Abstract
The feather is an important epidermal appendage, plays an important role in the life activities of avian specie, and has important economic value. Revealing the molecular regulation mechanism of feather growth has a significant meaning in studying adaptive evolution, physiology, and mating of avian species and also provides a theoretical reference for poultry breeding. In this study, the genome-wide association analysis (GWAS) of 358 ducks was based on primary feather length phenotypic data (28–60 d), length growth rates (LGRs), and maturity scores (60 d) to explore the genetic basis affecting feather growth and maturation. The results showed that, among the primary feather 1 to 5 in ducks, the mean LGR of primary feather 2 was the fastest, with the longest length. The primary feathers in males grew and matured slightly faster than in females. The mean maturity scores of primary feather 10∼7 were higher than primary feather 1 to 3 in ducks. GWAS further showed 116 SNPs associated with feather length traits. In addition, 2 candidate regions (Chr1: 127,407,230–127,524,879 bp and Chr21: 182,061,707–183,616,298 bp) were associated with LGR, which contain total 13 candidate genes (The extremely significant SNPs were mainly located in 2 genes: Chr1: REPS2 and Chr21: PTPRT). Four candidate regions (Chr1: 29,113,036–28,675,018 bp, Chr2: 18,253,612–149,111,290 bp, Chr15: 6,489,774 to 12,138,221 bp and Chr21: 6,578,021–8,472,904 bp) were associated with feather maturity, which contain total 24 candidate genes (The extremely significant SNPs were mainly located in 4 genes: Chr1: IMMP2L, DOCK4 and DDX10, Chr2: LDLRAD4). In conclusion, sex factors influence feather growth and maturity, and the genetic basis of the growth /maturity trait between different feathers is similar. REPS2, PTPRT genes, and IMMP2L, DOCK4, DDX10, and LDLRAD4 are important candidate genes that influence feather growth and maturity, respectively.
Key words: duck, primary feather, growth, maturity, GWAS
INTRODUCTION
Feathers are essential accessory organs of avian species and have various phenotypic variations, such as shapes, sizes, patterns, and pigmentation. The feathers provide conditions for avian species to fly, adapt to complex environments, protect the body's surface, avoid danger and go to mate (Hill and Montgomerie, 1994; CUERVO and MØLLER, 1999; Prum and Williamson, 2001; Norris et al., 2004; Leeson and Walsh, 2004a; Talloen et al., 2008; Kjaer and Bessei, 2013; Chen et al., 2015). Hence, feathers were often considered an ideal model for studying adaptive evolution, mating, physiological, and development of animals. Feathers also have significant economic value in the processing and manufacturing industry and were usually taken as an important industrial raw material for making quilts, clothes, sporting goods, etc. Besides that, feathers also play an essential role in poultry sexual identification (Chuong et al., 2012). The development of feathers is often a reflection of poultry individuals' growth and health conditions (Coon et al., 2016), influencing the appearance and consumers' choice of poultry products. Based on traditional consumption habits and specific requirements for the appearance of quality poultry, having neat, shiny, and plump feathers can improve their commercial value. If the feathers of poultries grow slowly, this will cause the time to market to be delayed, the cost of the breeding to increase, and the production efficiency reduction. Many recent studies have shown that feather growth traits are related to poultry production performance (Kalinowski et al., 2003; Khosravinia, 2009). Based on the above, feather growth and maturity traits are essential for developing biology selection criteria for poultry breeding. Hence, studying poultry feathers' growth and development rules and genetic mechanisms is necessary.
The heritability (h2) of feather growth traits has been widely assessed among poultry species, such as quail, chicken, and duck. Chambers (1990) firstly estimated the heritability of feather length traits of chicken and considered it at a moderate level (h2 = 0.3–0.5). Subsequently, Lou (1994) found that the heritabilities were 0.54 and 0.56 for the tail feather length of generations 1–2 and 1–3 broiler chickens, respectively. However, they also found that the heritabilities of the later generations were lower and were 0.32 and 0.33 for the tail feather length of generations 4–8 and 6–8, respectively. Rizzi et al. (1994) also found that the heritabilities were the following: 0.21 for primary remex 1; 0.23 for secondary remex length in pheasant (sire component). In follow-up studies, Hu et al. (1999) assessed the heritability of feather length in muscovy ducks. The results showed that the female's eighth primary feather was longer than the male's by 6% to 22% in the 10th wk. The heritability of feather length in male (h2 = 0.37) is higher than that in female (h2 = 0.14). Chen et al. (1995) assessed the heritability of feather length (FL20) in 20-wk-old Brown Tsaiya laying ducks. The heritability of FL20 was found to be low (h2 = 0.169). In other avian species, Gebhardt-Henrich et al. (1993) assessed the heritability of quail feather length in ad libitum and restricted feeding conditions. The results showed h2 = 0.59 ± 0.19 under restricted feeding and h2 = 0.53 ± 0.19 under ad libitum feeding of quail feather length.
Based on the above studies, the heritability estimates of feather growth traits are generally moderate or low in some poultry species. These traits are determined by abundant slightly or moderately effective loci, and these loci only contribute to limited genetic variance. At the same time, environmental factors also affect the phenotypes of such traits. In the traditional breeding methods, the phenotypic value of the target trait is directly measured, and the further decision to keep livestock and poultry is based on whether the phenotypic value is in line with the breeding goals. Phenotypic measurement is usually carried out under the target environment and in the critical period when traits are fully expressed, but the contribution of genotype and environment to the phenotype cannot be distinguished. Also, some phenotypic identification is difficult to be completely precise and reliable. Hence, applying traditional breeding methods is difficult to carry out accurate and efficient seed selection based on these traits as indicators. With the development of molecular breeding technology (e.g., marker-assisted selection [MAS] (Soller, 1994) and genomic selection [GS] (Meuwissen et al., 2001)), based on the use of high-density trait-related SNP (Single nucleotide polymorphism) markers covering the whole genome for selective breeding can effectively improve the accuracy of GEBV (Genomic estimated breeding value) estimation and accelerate genetic progress, especially for complex traits with low heritability and difficult to measure. The results of GWAS are the most commonly used prior biological information for GS and MAS (MacLeod et al., 2016; Abdollahi-Arpanahi et al., 2017; Hoff et al., 2019; Ma et al., 2019; Liu et al., 2020; Ye et al., 2020). Based on the P-value and/or marker effect in GWAS, it is easy to obtain the SNP's effect on a phenotypic trait. At present, molecular markers associated with reproductive (Chang et al., 2012; Huang et al., 2013), carcass (Wu et al., 2012), body size (Zhou et al., 2018), feather color (Xi et al., 2021), and eggshell (Liu et al., 2021) color traits have been discovered in ducks. However, less publications reported on ducks' QTLs or SNPs associated with feather growth traits. It is restricted in heredity improvement of duck feather growth traits for the above reasons, which is not conducive to the large-scale production of ducks.
Therefore, 400 ducks were raised in the same environment in this study. The length of primary feathers was measured at 28, 35, 40, 45, and 60-day-old, and the maturity trait of primary feathers was scored at 60-day-old. Finally, the primary feather length, LGRs, and maturity traits were computed for the correlation with genomic mutations by GWAS to screen candidate genes potentially causing differences in traits. This study provides a theoretical reference for studying feather growth and development, the genetic basis of avian species, and ducks' breeding.
