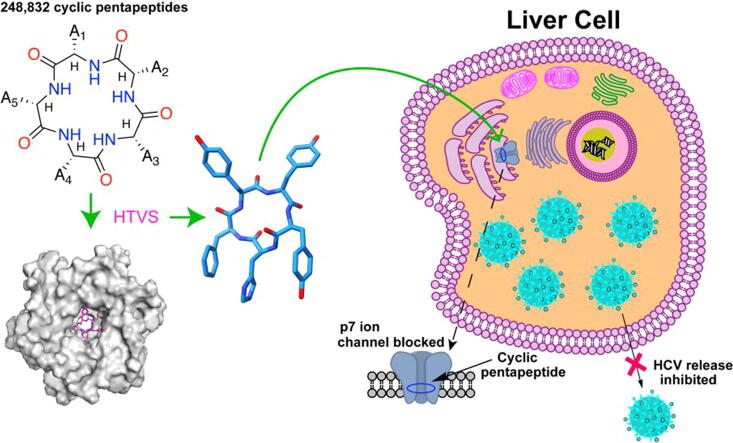
Graphical abstract
The top-hit penta-cyclopeptides targeting p7 channel are acquired using high-throughput virtual screening, and the efficacies of inhibiting HCV release are verified over strains of Jc1 or Jc1-S282T. The binding sites of the best inhibitor CP7 in the p7 channel lumen are deciphered using NMR and MD simulations. This channel blocking mechanism of inhibiting HCV release provides a novel strategy for eliminating HCV.
Keywords: Drug design, Cyclic penta-peptide, Channel blocker, P7, HCV, NMR, Molecular dynamics simulations
Abstract
The hepatitis C virus (HCV) p7 viroporin protein is essential for viral assembly and release, suggesting its unrealised potential as a target for HCV interventions. Several classes of small molecules that can inhibit p7 through allosteric mechanisms have shown low efficacy. Here, we used a high throughput virtual screen to design a panel of eight novel cyclic penta-peptides (CPs) that target the p7 channel with high binding affinity. Further examination of the effects of these CPs in viral production assays indicated that CP7 exhibits the highest potency against HCV among them. Moreover, the IC50 efficacy of CP7 in tests of strain Jc1-S282T suggested that this cyclopeptide could also effectively inhibit a drug-resistant HCV strain. A combination of nuclear magnetic resonance (NMR) spectroscopy and molecular dynamics (MD) simulations revealed that CP7 blocking activity relies on direct binding to the p7 channel lumen at the N-terminal bottleneck region. These findings thus present a promising anti-HCV cyclic penta-peptide targeting p7 viroporin, while also describing an alternative strategy for designing a new class of p7 channel blockers for strains resistant to direct-acting antiviral agents (DAA).
1. Introduction
Hepatitis C virus (HCV) is a positive sense single-stranded RNA virus that causes chronic and severe liver diseases in humans [1] and chimpanzees [2]. Approximately 58 million people worldwide currently live with HCV-related diseases, with an annual mortality rate of ∼290,000 people [3] according to recent studies by the World Health Organization. The introduction of highly effective and well-tolerated direct-acting antiviral agents (DAAs), such as sofosbuvir [4], elbasvir [5] and grazoprevir [6] has revolutionized therapeutic strategies for HCV in recent years. However, like all RNA viruses, HCV can develop resistance to these current drugs through rapid mutation, thus posing a serious threat to public health. For example, the resistance associated substitution (RAS) S282T mutation in the viral NS5B RNA polymerase decreases HCV susceptibility to sofosbuvir, the most essential composition of the DAAs [7]. Therefore, screening for novel targets and developing new drugs is an ongoing and essential process.
Viroporins, which are crucial for viral pathogenicity, have emerged as validated drug targets since the successful example of M2 proton channel as an anti-viral target against influenza A virus [8]. The HCV p7 viroporin is required for viral assembly and release of the viral particles [9]. In-frame deletions or point mutations of p7 reduced or ablated production of infectious viruses [10], [11]. Given its central role in infection, together with a wide array of compounds, including amantadine [12], rimantadine [13], BIT225 [14], and hexamethylene amiloride (HMA) [15], shown to inhibit its activity, p7 is a top potential target for anti-HCV drug development. However, these inhibitors generally show low efficacy [12], [16]. In addition, previous electron microscopy (EM) [17] and nuclear magnetic resonance (NMR) [18] results revealed that p7 has a hexameric funnel-like channel architecture that requires binding by six molecules of amantadine [18] or HMA [19] to the hydrophobic pockets on the lipid-facing side of the channel for successful allosteric inhibition of p7 ion transport. Thus, additional energy is likely required to deliver these inhibitor molecules to the binding pockets through the lipid bilayers.
To overcome these drawbacks, we previously proposed using cyclic tetra-peptides to block the p7 channel lumen [20]. However, the cyclic tetra-peptides with a limited panel of candidates exhibited only slightly lower (or even higher) binding free energies than those of small molecule inhibitors. Additionally, cyclic tetra-peptides are difficult to be synthesized due to the high intramolecular tensile forces, and no experimental evidence has yet demonstrated that cyclic peptides can indeed block the inner p7 channel as predicted. Here, we designed and tested a series of cyclic penta-peptides using high-throughput virtual screening (HTVS). Eight novel cyclic penta-peptides (CP) targeting the p7 channel were predicted to bind the p7 channel with high affinity. Further in vitro viral production assays showed that CP7 has the highest potency against HCV among them. Nuclear magnetic resonance (NMR) spectroscopy in conjunction with molecular dynamics (MD) simulations revealed that CP7 binds the interior channel of the p7, resembling the effects of a bottle cork. Furthermore, CP7 has been demonstrated its inhibition effect on the DAA-drug resistant mutant S282T, providing valuable insights into anti-HCV therapeutic development.
2. Materials and methods
2.1. High throughput virtual screening of cyclic penta-peptides targeting p7
For screening the potential cyclic penta-peptides inside the p7 protein channel, the combinatorial library of cyclic penta-peptides was generated using Discovery Studio 4.0 [21]. The twelve natural amino acids with relatively long sidechains were selected for building up the cyclic penta-peptides under the rule of permutation and combination. 248,832 cyclic penta-peptides were acquired and pre-optimised before virtual screening by Autodock Vina 1.2.2 [22]. The structure of the hexameric HCV p7-5a channel for docking was obtained from the Protein Data Bank (PDB ID: 2M6X) [18], which is also processed for computational screening. After the first round of semi-flexible docking method, 5146 potential hits were selected out with the cutoff of binding free energy < −6 kcal/mol. These 5146 conformations were input into p7-5a channel for a second-round screening using flexible-docking screening, the top 8 hits with good binding free energies and conformations were selected for further corresponding assays and calculations.
