Abstract
Whole-sporozoite (WSp) malaria vaccines induce protective immune responses in animal malaria models and in humans. A recent clinical trial with a WSp vaccine comprising genetically attenuated parasites (GAP) which arrest growth early in the liver (PfSPZ-GA1), showed that GAPs can be safely administered to humans and immunogenicity is comparable to radiation-attenuated PfSPZ Vaccine. GAPs that arrest late in the liver stage (LA-GAP) have potential for increased potency as shown in rodent malaria models. Here we describe the generation of four putative P. falciparum LA-GAPs, generated by CRISPR/Cas9-mediated gene deletion. One out of four gene-deletion mutants produced sporozoites in sufficient numbers for further preclinical evaluation. This mutant, PfΔmei2, lacking the mei2-like RNA gene, showed late liver growth arrest in human liver-chimeric mice with human erythrocytes, absence of unwanted genetic alterations and sensitivity to antimalarial drugs. These features of PfΔmei2 make it a promising vaccine candidate, supporting further clinical evaluation. PfΔmei2 (GA2) has passed regulatory approval for safety and efficacy testing in humans based on the findings reported in this study.
Subject terms: Diseases, Malaria
Introduction
Whole-sporozoite (WSp) malaria vaccines can induce strong protective immune responses in animal models of malaria and in humans1–4. WSp vaccines consist of whole parasites, i.e., metabolically active P. falciparum sporozoites (PfSPZ) that have the ability to migrate to and infect the human liver, but cannot transform into the symptomatic blood stage, and as such do not cause disease. The most advanced WSp vaccine candidate, the PfSPZ Vaccine, employs sporozoites that have been attenuated by radiation5. These sporozoites enter hepatocytes but are unable to replicate and thus abort development early in the liver. The PfSPZ Vaccine, consisting of cryopreserved, vialed radiation-attenuated sporozoites and produced by the biotech company Sanaria, Inc. (US), has entered phase 3 clinical development6,7. Sporozoite attenuation by genetic modification rather than by radiation, offers the advantage of a more homogeneous product, increased biosafety for sporozoite production, and potentially increased potency8,9. In a recent clinical study, human volunteers were immunized by intravenous injection with multiple doses of cryopreserved, vialed sporozoites of either the PfSPZ Vaccine or a WSp vaccine, consisting of genetically-attenuated parasites (GAP) that arrest growth soon after invasion of hepatocytes (early-arresting GAP; EA-GAP)10,11. This study showed that the EA-GAP vaccine, termed PfSPZ-GA1, was safe and induced immune responses that were comparable to those induced by the radiation-attenuated PfSPZ Vaccine10. Multiple clinical studies with the PfSPZ Vaccine in healthy volunteers from non-endemic areas (e.g., without prior exposure to malaria), have shown that this vaccine can induce levels of protection that are higher than those achieved with various subunit malaria vaccines (protein, peptide, or DNA-based)3,12,13. However, high sporozoite numbers are needed to achieve sufficient levels of efficacy. Moreover, the efficacy of the PfSPZ Vaccine appears to be lower in people living in areas of malaria endemicity compared to people with no previous history of malaria infection14,15, creating a need for highly potent WSp vaccines to achieve the 75% protection against clinical disease for >1 year as targeted in the malaria vaccine technology roadmap (https://www.malariavaccine.org/malaria-and-vaccines/malaria-vaccine-roadmap). Chemo-attenuated PfSPZ immunization studies suggest that the WSp vaccine efficacy may be improved by delaying the growth arrest in the liver and thereby broadening the antigen repertoire16. A study using a mouse malaria GAP has shown that attenuated sporozoites that arrest growth late in the liver (late-arresting GAP; LA-GAP) through deletion of a gene encoding a protein involved in fatty acid synthesis (Fabb/f), induced stronger protective immune responses than immunization with EA-GAP sporozoites17. The increased potency can most likely be explained by the exposure to an increased (liver- and blood-stage) antigen repertoire and/or biomass of LA-GAP liver-stages compared to those of EA-GAP. Unfortunately, removal of several fatty acid synthesis genes from the P. falciparum genome, results in the arrest of parasite growth in the mosquito preventing sporozoite formation18,19. These genes are therefore unsuitable targets for creating a P. falciparum LA-GAP for human use. To generate a P. falciparum LA-GAP, we selected in this study four additional genes (palm, hcs1, cbr, mei2) with a putative specific role in P. falciparum late liver-stage development, based on published phenotypes of rodent malaria mutants that lack the orthologous genes. We report the creation and analysis of four P. falciparum gene-deletion mutants that lack the palm, hcs1, cbr, or mei2 gene. We show that three gene-deletion mutants did not show the expected phenotype of sporozoite production, essential for further preclinical evaluation. Only the P. falciparum mutant lacking the mei2 gene (PfΔmei2) produced infective sporozoites that arrest growth late during development in the liver. We report results of preclinical evaluation of PfΔmei2, involving evaluation of late liver growth-arrest in cultured primary human hepatocytes and in chimeric mice with humanized liver and human red blood cells, genotype analysis by whole-genome sequencing and in-vitro drug-sensitivity testing of blood stages. Based on these preclinical studies, PfΔmei2 has passed regulatory approval for clinical evaluation of safety and efficacy as an LA-GAP vaccine (GA2).
Results
Selection of genes with a role during late liver-stage Plasmodium development
Gene-deletion studies have identified multiple proteins that play a role during late liver-stage development of rodent malaria parasites. Table 1 shows liver-stage growth phenotypes of published gene-deletion mutants that display a normal, wild-type-like blood-stage and sporozoite development but present defects in late liver-stage development. Most mutants do not display full growth arrest in the liver as revealed by ‘break-through’ blood infections, although with a (strong) increase in prepatency. Deletion of genes encoding apicoplast-located proteins involved in fatty acid synthesis results in the strongest attenuation phenotype, specifically in P. yoelii, with complete growth arrest. However, FASII-pathway genes, involved in the transformation of Acetyl-CoA into Acyl-ACP, are unsuitable targets for generation of P. falciparum LA-GAP because this pathway is essential for formation of viable sporozoites18. It can be assumed that genes encoding the apicoplast-located proteins of the pyruvate dehydrogenase (PDH) complex, which provides Acetyl-CoA, are also essential for P. falciparum sporozoite formation. Therefore, we excluded FASII-pathway genes for generation of P. falciparum LA-GAP. Four genes with a previously reported strong attenuation phenotype, palm, hcs1, cbr, and mei2, were selected for creating P. falciparum LA-GAPs. This selection was based on the absence of blood infections and/or compelling extension of the prepatent period after infection of mice with gene-deletion mutant sporozoites, while these sporozoites developed into late liver-stages in cultured hepatocytes (Table 1). The P. berghei PALM protein, whose exact function is unknown but plays a role during the final steps of P. berghei liver-stage merozoite formation20, has been shown to localize to the apicoplast20. HCS1, an ATP-dependent ligase that catalyzes biotin binding to biotin carboxylase, plays a role in adequate formation of P. berghei liver-stage merozoites21 and is expressed in the cytoplasm of liver-stages21. For CBR, a putative cytochrome-b5 oxidoreductase, a location in the food vacuole of blood-stages has been reported22. P. berghei parasites lacking CBR, showed a slight reduction in the size of mature liver-stages and in detachment of cultured hepatocytes containing maturing liver-stages23. The MEI2 protein is a member of a family of RNA-binding proteins containing an RNA recognition motif24. P. yoelii parasites lacking MEI2 develop into late liver schizonts but have a complete attenuation phenotype in highly susceptible BALB/cByJ mice infected with 50,000 sporozoites24 and only occasional breakthrough blood-infections were observed when >200,000 sporozoites were inoculated25. We confirmed this late liver-stage growth arrest and the strong attenuation phenotype in P. berghei by creating and characterizing P. berghei gene-deletion mutants lacking the mei2 gene (PbΔmei2; Supplementary Fig. 1). We also observed occasional breakthrough blood-infections when mice were inoculated with high numbers (2 × 105) of PbΔmei2 sporozoites. In one experiment we infected mice with 2 × 105 sporozoites of a PbΔmei2 line (PbΔmei2-breakthrough-a) that was collected from mice with a breakthrough blood-infection after the first infection with PbΔmei2 sporozoites. In this experiment we found that the percentage of mice with a ‘second’ breakthrough blood infection was comparable to that of mice with a ‘first’ breakthrough infection and the mice that became positive had again a strongly prolonged prepatent period, i.e., 5 of the 11 mice did develop a blood stage infection with a prepatent period of 9-10 days, whereas mice infected with wild-type parasites became blood-stage positive at day 4 or 5. These observations strongly indicate that the PbΔmei2-breakthrough-a parasites are not derived from parasites that had permanently switched to an efficient and Mei2-independent mechanism of liver stage development (Supplementary Fig. 1).
Table 1.
Published rodent malaria gene-deletion mutants with a ‘late-liver stage’ growth phenotype.
| berghei/yoelii gene ID | falciparum gene ID | product | blood infection | Extended prepatency | Ref. |
|---|---|---|---|---|---|
| PBANKA_1125100 | PF3D7_0626300 | 3-oxoacyl-acyl-carrier protein synthase I/II (FabB/FabF) | yes | yes | 70 |
| PY17X_1126500 | PF3D7_0626300 | 3-oxoacyl-acyl-carrier protein synthase I/II (FabB/FabF) | no | - | 71 |
| PBANKA_1229800 | PF3D7_0615100 | enoyl-acyl carrier reductase (ENR,FabI) | yes | yes | 72 |
| PY17X_1342900 | PF3D7_1323000 | beta-hydroxyacyl-ACP dehydratase (FabZ) | no | - | 71 |
| PBANKA_1410500 | PF3D7_1312000 | malonyl CoA-acyl carrier protein transacylase (FabD) | no | - | 23 |
| PBANKA_0308200 | PF3D7_0211400 | beta-ketoacyl-ACP synthase III (FabH) | yes | yes | 23 |
| PBANKA_0823800 | PF3D7_0922900 | 3-oxoacyl-[acyl-carrier-protein] reductase (FabG) | no | - | 23 |
| PY17X_0715100 | PF3D7_0815900 | dihydrolipoyl dehydrogenase, apicoplast (PDH E3) | no | - | |
| PBANKA_0923800 | PF3D7_1124500 | pyruvate dehydrogenase E1 component subunit | no | - | 73 |
| alpha (PDH E1α) | |||||
| PY17X_0934900 | PF3D7_1114800 | glycerol-3-phosphate dehydrogenase, putative (apiG3PDH) | no | - | 74 |
| PBANKA_0923800 | PF3D7_1114800 | glycerol-3-phosphate dehydrogenase, putative (apiG3PDH) | yes | yes | 73 |
| PY17X_1418400 | PF3D7_1318200 | glycerol-3-phosphate 1-O-acyltransferase (apiG3PAT) | no | - | 74 |
| PBANKA_0820900 | PF3D7_0920000 | long chain fatty acid elongation enzyme, putative (ELO3, ELO-A) | no | - | 23 |
| PBANKA_1357500 | PF3D7_1344600 | lipoyl synthase (LipA) | yes | yes | 23 |
| PBANKA_0707000 | PF3D7_0823600 | lipoate-protein ligase B (LipB) | yes | yes | 21 |
| PBANKA_1143400 | PF3D7_1367500 | NADH-cytochrome b5 reductase, putative (CBR) | yes | yes | 23 |
| PBANKA_0101100 | PF3D7_0602300 | liver merozoite formation protein (PALM) | Yes | Yes | 20 |
| PBANKA_0511000 | PF3D7_1026900 | biotin--protein ligase 1 (HCS1) | yes | yes | 21,23 |
| PY17X_1123700 | PF3D7_0623400 | MEI2-like RNA-binding protein (MEI2) | Yes | Yes | 24 |
| PBANKA_0214400 | PF3D7_0730300 | a transcription factor with AP2 domain(s) (AP2-L) | Yes | Yes | 75 |
| PBANKA_1024600 | PF3D7_1418100 | putative liver stage protein 1 (LISP1) | Yes | Yes | 76 |
| PBANKA_1003000 | PF3D7_0405300 | liver-specific protein 2 (LISP2; sequestrin) | Yes | Yes | 77,78 |
| PBANKA_0506500 | PF3D7_1022300 | ZIP domain-containing protein (ZIPCO) | Yes | Yes | 79 |
| PBANKA_0505000 | PF3D7_1020800 | dihydrolipoamide acyltransferase component E2 | yes | yes | 23 |
| PBANKA_1436200 | PF3D7_1221000 | histone-lysine N-methyltransferase, H3 lysine-4 specific | yes | yes | 23 |
| PBANKA_0304800 | PF3D7_0207400 | serine repeat antigen 7 | yes | yes | 80 |
| PBANKA_0404100 | PF3D7_0305600 | DNA-(apurinic or apyrimidinic site) endonuclease | yes | yes | 81 |
| PBANKA_1021500 | PF3D7_1421900 | copper transporter, putative (CTR2) | yes | yes | 82 |
Genes 1–7: Apicoplast-located proteins, involved in fatty acid synthesis (FAS II)–transformation of Acetyl-CoA into Acyl-ACP; Genes 8–12: Apicoplast-located proteins of the pyruvate dehydrogenase (PDH) complex, providing acetyl-CoA for fatty acid synthesis; Genes 13–17: (Putative) apicoplast-located proteins: not involved in the transformation of Acetyl-CoA into Acyl-ACP; Genes 16–19: genes selected for gene-deletion in P. falciparum.
P. falciparum mutants lacking the genes palm, hcs1 or cbr do not produce viable sporozoites
To create P. falciparum LA-GAP, independent mutants lacking the palm, hcs1, cbr, or mei2 genes (PfΔpalm, PfΔhcs1, PfΔcbr, PfΔmei2) were generated using established methods of CRISPR/Cas9 gene-editing26,27. Correct deletion of the genes was confirmed by diagnostic PCR and/or Southern blot analysis of digested genomic DNA (Supplementary Figs. 2–4; Fig. 1). These four mutants showed blood-stage growth rates comparable to wild-type P. falciparum NF54 (WT PfNF54) parasites. In vitro gametocyte production and oocyst numbers in An. stephensi mosquitoes fed with cultured gametocytes were in the range of those of WT PfNF54 (Supplementary Figs. 2–4; Fig. 1; Table 2). However, in mosquitoes infected with the PfΔpalm, PfΔhcs1, and PfΔcbr mutants, salivary gland sporozoites were either absent or their numbers were highly reduced. Midgut/hemocoel sporozoites were also absent or strongly reduced and most oocysts did not show clear signs of sporozoite formation, presenting an ‘empty’ or vacuolated appearance (Supplementary Fig. 5). These observations indicate a role of these three proteins in adequate sporozoite formation inside oocysts. Only PfΔmei2 mutants produced numbers of salivary gland sporozoites comparable to those observed for WT PfNF54 (Table 2).