RESULT AND ANALYSIS
Phenotype
Feather Growth
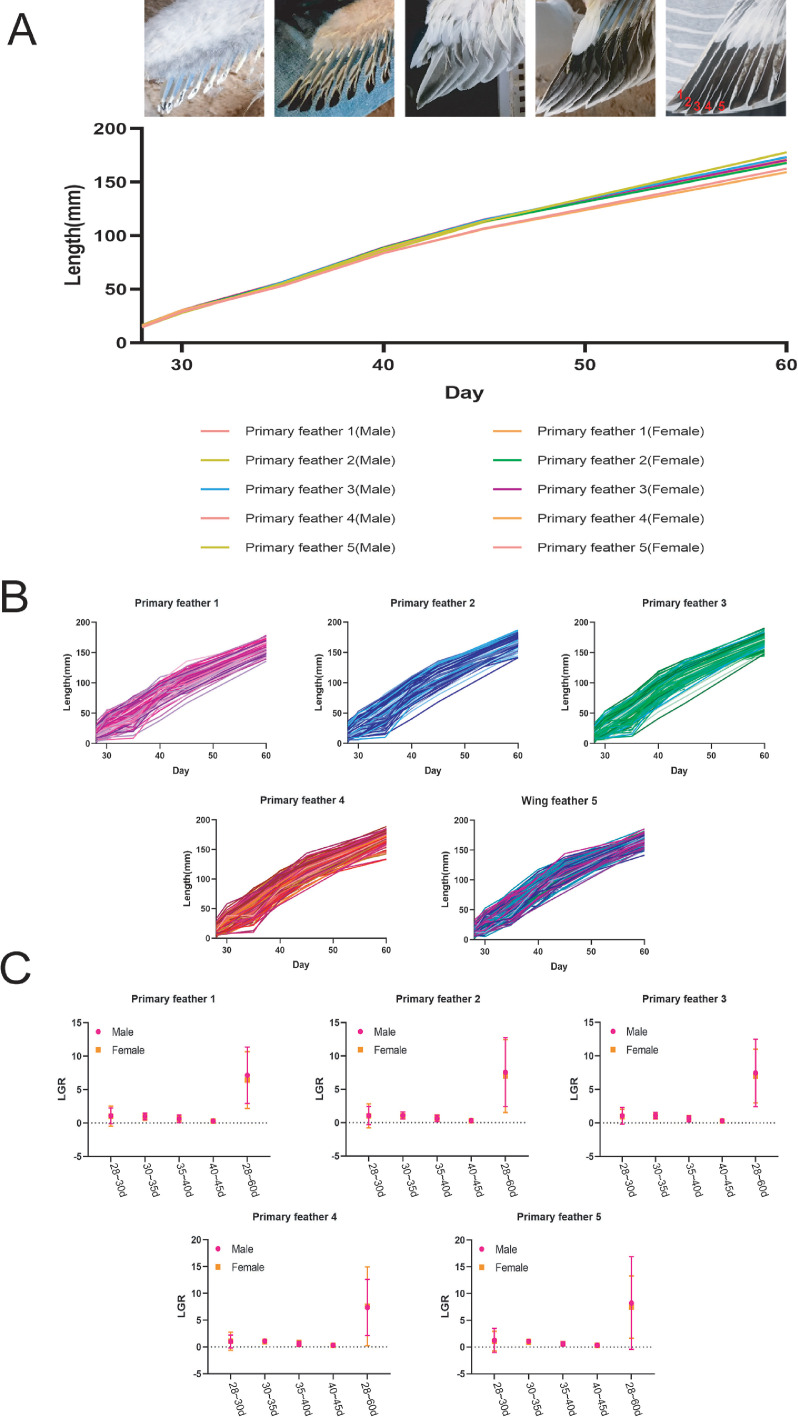
The descriptive statistics of growth traits of primary feathers 1 to 5 (Figure 1A) are shown in Figure 1 and Table S8, Supplementary Table S1, S2, and S3. The results showed that barbs and barbules had been formed at the top of the rachis of primary feather 1 to 5 (Refer to specific: primary feather 1, primary feather 2, primary feather 3, primary feather 4, and primary feather 5), and their lengths were both in the range of 15 to 16 mm at 28 d of old in ducks (Figures 1A and 1B, Table S8). Accompanying ducks grew older, and primary feathers were 1 to 5 lengthened (Figure 1A). Meanwhile, the average lengths between different categories of primary feathers were close at 28 to 40 d of age in ducks.
Figure 1.
Descriptive statistics of feather growth in ducks. (A) Growth curves of primary feather 1–5 in male and female populations, respectively. The growth curves were plotted using the average primary feather lengths for male or female populations at different growth stages. (B) Primary feather types group growth curves. The 5 small pictures documented the growth of primary feathers 1–5 from 20 to 60-day-old in each duck. (C) The LGRs of different primary feather types at 5 stages.
However, the average lengths gradually showed differences between these 5 categories of primary feathers at 45 to 60 days old in ducks (Figure 1A, Table S8). At 60-day-old ducks, the average length of primary feather 1∼5 was 159.43, 168.01, 170.68, 169.14, and 167.83 mm, respectively (Figures 1A and 1B, Table S8). Primary feather 2 has the length advantage, and primary feather 1 has the shortest length in the duck population at 45 to 60 days old. In addition, the standard deviation (S.D.) of primary feather 1 to 5 length of all ducks increased with the measuring age, the S.D. of primary feather length at 45 d was the largest, but the S.D. of feather length at 60 d was close to that of 28 d (Table S8), these results indicate that at 45 d of duck age, the difference in primary feather length between individuals was the largest.
Based on the intersexual comparison, the average lengths of primary feathers 1 to 5 of males were roughly the same as that of females at 28, 30, 35, and 40-day-old. The average length of primary feather 1 to 5 in males was longer than in females at 60-day-old ducks (Figure 1A and Table S9). Next, we compared the growth intensity of primary feathers 1 to 5 of different individuals at 5 stages (28–30-day-old, 30–35-day-old, 35–40-day-old, 40–45-day-old, and 45–60-day-old; Figure 1C). Here, LGR was used to reflect the intensity of primary feather growth. The results show that the mean LGR of primary feathers 1 to 5 in males was slightly higher than in females at 5 stages. The mean LGR of the primary feather 1 to 5 in males or females at 2 stages (28–30 d and 30–35 d) were similar, with mean LGR at 30–35 d>35–40 d>40–45 d. Throughout the observation phase (28–60 d), primary feathers 4 and 5 still had the highest mean LGR, and primary feather 1 had the lowest mean LGR value.
Then, correlation analysis also suggested that primary feathers length at 28 d was significantly associated with primary feathers length at 30, 40, 45, and 60 d of age (Table S1). One-way analysis of variance (ANOVA) based on sexes grouping also showed that sexes factors mainly affected primary feathers length at 28, 30, 35, and 60 d (P<0.05; Table S2).
