromb. Vasc. Biol. 2015;35:303–308. doi: 10.1161/ATVBAHA.114.303220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Browaeys R, Saelens W, Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods. 2020;17:159–162. doi: 10.1038/s41592-019-0667-5. [DOI] [PubMed] [Google Scholar]
- 72.McAndrews KM, et al. Identification of functional heterogeneity of carcinoma-associated fibroblasts with distinct IL6-mediated therapy resistance in pancreatic cancer. Cancer Discov. 2022;12:1580–1597. doi: 10.1158/2159-8290.CD-20-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Denhardt DT, Noda M, O’Regan AW, Pavlin D, Berman JS. Osteopontin as a means to cope with environmental insults: regulation of inflammation, tissue remodeling, and cell survival. J. Clin. Investig. 2001;107:1055–1061. doi: 10.1172/JCI12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McAllister SS, et al. Systemic endocrine instigation of indolent tumor growth requires osteopontin. Cell. 2008;133:994–1005. doi: 10.1016/j.cell.2008.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rowe GC, et al. PGC-1α induces SPP1 to activate macrophages and orchestrate functional angiogenesis in skeletal muscle. Circulation Res. 2014;115:504–517. doi: 10.1161/CIRCRESAHA.115.303829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li T, et al. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kumar V, et al. Single-cell atlas of lineage states, tumor microenvironment, and subtype-specific expression programs in gastric cancer. Cancer Discov. 2022;12:670–691. doi: 10.1158/2159-8290.CD-21-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hosaka K, et al. Pericyte-fibroblast transition promotes tumor growth and metastasis. Proc. Natl Acad. Sci. USA. 2016;113:E5618–E5627. doi: 10.1073/pnas.1608384113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Choi SH, et al. Tumour-vasculature development via endothelial-to-mesenchymal transition after radiotherapy controls CD44v6(+) cancer cell and macrophage polarization. Nat. Commun. 2018;9:5108. doi: 10.1038/s41467-018-07470-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ribatti D, Nico B, Crivellato E. The role of pericytes in angiogenesis. Int. J. Develop. Biol. 2011;55:261–268. doi: 10.1387/ijdb.103167dr. [DOI] [PubMed] [Google Scholar]
- 81.Wolock SL, Lopez R, Klein AM. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 2019;8:281–291.e289. doi: 10.1016/j.cels.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aibar S, et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hoadley KA, et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 2018;173:291–304.e296. doi: 10.1016/j.cell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marx V. Method of the year: spatially resolved transcriptomics. Nat. Methods. 2021;18:9–14. doi: 10.1038/s41592-020-01033-y. [DOI] [PubMed] [Google Scholar]
- 87.Bankhead P, et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017;7:16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen S, et al. Single-cell analysis reveals transcriptomic remodellings in distinct cell types that contribute to human prostate cancer progression. Nat. Cell Biol. 2021;23:87–98. doi: 10.1038/s41556-020-00613-6. [DOI] [PubMed] [Google Scholar]
- 89.Ma X, et al. Identification of a distinct luminal subgroup diagnosing and stratifying early stage prostate cancer by tissue-based single-cell RNA sequencing. Mol. Cancer. 2020;19:147. doi: 10.1186/s12943-020-01264-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li H, et al. Single-cell transcriptomic architecture unraveling the complexity of tumor heterogeneity in distal cholangiocarcinoma. Cell Mol. Gastroenterol. Hepatol. 2022;13:1592–1609 e1599. doi: 10.1016/j.jcmgh.2022.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peng J, et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019;29:725–738. doi: 10.1038/s41422-019-0195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lin W, et al. Single-cell transcriptome analysis of tumor and stromal compartments of pancreatic ductal adenocarcinoma primary tumors and metastatic lesions. Genome Med. 2020;12:80. doi: 10.1186/s13073-020-00776-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen Z, et al. Single-cell RNA sequencing highlights the role of inflammatory cancer-associated fibroblasts in bladder urothelial carcinoma. Nat. Commun. 2020;11:5077. doi: 10.1038/s41467-020-18916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sathe A, et al. Single-cell genomic characterization reveals the cellular reprogramming of the gastric tumor microenvironment. Clin. Cancer Res. 2020;26:2640–2653. doi: 10.1158/1078-0432.CCR-19-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Han L. et al. Pan-cancer single-cell analysis reveals the heterogeneity and plasticity of cancer-associated fibroblasts in the tumor microenvironment. Zenodo10.5281/zenodo.7095147, 2022. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the expression data can be obtained from the Gene Expression Omnibus, and the selected studies are listed in Supplementary Data 1. Analysis and visualization of the scRNA-seq datasets in this study can also be performed at https://gist-fgl.github.io/sc-caf-atlas/. Additionally, the integrated single-cell RNA sequencing matrix data that support the findings of this study are deposited in Gene expression Omnibus (accession No. GSE210347). Previously published scRNA-seq data reanalyzed here are available under accession codes GSE134355 (Normal data by Han et al.25), GSE141445 (Prostate cancer data by Chen et al.88), E-MTAB-8107 [https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-8107/sdrf] (Ovary/Breast/Colorectum cancer data by Qian et al.24), GSE157703 (Prostate cancer data by Ma et al.89), GSE131907 (Lung cancer data by Kim et al.26), GSE138709 (Intrahepatic cholangiocarcinoma data by Zhang et al.90), E-MTAB-6149 [https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-6149/sdrf], E-MTAB-6653 [https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-6653/sdrf] (Lung carcinomas data by Qian et al.24), CRA001160 (PDAC data by Peng et al.91), GSE154778 (PDAC data by Lin et al.92), HRA000212 (Bladder cancer data by Chen et al.93), HRA000686 (Thyroid cancer data by Luo et al.39). The gastric cancer data by Sathe et al.94 were downloaded from [http://dna-discovery.stanford.edu/download/1401/]. The raw sequencing data for the spatial transcriptome from this study have been deposited in the Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, under accession numbers (HRA003299 and HRA003300) that can be accessed at https://ngdc.cncb.ac.cn/gsa-human/. Source data are provided with this paper.
The bioinformatic analysis code has been uploaded into the github (https://github.com/Xiaxy-XuLab/PanCAF, 10.5281/zenodo.7095147)95.