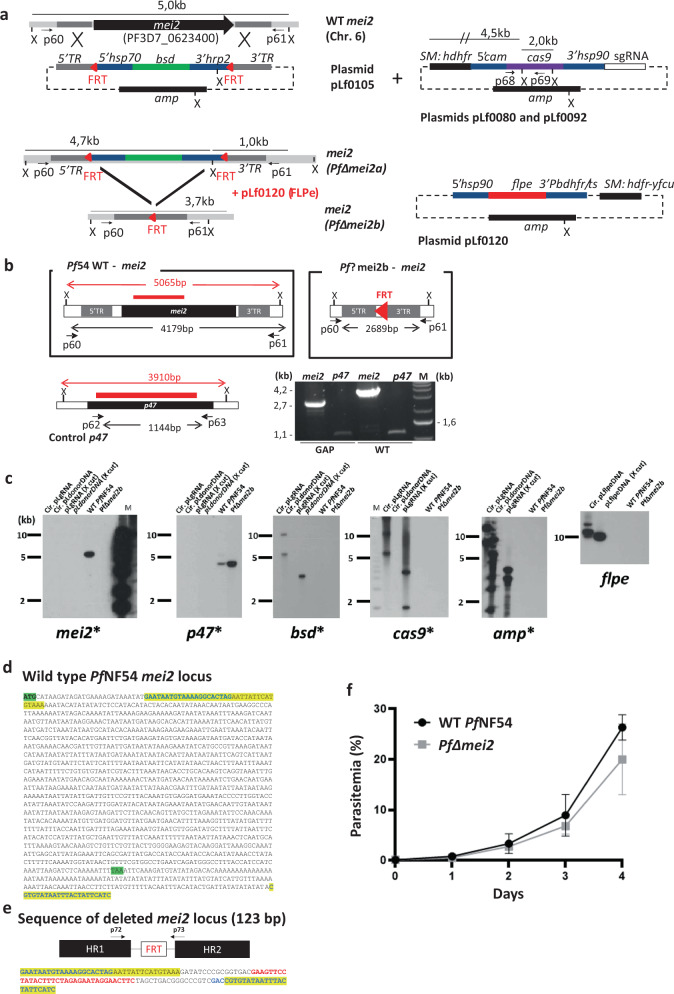
Fig. 1. Generation, genotyping, and blood-stage growth of PfΔmei2 (LA-GAP, GA2).
a Left: the mei2 (PF3D7_ 0623400) genomic locus on chromosome 6 (Chr. 6) of wild-type Pf NF54 (WT Pf NF54) and PfΔmei2 parasites before (PfΔmei2a) and after (PfΔmei2b) FLPe-mediated removal of the blasticidin-S-deaminase (bsd) selectable marker (SM). The donor plasmid pLf0105 to delete mei2 contains the bsd SM, flanked by two frt sites (red triangles) and mei2 targeting sequences (5′ TR and 3′ TR) for double cross-over integration. Primer pairs p60/p61 and PCR fragment size for diagnostic PCR are indicated (b); X (XmnI): restriction site used for Southern blot analyses (c). hsp70, heat shock protein 90; hrp2, histidine-rich protein II; amp, ampicillin. Right top: sgRNA plasmids (pLf0080, pLf0092) containing the human dihydrofolate reductase-thymidylate synthase (hdhfr) SM and the cas9 expression cassette. Cam, calmodulin. PCR primers (p68/p69) to amplify part of cas9, sizes of the sgRNA constructs after XmnI (X) digestion and mei2 and cas9 probes are indicated (c). Right bottom: construct pLf0120 with the hdhfr-yfcu SM and the flpe expression cassette. pbdhfr/ts,: P. berghei bifunctional dihydrofolate reductase-thymidylate synthase, putative. See Supplementary Table 1 for primers details. b WT Pf NF54 and PfΔmei2b genomic loci and the control gene p47 (coding sequence shown as black boxes). Shown are the 5′ and 3′ mei2 targeting regions (5′TR and 3′TR), used construct pLf0105 (a) and the frt site. PCR primers (in black) for amplifying mei2 (p60/p61) and p47 (p62/63), expected sizes of the full length mei2 and p47 genes and size of mei2 locus after mei2 deletion and removal of bsd SM cassette are shown. X (XmnI): restriction site used for Southern analysis (c). DNA probes used in Southern analyses (c) and sizes of digested DNA fragments recognized by the probes (mei2 and p47) are shown (in red). Red triangle: the 34 bp frt site in the PfΔmei2b genome after removal of the bsd SM cassette. PCR (right lower panel) analysis of WT Pf NF54 and PfΔmei2b genomic DNA confirms mei2 deletion (control: amplification of p47). Primer pairs: p60/61 for mei2 and p62/p63 for p47. See Supplementary Table 1 for primer details). M, molecular weight marker; 1 kb DNA ladder (Invitrogen). c Southern analysis of restricted genomic DNA from WT Pf NF54, PfΔmei2b, and plasmids used to delete mei2 (DNA digested with XmnI (X)). DNA-samples/lanes: (i) circular sgRNA plasmids (Cir.pLgRNA); (ii) circular donor DNA plasmid pLf0105 (pLΔmei2); (iii) XmnI-digested sgRNA plasmid (pLgRNA-X cut); (iv) XmnI-digested donor DNA plasmid; v) genomic WT Pf NF54 DNA; (vi) genomic PfΔmei2b DNA. Probes: part of mei2, p47 (control), cas9, bsd, amp and flpe (see a and b for probe location and expected fragment sizes). Hybridizations show correct mei2 deletion and absence of cas9, bsd, amp and flpe in PfΔmei2b. M, molecular weight marker; 1 kb DNA ladder (Invitrogen) labeled on the sides of the gels. d Sequence of the WT PfNF54 mei2 locus. Yellow: sequences present in the disrupted mei2 locus of PfΔmei2 (e); Green: start and stop codon of mei2. e The mei2 locus in the PfΔmei2 genome after mei2 deletion by integration of pLf0105 and FLPe-mediated removal of the bsd SM marker The mei2 targeting regions (HR1 and HR2) for double cross-over integration and the frt site are shown. In addition, the sequence of the PCR fragment of the mei2 locus is shown, amplified using primers p72/p73 (see Supplementary Table 1 for primer details). Yellow: sequences present in the mei2 locus of WT PfNF54 and PfΔmei2. Red: the 34 bp FRT sequence, flanked by 16 bp and 14 bp cloning restriction sites. f In vitro growth rate of PfΔmei2 and WT Pf NF154 asexual blood stages. Parasitemia (%) during a 4-day culture period (mean and s.d. of three cultures). Error bars represent standard deviation.
Table 2.
Gametocyte, oocyst, and sporozoite production of four P. falciparum gene-deletion mutants and wild-type WT PfNF54.
| Lines | Stage V Gam. (%)1 | Exflag./105 RBC2 | Infection rate (%)3 | Oocyst no.4 | Spor. no (×103)6 |
|---|---|---|---|---|---|
| Average (s.d) | Average (s.d.) | Average (s.d.) | Average (s.d.) | Average (s.d.) | |
| WT PfNF54 | 2.04 (0.88) | 804 (387) | 66 (39) | 92 (86) | 53 (51) |
| 0.8–3.7 (11 exp.) | 201–1459 (27 exp.) | 68–100 (34 exp.) | 21–247 (34 exp.) | 4–155 (34 exp.) | |
| PfΔpalm | 2.85 (1.34) | 1544 (116) | 75 (18) | 38 (27) | 0 |
| 1.9–3.8 (2 exp.) | 1462–1626 (2 exp.) | 56–93 (4 exp.) | 19–64 (4 exp.) | (4 exp.) | |
| PfΔcbr | 1.6 (0.6) | 440 (219) | 53 (45) | 65 (52) | 0 |
| 0.8–2.1 (4 exp.) | 147–660 (9 exp.) | 54–100 (13 exp.) | 22–156 (13 exp.) | (11 exp.) | |
| PfΔhcs1 | 2.6 (0.8) | 459 (133) | 92(14) | 89 (42) | 0 |
| 1.8–3.3 (3 exp.) | 365–554 (2 exp.) | 76–100 (3 exp.) | 63–138 (3 exp.) | (3 exp.) | |
| PfΔmei2 | 2.1 (0.6) | 288 (182) | 73 (32) | 63 (68) | 31 (26) |
| 1.8–2.8 (3 exp.) | 94–600 (9 exp.) | 23–100 (14 exp.) | 14–167 (13 exp.) | 10–84 (9 exp.) |
1Mean percentage of blood-stage parasites developing into gametocytes in vivo.
2Mean percentage of exflagellating males in vitro, 12–15 minutes after activation.
3percentage of infected mosquitoes at days 7–12 after feeding.
4Mean number of oocysts per mosquito (days 7–12).
5Mean number of sporozoites per mosquito (days 21).
s.d.: standard deviation. Second raw: range and number of experiments.
PfΔmei2 parasites develop into replicating liver stages but fail to produce red blood cell-infectious liver-stage merozoites
The growth arrest of PfΔmei2 liver stages was investigated using primary human hepatocytes. First, PfΔmei2 sporozoite infectivity and development was analyzed in vitro in cultures of cryopreserved primary human hepatocytes (from the company BioIVT) infected with isolated PfΔmei2 and WT PfNF54 sporozoites. A mean of 2,1% (s.d. 0.42) and 1.4% (s.d. 0.42) infected hepatocytes per well was observed for WT PfNF54 WT and PfΔmei2, respectively, at day 3 post infection (p.i.) (Fig. 2a). During in vitro liver-stage development (day 3, 5, 7, and 9 p.i.) the size and light-microscopy morphology of PfΔmei2 parasites was comparable to those of WT PfNF54 parasites and a strong increase in parasite size (Fig. 2b) and nuclear content (Fig. 2c) was observed in both WT PfNF54 and PfΔmei2 parasites. Liver-stages of WT PfNF54 and PfΔmei2 liver stages displayed similar staining patterns of the cytoplasmic marker HSP70 and of the parasitophorous vacuole marker EXP1. At days 7 and 9 p.i., liver-stages of WT PfNF54 and PfΔmei2 exhibited comparable staining of the merozoite surface protein 1 (MSP1) (Fig. 2c). We then confirmed wild-type-like hepatocyte invasion and development into late liver-stages of PfΔmei2 in cryopreserved primary human hepatocytes from the company Lonza (Supplementary Fig. 6) These analyses also showed that sporozoite infectivity and the size of PfΔmei2 liver stages (at day 3, 5, 7, and 9) were comparable to those of WT PfNF54 parasites. In addition, PfΔmei2 and WT PfNF54 liver stages displayed similar staining patterns with antibodies against the cytoplasmic marker HSP70 and the parasitophorous vacuole marker EXP1. All together, these results show that PfΔmei2 sporozoites can effectively infect cultures of human hepatocytes and are able to develop into large, replicating liver-stages that express merozoite-specific antigens.
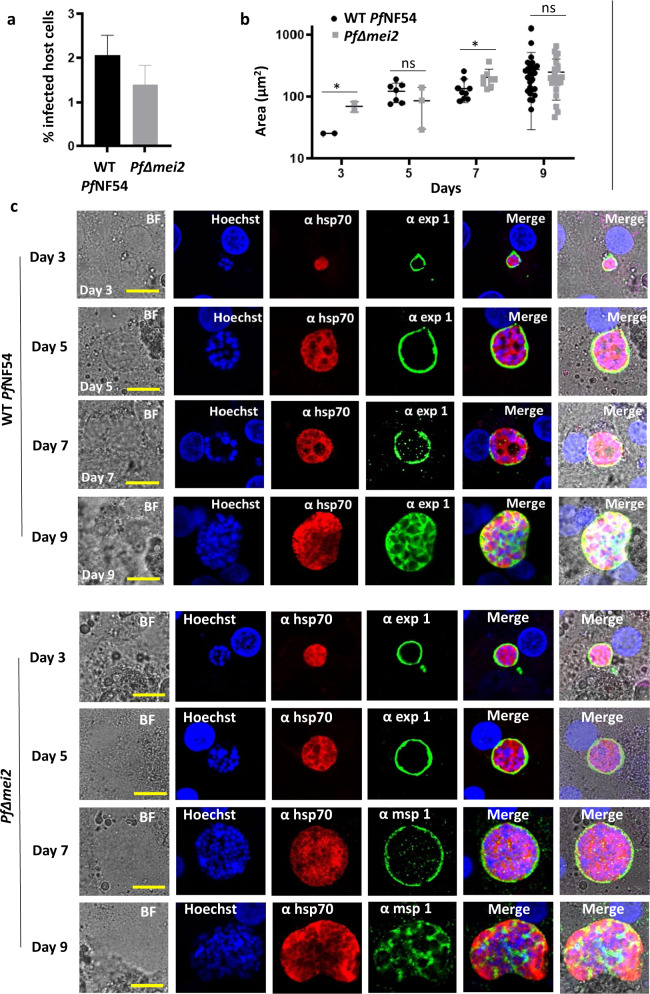
Fig. 2. PfΔmei2 liver-stage development in cultured human primary hepatocytes (BioIVT).
a Percentage of hepatocytes infected with WT PfNF54 and PfΔmei2 at day 3 post infection (p.i.) (p = 0.002; unpaired Mann–Whitney test). b Liver-stage size on day 3, 5, 7, and 9 p.i. (3-20 parasites measured in two wells). The average of the parasite’s cytoplasm at its greatest circumference using HSP70-positive area (μm2), s.d. and significances values are shown (unpaired Mann–Whitney test: *p < 0.05; ns: not significant). c Representative confocal microscopy images of liver stages on days 3, 5, 7, and 9 p.i. Upper panel WT PfNF54; lower panel PfΔmei2. Fixed hepatocytes were stained with the following antibodies: rabbit anti-PfHSP70 (α hsp70), mouse anti-PfEXP1 (α exp1), and anti-PfMSP1 (α msp1). Nuclei stained with Hoechst-33342. All pictures were recorded with standardized exposure/gain times; Alexa Fluor® 488 (green) 0.7 s; anti-IgG Alexa Fluor® 594 (red) 0.6 s; Hoechst (blue) 0.136 s; bright field (BF) 0.62 s (1× gain). Scale bar, 10 μm. Error bars represent standard deviation.