Feather Maturity
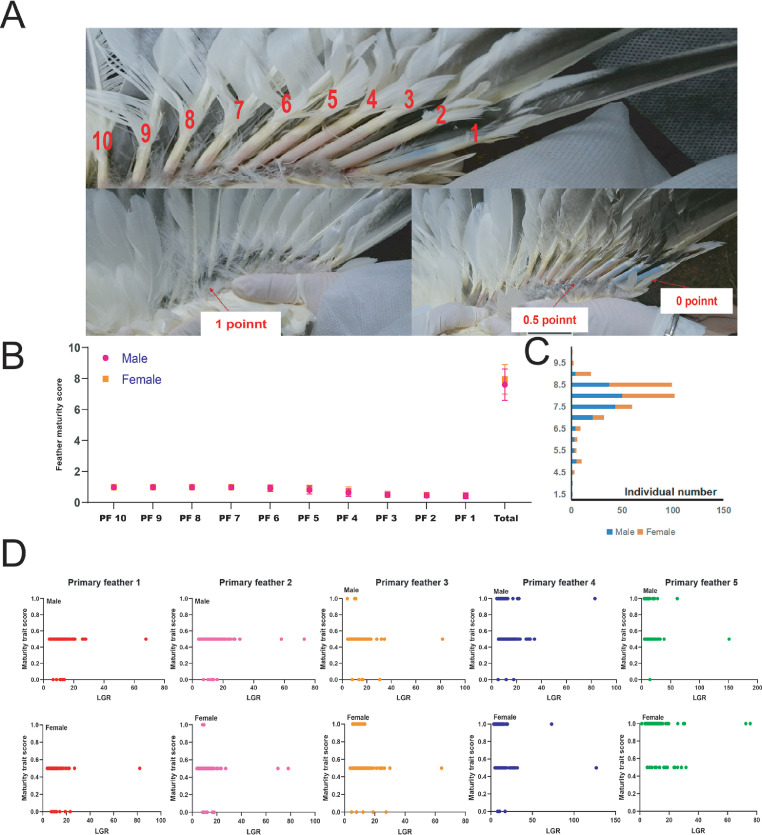
The descriptive statistics of feather maturity traits are shown in Figure 2, Table S3, and Table S4. In the 60-day-old duck population, individuals with a total maturity score of 8.5 to 10, 8.0 to 6, and 5.5 to 0 made up 35, 60, and 15%, respectively. The proportions of individuals with different full maturity scores were similar in male and female populations (Figure 2C). Meanwhile, the statistical results showed that primary feather 1 has the lowest maturity score, and primary feather 10 has the highest maturity score (Figure 2B). The above results suggested that the primary feathers 10 to 6 foremost matured, but the maturity of primary feathers 1 to 3 is inferior. Based on the intersexual comparison, the maturity scores of each primary feather category and total score in females were slightly higher than in males (Figure 2B). One-way ANOVA based on sexes grouping also showed that sexes factors mainly affected primary feather 3, 4, and 5 maturities at 60 d (Table S3). In addition, although we found differences in the primary feather maturity of different individuals or different types of feathers in the same individual, the maturity scores of primary feather 1 to 5 for most individuals were still centered around 0.5 (Figure 2D).
Figure 2.
Descriptive statistics for primary feather maturity traits. (A) These images show the grading criteria for feather maturity and the location of the main primary feather. (B) Maturity scores and overall scores for different types of feathers. (C) The number of individuals within the range of total feather maturity scores (D) Scatter plot of different feather LGR and maturity scores for each individual.
However, correlation analysis further showed that the LGRs of primary feather 3 and 4 significantly affected the maturity scores (P < 0.05), and that of primary feather 5 extremely significantly affected the maturity scores (P < 0.01; Table S4).
Quality Control and Population Structure
For 358 DNA samples, their OD260/280≥1.8, and OD260/230≥2.0 showed that all samples' quality was good. The above samples were used for whole-genome sequencing. After quality control, SNPs were kept for further analysis. The distribution of SNPs and principal component analysis (PCA) analysis is illustrated in Figure S1.
SNPs Associated With Primary Feather Length Traits
The GWAS results showed no clear signal peak in the length traits of primary feather 1 to 5 at 28, 30, 35, 40, 45, and 60-day-old ducks. Only a few SNPs passed the Bonferroni threshold. In results at some stages of ducks (Involved: length traits of Pf 1(primary feather 1) (40 d): 9 SNPs; Pf 1 (45 d): 30 SNPs; Pf 2 (28 d): 1 SNPs; Pf 2 (40 d): 4 SNPs; Pf 2 (45 d): 45 SNPs; Pf 3 (40 d): 1 SNPs; Pf 4 (60 d): 2 SNPs; Pf 5 (28 d): 17 SNPs and Pf 5 (60 d): 1 SNPs) (Figure S2). These SNPs are mainly located on chromosomes 1 and 2.
Thirty-eight potential candidate genes were annotated within all detected SNPs (54 SNP sites were involved). The details about significant SNPs are shown in Table S5. The SNP loci associated with different traits differ among these annotated SNPs. Among the above candidate genes, KIF18A, NMRK2, KIF1B, BAZ1B, CDK14, and STK32C genes are associated with the ATP binding signaling pathway (GO:0005524), KIF18A and KIF1B genes are associated with kinesin complex signaling pathway (GO:0005871). Notably, 1 common SNP located at position 24,764,847 bp on the KCND2 gene of Chr1, respectively, related to the length traits of primary feathers 1 and 2 at 45-day-old ducks.
Three Main Candidate Regions Distributed on Chromosome 1 and 21 Are Associated With LGRs of Primary Feather
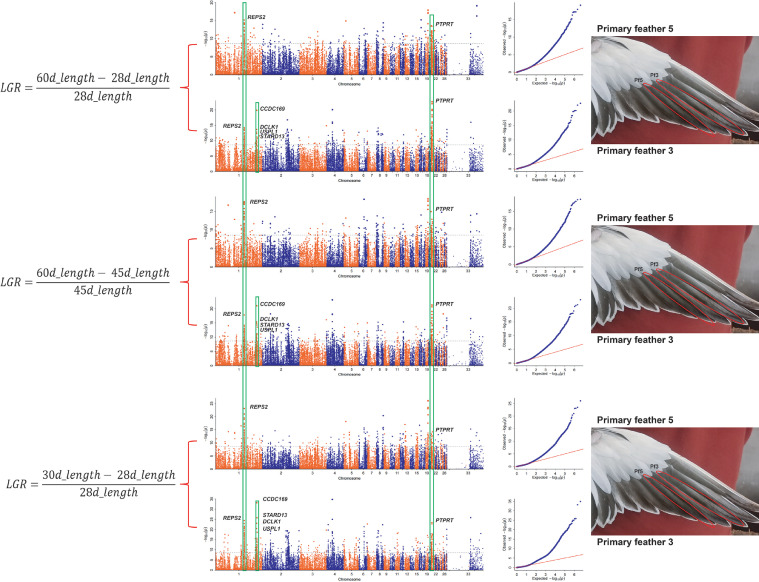
We performed GWAS based on LGRs of primary feathers 1 to 5 at 5 growth stages (The 5 stages included: 28–30 d, 30–35 d, 35–40 d, 45–60 d, and 28–60 d). The results showed that 2 significant signal peaks were observed on chromosomes 1 and 21, respectively, in GWAS results of LGRs of primary feather 3 and 5 at 3 stages (involved stages: 28–30 d, 45–60 d, and 28–60 d) (Figure 3). In addition, another signal peak located on chromosome 1 was found in the GWAS results of LGRs of primary feather 3 at the above 3 stages (Figure 3). In the results of LGRs at other stages, we did not find regular and significant signal peaks (Figure S3).
Figure 3.
GWAS of growth rate traits of primary feathers 3 and 5. On the left and right of the image, we indicate how the LGR is calculated and the position of primary feathers 3 and 5 on the duck primary, respectively.
The SNPs with significance (-Log10P > 8.39) were mainly distributed in a ∼118 kb region from 127,407,230 to 127,524,879 bp and a ∼1,555 kb region from 182,061,707 to 183,616,298 bp on chromosome 1, respectively. Then, other SNPs were distributed in a ∼146 kb region from 4,210,283 to 4,356,595 bp on chromosome 21.
Among these SNPs, 13 main candidate genes were annotated. The details about all significant SNPs on Chr.1 and 21 are shown in Table S6. Gene Ontology (GO) analysis further found that PLXNB2 and PLXNA4 genes were enriched in the nervous system formation and cell migration signaling pathways (GO:1902287, GO:0071526, GO:0030334, and GO:0002116), PTPRT and PLXNB2 genes were enriched in protein modifications and cell physiological activities signaling pathways (GO:0006470 and GO:0007156).