2.2. Modelling of cyclic penta-peptides and p7 channels
Since the infectious HCV genotype 5a (GT5a) is unavailable as a cell culture model, we used the Jc1 strain (genotype 2a, GT2a) in anti-HCV production assays to evaluate the inhibition effect of cyclopeptides. Therefore, besides the p7-5a model, a p7-2a (Jc1) model was originated from mutating the given residues on p7-5a. The modelling of cyclopeptides (CP1-CP8) were built up under Discovery Studio 4.0 [21] that the geometries were pre-optimised by molecular mechanics using the add-in force field.
2.3. Molecular docking of cyclic penta-peptides inside p7-5a and p7-2a channels
For deciphering the details of peptide binding sites inside the channel, the molecular docking of cyclic penta-peptides was conducted on HCV p7-5a and p7-2a. All peptides and targets were pre-optimised and the formats were converted from PDB to PDBQT files using ADFR suite [23]. For cyclic penta-peptides docking inside p7-5a and p7-2a, the grid box size was set to 22.2146 Å×27.9757 Å×23.3957 Å in order to cover the binding region (acquired from nuclear Overhauser effects (NOEs)) inside the p7-5a or p7-2a channel using local search and the centre of grid box is set to 13.2805 Å×4.3689 Å×13.2349 Å with spacing of 0.375 Å. The amount of computational effort (exhaustiveness) was set as 64 for considerably consistent performance. Molecular docking was performed using the Autodock Vina 1.2.2 [22] with Vina scoring force field. The best-fitted pose for each model was used for further analysis.
2.4. Molecular dynamics simulations of CP7 cyclo(YYYFH) with p7-5a and p7-2a
The MD simulations were performed using the Desmond package [24] and the force field OPLS2005 [25] in a bilayer with proper number of counter ions to balance the net charge of the system with 150 mM NaCl. The localisation of the membrane was defined using the Orientations of Proteins in Membranes (OPM) database [26]. p7-5a or p7-2a and CP7 were inserted into POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) bilayer containing the explicit water models (TIP3P) [27]. Nose-Hoover temperature coupling [28] and Martina-Tobias- Klein method [29] with isotropic scaling were utilised to control the simulation temperature (300 K) and atmospheric pressure (1 atm). The particle-mesh Ewald (PME) method [30] was used to calculate long-range electrostatic interactions with grid spacing of 0.8 Å. Van der Waals (VDW) and short-range electrostatic interactions were smoothly truncated at 9.0 Å. The system was equilibrated using the default membrane relax protocol before MD simulations provided in Desmond [24], which consists of a series of restrained minimisations to relax the system. The initial p7-5a or p7-2a channel coordinates for the MD simulation calculations were taken from Protein Data Bank (PDB: 2M6X) [31]. The backbone of p7-5a or p7-2a are restrained with 10 force constants. After minimisation and relaxation, the system was subject to 1 μs of normal pressure and temperature (NPT) production simulation for better showing the interaction of CP7 inside p7-5a or p7-2a channels at the timescale of microsecond.
2.5. Protein expression and purification
Because of the NMR feasibility of p7-5a, we next used p7-5a to characterize the interactions between p7 channel and CP7. The 2H, 15N labelled p7-5a protein was prepared as previously described [18]. In short, His9-trpLE-p7 was transformed into E. Coli BL21 (DE3) cells in the pMM-LR6 vector and grown in M9 minimal media prepared with D2O. Inclusion bodies were extracted and solubilised later in 6 M guanidine HCl, 50 mM Tris (pH 8.0), 200 mM NaCl, 1 % Triton X-100 (Buffer A). The His9-trpLE-p7 fusion protein was purified by immobilised Ni2+ affinity chromatography in Buffer A at room temperature and eluted from the Ni2+column in the same buffer with 400 mM imidazole. The eluate was cleaved at the site of a methionine residue by cyanogen bromide (CNBr) in 80 % formic acid solution. The sole p7 and fusion protein were then separated by reverse-phase chromatography using a preparative C18 column (Agilent Technologies). The NMR sample for NOE experiment was prepared and refolded by dialysis against the NMR buffer (25 mM MES, pH 6.5), containing 0.82 mM 2H, 15N labelled p7-5a and 8 mM CP7 with natural redundancy.
2.6. Circular dichroism spectroscopy
The p7 NMR samples were further characterized by circular dichroism (CD) spectroscopy using a J-715 circular dichroism spectropolarimeter. Wavelength scans were conducted from 180 to 260 nm. Experimental conditions were 20 μM of p7 in 0.1 % dodecylphosphorylcholine (DPC), 50 mM K-PO4, and pH 6.5 at 298 K.
2.7. Cell and medium
The cells were cultured as described previously [32]. Briefly, the hepatoma cell line (Huh 7.5.1) were maintained in complete medium consisting of Dulbecco's modified Eagle's medium (DMEM) supplemented with 10 % fetal bovine serum, 10 mM HEPES, 2 mM l-glutamine, 100 U of penicillin/ml, and 100 mg of streptomycin/ml (Invitrogen).
2.8. Plasmids, antibodies and viruses
pUC-Jc1 was constructed as previously [33]. A point mutation S282T in NS5B that is resistant to DAA-drug sofosbuvir was engineered into NS5B of HCV Jc1 strain (GT2a) by Mut Express® MultiS Fast Mutagenesis Kit (Vazyme) and verified by sequencing. Mouse monoclonal antibodies against HCV E2 (1C9) and NS3 were raised in our lab. The viruses were stored in Thermo Scientific Revco ULT and performed in BSL-2 facility at Institut Pasteur of Shanghai following the regulations.
2.9. In vitro transcription and transfection of HCV RNA
The Jc1 and Jc1-S282T plasmids were linearised by Xba I digestion. The linearised plasmids DNA were transcribed by MEGAscriptTM T7 Transcription Kit (Invitrogen) after purification. RNAs were delivered to cells by electroporation as described previously [34]. Briefly, Huh 7.5.1 cells were washed twice and resuspended in serum-free Opti-MEM (Invitrogen) at 1 × 107 cells per ml. 10 μg RNA mixed with 400 μL cells in a 4-mm cuvette, then pulsed by Bio-Rad Gene Pulser at 0.27 kV, 100 Ω and 950 μF. The pulsed cells were plated in a 10 cm dish (Corning).