Next, the liver-stage development of PfΔmei2 and WT PfNF54 parasites was analyzed in liver-chimeric humanized mice (FRG huHep mice) engrafted with high proportions of human red blood cells (RBC), injected intravenously (see Fig. 3a for a schematic representation of these experiments). The use of FRG huHep mice to mimic in vivo hepatocyte infection with P. falciparum sporozoites through to the blood-stage of infection has been previously described28,29. Briefly, mice were infected with 1 × 10(6) sporozoites of WT PfNF54 or PfΔmei2 followed by injection of human red blood cells at days 5 and 6 p.i. Blood was collected for qRT-PCR analysis at days 7 and 9 p.i., and mice were euthanized at day 9 p.i. for blood collection and cryopreservation. In WT PfNF54-infected mice an average of 1,4 × 10(10) 18 S copies per ml was detected at day 7 (range: 1,1 × 10(10) to 2,0 × 10(10)) which increased on day 9 to an average of 1,2 × 10(11) copies (range 7,0 × 10(10) to 1,6 × 10(11)). In the PfΔmei2-infected mice the average number of 18 S copies was much lower at day 7 (2,4 × 10(6); range 1 × 10(6) to 4,9 × 10(6)) and dropped 10-fold at day 9 in 6 mice (3,2 × 10(5); range 3,7 × 10(4) to 1,1 × 10(6)) while in one mouse the number of 18 S copies remained similar (5,8 × 10(6) at day 7 and 7,3 × 10(6) at day 9. (Fig. 3b). Combined these observations show the presence of replicating parasites in WT PfNF54-infected mice whereas in the PfΔmei2-infected mice replicating blood stages are absent. To determine if the WT PfNF54 or PfΔmei2 infected mice contained viable blood stage parasites, the cryopreserved blood of the FRG huHep mice collected at day 9 was cultured using standard in vitro culture conditions for blood-stage parasites. Samples from all WT PfNF54-infected mice showed a >0.1% parasitemia after 5 days of culture. In contrast, none of the cultures of blood samples of the PfΔmei2-infected mice became blood-stage positive up to 28 days, as determined by microscopy of Giemsa stained smears and 18 S qRT-PCR analyses (Fig. 3c, d). The absence of parasites in the cultures of PfΔmei2-infected mouse blood suggests that the low number of 18 S copies detected at days 7 and 9 in these mice most likely results from the presence of dead parasite material released from PfΔmei2-infected hepatocytes in the blood. Overall, our results show that PfΔmei2 sporozoites can effectively infect human hepatocytes in vitro, develop into large, replicating liver-stages that express merozoite-specific antigens but these maturing liver-stages fail to produce human RBC-infective merozoites in the livers of liver/blood-humanized mice.
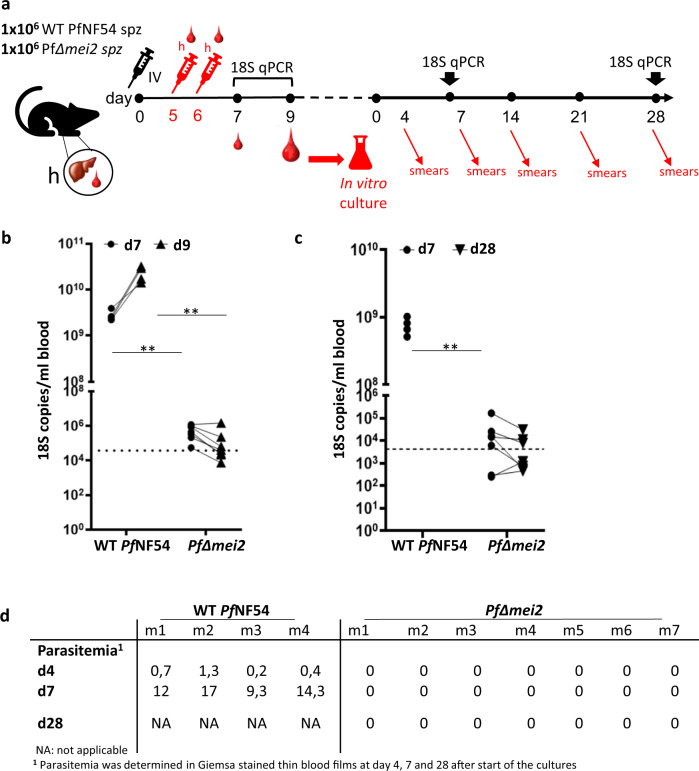
Fig. 3. Development of PfΔmei2 in FRG huHep mice containing human red blood cells.
a Timeline of experiments in liver-chimeric mice where mice were injected intravenously (IV) with 1 × 10(6) sporozoites on day 0 and then with human red blood cells (h) on days 5 and 6 prior to emergence of blood-stage parasites. Blood samples were taken on days 7 and 9 for qRT-PCR with day 9 samples used for 28-day in vitro culture of blood stages with subsequent parasitaemia readout by microscopy and qRT-PCR. b 18 S qPCR analysis of blood samples from FRG huHep mice on day 7 and 9 after infection with 1 × 10(6) sporozoites of PfΔmei2 (n = 7 mice) and WT PfNF54 (n = 4 mice). Significance values (unpaired Mann–Whitney test): **p < 0.001. Dotted line: the cutoff as used in controlled human malaria infection (CHMI) at OHSU of 5 parasites/ml, assuming 7400 18 S copies/per parasite. c Analysis of blood samples from FRG huHep mice for presence of blood-stage parasites by in vitro cultivation of blood stages. Cultures, maintained in a semi-automated shaker system, were monitored for blood-stages for 28 days by microscopy analysis of Giemsa-stained thin and thick blood smears (see d) and by 18 S qPCR. Significance values (unpaired Mann–Whitney test): **p < 0.001. Dotted line: the 10 parasites/ml cutoff used in CHMI at LUMC, assuming 4252 18 S copies per parasite. d Blood-stage parasites in cultured blood samples collected from FRG huHep mice (m) after infection with PfΔmei2 and WT PfNF54 sporozoites. Samples from in vitro cultures were analyzed at different days (d) for blood-stage parasitemia by microscopy (% of infected RBC).
PfΔmei2 genotyping by whole-genome sequencing confirms correct deletion of mei2 and indicates absence of major rearrangements or integration of heterologous DNA sequences
The promising results of late growth arrest of PfΔmei2 liver-stages warranted further characterization of PfΔmei2 parasites to obtain regulatory approval for clinical testing of this LA-GAP. Clinical evaluation involves safety testing, i.e., analysis of adverse events in humans resulting from injection of PfΔmei2 sporozoites or resulting from possible break-through blood infections, similar to procedures used in safety evaluation of the GA1 P. falciparum GAP10. Thorough identity testing and characterization of the genetic makeup of the parasites, in particular a confirmation of absence of unwanted genetic alterations which may impact the virulence of PfΔmei2 parasites, is crucial for regulatory approval of their clinical use. We therefore analyzed the presence of undesirable integration of plasmid DNA sequences that were used to generate PfΔmei2 and the presence of other genome rearrangements that might result from the genetic modification or mutations that may affect gene expression or drug-sensitivity. Four different plasmids with heterologous DNA sequences were used to create PfΔmei2, specifically, (i) a donor DNA plasmid containing two 34 bp frt (GAAGTTCCTATACTTTCTAGAGAATAGGAACTTC) sequences; (ii) two sgRNA plasmids containing the heterologous selectable-marker genes bsd, hdhfr and yfcu flanked by 3′ and 5′-utr sequences of P. falciparum-specific genes, employed to delete mei2 by double cross-over integration; (iii) a fourth plasmid, containing a yeast FLPe recombinase expression cassette, which was used to remove nearly all heterologous DNA from the donor DNA plasmid that is introduced into the genome for deletion of mei2, specifically the bsd selectable-marker cassette. These heterologous DNA sequences, located between the two frt sequences are excised in the presence of FLPe recombinase, leaving one frt sequence in the genome. This method of removal of heterologous DNA by FLPe recombinase is similar to that described for the generation of the GA110 that also contains a single frt site.
First, diagnostic PCR analysis and Southern blot analyses of digested genomic DNA were used to confirm the correct deletion of mei2 and subsequently the removal of the bsd selectable-marker cassette from the mei2 locus in PfΔmei2 by (Fig. 1b, c). In addition, PCR products that encompass the wild-type mei2 and deleted DNA regions were cloned and sequenced, revealing that (i) the expected gene-targeting had occurred at the mei2 locus resulting in deletion of mei2, ii) the bsd selection marker was absent in the mei2 locus, and (iii) only a single frt sequence was present in the mei2 locus (Fig. 1d, e). Next, the complete genome of PfΔmei2 was sequenced and single nucleotide polymorphisms (SNPs), insertions and deletions were analyzed by comparison with the reference 3D7 P. falciparum genome, which is a cloned line of PfNF54. This analysis showed that: (i) No reads were mapped on the mei2 coding sequence, while they mapped to the mei2 3′ and 5′ utr sequences, confirming correct deletion of mei2 and showing preservation of the flanking targeting sequences in the PfΔmei2 genome (Fig. 4a, b); (ii) None of the following plasmid sequences are present in the PfΔmei2 genome: cas9, ampicillin, the drug selection marker genes bsd, hdhfr, yfcu and the flpe recombinase gene (Fig. 4c); (iii) The endogenous 3′- and 5′utr sequences that were used in plasmids to drive gene expression were unaltered in the PfΔmei2 genome, showing the absence of unwanted recombination events in these genomic regions (Fig. 4d). These sequences comprised 5′utr sequences of calmodulin (PF3D7_1434200), Hsp70 (PF3D7_0818900), Hsp90 (PF3D7_0708400) and 3′utr sequences of Hpr2 (PF3D7_0831800); Hsp86 (PF3D7_0708400) and dhfr/ts (PF3D7_0417200); iv) No detectable rearrangements in the PfΔmei2 genome were observed by InDel analysis, using programs for identification of insertion and deletions; v) A total of 105 high-quality SNPs were identified in genes and only two of these 105 genes had a non-synonymous mutation: eukaryotic translation initiation factor 2 (PF3D7_0107600, A > C position 3421, amino acid N114 to H) and a Pfemp1 (PF3D7_0421100, A > T, position 914, amino acid Y350 to F). In conclusion, our analyses did not indicate the presence of unwanted integration of (heterologous) plasmid DNA sequences into the PfΔmei2 genome or the presence of genome rearrangements, except for the deletion of mei2, that may impact other phenotypic/virulence characteristics of PfΔmei2.
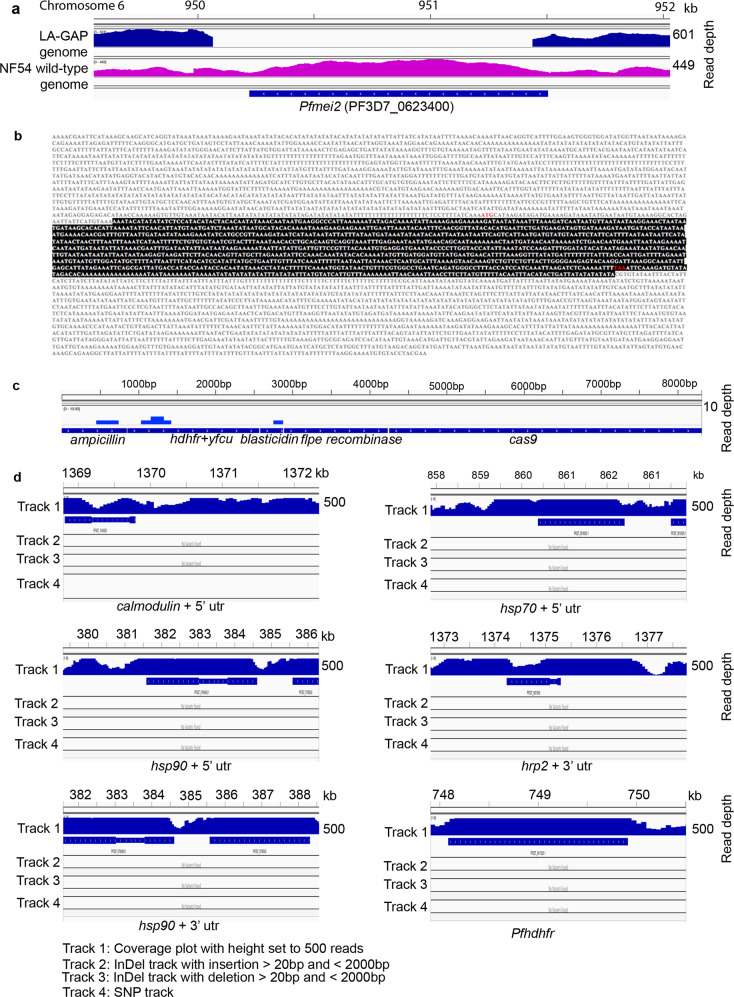
Fig. 4. PfΔmei2 genome sequence analysis.
a The mei2 locus of PfΔmei2. No sequence reads are mapped on the mei2 coding sequence while reads map in the mei2 up- and downstream regions. b Sequence of the mei2 locus of PfΔmei2. The mei2 flanking regions are unaltered and the expected mei2 deletion event is shown by the preservation of the mei2 targeting sequences (yellow). The mei2 coding sequence (in red) is absent (start and stop codon in green). c Uniquely mapped sequence read (Illimina reads) coverage of heterologous sequences used in the DNA constructs/plasmids for generation of PfΔmei2. None of the sequences (ampicilin, hdhfr-yfcu, blasticydin, flpe, and cas9) were mapped with sequence reads from the PfΔmei2 genome. d Read coverage, InDel, and SNP information of the following endogenous 5′- and 3′-utr sequences, used in DNA constructs/plasmids to drive gene expression. These sequences were unaltered in the PfΔmei2 genome. The regions were: 5′UTR: calmodulin, PF3D7_1434200; Hsp70, PF3D7_0818900; Hsp90, PF3D7_0708400; 3′UTR: Hpr2, PF3D7_0831800; Hsp90, PF3D7_0708400; and the dhfr/ts locus, PF3D7_0417200.