Four Main Candidate Regions Distributed on Chromosomes 1, 2, 15, and 18 Are Associated With Primary Feather Maturity Traits
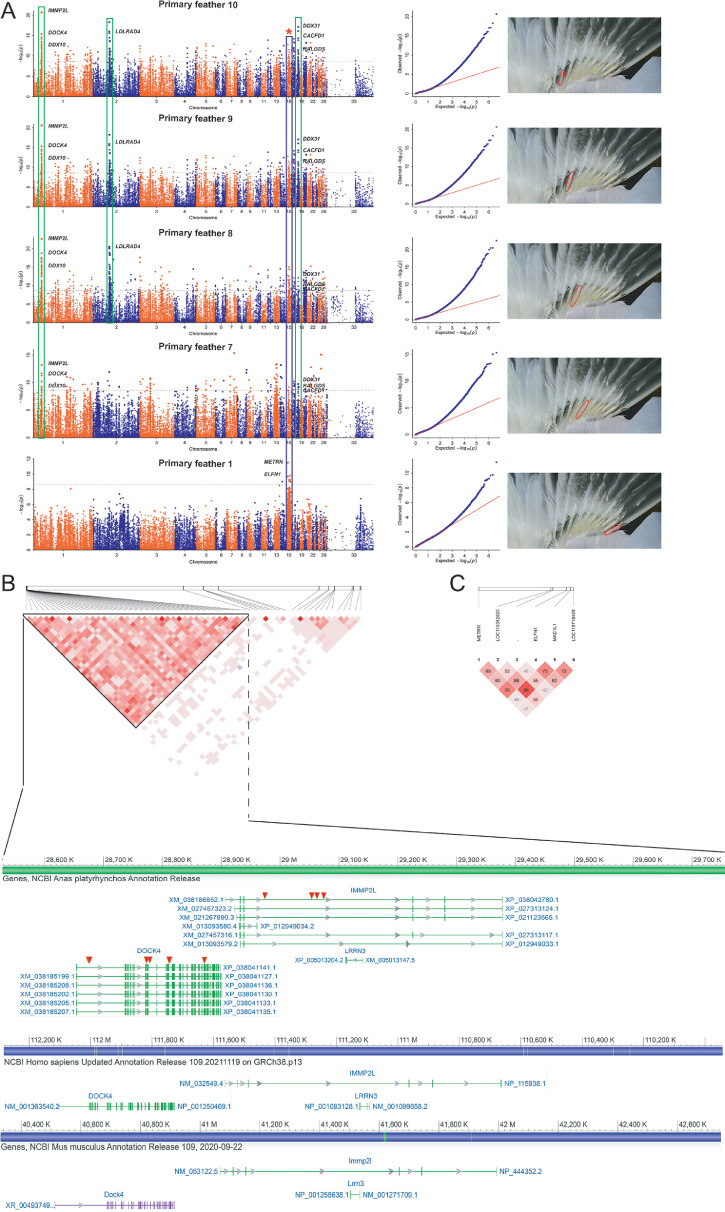
Next, we performed GWAS based on the maturity scores of primary feathers 1 to 10 at 60-day-old ducks. Three significant signal peaks were located on chromosomes 1, 2, and 15, respectively, and these signal peaks were observed in GWAS results of the maturity traits of primary feathers 10, 9, and 8 (Figure 4A). It is worth noting that these two signal peaks located on chromosome 1 and chromosome 15, respectively, were also found in GWAS results of the maturity traits of primary feathers 7 and 1 (Figure 4A). In addition, a significant signal peak was observed at the same position on chromosome 18 among the GWAS results of the maturity traits of primary feather 10 and 9 (Figure 4A). Among these 4 signal peaks, the signal peak on chromosome 1 is the most obvious (Figure 4A). In the results of maturity scores of primary feather 6 to 2, we did not find regular and significant signal peaks (Figure S4).
Figure 4.
GWAS of primary feather maturity traits. (A) The results of GWAS of primary feather 10, 9, 8, 7, and 1 maturity traits. (B) Linkage disequilibrium analysis and synteny analysis of some SNPs on chromosome 1 (C) Linkage disequilibrium analysis of some SNPs on chromosome 15.
The SNPs with significance (-Log10P > 8.39) were mainly distributed in a ∼43.8 kb region from 29,113,036 to 28,675,018 bp on chromosome 1 (primary feather 10: 29,113,036 ∼ 28,675,018 bp; primary feather 9: 29,108,773 ∼ 28,675,018 bp; primary feather 8: 29,074,033 ∼ 28,675,018 bp and primary feather 7: 29,074,033 ∼ 28,675,018 bp), in a ∼130,858 kb region from 18,253,612 to 149,111,290 bp on chromosome 2, in a ∼ 5,648 kb region from 6,489,774 to 12,138,221 bp on chromosome 15, and in a ∼ 1,894 kb region from 6,578,021 to 8,472,904 bp on chromosome 18.
The candidate region on chromosomes 1, 2, 15, and 18 harbored 15, 1, 5, and 3 genes. The details about all significant SNPs on Chr.1, 2, 15, and 18 are shown in Table S7. GO analysis further suggested that DOCK4, ITSN1, and RALGDS genes were significantly enriched in small GTPase mediated signal transduction (GO:0007264), positive regulation of GTPase activity (GO:0043547), and guanyl-nucleotide exchange factor activity (GO:0005085) signaling pathways. DDX31 and DDX10 genes were significantly enriched in ATP-dependent RNA helicase activity (GO:0004004), helicase activity, and RNA secondary structure unwinding (GO:0010501) signaling pathways.
Based on linkage disequilibrium (LD) analysis, we found that 11 common SNPs located in the IMMP2L and DOCK4 genes (The particular positions of the 11 SNPs on the IMMP2L or DOCK4 genes are kept in the sashimi plot of Figure 4B), respectively, have strong linkage characteristics (Figure 4B). The 6 SNPs on chromosome 15 also have certain linkage characteristics (Figure 4C). Synteny analysis showed that IMMP2L and DOCK4 genes are always neighbors and keep the same transcription direction in duck, human, and mouse genomes. Therefore, this fragment may be conserved.
Transcriptomes in Plumage Follicles During Feather Growth and Development Support Candidate Genes That Play a Role in Feather Follicles
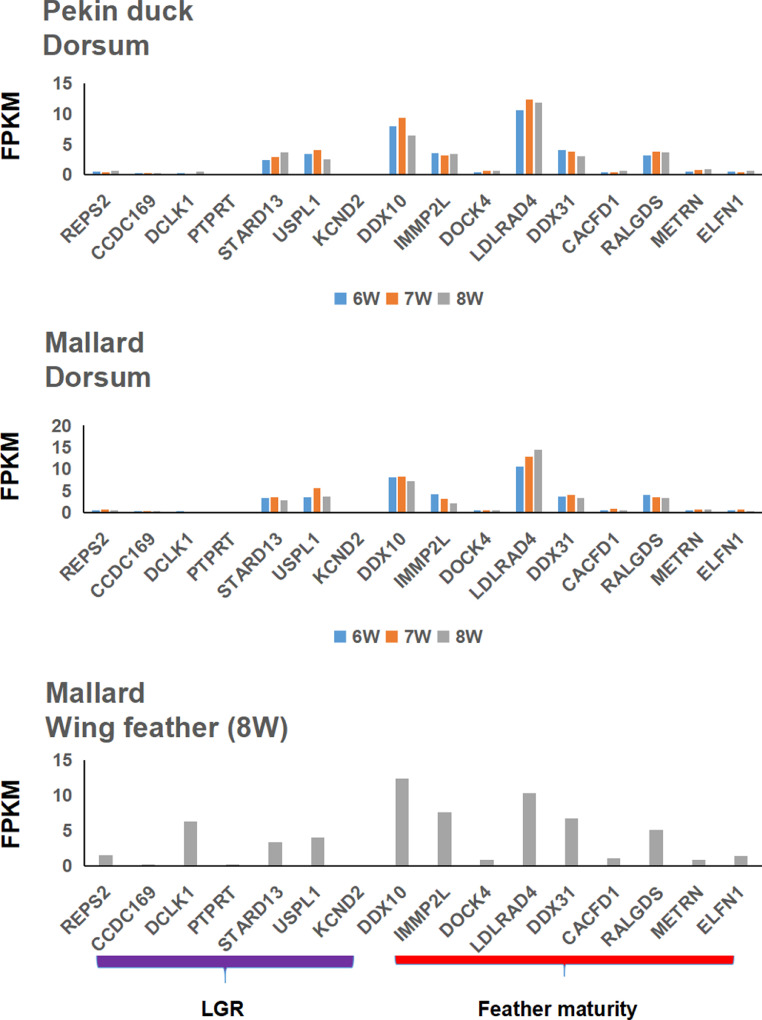
We focused on the expression patterns of 12 candidate genes related to LGR and feather maturity traits using the primary and dorsum feather follicle transcriptome data in Tianfu Nonghua duck and Pekin ducks. The transcriptomes of Tianfu Nonghua duck and Pekin duck set at 3 stages (6th, 7th, and 8th wk previously published in the database) showed 12 candidate genes expressed in primary and dorsum feather follicles at 3 stages. Still, the expression of these genes was lower in feather follicles. Meanwhile, the expression patterns of the above candidate genes in the dorsal feather follicle tissues of Tianfu Nonghua duck and Pekin duck were generally similar from wk 6 to wk 8. We also found that the expression pattern of the above genes in the primary feather follicles was identical to that in the dorsal feather follicles of an 8-wk-old Tianfu Nonghua duck (Figure 5).