2.10. Cellular viability and cytotoxicity assays
Cell viability and cytotoxicity were detected using Luminescent Cell Viability Assay reagent (Promega GS7570). Briefly, Huh 7.5.1 cells were seeded at a density of 40,000 cells per well in 48-well microplates and allowed to attach and grow overnight. The next day, the cell-culture media with serially diluted compounds in 0.5 % dimethyl sulfoxide(DMSO) or mock were added to the cells. After 12 h of incubation, cells were collected and lysed for the cell cytotoxicity assay.
2.11. Indirect immunofluorescence
Intracellular immuno-staining of HCV-infected cells was performed as described previously [34]. Briefly, the virus-infected cells were fixed, and stained with a human monoclonal anti-HCV E2 antibody (AR3A). Bound primary antibody was detected by Alexa Fluor 555-conjugated anti-human secondary antibodies (Thermo Fisher Scientific). Nuclei were stained with Hoechst dye.
2.12. Anti-HCV production assay of cyclic penta-peptides CP1–CP8
HCV Jc1 and Jc1-S282T fully infected Huh 7.5.1 cells were seeded in 24-well plate at a density of 80, 000 cells per well overnight. The supernatant was removed and the cells were washed with PBS for 3 times, then incubated with serially diluted compounds in 0.5 % DMSO for 6 h. The supernatant was collected, and the virus production was determined by focus-reduction assay. Dose-response curves and the half maximal inhibitory concentrations (IC50) of each compound were generated by non-linear fitting. IC50 was calculated as the concentration of inhibitor required for a 50 % reduction in the HCV production, as described previously [32].
2.13. Assignment of NOEs between CP7 cyclo(YYYFH) and p7-5a
Intermolecular NOEs between protein backbone amide and side-chain protons of ligand were assigned using a sample consisting of 0.82 mM 2H, 15N-p7-5a, 8 mM CP7 cyclo(YYYFH) with natural redundancy of 1H and deuterated d38-DPC (Avanti Polar Lipids). The sample was used to record a 15N-edited NOESY-TROSY(transverse relaxation optimized spectroscopy) (τNOE = 200 ms) on a 900 MHz Bruker spectrometer. This experiment allowed exclusive detection of NOEs between the exchangeable amide protons of p7-5a and the peptide protons (CP7). The strong NOE signals detected at ∼7.0 p.p.m between the p7-5a backbone amide and the side-chain protons of CP7 were assigned.
2.14. Chemistry
The cyclopeptides were synthesized by ChinaPeptides Company, Limited. For CP1: cyclo(DERyR) (Remark: y is d-Tyr) Step 1: Wash the reaction vessel with DCM. Bottom blow with nitrogen, then drain completely. Step 2: Weigh some 2-chlorotrityl chloride resin in the reaction vessel, swell the resin with dimethylformamide (DMF) (15 mL/g) for 30 min. Step 3: Weigh thrice mole Fmoc-l-Arg(Pbf)–OH in a test tube, dissolve Fmoc-amino acid in DMF/chloromethane (CM) (1: 1) (15 mL/g), transfer the solution into the reaction vessel above, add 10× N,N-diisopropylethylamine (DIEA), mixture for 30 min at room temperature with nitrogen. Step 4: Add 5 mL methanol into the reaction vessel and bottom blow for 10 min. Drain and wash with DMF (3×), dichloromethane (DCM) (3×), DMF (3×). Step 5: Drain then add 20 % piperidine (15 mL/g) to remove the Fmoc group. Bottom blow mixture for 10 min and 5 min. Wash with DMF (3×), DCM (3×), DMF (3×). Step 6: Take a little of resin, add in 2 drops of 25 % ninhydrin-alcohol solution and 1 drop of 20 % phenolic -alcohol solution, and then 1 drop of pyridine, to heat in 105 °C for 5 min, the colour changing into deep blue is positive reaction. Step 7: Place in 3× protected amino acid, 3×hexafluorophosphate benzotriazole tetramethyl uronium (HBTU)hydroxybenzotriazole (HOBT) (1 g), DIEA (2 mL), add in DMF to dissolve and then DCM (15 mL/g). React for 1 h. Step 8: Wash with DCM (15 mL/g) and DMF (15 mL/g) alternately for 3 times. Step 9: Take a little of resin, add in 2 drops of 25 % ninhydrin-alcohol solution and 1 drop of 20 % phenolic -alcohol solution, and then 1 drop of pyridine, to heat in 105 °C for 5 min, the color changing into deep blue is a positive reaction, heat in 105 °C for 5 min, no color changing is a negative reaction. Step 10: Repeat Step 5–9 to couple the other amino acids. Step 11: The method to wash resin after the last amino acid coupling and deprotection is below. Wash by the following reagents in turn: 2× DMF (10 mL/g), 2× methanol (10 mL/g), 2× DMF (10 mL/g), 2× DCM (10 mL/g). And then draw drying for 10 min. Step 12: Release the peptide from resin using 2,2,2-trifluoroethanol 30 % and DCM 70 % for 2 h. Step 13: Cleave the peptide using trifluoroacetic acid (TFA) 94.5 %, H2O 2.5 %,1,2-ethane dithiol (EDT) 2.5 %, triisopropyl silane (TIS) 1 % for 2 h. Step 14: Blow the cleavage solution drying with nitrogen gas as far as possible, and wash it 6 times with absolute aether, dry it in the air. Acquire the crude peptide in the end. For the syntheses of CP2-CP8, the procedures are the same except for the substrates.
2.15. Purification of cyclic penta-peptides
Dissolve the crude peptide with purified water. Purification condition is shown as follows, load 3 mL of sample at flow rate of 1 mL/min using 0.1 % TFA + 100 % H2O solution (buffer A) and 0.1 % TFA + 100 % acetonitrile (ACN) solution (buffer B) and equilibrate the preparative C18 column (Venusi MRC-ODS, 30 × 250 mm) at ratio of 90 % buffer A and 10 % buffer B for 5 min. The separation is performed under the gradient of 10 % buffer B to 80 % buffer B through 25 min. The purified solution is dried by lyophilisation to give the white-powder-form product. All peptides are greater than 95 % pure analyzed by HPLC.