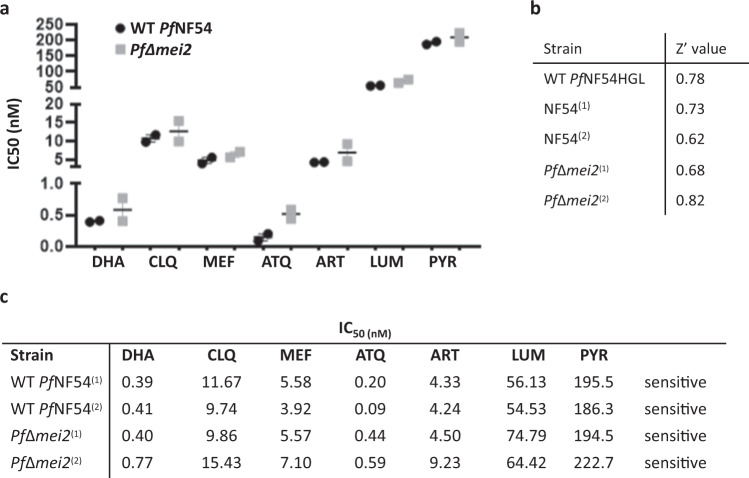
Drug sensitivity of PfΔmei2 blood-stage parasites is comparable to that of WT PfNF54 blood stages
An inherent safety feature of attenuated parasites is the ability to quickly cure any breakthrough blood infection with a variety of drugs. As noted above, the comparison of the genome sequences of PfΔmei2 and WT 3D7 P. falciparum revealed the presence of a relatively low number of SNP’s. Non-silent mutations in coding and non-coding sequences may affect genes that encode proteins involved in sensitivity/resistance of blood-stage parasites to commonly used antimalarials. We therefore determined drug sensitivity of PfΔmei2 and WT PfNF54 blood-stages to seven commonly used drugs for treating (experimental) malaria infections (dihydroartemisinin, chloroquine, mefloquine, atovaquone, artemisinin, lumefantrine and pyrimethamine). Blood-stages of both parasite lines were sensitive to all drugs tested, with IC50 values in the nanomolar range (Fig. 5). These results indicate that SNPs detected in PfΔmei2 did not impact on the sensitivity of PfΔmei2 blood-stages to the drugs tested in this study.
Fig. 5. Sensitivity of PfΔmei2 and WT PfNF54 blood stages to seven antimalarial drugs.
a Drug sensitivity of PfΔmei2 (gray) and WT PfNF54 (black) to seven antimalarial drugs was determined in an asexual Blood stage (ABS) SYBR Green drug assay. The graph shows the IC50 value with standard error of the mean (SEM) (see c below for the exact values). DHA dihydroartemisinin, CLQ chloroquine, MEF mefloquine, ATQ atovaquone, ART, artemisinin, LUM lumefantrine, PYR pyrimethamine. b Z’ values for each tested Plasmodium falciparum strain. The Table shows the Z’ values for each of the plates tested in the ABS replication assay. c IC50 values for each culture for the drugs shown in a. IC50 was determined using a four-parameter non-linear regression model using least-squares to find the best fit. Error bars represent the standard error of the mean.
Discussion
To generate P. falciparum GAPs with a late liver-stage growth arrest, we selected four genes for deletion based on published phenotypes of rodent malaria parasite mutants, lacking the equivalent orthologous genes. In contrast to the rodent parasite data, three out of these four genes (palm, hcs1, cbr) appear to be essential for the formation of P. falciparum sporozoites inside the oocyst. A comparable discrepancy in gene function between rodent malaria parasites and P. falciparum has been reported for genes encoding apicoplast-located proteins involved in the transformation of Acetyl-CoA into Acyl-ACP for generation of fatty acids by the FASII pathway18. There is no proof that the three proteins selected, PALM, HCS1 and CBR, play a direct role in this (part of the) FASII-pathway, although CBR may operate in fatty acid elongation30 and the rodent Plasmodium PALM localizes to the apicoplast, which would be compatible with a function in the FASII pathway20. However, additional studies are required to unravel the biochemical pathways in which these proteins take an indispensable role for formation of P. falciparum sporozoites
In contrast to palm, hcs1 and cbr, we found that P. falciparum GA2 lacking mei2 phenocopies P. yoelii24,25 and P. berghei mei2 gene-deletion mutants (this study). The rodent mei2 knock-out malaria parasite mutants develop similarly to wild-type parasites in the blood, in the mosquito and during part of their intra-hepatic development process, but their growth is arrested during replication inside hepatocytes. Recently, a similar phenotype of late liver growth arrest has also been described for a P. falciparum mei2 gene-deletion mutant by Goswami et al.31. We found a highly similar development of the PfΔmei2 parasites into replication-competent, late liver-stages and the absence of formation of merozoites that infect human red blood cells. The MEI2 (or PlasMei2) protein is a member of a family of RNA-binding proteins containing an RNA recognition motif and is only expressed in late-liver-stages where it has a granular cytoplasmic location31. Detailed observations on the development of P. falciparum liver-stages lacking the MEI2 protein showed aberrant formation of cytomeres, structures formed in maturing schizonts during the formation of the multiple daughter merozoites and impaired DNA replication and segregation31. Similar to what has been observed by Goswami et al.31, we provide evidence that these schizonts do not produce merozoites that are capable of infecting RBC. In standardized experiments using FRG huHep mice transfused with human RBC28,29, no evidence was found for development of blood-stage infections after injection of high numbers (1 × 10(6)) of PfΔmei2 sporozoites. These observations indicate a complete late-liver stage growth arrest of P. falciparum parasites lacking the MEI2 protein. However, occasional break-through blood infections were found in mice injected with more than 2 × 10(5) sporozoites of both P. yoelii28 and P. berghei mei2 gene-deletion mutants (this study). These occasional break-through blood infections of the rodent malaria mutants may be specific for the rodent malaria species which have a short period of full liver-stage development of only two days compared to a period of more than six days for P. falciparum. However, in the mouse studies with the rodent malaria parasites larger groups of mice were infected with sporozoites of the mei2 gene-deletion mutants compared to the number of FRG huHep mice injected with PfΔmei2 sporozoites. In addition, infectivity of P. falciparum sporozoites may be lower in the FRG huHep mouse model than in humans and therefore formal proof of complete attenuation of PfΔmei2, parasites awaits controlled safety studies in humans10.
The phenotype of growth arrest in the liver of PfΔmei2 is highly similar to the phenotype of P. falciparum mei2 gene-deletion mutant (P. falciparum mei2–) reported by Goswami et al.31. Both mutants have been generated by CRISPR/Cas9-mediated gene deletion. However, differences exist in the plasmids used for deleting mei2, the deleted sequence and in drug-selection of the mutants. All phenotype analyses of the P. falciparum mei2– were performed with a mutant that still contains the hdhfr selectable-marker cassette in the mei2- locus and no data is reported on whole-genome sequence analyses of the different P. falciparum mei2– mutants with or without the hdhfr selectable marker. CRISPR/Cas9 modification can make off target gene mutations, not only at or near the target site, but also far from the target site32,33, mutations that may affect the phenotype of P. falciparum mutants, for example impacting virulence features or drug sensitivity.
To obtain regulatory approval for clinical evaluation of PfΔmei2, we performed detailed studies to investigate whether genetic modification procedures did not cause unwanted genetic alterations, possibly impacting the virulence of the genetically modified parasites. Our genotype analyses, including whole-genome sequencing, did not show unwanted integration of (heterologous) plasmid DNA sequences or genome rearrangements in PfΔmei2 that may impact other phenotypic/virulence characteristics of PfΔmei2, except for the intentional deletion of mei2 from the PfΔmei2 genome. In addition, the sensitivity of PfΔmei2 blood-stages to seven antimalarial drugs showed IC50 values in the nanomolar range similar to those found for the parent WT PfNF54. Based on our observation, it can therefore be expected that the safety profile of growth-arrested PfΔmei2 sporozoites in humans is highly similar to sporozoites of PfSPZ-GA110, PfSPZ Vaccine1 or WT PfNF54 sporozoites administered under chloroquine prophylaxis34.
The results of the preclinical evaluation reported in this study have led to the regulatory approval of PfΔmei2 (named GA2) for use in human studies by the Gene Therapy Office of the Dutch Ministry of Infrastructure and the Environment in the Netherlands, licensed to the Leiden University Medical Centre (GGO IM-MV 20-018_000) and subsequent approval of the Central Committee on Research Involving Human Subjects (CCMO; NL75577.000.21). Safety and subsequent efficacy studies using the CHMI model have currently been initiated to provide the answer to the key question of whether attenuated parasites with a growth arrest late in the liver substantially increase potency compared to Wsp vaccines consisting of attenuated parasites with an early-growth arrest, similar to what has been found in rodent studies17 and is expected based on studies using chemo-attenuated PfSPZ immunization16. Moreover, parallel assessment of immunological samples obtained from individuals exposed to EA-GAP (GA1 and PfSPZ Vaccine) or LA-GAP GA2 will provide unprecedented insight into the immunology of the liver stage of human malaria, thus far a black box. Most importantly, the prospect of a highly efficacious malaria vaccine using cutting-edge molecular technology provides hope for regaining control for malaria control programs struggling with resistance.
Methods
Experimental animals (ethics statement): Leiden, LUMC (The Netherlands)
Animal experiments were granted with a license DEC12042 and 14207 by Competent Authority after an advice on the ethical evaluation by the Animal Experiments Committee Leiden and were performed in accordance with the Experiments on Animals Act (Wod, 2014), the applicable legislation in the Netherlands in accordance with the European guidelines (EU directive no. 2010/63/EU). Experiments were executed in a licensed establishment for experimental animals. Mice were housed in ventilated cages with autoclaved aspen woodchip, fun tunnel, wood chew block and nestlets (12:12 hour (h) light-dark cycle; 21 ± 2 °C; relative humidity of 55 ± 10%) and fed with a commercially-prepared autoclaved, dry rodent diet pellets and provided with water, both available ad libitum. Female OF1 and C57BL/6 mice (6–7 weeks; Charles River Laboratories, France) were used. Experiments involving generation of mutant parasite lines and phenotype analyses were performed using highly standardized and approved protocols that have been developed to reduce the number of animals and minimize suffering and distress. Mice were killed (cardiac puncture under isoflurane anesthesia or CO2) at a parasitemia of 2–5%, before malaria-associated symptoms occur. Humane endpoints: the animals/body condition was thoroughly examined daily. Animals are humanely sacrificed in case the following defined end points are reached: visible pain (abnormal posture and/or movement), abnormal behavior (isolation, abnormal reaction to stimuli, no food and water intake). If distress of the animals is observed by the animal caretakers, this will be reported to the investigators and according to the aforementioned criteria, the animals will be taken out of the experiment and euthanized. In all experiments no mice were euthanized before termination of the experiment and no mice died before meeting criteria for euthanasia.
Mosquitoes
Mosquitoes from a colony of Anopheles stephensi (line Nijmegen SDA500) were used. Larval stages were reared in water trays (at 28 ± 1 °C; relative humidity 80%). Adult females were transferred to incubators at 26 ± 0.2 °C (relative humidity of 80%) and were fed with 5% filter-sterilized glucose solution. For the transmission experiments, 3 to 5 day-old mosquitoes were used. Following infection, the P. berghei and P. falciparum, infected mosquitoes were maintained at 21 °C and 26 °C, respectively, at 80% relative humidity.
Parasites
For generation of the rodent malaria LA-GAP PbΔmei2, the P. berghei ANKA reference line 1868cl1 was used (line RMgm-1320; www.pberghei.eu) which contains the reporter genes mCherry and luciferase under control of the constitutive hsp70 and eef1α promoters, respectively, integrated into the neutral 230p gene locus (PBANKA_0306000). This line does not contain a drug-selectable marker. Production of PbΔmei2 and characterization of these parasites throughout their life cycle, including mosquito transmission, was performed under GMO permits IG 17-230_II-k en IG 17-135_III.
P. falciparum parasites NF54 strain35 was used as wild-type P. falciparum parasites (WT PfNF54). Parasites from the PfNF54 strain, and its derivative Pf3D7, are the most commonly used P. falciparum parasites in laboratory studies and in Controlled Human Malaria infections (CHMI;36). The complete genome sequences of Pf3D7 and PfNF54 have been published37,38. The parasites of PfNF54 and Pf3D7 have been deposited with the Malaria Research and Reference Reagent Resource Center (MR4; MRA-1000 and MRA-102), which was developed by the National Institute of Allergy and Infectious Diseases (NIAID) and is managed by the American Type Culture Collection (ATCC) (BEI Resources; https://www.beiresources.org/About/MR4.aspx). Parent parasites used for the generation of the PfΔmei2 and the other gene-deletion mutants were obtained from a characterized good manufacturing process (GMP) produced working cell bank of the WT PfNF5435, produced by Sanaria Inc39,40. WT PfNF54 is sensitive to the following antimalarial drugs: atovaquone/proguanil, arthemeter/lumefantrine and chloroquine41.
For cultivation of P. falciparum blood-stage parasites42, Fresh human serum and human red blood cells (RBC) were obtained from the Dutch National Blood Bank (Sanquin Amsterdam, the Netherlands; permission granted from donors for the use of blood products for malaria research and microbiology tested for safety). Production of genetically modified parasites and characterization of these parasites throughout their life cycle, including mosquito transmission, was performed under GMO permits IG 17-134_II-k en IG 17-135_III.