Figure 5.
Expression patterns of genes related to feather growth and candidate genes at 6th to 8th weeks in Tianfu Nonghua duck and Pekin duck.
DISCUSSION
Duck is an important poultry species, but we have known very little about the development and genetics of duck feathers for a long time. Compared with hair and wool in mammals, the structure of feathers is more complex, and more signal molecules may regulate the growth and maturity of feathers in avian species. Some molecular signaling pathways affecting avian feather formation have been reported. However, this cannot fully reveal the complex regulatory mechanism of feather growth and maturation in avian species and the individual differences. In this study, we preliminarily analyzed the regularity of growth and maturity of primary feathers in the duck population from 28 to 60-day-old and performed GWAS based on the feather growth and maturity trait phenotypic data to analyze further the genetic mechanisms regulating differences in feather growth and maturity among individuals.
First, our study found that the mean length differences between 5 categories of primary feathers (primary feather 1–5) were not obvious, and neither can length differences of primary feathers between sexes in the youth duck population (28 d). The growth rate of primary feathers was gradually accelerated, and the mean length differences between primary feathers 1–5 were more obvious as the age increased (28d–60 d). The primary feather 2 always has the advantage of length at 40, 45, and 60-day-old ducks. Also, males have longer primary feathers than females (60 d). The feather growth curve and LGR statistics showed that primary feather 2 has the fastest growth rate, and male feathers grow faster than females (40–60 d). Although there are differences in the growth rates of the 5 categories of primary feathers, the whole growth pattern of primary feathers of ducks was similar to that of Pekin, mule, muscovy, chicken, etc. (Szász et al., 1999; Leeson and Walsh, 2004b; Xie et al., 2020). In broilers, McDOUGALD and Keshavarz (1984) found that males have longer feathers than females. Although we have only measured the feather length at limited stages and cannot determine the fastest-growing period of primary feathers in the duck population, we preliminarily considered that sex factors might affect the length and growth rate of primary duck feathers, and initial length (28 d) may affect final length (60 d) of primary feathers. Previous classic case studies have revealed the sex-linked genetic basis of feather growth rate, supporting our conclusion from the side[4].
In subsequent GWAS, fewer SNPs associated with feather length were found. Among the 38 annotated main candidate genes, the KCND2 gene is closely related to the length traits of primary feathers 1 and 2 in 45-day-old ducks. Zhang et al. (2021) have found that the KCND2 variants are associated with global developmental delay. Tao et al. (2020) also found that some SNPs in the KCND2 gene are associated with bone development in neonatal sheep. Therefore, we further speculate that mutations within the KCND2 gene affect feather growth in ducks.
In the GWAS of LGRs, 3 significant candidate regions associated with LGRs of primary feather 5 and 3 at 3 stages (28–30 d, 45–60 d, and 28–60 d) were found, and we further got more candidate genes associated with feather LGRs. Among the 13 candidate genes, the REPS2 and PTPRT genes are very important, and many extremely significant SNPs are mainly located in these genes. Badway and Baleja (2011) suggested that Reps2 is cellular signaling and molecular trafficking nexus. In addition, previous studies have found that the EGF signaling pathway regulates the growth of avian feathers (Yue et al., 2012), and EGF signaling is also inhibited by a high expression level of REPS2 protein in some cell lines (Oosterhoff et al., 2005). Meanwhile, Lee (2015) found that PTPRT regulates the synaptic formation and neuronal development. Nervous tissue plays an important role in feather development (Yu et al., 2004). Combining the above results, we consider that primary feathers 3 and 5 may have a similar genetic basis for growth regulation, PTPRT and REPS2 are essential candidate genes.
Feather follicles are control centers for feather growth and regeneration. The feather follicle continuously delivers the substances needed for feather growth to the feather through the capillaries in the feather pulp and promotes the growth of the feather[2]. When the feather fully matures, the follicles degenerate, and the feather pith becomes hollow and transparent, which is an important indicator for judging whether the feather is mature. Based on the maturity scores of primary feather 1 to 10, we found that primary feather 10 to 5 matured earlier than primary feather 1 to 3, the feather follicles of primary feather 1 to 3 were still active and not degenerated, and primary feather 1 to 3 may not have finished growing. This may be due to the significantly shorter length and smaller primary feather 10 to 5 than primary feather 1 to 3. Lower maturity scores for primary feathers 1 and 2 may also be because the feather follicles provide the material needed for feather coloration or are related to feather wear and replacement (Yu et al., 2004). In addition, the maturity scores of primary feathers 10 to 5 for males were also higher than that of females, suggesting that male feathers grow faster than females. Therefore, we considered that the sexes also affect feather maturity traits.
The GWAS for maturity scores of primary feather 10 to 7 and 1 revealed that 4 important regions associated with feather maturity traits on Chr.1, 2, 15, and 18 contained 24 candidate genes. These results suggested that primary feather 10 to 7, 1 shared a similar genetic regulatory basis for feather maturity. Notably, 11 common SNPs were mapped to an important QTL region (IMMP2L-DOCK4) on Chromosome 1, and have strong linkage characteristics. Synteny analysis further showed that this fragment is relatively conserved in different species. Numerous studies have shown that the IMMP2L-DOCK4 fragment is a vital QTL, mutations which cause some congenital neurodevelopmental disorders and loss of sensory function (Maestrini et al., 2010). Bertelsen et al. (2014) suggested that the defective IMMP2L gene may lead to apoptosis due to a hyperactive mitochondrion. He et al. (2020) found that the IMMP2L mutation causes ovarian aging through the ROS-Wnt/β-catenin estrogen pathway. Zhou et al. (2011) also found that DOCK4 gene knockdown significantly increased apoptosis. The degeneration of the feather follicle is the process of cell senescence and apoptosis. In summary, these 2 genes are important candidate genes affecting feather maturity traits in ducks. In addition, some SNPs were also located within the DDX10 and LDLRAD4 genes. Deleting the DDX10 gene prohibits cell activities modulated by the MAPK pathway (Shi and Hao, 2019). MAPK signaling pathway is also important in regulating feather growth and development (Ji et al., 2021). Ito et al. (2020) found that downregulation of LDLRAD4 induces transforming growth factor β signaling, promoting cell proliferation and suppressing apoptosis. These results also suggest that DDX10 and LDLRAD4 genes play an important role in feather maturation.
Whether performing GWAS based on feather growth traits or maturity traits, we find many signal peaks distributed on different chromosomes with low correlation to the trait and are cluttered. The QQ plot also suggests that these SNPs with lower P-values may be associated with random drift and false-positive loci. Of course, we try to eliminate the influence of this factor on the association analysis through the mixed linear model as much as possible. However, we still do not rule out confounding bias effects (e.g., the influence of non-genetic factors on the trait itself, cryptic relatedness, and population stratification) for this result. Other researchers believe this phenomenon is related to many minor genes controlling quality traits (Bulik-Sullivan et al., 2015). Therefore, we did not perform a detailed analysis of these SNPs.