3. Results
3.1. Design of cyclic penta-peptides
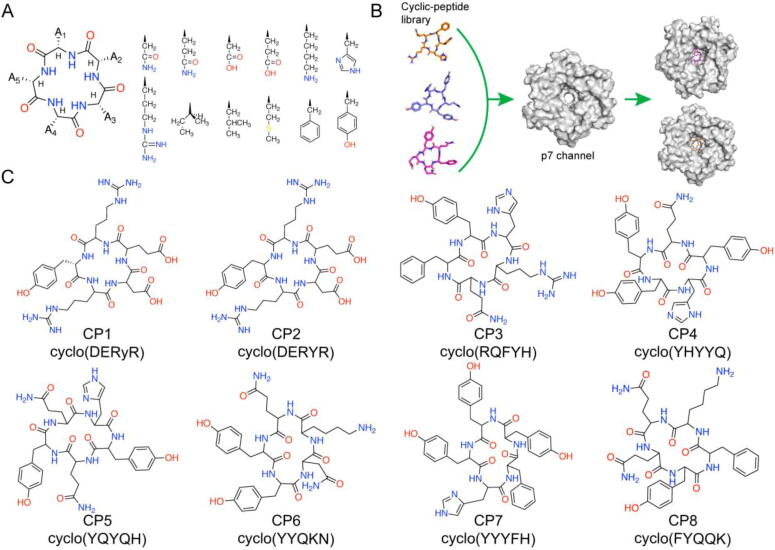
Analysis of the NMR structure of p7-5a (PDB: 2M6X) [18] reveals that the p7 pore radius is relatively large (3.9 Å at the narrowest conical region). Therefore, cyclic penta-, hexa- and hepta- peptides, which have the suitable size to fit into the p7 channel lumen, were initially docked into p7-5a and examined their respective binding modes. However, cyclic hexa- and hepta- peptides showed a high degree of flexibility and formed intramolecular hydrogen bonds, which resulted in random motion behavior and unpredictable peptide conformations in computational calculations. By contrast, cyclic penta-peptides showed higher stability and remained in a fixed position in simulations. We therefore generated a combinatorial cyclic penta-peptide library using different combinations to maximize the potential for interactions between cyclopeptides and the p7 channel (Fig. 1A). Twelve long-sidechain amino acids, including Asn, Gln, Asp, Glu, Lys, His, Arg, Ile, Leu, Met, Phe, and Tyr, were respectively incorporated at position A1–A5 and hence the theoretical diversity of the peptide library is 125 or 248,832.
Fig. 1.
Design of penta-cyclopeptides targeting p7. (A) Different combinations of 12 amino acid sidechains were introduced into the A1 ∼ A5 positions using a combinatorial method to generate a penta-cyclopeptide library. (B) High-throughput virtual screening (HTVS) of penta-cyclopeptides entry and binding within the p7-5a channel. (C) Eight positive candidates (CP1-CP8) were selected for synthesis and in vitro assays based on binding affinity and p7 conformations after HTVS docking and screening.
3.2. High-throughput virtual screening
The full library of cyclic penta-peptides was first generated by Discovery Studio 4.0 [21]. Docking simulations for the full library of penta-cyclopeptides were conducted with the p7-5a channel (PDB: 2M6X) [18] using via Autodock Vina 1.2.2 [22]. After first-round high-throughput virtual semi-flexible screening and second-round flexibility refinement (Fig. 1B), eight novel cyclic penta-peptides (CP1-CP8) were obtained with high binding affinity and capacity for stable channel blocking (Fig. 1C). More detailed information about molecular docking-based virtual screening is provided in Methods Section. The binding free energies of CP1-CP8 with p7-5a ranged from −6.481 to −8.160 kcal/mol (Table 1), substantially lower than that of small molecules and cyclic tetra-peptides [20].
Table 1.
Binding free energies of cyclic penta-peptides in the p7 channel.
| Binding free energies (kcal/mol) | p7-5a[a] | p7-2a[b] |
|---|---|---|
| CP1 | −6.653 | −6.863 |
| CP2 | −6.902 | −7.029 |
| CP3 | −7.577 | −8.098 |
| CP4 | −6.799 | −7.022 |
| CP5 | −7.005 | −7.019 |
| CP6 | −6.588 | −6.615 |
| CP7 | −8.160 | −8.147 |
| CP8 | −6.481 | −6.545 |
[a] Target p7-5a is used in the NMR. [b] Target p7-2a is used in the virus production assay.