Generation and genotyping of P. berghei PbΔmei2
The P. berghei mei2 (PBANKA_1122300) gene was deleted by standard methods of transfection43 using a gene-deletion plasmid (PbGEM-300555, pL2206) obtained from PlasmoGEM (Wellcome Trust Sanger Institute, UK; http://plasmogem.sanger.ac.uk)44. This construct is designed to replace the mei2 open reading frame (orf) by the hdhfr::yfcu selectable marker (SM) cassette by double cross-over homologous recombination. The SM cassette contains the hdhfr::yfcu flanked by the P. berghei eef1α promoter region and 3′ terminal sequence of pbdhfr. Before transfection, the construct was linearized by digesting with NotI. Parasites of line 1868cl1 were transfected with construct pL2206 (exp. 2834) and transformed parasites selected by positive selection with pyrimethamine43. Selected parasites were cloned by limiting dilution and cloned lines 2834cl1 and 2834cl2 were used for genotype analysis. Line 2834cl2 was further used to generate the gene-deletion mutant which is SM free. To remove the hdhfr::yfcu SM cassette from the genome of 2834cl2, the parasites were selected (negative selection) by treatment of infected mice with 5-fluorocytosine (5-FC)45. This treatment selects for parasites that have undergone homologous recombination between the two 3′-UTR of pbdhfr untranslated regions present in the integrated construct pL2206, flanking the hdhfr::yfcu cassette and thereby removing the SM46. Selection and cloning of the parasites resulted in the SM-free gene-deletion line PbΔmei2 (2834cl2m1cl1). Correct integration of the construct and deletion of the mei2 gene were confirmed by Southern blot analyses of Pulsed Field Gel (PFG)-separated chromosomes and diagnostic PCR analysis43. To show integration of the PlasmoGem construct containing the hdhfr::yfcu SM or removal of the hdhfr::yfcu SM by negative selection, the PFG-separated chromosomes were hybridized with a mixture of two probes: a probe recognizing the hdhfr gene and a control probe recognizing gene PBANKA_0508000 on chromosome 547. PCR primers for genotyping are listed in Supplementary Table 1.
Phenotyping of P. berghei PbΔmei2
The in vivo multiplication rate of asexual blood stages was determined during the cloning procedure of the different QC mutants48. Infection of ‘, For collection and counting of sporozoites from infected An. stephensi mosquitoes49 mosquito salivary glands were manually dissected (21 days after feeding). Salivary glands were collected in RPMI medium, homogenized and filtered (40 µm Falcon, Corning, NL). Free sporozoites were counted in a Bürker counting chamber using phase-contrast microscopy. The human-hepatoma cell line Huh750 was used for in vitro cultivation of liver-stages. Briefly, 5 × 10(4) isolated sporozoites were added to monolayers of Huh7 cells on coverslips in 24 well plates (with confluency of 80–90%) in complete RPMI-1640 medium supplemented with 10% (vol/vol) fetal bovine serum, 2% (vol/vol) penicillin-streptomycin, 1% (vol/vol) GlutaMAX (Invitrogen), and maintained at 37 °C with 5% CO2. At 24, 48 and 72 hours post infection (p.i.) nuclei were stained with Hoechst-33342 at a final concentration of 10 µM and live imaging of mCherry-expressing parasites was performed using a Leica fluorescence MDR microscope (×40 magnification). Pictures were recorded with a DC500 digital camera microscope using Leica LAS X software with the following exposure times: mCherry: 0.7 s and Hoechst 0.136 s (1× gain). Liver-stage parasite sizes were measured using Leica LAS X software by determining the area of the parasite at its greatest circumference using the mCherry-positive area (µm2).
To determine the attenuation phenotype of PbΔmei2, C57BL/6 mice were infected with 5 × 10(3), 5 × 10(4), or 2 × 10(5) sporozoites of WT or PbΔmei2. Isolated sporozoites, suspended in RPMI-1640 medium, were intravenously injected into the tail vein (200 μl per mouse). Parasite liver loads in live mice were quantified by real-time in vivo imaging51. Parasite liver loads were visualized and quantified by measuring luciferase activity of parasites in whole bodies of mice at 44, 56, and 65 h p.i using the IVIS Lumina II Imaging System (Perkin Elmer Life Sciences, Waltham, USA). D-luciferin was dissolved in PBS (100 mg/kg; Caliper Life Sciences, USA) and 60 µl injected subcutaneously in the neck. Measurements were performed within 8 min after the injection of D-luciferin. Quantitative analysis of the bioluminescence of whole bodies was performed by measuring the luminescence signal intensity (RLU; relative light units) using the ROI (region of interest) settings of the Living Image® 4.5.5 software. Mice were monitored for blood-stage infections by Giemsa-stained blood smears made at day 4 to 30 p.i. The prepatent period (measured in days after sporozoite challenge) is defined as the day when a blood-stage infection with a parasitemia of 0.5–2% is observed47.
Generation and genotyping of four P. falciparum gene-deletion mutants (see Supplementary Table 1 for primer sequences)
PfΔpalm: The palm gene (PF3D7_0602300) was deleted using a CRISPR/Cas9 approach where first a plasmid was introduced into parasites as an episome containing the cas9 gene52, CRISPR/Cas9 system in Plasmodium falciparum using the centromere plasmid53. This plasmid, pLf0086, contains the hdhfr SM (with the P. chabaudi dihydrofolate reductase thymidylate synthase gene promoter (PcDT; PCHAS_0728300) and the cas9 gene (with the P. falciparum heat shock protein 90 gene promoter the (PF3D7_0708400; Pfhsp90). To generate pLf0086, plasmid pLf0070 (pDC2-cam-Cas9-U6.2-hdhfr)54,55 was digested with BamHI to remove the sgRNA/U6 cassette. Re-circularized plasmid in the BamHI site was termed pLf0086. Transfection of WT PfNF54 parasites with plasmid pLf0086 was performed by the method of spontaneous plasmid uptake from plasmid-loaded RBC56. Transfected parasites were selected by treatment with the drug WR99210 (2.6 nM) for a period of two weeks (until parasites were detectable in Giemsa-stained thin blood films) to select for parasites containing the plasmid pLf0086 episomally (Exp.121). Subsequently, selected parasites were simultaneously transfected with two sgRNA/donor DNA plasmids, pLf0124, pLf0125. Both plasmids contain two homology regions (HR) targeting palm, a blasticidin-S-deaminase (bsd) SM cassette (with the P. falciparum hsp70 promoter; Pfhsp70; PF3D7_0818900) and a palm sgRNA cassette. Each plasmid contains a different sgRNA. To generate the palm targeting vectors, a basic plasmid pLf0103 was designed that contains the bsd SM cassette. To generate the bsd SM cassette, a gBlock was designed and ordered (https://eu.idtdna.com/pages), containing the bsd gene flanked by two 34 bp flippase recognition target (frt) sequences (GAAGTTCCTATTCTCTAGAAAGTATAGGAACTTC;57) (Supplementary Fig. 7). These sequences allow to recycle the SM cassette57. This fragment was cloned into the P. berghei transfection construct pL0034 (RMgm-687; www.pberghei.eu46;) using the restriction enzymes EcoRI/HindIII resulting in intermediate plasmid SKK159. The pfhsp70 promoter was obtained by PCR amplification (KOD Hot Start DNA Polymerase, Merck Millipore) using primers p1/p2 and cloned into the intermediate plasmid SKK159 using the restriction enzymes KpnI/XhoI resulting in the intermediate plasmid SKK160. Finally, the 3′utr of P. falciparum histidine rich protein 2 (Pfhrp2; PF3D7_0831800) was obtained by PCR amplification (KOD Hot Start DNA Polymerase, Merck Millipore) using primers p3/p4 and cloned into SKK160 using the restriction enzymes NotI/AvrII resulting in the final basic plasmid pLf0103. This construct contains additional restriction sites for introducing homology/targeting sequences to target any gene of interest, such as NaeI/SacII and ApaI/HindIII and for introducing the sgRNA/U6 cassettes, such as AatII/ BamHI (see below). Plasmid pLf0039 with the P. falciparum u6 RNA promoter (PF3D7_1341100) containing the BtgZI adaptor58 was used two generate two sgRNA expression cassettes for two intermediate plasmids containing sgRNA009 (pLf0110) and sgRNA010 (pLf0111). The guide sgRNA sequences for palm (sgRNA009 and sgRNA010) were identified using the Protospacer software (alpha version; https://sourceforge.net/projects/protospacerwb/files/Release/) and were amplified using the primers p5/p6 and p7/p8. These sgRNAs were selected based on the best off target hits score throughout the genome given by Protospacer and the total number of mismatches of the sgRNA with respect to the Protospacer adjacent motif site. Two 20 bp primer guide sgRNAs, surrounded by 15 bp vector-specific DNA necessary for InFusion cloning (HD Cloning Kit; Clontech), were annealed and used to replace the BtgZI adaptor58, resulting in intermediate plasmids pLf0100, pLf0101, that were digested with BlnI/ NruI for evaluation successful cloning and confirmed by Sanger sequencing using primers p9/p10. These constructs contain additional restriction sites for lifting the complete u6 cassette including the sgRNA, such as AatII/BamHI (see below). Next, two different sgRNA/donor DNA constructs, containing each of the sgRNA as well as the donor DNA sequences, were generated in multiple cloning steps resulting in pLf0124 and pLf0125. These constructs contain both the sgRNA expression cassettes and the bsd SM cassette. To generate the palm targeting vectors, plasmid pLf0103 was modified by introducing two HRs, HR1 and HR2, targeting palm. HR1 was amplified using primers P11/12 and HR2 with p13/p14 from WT PfNF54 genomic DNA. HR2 was cloned into pLf0103 using restriction sites ApaI/HindIII, resulting in intermediate plasmid F171. Subsequently, HR1 was cloned into F171 using NaeI/SacII, resulting in intermediate plasmid pLf0110 (F177). These plasmids are used to introduce sgRNA/U6 cassette from the intermediate plasmids pLf0100 and pLf0101, containing sgRNA009 and sgRNA010 respectively (using restriction sites AatII/ BamHI) to generate pLf0124 and pLf0125, respectively.
Parasites of Exp.121 were transfected with the two plasmids pLf0124 and pLf0125 (a mixture of 50 μg of each circular plasmid in 200 μl cytomix) using standard transfection methods59. Selection of transfected parasites was performed by applying double-positive drug pressure from day 3 until day 9 after transfection using the drugs WR99210 (2.6 nM) and Blasticidin (BSD, 5 µg/ml). On day 9 drug pressure was removed and parasites were maintained in drug-free medium until parasites were detectable in thin blood-smears (day 15 after transfection). Selected parasites were then grown without both drugs until the parasitemia reached over 10%, followed by a second BSD selection (5 µg/ml) for a period of 7 days, resulting in parasite population Exp.167 (PfΔpalm-1) and Exp.169 (PfΔpalm-2). After drug selection, diagnostic PCR42 was performed from material isolated from iRBC. Correct replacement of the palm gene with the bsd cassette in the parasites after the second BSD selection in PfΔpalm-1 and PfΔpalm-2 parasites was confirmed by long-range PCR amplification (LR-PCR) (primers P15/P16) and standard PCR amplification of the palm open reading frame (primers p17/p18) and the bsd SM cassette (primers p19/p20). The PCR fragments were amplified using KOD Hot Start Polymerase (Merck Millipore) following standard conditions with annealing temperatures of 50.5 and 51 °C for 25 s and an elongation step of 68 °C for 3 min.
PfΔcbr: The Pfcbr gene (PF3D7_1367500) was deleted in WT PfNF54 parasites by standard methods of CRISPR/Cas9 transfection58,59 using a sgRNA-expressing plasmid pLf0178, containing the cas9 expression cassette, guide-RNA expression cassette and an hdhfr SM cassette, in combination with a donor DNA plasmid pLf0179 that contains a bsd SM cassette (linked to gfp, and separated with skip peptide 2 A; bsd-2A-gfp) for positive selection and a yfcu SM cassette for negative selection. The sgRNA-expressing plasmid was generated as follows: pLf007054 was digested with BbsI and the sgRNA067 was selected using the CHOPCHOP webtool (https://chopchop.cbu.uib.no/)60 and subsequently cloned into the pLf0070 using primers p21/p22. In brief, the primers (100 µM each primer) were phosphorylated with T4 polynucleotide kinase (10 Units per reaction) during 30 min at 37 °C, followed by an annealing program of 5 min incubation at 94 °C and a ramp down to 25 °C at 5 °C per min, and subsequently ligated into the BbsI digested pLf0070 vector using T4 ligase (5 units) resulting in the plasmid pLf0178.
The donor DNA plasmid pLf0179 was generated to replace the Pfcbr open reading frame with the bsd SM cassette linked to gfp (bsd-2A-gfp). For generation of pLf0179, the HR1 and HR2 regions of Pfcbr were amplified from WT PfNF54 genomic DNA using primers p23/p24 and p25/p26. Fragments were digested with HindIII/ApaI and NheI/BamHI respectively and ligated into plasmid pLf0169 to obtain pLf0179. Plasmid pLf0169 was generated as follows: a new gBlock was designed and ordered (https://eu.idtdna.com/pages), containing the bsd-2A-gfp genes flanked by the two frt sequences. This fragment was cloned into the intermediate construct pL0f103 (see above) to replace the bsd cassette by bsd-2A-gfp using the restriction sites BsabI/AvrII, resulting in the intermediate plasmid pLf0165. A second intermediate plasmid F213 was created amplifying the complete yfcu SM cassette controlled by the pfhsp90 promoter and the 3′ terminator sequence from P. berghei dihydrofolate reductase thymidylate synthase gene (Pbdhfr/ts; PBANKA_0719300; 3′PcDT, PCHAS_0728300), from the existing vector pLf0003 (pHHT-FRT-Pf3657) using the primers p27/p28. The complete cassette was cloned into the vector pJET1.2/blunt (thermo scientific) using the restriction enzyme EcoRV. Finally the yfcu SM cassete from the F213 plasmid was subcloned into the vector pLf0165 using the restriction sites StuI for the pLf0165 and PvuII/StuI for the yfcu SM, resulting in the plasmid pLf0169.