Finally, we focused on the expression pattern of 12 candidate genes in feather follicles. Although the expression levels of some genes were low, these genes were all expressed in the dorsum and primary feather follicles of Tianfu Nonghua duck and Pekin duck at 3 stages. These genes may be indispensable and play a role in feather growth and development. However, these genes' specific functions and regulatory mechanisms still need further verification.
CONCLUSIONS
Sex factor may be an essential factor affecting primary feather growth and maturity. The genetic basis of growth and maturity traits of different categories of primary feathers are similar, but there are also some differences. In GWAS, the SNPs were mapped to REPS2, PTPRT genes, IMMP2L, DOCK4, LDLRAD4, and DDX10 genes, which were important candidate marker sites associated with feather growth and maturity, respectively.
MATERIALS AND METHODS
Animal Feeding and Phenotype Collection
In total, 400 one-day-old ducklings (Tianfu Nonghua duck) (200 males and 200 females) were provided by the waterfowl breeding farm at Sichuan Agricultural University, Sichuan, China. These ducks have been raised in a comfortable environment.
At 28, 35, 40, 45, and 60 d, we measured the length of primary feathers 1 to 5 of the duck population, respectively. Then, we calculated the LGRs of primary feather 1 to 5 at different stages (28–30 d, 30–35 d, 35–40 d, 45–60 d, and 28–60 d) based on the primary feather 1 to 5 lengths of 28, 35, 40, 45, and 60-day-old ducks. The LGR of the feather is a relative growth rate, which specifically refers to the percentage of feather length increase in a certain time range to the original length. The calculation formula is:
Next, at 60 d, the feather maturity trait of ten primary feathers was scored; the scoring standard is as follows:
-
•
Complete maturity (1 point): The feather pulp of a single primary feather is transparent, and the feather tube is clear.
-
•
Partial maturity (0.5 points): The feather pulp of the single main primary feather is pink, the umbilicus is milky white, and the root is pink.
-
•
Immaturity (0 points): The root of the single main primary feather is gray-black, and the umbilicus of the feather is gray-black.
The sum of the 10 primary feathers scores was statistically applied to the subsequent association analysis.
Sample Collection and DNA Extraction
Finally, the 358 blood samples of the duck population were obtained from primary veins and rapidly frozen to −20°C. Total genomic DNA was then extracted using a traditional phenol-chloroform protocol. All experiments with Tianfu Nonghua ducks were performed under the guidance of ethical regulation from the Institute of Animal Science, Sichuan Agricultural University.
Whole-Genome Resequencing
The quality and quantity of 358 DNA samples were examined using a NanoDrop device and agarose gel electrophoresis. After the examinations, standard procedures generated paired-end libraries for each eligible sample. The average insert size was 500 bp, and the average read length was 150 bp. All libraries were sequenced on an Illumina HiSeq 2500 platform.
Variant Discovery and Genotyping
The 150-bp paired-end raw reads were mapped to the reference genome (ZJU1.0) with Burrows-Wheeler alignment (BWA aln) using default parameters. We additionally performed local realignment using GATK to enhance the alignments in regions of InDel polymorphisms (McKenna et al., 2010). After mapping, SNP calling was performed using GATK 3.5 exclusively, and the output was further filtered using VCFtools 0.1.15 (Danecek et al., 2011). SNPs were filtered based on the following criteria: 1) SNPs had to have a minor allele frequency > 0.05 and a major allele frequency < 0.99; 2) the maximum missing rate was < 0.1; and 3) SNPs could only have 2 alleles.
Population Structure Analysis
PCA was performed based on all SNPs using the GCTA software further to analyze ducks' population structure (Yang et al., 2011). Then, PCA plots were plotted using the first and second principal components with the R 3.5.1 package.
GWAS
The GWAS used a mixed linear model program Emmax (Legarra et al., 2018). The first 3 principal component values (PCA eigenvectors) derived from whole-genome SNPs were set as a fixed effect to correct population stratification in the mixed model. Random effects were across all individual genome-wide SNP relation matrices of estimation. In addition, the effect of sex was used as a fixed condition in GWAS. QQman software in the R 3.5.1 package was used to draw Manhattan diagrams. Significance thresholds (P) were determined based on Bonferroni correction, and the calculation formula is P = 0.05/The total number of SNPs. Then, the screening condition of significant SNPs was -Log10P > 8.39 by calculation. Finally, the QQ plots were drawn to detect false positives due to population stratification. In the QQ plot, the ordinate was the observed SNP P-value, and the abscissa was the theoretical P-value determined using a chi-squared distribution.
LD Analysis
VCFtools were used to retrieve individual genotypes in the region of interest. LD analysis between the most significant SNPs (-Log10P > 8.39) in the candidate region was conducted by Plink, and a Locus zoom graph was generated by R 3.5.1. Haploview software was used to analyze the total LD in candidate regions and the haplotypes of four candidate SNPs (Barrett et al., 2005).
Gene Expression Analysis
Transcriptome data of wild duck hair follicles were obtained from the GSA database (Bio project: PRJCA004157). Then, clean data of each Illumina-seq transcriptome were mapped to the duck reference genome using HISAT 2.0 (Kim et al., 2015). Transcripts were spliced and quantified using Stringtie software (Pertea et al., 2015). The FPKM (Fragments per Kilobase Million) value quantifies the gene expression levels. After that, we focused on the expression levels of the candidate genes.
Functional Annotation
The functional annotation of candidate genes was completed using the online tool DAVID (https://david.ncifcrf.gov/), and the parameter is set to default. GO analysis divides the function of genes into 3 parts: cellular component (CC), molecular function (MF), and biological process (BP). Pathways with P-values<0.05 were retained.
Statistical Analysis
A normal distribution test was performed using SPSS 17.0 to check the primary feather length traits' distribution (all results are shown in Figure S5). Correlation and One-Way ANOVA analysis among different traits was done using SPSS software. Differences at P < 0.05 were considered significant.
ACKNOWLEDGMENTS
This work was supported by grants from the National Key R&D Program of China (2022YFF1000100), the National Natural Science Foundation of China (31872345 and 32072703) and the Key Technology Support Program of Sichuan Province (2021JDJQ0008).
Ethics approval and consent to participate: All experimental procedures that involved animal manipulation were approved by the Faculty Animal Care and Use Committee of Sichuan Agricultural University under permit no. DKY20170913.
Authors' contributions: The original idea for this study was conceived by Hehe Liu and Shengchao Ma, and Hehe Liu and Shengchao Ma designed the experimental methods. Shengchao Ma, Pengcheng Li, Yang Xi, Qian Xu, Jingjing Qi, Jianmei Wang, Jiwei Hu, Liang Li, Jiwen Wang, Hua He, Chunchun Han, and Lili Bai collected materials and studied. The manuscript was written by Shengchao Ma and edited by Hehe Liu and Yang Xi. Finally, all the authors read and approved the final manuscript.
Disclosures
The authors declare that they have no competing interests.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2022.102243.
Appendix. Supplementary materials
Figure S1 The PCA analysis of 358 individuals.
Figure S2 GWAS of primary feather length traits at 28, 30, 35, 40, 45, and 60-days-old of ducks.
Figure S3 GWAS of LGR of primary feather 1∼5 at 28, 30, 35, 40, 45, and 60-days-old of ducks.
Figure S4 GWAS of maturity scores of primary feather 6∼2 of ducks.
Figure S5 Tests for normal distribution of feather length trait measurements.
Table S1 Correlation analysis of primary feather length at different growth stages.
Table S2 One-way ANOVA for feather length based on sexes grouping.
Table S3 One-way ANOVA for feather maturity traits based on sexes grouping.