3.3. Anti-viral evaluation
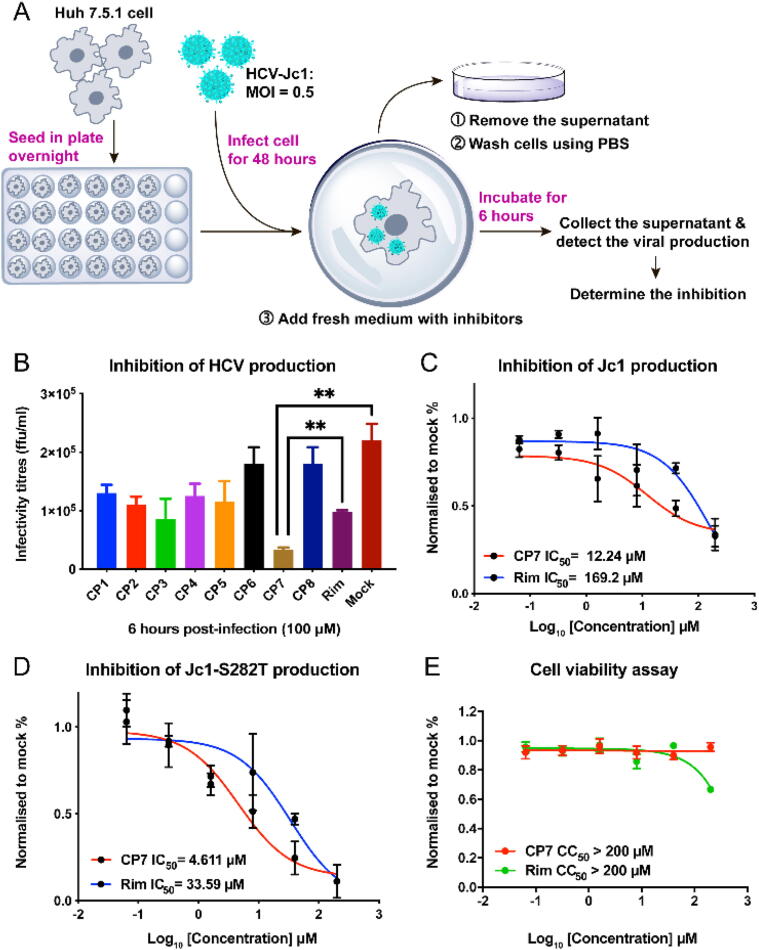
These eight novel cyclic penta-peptides (CP1-CP8) were then tested for their in vitro inhibitory effects on HCV production (Fig. 2A). As we mentioned in Materials and Methods, the available infectious Jc1 strain (GT2a) was used in the anti-HCV production assays. Briefly, virus-infected Huh 7.5.1 cells were treated with cyclopeptides for 6 h, then subsequent production of infectious viral particles was quantified by HCV titration [34]. While 100 μM of all eight of the candidate cyclic penta-peptides could significantly inhibit HCV viral production compared to that in the mock treatment, CP7 cyclo(YYYFH) treatment resulted in significantly lower HCV titres, indicating a better viral inhibition, than other cyclopeptides (Fig. 2B). Time-course evaluation of CP7 inhibition after 2 h, 4 h and 12 h post-infection further demonstrated its inhibitory effects on HCV production (Fig. S1A). Additionally, RT-qPCR-based quantification of intracellular RNA levels of subgenomic replicons showed no difference between CP7 (Fig. S1B), rimantadine (Rim), and mock treatments (Fig. S1C), indicating that CP7 could impair virus assembly and release but does not affect viral RNA replication.
Fig. 2.
Newly-designed CP7 inhibits HCV production with high efficacy. (A) The experimental scheme to test the inhibitory effects on viral production. Huh 7.5.1 cells infected by HCV-Jc1 at an MOI of 0.5 for 2 days were treated with 100 μM anti-viral cyclopeptides. Supernatants were collected after 6 h of incubation, and virus production was quantified by focus-reduction assay. (B) Inhibition of HCV production was determined for treatments of each candidate cyclic penta-peptide (CP1–CP8). (C) and (D) IC50 values of Rim (Rimantadine) and CP7 were determined, respectively. (E) After incubation with CP7 and Rim, cell viability and cytotoxicity were detected using Luminescent Cell Viability Assay reagent (Promega GS7570). (** denotes that p < 0.05, CP7 compared to Rim or Mock).
The 50 % maximal inhibition concentration (IC50) value of CP7 against Jc1 was 12.24 μM, considerably lower than that of Rim (Fig. 2C). Furthermore, the low HCV titres observed under exposure to CP7 aligned well with the calculations that showed CP7 had the lowest binding free energy to the p7-2a channel (modelled using PDB: 2M6X as a template, Fig. S2) (Table 1). By contrast, CP6 and CP8 showed the weakest binding and inhibitory effects on HCV. To test whether CP7 was effective against drug resistant strains, we generated a novel HCV strain, Jc1-S282T, harboring the known sofosbuvir resistance-inducing mutation S282T in the NS5B RNA polymerase (Fig. S3). The results of virus inhibition assays revealed an IC50 value of 4.61 μM for CP7 against Jc1-S282T (Fig. 2D), indicating that CP7 has high potency in inhibiting this drug resistant strain. Additionally, cell viability assays showed a half-maximum cytotoxicity concentration (CC50) of greater than 200 μM, indicating that CP7 has lower cell toxicity than that of Rim (Fig. 2E). Taken together, these strong inhibitory effects and low cytotoxicity of CP7 suggest its high potential for anti-HCV drug development.
3.4. Binding site of CP7 at the interior of p7 channel
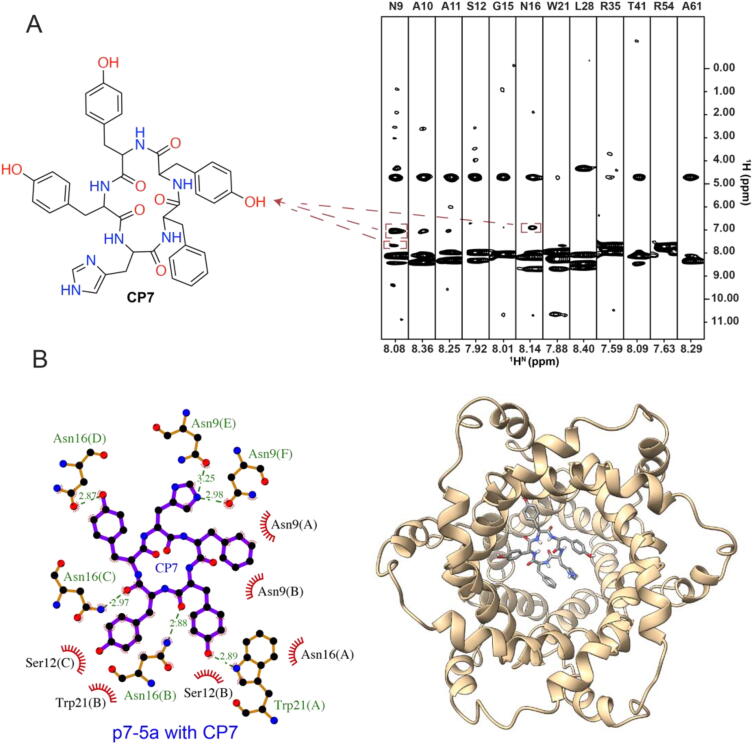
To determine the exact binding site of CP7 within the p7 channel, we carried out a protein–ligand nuclear Overhauser effect spectroscopy (NOESY) experiment. To this end, CP7 was added into a 15N-labelled and fully deuterated p7-5a sample to unambiguously measure the NOE signals between the protein backbone amide protons and CP7 sidechain protons. The NMR sample was examined using CD spectroscopy, showing typical α-helical characteristics (Fig. S4). The 15N-edited NOESY spectrum showed NOE crosspeaks between the CP7 aromatic protons and the amide protons of N9 and N16 in the p7 channel (Fig. 3A), which were incorporated as the experimental restraints in the molecular docking simulations of CP7 with the p7-5a or p7-2a channels. The final docking poses showed that the CP7 binding sites of p7-5a involved the N9, S12, N16, and W21 residues (Fig. 3B), while the corresponding binding sites in p7-2a involved residues H9, S12, C16, and Y21.