Transfection of WT PfNF54 parasites with plasmids pLf0178 and pLf0179 was performed by spontaneous plasmid uptake from plasmid-loaded red blood cells cultured59. Transgenic parasites were selected by applying ‘double’ positive selection 72 h after transfection with the drugs WR99210 (2.6 nM) and BSD (5 µg/ml) during 7 days. Subsequently, both drugs were removed from the cultures until thin blood-smears were parasite-positive, followed by applying negative selection by addition of 5-FC (1 µM) in order to eliminate parasites that retained the Donor DNA construct as episomal plasmid. Negative drug pressure in the cultures was maintained until thin blood-smears were parasite-positive. After negative selection parasites (Exp. 236) were harvested for genotyping by diagnostic PCR and Southern analysis58,59. To confirm the integration and the presence of the bsd-2A-gfp cassette, 5′-integration, 3′ integration, and bsd, PCRs were performed using the primers p29/p30, p31/p32 and p20/p33 respectively. In addition the absence of the pfcbr open reading frame was confirmed using the primers p34/p35. The PCR fragments were amplified using KOD Hot Start Polymerase (Merck Millipore) following standard conditions with annealing temperatures of 50, 55, 60 °C for 10 s and an elongation step of 68 °C. Southern blot analysis was performed with gDNA digested with EcoRI and NcoI (4 h at 37 °C) in order to confirm the deletion of Pfcbr. Digested DNA was hybridized with a probe targeting the Pfcbr HR2, amplified from WT PfNF54 genomic DNA by PCR using primers p25/p26, and the ampicillin probe (amp), was amplified using primers p36/p37.
PfΔhcs1: The Pfhcs1 gene (PF3D7_1026900) was deleted in WT PfNF54 parasites by standard methods of CRISPR/Cas9 transfection58,59 using two different sgRNA-expressing plasmids, containing the cas9 expression cassette, guide-RNA expression cassettes, and hdhfr SM cassette, in combination with donor DNA plasmid pLf0191 that contains a bsd-2A-gfp SM cassette. The two different sgRNA-expressing plasmids were generated as follows: pLf007054 was digested with BbsI and sgRNA074 and sgRNA075 (selected with CHOP-CHOP webtool) were cloned using primers p38/p39 and p40/p41, respectively, resulting in the plasmids pLf0193 and pLf0194. The donor DNA plasmid pLf0191 was generated to replace the Pfhcs1 open reading frame with a bsd-2A-gfpSM cassette flanked by two frt sequences. The HR1 and HR2 targeting regions of Pfhcs1 were amplified from WT PfNF54 genomic DNA using the primers P42/P43 and P44/P45 respectively (Supplementary Table 1). Fragments were digested with HindIII/AscI and SacII/NheI and ligated into plasmid pLf0169 to obtain pLf0191. Transfection of WT PfNF54 parasites with constructs pLf0191, pLf0193, and pLf0194 was performed by spontaneous plasmid uptake from plasmid-loaded red blood cells cultured59 and selection of PfΔhcs1 parasites was as described above for generation of PfΔcbr, resulting in the line PfΔhcs1 (Exp. 252). For genotyping PfΔhcs1 parasites, diagnostic PCR was performed. To confirm the integration and the presence of the bsd-2A-gfp cassette, 5′-integration, 3′ integration PCRs were performed using the primers p46/p47, p48/p49 respectively, additionally the absence of the pfhcs1 open reading frame was confirmed using the primers p50/p51. The PCR fragments were amplified using Phusion DNA Polymerase (NEB) following standard conditions with annealing temperatures of 58 °C for 30 s and an elongation step of 68 °C.
PfΔmei2: The mei2 gene (PF3D7_0623400) was deleted in WT PfNF54 parasites by standard methods of CRISPR/Cas9 transfection58 using a donor DNA plasmid pLf0105 and two different sgRNA-donor containing plasmids, pLf0080 and pLf0092, targeting the mei2 gene.
For generation of plasmid pLf0105, two homology regions targeting the mei2 gene were introduced in pLf0103. HR1 was amplified from WT PfNF54 genomic DNA using primers P52/P53 and HR2 with primers P54/P55. The PCR fragments were sequenced after TOPO TA (Invitrogen) subcloning and subsequently cloned into pLf0103 using restriction sites NaeI/SacII and ApaI/HindIII, resulting in plasmid pLf0105. To generate the two sgRNA-donor-containing vectors, plasmid pLf007054 was digested with BbsI and sgRNA030 and sgRNA032 were cloned, as described above, using primers P56/P57 and P58/P59, respectively, resulting in the plasmids pLf0080 and pLf0092. Transfection of WT PfNF54 (Leiden vial 9002 obtained from Nijmegen; NF54 54/329 4514) with constructs pLf0105, pLf0080 and pLf0092was performed by spontaneous plasmid uptake from plasmid-loaded RBC59. Selection of transfected WT PfNF54 parasites was performed by applying double positive selection (as described for selecting for generation of PfΔcbr) for a period of 6-19 days. After this treatment period with two drugs, cultures were maintained in drug-free medium until parasites were detectable in Giemsa-stained thin blood smears (a period of three weeks). Subsequently, parasites were treated for one week with BSD (5 μg/ml), resulting in parasite population Exp.151 (PfΔmei2a parasites; Fig. 1). Subsequently, selected parasites were cloned by limiting dilution. In order to remove the bsd SM cassette from the genome of PfΔmei2a, blood stage parasites of the uncloned population of PfΔmei2-a (Exp. 151) were transfected with plasmid pLf0120 that contains a flpe recombinase expression cassette (Fig. 1). To create plasmid pLf0120, we used plasmid pMV-FLPe57 (pLf0038) that contains a bsd SM cassette and an flpe recombinase expression cassette. We first replaced the bsd SM cassette of plasmid pMV-FLPe with the hdhfr-yfcu SM cassette of plasmid pLf003958 (HindIII/KpnI) to create pLf0120. The hdhfr-yfcu gene is flanked by the promoter of the Pfhsp86 and the Pbdhfr/ts short (0,5 kb) terminator sequences. The flpe gene is flanked by the promoter of the Pfhsp90 from PfDd2 strain (PfDd2_070012600;) and Pbdhfr/ts long (1 kb) terminator sequences. After transfection of PfΔmei2a blood stages with pLf0120 (as described above), cultures were treated for a period of six days (day 3–9) with WR99210 (2.6 nM), followed by a period of 2 weeks culture without WR99210 treatment. Subsequently, selected parasites were cloned by limiting dilution. DNA from iRBC was obtained from 10 ml cultures (parasitemia 3–10%). Diagnostic PCR was performed using primer pair (P60/P61). As a control, the gene p47 (PF3D7_1346800) was PCR amplified using primers 8428/8756 (P62/P63). Southern blot analysis was performed with gDNA digested with XmnI (4 h at 37 °C) in order to confirm the deletion of Pfmei2. Digested DNA was hybridized with probes targeting the Pfmei2 (a fragment of 539 bp of the mei2 coding sequence amplified using primers P64/P65, Pfp47 gene (PF3D7_1346800, as a control fragment of 3910 bp, amplified using primers P62/P66), ampicillin gene (amp probe; amplified using primers P36/P37), the bsd SM (amplified using primers P67/P20), the cas9 (amplified using primers P68/P69 and the flpe gene (amplified using primers P70/P71). In order to confirm the precise nature of the genetic deletion, a PCR product that encompass the mei2 gene-deletion region of PfΔmei2 was cloned and sequenced. This PCR product was obtained using primer pair P72/P73, cloned in pJET (Thermo Fisher Scientific) and sequenced using primers P74/P75.
Phenotyping of P. falciparum mutants PfΔpalm, PfΔhcs1 and PfΔcbr
The growth rate of asexual blood-stages (parasitemia) was monitored by determination of parasitemia in standard in vitro cultures (in a semi-automated shaker incubator system) for a period of 4 days with a starting parasitemia of 0.1%42. Parasitemia was determined by counting infected RBC in Giemsa-stained thin blood films in three independent experiments. Gametocyte production and exflagellation54 were quantified in gametocyte cultures.
For analysis of mosquito stages (oocysts and sporozoites) An. stephensi mosquitoes were infected with day 14 gametocyte cultures using the standard membrane feeding assay (SMFA)59,61. Oocysts and salivary gland sporozoites were counted at days 9 and day 21 p.i., respectively. For counting sporozoites, salivary glands from 10 to 15 mosquitoes were dissected, collected in 100 µl of PBS and homogenized using a grinder. Sporozoites were counted using a Bürker cell counter using phase-contrast microscopy.
Oocysts were analyzed in manually dissected midguts using a Leica MZ16 FA stereo-fluorescent microscope. The midguts were imaged with a Leica MZ camera at ×10 magnification using Leica LAS X software. Individual oocysts were observed under a Leica DM2500 light microscope and documented with at ×100 using Leica DC500 digital camera using Leica LAS X software. Sg-sporozoite numbers were analyzed in infected mosquitoes at day 18–21 p.i. For counting sporozoites, salivary glands from 30–0 mosquitoes were dissected and homogenized using a grinder in 100 μl of RPMI-1640 medium (pH 7.2) and sporozoites were analyzed in a Bürker cell counter using phase-contrast microscopy54.
Phenotyping of P. falciparum mutant PfΔmei2
The growth rate of asexual blood-stages and analysis of mosquito stages (oocysts and sporozoites) in An. stephensi mosquitoes were performed as described in the previous section for the other three gene-deletion mutants.
BioIVT hepatocytes: Analysis of the development of WT PfNF54 and PfΔmei2 parasites in primary human hepatocytes58 was performed as follows. Liver-stages of WT PfNF54 and PfΔmei2 were cultured in vitro using cryopreserved primary human hepatocytes obtained from BioIVT (Belgium) and thawed according to the instructions of the manufacturer. Cells were seeded at a density of 60,000 cells/well in a collagen-coated 96-well clear-bottomed black plate for 2 days. Medium was refreshed daily (hepatocyte medium: Williams’s E medium supplemented with 10% heat-inactivated fetal bovine serum, 2% penicillin-streptomycin, 1% fungizone, 0.1 lU/ml insulin, 1.6 μM dexamethasone). Per well, 7 × 104 freshly dissected WT PfNF54 and PfΔmei2 sporozoites were added to the hepatocyte monolayer. After a quick spin (10 min at 1900 g), the plate was incubated at 37 °C under 5% CO2. The medium was replaced with fresh hepatocyte culture medium 3 h p.i., and daily for 9 days thereafter. At days 3, 5, 7, and 9 p.i., hepatocytes were fixed with 4% paraformaldehyde in 1× PBS for 30 min. After fixation the wells were washed three times with 1× PBS and permeabilized with 300 μl of 0.5% triton in 1× PBS during 1 h and then blocked with 10% of FCS in 1× PBS for 1 h. Fixed cells were washed with 1× PBS and standard IFA was performed using antibodies against (1) the cytoplasmic protein PfHSP70 (PF3D7_0818900; rabbit anti-PfHSP70-PE/ATTO 594 conjugated primary antibody; 1:200 dilution of 100 μg/ml stock solution StressMarq, Biosciences, NL); (2) the plasma membrane surface protein MSP1 (PF3D7_0930300; mouse monoclonal antibody 1:1000 of 4.0 mg/ml stock solution obtained from The European Malaria Reagent Repository, Edinburgh, UK) (3) and the parasitophorous membrane protein EXP1 (PF3D7_1121600, mouse monoclonal antibody (1:200 of 4.0 mg/ml stock solution obtained from The European Malaria Reagent Repository, Edinburgh, UK). In addition, cells were stained with Hoechst-33342 for nuclear staining. Cells were mounted in Image-iT signal Enhancer (Invitrogen Thermofisher, USA) and examined with a SP8 Leica confocal microscope at ×100 magnification. Number of infected hepatocytes, antibody staining, and size of parasites were analyzed using ImageJ.
LONZA hepatocytes: Cryopreserved primary human hepatocytes purchased from Lonza Bioscience were thawed and seeded in μ-Slide 18 Well ibiTreat coverslip (IBIDI, Gräfelfing, Germany), pre-coated with rat-tail collagen I (BD Bioscience, USA) in William’s E medium (Gibco) supplemented with 10% fetal clone III serum (FCS, Hyclone), 100 u/mL penicillin and 100 ug/mL streptomycin (Gibco), 5 × 10−3 g/L human insulin (Sigma-Merck), 5 × 10−5 M hydrocortisone (Upjohn Laboratories SERB, France) at 37 °C in 5% CO2. The next day, cells were overlaid with matrigel (Corning) and medium was then renewed every 2 days. Three days later, sporozoites were isolated by aseptic hand dissection of salivary glands of PfΔmei2- and WT PfNF54-infected mosquitoes. Matrigel was removed from hepatocyte culture and 30,000 sporozoites were inoculated to cells before centrifugation at 560×g for 10 min at RT and further incubation at 37 °C, 5% CO2. Three hours later, infected cultures were covered with matrigel prior to addition of fresh cell culture medium. Medium was renewed every day, until cell fixation at the chosen times. Infected cultures were fixed with 4% paraformaldehyde for 15 min at room temperature and liver-stage parasites were immunostained with polyclonal anti-PfHSP70 murine serum prepared in the lab, anti-PfEXP1 (kindly provided by Pr Jude Przyborski) and revealed with anti-mouse IgG Alexa Fluor® 594 and anti-rabbit IgG Alexa-Fluor 488-conjugated, respectively (Invitrogen). DAPI was added to visualize nuclei. Parasite number and size were determined using a Cell Insight High Content Screening platform equipped with the Studio HCS software (Thermo Fisher Scientific) at the CELIS platform (ICM, La Pitié-Salpêtière, Paris). Graphs and statistical analysis were done using Prism 8.4.3 software. The areas of parasites were compared using the Mann–Whitney U test.