Table S4 Correlation analysis between LGRs and feather length traits.
Additional file 6: Table S5 The Annotation of SNPs associated with feather length traits.
Table S6 The annotation of SNPs associated with feather LGR on Chr. 1 and 21.
Additional file 8: Table S7 The annotation of SNPs associated with feather maturity traits.
Table S8 Phenotypic means of length traits of primary feathers.
Table S9 The average primary feather length of male and female duck populations at different stages.
REFERENCES
- Abdollahi-Arpanahi R., Morota G., Peñagaricano F. Predicting bull fertility using genomic data and biological information. J. Dairy Sci. 2017;100:9656–9666. doi: 10.3168/jds.2017-13288. [DOI] [PubMed] [Google Scholar]
- Badway J.A., Baleja J.D. Reps2: a cellular signaling and molecular trafficking nexus. Int. J. Biochem. Cell Biol. 2011;43:1660–1663. doi: 10.1016/j.biocel.2011.08.014. [DOI] [PubMed] [Google Scholar]
- Bertelsen B., Melchior L., Jensen L.R., Groth C., Glenthøj B., Rizzo R.…Tümer Z. Intragenic deletions affecting two alternative transcripts of the IMMP2L gene in patients with Tourette syndrome. Eur. J. Hum. Genet. 2014;22:1283–1289. doi: 10.1038/ejhg.2014.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan B.K., Loh P.R., Finucane H.K., Ripke S., Yang J., Patterson N.…Neale B.M. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J.C., Fsery B., Maller J.D.M.J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Chen C.F., Foley J., Tang P.C., Li A., Jiang T.X., Wu P.…Chuong C.M. Development, regeneration, and evolution of feathers. Annu. Rev. Anim. Biosci. 2015;3:169–195. doi: 10.1146/annurev-animal-022513-114127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo J.J., Møller A.P. Phenotypic variation and fluctuating asymmetry in sexually dimorphic feather ornaments in relation to sex and mating system. Biol. J. Linn. Soc. 1999;68:505–529. [Google Scholar]
- Chuong C.M., Randall V.A., Widelitz R.B., Wu P., Jiang T.X. Physiological regeneration of skin appendages and implications for regenerative medicine. Physiology. 2012;27:61–72. doi: 10.1152/physiol.00028.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon C.A., Garcia-Longoria L., Martin L.B., Magallanes S., de Lope F., Marzal A. Malaria infection negatively affects feather growth rate in the house sparrow Passer domesticus. J. Avian Biol. 2016;47:779–787. [Google Scholar]
- Chambers J.R. Genetics of growth and meat production in chicken. Poultr. Breed. Gen. 1990:599–643. ISBN 0-444-88557-9. [Google Scholar]
- Cheng Y.S., Rouvier R., Poivey J.P., Tai C. Genetic parameters of body weight, egg production and shell quality traits in the Brown Tsaiya laying duck. Gen. Select. Evolut. 1995;27:459–472. [Google Scholar]
- Chang M.T., Cheng Y.S., Huang M.C. The SNP genotypes of growth hormone gene associated with reproductive traits in Tsaiya ducks. Reprod. Domest. Ani. 2012;47:568–573. doi: 10.1111/j.1439-0531.2011.01918.x. [DOI] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A.…and 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt-Henrich S.G., Marks H.L. Heritabilities of growth curve parameters and age-specific expression of genetic variation under two different feeding regimes in Japanese quail (Coturnix coturnix japonica) Gen. Res. 1993;62:45–55. doi: 10.1017/s0016672300031554. [DOI] [PubMed] [Google Scholar]
- Hill G.E., Montgomerie R. Plumage colour signals nutritional condition in the house finch. Proc. R. Soc. Lond. B Biol. Sci. 1994;258:47–52. [Google Scholar]
- Hu Y.H., Poivey J.P., Rouvier R., Wang C.T., Tai C. Heritabilities and genetic correlations of body weights and feather length in growing Muscovy selected in Taiwan. Br. Poult. Sci. 1999;40:605–612. doi: 10.1080/00071669986972. [DOI] [PubMed] [Google Scholar]
- Hoff J.L., Decker J.E., Schnabel R.D., Seabury C.M., Neibergs H.L., Taylor J.F. QTL-mapping and genomic prediction for bovine respiratory disease in US Holsteins using sequence imputation and feature selection. BMC Genomics. 2019;20:1–15. doi: 10.1186/s12864-019-5941-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H.L., Huang L.T., Cheng Y.S. A novel SNP marker of ovalbumin gene in association with duck hatchability. Theriogenology. 2013;79:1218–1223. doi: 10.1016/j.theriogenology.2013.02.021. [DOI] [PubMed] [Google Scholar]
- He Q., Gu L., Lin Q., Ma Y., Liu C., Pei X.…Yang Y. The Immp2l mutation causes ovarian aging through ROS-Wnt/β-catenin-estrogen pathway: preventive effect of melatonin. Endocrinology. 2020;161:bqaa119. doi: 10.1210/endocr/bqaa119. [DOI] [PubMed] [Google Scholar]
- Ito Y., Nakajima K., Masubuchi Y., Kikuchi S., Okano H., Saito F.…Shibutani M. Downregulation of low-density lipoprotein receptor class A domain-containing protein 4 (Ldlrad4) in the liver of rats treated with nongenotoxic hepatocarcinogen to induce transforming growth factor β signaling promoting cell proliferation and suppressing apoptosis in early hepatocarcinogenesis. J. Appl. Toxicol. 2020;40:1467–1479. doi: 10.1002/jat.3998. [DOI] [PubMed] [Google Scholar]
- Ji G.G., Zhang M., Liu Y.F., Shan Y.J., Tu Y.J., Ju X.J.…Xie J.F. A gene co-expression network analysis of the candidate genes and molecular pathways associated with feather follicle traits of chicken skin. J. Ani. Breed. Genet. 2021;138:122–134. doi: 10.1111/jbg.12481. [DOI] [PubMed] [Google Scholar]
- Kjaer J.B., Bessei W. The interrelationships of nutrition and feather pecking in the domestic fowl. Arch. Geflügelk. 2013;77:1–9. [Google Scholar]
- Khosravinia H. Effect of the slow (K) or rapid (k+) feathering gene on carcass-related traits of broiler chickens selected for breast and thighs weight. Russ. J. Genet. 2009;45:98–104. [PubMed] [Google Scholar]
- Kalinowski A., Moran E.T., Jr, Wyatt C. Methionine and cystine requirements of slow-and fast-feathering male broilers from zero to three weeks of age. Poult. Sci. 2003;82:1423–1427. doi: 10.1093/ps/82.9.1423. [DOI] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeson S., Walsh T. Feathering in commercial poultry II. Factors influencing feather growth and feather loss. Worlds Poult. Sci. J. 2004;60:52–63. [Google Scholar]
- Leeson S., Walsh T. Feathering in commercial poultry I. Feather growth and composition. Worlds Poult. Sci. J. 2004;60:42–51. [Google Scholar]
- Lou, M. 1994. Genetic evaluation of the effects of divergent feathering selection and major feathering genes on growth performances and carcass traits in broiler chickens. University of Glasgow (United Kingdom).