Fig. 3.
Determination of CP7 binding site in the p7-5a channel using NMR. (A) Chemical structure of CP7 (left) and the representative strips (right) of a 3D 15N-edited NOESY-TROSY (200 ms, mixing time) spectrum acquired using 0.82 mM 2H-,15N-labelled p7-5a and 8 mM CP7. The amide protons of N9 and N16 showed NOE crosspeaks with CP7. (B) CP7 docked in the p7-5a channel using NOE constraints. Left: the binding sites, including residues N9, S12, N16 and W21, labelled in green (hydrogen bond) or black (hydrophobic) in the view of CP7 (purple) inside p7-5a. The letters in the parentheses are referred to the chain ID of p7-5a. Right: the top view of CP7 inside p7-5a. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5. MD simulations of CP7 and p7
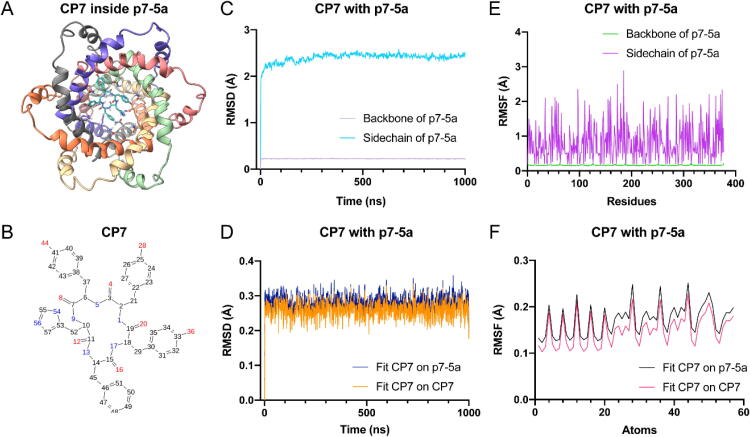
The binding stability and dynamics of CP7 inside p7-5a / p7-2a were further assessed using 1 μs MD simulations. In the presence of CP7, the root mean square deviation (RMSD) of p7-5a backbone was around 0.3 Å and ∼2 Å for sidechains. The root mean square of fluctuations (RMSF) of p7-5a and CP7 were ∼2 Å (p7-5a sidechains) and ∼0.2 Å (CP7 fitting on p7-5a or itself), respectively (Fig. 4). The interaction diagram of CP7 with p7-5a revealed that the hotspot residues involved in dynamic binding events mainly included N9, S12, N16, H17 and W21 (Fig. S5A), which was in agreement with the NOE results. Among these residues, N9 had the highest binding frequency. Furthermore, the interaction values of N9, S12, N16 and W21 were all greater than 0.5, which indicated that binding interactions with these sites in the p7-5a channel occurred in at least 50 % of the 1 μs simulations (Fig. S5B). The same results were obtained for p7-2a interactions with CP7, also with very low RMSD and RMSF (Fig. S6). A timeline plot of binding revealed that H9, S12, C16 and Y21 had the most frequent number of contacts (Fig. S7A), as well as the largest binding interaction values (Fig. S7B) between CP7 and p7-2a in 1 μs timelines. These results indicated that these interactions in the p7-2a channel remained stable over the MD trajectory.
Fig. 4.
Molecular dynamics simulation of CP7 inside p7-5a channel. (A) The docking complex of CP7 (blue) and p7-5a channel (rainbow). (B) Atom numberings of CP7. All the nitrogen atoms are labelled in blue and oxygen atoms are labelled in red. (C-D) Root-mean-square deviation (RMSD) plots of p7-5a in the p7-5a and CP7 complex against simulation time, showing system convergence and stability of simulations through 1 μs. Backbones of p7-5a (light purple) are extremely stable, while sidechains of p7-5a (cyan) are moderately stable in the presence of CP7 (C). Deviations of CP7 are within ∼0.3 Å inside p7-5a channel compared to its own coordinates (orange) and p7-5a protein coordinates (blue) (D). (E-F) Root Mean Square Fluctuations (RMSF) plots of the p7-5a and CP7 complex against simulation time, showing convergence and stability of simulations. Fluctuations of backbones and sidechains of p7-5a are ∼3 Å for every residue within 1 μs in the presence of CP7 (E). Fluctuations of CP7 are ∼0.2 Å for all the atoms of CP7 compared to its own coordinates (black) and p7-5a protein coordinates (red) (F). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Despite the high effectiveness of current DAAs against HCV, viral resistance has emerged rapidly leading to the urgent need for searching novel treatments. The essential activity and inhibition potential of HCV p7 channel make it an attractive target for antiviral inhibitor design. In this study, we designed a large library of cyclic penta-peptide inhibitors and performed the docking-based virtual screening to identify potential drug candidates targeting p7 channel. Instead of using the same amino acid for each position in cyclic tetra-peptides [20], here we used twelve natural amino acids with relatively long sidechains that were randomly input into cyclic penta-peptides. The long sidechains with different biochemical properties maximized the interactions between p7 and the cyclic penta-peptides. Therefore, the cyclic penta-peptide has a stronger binding affinity to the p7 channel. In contrast, the rigid structure of the cyclic tetra-peptide limits the flexibility of binding to p7 channels. The high intramolecular tension of the cyclic tetra-peptides also destabilizes themselves, resulting in a weaker binding free energy.
The primary hit rate from the high-throughput virtual screening was 2 %, which is relatively high due to the low energy threshold that was set to −6 kcal/mol for the primary hit selection. After elimination of unwanted modes of action in the second round, 8 hits remained. Of the eight selected cyclic penta-peptides, CP7 has been identified to be the most potent inhibitor of the p7 channel using anti-viral assays. CP7 is the least polar cyclic peptide containing four aromatic residues to inhibit HCV production, which is consistent to our previously quantitative structure-activity relationship (QSAR) analysis of small molecules, phenyl group contributes the most to inhibition effects [35]. We have further demonstrated that CP7 can specifically inhibit p7 activity with IC50 value of single-digit micromolar magnitude in a RAS-strain, indicating CP7 is promising to be further developed as a complementary therapy in cases of DAA failure.