Liver stage development of both PfΔmei2 and WT PfNF54 parasites were analyzed in liver-chimeric humanized mice (FRG huHep mice) purchased from Yecuris Corporation (Tualatin, OR) and housed at Oregon Health and Science University (OHSU) as per manufacturer’s recommendation. All studies were performed according to the regulations of the Institutional Animal Care and Use Committee (IACUC; protocol IP00002077). Female An. stephensi mosquitoes, aged 3–5 days, were infected with PfΔmei2 and WT PfNF54 at LUMC (Leiden, the Netherlands)54,59. 12 days after feeding, mosquitoes were shipped to OHSU. Sporozoites were isolated by salivary gland dissection from infected mosquitoes at day 16 p.i. at OHSU for infection of the FRG huHep mice. Isolated salivary gland sporozoite were run over glass wool to remove contaminating mosquito material and sporozoites enumerated by hemocytometer. FRG huHep mice (PfNF54 WT n = 4; PfΔmei2 n = 7) were infected by intravenous injection (retro-orbital injection) of 10(5) sporozoites in a 100 µl volume of phosphate buffered saline. Five days p.i., FRG huHep mice were injected intravenously with 400 µl freshly washed human type AB red blood cells (RBC; 70% hematocrit) supplemented with 0.035 mg/mouse clodronate (Formumax CloLip) and penicillin/streptomycin antibiotic (50 units penicillin, 50 µg streptomycin/ml, Sigma). The following day, FRG huHep mice were injected intraperitoneally with 700 µl freshly washed human type AB RBC (70% hematocrit). At 7 and 9 p.i. 100 µl of blood was collected into 2 ml NucliSens buffer (Biomerieux) for qRT-PCR analysis. At day 9 p.i. mice were euthanized and blood collected for cryopreservation in glycerolyte. To analyze the presence of blood stages in blood samples collected from the FRG huHep mice (day 7, 9) 18 S qRT-PCR62 quantification of blood-stage parasites was performed. In addition to the qRT-PCR analyses, the cryopreserved blood of all FRG huHep mice (collected at day 9) was used for in vitro cultivation of parasites, performed at the LUMC (Leiden, the Netherlands), to assess the presence/absence of blood stage parasites. For these experiments, a standard protocol for in vitro cultivation of blood stages was used58,59. Cultures, maintained in a semi-automated shaker system, were monitored for blood stage parasites for a period of 28 days by analyzing Giemsa stained thin and thick blood smears and 18 S qRT-PCR and fresh RBC (100 µl packed RBC in 1 ml culture medium) were added to the cultures weekly. Briefly, total DNA was isolated from blood samples using QIAamp DNA Blood Mini Kit (Qiagen, NL) in accordance with the manufacturer’s instructions, and a one-step qPCR Hotstar mastermix (Qiagen, NL) assay specific for the 18 S ribosomal subunit of P. falciparum was used to quantify the presence of parasites (forward primer: 5′-CCGACTAGGTGTTGGATGAAAGTGTTAA-3′; reverse primer: 5′-AACCCAAAGACTTTGATTTCTCATAA-3′; Probe: 5′-FAM- CTTTCGAGGTGACTTTTAGAT- MGB (quencher)63; cycling profile: 15 min at 95 °C followed by 45 cycles of 30 sec at 95 °C, 20 sec at 60 °C and 30 sec at 72 °C. The resulting CT values were compared to a standard curve of a known quantity of 18 S plasmid spanning 100 to 10(8) copies. To determine positive/negative parasite samples, the same cutoff as is used in controlled human malaria infection (CHMI) of 5 parasites/ml, assuming 7400 18 s copies/parasite (at OHSU) and 5 parasites/ml, assuming 4252 18 s copies/parasite.
Uncropped and unprocessed scans of gels and blots from (supplementary) Figs. are shown in Supplementary Figs. 8–13.
Whole-genome sequencing of PfΔmei2
Genomic DNA of PfΔmei2 was isolated from blood stage-infected RBC obtained from 10 ml cultures (parasitemia 3–10%). RBC were pelleted by centrifugation (1150 g; 5 min.) and subsequently lysed with 5–10 ml of cold (4 °C) erythrocyte lysis buffer (10× stock solution 1.5 M NH4Cl, 0.1 M KHCO3, 0.01 M Na2EDTA; pH 7.4). Pelleted parasites were treated with RNAse and proteinase-K before genomic DNA isolation by standard phenol-chloroform methods. Whole-genome sequencing was performed at the King Abdullah University of Science and Technology (KAUST, Thuwal, Saudi Arabia Prof. Arnab Pain). Genomic DNA quantification was carried out using Qubit-based colorimetric assay (Invitrogen). A total of 200 ng of DNA was used for DNA library preparation using NebNext Ultra II DNA library prep kit for Illumina (NEB). Upon library quantification and size verification, DNA library sequencing was carried out on a MiSeq platform (Illumina) that produced 2 × 150 bp paired-end reads. The quality of the raw reads was assessed using FATSQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Low-quality reads and Illumina adaptors sequences from the end of the reads were removed using Trimmomatic64. Quality trimmed reads were mapped to P. falciparum 3D7 reference genome (release 40 in PlasmoDB- http://www.plasmoddb.org) using BWA (version 0.7.17)65. Read pairing information, flag, and duplicate reads were removed using Picard’s CleanSam, FixMateInformation, and MarkDuplicates tools. The full genome sequence has been uploaded in the European Nucleotide Archive. Project ID has been created in ENA with the study accession number PRJEB40003 (Public release date set to 25/08.2021). SNPs were called using the genome analysis tool kit (GATK) best practices pipeline66. Identified SNPs were filtered using vcftools to keep high-quality SNP with the quality score (Q) ≥ 30 and depth (d) ≥50. A total of 105 high-quality SNPs were identified. SNPs were annotated and their effect on coding sequences of genes was done via snpEFF.
For insertion and deletion (InDel) identification, raw gapped alignment were realigned was using GATK RealignerTargetCreator and IndelRealigner tools. Insertion between 20 and 2000 bp and deletion between 20 and 2000 bp were identified using GATK’s SelectVariants tool. Variants were tagged with quality score (Q) ≥ 30 and depth (d) ≥50 tagged using vcfannotate option and only InDels with a quality score (Q) ≥ 30 and depth (d) ≥100 were filtered for further analysis. Confirmation of absence of the gene sequences of cas9, ampicillin, blasticidin, hdhfr, yfcu, flpe recombinase was determined by concatenating the DNA sequence of these genes into a single fasta file and mapping the quality trimmed reads to the indexed sequences using HISAT2 (V 2.1.067). Read coverage was visualized using Integrative Genome Viewer browser68.
Drug-sensitivity testing of PfΔmei2
Drug sensitivity was established in standard drug-sensitivity assays69. Comparative drug sensitivity testing was performed for three P. falciparum lines (NF54-HGL, WT PfNF54, and PfΔmei2) with 7 different compounds in the asexual blood stage (ABS) replication assay. As assay reference, NF54-ΔPf47-5′ hsp70-GFP-Luc69 (hereafter: NF54-HGL), a transgenic strain of P. falciparum that stably expresses a GFP-Luciferase fusion protein under control of the constitutive P. falciparum hsp70 promoter, was used. In the assay parasites from asynchronous asexual blood stage cultures were diluted to achieve a parasitemia of 0.8% in 3% hematocrit in RPMI-1640 medium supplemented with 10% human serum. 30 µl of diluted parasites were combined with 30 µl of diluted compound in a 384-well microtiter plate. Following 72 hour incubation at 37 °C, 3% O2, 4% CO2, relative parasitemia was determined through a SYBR Green assay. Compounds were dissolved in DMSO to a stock concentration of 10 mM and diluted in DMSO and then in RPMI-1640 medium to reach a final DMSO concentration of 0.1% for all conditions tested. Compounds were tested at 9 dilutions in 0.5 log steps from 10 µM to 1 nM with duplicate wells on each plate. Dihydroartemisinin (DHA) was taken along as a reference compound from 100 pM to 1 µM. Plate controls included MAX (0.1% DMSO) and MIN (1 µM DHA) with 14 replicates each per plate. Relative parasitemia was normalized to the MIN and MAX controls. IC50 values were determined using a four-parameter non-linear regression model using least-squares to find the best fit using the Prism software package (GraphPad Software, San Diego). For each of the plates, Z’ values were calculated based on the MIN and MAX controls.
Statistical analysis
Statistical analyses were performed using Mann–Whitney test with the GraphPad Prism software package 9 (GraphPad Software, Inc). A P value <0.05 was considered significant.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We acknowledge the critical contribution of the late Dr Shahid Khan to the design of these studies. We are grateful to Pr Jude Przyborski for the anti- P. falciparum EXP-1 antibody. This work was supported by LUMC internal funds. The funders had no role in design, analysis, or reporting of these studies. CM-M was, in part, supported by Colciencias Ph.D. fellowship (Call 568 from 2012 Resolution 01218 Bogotá, Colombia) and LUF project grant, Den Dulk-Moermans Fonds (grant reference number: W19374-2-32, 2019-2021). A.O. is supported by a Skim Latihan Akademik IPTA-SLAI (Ministry of Higher Education, Malaysia). A.P. acknowledges the Faculty Baseline funding (BAS/1/1020-01-01) from King Abdullah University of Science and Technology (KAUST).
Author contributions
Study concept and design: B.F., C.M., A.M., M.P., V.H., M.R., C.J. Acquisition of data and technical support: F.G., C.M., S.M., J.R., H.K., E.B., E.W., V.S., Y.M., A.O., S.K., R.S., D.M., (generation and characterization of the transgenic P. berghei and falciparum parasites); T.M., M.A., S.M., L.F. (FRG liver and blood humanized mice experiments); A.S. (sequencing), A.H. (drug assay). Analysis and interpretation: B.F., C.M., A.M., M.P., K.D., V.H., A.P., B.W., M.R., C.J. Drafting of the manuscript: B.F., C.M., C.J. Critical revision of the manuscript: B.F., C.M., Y.M., A.M., M.P., K.D., V.H., A.P., B.W., M.R., C.J. Obtained funding: M.R., C.J.
Data availability
Parasite whole-genome sequences have been deposited in the repository https://www.ebi.ac.uk/ena/browser/view/PRJEB40003; the sequence file is accessible under accession number ERR4620262. The P. berghei mutant line PbΔmei2 has been deposited in the repository https://www.pberghei.eu/index.php?rmgm=4937.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Blandine Franke-Fayard, Email: bfranke@lumc.nl.
Meta Roestenberg, Email: M.Roestenberg@lumc.nl.
Chris J. Janse, Email: c.j.janse@lumc.nl
Supplementary information
The online version contains supplementary material available at 10.1038/s41541-022-00558-x.