- Liu A., Lund M.S., Boichard D., Karaman E., Fritz S., Aamand G.P.…Su G. Improvement of genomic prediction by integrating additional single nucleotide polymorphisms selected from imputed whole genome sequencing data. Heredity. 2020;124:37–49. doi: 10.1038/s41437-019-0246-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Hu J., Guo Z., Fan W., Xu Y., Liang S.…Hou S. A single nucleotide polymorphism variant located in the cis-regulatory region of the ABCG2 gene is associated with mallard egg colour. Mol. Ecol. 2021;30:1477–1491. doi: 10.1111/mec.15785. [DOI] [PubMed] [Google Scholar]
- Lee J.R. Protein tyrosine phosphatase PTPRT as a regulator of synaptic formation and neuronal development. BMB Rep. 2015;48:249. doi: 10.5483/BMBRep.2015.48.5.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legarra A., Ricard A., Varona L. GWAS by GBLUP: single and multimarker EMMAX and Bayes factors, with an example in detection of a major gene for horse gait. G3: Genes Genomes Genet. 2018;8:2301–2308. doi: 10.1534/g3.118.200336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen T.H., Hayes B.J., Goddard M. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157:1819–1829. doi: 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod I.M., Bowman P.J., Vander Jagt C.J., Haile-Mariam M., Kemper K.E., Chamberlain A.J.…Goddard M.E. Exploiting biological priors and sequence variants enhances QTL discovery and genomic prediction of complex traits. BMC Gen. 2016;17:1–21. doi: 10.1186/s12864-016-2443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P., Lund M.S., Aamand G.P., Su G. Use of a Bayesian model including QTL markers increases prediction reliability when test animals are distant from the reference population. J. Dairy Sci. 2019;102:7237–7247. doi: 10.3168/jds.2018-15815. [DOI] [PubMed] [Google Scholar]
- McDougald L.R., K.A.V.O.U.S. Keshavarz The effect of polyether, ionophorous anticoccidial drugs on feather growth in genetically slow-feathering broilers. Poult. Sci. 1984;63:1322–1326. doi: 10.3382/ps.0631322. [DOI] [PubMed] [Google Scholar]
- Maestrini E., Pagnamenta A.T., Lamb J.A., Bacchelli E., Sykes N.H., Sousa I.…Monaco A.P. High-density SNP association study and copy number variation analysis of the AUTS1 and AUTS5 loci implicate the IMMP2L–DOCK4 gene region in autism susceptibility. Mol. Psychiatr. 2010;15:954–968. doi: 10.1038/mp.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A.…DePristo M.A. The genome analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris D.R., Marra P.P., Montgomerie R., Kyser T.K., Ratcliffe L.M. Reproductive effort, molting latitude, and feather color in a migratory songbird. Science. 2004;306:2249–2250. doi: 10.1126/science.1103542. [DOI] [PubMed] [Google Scholar]
- Oosterhoff J.K., Kühne L.C., Grootegoed J.A., Blok L.J. EGF signalling in prostate cancer cell lines is inhibited by a high expression level of the endocytosis protein REPS2. Int. J. Cancer. 2005;113:561–567. doi: 10.1002/ijc.20612. [DOI] [PubMed] [Google Scholar]
- Prum R.O., Williamson S. Theory of the growth and evolution of feather shape. J. Exp. Zool. 2001;291:30–57. doi: 10.1002/jez.4. [DOI] [PubMed] [Google Scholar]
- Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzi R., Cerolini S., Mantovani C., Pagnacco G., Mangiagalli M.G., Cavalchini L.G. Heritabilities and genetic correlations of conformation and plumage characteristics in pheasant (Phasianus colchicus) Poult. Sci. 1994;73:1204–1210. doi: 10.3382/ps.0731204. [DOI] [PubMed] [Google Scholar]
- Soller M. Marker assisted selection-an overview. Anim. Biotechnol. 1994;5:193–207. [Google Scholar]
- Szász S., Bogenfürst F., Varju M. Development of the fourth primary wing feathers in three types of duck. Acta Agraria Kaposváriensis. 1999;3:239–245. [Google Scholar]
- Shi J.H., Hao Y.J. DDX10 overexpression predicts worse prognosis in osteosarcoma and its deletion prohibits cell activities modulated by MAPK pathway. Biochem. Biophys. Res. Commun. 2019;510:525–529. doi: 10.1016/j.bbrc.2019.01.114. [DOI] [PubMed] [Google Scholar]
- Talloen W., Lens L., Van Dongen S., Matthysen E. Feather development under environmental stress: lead exposure effects on growth patterns in Great Tits Parus major. Bird Study. 2008;55:108–117. [Google Scholar]
- Tao L., He X.Y., Pan L.X., Wang J.W., Gan S.Q., Chu M.X. Genome-wide association study of body weight and conformation traits in neonatal sheep. Anim. Genet. 2020;51:336–340. doi: 10.1111/age.12904. [DOI] [PubMed] [Google Scholar]
- Wu Y., Pi J.S., Pan A.L., Pu Y.J., Du J.P., Shen J.…Zhang J.R. An SNP in the MyoD1 gene intron 2 associated with growth and carcass traits in three duck populations. Biochem. Genet. 2012;50:898–907. doi: 10.1007/s10528-012-9530-4. [DOI] [PubMed] [Google Scholar]
- Xi Y., Xu Q., Huang Q., Ma S., Wang Y., Han C.…Li L. Genome-wide association analysis reveals that EDNRB2 causes a dose-dependent loss of pigmentation in ducks. BMC Gen. 2021;22:1–11. doi: 10.1186/s12864-021-07719-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W.Y., Pan N.X., Zeng H.R., Yan H.C., Wang X.Q., Gao C.Q. Comparison of nonlinear models to describe the feather growth and development curve in yellow-feathered chickens. Animal. 2020;14:1005–1013. doi: 10.1017/S1751731119003082. [DOI] [PubMed] [Google Scholar]
- Ye S., Li J., Zhang Z. Multi-omics-data-assisted genomic feature markers preselection improves the accuracy of genomic prediction. J. Anim. Sci. Biotechnol. 2020;11:1–12. doi: 10.1186/s40104-020-00515-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z., Jiang T.X., Wu P., Widelitz R.B., Chuong C.M. Sprouty/FGF signaling regulates the proximal–distal feather morphology and the size of dermal papillae. Dev. Biol. 2012;372:45–54. doi: 10.1016/j.ydbio.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Yue Z., Wu P., Wu D.Y., Mayer J.A., Medina M.…Chuong C.M. The developmental biology of feather follicles. Int. J. Dev. Biol. 2004;48:181. doi: 10.1387/ijdb.031776my. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Li M., Cheng H., Fan W., Yuan Z., Gao Q.…Jiang Y. An intercross population study reveals genes associated with body size and plumage color in ducks. Nat. Commun. 2018;9:1–10. doi: 10.1038/s41467-018-04868-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Tachtsidis G., Schob C., Koko M., Hedrich U.B., Lerche H.…Bähring R. KCND2 variants associated with global developmental delay differentially impair Kv4. 2 channel gating. Hum. Mol. Genet. 2021;30:2300–2314. doi: 10.1093/hmg/ddab192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Opalinska J., Sohal D., Yu Y., Mo Y., Bhagat T.…Verma A. Aberrant epigenetic and genetic marks are seen in myelodysplastic leukocytes and reveal Dock4 as a candidate pathogenic gene on chromosome 7q. J. Biol. Chem. 2011;286:25211–25223. doi: 10.1074/jbc.M111.235028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 The PCA analysis of 358 individuals.
Figure S2 GWAS of primary feather length traits at 28, 30, 35, 40, 45, and 60-days-old of ducks.
Figure S3 GWAS of LGR of primary feather 1∼5 at 28, 30, 35, 40, 45, and 60-days-old of ducks.
Figure S4 GWAS of maturity scores of primary feather 6∼2 of ducks.
Figure S5 Tests for normal distribution of feather length trait measurements.
Table S1 Correlation analysis of primary feather length at different growth stages.
Table S2 One-way ANOVA for feather length based on sexes grouping.
Table S3 One-way ANOVA for feather maturity traits based on sexes grouping.
Table S4 Correlation analysis between LGRs and feather length traits.
Additional file 6: Table S5 The Annotation of SNPs associated with feather length traits.
Table S6 The annotation of SNPs associated with feather LGR on Chr. 1 and 21.
Additional file 8: Table S7 The annotation of SNPs associated with feather maturity traits.
Table S8 Phenotypic means of length traits of primary feathers.
Table S9 The average primary feather length of male and female duck populations at different stages.