We further characterized the CP7 binding site using NMR technology and verified that CP7 indeed binds to the channel lumen to restrict downstream viral production in combination with anti-viral assays. This direct binding to the interior of HCV p7 is distinct from the known small molecule inhibitors, amantadine and HMA [18], [19], that bind to the hydrophobic pockets on the channel periphery and inhibit channel conduction via long-range conformational changes in the p7 bottleneck region. These small molecules act as allosteric inhibitors to stabilize p7 channel in a closed conformation, while CP7 showed an unprecedented ‘bottle cork’ inhibition mechanism of p7 channel to inhibit the production of virus. MD simulations of p7 with this top hit further explained the key and stable interactions of different peptide side-chains with critical residues in p7 N-terminal bottleneck region. The key determinants of CP7 in preventing the channel opening are hydrophobic interactions and hydrogen bonds. In contrast, the central interaction of p7 allosteric inhibitors is hydrophobic interaction. Previous study demonstrated that deleting p7 N-terminal domain (NTD) or introducing H9W and S12W mutations can disable the release processes of HCV viral particles [36], indicating the NTD is a key mediator for p7 activity, which is in accordance with our discovery that blocking NTD of p7 channel for inhibiting HCV infection.
Cyclic peptide-based drugs with several advantageous biochemical and therapeutic properties have increasingly attracted attention in the past two decades [37], [38]. Previous literature showed that cyclic peptides usually inhibit receptor or enzyme activity, and modulate protein–protein or protein-RNA interactions [38]. A recent study using molecular modeling showed that a small cyclic peptide, PnCS1, binds to the pore region of the CaV3.1 channel and blocks the conduction pathway of the channel [39]. Our NMR characterization of CP7 and p7 viroporin for the first time experimentally revealed the structural basis of blocking the channel lumen by cyclic peptides, which enhances our understanding of the molecular mechanism for cyclic peptide inhibitions and further provides mechanistic insights into therapeutic development strategies of channel blockers.
Collectively, our study of cyclopeptide-based inhibition of the p7 inner channel provides guiding principles for designing a new class of anti-HCV drugs.
Funding
This Work was supported by National Natural Science Foundation of China, Grant No 21772146 (S.W.) and National Key R&D Program of China, grant number 2017YFA0504804 (B.O.), Key Research Program of Frontier Sciences, CAS, grant number QYZDB-SSW-SMC043 (B.O.), the Chinese Academy of Sciences (CAS) Major Science and Technology Infrastructure Open Research Projects (B.O.).
CRediT authorship contribution statement
Shukun Wei: Data curation, Methodology, Software, Validation, Formal analysis. Chaolun Liu: Data curation. Lingyu Du: Data curation, Methodology. Bin Wu: Methodology. Jin Zhong: Writing – review & editing. Yimin Tong: Investigation, Writing – original draft, Supervision. Shuqing Wang: Conceptualization, Validation, Investigation, Writing – original draft, Writing – review & editing, Supervision, Funding acquisition. Bo OuYang: Conceptualization, Validation, Investigation, Writing – original draft, Writing – review & editing, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the staffs from Nuclear Magnetic Resonance System/Mass Spectrometry System/Database and Calculation Analysis System at National Facility for Protein Science in Shanghai, Zhangjiang Laboratory (NFPS, ZJLab), High Performance Calculation (HPC) Platform at Centre for Excellence of Molecular Cell Science (CEMCS) of Chinese Academy of Sciences (CAS), China for their instrument support and technical assistance in data collection and analysis.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.10.035.
Contributor Information
Yimin Tong, Email: ymtong@sibs.ac.cn.
Shuqing Wang, Email: wangshuqing@tmu.edu.cn.
Bo OuYang, Email: ouyang@sibcb.ac.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Mohd Hanafiah K., Groeger J., Flaxman A.D., Wiersma S.T. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57:1333–1342. doi: 10.1002/hep.26141. [DOI] [PubMed] [Google Scholar]
- 2.Bukh J. A critical role for the chimpanzee model in the study of hepatitis C. Hepatology. 2004;39:1469–1475. doi: 10.1002/hep.20268. [DOI] [PubMed] [Google Scholar]
- 3.WHO, Hepatitis C Key Facts. 2021; https://www.who.int/news-room/fact-sheets/detail/hepatitis-c. Accessed 27th July, 2021.
- 4.Cholongitas E., Papatheodoridis G.V. Sofosbuvir: a novel oral agent for chronic hepatitis C. Ann Gastroenterol. 2014;27:331–337. [PMC free article] [PubMed] [Google Scholar]
- 5.Lee C. Discovery of hepatitis C virus NS5A inhibitors as a new class of anti-HCV therapy. Arch Pharm Res. 2011;34:1403–1407. doi: 10.1007/s12272-011-0921-6. [DOI] [PubMed] [Google Scholar]
- 6.Chen S.H., Tan S.L. Discovery of small-molecule inhibitors of HCV NS3-4A protease as potential therapeutic agents against HCV infection. Curr Med Chem. 2005;12:2317–2342. doi: 10.2174/0929867054864769. [DOI] [PubMed] [Google Scholar]
- 7.Malandris K., Kalopitas G., Theocharidou E., Germanidis G. The Role of RASs /RVs in the Current Management of HCV. Viruses. 2021;13(10):2096. doi: 10.3390/v13102096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.OuYang B., Chou J.J. The minimalist architectures of viroporins and their therapeutic implications. Biochim Biophys Acta. 2014;1838:1058–1067. doi: 10.1016/j.bbamem.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steinmann E., Penin F., Kallis S., Patel A.H., Bartenschlager R., et al. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 2007;3:e103. doi: 10.1371/journal.ppat.0030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakai A., Claire M.S., Faulk K., Govindarajan S., Emerson S.U., et al. The p7 polypeptide of hepatitis C virus is critical for infectivity and contains functionally important genotype-specific sequences. Proc Natl Acad Sci USA. 2003;100:11646–11651. doi: 10.1073/pnas.1834545100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones C.T., Murray C.L., Eastman D.K., Tassello J., Rice C.M. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J Virol. 2007;81:8374–8383. doi: 10.1128/JVI.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffin S.D., Beales L.P., Clarke D.S., Worsfold O., Evans S.D., et al. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug. Amantadine FEBS Lett. 2003;535:34–38. doi: 10.1016/s0014-5793(02)03851-6. [DOI] [PubMed] [Google Scholar]