References
- 1.Richie TL, et al. Progress with Plasmodium falciparum sporozoite (PfSPZ)-based malaria vaccines. Vaccine. 2015;33:7452–7461. doi: 10.1016/j.vaccine.2015.09.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hollingdale MR, Sedegah M. Development of whole sporozoite malaria vaccines. Expert Rev. Vaccines. 2017;16:45–54. doi: 10.1080/14760584.2016.1203784. [DOI] [PubMed] [Google Scholar]
- 3.Duffy PE, Patrick Gorres J. Malaria vaccines since 2000: progress, priorities, products. NPJ Vaccines. 2020;5:48. doi: 10.1038/s41541-020-0196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goh YS, McGuire D, Rénia L. Vaccination with sporozoites: models and correlates of protection. Front. Immunol. 2019;10:1227. doi: 10.3389/fimmu.2019.01227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffman SL, et al. Protection of humans against malaria by immunization with radiation-attenuated Plasmodium falciparum sporozoites. J. Infect. Dis. 2002;185:1155–1164. doi: 10.1086/339409. [DOI] [PubMed] [Google Scholar]
- 6.Sissoko, M. S. et al. Safety and efficacy of a three-dose regimen of Plasmodium falciparum sporozoite vaccine in adults during an intense malaria transmission season in Mali: a randomised, controlled phase 1 trial. Lancet. Infect. Dis.10.1016/s1473-3099(21)00332-7 (2021). [DOI] [PMC free article] [PubMed]
- 7.Oneko M, et al. Safety, immunogenicity and efficacy of PfSPZ Vaccine against malaria in infants in western Kenya: a double-blind, randomized, placebo-controlled phase 2 trial. Nat. Med. 2021;27:1636–1645. doi: 10.1038/s41591-021-01470-y. [DOI] [PubMed] [Google Scholar]
- 8.Goswami D, Minkah NK, Kappe SHI. Designer parasites: genetically engineered plasmodium as vaccines to prevent malaria infection. J. Immunol. 2019;202:20–28. doi: 10.4049/jimmunol.1800727. [DOI] [PubMed] [Google Scholar]
- 9.Bijker EM, et al. Novel approaches to whole sporozoite vaccination against malaria. Vaccine. 2015;33:7462–7468. doi: 10.1016/j.vaccine.2015.09.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roestenberg, M. et al. A double-blind, placebo-controlled phase 1/2a trial of the genetically attenuated malaria vaccine PfSPZ-GA1. Sci. Transl. Med.12, 10.1126/scitranslmed.aaz5629 (2020). [DOI] [PubMed]
- 11.van Schaijk, B. C. et al. A genetically attenuated malaria vaccine candidate based on P. falciparum b9/slarp gene-deficient sporozoites. eLife3, 10.7554/eLife.03582 (2014). [DOI] [PMC free article] [PubMed]
- 12.Doolan DL, Hoffman SL. The complexity of protective immunity against liver-stage malaria. J. Immunol. 2000;165:1453–1462. doi: 10.4049/jimmunol.165.3.1453. [DOI] [PubMed] [Google Scholar]
- 13.Chatterjee D, Cockburn IA. The challenges of a circumsporozoite protein-based malaria vaccine. Expert Rev. Vaccines. 2021;20:113–125. doi: 10.1080/14760584.2021.1874924. [DOI] [PubMed] [Google Scholar]
- 14.Jongo SA, et al. Safety, immunogenicity, and protective efficacy against controlled human malaria infection of plasmodium falciparum sporozoite vaccine in Tanzanian adults. Am. J. Trop Med. Hyg. 2018;99:338–349. doi: 10.4269/ajtmh.17-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sissoko MS, et al. Safety and efficacy of PfSPZ Vaccine against Plasmodium falciparum via direct venous inoculation in healthy malaria-exposed adults in Mali: a randomised, double-blind phase 1 trial. Lancet Infect. Dis. 2017;17:498–509. doi: 10.1016/S1473-3099(17)30104-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mwakingwe-Omari A, et al. Two chemoattenuated PfSPZ malaria vaccines induce sterile hepatic immunity. Nature. 2021;595:289–294. doi: 10.1038/s41586-021-03684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butler NS, et al. Superior antimalarial immunity after vaccination with late liver stage-arresting genetically attenuated parasites. Cell Host. Microbe. 2011;9:451–462. doi: 10.1016/j.chom.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Schaijk BC, et al. Type II fatty acid biosynthesis is essential for Plasmodium falciparum sporozoite development in the midgut of Anopheles mosquitoes. Eukaryot. Cell. 2014;13:550–559. doi: 10.1128/EC.00264-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biddau M, et al. Plasmodium falciparum LipB mutants display altered redox and carbon metabolism in asexual stages and cannot complete sporogony in Anopheles mosquitoes. Int. J. Parasitol. 2021;51:441–453. doi: 10.1016/j.ijpara.2020.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haussig JM, Matuschewski K, Kooij TW. Inactivation of a Plasmodium apicoplast protein attenuates formation of liver merozoites. Mol. Microbiol. 2011;81:1511–1525. doi: 10.1111/j.1365-2958.2011.07787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dellibovi-Ragheb TA, et al. Host biotin is required for liver stage development in malaria parasites. Proc. Natl Acad. Sci. USA. 2018;115:E2604–e2613. doi: 10.1073/pnas.1800717115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ostera G, et al. Plasmodium falciparum: food vacuole localization of nitric oxide-derived species in intraerythrocytic stages of the malaria parasite. Exp. Parasitol. 2008;120:29–38. doi: 10.1016/j.exppara.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stanway RR, et al. Genome-scale identification of essential metabolic processes for targeting the plasmodium liver stage. Cell. 2019;179:1112–1128. doi: 10.1016/j.cell.2019.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dankwa DA, Davis MJ, Kappe SHI, Vaughan AM. A Plasmodium yoelii Mei2-like rna binding protein is essential for completion of liver stage schizogony. Infect. Immun. 2016;84:1336–1345. doi: 10.1128/IAI.01417-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaughan, A. M. et al. A plasmodium parasite with complete late liver stage arrest protects against preerythrocytic and erythrocytic stage infection in mice. Infect. Immun.86, 10.1128/iai.00088-18 (2018). [DOI] [PMC free article] [PubMed]
- 26.Ghorbal M, et al. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat. Biotechnol. 2014;32:819–821. doi: 10.1038/nbt.2925. [DOI] [PubMed] [Google Scholar]
- 27.Lee MCS, Lindner SE, Lopez-Rubio JJ, Llinás M. Cutting back malaria: CRISPR/Cas9 genome editing of Plasmodium. Brief. Funct. Genomics. 2019;18:281–289. doi: 10.1093/bfgp/elz012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaughan AM, et al. Complete Plasmodium falciparum liver-stage development in liver-chimeric mice. J. Clin. Investig. 2012;122:3618–3628. doi: 10.1172/JCI62684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sack BK, et al. Humoral protection against mosquito bite-transmitted Plasmodium falciparum infection in humanized mice. NPJ Vaccines. 2017;2:27. doi: 10.1038/s41541-017-0028-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mi-Ichi F, Kita K, Mitamura T. Intraerythrocytic Plasmodium falciparum utilize a broad range of serum-derived fatty acids with limited modification for their growth. Parasitology. 2006;133:399–410. doi: 10.1017/S0031182006000540. [DOI] [PubMed] [Google Scholar]
- 31.Goswami, D. et al. A replication-competent late liver stage-attenuated human malaria parasite. JCI Insight.10.1172/jci.insight.135589 (2020). [DOI] [PMC free article] [PubMed]
- 32.Ledford H. CRISPR gene editing in human embryos wreaks chromosomal mayhem. Nature. 2020;583:17–18. doi: 10.1038/d41586-020-01906-4. [DOI] [PubMed] [Google Scholar]
- 33.Höijer I, et al. CRISPR-Cas9 induces large structural variants at on-target and off-target sites in vivo that segregate across generations. Nat. Commun. 2022;13:627. doi: 10.1038/s41467-022-28244-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Odedra A, McCarthy JS. Safety considerations for malaria volunteer infection studies: a mini-review. Am. J. Trop Med. Hyg. 2020;102:934–939. doi: 10.4269/ajtmh.19-0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ponnudurai T, Leeuwenberg AD, Meuwissen JH. Chloroquine sensitivity of isolates of Plasmodium falciparum adapted to in vitro culture. Trop. Geogr. Med. 1981;33:50–54. [PubMed] [Google Scholar]
- 36.Stanisic, D. I., McCarthy, J. S. & Good, M. F. Controlled human malaria infection: applications, advances, and challenges. Infect. Immun.86, 10.1128/IAI.00479-17 (2018). [DOI] [PMC free article] [PubMed]
- 37.Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moser KA, et al. Strains used in whole organism Plasmodium falciparum vaccine trials differ in genome structure, sequence, and immunogenic potential. Genome Med. 2020;12:6. doi: 10.1186/s13073-019-0708-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoffman SL, et al. Development of a metabolically active, non-replicating sporozoite vaccine to prevent Plasmodium falciparum malaria. Hum. Vaccines. 2010;6:97–106. doi: 10.4161/hv.6.1.10396. [DOI] [PubMed] [Google Scholar]
- 40.Epstein JE, et al. Live attenuated malaria vaccine designed to protect through hepatic CD8(+) T cell immunity. Science. 2011;334:475–480. doi: 10.1126/science.1211548. [DOI] [PubMed] [Google Scholar]
- 41.Teirlinck AC, et al. NF135.C10: a new Plasmodium falciparum clone for controlled human malaria infections. J. Infect. Dis. 2013;207:656–660. doi: 10.1093/infdis/jis725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mogollon CM, et al. Rapid generation of marker-free p. falciparum fluorescent reporter lines using modified CRISPR/Cas9 constructs and selection protocol. PLoS One. 2016;11:e0168362. doi: 10.1371/journal.pone.0168362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Janse CJ, et al. High efficiency transfection of Plasmodium berghei facilitates novel selection procedures. Mol. Biochem. Parasitol. 2006;145:60–70. doi: 10.1016/j.molbiopara.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 44.Schwach F, et al. PlasmoGEM, a database supporting a community resource for large-scale experimental genetics in malaria parasites. Nucleic Acids Res. 2015;43:D1176–D1182. doi: 10.1093/nar/gku1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orr RY, Philip N, Waters AP. Improved negative selection protocol for Plasmodium berghei in the rodent malarial model. Malar. J. 2012;11:103. doi: 10.1186/1475-2875-11-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Braks JA, Franke-Fayard B, Kroeze H, Janse CJ, Waters AP. Development and application of a positive-negative selectable marker system for use in reverse genetics in Plasmodium. Nucleic Acids Res. 2006;34:e39. doi: 10.1093/nar/gnj033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salman AM, et al. Generation of transgenic rodent malaria parasites expressing human malaria parasite proteins. Methods Mol. Biol. 2015;1325:257–286. doi: 10.1007/978-1-4939-2815-6_21. [DOI] [PubMed] [Google Scholar]
- 48.Spaccapelo R, et al. Plasmepsin 4-deficient Plasmodium berghei are virulence attenuated and induce protective immunity against experimental malaria. Am. J. Pathol. 2010;176:205–217. doi: 10.2353/ajpath.2010.090504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Othman AS, et al. OX40 stimulation enhances protective immune responses induced after vaccination with attenuated malaria parasites. Front. Cell. Infect. Microbiol. 2018;8:247. doi: 10.3389/fcimb.2018.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fougere A, et al. Variant exported blood-stage proteins encoded by plasmodium multigene families are expressed in liver stages where they are exported into the parasitophorous vacuole. PLoS Pathog. 2016;12:e1005917. doi: 10.1371/journal.ppat.1005917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Annoura T, Chevalley S, Janse CJ, Franke-Fayard B, Khan SM. Quantitative analysis of Plasmodium berghei liver stages by bioluminescence imaging. Methods Mol. Biol. 2013;923:429–443. doi: 10.1007/978-1-62703-026-7_30. [DOI] [PubMed] [Google Scholar]
- 52.Payungwoung T, et al. CRISPR/Cas9 system in Plasmodium falciparum using the centromere plasmid. Parasitol. Int. 2018;67:605–608. doi: 10.1016/j.parint.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 53.Shinzawa N, et al. Improvement of CRISPR/Cas9 system by transfecting Cas9-expressing Plasmodium berghei with linear donor template. Commun. Biol. 2020;3:426. doi: 10.1038/s42003-020-01138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marin-Mogollon C, et al. Chimeric Plasmodium falciparum parasites expressing Plasmodium vivax circumsporozoite protein fail to produce salivary gland sporozoites. Malar. J. 2018;17:288. doi: 10.1186/s12936-018-2431-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lim MY, et al. UDP-galactose and acetyl-CoA transporters as Plasmodium multidrug resistance genes. Nat. Microbiol. 2016;1:16166. doi: 10.1038/nmicrobiol.2016.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deitsch K, Driskill C, Wellems T. Transformation of malaria parasites by the spontaneous uptake and expression of DNA from human erythrocytes. Nucleic Acids Res. 2001;29:850–853. doi: 10.1093/nar/29.3.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Schaijk BC, Vos MW, Janse CJ, Sauerwein RW, Khan SM. Removal of heterologous sequences from Plasmodium falciparum mutants using FLPe-recombinase. PLoS One. 2010;5:e15121. doi: 10.1371/journal.pone.0015121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marin-Mogollon C, et al. A P. falciparum NF54 reporter line expressing mcherry-luciferase in gametocytes, sporozoites, and liver-stages. Front. Cell. Infect. Microbiol. 2019;9:96. doi: 10.3389/fcimb.2019.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miyazaki S, et al. Generation of novel plasmodium falciparum NF135 and NF54 lines expressing fluorescent reporter proteins under the control of strong and constitutive promoters. Front. Cell. Infect. Microbiol. 2020;10:270. doi: 10.3389/fcimb.2020.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016;44:W272–W276. doi: 10.1093/nar/gkw398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ponnudurai T, et al. Infectivity of cultured Plasmodium falciparum gametocytes to mosquitoes. Parasitology. 1989;98:165–173. doi: 10.1017/s0031182000062065. [DOI] [PubMed] [Google Scholar]
- 62.Seilie AM, et al. Beyond blood smears: qualification of plasmodium 18S rRNA as a biomarker for controlled human malaria infections. Am. J. Trop. Med. Hyg. 2019;100:1466–1476. doi: 10.4269/ajtmh.19-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nijhuis RHT, van Lieshout L, Verweij JJ, Claas ECJ, Wessels E. Multiplex real-time PCR for diagnosing malaria in a non-endemic setting: a prospective comparison to conventional methods. Eur. J. Clin. Microbiol. Infect. Dis. 2018;37:2323–2329. doi: 10.1007/s10096-018-3378-4. [DOI] [PubMed] [Google Scholar]
- 64.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vos MW, et al. A semi-automated luminescence based standard membrane feeding assay identifies novel small molecules that inhibit transmission of malaria parasites by mosquitoes. Sci. Rep. 2015;5:18704. doi: 10.1038/srep18704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Annoura, T. et al. Assessing the adequacy of attenuation of genetically modified malaria parasite vaccine candidates. Vaccine10.1016/j.vaccine.2012.02.010 (2012). [DOI] [PubMed]
- 71.Vaughan AM, et al. Type II fatty acid synthesis is essential only for malaria parasite late liver stage development. Cell Microbiol. 2009;11:506–520. doi: 10.1111/j.1462-5822.2008.01270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu M, et al. The fatty acid biosynthesis enzyme FabI plays a key role in the development of liver-stage malarial parasites. Cell Host Microbe. 2008;4:567–578. doi: 10.1016/j.chom.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nagel A, et al. A new approach to generate a safe double-attenuated Plasmodium liver stage vaccine. Int. J. Parasitol. 2013;43:503–514. doi: 10.1016/j.ijpara.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 74.Lindner SE, et al. Enzymes involved in plastid-targeted phosphatidic acid synthesis are essential for Plasmodium yoelii liver-stage development. Mol. Microbiol. 2014;91:679–693. doi: 10.1111/mmi.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Iwanaga S, Kaneko I, Kato T, Yuda M. Identification of an AP2-family protein that is critical for malaria liver stage development. PLoS One. 2012;7:e47557. doi: 10.1371/journal.pone.0047557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ishino T, et al. LISP1 is important for the egress of Plasmodium berghei parasites from liver cells. Cell Microbiol. 2009;11:1329–1339. doi: 10.1111/j.1462-5822.2009.01333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kumar H, et al. Protective efficacy and safety of liver stage attenuated malaria parasites. Sci. Rep. 2016;6:26824. doi: 10.1038/srep26824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Orito Y, et al. Liver-specific protein 2: a Plasmodium protein exported to the hepatocyte cytoplasm and required for merozoite formation. Mol. Microbiol. 2013;87:66–79. doi: 10.1111/mmi.12083. [DOI] [PubMed] [Google Scholar]
- 79.Sahu T, et al. ZIPCO, a putative metal ion transporter, is crucial for Plasmodium liver-stage development. EMBO Mol. Med. 2014;6:1387–1397. doi: 10.15252/emmm.201403868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Putrianti ED, et al. The plasmodium serine-type SERA proteases display distinct expression patterns and non-essential in vivo roles during life cycle progression of the malaria parasite. Cell Microbiol. 2010;12:725–739. doi: 10.1111/j.1462-5822.2009.01419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Verma N, Shukla H, Tiwari A, Mishra S, Habib S. Plasmodium Ape1 is a multifunctional enzyme in mitochondrial base excision repair and is required for efficient transition from liver to blood stage infection. DNA Repair. 2021;101:103098. doi: 10.1016/j.dnarep.2021.103098. [DOI] [PubMed] [Google Scholar]
- 82.Kenthirapalan S, Waters AP, Matuschewski K, Kooij TW. Functional profiles of orphan membrane transporters in the life cycle of the malaria parasite. Nat. Commun. 2016;7:10519. doi: 10.1038/ncomms10519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Parasite whole-genome sequences have been deposited in the repository https://www.ebi.ac.uk/ena/browser/view/PRJEB40003; the sequence file is accessible under accession number ERR4620262. The P. berghei mutant line PbΔmei2 has been deposited in the repository https://www.pberghei.eu/index.php?rmgm=4937.