- 13.Griffin S., Stgelais C., Owsianka A.M., Patel A.H., Rowlands D., et al. Genotype-dependent sensitivity of hepatitis C virus to inhibitors of the p7 ion channel. Hepatology. 2008;48:1779–1790. doi: 10.1002/hep.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luscombe C.A., Huang Z., Murray M.G., Miller M., Wilkinson J., et al. A novel Hepatitis C virus p7 ion channel inhibitor, BIT225, inhibits bovine viral diarrhea virus in vitro and shows synergism with recombinant interferon-alpha-2b and nucleoside analogues. Antiviral Res. 2010;86:144–153. doi: 10.1016/j.antiviral.2010.02.312. [DOI] [PubMed] [Google Scholar]
- 15.Premkumar A., Wilson L., Ewart G.D., Gage P.W. Cation-selective ion channels formed by p7 of hepatitis C virus are blocked by hexamethylene amiloride. FEBS Lett. 2004;557:99–103. doi: 10.1016/s0014-5793(03)01453-4. [DOI] [PubMed] [Google Scholar]
- 16.Pavlovic D., Neville D.C., Argaud O., Blumberg B., Dwek R.A., et al. The hepatitis C virus p7 protein forms an ion channel that is inhibited by long-alkyl-chain iminosugar derivatives. Proc Natl Acad Sci USA. 2003;100:6104–6108. doi: 10.1073/pnas.1031527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luik P., Chew C., Aittoniemi J., Chang J., Wentworth P., Jr, et al. The 3-dimensional structure of a hepatitis C virus p7 ion channel by electron microscopy. Proc Natl Acad Sci USA. 2009;106:12712–12716. doi: 10.1073/pnas.0905966106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.OuYang B., Xie S., Berardi M.J., Zhao X., Dev J., et al. Unusual architecture of the p7 channel from hepatitis C virus. Nature. 2013;498:521–525. doi: 10.1038/nature12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao L., Wang S., Du L., Dev J., Zhou L., et al. Structural basis of interaction between the hepatitis C virus p7 channel and its blocker hexamethylene amiloride. Protein Cell. 2016;7:300–304. doi: 10.1007/s13238-016-0256-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pang S., Zhao R., Wang S., Wang J. Cyclopeptides design as blockers against HCV p7 channel in silico. Mol Simul. 2019;45:1419–1425. [Google Scholar]
- 21.Dassault Systèmes BIOVIA, Discovery Studio Modeling Environment, Release 4.0, San Diego: Dassault Systèmes. 2013; https://www.3dsbiovia.com/. Accessed 20th January, 2021.
- 22.Eberhardt J., Santos-Martins D., Tillack A.F., Forli S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J Chem Inf Model. 2021;61:3891–3898. doi: 10.1021/acs.jcim.1c00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ravindranath P.A., Forli S., Goodsell D.S., Olson A.J., Sanner M.F. AutoDockFR: Advances in Protein-Ligand Docking with Explicitly Specified Binding Site Flexibility. PLoS Comput Biol. 2015;11:e1004586. doi: 10.1371/journal.pcbi.1004586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bowers KJ, Chow E, Xu H, Dror RO, Eastwood MP, et al. In: Proceedings of the 2006 ACM/IEEE conference on Supercomputing, Association for Computing Machinery, Tampa, Florida, 2006. pp. 84.
- 25.Jorgensen W.L., Tirado-Rives J. The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J Am Chem Soc. 1988;110:1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 26.Lomize M.A., Lomize A.L., Pogozheva I.D., Mosberg H.I. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 27.Price D.J., Brooks C.L., 3rd A modified TIP3P water potential for simulation with Ewald summation. J Chem Phys. 2004;121:10096–10103. doi: 10.1063/1.1808117. [DOI] [PubMed] [Google Scholar]
- 28.Hoover Canonical dynamics: Equilibrium phase-space distributions. Phys Rev A. 1985;31(3):1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 29.Deng M. Klein, Structure and dynamics of bipolarons in liquid ammonia. Phys Rev Lett. 1992;68(16):2496–2499. doi: 10.1103/PhysRevLett.68.2496. [DOI] [PubMed] [Google Scholar]
- 30.Petersen H.G. Accuracy and efficiency of the particle mesh Ewald method. J Chem Phys. 1995;103:3668. [Google Scholar]
- 31.Bernstein F.C., Koetzle T.F., Williams G.J., Meyer E.F., Jr., Brice M.D., et al. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 32.Tong Y., Li Q., Li R., Xu Y., Pan Y., et al. A Novel Approach To Display Structural Proteins of Hepatitis C Virus Quasispecies in Patients Reveals a Key Role of E2 HVR1 in Viral Evolution. J Virol. 2020;94:e00622–e00720. doi: 10.1128/JVI.00622-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pietschmann T., Kaul A., Koutsoudakis G., Shavinskaya A., Kallis S., et al. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc Natl Acad Sci USA. 2006;103:7408–7413. doi: 10.1073/pnas.0504877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong J., Gastaminza P., Cheng G., Kapadia S., Kato T., et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei S., Hu X., Du L., Zhao L., Xue H., et al. Inhibitor Development against p7 Channel in Hepatitis C Virus. Molecules. 2021;26(5):1350. doi: 10.3390/molecules26051350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scull M.A., Schneider W.M., Flatley B.R., Hayden R., Fung C., et al. The N-terminal Helical Region of the Hepatitis C Virus p7 Ion Channel Protein Is Critical for Infectious Virus Production. PLoS Pathog. 2015;11:e1005297. doi: 10.1371/journal.ppat.1005297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi J.S., Joo S.H. Recent Trends in Cyclic Peptides as Therapeutic Agents and Biochemical Tools. Biomol Ther (Seoul) 2020;28:18–24. doi: 10.4062/biomolther.2019.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H., Chen S. Cyclic peptide drugs approved in the last two decades (2001–2021) RSC Chem Biol. 2022;3:18–31. doi: 10.1039/d1cb00154j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Depuydt A.S., Rihon J., Cheneval O., Vanmeert M., Schroeder C.I., et al. Cyclic Peptides as T-Type Calcium Channel Blockers: Characterization and Molecular Mapping of the Binding Site. ACS Pharmacol Transl Sci. 2021;4:1379–1389. doi: 10.1021/acsptsci.1c00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.